
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 22 March 2021
Sec. Statistical Genetics and Methodology
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.652315
This article is part of the Research TopicApplication of Novel Statistical and Machine-learning Methods to High-dimensional Clinical Cancer and (Multi-)Omics dataView all 11 articles
 Liye Zhou1
†
Liye Zhou1
† Zhifei Guo1
†
Zhifei Guo1
† Bijue Wang1
Bijue Wang1 Yongqing Wu2
Yongqing Wu2 Zhi Li3
Zhi Li3 Hongmei Yao4
Hongmei Yao4 Ruiling Fang2
Ruiling Fang2 Haitao Yang5
Haitao Yang5 Hongyan Cao2,6*
Hongyan Cao2,6* Yuehua Cui7*
Yuehua Cui7*Heart failure with preserved ejection fraction (HFpEF) has become a major health issue because of its high mortality, high heterogeneity, and poor prognosis. Using genomic data to classify patients into different risk groups is a promising method to facilitate the identification of high-risk groups for further precision treatment. Here, we applied six machine learning models, namely kernel partial least squares with the genetic algorithm (GA-KPLS), the least absolute shrinkage and selection operator (LASSO), random forest, ridge regression, support vector machine, and the conventional logistic regression model, to predict HFpEF risk and to identify subgroups at high risk of death based on gene expression data. The model performance was evaluated using various criteria. Our analysis was focused on 149 HFpEF patients from the Framingham Heart Study cohort who were classified into good-outcome and poor-outcome groups based on their 3-year survival outcome. The results showed that the GA-KPLS model exhibited the best performance in predicting patient risk. We further identified 116 differentially expressed genes (DEGs) between the two groups, thus providing novel therapeutic targets for HFpEF. Additionally, the DEGs were enriched in Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways related to HFpEF. The GA-KPLS-based HFpEF model is a powerful method for risk stratification of 3-year mortality in HFpEF patients.
Heart failure (HF) is the leading cause of death and disability worldwide among older adults (Manolis et al., 2019). Over 50% of patients with HF exhibit heart failure with preserved ejection fraction (HFpEF; Komajda et al., 2011; Rich et al., 2018), and the prevalence of HFpEF is increasing relative to heart failure with reduced ejection fraction (HFrEF) at an alarming rate of 1% per year (Monika et al., 2018). HFpEF is a heterogeneous syndrome that contributes to abnormal cardiac structure or function, seriously endangering human health (Antlanger et al., 2017; Garg et al., 2017). HFpEF patients have a poor prognosis, and the 5-year mortality rate of HFpEF is as high as 50% (Shah et al., 2017). While the mortality rate of HFrEF has significantly decreased over the past few years because of specific HFrEF treatments (Loh et al., 2013), no effective treatment has been identified for HFpEF patients (Shah et al., 2014). Arguably, with an aging population worldwide, the emerging epidemic of HFpEF requires urgent attention to determine methods for faster disease risk assessment and to predict clinical outcomes to guide therapy, monitoring, and patient management.
While numerous risk assessment models have been developed in cohorts with HFrEF or a mixture of HFrEF and HFpEF, risk prediction in HFpEF patients has been less studied (Thorvaldsen et al., 2017; Angraal et al., 2020). This may be associated with the poor prognostic factors used to predict HFpEF patients (Kanda et al., 2018). The existing risk assessment models for HFpEF are predominantly based on clinical phenotype data, such as baseline demographic and clinical data and electrocardiographic, echocardiographic, and laboratory testing data (Komajda et al., 2011; Thorvaldsen et al., 2017; Rich et al., 2018; Angraal et al., 2020). Unfortunately, these models constructed using clinical phenotypic data have low sensitivity or specificity, and patients are likely to be misdiagnosed. No model has gained widespread acceptance to date. The estimate of an HFpEF patient’s prognosis in daily practice is still mainly based on the experience of clinicians (Ferrero et al., 2015; Thorvaldsen et al., 2017; Manolis et al., 2019). A great need exists to develop an effective risk model for HFpEF to aid in the design of future clinical trials.
With advances in sequencing and computer technology, high throughput expression data can be extracted without limits. Genomic measures of gene expression offer rich information about the underlying disease mechanism and have provided new possibilities of using these molecular data to understand the disease gene function and further predict disease outcomes (Haring and Wallaschofski, 2012). Based on the expression data, great efforts have been devoted to disease classification, clinical outcome prediction, and the identification of genes with potential therapeutic molecular signatures (Penney et al., 2011; Khan et al., 2012; Vargas and Lima, 2013; Wang et al., 2019). HFpEF is a complicated clinical syndrome with high molecular heterogeneity and diverse manifestations (Shah et al., 2015) and is further complicated with a potentially nonlinear relationship between genes and the clinical outcome. Thus, conventional generalized linear models (e.g., logistic regression) are poor choices for risk prediction. Advanced statistical techniques and machine learning methods show great potential in improving the classification performance over conventional statistical tools through the nonlinear effects of variables to achieve accurate prediction (Angraal et al., 2020) and should be studied for HFpEF prediction.
The purpose of this work is to evaluate six different risk stratification models and to predict the survival risk of HFpEF patients based on gene expression profiles using data from a high-quality epidemiologic study, the Framingham Heart Study (FHS). We applied five advanced machine learning methods [i.e., kernel partial least squares based on the genetic algorithm (GA-KPLS), random forest (RF), the least absolute shrinkage and selection operator (LASSO), ridge regression (RR), support vector machine (SVM), and a conventional logistic regression model (Logit)] to build an optimal risk stratification model. Identification of patients with a high risk of HFpEF will be helpful for targeted interventions and clinical trials to further improve the survival of HFpEF patients.
The FHS data used in this study included clinical, survival, and expression data downloaded from dbGAP (study accession: phs000007, http://dbgap.ncbi.nlm.nih.gov). The FHS has recruited participants from Framingham, MA, United States, to undergo biennial examinations to investigate cardiovascular disease and its risk factors since 1948 (Oppenheimer, 2005). Offspring (and their spouses) and adult grandchildren of the original cohort of participants were recruited into the second- and third-generation cohorts in 1971 and 2002, respectively (Yao et al., 2015). In this study, the clinical and gene expression data were obtained from the offspring cohort who (i) attended the eighth examination cycle conducted between 2005 and 2008 and (ii) had both clinical and gene expression profiles.
According to the guidelines of the European Society of Cardiology (McMurray et al., 2018), patients were diagnosed with HFpEF using the following four conditions: (1) typical signs or symptoms of HF, (2) B-type natriuretic peptide >35 pg/ml and/or N-terminal-pro hormone B-type natriuretic peptide >125 pg/ml, (3) left ventricular ejection fraction >50%; and (4) structural HF (left ventricular hypertrophy/left atrial enlargement) and/or diastolic dysfunction. We excluded patients with valvular stroma and/or hypertrophic cardiomyopathy, resulting in inclusion of 172 HFpEF patients (103 males and 69 females). Patients whose 3-year survival status was unknown were filtered out by design (Fransen et al., 2011). Finally, 149 individuals (91 males and 58 females) who had full survival information after 3 years were included in the study.
The expression data contained 17,873 gene expression probes. We mapped these probes to genes following the annotation from the Affymetrix Human Exon 1.0 ST GeneChip platform, which yielded 17,358 genes. The gene expression data were log 2 (x + 1) transformed and then standardized (Cheerla and Gevaert, 2017). A variable screening procedure called as sure independence screening was applied to reduce the gene expression dimensionality from an ultra-high to a moderate scale, with a binary response defined as a “good outcome” or “poor outcome” for each individual. Following the sure independence screening criterion {i.e., keeping d = [2n/log(n)] features; Fan and Lv, 2008}, the top 137 features were retained for further analysis.
The clinical outcome was defined as a good or poor outcome based on patients’ survival status. The good-outcome group had event-free survival for at least 3 years [survival time was measured from the time of admission for HFpEF diagnosis to the time of last follow-up (2011) or time of death from cardiovascular disease]. The poor-outcome group included patients who died because of cardiovascular disease during the 3-year period. We further explored the differentially expressed genes (DEGs) between the good-outcome and poor outcome groups using significance analysis of microarrays (Tusher et al., 2001) and then conducted Gene Ontology (GO) enrichment analysis and the Kyoto Encyclopedia of the Genes and Genomes (KEGG) pathway analysis based on the DEGs using KOBAS software1 (Ai and Kong, 2018).
The kernel partial least squares method can map the original data points from the original input space RN into a high-dimensional feature space ℱ, and therefore, original data that cannot be linearly separated in RN can be separated in ℱ (Rosipal and Trejo, 2002), which improves the classification performance to achieve accurate prediction. A genetic algorithm (GA) is an optimization method based on the genetic mechanism of “survival of the fittest.” In this study, we used a Gaussian kernel function to construct the kernel matrix for gene expression data and then used the genetic algorithm to optimize the Gaussian kernel function parameter σ. The Gaussian kernel function is given as . For the details of the method, readers are referred to Yang et al. (2020). Because we only used gene expression data for prediction, the only parameter that needed to be optimized was the kernel bandwidth σ.
Ridge regression and LASSO fit prediction models by shrinkage or regularization of the regression coefficients (Frank and Friedman, 1993; Tibshirani, 1996). The LASSO method can shrink some coefficients to exactly zero. Both models were developed to minimize prediction errors. For the LASSO and RR methods, the optimal tuning parameter λ was chosen by 10-fold cross-validation over a grid of 100 λ values. The RR and LASSO methods were performed using the R glmnet package.
The SVM method was developed to solve high-dimensional classification problems (Furey et al., 2000) and was performed using the R e1071 package. The radial basis kernel function was used in the SVM.
An RF uses the bootstrap method to extract n samples from the original data and generate B classification trees. These B trees constitute a random forest. Each observation’s predictive result is determined by a majority vote; the overall prediction is the most commonly occurring class among the B classification trees (Austin et al., 2013). The RF method was performed using the randomForest package in R. All parameter values were set using the default.
In our study, the original data were divided into two non-overlapping data sets: modeling data and external testing data. We randomly selected modeling data and external testing data at a ratio of 80:20. The modeling set was used to train the prediction model, and the testing set was used to evaluate the prediction performance. The entire process of randomly selecting the modeling and testing data was repeated 1,000 times to increase the stability and repeatability of the results.
We used multiple evaluation criteria to evaluate the predictive performances of the six models, including the area under the curve (AUC), sensitivity (Se), specificity (Sp), accuracy (ACC), Youden index, G-means, and Matthews correlation coefficient (MCC). The MCC and AUC were mainly used to evaluate the model performance because they are more comprehensive evaluation criteria. We employed one-way ANOVA, followed by Dunnett’s multiple-comparison test, to compare the performance of the GA-KPLS and the five other models (RF, LASSO, RR, Logit, and SVM). Statistical significance was indicated by a value of p < 0.05.
At the end of the 3-year period, 42 patients (28.19%) met the study endpoint of cardiovascular disease-related death, and 107 patients (71.81%) had survived. There were 91 males (61.07%) and 58 females (38.93%). The average age was 75.02 (±8.02) years old. Table 1 shows the baseline condition of both groups, patients with good outcomes, and those with poor outcomes. There was no significant difference in age, gender, comorbidities, vital signs, or laboratory data (except for systolic blood pressure) between the two groups.
We compared the classification performance of the six models: GA-KPLS, RF, LASSO, RR, SVM, and Logit. The evaluation index of the six models was summarized as the average value obtained by repeating the data partition 1,000 times. Table 2 shows the prediction results of the six models. As shown in the table, the GA-KPLS model exhibited the best performance in nearly all the criteria except for specificity. This finding clearly demonstrates the superior performance of the GA-KPLS model. To further display the prediction results, we chose the evaluation criterion AUC to demonstrate the performance obtained by 1,000 random splits (see Figure 1). The AUC of the GA-KPLS model was significantly different from those of the RF, LASSO, RR, Logit, and SVM models, indicating the superior performance of the GA-KPLS model over the other models. It is interesting to note that the performance of the SVM model was quite similar to that of the GA-KPLS model. Based on the results, we concluded that the risk prediction model constructed by the GA-KPLS method had the best performance and can provide a methodological reference to assess the risk of HFpEF.
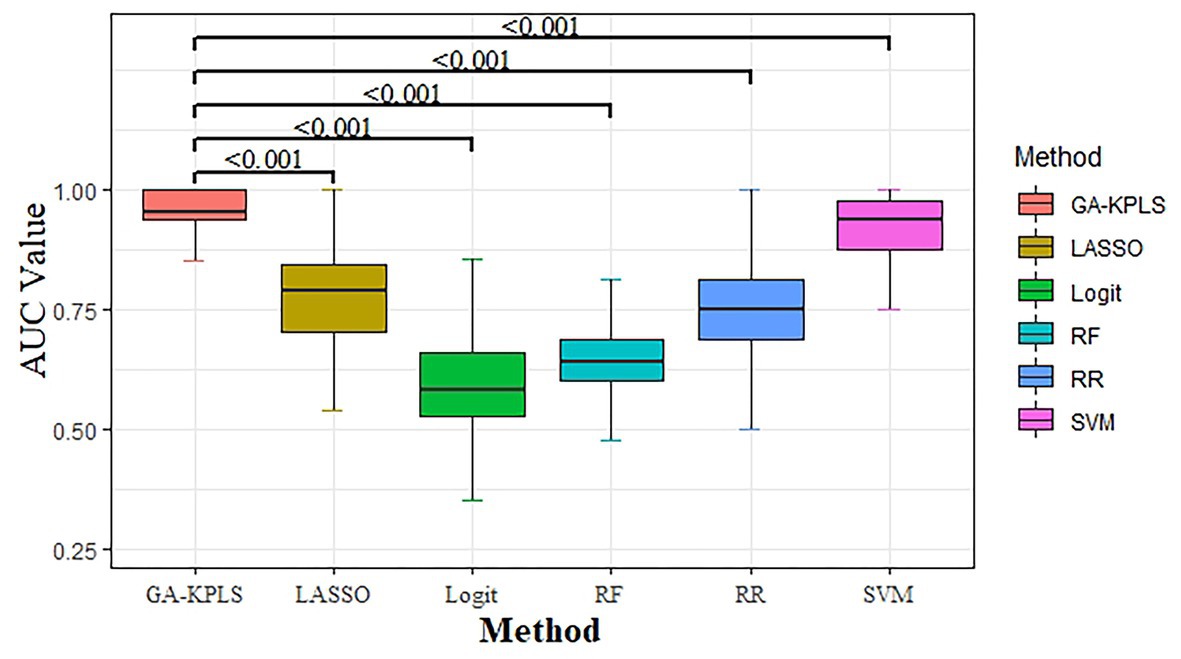
Figure 1. Boxplot of the area under the curve (AUC) values for the six different models (based on 1,000 random splits). The y-axis represents the AUC value. Values of p were obtained using Dunnett’s multiple-comparison test.
To demonstrate the clinical significance of identifying high-risk patients, we selected the prediction result of one random split with 120 training samples and 29 testing samples, which gave an MCC = 0.920 (close to MCCmean = 0.921). The Kaplan-Meier curves based on the original and predicted data yielded significantly different survival probabilities (p < 0.0001). Figure 2 shows the survival curves of the two groups. The left panel shows the survival curve from the original data, and the right panel shows the survival curve based on the newly predicted risk group with the GA-KPLS method. The prediction method exhibited good performance because the survival curves using the original and predicted values were very similar. To predict a future event, all the data can be used as the training set, and then the risk group status can be predicted based on measured gene expression data.
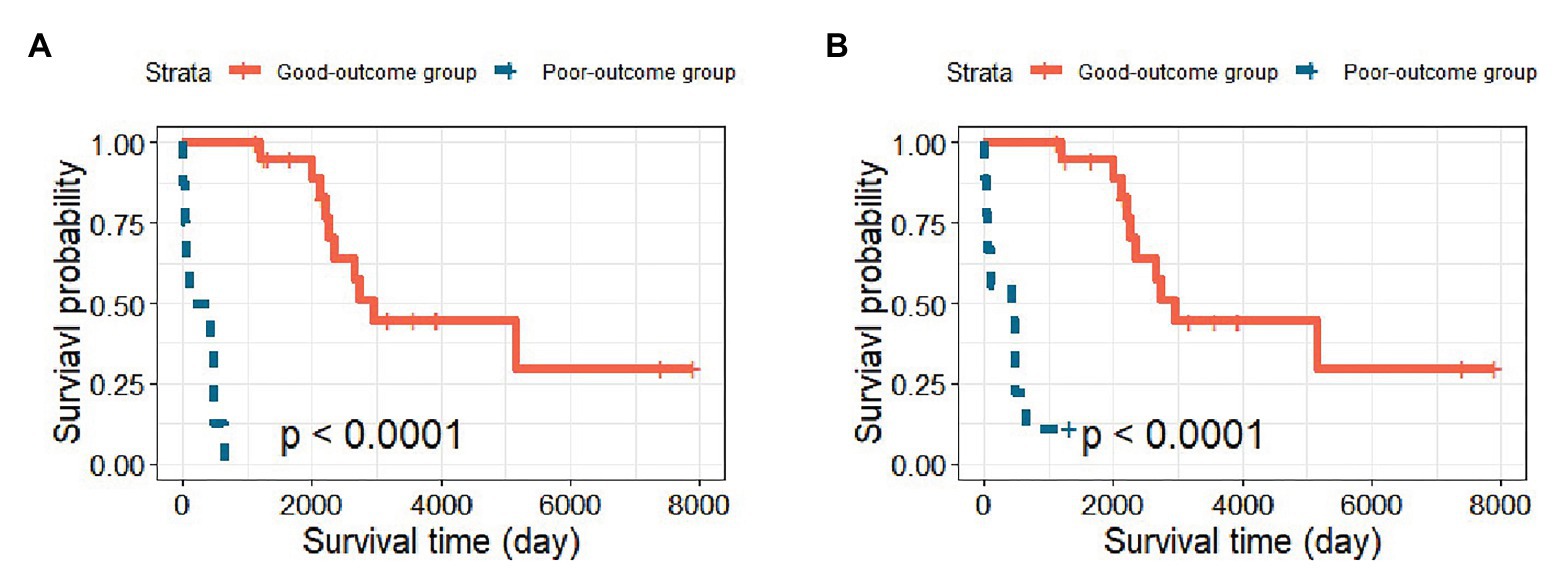
Figure 2. Kaplan-Meier survival curves of the good-outcome and poor-outcome groups. (A) The survival curve including the original 29 patients in the testing cohort and (B) the survival curve based on the predicted survival outcomes using the GA-KPLS method.
We treated the good-outcome group as the control group to identify DEGs. Of a total of 137 top genes, 116 DEGs were identified based on a threshold value of q < 0.05, among which 70 genes were upregulated and 46 were downregulated. The significant features of gene expression are shown in a heat map (see Figure 3). A block-like structure can be observed between the good-outcome and poor-outcome groups.
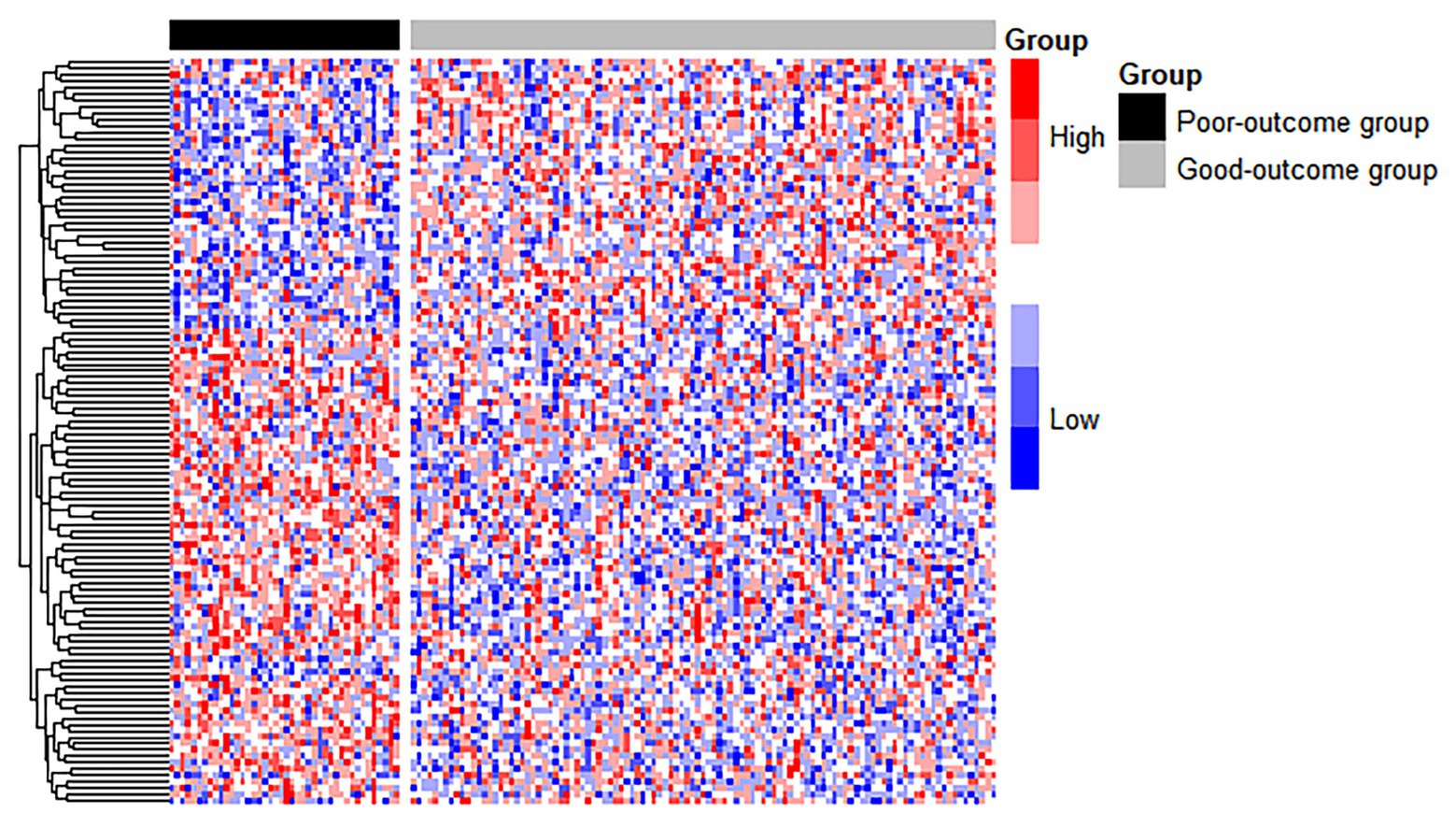
Figure 3. The heatmap of DEGs between the good-outcome and poor-outcome groups. Each column represents a patient, and each row represents a gene. Patients labeled with the black bar are poor-outcome samples, and those with the gray bar are good-outcome samples.
Among the 116 DEGs, the TRAF3IP2, C1QTNF9, TECRL, and Eph genes have been reported to be associated with HF. TRAF3IP2 is an upstream regulator of multiple proinflammatory pathways. TRAF3IP2 overexpression may activate IKK/NF-B, p38 MAPK, and JNK/AP-1 and induce proinflammatory cytokines, leading to cardiac fibrosis and contractile dysfunction (Yariswamy et al., 2016). C1QTNF9 (CTRP9) is an important member of the CTRP protein family. Appari et al. (2016) found that C1QTNF9 knock-out mice were protected from left ventricular dilatation and contractile dysfunction; however, C1QTNF9 overexpression promoted ventricular remodeling and systolic dysfunction. TECRL was recently suggested to play a key role in the electrical activity of the heart. TECRL affects the electrical conduction system of the heart by causing mutations in a calcium-processing protein, which eventually leads to arrhythmia (Perry and Vandenberg, 2016). The Eph/ephrin receptor ligand comprises the largest family of receptor tyrosine kinases and affects the behavior of cells mainly by activating signal transduction pathways. Eph/ephrin expression may lead to phenotypic changes in the vascular endothelium during inflammation, causing inflammatory cells to enter the interstitial tissue from the vascular space (Coulthard et al., 2012).
The role of DUSP1 is controversial, as both anti-inflammatory and pro-atherosclerotic actions have been suggested (Hahn et al., 2014). Auger-Messier et al. (2013) suggested that the disruption of DUSP1 promoted p38 MAPK activity, which could reduce cardiac contractility and calcium handling; thus, DUSP1 could be a target gene for prevention of HF. In addition, LHFPL2 and SNX24 are associated with coronary artery disease (Lin et al., 2013; Shendre et al., 2017). HIST1H4B is associated with the immune process (Zhang et al., 2019). OXER1 is involved in the inflammatory response of the disease (Dattilo et al., 2015). The empirical evidence suggests the importance of the identified DEGs associated with HFpEF.
To further investigate the functional relevance of the DEGs, we performed GO enrichment and KEGG pathway analyses. The DEGs were significantly enriched in 12 GO terms, with a corrected value of p < 0.05. GO terms comprised three categories: biological process, cell component, and molecular function. Figure 4 shows all significant GO terms. The most significantly enriched GO terms were plasma membrane (corrected value of p = 2.67E−07), G protein-coupled receptor signaling pathway (corrected value of p = 3.06E−04), and protein binding (corrected value of p = 3.06E−04). The plasma membrane plays important roles in maintaining homeostasis, cell material exchange, and information transmission (Lutz et al., 2003; Wang et al., 2017). The G protein-coupled receptor signaling pathway mediates cardiac functions, such as those of inotropy and vasodilation in peripheral vessels, participates in the occurrence and development of HF and may serve as the molecular underpinning for future HF therapeutics (Wang et al., 2018; Altamish et al., 2020). Protein binding, including fatty acid-binding proteins, has been related to cardiac alterations, e.g., systolic and diastolic cardiac dysfunction (Rodriguez-Calvo et al., 2017). In the KEGG analysis, the olfactory transduction pathway was identified, with a corrected value of p < 0.05. The olfactory system uses G protein-coupled receptors to accomplish its vital task (Ronnett and Moon, 2002).
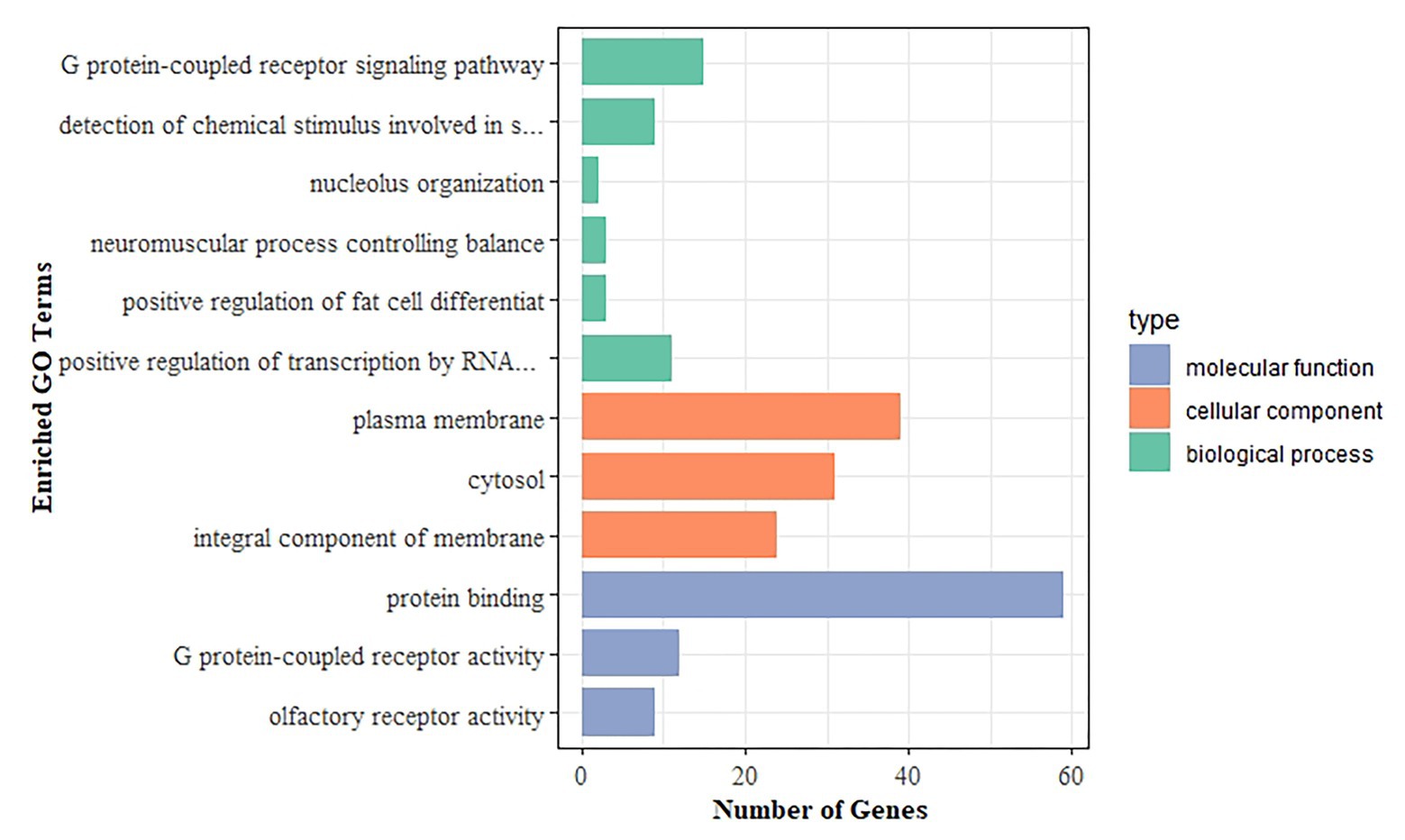
Figure 4. Gene Ontology (GO) enrichment analysis of DEGs. The x-axis shows the number of genes, and the y-axis indicates the GO terms. Bars with different colors correspond to different GO categories, with green representing biological process, orange representing cellular component, and blue representing molecular function.
Accurately predicting disease outcomes are essential for patient-centered care, both for making treatment decisions and monitoring the quality of health care (Angraal et al., 2020). Using the gene expression data of HFpEF patients, this study explored five machine learning methods and one conventional logistic regression model to predict the survival status of patients with HFpEF. The GA-KPLS based HFpEF model could predict patient survival status with high accuracy. Furthermore, the identification of molecular markers (i.e., DEGs) of HFpEF may lead to the development of novel targeted therapies.
The ability to assess survival outcomes of patients with cardiovascular diseases has great clinical value in an era with multiple treatment options. Although previous studies have devoted great effort to predicting clinical outcomes of HF patients, the current study has several unique merits. There are many studies being conducted to predict HF. However, few studies are focused on HFpEF. By evaluating six models, we showed that the GA-KPLS model using gene expression data may be a powerful and highly accurate prediction model of survival status in HFpEF patients. A prediction model using gene expression data can be an alternative means to the currently used models based on clinical data, such as the Enhanced Feedback for Effective Cardiac (EFFECT) study risk scores (Thorvaldsen et al., 2017) and Meta-Analysis Global Group in Chronic Heart Failure (MAGGIC) scores (Pocock et al., 2013).
Second, because of the highly heterogenous nature of HFpEF, a consensus has not been reached on which predictors can be used to reliably predict HFpEF. We demonstrated that gene expression can be used to predict HFpEF survival status with high accuracy using the GA-KPLS prediction model. With the availability of increasing types of omics data (e.g., copy number variants, microRNAs, and epigenetic data), we can further improve the prediction accuracy by integrating different data sources with the GA-KPLS model. Our study illustrates the development of new machine learning methods for HFpEF risk prediction by integrating different omics data types.
Current studies have focused on single or multiple clinical indicators to identify patients at high risk for HFpEF. However, most methods can only achieve an AUC of 0.7, which is unrealistic for application in clinical practice (Kanda et al., 2018; Shen et al., 2020). Many researchers have also used statistical methods to construct stratification models such as Cox proportional hazards models and logistic regression models. However, these methods fail to capture the nonlinear relationship between predictors and the disease outcome (Komajda et al., 2011; Rich et al., 2018; Angraal et al., 2020). In contrast, the GA-KPLS model uses the advantage of kernel functions to extract nonlinear relationships between genomic features and survival outcomes, hence achieving more accurate predictions than its counterparts.
Risk prediction in HFpEF patients using the GA-KPLS model may (1) serve to motivate patients to adhere to recommended treatments and lifestyle modifications (Oktay et al., 2013); (2) help clinicians to make treatment decisions, especially for high-risk groups of patients who may progress to circulatory failure when administered routine clinical therapeutics, and these patients may have the opportunity to undergo active therapeutic interventions such as mechanical circulatory assistance, heart transplantation, or new trials (Wang et al., 2019); and (3) help to inform the design of future HFpEF clinical trials.
However, our study had some limitations. First, because of the lack of additional external data on HFpEF, we cannot validate our findings in another data set. Second, we focused on gene expression data in our study. As lifestyle is an important risk factor for HF, further research should be performed to predict HFpEF risk by integrating both clinical and genomic data to improve the prediction performance because potential interactions may exist between these factors. Third, the HFpEF data set is imbalanced, with a ratio of 28:72 between the poor-outcome and good-outcome groups. However, the GA-KPLS and SVM methods performed well, with high sensitivity and specificity. If either low sensitivity or specificity becomes a concern, the SMOTE algorithm can be applied (Chawla et al., 2002), which is designed to handle prediction with imbalanced data.
In conclusion, the GA-KPLS-based HFpEF prediction model using gene expression data represents a valuable tool to improve the prognosis of HFpEF patients with different risk levels. The discovered transcriptional biomarkers of HFpEF provide new insight to the understanding the complex mechanism of HFpEF, leading to the development of novel targeted therapies for HFpEF. It is expected that integrating multi-omics and clinical data can further improve HFpEF outcome prediction, leading to the development of targeted, adaptive, and precision treatment of HFpEF patients with different risk levels.
The data analyzed in this study is subject to the following licenses/restrictions: The data analyzed in this study require NIH approval through the dbGap website. Requests to access these datasets should be directed to http://dbgap.ncbi.nlm.nih.gov.
LZ and ZG performed the study and drafted the manuscript. BW, YW, ZL, RF, and HtY participated in the data processing and analysis. HY provided the clinical interpretation. HC and YC conceived of the idea and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from NSFC (71403156 and 81872717), the Applied Basic Research Program of Shanxi Province (201901D111204), the China Scholarship Council (201908140151), a fund from the Provincial Education Department of Hebei Province (ZD2018022), and a fund from Michigan State University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the two reviewers for their insightful comments that helped us to greatly improve our manuscript. The Framingham Heart Study was conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (contract no. N01-HC-25195, HHSN268201500001I, and 75N92019D00031). This manuscript was not prepared in collaboration with the investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or the NHLBI. We thank Lisa Kreiner, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Ai, C., and Kong, L. (2018). CGPS: a machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 45, 489–504. doi: 10.1016/j.jgg.2018.08.002
Altamish, M., Samuel, V. P., Dahiya, R., Singh, Y., Deb, P. K., Bakshi, H. A., et al. (2020). Molecular signaling of G-protein-coupled receptor in chronic heart failure and associated complications. Drug Dev. Res. 81, 23–31. doi: 10.1002/ddr.21627
Angraal, S., Mortazavi, B. J., Gupta, A., Khera, R., Ahmad, T., Desai, N. R., et al. (2020). Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction. JACC Heart Fail. 8, 12–21. doi: 10.1016/j.jchf.2019.06.013
Antlanger, M., Aschauer, S., Kopecky, C., Hecking, M., Kovarik, J. J., Werzowa, J., et al. (2017). Heart failure with preserved and reduced ejection fraction in hemodialysis patients: prevalence, disease prediction and prognosis. Kidney Blood Press. Res. 42, 165–176. doi: 10.1159/000473868
Appari, M., Breitbart, A., Brandes, F., Szaroszyk, M., and Heineke, J. (2016). C1q-TNF-related protein-9 promotes cardiac hypertrophy and failure. Circ. Res. 120, 66–77. doi: 10.1161/CIRCRESAHA.116.309398
Auger-Messier, M., Accornero, F., Goonasekera, S. A., Bueno, O. F., Lorenz, J. N., Van Berlo, J. H., et al. (2013). Unrestrained p38 MAPK activation in Dusp1/4 double-null mice induces cardiomyopathy. Circ. Res. 112, 48–56. doi: 10.1161/CIRCRESAHA.112.272963
Austin, P. C., Tu, J. V., Ho, J. E., Levy, D., and Lee, D. S. (2013). Using methods from the data-mining and machine-learning literature for disease classification and prediction: a case study examining classification of heart failure subtypes. J. Clin. Epidemiol. 66, 398–407. doi: 10.1016/j.jclinepi.2012.11.008
Chawla, N., Bowyer, K. W., Hall, L. O., and Kegelmeyer, W. P. (2002). SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357. doi: 10.1613/jair.953
Cheerla, N., and Gevaert, O. (2017). MicroRNA based pan-Cancer diagnosis and treatment recommendation. BMC Bioinform. 18:32. doi: 10.1186/s12859-016-1421-y
Coulthard, M. G., Morgan, M., Woodruff, T. M., Arumugam, T. V., Taylor, S. M., Carpenter, T. C., et al. (2012). Eph/Ephrin signaling in injury and inflammation. Am. J. Pathol. 181, 1493–1503. doi: 10.1016/j.ajpath.2012.06.043
Dattilo, M., Neuman, I., Muñoz, M., Maloberti, P., and Cornejo Maciel, F. (2015). OxeR1 regulates angiotensin II and cAMP-stimulated steroid production in human H295R adrenocortical cells. Mol. Cell. Endocrinol. 408, 38–44. doi: 10.1016/j.mce.2015.01.040
Fan, J., and Lv, J. (2008). Rejoinder: sure independence screening for ultrahigh dimensional feature space. J. R. Stat. Soc. Series B Stat. Methodol. 70, 849–911. doi: 10.1111/j.1467-9868.2008.00674.x
Ferrero, P., Iacovoni, A., D’Elia, E., Vaduganathan, M., Gavazzi, A., and Senni, M. (2015). Prognostic scores in heart failure - critical appraisal and practical use. Int. J. Cardiol. 188, 1–9. doi: 10.1016/j.ijcard.2015.03.154
Frank, L. E., and Friedman, J. H. (1993). A statistical view of some chemometrics regression tools. Technometrics 35, 109–135. doi: 10.1080/00401706.1993.10485033
Fransen, J., Popa-Diaconu, D., Hesselstrand, R., Carreira, P., Valentini, G., and Beretta, L. (2011). Clinical prediction of 5-year survival in systemic sclerosis: validation of a simple prognostic model in EUSTAR centres. Ann. Rheum. Dis. 70, 1788–1792. doi: 10.1136/ard.2010.144360
Furey, T. S., Cristianini, N., Duffy, N., Bednarski, D. W., Schummer, M., and Haussler, D. (2000). Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 16, 906–914. doi: 10.1093/bioinformatics/16.10.906
Garg, A., Virmani, D., Agrawal, S., Agarwal, C., Sharma, A., Stefanini, G., et al. (2017). Clinical application of biomarkers in heart failure with a preserved ejection fraction: a review. Cardiology 136, 192–203. doi: 10.1159/000450573
Hahn, R. T., Hoppstädter, J., Hirschfelder, K., Hachenthal, N., Diesel, B., Kessler, S. M., et al. (2014). Downregulation of the glucocorticoid-induced leucine zipper (GILZ) promotes vascular inflammation. Atherosclerosis 234, 391–400. doi: 10.1016/j.atherosclerosis.2014.03.028
Haring, R., and Wallaschofski, H. (2012). Diving through the “-omics”: the case for deep Phenotyping and systems epidemiology. OMICS 16, 231–234. doi: 10.1089/omi.2011.0108
Kanda, T., Uematsu, M., Fujita, M., Iida, O., Masuda, M., Okamoto, S., et al. (2018). A novel predictor of clinical outcomes in patients with heart failure with preserved left-ventricular ejection fraction: a pilot study. Heart Vessel. 33, 1490–1495. doi: 10.1007/s00380-018-1211-8
Khan, J., Wei, J. S., and Greer, B. T. (2012). Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 64, 6883–6891. doi: 10.1158/0008-5472.CAN-04-0695
Komajda, M., Carson, P. E., Hetzel, S., McKelvie, R., McMurray, J., Ptaszynska, A., et al. (2011). Factors associated with outcome in heart failure with preserved ejection fraction: findings from the Irbesartan in heart failure with preserved ejection fraction study (I-PRESERVE). Circ. Heart Fail. 4, 27–35. doi: 10.1161/CIRCHEARTFAILURE.109.932996
Lin, Y. J., Chang, J. S., Liu, X., Lin, T. H., Huang, S. M., Liao, C. C., et al. (2013). Sorting nexin 24 genetic variation associates with coronary artery aneurysm severity in Kawasaki disease patients. Cell Biosci. 3:44. doi: 10.1186/2045-3701-3-44
Loh, J. C., Creaser, J., Rourke, D. A., Livingston, N., Harrison, T. K., Vandenbogaart, E., et al. (2013). Temporal trends in treatment and outcomes for advanced heart failure with reduced ejection fraction from 1993-2010: findings from a university referral center. Circ. Heart Fail. 6, 411–419. doi: 10.1161/CIRCHEARTFAILURE.112.000178
Lutz, S., Mura, R. A., Hippe, H. J., Tiefenbacher, C., and Niroomand, F. (2003). Plasma membrane-associated nucleoside diphosphate kinase (nm23) in the heart is regulated by beta-adrenergic signaling. Br. J. Pharmacol. 140:1019. doi: 10.1038/sj.bjp.0705527
Manolis, A. S., Manolis, A. A., Manolis, T. A., and Melita, H. (2019). Sudden death in heart failure with preserved ejection fraction and beyond: an elusive target. Heart Fail. Rev. 24, 847–866. doi: 10.1007/s10741-019-09804-2
McMurray, J. J., Adamopoulos, S., Anker, S. D., Auricchio, A., Böhm, M., Dickstein, K., et al. (2018). ESC guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: the task force for the diagnosis and treatment of acute and chronic heart failure 2012 of the European Society of Cardiology. Developed in collaboration with the heart. Eur. Heart J. 33, 1787–1847. doi: 10.1093/eurheartj/ehs104
Monika, R., Arantxa, B. A., Vanessa, V. E., Marc, V. B., and Blanche, S. (2018). Pathophysiological understanding of HFpEF: microRNAs as part of the puzzle. Cardiovasc. Res. 114, 782–793. doi: 10.1093/cvr/cvy049
Oktay, A. A., Rich, J. D., and Shah, S. J. (2013). The emerging epidemic of heart failure with preserved ejection fraction. Curr. Heart Fail. Rep. 10, 401–410. doi: 10.1007/s11897-013-0155-7
Oppenheimer, G. M. (2005). Becoming the Framingham study 1947-1950. Am. J. Public Health 95, 602–610. doi: 10.2105/AJPH.2003.026419
Penney, K. L., Sinnott, J. A., Fall, K., Pawitan, Y., Hoshida, Y., Kraft, P., et al. (2011). mRNA expression signature of Gleason grade predicts lethal prostate cancer. J. Clin. Oncol. 29, 2391–2396. doi: 10.1200/JCO.2010.32.6421
Perry, M. D., and Vandenberg, J. I. (2016). TECRL: connecting sequence to consequence for a new sudden cardiac death gene. EMBO Mol. Med. 8, 1364–1365. doi: 10.15252/emmm.201606967
Pocock, S. J., Ariti, C. A., McMurray, J. J., Maggioni, A., Køber, L., Squire, I. B., et al. (2013). Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur. Heart J. 34, 1404–1413. doi: 10.1093/eurheartj/ehs337
Rich, J. D., Burns, J., Freed, B. H., Maurer, M. S., Burkhoff, D., and Shah, S. J. (2018). Meta-analysis Global Group in Chronic (MAGGIC) heart failure risk score: validation of a simple tool for the prediction of morbidity and mortality in heart failure with preserved ejection fraction. J. Am. Heart Assoc. 7:e009594. doi: 10.1161/JAHA.118.009594
Rodriguez-Calvo, R., Girona, J., Alegret, J. M., Bosquet, A., Ibarretxe, D., and Masana, L. (2017). Role of the fatty acid binding protein 4 in heart failure and cardiovascular disease. J. Endocrinol. 233, R173–R184. doi: 10.1530/JOE-17-0031
Ronnett, G. V., and Moon, C. (2002). G proteins and olfactory signal transduction. Annu. Rev. Physiol. 64, 189–222. doi: 10.1146/annurev.physiol.64.082701.102219
Rosipal, R., and Trejo, L. J. (2002). Kernel partial least squares regression in reproducing kernel Hilbert space. J. Mach. Learn. Res. 2, 97–123. doi: 10.1162/15324430260185556
Shah, S. J., Katz, D. H., and Deo, R. C. (2014). Phenotypic spectrum of heart failure with preserved ejection fraction. Heart Fail. Clin. 10, 407–418. doi: 10.1016/j.hfc.2014.04.008
Shah, S. J., Katz, D. H., Selvaraj, S., Burke, M. A., Yancy, C. W., Gheorghiade, M., et al. (2015). Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 131, 269–279. doi: 10.1161/CIRCULATIONAHA.114.010637
Shah, K. S., Xu, H., Matsouaka, R. A., Bhatt, D. L., Heidenreich, P. A., Hernandez, A. F., et al. (2017). Heart failure with preserved, borderline, and reduced ejection fraction: 5-year outcomes. J. Am. Coll. Cardiol. 70, 2476–2486. doi: 10.1016/j.jacc.2017.08.074
Shen, L., Jhund, P. S., Anand, I. S., Carson, P. E., Desai, A. S., Granger, C. B., et al. (2020). Developing and validating models to predict sudden death and pump failure death in patients with heart failure and preserved ejection fraction. Clin. Res. Cardiol. doi: 10.1007/s00392-020-01786-8 [Epub ahead of print]
Shendre, A., Wiener, H., Irvin, M. R., Zhi, D., Limdi, N. A., Overton, E. T., et al. (2017). Admixture mapping of subclinical atherosclerosis and subsequent clinical events among African Americans in 2 large cohort studies. Circ. Cardiovasc. Genet. 10:e001569. doi: 10.1161/CIRCGENETICS.116.001569
Thorvaldsen, T., Claggett, B. L., Shah, A., Cheng, S., Agarwal, S. K., Wruck, L. M., et al. (2017). Predicting risk in patients hospitalized for acute decompensated heart failure and preserved ejection fraction. Circ. Heart Fail. 10:e003992. doi: 10.1161/CIRCHEARTFAILURE.117.003992
Tibshirani, R. J. (1996). Regression shrinkage and selection via the LASSO. J. R. Stat. Soc. Ser. B Methodol. 73, 273–282.
Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98, 5116–5121. doi: 10.1073/pnas.091062498
Vargas, J. D., and Lima, J. A. (2013). Coronary artery disease: a gene-expression score to predict obstructive CAD. Nat. Rev. Cardiol. 10, 243–244. doi: 10.1038/nrcardio.2013.50
Wang, J., Gareri, C., and Rockman, H. A. (2018). G-protein–coupled receptors in heart disease. Circ. Res. 123, 716–735. doi: 10.1161/CIRCRESAHA.118.311403
Wang, Y., Wilson, C., Cartwright, E. J., and Lei, M. (2017). Plasma membrane Ca2+ -ATPase 1 is required for maintaining atrial Ca2+ homeostasis and electrophysiological stability in the mouse. J. Physiol. 595, 7383–7398. doi: 10.1113/JP274110
Wang, Q., Xu, M., Sun, Y., Chen, J., and Yang, W. (2019). Gene expression profiling for diagnosis of triple-negative breast cancer: a Multicenter, retrospective cohort study. Front. Oncol. 9:1576. doi: 10.3389/fonc.2019.01576
Yang, H., Cao, H., He, T., Wang, T., and Cui, Y. (2020). Multilevel heterogeneous omics data integration with kernel fusion. Brief. Bioinform. 21, 156–170. doi: 10.1093/bib/bby115
Yao, C., Chen, B. H., Joehanes, R., Otlu, B., Zhang, X., Liu, C., et al. (2015). Integromic analysis of genetic variation and gene expression identifies networks for cardiovascular disease phenotypes. Circulation 131, 536–549. doi: 10.1161/CIRCULATIONAHA.114.010696
Yariswamy, M., Yoshida, T., Valente, A. J., Kandikattu, H. K., Sakamuri, S. S. V. P., Siddesha, J. M., et al. (2016). Cardiac-restricted overexpression of TRAF3 interacting protein 2 (TRAF3IP2) results in spontaneous development of myocardial hypertrophy, fibrosis, and dysfunction. J. Biol. Chem. 291:19425. doi: 10.1074/jbc.M116.724138
Keywords: risk prediction, kernel partial least squares, genetic algorithm, heart failure with preserved ejection fraction, machine learning
Citation: Zhou L, Guo Z, Wang B, Wu Y, Li Z, Yao H, Fang R, Yang H, Cao H and Cui Y (2021) Risk Prediction in Patients With Heart Failure With Preserved Ejection Fraction Using Gene Expression Data and Machine Learning. Front. Genet. 12:652315. doi: 10.3389/fgene.2021.652315
Received: 12 January 2021; Accepted: 02 March 2021;
Published: 22 March 2021.
Edited by:
Chao Xu, University of Oklahoma Health Sciences Center, United StatesReviewed by:
Zhongshang Yuan, Shandong University, ChinaCopyright © 2021 Zhou, Guo, Wang, Wu, Li, Yao, Fang, Yang, Cao and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyan Cao, Y2FvaHlAc3htdS5lZHUuY24=; Yuehua Cui, Y3VpeUBtc3UuZWR1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.