
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet., 10 February 2021
Sec. Computational Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.624820
This article is part of the Research TopicMachine Learning Used in Biomedical Computing and Intelligence Healthcare, Volume IView all 11 articles
 Xi Wang1,2
Xi Wang1,2 Bin-bin Li1,2*
Bin-bin Li1,2*Head and neck tumors are the sixth most common neoplasms. Multiomics integrates multiple dimensions of clinical, pathologic, radiological, and biological data and has the potential for tumor diagnosis and analysis. Deep learning (DL), a type of artificial intelligence (AI), is applied in medical image analysis. Among the DL techniques, the convolution neural network (CNN) is used for image segmentation, detection, and classification and in computer-aided diagnosis. Here, we reviewed multiomics image analysis of head and neck tumors using CNN and other DL neural networks. We also evaluated its application in early tumor detection, classification, prognosis/metastasis prediction, and the signing out of the reports. Finally, we highlighted the challenges and potential of these techniques.
Head and neck tumors are the sixth most common neoplasms (529,000 new cases annually) and cause 350,000 cancer-related deaths each year (Ferlay et al., 2015; Fidler et al., 2017). Accurate diagnosis and analysis, especially histologic, radiologic, and biological findings, are crucial for therapeutic efficacy and prognosis prediction in precision medicine. A histologic section typically contains 106–107 cells and provides information on cell numbers and the tumor microenvironment (Koelzer et al., 2017). Radiological images contain 50–5,000 quantitative features (Limkin et al., 2017). Therefore, pathologists and radiologists must spend considerable time and effort on the qualitative and quantitative analyses of cell subsets and biomarker expression in a series of images. Also, inter- and intraobserver variations caused by subjective evaluation are inevitable in clinical practice.
Artificial intelligence (AI) was developed in the 1950s (Bini, 2018). The term big data was first proposed by the National Aeronautics and Space Administration in 1997 because a dataset is too large to be easily manipulated and managed. Big data refers to extra huge amounts of data integration, storage, analysis, and reuse of various forms of data, such as audio, video, and images. Big data is aimed at generating a large amount of information to assist decision-making and estimate outcomes, at a lower cost in time and labor (Conway et al., 2018). Computer algorithms and well-integrated data are critical for decoding medical big data. Because radiologic images are digitalized, no additional processing is required. Hung et al. (2020) used clinical big data from the SEER database to predict the survival time of patients of oral tumor by machine learning algorithms in 2020. For pathologic diagnosis, the first major step in adopting deep learning (DL) is to use digital whole-slide imaging (WSI) in routine practice (Jeyaraj and Samuel Nadar, 2019). WSI is non-inferior to traditional microscopy for clinical diagnosis (Halicek et al., 2019).
Machine learning (ML), a type of AI, refers to a computer software performing a task by being exposed to the manually crafted features of representative data (Niel and Bastard, 2019). Head and neck tumors are diverse in histology, in the pattern of underlying genetic alterations, and in metabolic signatures, which need a new method to reveal the sophisticated features. An evolution of ML—DL (Helm et al., 2020)—was first applied to the analysis of pathologic images of the head and neck in 2017 (Lu et al., 2017). Several new theories and methods have arisen to facilitate the application of DL in precision medicine, such as backpropagation and multiple layers in the convolution network. The main beauty of DL is to get rid of the handcrafted features and the end-to-end learning procedure. In the same year, DL was applied to radiomics image segmentation of head and neck tumors (Ibragimov and Xing, 2017). As a result of the improvements of computer algorithms and computational pathology, DL now facilitates the identification of benign and malignant tumors, grading of malignant tumors, and prognosis prediction.
Here, we outlined the application of DL algorithms in multiomics to diagnose and analyze head and neck tumors. Because pathological diagnosis of tumors is the gold standard, the application of DL in pathomics is emphasized in the diagnosis section and radiomics in the prognosis section. Finally, we review the challenges and prospects of DL in multiomics diagnosis and analysis.
The term “multiomics” in medicine refers to the combination of multiple sources of information (genomics, transcriptomics, proteomics, metabolomics, radiomics, and pathomics) to provide a deeper understanding of the tumor pathogenesis and lesion nature (Mars et al., 2020; Ye Y. et al., 2020; Figure 1). A schematic representation of a synergetic integration of multiomics data is shown in Figure 1. DL techniques have already been applied for multiomics analysis in various tumors. The identification of tumor origin and essential gene is critical for molecular targeted therapies and accurate treatment and lays the foundation to reveal changes in oncogenic mutation by liquid biopsy. Actually, multiomics is heterogeneous data which is difficult to be comprehensively analyzed. However, the DL network takes this challenge into an opportunity. DL-based multiomics analysis has allowed to classify groups of patients based on a more individual scale in the era of precision medicine. A timeline demonstrating the researches of DL in tumor diagnosis and multiomics analysis is shown in Supplementary Figure 1. Identifying robust survival subgroups of head and neck squamous cell carcinoma (HNSCC) will significantly improve patient outcome. Zhao et al. (2020) established a DL-based disease progression model on 86 HNSCC patients’ data using methylation data, RNA sequencing (RNA-Seq), and miRNA sequencing (miRNA-Seq) from The Cancer Genome Atlas (TCGA). The results of the autoencoder DL model demonstrated that patients were classified into two subgroups with a significant difference in progression-free survival (PFS). The predictability of this model was validated using three independent cohorts. The different biological origin of the tumor tissue has distinct clinical behavior. In practical clinical situations, it is difficult to distinguish between poorly differentiated carcinoma and metastatic carcinomas. Jiao et al. (2020) constructed a multiclass deep learning/neural network (DNN) model to integrate the whole genome sequence and pathomics data to shed light on a comprehensive view of the histological origin of the tumor cells. They evaluated three features, namely mutation distribution, mutation type, and driver gene/pathway. The classifier achieved predictive accuracies of 91% in 24 types of tumors.
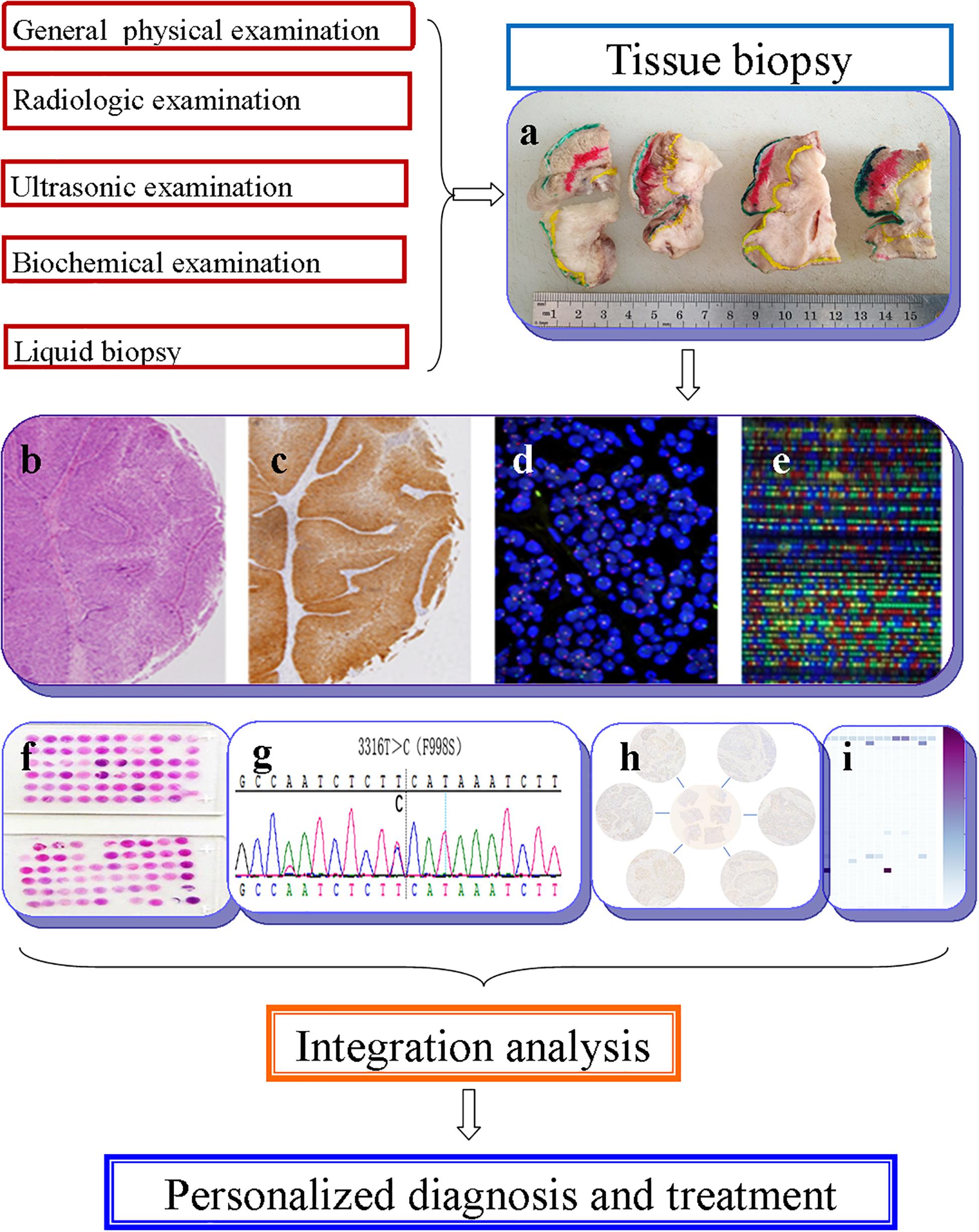
Figure 1. Schematic representation of a synergetic integration of multiomics data. (a) Biopsy specimens from head and neck tumor tissue. (b) Pathological data. (c) Tumor protein data. (d) Chromosomal data. (e) Gene microarray data. (f) Tissue microarray data. (g) Gene mutation data. (h) mRNA expression data. (i) Methylation data.
Artificial neural network models have been used to investigate the relationship between the symptoms of oral cancer and its prognosis (Tseng et al., 2015). Phillips et al. (2019) used DL models to detect pigmented dermoscopic images, thus improving the accuracy of early melanoma diagnosis. Clinically, it is difficult to differentiate ameloblastomas from keratocystic odontogenic tumors depending only on X-ray. CNN can assist in the diagnosis of ameloblastoma and keratocystic odontogenic tumors based on transfer learning. The sensitivity, specificity, and accuracy were 81.8, 83.3, and 83.0%, respectively. Interestingly, the model performed consistently well, just like skilled experts (Poedjiastoeti and Suebnukarn, 2018). In addition to X-ray research, Fu et al. (2020) used clinical photographic images to predict the early occurrence of oral tumor through a cascaded CNN model. After training by 1,469 samples, the sensitivity and specificity reached 94.9 and 88.7%, separately. This study also provided a non-invasive and highly efficient perspective on oral tumor detection. It was also possible to start providing early treatment immediately. In IDH1 wild-type glioblastomas, methylation modification had a great influence on chemotherapy response and prognosis. Le et al. (2020) used a radiomics-based eXtreme Gradient Boosting (XGBoost) model to predict the IDH1 wild-type patients with O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status. Nine robust radiomics features were selected based on the F score to improve the diagnosis of MGMT methylation status in IDH1 wild-type glioblastomas and predict patient prognosis. Sulfation of the protein S site is an important posttranscriptional modification, which plays a vital role in signal transduction, transcriptional regulation, and cell apoptosis. However, traditional experiments for its biological functions were not timely due to its rapid degradation. Do et al. (2020) used a DL network to predict protein phosphorylation S site based on the proteomics data. The DL network was also used to predict the function of fertility-related proteins in infertility patients and paved the way for a better understanding of the function of fertility proteins (Le, 2019). Therefore, the integration of DL, image analysis, and big data enables the evaluation of tumor biological behavior and, hence, facilitates diagnosis, personalized treatment, and survival prediction.
The global incidence of head and neck cancer is 1.3 million annually. The risk factors for head and neck tumors are chewing tobacco, local irritation, smoking, alcohol abuse, human papillomavirus infection, etc. It is necessary to monitor the occurrence of oral cancer in high-risk groups. Early diagnosis could reduce the mortality rate to 70% at present (Erickson et al., 2018). Also, DL could enable regular follow-up of high-risk groups. Moreover, DL methods can be applied not only to low-level tasks (e.g., recognition, detection, and segmentation) but also to more advanced tasks (e.g., selection of the optimal treatment and prediction of prognosis).
As we know, routine tissue biopsies are invasive. Although it is safe, some risks may be brought in rare cases and non-invasive biopsy comes into being. In non-invasive modalities, a large number of images appeared combined with training of DL networks based on oral clinical examinations and histological findings, which would assist in the evaluation of precancerous and cancerous lesions. The human eyes and cameras capture three color channels—red, green, and blue. Hyperspectral imaging involves multiple wavelengths, enabling the identification of cancerous and normal tissue by optical biopsy. Halicek et al. (2017) trained a CNN to identify hyperspectral images of squamous cell carcinoma (SCC). The reported accuracy, sensitivity, and specificity of the training set were 81, 81, and 80%, respectively. The hypercube contained 91 spectral bands, ranging from 450 to 900 nm with a 5-nm spectral sampling interval. Similarly, confocal laser endomicroscopy (CLE) allows real-time visualization of epithelium in vivo and enables early diagnosis of oral cancer and prediction of the prognosis. In 2007, Soo et al. reported the application of CLE for the diagnosis of oral SCC (OSCC) (Thong et al., 2007). Subsequently, Nathan et al. applied CLE to detect head and neck precancerous lesions; the sensitivity and specificity for the diagnosis of oral epithelial dysplasia were 85.7 and 80.0%, respectively (Moore et al., 2016). Aubreville et al. (2017) proposed an automatic framework for the application of CLE to detect cancerous lesions by CNN. In the proteomics research of head and neck tumors, Ni et al. (2015) used artificial neural networks to screen out proteins which were related to lymph node metastasis using the proteins extracted from the saliva of OSCC patients.
Radiomics is also used as one of the non-invasive clinical examinations. Ren et al. (2018) used the least absolute shrinkage and selection operator (LASSO) logistic regression to extract features from magnetic resonance images (MRI) of head and neck SCC to predict the histological grade before surgery. Subsequently, the same method was applied in floor-of-the-mouth and tongue SCC by Ren et al. (2020). Computed tomography (CT) can also be used to predict the histological classification before surgery by kernel principal component analysis (KPCA) and the random forest classifier (Wu et al., 2019). Mukherjee et al. (2020) performed principal component analysis and regularized regression to predict tumor grade, extracapsular spread, perineural invasion, lymphovascular invasion, and human papillomavirus infection status. The accuracy, sensitivity, and specificity of the model were 0.72, 0.83, and 0.48, respectively. DL is also applied in radiomics. Ye J. et al. (2020) used a CNN model for histological classification of head and neck tumors; the accuracy, sensitivity, and specificity were 0.79, 0.71, and 0.85, respectively. The utility of AI for the analysis of head and neck pathologic sections and radiologic images is summarized in Tables 1, 2.
Deep learning is suitable for digital pathology (DP)-related image analysis tasks, such as detection (e.g., lymphocyte), segmentation (e.g., nuclei and epithelium), and classification (e.g., the tumor subclass). Figures 2, 3 demonstrate an example of epithelial segmentations on WSI images and an example of segmentation of nuclei in a cell layer on WSI images. Different from ML, which classifies handcrafted features (Das et al., 2018), DL takes an agnostic approach by combining feature extraction and the interest region analysis.
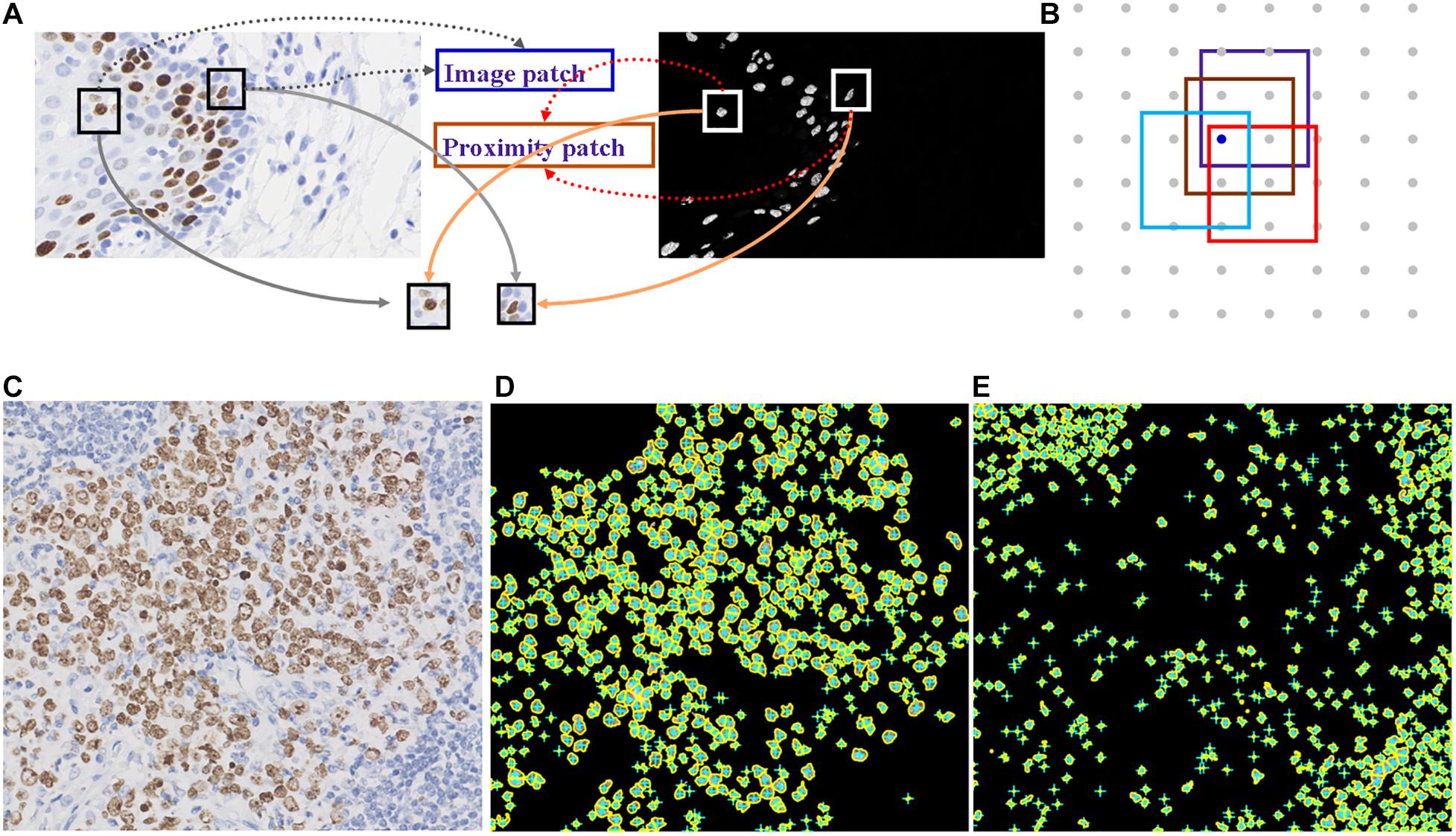
Figure 2. Example of segmentation of nuclei in a cell layer on WSI images. (A,B) Illustrations of nucleus segmentation based on the DL model. (C) The original image of immunohistochemical staining. (D) Nucleus segmentation for immunohistochemistry-positive cells. (E) Nucleus segmentation for immunohistochemistry-negative cells.
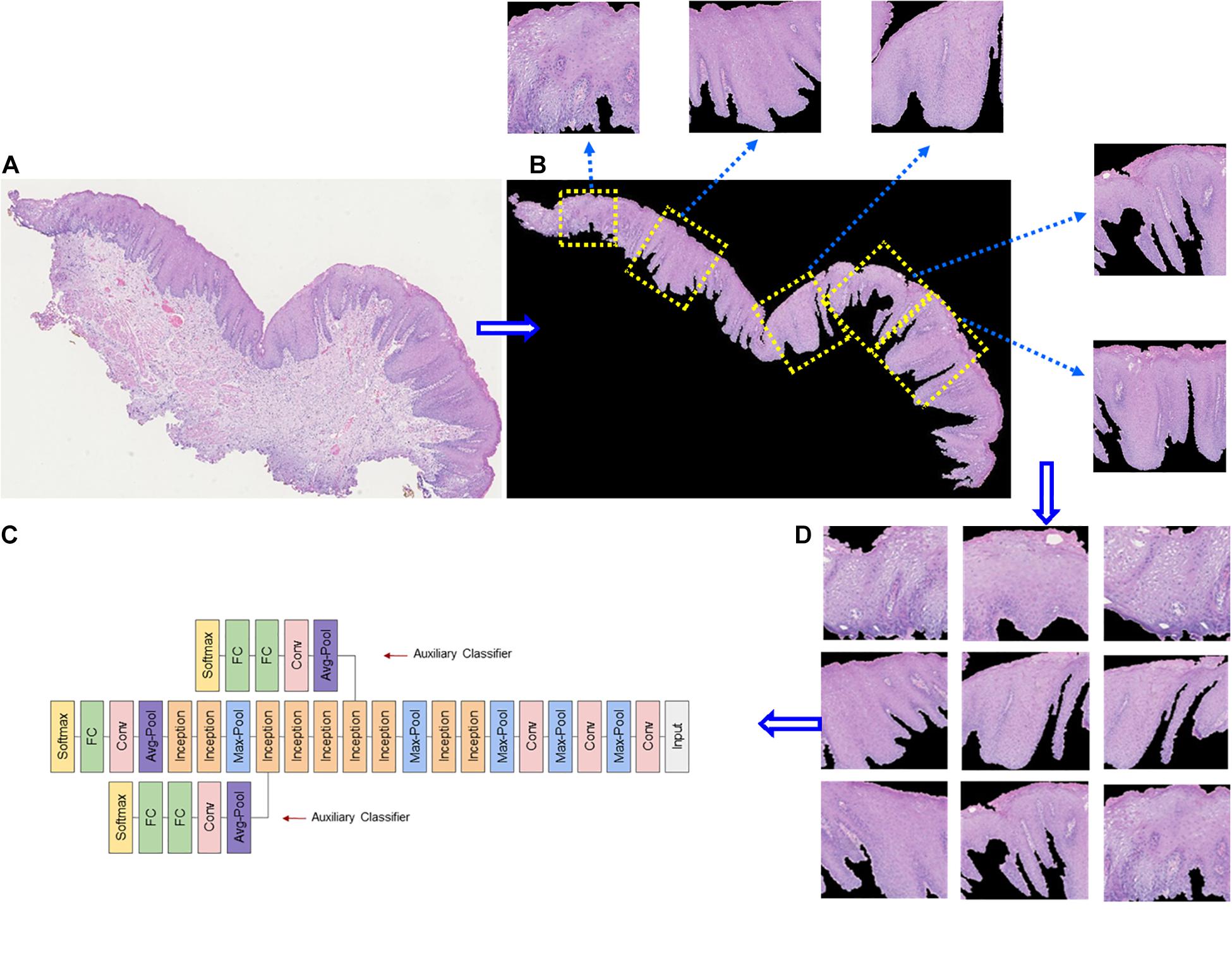
Figure 3. Example of epithelial segmentations on WSI images. (A) The original image. (B) Black curves indicate the segmented boundary of the epithelium. (C) Input patches for training the DL network. (D) Schematic of a deep learning framework.
In head and neck tumor diagnosis, the morphology of heterogeneous cell types needs to be evaluated. This can be formulated as a pixel-wise detection task. The detection tasks frequently align with the classification tasks, and the algorithms learn the weighted parameters of the feature map. The algorithms map clusters of similar features to the output labels. The workflow for DL approaches in digital pathology is shown in Figure 4. In traditional ML, the workflow is comprised of two steps: detection and classification. For instance, Lewis et al. (2014) developed an approach to quantify automatically the morphologic features used for the classification of aggressive or indolent p16-positive oropharyngeal SCC. A cluster cell graph was generated to evaluate the spatial distribution of mitotic cells, and a random forest (RF) decision tree and SVM were used to classify features. The accuracy of the model was 87.5% (140 patients). However, it may not be applicable to other situations because of the small training dataset and the overfitting problem. Moreover, the accuracy of DL is unsatisfactory. Several proposed DL models for detecting head and neck tumors overcome the abovementioned shortcomings. Aubreville et al. (2017) trained a DL model to detect an image patch from doubtful OSCC cases. Overall image recognition had an area under the curve (AUC) of 0.96 and a mean accuracy of 88.3% (sensitivity 86.6% and specificity 90%).
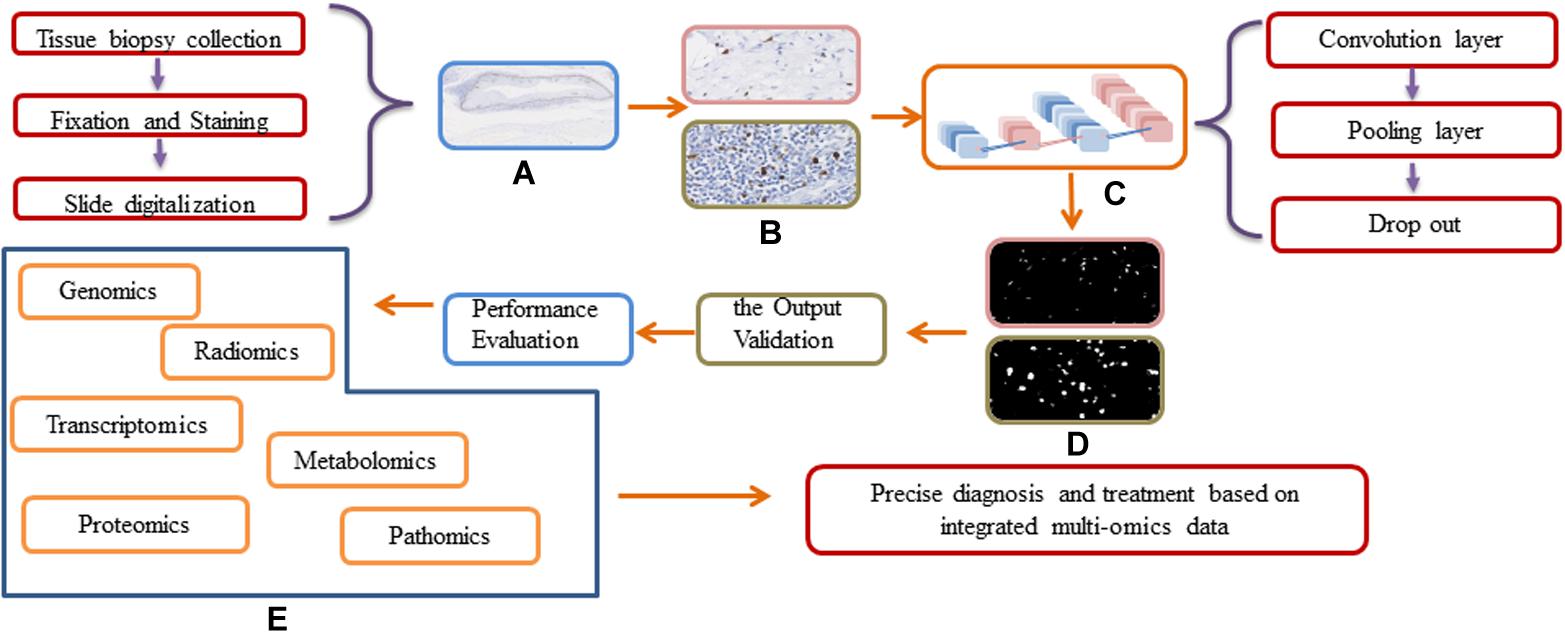
Figure 4. Workflow and general framework for DL approaches in digital pathology. (A) The original WSI image. (B) Different resolution of WSI images before input the DL network. (C) The Schematic of a deep learning framework. (D) The output of different resolution of WSI images. (E) The integration of multi-omics data.
Halicek et al. trained a deep CNN to identify surgical margins accurately in hyperspectral images. Additionally, an end-to-end DL network can simultaneously detect and enumerate mitotic cells (Jimenez and Racoceanu, 2019). In the above reports, DL was only used for dimension reduction or feature extraction. It also may be a classifier to perform classification (Boldrini et al., 2019). Usually, the end-to-end DL approach performed better than none end-to-end learning. However, as for pathomics, one end-to-end DL model cannot perform multiple tasks simultaneously. Lei et al. (2019) trained a convolutional neural network by DL to extract mitosis features automatically and proposed a network to determine the location of all mitotic cells. This approach showed an unexpectedly high accuracy in the International Conference on Pattern Recognition (ICPR2012) mitosis detection test dataset. The remaining challenges include accurate identification and enumeration of mitotic cells in two-dimensional (2D) digital histology images. The imaging of three-dimensional (3D) tissues in 2D results in loss of spatial information. Radiomics enables enhanced 3D assessment of tumor growth by quantifying changes in tumor cellularity and angiogenesis. Radiomics analysis also shows potential for the accurate quantification of heterogeneity and outcome prediction.
Microscopic cell structure recognition is emphasized in pathomics. A common strategy for detecting cells or nuclei is to train a CNN classifier as a pixel classifier, in which a patch centered on the object of interest is used to train the network under supervised conditions. Trained CNN models typically comprise two classifiers (yes or no) and can be applied to WSI in a sliding window to detect all histological components of interest and output a probability map, where each pixel is transferred to a probable value. Therefore, in principle, the target objects can be located by finding a local maximum in the generated probability map. Fully convolutional networks can share calculations on sliding windows. After completing nuclear or mitotic detection tasks, it begins counting or extracting quantitative indicators in WSI. The algorithm is built on mapping an input image patch to a density map, which is used to estimate the number of cells in the original image.
Deep learning also plays an important role in the analysis of tumor microenvironment characteristics (TMC). The crucial step in TMC analysis is segmenting different types of tissue and cell structures in pathology images. Tumor cells can be classified into parenchymal and stromal cells. Niranjan and Sarathy (2018) reported the ratio of tumor to stroma (TSR) as a reliable histologic predictor of overall survival and outcome in OSCC. In a cohort of 60 OSCC patients, the 3-year overall survival (OS) and disease-free survival (DFS) rates of patients with >50% intratumor stroma had been shown to be better than the patients with <50% intratumor stroma.
The segmentation task is more difficult than mitosis detection because parenchyma segmentation can be labeled by experts at lower magnifications. However, stroma (e.g., lymphocytes, macrophages, fibroblasts, etc.) must be analyzed at high magnification. Indeed, × 40 magnification performed better than × 20 magnification for nucleus segmentation. By contrast, epithelium segmentation is typically more precise by experts at × 20 than at 40 × magnification, as indicated by a higher accuracy and F score (Janowczyk and Madabhushi, 2016). To remedy this drawback, the fully convolutional network (FCN) and UNet were designed to accept discretional size as an input and product proportionate-sized outputs by removing all fully connected layers and introducing unsampled layers to offset the shortcomings of downsampling in CNN (Zhou et al., 2019). Considering that head and neck tumors are heterogeneous and complex, segmentation may involve varied anonymous anaplastic cells and then can be achieved by data augmentation. Halicek et al. (2018) trained a CNN to segment the tumor and normal tissue of OSCC with 81% accuracy, 84% sensitivity, and 77% specificity. The sensitivity and specificity of FCN for cervical tumor segmentation on 3D FDG-PET images were 88 and 98%, which were markedly superior to CNN. Unfortunately, FCN has not been used for segmentation of pathologic images of head and neck tumors. Moreover, tumor segmentation accuracy is associated with loss function. Now, the well-known loss function is cross-entropy loss. A new loss function, class-wise DSC loss, for training the segmentation network of colonoscopy pathology images was presented by Feng et al. (2020).
The high heterogeneity and complexity of head and neck tumor pathology images hamper the prediction of outcomes only by TNM stage. In recent years, more and more scholars have been interested in the potential of DL networks for predicting postoperative outcomes. The applications of radiomics to predict overall survival, biomarker status, recurrence, distant or local metastasis, and lymph node metastasis are summarized in Table 2. Tixier et al. used the Genomica software to analyze PET and transcriptomics data of 45 patients with locally advanced head and neck cancer. They applied a fuzzy locally adaptive Bayesian (FLAB) algorithm to assess the associations between radiomics features (a total of 28 image biomarker standardization initiative-compliant radiomic features) (Zwanenburg et al., 2020) and alterations of biological pathways (e.g., extracellular matrix organization, cell cycle, signal transduction, cell cycle, etc.). The results demonstrated that FDG-PET radiomic features were associated with cell cycle, DNA repair, extracellular matrix organization, immune system, metabolism, and signal transduction pathways, providing a thorough understanding of genetic mutations and minimizing the costs (Tixier et al., 2020). Zhu et al. (2019) integrated the genome-wide multiomics data of 126 patients with head and neck SCC with CT imaging data and found the significant association between genomic characteristics and CT features. The use of DL together with sophisticated biomarkers can significantly improve prognostic and predictive accuracy. Subsequently, the DL-extracted imaging features of morphology structure on digitized H&E-stained tissue sections have been used for risk stratification of head and neck tumor patients. Patients with p16-positive human papillomavirus-related oropharyngeal SCC have a more favorable prognosis than those negative for P16 (Ali et al., 2013). Lewis et al. (2014) used a typical ML approach (the random forest decision tree) to extract nuclear morphologic features and predict progression. Before the advent of DL, improvement of prognosis was evaluated by multifactor analysis, conventional logistic regression, and Cox analysis in traditional ML models. However, the absence of a decision rule and linear combinations of covariates hampered the prediction of outcomes. DL-based survival prediction has improved predictive accuracy and, together with nonlinear algorithms, will facilitate precision medicine. Therefore, it is suitable for predicting the survival of inpatients (Tan et al., 2016). Tseng et al. constructed a DNN to predict the survival of patients with oral tumors using clinical variables and histopathological features. It was suggested that the DNN model established by data mining was superior to logistic regression in terms of both training accuracy and cross-validation accuracy. Brennan et al. (2017) used an unsupervised cluster analysis method to interpret the genomics and epigenetics data of morphologically atypical head and neck SCC and found CpG island methyl groups in atypical SCC. Therefore, novel prognostic factors, such as genetic mutations and molecular markers, combined with clinicopathologic and radiologic features and a multi-nonlinear DL network would yield optimal results.
Proteomics and transcriptome have also been used to study lymph node and distant metastasis and recurrence of SCC patients. Onken et al. (2014) used an unsupervised clustering algorithm to extract transcriptome signature predicting distant metastasis in oral tumor over four SCC datasets. Xu et al. (2014) applied a ML approach called maximum relevance minimum redundancy algorithm to a set of transcriptome data generated from papillary carcinoma and anaplastic carcinoma for differential diagnoses. The lung is the most common site of distant metastasis of OSCC. Primary SCC can also occur in the lung. Through supervised learning and analysis of proteomic data, Bohnenberger et al. (2018) found the vital difference of protein characteristics between lung metastatic head and neck SCC and primary lung SCC. Their data provided reference information for the origin of lung SCC. Carnielli et al. (2018) used histological morphology-oriented proteomics analysis of the protein expression in tumor islands and stroma to forecast the possibility of tumor recurrence and lymph node metastasis. Six ML approaches were used by Kaddi and Wang (2017) to analyze proteomics and transcriptome data, including KNN, SVM, naive Bayes, DT, AdaBoost, and RF. It was shown that the prognostic model based on both transcriptome and proteomics data had better predictive performance than transcriptomics or proteomics alone.
Zhang et al. (2017) developed MDNet, which generated pathological reports by directly mapping pathology images with simultaneous retrieval of pathology images according to symptom descriptions. MDNet added a language network to the original image model. Integration of a language model with the multiscale features proposed by the image model allowed the identification of critical image features and enabled the direct mapping from words to pixels. Changes in the size or density of nuclei and epithelial thickness may indicate neoplastic invasion. However, these discriminant imaging features were not directly supported to generate a diagnostic report. MDNet allowed direct multimodal mapping from medical images and diagnostic reports. Mimicking diagnosis by pathologists, long short-term memory (LSTM) networks were used to generate semantic information as a language model. The LSTM was a representative gated RNN that controlled the forget gate and input gate to emphasize or forget some weights. It could reduce the problem of multiple layers and vanished gradient multilayers from input to output.
Radiogenomics is a computational nomenclature which identifies correlations between radiomics imaging features and genomics or proteomics data. These imaging feature correlations can be used to predict a tumor’s molecular profile in clinical radiomics data (West and Rosenstein, 2010). Radiogenomics has two goals: i) discover the patients who are more likely to develop radiotherapy complications based on molecular data and (ii) analyze the targeted molecular pathway responsible for radiotoxicity in radiation-induced normal tissue (Kerns et al., 2014). Postoperative radiotherapy is an effective treatment for head and neck tumors. The existence of radiosensitivity and radioresistance may be related to genetic factors partially. The remaining differences between individuals were caused by differences in treatment (radiation dose), physical habits, and random factors (Rattay and Talbot, 2014). Werbrouck et al. (2009) reported that the DNA repair genes XRCC3 and Ku70 were connected to the intensity of dysphagia after radiotherapy in H&N tumor in 2009. For the study of postradiotherapy mucositis, Yang et al. (2020) sequenced and located the gene expression in 1,497 patients with postoperative radiotherapy. They found that 64 target genes were enriched in the process of telomerase regulation, which confirmed the importance of telomere function in the development of radiation-induced adverse reactions. The combination of PET-based spatial radiation features and sequencing data provided a new perspective for further revealing the spatial heterogeneity of tumors (Clasen et al., 2020). Furthermore, the predictive analysis of gene expression and cellular and molecular expression can be provided from a non-invasive point of view, based on the radiological characteristics and gene differential expression data of head and neck tumors obtained from the TCGA and TCIA databases (Katsoulakis et al., 2020).
AI is highly dependent on a robust and large database, but the database of pathological slides of head and neck tumors has not been established yet. Apart from the hardware needed to set up the database, setting up an autoprocessed image database is also needed. When there were images captured from clinical cases, the database could have the images with their properties at the same time, which would help in further analysis. As time goes on, the database could grow by itself (Ibrahim et al., 2020). The low-quality images are also a problem for DL analysis. According to a jointed framework proposed by Chen J. et al. (2020), a novel transfer learning strategy called channel fusion transfer learning and a deep super-resolution framework called SRFBN+ were dedicated to generating higher-resolution slice images with lower-resolution ones as input. The most successful application of DL in medical image analysis has been in supervised learning. On the other hand, the rarity of pathologists added the extra difficulties in data cleaning and labeling, while the high heterogeneity of head and neck tumors means that many rare tumors need to be accurately labeled.
A crucial step is to avoid subjective and sample biases in the training sets as the quality of the output depends on the quality of the input data (Oakden-Rayner, 2020). So, establishing a unified standard to normalize the image input in the network by multi-institution datasets can not only reduce the bias from the samples and the bias caused by inconsistent diagnostic from the physicians but also fully fit and train the model to reduce overfitting and reduce to a maximum the highly opaque nature of medical image (Martorell-Marugán et al., 2019). However, current DL algorithms are mainly trained on a small dataset from a single center (Jiang et al., 2020). The limited availability of well-characterized and adequately stored clinical tumor and non-tumor samples is a major challenge in proteomics and genomics researches (Matta et al., 2010).
For the algorithms themselves, the tendency has been to propose new algorithms rather than optimize those already used, leading to the conclusion that there is no improvement of some subdomain algorithms. In addition, due to the limitations of, for instance, data and computational power, the improvement of algorithms must take into account various trade-offs. Additionally, some studies used a non-open-source code or a non-open-source model, such as an in-house developed model, hampering model verification in other types of tumors (Parmar et al., 2015; Leijenaar et al., 2018). A flowchart demonstrating the relationship for the subsection of difficulties and expectation of DL in tumor diagnosis and multiomics analysis is shown in Supplementary Figure 2.
The lack of unified innovation evaluation standards in AI has led to some exaggeration of the improvements achieved. This can be overcome by a variety of methods, e.g., an open-source or source model. Unifying evaluation standards is difficult but is possible for some mature domains. The relevant data management domains are as follows: (i) administrative standards, (ii) patient privacy protection standards, and (iii) intellectual property protection standards. The establishment of data management standards would allow access to diverse anonymized imaging datasets. Technical standardization cannot resolve all of the issues described in this review. The use of different image normalization or style conversion methods (e.g., rotating, cropping, zooming, and image histogram-based modifications) for preprocessing could overcome the technical obstacles.
The architectures of CNNs have been especially powerful for computer vision, particularly in image interpretation and procession. WSI combined with DL algorithms for tumor detection, classification, and prognosis prediction has played an ever-increasing part in supporting pathologists in clinical assessments. The main components of CNN are convolutional layers and pooling layers. Although CNN has advantages in the processing of object detection, it has notable drawbacks: (1) both the training and the detection process is considerably time-consuming and (2) the normalization method would lead to lose some discriminative details. FCN is suitable for image segmentation at the pixel level. It consists of convolution and deconvolution layers, which can accept input images of any size and retain the spatial information of the original input lines. The major disadvantages of FCN may be that it is noisy and contains redundant information, requiring a huge number of reliable samples. To overcome the issues mentioned above, more novel architectures (e.g., UNet++, SegNet, and ENet) based on FCN or CNN have been proposed for image segmentation. Pan et al. (2020) proposed a DL model based on the architectures of FCN to automatically recognize lymph node metastasis of esophageal SCC. Compared with previous studies focused on the isolated tasks in the analysis of pathology and radiology images, the integration of independent DL models into a general model would be beneficial (Wang et al., 2019). It was also anticipated that biological pathways and gene regulation networks would be incorporated into prediction models, improving their performance and interpretability. For multimodal learning, collecting data from the required modalities simultaneously could be problematic. A slight disturbance to the inputs of multimodal can influence the stability of CNN. Lin et al. (2020) trained a multiscale activity transition network to provide an activity state pyramid consisting of multiscale recurrent neural networks to capture the accurate feature of input. Transfer learning is frequently used and is an effective pretraining strategy. The fusion of different modal representations is the key point of a multimodal task. Specific fusion operations are based on an attention mechanism or bilinear pooling. In practice, fusion operations are often diverse and complicated (Mormont et al., 2020).
Now, DL algorithms still have several difficulties of integrating multiomics data or various sources of information such as pathology images and electronic medical records. The use of DL to accomplish simple tasks can yield useful results. Furthermore, complex datasets, abundant neural network architecture, and adequate DL methods are anticipated to provide useful information for precision medicine. Pathomics and radiomics are crucial components of multiomics, which also include genomics, transcriptomics, proteomics, and metabolomics information. Although there are still some limitations that restricted the direct clinical usage of multiomics analysis, there is still an increasing effort in solving the drawbacks to provide promising applications. The increasing number of omics datasets is fuelling the quantitative analysis of biological specimens at the gene, cell, and tissue levels. It will generate novel hypotheses on the molecular mechanisms of tumor development and progression for guiding precise diagnosis and treatment.
BL: conceptualization, writing—review and editing, supervision, and funding acquisition. XW: formal analysis, data curation, and writing—original draft. Both authors contributed to the article and approved the submitted version.
This study received funding from the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-038).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.624820/full#supplementary-material
Supplementary Figure 1 | A timeline demonstrating the DL researches in tumor diagnosis and multi-omics analysis.
Supplementary Figure 2 | A flow chart demonstrating the difficulties and expectation of DL in tumor diagnosis and multi-omics analysis.
Ali, S., Lewis, J., and Madabhushi, A. (2013). “Spatially aware cell cluster(spACCl) graphs: predicting outcome in oropharyngeal p16+ tumors,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. MICCAI 2013. Lecture Notes in Computer Science, Vol. 8149, eds K. Mori, I. Sakuma, Y. Sato, C. Barillot and N. Navab (Berlin: Springer), 412–419. doi: 10.1007/978-3-642-40811-3_52
Aubreville, M., Knipfer, C., Oetter, N., Jaremenko, C., Rodner, E., Denzler, J., et al. (2017). Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Sci. Rep. 7:11979. doi: 10.1038/s41598-017-12320-8
Bagher-Ebadian, H., Lu, M., Siddiqui, F., Ghanem, A. I., Wen, N., Wu, Q., et al. (2020). Application of radiomics for the prediction of HPV status for patients with head and neck cancers. Med. Phys. 47, 563–575. doi: 10.1002/mp.13977
Beaumont, J., Acosta, O., Devillers, A., Palard-Novello, X., Chajon, E., de Crevoisier, R., et al. (2019). Voxel-based identification of local recurrence sub-regions from pre-treatment PET/CT for locally advanced head and neck cancers. EJNMMI Res. 9:90. doi: 10.1186/s13550-019-0556-z
Bini, S. A. (2018). Artificial intelligence, machine learning, deep learning, and cognitive computing: What do these terms mean and how will they impact health care? J. Arthroplasty 33, 2358–2361. doi: 10.1016/j.arth.2018.02.067
Bogowicz, M., Jochems, A., Deist, T. M., Tanadini-Lang, S., Huang, S. H., Chan, B., et al. (2020). Privacy-preserving distributed learning of radiomics to predict overall survival and HPV status in head and neck cancer. Sci. Rep. 10:4542. doi: 10.1038/s41598-020-61297-4
Bogowicz, M., Tanadini-Lang, S., Guckenberger, M., and Riesterer, O. (2019a). Combined CT radiomics of primary tumor and metastatic lymph nodes improves prediction of loco-regional control in head and neck cancer. Sci. Rep. 9:15198. doi: 10.1038/s41598-019-51599-7
Bogowicz, M., Tanadini-Lang, S., Veit-Haibach, P., Pruschy, M., Bender, S., Sharma, A., et al. (2019b). Perfusion CT radiomics as potential prognostic biomarker in head and neck squamous cell carcinoma. Acta Oncol. 58, 1514–1518. doi: 10.1080/0284186x.2019.1629013
Bohnenberger, H., Kaderali, L., Ströbel, P., Yepes, D., Plessmann, U., Dharia, N. V., et al. (2018). Comparative proteomics reveals a diagnostic signature for pulmonary head-and-neck cancer metastasis. EMBO Mol. Med. 10:e8428. doi: 10.15252/emmm.201708428
Boldrini, L., Bibault, J. E., Masciocchi, C., Shen, Y., and Bittner, M. I. (2019). Deep learning: a review for the radiation oncologist. Front. Oncol. 9:977. doi: 10.3389/fonc.2019.00977
Brennan, K., Koenig, J. L., Gentles, A. J., Sunwoo, J. B., and Gevaert, O. (2017). Identification of an atypical etiological head and neck squamous carcinoma subtype featuring the CpG island methylator phenotype. EBioMedicine 17, 223–236. doi: 10.1016/j.ebiom.2017.02.025
Carnielli, C. M., Macedo, C. C. S., De Rossi, T., Granato, D. C., Rivera, C., Domingues, R. R., et al. (2018). Combining discovery and targeted proteomics reveals a prognostic signature in oral cancer. Nat. Commun. 9:3598. doi: 10.1038/s41467-018-05696-2
Chen, J., Ying, H., Liu, X., Gu, J., Feng, R., Chen, T., et al. (2020). A transfer learning based super-resolution microscopy for biopsy slice images: the joint methods perspective. IEEE/ACM Trans. Comput. Biol. Bioinform. doi: 10.1109/tcbb.2020.2991173 [Epub ahead of print].
Chen, L., Wang, H., Zeng, H., Zhang, Y., and Ma, X. (2020). Evaluation of CT-based radiomics signature and nomogram as prognostic markers in patients with laryngeal squamous cell carcinoma. Cancer Imaging 20:28. doi: 10.1186/s40644-020-00310-5
Chen, L., Zhou, Z., Sher, D., Zhang, Q., Shah, J., Pham, N. L., et al. (2019). Combining many-objective radiomics and 3D convolutional neural network through evidential reasoning to predict lymph node metastasis in head and neck cancer. Phys. Med. Biol. 64:075011. doi: 10.1088/1361-6560/ab083a
Clasen, K., Leibfarth, S., Hilke, F. J., Admard, J., Winter, R. M., Welz, S., et al. (2020). PET/MRI and genetic intrapatient heterogeneity in head and neck cancers. Strahlenther. Onkol. 196, 542–551. doi: 10.1007/s00066-020-01606-y
Conway, D. I., Purkayastha, M., and Chestnutt, I. G. (2018). The changing epidemiology of oral cancer: definitions, trends, and risk factors. Br. Dent. J. 225, 867–873. doi: 10.1038/sj.bdj.2018.922
Cozzi, L., Franzese, C., Fogliata, A., Franceschini, D., Navarria, P., Tomatis, S., et al. (2019). Predicting survival and local control after radiochemotherapy in locally advanced head and neck cancer by means of computed tomography based radiomics. Strahlenther. Onkol. 195, 805–818. doi: 10.1007/s00066-019-01483-0
Das, D. K., Bose, S., Maiti, A. K., Mitra, B., Mukherjee, G., and Dutta, P. K. (2018). Automatic identification of clinically relevant regions from oral tissue histological images for oral squamous cell carcinoma diagnosis. Tissue Cell 53, 111–119. doi: 10.1016/j.tice.2018.06.004
Do, D., Le, T., Le, T. Q. T., and Le, N. Q. K. (2020). Using deep neural networks and biological subwords to detect protein S-sulfenylation sites. Brief. Bioinformatics. doi: 10.1093/bib/bbaa128 [Epub ahead of print].
Erickson, B. J., Korfiatis, P., Kline, T. L., Akkus, Z., Philbrick, K., and Weston, A. D. (2018). Deep learning in radiology: Does one size fit all? J. Am. Coll. Radiol. 15, 521–526. doi: 10.1016/j.jacr.2017.12.027
Feng, R., Liu, X., Chen, J., Chen, D. Z., Gao, H., and Wu, J. (2020). A deep learning approach for colonoscopy pathology WSI analysis: accurate segmentation and classification. IEEE J. Biomed. Health Inform. doi: 10.1109/jbhi.2020.3040269 [Epub ahead of print].
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386. doi: 10.1002/ijc.29210
Fidler, M. M., Bray, F., Vaccarella, S., and Soerjomataram, I. (2017). Assessing global transitions in human development and colorectal cancer incidence. Int. J. Cancer 140, 2709–2715. doi: 10.1002/ijc.30686
Fu, Q., Chen, Y., Li, Z., Jing, Q., Hu, C., Liu, H., et al. (2020). A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: a retrospective study. EClinicalMedicine 27:100558. doi: 10.1016/j.eclinm.2020.100558
Ger, R. B., Zhou, S., Elgohari, B., Elhalawani, H., Mackin, D. M., Meier, J. G., et al. (2019). Radiomics features of the primary tumor fail to improve prediction of overall survival in large cohorts of CT- and PET-imaged head and neck cancer patients. PLoS One 14:e0222509. doi: 10.1371/journal.pone.0222509
Halicek, M., Little, J. V., Wang, X., Chen, A. Y., and Fei, B. (2019). Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks. J. Biomed. Opt. 24, 1–9. doi: 10.1117/1.Jbo.24.3.036007
Halicek, M., Little, J. V., Wang, X., Patel, M., Griffith, C. C., Chen, A. Y., et al. (2018). Tumor margin classification of head and neck cancer using hyperspectral imaging and convolutional neural networks. Proc. SPIE Int. Soc. Opt. Eng. 10576:1057605. doi: 10.1117/12.2293167
Halicek, M., Lu, G., Little, J. V., Wang, X., Patel, M., Griffith, C. C., et al. (2017). Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J. Biomed. Opt. 22:60503. doi: 10.1117/1.Jbo.22.6.060503
Helm, J. M., Swiergosz, A. M., Haeberle, H. S., Karnuta, J. M., Schaffer, J. L., Krebs, V. E., et al. (2020). Machine learning and artificial intelligence: definitions, applications, and future directions. Curr. Rev. Musculoskelet. Med. 13, 69–76. doi: 10.1007/s12178-020-09600-8
Hung, M., Park, J., Hon, E. S., Bounsanga, J., Moazzami, S., Ruiz-Negrón, B., et al. (2020). Artificial intelligence in dentistry: harnessing big data to predict oral cancer survival. World J. Clin. Oncol. 11, 918–934. doi: 10.5306/wjco.v11.i11.918
Ibragimov, B., and Xing, L. (2017). Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med. Phys. 44, 547–557. doi: 10.1002/mp.12045
Ibrahim, A., Gamble, P., Jaroensri, R., Abdelsamea, M. M., Mermel, C. H., Chen, P. C., et al. (2020). Artificial intelligence in digital breast pathology: techniques and applications. Breast 49, 267–273. doi: 10.1016/j.breast.2019.12.007
Janowczyk, A., and Madabhushi, A. (2016). Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inform. 7:29. doi: 10.4103/2153-3539.186902
Jeyaraj, P. R., and Samuel Nadar, E. R. (2019). Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. J. Cancer Res. Clin. Oncol. 145, 829–837. doi: 10.1007/s00432-018-02834-7
Jiang, Y., Yang, M., Wang, S., Li, X., and Sun, Y. (2020). Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 40, 154–166. doi: 10.1002/cac2.12012
Jiao, W., Atwal, G., Polak, P., Karlic, R., Cuppen, E., Danyi, A., et al. (2020). A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat. Commun. 11:728. doi: 10.1038/s41467-019-13825-8
Jimenez, G., and Racoceanu, D. (2019). Deep learning for semantic segmentation vs. classification in computational pathology: application to mitosis analysis in breast cancer grading. Front. Bioeng. Biotechnol. 7:145. doi: 10.3389/fbioe.2019.00145
Kaddi, C. D., and Wang, M. D. (2017). Models for predicting stage in head and neck squamous cell carcinoma using proteomic and transcriptomic data. IEEE J. Biomed. Health Inform. 21, 246–253. doi: 10.1109/jbhi.2015.2489158
Katsoulakis, E., Yu, Y., Apte, A. P., Leeman, J. E., Katabi, N., Morris, L., et al. (2020). Radiomic analysis identifies tumor subtypes associated with distinct molecular and microenvironmental factors in head and neck squamous cell carcinoma. Oral Oncol. 110:104877. doi: 10.1016/j.oraloncology.2020.104877
Keek, S., Sanduleanu, S., Wesseling, F., de Roest, R., van den Brekel, M., van der Heijden, M., et al. (2020). Computed tomography-derived radiomic signature of head and neck squamous cell carcinoma (peri)tumoral tissue for the prediction of locoregional recurrence and distant metastasis after concurrent chemo-radiotherapy. PLoS One 15:e0232639. doi: 10.1371/journal.pone.0232639
Kerns, S. L., Ostrer, H., and Rosenstein, B. S. (2014). Radiogenomics: using genetics to identify cancer patients at risk for development of adverse effects following radiotherapy. Cancer Discov. 4, 155–165. doi: 10.1158/2159-8290.cd-13-0197
Kim, D. W., Lee, S., Kwon, S., Nam, W., Cha, I. H., and Kim, H. J. (2019). Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 9:6994. doi: 10.1038/s41598-019-43372-7
Koelzer, V. H., Sokol, L., Zahnd, S., Christe, L., Dawson, H., Berger, M. D., et al. (2017). Digital analysis and epigenetic regulation of the signature of rejection in colorectal cancer. Oncoimmunology 6:e1288330. doi: 10.1080/2162402x.2017.1288330
Kwan, J. Y. Y., Su, J., Huang, S. H., Ghoraie, L. S., Xu, W., Chan, B., et al. (2018). Radiomic biomarkers to refine risk models for distant metastasis in HPV-related oropharyngeal carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 102, 1107–1116. doi: 10.1016/j.ijrobp.2018.01.057
Le, N. (2019). Fertility-GRU: identifying fertility-related proteins by incorporating deep-gated recurrent units and original position-specific scoring matrix profiles. J. Proteome Res. 18, 3503–3511. doi: 10.1021/acs.jproteome.9b00411
Le, N., Do, D., Chiu, F., Yapp, E., Yeh, H., and Chen, C. (2020). XGBoost improves classification of MGMT promoter methylation status in IDH1 wildtype glioblastoma. J. Pers. Med. 10:128. doi: 10.3390/jpm10030128
Leger, S., Zwanenburg, A., Pilz, K., Zschaeck, S., Zöphel, K., Kotzerke, J., et al. (2019). CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother. Oncol. 130, 10–17. doi: 10.1016/j.radonc.2018.07.020
Lei, H., Liu, S., Xie, H., Kuo, J. Y., and Lei, B. (2019). “An improved object detection method for mitosis detection,” in Proceedings of the 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, 130–133. doi: 10.1109/embc.2019.8857343
Leijenaar, R. T., Bogowicz, M., Jochems, A., Hoebers, F. J., Wesseling, F. W., Huang, S. H., et al. (2018). Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br. J. Radiol. 91:20170498. doi: 10.1259/bjr.20170498
Lewis, J. S. Jr., Ali, S., Luo, J., Thorstad, W. L., and Madabhushi, A. (2014). A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am. J. Surg. Pathol. 38, 128–137. doi: 10.1097/pas.0000000000000086
Limkin, E. J., Sun, R., Dercle, L., Zacharaki, E. I., Robert, C., Reuzé, S., et al. (2017). Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 28, 1191–1206. doi: 10.1093/annonc/mdx034
Lin, B., Deng, S., Gao, H., and Yin, J. (2020). A Multi-scale activity transition network for data translation in EEG signals decoding. IEEE/ACM Trans. Comput. Biol. Bioinform. doi: 10.1109/tcbb.2020.3024228 [Epub ahead of print].
Liu, T., Zhou, S., Yu, J., Guo, Y., Wang, Y., Zhou, J., et al. (2019). Prediction of lymph node metastasis in patients with papillary thyroid carcinoma: a radiomics method based on preoperative ultrasound images. Technol. Cancer Res. Treat. 18:1533033819831713. doi: 10.1177/1533033819831713
Lu, C., Lewis, J. S., Dupont, W. D., Plummer, W. D., Janowczyk, A., and Madabhushi, A. (2017). An oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier of nuclear morphology can risk stratify patients for disease-specific survival. Mod. Pathol. 30, 1655–1665. doi: 10.1038/modpathol.2017.98
Lv, W., Ashrafinia, S., Ma, J., Lu, L., and Rahmim, A. (2020). Multi-level multi-modality fusion radiomics: application to PET and CT imaging for prognostication of head and neck cancer. IEEE J. Biomed. Health Inform. 24, 2268–2277. doi: 10.1109/jbhi.2019.2956354
Mars, R. A. T., Yang, Y., Ward, T., Houtti, M., Priya, S., Lekatz, H. R., et al. (2020). Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 1821460–1473.e17. doi: 10.1016/j.cell.2020.08.007
Martens, R. M., Koopman, T., Noij, D. P., Pfaehler, E., Übelhör, C., Sharma, S., et al. (2020). Predictive value of quantitative (18)F-FDG-PET radiomics analysis in patients with head and neck squamous cell carcinoma. EJNMMI Res. 10:102. doi: 10.1186/s13550-020-00686-2
Martorell-Marugán, J., Tabik, S., Benhammou, Y., del Val, C., Zwir, I., Herrera, F., et al. (2019). “Deep learning in Omics data analysis and precision medicine,” in Computational Biology, ed. H. Husi (Brisbane: Codon Publications).
Matta, A., Ralhan, R., DeSouza, L. V., and Siu, K. W. (2010). Mass spectrometry-based clinical proteomics: head-and-neck cancer biomarkers and drug-targets discovery. Mass Spectrom. Rev. 29, 945–961. doi: 10.1002/mas.20296
Mes, S. W., van Velden, F. H. P., Peltenburg, B., Peeters, C. F. W., Te Beest, D. E., van de Wiel, M. A., et al. (2020). Outcome prediction of head and neck squamous cell carcinoma by MRI radiomic signatures. Eur. Radiol. 30, 6311–6321. doi: 10.1007/s00330-020-06962-y
Moore, C., Mehta, V., Ma, X., Chaudhery, S., Shi, R., Moore-Medlin, T., et al. (2016). Interobserver agreement of confocal laser endomicroscopy for detection of head and neck neoplasia. Laryngoscope 126, 632–637. doi: 10.1002/lary.25646
Mormont, R., Geurts, P., and Maree, R. (2020). Multi-task pre-training of deep neural networks for digital pathology. IEEE J. Biomed. Health Inform. doi: 10.1109/jbhi.2020.2992878 [Epub ahead of print].
Mukherjee, P., Cintra, M., Huang, C., Zhou, M., Zhu, S., Colevas, A. D., et al. (2020). CT-based radiomic signatures for predicting histopathologic features in head and neck squamous cell carcinoma. Radiol. Imaging Cancer 2:e190039. doi: 10.1148/rycan.2020190039
Ni, Y. H., Ding, L., Hu, Q. G., and Hua, Z. C. (2015). Potential biomarkers for oral squamous cell carcinoma: proteomics discovery and clinical validation. Proteomics Clin. Appl. 9, 86–97. doi: 10.1002/prca.201400091
Niel, O., and Bastard, P. (2019). Artificial intelligence in nephrology: core concepts, clinical applications, and perspectives. Am. J. Kidney Dis. 74, 803–810. doi: 10.1053/j.ajkd.2019.05.020
Niranjan, K. C., and Sarathy, N. A. (2018). Prognostic impact of tumor-stroma ratio in oral squamous cell carcinoma - A pilot study. Ann. Diagn. Pathol. 35, 56–61. doi: 10.1016/j.anndiagpath.2018.05.005
Oakden-Rayner, L. (2020). Exploring large-scale public medical image datasets. Acad. Radiol. 27, 106–112. doi: 10.1016/j.acra.2019.10.006
Onken, M. D., Winkler, A. E., Kanchi, K. L., Chalivendra, V., Law, J. H., Rickert, C. G., et al. (2014). A surprising cross-species conservation in the genomic landscape of mouse and human oral cancer identifies a transcriptional signature predicting metastatic disease. Clin. Cancer Res. 20, 2873–2884. doi: 10.1158/1078-0432.Ccr-14-0205
Pan, Y., Sun, Z., Wang, W., Yang, Z., Jia, J., Feng, X., et al. (2020). Automatic detection of squamous cell carcinoma metastasis in esophageal lymph nodes using semantic segmentation. Clin. Transl. Med. 10:e129. doi: 10.1002/ctm2.129
Parmar, C., Leijenaar, R. T., Grossmann, P., Rios Velazquez, E., Bussink, J., Rietveld, D., et al. (2015). Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci. Rep. 5:11044. doi: 10.1038/srep11044
Phillips, M., Marsden, H., Jaffe, W., Matin, R. N., Wali, G. N., Greenhalgh, J., et al. (2019). Assessment of accuracy of an artificial intelligence algorithm to detect melanoma in images of skin lesions. JAMA Netw. Open 2:e1913436. doi: 10.1001/jamanetworkopen.2019.13436
Poedjiastoeti, W., and Suebnukarn, S. (2018). Application of convolutional neural network in the diagnosis of jaw tumors. Healthc. Inform. Res. 24, 236–241. doi: 10.4258/hir.2018.24.3.236
Rattay, T., and Talbot, C. J. (2014). Finding the genetic determinants of adverse reactions to radiotherapy. Clin. Oncol. 26, 301–308. doi: 10.1016/j.clon.2014.02.001
Ren, J., Qi, M., Yuan, Y., and Tao, X. (2020). Radiomics of apparent diffusion coefficient maps to predict histologic grade in squamous cell carcinoma of the oral tongue and floor of mouth: a preliminary study. Acta Radiol. doi: 10.1177/0284185120931683 [Epub ahead of print].
Ren, J., Tian, J., Yuan, Y., Dong, D., Li, X., Shi, Y., et al. (2018). Magnetic resonance imaging based radiomics signature for the preoperative discrimination of stage I-II and III-IV head and neck squamous cell carcinoma. Eur. J. Radiol. 106, 1–6. doi: 10.1016/j.ejrad.2018.07.002
Shaban, M., Khurram, S. A., Fraz, M. M., Alsubaie, N., Masood, I., Mushtaq, S., et al. (2019). A novel digital score for abundance of tumour infiltrating lymphocytes predicts disease free survival in oral squamous cell carcinoma. Sci. Rep. 9:13341. doi: 10.1038/s41598-019-49710-z
Tan, M. S., Tan, J. W., Chang, S.-W., Yap, H. J., Abdul Kareem, S., and Zain, R. B. (2016). A genetic programming approach to oral cancer prognosis. PeerJ 4:e2482. doi: 10.7717/peerj.2482
Thong, P. S., Olivo, M., Kho, K. W., Zheng, W., Mancer, K., Harris, M., et al. (2007). Laser confocal endomicroscopy as a novel technique for fluorescence diagnostic imaging of the oral cavity. J. Biomed. Opt. 12:014007. doi: 10.1117/1.2710193
Tixier, F., Cheze-le-Rest, C., Schick, U., Simon, B., Dufour, X., Key, S., et al. (2020). Transcriptomics in cancer revealed by positron emission tomography radiomics. Sci. Rep. 10:5660. doi: 10.1038/s41598-020-62414-z
Tosado, J., Zdilar, L., Elhalawani, H., Elgohari, B., Vock, D. M., Marai, G. E., et al. (2020). Clustering of largely right-censored oropharyngeal head and neck cancer patients for discriminative groupings to improve outcome prediction. Sci. Rep. 10:3811. doi: 10.1038/s41598-020-60140-0
Tran, W. T., Suraweera, H., Quaioit, K., Cardenas, D., Leong, K. X., Karam, I., et al. (2019). Predictive quantitative ultrasound radiomic markers associated with treatment response in head and neck cancer. Future Sci OA 6:FSO433. doi: 10.2144/fsoa-2019-0048
Tseng, W. T., Chiang, W. F., Liu, S. Y., Roan, J., and Lin, C. N. (2015). The application of data mining techniques to oral cancer prognosis. J. Med. Syst. 39:59. doi: 10.1007/s10916-015-0241-3
Wang, S., Yang, D. M., Rong, R., Zhan, X., Fujimoto, J., Liu, H., et al. (2019). Artificial intelligence in lung cancer pathology image analysis. Cancers 11, 1673–1689. doi: 10.3390/cancers11111673
Werbrouck, J., De Ruyck, K., Duprez, F., Veldeman, L., Claes, K., Van Eijkeren, M., et al. (2009). Acute normal tissue reactions in head-and-neck cancer patients treated with IMRT: influence of dose and association with genetic polymorphisms in DNA DSB repair genes. Int. J. Radiat. Oncol. Biol. Phys. 73, 1187–1195. doi: 10.1016/j.ijrobp.2008.08.073
West, C., and Rosenstein, B. S. (2010). Establishment of a radiogenomics consortium. Radiother. Oncol. 94, 117–118. doi: 10.1016/j.radonc.2009.12.007
Witjes, M. J., Ilgner, J. F., Wong, B. J. F., El-Deiry, M. W., Chen, A. Y., Griffith, C. C., et al. (2018). “Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks,” in Proceedings of the Optical Imaging, Therapeutics, and Advanced Technology in Head and Neck Surgery and Otolaryngology, San Francisco, CA.
Wu, W., Ye, J., Wang, Q., Luo, J., and Xu, S. (2019). CT-based radiomics signature for the preoperative discrimination between head and neck squamous cell carcinoma grades. Front. Oncol. 9:821. doi: 10.3389/fonc.2019.00821
Xu, Y., Deng, Y., Ji, Z., Liu, H., Liu, Y., Peng, H., et al. (2014). Identification of thyroid carcinoma related genes with mRMR and shortest path approaches. PLoS One 9:e94022. doi: 10.1371/journal.pone.0094022
Yang, D. W., Wang, T. M., Zhang, J. B., Li, X. Z., He, Y. Q., Xiao, R., et al. (2020). Genome-wide association study identifies genetic susceptibility loci and pathways of radiation-induced acute oral mucositis. J. Transl. Med. 18:224.
Ye, J., Luo, J., Xu, S., and Wu, W. (2020). One-slice CT image based kernelized radiomics model for the prediction of low/mid-grade and high-grade HNSCC. Comput. Med. Imaging Graph 80:101675. doi: 10.1016/j.compmedimag.2019.101675
Ye, Y., Zhang, Z., Liu, Y., Diao, L., and Han, L. (2020). A multi-omics perspective of quantitative trait loci in precision medicine. Trends Genet. 36, 318–336. doi: 10.1016/j.tig.2020.01.009
Yu, M., Yan, H., Xia, J., Zhu, L., Zhang, T., Zhu, Z., et al. (2019). Deep convolutional neural networks for tongue squamous cell carcinoma classification using Raman spectroscopy. Photodiagnosis Photodyn. Ther. 26, 430–435. doi: 10.1016/j.pdpdt.2019.05.008
Yuan, Y., Ren, J., Shi, Y., and Tao, X. (2019). MRI-based radiomic signature as predictive marker for patients with head and neck squamous cell carcinoma. Eur. J. Radiol. 117, 193–198. doi: 10.1016/j.ejrad.2019.06.019
Zhai, T. T., Langendijk, J. A., van Dijk, L. V., van der Schaaf, A., Sommers, L., Vemer-van den Hoek, J. G. M., et al. (2020). Pre-treatment radiomic features predict individual lymph node failure for head and neck cancer patients. Radiother. Oncol. 146, 58–65. doi: 10.1016/j.radonc.2020.02.005
Zhang, Z., Xie, Y., Xing, F., McGough, M., and Yang, L. (2017). “MDNet: a semantically and visually interpretable medical image diagnosis network,” in Proceedings of the IEEE Conference on Computer Vision Pattern Recognition, Honolulu, HI, 3549–3557.
Zhao, Z., Li, Y., Wu, Y., and Chen, R. (2020). Deep learning-based model for predicting progression in patients with head and neck squamous cell carcinoma. Cancer Biomark. 27, 19–28. doi: 10.3233/cbm-190380
Zhou, Z., Chen, L., Sher, D., Zhang, Q., Shah, J., Pham, N. L., et al. (2018). Predicting lymph node metastasis in head and neck cancer by combining many-objective radiomics and 3-dimensioal convolutional neural network through evidential reasoning. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2018, 1–4. doi: 10.1109/embc.2018.8513070
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., and Liang, J. (2019). UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging. 39, 1856–1867. doi: 10.1109/tmi.2019.2959609
Zhu, Y., Mohamed, A. S. R., Lai, S. Y., Yang, S., Kanwar, A., Wei, L., et al. (2019). Imaging-genomic study of head and neck squamous cell carcinoma: associations between radiomic phenotypes and genomic mechanisms via integration of the cancer genome atlas and the cancer imaging archive. JCO Clin. Cancer Inform. 3, 1–9. doi: 10.1200/cci.18.00073
Keywords: artificial intelligence, deep learning, head and neck tumors, diagnosis, multi-omics
Citation: Wang X and Li B-b (2021) Deep Learning in Head and Neck Tumor Multiomics Diagnosis and Analysis: Review of the Literature. Front. Genet. 12:624820. doi: 10.3389/fgene.2021.624820
Received: 01 November 2020; Accepted: 07 January 2021;
Published: 10 February 2021.
Edited by:
Ying Li, Zhejiang University, ChinaReviewed by:
Yuqi Zhao, Jackson Laboratory, United StatesCopyright © 2021 Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin-bin Li, a3FsaWJpbmJpbkBiam11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.