- 1Department of Biological Sciences, Smith College, Northampton, MA, United States
- 2Program in Organismic and Evolutionary Biology, University of Massachusetts Amherst, Amherst, MA, United States
Introduction
Microbial eukaryotes, i.e., protists, represent the bulk of eukaryotic diversity in terms of species diversity and biomass. Protists are globally distributed in all ecosystems and play important roles in food webs and nutrient cycles. To date it remains enigmatic how protist diversity is generated, especially in lineages with large populations in ecosystems without apparent dispersal barriers (i.e., many marine species, species that encyst). We argue that epigenetic processes, such as chromatin modification and/or regulation by small non-protein-coding RNAs (npc-RNAs) that rapidly modify genomes and gene expression states, play important roles in driving phenotypic plasticity, differential adaptation and ultimately diversification of protists. Our argument is based on two recent developments in epigenetic research: (1) it is now clear that epigenetic processes were present in the last eukaryotic common ancestor (LECA) and are widespread across eukaryotes, and (2) numerous studies have demonstrated that at least some epigenetic marks can be inherited across generations. Given this, we suggest to combine morphometrics, genomics, and epigenomics for research on adaptability and diversification in microbial eukaryotes.
Diversity of Microbial Eukaryotes
Many lineages of protists have a tremendous species diversity, which is reflected in a wide variety of morphologies and ecological functions (e.g., Adl et al., 2019). In addition, research of the last two decades has unearthed a large amount of cryptic diversity, suggesting a decoupling of morphological and molecular evolution (e.g., Katz et al., 2005; Šlapeta et al., 2005; Darling and Wade, 2008; Oliverio et al., 2014). Protists occur globally in all ecosystems, and while some species are endemic to certain areas, others have a cosmopolitan distribution and vast population sizes (e.g., Ryšánek et al., 2015; Faure et al., 2019). Large-scale barcoding studies revealed that some closely related cryptic species are able to co-occur in close biogeographical proximity (e.g., Amato et al., 2007; Weiner et al., 2014; Badger et al., 2017; Tucker et al., 2017). In addition, protists show a variety of complex life cycles, sometimes alternating sexual and asexual generations (e.g., Grell, 1973; Parfrey et al., 2008). Given these characteristics, the enormous species diversity is perhaps not surprising. However, which (molecular/epigenetic) mechanisms allow for speciation in microbes, especially in habitats with seemingly unlimited dispersal potential, remains unresolved. Several groups have hypothesized that differential adaptation to environmental factors may be the underlying driver for diversification in sympatry (e.g., Ryšánek et al., 2016; Irwin et al., 2017; Škaloud et al., 2019). However, for gene flow between populations to be overcome, mechanisms leading to the establishment of reproductive isolation would have to be fast and efficient. We argue that in order for rapid diversification to be achieved, epigenetic processes that regulate gene activity and that may be influenced by the environment play important roles in establishing phenotypic plasticity; if the epigenetic marks are inherited across generations—what we refer to as “epigenetic assimilation”—they can provide a fitness advantage to members of the population and ultimately lead to differential adaptation that drives speciation (Figure 1).
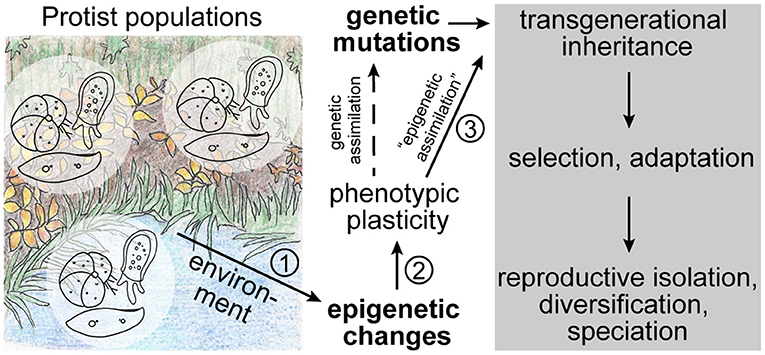
Figure 1. Theoretical sequence of events in ecological speciation driven by epigenetics. In addition to genetic mutations, naturally occurring protist populations experience epigenetic modifications that may be stochastic or be triggered by the environment. These modifications can lead to phenotypic plasticity in the population through changes in genome structure or gene expression states. If the epigenetic modification is followed by a genetic mutation, it may be fixed in the genome through genetic assimilation. However, if the epigenetic mark itself is stably inherited (i.e., “epigenetically assimilation”) across generations, it may represent a selectable advantage that can lead to an increase in fitness of the population and ultimately to adaptation, diversification, and speciation without changes to the genome. The numbers indicate the most critical steps in this sequence of events that we discuss throughout the text.
Epigenetics in Microbial Eukaryotes
A prerequisite for our model of epigenetics as driver of ecological speciation in protists (Figure 1) to be valid is the widespread existence of epigenetic phenomena in microbial eukaryotes. Eukaryotic epigenetics comprises processes such as chromatin and DNA modifications and regulation by npc-RNAs (e.g., Razin and Riggs, 1980; Ng and Bird, 1999; Shabalina and Koonin, 2008), that are thought to have evolved originally for mediating genome conflict between mobile genetic elements and host genomes (Lisch, 2009; Fedoroff, 2012). The effects of epigenetics include, among others, gene activation or silencing, and altering genome structures through DNA elimination or polyploidization (e.g., Liu and Wendel, 2003; Bernstein and Allis, 2005). Most epigenetic research has focused on animals and plants, yet it was recently confirmed that the basic epigenetic gene toolkit was present in LECA and is now widespread throughout the eukaryotic tree of life (Aravind et al., 2014; Weiner et al., 2020). This highlights the importance of epigenetics for the functioning of eukaryotic genomes. For the majority of protists, however, knowledge on their epigenetics remains limited, mostly because many are uncultivable and annotated reference genomes are lacking. What is known so far mostly stems from research on model organisms, such as ciliates (Alveolata) and human pathogens [e.g., Plasmodium (Alveolata) and Trypanosoma (Excavata)].
In ciliates, which contain both a germline and somatic nucleus within one cell, epigenetics plays key roles in distinctions between the two genomes and in elimination of DNA during the development of a new somatic nucleus during reproduction (e.g., Chicoine and Allis, 1986; Jahn and Klobutcher, 2002; Chalker et al., 2013; Pilling et al., 2017). Small npc-RNAs, such as “scan RNAs” and “macronuclear RNAs,” bind to homologous regions in the genome or direct histone modifications (e.g., H3K9 methylation) in those regions to mark them for either retention or elimination (Chen et al., 2014; Swart et al., 2014). Similarly, npc-RNAs, so-called “template RNAs,” were found to be involved in the reordering of scrambled genes in some ciliates (e.g., Garnier et al., 2004; Nowacki et al., 2011). Another phenomenon of genome dynamics that is likely driven by epigenetics is the determination of ploidy levels throughout the life cycle. Many protist lineages, such as some Foraminifera (Rhizaria), ciliates and Amoebozoa have been observed to undergo significant changes in ploidy, sometimes containing thousands of copies of the genome that later are eliminated again (Parfrey et al., 2008; Bellec and Katz, 2012; Goodkov et al., 2020). In the case of ciliates, research suggested that RNA interference, which is part of the “epigenetic toolkit,” is driving these changes (Heyse et al., 2010).
In addition to these large-scale modifications to the genome architecture, epigenetic processes are involved in changes to the morphology or physiology of protists. This is especially prevalent in parasites, in which epigenetics controls virulence and cell differentiation through regulation of gene expression and thus plays an important role in host-pathogen interaction (e.g., Croken et al., 2012; Gomez-Diaz et al., 2012). For example, the formation of cysts (an important life cycle stage for host infection) in Toxoplasma (Apicomplexa), Acanthamoeba (Amoebozoa), and Giardia (Excavata) is driven at least partly by epigenetic mechanisms such as histone acetylation and methylation (e.g., H3K18 acetylation and H3R17 methylation in Toxoplasma; Saksouk et al., 2005; Dixon et al., 2010; Sonda et al., 2010; Moon et al., 2017; Lagunas-Rangel and Bermudez-Cruz, 2019). Antigenic variation, a strategy used by many pathogens (e.g., Trypanosoma brucei, Giardia lamblia, Giardia doudenalis, and Plasmodium falciparum) to avoid the host immune system, also is achieved through epigenetic regulation of gene expression (Kulakova et al., 2006; Elias and Faria, 2009; Juarez-Reyes and Castano, 2019; Lagunas-Rangel and Bermudez-Cruz, 2019). Their genomes contain many genes for surface proteins and the timing of gene expression is at least partly epigenetically regulated, e.g., through histone methylation (H3K4) or acetylation (H3K9) of the var genes in Plasmodium falciparum (e.g., Freitas-Junior et al., 2005; Guizetti and Scherf, 2013; Duffy et al., 2014). In this way, pathogenic protists are able to rapidly react and adapt to a changing host environment.
A further aspect of cell physiology that seems to involve epigenetics is mating type determination in ciliates (e.g., Pilling et al., 2017). While most ciliate species have different mating types, their number varies greatly (up to a 100; Phadke and Zufall, 2009) and so do the molecular mechanisms for mating type determination (e.g., Orias et al., 2017). For Paramecium tetraurelia it could be shown that the difference between its two mating types lies in the presence/absence of a transmembrane protein, whose expression is regulated by “scan RNAs” (Singh et al., 2014).
Despite the fact that details on the exact molecular processes and the genes/enzymes involved often remain scarce, the above-mentioned examples highlight the ubiquity and importance of epigenetics in the life histories of microbial eukaryotes.
The Potential Role of Epigenetics in Driving Adaptation and Diversification
Ecological speciation through differential adaptation to environmental factors may be a plausible explanation for diversification in protists considering their often-large population sizes and wide biogeographic distribution. Here, we focus on the role of epigenetics in these events, yet we acknowledge that bottlenecks and drift likely are also important drivers of diversity in protists, especially in lineages with small populations and restricted distribution. However, the effects of these population genetic phenomena on epigenetics remain largely unknown.
In order to elucidate the interactions between ecology, epigenetics, and evolution that are the basis of our suggested model, special consideration has to be placed on the following questions (Figure 1): (1) does the environment trigger epigenetic variations, (2) can epigenetic modifications lead to phenotypic plasticity, and (3) are environmentally acquired epigenetic marks stably inherited to establish reproductive isolation and speciation? Over the last few years research efforts investigating these interactions have rapidly increased, yet so far mostly focusing on multicellular model species (e.g., Smith and Ritchie, 2013; Vogt, 2017; Boskovic and Rando, 2018; Perez and Lehner, 2019).
The notion that the environment influences epigenetic modifications is by now well-established. Many studies have focused on the effects of stress, toxin exposure or nutrition on epigenetic marks (e.g., Yaish et al., 2011; Collotta et al., 2013; Tiffon, 2018; Weyrich et al., 2019), and research on natural non-model systems showed epigenetic variability in populations across ecological gradients (e.g., Foust et al., 2016; Mcnew et al., 2017; Johnson and Kelly, 2020; Wogan et al., 2020). These studies usually focus on patterns of DNA methylation as this epigenetic modification is better understood and easy to analyze through bisulfite sequencing methods (e.g., Meissner et al., 2005; Smallwood et al., 2014). To our knowledge, few data exist on similar studies of protists, yet we argue that due to their ubiquitous occurrence across a wide range of environments, protist populations hold great promise for investigating environmental effects on epigenetic variation.
Elucidating the influence of epigenetics on phenotypic plasticity is more challenging as it can be difficult to rule out underlying genetic influences. However, recently progress has been made, mostly through experimental modifications to epigenetic marks on DNA or histones and the investigation of subsequent effects on the phenotype (e.g., Kronholm et al., 2016; Verhoeven et al., 2016). Research on a natural system was able to show that epigenetic modifications were more likely than genetic variability to have shaped the behavioral reproductive isolation in fish species (Smith et al., 2016). Similarly, epigenetic mechanisms were found to be responsible for phenotypic plasticity in asexual lineages allowing them to respond to environmental fluctuations (Castonguay and Angers, 2012). A further striking example of rapid phenotypic plasticity induced by epigenetics can be found in protist lineages that use antigenic variation through epigenetically regulated changes in gene expression to adjust to changing environments (see above).
For epigenetics to act as driver of speciation, it is important that epigenetic marks are stably inherited across generations, at least until reproductive isolation is established and/or genetic assimilation has occurred (Figure 1; discussed in Rey et al., 2016). The stable inheritance of epigenetic marks has long been debated as they were assumed to be eliminated during reproduction and only affect the current generation (discussed in: Richards, 2006), a view that is in line with the concepts of the modern synthesis and the rejection of the idea that acquired traits can be passed down to future generations (discussed in: Jablonka and Lamb, 2008; Bonduriansky, 2012). However, in recent years, examples of soft inheritance through transgenerational inheritance of epigenetic marks became more numerous (e.g., Richards, 2006; Bond and Finnegan, 2007; Perez and Lehner, 2019). Again, important examples can be found among protists, such as ciliates, in which acquired changes to the morphology or physiology, such as doublet morphology and mating types, are inherited to progeny without changes in the underlying nucleotide sequence (e.g., Pilling et al., 2017; Neeb and Nowacki, 2018). In addition, experimental evolution on the unicellular algae Chlamydomonas (Archaeplastida) showed that epigenetic variation is stably inherited across generations and thus influences adaptability of the organism (Kronholm et al., 2017).
Conclusion
In recent years, a large amount of research has been published that focuses on the role of epigenetics in ecological speciation. It has been shown that environmentally induced epigenetic modifications can lead to differential gene expression and phenotypic plasticity. If these epigenetic marks are stably inherited across generations (“epigenetic assimilation”) and increase the fitness of the population, they could be substrate for selection and thus represent a first step toward ecological speciation (Figure 1).
While detailed information on the molecular processes of epigenetics in microbial eukaryotes remains scarce, its prominent role in shaping genome dynamics and driving phenotypic plasticity even across generations makes it likely that epigenetics is involved in generating their tremendous diversity. This, as well as their short generation times, make protists interesting model systems for studying the influence of epigenetics on adaptation and speciation. The model of ecological speciation driven by epigenetics presented here is consistent with the idea of rapid diversification in lineages with large population sizes and therefore weak genetic drift. Recent improvements in the sensitivity of high-throughput sequencing techniques to sequence the genomes, transcriptomes, and epigenomes of non-model microbes make this an exciting time to combine molecular, morphological, and epigenetic approaches for elucidating the origin of species diversity and a species' response to changing environmental conditions.
Author Contributions
AW and LK developed the ideas and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
LK was supported by grants from the National Institute of Health (grant number R15HG010409) and the National Science Foundation (grant numbers OCE-1924570, DEB-1651908, and DEB-1541511).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the editors for organizing the Frontiers Research Topic Gene Regulation as a Driver of Adaptation and Speciation and two reviewers for helpful comments on our manuscript. We thank Eleanor Goetz and Caitlin Timmons for the drawings in the figure.
References
Adl, S. M., Bass, D., Lane, C. E., Lukes, J., Schoch, C. L., Smirnov, A., et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119. doi: 10.1111/jeu.12691
Amato, A., Kooistra, W. H., Ghiron, J. H. L., Mann, D. G., Pröschold, T., and Montresor, M. (2007). Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 158, 193–207. doi: 10.1016/j.protis.2006.10.001
Aravind, L., Burroughs, A. M., Zhang, D. P., and Iyer, L. M. (2014). Protein and DNA modifications: evolutionary imprints of bacterial biochemical diversification and geochemistry on the provenance of eukaryotic epigenetics. Cold Spring Harb. Perspect. Biol. 6:a016063. doi: 10.1101/cshperspect.a016063
Badger, M., Tucker, S. J., Grattepanche, J. D., and Katz, L. A. (2017). Rapid turnover of ciliate community members in New England tide pools. Aquat. Microb. Ecol. 80, 43–54. doi: 10.3354/ame01839
Bellec, L., and Katz, L. A. (2012). Analyses of chromosome copy number and expression level of four genes in the ciliate Chilodonella uncinata reveal a complex pattern that suggests epigenetic regulation. Gene 504, 303–308. doi: 10.1016/j.gene.2012.04.067
Bernstein, E., and Allis, C. D. (2005). RNA meets chromatin. Genes Dev. 19, 1635–1655. doi: 10.1101/gad.1324305
Bond, D. M., and Finnegan, E. J. (2007). Passing the message on: inheritance of epigenetic traits. Trends Plant Sci. 12, 211–216. doi: 10.1016/j.tplants.2007.03.010
Bonduriansky, R. (2012). Rethinking heredity, again. Trends Ecol. Evol. 27, 330–336. doi: 10.1016/j.tree.2012.02.003
Boskovic, A., and Rando, O. J. (2018). Transgenerational epigenetic inheritance. Ann. Rev. Genet. 52, 21–41. doi: 10.1146/annurev-genet-120417-031404
Castonguay, E., and Angers, B. (2012). The key role of epigenetics in the persistence of asexual lineages. Genet. Res. Int. 2012:534289. doi: 10.1155/2012/534289
Chalker, D. L., Meyer, E., and Mochizuki, K. (2013). Epigenetics of ciliates. Cold Spring Harb. Perspect. Biol. 5:a017764. doi: 10.1101/cshperspect.a017764
Chen, X., Bracht, J. R., Goldman, A. D., Dolzhenko, E., Clay, D. M., Swart, E. C., et al. (2014). The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 158, 1187–1198. doi: 10.1016/j.cell.2014.07.034
Chicoine, L. G., and Allis, C. D. (1986). Regulation of histone acetylation during macronuclear differentiation in Tetrahymena: evidence for control at the level of acetylation and deacetylation. Dev. Biol. 116, 477–485. doi: 10.1016/0012-1606(86)90148-X
Collotta, M., Bertazzi, P. A., and Bollati, V. (2013). Epigenetics and pesticides. Toxicology 307, 35–41. doi: 10.1016/j.tox.2013.01.017
Croken, M. M., Nardelli, S. C., and Kim, K. (2012). Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol. 28, 202–213. doi: 10.1016/j.pt.2012.02.009
Darling, K. F., and Wade, C. M. (2008). The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes. Mar. Micropaleontol. 67, 216–238. doi: 10.1016/j.marmicro.2008.01.009
Dixon, S. E., Stilger, K. L., Elias, E. V., Naguleswaran, A., and Sullivan, W. J. (2010). A decade of epigenetic research in Toxoplasma gondii. Mol. Biochem. Parasitol. 173, 1–9. doi: 10.1016/j.molbiopara.2010.05.001
Duffy, M. F., Selvarajah, S. A., Josling, G. A., and Petter, M. (2014). Epigenetic regulation of the Plasmodium falciparum genome. Brief. Funct. Genom. 13, 203–216. doi: 10.1093/bfgp/elt047
Elias, M. C., and Faria, M. (2009). Are there epigenetic controls in Trypanosoma cruzi? Ann. N. Y. Acad. Sci. 1178, 285–290. doi: 10.1111/j.1749-6632.2009.05008.x
Faure, E., Not, F., Benoiston, A.-S., Labadie, K., Bittner, L., and Ayata, S.-D. (2019). Mixotrophic protists display contrasted biogeographies in the global ocean. ISME J. 13, 1072–1083. doi: 10.1038/s41396-018-0340-5
Fedoroff, N. V. (2012). Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338, 758–767. doi: 10.1126/science.338.6108.758
Foust, C. M., Preite, V., Schrey, A. W., Alvarez, M., Robertson, M. H., Verhoeven, K. J., et al. (2016). Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol. Ecol. 25, 1639–1652. doi: 10.1111/mec.13522
Freitas-Junior, L. H., Hernandez-Rivas, R., Ralph, S. A., Montiel-Condado, D., Ruvalcaba-Salazar, O. K., Rojas-Meza, A. P., et al. (2005). Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121, 25–36. doi: 10.1016/j.cell.2005.01.037
Garnier, O., Serrano, V., Duharcourt, S., and Meyer, E. (2004). RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 24, 7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004
Gomez-Diaz, E., Jorda, M., Angel Peinado, M., and Rivero, A. (2012). Epigenetics of host-pathogen interactions: the road ahead and the road behind. PLoS Pathog. 8:e1003007. doi: 10.1371/journal.ppat.1003007
Goodkov, A. V., Berdieva, M. A., Podlipaeva, Y. I., and Demin, S. Y. (2020). The chromatin extrusion phenomenon in Amoeba proteus cell cycle. J. Eukaryot. Microbiol. 67, 203–208. doi: 10.1111/jeu.12771
Guizetti, J., and Scherf, A. (2013). Silence, activate, poise and switch! mechanisms of antigenic variation in Plasmodium falciparum. Cell. Microbiol. 15, 718–726. doi: 10.1111/cmi.12115
Heyse, G., Jonsson, F., Chang, W. J., and Lipps, H. J. (2010). RNA-dependent control of gene amplification. Proc. Natl. Acad. Sci. U.S.A. 107, 22134–22139. doi: 10.1073/pnas.1009284107
Irwin, N. A., Sabetrasekh, M., and Lynn, D. H. (2017). Diversification and phylogenetics of mobilid peritrichs (ciliophora) with description of Urceolaria parakorschelti sp. nov. Protist 168, 481–493. doi: 10.1016/j.protis.2017.07.003
Jablonka, E., and Lamb, M. J. (2008). The epigenome in evolution: beyond the modern synthesis. Becmhuk BOTuC, 12, 242–254.
Jahn, C. L., and Klobutcher, L. A. (2002). Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56, 489–520. doi: 10.1146/annurev.micro.56.012302.160916
Johnson, K. M., and Kelly, M. W. (2020). Population epigenetic divergence exceeds genetic divergence in the Eastern oyster Crassostrea virginica in the Northern Gulf of Mexico. Evol. Appl. 13, 945–959. doi: 10.1111/eva.12912
Juarez-Reyes, A., and Castano, I. (2019). Chromatin architecture and virulence-related gene expression in eukaryotic microbial pathogens. Curr. Genet. 65, 435–443. doi: 10.1007/s00294-018-0903-z
Katz, L. A., Mcmanus, G. B., Snoeyenbos-West, O. L. O., Griffin, A., Pirog, K., Costas, B., et al. (2005). Reframing the “Everything is everywhere” debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquat. Microb. Ecol. 41, 55–65. doi: 10.3354/ame041055
Kronholm, I., Bassett, A., Baulcombe, D., and Collins, S. (2017). Epigenetic and genetic contributions to adaptation in Chlamydomonas. Mol. Biol. Evol. 34, 2285–2306. doi: 10.1093/molbev/msx166
Kronholm, I., Johannesson, H., and Ketola, T. (2016). Epigenetic control of phenotypic plasticity in the filamentous fungus Neurospora crassa. Genes Genom. Genet. 6, 4009–4022. doi: 10.1534/g3.116.033860
Kulakova, L., Singer, S. M., Conrad, J., and Nash, T. E. (2006). Epigenetic mechanisms are involved in the control of Giardia lamblia antigenic variation. Mol. Microbiol. 61, 1533–1542. doi: 10.1111/j.1365-2958.2006.05345.x
Lagunas-Rangel, F. A., and Bermudez-Cruz, R. M. (2019). Epigenetics in the early divergent eukaryotic Giardia duodenalis: an update. Biochimie 156, 123–128. doi: 10.1016/j.biochi.2018.10.008
Lisch, D. (2009). Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 60, 43–66. doi: 10.1146/annurev.arplant.59.032607.092744
Liu, B., and Wendel, J. F. (2003). Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 29, 365–379. doi: 10.1016/S1055-7903(03)00213-6
Mcnew, S. M., Beck, D., Sadler-Riggleman, I., Knutie, S. A., Koop, J. A., Clayton, D. H., et al. (2017). Epigenetic variation between urban and rural populations of Darwin's finches. BMC Evol. Biol. 17:183. doi: 10.1186/s12862-017-1025-9
Meissner, A., Gnirke, A., Bell, G. W., Ramsahoye, B., Lander, E. S., and Jaenisch, R. (2005). Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877. doi: 10.1093/nar/gki901
Moon, E. K., Hong, Y., Lee, H. A., Quan, F. S., and Kong, H. H. (2017). DNA Methylation of gene expression in Acanthamoeba castellanii encystation. Korean J. Parasitol. 55, 115–120. doi: 10.3347/kjp.2017.55.2.115
Neeb, Z. T., and Nowacki, M. (2018). RNA-mediated transgenerational inheritance in ciliates and plants. Chromosoma 127, 19–27. doi: 10.1007/s00412-017-0655-4
Ng, H. H., and Bird, A. (1999). DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9, 158–163. doi: 10.1016/S0959-437X(99)80024-0
Nowacki, M., Shetty, K., and Landweber, L. F. (2011). RNA-Mediated epigenetic programming of genome rearrangements. Annu. Rev. Genom. Hum. Genet. 12, 367–389. doi: 10.1146/annurev-genom-082410-101420
Oliverio, A., Lahr, D. J. G., Nguyen, T., and Katz, L. A. (2014). Cryptic diversity within morphospecies of testate (shelled) amoebae in New England bogs and fens. Protist 165, 196–207. doi: 10.1016/j.protis.2014.02.001
Orias, E., Singh, D. P., and Meyer, E. (2017). Genetics and epigenetics of mating type determination in Paramecium and Tetrahymena. Annu. Rev. Microbiol. 71, 133–156. doi: 10.1146/annurev-micro-090816-093342
Parfrey, L. W., Lahr, D. J. G., and Katz, L. A. (2008). The dynamic nature of eukaryotic genomes. Mol. Biol. Evol. 25, 787–794. doi: 10.1093/molbev/msn032
Perez, M. F., and Lehner, B. (2019). Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143–151. doi: 10.1038/s41556-018-0242-9
Phadke, S. S., and Zufall, R. A. (2009). Rapid diversification of mating systems in ciliates. Biol. J. Linn. Soc. 98, 187–197. doi: 10.1111/j.1095-8312.2009.01250.x
Pilling, O. A., Rogers, A. J., Gulla-Devaney, B., and Katz, L. A. (2017). Insights into transgenerational epigenetics from studies of ciliates. Eur. J. Protistol. 61, 366–375. doi: 10.1016/j.ejop.2017.05.004
Razin, A., and Riggs, A. D. (1980). DNA Methylation and gene-function. Science 210, 604–610. doi: 10.1126/science.6254144
Rey, O., Danchin, E., Mirouze, M., Loot, C., and Blanchet, S. (2016). Adaptation to global change: a transposable element-epigenetics perspective. Trends Ecol. Evol. 31, 514–526. doi: 10.1016/j.tree.2016.03.013
Richards, E. J. (2006). Inherited epigenetic variation–revisiting soft inheritance. Nat. Rev. Genet. 7, 395–401. doi: 10.1038/nrg1834
Ryšánek, D., Holzinger, A., and Škaloud, P. (2016). Influence of substrate and pH on the diversity of the aeroterrestrial alga Klebsormidium (Klebsormidiales, Streptophyta): a potentially important factor for sympatric speciation. Phycologia 55, 347–358. doi: 10.2216/15-110.1
Ryšánek, D., Hrčková, K., and Škaloud, P. (2015). Global ubiquity and local endemism of free-living terrestrial protists: phylogeographic assessment of the streptophyte alga Klebsormidium. Environ. Microbiol. 17, 689–698. doi: 10.1111/1462-2920.12501
Saksouk, N., Bhatti, M. M., Kieffer, S., Smith, A. T., Musset, K., Garin, J., et al. (2005). Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25, 10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005
Shabalina, S. A., and Koonin, E. V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587. doi: 10.1016/j.tree.2008.06.005
Singh, D. P., Saudemont, B., Guglielmi, G., Arnaiz, O., Gout, J. F., Prajer, M., et al. (2014). Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature 509, 447–452. doi: 10.1038/nature13318
Škaloud, P., Škaloudová, M., Doskočilová, P., Kim, J. I., Shin, W., and Dvorák, P. (2019). Speciation in protists: spatial and ecological divergence processes cause rapid species diversification in a freshwater chrysophyte. Mol. Ecol. 28, 1084–1095. doi: 10.1111/mec.15011
Šlapeta, J., Moreira, D., and López-García, P. (2005). The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. R. Soc. Lond. B Biol. Sci. 272, 2073–2081. doi: 10.1098/rspb.2005.3195
Smallwood, S. A., Lee, H. J., Angermueller, C., Krueger, F., Saadeh, H., Peat, J., et al. (2014). Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820. doi: 10.1038/nmeth.3035
Smith, G., and Ritchie, M. G. (2013). How might epigenetics contribute to ecological speciation? Curr. Zool. 59, 686–696. doi: 10.1093/czoolo/59.5.686
Smith, T. A., Martin, M. D., Nguyen, M., and Mendelson, T. C. (2016). Epigenetic divergence as a potential first step in darter speciation. Mol. Ecol. 25, 1883–1894. doi: 10.1111/mec.13561
Sonda, S., Morf, L., Bottova, I., Baetschmann, H., Rehrauer, H., Caflisch, A., et al. (2010). Epigenetic mechanisms regulate stage differentiation in the minimized protozoan Giardia lamblia. Mol. Microbiol. 76, 48–67. doi: 10.1111/j.1365-2958.2010.07062.x
Swart, E. C., Wilkes, C. D., Sandoval, P. Y., Arambasic, M., Sperling, L., and Nowacki, M. (2014). Genome-wide analysis of genetic and epigenetic control of programmed DNA deletion. Nucleic Acids Res. 42, 8970–8983. doi: 10.1093/nar/gku619
Tiffon, C. (2018). The impact of nutrition and environmental epigenetics on human health and disease. Int. J. Mol. Sci. 19:3425. doi: 10.3390/ijms19113425
Tucker, S. J., Mcmanus, G. B., Katz, L. A., and Grattepanche, J. D. (2017). Distribution of abundant and active planktonic ciliates in coastal and slope waters off New England. Front. Microbiol. 8:2178. doi: 10.3389/fmicb.2017.02178
Verhoeven, K. J. F., Vonholdt, B. M., and Sork, V. L. (2016). Epigenetics in ecology and evolution: what we know and what we need to know. Mol. Ecol. 25, 1631–1638. doi: 10.1111/mec.13617
Vogt, G. (2017). Facilitation of environmental adaptation and evolution by epigenetic phenotype variation: insights from clonal, invasive, polyploid, and domesticated animals. Environ. Epigenet. 3:dvx002. doi: 10.1093/eep/dvx002
Weiner, A. K., Cerón-Romero, M. A., Yan, Y., and Katz, L. A. (2020). Phylogenomics of the epigenetic toolkit reveals punctate retention of genes across eukaryotes. Genome Biol. Evol. 12, 2196–2210. doi: 10.1093/gbe/evaa198
Weiner, A. K., Weinkauf, M. F., Kurasawa, A., Darling, K. F., Kucera, M., and Grimm, G. W. (2014). Phylogeography of the tropical planktonic foraminifera lineage Globigerinella reveals isolation inconsistent with passive dispersal by ocean currents. PLoS ONE 9:e92148. doi: 10.1371/journal.pone.0092148
Weyrich, A., Lenz, D., and Fickel, J. (2019). Environmental change-dependent inherited epigenetic response. Genes 10:4. doi: 10.3390/genes10010004
Wogan, G. O. U., Yuan, M. L., Mahler, D. L., and Wang, I. J. (2020). Genome-wide epigenetic isolation by environment in a widespread Anolis lizard. Mol. Ecol. 29, 40–55. doi: 10.1111/mec.15301
Keywords: epigenetics, adaptation, speciation, chromatin modification, non-protein-coding RNA, protists
Citation: Weiner AKM and Katz LA (2021) Epigenetics as Driver of Adaptation and Diversification in Microbial Eukaryotes. Front. Genet. 12:642220. doi: 10.3389/fgene.2021.642220
Received: 15 December 2020; Accepted: 15 February 2021;
Published: 16 March 2021.
Edited by:
Ekaterina Shelest, German Centre for Integrative Biodiversity Research (iDiv), GermanyReviewed by:
Rosa Maria Bermudez-Cruz, Instituto Politécnico Nacional de México (CINVESTAV), MexicoDouglas Chalker, Washington University in St. Louis, United States
Copyright © 2021 Weiner and Katz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnes K. M. Weiner, YXdlaW5lckBzbWl0aC5lZHU=; orcid.org/0000-0002-9917-5235; Laura A. Katz, bGthdHpAc21pdGguZWR1; orcid.org/0000-0002-9138-4702
 Agnes K. M. Weiner
Agnes K. M. Weiner Laura A. Katz
Laura A. Katz