
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 25 March 2021
Sec. Genetics of Common and Rare Diseases
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.630359
This article is part of the Research Topic Genetics of Complex Traits & Diseases from Under-Represented Populations View all 11 articles
 Zixian Wang1,2,3,4†
Zixian Wang1,2,3,4† Hui Chen1,2,3†
Hui Chen1,2,3† Min Qin1,2,3
Min Qin1,2,3 Chen Liu5
Chen Liu5 Qilin Ma6
Qilin Ma6 Xiaoping Chen7
Xiaoping Chen7 Ying Zhang8
Ying Zhang8 Weihua Lai2
Weihua Lai2 Xiaojuan Zhang2*
Xiaojuan Zhang2* Shilong Zhong1,2,3,4*
Shilong Zhong1,2,3,4*Plasma lipids have been at the center stage of the prediction and prevention strategies for cardiovascular diseases (CVDs), and novel lipidomic traits have been recognized as reliable biomarkers for CVD risk prediction. The mitochondria serve as energy supply sites for cells and can synthesize a variety of lipids autonomously. Therefore, investigating the relationships between mitochondrial single nucleotide polymorphism (SNPs) and plasma lipidomic traits is meaningful. Here, we enrolled a total of 1,409 Han Chinese patients with coronary artery disease from three centers and performed linear regression analyses on the SNPs of mitochondrial DNA (mtDNA) and lipidomic traits in two independent groups. Sex, age, aspartate aminotransferase, estimated glomerular filtration rate, antihypertensive drugs, hypertension, and diabetes were adjusted. We identified three associations, namely, D-loopm.16089T>C with TG(50:4) NL-16:0, D-loopm.16145G>A with TG(54:5) NL-18:0, and D-loopm.16089T>C with PC(16:0_16:1) at the statistically significant threshold of FDR < 0.05. Then, we explored the relationships between mitochondrial genetic variants and traditional lipids, including triglyceride, total cholesterol (TC), low-density lipoprotein cholesterol (LDLC), and high-density lipoprotein cholesterol. Two significant associations were found, namely MT-ND6m.14178T>C with TC and D-loopm.215A>G with LDLC. Furthermore, we performed linear regression analysis to determine on the SNPs of mtDNA and left ventricular ejection fraction (LVEF) and found that the SNP D-loopm.16145G>A was nominally significantly associated with LVEF (P = 0.047). Our findings provide insights into the lipidomic context of mtDNA variations and highlight the importance of studying mitochondrial genetic variants related to lipid species.
Coronary artery disease (CAD) is one of the greatest threats to human health (Virani et al., 2020). Lipid metabolism plays a central role in the development of cardiovascular diseases (CVDs) and is considered a therapeutic intervention target for CVDs (Ference et al., 2017; Ouchi et al., 2019; Echouffo-Tcheugui et al., 2020). Lipidomic traits can be used independently in predicting long-term CVD risk and adverse clinical outcome. The application of lipidomic traits for risk prediction is more comprehensive than traditional lipids, namely triglycerides (TRIG), total cholesterol (TC), low-density lipoprotein (LDLC), and high-density lipoprotein cholesterol (HDLC) (Razquin et al., 2018; Cadby et al., 2020; Poss et al., 2020).
Mitochondria serve as energy supply sites for cells and can synthesize a variety of lipids autonomously, such as phosphatidylglycerol, cardiolipin, and phosphatidic acid (Horvath and Daum, 2013). Mitochondrial dysfunction can cause various diseases, including CVDs (cardiomyopathy, heart failure, and arrhythmia) (Marín-García and Goldenthal, 2002; Yang et al., 2014; Wahbi et al., 2015). Human mitochondrial DNA (mtDNA) is in a circular double-stranded state and approximately 16.5 kb in length and encodes 37 genes (Sobenin et al., 2012). The replication of mtDNA starts with the displacement loop (D-loop) and contains a transcript promoter adjacent to the D-loop (Sharma and Sampath, 2019). Although the exact function of the D-loop is unclear, this region has a high sequence variability (Ingman et al., 2000). The sequences outside the D-loop region are all exons and encode genes, and thus mtDNA mutations are more likely to occur in the coding region than autosomal DNA mutations and directly affect the physiological functions of the genes encoded by them. Therefore, identifying the associations between mitochondrial genetic patterns and lipid species can provide a deeper understanding of lipid metabolism and its biological links to clinical features and disease progression.
Here, we performed large-scale association analyses on mtDNA variations and lipidomic traits in a Han Chinese population to obtain insights into the relationships between mitochondrial genetic variants and plasma lipidomic traits in CAD patients. We recruited 1,409 subjects with CAD from three centers and used multiple linear regression to explore the relationships between the 42 single nucleotide polymorphisms (SNPs) of mtDNA and 309 lipid species in two independent groups. Moreover, we investigated the relationships between mtDNA variations and the plasma profiling of traditional lipids, including TRIG, TC, LDLC, and HDLC to search more SNPs affecting these lipids. We then explored the effects of mitochondrial genetic variants on heart function by using linear regression to analyze the variations in mtDNA with left ventricular ejection fraction (LVEF). Our findings may provide novel insights into the lipidomic context of mtDNA variations and provide a potential target for pharmacological intervention and optimized treatment of patients with CAD.
Two groups were studied, and the Group I and Group II were mainly grouped according to the time of enrolment. Subjects of the Group I were sequentially enrolled from Guangdong Provincial People’s Hospital between January 2010 and December 2013. The samples in Group I were obtained from patients who had complete baseline characteristics and genetic data in our previous study (Liu et al., 2019). Group II was a multicenter cohort including patients from three centers (Guangdong Provincial People’s Hospital, Xiangya Hospital, Center South University, and the First Affiliated Hospital, Sun Yat-sen University) from September 2017 to October 2018 to illustrate the repeatability and representation of the results from the Group I. All patients enrolled in this study had taken statin lipid-lowering drugs, diagnosed with CAD through coronary angiography, and had undergone percutaneous coronary intervention (PCI). The exclusion criteria in Group II were the same as those used in the previous study, namely, (1) age < 18 years or >80 years; (2) renal insufficiency [defined as serum creatinine concentration > two times the upper limit of normal (230 μmol/L) and renal transplantation or dialysis]; (3) hepatic insufficiency [defined as serum transaminase concentration > two times the upper limit of normal (80 U/L) or diagnosis of cirrhosis]; (4) pregnancy or lactating status; (5) advanced cancer or hemodialysis; (6) history of thyroid problems and the use of antithyroid drugs or thyroid hormone medication; and (7) incomplete information about cardiovascular events during follow-up.
Our study was approved by the Medical Ethical Review Committee of Guangdong General Hospital (Nos. GDREC2010137 and GDREC2017071H) and conducted in accordance with the Declaration of Helsinki. Informed consent (Nos. 20100910 and 20170211) was obtained each participant enrolled in the study. Baseline information, such as demographics, medical history, biochemical measurements, and medication, were all obtained from the hospital information database.
The lipidomic data of the two groups were obtained through widely targeted lipidomic profiling. Details were described in our previous study (Qin et al., 2020), see Supplementary Material. In general, 667 lipid species were detected in the Group I, which contained 14 categories, namely, monoglyceride (MG), cholesteryl esters (CE), diacylglycerol (DG), triacylglycerol (TG), phosphatidic acids (PA), phosphatidylcholines (PC), phosphatidylglycerol (PG), phosphatidylserines (PS), phosphatidylethanolamines (PE), lysophosphatidic acids (LPA), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), hemolytic serine (LPS), and ceramides (Cer). 687 lipid species were annotated in Group II, and a total of 309 lipid species were detected in both groups. For lipidomic analyses, raw signals with more than half of missing rate in the QC samples (those with zero ion intensity) were removed. Missing lipidomic data were imputed by replacing the missing value with a minimum value of the lipid species quantified. The lipidomic data after the batch effect correction by Quality Control–based Robust LOESS (Locally Estimated Scatterplot Smoothing) Signal Correction (QC-RLSC) algorithm was then scaled by pareto scaling, which utilized the square root of the standard deviation as the scaling factor (van den Berg et al., 2006).
For Group II, each eligible patient fasted for at least 8 h. This procedure was performed to minimize the influence of nutrition on lipid species levels. Blood samples were collected using ethylenediaminetetraacetic acid-coated tubes. Whole blood samples were separated into plasma and hemocytes within 2 h through centrifugation at 2,095 g for 10 min at 4°C and then stored at -80°C for further analysis. DNA was extracted from the hemocyte samples for genotyping with a TGuideM16 automatic nucleic acid extractor (Cat. No. OSE-M16) and genomic DNA extraction kit (Cat. No. OSR-M102; TIANGEN). Concentration quantification and electrophoresis were conducted for DNA QC, and qualified DNA was genotyped using an Illumina Infinium global screening array (GSA) bead chip. In brief, the 140 SNPs of mtDNA were genotyped for the two groups. We combined the mitochondrial genetic data of both groups, and the exclusion criteria were as follows: (1) maximum per-person missing rate > 0.05; (2) maximum per-SNP missing rate > 0.05; (3) Hardy–Weinberg disequilibrium P value < 1e-6; and (4) minor allele frequency < 0.01.
Linear regression analyses were conducted at each qualified SNPs of mtDNA for the levels of 309 lipid species presented in Group I and Group II. Sex, age, aspartate aminotransferase (AST), estimated glomerular filtration rate (eGFR), antihypertensive drugs (including β-blockers, angiotensin converting enzyme inhibitors, calcium channel blockers, and proton pump inhibitors), hypertension, and diabetes mellitus were adjusted. All association analyses were conducted using PLINK v2.0 software (Chang et al., 2015).
We used an inverse-variance weighted method meta-analysis, which was based on the effect sizes, and the standard errors adjusting for genomic control were used to combine the association results for the two groups. The heterogeneities in each association between the datasets were tested using the Cochran’s Q test. The analyses above were conducted using METAL software (Willer et al., 2010). Only the associations that met the following conditions were presented: (1) P-value of both groups < 0.05 and (2) the P-value of heterogeneity of effects between the two groups > 0.001. The results of meta-analysis were adjusted to account for multiple testing by the Benjamini-Hochberg false discovery rate (FDR) method and associations were considered statistically significant at FDR < 0.05. All analyses were performed in R v3.6.11.
To explore the influence of mitochondrial genetic variants with traditional lipids and heart function, multiple linear regression of the qualified SNPs of mtDNA to the traditional lipids (TRIG, TC, LDLC, and HDLC), and the heart function (LVEF) were conducted with adjusting for confounding factors including sex, age, AST, eGFR, antihypertensive drugs, hypertension, and diabetes mellitus. Benjamini-Hochberg FDR was used in correcting the number of the SNPs of mtDNA for multiple hypothesis testing, and statistical analyses were performed using PLINK v2.0 and R v3.6.1.
This study enrolled 1,409 Han Chinese patients with CAD who were undergoing PCI therapy. A total of 914 subjects were included in Group I (62.90 ± 10.06 years old), and 495 subjects from three centers (353 patients from Guangdong Provincial People’s Hospital, 138 patients from Xiangya Hospital, Center South University, and 4 patients from the First Affiliated Hospital, Sun Yat-sen University) were included in Group II (61.98 ± 9.81 years old). All the subjects had complete mitochondrial genetic, lipidomic, and traditional lipids data. The average TRIG levels were 1.61 ± 1.13 and 1.84 ± 1.90 mmol/L for Group I and Group II, respectively. The average TC levels were 4.25 ± 1.09 and 4.28 ± 1.78 for Group I and Group II. The average LDLC levels were 2.56 ± 0.90 and 2.70 ± 0.97 mmol/L for Group I and Group II. The average HDLC levels were 0.96 ± 0.25 and 0.99 ± 0.25 mmol/L for Group I and Group II. The detailed demographic characteristics are presented in Table 1.
As indicated in the Materials and methods section, the 42 SNPs of mtDNA, including nine SNPs in the D-loop region, two SNPs on the 12S rRNA, two SNPs on the 16S rRNA, one SNP on the tRNA, and 28 SNPs in the exon region met the quality control criteria for merged genotype data. In the lipidomic data, 309 plasma lipid species, including 7 CE, 23 DG, 172 TG, 50 PC, 18 LPC, 19 LPE, 1 LPS, and 19 Cer were included for the association analyses conducted on each group.
Primary association analyses were conducted using the 42 SNPs of mtDNA with 309 lipid species in the two groups. The association results were merged through meta-analysis. P-values in the two groups are below 0.05, and no significant heterogeneity between them was observed (see section “Materials and Methods”; Table 2). In total, we identified seven associations of mitochondrial genetic variants with lipid species (P < 0.05). Three associations were statistically significant after FDR correction (Table 2 and Figure 1). m.16089T > C (T > C) in D-loop was associated with the increased level of TG(50:4) NL-16:0 (P = 0.00035, FDR = 0.01462), m.16145G > A (A > G) in D-loop was associated with the decreased level of TG(54:5) NL-18:0 (P = 0.00111, FDR = 0.04670), and m.16089T > C (T > C) in D-loop was associated with the increased level of PC(16:0_16:1) (P = 0.00113, FDR = 0.04759). Four of them were in nominally significant associations (P < 0.05, but FDR > 0.05). Specifically, m.15924A > G (A > G) in tRNA-Thr was nominally significantly associated with the increased level of TG(58:9) NL-20:4 (P = 0.00256, FDR = 0.10756), m.16217T > C (T > C) in D-loop was nominally significantly associated with the increased levels of Cer(m18:1/26:0) (P = 0.00401, FDR = 0.16859), and TG(58:3) NL-18:2 (P = 0.00567, FDR = 0.23801), and m.6680T > C (T > C) in MT-CO1 was nominally significantly associated with the increased level of CE(18:1) (P = 0.00749, FDR = 0.20076).
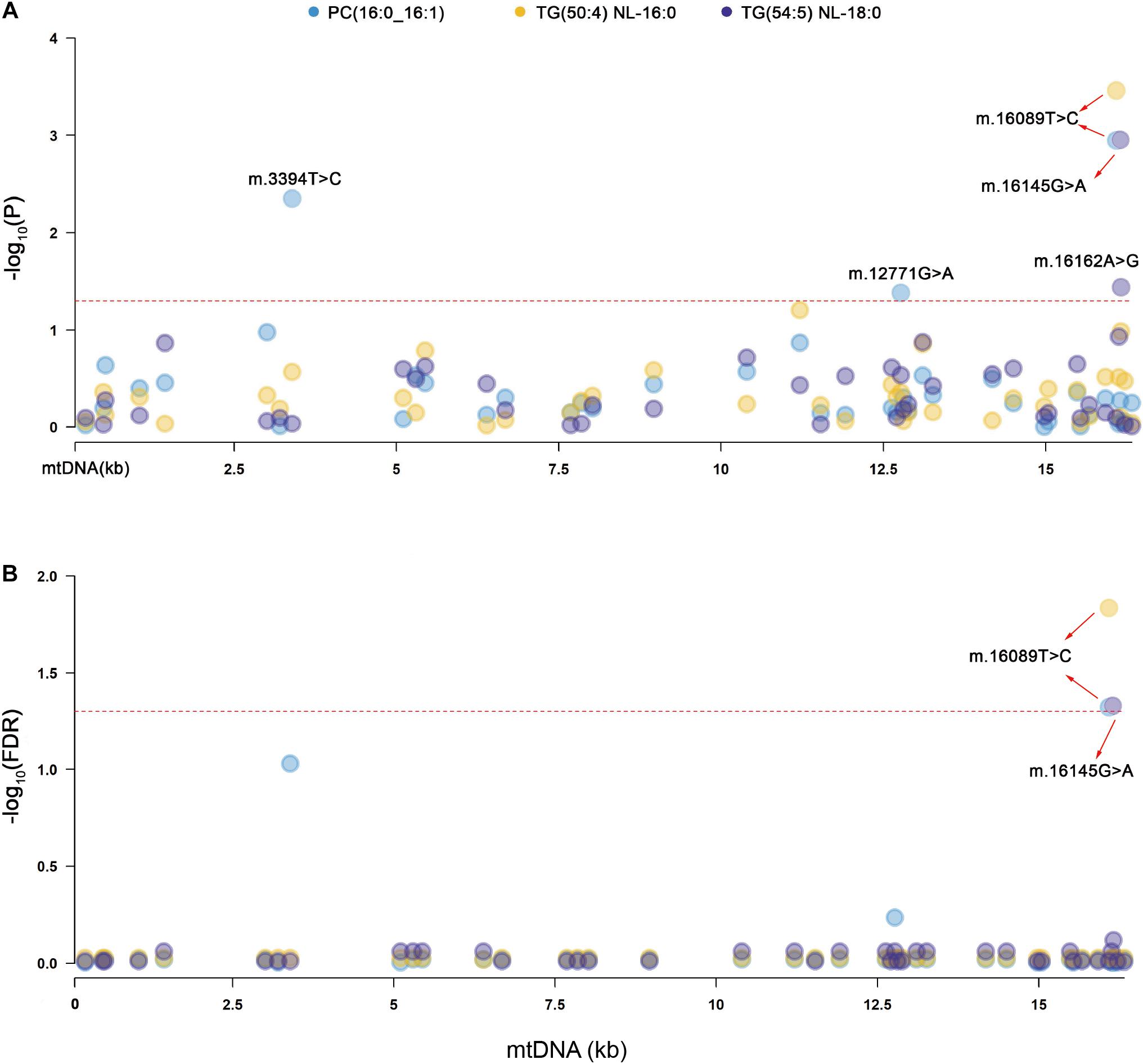
Figure 1. Manhattan plots for the significant associations of mitochondrial genetic variants with lipid species. PC, phosphatidylcholines, TG, triacylglycerol. (A) The red dashed line indicates the threshold of nominal significance on lipid species (P < 0.05). (B) The red dashed line indicates the threshold of statistical significance on lipid species after corrected (FDR < 0.05).
Moreover, we performed association analyses between the 42 SNPs of mtDNA and four traditional lipids to explore the relationships of mtDNA variants and traditional lipids (TRIG, TC, LDLC, and HDLC), using a merged genotype and lipids data (N = 1409). No SNP was statistically significantly associated with TRIG. The results of potential associations (P < 0.05) were presented in Table 3. After FDR corrections, two associations were considered statistically significant (FDR < 0.05). m.14178T > C (T > C) in MT-ND6 was associated with the increased level of TC, and m.215A > G (A > G) in D-loop was associated with the increased level of LDLC (Figure 2). All 42 SNPs with the four lipids were presented in Supplementary Tables 1–4.
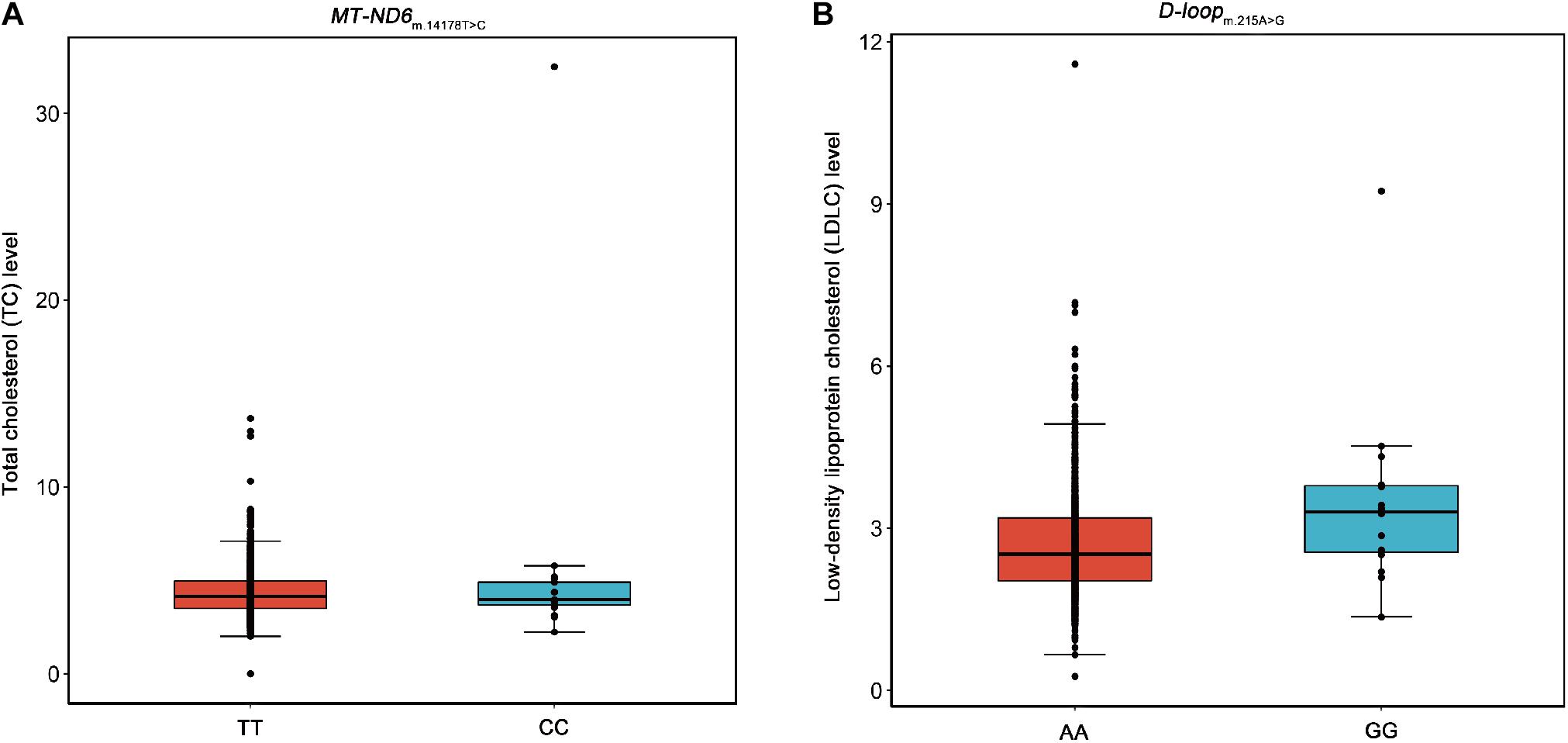
Figure 2. Box plots for the SNPs m.14178T > C and m.215A > G against the levels of TC and LDLC. Five lines from top to bottom for each box were upper extreme, upper quartile, median, lower quartile, and lower extreme. Whiskers were drawn between upper extreme and upper quartile, lower quartile and lower extreme, and the dots above upper extreme or below lower extreme were outliers. (A) The SNP m.14178T > C (T > C) in MT-ND6 against the level of TC. (B) The SNP m.215A > G (A > G) in D-loop against the level of LDLC.
We performed further analyses to explore the relationships between the SNPs of mtDNA and heart function. LVEF is a continuous variable that can be used as an index for evaluating heart function, and a low LVEF value implies poor heart function. A total of 1,151 subjects with LVEF data were available in linear regression analyses, whose LVEF(%) was 59.92 ± 11.47 (mean ± standard deviation). Three SNPs were nominally significantly associated with LVEF (P < 0.05 but FDR > 0.05). m.1048C > T (C > T) in MT-RNR1 was nominally significantly associated with decreased LVEF, m.12705C > T (T > C) in MT-ND5 was nominally significantly associated with decreased LVEF, and m.16145G > A (G > A) in D-loop was nominally significantly associated with decreased LVEF (Table 4). The SNP m.16145G > A showed a potential association with LVEF (P < 0.05) among the SNPs that were statistically significantly associated with lipid species (FDR < 0.05). All 42 associations were presented in Supplementary Table 5.
We performed large-scale association analyses between mitochondrial genetic variants and lipidomic traits in a Chinese cohort with CAD. We identified two SNPs (m.16089T > C and m.16145G > A) of mtDNA that were statistically significantly associated with three lipid species (FDR < 0.05) in two independent groups. Then, we performed linear regression to identify the relationships of the SNPs of mtDNA with traditional lipids, and heart function to explore the influence of mitochondrial genetic variants with clinical manifestations. We found that two SNPs (m.14178T > C and m.215A > G) were statistically significantly associated with TC and LDLC (FDR < 0.05), and three SNPs (m.1048C > T, m.12705C > T, and m.16145G > A) were nominally significantly associated with LVEF (P < 0.05; Figure 3). The results may provide a potential theoretical basis for further explaining the large difference in the levels of lipids and poor prognosis in patients with CAD and provide potential drug intervention targets for precision medicine in patients with CAD.
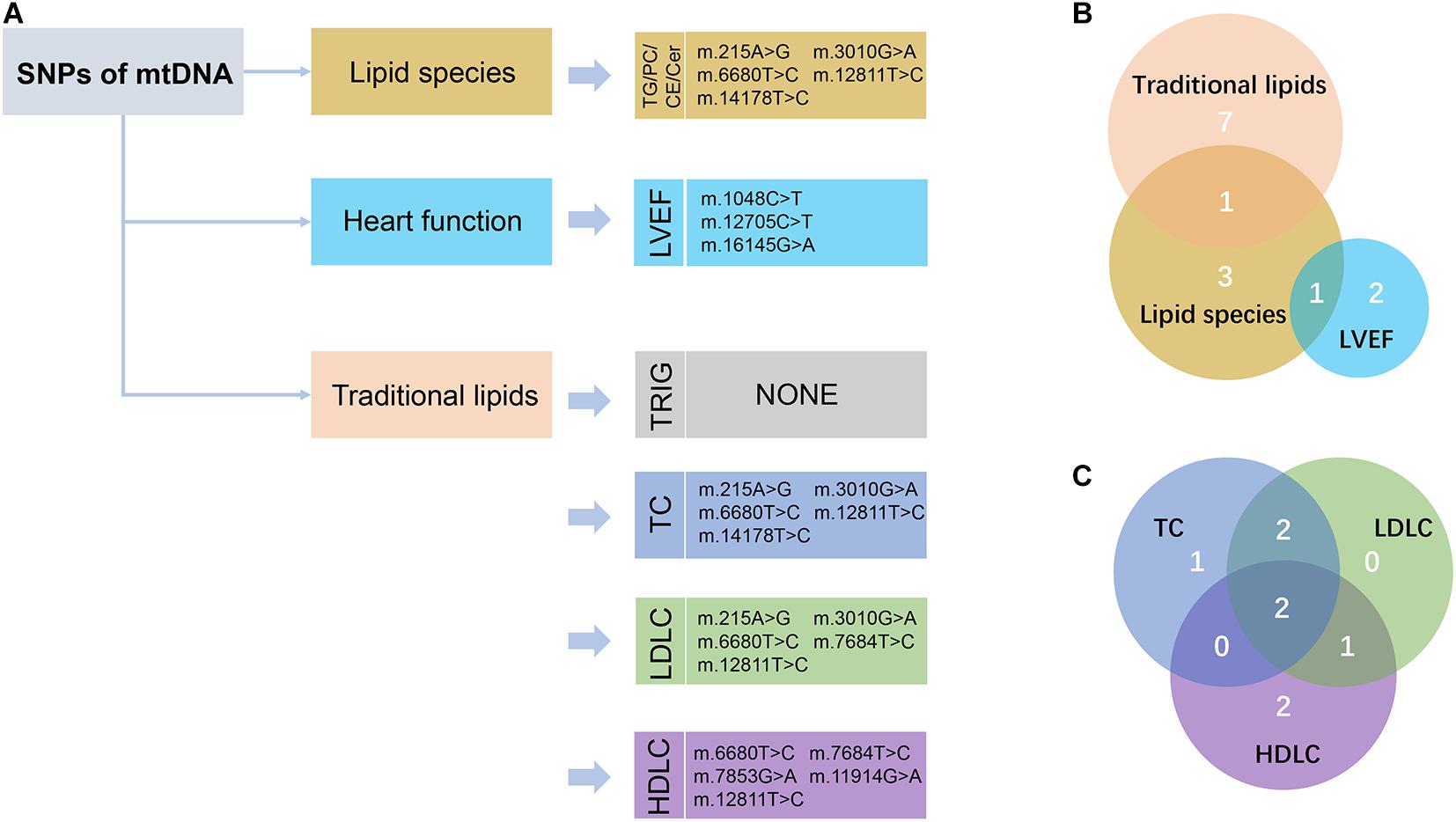
Figure 3. Schematic view of the association analyses results. (A) The nominally significant (P < 0.05) SNPs of mtDNA with lipid species, traditional lipids, and heart function were presented. TG, triacylglycerol; PC, phosphatidylcholines; CE, cholesteryl esters; Cer, ceramides; LVEF, left ventricular ejection fraction; TRIG, triglyceride; TC, total cholesterol; LDLC, low-density lipoprotein cholesterol; HDLC, high-density lipoprotein cholesterol. (B) Venn diagram showed the intersection of these nominally significant SNPs in lipid species, traditional lipids, and LVEF. (C) Venn diagram showed the intersection of the nominally significant SNPs in the traditional lipids, including TC, LDLC, and HDLC.
m.16145G > A (G > A) associated with the increased level of TG(54:5) NL-18:0 but with the decreased level of LVEF were found in this study (Figure 4). This result suggests that the variation at m.16145G > A may bring adverse effect to patients with CAD. Interestingly, the SNPs of mtDNA that are statistically significantly related to the traditional lipids (FDR < 0.05) showed no significant correlation with LVEF (P > 0.05). The variation at the m.16145G > A was not statistically significantly related to the traditional lipids, suggesting the importance of studying mitochondrial genetic variants related to specific lipid species.
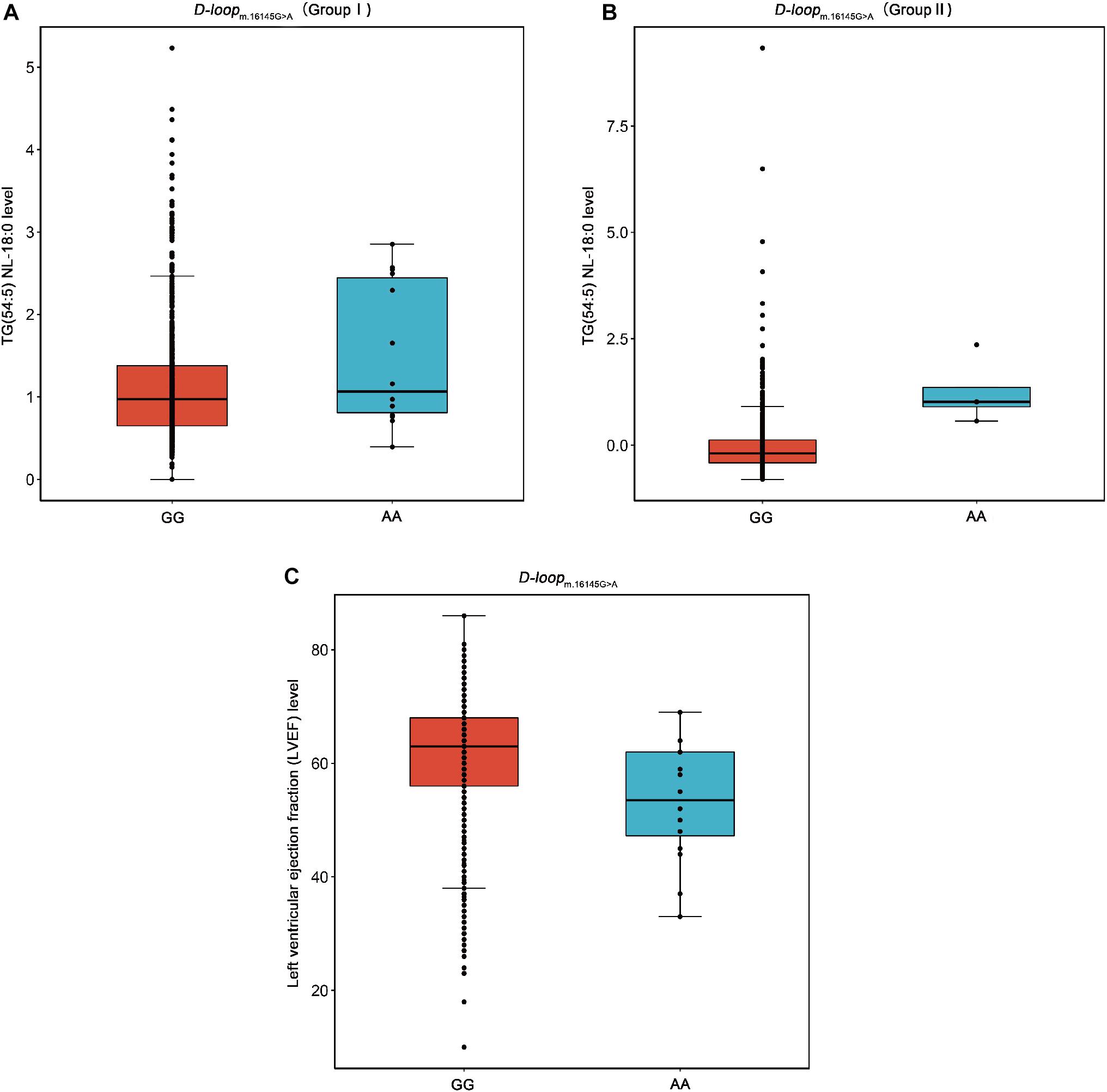
Figure 4. Box plots for the SNP m.16145G > A against the levels of TG(54:5) NL-18:0 and LVEF. TG, triacylglycerol; LVEF, left ventricular ejection fraction. (A) The SNP m.16145G > A (G > A) in D-loop against the level of TG(54:5) NL-18:0 in the Group I. (B) The SNP m.16145G > A (G > A) in D-loop against the level of TG(54:5) NL-18:0 in the Group II. (C) The SNP m.16145G > A (G > A) in D-loop against the level of LVEF.
The SNPs of mtDNA were significantly associated with the level of lipid species (FDR < 0.05) in the D-loop. The D-loop region is a non-coding region but accumulates more mutations under increased oxidative stress than the other regions of mtDNA (Clayton, 2000). The initiation site for heavy chain replication and the promoter for heavy and light chain transcription are contained in this region (Chen et al., 2012). Therefore, the SNPs in the region of the D-loop may contain essential transcription and replication elements (Sharma et al., 2005). For example, the SNP m.16189T > C in D-loop is associated with vascular pathologies such as stroke and CAD (Liou et al., 2004; Mueller et al., 2011). We speculate that the variants in the D-loop region may affect the metabolism of lipid species, thereby affecting the cardiovascular prognosis.
By analyzing the correlation between mitochondrial genetic variation and plasma TC concentration, we found that m.14178T > C (T > C) in MT-ND6 could significantly increase TC level (FDR < 0.001). The variation at m.14178T > C is a missense mutation that can change the encoded amino acid (T > C, isoleucine > valine), and MT-ND6 is a mitochondrial gene that codes the NADH-ubiquinone oxidoreductase chain 6 protein (Shao et al., 2008). NADH is a reduced form of nicotinamide adenine dinucleotide, which is a cofactor in the metabolic center. NADH plays an important role in energy-producing functions of the mitochondria (Stein and Imai, 2012). Therefore, variations in MT-ND6 may cause energy imbalance and metabolic diseases. A recent study reported that variants in MT-ND6 can significantly increase the severity of cardiomyopathy (McManus et al., 2019). Our study found that the T to C base in MT-ND6 m.14178T > C was associated with the increased level of plasma TC, thus possibly contributing to the occurrence and development of atherosclerosis. However, the causal relationship between this mutation and atherosclerosis is still unclear, and further research is needed.
Furthermore, we identified three SNPs that were nominally significantly associated with the level of LVEF (P < 0.05 but FDR > 0.05). The m.1048C > T in MT-RNR1 was the most significant SNP of the results. MT-RNR1 is an RNA gene that encodes 12S rRNA, which is mainly involved in the regulation of insulin sensitivity and metabolic balance (Lee et al., 2015). Previous studies reported that the SNP m.1555A > G in MT-RNR1 was associated with atherosclerosis (Sobenin et al., 2012, 2013). We found that the mutation in this gene could affect heart function, suggesting that MT-RNR1 may play an important role in the development of CAD. However, its relationship with LVEF level needs further analysis, and a large sample size is needed for the validation of this role.
Our present study has several limitations. Although we enrolled approximately 1,400 participants from three centers, a larger central cohort is needed for fully understanding the relationships between mitochondrial genetic variants and lipidomic profiling. Besides, the patients enrolled in this study were almost treated with statins for lipid-lowering medications, which may influence the plasma levels of cholesterol-derived lipids. Moreover, mitochondrial genetic data were obtained using GSA bead chips, and the number of SNPs detected was relatively small in this study. Increasing the number of SNPs and performing full mitochondrial sequencing could solve this problem in the future. Furthermore, our study can only provide the correlation, and the causality may need to be combined with causality analysis or other approaches, such as Mendelian randomization or molecular biology verification in the future.
We performed large-scale association analyses in a Han Chinese population with CAD to explore the relationships between mitochondrial genetic variants and lipidomic profiling. We identified three statistically significant associations (FDR < 0.05), namely, m.16089T > C with TG(50:4) NL-16:0, m.16145G > A with TG(54:5) NL-18:0, and m.16089T > C with PC(16:0_16:1). Then, we further investigated the relationships between mitochondrial genetic variants and traditional lipids. Two statistically significant associations were found (FDR < 0.05), namely, m.14178T > C with TC and m.215A > G with LDLC. Furthermore, we performed association analyses on the SNPs of mtDNA and LVEF. We found that the association between the SNP m.16145G > A and LVEF was nominally significant (P = 0.047). These findings highlight the importance of studying mitochondrial genetic variants related to lipid species and could provide a potential target for pharmacological intervention and optimized treatment of patients with CAD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: EMBL-EBI (Project: PRJEB42554; Analyses: ERZ1714343).
The studies involving human participants were reviewed and approved by the Medical Ethical Review Committee of Guangdong Provincial People’s Hospital (Nos. GDREC2010137 and GDREC2017071H) and conducted according to the Declaration of Helsinki. Informed consent (Nos. 20100910 and 20170211) was obtained from all individual participants included in the study. The patients/participants provided their written informed consent to participate in this study.
ZW designed the study, performed the data analysis, and wrote the manuscript. HC participated in patient recruitment, performed the experiment, and revised the manuscript. MQ performed the experiment and the data analysis. CL, QM, and XC curated the data. YZ made the investigation. WL provided the resources. XZ and SZ designed the study, leaded the study, and provided the resources. All authors reviewed and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was funded by the National Natural Science Foundation of China [No. 81872934 (Principle investigators: Shilong Zhong), 81673514 (Principle investigators: Shilong Zhong); http://www.nsfc.gov.cn/], the National Key Research and Development Program [No. 2017YFC0909301 (Principle investigators: Shilong Zhong); https://service.most.gov. cn/index/], and the Key Research and Development Program of Guangdong Province, China [2019B020229003 (Principle investigators: Shilong Zhong); http://pro.gdstc.gd.gov.cn/egrantweb/]. No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.630359/full#supplementary-material
CAD, coronary artery disease; SNP, single nucleotide polymorphism; mtDNA, mitochondrial DNA; TRIG, triglyceride; TC, total cholesterol; LDLC, low-density lipoprotein cholesterol; HDLC, high-density lipoprotein cholesterol; LVEF, left ventricular ejection fraction; CVD, cardiovascular disease; D-loop, displacement loop; PCI, percutaneous coronary intervention; MG, monoglyceride; CE, cholesteryl esters; DG, diacylglycerol; TG, triacylglycerol; PA, phosphatidic acids; PC, phosphatidylcholines; PG, phosphatidylglycerol; PS, phosphatidylserines; PE, phosphatidylethanolamines; LPA, lysophosphatidic acids; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPS, hemolytic serine; Cer, ceramides; GSA, global screening array; AST, aspartate aminotransferase; eGFR, estimated glomerular filtration rate; FDR, false discovery rate.
Cadby, G., Melton, P. E., McCarthy, N. S., Giles, C., Mellett, N. A., Huynh, K., et al. (2020). Heritability of 596 lipid species and genetic correlation with cardiovascular traits in the Busselton Family Heart Study. J. Lipid Res. 61, 537–545. doi: 10.1194/jlr.RA119000594
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8
Chen, J. B., Yang, Y. H., Lee, W. C., Liou, C. W., Lin, T. K., Chung, Y. H., et al. (2012). Sequence-based polymorphisms in the mitochondrial D-loop and potential SNP predictors for chronic dialysis. PLoS One 7:e41125. doi: 10.1371/journal.pone.0041125
Clayton, D. A. (2000). Transcription and replication of mitochondrial DNA. Hum. Reprod. 15(Suppl. 2), 11–17. doi: 10.1093/humrep/15.suppl_2.11
Echouffo-Tcheugui, J. B., Jain, M., and Cheng, S. (2020). Breaking through the surface: more to learn about lipids and cardiovascular disease. J. Clin. Invest. 130, 1084–1086. doi: 10.1172/jci134696
Ference, B. A., Ginsberg, H. N., Graham, I., Ray, K. K., Packard, C. J., Bruckert, E., et al. (2017). Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 38, 2459–2472. doi: 10.1093/eurheartj/ehx144
Horvath, S. E., and Daum, G. (2013). Lipids of mitochondria. Prog. Lipid Res. 52, 590–614. doi: 10.1016/j.plipres.2013.07.002
Ingman, M., Kaessmann, H., Pääbo, S., and Gyllensten, U. (2000). Mitochondrial genome variation and the origin of modern humans. Nature 408, 708–713. doi: 10.1038/35047064
Lee, C., Zeng, J., Drew, B. G., Sallam, T., Martin-Montalvo, A., Wan, J., et al. (2015). The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21, 443–454. doi: 10.1016/j.cmet.2015.02.009
Liou, C. W., Lin, T. K., Huang, F. M., Chen, T. L., Lee, C. F., Chuang, Y. C., et al. (2004). Association of the mitochondrial DNA 16189 T to C variant with lacunar cerebral infarction: evidence from a hospital-based case-control study. Ann. N. Y. Acad. Sci. 1011, 317–324. doi: 10.1007/978-3-662-41088-2_31
Liu, Y., Ma, H., Zhu, Q., Zhang, B., Yan, H., Li, H., et al. (2019). A genome-wide association study on lipoprotein (a) levels and coronary artery disease severity in a Chinese population. J. Lipid Res. 60, 1440–1448. doi: 10.1194/jlr.P091009
Marín-García, J., and Goldenthal, M. J. (2002). Understanding the impact of mitochondrial defects in cardiovascular disease: a review. J. Card. Fail. 8, 347–361. doi: 10.1054/jcaf.2002.127774
McManus, M. J., Picard, M., Chen, H. W., de Haas, H. J., Potluri, P., Leipzig, J., et al. (2019). Mitochondrial DNA variation dictates expressivity and progression of nuclear DNA mutations causing cardiomyopathy. Cell Metab. 29, 78–90.e5. doi: 10.1016/j.cmet.2018.08.002
Mueller, E. E., Eder, W., Ebner, S., Schwaiger, E., Santic, D., Kreindl, T., et al. (2011). The mitochondrial T16189C polymorphism is associated with coronary artery disease in Middle European populations. PLoS One 6:e16455. doi: 10.1371/journal.pone.0016455
Ouchi, Y., Sasaki, J., Arai, H., Yokote, K., Harada, K., Katayama, Y., et al. (2019). Ezetimibe lipid-lowering trial on prevention of atherosclerotic cardiovascular disease in 75 or older (EWTOPIA 75): a randomized, controlled trial. Circulation 140, 992–1003. doi: 10.1161/circulationaha.118.039415
Poss, A. M., Maschek, J. A., Cox, J. E., Hauner, B. J., Hopkins, P. N., Hunt, S. C., et al. (2020). Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J. Clin. Invest. 130, 1363–1376. doi: 10.1172/jci131838
Qin, M., Zhu, Q., Lai, W., Ma, Q., Liu, C., Chen, X., et al. (2020). Insights into the prognosis of lipidomic dysregulation for death risk in patients with coronary artery disease. Clin. Transl. Med. 10:e189. doi: 10.1002/ctm2.189
Razquin, C., Liang, L., Toledo, E., Clish, C. B., Ruiz-Canela, M., Zheng, Y., et al. (2018). Plasma lipidome patterns associated with cardiovascular risk in the PREDIMED trial: a case-cohort study. Int. J. Cardiol. 253, 126–132. doi: 10.1016/j.ijcard.2017.10.026
Shao, L., Martin, M. V., Watson, S. J., Schatzberg, A., Akil, H., Myers, R. M., et al. (2008). Mitochondrial involvement in psychiatric disorders. Ann. Med. 40, 281–295. doi: 10.1080/07853890801923753
Sharma, H., Singh, A., Sharma, C., Jain, S. K., and Singh, N. (2005). Mutations in the mitochondrial DNA D-loop region are frequent in cervical cancer. Cancer Cell Int. 5:34. doi: 10.1186/1475-2867-5-34
Sharma, P., and Sampath, H. (2019). Mitochondrial DNA integrity: role in health and disease. Cells 8:100. doi: 10.3390/cells8020100
Sobenin, I. A., Sazonova, M. A., Postnov, A. Y., Bobryshev, Y. V., and Orekhov, A. N. (2012). Mitochondrial mutations are associated with atherosclerotic lesions in the human aorta. Clin. Dev. Immunol. 2012:832464. doi: 10.1155/2012/832464
Sobenin, I. A., Sazonova, M. A., Postnov, A. Y., Bobryshev, Y. V., and Orekhov, A. N. (2013). Changes of mitochondria in atherosclerosis: possible determinant in the pathogenesis of the disease. Atherosclerosis 227, 283–288. doi: 10.1016/j.atherosclerosis.2013.01.006
Stein, L. R., and Imai, S. (2012). The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol. Metab. TEM 23, 420–428. doi: 10.1016/j.tem.2012.06.005
van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K., and van der Werf, M. J. (2006). Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7:142. doi: 10.1186/1471-2164-7-142
Virani, S. S., Alonso, A., Benjamin, E. J., Bittencourt, M. S., Callaway, C. W., Carson, A. P., et al. (2020). Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation 141, e139–e596. doi: 10.1161/cir.0000000000000757
Wahbi, K., Bougouin, W., Béhin, A., Stojkovic, T., Bécane, H. M., Jardel, C., et al. (2015). Long-term cardiac prognosis and risk stratification in 260 adults presenting with mitochondrial diseases. Eur. Heart J. 36, 2886–2893. doi: 10.1093/eurheartj/ehv307
Willer, C. J., Li, Y., and Abecasis, G. R. (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. doi: 10.1093/bioinformatics/btq340
Keywords: mitochondrial DNA, polymorphisms, lipidomic, association analyses, coronary artery disease
Citation: Wang Z, Chen H, Qin M, Liu C, Ma Q, Chen X, Zhang Y, Lai W, Zhang X and Zhong S (2021) Associations of Mitochondrial Variants With Lipidomic Traits in a Chinese Cohort With Coronary Artery Disease. Front. Genet. 12:630359. doi: 10.3389/fgene.2021.630359
Received: 17 November 2020; Accepted: 08 March 2021;
Published: 25 March 2021.
Edited by:
Segun Fatumo, University of London, United KingdomReviewed by:
Corey Giles, Curtin University, AustraliaCopyright © 2021 Wang, Chen, Qin, Liu, Ma, Chen, Zhang, Lai, Zhang and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojuan Zhang, emhhbmd4anRqQDE2My5jb20=; Shilong Zhong, Z2RwaF96aG9uZ3NsQGdkLmdvdi5jbg==; emhvbmdzbEBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.