
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 03 May 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.613197
 Dan Li1
Dan Li1 Jianhui Yang1
Jianhui Yang1 Yong Yang2
Yong Yang2 Jianxin Liu1,3
Jianxin Liu1,3 Hui Li1
Hui Li1 Rongfei Li1
Rongfei Li1 Chunya Cao1
Chunya Cao1 Liping Shi1
Liping Shi1 Weihua Wu1,3*
Weihua Wu1,3* Kai He1,3*
Kai He1,3*MicroRNAs (miRNAs) belong to a class of non-coding RNAs that suppress gene expression by complementary oligonucleotide binding to the sites in target messenger RNAs. Numerous studies have demonstrated that miRNAs play crucial role in virtually all cellular processes of both plants and animals, such as cell growth, cell division, differentiation, proliferation and apoptosis. The study of rice MIR168a has demonstrated for the first time that exogenous plant MIR168a influences cholesterol transport in mice by inhibiting low-density lipoprotein receptor adapter protein 1 expression. Inspired by this finding, the cross-kingdom regulation of plant-derived miRNAs has drawn a lot of attention because of its capability to provide novel therapeutic agents in the treatment of miRNA deregulation-related diseases. Notably, unlike mRNA, some plant miRNAs are robust because of their 3′ end modification, high G, C content, and the protection by microvesicles, miRNAs protein cofactors or plant ingredients. The stability of these small molecules guarantees the reliability of plant miRNAs in clinical application. Although the function of endogenous miRNAs has been widely investigated, the cross-kingdom regulation of plant-derived miRNAs is still in its infancy. Herein, this review summarizes the current knowledge regarding the anti-virus, anti-tumor, anti-inflammatory, anti-apoptosis, immune modulation, and intestinal function regulation effects of plant-derived miRNAs in mammals. It is expected that exploring the versatile role of plant-derived miRNAs may lay the foundation for further study and application of these newly recognized, non-toxic, and inexpensive plant active ingredients.
MicroRNAs (miRNAs) are a category of endogenous, highly conserved, 18-24 nucleotides single stranded RNAs. By binding to the 3′-untranslated region of various target genes, miRNAs play a crucial role in cell growth, cell division, differentiation, proliferation, and apoptosis (Chen et al., 2006; Guo et al., 2010). It is believed that lin-4 is the first discovered miRNA, which targets lin-14 mRNA in Caenorhabditis elegans (Lee et al., 1993). Thereafter, tens of thousands of miRNAs have been identified from mammals, plants and microorganisms.
Mechanisms of miRNA gene transcription and maturation were mainly revealed using Caenorhabditis elegans (Knight and Bass, 2001), Drosophila melanogaster (Hammond et al., 2000) and human cell lines (Lee et al., 2002). Although no homologs of Drosha and DGCR8/Pasha have been found in plants, RNase III Dicer, which is responsible for processing pre-miRNAs into mature miRNAs, is highly conserved in plants and animals (Kim, 2005). It is reported that the miRNA in plants and animals were processed in a similar manner (Reinhart et al., 2002). Normally, mature animal miRNAs drive from pri-miRNA, which is transcribed by RNA polymerase II/III from miRNA genes. Then these longer, hairpin loop-containing pri-miRNA is cleaved by microprocessor complex to generates about 70 nucleotides stem loop pre-miRNA. After transport from nucleus into the cytoplasm, pre-miRNA is further cleaved by RNAse III enzyme dicer and transactivation-response RNA-binding protein to produce mature miRNAs (Akbari Kordkheyli et al., 2019). Studies have revealed that miRNAs can regulate the translation of over 60% of protein-coding genes in animals (Esteller, 2011). Some miRNAs target specific gene, while, crucial miRNAs may mediate the expression of hundreds of genes simultaneously. The recent discovery and characterization of miRNAs not only add the rapidly growing repository of the pathogenic mechanisms of diseases, but also offer opportunities to applying these small molecules to therapy various disorders. Traditionally, the chemical compounds in herb or plant extracts were considered as the main active ingredients, and responsible for the observed pharmacological activities. Recently, people begin to notice that nutrients and bioactive compound, such as flavonoids, alkaloids and diterpenoids could regulate the crosstalk of miRNA, lncRNA and circRNA, and exert pro-apoptosis, anti-inflammation, anti-proliferation, anti-atherosclerosis and anti-infection activities (Bruna et al., 2017; Dong et al., 2019). Specifically, medicinal plants derived miRNAs are being recognized as a class of new bioactive compounds in nowadays. For instance, as one of the most valuable herb medicines in China, Panax ginseng C. A. Mey (Panax ginseng) is well known for its regulation of immune function, anti-tumor, cardiovascular protection, and cognitive improvement activities. Using Illumina HiSeq platform, Wang et al. (2019) identified 298 known and 3,500 possible novel miRNAs in Panax ginseng. Bioinformatics analysis showed that 2686 miRNAs in ginseng may regulate 50992 potential human genes, which involving 296 signaling pathways. Among 24 significantly enriched pathways, 3 were tightly associated with bladder cancer, small cell lung cancer, and cancer cell metabolism. Four significantly enriched pathways were related to signal transduction pathway, including Hedgehog, TGF-β, Hippo and neuroactive ligand receptor pathway. The results suggested Panax ginseng miRNAs are responsible, at least partially, for the clinical efficacy of Panax ginseng. Moreover, as a family of versatile molecular regulators, miRNAs from plant not only regulate host-gene expression, importantly, by dietary intake, exogenous plant miRNAs also affect the physiological and pathological conditions of mammals. Shahid et al. (2018) reported that while parasitizing Arabidopsis thaliana Heynh, Cuscuta campestris haustoria affects the growth and pathogenesis of Arabidopsis thaliana by inducing many 22 nt miRNAs, some of them could reduce mRNAs accumulation, stimulate secondary siRNA production, or promote mRNA cleavage of the host. Zhang et al. (2012) have demonstrated for the first time that through food intake, exogenous plant MIR168a can be detected in the sera and tissues of animals, and served as signaling molecules in intercellular communication. After feeding mice with total RNA extracted from fresh rice, or synthetic MIR168a, or synthetic methylated MIR168a, the levels of MIR168a in mouse serum and liver were all elevated 3 h after feeding. By binding at nucleotide sequence CCAAGCG, located on the open reading frame of low-density lipoprotein receptor adapter protein 1 (LDLRAP1), MIR168a inhibits LDLRAP1 expression, and thereby decreases LDL removal from mouse plasma. This pioneering work indicating that ingestion of plants containing high-content of functional miRNAs might be a promising way for clinical applications, because deregulation of miRNAs is involved in the pathogenesis of many human diseases such as cancers, immunodeficiency, cardiovascular diseases, and, with the new data emerging, lung diseases (Boateng and Krauss-Etschmann, 2020). More importantly, rapidly accumulating data have implicated both endogenous and dietary exogenous miRNAs regulate gene expression mainly at the post-transcriptional level via direct cleavage of transcripts (Sanchita et al., 2018; Ren et al., 2019). The high stability of plant miRNAs lays the foundation for their therapeutic application and these exogenous can survive in animals and human gastrointestinal tract (Liang et al., 2014). For instance, after drank watermelon juice, 10 miRNAs can be detected at high basal levels in human plasma among the selected 16 miRNAs (Liang et al., 2015). Unlike mRNA, plant miRNAs are robust during homogenization, mammalian circulation and even boiling processes for the following reasons (Xie et al., 2016): (1) some plant miRNAs are 2′-O-methylated on their 3′ end, this modification has a protective effect on miRNA stability. (2) The primary sequence (like high G,C content), structure of miRNA and absence of RNases digestion motifs also contribute to the stability of miRNAs (Yang et al., 2018). (3) the miRNAs protein cofactors like high-density lipoproteins (Vickers et al., 2014), and Argonaute proteins prevent them from decay. (4) herb ingredients of plant metabolites not only create a favorable environment for miRNAs but also exert RNase inhibitory activities (Xie and Melzig, 2018; Vora et al., 2019). (5) During transportation, plant miRNAs are packaged into microvesicles or exosomes, which protect them against degradation. Moreover, the plant miRNA-like miRNAs, mimicked from mammalian tumor suppressor miRNAs by methylation at 2′position of the ribose of the 3′terminal nucleotide, effectively reduced tumor burden in ApcMin/ + mice of colon cancer model (Mlotshwa et al., 2015). This observation has led to the idea that apart from plant derived therapeutic miRNAs, functional miRNAs could be produced by edible plants using bioengineering to meet the increasing demand of therapeutic miRNAs in future.
In 2018, Wang et al. (2018) have reviewed several studies concerning plant miRNAs regulate gene expression in animals (mainly honey bee), and the effect of hosts miRNAs on pathogens. Zhao et al. (2018) have highlighted some discoveries of both intra- and inter-kingdom communication of plant and animal miRNAs. Because current study for plant miRNAs is advancing rapidly, it is necessary to provide timely updates. This paper summarizes the plant-derived miRNAs with cross-kingdom activity in mammals (Table 1), and reviews the updated evidences regarding potential therapeutic benefit of miRNA cross-kingdom activity, like anti-virus, anti-cancer, anti-inflammation, and immune modulation, etc. (Figure 1). The current review may facilitate the recognition of a more profound understanding of the therapeutic value of plant miRNAs, and the application of these new natural resources.
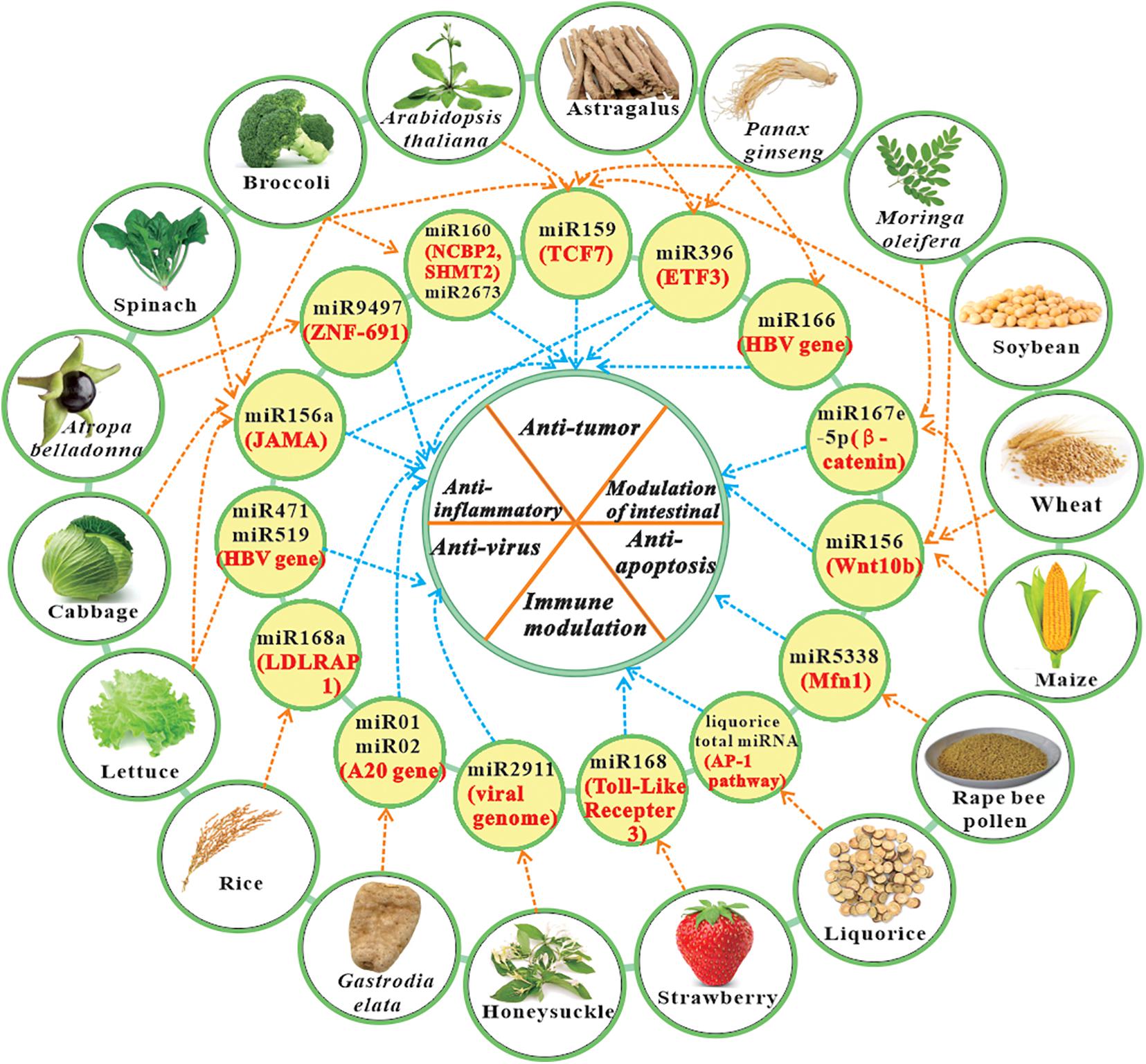
Figure 1. The representative plant-derived miRNAs with cross-kingdom bioactivity and their role in cross-kingdom communications. Note: The information in yellow circle contains miRNAs derived from specific plants (black text) and their main targets (red text). Some conservative miRNA family can be found in many different plant species. Other information like miRNA sequence and references can be found in Table 1. ETF3, Endothelial transcription factor 3; HBV, Hepatitis B virus; JAMA, Junctional adhesion molecule A; LDLRAP 1, Low-density lipoprotein receptor adapter protein 1; Mfn1, Mitochondrial fusion requires fusion protein 1; TCF7, Transcription factor 7; ZNF-691, zinc-finger transcription factor 691.
The relationship between miRNAs and viral replication is complex and not completely understood. Some miRNAs could target viral genomes and proteins to suppress virus replication (Wen et al., 2013). While, other miRNAs, like gga-miR-142-5p (Ouyang et al., 2018), miRNA-16-5p (Duan et al., 2020), may inhibit host defense and enhance virus replication and infection (Li and Zheng, 2020). There is evidence that miRNAs exert antiviral effects on virus infection by modulation of certain immune and antiviral pathways, such as the janus kinase-signal transducer and activator of transcription (JAK-STAT), toll-like receptor, and mitogen-activated protein kinase signaling pathways (Lin et al., 2016; Fu et al., 2018).
Facing the novel Coronavirus SARS-CoV-2 pandemic, Chauhan et al. (2020) have indicated that using miRNAs that can binding to spike protein or the genome of SARS-CoV 2 is a promising way to fight back against this viral disease. Lonicera japonica Thunb (honeysuckle, HS)-encoded atypical miRNA2911 is considered as the first active plant miRNA identified in Traditional Chinese Medicine (TCM), which inhibiting the replication of influenza A viruses, including H1N1, H5N1 and H7N9. Three days gavage of HS decoction significantly elevated the MIR2911 level in mouse peripheral blood and lung. Besides, synthetic MIR2911 gavage administration reduced the viral infection-induced weight loss and even mortality in mouse. The antiviral activity of MIR2911 may attributed to its ability to bind to the nucleotide sequences of H1N1 virus-encoded PB2 and NS1 genes (Zhou et al., 2015). The authors further demonstrated that MIR2911 in honeysuckle decoction (HD) had 28 binding sites in the SARSCoV-2 genome and was able to inhibit virus replication and accelerate the negative conversion of infected patients. In vitro, exosomal-MIR2911 inhibited 93% of viral replication. Patients who intake 30 g dried honeysuckle (MIR 2911 level: 10.5 pmol) per day for 7 days exhibited a dramatically higher negative conversion rate compared with TCM mixture control group. Besides, HD-MIR2911 also reduced the time taken to become SARS-CoV-2 PCR-negative in both male and female patients (Zhou et al., 2020). Zhang et al. (2019) generated two plant-derived small silencing RNAs (amiR471 and amiR519) in edible Lactuca sativa L. var. ramosa Hort (lettuce) and found that these miRNAs could inhibit HBsAg expression. Using HBsAg–/+ transgenic mice, the authors further revealed that amiR519 lettuce decoction decreased both the fat droplets and immune-associated factors, such as TNF-α, IL-6, CCL5 and IFN-γ in mice liver. Long-term feeding with amiRNA-containing decoction showed no toxicological effects but alleviated liver injury in HBsAg–/+ transgenic mice caused by persistent HBsAg expression. Using bioinformatics approach, Perez-Quintero et al. (2010) have shown that plant miRNAs, including miRNA families of 156, 395, 159, 166, and 160 can target the coding regions of RNA polymerases, cylindrical inclusion proteins, capsid proteins, and nuclear inclusion body proteins in plant-infecting viruses. Due to a higher matching percentage for plants miRNAs to plant viruses than animal and other source of miRNAs, plant miRNAs target plant-infecting viruses more efficiently. Similarly, the following study on screening conserved plant miRNAs against the hepatitis B and C viruses showed that five plant conserved miRNAs: miR-166, 169, 172, 390 and 399 were quite likely to bind to the six HBV gene sequences. While, miR-156/157, 166, 169, 172 and 390 were predicted to target the six HCV gene sequences (Barozai and Din, 2017).
The anti-tumor effects of plant-derived miRNAs were mainly achieved by down-regulating the critical targets or pathways in tumorigenesis, and exerting tumor-suppressor function (like miRNA34). By sequence analysis, four common miRNAs in edible plants were found to be perfect match with 22 human transcripts. These plant miRNAs targeted genes are involved in muscle contraction and tumor suppression, indicating that food miRNAs may altering our body functions by regulating human genome (Kashani et al., 2019). Recently, microvesicles purified from Moringa oleifera seeds increased apoptosis levels of Jurkat and Hela cells. BCL2 protein expression and mitochondrial membrane potential of the two cell lines were also significantly downregulated after treatment (Potestà et al., 2020). The extracts of Camptotheca acuminata Decne (C. acuminate) have been used for thousands of years in China and Indian for the treatment of various cancers. Thirty-three stable novel miRNAs in C. acuminate were found to regulate 152 target genes in human. These predicted target genes are significantly associated with vital cell functions like focal adhesion, lipolysis in adipocytes and mTOR pathways. Thus, miRNAs in C. acuminate have great potential in modulation of cancer pathways (Kumar et al., 2017). Pirrò et al. (Stefano et al., 2016) have identified 94 conserved and 2 novel miRNAs in Moringa oleifera. Bioinformatics prediction have revealed that these miRNAs may regulate apoptosis, cell cycle and protein degradation related human genes. Additionally, transfection of Moringa oleifera mol-mir168a to HepG2 cells significantly decreased SIRT1 protein level, suggesting a key role of mol-mir168a in modulation of cell cycle, apoptosis, and inflammation in human. By profiling small RNAs in the sera of 42 breast cancer patients, Chin et al. (2016) found that the serum level of plant miR159 (present in Arabidopsis thaliana Heynh, Glycine max Merr and Brassica oleracea L. var. capitata) is inversely correlated with breast cancer incidence and progression. Oral intake of miR159 at the concentration of 25 mg/kg for 16 days dramatically reduced tumor growth in tumor xenograft models. In addition, the expression of transcription factor 7 (TCF7) and potent oncogene MYC were significantly down-regulated. Mechanism study revealed that synthetic miR159 had the potential to inhibit the growth of breast cancer cells by aiming the region of nucleotide 1445-1470 and 1842-1870 in 3′UTR of TCF7, then resulting in the decrease of MYC protein levels. Similarly, Glycine max Merr (soybean)-derived Gma-miR159a, accounts for 83.3% soybean miRNA families, was also reported to promote the apoptosis of Caco-2 cells by inhibiting TCF7 expression (Liu et al., 2020). A controlled intervention study involving 19 volunteers showed that two weeks administration of 80 g Brassica oleracea L (broccoli) significantly increased the serum level of miR160, miR2673 in a dose and time-dependent manner. Transfection of miR160 mimic significantly downregulated CDC6, POL2RF, PYCR1, MTHFD2, NCBP2, SHMT2; CELSR3 and FKBP4 expression in cancer cell lines. Among them, NCBP2 and SHMT2 were significantly reduced at mRNA and protein level. Thus, except for the chemical compounds, Brassica miRNAs may also attribute to its cancer prevention function (Pastrello et al., 2016). Junctional adhesion molecule A (JAMA) is a transmembrane protein and plays crucial role in cell invasion, platelet aggregation and lymphocyte adhesion. Previous research has identified 84 conserved miRNAs and 184 novel miRNAs from broccoli, and noticed that miR156a is the most highly expressed miRNA in flower and leaf (Tian, 2014). By binding to the JAMA mRNA 3′-UTR region, miR156a inhibited JAMA protein expression and result to down-regulation of Vimentin, up-regulation of E-cadherin in human nasopharyngeal carcinoma cell lines CNE2 and HONE1. These effects were also accompanied by a remarkable reduction of invasion ability of CNE2 and HONE1 cell. MiRNA171 is a ubiquitous and conserved plant miRNA. Recently, it is shown that miRNA171 significantly decreased G protein subunit alpha 12 mRNA and protein levels of human embryonic kidney 293 cells, and then modulated mTOR pathway, one of downstream signaling factors of GNA12. Thus, miRNA171 may be of great value in the treatment of GNA12 over-expressed carcinomas (Gismondi et al., 2021).
The anti-tumor effect of human miRNA34 families is different from the above-mentioned miRNAs. Human miRNA34 families are well known as tumor-suppressor miRNAs, play an important role in apoptotic cellular mechanisms by repressing BCL2 mRNA (Dang et al., 2013). A recent study showed that Olea europaea Linn miRNAs, oeu-sR20, oeu-sR27 and oeu-sR34 are homologous to human miRNA34 in cross-kingdom regulation of tumorigenesis. Like hsa-miR34a, transfect Olea europaea miRNAs mimics to THP1 and Jurkat cells affects cell viability and apoptosis via significantly decrease SIRT1 and BCL2 proteins expression (Minutolo et al., 2018). These results indicate Olea europaea miRNAs have great potential to be used as a novel, natural non-toxic, anticancer drug.
MicroRNAs have emerged as an important regulator in the mammalian immune system, as they participate in various critical events, such as development, homeostasis, and regulation of multiple pathways. For instance, Glycyrrhiza uralensis Fisch (Liquorice) miRNA induced obvious proliferation and aggregation of human peripheral blood mononuclear cells (PBMC). Moreover, the proportion of HLA-DR+ cells was significantly increased from 16.7% to 68.1%, which indicating that liquorice miRNAs improve immune function by stimulate PBMC growth (Shao et al., 2015). Later study further demonstrated that immune modulation activity of glycyrrhiza miRNA was related to its effect on the regulation of genes involved in T cell differentiation, inflammation and apoptosis. Transfection of glycyrrhiza miRNA to PBMC inhibited Th2 differentiation by inhibiting AP-1 signaling pathway. Besides, the down-regulated NF-κB, Bax and p53 expression proves its anti-inflammation and anti-apoptosis activity (Xiang et al., 2017). Cavalieri et al. (2016) have suggested that dietary plant miRNAs may reduce chronic diseases by regulating inflammation response. FvmiR168 interacts with dendritic cells (DCs) by binding to the ectodomain of TLR3, leading to the reduction of cytokines release and inhibition of T cell proliferation. Injection of “cocktail” plant sRNA to experimental autoimmune encephalomyelitis (EAE) mice significantly reduced disease onset and maximum EAE score. Besides, plant sRNA-treated mice had less CD11c infiltrates in spinal cord areas. These were attributed to the reduction of serious inflammatory cytokines by plant sRNA via down-regulating the expression of master regulators of pro-inflammatory including Tbet, Rorc and IL-10 master regulator FoxP3. JAK-STAT pathway regulates cell growth, proliferation, differentiation, and apoptosis, and thus critical to immune and inflammatory responses (Jin et al., 2018). By analysis of 410 human plasma small RNA sequencing data, Liu et al. (2017) have found that high relative amount of plant miRNA peu-MIR2910, which conserved in maize, melon, sorghum, Solanum lycopersicum Mill (tomato), and tea, can be detected in the plasma samples. Moreover, two seed sequences of miR2910 are the same as human miR-4715-5p and miR-4259. The authors further explored that miR2910 may target on 5’UTR of LIMA1, and the CDS of a group of genes like SPRY4, CTNND1, LBX1, STK38, etc. Since SPRY4 is involved in JAK-STAT signaling pathway, it is possible that plant miR2910 can be applied for the treatment of various diseases resulted from deregulation of the JAK-STAT pathway.
MicroRNAs exert significant impact on inflammatory process by targeting proteins that response for transmit of intracellular signals, and by reducing mRNAs that translate cytokines. Sharma et al. (2017) have identified thirty-four mature Curcuma longa L. Among them, clo-mir-14 showed sequence homology with human miRNA-4693-5p. A total of twenty human genes such as regulator of G-protein signaling 13, adenylate cyclase 9, gap junction protein A1, calcitonin receptor, etc. were predicted as target of clo-mir-14, indicating that clo-mir-14 may evoke the cellular immune response during inflammation of Rheumatoid Arthritis. By intranasal delivery, Astragalus membranaceus Moench (Astragalus)-derived miR-396 showed about ten times higher expression in the spleen, peripheral blood and lung tissue of asthmatic mice compared with internal reference gene. After seven times consecutive treatment with 20 μg miR-396 mimics, the levels of IL-5, IL-13 cytokines were significantly reduced compared to asthmatic group, moreover, the expression of the master transcription factor GATA-3 of Th2 cells was also down-regulated by miR-396 treatment (Shen et al., 2019). As a precious Chinese herbal medicine, Gastrodia elata Bl (GE) possesses anti-convulsant, neuroprotection, anti-inflammatory and immune regulation effects. By Illumina high-throughput sequencing, 5718 miRNAs were discovered from GE, of which 52 were new GE miRNAs. Function prediction showed that Gas-mi R01 and Gas-mi R02 might interfere with three key genes: A20, TNF-α and IκBα. Transfect Gas-mi R01 and Gas-mi R02 mimics to 293T cell significantly inhibited the expression of A20, which is an important feedback regulator of NF-kB-driven gene. Additionally, after gavage GE decoction to mice, high levels of Gas-mi R01 and Gas-mi R02 could be detected in mice serum, liver, brain, kidney, spleen, and reduced A20 expression (Zhou, 2018; Xia et al., 2020). These results suggested that GE miRNAs are a group of active ingredients in GE and may responsible for the well know anti-inflammatory and immune regulation efficiency of Gastrodia elata. Except for the cholesterol metabolism modulation role of MIR168a (Zhang et al., 2012), MIR156a was also reported to exert beneficial effects on cardiovascular disease (CVD). In a controlled intervention study, after eating 500 g lettuce, miR156a was stably presents in human serum and reached its peak value one to three hours after ingestion. Moreover, CVD patients have exhibited a decreased MIR156a level in serum and blood vessel. Plant MIR156a combines with 3′-UTR of JAMA mRNA to suppress the junction JAMA expression and mediate the cleavage of JAMA mRNA, thus inhibiting inflammatory-induced monocytes adhesion in CVD patients (Hou et al., 2018). Aba miRNA 9497 derived from Atropa belladonna L, has a highly homologous sequence to miRNA-378 of Homo sapiens. Avsar et al. (2020) have demonstrated for the first time that both aba miRNA 9497 and miRNA-378 could downregulate the mRNA expression of human CNS-enriched ZNF-691 by binding to 3′-UTR of this gene. The authors speculated that the neurotoxic actions of Atropa belladonna may partially results from the modulation role of ZNF-691 by aba-miRNA-9497. Since ZNF-691 has been involved in many neuropathological roles such as inflammation, targeting ZNF-691 by aba miRNA 9497 should be of great interest as the crucial role of inflammation in various diseases.
Apoptosis is a cell death process occurring when cells face irreversible stress, and its deregulation in humans will result in various proliferative and degenerative diseases. Normally, proteins of anti-apoptotic family members (BCL2, MCL1) are binding to Beclin 1, an important autophagy-promoting member, to maintain cellular homeostasis. Cell stress can cause the disassociation of these two kinds of proteins, thus promoting autophagy and inhibiting apoptosis, respectively. Many miRNAs regulate autophagy and apoptosis signals by modulating the above-mentioned protein-protein interactions (Xu et al., 2012). Using testosterone propionate induced benign prostatic hyperplasia (BPH) rat model, Chen et al. (2018) have shown that miRNA derived from the pollen of Brassica napus L (rape bee) could cross the blood-prostate barrier and entry prostate. After gavage fed rape bee pollen to BPH rats at the concentration of 6.39 g/kg for 3 weeks, the abundance of seven plant miRNAs such as miR894, miR5338, miR3440-5p, miR2878-5p, miR7754-5p, miR5015, and iR7731-3p were elevated in the prostate gland of rats, accompanied by a decreased posterior lobes index compared with the model group. Further study has revealed that the most enriched miRNA in rat’s prostate by rape bee pollen treatment is miR5338, which prevents BPH by decrease Mfn1 expression and inhibit mitochondrial fusion, cell apoptosis via Bcl-2 pathway.
The intestine is not only essential for digestion and absorption, but also crucial for secretion and immunity, as well as mediating various bile acids or microbiota involved signaling pathways. Recent studies have revealed that plant MIR167e-5p elicits cross-kingdom effects on the proliferation of enterocyte. MIR167e-5p, a conserved and highly expressed miRNA in Moringa oleifera Lam and Zea mays L (maize), significantly reduced IPEC-J2 and Caco-2 cell proliferation in a dose-and time-dependent manner. This event was achieved by downregulating the mRNA and protein levels of β-catenin, which is a crucial target in intestinal epithelium development (Li et al., 2019a). The same group further demonstrated that MIR156, a common plant miRNA can be found in soybean, Triticum aestivum L (wheat) and maize, regulates intestinal development and health in vitro and in vivo. MIR156 may bind to the 3′-UTR sequence of Wnt10b, then suppresses the proliferation of IPEC-J2 cells. In mice study, MIR156-containing maize diet administration significantly reduced the villus height:crypt depth ratio in mice duodenum, accompanied with down-regulated expression of Wnt10b and β-catenin. Additionally, 7 days treatment of 300 pmol synthetic MIR156 significantly decreased the mRNA levels of Cdx2, Wnt10b, β-catenin and cyclin D1 in mice intestine (Li et al., 2019b). These studies provide evidences that plant miRNA156 and miRNA167e-5p could regulate Wnt/β-catenin pathway to maintain intestinal epithelium homeostasis and prevent colitis.
The discovery of miRNAs and plant miRNA cross-kingdom regulation in Homo sapiens has generated considerable excitement. According to the latest information in miRNA database 22, there are 38589 mature miRNAs from nearly 300 species (miRBase 22 release, October, 2018). Plant miRNAs are crucial for many aspects of plant biology, such as leaf morphogenesis, flowering time, stress response and signal transduction. Zhang H. et al. (2016) have investigated the target genes of 25 miRNAs in Arabidopsis thaliana in both human and Arabidopsis. The results showed that Arabidopsis miRNAs may exert similar functions such as stress response and ion transport in both human and plant, suggesting a key role of exogenous plant miRNA on physiological function of human body. According to the sequence similarity, miRNAs can be grouped into different families. In Solanum tuberosum L (potato), 982 miRNAs have been classified in 71 miRNA families, and as the largest family, miR1128 family have 141 members (Mian, 2014). The specific miRNA can modulate multiple gene targets, for instance, miR168-5p, one variants of the plant miRNA168a, is wide distributed in 28 plants species of 15 plants families and has the potential to regulate 123 genes of human transcriptome (Javed et al., 2017).
Plant miRNAs seem to be a promising alternative for further preclinical use, due to their diverse expression patterns and the ability to regulate many development and physiology process. Besides, widespread deregulation of miRNA expression can be found in many human diseases (Lina et al., 2020; Mansoor et al., 2020). Strategies to target miRNAs in human disease involving two aspects, one is to inhibit miRNA function use antisense oligonucleotides or “miRNA sponges.” The other is restoring miRNA function by epigenetic drugs, miRNA virus delivery system or ingestion plants/herb medicine that rich in specific miRNA (Esteller, 2011). Thus, using abundant, easy to obtain exogenous miRNA would be a promising choice and new alternative way for human diseases treatment. Plant derived nanoparticles which mainly contains miRNA, lipids, and proteins have been demonstrated to function as natural therapeutical agents for cancer, colitis, and inflammatory diseases (Zhang M. et al., 2016). Lukasik et al. (2018) reported that five plant food-derived miRNAs, including miR156a, miR166a, miR157a, miR168a, and miR172a can be found in breast milk samples of healthy volunteers. The bioinformatics results indicated that these plant miRNAs may interact with targets involving protein transport, immune processes, and signal transduction, etc. in faints. As for the regulating mechanisms, accumulating evidences demonstrated that plant derived miRNAs exert their activities both directly and indirectly. For instance, in plants and animals, miRNAs regulate gene expression by recognizing a complementary sequence, which is located in 3′UTR, ORF (Zhang et al., 2018), or 5′UTR of the targeted mRNA, with the assistance of RNA-induced silencing complex. This combination generally results in translation suppression or mRNA decapping and 5′-to-3′ decay. Besides, most plants and few animal miRNAs are highly complementary to their target mRNA, leading to endonucleolytic cleavage of their mRNA targets (Ameres and Zamore, 2013). Intriguingly, a recent study has showed that plant-derived exosome miRNAs interact with gut microbiota and indirectly maintain gut health. MiR7267-3p binds to the “TGGCTGGA” region of ycnE mRNA which encoding Lactobacillus rhamnosus monooxygenase, and resulted to increased indole-3-carboxaldehyde level. Subsequently, aryl hydrocarbon receptor-mediated pathway was activated, leading to the expression of IL-22 and evoking antimicrobial immunity and tissue repair responses in mice (Teng et al., 2018).
Among plant-derived miRNAs, medicinal herb derived-miRNAs have drawn a lot of attention because these active ingredients can be absorbed through diets, and most people in developing countries are still rely upon the herb medicine for the treatment of diseases. A large-scale analysis has discovered thousands of small RNAs derived from 10 TCM herbs and most of these miRNAs can be delivered to human blood cells and tissues after oral administration of herbal decoctions (Huang et al., 2019). One question is to what extent herb miRNAs contribute to the known activities of individual herb? Compared with miRNA alone, administration miRNA together with plant extracts showed higher stability and delayed kinetics curve (Zhang et al., 2019). Notably, studies have indicated that therapeutic effects of herbal medicines may attributed to both the chemical compounds and miRNAs in herb (Hamed et al., 2018). For instance, Pastrello et al. (2016) compared the anticancer effects of the main compounds and miRNA in broccoli. It turned out that among the tested 11 genes, 6 were down-regulated solely by miR160, one down-regulated by both miR160 and sulforaphane, and one by miR160 and the active compound in broccoli (3,3-diindolylmethane). Therefore, the interaction between chemical compounds and miRNAs in plant should be considered in future clinical application.
Since the study of cross-kingdom regulation of exogenous plant miRNAs is currently at an early stage, controversy exists regarding the detection and effectiveness of consumed plant miRNA, as well as the repeatability of the published results. In some studies, plant miRNAs were not detected or the levels of target miRNAs were no higher than the background (Mico et al., 2016). Generally, the basis for clinical use of exogenous miRNAs is their exceptional stability toward harsh gastrointestinal environment. Due to the various sequence, structure and binding lipoproteins, miRNAs exert different stabilities, which may fundamentally influence their in vivo activity (Zhang et al., 2019). For example, unlike other plant miRNAs, MIR2911 from honeysuckle decoction is resistant to RNase treatment or even boiling process, because of its unique sequence and high GC content (Zhou et al., 2015). Intriguingly, sequence variance not only affects the stability of plant miRNA, but also determines its bioactivity. Recently, study of silencing efficacy of plant variants of miR168a on human LDLRAP1 has demonstrated that although predicted energy scores and miRNA-mRNA duplex structures are similar among rice, tomato and apple miR168a variants, a single nucleotide difference at position 14 lead to the formation of G:U wobble in tomato and apple miRNA-mRNA duplex, results to the significantly decreased LDLRAP1 expression-regulation activity of tomato and apple miR168a variants (Lang et al., 2019). Tripathi et al. (2018) have also found that miRNAs from two tomato varieties showed completely different response to tomato leaf curl virus infections. This reminds us that to ensure the repeatability of previously reported work, plant materials and experimental conditions should be strictly followed. Moreover, it is demonstrated that the concentration of miRNA should higher than 100 copies per cell to effectively suppress the target-containing transcripts (Brown et al., 2006). Therefore, ingestion of insufficient concentration of miRNA, poor bioavailability (Huang et al., 2018), and degradation could all lead to the reduced effectiveness of plant-derived miRNAs. Lastly, improper handling, incorrect selection of endogenous references and contamination of dietary miRNAs may also lead to false results.
Taken together, further efforts should be devoted to two aspects, one is to discovery more novel plant miRNAs and validate their function in human disease treatment, because many bioinformatic predicted miRNA targets were invalid when it comes to in vivo. The other is to elucidate the mechanisms and any potential risk of these new active therapeutic substances. The versatile cross-kingdom regulation of plant-derived miRNAs summarized in present study, may lay the foundation for further investigation and application of these newly recognized, non-toxic, and inexpensive plant active ingredients.
DL, JY, and YY did the investigation, resources, and data curation. DL, JL, and HL did the writing and draft preparation. RL, CC, and KH revised and edited. LS did the visualization. WW and KH did the supervision and discussion. All authors have read and agreed to the published version of the manuscript.
This work was financially supported by Natural Science Foundation of Hunan Province (2019JJ50419), Scientific Research Fund of Hunan Provincial Education Department (18A490, 18A488), Young backbone teachers in Hunan Province (2020-379), Doctoral research Fund of Hunan university of medicine (2018-3), and Training program for young scholars of Hunan university of medicine (2019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
BPH, Benign prostatic hyperplasia; DCs, Dendritic cells; EAE, Experimental autoimmune encephalomyelitis; GE, Gastrodia elata; HD, Honeysuckle decoction; JAK-STAT, Janus kinase-signal transducer and activator of transcription; JAMA, Junctional adhesion molecule A; LDLRAP1, Low-density lipoprotein receptor adapter protein 1; miRNAs, MicroRNAs; PBMC, Peripheral blood mononuclear cells; TCF7, Transcription factor 7; TCM, Traditional Chinese Medicine.
Abla, M., Sun, H., Li, Z., Wei, C., Gao, F., Zhou, Y., et al. (2019). Identification of miRNAs and Their Response to Cold Stress in Astragalus Membranaceus. Biomol. 9, 1–20. doi: 10.3390/biom9050182
Akbari Kordkheyli, V., Khonakdar Tarsi, A., Mishan, M. A., Tafazoli, A., Bardania, H., Zarpou, S., et al. (2019). Effects of quercetin on microRNAs: a mechanistic review. J. Cell Biochem. 120, 12141–12155. doi: 10.1002/jcb.28663
Ameres, S. L., and Zamore, P. D. (2013). Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488. doi: 10.1038/nrm3611
Avsar, B., Zhao, Y., Li, W., and Lukiw, W. J. (2020). Atropa belladonna Expresses a microRNA (aba-miRNA-9497) Highly Homologous to Homo sapiens miRNA-378 (hsa-miRNA-378); both miRNAs target the 3’-Untranslated Region (3’-UTR) of the mRNA Encoding the Neurologically Relevant, Zinc-Finger Transcription Factor ZNF-691. Cell Mol. Neurobiol. 40, 179–188. doi: 10.1007/s10571-019-00729-w
Barozai, M. Y. K., and Din, M. (2017). “Initial screening of plant most conserved MicroRNAs targeting infectious viruses: HBV and HCV,” in 14th International Bhurban Conference on Applied Sciences and Technology (IBCAST) (New York: IEEE), doi: 10.1109/IBCAST.2017.7868055
Boateng, E., and Krauss-Etschmann, S. (2020). miRNAs in Lung Development and Diseases. Int. J. Mol. Sci. 21:2765. doi: 10.3390/ijms21082765
Brown, B. D., Venneri, M. A., Zingale, A., Sergi Sergi, L., and Naldini, L. (2006). Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 12, 585–591. doi: 10.1038/nm1398
Bruna, Q., Bruna, R., Graziela, D., Silvia, C., and Marcelo, R. (2017). Nutrimiromics: role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 9:1168. doi: 10.3390/nu9111168
Cavalieri, D., Rizzetto, L., Tocci, N., Rivero, D., Asquini, E., Si-Ammour, A., et al. (2016). Plant microRNAs as novel immunomodulatory agents. Sci. Rep. 6:25761. doi: 10.1038/srep25761
Chauhan, N., Jaggi, M., Chauhan, S. C., and Yallapu, M. M. (2020). COVID-19: fighting the invisible enemy with microRNAs. Expert Rev. Anti Infect. Ther. 19, 137–145. doi: 10.1080/14787210.2020.1812385
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Chen, X., Wu, R. Z., Zhu, Y. Q., Ren, Z. M., Tong, Y. L., Yang, F., et al. (2018). Study on the inhibition of Mfn1 by plant-derived miR5338 mediating the treatment of BPH with rape bee pollen. BMC Complem. Altern. M. 18:38. doi: 10.1186/s12906-018-2107-y
Chin, A. R., Fong, M. Y., Somlo, G., Wu, J., Swiderski, P., Wu, X., et al. (2016). Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 26, 217–228. doi: 10.1038/cr.2016.13
Dang, Y., Luo, D., Rong, M., and Chen, G. (2013). Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One 8:e61054. doi: 10.1371/journal.pone.0061054
Dong, Y., Chen, H., Gao, J., Liu, Y., and Wang, J. (2019). Bioactive Ingredients in Chinese Herbal Medicines that Target Non-coding RNAs: promising New Choices for Disease Treatment. Front. Pharmacol. 10:515. doi: 10.3389/fphar.2019.00515
Duan, X., Zhao, M., Wang, Y., Li, X., Cao, H., and Zheng, S. J. (2020). Epigenetic Upregulation of Chicken MicroRNA-16-5p Expression in DF-1 Cells following Infection with Infectious Bursal Disease Virus (IBDV) Enhances IBDV-Induced Apoptosis and Viral Replication. J. Virol. 94, 1–50. doi: 10.1128/JVI.01724-19
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi: 10.1038/nrg3074
Fu, M., Wang, B., Chen, X., He, Z., Wang, Y., Li, X., et al. (2018). MicroRNA gga-miR-130b Suppresses Infectious Bursal Disease Virus Replication via Targeting of the Viral Genome and Cellular Suppressors of Cytokine Signaling 5. J. Virol. 92, e1646–e1617. doi: 10.1128/JVI.01646-17
Gismondi, A., Nanni, V., Monteleone, V., Colao, C., Di Marco, G., and Canini, A. (2021). Plant miR171 modulates mTOR pathway in HEK293 cells by targeting GNA12. Mol. Biol. Rep. 48, 435–449. doi: 10.1007/s11033-020-06070-6
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Hamed, M., Aria, M., Amirhossein, S., Naser, Z., Javid, S. N., Bahman, R., et al. (2018). MicroRNA: a novel target of curcumin in cancer therapy. J. Cell Physiol. 233, 1–13. doi: 10.1002/jcp.26055
Hammond, S. M., Bernstein, E., Beach, D., and Hannon, G. J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. doi: 10.1038/35005107
Hou, D., He, F., Ma, L., Cao, M., Zhou, Z., Wei, Z., et al. (2018). The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J. Nutr. Biochem. 57, 197–205. doi: 10.1016/j.jnutbio.2018.03.026
Huang, F., Du, J., Liang, Z., Xu, Z., Xu, J., Zhao, Y., et al. (2019). Large-scale analysis of small RNAs derived from traditional Chinese herbs in human tissues. Sci. China Life Sci. 62, 321–332. doi: 10.1007/s11427-018-9323-5
Huang, H., Davis, C. D., and Wang, T. T. Y. (2018). Extensive Degradation and Low Bioavailability of Orally Consumed Corn miRNAs in Mice. Nutrients 10, 1–13. doi: 10.3390/nu10020215
Javed, M., Solanki, M., Sinha, A., and Shukla, L. I. (2017). Position Based Nucleotide Analysis of miR168 Family in Higher Plants and its Targets in Mammalian Transcripts. Microrna 2, 136–142.
Jin, W. N., Ducruet, A. F., Liu, Q., Shi, S. X., Waters, M., Zou, M., et al. (2018). Activation of JAK/STAT3 restores NK-cell function and improves immune defense after brain ischemia. FASEB J. 32, 2757–2767. doi: 10.1096/fj.201700962R
Kashani, B., Hasani Bidgoli, M., Motahari, S. A., Sedaghat, N., and Modarressi, M. H. (2019). You are what you eat: sequence analysis reveals how plant microRNAs may regulate the human genome. Comput. Biol. Med. 106, 106–113. doi: 10.1016/j.compbiomed.2019.01.020
Kim, V. N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Bio. 6, 376–385. doi: 10.1038/nrm1644
Knight, S. W., and Bass, B. L. (2001). A Role for the RNase III Enzyme DCR-1 in RNA Interference and Germ Line Development in Caenorhabditis elegans. Science 293, 2269–2271. doi: 10.1126/science.1062039
Kumar, D., Kumar, S., Ayachit, G., Bhairappanavar, S. B., Ansari, A., Sharma, P., et al. (2017). Cross-Kingdom Regulation of Putative miRNAs Derived from Happy Tree in Cancer Pathway: a Systems Biology Approach. Int. J. Mol. Sci. 18:1191. doi: 10.3390/ijms18061191
Lang, C., Karunairetnam, S., Lo, K. R., Kralicek, A. V., Crowhurst, R. N., Gleave, A. P., et al. (2019). Common Variants of the Plant microRNA-168a Exhibit Differing Silencing Efficacy for Human Low-Density Lipoprotein Receptor Adaptor Protein 1 (LDLRAP1). Microrna 8, 166–170. doi: 10.2174/2211536608666181203103233
Lee, R. C., Feinbaum, R. L., and Ambros, V. R. (1993). The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to lin-14. Cell 75, 861–874. doi: 10.1016/0092-8674(93)90529-Y
Lee, Y., Jeon, K., Lee, J. T., Kim, S., and Kim, V. N. (2002). MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21, 4663–4670. doi: 10.1093/emboj/cdf476
Li, J., and Zheng, S. J. (2020). Role of MicroRNAs in Host Defense against Infectious Bursal Disease Virus (IBDV) Infection: a Hidden Front Line. Viruses 12:543. doi: 10.3390/v12050543
Li, M., Chen, T., He, J. J., Wu, J. H., Luo, J. Y., Ye, R. S., et al. (2019a). Plant MIR167e-5p Inhibits Enterocyte Proliferation by Targeting β-Catenin. Cells 8, 1385–1391. doi: 10.3390/cells8111385
Li, M., Chen, T., Wang, R., Luo, J. Y., He, J. J., Ye, R. S., et al. (2019b). Plant MIR156 regulates intestinal growth in mammals by targeting the Wnt/β-catenin pathway. Am. J. Physiol-Cell Ph. 317, 434–448. doi: 10.1152/ajpcell.00030.2019
Liang, G. F., Zhu, Y. L., Sun, B., Shao, Y. H., Jing, A. H., Wang, J. H., et al. (2014). Assessing the survival of exogenous plant microRNA in mice. Food Sci. Nutr. 2, 380–388. doi: 10.1002/fsn3.113
Liang, H., Zhang, S., Fu, Z., Wang, Y., Wang, N., Liu, Y., et al. (2015). Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J. Nutr. Biochem. 26, 505–512. doi: 10.1016/j.jnutbio.2014.12.002
Lin, J., Xia, J., Zhang, K., and Yang, Q. (2016). Genome-wide profiling of chicken dendritic cell response to infectious bursal disease. BMC Genomics. 17:878. doi: 10.1186/s12864-016-3157-5
Lina, T. M., Raffaella, F., Luigi, B., Clorinda, A., Noemy, S., Andrea, C., et al. (2020). The Role of microRNAs, Long Non-coding RNAs, and Circular RNAs in Cervical Cancer. Front. Oncol. 10:150. doi: 10.3389/fonc.2020.00150
Liu, J., Wang, F., Weng, Z., Sui, X., Fang, Y., Tang, X., et al. (2020). Soybean-derived miRNAs specifically inhibit proliferation and stimulate apoptosis of human colonic Caco-2 cancer cells but not normal mucosal cells in culture. Genomics 112, 2949–2958. doi: 10.1016/j.ygeno.2020.05.011
Liu, Y. C., Chen, W. L., Kung, W. H., and Huang, H. D. (2017). Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genomics. 18:112. doi: 10.1186/s12864-017-3502-3
Lukasik, A., Brzozowska, I., Zielenkiewicz, U., and Zielenkiewicz, P. (2018). Detection of Plant miRNAs Abundance in Human Breast Milk. Int. J. Mol. Sci. 19:37. doi: 10.3390/ijms19010037
Mansoor, S. R., Ghasemi-Kasman, M., and Yavarpour-Bali, H. (2020). The role of microRNAs in multiple sclerosis. Int. Rev. Immunol. 30, 1–15. doi: 10.1080/08830185.2020.1826474
Mian, G. (2014). Identification of microRNAs in six solanaceous plants and their potential link with phosphate and mycorrhizal signaling. J. Integr. Plant Biol. 12, 1164–1178. doi: 10.1111/jipb.12233
Mico, V., Martin, R., Lasuncion, M. A., Ordovas, J. M., and Daimiel, L. (2016). Unsuccessful Detection of Plant MicroRNAs in Beer, Extra Virgin Olive Oil and Human Plasma After an Acute Ingestion of Extra Virgin Olive Oil. Plant Food. Hum. Nutr. 71, 102–108. doi: 10.1007/s11130-016-0534-9
Minutolo, A., Potestà, M., Gismondi, A., Pirrò, S., Cirilli, M., Gattabria, F., et al. (2018). Olea europaea small RNA with functional homology to human miR34a in cross-kingdom interaction of anti-tumoral response. Sci. Rep. 8:12413. doi: 10.1038/s41598-018-30718-w
Mlotshwa, S., Pruss, G. J., MacArthur, J. L., Endres, M. W., Davis, C., Hofseth, L. J., et al. (2015). A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 25, 521–524. doi: 10.1038/cr.2015.25
Ouyang, W., Qian, J., Pan, Q. X., Wang, J. Y., Xia, X. X., Wang, X. L., et al. (2018). gga-miR-142-5p attenuates IRF7 signaling and promotes replication of IBDV by directly targeting the chMDA5’s 3’ untranslated region. Vet. Microbiol. 221, 74–80. doi: 10.1016/j.vetmic.2018.05.018
Pastrello, C., Tsay, M., McQuaid, R., Abovsky, M., Pasini, E., Shirdel, E., et al. (2016). Circulating plant miRNAs can regulate human gene expression in vitro. Sci. Rep. 6:32773. doi: 10.1038/srep32773
Peng, T., Lv, Q., Zhang, J., Li, J., Du, Y., and Zhao, Q. (2011). Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J. Exp. Bot. 62, 4943–4954. doi: 10.1093/jxb/err205
Perez-Quintero, A. L., Neme, R., Zapata, A., and Lopez, C. (2010). Plant microRNAs and their role in defense against viruses: a bioinformatics approach. BMC Plant Biol. 10:138. doi: 10.1186/1471-2229-10-138
Potestà, M., Roglia, V., Fanelli, M., Pietrobono, E., Gismondi, A., Vumbaca, S., et al. (2020). Effect of microvesicles from Moringa oleifera containing miRNA on proliferation and apoptosis in tumor cell lines. Cell Death Discov. 6:43. doi: 10.1038/s41420-020-0271-6
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Ren, W., Wu, S., Wu, Y., Liu, T., Zhao, X., and Li, Y. (2019). MicroRNA-196a/-196b regulate the progression of hepatocellular carcinoma through modulating the JAK/STAT pathway via targeting SOCS2. Cell Death. Dis. 10:333. doi: 10.1038/s41419-019-1530-4
Sanchita, Trivedi, R., Asif, M. H., and Trivedi, P. K. (2018). Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation. RNA Biol. 15, 1433–1439. doi: 10.1080/15476286.2018.1551693
Shahid, S., Kim, G., Johnson, N. R., Wafula, E., Wang, F., Coruh, C., et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85. doi: 10.1038/nature25027
Shao, H.-W., He, M., Chen, J.-X., Chen, H., Xiang, J., and Huang, S.-L. (2015). Extraction of miRNA from Glycyrrhiza uralensis Decoction and Its Effect on Immune Cells. J. Chin. Med. Mater. 38, 1449–1453. doi: 10.13863/j.issn1001-4454.2015.07.029
Sharma, A., Sahu, S., Kumari, P., Gopi, S. R., Malhotra, R., and Biswas, S. (2017). Genome-wide identification and functional annotation of miRNAs in anti-inflammatory plant and their cross-kingdom regulation in Homo sapiens. J. Biomol. Struct. Dyn. 35, 1389–1400. doi: 10.1080/07391102.2016.1185381
Shen, C.-B., Yu, L., Gu, Y. N., Wang, S., and Lu, L. (2019). Inhibited expression of GATA-3 on Th2 cells transfect Astragalus-derived miR-396 of asthmatic mice in vivo. Chin. J. Immunol. 35, 3001–3007. doi: 10.3969/j.issn.1000-484X.2019.24.011
Stefano, P., Letizia, Z., Maurice, K., Carla, M., Antonella, M., Marina, P., et al. (2016). MicroRNA from Moringa oleifera: identification by High Throughput Sequencing and Their Potential Contribution to Plant Medicinal Value. PloS One 11:e0149495. doi: 10.1371/journal.pone.0149495
Teng, Y., Ren, Y., Sayed, M., Hu, X., Lei, C., Kumar, A., et al. (2018). Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe. 24, 637–652e638. doi: 10.1016/j.chom.2018.10.001
Tian, Y.-H. (2014). miR156a Targets Mammalian Junctional Adhesion Molecule-A to Repress Epithelial-Mesenchymal Transition in Human Nasopharyngeal Cancer. Ph. D. Thesis. China: Southern Medical University., doi: 10.7666/d.Y2618423.
Tripathi, A., Goswami, K., Tiwari, M., Mukherjee, S. K., and Sanan-Mishra, N. (2018). Identification and comparative analysis of microRNAs from tomato varieties showing contrasting response to ToLCV infections. Physiol. Mol. Biol. Pla. 24, 185–202. doi: 10.1007/s12298-017-0482-3
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2014). Correction: corrigendum: microRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 17, 104–104. doi: 10.1038/ncb3074
Vora, J., Patel, S., Sinha, S., Sharma, S., Srivastava, A., Chhabria, M., et al. (2019). Molecular docking, QSAR and ADMET based mining of natural compounds against prime targets of HIV. J. Biomol. Struct. Dyn. 37, 131–146. doi: 10.1080/07391102.2017.1420489
Wang, W., Liu, D., Zhang, X., Chen, D., Cheng, Y., and Shen, F. (2018). Plant MicroRNAs in Cross-Kingdom Regulation of Gene Expression. Int. J. Mol. Sci. 19, 1–12. doi: 10.3390/ijms19072007
Wang, Y., Peng, M., Chen, Y., Wang, W., He, Z., Yang, Z., et al. (2019). Analysis of Panax ginseng miRNAs and Their Target Prediction Based on High-Throughput Sequencing. Planta Med. 85, 1168–1176. doi: 10.1055/a-0989-7302
Wen, B. P., Dai, H. J., Yang, Y. H., Zhuang, Y., and Sheng, R. (2013). MicroRNA-23b inhibits enterovirus 71 replication through downregulation of EV71 VPl protein. Intervirology 56, 195–200. doi: 10.1159/000348504
Xia, C., Zhou, H., Xu, X., Jiang, T., Li, S., Wang, D., et al. (2020). Identification and Investigation of miRNAs From Gastrodia elata Blume and Their Potential Function. Front. Pharmacol. 11, 1–12. doi: 10.3389/fphar.2020.542405
Xiang, J., Huang, J. C., Xu, C., He, M., Xu, P. C., Zhang, D., et al. (2017). Effect of miRNA from Glycyrrhiza uralensis decoction on gene expression of human immune cells. China J. Chin. Mater. Med. 42, 1752–1756. doi: 10.19540/j.cnki.cjcmm.2017.0068
Xie, W., and Melzig, M. F. (2018). The Stability of Medicinal Plant microRNAs in the Herb Preparation Process. Molecules 23:919. doi: 10.3390/molecules23040919
Xie, W., Weng, A., and Melzig, M. F. (2016). MicroRNAs as New Bioactive Components in Medicinal Plants. Planta Med. 82, 1153–1162. doi: 10.1055/s-0042-108450
Xu, J., Wang, Y., Tan, X., and Jing, H. (2012). MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 8, 873–882. doi: 10.4161/auto.19629
Yang, J., Elbaz-Younes, I., Primo, C., Murungi, D., and Hirschi, K. D. (2018). Intestinal permeability, digestive stability and oral bioavailability of dietary small RNAs. Sci. Rep. 8:10253. doi: 10.1038/s41598-018-28207-1
Zhang, H., Li, Y., Liu, Y., Liu, H., Wang, H., Jin, W., et al. (2016). Role of plant MicroRNA in cross-species regulatory networks of humans. BMC Syst. Biol. 10:60. doi: 10.1186/s12918-016-0292-1
Zhang, K., Zhang, X., Cai, Z., Zhou, J., Cao, R., Zhao, Y., et al. (2018). A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 25, 1019–1027. doi: 10.1038/s41594-018-0136-3
Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L., Zhang, Y., et al. (2012). Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126. doi: 10.1038/cr.2011.158
Zhang, M., Viennois, E., Xu, C., and Merlin, D. (2016). Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 4:e1134415. doi: 10.1080/21688370.2015.1134415
Zhang, S., Sang, X., Hou, D., Chen, J., Gu, H., Zhang, Y., et al. (2019). Plant-derived RNAi therapeutics: a strategic inhibitor of HBsAg. Biomaterials 210, 83–93. doi: 10.1016/j.biomaterials.2019.04.033
Zhao, Y., Cong, L., and Lukiw, W. J. (2018). Plant and Animal microRNAs (miRNAs) and Their Potential for Inter-kingdom Communication. Cell. Mol. Neurobiol. 38, 133–140. doi: 10.1007/s10571-017-0547-4
Zhou, H.-X. (2018). Identification and Analysis of Gastrodia elata mi RNA and Preliminary Studies on Its Cross-kingdom Regulation. Ph. D. Thesis. China: Zhejiang Sci-Tech Universit.
Zhou, L. K., Zhou, Z., Jiang, X. M., Zheng, Y., Chen, X., Fu, Z., et al. (2020). Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 6:54. doi: 10.1038/s41421-020-00197-3
Keywords: microRNA, plant, cross-kingdom regulation, gene expression, activity
Citation: Li D, Yang J, Yang Y, Liu J, Li H, Li R, Cao C, Shi L, Wu W and He K (2021) A Timely Review of Cross-Kingdom Regulation of Plant-Derived MicroRNAs. Front. Genet. 12:613197. doi: 10.3389/fgene.2021.613197
Received: 22 October 2020; Accepted: 12 April 2021;
Published: 03 May 2021.
Edited by:
Jianping Wang, University of Florida, United StatesReviewed by:
Clicia Grativol, State University of the North Fluminense Darcy Ribeiro, BrazilCopyright © 2021 Li, Yang, Yang, Liu, Li, Li, Cao, Shi, Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihua Wu, d3doODE1QGhvdG1haWwuY29t; Kai He, aGVrYWk2OUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.