
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 29 April 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.675498
Gliomas are the most common and malignant primary brain tumors. Various hallmarks of glioma, including sustained proliferation, migration, invasion, heterogeneity, radio- and chemo-resistance, contribute to the dismal prognosis of patients with high-grade glioma. Dysregulation of cancer driver genes is a leading cause for these glioma hallmarks. In recent years, a new mechanism of post-transcriptional gene regulation was proposed, i.e., “competing endogenous RNA (ceRNA).” Long non-coding RNAs, circular RNAs, and transcribed pseudogenes act as ceRNAs to regulate the expression of related genes by sponging the shared microRNAs. Moreover, coding RNA can also exert a regulatory role, independent of its protein coding function, through the ceRNA mechanism. In the latest glioma research, various studies have reported that dysregulation of certain ceRNA regulatory networks (ceRNETs) accounts for the abnormal expression of cancer driver genes and the establishment of glioma hallmarks. These achievements open up new avenues to better understand the hidden aspects of gliomas and provide new biomarkers and potential efficient targets for glioma treatment. In this review, we summarize the existing knowledge about the concept and logic of ceRNET and highlight the emerging roles of some recently found ceRNETs in glioma progression.
Gliomas are the most common and malignant primary brain tumors, accounting for about 30% of all primary brain tumors and 80% of malignant ones (Weller et al., 2015). The origin of gliomas is thought to be from neuroglial stem or progenitor cells. Based on morphological similarities to the neuroglial cells of normal brain, the World Health Organization (WHO) 2007 classification system categorizes gliomas into astrocytomas, oligodendrogliomas, mixed oligoastrocytic gliomas, or ependymomas and into I–IV grades with grades I and II and grades III and IV considered low- and high-grade gliomas, respectively (Louis et al., 2007). Patients with high-grade glioma, such as glioblastoma (GBM, a grade IV astrocytoma), have a median survival time of only 15 months after initial diagnosis (Jemal et al., 2010; Rynkeviciene et al., 2019). Research over the past decade using advanced sequencing technologies has unraveled molecular alterations or biomarkers underlying gliomas, which updated our understanding of glioma’s biology and resulted in a new classification system (Louis et al., 2016) with more precision for gliomas. This system integrated histology and molecular biomarkers, including IDH (encoding isocitrate dehydrogenase) mutation and 1p/19q-codeletion status (Louis et al., 2016). Despite these progresses, as well as progress in surgical resection, radiotherapy, and chemotherapy, the prognosis for patients with high-grade gliomas remains dismal (Dong and Cui, 2020). Besides that, some low-grade gliomas can develop into secondary high-grade ones after surgical resection, radiotherapy, or chemotherapy (Hamisch et al., 2017). A better understanding of the molecular mechanisms of gliomagenesis is urgently needed to develop potential new biomarkers and therapeutic strategies for improved treatments.
Dysregulation of oncogenes (e.g., RAS, PIK3CA, and MYC) and/or tumor-suppressive genes (e.g., PTEN, TP53, and RB1) leads to cell transformation (Singh et al., 2002; van’t Veer et al., 2002; Ballestar and Esteller, 2008; Liu et al., 2021). Enormous efforts have been devoted to illustrating the dysregulation mechanisms of these cancer driver genes at the transcriptional and post-transcriptional levels. Noticeably, more than 75% of the human genome can generate RNA transcripts, of which only approximately 2% are messenger RNAs (mRNAs) that contain cancer driver genes, and the majority of the rest are noncoding RNAs including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), transcribed pseudogenes, and circular RNAs (circRNAs) (Hangauer et al., 2013; Abdollahzadeh et al., 2019). These noncoding RNAs did not draw attention until in the recent years, and our understanding of their function is still in infancy and requires more research. In 2011, Salmena et al. (2011) proposed that mRNAs, lnRNAs, and transcribed pseudogenes regulate each other via acting as competing endogenous RNAs (ceRNAs) to compete for binding of shared miRNAs. This milestone conception of ceRNA implies that all of the above-mentioned types of RNA transcripts, even protein-coding mRNAs themselves, can perform post-transcriptional regulation and constitute ceRNA regulatory networks (ceRNETs), which may profoundly affect the expression of cancer driver genes and promote tumorigenesis.
Examples of ceRNA crosstalk have been described in the latest research on glioma. Indeed the dysregulation of ceRNETs between different kinds of RNAs contributes to the establishment of the hallmarks of different subtypes of gliomas, suggesting the important roles of ceRNETs in the development of gliomas. Therefore, understanding this novel language of RNA crosstalk will give a new insight into gene regulatory networks, open a new window to better understand the hidden and complex aspects of gliomas, and provide a new way to find specific biomarkers and potential efficient therapeutic targets for the diagnosis and treatment of gliomas. In this review, we first introduce the pieces of knowledge of ceRNA hypothesis, particularly highlighting their building blocks including miRNAs, mRNAs, lncRNAs, circRNAs, and transcribed pseudogenes as well as the logic for effective ceRNA crosstalk. Then, we specifically discuss the latest discoveries of distinct ceRNETs in glioma research.
miRNAs are small single-stranded RNAs (approximately 22 nucleotides) that play key roles in ceRNA crosstalk. They bind to miRNA response elements (MREs) on target RNAs through sequence complementarity, which reduces the stability of targets or restricts their translation. MREs can be found in 5’ untranslated regions (5’ UTRs), coding sequences, and especially 3’ untranslated regions (3’ UTRs) of various types of RNA transcripts, such as mRNAs, lncRNAs, transcribed pseudogenes, and circRNAs. Most miRNAs pair imperfectly with their targets, and pairing between miRNA (two to eight nucleotides, especially six or seven nucleotides from the 5’ end of miRNA) and 3’ UTR of target mRNA is often crucial. Each miRNA can regulate up to thousands of target RNAs, and miRNAs can act in a combinatorial manner if a target RNA has multiple different MREs. miRNA-mediated regulation is estimated to affect a large proportion of human transcriptome, which makes miRNA an important modulator in numerous diseases, including various types of cancers.
The above-mentioned miRNA → RNAs regulation model has been updated by introducing the concept of ceRNAs. As shown in Figure 1, RNA transcripts that share the same MREs can regulate each other’s expression by competing for the same pool of miRNAs and thus modulating the availability of miRNAs. A new miRNA-mediated regulation model is RNAs ↔ miRNAs ↔ RNAs based on the concept of ceRNA. This means that miRNAs no longer act only as active regulators but that they are also passively regulated by their target RNAs. In addition, even a few miRNAs and related target RNAs can generate a complex ceRNET. Studying the larger interconnected ceRNET, rather than isolated ceRNA pair interactions, may generate deeper insight into ceRNA-mediated gene regulation in a setting that is closer to physiological conditions.
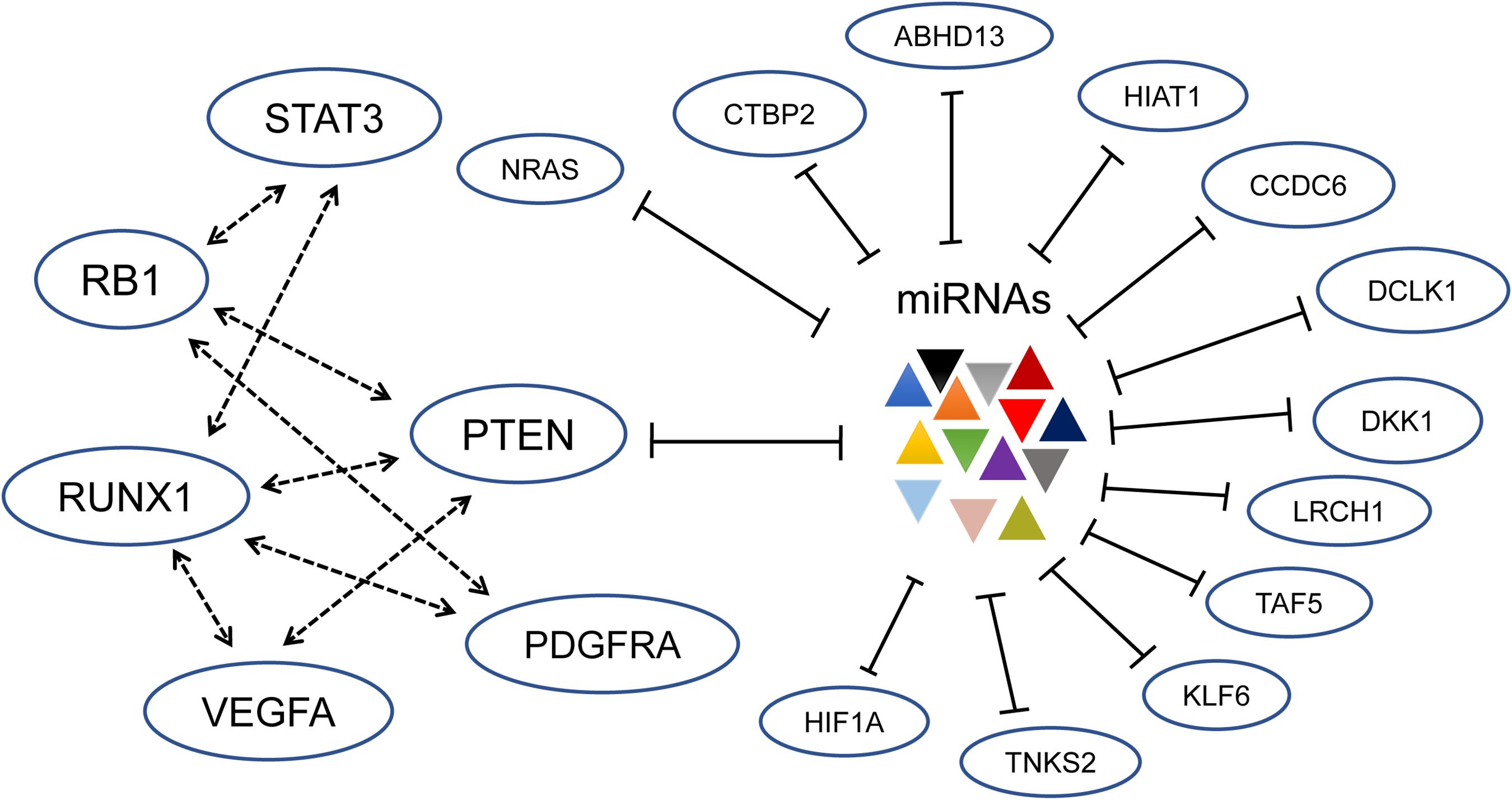
Figure 1. Established driver genes of gliomagenesis constitute a complex ceRNET; especially the ceRNAs for PTEN are shown (Sumazin et al., 2011). Through shared miRNA, the six cancer driver genes including PTEN, RB1, STAT3, PDGFRA, RUNX1, and VEGFA regulate each other. At the same time, other genes constitute a subnetwork and interact with PTEN. The dotted arrows represent activation, and the lines with blunt ends indicate inhibition.
The central dogma of molecular biology is that information on the DNA is transcribed into mRNAs that, in turn, are used as templates for protein synthesis. There are approximately 20,000 protein-coding genes in the human genome (Baltimore, 2001). Many of them, including cancer driver genes, harbor MREs, making their expression profoundly affected by the cognate miRNAs (Friedman et al., 2009). The finding of ceRNA implies that mRNAs possess a regulatory function independent of protein coding function. As a result, the ceRNA activity of mRNAs may confer them independent and even opposite roles to their encoded protein in a process such as tumorigenesis (Salmena et al., 2011). In this scenario, gross genomic losses or amplifications that commonly happened in cancers could potentially affect the function of ceRNAs in these regions and interrupt the related ceRNET. In addition, gene loss events should be distinguished with point mutations, as the former lose both protein-coding sequence and MREs, while the latter lose protein function but retain ceRNA function. The potential role of mRNA-mediated ceRNET has been confirmed in glioma (Sumazin et al., 2011).
lncRNAs are a large variety of RNA transcripts longer than 200 nucleotides without protein-coding capacity but with a similar structure to mRNAs since they typically have a 5’ m7G cap and 3’ poly (A) tail (Ulitsky and Bartel, 2013). The expression of lncRNAs is more tissue specific and dynamic, suggesting that they have distinct biological roles (Deveson et al., 2017). Dysregulation of cancer-related lncRNAs plays important roles in tumorigenesis, and an increasing amount of lncRNAs has been linked to gliomagenesis (Kiang et al., 2015; Peng et al., 2018; Rynkeviciene et al., 2019). lncRNAs perform their regulatory function through interacting with DNA, mRNAs, other non-coding RNAs, and proteins, covering almost all aspects of gene expression regulation including chromatin modification, transcription, post-transcription, and translation (Wang and Chang, 2011). Functionally, lncRNAs can act as signals, decoys, guides, scaffolds, and sponges. Particularly, the miRNA sponging function of lncRNAs that inhibits miRNAs makes lncRNA an important active player in the ceRNET (Xia et al., 2014; Greco et al., 2019; Wang L. et al., 2019; Ebrahimpour et al., 2021).
CircRNAs are a class of endogenous non-coding RNA without a 5’ m7G cap and 3’ poly (A) tail structure and are formed by the circularization of pre-RNAs via back splicing (Li X. et al., 2018; Kristensen et al., 2019). They are widely present in a variety of human cells. Because of lack of exposed 5’ and 3’ ends, circRNAs are more stable than linear RNAs in terms of resistance to degradation by exonucleases or RNase R (Suzuki et al., 2006). Thus, circRNAs are stable in human body fluids, including blood and saliva, making them suitable biomarkers for diagnosis of cancer (Bahn et al., 2015; Memczak et al., 2015). CircRNAs carry out their function through a variety of mechanisms, such as acting as ceRNAs, interacting with RNA-binding proteins, alternative splicing, and translation (Li X. et al., 2018; Kristensen et al., 2019). Due to the high stability of circRNAs, the sponge effect of circRNAs on miRNAs has been appreciated. Rybak-Wolf et al. (2015) revealed that circRNAs are highly abundant in the mammalian brain, dynamically expressed, and conserved among human, mouse, and Drosophila. Furthermore, Song et al. (2016) developed a computational tool called UROBORUS to detect circRNAs in total RNA-seq data and found that more than 476 circRNAs were differentially expressed in control brain tissues and gliomas. Increasing recent reports have illustrated the crucial role of dysregulated circRNAs in gliomagenesis, showing great potential as valuable diagnostic and therapeutic biomarkers (Jin et al., 2018; Ding et al., 2020; Long et al., 2020).
Pseudogenes are genomic loci similar to the known genes but lost their protein coding ability as a result of premature stop codons, deletions, insertions, or frameshift mutations (D’Errico et al., 2004). Therefore, they were considered as “non-functional,” “junk,” or “genomic fossils,” until recently their roles in various biological processes have been illustrated. Genomic sequencing analyses showed a huge number of pseudogenes (∼19,000) in humans, and many of them are transcribed and well-conserved (Pink et al., 2011). The ENCODE project further revealed that the transcription of some pseudogenes is tissue specific or constitutive (Pei et al., 2012). Mechanically, transcribed pseudogenes regulate the expression of target genes by the generation of endogenous small interference RNAs (siRNAs) (Watanabe et al., 2008) and antisense transcripts (Zhou et al., 1992) or acting as ceRNAs (Poliseno et al., 2010). Since pseudogenes are highly similar to their ancestral protein-coding genes, they can actively compete for the same pool of miRNAs through shared MREs (An et al., 2017). Poliseno et al. (2010) firstly reported that pseudogene PTENP1 derepresses the expression of tumor-suppressor gene PTEN through competing for PTEN-targeting miRNAs in prostate cancer cells and colon carcinoma cells, therefore exerting a tumor-suppressive role. Furthermore, they extended their analysis to other cancer driver genes with pseudogenes, such as oncogene KRAS and its pseudogene KRAS1P. Thereafter, the ceRNA function of more pseudogenes is revealed in various cancers, including breast cancer and gliomas (Shi X. et al., 2016; Li et al., 2017b; Wang Y. et al., 2019).
The effectiveness and result of crosstalk between ceRNAs are regulated by multiple factors, including relative concentration and subcellular localization of the ceRNAs and miRNAs, number of shared MREs, and miRNA-ceRNA binding affinity (Salmena et al., 2011; Sanchez-Mejias and Tay, 2015). Both mathematical models and experimental models support that ceRNA crosstalk conform to a titration mechanism that is sensitive to the relative abundance of miRNA/target RNAs and often exhibit a threshold-like manner (Buchler and Louis, 2008). Alterations of the ceRNA levels should be large enough to overcome or relieve suppression on competing ceRNAs by the miRNAs. Similarly, absent expression or overexpression of shared miRNAs will abolish ceRNA competition. When the levels of miRNA and ceRNAs are near equimolar, optimal ceRNA crosstalk is expected to happen, in which one ceRNA has the biggest effect on its ceRNA partners (Mullokandov et al., 2012; Bosia et al., 2013). Subcellular localization influences ceRNA’s accessibility to miRNAs. Not all miRNAs are present everywhere and at all times, and many RNA-binding proteins can profoundly affect the localization or compartmentation of RNA transcripts through mechanisms including phase separation (Venables et al., 2009; Fox et al., 2018; Ries et al., 2019). The number of shared miRNAs among ceRNAs is important for effective ceRNA crosstalk; the more shared, the deeper the communication (Ala et al., 2013; Figliuzzi et al., 2013). MREs on ceRNAs are not equal. Although two MREs can bind the same miRNA, their partially different nucleotide composition contributes to the distinct binding affinity between miRNAs and ceRNAs (Salmena et al., 2011). The nonreciprocal competing effect between partially and perfectly complementary ceRNAs was predicted computationally and validated experimentally in cultured human cells using synthetic gene circuits (Yuan et al., 2015). Collectively, the above-mentioned factors should be considered when studying ceRNAs and especially assigning their contribution to specific human diseases, which may facilitate the translation of research results to clinical application.
Sumazin et al. (2011) presented a pioneer and comprehensive study of mRNA–mRNA crosstalk through shared miRNAs in GBM. Using computational tools, an extensive ceRNET, consisting of about 7,000 genes and more than 248,000 miRNAs, is constructed. Further biochemical assays in cell lines confirmed that established drivers of tumor initiation and subtype implementation are indeed regulated by this ceRNET, including PTEN, RB1, STAT3, PDGFRA, RUNX1, and VEGFA (Figure 1). Specifically, they focused on 13 genes, including ABHD13, CCDC6, CTBP2, NRAS, and RB1, and confirmed that these genes can act through ceRNA mechanism to regulate the expression of PTEN and vice versa (Figure 1). The overexpression of PETN 3’ UTR increases the expression of 13 ceRNAs, elevates PTEN protein level, and decreases the growth rates of glioma cells, while knockdown of each of the 13 genes can reduce PTEN 3’ UTR luciferase expression and significantly promote glioma cell growth. The silencing effect mediated by the ceRNA mechanism is comparable to that of siRNA-mediated PTEN silencing. Moreover, PTEN composes a subnetwork with the known drivers of glioma tumorigenesis and GBM subtypes, i.e., RB1, STAT3, PDGFRA, RUNX1, and VEGFA. The ectopic expression of the 3’ UTRs of genes in this subnetwork can upregulate the expression of the other genes. Therefore, the ceRNA mechanism provides a way for these cancer drivers to be coordinately expressed through a shared miRNA pool, which is implicated in high-grade gliomagenesis (Chow et al., 2011).
The ceRNET concept is one of the hot research topics in recent years, and reports of lncRNA- or circRNA-mediated ceRNETs in glioma research are increasing fast using computational or experimental methods (Wu and Qian, 2019; Zhu et al., 2020c). For example, Zhu et al. (2020c) constructed a comprehensive lncRNA–miRNA–mRNA ceRNET consisting of 61 lncRNAs, 12 miRNAs, and 92 mRNAs through a computational method. Here we will discuss and highlight several lately discovered ceRNETs in the following sections. More ceRNETs are summarized in Figures 2, 3 and Tables 1, 2.
lncRNA X-inactive specific transcript (XIST), located on chromosome Xq13.2, is frequently diagnosed in various cancers, including gastric cancer, lung cancer, and glioma (Wang Y.P. et al., 2020). Acting as a ceRNA, at least five miRNAs (e.g., miR-133a, miR-126, miR-137, miR-204-5p, and miR-329-3p) have been identified in the XIST-mediated ceRNETs that affect multiple hallmarks of glioma progression, including proliferation, apoptosis, migration, EMT, and angiogenesis (Figure 2 and Table 1).
Wang et al. (2017) reported that XIST promotes gliomagenesis through the XIST/miR-137/Rac1 regulatory axis. Specifically, the expression of XIST and miR-137 is significantly up- and down-regulated in glioma tissues, respectively. Overexpression of XIST promotes the proliferation of glioma cells, which can be reversed by miR-137 overexpression (Wang et al., 2017). Ract1 (Ras-related C3 botulinum toxin substrate1) is a member of the Rho family that belongs to the Ras superfamily of GTPases (Coso et al., 1995). It plays a crucial role in the regulation of proliferation, differentiation, and apoptosis of tumor cells and is abnormally expressed in several cancer types, including non-small cell lung cancer (Zhou et al., 2016), breast cancer (Algayadh et al., 2016), and sarcoma (Manara et al., 2016). Interestingly, through sponging miR-137, XIST regulates glioma angiogenesis by regulating FOXC1 (forkhead box C1) expression (Yu et al., 2017). Yu et al. (2017) found that XIST is upregulated in endothelial cells in a blood–tumor–barrier model in vitro. FOXC is a transcription factor of the forkhead box family, and it promotes glioma angiogenesis by activating the expression of CXCR7 [chemokine (C–X–C motif) receptor 7b] (Yu et al., 2017).
In 2020, four groups reported novel XIST-mediated ceRNETs that promote glioma progression, i.e., XIST/miR-133a/SOX4 (Luo et al., 2020), XIST/miR-126/IRS1 (Cheng et al., 2020), XIST/miR-204-5p/Bcl-2 (Shen et al., 2020), and XIST/miR-329-3p/CREB1 (Wang Y.P. et al., 2020). Luo et al. (2020) reported that XIST/miR-133a/SOX4 ceRNET regulates the proliferation, invasion, and EMT of glioma. Sox4 is a member of the Sox (SRY-related HMG-box) family of transcription factors and is involved in cell differentiation and proliferation (Tiwari et al., 2013). Cheng et al. (2020) reported that XIST/miR-126/IRS1 ceRNET regulates cell viability, migration, invasion, glucose metabolism, and resistance to apoptosis in glioma cells. IRS1 (insulin receptor substrate 1) is a key target of the insulin receptor tyrosine kinase involved in hormonal control of metabolism (Shah et al., 2004). Furthermore, Cheng et al. (2020) demonstrated that IRS1 promotes glioma progression by activating the PI3K/AKT pathway. Shen et al. (2020) reported that XIST/miR-204-5p/Bcl-2 ceRNET regulates the proliferation, migration, invasion, and apoptosis of glioma cells. XIST can regulate a variety of apoptosis-related genes, including Bax, caspase 3, caspase 9, and Bcl-2; however, only Bcl-2 had been shown to be a direct target of miR-204-5p and was only tested in their study (Shen et al., 2020). Wang Y.P. et al. (2020) reported that XIST/miR-329-3p/CREB1 ceRNET regulates proliferation, invasion, apoptosis, and radiosensitivity in glioma. CREB1 (cAMP response element binding protein 1) is a member of the leucine zipper with a basic domain (bZip) family of transcription factors and regulates responses to a variety of growth factors and stress signals (Wang et al., 2016). In addition to the above-mentioned studies with known competing endogenous mRNAs for XIST, XIST can also sponge other miRNAs to affect glioma progression, such as miR-152 (Yao et al., 2015), yet the downstream mRNA targets remain unknown.
Collectively, these results clearly indicate that lncRNA XIST plays key roles in gliomagenesis by targeting multiple miRNAs and, in turn, de-represses the expression of multiple cancer driver genes. Therefore, XIST-mediated ceRNETs could be potential diagnostic and prognostic biomarkers and therapeutic targets.
Mu et al. (2020) identified 183 lncRNAs that were significantly differentially expressed in the glioma samples of patients compared with normal control and further investigated the function of lncRNA BCYRN1 (brain cytoplasmic RNA 1, also called BC200) which was the most downregulated one. Functionally, they showed that BCYRN1 overexpression can repress the proliferation and migration of glioma cells, while its knockdown has opposite effects. Mechanically, BCYRN1 acts as a ceRNA to impede gliomagenesis by sponging miR-619-5p to regulate the expression of CUEDC2 (CUE domain-containing protein 2) and PTEN/AKT/p21 pathway (Mu et al., 2020). CUEDC2 is an adapter protein with a CUE domain that is involved in regulating protein stability via ubiquitination of specific substrates (Shih et al., 2003). The PTEN/AKT/p21 pathway acts on downstream targets of CUEDC2 to mediate the anti-tumor effect of BCYRN1 (Mu et al., 2020).
In contrast, Liu et al. (2015) showed that BCYRN1 is significantly downregulated during genotoxic stress-induced necrosis in human glioma cell lines, implying an oncogenic function of lncRNA. In 2020, the same group reported that BCYRN1 functions as an oncogene and promotes proliferation, invasion, and migration (Yu et al., 2020). Mechanically, BCYRN1 sponges endogenous tumor suppressor miR-125a-5p to de-depress the expression of TAZ (transcriptional coactivator with PDZ-binding motif). TAZ has been shown to regulate mesenchymal differentiation in GBM, i.e., TAZ is required for self-renewal, invasion, and tumor formation of mesenchymal glioma stem cells (Bhat et al., 2011). Future studies could reconcile the difference between these studies and shed light on the exact role of BCYRN1-mediated ceRNETs in glioma progression.
Long et al. (2020) demonstrated in glioma cells that circPOSTN (has_circ_0030018), as a ceRNA, is involved in the stimulation of cell growth and aerobic glycolysis and inhibition of apoptosis by upregulating the mRNA TPX2 (targeting protein for Xenopus kinesin-like protein 2) through sponging miR-361-5p (Figure 3 and Table 2). CircPOSTN (has_circ_0030018), located at chr13:38136718–38161065 (2,656 nucleotides), was screened by high-throughput circRNA microarray to be upregulated in glioma tissues compared with normal tissues. A high level of circPOSTN was significantly associated with larger tumor size, higher WHO grades, and shorter overall survival (Yang Y. et al., 2019). In the study of Long et al. (2020), the effect of circPOSTN on apoptosis, proliferation, and aerobic glycolysis is mitigated by silencing miR-361-5p. miR-361-5p is a tumor suppressor in multiple types of cancers, including prostate cancer (Liu et al., 2014), cutaneous squamous cell carcinoma (Kanitz et al., 2012), hepatocellular carcinoma (Sun et al., 2016a), non-small cell lung cancer (Chen et al., 2016), and breast cancer (Cao et al., 2016). Zhang et al. (2017) reported that miR-361-5p inhibits the migration, invasion, and epithelial–mesenchymal transition of glioma cells via regulating the Twist1/Bmi-1 signaling axis. Whether Twist1 is targeted by circPOSTN remains unknown, while Long et al. (2020) found that TPX2 acts as a downstream target of miR-361-5p/circPOSTN in glioma cells. Depletion of circPOSTN or TPX2 significantly suppresses cell proliferation and aerobic glycolysis while promoting the apoptosis of glioma cells (Long et al., 2020). TPX2 is a cell cycle-regulated nuclear protein that functions in proliferation and mitotic spindle assembly (Heidebrecht et al., 1997; Kufer et al., 2002). As an oncogene, TPX2 is involved in multiple cancers, including gastric cancer (Tomii et al., 2017), colon cancer (Wei et al., 2013), lung squamous cell carcinoma (Ma et al., 2006), pancreatic cancer (Ludwig et al., 2017), and prostate cancer (Zou et al., 2018). In glioma cells, TPX2 promotes cell proliferation and invasion by activating the AKT signaling pathway (Gu et al., 2016). On the other hand, Yang Y. et al. (2019) showed that CircPOSTN promotes cell growth and invasion by sponging miR-1205, yet the targets of miR-1205 have not been explored. Therefore, these results illustrate that CircPOSTN play a crucial role in the progression and invasion of gliomas as a miRNA sponger and may be a useful new prognostic biomarker and therapeutic target for gliomas.
Hsa_circ_0082374 was screened by Peng et al. (2019) in a circRNA microarray analysis of glioma and matched normal brain tissues as the most up-regulated one among the top 20 up-regulated circRNAs. It locates at chr7:129948146–129964020 and is named circCPA4 as it was assumed to be derived from carboxypeptidaseA4 (CPA4) according to the human reference genome (GRCh47/hg19). A high level of circCPA4 correlates with a poor prognosis of glioma, and the knockdown of it impedes cell proliferation and invasion in glioma. Mechanically, circCPA4 acts as a ceRNA and sponges miR-let-7 to derepress the expression of CPA4 (Peng et al., 2019). CPA4 is a member of the metallocarboxypeptidase family and may be involved in the regulation of peptide hormone activity and hormone-regulated cell proliferation and differentiation (Huang et al., 1999; Tanco et al., 2010). The expression of CPA4 is elevated in multiple types of cancer tissues of patients, such as gastric cancer (Sun et al., 2016b), pancreatic cancer (Sun et al., 2015), breast cancer (Handa et al., 2019), lung cancer (Sun et al., 2016c), and esophageal squamous cell carcinoma (Sun L. et al., 2017), and can be used as a potential diagnostic and prognostic biomarker as well as a therapeutic target.
Zhang Y. et al. (2020) further explored the circCPA4 function in glioma cells and found that suppression of the circRNA inhibits tumor cell proliferation, migration, and invasion while promoting cell apoptosis and radiosensitivity in vitro and repressing tumor growth in vivo. They pointed out that a higher expression of circCPA4 is positively correlated with tumor size, WHO grade, and poor prognosis in patients. Differently from Peng et al. (2019) in the mechanism, Zhang Y. et al. (2020) revealed that circCPA4 sponges MEF2D (myocyte enhancer factor 2D)-targeting miR-760 to promote glioma progress. Knockdown of miR-760 can reverse the antitumor effects mediated by the suppression of circCPA4. MiR-760 is a well-identified tumor-suppressive miRNA that functions in many types of cancers, including hepatocellular carcinoma (Tian et al., 2018), breast cancer (Han et al., 2016), and non-small cell lung cancer (Zhu et al., 2019), via regulating the malignant properties of tumor, such as cell proliferation, apoptosis, migration, and drug resistance. Then, Zhang Y. et al. (2020) confirmed that MEF2D is targeted by miR-760 glioma cells. MEF2D is a transcription factor of the myocyte-specific enhancer factor 2 (MEF2) family involved in the regulation of the differentiation and development of muscle and neuronal cells (McKinsey et al., 2002). Interestingly, miR-760 and MEF2D are also involved in lncRNA LOC730100- and lncRNA DLEU1-mediated ceRNETs in glioma cells, respectively (Feng et al., 2019; Li Q. et al., 2019), implying the potential crosstalk between the circCPA4 and the lncRNAs. These results indicate the importance and complexity of circCPA4-mediated ceRNETs in gliomagenesis, providing potential biomarkers and targets for glioma treatment.
Circular RNA circ_0001946 (also known as CDR1as and CiRS-7) that derives from chrX:139865339–139866824 is involved in the progression of multiple cancer types, such as esophageal squamous cell cancer (Fan et al., 2019), colorectal cancer (Deng et al., 2020), lung adenocarcinoma (Yao et al., 2019), and glioblastoma (GBM) (Li and Diao, 2019). Li and Diao (2019) revealed that circ_0001946 suppresses GBM progression by activating the expression of CDR1 through sponging miR-671-5p. Circ_0001946 and its competing mRNA CDR1 can inhibit the proliferation, migration, and invasion and promote the apoptosis of GBM cells, while miR-671-5p has the opposite effect. Microarray analyses showed that circ_0001946 and CDR1 were down-regulated in GBM, while miR-671-5 was up-regulated. The genomic region containing miR-671-5p gene is frequently amplified in GBM (Barbagallo et al., 2016), and its regulatory role on circ_0001946 and CDR1 expression was proven by Hansen et al. (2011) earlier using HEK293 cells derived from human embryonic kidney cells. CDR1 (cerebellar degeneration-related autoantigen 1) is encoded by the CDR34 gene and is required for neuronal–glial functions. Inhibition of CDR1 expression leads to the loss of differentiation of neural cells and neoplastic transformation (Chen et al., 1990; Satoh and Yamamura, 2004). These results suggest that stimulating the circ_0001946/miR-671-5p/CDR1 axis may be a potential therapeutic strategy for GBM treatment.
CircMMP9 (hsa_circ_0001162) was screened as the circRNA with the greatest differential expression in the GBM tissues compared with the adjacent normal brain tissues in a microarray analysis performed by Wang et al. (2018). It is derived from exons 12 and 13 of MMP9 (matrix metalloproteinase-9), with 328 nucleotides in length. Overexpression of circMMP9 promotes the proliferation, migration, and invasion of GBM cells through sponging miR-124 (Wang et al., 2018). Thereafter, the oncogenic effect of circMMP9 was demonstrated in osteosarcoma (Pan et al., 2019) and oral squamous cell carcinoma (Xia et al., 2019). Cyclin-dependent kinase 4 (CDK4) and aurora kinase A (AURKA) are two downstream targets of miR-124/circMMP9 in GBM cells (Wang et al., 2018). Furthermore, Wang et al. (2018) found that eukaryotic initiation factor 4A3 (EIF4A3) binds to the MMP9 mRNA transcript to induce circMMP9 cyclization, which improves the circMMP9 level in GBM. EIF4A3 is a component of the exon junction complex involved in exon splicing (Chan et al., 2004). The expression of EIF4A3 shows prognostic significance in Chinese Glioma Genome Atlas but not The Cancer Genome Atlas (TCGA) database, which may be caused by the difference in sample size and ethnicity between the two data sets (Wei et al., 2020).
circPITX1 (hsa-circ-0074026) is another circRNA found to be up-regulated in GBM tissues compared with the noncancerous controls in the microarray analysis performed by Wang et al. (2018). It locates in chr5:134363423–134365011, with 2,383 bp in length. Recently, four groups reported different circPITX1-mediated ceRNETs in glioma cells, i.e., circPITX1/miR-518a-5p/IL17RD (Zhan et al., 2019), circPITX1/miR-1304/ERBB4 (Chen M. et al., 2020), circPITX1/miR-379-5p/mitogen-activated protein kinase 2 (MAP3K2) (Lv et al., 2019), and circPITX1/miR-329-3p/NIMA-related kinase 2 (NEK2) (Guan et al., 2020).
Zhan et al. (2019) further confirmed that circPITX1 is upregulated in cancerous tissues of 52 patients and four glioma cell lines, which is correlated with the patient’s tumor size and WHO grade. Through gain- and loss-of-function assays, they demonstrated that circPITX1 can promote the growth, migration, invasion, and survival of glioma cells. Mechanically, Zhan et al. (2019) proposed that circPITX1 promotes IL17RD (interleukin 17 receptor D) expression by sponging miR-518a-5p. miR-518a-5p also plays a tumor-suppressive role in colorectal cancer (Rubie et al., 2014), diffuse large B cell lymphoma (Huang et al., 2021), and gastrointestinal stromal tumor (Shi Y. et al., 2016) but an oncogenic role in ovarian cancer (Zhang N. et al., 2020). IL17RD can interact with the IL-17 receptor and mediates IL-17 signaling (Rong et al., 2009). The oncogenic role of IL17RD has been shown in colon cancer (Pekow et al., 2017) and colorectal cancer (Yu et al., 2019).
Similar to the study of Zhan et al. (2019); Chen M. et al. (2020) observed a clinical significance of circPITX1 in larger tumor size and higher WHO grade of patients and an inhibitory effect of circPITX1 knockdown on the proliferation, migration, invasion, and survival of glioma cells. Mechanically, they proposed that circPITX1 regulates ERBB4 expression to promote glioma progression by sponging miR-1304. ERBB4 (HER4) belongs to the epidermal growth factor (EGF)/ERBB family of receptor tyrosine kinases, which also includes the EGF receptor (EGFR/HER1/ERBB1), ERBB2 (HER2/Neu), and ERBB3 (HER3) (Qiu et al., 2008). The abnormal expression of each ERBB is associated with many human cancers (Hynes and Lane, 2005).
Lv et al. (2019) performed circPITX1 knockdown experiments and observed reduced proliferation and increased apoptosis of GBM cells. Mechanically, they proposed that circPITX1 promotes GBM progression by sponging miR-379-5p to increase the expression of MAP3K2 (Lv et al., 2019). The effects of circPITX1 knockdown on the proliferation and apoptosis of GBM cells can be rescued partly by upregulating MAP3K2. MAP3K2 is a member of the serine/threonine protein kinase family (Mazur et al., 2014). It preferentially activates other kinases of the MAP kinase signaling pathway and is frequently overexpressed in multiple human cancers including non-small cell lung cancer (Yu et al., 2015), hepatocellular carcinoma (Shi et al., 2020), cutaneous melanoma (Chen et al., 2019), and breast cancer (Wu et al., 2017).
Guan et al. (2020) reported that the down-regulation of circPITX1 leads to reduced viability, glycolysis, colony formation, and radioresistance of glioma cells in vitro and suppresses tumor growth in vivo. Mechanically, Guan et al. (2020) proposed that circPITX1 promotes NEK2 expression by sponging miR-329-3p. Depletion of miR-329-3p reverses the inhibitory effects of circPITX1 knockdown on glycolysis and radioresistance. It has been shown that miR-329-3p plays a tumor-suppressive role in multiple cancers, including non-small cell lung cancer, osteosarcoma, cervical cancer, and hepatocellular carcinoma, through regulating the proliferation and migration of tumor cells (Chang et al., 2017). Furthermore, Guan et al. (2020) demonstrate that NEK2 acts downstream of miR-329-3p/circPITX1 to affect the glycolysis and radiosensitivity of glioma cells. NEK2 is a conserved centrosome kinase of the NIMA-related kinase family, with abnormal expression in a wide variety of human cancers (Hayward and Fry, 2006). NEK2 is widely upregulated in gliomas and associated with WHO grades, proliferation, and prognosis in malignant gliomas (Liu et al., 2017). An earlier study by Ye et al. (2018) showed that NEK2 is overexpressed in the glioma tissues and is targeted by miR-128 involved in regulating the apoptosis of glioma cells.
Collectively, these studies solidly confirmed the crucial roles of circPITX1-mediated ceRNETs in the regulation of multiple hallmarks of gliomas, including proliferation, migration, invasion, apoptosis, and treatment resistance. Considering that circPITX1 knockdown or overexpression has no side effect on the cell proliferation and apoptosis of normal human astrocytes (Chen M. et al., 2020), targeting circPITX1 may be a valuable and promising strategy for glioma treatment.
The finding of pseudogene-mediated ceRNETs laid the foundation for the establishment of the ceRNA hypothesis (Poliseno et al., 2010; Salmena et al., 2011), yet the description of this type of ceRNETs in glioma is less than that of lncRNA- and circRNA-mediated ceRNETs. PTEN pseudogene-1 (PTENP1), the pseudogene of PTEN, acts through ceRNA mechanism to inhibit cancer progression in prostate cancer (Poliseno et al., 2010) and breast cancer (Gao et al., 2019). Although PTENP1 is also found to be involved in regulating the proliferation and invasion of glioma cells, whether its anti-tumor effect is mediated by ceRNA mechanism has not been reported (Hu et al., 2019).
Three annexin A2 pseudogenes, including ANXA2P1, ANXA2P2, and ANXA2P3, are significantly upregulated, along with their parent gene annexin A2 (ANXA2), which is correlated with poor survival outcome of glioma patients (Li et al., 2017b). Whether the co-expression of the pseudogenes (ANXA2P1, ANXA2P2, and ANXA2P) and parent gene (ANXA2) is caused by shared miRNA(s) or ceRNET(s) remain unknown. Yu et al. (2019) identified five pseudogenes correlating with glioma survival from the TCGA dataset and established a complex ceRNET consisting of three pseudogenes (ANXA2P2, EEF1A1P9, and FER1L4), 72 microRNAs, and 322 targeted genes (Wang Y. et al., 2019). However, the computational ceRNET and each ceRNA pair need further experimental validation. Du et al. (2020) reported an experimentally validated ceRNET involving ANXA2P2, in which ANXA2P2 functions as a ceRNA to regulate the expression of lactate dehydrogenase A (LDHA) by sponging miR-9 in GBM (Du et al., 2020). The ANXA2P2/miR-9/LDHA ceRNET regulates glucose metabolism, proliferation, and apoptosis of GBM (Du et al., 2020). LDHA catalyzes the final step of aerobic glycolysis and is abnormally expressed in many human cancers (Li et al., 2017a).
Wang S. et al. (2020) reported a new pseudogene-mediated ceRNET in glioma, protein disulfide isomerase family A member 3 pseudogene 1 (PDIA3P1)/miR-124-3p/RELA. PDIA3P1 is a 2,099-bp fragment mapped to chromosome 1q21.1. High PDIA3P1 expression is correlated with EMT, extracellular matrix disassembly, and angiogenesis and can promote the migration and invasion of glioma cells (Wang S. et al., 2020). Mechanically, PDIA3P1 sponges miR-124-3p to upregulate RELA expression and, in turn, activates the downstream NF-κB pathway, which promotes a highly invasive mesenchymal (MES) transition of glioma cells (Wang S. et al., 2020). RELA (v-rel avian reticuloendotheliosis viral oncogene homolog A) gene encodes the major component of the NF-κB complex (Hashimoto et al., 2011). Intriguingly, hypoxia inducible factor 1 (HIF1) upregulates the transcription of PDIA3P1 by directly binding its promoter, linking hypoxia to MES transition (Wang S. et al., 2020). In addition, the ceRNA function of PDIA3P1 has also been reported in oral squamous cell carcinoma (Sun C.C. et al., 2017) as well as its oncogenic effect in hepatocellular carcinoma (Kong et al., 2017). These results underscore the importance of PDIA3P1-mediated ceRNET in cancer progression and suggest it as a promising prognostic biomarker and therapeutic target in glioma treatment.
Despite the improved surgical resection, radiotherapy, and chemotherapy, the prognosis for patients with high-grade gliomas is still poor. Understanding the molecular mechanisms underlying gliomagenesis is urgently needed to “break the ice.” Formally proposed in 2011 (Salmena et al., 2011), the ceRNA hypothesis opens up new avenues for basic cancer research, including glioma (Di Palo et al., 2020; Liu Z. et al., 2020; Wang S. et al., 2020). Research in the past decade have shed light on the dysregulated ceRNETs consisting of coding and non-coding RNAs (miRNAs, lncRNAs, circRNAs, and transcribed pseudogenes) in gliomas. As we have discussed here, various hallmarks of gliomas, such as cell proliferation, growth, invasion, EMT, apoptosis, angiogenesis, chemo-resistance, and radio-resistance, are associated with dysregulated ceRNETs. Illustration of the different nature of the RNA interaction within the ceRNETs will provide new insights into the initiation and progression of gliomas and therefore novel biomarkers for the diagnosis, prognosis, and targets for glioma treatment.
LC wrote the manuscript. RL drew the cartoon figures. WL collected the articles. HC and QL provided the idea and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (81872071), the Natural Science Foundation of Chongqing (cstc2019jcyj-zdxmX0033), the Fundamental Research Funds for the Central Universities (XYDS201912), and the Chongqing University Innovation Team Building Program (CXTDX201601010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdollahzadeh, R., Daraei, A., Mansoori, Y., Sepahvand, M., Amoli, M. M., and Tavakkoly-Bazzaz, J. (2019). Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J. Cell. Physiol. 234, 10080–10100. doi: 10.1002/jcp.27941
Ala, U., Karreth, F. A., Bosia, C., Pagnani, A., Taulli, R., Léopold, V., et al. (2013). Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl. Acad. Sci. U.S.A 110, 7154–7159. doi: 10.1073/pnas.1222509110
Algayadh, I. G., Dronamraju, V., and Sylvester, P. W. (2016). Role of Rac1/WAVE2 signaling in mediating the inhibitory effects of γ-tocotrienol on mammary cancer cell migration and invasion. Biol. Pharmaceutical Bull. 39, 1974–1982. doi: 10.1248/bpb.b16-00461
An, Y., Furber, K. L., and Ji, S. (2017). Pseudogenes regulate parental gene expression via ceRNA network. J. Cell. Mol. Med. 21, 185–192. doi: 10.1111/jcmm.12952
Bahn, J. H., Zhang, Q., Li, F., Chan, T.-M., Lin, X., Kim, Y., et al. (2015). The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 61, 221–230. doi: 10.1373/clinchem.2014.230433
Ballestar, E., and Esteller, M. (2008). Chapter 9 epigenetic gene regulation in cancer. Adv. Genet. 61, 247–267. doi: 10.1016/s0065-2660(07)00009-0
Barbagallo, D., Condorelli, A., Ragusa, M., Salito, L., Sammito, M., Banelli, B., et al. (2016). Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget 7, 4746–4759. doi: 10.18632/oncotarget.6621
Bhat, K. P. L., Salazar, K. L., Balasubramaniyan, V., Wani, K., Heathcock, L., Hollingsworth, F., et al. (2011). The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 25, 2594–2609. doi: 10.1101/gad.176800.111
Bian, A., Wang, Y., Liu, J., Wang, X., Liu, D., Jiang, J., et al. (2018). Circular RNA complement factor H (CFH) promotes glioma progression by sponging miR-149 and regulating AKT1. Med. Sci. Monit. 24, 5704–5712. doi: 10.12659/msm.910180
Bosia, C., Pagnani, A., and Zecchina, R. (2013). Modelling competing endogenous RNA networks. PLoS One 8:e66609. doi: 10.1371/journal.pone.0066609
Buchler, N. E., and Louis, M. (2008). Molecular titration and ultrasensitivity in regulatory networks. J. Mol. Biol. 384, 1106–1119. doi: 10.1016/j.jmb.2008.09.079
Cai, J., Zhang, J., Wu, P., Yang, W., Ye, Q., Chen, Q., et al. (2018). Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-κB pathway. J. Neuro Oncol. 140, 225–236. doi: 10.1007/s11060-018-2951-0
Cao, Z.-G., Huang, Y.-N., Yao, L., Liu, Y.-R., Hu, X., Hou, Y.-F., et al. (2016). Positive expression of miR-361-5p indicates better prognosis for breast cancer patients. J. Thorac. Dis. 8, 1772–1779. doi: 10.21037/jtd.2016.06.29
Chan, C. C., Dostie, J., Diem, M. D., Feng, W., Mann, M., Rappsilber, J., et al. (2004). eIF4A3 is a novel component of the exon junction complex. RNA 10, 200–209. doi: 10.1261/rna.5230104
Chang, Y. H., Yin, F., Fan, G. F., and Zhao, M. (2017). Down-regulation of miR-329-3p is associated with worse prognosis in patients with cervical cancer. Eur. Rev. Med. Pharmacol. Sci. 21, 4045–4049.
Chen, C., Deng, L., Nie, D.-K., Jia, F., Fu, L.-S., Wan, Z.-Q., et al. (2020). Circular RNA Pleiotrophin promotes carcinogenesis in glioma via regulation of microRNA-122/SRY-box transcription factor 6 axis. Eur. J. Cancer Prevent. 29, 165–173. doi: 10.1097/cej.0000000000000535
Chen, J., Liu, T., Wang, H., Wang, Z., Lv, Y., Zhao, Y., et al. (2020). Elevation in the expression of circ_0079586 predicts poor prognosis and accelerates progression in glioma via interactions with the miR-183-5p/MDM4 signaling pathway. Onco. Targets Ther. 13, 5135–5143. doi: 10.2147/ott.s234758
Chen, M., Liu, X., Xie, P., Wang, P., Liu, M., Zhan, Y., et al. (2020). Circular RNA circ_0074026 indicates unfavorable prognosis for patients with glioma and facilitates oncogenesis of tumor cells by targeting miR-1304 to modulate ERBB4 expression. J. Cell. Physiol. 235, 4688–4697. doi: 10.1002/jcp.29347
Chen, W., Wang, J., Liu, S., Wang, S., Cheng, Y., Zhou, W., et al. (2016). MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J. Exp. Clin. Cancer Res. 35:76.
Chen, X., Gao, J., Yu, Y., Zhao, Z., and Pan, Y. (2019). LncRNA FOXD3-AS1 promotes proliferation, invasion and migration of cutaneous malignant melanoma via regulating miR-325/MAP3K2. Biomed. Pharmacother. 120:109438. doi: 10.1016/j.biopha.2019.109438
Chen, X., Li, D., Gao, Y., Tang, W., Iw, L., Cao, Y., et al. (2018). Long intergenic noncoding RNA 00152 promotes glioma cell proliferation and invasion by interacting with MiR-16. Cell. Physiol. Biochem. 46, 1055–1064. doi: 10.1159/000488836
Chen, Y. T., Rettig, W. J., Yenamandra, A. K., Kozak, C. A., Chaganti, R. S., Posner, J. B., et al. (1990). Cerebellar degeneration-related antigen: a highly conserved neuroectodermal marker mapped to chromosomes X in human and mouse. Proc. Natl. Acad. Sci. 87, 3077–3081. doi: 10.1073/pnas.87.8.3077
Chen, Z., and Duan, X. (2018). hsa_circ_0000177-miR-638-FZD7-Wnt signaling cascade contributes to the malignant behaviors in glioma. DNA Cell Biol. 37, 791–797. doi: 10.1089/dna.2018.4294
Cheng, Z., Luo, C., and Guo, Z. (2020). LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell. Biochem. 121, 2170–2183. doi: 10.1002/jcb.29440
Chow, L. M., Endersby, R., Zhu, X., Rankin, S., Qu, C., Zhang, J., et al. (2011). Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19, 305–316. doi: 10.1016/j.ccr.2011.01.039
Coso, O. A., Chiariello, M., Yu, J.-C., Teramoto, H., Crespo, P., Xu, N., et al. (1995). The small GTP-binding proteins Rac1 and Cdc42regulate the activity of the JNK/SAPK signaling pathway. Cell 81, 1137–1146. doi: 10.1016/s0092-8674(05)80018-2
Cui, B., Li, B., Liu, Q., and Cui, Y. (2017). lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J. Cell. Biochem. 118, 4548–4557. doi: 10.1002/jcb.26116
Deng, Z., Li, X., Wang, H., Geng, Y., Cai, Y., Tang, Y., et al. (2020). Dysregulation of CircRNA_0001946 contributes to the proliferation and metastasis of colorectal cancer cells by targeting microRNA-135a-5p. Front. Genet. 11:357. doi: 10.3389/fgene.2020.00357
D’Errico, I., Gadaleta, G., and Saccone, C. (2004). Pseudogenes in metazoa: origin and features. Br. Funct. Geno. Proteomic. 3, 157–167. doi: 10.1093/bfgp/3.2.157
Deveson, I. W., Hardwick, S. A., Mercer, T. R., and Mattick, J. S. (2017). The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 33, 464–478. doi: 10.1016/j.tig.2017.04.004
Di Palo, A., Siniscalchi, C., Mosca, N., Russo, A., and Potenza, N. (2020). A novel ceRNA regulatory network involving the long non-coding antisense RNA SPACA6P-AS, miR-125a and its mRNA targets in hepatocarcinoma cells. Int. J. Mol. Sci. 21:5068. doi: 10.3390/ijms21145068
Ding, X., Yang, L., Geng, X., Zou, Y., Wang, Z., Li, Y., et al. (2020). CircRNAs as potential biomarkers for the clinicopathology and prognosis of glioma patients: a meta-analysis. BMC Cancer 20:1005. doi: 10.1186/s12885-020-07446-4
Dong, Z., and Cui, H. (2020). The emerging roles of RNA modifications in glioblastoma. Cancers 12:736. doi: 10.3390/cancers12030736
Du, P., Liao, Y., Zhao, H., Zhang, J., Muyiti, K., and Mu, K. (2020). ANXA2P2/miR-9/LDHA axis regulates Warburg effect and affects glioblastoma proliferation and apoptosis. Cell. Signal. 74:109718. doi: 10.1016/j.cellsig.2020.109718
Duan, X., Liu, D., Wang, Y., and Chen, Z. (2018). Circular RNA hsa_circ_0074362 promotes glioma cell proliferation, migration, and invasion by attenuating the inhibition of miR-1236-3p on HOXB7 expression. DNA Cell Biol. 37, 917–924. doi: 10.1089/dna.2018.4311
Ebrahimpour, A., Sarfi, M., Rezatabar, S., and Tehrani, S. S. (2021). Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma. Mol. Cell. Biochem. Epub ahead of print
Fan, L., Cao, Q., Liu, J., Zhang, J., and Li, B. (2019). Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol. Cancer 18:16.
Feng, L., He, M., Rao, M., Diao, J., and Zhu, Y. (2019). Long noncoding RNA DLEU1 aggravates glioma progression via the miR-421/MEF2D axis. Onco. Targets Ther. 12, 5405–5414. doi: 10.2147/ott.s207542
Figliuzzi, M., Marinari, E., and De Martino, A. (2013). MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys. J. 104, 1203–1213. doi: 10.1016/j.bpj.2013.01.012
Fox, A. H., Nakagawa, S., Hirose, T., and Bond, C. S. (2018). Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 43, 124–135. doi: 10.1016/j.tibs.2017.12.001
Friedman, R. C., Farh, K. K.-H., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Geno. Res. 19, 92–105. doi: 10.1101/gr.082701.108
Gao, F., Du, Y., Zhang, Y., Ren, D., Xu, J., and Chen, D. (2020). Circ-EZH2 knockdown reverses DDAH1 and CBX3-mediated cell growth and invasion in glioma through miR-1265 sponge activity. Gene 726:144196. doi: 10.1016/j.gene.2019.144196
Gao, X., Qin, T., Mao, J., Zhang, J., Fan, S., Lu, Y., et al. (2019). PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J. Exp. Clin. Cancer Res. CR 38:256.
Greco, S., Gaetano, C., and Martelli, F. (2019). Long noncoding competing endogenous RNA networks in age-associated cardiovascular diseases. Int. J. Mol. Sci. 20:3079. doi: 10.3390/ijms20123079
Gu, J. J., Zhang, J. H., Chen, H. J., and Wang, S. S. (2016). TPX2 promotes glioma cell proliferation and invasion via activation of the AKT signaling pathway. Oncol. Lett. 12, 5015–5022. doi: 10.3892/ol.2016.5371
Guan, Y., Cao, Z., Du, J., Liu, T., and Wang, T. (2020). Circular RNA circPITX1 knockdown inhibits glycolysis to enhance radiosensitivity of glioma cells by miR-329-3p/NEK2 axis. Cancer Cell Int. 20:80.
Hamisch, C., Ruge, M., Kellermann, S., Kohl, A.-C., Duval, I., Goldbrunner, R., et al. (2017). Impact of treatment on survival of patients with secondary glioblastoma. J. Neuro Oncol. 133, 309–313. doi: 10.1007/s11060-017-2415-y
Han, M. l., Wang, F., Gu, Y. t., Pei, X.-h, Ge, X., and Guo, G.-c, et al. (2016). MicroR-760 suppresses cancer stem cell subpopulation and breast cancer cell proliferation and metastasis: By down-regulating NANOG. Biomed. Pharmacother. 80, 304–310. doi: 10.1016/j.biopha.2016.03.024
Handa, T., Katayama, A., Yokobori, T., Yamane, A., Fujii, T., Obayashi, S., et al. (2019). Carboxypeptidase A4 accumulation is associated with an aggressive phenotype and poor prognosis in triple-negative breast cancer. Int. J. Oncol. 54, 833–844.
Hangauer, M. J., Vaughn, I. W., and McManus, M. T. (2013). Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9:e1003569. doi: 10.1371/journal.pgen.1003569
Hansen, T. B., Wiklund, E. D., Bramsen, J. B., Villadsen, S. B., Statham, A. L., Clark, S. J., et al. (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422. doi: 10.1038/emboj.2011.359
Hashimoto, R., Ohi, K., Yasuda, Y., Fukumoto, M., Yamamori, H., Takahashi, H., et al. (2011). Variants of the RELA gene are associated with schizophrenia and their startle responses. Neuropsychopharmacology 36, 1921–1931. doi: 10.1038/npp.2011.78
Hayward, D. G., and Fry, A. M. (2006). Nek2 kinase in chromosome instability and cancer. Cancer Lett. 237, 155–166. doi: 10.1016/j.canlet.2005.06.017
He, Q., Zhao, L., Liu, X., Zheng, J., Liu, Y., Liu, L., et al. (2019). MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4 expression change. J. Exp. Clin. Cancer Res. CR 38:9.
He, Q., Zhao, L., Liu, Y., Liu, X., Zheng, J., Yu, H., et al. (2018). circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol. Ther. Nucleic Acids 10, 331–348. doi: 10.1016/j.omtn.2017.12.014
He, Z., Long, J., Yang, C., Gong, B., Cheng, M., Wang, Q., et al. (2020). LncRNA DGCR5 plays a tumor-suppressive role in glioma via the miR-21/Smad7 and miR-23a/PTEN axes. Aging (Albany NY) 12, 20285–20307. doi: 10.18632/aging.103800
He, Z., Ruan, X., Liu, X., Zheng, J., Liu, Y., Liu, L., et al. (2019). FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J. Exp. Clin. Cancer Res. CR 38:65.
Heidebrecht, H. J., Buck, F., Steinmann, J., Sprenger, R., Wacker, H. H., and Parwaresch, R. (1997). p100: a novel proliferation-associated nuclear protein specifically restricted to cell cycle phases S, G2, and M. Blood 90, 226–233. doi: 10.1182/blood.v90.1.226
Hu, S., Xu, L., Li, L., Luo, D., Zhao, H., Li, D., et al. (2019). Overexpression of lncRNA PTENP1 suppresses glioma cell proliferation and metastasis in vitro. Onco. Targets Ther. 12, 147–156. doi: 10.2147/ott.s182537
Hu, T., Wang, F., and Han, G. (2020). LncRNA PSMB8-AS1 acts as ceRNA of miR-22-3p to regulate DDIT4 expression in glioblastoma. Neurosci. Lett. 728:134896. doi: 10.1016/j.neulet.2020.134896
Huang, H., Reed, C. P., Zhang, J. S., Shridhar, V., Wang, L., and Smith, D. I. (1999). Carboxypeptidase A3 (CPA3): a novel gene highly induced by histone deacetylase inhibitors during differentiation of prostate epithelial cancer cells. Cancer Res. 59, 2981–2988.
Huang, Q., Zhang, F., Fu, H., and Shen, J. (2021). Epigenetic regulation of miR-518a-5p-CCR6 feedback loop promotes both proliferation and invasion in diffuse large B cell lymphoma. Epigenetics 16, 28–44. doi: 10.1080/15592294.2020.1786317
Huang, W., Shi, Y., Han, B., Wang, Q., Zhang, B., Qi, C., et al. (2020). LncRNA GAS5-AS1 inhibits glioma proliferation, migration, and invasion via miR−106b-5p/TUSC2 axis. Hum. Cell 33, 416–426. doi: 10.1007/s13577-020-00331-z
Hynes, N. E., and Lane, H. A. (2005). ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354. doi: 10.1038/nrc1609
Jemal, A., Siegel, R., Xu, J., and Ward, E. (2010). Cancer statistics, 2010. CA Cancer J. Clin. 60, 277–300. doi: 10.3322/caac.20073
Jin, C., Zhao, J., Zhang, Z. P., Wu, M., Li, J., Liu, B., et al. (2021). CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10. Mol. Oncol. 15, 596–622. doi: 10.1002/1878-0261.12830
Jin, P., Huang, Y., Zhu, P., Zou, Y., Shao, T., and Wang, O. (2018). CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem. Biophys. Res. Commun. 503, 1570–1574. doi: 10.1016/j.bbrc.2018.07.081
Jin, T., Liu, M., Liu, Y., Li, Y., Xu, Z., He, H., et al. (2020). Lcn2-derived circular RNA (hsa_circ_0088732) inhibits cell apoptosis and promotes EMT in Glioma via the miR-661/RAB3D Axis. Front. Oncol. 10:170. doi: 10.3389/fonc.2020.00170
Kanitz, A., Imig, J., Dziunycz, P. J., Primorac, A., Galgano, A., Hofbauer, G. F. L., et al. (2012). The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PLoS One 7:e49568. doi: 10.1371/journal.pone.0049568
Kiang, K. M.-Y., Zhang, X.-Q., and Leung, G. K.-K. (2015). Long non-coding RNAs: the key players in glioma pathogenesis. Cancers 7, 1406–1424. doi: 10.3390/cancers7030843
Kong, Y., Zhang, L., Huang, Y., He, T., Zhang, L., Zhao, X., et al. (2017). Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 407, 76–83. doi: 10.1016/j.canlet.2017.07.031
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691.
Kufer, T. A., Silljé, H. H., Körner, R., Gruss, O. J., Meraldi, P., and Nigg, E. A. (2002). Human TPX2 is required for targeting aurora-a kinase to the spindle. J. Cell Biol. 158, 617–623. doi: 10.1083/jcb.200204155
Lei, B., and Huang, Y. (2019). Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. 120, 6698–6708. doi: 10.1002/jcb.27966
Li, C., Liu, Y., Lv, Z., Zheng, H., Li, Z., Zhang, J., et al. (2021). Circular RNA circHECTD1 facilitates glioma progression by regulating the miR-296-3p/SLC10A7 axis. J. Cell. Physiol.
Li, F., Ma, K., Sun, M., and Shi, S. (2018). Identification of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am. J. Transl. Res. 10, 1373–1386.
Li, G., Huang, M., Cai, Y., Yang, Y., Sun, X., and Ke, Y. (2019). Circ-U2AF1 promotes human glioma via derepressing neuro-oncological ventral antigen 2 by sponging hsa-miR-7-5p. 234, 9144–9155. doi: 10.1002/jcp.27591
Li, G., Yang, H., Han, K., Zhu, D., Lun, P., and Zhao, Y. (2018). A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem. Biophys. Res. Commun. 498, 254–261. doi: 10.1016/j.bbrc.2018.01.076
Li, G.-F., Li, L., Yao, Z.-Q., and Zhuang, S.-J. (2018). Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem. Biophys. Res. Commun. 499, 765–771. doi: 10.1016/j.bbrc.2018.03.219
Li, L., Kang, L., Zhao, W., Feng, Y., Liu, W., Wang, T., et al. (2017a). miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 400, 89–98. doi: 10.1016/j.canlet.2017.04.034
Li, Q., Lu, J., Xia, J., Wen, M., and Wang, C. (2019). Long non-coding RNA LOC730100 enhances proliferation and invasion of glioma cells through competitively sponging miR-760 from FOXA1 mRNA. Biochem. Biophys. Res. Commun. 512, 558–563. doi: 10.1016/j.bbrc.2019.03.124
Li, S., Zou, H., Shao, Y.-Y., Mei, Y., Cheng, Y., Hu, D.-L., et al. (2017b). “Pseudogenes of annexin A2, novel prognosis biomarkers for diffuse gliomas,” Oncotarget 8, 106962–106975. doi: 10.18632/oncotarget.22197
Li, X., and Diao, H. (2019). Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J. Cell. Physiol. 234, 13807–13819. doi: 10.1002/jcp.28061
Li, X., Yang, L., and Chen, L.-L. (2018). The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71, 428–442. doi: 10.1016/j.molcel.2018.06.034
Li, Z., Xu, C., Ding, B., Gao, M., Wei, X., and Ji, N. (2017c). Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J. Neurooncol. 134, 19–28. doi: 10.1007/s11060-017-2498-5
Liao, Y., Shen, L., Zhao, H., Liu, Q., Fu, J., Guo, Y., et al. (2017). LncRNA CASC2 interacts with miR-181a to modulate glioma growth and resistance to TMZ through PTEN pathway. J. Cell. Biochem. 118, 1889–1899. doi: 10.1002/jcb.25910
Liu, D., Tao, T., Xu, B., Chen, S., Liu, C., Zhang, L., et al. (2014). MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6). Biochem. Biophys. Res. Commun. 445, 151–156. doi: 10.1016/j.bbrc.2014.01.140
Liu, H., Liu, B., Hou, X., Pang, B., Guo, P., Jiang, W., et al. (2017). Overexpression of NIMA-related kinase 2 is associated with poor prognoses in malignant glioma. J. Neuro Oncol. 132, 409–417. doi: 10.1007/s11060-017-2401-4
Liu, N. Z., Li, T., Liu, C. M., Liu, F. R., and Wang, Y. X. (2020). Hsa_circ_0000337 promotes proliferation, migration and invasion in glioma by competitively binding miRNA-942-5p and thus upregulates MAT2A. Eur. Rev. Med. Pharmacol. Sci. 24, 12251–12257.
Liu, Q., Sun, S., Yu, W., Jiang, J., Zhuo, F., Qiu, G., et al. (2015). Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells. J. Neurooncol. 122, 283–292. doi: 10.1007/s11060-015-1718-0
Liu, R., Shi, P., Wang, Z., Yuan, C., and Cui, H. (2021). Molecular mechanisms of MYCN dysregulation in cancers. Front. Oncol. 10:625332. doi: 10.3389/fonc.2020.625332
Liu, Z., Guo, S., Sun, H., Bai, Y., Song, Z., and Liu, X. (2020). Circular RNA CircHIPK3 elevates CCND2 expression and promotes cell proliferation and invasion through miR-124 in Glioma. Front. Genet. 11:1013. doi: 10.3389/fgene.2020.01013
Long, N., Chu, L., Jia, J., Peng, S., Gao, Y., Yang, H., et al. (2020). CircPOSTN/miR-361-5p/TPX2 axis regulates cell growth, apoptosis and aerobic glycolysis in glioma cells. Cancer Cell Int. 20:374.
Louis, D. N., Ohgaki, H., Wiestler, O. D., Cavenee, W. K., Burger, P. C., Jouvet, A., et al. (2007). The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109. doi: 10.1007/978-94-007-1399-4_10
Louis, D. N., Perry, A., Reifenberger, G., von Deimling, A., Figarella-Branger, D., Cavenee, W. K., et al. (2016). The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820. doi: 10.1007/s00401-016-1545-1
Ludwig, R., Teran, F. J., Teichgraeber, U., and Hilger, I. (2017). Nanoparticle-based hyperthermia distinctly impacts production of ROS, expression of Ki-67, TOP2A, and TPX2, and induction of apoptosis in pancreatic cancer. Int. J. Nanomed. 12, 1009–1018. doi: 10.2147/ijn.s108577
Luo, C., Quan, Z., Zhong, B., Zhang, M., Zhou, B., Wang, S., et al. (2020). lncRNA XIST promotes glioma proliferation and metastasis through miR-133a/SOX4. Exp. Ther. Med. 19, 1641–1648.
Lv, X., Wang, M., Qiang, J., and Guo, S. (2019). Circular RNA circ-PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR-379–5p/MAP3K2 axis. Eur. J. Pharmacol. 863, 172643. doi: 10.1016/j.ejphar.2019.172643
Ma, Y., Lin, D., Sun, W., Xiao, T., Yuan, J., Han, N., et al. (2006). Expression of targeting protein for xklp2 associated with both malignant transformation of respiratory epithelium and progression of squamous cell lung cancer. Clin. Cancer Res. 12, 1121–1127. doi: 10.1158/1078-0432.ccr-05-1766
Manara, M. C., Terracciano, M., Mancarella, C., Sciandra, M., Guerzoni, C., Pasello, M., et al. (2016). CD99 triggering induces methuosis of Ewing sarcoma cells through IGF-1R/RAS/Rac1 signaling. Oncotarget 7, 79925–79942. doi: 10.18632/oncotarget.13160
Mazur, P. K., Reynoird, N., Khatri, P., Jansen, P. W. T. C., Wilkinson, A. W., Liu, S., et al. (2014). SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287. doi: 10.1038/nature13320
McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002). MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27, 40–47. doi: 10.1016/s0968-0004(01)02031-x
Memczak, S., Papavasileiou, P., Peters, O., and Rajewsky, N. (2015). Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10:e0141214. doi: 10.1371/journal.pone.0141214
Meng, Q., Li, S., Liu, Y., Zhang, S., Jin, J., Zhang, Y., et al. (2019). Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR-421/SP1/VEGFA Axis. Mol. Ther. Nucleic Acids 17, 669–677. doi: 10.1016/j.omtn.2019.06.022
Mu, M., Niu, W., Zhang, X., Hu, S., and Niu, C. (2020). LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene 39, 6879–6892. doi: 10.1038/s41388-020-01466-x
Mullokandov, G., Baccarini, A., Ruzo, A., Jayaprakash, A. D., Tung, N., Israelow, B., et al. (2012). High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 9, 840–846. doi: 10.1038/nmeth.2078
Mutalifu, N., Du, P., Zhang, J., Akbar, H., Yan, B., Alimu, S., et al. (2020). Circ_0000215 increases the expression of CXCR2 and promoted the progression of glioma cells by sponging miR-495-3p. Technol. Cancer Res. Treatment 19:1533033820957026.
Pan, G., Hu, T., Chen, X., and Zhang, C. (2019). Upregulation Of circMMP9 promotes osteosarcoma progression via targeting miR-1265/CHI3L1 Axis. Cancer Manag. Res. 11, 9225–9231. doi: 10.2147/cmar.s226264
Pei, B., Sisu, C., Frankish, A., Howald, C., Habegger, L., Mu, X. J., et al. (2012). The GENCODE pseudogene resource. Geno. Biol. 13:R51.
Pekow, J., Meckel, K., Dougherty, U., Huang, Y., Chen, X., Almoghrabi, A., et al. (2017). miR-193a-3p is a key tumor suppressor in ulcerative colitis–associated colon cancer and promotes carcinogenesis through upregulation of IL17RD. Clin. Cancer Res. 23, 5281–5291. doi: 10.1158/1078-0432.ccr-17-0171
Peng, H., Qin, C., Zhang, C., Su, J., Xiao, Q., Xiao, Y., et al. (2019). circCPA4 acts as a prognostic factor and regulates the proliferation and metastasis of glioma. J. Cell. Mol. Med. 23, 6658–6665. doi: 10.1111/jcmm.14541
Peng, Z., Liu, C., and Wu, M. (2018). New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer 17:61.
Pink, R. C., Wicks, K., Caley, D. P., Punch, E. K., Jacobs, L., and Francisco Carter, D. R. (2011). Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 17, 792–798. doi: 10.1261/rna.2658311
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., and Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi: 10.1038/nature09144
Qiu, C., Tarrant, M. K., Choi, S. H., Sathyamurthy, A., Bose, R., Banjade, S., et al. (2008). Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase. Structure 16, 460–467. doi: 10.1016/j.str.2007.12.016
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m6A enhances the phase separation potential of mRNA. Nature 571, 424–428. doi: 10.1038/s41586-019-1374-1
Rong, Z., Wang, A., Li, Z., Ren, Y., Cheng, L., Li, Y., et al. (2009). IL-17RD (Sef or IL-17RLM) interacts with IL-17 receptor and mediates IL-17 signaling. Cell Res. 19, 208–215. doi: 10.1038/cr.2008.320
Rubie, C., Kruse, B., Frick, V. O., Kölsch, K., Ghadjar, P., Wagner, M., et al. (2014). Chemokine receptor CCR6 expression is regulated by miR-518a-5p in colorectal cancer cells. J. Trans. Med. 12:48. doi: 10.1186/1479-5876-12-48
Rybak-Wolf, A., Stottmeister, C., Glažar, P., Jens, M., Pino, N., Giusti, S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885. doi: 10.1016/j.molcel.2015.03.027
Rynkeviciene, R., Simiene, J., Strainiene, E., Stankevicius, V., Usinskiene, J., Miseikyte Kaubriene, E., et al. (2019). Non-coding RNAs in Glioma. Cancers 11:17.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, and Pier, P. (2011). A ceRNA Hypothesis: the rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Sanchez-Mejias, A., and Tay, Y. (2015). Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J. Hematol. Oncol. 8:30.
Sang, J., Li, X., Lv, L., Zhang, C., Zhang, X., and Li, G. (2021). Circ-TOP2A acts as a ceRNA for miR-346 and contributes to glioma progression via the modulation of sushi domain-containing 2. Mol. Med. Rep. 23:1.
Satoh, J.-i, and Yamamura, T. (2004). Gene expression profile following stable expression of the cellular prion protein. Cell. Mol. Neurobiol. 24, 793–814. doi: 10.1007/s10571-004-6920-0
Shah, O. J., Wang, Z., and Hunter, T. (2004). Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14, 1650–1656. doi: 10.1016/j.cub.2004.08.026
Shen, J., Xiong, J., Shao, X., Cheng, H., Fang, X., Sun, Y., et al. (2020). Knockdown of the long noncoding RNA XIST suppresses glioma progression by upregulating miR-204-5p. J. Cancer. 11, 4550–4559. doi: 10.7150/jca.45676
Shi, X., Liu, T.-T., Yu, X.-N., Balakrishnan, A., Zhu, H.-R., Guo, H.-Y., et al. (2020). microRNA-93-5p promotes hepatocellular carcinoma progression via a microRNA-93-5p/MAP3K2/c-Jun positive feedback circuit. Oncogene 39, 5768–5781. doi: 10.1038/s41388-020-01401-0
Shi, X., Nie, F., Wang, Z., and Sun, M. (2016). Pseudogene-expressed RNAs: a new frontier in cancers. Tumor Biol. 37, 1471–1478. doi: 10.1007/s13277-015-4482-z
Shi, Y., Gao, X., Hu, Q., Li, X., Xu, J., Lu, S., et al. (2016). PIK3C2A is a gene-specific target of microRNA-518a-5p in imatinib mesylate-resistant gastrointestinal stromal tumor. Laborat. Investigat. 96, 652–660. doi: 10.1038/labinvest.2015.157
Shih, S. C., Prag, G., Francis, S. A., Sutanto, M. A., Hurley, J. H., and Hicke, L. (2003). A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. Embo. J. 22, 1273–1281. doi: 10.1093/emboj/cdg140
Singh, D., Febbo, P. G., Ross, K., Jackson, D. G., Manola, J., Ladd, C., et al. (2002). Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1, 203–209. doi: 10.1016/s1535-6108(02)00030-2
Song, X., Zhang, N., Han, P., Moon, B. S., Lai, R. K., Wang, K., et al. (2016). Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 44:e87. doi: 10.1093/nar/gkw075
Sumazin, P., Yang, X., Chiu, H.-S., Chung, W.-J., Iyer, A., Llobet-Navas, D., et al. (2011). An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147, 370–381. doi: 10.1016/j.cell.2011.09.041
Sun, C. C., Zhang, L., Li, G., Li, S. J., Chen, Z. L., Fu, Y. F., et al. (2017). The lncRNA PDIA3P interacts with miR-185-5p to modulate oral squamous cell carcinoma progression by targeting cyclin D2. Mol. Ther. Nucleic Acids 9, 100–110. doi: 10.1016/j.omtn.2017.08.015
Sun, J. J., Chen, G. Y., and Xie, Z. T. (2016a). MicroRNA-361-5p inhibits cancer cell growth by targeting CXCR6 in hepatocellular carcinoma. Cell. Physiol. Biochem. 38, 777–785. doi: 10.1159/000443033
Sun, L., Burnett, J., Guo, C., Xie, Y., Pan, J., Yang, Z., et al. (2015). CPA4 is a promising diagnostic serum biomarker for pancreatic cancer. Am. J. Cancer Res. 6, 91–96.
Sun, L., Cao, J., Guo, C., Burnett, J., Yang, Z., Ran, Y., et al. (2017). Associations of carboxypeptidase 4 with ALDH1A1 expression and their prognostic value in esophageal squamous cell carcinoma. Dis. Esophagus 30, 1–5. doi: 10.1093/dote/dox011
Sun, L., Guo, C., Yuan, H., Burnett, J., Pan, J., Yang, Z., et al. (2016b). Overexpression of carboxypeptidase A4 (CPA4) is associated with poor prognosis in patients with gastric cancer. Am. J. Transl. Res. 8, 5071–5075.
Sun, L., Wang, Y., Yuan, H., Burnett, J., Pan, J., Yang, Z., et al. (2016c). CPA4 is a novel diagnostic and prognostic marker for human non-small-cell lung cancer. J. Cancer 7, 1197–1204. doi: 10.7150/jca.15209
Sun, S.-L., Shu, Y.-G., and Tao, M.-Y. (2020). LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol. Cell. Biochem. 468, 69–82. doi: 10.1007/s11010-020-03712-y
Sun, W. Y., Lu, Y. F., Cai, X. L., Li, Z. Z., Lv, J., Xiang, Y. A., et al. (2020). Circ-ABCB10 acts as an oncogene in glioma cells via regulation of the miR-620/FABP5 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 6848–6857.
Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., and Mayeda, A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34:e63. doi: 10.1093/nar/gkl151
Tanco, S., Zhang, X., Morano, C., Avilés, F. X., Lorenzo, J., and Fricker, L. D. (2010). Characterization of the substrate specificity of human carboxypeptidase A4 and implications for a role in extracellular peptide processing. J. Biol. Chem. 285, 18385–18396. doi: 10.1074/jbc.m109.060350
Tao, W., Jia, Z., Mengshi, W., Wei, L., and Feng, L. (2020). Circular RNA circFANCL motivates the glioma progression via the action on the miR-337-3p/HMGB1 signal axis. Minerva Med. Epub ahead of print
Tian, T., Fu, X., Lu, J., Ruan, Z., Nan, K., Yao, Y., et al. (2018). MicroRNA-760 inhibits doxorubicin resistance in hepatocellular carcinoma through regulating notch1/Hes1-PTEN/Akt signaling pathway. J. Biochem. Mol. Toxicol. 32:e22167. doi: 10.1002/jbt.22167
Tiwari, N., Tiwari, Vijay, K., Waldmeier, L., Balwierz, Piotr, J., et al. (2013). Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783. doi: 10.1016/j.ccr.2013.04.020
Tomii, C., Inokuchi, M., Takagi, Y., Ishikawa, T., Otsuki, S., Uetake, H., et al. (2017). TPX2 expression is associated with poor survival in gastric cancer. World J. Surg. Oncol. 15:14.
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics. evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
van’t Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A. M., Mao, M., et al. (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature, 415, 530–536.
Venables, J. P., Klinck, R., Koh, C., Gervais-Bird, J., Bramard, A., Inkel, L., et al. (2009). Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16, 670–676.
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, L., Cho, K. B., Li, Y., Tao, G., Xie, Z., and Guo, B. (2019). Long noncoding RNA (lncRNA)-mediated competing endogenous rna networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 20:5758. doi: 10.3390/ijms20225758
Wang, R., Zhang, S., Chen, X., Li, N., Li, J., Jia, R., et al. (2018). EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer 17:166.
Wang, S., Qi, Y., Gao, X., Qiu, W., Liu, Q., Guo, X., et al. (2020). Hypoxia-induced lncRNA PDIA3P1 promotes mesenchymal transition via sponging of miR-124-3p in glioma. Cell Death Dis. 11:168.
Wang, X., and Zhu, Y. (2021). Circ_0000020 elevates the expression of PIK3CA and facilitates the malignant phenotypes of glioma cells via targeting miR-142-5p. Cancer Cell Int. 21:79.
Wang, Y., Liu, X., Guan, G., Xiao, Z., Zhao, W., and Zhuang, M. (2019). Identification of a five-pseudogene signature for predicting survival and its ceRNA network in glioma. Front. Oncol.:9. doi: 10.3389/fonc.2019.01059
Wang, Y. P., Li, H. Q., Chen, J. X., Kong, F. G., Mo, Z. H., Wang, J. Z., et al. (2020). Overexpression of XIST facilitates cell proliferation, invasion and suppresses cell apoptosis by reducing radio-sensitivity of glioma cells via miR-329-3p/CREB1 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 3190–3203.
Wang, Y.-W., Chen, X., Ma, R., and Gao, P. (2016). Understanding the CREB1-miRNA feedback loop in human malignancies. Tumor. Biol. 37, 8487–8502. doi: 10.1007/s13277-016-5050-x
Wang, Z., Yuan, J., Li, L., Yang, Y., Xu, X., and Wang, Y. (2017). Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am. J. Transl. Res. 9, 1845–1855.
Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., et al. (2008). Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543. doi: 10.1038/nature06908
Wei, P., Zhang, N., Xu, Y., Li, X., Shi, D., Wang, Y., et al. (2013). TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J. Trans. Med. 11:313. doi: 10.1186/1479-5876-11-313
Wei, Y., Lu, C., Zhou, P., Zhao, L., Lyu, X., Yin, J., et al. (2020). EIF4A3-induced circular RNA ASAP1(circASAP1) promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1/2 signaling. Neuro Oncol.
Weller, M., Wick, W., Aldape, K., Brada, M., Berger, M., Pfister, S. M., et al. (2015). Glioma. Nat. Rev. Dis. Primers 1:15017.
Wu, J., Li, W.-z, Huang, M.-l, Wei, H.-l, Wang, T., Fan, J., et al. (2017). Regulation of cancerous progression and epithelial-mesenchymal transition by miR-34c-3p via modulation of MAP3K2 signaling in triple-negative breast cancer cells. Biochem. Biophys. Res. Commun. 483, 10–16. doi: 10.1016/j.bbrc.2017.01.023
Wu, Y., and Qian, Z. (2019). Long non-coding RNAs (lncRNAs) and microRNAs regulatory pathways in the tumorigenesis and pathogenesis of glioma. Discov. Med. 28, 129–138.
Xia, B., Hong, T., He, X., Hu, X., and Gao, Y. (2019). A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Trans. 28, 1614–1623. doi: 10.1177/0963689719875409
Xia, L., Yi, F., Zhai, X., and Zhang, M. (2020). [Circular RNA homeodomain-interacting protein kinase 3 (circHIPK3) promotes growth and metastasis of glioma cells by sponging miR-124-3p]. Xi Bao yu fen zi mian yi xue za zhi = Chin. J. Cell. Mol. Immunol. 36, 609–615.
Xia, T., Liao, Q., Jiang, X., Shao, Y., Xiao, B., Xi, Y., et al. (2014). Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 4:6088.
Xie, Z., Li, X., Chen, H., Zeng, A., Shi, Y., and Tang, Y. (2019). The lncRNA-DLEU2/miR-186-5p/PDK3 axis promotes the progress of glioma cells. Am. J. Transl. Res. 11, 4922–4934.
Xu, D., Yu, J., Gao, G., Lu, G., Zhang, Y., and Ma, P. (2018). LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci. Rep. 38:BSR20171664.
Xu, H., Zhang, Y., Qi, L., Ding, L., Jiang, H., and Yu, H. (2018). NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front. Mol. Neurosci. 11:225. doi: 10.3389/fnmol.2018.00225
Xu, W., Xue, R., Xia, R., Liu, W. W., Zheng, J. W., Tang, L., et al. (2020). Sevoflurane impedes the progression of glioma through modulating the circular RNA has_circ_0012129/miR-761/TGIF2 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 5534–5548.
Yang, M., Li, G., Fan, L., Zhang, G., Xu, J., and Zhang, J. (2019). Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem. Biophys. Res. Commun. 508, 980–985. doi: 10.1016/j.bbrc.2018.12.052
Yang, Y., Zhang, Y., Chen, B., Ding, L., Mu, Z., and Li, Y. (2019). Elevation of circular RNA circ-POSTN facilitates cell growth and invasion by sponging miR-1205 in glioma. J. Cell. Biochem. 120, 16567–16574. doi: 10.1002/jcb.28916
Yang, Z., Li, C., Fan, X. Y., and Liu, L. J. (2020). Circular RNA circ_0079593 promotes glioma development through regulating KPNA2 expression by sponging miR-499a-5p. Eur. Rev. Med. Pharmacol. Sci. 24, 1288–1301.
Yao, Y., Hua, Q., Zhou, Y., and Shen, H. (2019). CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin signaling pathway. Biomed. Pharmacother. 111, 1367–1375. doi: 10.1016/j.biopha.2018.12.120
Yao, Y., Ma, J., Xue, Y., Wang, P., Li, Z., Liu, J., et al. (2015). Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 359, 75–86. doi: 10.1016/j.canlet.2014.12.051
Ye, J., Zhu, J., Chen, H., Qian, J., Zhang, L., Wan, Z., et al. (2020). A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int. J. Cancer 146, 248–261. doi: 10.1002/ijc.32483
Ye, Y., Zhi, F., Peng, Y., and Yang, C. C. (2018). MiR-128 promotes the apoptosis of glioma cells via binding to NEK2. Eur. Rev. Med. Pharmacol. Sci. 22, 8781–8788.
Yin, H., and Cui, X. (2020). Knockdown of circHIPK3 facilitates temozolomide sensitivity in glioma by regulating cellular behaviors through miR-524-5p/KIF2A-mediated PI3K/AKT pathway. Cancer Biother. Radiopharm.
Yu, H., Xue, Y., Wang, P., Liu, X., Ma, J., Zheng, J., et al. (2017). Knockdown of long non-coding RNA XIST increases blood–tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis 6, e303–e303.
Yu, H.-M., Wang, C., Yuan, Z., Chen, G.-L., Ye, T., and Yang, B.-W. (2019). LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR-193a-3p. Cell Prolif. 52:e12526. doi: 10.1111/cpr.12526
Yu, J., Tan, Q., Deng, B., Fang, C., Qi, D., and Wang, R. (2015). The microRNA-520a-3p inhibits proliferation, apoptosis and metastasis by targeting MAP3K2 in non-small cell lung cancer. Am. J. Cancer Res. 5, 802–811.
Yu, W., Xiang, D., Jia, H., He, X., Sheng, J., Long, Y., et al. (2020). The lncRNA BCYRN1 functions as an oncogene in human glioma by downregulating miR-125a-5p in vitro. Cancer Manag. Res. 12, 1151–1161. doi: 10.2147/cmar.s227327
Yuan, Y., Liu, B., Xie, P., Zhang, M. Q., Li, Y., Xie, Z., et al. (2015). Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit. Proc. Natl. Acad. Sci.U.S.A. 112, 3158–3163. doi: 10.1073/pnas.1413896112
Zhan, L., Mu, Z., Yang, M., Zhang, T., Li, H., and Qian, L. (2019). Elevation of circ-PITX1 upregulates interleukin 17 receptor D expression via sponging miR-518a-5p and facilitates cell progression in glioma. J. Cell. Biochem. 120, 16495–16502. doi: 10.1002/jcb.28868
Zhang, H. Y., Zhang, B. W., Zhang, Z. B., and Deng, Q. J. (2020). Circular RNA TTBK2 regulates cell proliferation, invasion and ferroptosis via miR-761/ITGB8 axis in glioma. Eur. Rev. Med. Pharmacol. Sci. 24, 2585–2600.
Zhang, N., Jin, Y., Hu, Q., Cheng, S., Wang, C., Yang, Z., et al. (2020). Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. 13:64.
Zhang, S., Qin, W., Yang, S., Guan, N., Sui, X., and Guo, W. (2020). Circular RNA SFMBT2 inhibits the proliferation and metastasis of glioma cells through mir-182-5p/Mtss1 pathway. Technol. Cancer Res. Treatment 19:1533033820945799.
Zhang, W., and Xu, C. (2020). Circ-ELF2 acts as a competing endogenous RNA to facilitate glioma cell proliferation and aggressiveness by targeting MiR-510-5p/MUC15 signaling. 13, 10087–10096. doi: 10.2147/ott.s275218
Zhang, X., Wei, C., Li, J., Liu, J., and Qu, J. (2017). MicroRNA-361-5p inhibits epithelial-to-mesenchymal transition of glioma cells through targeting Twist1. Oncol. Rep. 37, 1849–1856. doi: 10.3892/or.2017.5406
Zhang, Y., Cai, Z., Liang, J., Chai, E., Lu, A., and Shang, Y. (2020). CircCPA4 promotes the malignant phenotypes in glioma via miR-760/MEF2D Axis. Neurochem. Res. 45, 2903–2913. doi: 10.1007/s11064-020-03139-3
Zheng, D., Chen, D., Lin, F., Wang, X., Lu, L., Luo, S., et al. (2020). LncRNA NNT-AS1 promote glioma cell proliferation and metastases through miR-494-3p/PRMT1 axis. Cell Cycle 19, 1621–1631. doi: 10.1080/15384101.2020.1762037
Zheng, J., Liu, X., Xue, Y., Gong, W., Ma, J., Xi, Z., et al. (2017). TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J. Hematol. Oncol. 10:52.
Zheng, S.-Q., Qi, Y., Wu, J., Zhou, F.-L., Yu, H., Li, L., et al. (2019). CircPCMTD1 acts as the sponge of miR-224-5p to promote glioma progression. Front. Oncol. 9:398. doi: 10.3389/fonc.2019.00398
Zhou, B.-S., Beidler, D. R., and Cheng, Y.-C. (1992). Identification of antisense RNA transcripts from a human DNA Topoisomerase I pseudogene. Cancer Res. 52, 4280–4285.
Zhou, H., Zhang, Y., Lai, Y., Xu, C., and Cheng, Y. (2020). Circ_101064 regulates the proliferation, invasion and migration of glioma cells through miR-154–5p/PIWIL1 axis. Biochem. Biophys. Res. Commun. 523, 608–614.
Zhou, X.-Y., Liu, H., Ding, Z.-B., Xi, H.-P., and Wang, G.-W. (2020). lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics 112, 1021–1029.
Zhou, Y., Liao, Q., Han, Y., Chen, J., Liu, Z., Ling, H., et al. (2016). Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. J. Cancer 7, 2100–2109. doi: 10.7150/jca.16198
Zhu, F., Cheng, C., Qin, H., Wang, H., and Yu, H. (2020a). A novel circular RNA circENTPD7 contributes to glioblastoma progression by targeting ROS1. Cancer Cell Int. 20:118.
Zhu, J., Gu, W., and Yu, C. (2020b). MATN1-AS1 promotes glioma progression by functioning as ceRNA of miR-200b/c/429 to regulate CHD1 expression. Cell Prolif. 53:e12700.
Zhu, J., Wang, H., Huang, Y. Q., Song, W., Li, Y. F., Wang, W. J., et al. (2020c). Comprehensive analysis of a long non-coding RNA-associated competing endogenous RNA network in glioma. Oncol. Lett. 20:63.
Zhu, L., Xue, F., Cui, Y., Liu, S., Li, G., Li, J., et al. (2019). miR-155-5p and miR-760 mediate radiation therapy suppressed malignancy of non-small cell lung cancer cells. BioFactors 45, 393–400. doi: 10.1002/biof.1500
Keywords: ceRNET, ceRNA, glioma, ncRNA, cancer driver dysregulation, glioblastoma
Citation: Cen L, Liu R, Liu W, Li Q and Cui H (2021) Competing Endogenous RNA Networks in Glioma. Front. Genet. 12:675498. doi: 10.3389/fgene.2021.675498
Received: 03 March 2021; Accepted: 22 March 2021;
Published: 29 April 2021.
Edited by:
Nicoletta Potenza, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Nicola Mosca, Fondazione Policlinico Universitario A. Gemelli, Scientific Institute for Research, Hospitalization and Healthcare, ItalyCopyright © 2021 Cen, Liu, Liu, Li and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianqian Li, c2lzc2kxMDBAdmlwLnNpbmEuY29t; Hongjuan Cui, aGN1aUBzd3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.