- School of Biological Science and Technology, University of Jinan, Jinan, China
Phospholipase C (PLC) is one of the main hydrolytic enzymes in the metabolism of phosphoinositide and plays an important role in a variety of signal transduction processes responding to plant growth, development, and stress. Although the characteristics of many plant PLCs have been studied, PLC genes of maize have not been comprehensively identified. According to the study, five phosphatidylinositol-specific PLC (PI-PLC) and six non-specific PLC (NPC) genes were identified in maize. The PI-PLC and NPC genes of maize are conserved compared with homologous genes in other plants, especially in evolutionary relationship, protein sequences, conserved motifs, and gene structures. Transient expression of ZmPLC-GFP fusion protein in Arabidopsis protoplast cells showed that ZmPLCs are multi-localization. Analyses of transcription levels showed that ZmPLCs were significantly different under various different tissues and abiotic stresses. Association analysis shown that some ZmPLCs significantly associated with agronomic traits in 508 maize inbred lines. These results contribute to study the function of ZmPLCs and to provide good candidate targets for the yield and quality of superior maize cultivars.
Introduction
Phospholipids are important basic structural components of biological membranes and also as key signaling components responding to the plant development and various environmental stresses (Pokotylo et al., 2013). Phospholipids could be degraded into various products, such as diacylglycerol (DAG), phosphatidic acid (PA), free fatty acids (FFAs), and lysophospholipids (LPLs) by phospholipases, which include phospholipase C (PLC), phospholipase D (PLD), and phospholipase A (PLA) (Tuteja and Sopory, 2008; Hong et al., 2016). Among them, PLC is recognized as an important lipid hydrolase in animals and plants and has a profound effect on membrane lipid remodeling and intracellular signaling (Meldrum et al., 1991).
Based on different substrate affinities and cellular functions, plant PLCs have two different types: phosphatidylinositol-specific PLCs (PI-PLCs) and phosphatidylcholine-PLC (PC-PLC) (Kocourková et al., 2011; Pokotylo et al., 2014). PI-PLC hydrolyzes phosphoinositides to produce inositol 1,4,5-trisphosphate (IP3) and DAG, which may function as the second messengers (Berridge, 1987; Meldrum et al., 1991). IP3 could be quickly synthesized into hexakisphosphate (IP6) and trigger Ca2+ influx, while DAG could be phosphorylated by DAG kinase (DGK) and transformed into PAs (Wang et al., 2006). Unlike PI-PLC, PC-PLC, also known as non-specific PLC (NPC), preferentially hydrolyzes the common membrane phospholipids, for example, PC, phosphatidylethanolamine (PE), and phosphatidylserine (PS) (Kocourková et al., 2011).
In plants, two types of PLCs were composed of many gene members. For example, Arabidopsis contains nine PI-PLCs and six NPCs, whereas there are four PI-PLCs and five NPCs in rice, respectively (Nakamura et al., 2005; Zheng et al., 2012; Singh et al., 2013). In general, a typical plant PI-PLC enzyme structurally contains two domains at least, catalytic PI-PLC-X domain and PI-PLC-Y domain, which are necessary for PI-PLC to function as phosphoesterase (Hicks et al., 2008). Furthermore, the X and Y domains could form together a distorted triose phosphate isomerase (TIM) barrel structure, which contains the active-site residues (Chen et al., 2011). In addition, PI-PLCs have a C-terminal Ca2+/phospholipid binding C2 domain and an N-terminal EF hand domain involved in calcium binding (Chen et al., 2011). Some plant NPCs contain a putative signal peptide at the N-terminus, while all NPCs have a phosphoesterase domain, which is necessary for the function of esterase, such as NPCs and acid phosphatases (Wimalasekera et al., 2010). Generally, the phosphoesterase domain contains two highly conserved motifs, ENRSFDxxxG and TxPNR, and two other invariable motifs, DExxGxxDHV, and GxRVPxxxxxP (Pokotylo et al., 2013).
Members of the plant PLC family play important roles in various biological processes, for example, plant growth, development, and stress response. AtPI-PLC2 is required for female gametogenesis and embryo development in Arabidopsis, and loss of AtPI-PLC2 resulted in defective male and female gametophyte development (Li et al., 2015; Di Fino et al., 2017). Similarly, AtNPC2 and AtNPC6 are involved in gametophyte and embryo development and glycerolipid metabolism in the flower buds (Ngo et al., 2018), while AtNPC3 and AtNPC4 have an important regulatory role in root development (Wimalasekera et al., 2010). In addition, plant PLCs have also been confirmed to participate in a variety of tolerances to abiotic or biotic stresses. Arabidopsis AtPI-PLC members, except AtPI-PLC2, could be induced under various abiotic stresses such as salinity, drought, and cold (Tasma et al., 2008). Overexpression of Brassica napus PI-PLC2 in canola induces significant changes in the expression of stress-related genes and enhances drought tolerance (Georges et al., 2009). Overexpression of ZmPI-PLC1 enhanced the grain yield of maize under drought conditions, while suppression of ZmPI-PLC1 had an opposite effect (Wang et al., 2008). Knockout of AtNPC4 in Arabidopsis could increase sensitivity to salt stress in root elongation, seedling biomass, and seed germination, while AtNPC5 expression could be significantly upregulated under salt stress and positively regulate the development of lateral root under salt stress (Kocourková et al., 2011).
In this research, the PLC-encoding genes including PI-PLC and NPC were identified in the maize genome. The ZmPLCs have been analyzed in detail, including phylogenetic relationships, gene structures, conserved motifs, and subcellular localization. The transcription levels of the ZmPLCs were determined by qRT-PCR in different tissues and various abiotic stresses. In addition, the ZmPLC genes with quantitative trait loci (QTLs) associated with agronomic traits. Furthermore, the results presented herein provide valuable clues for studying the functions of ZmPLCs in response to the growth, development, and stress responses of maize.
Materials and Methods
Genome-Wide Identification of the PLC Genes in Maize
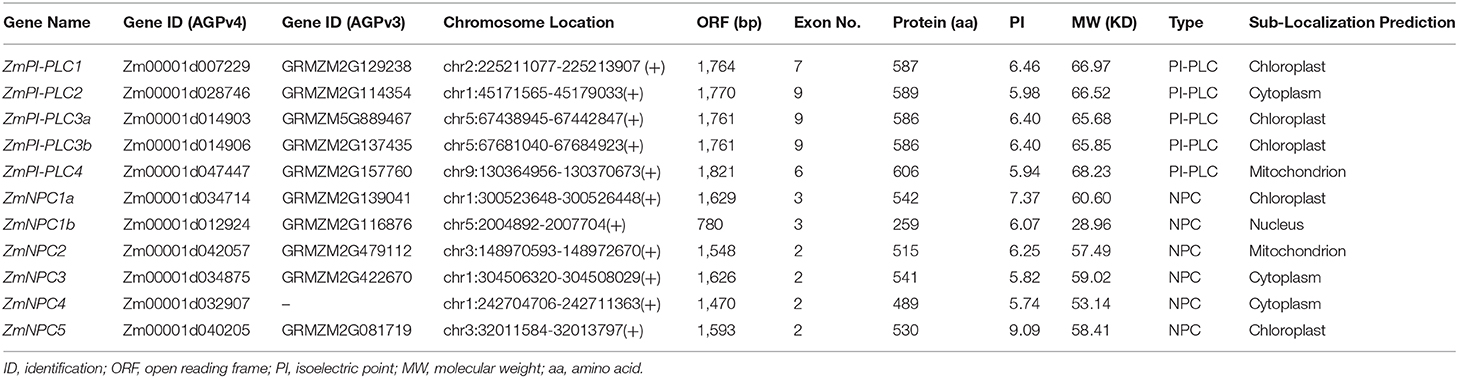
To identify putative PLC proteins, the hidden Markov models (HMMs) of the two characteristic domains of a PLC protein from PFam (http://pfam.sanger.ac.uk/), PI-PLC-X (PF00388), and PI-PLC-X (PF00387) were used as query sequences in local HMM-based searches, setting E-values < 0.01. In addition, to identify ZmPLC proteins that might have been missed through HMM searching, ZmPLC sequences were further identified using the previously reported Arabidopsis PLC protein sequences from the Maize Genome Database (https://www.maizegdb.org) and Phytozomev12.0 (http://www.phytozome.net) under the E-value cutoff 0.1. The matched sequences were subjected to SMART (http://smart.embl.de/) analyses to detect the presence and number of the PLC domain. Therefore, 11 independent ZmPLC genes were identified in maize. The chromosomal location image was mapped by MapInspect software. The ExPASy (https:// web.expasy.org/protparam/) was performed to calculate the molecular weight (MW) and the theoretical isoelectric point (PI).
Phylogenetic Analysis and Synteny Analysis
The multi-species PLC sequences, including 11 ZmPLCs from Zea mays, 15 AtPLCs from Arabidopsis thaliana, nine OsPLCs from Oryza sativa, 22 GhPLCs from Gossypium hirsutum, eight BdPLCs from Brachypodium distachyon, 10 SbPLCs from Sorghum bicolor, and 19 GmPLCs from Glycine max (Supplementary File 1), were constructed into a phylogenetic tree using the neighbor-joining (NJ) method in MEGA7.1 (Tamura et al., 2011). ZmPLCs were named basing on the phylogenetic relationship with AtPLCs and OsPLCs. Syntenic gene pairs among Zea mays and between Arabidopsis thaliana and Oryza sativa were identified using the TBtools (Chen et al., 2020).
Analyses of Gene Structures, cis-Acting Elements, and Motifs
The exon/intron structures of PLCs were analyzed by Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/). The genomic sequences 2,000 bp upstream of ZmPLCs predicted the cis-acting elements using PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/?tdsourcetag=s_pcqq_aiomsg). The conserved motifs were analyzed with MEME (http://meme.sdsc.edu/meme4_3_0/intro.html).
Subcellular Localization of ZmPLCs
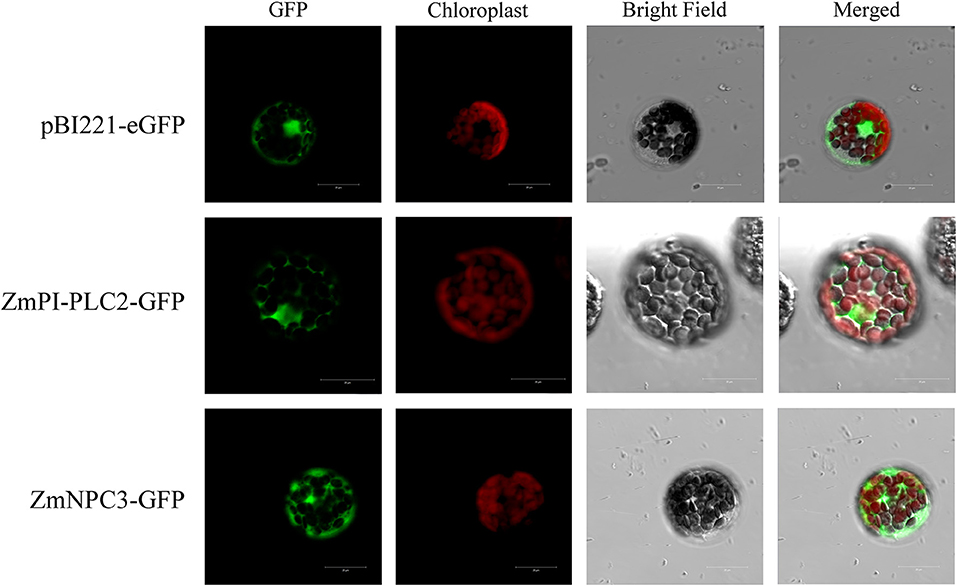
In order to confirm the subcellular localization, the coding sequences of selected ZmPLC genes were amplified using gene-specific primers. Then, ZmPLCs minus the stop codons were cloned and inserted into pBI221:eGFP, and the corresponding expression vectors were introduced into Arabidopsis protoplasts. The green fluorescent protein (GFP) fluorescence was excited with a confocal laser scanning microscope LSM 800 (Zeiss).
Plant Growth Conditions and Treatments
The maize inbred line W22 (from Huazhong Agricultural University) was used for all the experimental treatments. The seeds were sterilized with 70% ethanol for 5 min and then washed three times with sterile water. The seedlings were grown in a Hoagland solution in a greenhouse with a regime of 16 h light/8 h dark and at 28°C. When the maize seedlings were raised to the three-leaf stage, seedlings were selected for stress treatments according to Lin et al. (2014), including 20% PEG 6000 for drought stress, 200 mM NaCl for salt stress, 4°C for cold stress, and 20 μmol/L Cu2+ for heavy metal stress. The leaves were collected at different points in time after treatment. Each treatment consisted of three replicates. Adult plants were grown in the field, and then all organs and tissues were harvested. The root was harvested at the three-leaf stage; the stem and leaf were collected at five fully extended leaves; the silk, cob, and anther were harvested at 13 extended leaves; the kernel was harvested at 10 days after pollination (DAP). All materials were immediately frozen in liquid nitrogen after harvesting and stored at −80°C prior to RNA isolation.
Expression Analysis and Quantitative Real-Time PCR
For quantitative real-time PCR, total RNAs were extracted from various maize tissues with the RNAprep Pure Plant Kit (Tiangen, Beijing, China). According to supplier instructions, total cDNA was synthesized using PrimeScriptTM RT Reagent Kit with gDNA Eraser (Tiangen, Beijing, China). The Primer Premier 5.0 was used to design the primers for qRT-PCR (Supplementary Table 1), and the maize TUB-ribosylation factor was selected as an internal control. The reaction was performed on Bio-Rad CFX ConnectTM using SYBR-Green to detect gene expression levels. For all qRT-PCR analyses, triplicate biological samples were collected. Data were analyzed using Bio-Rad CFX Manager software.
Candidate Gene-Based Association Mapping of ZmPLC Family Members in Maize
Regional association tests between the single-nucleotide polymorphisms (SNPs) of candidate genes and the 17 traits agronomic, including plant height, ear height, tassel branch number, ear diameter, 100 grain weight, silking time, heading date, leaf number above ear, tassel main axis length, ear length, kernel width, cob weight, pollen shed, kernel weight, kernel number per row, ear leaf width, ear leaf length, were conducted in the 508 inbred lines (Fu et al., 2013). The genotype and phenotypes of the association panel were detected by two genotyping platforms, resulting in 550,000 high-quality SNPs (Yang et al., 2011; Li et al., 2013), and only SNPs within the range of 100 kb upstream and downstream of candidate genes were used. The association analysis was estimated using a mixed linear model (MLM) incorporated in TASSEL V5.0 (Bradbury et al., 2007). P ≤ 0.05 was considered the significance threshold.
Results
Identification and Characterization of PLCs in Maize
Eleven ZmPLCs, including five PI-PLC and six NPC sequences, were identified in the maize genome according to their domain structures (Supplementary Figure 1A). The ZmPI-PLC group contains five members: ZmPI-PLC1, ZmPI-PLC2, ZmPI-PLC3a, ZmPI-PLC3b, and ZmPI-PLC4. The ZmNPC group has six members: ZmNPC1a, ZmNPC1b, ZmNPC2, ZmNPC3, ZmNPC4, and ZmNPC5. Domain analysis showed that the five ZmPI-PLCs contained the catalytic PI-PLC-X, PI-PLC-Y domain, and Ca2+/phospholipid binding C2 domain, whereas an EF hand-like motif was found only in ZmPI-PLC4 (Supplementary Figure 1A). Six ZmNPCs had a phosphoesterase domain, which contains two highly conserved motifs, ENRSFDxxxG and TxPNR, and two invariable motifs, DExxGxxDHV and GxRVPxxxxxP (Supplementary Figure 1B). The gene identification (ID), gene name, open reading frame size, exon number, length, molecular weight, and isoelectric point of ZmPLCs were shown in Table 1. Especially, the number of amino acids (aa) in ZmPLCs of maize is comparable, with ZmPI-PLCs ranging from 586 to 606 aa and ZmNPCs ranging from 489 to 542 aa (ZmNPC1b is the only exception with 259 aa) (Table 1).
Phylogenetic Relationship, Gene Structure, and Conserved Motifs of ZmPLCs in Maize
To investigate the phylogenetic relationship of PLC among different species, a phylogenetic tree consisting of 11 ZmPLCs, 15 AtPLCs, nine OsPLCs, 19 GmPLCs, eight BdPLCs, 10 SbPLCs, and 22 GhPLCs was constructed using the NJ method (Figure 1). A total of 94 PLC protein sequences were classified into two subfamilies, named PI-PLC and NPC, based on the differences in the domain and phylogenetic relationships (Figure 1). The phylogenetic analysis showed that ZmPLCs shared high homology with those from other plants (Figure 1), especially ZmPI-PLC1, ZmPI-PLC2, ZmPI-PLC3a, and ZmPI-PLC3b made a separate small clade, while ZmPLC4 fell apart (Figure 1). While ZmNPC1a and ZmNPC1b; ZmNPC2; ZmNPC5; ZmNPC3 and ZmNPC4 made a separate small clade (Figure 1).
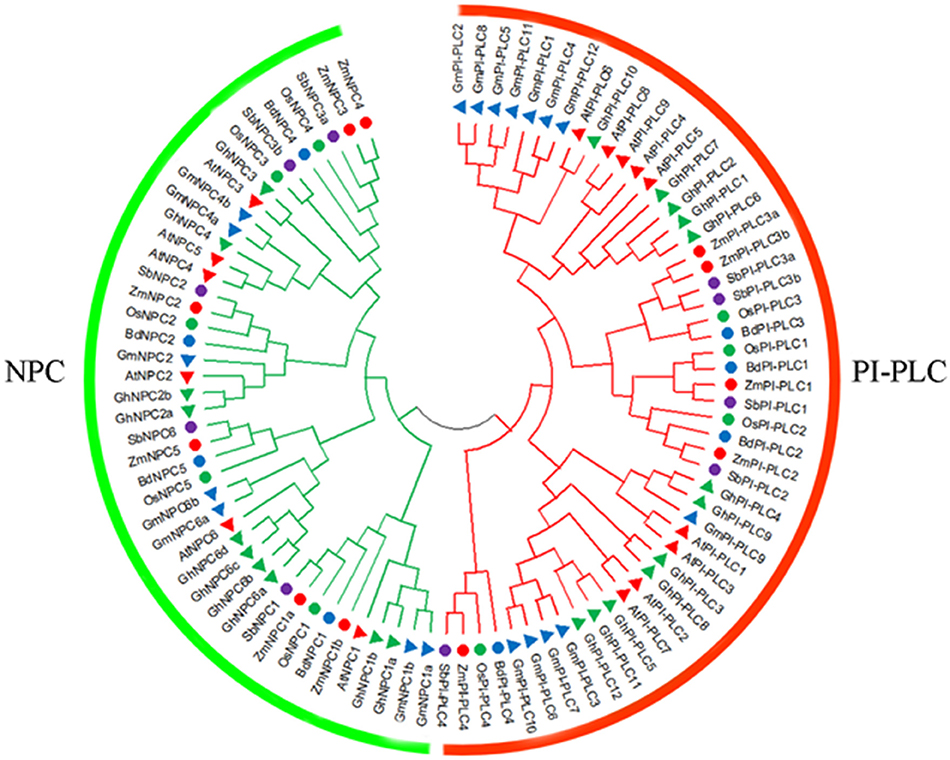
Figure 1. Phylogenetic analysis of the PLC family. The phylogenetic tree was made by MEGA 7.1 software using the neighbor-joining (NJ) method with bootstrapping analysis. The analysis involved 94 amino acid sequences from various plants, including Arabidopsis thaliana (At), Glycine max (Gm), Oryza sativa (Os), Sorghum bicolor (Sb), Gossypium hirsutum (Gh), Brachypodium distachyon (Bd).
Corresponding to the evolutionary relationship, analysis of the exon/intron structures of the PLC genes revealed that these genes are also divided into two different types: PI-PLC with an exon-rich clade (≥5 exons per gene) generally containing 5–10 exons, while NPC with an exon-poor clade (≤5 exons per gene) containing 1–5 exons (Figure 2). It is worth noting that a similar exon/intron pattern exists in each clade, for instance, most NPC1 genes contain three exons, and the vast majority of PI-PLC1 genes have seven exons (Figure 2).

Figure 2. Phylogenetic relationships, gene structures, and conserved protein motifs of maize PLC family. Green boxes indicate exons, and black lines indicate introns. The motifs, numbered 1–10, are displayed in different colored boxes. The length of the protein can be estimated using the scale at the bottom.
To further study the characteristic regions of the PLC proteins, a total of 10 conserved motifs were identified in PLCs using the online MEME tool (Figure 2). Interestingly, the members of the same clade usually had similar structures and lengths in terms of domain. The group PI-PLC members had the conserved motifs 1, 2, 4, 5, 7, and 9, which are PI-PLC-X, PI-PLC-Y, and Ca2+/phospholipid binding C2 domain. While the conserved motifs 3, 6, 8, 9, and 10, which were annotated as the phosphoesterase domain, were found in group NPC family (Figure 2).
Chromosomal Location of the ZmPLCs and Synteny Analysis of PLCs Among Several Different Species
According to the data of the gene locus, 11 ZmPLC genes were found in five different chromosomes (Table 1, Figure 3A). ZmPI-PLC2, ZmNPC1a, ZmNPC3, and ZmNPC4 are localized on chromosome 1, ZmPI-PLC1 and ZmPI-PLC4 are distributed on chromosomes 2 or 9, and ZmNPC2 and ZmNPC5 are located on chromosomes 3, while chromosomes 5 contained ZmNPC1b, ZmPI-PLC3a, and ZmPI-PLC3b (Figure 3A).
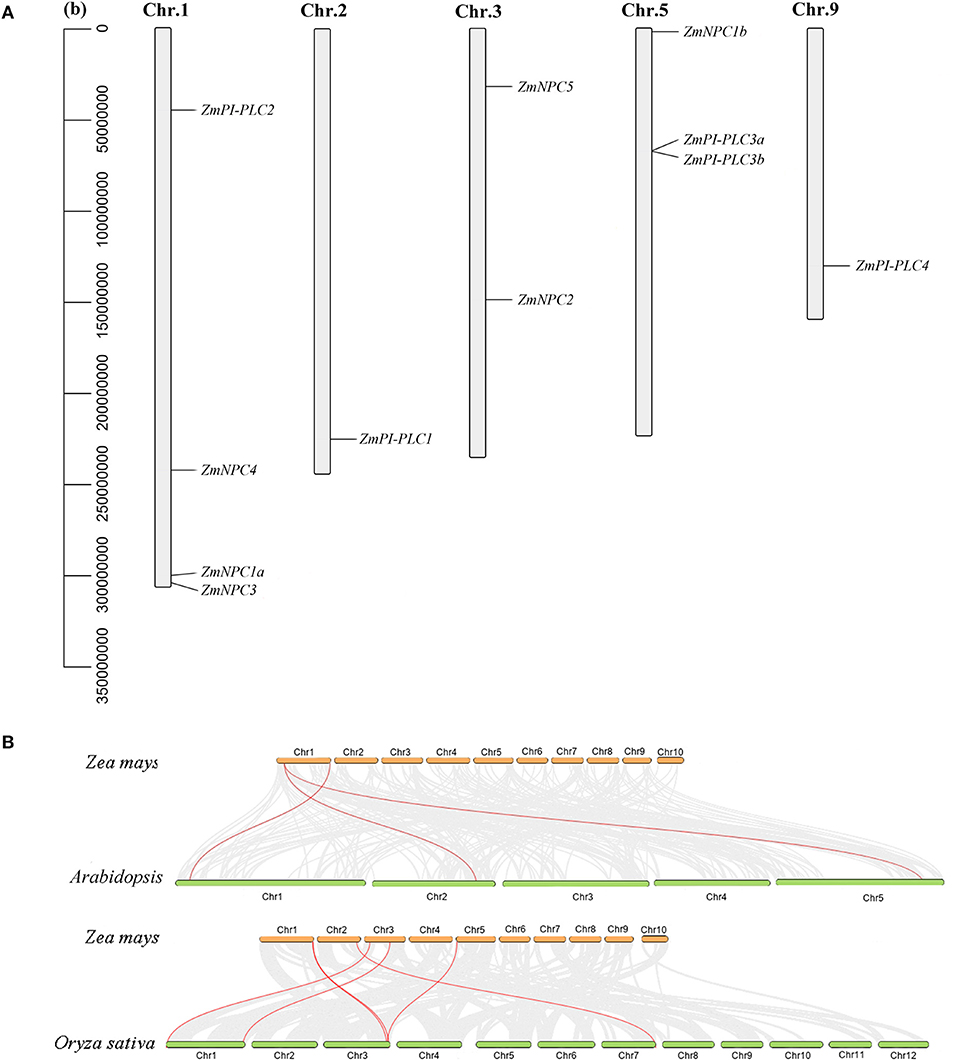
Figure 3. The chromosomal distribution of ZmPLC genes (A) and synteny analysis between ZmPLCs and PLCs from Arabidopsis or rice (B). The number of the chromosome is shown on each chromosome. Gray lines in the background indicate the collinear blocks within maize and other plant genomes, while the red lines highlight the syntenic PLC gene pairs.
To understand more about the phylogeny of ZmPLC genes family, syntenic analysis was performed between maize and two other plant species, including Arabidopsis thaliana and Oryza sativa (Figure 3B). There were three ZmPLC genes that were synchronized with those in Arabidopsis thaliana (Figure 3B, Supplementary Table 2). The comparative syntenic maps of maize associated with rice were analyzed further, and six out of 11 ZmPLC genes had collinear genes in rice (Figure 3B, Supplementary Table 2), indicating that these genes may be derived from a common ancestor.
Analysis of cis-Elements in the Promoters of ZmPLCs
To study the expression regulation patterns and the potential function of ZmPLCs, putative cis-elements on promoter regions were searched in the Plant CARE database. Cis-elements related to developmental processes, such as the meristem expression (CAT-box), were found in the promoter regions of ZmPI-PLC4, ZmNPC1a, ZmNPC1b, ZmNPC2, ZmNPC3, and ZmNPC5, suggesting that ZmPLC genes play important roles in differentiation (Figure 4). The hormone-responsive elements, including abscisic acid (ABA) responsive elements (ABREs), MeJA-responsive elements (TGACG-motif and CGTCA-motif), salicylic acid-responsive element (TCA-element and SARE), auxin-responsive element (TGA-element and AuxRR-core), and gibberellin-responsive element (TATC-box, P-box, and GARE-motif), were also found in some ZmPLC gene promoters (Figure 4), showing that ZmPLC genes may play a role in growth and development. Furthermore, stress-responsive elements, such as low-temperature responsive (LTR) element, drought-responsive element, anaerobic induction responsive element (ARE), and defense and stress responsive element (AT-rich and TC-rich repeats), were observed in some promoters of ZmPLC genes. For example, LTR elements were detected in ZmPI-PLC1/2/3a/3b and ZmNPC3/4/5 gene promoters (Figure 4), while drought-responsive elements were found in the promoters of ZmPI-PLC1/2 and ZmNPC1a/1b/5 (Figure 4).
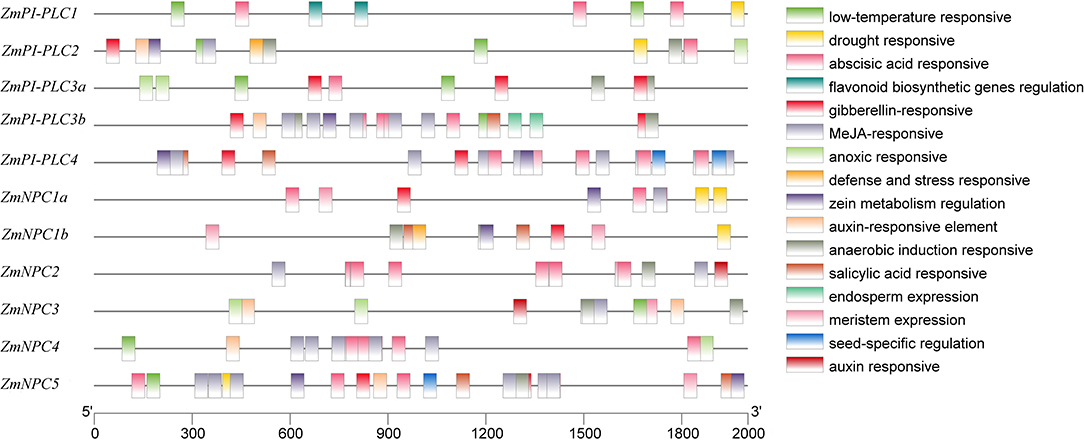
Figure 4. Putative regulatory cis-elements in ZmPLC family gene promoters. Different cis-elements are represented with different color box.
Expression Profiles of ZmPLCs in Different Tissues and Developmental Stages of Maize
To study the roles of ZmPLCs in growth and development of maize, qRT-PCR was used to determine the expression profiles of the 11 ZmPLC mRNAs in different tissues, including root, stem, leaf, silk, immature cob, anther, tassel, kernel (10 DAP) (Figure 5). The results showed that ZmPLCs were significantly different under various different tissues. For example, ZmPI-PLC1 was more highly expressed in silk; ZmPI-PLC2 and ZmNPC1a were more highly expressed in the leaf than in other tissues; ZmPI-PLC3a and ZmPI-PLC3b had similar expression patterns with relatively lower expressions in anther, while ZmNPC2, ZmNPC3, and ZmNPC4 had relatively high expressions in anther (Figure 5). Except for the relatively high expression in root, ZmPI-PLC4 and ZmNPC1b showed low expression in others organs (Figure 5), and ZmNPC5 was relatively highly expressed in 10 DAP of kernel (Figure 5). These results indicated that ZmPLCs may have important functions in different developmental stages and tissues of maize.
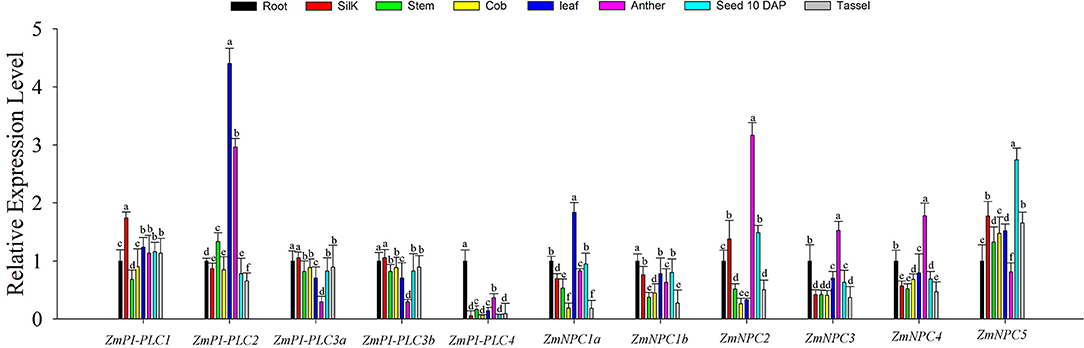
Figure 5. Tissue-specific expression patterns of the 11 ZmPLC genes in maize root, stem, leaf, silk, immature cob, anther, and kernel [10 days after pollination (DAP)] by using qRT-PCR. The expression levels were normalized to that of TUB. Bars represent SE (n = 3). The entire experiment was repeated three times. Different letters indicate values that departed significantly from those of root.
Expression Profiling of ZmPLCs in Response to Various Stresses
To verify the expression changes of ZmPLCs under stresses, the expression profiles were examined by qRT-PCR under different abiotic stress treatments, including 20% PEG6000, 200 mM NaCl, 4°C, and 20 μmol/L Cu2+ (Figure 6). The results show that ZmPLC genes, except ZmNPC4 and ZmNPC5, were induced by 20% PEG 6000, peaking at 6 or 12 h after treatments, and then reduced to the lowest level at 48 h (Figure 6A). ZmPI-PLC1, ZmPI-PLC3a, ZmNPC1a, ZmNPC1b, ZmNPC4, and ZmNPC5 were rapidly induced by high salinity, and ZmPI-PLC3b, ZmPI-PLC4, and ZmNPC3 mRNAs were almost unaffected; however, the expressions of ZmPI-PLC2 and ZmNPC2 were suppressed (Figure 6B). The expressions of ZmPI-PLC1, ZmPI-PLC2, ZmNPC1b, ZmNPC2, ZmNPC3, ZmNPC4, and ZmNPC5 were decreased after cold treatment and then reached the lower level at 12 h, while ZmPI-PLC3a, ZmPI-PLC3b, and ZmNPC1a mRNAs were induced with cold treatment, then still relatively high level at 12 h (Figure 6C). Under 20-μmol/L Cu2+ treatments, ZmPI-PLC3a, ZmPI-PLC3b, ZmPI-PLC4, ZmNPC2, and ZmNPC3 mRNAs were suppressed within 48 h, while ZmPI-PLC1, ZmPI-PLC2, ZmNPC1a, ZmNPC1b, ZmNPC2, and ZmNPC5 were induced within 48 h and peaked at 3 or 6 h after treatment (Figure 6D). These results show that ZmPLCs genes might play important roles in response to various abiotic stresses.
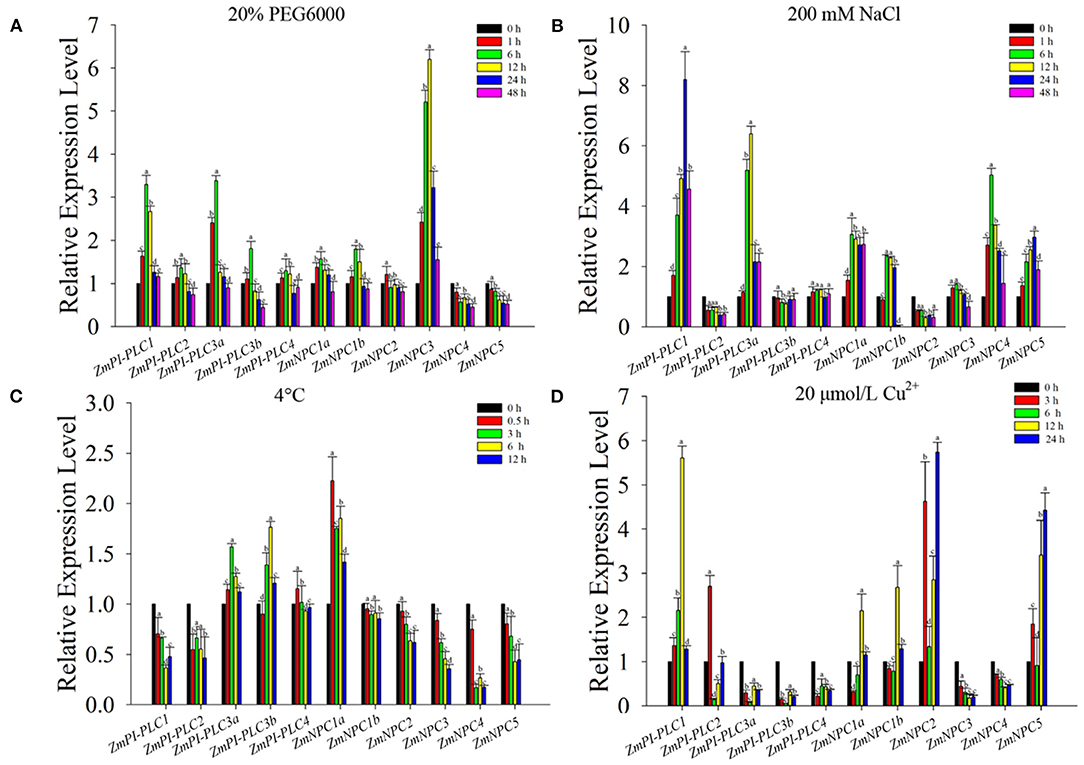
Figure 6. Quantitative real-time PCR analysis of ZmPLCs expression under stress treatments: (A) 20% PEG 6000, (B) 200 mM NaCl, (C) 4°C, (D) 20 μmol/L Cu2+. The expression levels were normalized to that of TUB. Bars represent SE (n = 3). The entire experiment was repeated three times. Different letters indicate values that departed significantly from those of without stress.
Subcellular Localization of ZmPLC Proteins
Previously, it was reported that the substrates of PLCs, PI4P and PI(4,5)P2, are located in the plasma membrane (Munnik et al., 1998), so it is speculated that the subcellular localization of PLCs should also be in the plasma membrane; however, the sub-localization prediction shows that ZmPLCs may be multi-localized (Table 1). In order to confirm the subcellular localization, two randomly selected ZmPLCs, ZmPI-PLC2 and ZmNPC3, were transiently expressed in A. thaliana mesophyll protoplasts to analyze subcellular localization by fusing to the N-terminus of GFP. In Arabidopsis leaf protoplasts, the fluorescence of the vector control was distributed throughout the nucleus and cytoplasm (Figure 7); however, the signals of ZmPI-PLC2 and ZmNPC3 were only observed in the cytoplasm (Figure 7), showing that ZmPI-PLC2 and ZmNPC3 are located in the cytosol.
Regional Association Mapping of ZmPLCs for Agronomic Traits in Maize
To understand the possible functions of the ZmPLCs, an analysis of agronomy -related trait QTLs from 508 maize inbred lines was performed. There were significant correlations between all 11 ZmPLC genes and more than one agronomic trait at the P ≤ 0.05 level, such as kerner number per row, 100 grain weight, cob weight, and so on (Figure 8). At P ≤ 0.01, 10 ZmPLC genes were identified to be related to some important agronomic traits (Supplementary Table 3). For example, ZmPI-PLC1 was significantly correlated with ear height, ear leaf length, ear length, kerner number per row, plant height, pollen shed, and tassel branch number (Supplementary Table 3, Figure 8A); ZmPI-PLC4 significantly affected heading date, pollen shed, 100 grain weight, silking time, ear leaf width, cob weight, kerner number per row, and so on (Supplementary Table 3, Figure 8E); ZmNPC1a significantly affected ear leaf length, kerner number per row, pollen shed, and plant height (Supplementary Table 3, Figure 8F).
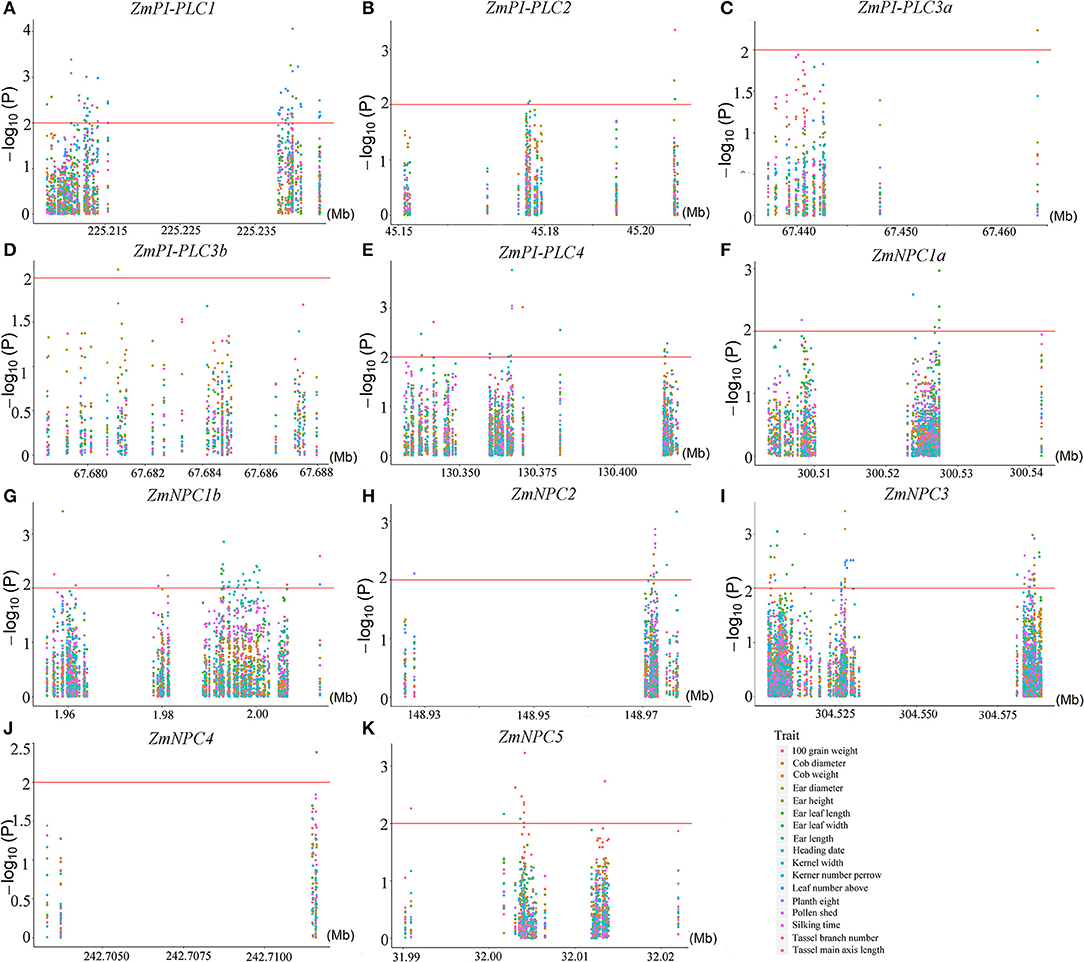
Figure 8. Regional correlation analysis between ZmPLC genes with the agronomic traits in maize. (A–K) show the correlation between ZmPLC genes with the agronomic traits, respectively. Local Manhattan plot (top) and linkage disequilibrium heatmap (bottom) surrounding the SNPs for agronomic traits on the range of 100 kb upstream/downstream of ZmPLC genes. An logarithm of the odds (LOD) > 2 indicates that a SNP is significantly related to kernel-related traits.
Discussion
ZmPLC Gene Identification and Evolutionary Analysis in Maize
At present, PLC genes and their functions in many plants have been studied and reported, such as Arabidopsis, rice, and soybean (Wang et al., 2015; Zhang et al., 2018). It was reported that nine, 15, and 19 PLC genes were found in rice (Singh et al., 2013), Arabidopsis (Zheng et al., 2012), and cotton (Zhang et al., 2018), respectively. In this study, 11 ZmPLCs, five PI-PLCs, and six NPC members were identified according to the domain structure and phylogenetic analysis (Supplementary Figure 1, Figure 2). Based on the phylogenetic analysis, although PLCs from closely related species made a phylogenetic clade due to different genetic relationships (Figure 1), maize PI-PLCs made a phylogenetic clade with dicots PI-PLCs with very low bootstrap; however, maize NPCs fall in the same clade with dicot NPCs with a relative high bootstrap (Figure 1), suggesting that PLCs from different species diversify during the course of evolution even after being originated from a common ancestor. In addition, there are generally more PLCs in dicots than monocots (Zheng et al., 2012; Zhang et al., 2018), especially PI-PLC, which indicates that the natural selection of the PLC genes is a variant for different plant species. Domain and motif analysis of the PI-PLC protein sequences in maize with homologs from other plants contained the conserved and typical PI-PLC-X and PI-PLC-Y catalytic domains and C2 domain (Supplementary Figure 1B, Figure 2). It is worth noting that an EF hand-like motif was only found in ZmPI-PLC4 (Supplementary Figure 1A). Similarly, all the NPC members harbored a highly conserved phosphoesterase domain (Supplementary Figure 1A). The gene structure of PLCs from different species could be distinguished into two types: exon-rich (PI-PLC) and exon-poor (NPC) (Figure 2). Genes in the same branch usually have similar exon–intron structures, whereas the gene structures differed markedly among the different branches (Figure 2). Furthermore, there are similar results in the motif compositions of the PLC protein (Figure 2). Synteny analysis showed that the number of PLC homologous genes between maize and rice were more than those between maize and Arabidopsis, which indicating that PLC has a higher homology among related species (Figure 3). These findings indicated that highly conserved PLC from different plants might have a similar function in the evolution of plants.
Functional Analysis of the ZmPLCs Response to the Development and Abiotic Stress in Maize
The cis-regulatory elements in the promoter play an important role in regulating the expression patterns of the genes (Vedel and Scotti, 2011). According to analysis of the cis-acting elements in ZmPLC promoter region, the ZmPLC promoters contain various element responses to plant development and stresses, for example, light-responsive elements, ABA-response elements, MeJA-responsive elements, LTR element, and so on (Figure 4). These results mean that ZmPLC genes could be involved not only in the regulation of the growth and development of maize but also in various stress responses.
The expression patterns of PLC genes have already been studied in different tissues and developments of many plants, such as Arabidopsis, rice, soybean, and so on. In this study, the expression profiles of ZmPLC genes were validated using qRT-PCR under different tissues. All ZmPLC genes were expressed in our tested tissues, while the expression abundance of different ZmPLC genes is significantly different in tissues (Figure 5). For example, the transcript levels of ZmPI-PLC2 were relatively high in all organs, especially leaf and anther (Figure 5). Interestingly, AtPI-PLC2, involved in reproductive organ development, was also highly expressed in leaves, stems, roots, and flowers (Tasma et al., 2008; Li et al., 2015). OsPLC genes from rice are also expressed differentially during reproductive developmental phases including stages of panicle and seed development (Singh et al., 2013). Furthermore, the regional association analysis between 11 ZmPLCs and 17 agronomic traits in maize indicated that some ZmPLC genes are significantly correlated with many development-related traits (Figure 8). These results suggested that ZmPLC genes have important functions in the regulation of growth and development in maize and could act as important candidate genes to improve maize agronomic traits for breeding.
Many previous studies have reported that PLC members are involved in various abiotic stress-triggered signaling transductions in many plant species. For example, AtPI-PLC3, AtPI-PLC9, and AtNPC1 genes play an important function in regulating heat tolerance (Gao et al., 2014; Krčková et al., 2015), whereas AtNPC4 and AtNPC5 genes could respond to salt stress (Kocourková et al., 2011; Peters et al., 2014). OsPI-PLC4 plays a positive role in osmotic stress response (Deng et al., 2019). Overexpression of maize PI-PLC1 could enhance drought tolerance of transgenic plants (Wang et al., 2008). In this study, the expression of ZmPLC genes showed different changes under different stresses; however, they presented their own characteristics (Figure 6). For example, all maize PI-PLC genes were induced by osmotic treatments, while only ZmPI-PLC1 and ZmPI-PLC3a were rapidly induced by high salinity (Figures 6A,B). ZmPI-PLC3a and ZmPI-PLC3b were upregulated by cold treatment and suppressed by copper ion treatment (Figures 6C,D). ZmNPC4 and ZmNPC5 were downregulated by osmotic treatments; however, they were rapidly induced by high salinity (Figure 6B). The different expression changes of ZmPLCs under different stresses may be related to the cis-acting elements in their promoters; for example, the cis-regulatory elements including MBS (MYB binding site), ABRE, and defense and stress responsive element (AT-rich and TC-rich repeats) are known to regulate various stress responses (Abe et al., 2003; Narusaka et al., 2003). In this study, the LTR could be detected in ZmPI-PLC1/2/3a/3b and ZmNPC3/4/5 gene promoters (Figure 4), and ZmPI-LC3a and ZmPI-PLC3b mRNAs were induced by cold treatments.
The substrates of PLCs, phosphoinositides, PC, or PE, are mainly distributed in the plasma membrane (Munnik et al., 1998), so it is speculated that the subcellular localization of PLCs should also be in the plasma membrane. However, the sub-localization from many plants showed that PLCs maybe multi-localized. For example, Arabidopsis AtPLC9 and soybean GmPLC10-GFP fluorescence were located in the plasma membrane (Zheng et al., 2012), OsPLC1 and OsPLC4 were distributed throughout the cytoplasm and nucleus, while OsNPC3 protein might be localized in the chloroplast/plastids (Singh et al., 2013). DAG derived from PI-PLC or NPC activities can be phosphorylated to PA by DGK, while DAG could act as a substrate to produce various lipid species and also significantly affect properties of cell membranes as sites of crucial cell activity (Darwish et al., 2009; Hong et al., 2016). On the other hand, PI-PLC hydrolyzes PtdIns(4,5)P2 into Ins(1,4,5)P3, and Ins(1,4,5)P3 is synthesized into InsP6, which could bind to IP6 receptors and lead to release of Ca2+ (Hong et al., 2016). Ca2+ and PA could act as a signaling molecule to play vital roles in various signaling pathways such as plant development, hormone signaling, and abiotic or biotic stresses to produce favorable response to plants (Xue et al., 2007). There is a possibility that the diverse localization of plant PLCs implies that they might take part in different cellular processes during development and abiotic stress.
Conclusions
In this study, 11 ZmPLCs were identified by genome-wide analysis. The results of evolution, gene structures, and motifs of PLCs indicated that ZmPLCs were highly conserved compared with their homologous genes from other plants. Additionally, the cis-elements, expression profiles, and association analysis between ZmPLC genes and agronomic traits for ZmPLC genes were also analyzed, which showed that ZmPLC genes may have important functions in regulating development and various stresses. Taken together, these results provide useful information for further study of the roles of ZmPLCs in plant development and environmental stress conditions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JZ and HL wrote the paper. JZ, YZ, and JL performed the experiments. JZ contributed to the data analysis. All authors read and approved the manuscript.
Funding
This study was funded by grants from the National Natural Science Foundation of China (31901497) and the scientific research leaders studio of Jinan (2019GXRC052).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to Dr. Jianbing Yan (Huazhong Agricultural University, China) for providing maize inbred line W22.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.611414/full#supplementary-material
Supplementary File 1. List of the PLC sequences from multi-species in this study.
Supplementary Figure 1. Schematic domain structures of ZmPLC (A) and Multiple sequence alignments of ZmPLCs (B). Protein sequences of ZmPLC were aligned by Bioedit. The conserved and characteristic domains and sequences of PI-PLCs and NPCs have been marked with colored box and red underline, respectively.
Supplementary Table 1. Primers and their sequences used in this study.
Supplementary Table 2. Synteny blocks of PLC genes between maize and two representative plant species.
Supplementary Table 3. The ZmPLC genes list associated with agronomic traits in maize. The red mark indicates the significant value after Bonferroni correction (p < 0.05/n).
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Berridge, M. J. (1987). Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu. Rev. Biochem. 56, 159–193. doi: 10.1146/annurev.bi.56.070187.001111
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Snyder, C. L., Greer, M. S., and Weselake, R. J. (2011). Biology and biochemistry of plant phospholipases. Crit. Rev. Plant Sci. 30, 239–258. doi: 10.1080/07352689.2011.572033
Darwish, E., Testerink, C., Khalil, M., El-Shihy, O., and Munnik, T. (2009). Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol. 50, 986–997. doi: 10.1093/pcp/pcp051
Deng, X. J., Yuan, S., Cao, H. S., Lam, S. M., Shui, G. H., Hong, Y. Y., et al. (2019). Phosphatidylinositol-hydrolyzing phospholipase C4 modulates rice response to salt and drought. Plant Cell Environ. 42, 536–548. doi: 10.1111/pce.13437
Di Fino, L. M., D'Ambrosio, J. M., Tejos, R., Wijk, R., Lamattina, L., Munnik, T., et al. (2017). Arabidopsis phosphatidylinositol-phospholipase C2 (PLC2) is required for female gametogenesis and embryo development. Planta 245, 717–728. doi: 10.1007/s00425-016-2634-z
Fu, J., Cheng, Y., Linghu, J., Yang, X., Kang, L., Zhang, Z., et al. (2013). RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun. 4:2832. doi: 10.1038/ncomms3832
Gao, K., Liu, Y. L., Li, B., Zhou, R. G., Sun, D. Y., and Zheng, S. Z. (2014). Arabidopsis thaliana phosphoinositide-specific phospholipase C isoform 3 (AtPLC3) and AtPLC9 have an additive effect on thermotolerance. Plant Cell Physiol. 55, 1873–1883. doi: 10.1093/pcp/pcu116
Georges, F., Das, S., Ray, H., Bock, C., Nokhrina, K., Kolla, V. A., et al. (2009). Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant Cell Environ. 32, 1664–1681. doi: 10.1111/j.1365-3040.2009.02027.x
Hicks, S. N., Jezyk, M. R., Gershburg, S., Seifert, J. P., Harden, T. K., and Sondek, J. (2008). General and versatile autoinhibition of PLC isozymes. Mol. Cell 31, 383–394. doi: 10.1016/j.molcel.2008.06.018
Hong, Y., Zhao, J., Guo, L., Kim, S. C., Deng, X., Wang, G., et al. (2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62, 55–74. doi: 10.1016/j.plipres.2016.01.002
Kocourková, D., Krcková, Z., Pejchar, P., Veselková, S., Valentová, O., Wimalasekera, R., et al. (2011). The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J. Exp. Bot. 62, 3753–3763. doi: 10.1093/jxb/err039
Krčková, Z., Brouzdová, J., Daněk, M., Kocourková, D., Rainteau, D., Ruelland, E., et al. (2015). Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress. Front. Plant Sci. 6:928. doi: 10.3389/fpls.2015.00928
Li, H., Peng, Z., Yang, X., Wang, W., Fu, J., Wang, J., et al. (2013). Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, 43–50. doi: 10.1038/ng.2484
Li, L., He, Y., Wang, Y., Zhao, S., Chen, X., Ye, T., et al. (2015). Arabidopsis PLC2 is involved in auxin modulated reproductive development. Plant J. 84, 504–515. doi: 10.1111/tpj.13016
Lin, Y., Zhang, C., Lan, H., Gao, S., Liu, H., Liu, J., et al. (2014). Validation of potential reference genes for qpcr in maize across abiotic stresses, hormone treatments, and tissue types. PLoS ONE 9:e95445. doi: 10.1371/journal.pone.0095445
Meldrum, E., Parker, P. J., and Carozzi, A. (1991). The PtdIns-PLC superfamily and signal transduction. Biochim. Biophys. Acta. 1092, 49–71. doi: 10.1016/0167-4889(91)90177-Y
Munnik, T., Irvine, R. F., and Musgrave, A. (1998). Phospholipid signaling in plants. Biochim. Biophys. Acta 1389, 222–272. doi: 10.1016/S0005-2760(97)00158-6
Nakamura, Y., Awai, K., Masuda, T., Yoshioka, Y., Takamiya, K. I., and Ohta, H. (2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280, 7469–7476. doi: 10.1074/jbc.M408799200
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Ngo, A. H., Lin, Y. C., Liu, Y. C., Gutbrod, K., Peisker, H., Dörmann, P., et al. (2018). A pair of nonspecific phospholipases C, NPC2 and NPC6, are involved in gametophyte development and glycerol lipid metabolism in Arabidopsis. New Phytol. 219, 163–175. doi: 10.1111/nph.15147
Peters, C., Kim, S. C., Devaiah, S., Li, M., and Wang, X. (2014). Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 37, 2002–2013. doi: 10.1111/pce.12334
Pokotylo, I., Kolesnikov, Y., Kravets, V., Zachowski, A., and Ruelland, E. (2014). Plant phosphoinositide-dependent phospholipases C: variations around a canonical theme. Biochimie 96, 144–157. doi: 10.1016/j.biochi.2013.07.004
Pokotylo, I., Pejchar, P., Potocký, M., Kocourková, D., Krčková, Z., Ruelland, E., et al. (2013). The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog. Lipid. Res. 52, 62–79. doi: 10.1016/j.plipres.2012.09.001
Singh, A., Kanwar, P., Pandey, A., Tyagi, A. K., Sopory, S. K., Kapoor, S., et al. (2013). Comprehensive genomic analysis and expression profiling of phospholipase C gene family during abiotic stresses and development in rice. PLoS ONE 8:e62494. doi: 10.1371/journal.pone.0062494
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tasma, I. M., Brendel, V., Whitham, S. A., and Bhattacharyya, M. K. (2008). Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 46, 627–637. doi: 10.1016/j.plaphy.2008.04.015
Tuteja, N., and Sopory, S. K. (2008). Plant signaling in stress: G-protein coupled receptors,heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal. Behav. 3, 79–86. doi: 10.4161/psb.3.2.5303
Vedel, V., and Scotti, I. (2011). Promoting the promoter. Plant Sci. 180, 182–189. doi: 10.1016/j.plantsci.2010.09.009
Wang, C. R., Yang, A. F., Yue, G. D., Gao, Q., Yin, H. Y., and Zhang, J. R. (2008). Enhanced expression of phospholipase C 1 (ZmPLC1) improves drought tolerance in transgenic maize. Planta 227, 1127–1140. doi: 10.1007/s00425-007-0686-9
Wang, F. W., Deng, Y., Zhou, Y. G., Dong, J. Y., Chen, H., Dong, Y. Y., et al. (2015). Genome-wide analysis and expression profiling of the phospholipase C gene family in soybean (Glycine max). PLoS ONE 10:e0138467. doi: 10.1371/journal.pone.0138467
Wang, X. M., Devaiah, S. P., Zhang, W. H., and Welti, R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid. Res. 45, 250–278. doi: 10.1016/j.plipres.2006.01.005
Wimalasekera, R., Pejchar, P., Holk, A., Martinec, J., and Scherer, G. F. (2010). Plant phosphatidylcholine-hydrolyzing phospholipases C NPC3 and NPC4 with roles in root development and brassinolide signaling in Arabidopsis thaliana. Mol. Plant 3, 610–625. doi: 10.1093/mp/ssq005
Xue, H. W., Chen, X., and Li, G. (2007). Involvement of phospholipid signaling in plant growth and hormone effects. Curr. Opin. Plant Biol. 10, 483–489. doi: 10.1016/j.pbi.2007.07.003
Yang, X. H., Gao, S. B., Xu, S. T., Zhang, Z. X., Prasanna, B. M., Li, L., et al. (2011). Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 28, 511–526. doi: 10.1007/s11032-010-9500-7
Zhang, B., Wang, Y. M., and Liu, J. Y. (2018). Genome-wide identification and characterization of phospholipase C gene family in cotton (Gossypium spp.). Sci. China Life Sci. 61, 88–99. doi: 10.1007/s11427-017-9053-y
Keywords: maize, PLC, genome-wide, expression pattern, stress, association analysis
Citation: Zhu J, Zhou Y, Li J and Li H (2021) Genome-Wide Investigation of the Phospholipase C Gene Family in Zea mays. Front. Genet. 11:611414. doi: 10.3389/fgene.2020.611414
Received: 29 September 2020; Accepted: 26 November 2020;
Published: 12 January 2021.
Edited by:
Jian Ma, Sichuan Agricultural University, ChinaReviewed by:
Jun Ma, China Agricultural University, ChinaXiao-Wei Zhang, Chinese Academy of Agricultural Sciences, China
Copyright © 2021 Zhu, Zhou, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, YmlvX2xpaEB1am4uZWR1LmNu
†These authors have contributed equally to this work
 Jiantang Zhu
Jiantang Zhu Yuanyuan Zhou
Yuanyuan Zhou Jiale Li
Jiale Li Hui Li
Hui Li