- 1Biocontrol Engineering Research Center of Plant Disease and Pest, Biocontrol Engineering Research Center of Crop Disease and Pest, Yunnan University, Kunming, China
- 2School of Life Sciences, Yunnan University, Kunming, China
- 3School of Ecology and Environmental Science, Yunnan University, Kunming, China
- 4Wenshan Branch of Yunnan Tobacco Company, Wenshan, China
Melatonin plays key roles in development and confers stress tolerance to plants. Serotonin N-acetyltransferase (SNAT) is either the enzyme involved in the last step or the penultimate enzyme of phytomelatonin biosynthesis. To date, SNAT genes have not been characterized in tobacco (Nicotiana tabacum), an economically important plant species. The sequence of the Acetyltransf_7 conserved domain was used as a query sequence, and 12 NtSNAT candidate genes were in turn identified in the genome of tobacco. These NtSNATs could be divided into two groups based on the phylogenetic tree. NtSNAT1 and NtSNAT2 clustered together with the other typical SNATs, but the other 10 NtSNATs separately clustered outside of the typical SNATs. These 10 NtSNATs have only motif 1, whereas representative SNATs, such as NtSNAT1 and NtSNAT2 or a SNAT from cyanobacteria, have five motifs. In addition, NtSNAT1 and NtSNAT2 are highly homologous to the characterized OsSNAT1, 62.95 and 71.36%, respectively; however, the homology between the other 10 NtSNAT genes and OsSNAT1 is low. Concomitantly, it is hypothesized that NtSNAT1 and NtSNAT2 are the homolog of SNATs, whereas the other 10 candidates could be considered NtSNAT-like genes. Furthermore, both Nicotiana tomentosiformis and Nicotiana sylvestris, two diploid ancestor species of N. tabacum, have two SNAT candidates; therefore, it is speculated that gene rearrangement or deletion during the process of genomic stabilization after whole-genome duplication or polyploidization led to the preservation of NtSNAT1 and NtSNAT2 during the evolution of tobacco from the ancestral diploid to the allotetraploid. NtSNAT and NtSNAT-like genes were differentially expressed in all organs under different stress conditions, indicating that these genes potentially associated with plant growth and development and stress resistance. Under different stress conditions, the expression of NtSNAT1 was significantly upregulated upon high-temperature and cadmium stresses, while the expression of NtSNAT2 did not significantly increase under any of the tested stress treatments. These results provide valuable information for elucidating the evolutionary relationship of SNAT genes in tobacco and genetic resources for improving tobacco production in the future.
Introduction
Melatonin (N-acetyl-5-methoxytryptamine) is an evolutionarily conserved pleiotropic molecule that exists ubiquitously in living organisms (Tan et al., 2011; Lee et al., 2014a; Kanwar et al., 2018). Melatonin is considered to be a growth regulator and is involved in regulating numerous plant biological processes, including seed germination, rooting, flowering, senescence, and photosynthesis (Chen et al., 2009; Kolář et al., 2010; Wang et al., 2012; Hernández et al., 2015; Wei et al., 2015; Zhang et al., 2017; Arnao and Hernández-Ruiz, 2019; Turk and Genisel, 2019). In addition to its roles in plant development, melatonin is also involved in the tolerance of plants to a range of biotic and abiotic stresses, such as pathogen infection, drought, high temperature, cold, and salinity (Tal et al., 2011; Byeon et al., 2014; Lee et al., 2014a; Shi et al., 2015a; Zhang et al., 2015; Arnao and Hernández-Ruiz, 2017; Zhao D. et al., 2019).
Melatonin is synthesized from tryptophan through four distinct reaction steps. A total of six enzymes, tryptophan decarboxylase (TDC), TPH, tryptamine 5-hydroxylase (T5H), serotonin N-acetyltransferase (SNAT), acetylserotonin-O-methyltransferase (ASMT), and caffeic acid O-methyltransferase (COMT), participate in the synthesis of melatonin in plants, among which SNAT is either the penultimate enzyme or the last step enzyme involved in melatonin biosynthesis (Back et al., 2016; Hardeland, 2016; Wang et al., 2017; Yu et al., 2018). Since the first plant SNAT gene was identified and cloned in rice, homologous SNAT genes in other species, such as cyanobacteria, alga laver, Arabidopsis thaliana, grapevine and apple, also have been identified, found to be present at even higher frequencies than the frequency in rice, and enzymatically characterized (Byeon et al., 2013, 2015, 2016b; Wang et al., 2017; Yu et al., 2019).
SNATs have been reported to play a regulatory role in maintaining the steady-state level of melatonin, and there is a relationship between SNATs and the response to abiotic stress (Park et al., 2014). A growing body of evidence indicates that different plant SNATs have different thermophilic properties: a SNAT protein of cyanobacteria presented increased catalytic activity at 70°C, a SNAT protein of loblolly pine presented increased catalytic activity at 55°C, and a SNAT protein of apple presented increased catalytic activity at 35°C. The heat resistance of SNATs shows that it functions in the heat stress response (Byeon et al., 2013, 2016b; Kang et al., 2013; Park et al., 2014; Yu et al., 2019). Additionally, transgenic Arabidopsis ectopically expressing MzSNAT5 presented elevated melatonin levels, thus resulting in enhanced drought tolerance (Wang et al., 2017). Low melatonin production by suppression of SNATs in rice causes stunted seedling growth together with yield penalty, increased abiotic stress susceptibility, and increased coleoptile growth under anoxic conditions (Byeon and Back, 2016). In addition to these abiotic defense responses, inhibition of the GhSNAT1 melatonin biosynthesis-relate genes reduced the resistance of cotton inoculated with pathogenic bacteria (Li et al., 2019). Hence, SNATs play an imperative role in the plant response to biotic and abiotic stresses.
Tobacco (Nicotiana tabacum) is an allotetraploid (2n = 48) that originated from chromosome doubling after an intraspecific hybridization event between Nicotiana tomentosiformis (2n = 24) and Nicotiana sylvestris (2n = 24) (Lim et al., 2004). Tobacco is an essential commercial crop species within the Solanaceae family. Tobacco is cultivated in more than 120 countries contributes substantially to the economic development of countries worldwide (Wang and Bennetzen, 2015; Tong et al., 2019). However, tobacco is often affected by various biotic and abiotic stresses during its growth and development, such as drought, pathogens, cold, high temperature, and heavy metals (Cho and Hong, 2006; Liu et al., 2011; Ma et al., 2014). These stresses cause stunted growth, senescence, reduce yields, and even death (Debnath et al., 2019). A large number of studies have confirmed that melatonin plays a pivotal role in plant growth and development and the response to biotic or abiotic stress (Hernández-Ruiz et al., 2004; Shi et al., 2015a,b; Yu et al., 2018; Zhao L. et al., 2019). However, numerous important biological pathways and gene families, including SNAT family members responsible for melatonin biosynthesis, remain unexplored in tobacco due to the lack of fully annotated reference genomes (Yu et al., 2020).
To further explore the roles of SNAT genes in tobacco, we used tobacco genomic data and performed a genome-wide investigation of the SNAT gene family. The phylogenetic relationships, sequence features, gene structures, and protein motifs of these NtSNAT genes were analyzed. Moreover, the functional diversity of SNATs in tobacco was studied through the analysis of tissue-specific expression patterns and stress responses. Our results provide a reference for the identification of tobacco SNAT gene function and facilitate further work on improving the stress resistance of tobacco.
Materials and Methods
Tobacco SNAT Gene Sequence Retrieval and Gene Identification
To identify the tobacco SNAT candidates, the hidden Markov model (HMM) profile of the SNAT conserved domain (Pfam13508) was used as a query to search the genomic databases of N. tabacum (tobacco), N. tomentosiformis, N. sylvestris, and Rhodospirillum rubrum by the Markov model-based HMMER program. R. rubrum is the first photosynthetic α-proteobacterium indicated to synthesize melatonin (Manchester et al., 1995). All the candidate sequences were analyzed via the PFAM1 and SMART2 databases (Zhao Y. et al., 2019), and proteins without a typical SNAT conserved domain were removed. Chloroplast transit signal peptides were identified via ChloroP3 (Byeon et al., 2016a). The molecular weight (MW) and isoelectric point (pI) of each protein sequence were calculated using the online tool ExPASY4 (Liu et al., 2017).
Distribution of Conserved Domains and Analysis of NtSNAT Structure
Conserved domains within NtSNAT genes were identified via the NCBI database5. Exon-intron structures were determined according to the alignments of their transcribed sequences with corresponding genomic sequences, and a diagram was generated with the online Gene Structure Display Server6. All the conserved motifs of the SNATs and NtSNAT proteins were subsequently identified by the MEME program7.
Sequence Alignment and Phylogenetic Tree Construction
The deduced protein sequences of the NtSNATs were aligned with the sequence of OsSNAT1 (AK059369) using ClustalX (2.0.9). We performed phylogenetic analyses based on the neighbor-joining method using MEGA 7.0 software. Except for these of the SNATs identified in Nicotiana, the sequences of other putative SNATs were retrieved from the NCBI database (Supplementary Table S1). Bootstrap analysis was performed using 1,000 resampling replications, and branch lengths were assigned through pairwise calculations of the genetic distances.
Plant Growth Conditions and Stress Treatments
Seeds of tobacco (K326 cultivar) were germinated in growth media consisting of a mixture of vermiculite and humus (v:v = 1:2) in polystyrene, square dishes. The seedlings were grown in a greenhouse at a day/night temperature of 25/18°C, an air humidity of 50–60%, and a photoperiod consisting of 16 h light/8 h dark; the light intensity was 100 μmol/m2/s. For heat-stress treatment, a group of 6 week-old tobacco seedlings was treated at 45°C in a growth chamber for 3, 6, and 9 h. For cold stress treatment, the seedlings were placed in a 4°C incubator for durations of 3, 6, and 24 h. The drought stress treatment was stopped, after which the seedlings were watered for 1, 4, and 7 days. For cadmium treatment, the cadmium concentration was determined on the basis of 10 mg/kg mixed soil, and samples were taken at 1, 4, and 7 days. The photoperiod and humidity of all the stress treatments were the same as those of the growth conditions described above. The conditions of the control treatment were the same those during seedling growth. Each treatment involved three biological replicates, and all the samples were immediately frozen in liquid nitrogen and stored at −80°C until analysis.
Isolation of Total RNA and Reverse Transcription
Total RNA was extracted from tobacco leaves, stems, flowers, fruits, and roots using an RNAprep Pure Plant Kit (TIANGEN). First-strand cDNA was synthesized via a cDNA synthesis kit. Gene-specific primers were designed against the genome sequence of tobacco present in the NCBI database. qRT-PCRs assay were performed in a 10.0 μl reaction volume using Super Real PreMix Plus (TIANGEN), and the actin gene was used as an internal control. All the primers used for qRT-PCR-based analysis are presented in Supplementary Table S2. Expression data were calculated using the 2–Δ Δ CT method, and the actin gene was used as a reference for the expression analysis of the NtSNAT genes in tobacco. All the results were generated via six samples: three biological replicates and three technical replicates. The 2–Δ Δ CT values were subsequently used to draw heatmaps via MeV 4.9 and GraphPad Prism 5 software.
Statistical Analysis
All the data were statistically analyzed using SPSS 23.0 statistical software. To investigate the expression differences of SNAT genes in all the samples, the T-test was used. Single asterisk (∗) indicate significant differences, at p ≤ 0.05, and double asterisks (∗∗) indicate extremely significant differences, at p ≤ 0.01.
Results
Primary Identification of NtSNAT Genes in the Tobacco Genome
Name searches and HMM analysis revealed a total of 12 candidate SNAT genes in tobacco. For convenience, we named the NtSNAT genes according to their MW. Details concerning the gene name, locus name, open reading frame (ORF) length, exon and intron numbers, protein length, MW, pI, and chloroplast transit signal peptide are listed in Table 1 and Supplementary Table S3. The MWs of the predicted NtSNAT proteins ranged from 20.4 to 34.9 kDa. In addition, the pIs ranged from 5.25 to 9.28, the ORF lengths ranged from 525 to 906, and protein lengths ranged from 174 to 301 amino acids (aa). A chloroplast transit signal peptide was identified in five of the candidate genes: NtSNAT1, NtSNAT2, NtSNAT3, NtSNAT8, and NtSNAT9. To examine the structural features of the NtSNAT genes, the exon/intron configurations of NtSNAT genes in the tobacco plants were compared. Sequence analysis revealed that introns were present in the coding DNA sequences (CDSs) of these genes, except in NtSNAT7 and NtSNAT12, and the number of introns varied from 2 to 8 (Figure 1). On the basis of the established SNAT identification standard, for the two diploid ancestors of tobacco, N. tomentosiformis, and N. sylvestris, each have two SNAT genes.
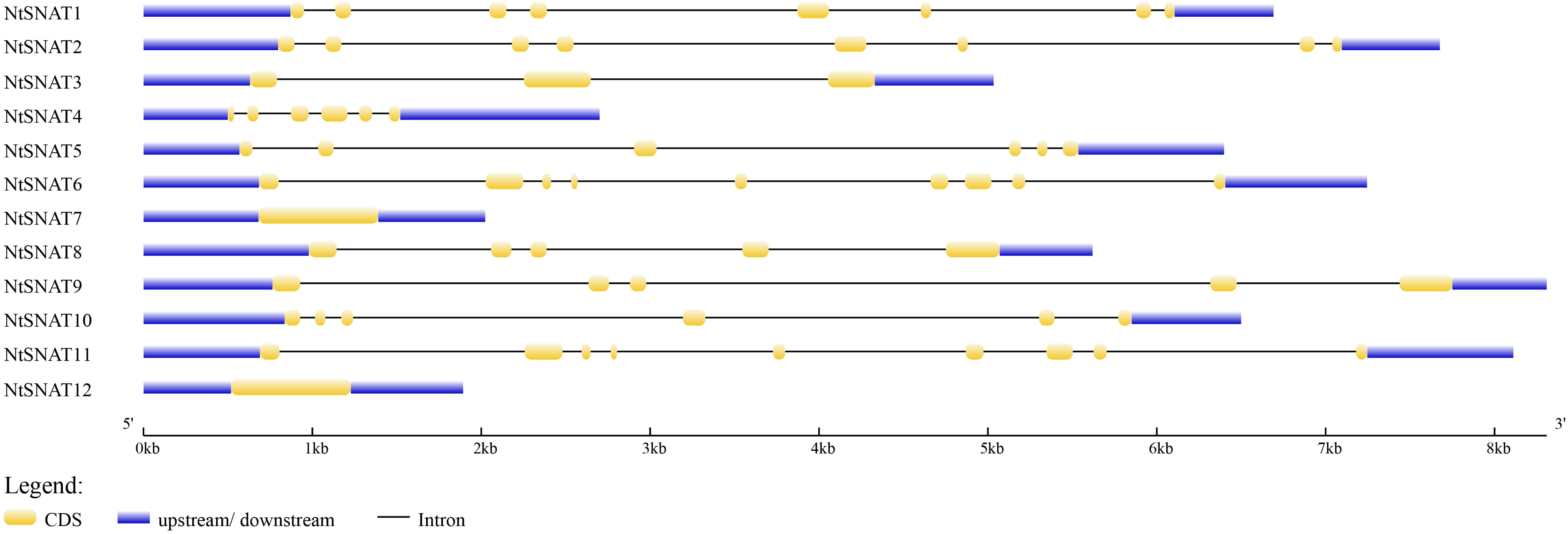
Figure 1. Exon-intron configurations of NtSNAT genes in tobacco plants. The introns and exons are drawn to scale together with the complete coding regions of their respective genes. The boxes indicate the exons, and lines indicate the introns.
Multiple Sequence Alignment of NtSNATs and the Characterized Rice OsSNAT1 Suggests NtSNAT1 and NtSNAT2 Are Authentic Tobacco SNATs
Sequence alignment of the NtSNAT proteins is shown in Figure 2. The results showed that NtSNAT1 and NtSNAT2 are highly homologous to the identified OsSNAT1 (62.95 and 71.36%, respectively), while several other candidate genes have low homology with OsSNAT1. Therefore, we preliminarily infer that NtSNAT1 and NtSNAT2 are homologs of SNATs and that the other 10 members belong to another subgroup. It was found that aa 169–242 of NtSNAT1 and NtSNAT2 compose conserved domains according to NCBI conserved domain analysis. Each of these two proteins has a GNAT functional domain with a coenzyme-A-binding site, and it is suggested that these two proteins could be tobacco SNATs. In addition, the motif 1 conserved region, which may have important functions, was detected in these proteins.
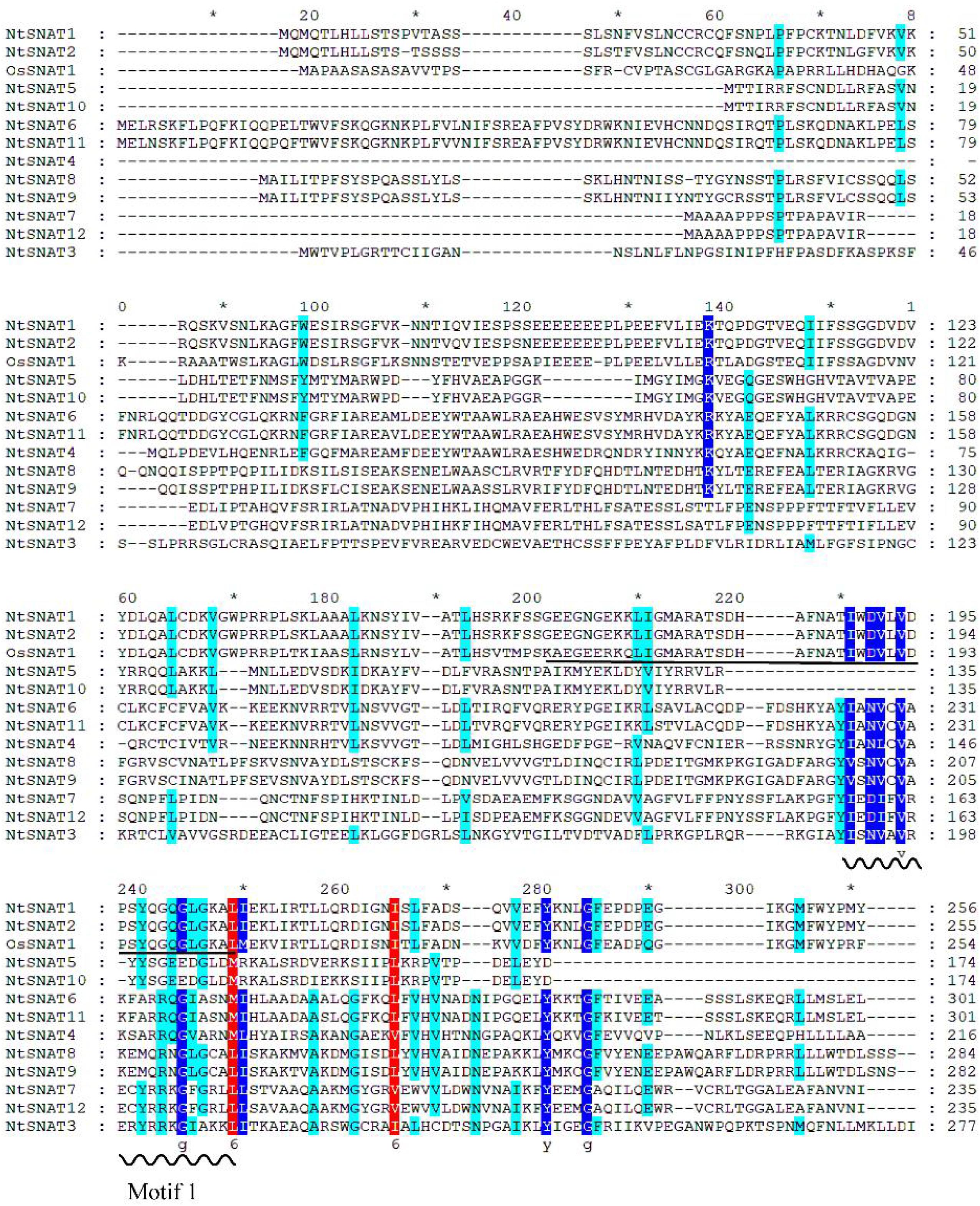
Figure 2. Multiple sequence alignment of OsSNAT1 and NtSNAT proteins. The wavy line in the figure indicates conserved motif 1, and the conserved acetyl-CoA binding motif is underlined. The conserved amino acid residues are indicated by colored shading. The asterisk “*” represent the length of every twenty amino acids (including blanks inside).
Further Validation of the NtSNAT1 and NtSNAT2 Members of NtSNAT Family Based on Phylogenetic Tree and Motif Analysis
For the phylogenetic tree, a BLAST search was performed using the rice OsSNAT1 amino acid sequence. SNAT homologs were found in various taxa, including bacteria, algae, mosses, ferns, gymnosperms, and angiosperms (Figure 3A). These homologous genes not only have typical SNAT conserved domains but also complete conserved motifs. The phylogenetic tree indicated that the NtSNAT and SNAT genes from the included species could be divided into two clades. NtSNAT1 and NtSNAT2 could be classified as typical SNATs, while the remaining 10 NtSNATs clustered together separately. Additionally, four SNATs from two diploid ancestors of tobacco, N. tomentosiformis and N. sylvestris, clustered into clade I together with the typical SNATs. Protein motifs are often used to predict protein function. Twelve NtSNAT and twenty-three SNAT genes were analyzed according to their conserved motifs (Figure 3B and Supplementary Figure S1). The results showed that motif 1 was the most widely distributed and was present in all the members. However, except in R. rubrum, motif 2, motif 3, motif 4, and motif 5 were present in all the members of clade I. The SNAT genes in clade II contained only motif 1. Therefore, it was further speculated that NtSNAT1 and NtSNAT2 were likely tobacco SNATs, and the other 10 candidates were considered NtSNAT-like genes that belonged to another subgroup. Each of the two ancestor diploids has two possible SNAT homologs. During the evolution from diploids to allotetraploids, there should be eight NtSNAT homologs in allotetraploid tobacco under normal circumstances, but only two SNAT homologs were identified in tobacco. These results suggest that the current typical SNATs in tobacco may have arisen via gene loss during the process of genomic stabilization after the occurrence of polyploidization or whole-genome duplication. Moreover, motif 1, motif 2, motif 3, and motif 4 were present at the C-terminus, suggesting important biological functions of the C-terminus for members of the SNAT gene family.
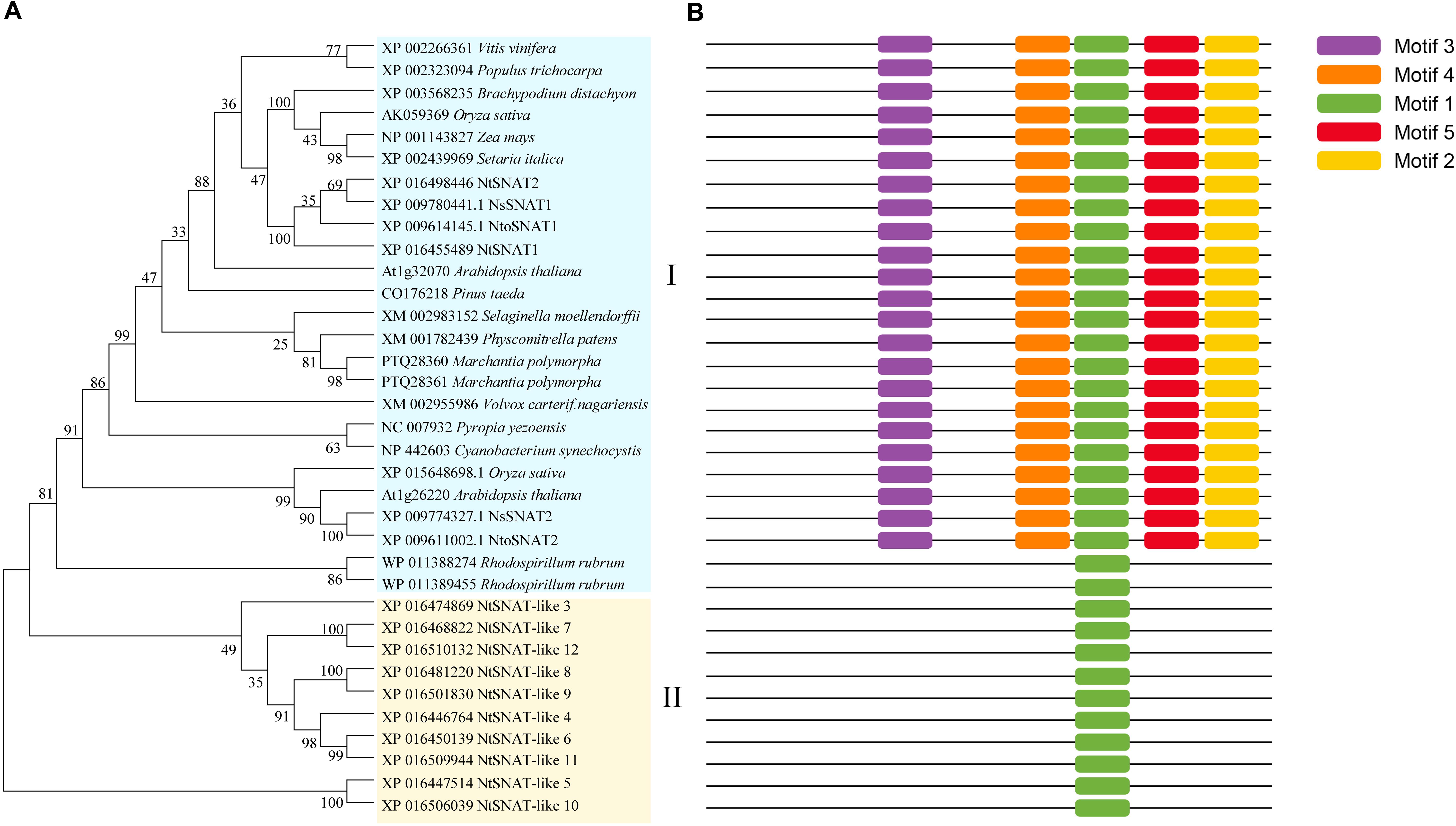
Figure 3. Phylogenetic tree of NtSNATs and other SNATs (A) and corresponding conserved motifs for each gene (B). Different motifs are highlighted with different colored boxes with numbers 1–5.
Organ-Specific Expression Analysis of NtSNAT and NtSNAT-Like Genes
Expression profiling provides useful clues about gene function. To examine the expression patterns of the candidate NtSNAT genes, we evaluated their expression levels in the roots, stems, young leaves, flowers, and fruits (Figure 4). The results revealed that NtSNAT and NtSNAT-like genes were expressed in all of the tested organs. Among them, the expression of NtSNAT1, NtSNAT2, and NtSNAT-like 10 in the leaves was significantly higher than that in other organs. In addition, NtSNAT-like 3 and NtSNAT-like 5 were expressed at relatively high levels in the flowers. Similarly, NtSNAT-like 6, NtSNAT-like 8, NtSNAT-like 9, NtSNAT-like 11, and NtSNAT-like 12 were highly expressed in the fruits. The expression of NtSNAT-like 4 was extremely low in the stems and fruits, and this gene was expressed mainly in the roots. Last, the expression of NtSNAT-like 7 was significantly higher in the stems than in the other organs. And these genes related Gene Ontology terms were listed in Supplementary Table S4, further indicating the functions of 12 SNAT genes in different organs of tobacco.
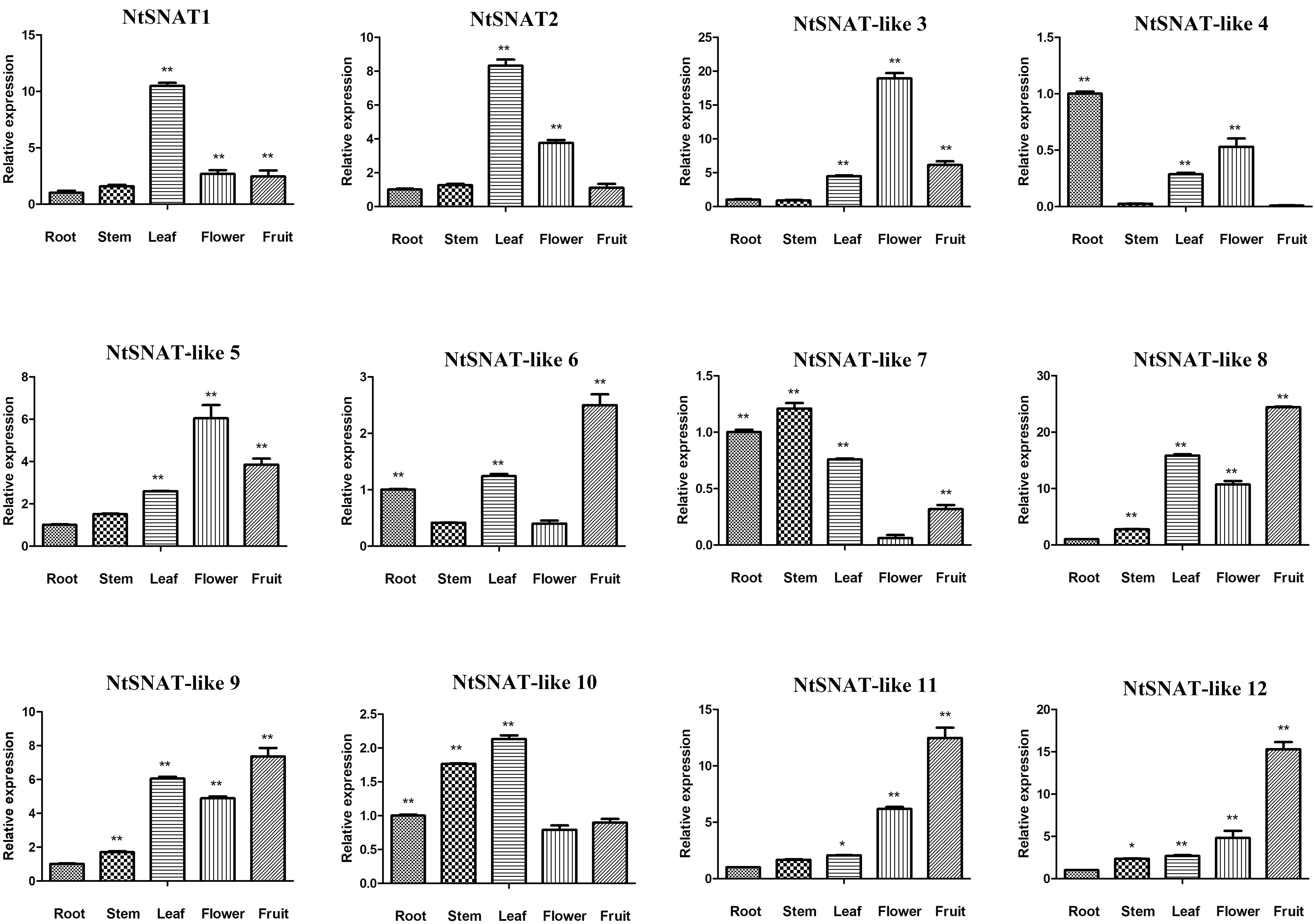
Figure 4. Expression profile of NtSNAT candidates in different tobacco organs based on qRT-PCR results. Single asterisk (*) indicates significant differences at p ≤ 0.05, and double asterisks (**) indicate extremely significant differences at p ≤ 0.01.
Expression Profiles of NtSNAT and NtSNAT-Like Genes Under Different Stress Conditions
Under various stress conditions, it is evident that the expression of NtSNAT and NtSNAT-like genes was induced to a greater extent when the plants were subjected to abiotic stress, including heat, cold, cadmium, and drought than when they were under no stress (Figure 5). Furthermore, the expression of most of these genes was highly variable. The expression of six NtSNAT genes, NtSNAT1, NtSNAT-like 3, NtSNAT-like 5, NtSNAT-like 6, NtSNAT-like 10, and NtSNAT-like 11, dramatically increased in response to high-temperature conditions. Under cadmium stress, the expression of the NtSNAT1, NtSNAT-like 3, NtSNAT-like 10, NtSNAT-like 11, and NtSNAT-like 12 genes strongly increased. When the plants were under drought stress, the expression of only three genes, NtSNAT-like 3, NtSNAT-like 8, and NtSNAT-like 9, was significantly upregulated, whereas the NtSNAT-like 7, NtSNAT-like 8, NtSNAT-like 10, NtSNAT-like 11, and NtSNAT-like 12 genes responded strongly to cold stress. Furthermore, the expression of three of these genes (NtSNAT2, NtSNAT-like 4, and NtSNAT-like 9) did not significantly increase under all the tested stresses.
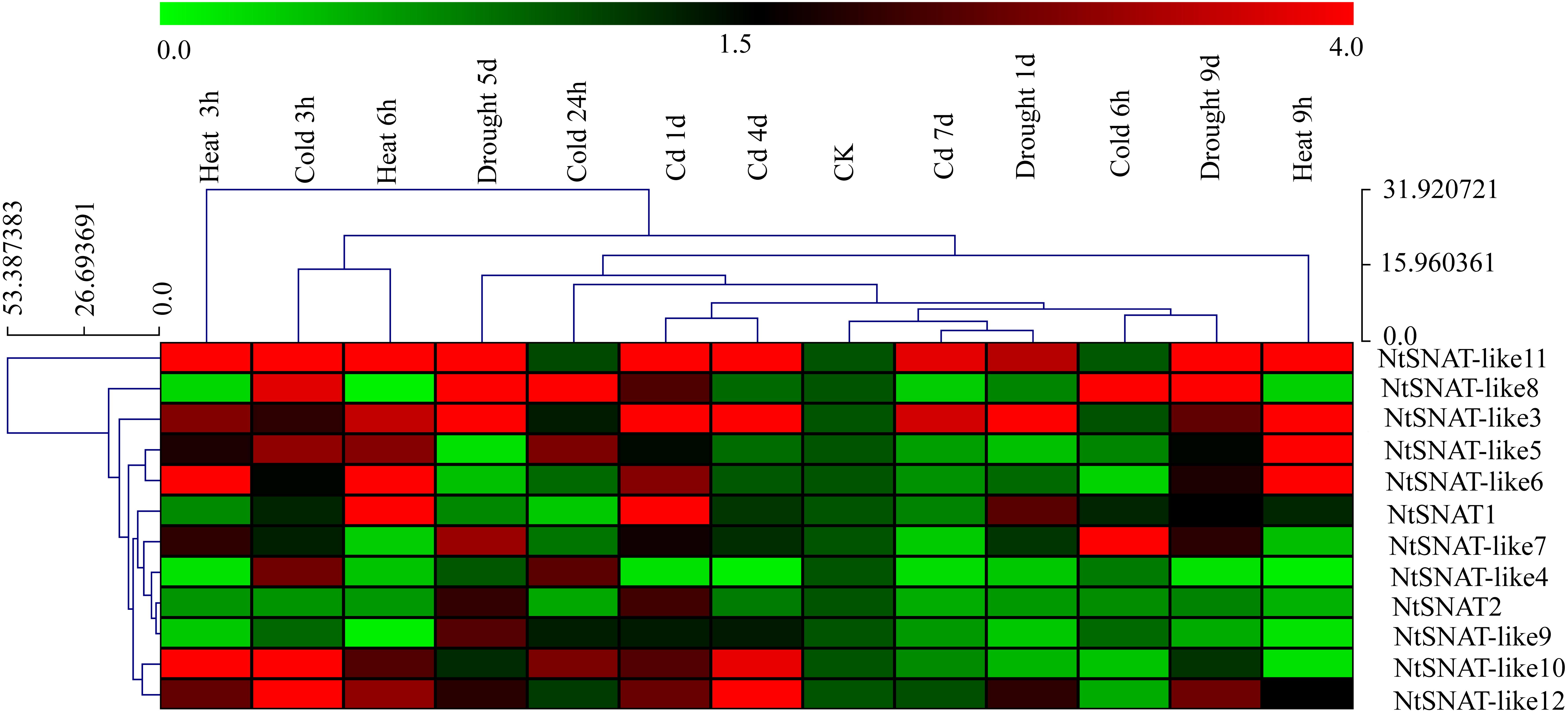
Figure 5. Hierarchical cluster display of expression profiles of NtSNAT and NtSNAT-like genes in tobacco leaves in response to drought, heat, cold, and cadmium (Cd) treatments. The blocks with colors indicate low (green) or high (red) transcript accumulation.
Discussion
The SNAT family belongs to the GNAT superfamily, and the members of the SNAT family play a critical role in regulating the accumulation of melatonin (Dyda et al., 2000; Lee et al., 2015). SNAT genes have been identified in various plant species (Byeon et al., 2016b; Yu et al., 2019). Twelve candidate SNAT genes in tobacco were recognized. When aligned with the defined OsSNAT1 amino acid sequence (Kang et al., 2013), NtSNAT1 and NtSNAT2 were highly homologous to OsSNAT1 (62.95 and 71.36%, respectively), while several other NtSNAT genes were found to have low homology with OsSNAT1. Hence, we preliminarily determined that NtSNAT1 and NtSNAT2 might be tobacco SNATs. Additionally, among the 12 candidate NtSNAT genes, only NtSNAT1 and NtSNAT2 have five conserved motifs, which is consistent with the findings of typical SNATs. Moreover, NtSNAT1 and NtSNAT2 clustered together with the classic SNATs, while the rest of the NtSNATs clustered separately from the identified SNATs. Therefore, these findings further confirmed our speculation.
Previous studies have shown that plant SNATs are vertically transferred to descendants through endosymbiosis (Kang et al., 2013; Lee et al., 2014b; Byeon et al., 2015; Wang et al., 2020). The evolution of SNAT genes from cyanobacteria to higher plants verified that NtSNAT1 and NtSNAT2 indeed evolved vertically through endosymbiosis. From bacteria to higher plants, SNAT proteins are ubiquitous, which means that SNAT proteins may have evolved in the early stages of biological history. Aside from cyanobacteria, we also identified SNAT homologous genes from the purple non-sulfur bacteria R. rubrum, which has been proven to be able to synthesize melatonin (Tilden et al., 1997; Tan et al., 2013). Interestingly, R. rubrum is one of the most ancestral species of living organisms and is the first photosynthetic α-proteobacterium shown to synthesize melatonin (Manchester et al., 1995), despite only motif 1 being present in the SNATs of R. rubrum. These facts, when taken together with all the tested species of common SNAT genes containing conserved motif 1, suggest that motif 1 is probably the core factor of SNAT genes.
Whole-genome duplication or polyploidization is an important driver of adaptation and speciation in plants (Hovav et al., 2008; Liu et al., 2019). In the polyploid genome, there are a large number of duplicated genes and duplicated genes from different diploid ancestors. Due to rearrangement or deletion of these duplicated genes, there are approximately three different fates of a polyploid genome, namely, subfunctionalization, pseudogenization or functional diversification (Adams et al., 2003; Hovav et al., 2008; Sojli et al., 2020). Therefore, it is speculated that the evolution of tobacco from being diploid (the two ancestral species of which were N. tomentosiformis and N. sylvestris) to allotetraploid may have caused the rearrangement or deletion of different genes due to gene duplication. In the present investigation, each of the two ancestor diploids has two possible SNATs; however, only two typical SNATs, NtSNAT1 and NtSNAT2, were identified in tobacco. These results suggest that the typical SNATs currently in tobacco may have arisen by gene loss during the process of genomic stabilization following polyploidization or whole-genome duplication. Ten additional NtSNAT-like genes branching independently from the typical SNATs were identified, and the proteins encoded by these genes had markedly fewer motifs than NtSNAT1 and NtSNAT2 did or the representative SNAT from cyanobacteria did. Therefore, motif 1 has been stable throughout the evolutionary process.
To explore the possible functional differences of NtSNAT and NtSNAT-like genes, their expression patterns in different tissues and under different stresses were determined. The results demonstrated different types of expression patterns among these genes. With respect to the expression in different organs, 2 NtSNAT and 10 NtSNAT-like genes were expressed in the roots, stems, leaves, flowers, and fruits, indicating that these genes may have potential effects in vegetative and reproductive growth. Moreover, the high expression of NtSNAT1 and NtSNAT2 in the leaves indicates that these two genes could be pivotal in leaf growth and development. In addition, studies have shown that SNATs play an important role in the process of plant stress resistance (Wang et al., 2017). The SNAT enzyme is involved in the biosynthesis of melatonin, which is reported to regulate the thermotolerance of many plant species. For example, the cyanobacteria SNAT gene was proven to be involved in melatonin in response to high temperature (Byeon et al., 2013). Under heat-stress conditions, SlSNAT interacts with HSP40 to maintain melatonin levels, thereby increasing the heat resistance of tomato plants (Wang et al., 2020). In this study, NtSNAT1, NtSNAT-like 3, NtSNAT-like 5, NtSNAT-like 6, and NtSNAT-like 10 responded to heat stress at different heat treatment time points, indicating that these genes may have potential effects in increasing heat resistance. Similarly, Lee and Back (2017) showed that overexpression of OsSNAT in rice can significantly increase plant resistance to cadmium stress and senescence. In this paper, under cadmium-stress conditions, NtSNAT1, NtSNAT-like 3, NtSNAT-like 10, NtSNAT-like 11, and NtSNAT-like 12 were found to respond to cadmium stress at different time points, suggesting that these genes potentially associated with increasing plant tolerance to cadmium. Notably, the expression of NtSNAT2, a representative SNAT gene, was not significantly upregulated compared with that in the control group under all the stress treatments. Accordingly, it is speculated that NtSNAT2 may not respond during the actual stress time and may be expressed at other time points. The exact role of NtSNAT1 and NtSNAT2 and the catalytic activities of their encoded proteins require further study.
Conclusion
Serotonin N-acetyltransferase is a key enzyme in the melatonin biosynthesis pathway. NtSNAT1 and NtSNAT2, together with 10 additional NtSNAT-like genes, were identified as candidate genes for improving tobacco production. Among the five motifs typically present within SNATs, motif 1 is indispensable for melatonin biosynthesis. During the evolutionary process through which tobacco changed from being diploid to allotetraploid, NtSNAT1 and NtSNAT2 were retained due to gene rearrangement or deletion during genome stabilization after whole-genome duplication and polyploidization. NtSNAT1 potentially associated with regulating plant growth and development and increasing plant tolerance to stress.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
DZ and SC designed the experiments. JZ designed and carried out the experiments and wrote the manuscript. ZY, RZ, and ZM participated and analyzed the data from the experiments. All the authors reviewed and approved the manuscript in its final form.
Funding
This work was supported financially by the National Natural Science Foundation of China (Nos. 31960082 and 81560622) and the Key Project of China National Tobacco Corporation’s Yunnan Provincial Company Science and Technology Plan (201853010020082).
Conflict of Interest
HY and TX was employed by the Wenshan Branch of Yunnan Tobacco Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.591984/full#supplementary-material
Supplementary Figure 1 | Conserved amino acid sequences of 12 motifs.
Supplementary Table 1 | Accession number and source organism of 35 SNATs from bacteria, algae, mosses, ferns, gymnosperms, and angiosperms.
Supplementary Table 2 | Primers used for the qRT-PCR assay of 12 candidate NtSNATs.
Supplementary Table 3 | Full amino acid sequences of 12 candidate NtSNATs.
Supplementary Table 4 | NtSNAT and NtSNAT-like genes and their related Gene Ontology (GO) terms.
Footnotes
- ^ http://pfam.xfam.org/
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.cbs.dtu.dk/services/Chlorop/
- ^ http://web.expas-y.org/compute_pi
- ^ https://www.ncbi.nlm.nih.gov/Strucure/cd-d/Wrpsb.cgi
- ^ http://www.cbi.pku.edu.cn/resource/index.htm
- ^ http://meme-suite.org/tools/meme
References
Adams, K. L., Richard, C., Ryan, P., and Wendel, J. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. U.S.A. 100, 4649–4654. doi: 10.1073/pnas.0630618100
Arnao, M. B., and Hernández-Ruiz, J. (2017). Melatonin and its relationship to plant hormones. Ann. Bot. 121, 195–207. doi: 10.1093/aob/mcx114
Arnao, M. B., and Hernández-Ruiz, J. (2019). Melatonin: a new plant hormone and/or a plant master regulator? Trends Plant Sci. 24, 38–48. doi: 10.1016/j.tplants.2018.10.010
Back, K., Tan, D. X., and Reiter, R. J. (2016). Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 61, 426–437. doi: 10.1111/jpi.12364
Byeon, Y., and Back, K. (2014). Melatonin synthesis in rice seedlings in vivo is enhanced at high temperatures and under dark conditions due to increased serotonin N-acetyltransferase and N-acetylserotonin methyltransferase activities. J. Pineal Res. 56, 189–195. doi: 10.1111/jpi.12111
Byeon, Y., and Back, K. (2016). Low melatonin production by suppression of either serotonin N-acetyltransferase or N-acetylserotonin methyltransferase in rice causes seedling growth retardation with yield penalty, abiotic stress susceptibility, and enhanced coleoptile growth under anoxi. J. Pineal Res. 60, 348–359. doi: 10.1111/jpi.12317
Byeon, Y., Lee, H., Choi, D. W., and Back, K. (2015). Chloroplast-encoded serotonin N-acetyltransferase in the red alga Pyropia yezoensis: gene transition to the nucleus from chloroplasts. J. Exp. Bot. 66, 709–717. doi: 10.1093/jxb/eru357
Byeon, Y., Lee, H. Y., and Back, K. (2016a). Cloning and characterization of the serotonin N–acetyltransferase–2 gene (SNAT2) in rice (Oryza sativa). J. Pineal Res. 61, 198–207. doi: 10.1111/jpi.12339
Byeon, Y., Lee, H. J., Lee, H. Y., and Back, K. (2016b). Cloning and functional characterization of the Arabidopsis N-acetylserotonin O-methyltransferase responsible for melatonin synthesis. J. Pineal Res. 60, 65–73. doi: 10.1111/jpi.12289
Byeon, Y., Lee, K., Park, Y. I., Park, S., and Back, K. (2013). Molecular cloning and functional analysis of serotonin N-acetyltransferase from the cyanobacterium Synechocystis sp. PCC 6803. J. Pineal Res. 55, 371–376. doi: 10.1111/jpi.12080
Chen, Q., Qi, W. B., Reiter, R. J., Wei, W., and Wang, B. M. (2009). Exogenously applied melatonin stimulates root growth and raises endogenous indoleacetic acid in roots of etiolated seedlings of Brassica juncea. J. Plant Physiol. 166, 324–328. doi: 10.1016/j.jplph.2008.06.002
Cho, E. K., and Hong, C. B. (2006). Over-expression of tobacco NtHSP70-1 contributes to drought-stress tolerance in plants. Plant Cell Rep. 25, 349–358. doi: 10.1007/s00299-005-0093-2
Debnath, B., Islam, W., Li, M., Sun, Y., Lu, X., and Mitra, S. (2019). Melatonin mediates enhancement of stress tolerance in plants. Int. J. Mol. Sci. 20:1040. doi: 10.3390/ijms20051040
Dyda, F., Klein, D. C., and Hickman, A. B. (2000). GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Bioph. Biomol. 29, 81–103. doi: 10.1146/annurev.biophys.29.1.81
Hardeland, R. (2016). Melatonin in plants-diversity of levels and multiplicity of functions. Front. Plant Sci. 7:198. doi: 10.3389/fpls.2016.00198
Hernández, I. G., Gomez, F. J. V., Cerutti, S., Arana, M. V., and Silva, M. F. (2015). Melatonin in Arabidopsis thaliana acts as plant growth regulator at low concentrations and preserves seed viability at high concentrations. Plant Physiol. Biochem. 94, 191–196. doi: 10.1016/j.plaphy.2015.06.011
Hernández-Ruiz, J., Cano, A., and Arnao, M. B. (2004). Melatonin: a growth-stimulating compound present in lupin tissues. Planta 220, 140–144. doi: 10.1007/s00425-004-1317-3
Hovav, R., Udall, J. A., Chaudhary, B., Rapp, R., Flagel, L., and Wendel, J. F. (2008). Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. U.S.A. 105, 6191–6195. doi: 10.1073/pnas.0711569105
Kang, K., Lee, K., Park, S., Byeon, Y., and Back, K. (2013). Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 55, 7–13. doi: 10.1111/jpi.12011
Kanwar, M. K., Yu, J., and Zhou, J. (2018). Phytomelatonin: recent advances and future prospects. J. Pineal Res. 65:e12526. doi: 10.1111/jpi.12526
Kolář, J., Johnson, C. H., and Macháčková, I. (2010). Exogenously applied melatonin (N-acetyl-5-methoxytryptamine) affects flowering of the short-day plant Chenopodium rubrum. Physiol. Plant 118, 605–612. doi: 10.1034/j.1399-3054.2003.00114.x
Lee, H. Y., Byeon, Y., and Back, K. (2014a). Melatonin as a signal molecule triggering defense responses against pathogen attack in Arabidopsis and tobacco. J. Pineal Res. 57, 262–268. doi: 10.1111/jpi.12165
Lee, H. Y., Byeon, Y., Lee, K., Lee, H., and Back, K. (2014b). Cloning of Arabidopsis SNAT and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations. J. Pineal Res. 57, 418–426. doi: 10.1111/jpi.12181
Lee, H. Y., Byeon, Y., Tan, D. X., Reiter, R. J., and Back, K. (2015). Arabidopsis serotonin N-acetyltransferase knockout mutant plants exhibit decreased melatonin and salicylic acid levels resulting in susceptibility to an avirulent pathogen. J. Pineal Res. 58, 291–299. doi: 10.1111/jpi.12214
Lee, K., and Back, K. (2017). Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 62:e12392. doi: 10.1111/jpi.12392
Li, C., He, Q., Zhang, F., Yu, J. W., Li, C., and Zhao, T. L. (2019). Melatonin enhances cotton immunity to Verticillium wilt via manipulating lignin and gossypol biosynthesis. Plant J. 100, 784–800. doi: 10.1111/tpj.14477
Lim, K. Y., Matyasek, R., Kovarik, A., and Leitch, A. (2004). Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 82, 599–606. doi: 10.1111/j.1095-8312.2004.00344.x
Liu, H. M., Ekrt, L., Kouteck, P., Pellicer, J., Hidalgo, O., and Marquardt, J. (2019). Polyploidy does not control all: lineage-specific average chromosome length constrains genome size evolution in ferns. J. Syst. Evol. 57, 418–430. doi: 10.1111/jse.12525
Liu, W., Zhao, D., Zheng, C., Chen, C., Xin, P., and Yuan, C. (2017). Genomic analysis of the ASMT gene Family in Solanum lycopersicum. Molecules 22:1984. doi: 10.3390/molecules22111984
Liu, H., Zhou, X., Dong, N., Liu, X., Zhang, H., and Zhang, Z. (2011). Expression of a wheat MBY gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Funct. Integ. Genom. 11, 431–443. doi: 10.1007/s10142-011-0228-1
Ma, Y., Zhang, L., Zhang, J., Chen, J., Wu, T., Zhu, S., et al. (2014). Expressing a Citrus ortholog of Arabidopsis ERF1 enhanced cold-tolerance in tobacco. Sci. Hortic. 174, 65–76. doi: 10.1016/j.scienta.2014.05.009
Manchester, L. C., Poeggeler, B., Alvares, F. L., Ogden, G. B., and Reiter, R. J. (1995). Melatonin immunoreactivity in the photosynthetic prokaryote Rhodospirillum rubrum: implications for an ancient antioxidant system. Cell. Mol. Biol. Res. 41, 391–395. doi: 10.1016/0008-8749(95)80021-A
Park, S., Byeon, Y., Lee, H. Y., Kim, Y. S., Ahn, T., and Back, K. (2014). Cloning and characterization of a serotonin N-acetyltransferase from a gymnosperm, loblolly pine (Pinus taeda). J. Pineal Res. 57, 348–355. doi: 10.1111/jpi.12174
Shi, H., Chen, Y., Tan, D. X., Reiter, R. J., Chan, Z., and He, C. (2015a). Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 59, 102–108. doi: 10.1111/jpi.12244
Shi, H., Qian, Y., Tan, D. X., Reiter, R. J., and He, C. (2015b). Melatonin induces the transcripts of CBF/DREB1s and their involvement in both abiotic and biotic stresses in Arabidopsis. J. Pineal Res. 59, 334–342. doi: 10.1111/jpi.12262
Sojli, L., Sehyun, C., Jeon, D., Donghyun, J., Yuna, K., and Chang, K. (2020). Evolutionary impact of whole genome duplication in Poaceae family. J. Crop Sci. Biotechnol. 57, doi: 10.1007/s12892-020-00049-2
Tal, O., Haim, A., Harel, O., and Gerchman, Y. (2011). Melatonin as an antioxidant and its semi-lunar rhythm in green macroalga Ulva sp. J. Exp. Bot. 62, 1903–1910. doi: 10.1093/jxb/erq378
Tan, D. X., Hardeland, R., Manchester, L. C., Korkmaz, A., Ma, S., Rosales-Corral, S., et al. (2011). Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. J. Exp. Bot. 63, 577–597. doi: 10.1093/jxb/err256
Tan, D. X., Manchester, L. C., Liu, X., Rosales-Corral, S. A., Acuna-Castroviejo, D., and Reiter, R. J. (2013). Mitochondria and chloroplasts as the original sites of melatonin synthesis: a hypothesis related to melatonin’s primary function and evolution in eukaryotes. J. Pineal Res. 54, 127–138. doi: 10.1111/jpi.12026
Tilden, A. R., Becker, M. A., Amma, L. L., Arciniega, J., and McGaw, A. K. (1997). Melatonin production in an aerobic photosynthetic bacterium: an evolutionarily early association with darkness. J. Pineal Res. 22, 102–106. doi: 10.1111/j.1600-079X.1997.tb00310.x
Tong, Z., Zhou, J., Xiu, Z., Jiao, F., Hu, Y., Zheng, F., et al. (2019). Construction of a high-density genetic map with whole genome sequencing in Nicotiana tabacum L. Genomics 112, 2028–2033. doi: 10.1016/j.ygeno.2019.11.015
Turk, H., and Genisel, M. (2019). Melatonin-related mitochondrial respiration responses are associated with growth promotion and cold tolerance in plants. Cryobiology 92, 76–85. doi: 10.1016/j.cryobiol.2019.11.006
Wang, L., Feng, C., Zheng, X., Guo, Y., Zhou, F., Shan, D., et al. (2017). Plant mitochondria synthesize melatonin and enhance the tolerance of plants to drought stress. J. Pineal Res. 63:e12429. doi: 10.1111/jpi.12429
Wang, P., Yin, L., Liang, D., Li, C., Ma, F., and Yue, Z. (2012). Delayed senescence of apple leaves by exogenous melatonin treatment: toward regulating the ascorbate-glutathione cycle. J. Pineal Res. 53, 11–20. doi: 10.1111/j.1600-079x.2011.00966.x
Wang, X., and Bennetzen, J. L. (2015). Current status and prospects for the study of Nicotiana genomics, genetics, and nicotine biosynthesis genes. Mol. Genet. Genomics 290, 11–21. doi: 10.1007/s00438-015-0989-7
Wang, X., Zhang, H., Xie, Q., Liu, Y., Lv, H., Bai, R., et al. (2020). SlSNAT interacts with HSP40, a molecular chaperone, to regulate melatonin biosynthesis and promote thermotolerance in tomato. Plant Cell Physiol. 61, 909–921. doi: 10.1093/pcp/pcaa018
Wei, W., Li, Q. T., Chu, Y. N., Reiter, R. J., Yu, X. M., Zhu, D. H., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. doi: 10.1093/jxb/eru392
Yu, S., Kakar, K., Yang, Z., Nawaz, Z., Lin, S., Guo, Y., et al. (2020). Systematic study of the stress-responsive Rboh gene family in Nicotiana tabacum: genome-wide identification, evolution and role in disease resistance. Genomics 112, 1404–1418. doi: 10.1016/j.ygeno.2019.08.010
Yu, Y., Bian, L., Jiao, Z., Yu, K., Wan, Y., Zhang, G., et al. (2019). Molecular cloning and characterization of a grapevine (Vitis vinifera L.) serotonin N-acetyltransferase (VvSNAT2) gene involved in plant defense. BMC Genomics 20:880. doi: 10.1186/s12864-019-6085-3
Yu, Y., Lv, Y., Shi, Y., Li, T., Chen, Y., Zhao, D., et al. (2018). The role of phyto-melatonin and related metabolites in response to stress. Molecules 23:1887. doi: 10.3390/molecules23081887
Zhang, H. J., Sun, Q. Q., Cao, Y. Y., Li, X., Zhao, B., Wu, P., et al. (2017). Proteomic analysis reveals a role of melatonin in promoting cucumber seed germination under high salinity by regulating energy production. Sci. Rep. 7:503. doi: 10.1038/s41598-017-00566-1
Zhang, N., Sun, Q., Zhang, H., Cao, Y., Weeda, S., Ren, S., et al. (2015). Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 66, 647–656. doi: 10.1093/jxb/eru336
Zhao, D., Yu, Y., Shen, Y., Liu, Q., Zhao, Z., Sharma, R., et al. (2019). Melatonin synthesis and function: evolutionary history in animals and plants. Front. Endocrinol. 10:249. doi: 10.3389/fendo.2019.00249
Zhao, L., Chen, L., Gu, P., Zhan, X., Zhang, Y., Hou, C., et al. (2019). Exogenous application of melatonin improves plant resistance to virus infection. Plant Pathol. 68, 1287–1295. doi: 10.1111/ppa.13057
Keywords: Nicotiana tabacum, melatonin, serotonin N-acetyltransferase, tissue-specific expression, abiotic stress
Citation: Zhang J, Yao Z, Zhang R, Mou Z, Yin H, Xu T, Zhao D and Chen S (2020) Genome-Wide Identification and Expression Profile of the SNAT Gene Family in Tobacco (Nicotiana tabacum). Front. Genet. 11:591984. doi: 10.3389/fgene.2020.591984
Received: 05 August 2020; Accepted: 30 September 2020;
Published: 27 October 2020.
Edited by:
Rong Zhou, Aarhus University, DenmarkReviewed by:
Tanweer Kumar, Lanzhou University, ChinaJianghua Chen, Xishuangbanna Tropical Botanical Garden (CAS), China
Copyright © 2020 Zhang, Yao, Zhang, Mou, Yin, Xu, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dake Zhao, emhhb2RrMjAxMkB5bnUuZWR1LmNu; Suiyun Chen, Y2hlbnN1aXl1bkB5bnUuZWR1LmNu
 Jiemei Zhang1,2
Jiemei Zhang1,2 Renjun Zhang
Renjun Zhang Dake Zhao
Dake Zhao