- 1Davidson School of Chemical Engineering, Purdue University, West Lafayette, IN, United States
- 2Purdue University Center for Cancer Research, Purdue University, West Lafayette, IN, United States
Developmental toxicity and its affiliation to long-term health, particularly neurodegenerative disease (ND) has attracted significant attentions in recent years. There is, however, a significant gap in current models to track longitudinal changes arising from developmental toxicity. The advent of induced pluripotent stem cell (iPSC) derived neuronal culture has allowed for more complex and functionally active in vitro neuronal models. Coupled with recent progress in the detection of ND biomarkers, we are equipped with promising new tools to understand neurotoxicity arising from developmental exposure. This review provides a brief overview of current progress in neuronal culture derived from iPSC and in ND markers.
Introduction
Developmental exposure to environmental chemicals, such as heavy metal [e.g., lead (Bellinger et al., 1987), manganese (Lynam et al., 1999), mercury (Falluel-Morel et al., 2007)] and organic chemicals (Thiruchelvam et al., 2002) (e.g., pesticides, herbicide, and industrial surfactants) has been associated with various neurodevelopmental and neurodegenerative diseases (NDs) in the past decades and attracted significant attention in the scientific area. These chemicals have pervaded nearly all parts of our environment, including the food chain (Watras et al., 1998), drinking water (Remoundaki et al., 2016), and atmosphere (Lynam et al., 1999) due to decades of industrial and commercial use. Chronic exposure to low-doses of environmental chemicals does not necessarily illicit immediate response in the exposed population but can have significant bearings on shaping the health of individuals later in life (Lee et al., 2003; Wang et al., 2015). Among different exposure windows, the developmental window has been considered the most sensitive to chemical exposure, which has led to the establishment of the Developmental Origins of Health and Disease (DOHaD) hypothesis. The DOHaD hypothesis postulates that the developmental and growth window presents a critical period of time where exposure to certain environmental chemicals can impose significant consequences on an individual’s short and long-term health (Barker, 2007). The attempts to elucidate toxicity arising from developmental exposure, however, have been very challenging because of the latent time between exposure and disease on-set; as well as the lack of established biomarkers that “record” past exposure events and trigger disease on-set later in life.
Longitudinal epidemiology studies have been commonly used to establish the connection between developmental exposure and disease onset later in life. For example, developmental exposure to lead (Pb) can result in an increased risk of developing learning disabilities in exposed children (Needleman et al., 1979) and NDs such as Alzheimer’s disease later in life as suggested in rodent models (Eid et al., 2016). Exposure to organic pesticides such as the herbicide paraquat and fungicide maneb during the developmental stage can cause a decrease of striatal dopamine production and impairment of locomotor activity which aligns with Parkinson’s disease phenotype (Thiruchelvam et al., 2002). Unfortunately, the majority of studies provide limited mechanistic insights into how toxicity affects cells. Various animal models have also been adopted, including rodent (Filipov et al., 2007; Eid et al., 2016), fish (Weber et al., 2013; Wirbisky et al., 2016), and monkey (Lasky et al., 2005) models. These have had success in assessing survival toxicity (Gad, 2014) and damage to reproductive tissues (Song et al., 2014) but have had only limited success for assessing neurotoxicity because of the vast difference between human and non-primate brains. For example, rodent models are extremely popular for the study of cancer (Ding et al., 2011) and psychiatric (Majdak et al., 2016) diseases; however, it has become increasingly clear that rodent models do not necessarily recapitulate human brain function as accurately as previously believed and thus may not be an ideal surrogate system for studying neurotoxicity arising from environmental exposures (Hodge et al., 2019). Although rodent and human brains share a conserved structure, RNA-seq studies have revealed that the expression of ion channels, neurotransmitter synthetase, and neurotransmitter receptors varies greatly between species (Hodge et al., 2019). Uncertainty in the accuracy of animal models for studying brain-related diseases has led to the popularization of human stem cells, including embryonic (ESC) and induced pluripotent stem cells (iPSCs) as an in vitro system for assessing neurotoxicity. In contrast to ESC, which must be collected from blastocysts, iPSCs are derived from fibroblasts of human patients. They are easier to collect; account for genetic variations among human patients and thus are becoming a preferred cell model in studying neurotoxicity. iPSCs have been differentiated into various lineages to partially mimic human organs since their debut in 2006 (Takahashi and Yamanaka, 2006). Of particular interest in assessing environmental exposure, iPSCs can be differentiated into neural stem cells (NSCs) (D’Aiuto et al., 2014) and later glutaminergic (Anderson et al., 2015), dopaminergic (Suzuki et al., 2017), and GABAergic (Lin et al., 2015) neurons that mimic part of the brain functions. Here, we will review the recent progress in using human iPSCs as a cell culture model to study neurotoxicity arising from developmental exposure.
Towards Recaptulating (Part of) Brain Complexity in Human Cell Culture
The brain is a complex organ and thus understandably difficult to reconstruct in vitro given its composition, structural, and functional complexity. However, significant progress has been made in recent years enabling partial recapitulation of a human brain in a culture dish.
Composition Complexity
The human brain consists of various cell types, including neurons (Lake et al., 2016), astrocytes (Lewis et al., 2010), and microglia (Hickey and Kimura, 1988). To address the challenge in composition complexity, various protocols have been developed for incorporating multiple cell types via co-culture systems [i.e., neuron- astrocyte (Odawara et al., 2014; Kuijlaars et al., 2016) and neuron- microglia (Haenseler et al., 2017)] differentiated from iPSC and neural progenitor cells (NPCs). Figure 1 illustrates some commonly used co-culturing techniques that can be used to reconstruct brain-like tissues on a dish. These co-culture systems facilitate the reconstruction of complex neural systems resembling neural development (Kuijlaars et al., 2016) or mimicking inflammatory responses that are provoked in NDs such as Alzheimer’s and Parkinson’s disease (McGeer et al., 1988; Sapp et al., 2001). It is worth noting that not all cells found in the brain are from the same stem cell lineage. For example, microglial cells in the brain result from the differentiation of mesodermal progenitors (Murabe and Sano, 1982), rather than the neuroectoderm, as is the case with neurons (Jiang et al., 2003). The combination of multiple types of cells in vitro has arisen as a critical tool in the study of brain development, particularly in the study of NDs, where the interaction between multiple cell types is crucial for disease pathology (Di Malta et al., 2012; Heneka et al., 2013).
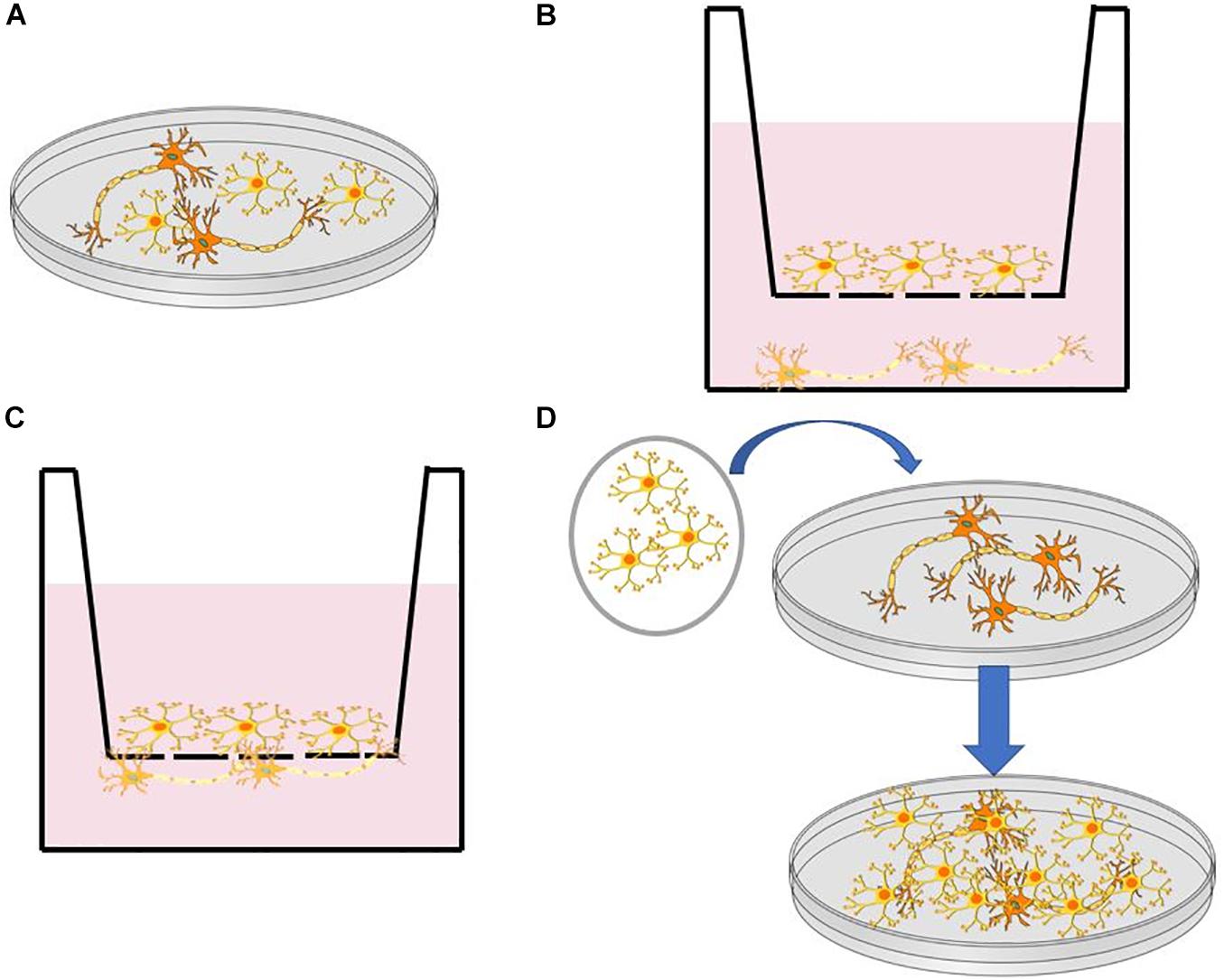
Figure 1. (A) Direct co-culture in a dish. (B) Indirect co-culture using a transwell insert with a semipermeable membrane. (C) Direct co-culture through a semipermeable membrane in a transwell dish. (D) Transfer of cell monolayer to established culture through a membrane such as PVDF.
Structure Complexity
Significant advances in cell culturing approaches have occurred in recent years that enabled the transition from 2D to 3D cell culture to reconstruct the architecture of the brain. Compared to 3D culture, 2D cell cultures are typically easier to perform and more compatible with imaging-based analysis, including neurite morphology analysis and live-cell tracing (Shin et al., 2018). 3D culture, however, takes advantage of the self-assembly that iPSCs undergo when grown in a 3D environment (Lancaster et al., 2013). This self-assembly causes iPSCs to form distinct cell types native to specific areas of the brain, such as the hippocampus, ventral forebrain, and cerebral cortex (Lancaster et al., 2017; Quadrato et al., 2017). Using this method, researchers can create brain organoids that contain various discrete but interdependent brain regions (Lancaster et al., 2013). This structure complexity, particularly the development and interaction of different brain regions, can produce organoids with active neural networks capable of firing in response to stimuli (Giandomenico et al., 2019). 3D organoids provide an ideal testing platform for drug discovery (Phan et al., 2019), modeling neurological diseases (Lancaster et al., 2013), and assessing environmental toxin exposure (Forsythe et al., 2018). While early brain organoids showed significant heterogeneity (Camp et al., 2015), brain organoids generated via more recent protocols have improved reproducibility (Velasco et al., 2019; Nickels et al., 2020) but still require further optimization.
Functional Connectivity
Although iPSCs can differentiate into neurons and express the mature neuronal markers such as MAP2 and NeuN, they do not necessarily exhibit the correct electrophysiological properties, hampering their uses in assessing neural circuitry activities. Researchers have developed simplified protocols for differentiating neuronal networks that are electro-physiologically active in 2D (Gunhanlar et al., 2018) and 3D (Paşca et al., 2015; Birey et al., 2017). These protocols typically include cAMP in their culture to facilitate the establishment of neuronal connectivity (Kang et al., 2017; Gunhanlar et al., 2018). With the initial claim of success, the prevalence, composition, properties, and relevance of these established neuronal connections remain to be tested by time. The occurrence of in vivo-like complex neuronal activity and functional circuitry remains elusive and more in-depth analysis will be crucial for unraveling the connectivity in cultures, particularly organoids, to understand the impact of genetic and environmental chemical perturbations on human synaptogenesis, neuronal activity, and network function.
Mimicking Developmental Exposure Using Stem Cell Models
Stem cells can undergo differentiation and maturation in a similar manner compared to animal models, providing multiple assessment windows for studying neurotoxicity. Human brains contain many unique features that define cognition (Lui et al., 2011). On average, 86.1 billion neurons can be found in the brain and spinal cord of an adult male and 16.34 billion of these are located in the cerebral cortex (Azevedo et al., 2009; Herculano-Houzel et al., 2016). 80% of the neurons in the cerebral cortex are thought to be excitatory glutaminergic neurons which were differentiated in the ventricular zone and sub-ventricular zone of the cortical wall during prenatal development from 50 postconceptional days (pcd) to 24 postconceptional weeks (pcw) at a rate of ∼3.86 million neurons per hour (Workman et al., 2013). Neurogenesis occurs in the spinal cord and brain stem beginning at 32 pcd followed by cerebral cortex (O’Rahilly, 2006). Most of this neuron generation is completed before birth. Neocortical interneurons and inhibitory GABAergic neurons are two of the few neuron types that are known to undergo differentiation after birth (Sanai et al., 2011; Radonjić et al., 2014). Although most neurons have completed differentiation before birth, the neuronal network is yet to be established. Recent studies have revealed that synaptogenesis (Peter, 1979; Kang et al., 2011), myelination (Miller et al., 2012), and synaptic pruning (Petanjek et al., 2011) are still ongoing until 20 years of age. Figure 2 summarizes the neuronal developmental processes at different time windows in the human brain matched to various stages of stem cell differentiation. Specifically, the procedure of generating mature neurons from NPCs can be divided into two sequential steps, namely, differentiation followed by maturation. During the differentiation step, retinoic acid or bone morphogenetic protein is used to mediate neuronal differentiation signaling pathways (Cazillis et al., 2006). After that, brain-derived neurotrophic factor (BDNF) and dibutyryl cyclic adenosine monophosphate (db-cAMP) are added to facilitate neuron maturation and promote synaptogenesis (Kang et al., 2017; Gunhanlar et al., 2018). The initial differentiation stage is completed after 8 days but it can take up to 6–8 weeks for neurons to mature and develop proper electrophysiological functions (Gunhanlar et al., 2018).
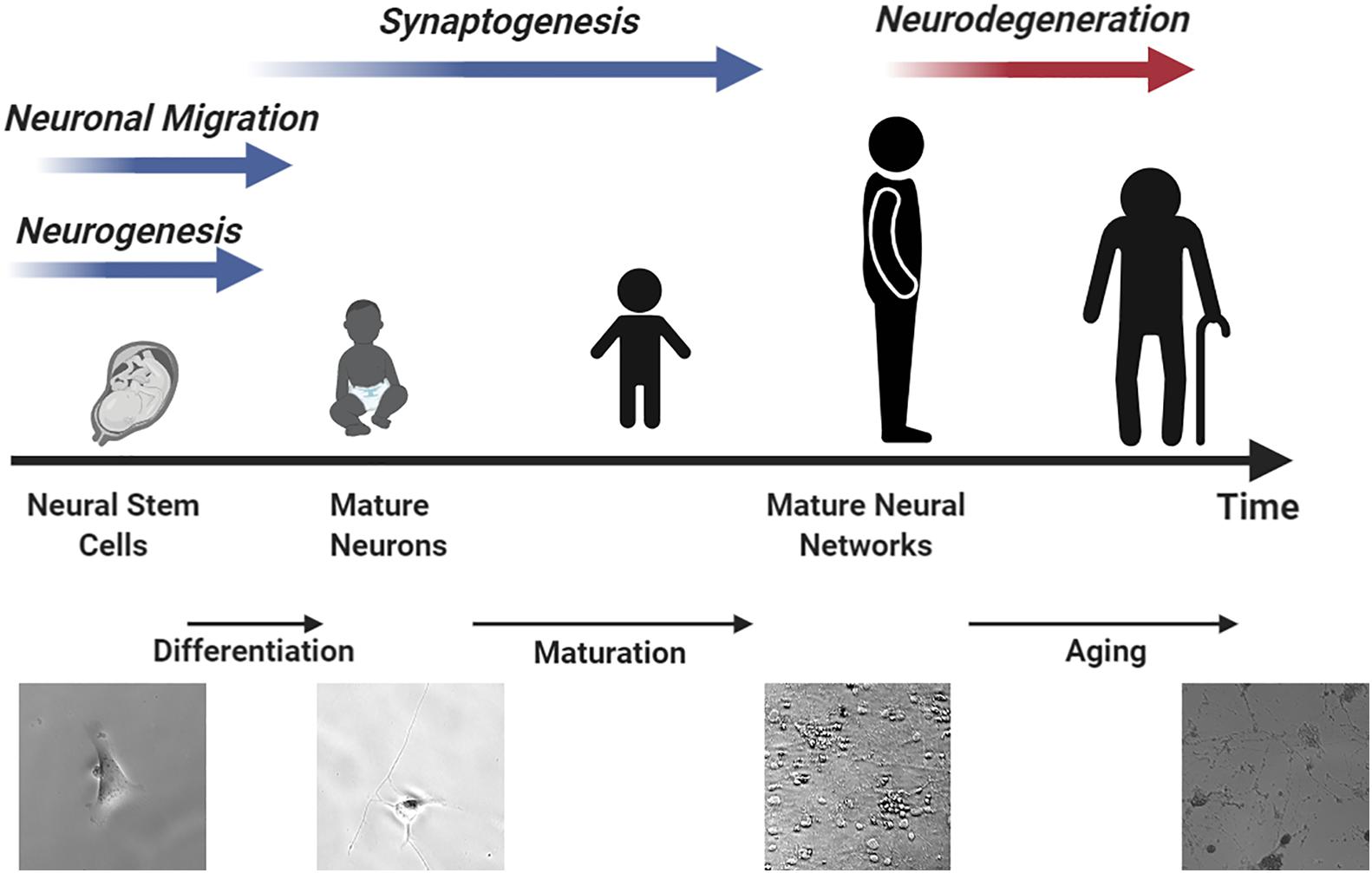
Figure 2. An illustration of neurodevelopment processes matched to different stem cell differentiation time windows resembling human brain development.
Assess Neurotoxicity Using Cell Cultur Model
Compared to animal models, cell culture presents several advantages for assessing neurotoxicity, including homogeneity in cell identity, compatibility with long-term monitoring, and controllable dosing. Several disadvantages remain, most notably the inability to perform behavior and aging related assessments. We will thus focus on summarizing molecular markers that have been previously established to facilitate the assessment of neurotoxicity.
Disease Biomarkers
Cell viability and proliferation are conventionally used as the starting point for assessing neurotoxicity. Although insightful for acute toxicity, limited knowledge can be gained regarding the chronic effects of neurotoxicity arising from environmental exposures. Early-stage disease biomarkers with established correlations of disease on-set thus play a critical role in assessing neurotoxicity. Several good reviews exist summarizing prevalent early-stage biomarkers that have been used to assess ND risks (Molinuevo et al., 2018; Ehrenberg et al., 2020). For example, Aβ42/Aβ40 ratio (Doecke et al., 2020) and phosphorylated tau p-Thr181 tau (Thijssen et al., 2020) and p-Thr205 tau (Barthélemy et al., 2020)) can be used to assist the diagnosis and prognosis of AD. α-synuclein and α-synuclein phosphorylation can be used in PD for similar purposes (Wang et al., 2012; Lin et al., 2017). These biomarkers can be visualized in cells exposed to environmental chemicals using either immunofluorescence or immunohistology methods, or quantified via ELISA, providing a robust approach to connect environmental exposure with neurodevelopmental and NDs.
Neuronal Activity
Recent expansion of toolsets to conduct electrophysiology measurements have further expanded our capability to probe neural circuit activities related to environmental exposure. Synaptic and ion channel activities, membrane potential, and action potential can be conventionally recorded using patch-clamp and multi-electrode array (MEA). These approaches have been primarily used to record changes in neural circuits of animal brain slices (Cholanian et al., 2017), cultured primary cells (Cannady et al., 2017; Dunn et al., 2018), and cell cultures (James et al., 2017) responding to various environmental chemical exposures. Neurons derived from iPSCs can establish electro-physiologically mature neuronal networks that closely resemble mature primary neurons (Gunhanlar et al., 2018). Recently, patch-clamp and MEA have been applied to cortical neurons derived from iPSCs of AD patient and healthy control; and demonstrated that AD neuronal cultures exhibit increased spontaneous firing, slow oscillatory events, and hypersynchronous circuit activity (Ghatak et al., 2020). Furthermore, various imaging probes, including calcium indicators, i.e., Fura-2 (Tsien et al., 1982; Grynkiewicz et al., 1985) and GCaMP (Chen et al., 2013; Yang et al., 2018); membrane potential probes, i.e., ASAP2 (Yang et al., 2016; Chamberland et al., 2017) and Voltron (Abdelfattah et al., 2019); and neurotransmitter probes, i.e., iGluSnFR (Marvin et al., 2013) and dLight (Patriarchi et al., 2018) can be introduced to neuronal cultures to monitor neural circuit activity in situ. These measurements collectively provide a viable approach to characterize abnormal circuitry activity that are known to be altered in various neurodevelopmental and neurodegenerative conditions. For example, AD neuronal cultures show oscillatory events and hypersynchronous network activity compared to their wild-type isogenic controls (Ghatak et al., 2020). Furthermore, neurons derived from iPSCs of Autism Spectrum Disorders (ASD) have significantly altered glutamate neurotransmitter release and reduced spontaneous firing rate (Russo et al., 2018).
Epigenetic Changes
Epigenetic modifications account for inheritable changes in chromatin that are not accounted for by DNA mutations. Epigenetic modifications have been increasingly recognized as viable markers for various neurological diseases and are being actively researched. Among different epigenetic modifications, DNA methylation, such as cytosine methylation (5mC), hydroxymethylation (5hmC), and histone acetylation have attracted the most significant attention. Mutations in DNA and histone methyltransferase have been identified as risk factors in autism (Satterstrom et al., 2020). In addition, abnormal alterations in H3K9ac and H3K27ac were identified as the major distinctions between aging and AD brains (Nativio et al., 2020). Furthermore, CpH hypomethylation is accelerated in AD brains compared to normal aging ones (Li et al., 2019). Abnormal DNA methylation of ASCC1 and SLC7A11 has also been linked to PD (Vallerga et al., 2020). Similar methylation features were also revealed in iPSC models reprogrammed from patients iPSCs (Fetahu et al., 2019). Interestingly, environmental exposure of neuronal cells can also lead to major changes in DNA methylation, histone methylation, and acetylation. This suggests that epigenetic mechanisms may serve as a potential bridging marker between environmental exposure and neurological conditions (Pavanello et al., 2009; Luo et al., 2014; Wirbisky-Hershberger et al., 2017; Lin et al., 2020). The detection of epigenetic changes by protein-based fluorescent probes allows for real-time analysis of the changes to the global epigenome levels. So far protein probes have been developed for binding and visualization of both DNA methylation and histone modifications (Hendrich and Bird, 1998; Lungu et al., 2017; Sanchez et al., 2017; Sanchez et al., 2019). Further development of nanobody fragments have provided an additional domain that allows for high specificity and affinity for selected epigenetic targets (Hattori et al., 2016; Jullien et al., 2016). Accurate understanding and quantification of epigenetic changes that proceed chemically induced disease development could provide a system that can detect neurological conditions at an earlier time point.
Conclusion
In closing, patient derived iPSCs offer a promising platform to assess neurotoxicity given its ability to partially recapitulate the complexity and functionality of in vivo neurological structures. The advent and exploration of both 2D and 3D iPSC systems provide great promise for the examination of environmental exposure effects on adolescent brains. In addition, these cell assemblies provide a platform that more closely matches the developmental timeline and disease phenotypes of a human brain in comparison to rodent models. The application of environmental chemicals to iPSC is in its early stages, but provides a clear opportunity to understand the mechanisms that affect the human brain when exposed to heavy metals and pesticides. The use of phenotypic assays that quantify ND risk factors, neuron activity, and epigenetic changes can be used to elucidate the molecular mechanisms that are perturbed by environmental exposure. With the application of the numerous live-cell compatible techniques to human iPSCs, we are at an optimal position to understand the biological response to chronic toxin exposure and how it informs long-term disease development.
Author Contributions
All authors have contributed in planning and writing the review.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdelfattah, A. S., Kawashima, T., Singh, A., Novak, O., Liu, H., Shuai, Y., et al. (2019). Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704. doi: 10.1126/science.aav6416
Anderson, G. W., Deans, P. J. M., Taylor, R. D. T., Raval, P., Chen, D., Lowder, H., et al. (2015). Characterisation of neurons derived from a cortical human neural stem cell line CTX0E16. Stem Cell Res. Ther. 6:149.
Azevedo, F. A. C., Carvalho, L. R. B., Grinberg, L. T., Farfel, J. M., Ferretti, R. E. L., Leite, R. E. P., et al. (2009). Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541. doi: 10.1002/cne.21974
Barker, D. J. (2007). The origins of the developmental origins theory. J. Intern. Med. 261, 412–417. doi: 10.1111/j.1365-2796.2007.01809.x
Barthélemy, N. R., Li, Y., Joseph-Mathurin, N., Gordon, B. A., Hassenstab, J., Benzinger, T. L. S., et al. (2020). A soluble phosphorylated tau signature links tau., amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat. Med. 26, 398–407. doi: 10.1038/s41591-020-0781-z
Bellinger, D., Leviton, A., Waternaux, C., Needleman, H., and Rabinowitz, M. (1987). Longitudinal analyses of prenatal and postnatal lead exposure and early cognitive development. N. Engl. J. Med. 316, 1037–1043. doi: 10.1056/nejm198704233161701
Birey, F., Andersen, J., Makinson, C. D., Islam, S., Wei, W., Huber, N., et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. doi: 10.1038/nature22330
Camp, J. G., Badsha, F., Florio, M., Kanton, S., Gerber, T., Wilsch-Bräuninger, M., et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U.S.A. 112, 15672–15677. doi: 10.1073/pnas.1520760112
Cannady, R., McGonigal, J. T., Newsom, R. J., Woodward, J. J., Mulholland, P. J., and Gass, J. T. (2017). Prefrontal Cortex K(Ca)2 channels regulate mGlu(5)-Dependent plasticity and extinction of alcohol-seeking behavior. J. Neurosci. 37, 4359–4369. doi: 10.1523/jneurosci.2873-16.2017
Cazillis, M., Rasika, S., Mani, S., Gressens, P., and Leliévre, V. (2006). In vitro induction of neural differentiation of embryonic stem (ES) cells closely mimics molecular mechanisms of embryonic brain development. Pediatr. Res. 59, 48–53.
Chamberland, S., Yang, H. H., Pan, M. M., Evans, S. W., Guan, S., Chavarha, M., et al. (2017). Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 6:e25690.
Chen, T.-W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354
Cholanian, M., Wealing, J., Levine, R. B., and Fregosi, R. F. (2017). Developmental nicotine exposure alters potassium currents in hypoglossal motoneurons of neonatal rat. J. Neurophysiol. 117, 1544–1552. doi: 10.1152/jn.00774.2016
D’Aiuto, L., Zhi, Y., Kumar Das, D., Wilcox, M. R., Johnson, J. W., McClain, L., et al. (2014). Large-scale generation of human iPSC-derived neural stem cells/early neural progenitor cells and their neuronal differentiation. Organogenesis 10, 365–377. doi: 10.1080/15476278.2015.1011921
Di Malta, C., Fryer, J. D., Settembre, C., and Ballabio, A. (2012). Astrocyte dysfunction triggers neurodegeneration in a lysosomal storage disorder. Proc. Natl. Acad. Sci. U.S.A. 109, E2334–E2342.
Ding, Z., Wu, C.-J., Chu, G. C., Xiao, Y., Ho, D., Zhang, J., et al. (2011). SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273. doi: 10.1038/nature09677
Doecke, J. D., Pérez-Grijalba, V., Fandos, N., Fowler, C., Villemagne, V. L., Masters, C. L., et al. (2020). Total Aβ42/Aβ40 ratio in plasma predicts amyloid-PET status, independent of clinical AD diagnosis. Neurology 94, e1580–e1591.
Dunn, A. R., Neuner, S. M., Ding, S., Hope, K. A., O’Connell, K. M. S., and Kaczorowski, C. C. (2018). Cell-type-specific changes in intrinsic excitability in the subiculum following learning and exposure to novel environmental contexts. eNeuro 5:ENEURO.0484-18.2018. doi: 10.1523/ENEURO.0484-18.2018
Ehrenberg, A. J., Khatun, A., Coomans, E., Betts, M. J., Capraro, F., Thijssen, E. H., et al. (2020). Relevance of biomarkers across different neurodegenerative diseases. Alzheimers Res. Ther. 13:56.
Eid, A., Bihaqi, S. W., Renehan, W. E., and Zawia, N. H. (2016). Developmental lead exposure and lifespan alterations in epigenetic regulators and their correspondence to biomarkers of Alzheimer’s disease. Alzheimer’s Dement. 2, 123–131. doi: 10.1016/j.dadm.2016.02.002
Falluel-Morel, A., Sokolowski, K., Sisti, H. M., Zhou, X., Shors, T. J., and DiCicco-Bloom, E. (2007). Developmental mercury exposure elicits acute hippocampal cell death., reductions in neurogenesis., and severe learning deficits during puberty. J. Neurochem. 103, 1968–1981. doi: 10.1111/j.1471-4159.2007.04882.x
Fetahu, I. S., Ma, D., Rabidou, K., Argueta, C., Smith, M., Liu, H., et al. (2019). Epigenetic signatures of methylated DNA cytosine in Alzheimer’s disease. Sci. Adv. 5:eaaw2880. doi: 10.1126/sciadv.aaw2880
Filipov, N. M., Stewart, M. A., Carr, R. L., and Sistrunk, S. C. (2007). Dopaminergic toxicity of the herbicide atrazine in rat striatal slices. Toxicology 232, 68–78. doi: 10.1016/j.tox.2006.12.007
Forsythe, S. D., Devarasetty, M., Shupe, T., Bishop, C., Atala, A., Soker, S., et al. (2018). Environmental toxin screening using human-derived 3d bioengineered liver and cardiac organoids. Front. Public Health 6:103. doi: 10.3389/fpubh.2018.00103
Gad, S. C. (2014). “Chapter 2 - Rodents model for toxicity testing and biomarkers,” in Biomarkers in Toxicology, ed. R. C. Gupta (Boston, MA: Academic Press), 7–69. doi: 10.1016/b978-0-12-404630-6.00002-6
Ghatak, S., Dolatabadi, N., Gao, R., Wu, Y., Scott, H., Trudler, D., et al. (2020). NitroSynapsin ameliorates hypersynchronous neural network activity in Alzheimer hiPSC models. Mol. Psychiatry doi: 10.1038/s41380-020-0776-7 [Epub ahead of print].
Giandomenico, S. L., Mierau, S. B., Gibbons, G. M., Wenger, L. M. D., Masullo, L., Sit, T., et al. (2019). Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 22, 669–679. doi: 10.1038/s41593-019-0350-2
Grynkiewicz, G., Poenie, M., and Tsien, R. Y. (1985). A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450.
Gunhanlar, N., Shpak, G., van der Kroeg, M., Gouty-Colomer, L. A., Munshi, S. T., Lendemeijer, B., et al. (2018). A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells. Mol. Psychiatry 23, 1336–1344. doi: 10.1038/mp.2017.56
Haenseler, W., Sansom, S. N., Buchrieser, J., Newey, S. E., Moore, C. S., Nicholls, F. J., et al. (2017). A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Rep. 8, 1727–1742. doi: 10.1016/j.stemcr.2017.05.017
Hattori, T., Lai, D., Dementieva, I. S., Montano, S. P., Kurosawa, K., Zheng, Y. P., et al. (2016). Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation. Proc. Natl. Acad. Sci. U.S.A. 113, 2092–2097. doi: 10.1073/pnas.1522691113
Hendrich, B., and Bird, A. (1998). Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18, 6538–6547. doi: 10.1128/mcb.18.11.6538
Heneka, M. T., Kummer, M. P., Stutz, A., Delekate, A., Schwartz, S., Vieira-Saecker, A., et al. (2013). NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678. doi: 10.1038/nature11729
Herculano-Houzel, S., Kaas, J. H., and de Oliveira-Souza, R. (2016). Corticalization of motor control in humans is a consequence of brain scaling in primate evolution. J. Comp. Neurol. 524, 448–455. doi: 10.1002/cne.23792
Hickey, W. F., and Kimura, H. (1988). Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science 239, 290–292. doi: 10.1126/science.3276004
Hodge, R. D., Bakken, T. E., Miller, J. A., Smith, K. A., Barkan, E. R., Graybuck, L. T., et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68.
James, T. F., Nenov, M. N., Tapia, C. M., Lecchi, M., Koshy, S., Green, T. A., et al. (2017). Consequences of acute Na(v)1.1 exposure to deltamethrin. Neurotoxicology 60, 150–160. doi: 10.1016/j.neuro.2016.12.005
Jiang, Y., Henderson, D., Blackstad, M., Chen, A., Miller, R. F., and Verfaillie, C. M. (2003). Neuroectodermal differentiation from mouse multipotent adult progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11854–11860. doi: 10.1073/pnas.1834196100
Jullien, D., Vignard, J., Fedor, Y., Bery, N., Olichon, A., Crozatier, M., et al. (2016). Chromatibody., a novel non-invasive molecular tool to explore and manipulate chromatin in living cells. J. Cell Sci. 129, 2673–2683. doi: 10.1242/jcs.183103
Kang, H. J., Kawasawa, Y. I., Cheng, F., Zhu, Y., Xu, X., Li, M., et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489.
Kang, S., Chen, X., Gong, S., Yu, P., Yau, S., Su, Z., et al. (2017). Characteristic analyses of a neural differentiation model from iPSC-derived neuron according to morphology., physiology., and global gene expression pattern. Sci. Rep. 7:12233.
Kuijlaars, J., Oyelami, T., Diels, A., Rohrbacher, J., Versweyveld, S., Meneghello, G., et al. (2016). Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci. Rep. 6:36529.
Lake, B. B., Ai, R., Kaeser, G. E., Salathia, N. S., Yung, Y. C., Liu, R., et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. doi: 10.1126/science.aaf1204
Lancaster, M. A., Corsini, N. S., Wolfinger, S., Gustafson, E. H., Phillips, A. W., Burkard, T. R., et al. (2017). Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666. doi: 10.1038/nbt.3906
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379.
Lasky, R. E., Luck, M. L., Parikh, N. A., and Laughlin, N. K. (2005). The effects of early lead exposure on the brains of adult rhesus monkeys: a volumetric MRI study. Toxicol. Sci. 85, 963–975. doi: 10.1093/toxsci/kfi153
Lee, Y. L., Pai, M. C., Chen, J. H., and Guo, Y. L. (2003). Central neurological abnormalities and multiple chemical sensitivity caused by chronic toluene exposure. Occup. Med. 53, 479–482. doi: 10.1093/occmed/kqg095
Lewis, N. E., Schramm, G., Bordbar, A., Schellenberger, J., Andersen, M. P., Cheng, J. K., et al. (2010). Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat. Biotechnol. 28, 1279–1285. doi: 10.1038/nbt.1711
Li, P., Marshall, L., Oh, G., Jakubowski, J. L., Groot, D., He, Y., et al. (2019). Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer’s disease pathology and cognitive symptoms. Nat. Commun. 10:2246.
Lin, C.-H., Yang, S.-Y., Horng, H.-E., Yang, C.-C., Chieh, J.-J., Chen, H.-H., et al. (2017). Plasma α-synuclein predicts cognitive decline in Parkinson’s disease. J. Neurol.Neurosurg. Psychiatry 88, 818–824. doi: 10.1136/jnnp-2016-314857
Lin, L., Xie, J., Sanchez, O. F., Bryan, C., Freeman, J., and Yuan, C. (2020). Low dose lead exposure induces alterations on heterochromatin hallmarks persisting through SH-SY5Y cell differentiation. Chemosphere 264:128486. doi: 10.1016/j.chemosphere.2020.128486
Lin, L., Yuan, J., Sander, B., and Golas, M. M. (2015). In vitro differentiation of human neural progenitor cells into striatal GABAergic neurons. Stem Cells Transl. Med. 4, 775–788. doi: 10.5966/sctm.2014-0083
Lui, J. H., Hansen, D. V., and Kriegstein, A. R. (2011). Development and evolution of the human neocortex. Cell 146, 18–36. doi: 10.1016/j.cell.2011.06.030
Lungu, C., Pinter, S., Broche, J., Rathert, P., and Jeltsch, A. (2017). Modular fluorescence complementation sensors for live cell detection of epigenetic signals at endogenous genomic sites. Nat. Commun. 8:649.
Luo, M., Xu, Y., Cai, R., Tang, Y., Ge, M.-M., Liu, Z.-H., et al. (2014). Epigenetic histone modification regulates developmental lead exposure induced hyperactivity in rats. Toxicol. Lett. 225, 78–85. doi: 10.1016/j.toxlet.2013.11.025
Lynam, D. R., Roos, J. W., Pfeifer, G. D., Fort, B. F., and Pullin, T. G. (1999). Environmental effects and exposures to manganese from use of methylcyclopentadienyl manganese tricarbonyl (MMT) in gasoline. Neurotoxicology 20, 145–150.
Majdak, P., Ossyra, J. R., Ossyra, J. M., Cobert, A. J., Hofmann, G. C., Tse, S., et al. (2016). A new mouse model of ADHD for medication development. Sci. Rep. 6:39472.
Marvin, J. S., Borghuis, B. G., Tian, L., Cichon, J., Harnett, M. T., Akerboom, J., et al. (2013). An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170. doi: 10.1038/nmeth.2333
McGeer, P. L., Itagaki, S., Boyes, B. E., and McGeer, E. G. (1988). Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology 38, 1285–1285. doi: 10.1212/wnl.38.8.1285
Miller, D. J., Duka, T., Stimpson, C. D., Schapiro, S. J., Baze, W. B., McArthur, M. J., et al. (2012). Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 16480–16485. doi: 10.1073/pnas.1117943109
Molinuevo, J. L., Ayton, S., Batrla, R., Bednar, M. M., Bittner, T., Cummings, J., et al. (2018). Current state of Alzheimer’s fluid biomarkers. Acta Neuropathol. 136, 821–853.
Nativio, R., Lan, Y., Donahue, G., Sidoli, S., Berson, A., Srinivasan, A. R., et al. (2020). An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 52, 1024–1035. doi: 10.1038/s41588-020-0696-0
Needleman, H. L., Gunnoe, C., Leviton, A., Reed, R., Peresie, H., Maher, C., et al. (1979). Deficits in psychologic and classroom performance of children with elevated dentine lead levels. N. Engl. J. Med. 300, 689–695. doi: 10.1056/nejm197903293001301
Nickels, S. L., Modamio, J., Mendes-Pinheiro, B., Monzel, A. S., Betsou, F., and Schwamborn, J. C. (2020). Reproducible generation of human midbrain organoids for in vitro modeling of Parkinson’s disease. Stem Cell Res. 46:101870. doi: 10.1016/j.scr.2020.101870
Odawara, A., Saitoh, Y., Alhebshi, A. H., Gotoh, M., and Suzuki, I. (2014). Long-term electrophysiological activity and pharmacological response of a human induced pluripotent stem cell-derived neuron and astrocyte co-culture. Biochem. Biophys. Res. Commun. 443, 1176–1181. doi: 10.1016/j.bbrc.2013.12.142
O’Rahilly, F. M. (2006). “Embryonic Staging,” in The Embryonic Human Brain: An Atlas of Developmental Stages, 3rd Edn, ed. John Wiley & Sons (Hoboken, NJ: Wiley), 11–13. doi: 10.1002/0471973084.ch4
Paşca, A. M., Sloan, S. A., Clarke, L. E., Tian, Y., Makinson, C. D., Huber, N., et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678. doi: 10.1038/nmeth.3415
Patriarchi, T., Cho, J. R., Merten, K., Howe, M. W., Marley, A., Xiong, W.-H., et al. (2018). Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360:eaat4422. doi: 10.1126/science.aat4422
Pavanello, S., Bollati, V., Pesatori, A. C., Kapka, L., Bolognesi, C., Bertazzi, P. A., et al. (2009). Global and gene-specific promoter methylation changes are related to anti-B [a] PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int. J. Cancer 125, 1692–1697. doi: 10.1002/ijc.24492
Petanjek, Z., Judaš, M., Šimić, G., Rašin, M. R., Uylings, H. B. M., Rakic, P., et al. (2011). Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. U.S.A. 108, 13281–13286. doi: 10.1073/pnas.1105108108
Peter, R. H. (1979). Synaptic density in human frontal cortex — Developmental changes and effects of aging. Brain Res. 163, 195–205. doi: 10.1016/0006-8993(79)90349-4
Phan, N., Hong, J. J., Tofig, B., Mapua, M., Elashoff, D., Moatamed, N. A., et al. (2019). A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun. Biol. 2:78.
Quadrato, G., Nguyen, T., Macosko, E. Z., Sherwood, J. L., Min Yang, S., Berger, D. R., et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53. doi: 10.1038/nature22047
Radonjić, N. V., Ayoub Albert, E., Memi, F., Yu, X., Maroof, A., Jakovcevski, I., et al. (2014). Diversity of cortical interneurons in primates: the role of the dorsal proliferative niche. Cell Rep. 9, 2139–2151. doi: 10.1016/j.celrep.2014.11.026
Remoundaki, E., Vasileiou, E., Philippou, A., Perraki, M., Kousi, P., Hatzikioseyian, A., et al. (2016). Groundwater deterioration: the simultaneous effects of intense agricultural activity and heavy metals in soil. Proc. Eng. 162, 545–552. doi: 10.1016/j.proeng.2016.11.099
Russo, F. B., Freitas, B. C., Pignatari, G. C., Fernandes, I. R., Sebat, J., Muotri, A. R., et al. (2018). Modeling the interplay between neurons and astrocytes in autism using human induced pluripotent stem cells. Biol. Psychiatry 83, 569–578. doi: 10.1016/j.biopsych.2017.09.021
Sanai, N., Nguyen, T., Ihrie, R. A., Mirzadeh, Z., Tsai, H.-H., Wong, M., et al. (2011). Corridors of migrating neurons in the human brain and their decline during infancy. Nature 478, 382–386. doi: 10.1038/nature10487
Sanchez, O. F., Mendonca, A., Carneiro, A. D., and Yuan, C. L. (2017). Engineering recombinant protein sensors for quantifying histone acetylation. ACS Sens. 2, 426–435. doi: 10.1021/acssensors.7b00026
Sanchez, O. F., Mendonca, A., Min, A., Liu, J. C., and Yuan, C. L. (2019). Monitoring histone methylation (H3K9me3) changes in live cells. ACS Omega 4, 13250–13259. doi: 10.1021/acsomega.9b01413
Sapp, E., Kegel, K. B., Aronin, N., Hashikawa, T., Uchiyama, Y., Tohyama, K., et al. (2001). Early and progressive accumulation of reactive microglia in the Huntington disease brain. J. Neuropathol. Exp. Neurol. 60, 161–172. doi: 10.1093/jnen/60.2.161
Satterstrom, F. K., Kosmicki, J. A., Wang, J., Breen, M. S., De Rubeis, S., An, J. Y., et al. (2020). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568.e23–584.e23.
Shin, H. Y., Pfaff, K. L., Davidow, L. S., Sun, C., Uozumi, T., Yanagawa, F., et al. (2018). Using automated live cell imaging to reveal early changes during human motor neuron degeneration. eNeuro 5:ENEURO.0001-18.2018.
Song, Y., Jia, Z. C., Chen, J. Y., Hu, J. X., and Zhang, L. S. (2014). Toxic effects of atrazine on reproductive system of male rats. Biomed. Environ. Sci. 27, 281–288.
Suzuki, S., Akamatsu, W., Kisa, F., Sone, T., Ishikawa, K.-I., Kuzumaki, N., et al. (2017). Efficient induction of dopaminergic neuron differentiation from induced pluripotent stem cells reveals impaired mitophagy in PARK2 neurons. Biochem. Biophys. Res. Commun. 483, 88–93. doi: 10.1016/j.bbrc.2016.12.188
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Thijssen, E. H., La Joie, R., Wolf, A., Strom, A., Wang, P., Iaccarino, L., et al. (2020). Diagnostic value of plasma phosphorylated tau181 in Alzheimer’s disease and frontotemporal lobar degeneration. Nat. Med. 26, 387–397.
Thiruchelvam, M., Richfield, E. K., Goodman, B. M., Baggs, R. B., and Cory-Slechta, D. A. (2002). Developmental exposure to the pesticides paraquat and maneb and the Parkinson’s Disease phenotype. NeuroToxicology 23, 621–633. doi: 10.1016/s0161-813x(02)00092-x
Tsien, R. Y., Pozzan, T., and Rink, T. J. (1982). Calcium homeostasis in intact lymphocytes: cytoplasmic free calcium monitored with a new., intracellularly trapped fluorescent indicator. J. Cell Biol. 94, 325–334. doi: 10.1083/jcb.94.2.325
Vallerga, C. L., Zhang, F., Fowdar, J., McRae, A. F., Qi, T., Nabais, M. F., et al. (2020). Analysis of DNA methylation associates the cystine–glutamate antiporter SLC7A11 with risk of Parkinson’s disease. Nat. Commun. 11:1238.
Velasco, S., Kedaigle, A. J., Simmons, S. K., Nash, A., Rocha, M., Quadrato, G., et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. doi: 10.1038/s41586-019-1289-x
Wang, Y., Shi, M., Chung, K. A., Zabetian, C. P., Leverenz, J. B., Berg, D., et al. (2012). Phosphorylated α-synuclein in Parkinson’s disease. Sci. Transl. Med. 4:121ra120.
Wang, Z., Zheng, Y., Zhao, B., Zhang, Y., Liu, Z., Xu, J., et al. (2015). Human metabolic responses to chronic environmental polycyclic aromatic hydrocarbon exposure by a metabolomic approach. J. Proteome Res. 14, 2583–2593. doi: 10.1021/acs.jproteome.5b00134
Watras, C. J., Back, R. C., Halvorsen, S., Hudson, R. J. M., Morrison, K. A., and Wente, S. P. (1998). Bioaccumulation of mercury in pelagic freshwater food webs. Sci. Total Environ. 219, 183–208. doi: 10.1016/s0048-9697(98)00228-9
Weber, G. J., Sepúlveda, M. S., Peterson, S. M., Lewis, S. S., and Freeman, J. L. (2013). Transcriptome alterations following developmental atrazine exposure in zebrafish are associated with disruption of neuroendocrine and reproductive system function., cell cycle., and carcinogenesis. Toxicol. Sci. 132, 458–466. doi: 10.1093/toxsci/kft017
Wirbisky, S. E., Weber, G. J., Sepúlveda, M. S., Lin, T.-L., Jannasch, A. S., and Freeman, J. L. (2016). An embryonic atrazine exposure results in reproductive dysfunction in adult zebrafish and morphological alterations in their offspring. Sci. Rep. 6:21337.
Wirbisky-Hershberger, S. E., Sanchez, O. F., Horzmann, K. A., Thanki, D., Yuan, C. L., and Freeman, J. L. (2017). Atrazine exposure decreases the activity of DNMTs., global DNA methylation levels., and dnmt expression. Food Chem. Toxicol. 109, 727–734. doi: 10.1016/j.fct.2017.08.041
Workman, A. D., Charvet, C. J., Clancy, B., Darlington, R. B., and Finlay, B. L. (2013). Modeling transformations of neurodevelopmental sequences across Mammalian species. J. Neurosci. 33, 7368–7383. doi: 10.1523/jneurosci.5746-12.2013
Yang, H. H., St-Pierre, F., Sun, X., Ding, X., Lin, M. Z., and Clandinin, T. R. (2016). Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell 166, 245–257. doi: 10.1016/j.cell.2016.05.031
Keywords: IPSC, epigenetics, neurodegenerative disease, developmental exposure, organoid
Citation: Xie J, Wettschurack K and Yuan C (2020) Review: In vitro Cell Platform for Understanding Developmental Toxicity. Front. Genet. 11:623117. doi: 10.3389/fgene.2020.623117
Received: 29 October 2020; Accepted: 03 December 2020;
Published: 23 December 2020.
Edited by:
Yanqiang Li, Boston Children’s Hospital and Harvard Medical School, United StatesReviewed by:
Shuo Zhang, University of Pennsylvania, United StatesLu Li, Sun Yat-sen University, China
Copyright © 2020 Xie, Wettschurack and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongli Yuan, Y3l1YW5AcHVyZHVlLmVkdQ==
 Junkai Xie
Junkai Xie Kyle Wettschurack
Kyle Wettschurack Chongli Yuan
Chongli Yuan