- 1Bio-Tech Research Center, Shandong Academy of Agricultural Sciences, Shandong Provincial Key Laboratory of Crop Genetic Improvement, Ecology and Physiology, Jinan, China
- 2College of Life Science, Shandong Normal University, Jinan, China
The NAC transcription factor (TF) is one of the most significant TFs in plants and is widely involved in plant growth, development, and responses to biotic and abiotic stresses. To date, there are no systematic studies on the NAC family in peanuts. Herein, 132 AhNACs were identified from the genome of cultivated peanut, and they were classified into eight subgroups (I–VIII) based on phylogenetic relationships with Arabidopsis NAC proteins and their conserved motifs. These genes were unevenly scattered on all 20 chromosomes, among which 116 pairs of fragment duplication events and 1 pair of tandem duplications existed. Transcriptome analysis showed that many AhNAC genes responded to drought and abscisic acid (ABA) stresses, especially most of the members in groups IV, VII, and VIII, which were expressed at larger differential levels under polyethylene glycol (PEG) and/or ABA treatment in roots or leaves. Furthermore, 20 of them selected in response to PEG and ABA treatment were evaluated by quantitative real-time polymerase chain reaction. The results showed that these genes significantly responded to drought and ABA in roots and/or leaves. This study was helpful for guiding the functional characterization and improvement of drought-resistant germplasms in peanuts.
Introduction
The NAC transcription factor (TF) is one of the most abundant types of TFs in plants; its designated name comes from the homologous proteins encoded by NAM, ATAF1/ATAF2, and CUC2 (Souer et al., 1996). NAC TFs have similar N-terminal structures (Aida et al., 1997), which consist of five subdomains (A–E), and usually contain approximately 160 amino acid residues. Among them, subdomains A, C, and D are highly conserved. The nuclear localization signal that involves the recognition process of the NAC TF localizes to the C and D subdomains. Although the E subdomain has lower conservation, it may participate in developmental regulation and cooperation with the D domain in a tissue-specific manner. The C-terminus of the NAC protein contains a transcriptional activation region, which has higher diversity, and some repeated amino acids, such as serine, threonine, and proline, are enriched in this region (Tran et al., 2010; Wang and Dane, 2013; Kim et al., 2016; Mathew and Agarwal, 2018).
In angiosperms, the NAC TF family has large numbers of members, for example, 117 NAC members in Arabidopsis (Ooka et al., 2003), 151 in rice (Ooka et al., 2003), 152 in soybean (Le et al., 2011) and corn (Shiriga et al., 2014), and 110 members in potato (Singh et al., 2013). NAC proteins regulate multiple biological processes in plants, including seed and embryo development (Duval et al., 2002; Sperotto et al., 2009), shoot tip meristem formation (Kim S.G. et al., 2007), fiber development (Ko et al., 2007), leaf senescence (Guo and Gan, 2006), cell division (Kim et al., 2006), and lateral root growth (Xie et al., 2000). Additionally, many NAC genes participate in the response to abiotic stresses in plants, such as drought, salinity, cold, and water logging (Hu et al., 2006; Jeong et al., 2010; Nuruzzaman et al., 2012). In Arabidopsis, overexpressing the NAC genes ANAC019, ANAC055, and ANAC072 enhanced tolerance to drought in transgenic lines (Tran et al., 2004). Overexpression of the AtNAC2 gene improved the salt tolerance of transgenic plants (He et al., 2005). The NAC TF gene TaNAC2 from wheat also responded to drought, salt, cold, and abscisic acid (ABA) stress. Overexpressing TaNAC2 in Arabidopsis improved their tolerance to these stresses (Mao et al., 2012). Overexpression of the SNAC1, OsNAC6, OsNAC5, OsNAC45, and OsNAC63 genes in rice can significantly increase drought and salt tolerance (Hu et al., 2006; Nakashima et al., 2007; Song et al., 2011; Trapnell et al., 2012). GmNAC11 and GmNAC20 in soybean are involved in the response to low temperature, and their overexpression can enhance the low-temperature tolerance of transgenic soybean plants (Hao et al., 2011).
Cultivated peanut (Arachis hypogaea) is a major oilseed crop and cash crop widely cultivated worldwide; its yield per unit area is higher than those of other oilseed crops. In China, peanut is always planted in arid and semiarid hilly regions because of its higher tolerance to drought and barren conditions. The growth period of peanuts is exceptionally susceptible to drought stress, which affects the yield and quality of peanuts (Yan et al., 2007). Recently, the functions of several peanut NAC genes in abiotic stress tolerance were investigated. Overexpression of the AhNAC2 (renamed AhNAC53 in our article) gene in Arabidopsis significantly improved drought and salt resistance (Liu et al., 2011, 2014). Overexpression of AhNAC3 (renamed AhNAC12 in our article) in tobacco increased the drought resistance of transgenic tobacco (Liu et al., 2013). Transgenic tobacco plants with AhNAC4 (renamed AhNAC53 in our article) transcripts enhanced their drought resistance by regulating stomatal opening and closing, reducing transpiration, and improving water use efficiency (Tang et al., 2017). However, to date, there has been no systematic study about the NAC family in peanut. In this article, 132 NAC genes were identified from the genome of cultivated peanut and were classified into eight main groups according to their phylogenetic relationships. Additionally, their gene structure, chromosome location, and conserved motifs were analyzed using bioinformatics methods. Furthermore, the function of these AhNAC genes in responding to drought stress was predicted by a genome-wide survey, and their expression patterns after treatment with drought and ABA were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). This study will help elucidate the functions of these NAC genes in the drought response and in the molecular breeding of drought resistance in peanut.
Materials and Methods
Identification of NAC TFs in Peanut
The peanut protein sequences were downloaded from the peanut library1. The HMM file of the NAM domain (PF02365) was retrieved from the Pfam database2 and was used to search the NAC family proteins with an e value less than 0.001 in the peanut protein database by HMMER 3.0 local software. Subsequently, some incomplete and redundant amino acid sequences were deleted, and the possible AhNACs were confirmed by BLASTP and CDD programs. The number of amino acids, molecular weight (MW), and theoretical isoelectric point of each NAC protein sequence were calculated using ExPASy3.
Sequence Alignment and Phylogenetic Analysis of AhNAC TFs
Arabidopsis NAC protein sequences were downloaded from the Arabidopsis Information Resource4, and sequence alignment of NAC TFs in peanut and Arabidopsis was performed by the MUSCLE method (Kumar et al., 2016). Subsequently, an unrooted phylogenetic tree was established by MEGA 7 using the neighbor-joining (NJ) method with a p-distance model and 1,000 bootstrap repeats (Kumar et al., 2018).
Chromosome Location and Gene Duplication of AhNAC Genes
The position of each AhNAC gene and the sizes of every peanut chromosome were extracted from the gff3 annotation files in the peanut library, and the chromosome distribution of genes was visualized using MapChart software (Voorrips, 2002). The duplication analysis of AhNAC genes considered the following two aspects: (1) the length of the shorter aligned sequence covered more than 70% of the longer sequence; and (2) the identity between two aligned sequences was greater than 70% (Gu et al., 2002; Yang et al., 2008). AhNAC genes were aligned with the whole peanut genome by local BLAST software, and gene duplications were then identified by the MCScanX (Wang et al., 2012) algorithm with the default setting (e ≤ 1e-10). A gene duplication diagram was drawn using Circos software (Krzywinski et al., 2009).
Gene Structure and Conserved Motif Analyses
The motif analysis tool MEME5 was used to analyze the conserved motifs of the NAC family according to any number of repetitions and parameters with a maximum motif number of 50. The gene structures were assessed with the Gene Structure Display Server6. The MEME theme was further annotated with InterPro7. The transmembrane (TM) domain was analyzed with TMHMM Server v. 2.08
Promoter Analysis of NAC Family Genes
The 2,000-base-pair (bp) sequences upstream of the start codon ATG of AhNAC genes were retrieved from the peanut genome using TB tools software (Chen et al., 2020) and were submitted to the online software PlantCARE9 for the analysis of cis-acting elements. The draft of the element distribution on each promoter was sketched using TBtools.
Expression Profiles Under Different Stresses Using RNA-Seq Data
The Illumina RNA-seq data (PRJNA553073) derived from roots and leaves of the peanut variety Fenghua No. 1 (FH1) were downloaded from the Sequence Read Archive10. The seedlings of FH1 used for sequencing were cultivated in 1/2 MS0 medium at room temperature for 10 days and were then treated with 20% polyethylene glycol (PEG) 6000 or 20 mg/L ABA for 24 h. RNA-seq data were analyzed by Hisat2, Samtools, and Cufflinks to obtain FPKM values. A heat map showing tissue-specific expression profiles (log2FPKM values) was made using TBtools.
RNA Extraction and qRT-PCR Analysis
The FH1 seedlings were cultivated in nutrient soil at room temperature for 10 days and irrigated twice daily using Hoagland solution during seedling growth. Then, the seedlings were treated with Hoagland solution supplemented with 20% PEG6000 or 20 mg/L ABA, and their leaves and roots after treatment for 0 (control, CK) and 24 h were picked and stored at −80°C. Total RNA was extracted using a Quick RNA Isolation Kit 3.0 (Huayueyang Biotechnology, Beijing, China) according to the manufacturer’s protocol and was then digested with RNase-free DNase I (TaKaRa, Dalian, Liaoning Sheng, China). The quality and quantity of RNA were determined by agarose gel electrophoresis and ultraviolet spectrophotometry (BioPhotometer Plus, Eppendorf, Germany). First-strand cDNA was synthesized by the PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, Liaoning Sheng, China).
The relative expression levels of AhNAC genes under drought and ABA stress conditions were analyzed by qRT-PCR using the ACTIN7 gene as an internal control and were calculated using the 2–ΔΔCt method. Primer Premier 5.0 software was used to design qRT-PCR primers, and PCR was performed with the following steps: first, predenaturing at 95°C for 10 min, then 40 cycles of 95°C for 15 s and 60°C for 1 min, and finally 95°C for 30 s and 60°C for 15 s. The experiments were performed in three biological replicates, with technical triplicates per biological repeat. All primer sequences are shown in Supplementary Table 1.
Results
Identification of the AhNAC Gene Family
A total of 196 NAC sequences were retrieved after searching the peanut protein database using the NAM conserved domain (Pfam: PF02365) by the local HMMER 3.0 software. Among them, some sequences with incomplete NAM domains or with e values greater than 1e–3 were excluded by BLASTP and CDD verification. Ultimately, 132 NAC TFs in peanut with the canonical NAM domain were identified and named AhNAC1 to AhNAC132 according to their physical locations on the chromosome.
The AhNAC genes vary in length from 535 bp (AhNAC93) to 9,732 bp (AhNAC53), and their coding proteins range from 127 (AhNAC1) to 740 (AhNAC122) amino acids. The MWs of AhNACs were between 15.2 kDa (AhNAC1) and 83.19 kDa (AhNAC122), with the majority of them ranging from 20 to 50 kDa in peanut. Their average MW was 40.08 kDa. The predicted pI ranged from 4.67 (AhNAC123) to 9.77 (AhNAC56), with an average of 6.85, among which 60 AhNACs had pI > 7, and 72 AhNACs had pI < 7 (Supplementary Table 2).
Phylogenetic Analysis and Classification of NAC Genes
To explore the evolutionary relationship between AhNAC TFs, an unrooted phylogenetic tree was constructed using 132 AhNAC proteins in peanut and 117 AtNACs from Arabidopsis (Supplementary Table 3). According to the clustering result of NACs from Arabidopsis thaliana and rice in the article by Ooka et al. (2003), all members of NAC TF mentioned previously were divided into eight groups, the NAM/NAC1 (group I), OsNAC7 (group II), ANAC11 (group III), SENU5/NAP/AtNAC3/ATAF (group IV), ONAC22/TERN (group V), OsNAC8/TIP/ANAC1(group VI), ONAC3(group VII), and NAC2/ANAC63 (group VIII) groups (Ooka et al., 2003); the largest group (group VII) consists of 42 proteins, including 27 peanut NACs, and the smallest (group V) has 17 proteins, including seven NAC members of peanut (Figure 1). The other groups from most to least had 40 (23 AhNACs, group IV), 37 (15 AhNACs, group VI), 36 (21 AhNACs, group I), 33 (16 AhNACs, group VIII), 24 (11 AhNACs, group II), and 20 (12 AhNACs, group III) members.
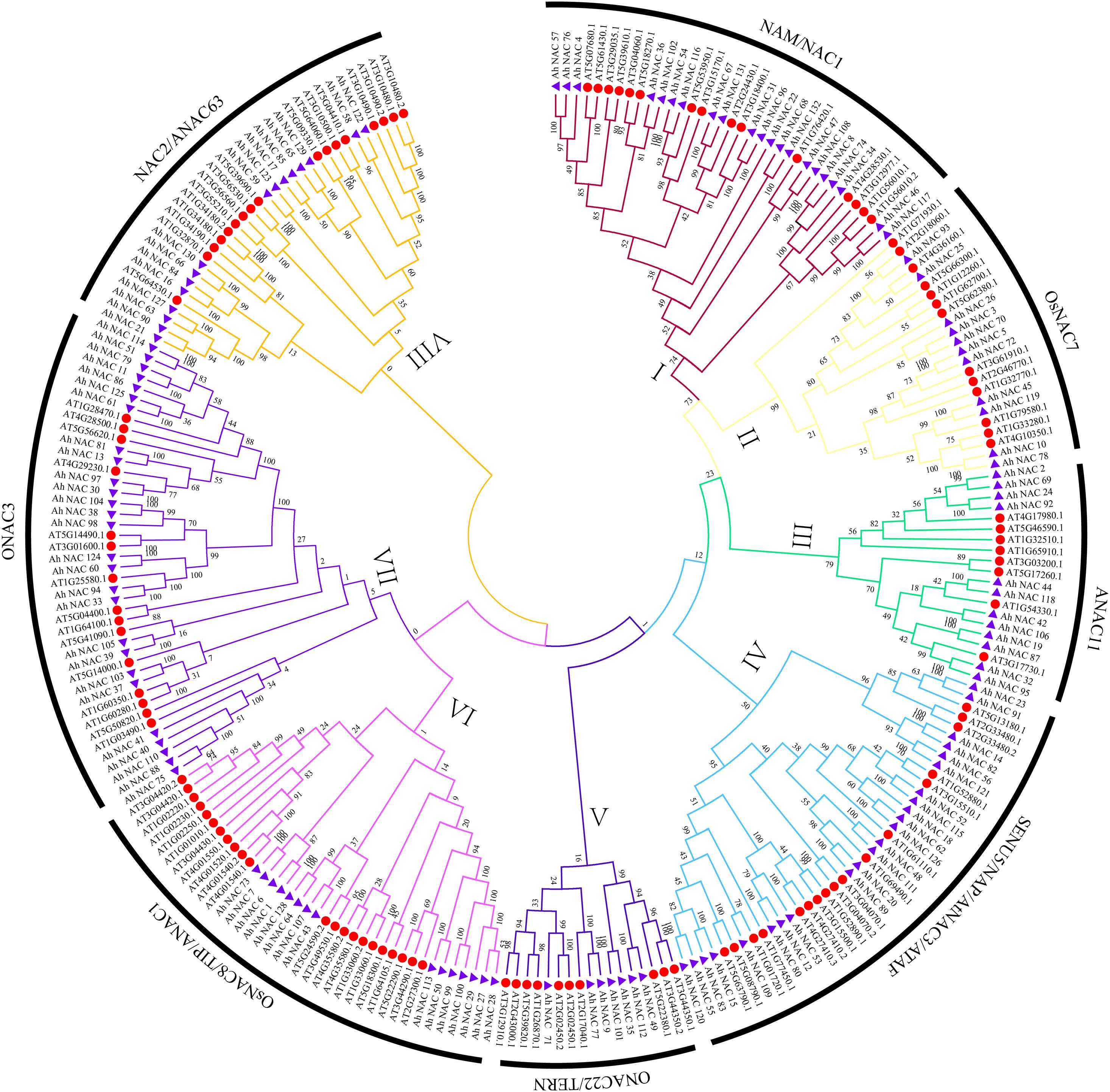
Figure 1. An unrooted phylogenetic tree representing the relationships among the NAC proteins of peanut and Arabidopsis. All full-length protein sequences were aligned by MUSCLE, and the tree was generated using the neighbor-joining (NJ) method in MEGA7 software. The I–VIII subgroups are represented by different colors, the purple triangles represent NAC proteins from peanut, and the red circles indicate the Arabidopsis protein.
Previous studies on NAC TFs indicated that genes clustered in one subgroup might have similar functions (Tran et al., 2009; Nuruzzaman et al., 2010; Le et al., 2011; Ahmad et al., 2018; Liu et al., 2019). It is worth noting that a variety of representative abiotic stress–related genes appeared in group IV. For example, the expression of the Arabidopsis RD26 gene (AT4G27410.2) was induced by drought-related high salinity (Fujita et al., 2004). Overexpression of ATAF1 (AT1G01720.1) in Arabidopsis increased plant sensitivity to ABA, salt, and oxidative stresses (Wu et al., 2009). Our results found that there were 23 AhNAC members in group IV, of which 17 Arabidopsis NACs were divided into three subclades. AhNAC53 clustered together with Arabidopsis AT1G52890.1, AT3G15800.1, and AT4G27410.2, and their sequence similarities were 57.82%, 69.37%, and 76.79%, respectively. AhNAC12 and AhNAC80 also belonged to this subclade and had higher homology with AhNAC53, and their sequence identities were 60% and 60.36%, respectively. AhNAC80 had 60.98% sequence similarity with Arabidopsis ANAC55 (AT3G15500.1) in the same group. Overexpression of AtNAC2 (AT3G15510.1) in Arabidopsis can improve resistance to drought by stimulating secondary root development under drought conditions (He et al., 2005). AhNAC62, AhNAC126, AhNAC18, AhNAC115, AhNAC52, AT1G52880.1, and ATNAC2 were clustered into the same subclade. Among them, AhNAC62 and 126 are orthologous genes from the A and B genomes and have 85.50% and 71.20% sequence similarity with ATNAC2, respectively. Some NACs with a TM motif 1-like (NIL) of Arabidopsis, including AT1G33060 (NTL2), AT3G44290 (NTL4), AT3G49530 (NTL6), AT2G27300 (NTL8), AT4G35580 (NTL9), AT1G01010 (NTL10), AT4G01540 (NTL12), and AT4G01550 (NTL13), were clustered in group VI. Peanut AhNACs (1, 6, 7, 27, 28, 29, 43, 50, 64, 73, 99, 100, 107, 113, and 128) gathered in this group, and AhNAC64, 128, and AhNAC107 had higher sequence similarity with NTL2 and NTL9, respectively. In group VIII, the homologous genes AhNAC127 and AhNAC63 from the A and B genomes were clustered in the same branch as Arabidopsis XND1 (AT5G64530.1), and both proteins had 70.16% sequence similarity with XND1. XND1 could indirectly affect aquaporin function to reduce its tolerance to drought stress (Tang et al., 2018). The other members in this subclade, AhNAC21 and AhNAC90, also had 65.1% and 65.63% sequence identity with XND1 (Supplementary Table 4).
Chromosomal Mapping and Duplication Analysis of the Peanut NAC Gene
To confirm the chromosome distribution of AhNAC genes, a physical map was constructed with MapChart. It appeared that 132 NAC genes were unevenly mapped on all 20 chromosomes, half of which were derived from the A genome (Chr 1–10) and the other half from the B genome (Chr 11–20, Figure 2). Most genes were present on chromosomes 3 and 13 belonging to the A and B genomes, respectively, accounting for 9.85% (13 genes) and 11.36% (15 genes) of the total gene numbers, and the fewest genes were scattered on chromosome 4 from the A genome and chromosome 14 of the B genome, accounting for 1.52% (2 genes) and 0.76% (1 gene), respectively. Some corresponding chromosomes from the A/B genome, such as chromosomes 2 and 12 (4 genes), 8 and 18 (11 genes), 9 and 19 (3 genes), and 10 and 20 (8 genes), had equal numbers of genes (Figure 2).
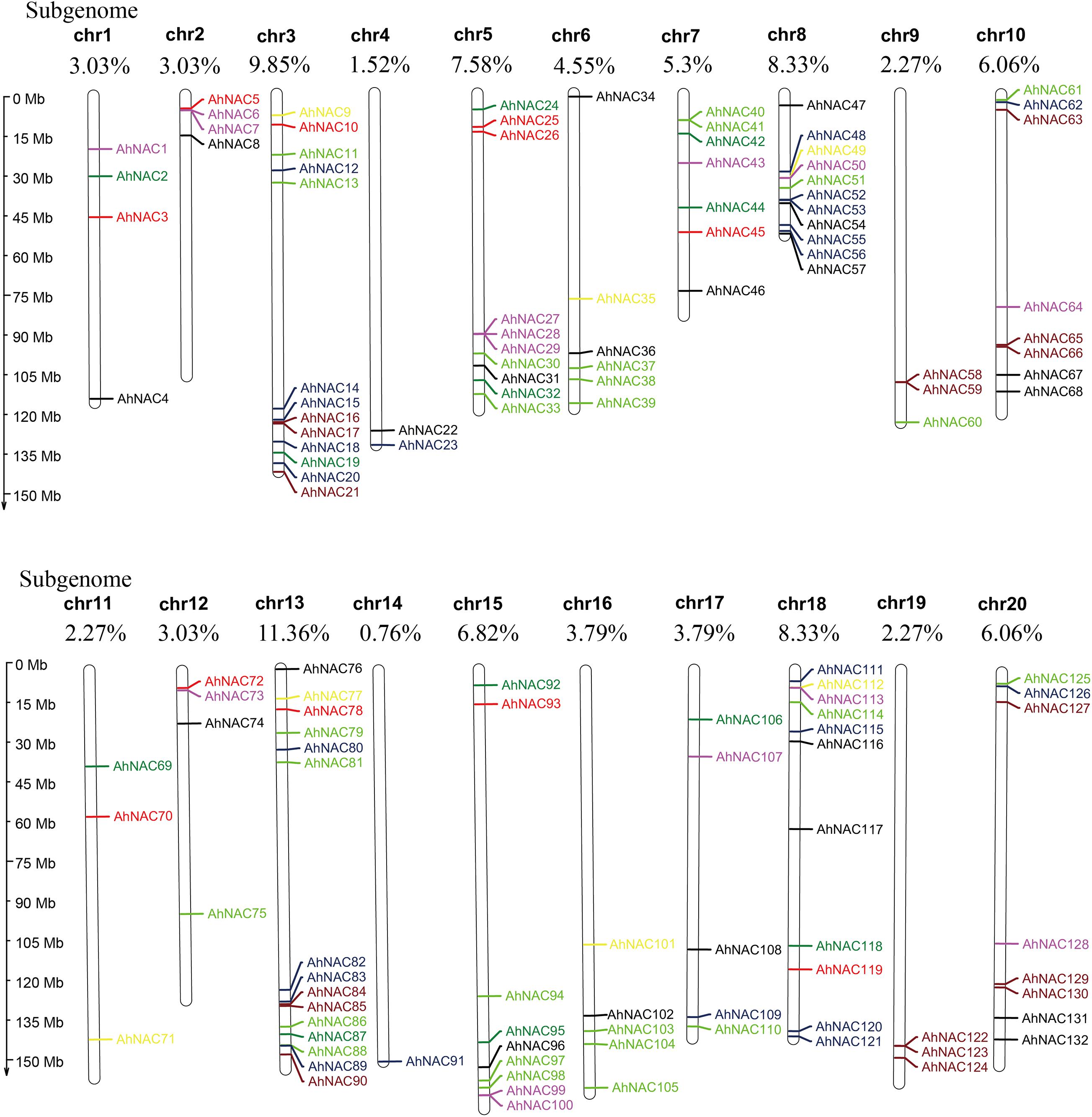
Figure 2. Chromosomal physical map showing the uneven distribution of AhNAC genes on each chromosome. Different colors indicate the different subgroups. The chromosome length is shown to the left of the map, and the serial numbers of chromosomes (chr1–chr20) and the distribution frequencies are shown on the top of each chromosome. Black represents group I, red represents group II, green represents group III, blue–black represents group IV, yellow represents group V, purple represents group VI, light green represents group VII, and brown represents group VIII.
A collinearity analysis by MCScanX software found that among 132 AhNACs, 116 pairs of segmental duplication genes were identified; these were divided into eight groups according to the phylogenetic tree (Figure 3). Maximum segmental duplication events occurred in group VII (31 pairs), followed by group IV (28 pairs), whereas groups V and VI (5 pairs and 3 pairs) showed far fewer segmental duplication events. Only one pair of tandem duplication genes, AhNAC28 and AhNAC29, was detected in group VI (Figure 3B).
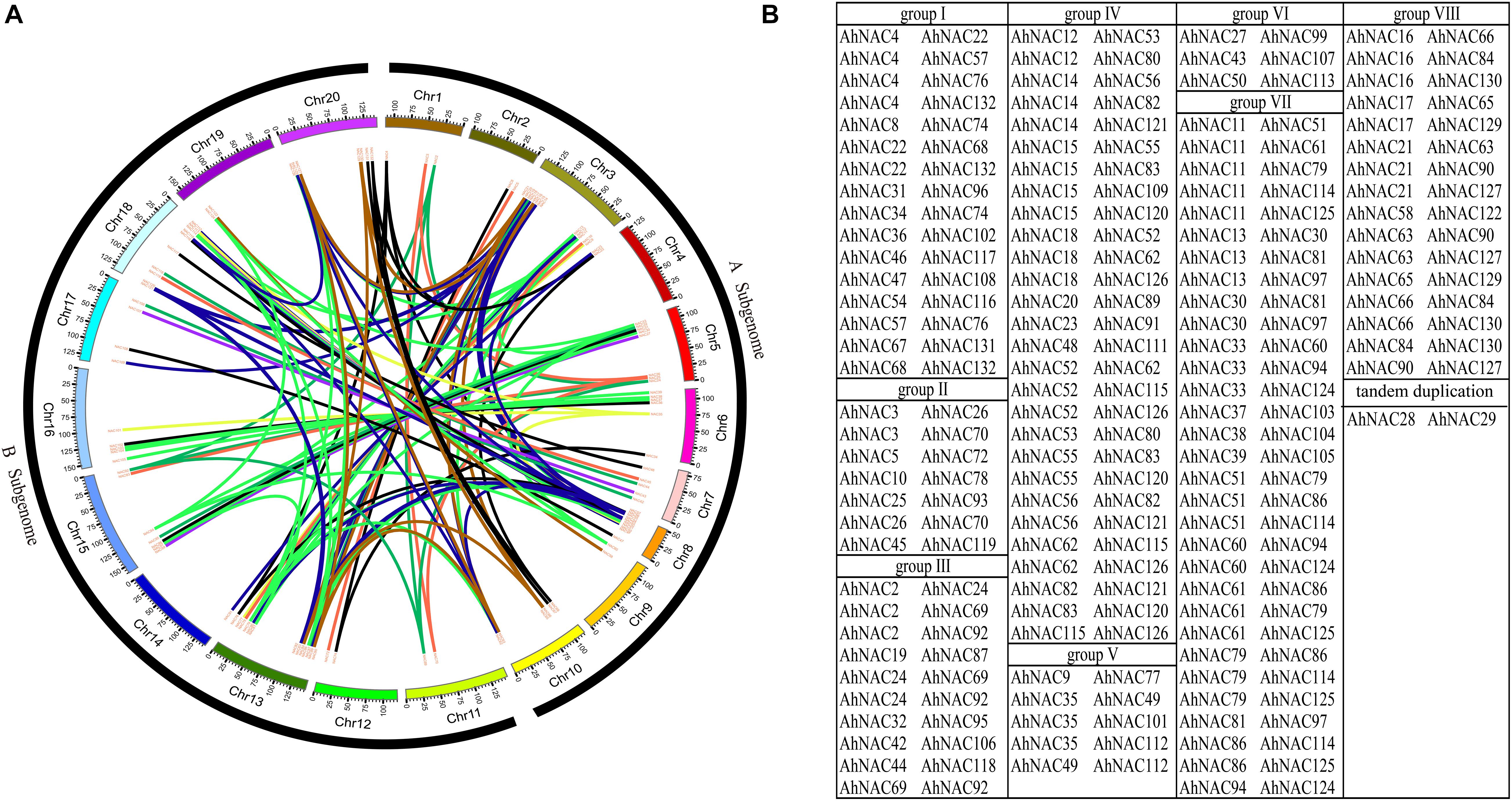
Figure 3. Collinearity analysis results indicating that a number of duplication events existed in AhNAC genes. (A) The colinear relationship among AhNAC genes is shown as a circle map. The red line indicates the duplication events between pairs, and the different-colored bars represent the different chromosomes. (B) The statistics of duplicated gene pairs according to their assigned groups. Black represents group I, red represents group II, green represents group III, blue–black represents group IV, yellow represents group V, purple represents group VI, light green represents group VII, and brown represents group VIII.
Conserved Motifs and Gene Structure of AhNAC
To further investigate the features of AhNACs, conserved motifs were verified by searching the MEME database, and homologous relationships among their coding proteins were constructed according to the motif composition. A total of 20 motifs were identified in all AhNAC members. The AhNAC gene family was classified into eight groups (I–VIII), which was similar to the unrooted phylogenetic tree of NAC proteins (Figure 4A). Group VIII was further divided into two subgroups, A and B, because of a lack of motifs 7 or 12 at the N-terminus and different lengths and motifs at the C-terminus. Almost every AhNAC had motifs 1, 3, 5, and 6, which were considered the NAC domains through InterProScan searching. Most of the AhNAC proteins also contained motifs 2, 4, and 7, which included all members of groups I–VI and group VIIIB and two members of group VII. Sequence analysis found that motifs 1 and 4, 2 and 7, 3 and 5, and 6 corresponded to subdomains A to E localized at the N-terminus of NAC proteins (Supplementary Table 5) (Diao et al., 2018; Liu et al., 2019). The majority of AhNACs in group VII also had motifs 8, 9, and 10, and almost half of the AhNACs in group VI had motif 16 at the N-terminus. The C-termini of AhNACs were less conserved, even though in the same group, they had different motif compositions. There were almost no new motifs found at the C-termini of groups I, II, and V members. Motifs 10, 16, and 18 appeared in some members of group III, and motif 15 appeared in three members of group IV. Nearly half of the members in group VI contained motifs 11, 12, and 16, and four members in group VII had motif 19 at their C-termini (Figure 4B). Analysis of the TM domains of the AhNACs at the C-terminus revealed that AhNAC7, 43, 50, 64, 73, 107, 113, and 128 of group VI all had one TM domain, and AhNAC19, 42, 87, and 106 in group III and AhNAC59 and 123 in group VIII contained two TM domains, whereas the other members had no TM structures at the C-terminus (Supplementary Table 6). Most closely related members of the phylogenetic tree showed a characteristic motif with the same arrangement and location, suggesting that NAC members clustered in the same subgroup may have similar biological functions.
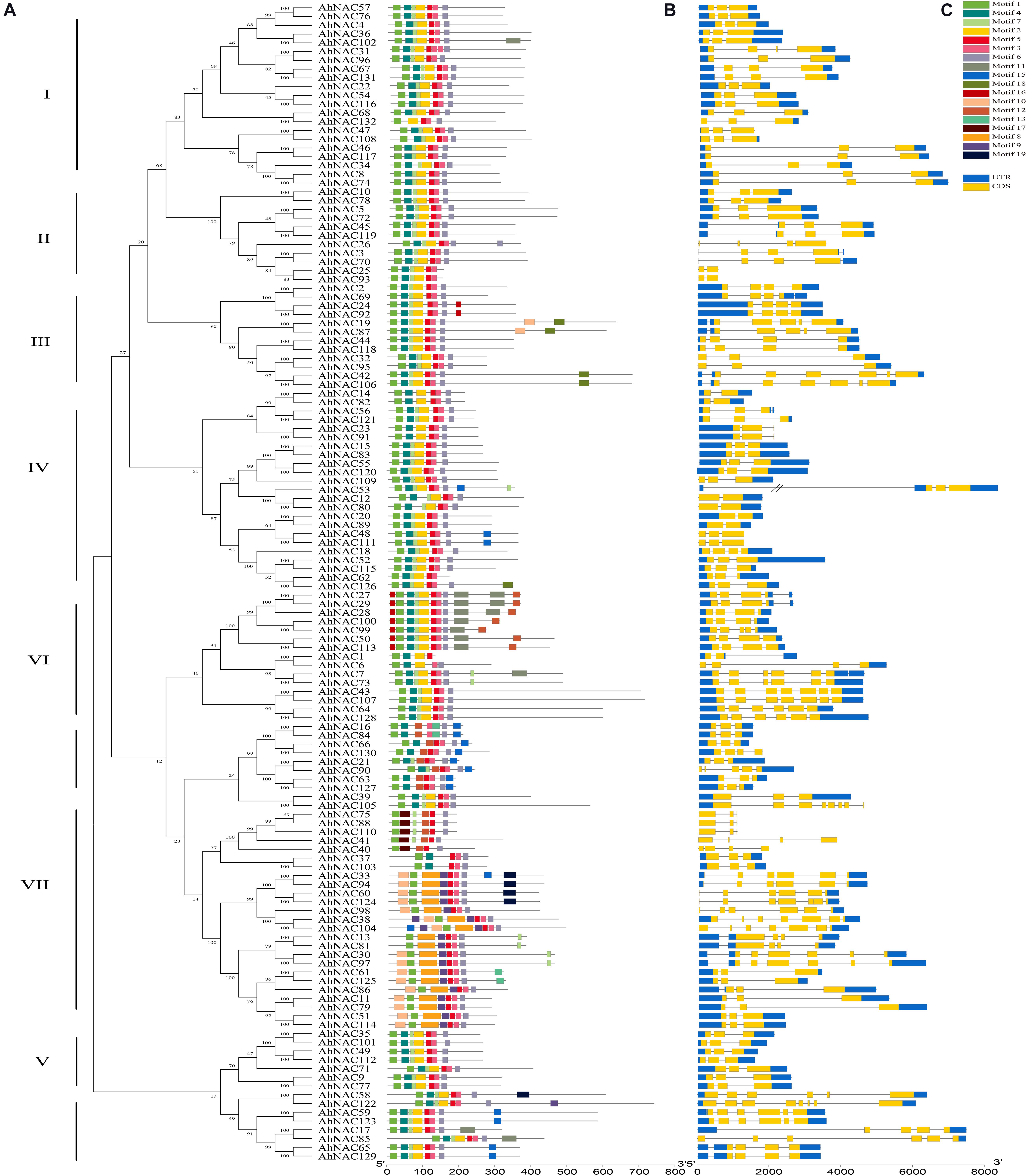
Figure 4. Analysis of the conserved motifs of AhNAC family members and their coding gene structures. (A) The phylogenetic tree of peanut AhNACs was constructed in MEGA7 using the NJ method. (B) Schematic diagrams of putative conserved motifs of 132 AhNAC proteins. The conserved motifs were found using the MEME tool and are shown by different colored boxes. (C) Sketch map of the exon–intron structures of 132 AhNAC genes. The blue and yellow boxes indicate the regions of the 5′- and 3′-UTRs and the exons, respectively, and the black lines indicate the introns in genes.
A sketch of the exon–intron structure of each AhNAC gene is displayed in Figure 4C. The numbers and lengths of exons in most AhNAC genes were essentially in accord with the classification of the phylogenetic relationship of the AhNAC family. In groups I and V, apart from AhNAC31, AhNAC67, and AhNAC131, the other genes had three exons. There were two to four exons in groups II and IV, but the genes in different groups had largely distinct sizes and arrays of introns, especially AhNAC53, which contained a large intron in its 5’ UTR. In groups III and VII, most AhNAC genes had three to four exons, and the others had six to seven exons, whereas these genes displayed diverse exon–intron structures. In group VI, the exon numbers of AhNAC genes were different from 3 to 7. The gene structures between groups VIIIA and VIIIB showed many variant patterns; most genes in VIIIA had three exons and relatively shorter introns, whereas the VIIIB genes had more than five exons and some longer introns. These results indicated the diversity of the AhNAC gene structure (Figure 4C).
Expression Patterns of AhNAC Genes Treated With Different Stresses
In order to discover the roles of the AhNAC genes, the expression profiles were summarized by analyzing the RNA-Seq data (PRJNA553073) of the roots and leaves under PEG and ABA treatment, and heat maps of the relative expression levels were established using TBtools. The results showed that in either roots or leaves, the expression levels of most genes in groups I and II did not change significantly under the ABA and PEG treatments. However, AhNAC47, AhNAC68, and AhNAC132 of group I in leaves were upregulated under PEG treatment, whereas AhNAC54 was downregulated by PEG and induced by ABA; in roots, the expression levels of six genes (AhNAC22, 31, 36, 96, 102, and 108) of group I were improved by the treatment of ABA, whereas only AhNAC47 was induced by PEG, and AhNAC54, 57, 76, and 108 were downregulated. In group II, AhNAC3 and its homologous gene AhNAC70 had different response patterns to treatment in roots and leaves; their expression was reduced in leaves treated with PEG, whereas in roots, AhNAC3 and AhNAC70 were induced by ABA and PEG, respectively; the expression levels of AhNAC25 and 93 were enhanced by treatment with both PEG and ABA (Figure 5, Supplementary Table 7).
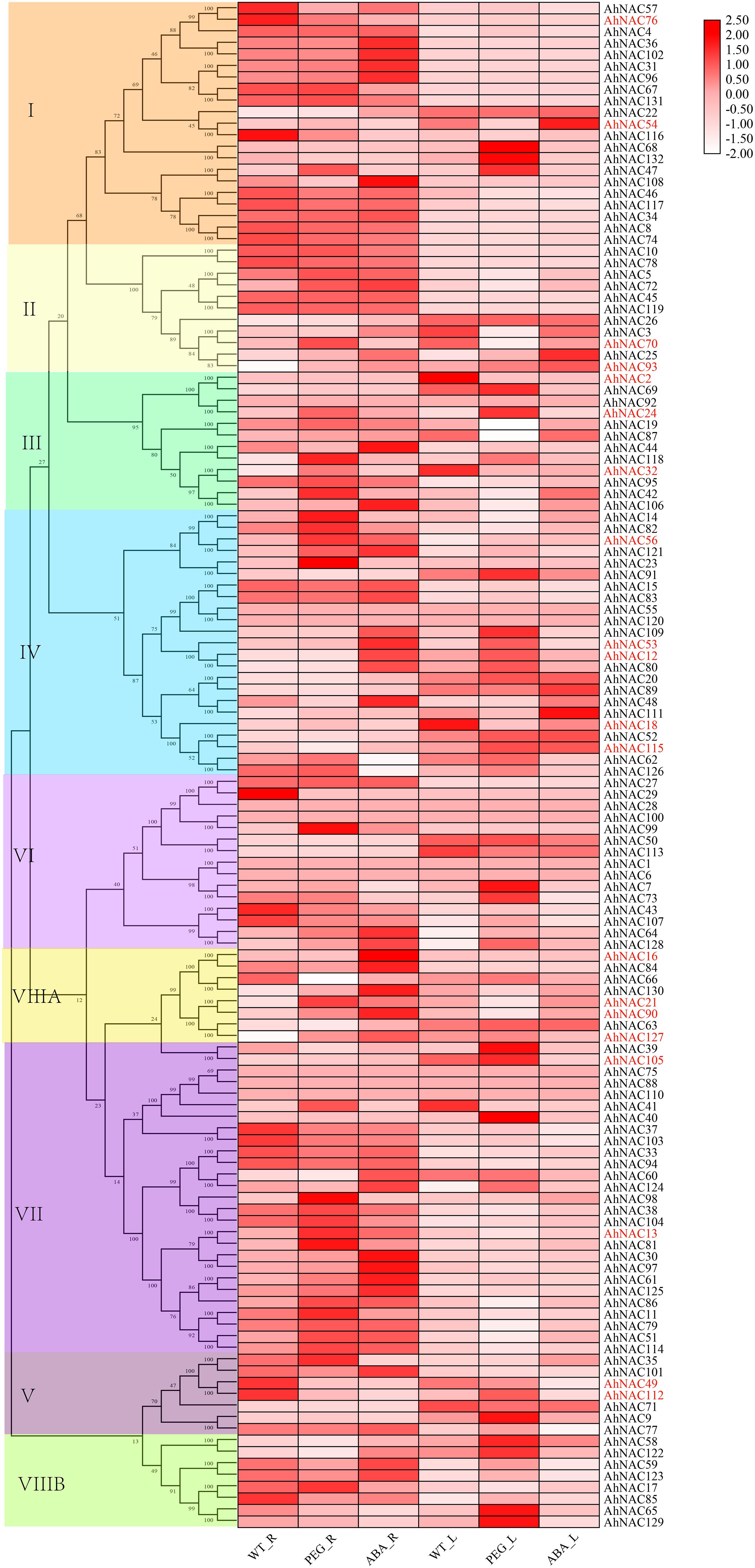
Figure 5. Heat map presenting the expression patterns of AhNAC genes in roots and leaves treated with PEG and ABA. The relative expression values were transformed by log2 of FPKM values and are displayed as colored boxes from light pink to red, and genes selected by qPCR are indicated in red font.
In leaves, most of the genes in groups VI, VII, and VIII were less affected by PEG and ABA treatment, but AhNAC7, 64, 73, and 128 in group VI; AhNAC39, 40, 105, and 124 in group VII; and AhNAC58, 65, 122, and 129 in group VIIIB were all significantly upregulated by PEG treatment. Few members in groups III and V responded to ABA, and six genes (AhNAC2, 19, 32, and 87 downregulated, and AhNAC92 and 118 expressed in the opposite manner) of group III and two genes (AhNAC9 and 112) of group V were affected by PEG. In group IV, most genes responded to PEG treatment; for example, AhNAC12, 53, 62, 80, 91, and 109 were upregulated by PEG treatment; only three genes (AhNAC48, 89, and 111) were upregulated under ABA treatment; and AhNAC20, 52, and 115 significantly responded to both conditions (Figure 5, Supplementary Table 7).
In roots, the subclade members with higher homology in group IV, including AhNAC14, 23, 56, 82, and 121, were more responsive to PEG treatment, and only AhNAC56 and 121 were upregulated under ABA treatment, whereas the higher homologous AhNACs (AhNAC12, 53, 80, and 109) in another subclade and AhNAC48 were markedly upregulated under ABA treatment. Many genes in group VII also responded to PEG and ABA treatment; for example, six genes (AhNAC13, 51, 61, 86, 114 and 125) were upregulated under PEG and ABA treatment, and AhNAC37 and AhNAC103 showed the opposite trend; another six genes (AhNAC11, 38, 41, 81, 98, and 104) were induced only by PEG treatment, and four genes (AhNAC30, 60, 97, and 124) were upregulated by ABA treatment. Almost all of the genes in group VIIIA responded to ABA and PEG, among which AhNAC21, 90, 127, and AhNAC130 were upregulated, and only AhNAC66 was downregulated; the expression levels of AhNAC16 and 84 were improved under ABA treatment. In the remaining groups, the minority of genes were affected by PEG and ABA. AhNAC32, 42, 92, and 118 in group III; AhNAC35 in group V; AhNAC99 in group VI; and AhNAC17 in group VIIIB showed upregulated trends under PEG treatment, and AhNAC44 and 106 in group III, AhNAC101 in group V, AhNAC64 and 128 in group VI, and AhNAC59, 122, and 123 in group VIIIB were upregulated under ABA treatment. The genes showing decreased expression under PEG and ABA treatment included AhNAC49 and 112 in group V, AhNAC29, 43, and 107 in group VI and AhNAC85 in group VIIIB (Figure 5, Supplementary Table 7).
To verify the expression profiles of some AhNACs that responded to PEG and ABA treatment, 20 AhNAC genes, of which AhNAC54 and 76 were from group I; AhNAC70 and 93 were from group II; AhNAC2, 21, and 56 were from group III; AhNAC12, 18, 53, 56, and 115 were from group IV; AhNAC49 and 112 were from group V; AhNAC13, 105 were from group VII; and AhNAC16, 21, 90, and 127 were from group VIII, were selected for qRT-PCR analysis. The results showed that the majority of genes analyzed by the two methods had similar expression patterns in response to ABA and PGE treatment, especially in roots. However, some genes without obvious responses to ABA and/or PEG in the RNA-seq data showed inconsistent patterns when analyzed by qRT-PCR. For example, the qRT-PCR results showed that AhNAC13, 21, 76, 90, and 127 were downregulated, and AhNAC16 was upregulated significantly by both ABA and PEG treatment, and the expression of AhNAC53 was induced by not only PEG but also ABA in leaves; in roots, AhNAC18 was markedly upregulated under both ABA and PEG treatment. The response patterns of AhNAC56 and 115 to ABA and PEG in roots and leaves found by qRT-PCR were inconsistent with the RNA-seq data. The qRT-PCR results showed that the expression level of AhNAC56 significantly increased under ABA treatment and decreased with PEG treatment in both roots and leaves, and AhNAC115 was only markedly induced by ABA in roots and was repressed by ABA in leaves and by PEG in the two organs. It was inferred that the samples from different picking times used for detection might lead to discrepancies in the results (Figure 6, Supplementary Table 8).
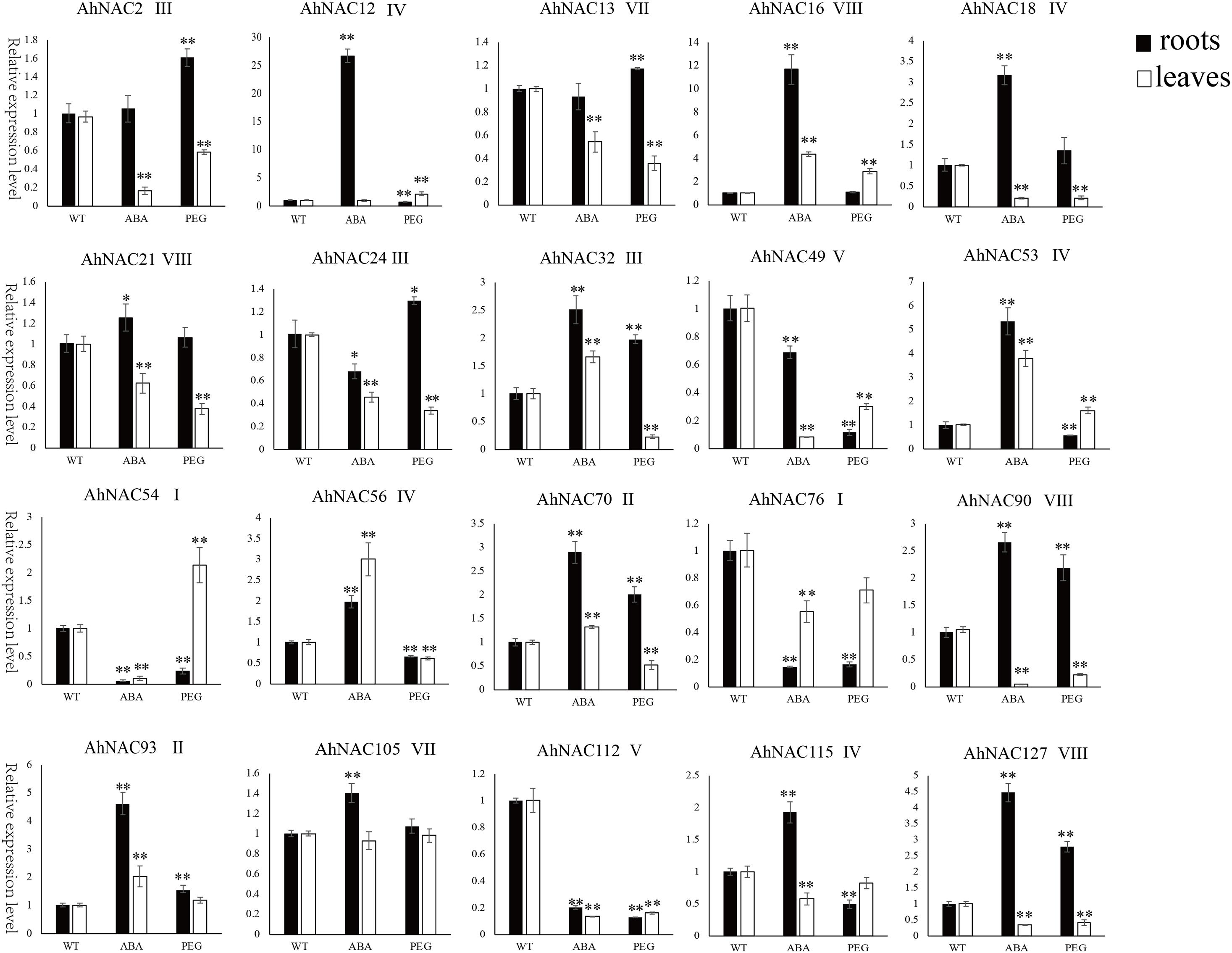
Figure 6. The expression patterns of 20 AhNAC genes in roots and leaves after PEG and ABA treatment were verified by qRT-PCR. The relative expression levels of AhNAC genes in untreated peanut (WT) and PEG and ABA treated for 24 h were compared. Data were calculated from three biological replicates, and the significance of variants between the control and treatment groups was analyzed by one-way ANOVA. Significant differences at the levels of ∗p < 0.05 and ∗∗p < 0.01.
Discussion
NAC TF is one of the largest plant-specific transcriptional regulators that play critical roles in stress responses and various developmental processes. Previous studies have identified NAC families in many plant species. However, there is still no detailed information available on the NAC family in peanut. Here, we identified 132 AhNAC genes from the peanut genome, which were classified into eight groups in accordance with the clusters of Arabidopsis AtNACs. The average MW of AhNACs (40.08 kDa) falls in the MW range (40–55 kDa) of the NAC proteins from 160 plant species (Mohanta et al., 2020). In addition, 116 pairs of segmental duplications and one pair of tandem duplications occurred, resulting in the large AhNAC gene family in the peanut genome. Mohanta et al. reported more AhNAC members (162) and larger duplicated events (161) in the genome of A. hypogaea. Similarly, there were multiple family members derived from a great quantity of duplicated events in some species of dicots and monocots (Ooka et al., 2003; Mohanta et al., 2020), and more duplication events probably occurred in these species after differentiation from their earliest ancestors. Several studies have indicated that the NAC family undergoes two discontinuous expansion processes during species evolution: the first occurred in land plants diverged from other streptophytes, whereas the second took place in flowering plants after their divergence from other vascular plants (Zhu et al., 2012; Maugarny-Calès et al., 2016; Mohanta et al., 2020). Their establishment in early diverged land plants was conserved during subsequent plant evolution, although many gene duplications and losses occurred (Zhu et al., 2012). Gene duplications are considered to be one of the primary driving forces in the evolution of genomes and gene families and are the foundation of producing new genes and new functions (Bowers et al., 2003; Moore and Purugganan, 2003). In peanut, these duplicated genes were observed in all groups, with more duplicated events in groups VII and IV. Multiple members of these two groups participated in responding to abiotic stresses. Domestication of A. hypogaea took place ≈4,500 years ago, and the modern cultivated tetraploid improved many agronomic traits but lost some resistance traits of wild relatives (Yin et al., 2019). This finding is probably due to the selective pressure conferred by environmental cues to facilitate the growth and development of plants. The peanut living environment is changeable and affected by the environment. The expansion of the NAC TF family might be beneficial for adaptability to the environment during peanut domestication.
Most of the NAC proteins that have been studied to date are involved in the responses to abiotic and biotic stress and the regulation of developmental processes (Pei et al., 2013; Kim S.G. et al., 2007; Kim et al., 2008; Li et al., 2016; Shahnejat-Bushehri et al., 2016). Under adverse environments, several NAC members of group IV in Arabidopsis, such as ANAC019, ANAC055, ANAC072 (RD26), and ATAF1 (ANAC002), participate in the stress response (Fujita et al., 2004; Wu et al., 2009). Arabidopsis RD26 is involved in the response to drought and high salinity in an ABA-dependent manner (Fujita et al., 2004). ANAC019 acts as an upstream regulator to induce the expression of several key drought-induced genes, including DREB2A, DREB2B, ARF2, MYB21, and MYB24, synergistically promoting both drought response and flower development (Sukiran et al., 2019). Overexpression of ATAF1 in Arabidopsis increased plant sensitivity to ABA, salt, and oxidative stresses (Wu et al., 2009). Recent studies have shown that overexpression of the ATAF1 gene of Arabidopsis enhances the transcription of the stress-related genes OsLEA3, OsSalT1, and OsPM1 and improves salt tolerance in rice (Liu et al., 2016). In this study, many AhNAC genes, especially the members of group IV, responded to ABA treatment and/or PEG treatment, implying that they were associated with regulating the stress response and ABA signaling. For instance, peanut AhNAC109 was significantly induced in roots by ABA, which has 68.39% sequence similarity to ATAF1 from Arabidopsis. Shao et al. found that AhNAC1 (renamed AhNAC109 in this study) played a role in seed development and response to abiotic stress (Shao et al., 2008; Wu et al., 2009). AhNAC53 (known as AhNAC2 in the study of Liu et al.) has 50% identity with RD26, which had lower water losses under drought stress and higher chlorophyll contents under salt stress in transgenic leaves, and its overexpression lines in Arabidopsis were more sensitive to ABA in root growth, seed emergence, and stomatal closure than those in the wild-type Arabidopsis (Liu et al., 2011, 2014; Tang et al., 2017). AhNAC12 (known as AhNAC3 in the study of Liu et al.) was verified to have higher expression under ABA treatment in roots and was significantly upregulated under PEG treatment in leaves. AhNAC12 and AhNAC80, as well as AhNAC53, were clustered in the same subclades, and their sequence similarities reached 60% and 60.36%, respectively. A previous study confirmed that overexpressing AhNAC3 (i.e., AhNAC12) in tobacco could activate the SOD, P5SC, LEA, and ERD10C genes and enhance the drought tolerance of transgenic tobacco (Liu et al., 2013). Taken together, it was speculated that these genes, which are evolutionarily close, might play similar functions in abiotic stress responses; however, their regulatory roles and mechanisms in response to stress need to be explored in subsequent research.
Some NIL genes were found to respond to different abiotic stresses differentially in plants. In Arabidopsis, 13 NAC members have α-helix TM motifs in the C-terminal region, and most of them are upregulated under stress conditions, suggesting their possible involvement in stress responses (Kim S.Y. et al., 2007; Wang and Dane, 2013). The expression levels of NTL1 and NTL11 and NTL4 and NTL7 were upregulated specifically by heat (37°C) and cold (4°C), respectively, and NTL6 expression was dramatically induced by NaCl. Notably, some NTLs, such as NTL2 and NTL3, were broadly influenced by cold, drought, and NaCl (Kim S.G. et al., 2007). In rice, eight genes clustered in the TIP subgroup showed upregulation by abiotic stress, among which three genes (Os01g15640, Os06g01230, and Os08g06140) share high similarity with Arabidopsis NTL6 (Nuruzzaman et al., 2010). Os02g57650, another TIP subgroup gene, was highly induced in the roots and leaves of rice under severe drought stress (Nuruzzaman et al., 2012). In our analysis of phylogenetic relationships, eight AhNACs (AhNAC7, 43, 50, and 64 and their orthologous AhNAC73, 107, 113, and 128) with one TM motif at the C-terminus, along with major Arabidopsis NTLs, were clustered in the OsNAC8/TIP/ANAC1 group (group VI) (Supplementary Table 6). Our RNA-Seq analysis also revealed that six of those genes were affected by PEG and/or ABA in roots and/or leaves. AhNAC64 and 128 were induced by PEG and ABA in both roots and leaves, and AhNAC7 and 73 were upregulated only in leaves under PEG treatment, whereas AhNAC43 and 107 were downregulated in roots treated with both PEG and ABA. These six genes were found to be increased at higher levels in PEG-treated roots than in untreated roots in the RNA-Seq data (accession no. SRP093341) released by Zhao et al. (2018). Notably, the response patterns to PEG of AhNAC43 and 107 and AhNAC7 and 73 were obviously inconsistent with our analysis (Supplementary Table 9). This might be attributed to the difference in sampling time under drought conditions. Root samples in the study of Zhao et al. were collected at 0, 6, 12, 18, 24, and 48 h poststress and were pooled for RNA-Seq, whereas in our study, sampling times before 24 h and at 48 h were absent. Some NAC genes respond rapidly to abiotic stress and increase their expression to high levels. For instance, the expression of ONAC095 in rice leaves was significantly and rapidly upregulated within 3 and 6 h by dehydration and ABA, showing 2.2- to 3.9- and 3.3-fold increases, respectively (Huang et al., 2016). AtNTL6 was expressed within 10 min after cold treatment and continued to increase, reaching a peak at 18 h (Seo et al., 2010). Therefore, the expression profiles of the AhNACs involved in the shock response to drought and ABA might be missed in the current study.
Roots are important organs for the uptake of water and nutrients. The ability of plants to adapt to rhizosphere drought stress requires the reprogramming of root growth and development (Lee et al., 2017). Recent studies have shown that improvement of the root system, such as longer roots and larger root diameters, can improve crop yield under water-limited conditions (Ghanem et al., 2011; Meister et al., 2014; Rogers and Benfey, 2015). Several NAC TFs have also been shown to promote root growth in some crop species and Arabidopsis. Overexpression of AtNAC2 in transgenic Arabidopsis plants resulted in the promotion of lateral root development (He et al., 2005). GmNAC20 and GmNAC4 also have roles in promoting lateral root formation in soybean (Hao et al., 2011; Quach et al., 2014). OsNAC5 transgenic rice could upregulate GLP, PDX, MERI5, and O-methyltransferase genes related to root growth and development, resulting in larger roots and thicker root diameters and higher yields under drought conditions (Jeong et al., 2013). Rice OsNAC6 also mediates root structural adaptations for drought tolerance (Lee et al., 2017). Overexpressing OsNAC9 altered its root architecture, involving an enlarged stele and aerenchyma, which enhanced the drought-resistance phenotype (Redillas et al., 2012). Peanuts AhNAC62 and 126 had higher homology with AtNAC2, and their expression levels in roots were significantly induced by PEG; AhNAC53 and 109, markedly induced by ABA in roots, had higher sequence similarity with OsNAC5 and OsNAC6, GmNAC20, GmNAC4 and OsNAC9. Whether they had similar functions in regulating root development under drought conditions or some functional redundancy needs to be further investigated.
Conclusion
In our study, a comprehensive in silico analysis including evolutionary, chromosomal location, gene structure, and regulatory elements of the NAC gene family in peanut was performed. A total of 132 AhNAC genes were identified and classified into eight groups. Their differential expression patterns under PEG and ABA treatment were analyzed. The results showed that many AhNAC genes were involved in the response to ABA and/or PEG in roots and leaves, especially the majority of genes in groups IV, VII, and VIII. These findings could be helpful for characterizing the genes’ functions and guiding the breeding of novel drought-resistant germplasms in peanuts.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LS designed and conceived the research. PL and LS wrote the manuscript. PL, ZP, and CM analyzed the data. PL, PX, GT, and JZ performed the experiments. LS and SW revised the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by the Major Basic Research Project of the Natural Science Foundation of Shandong Province (2018GHZ007), the National Key R&D Program of China (2018YFD1000906), and the Shandong Province Germplasm Innovation (2019LZGC017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.630292/full#supplementary-material
Footnotes
- ^ https://www.peanutbase.org/search/gene/
- ^ http://pfam.xfam.org/
- ^ https://www.expasy.org/
- ^ http://www.arabidopsis.org/
- ^ http://meme-suite.org/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://www.ebi.ac.uk/interpro/
- ^ http://www.cbs.dtu.dk/services/TMHMM/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/
- ^ https://www.ncbi.nlm.nih.gov/sra/
References
Ahmad, M., Yan, X., Li, J., Yang, Q., Jamil, W., Teng, Y., et al. (2018). Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 18:214. doi: 10.1186/s12870-018-1427-x
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Diao, W. P., John, S., Wang, S. B., Liu, J. B., Pan, B. G., Guo, G. J., et al. (2018). Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum L.): chromosome location, phylogeny, structure, expression patterns, Cis-elements in the promoter, and interaction network. Int. J. Mol. Sci. 19, 1048–1061. doi: 10.3390/ijms19041028
Duval, M., Hsieh, T. F., Kim, S. Y., and Thomas, T. L. (2002). Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248. doi: 10.1023/a:1016028530943
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Ghanem, M. E., Hichri, I., Smigocki, A. C., Albacete, A., Fauconnier, M. L., Diatloff, E., et al. (2011). Root-targeted biotechnology to mediate hormonal signalling and improve crop stress tolerance. Plant Cell Rep. 30, 807–823. doi: 10.1007/s00299-011-1005-2
Gu, Z., Cavalcanti, A., Chen, F. C., Bouman, P., and Li, W. H. (2002). Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262. doi: 10.1093/oxfordjournals.molbev.a004079
Guo, Y. F., and Gan, S. S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. doi: 10.1111/j.1365-313X.2011.04687.x
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Hu, H. H., Dai, M. Q., Yao, J. L., Xiao, B. Z., Li, X. H., Zhang, Q. F., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U. S. A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Huang, L., Hong, Y., Zhang, H., Li, D., and Song, F. (2016). Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 16:203. doi: 10.1186/s12870-016-0897-y
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Do Choi, Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jeong, J. S., Kim, Y. S., Redillas, M. C. F. R., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Kim, H. J., Nam, H. G., and Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kim, S. G., Kim, S. Y., and Park, C. M. (2007). A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226, 647–654. doi: 10.1007/s00425-007-0513-3
Kim, S. G., Lee, A. K., Yoon, H. K., and Park, C. M. (2008). A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 55, 77–88. doi: 10.1111/j.1365-313X.2008.03493.x
Kim, S. Y., Kim, S. G., Kim, Y. S., Seo, P. J., Bae, M., Yoon, H. K., et al. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 35, 203–213. doi: 10.1093/nar/gkl1068
Kim, Y. S., Kim, S. G., Park, J. E., Park, H. Y., Lim, M. H., Chua, N. H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Ko, J. H., Yang, S. H., Park, A. H., Lerouxel, O., and Han, K. H. (2007). ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 50, 1035–1048. doi: 10.1111/j.1365-313X.2007.03109.x
Krzywinski, M. I., Schein, J. E., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276. doi: 10.1093/dnares/dsr015
Lee, D. K., Chung, P. J., Jeong, J. S., Jang, G., Bang, S. W., Jung, H., et al. (2017). The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 15, 754–764. doi: 10.1111/pbi.12673
Li, X. Y., Li, X. L., Li, M. J., Yan, Y. C., Liu, X., and Li, L. (2016). Dual function of NAC072 in ABF3-mediated ABA-responsive gene regulation in Arabidopsis. Front. Plant Sci. 7:1075. doi: 10.3389/fpls.2016.01075
Liu, M. Y., Ma, Z. T., Sun, W. J., Huang, L., Wu, Q., Tang, Z. Z., et al. (2019). Genome-wide analysis of the NAC transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Genomics 20:113. doi: 10.1186/s12864-019-5500-0
Liu, X., Hong, L., Li, X. Y., Yao, Y., Hu, B., and Li, L. (2011). Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci. Biotechnol. Biochem. 75, 443–450. doi: 10.1271/bbb.100614
Liu, X., Liu, S., Wu, J. L., Zhang, B. Y., Li, X. Y., Yan, Y. C., et al. (2013). Overexpression of Arachis hypogaea NAC3 in tobacco enhances dehydration and drought tolerance by increasing superoxide scavenging. Plant Physiol. Biochem. 70, 354–359. doi: 10.1016/j.plaphy.2013.05.018
Liu, X., Zhang, B. Y., Hong, L., Su, L. C., Liang, X. Q., Li, X. Y., et al. (2014). Molecular characterization of Arachis hypogaea NAC 2 (AhNAC2) reveals it as a NAC-like protein in peanut. Biotechnol. Biotechnol. Equip. 24, 2066–2070. doi: 10.2478/v10133-010-0085-4
Liu, Y. C., Sun, J., and Wu, Y. R. (2016). Arabidopsis ATAF1 enhances the tolerance to salt stress and ABA in transgenic rice. J. Plant Res. 129, 955–962. doi: 10.1007/s10265-016-0833-0
Mao, X. G., Zhang, H. Y., Qian, X. Y., Li, A., Zhao, G. Y., and Jing, R. L. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Mathew, I. E., and Agarwal, P. (2018). May the fittest protein evolve: favoring the plant-specific origin and expansion of NAC transcription factors. Bioessays 40:e1800018. doi: 10.1002/bies.201800018
Maugarny-Calès, A., Gonçalves, B., Jouannic, S., Melkonian, M., Ka-Shu Wong, G., and Laufs, P. (2016). Apparition of the NAC transcription factors predates the emergence of land plants. Mol. Plant 9, 1345–1348. doi: 10.1016/j.molp.2016.05.016
Meister, R., Rajani, M. S., Ruzicka, D., and Schachtman, D. P. (2014). Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 19, 779–788. doi: 10.1016/j.tplants.2014.08.005
Mohanta, T. K., Yadav, D., Khan, A., Hashem, A., Tabassum, B., Khan, A. L., et al. (2020). Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS One 15:e0231425. doi: 10.1371/journal.pone.0231425
Moore, R. C., and Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U. S. A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Nuruzzaman, M., Sharoni, A. M., Satoh, K., Moumeni, A., Venuprasad, R., Serraj, R., et al. (2012). Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing aday selection (drought tolerant) and IR64. Mol. Genet. Genomics 287, 389–410. doi: 10.1007/s00438-012-0686-8
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Pei, H., Ma, N., Tian, J., Luo, J., Chen, J., Li, J., et al. (2013). An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. 163, 775–791. doi: 10.1104/pp.113.223388
Quach, T. N., Tran, L. S., Valliyodan, B., Nguyen, H. T., Kumar, R., Neelakandan, A. K., et al. (2014). Functional analysis of water stress-responsive soybean GmNAC003 and GmNAC004 transcription factors in lateral root development in Arabidopsis. PLoS One 9:e84886. doi: 10.1371/journal.pone.0084886
Redillas, M. C. F. R., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Rogers, E. D., and Benfey, P. N. (2015). Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotechnol. 32, 93–98. doi: 10.1016/j.copbio.2014.11.015
Seo, P. J., Kim, M. J., Song, J. S., Kim, Y. S., Kim, H. J., and Park, C. M. (2010). Proteolytic processing of an Arabidopsis membrane-bound NAC transcription factor is triggered by cold-induced changes in membrane fluidity. Biochem. J. 427, 359–367. doi: 10.1042/bj20091762
Shahnejat-Bushehri, S., Tarkowska, D., Sakuraba, Y., and Balazadeh, S. (2016). Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2:16013. doi: 10.1038/nplants.2016.13
Shao, F. X., Liu, Z. J., Wei, L. Q., Cao, M., and Bi, Y. P. (2008). Cloning and sequence analysis of a novel NAC-like gene AhNAC1 in peanut (Arachis hypogaea). Acta Bot. Boreali Occidential Sin. 28, 1929–1934. doi: 10.3321/j.issn:1000-4025.2008.10.001
Shiriga, K., Sharma, R., Kumar, K., Yadav, S. K., Hossain, F., and Thirunavukkarasu, N. (2014). Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2, 407–417. doi: 10.1016/j.mgene.2014.05.001
Singh, A. K., Sharma, V., Pal, A. K., Acharya, V., and Ahuja, P. S. (2013). Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20, 403–423. doi: 10.1093/dnares/dst019
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., and Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–345. doi: 10.1007/s00425-011-1403-2
Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/s0092-8674(00)81093-4
Sperotto, R. A., Ricachenevsky, F. K., Duarte, G. L., Boff, T., Lopes, K. L., Sperb, E. R., et al. (2009). Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230, 985–1002. doi: 10.1007/s00425-009-1000-9
Sukiran, N. L., Ma, J. C., Ma, H., and Su, Z. (2019). ANAC019 is required for recovery of reproductive development under drought stress in Arabidopsis. Plant Mol. Biol. 99, 161–174. doi: 10.1007/s11103-018-0810-1
Tang, G. Y., Shao, F. X., Xu, P. L., Shan, L., and Liu, Z. J. (2017). Overexpression of a peanut NAC gene, AhNAC4, confers enhanced drought tolerance in tobacco. Russ. J. Plant Physiol. 64, 525–535. doi: 10.1134/s1021443717040161
Tang, N., Shahzad, Z., Lonjon, F., Loudet, O., Vailleau, F., and Maurel, C. (2018). Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis. Nat. Commun. 9, 3884–3884. doi: 10.1038/s41467-018-06430-8
Tran, L. S., Nishiyama, R., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1, 32–39. doi: 10.4161/gmcr.1.1.10569
Tran, L. S., Quach, T. N., Guttikonda, S. K., Aldrich, D. L., Kumar, R., Neelakandan, A., et al. (2009). Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics 281, 647–664. doi: 10.1007/s00438-009-0436-8
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498. doi: 10.1105/tpc.104.022699
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Voorrips, E. R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Z. Y., and Dane, F. (2013). NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway. Acta Physiol. Plant. 35, 1397–1408. doi: 10.1007/s11738-012-1195-4
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290. doi: 10.1038/cr.2009.108
Xie, Q., Frugis, G., Colgan, D., and Chua, N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. doi: 10.1101/gad.852200
Yan, M. L., Li, X. D., Lin, Y. J., Wang, L. L., and Zhou, L. Y. (2007). Effects of drought during seedling stage on physiological traits, yield and quality of different peanut cultivars. Acta Agron. Sin. 33. 113–119.
Yang, S. H., Zhang, X. H., Yue, J. X., Tian, D. C., and Chen, J. Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280, 187–198. doi: 10.1007/s00438-008-0355-0
Yin, D. M., Ji, C. M., Song, Q. X., Zhang, W. K., Zhang, X. G., Zhao, K. K., et al. (2019). Comparison of Arachis monticola with diploid and cultivated tetraploid genomes reveals asymmetric subgenome evolution and improvement of peanut. Adv. Sci. (Weinh.) 7:1901672. doi: 10.1002/advs.201901672
Zhao, X., Li, C., Wan, S., Zhang, T., Yan, C., and Shan, S. (2018). Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 45, 119–131. doi: 10.1007/s11033-018-4145-4
Keywords: peanut, NAC transcription factor, gene family, abiotic stress, expression analysis
Citation: Li P, Peng Z, Xu P, Tang G, Ma C, Zhu J, Shan L and Wan S (2021) Genome-Wide Identification of NAC Transcription Factors and Their Functional Prediction of Abiotic Stress Response in Peanut. Front. Genet. 12:630292. doi: 10.3389/fgene.2021.630292
Received: 17 November 2020; Accepted: 08 February 2021;
Published: 09 March 2021.
Edited by:
Madhav P. Nepal, South Dakota State University, United StatesReviewed by:
Hui Song, Qingdao Agricultural University, ChinaTie Liu, University of Florida, United States
Copyright © 2021 Li, Peng, Xu, Tang, Ma, Zhu, Shan and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Shan, c2hsZWkxMDI1QHNpbmEuY29t; Shubo Wan, d2Fuc2h1Ym8yMDE2QDE2My5jb20=
 Pengxiang Li
Pengxiang Li Zhenying Peng
Zhenying Peng Pingli Xu
Pingli Xu Guiying Tang
Guiying Tang Changle Ma
Changle Ma Jieqiong Zhu1,2
Jieqiong Zhu1,2 Lei Shan
Lei Shan