- 1Department of Vip center, School and Hospital of Stomatology, Cheeloo College of Medicine, Shandong University & Shandong Key Laboratory of Oral Tissue Regeneration & Shandong Engineering Laboratory for Dental Materials and Oral Tissue Regeneration, Jinan, China
- 2Endodontics, Faculty of Dentistry, The University of Hong Kong, Hong Kong, Hong Kong
- 3Department of Breast Surgery, Qilu Hospital of Shandong University, Jinan, China
- 4Department of Stomatology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 5Department of Oral Pathology, School of Stomatology, Zhengzhou University, Zhengzhou, China
Osteo/odontogenic differentiation is a key process of human stem cells from apical papilla (SCAP) in tooth root development. Emerging evidence indicates microRNAs (miRNAs) play diverse roles in osteogenesis. However, their functions in osteo/odontogenic differentiation of SCAP require further elucidation. To investigate the role of miRNA in SCAP osteo/odontogenic differentiation and underlying mechanisms, miRNA microarray analysis was performed to screen differentially expressed miRNAs between control and osteo/odontogenic-induced group. Quantitative real-time PCR (qRT-PCR) and western blot were used to detected osteo/odontogenic differentiation-related markers and possible signaling pathway SCAP-associated genes. Alizarin Red Staining (ARS) were applied to evaluated osteogenic capacity. The results showed that miR-497-5p increased during SCAP osteo/odontogenic differentiation. Overexpression of miR-497-5p enhanced the osteo/odontogenic differentiation of SCAP, whereas downregulation of miR-497-5p elicited the opposite effect, thus suggesting that miR-497-5p is a positive regulator of the osteo/odontogenic differentiation of SCAP. Bioinformatic analysis and dual luciferase reporter assay identified that SMAD specific E3 ubiquitin protein ligase 2 (Smurf2) is a direct target of miR-497-5p. Further study demonstrated that Smurf2 negatively regulates SCAP osteo/odontogenic differentiation, and silencing Smurf2 could block the inhibitory effect of the miR-497-5p inhibitor. Meanwhile, pathway detection manifested that miR-497-5p promotes osteo/odontogenic differentiation via Smad signaling pathway. Collectively, our findings demostrate that miR-497-5p promotes osteo/odontogenic differentiation of SCAP via Smad signaling pathway by targeting Smurf2.
Introduction
Stem cells from apical papilla (SCAP) were first isolated and characterized from the apical papilla of human immature third molar by Sonoyama et al. (2006, 2008). This indicates that SCAP play a vital role in the development of tooth root. Similar to postnatal mesenchymal stem cells (MSCs), SCAP possess self-renewal and multidirectional differentiation potential (Abe et al., 2008; Sonoyama et al., 2008; Bakopoulou et al., 2011; Koutsoumparis et al., 2018). Compared with dental pulp stem cells (DPSCs), SCAP have stronger proliferation ability, cell migration, and telomere activity, and are considered to be a valuable source of postnatal MSCs for dental tissue engineering (Sonoyama et al., 2006; Huang et al., 2010; Na et al., 2016; Chrepa et al., 2017). The osteo/odontogenic differentiation of SCAP is prerequisite in pulp-dentine regeneration. Therefore, how to effectively promote the osteogenic/odontogenic differentiation of SCAP has become a key issue. Our previous studies have demonstrated that many factors could regulate osteo/odontogenic differentiation of SCAP (Yang et al., 2012; Wan et al., 2016; Liu et al., 2019a). However, the molecular mechanisms underlying the osteo/odontogenic differentiation in SCAP remains unknown.
MicroRNAs (miRNAs) are a class of small single-stranded non-coding RNAs. They function as post-transcriptional regulators. MiRNAs bind to 3′-untranslated regions (3′-UTR) of the target mRNA and modulate the expression of target genes through degrading target mRNAs or inhibiting their translation. MiRNAs have been implicated in various biological processes, including cell proliferation, differentiation, apoptosis, and carcinogenesis (Hammond et al., 2001; Bartel, 2004; Lindsay, 2008; Julia et al., 2009). A number of miRNAs were reported to regulate osteogenic differentiation in mesenchymal stem cells (Chen et al., 2017; Tang et al., 2018). Accumulating evidence revealed that miRNAs participated in osteogenic differentiation of dental stem cells. MiR-214 down-regulated the osteogenic differentiation of periodontal ligament stem cells (PDLSCs) by targeting transcription factor 4 (Yao et al., 2017). MiR-508-5p suppressed the osteogenesis of human dental pulp stem cells through inhibiting glycoprotein non-metastatic melanoma protein B (Liu et al., 2019b). NOTCH activation inhibited osteogenic differentiation of SCAP and promoted the expression of miR-34a, while miR-34a suppressed Notch signaling by targeting NOTCH2 and HES1 (Sun et al., 2014). To date, there have been few studies on miRNA regulating osteo/odontogenic differentiation of SCAP and their molecular mechanisms remained still unclear. Thus, we hypothesized that certain miRNAs could positively regulate the osteo/odontogenic differentiation of SCAP, which could be used as a novel target for regulating dental tissue regeneration.
In this study, a microarray was applied to investigate the miRNA expression profiles of SCAP during osteo/odontogenic differentiation. qRT-PCR was performed to verify gene expression. Bioinformatics and qRT-PCR analysis demonstrated miR-497-5p was up-regulated during osteogenic differentiation. Further study confirmed that miR-497-5p increased the osteo/odontogenic differentiation of SCAP. Target gene prediction and dual luciferase reporter assay showed that Smurf2, a negative regulator of osteogenesis, was a direct target of miR-497-5p. The Smad signaling pathway was involved in the osteo/odontogenic differentiation of SCAP. Our study suggested miR-497-5p could suppress Smurf2 and promote osteo/odontogenic differentiation of SCAP through Smad signaling pathway.
Materials and Methods
Cell Culture
Human apical papilla tissues were obtained from immature third permanent molars extracted from patients aged 16–20 years for orthodontic reason at the School and Hospital of Stomatology, Shandong University. This project was approved by the Ethics Committee of the School and Hospital of Stomatology, Shandong University. The consent was obtained from the patients or their parents. The apical papilla was separated from the third molar and digested with 3 mg/mL Collagenase Type I (Solarbio, Beijing, China) and 4 mg/mL Dispase (Roche, Indianapolis, IN, United States) at 37°C for 1 h. Then cells were cultured in α-minimum essential medium (HyClone, Logan, UT, United States) supplemented with 20% fetal bovine serum (FBS) (Gibco, Grand Island, NY, United States), 100 U/mL penicillin and 100 μg/mL streptomycin (HyClone) incubated at 37°C in a 5% CO2 atmosphere. The cells from passage 2–5 were used for subsequent experiments.
Human embryonic kidney 293T (HEK 293T) cells were cultured in DMEM medium containing 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin.
Flow Cytometry
SCAP were characterized by flow cytometry. A total of 8 × 105 cells were collected and incubated respectively with monoclonal antibodies specific for STRO-1 (R&D Systems, Minneapolis, MN, United States), CD24, CD146, CD34 and CD45 (BD Biosciences, Franklin Lakes, NJ, United States) for 30 min at room temperature. Expression profiles were analyzed by BD FACSCalibur flow cytometer (BD Biosciences).
Cell Counting Kit-8 (CCK-8) Analysis
The proliferation ability of SCAP was examined using CCK-8 (MedChemExpress, Monmouth Junction, NJ, United States). Cells were seeded in 96-well plates at a density of 3 × 103 cells/well. The cells were treated with CCK-8 reagent on days 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 and incubated at 37°C for 2 h. Then, the absorbance was measured at 450 nm using a microplate reader.
Osteogenic Differentiation
For osteogenic differentiation, after the SCAP reached 80% confluence, the medium was changed to osteogenic medium containing 10 mM β-glycerolphosphate (Sigma-Aldrich, St. Louis, MO, United States), 10 nM dexamethasone (Sigma-Aldrich) and 50 μg/mL L-ascorbic acid (Sigma-Aldrich). The osteogenic medium was changed every 3 days.
Alkaline Phosphatase (ALP) Activity Assay
SCAP were cultured in osteogenic medium for 7 days. Then cells were washed with PBS and lysed with radioimmunoprecipitation (RIPA) lysis buffer (Solarbio) on ice for 30 min. After sonication and centrifugation, the supernatants were obtained and ALP activity was measured using an ALP activity assay kit (Jiancheng, Nanjing, China), according to the manufacturer’s instructions.
ALP Staining and Alizarin Red Staining (ARS)
SCAP were grown in osteogenic medium. On day 7, cells were fixed with 4% paraformaldehyde, followed by washing with PBS. ALP staining was performed using an ALP staining kit (Solarbio), according to manufacturer’s protocol. For Alizarin Red staining, after 21 days induction, cells were fixed with 4% paraformaldehyde and stained with 2% Alizarin red (Sigma-Aldrich) with pH 4.2 for 30 min. Then, cells were rinsed thrice with double distilled water to remove the unbound dye and visualized under a microscope. For quantitative assay, the alizarin red was dissolved in 10% cetylpyridinium chloride (Sigma-Aldrich) for 30 min. The absorbance was detected at 560 nm and the results were normalized to the total protein content.
MiRNA Microarray and Identification of Differentially Expressed Genes
SCAP treated with osteogenic induction for 7 days were served as experimental group (n = 3), while the untreated cells were control group (n = 3). Total RNA was extracted and analyzed miRNAs expression using microarray (Affymetrix miRNA 4.0 Array). To determine differentially expressed genes, the R package “limma” was used (Ritchie et al., 2015). MiRNAs (fold change ≥2.0 or ≤0.5, P value <0.05, compared with control group) were considered to be statistically significant. The corresponding heatmaps were drawn by R package “pheatmap” (Wang et al., 2019).
Cell Transfection
Synthetic miR-497-5p mimics/inhibitor, siRNA targeting Smurf2 (siSmurf2), and their negative control (NC) were purchased from GenePharma. SCAP were transfected with these oligonucleotides using LipofectamineTM 2000 (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. For osteo/odontogenic induction, the medium was changed to osteogenic medium after 24 h transfection.
Dual Luciferase Reporter Assay
TargetScan, miRPathDB, starBase and PiTA database were used to predict potential targets of miR-497-5p. The dual luciferase reporter assay was performed to determine whether miR-497-5p could directly regulate Smurf2. A Smurf2 3′-UTR reporter vector was synthetized by BioSune Biotechnology. The wild type (WT) and mutant-type (Mut) plasmid was termed Smurf2-WT and Smurf2-Mut, respectively. HEK293T cells were cultured in a 96-well plate. Then Smurf2-WT or Smurf2-Mut was co-transfected with miR-497-5p mimics or its negative control into HEK293T cells. After 48 h of incubation, firefly and renilla luciferase activities were analyzed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States). Renilla luciferase activity was used as an internal control.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from osteogenic induced cells using RNAiso Plus (Takara, Shiga, Japan). Complementary DNA (cDNA) was synthesized with a Reverse Transcription System (Takara) following the manufacturer’s protocol. qRT-PCR was performed on by Roche LightCycler§ 480 (Roche, Mannheim, Germany) using SYBR Premix Ex Taq (Takara). Relative expression of mRNA or miRNA were evaluated, GAPDH or U6 was used as an endogenous normalization control. The primers used for amplification were listed in the Supplementary Table 1.
Western Blot Analysis
After 7 days of osteogenic induction, the transfected SCAP were harvested and lysed with RIPA lysis buffer containing 1% phenylmethylsulfonyl fluoride (PMSF) (Beyotime, Shanghai, China) on ice. Total protein concentrations were measured by the BCA Protein Assay Kit (Solarbio). The proteins in all samples were separated by SDS-PAGE gels, and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, United States). After blocking in 5% skim milk for 1 h at room temperature, the membranes were incubated with primary antibodies overnight at 4°C. Primary antibodies included DSPP (Santa Cruz Biotechnology, Santa Cruz, CA, United States), Collagen I (Wanlei, Shenyang, China), ALP (Abcam, Cambridge, MA, United Kingdom), Runx2, OSX (Abcam), OPN (Abcam), SNIP1 (Abcam), Smurf1 (Proteintech Group, Chicago, IL, United States), Smurf2, Smad2, Smad3, Smad4 (Cell Signaling Technology), and GAPDH (Proteintech Group). After that, the membranes were incubated with horseradish peroxidase–conjugated anti-mouse or anti-rabbit secondary antibodies (Proteintech Group) at room temperature for 1 h. Protein bands were visualized using the enhanced chemiluminescence (Millipore) and the protein levels were quantified using the ImageJ software.
Statistical Analysis
Data are presented as the mean ± SD from at least three independent experiments. The t-test, one-way and two-way ANOVA were used for statistical analysis. P < 0.05 was considered statistically significant. All statistical analyses were performed using the GraphPad Prism 6.
Results
Characteristics of SCAP and Osteogenic Differentiation
Human SCAP was isolated and adhered to the culture dish with spindle-liked morphology (Figures 1A,B). CCK-8 assayed the proliferation of SCAP including the incubation, logarithmic growth, and plateau periods. The growth curve was shown in Figure 1C. Flow cytometry analysis showed that cell surface markers of SCAP were positive for STRO-1 (31.8%), CD24 (23.2%) and CD146 (99.5%), and negative for CD34 (0.27%) and CD45 (0.16%) (Figure 1D). After osteogenic induction, the ALP activity increased significantly compared with control group (Figure 1E). ALP staining and Alizarin red staining showed a clear bluish violet stains and mineralized nodules, respectively (Figure 1F). The mRNA expression of specific markers, including Dentin sialophosphoprotein (DSPP), Collagen type I (Collagen I), Alkaline phosphatase (ALP), Runt-related transcription factor 2 (Runx2) and Bone sialoprotein (BSP), were upregulated after the osteogenic induction (Figure 1G). These results revealed that SCAP has a good proliferation ability and osteogenic differentiation potential.
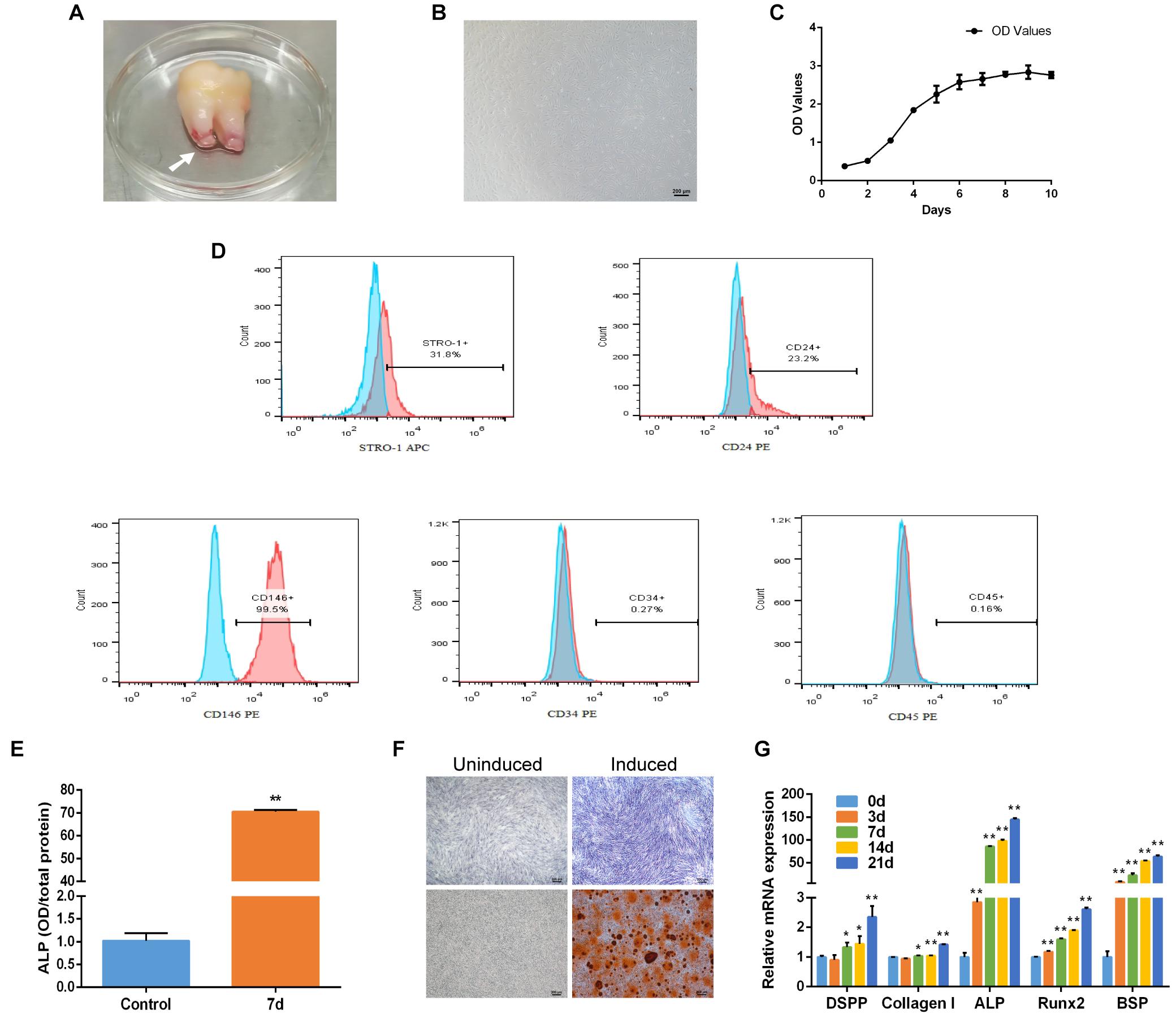
Figure 1. Characteristics and osteogenic differentiation of SCAP. (A) Human apical papilla (white arrow) attached to the apex of extracted immature third molar. (B) The morphology of SCAP. (C) Growth curve gained according to CCK-8 method. (D) Cell surface markers of SCAP displayed by flow cytometry. (E) ALP activity at day 7 of osteogenic differentiation. (F) ALP staining on day 7 and Alizarin red staining on day 21 after osteogenic induction. (G) Osteogenic specific genes examined by qRT-PCR at different time points. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
Increased Expression of miR-497-5p During Osteo/Odontogenic Differentiation of SCAP
MiRNA microarray analysis showed that miRNAs differentially expressed between osteo/odontogenic induction SCAP and control group. Heatmap revealed that the expression levels of 45 miRNAs significantly changed, with 24 miRNAs up-regulated and 21 down-regulated compared to those in the control group (Figure 2A). To verify the microarray data, five miRNAs from differentially expressed miRNAs were selected and detected using qRT-PCR. The results showed that all five miRNAs were significantly changed and were consistent with microarray data (Figure 2B). In order to screen for the most differentially expressed miRNA in osteo/odontogenic differentiation, five miRNAs were overexpressed by transfecting miRNAs mimics and the protein expression of osteo/odontogenic differentiation-related genes (DSPP, Collagen I, ALP, Runx2, and OPN) was detected using western blot. We found these markers were increased significantly in miR-497-5p group compared to those in the NC group (Figures 2C,D). To further observe the phase changes of miR-497-5p in the SCAP osteo/odontogenic differentiation, we determined its expression and found that miR-497-5p increased on the day 7 and remained high level until day 21 (Figure 2E), indicating that miR-497-5p might promote the osteo/odontogenic differentiation of SCAP.
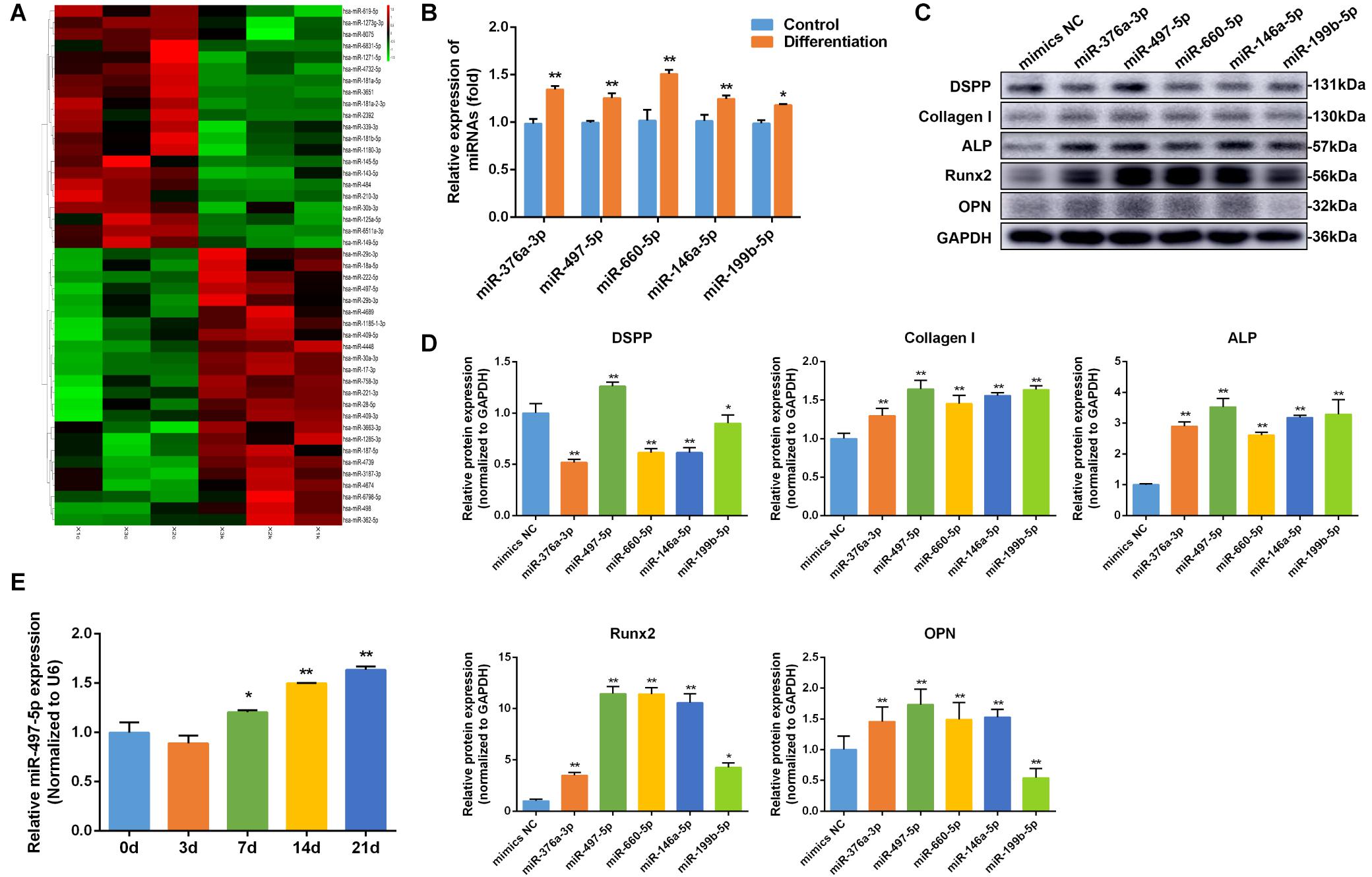
Figure 2. miR-497-5p was upregulated during osteo/odontogenic differentiation of SCAP. (A) Heat map of the differentially expressed miRNAs in differentiated SCAP. Red indicates up-regulated miRNAs and green indicates down-regulated miRNAs. (B) Expression levels of miR-376a-3p, miR-497-5p, miR-660-5p, miR-146a-5p, miR-199b-5p in differentiated SCAP examined by qRT-PCR. (C,D) The expression of osteo/odontogenic differentiation related proteins in overexpressed miR-376a-3p, miR-497-5p, miR-660-5p, miR-146a-5p, miR-199b-5p examined by western blot. (E) Time course of miR-497-5p expression during osteogenic differentiation of SCAP. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
MiR-497-5p Promoted Osteo/Odontogenic Differentiation in SCAP
To determine whether miR-497-5p regulated osteo/odontogenic differentiation, miR-497-5p mimics was transfected into SCAP and the cells were cultured in osteogenic medium. The levels of miR-497-5p were elevated by approximately 6,000-fold compared to those in the NC group, as confirmed by qRT-PCR after 24 h of transfection, which indicated high transfection efficiency (Figure 3A). The overexpression of miR-497-5p enhanced the ALP activity and the mRNA expression of osteo/odontogenic specific genes, including DSPP, Collagen I, ALP, Runx2 and Osterix (OSX), compared to that of negative control (Figures 3B,C). Consistently, the protein levels of DSPP, Collagen I, ALP, Runx2, OSX, OPN were upregulated in miR-497-5p overexpression group (Figure 3D). The Alizarin red staining also showed enhanced osteogenic capacity of SCAP (Figure 3E). These findings suggested that miR-497-5p might be a promoter for osteo/odontogenic differentiation of SCAP.
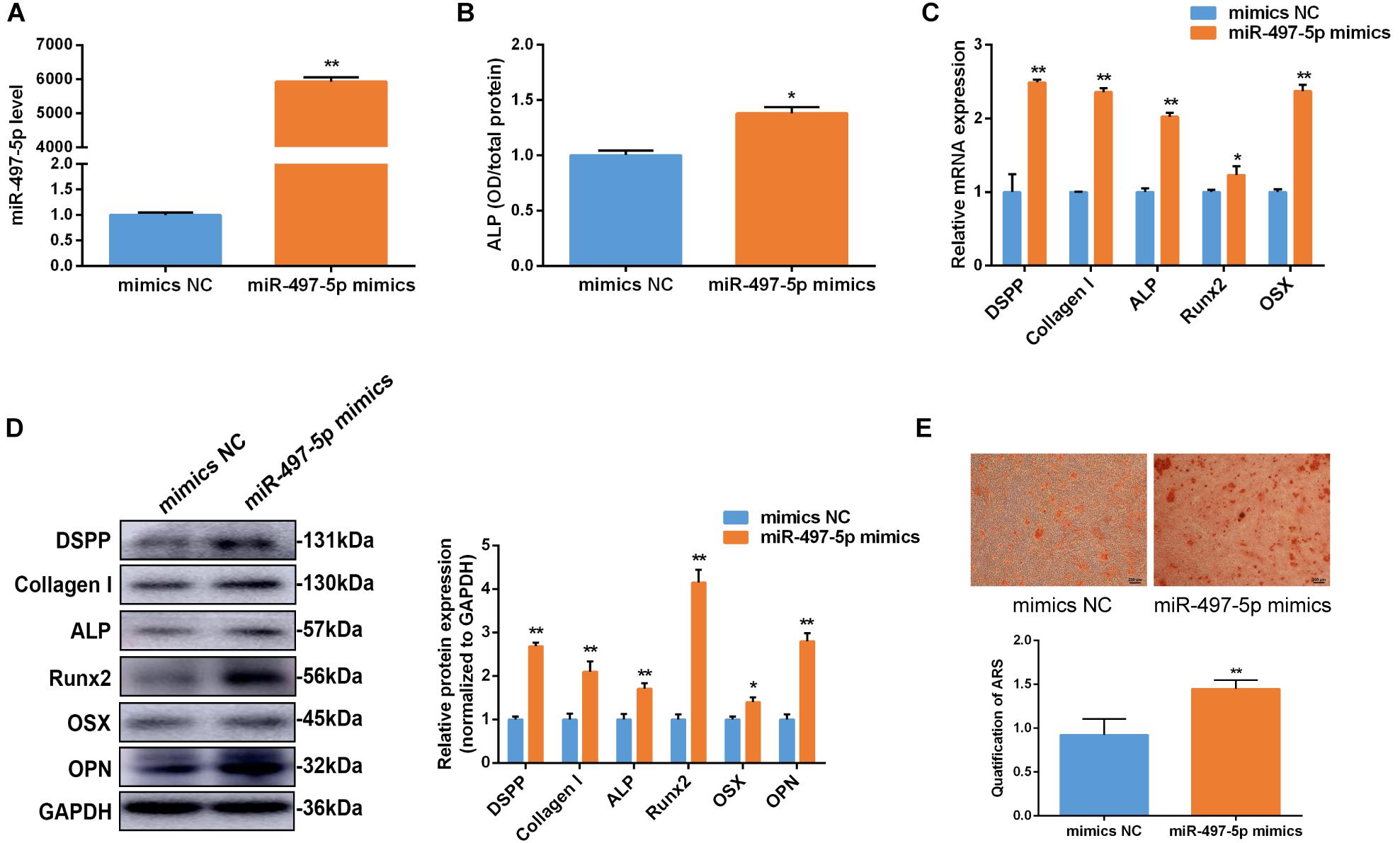
Figure 3. miR-497-5p promoted osteo/odontogenic differentiation in SCAP. (A) miR-497-5p expression assessed by qRT-PCR in SCAP transfected with miR-497-5p mimics. (B) ALP activity measured on day 7 of osteogenic differentiation. (C,D) mRNA and protein expression of osteo/odontogenic markers after miR-497-5p overexpression. (E) Alizarin red staining and quantitative assay after miR-497-5p overexpression. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
Inhibition of miR-497-5p Suppressed Osteo/Odontogenic Differentiation in SCAP
To further clarify the effect of miR-497-5p on the osteo/odontogenic differentiation, miR-497-5p inhibitor was transfected into SCAP and the cells were cultured in osteogenic medium. The inhibition efficiency of miR-497-5p was detected (Figure 4A). Consequently, the ALP activity was suppressed (Figure 4B). The osteo/odontogenic specific markers (DSPP, Collagen I, ALP, Runx2, OSX, and OPN) were remarkably down-regulated compared to their negative controls (Figures 4C,D). The Alizarin red staining reflected the same effects (Figure 4E). All the results thus suggest that down-regulating miR-497-5p could suppress the osteo/odontogenic differentiation of SCAP.
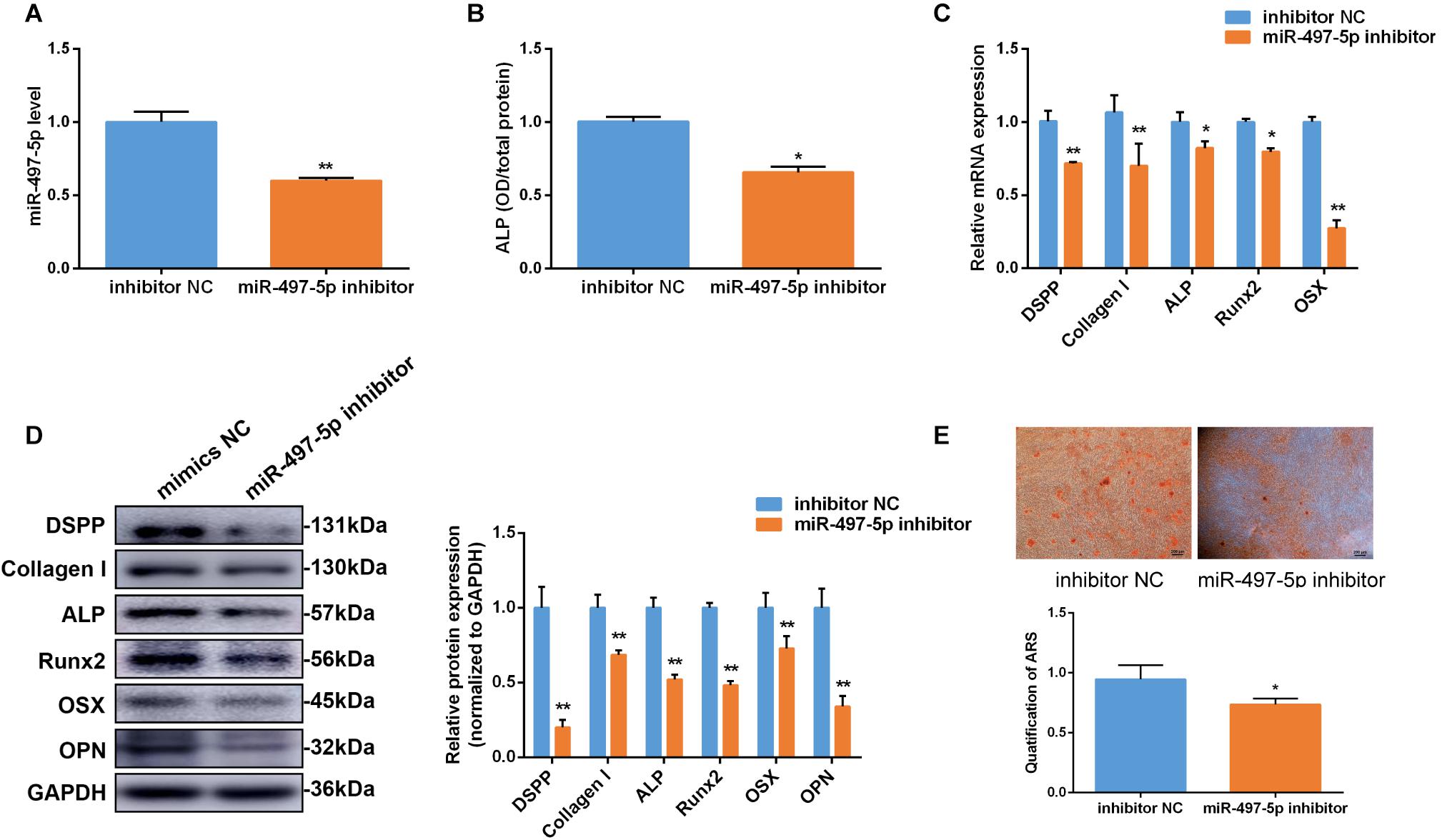
Figure 4. Inhibition of miR-497-5p suppress osteo/odontogenic differentiation in SCAP. (A) miR-497-5p expression assessed by qRT-PCR in SCAP transfected with miR-497-5p inhibitor. (B) ALP activity measured on day 7 of osteogenic differentiation. (C,D) mRNA and protein expression of osteo/odontogenic markers examined after miR-497-5p suppression. (E) Alizarin red staining and quantitative assay after miR-497-5p suppression. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
miR-497-5p Directly Targeted Smurf2
To reveal the probable molecular mechanism by which miR-497-5p mediates osteo/odontogenic differentiation of SCAP, TargetScan, miRPathDB, starBase and PiTA were used to predict potential targets of miR-497-5p (Figure 5A). It was discovered that three osteogenic-associated potential target genes including SMAD specific E3 ubiquitin protein ligase 1 (Smurf1), Smurf2 and Smad nuclear interacting protein 1 (SNIP1), contain a miR-497b-5p binding site in their 3′-UTRs (Supplementary Figure 1). After miR-497-5p overexpressed or suppressed, three target genes were detected. The most significant change was observed in the mRNA and protein levels of Smurf2 (Figures 5B,C). To further confirm whether miR-497-5p directly targets Smurf2, we constructed dual luciferase reporter vectors containing the Smurf2 3′UTR wild type (WT) or mutated sequence (Mut) (Figure 5D). The dual luciferase vector and miR-497-5p mimics (or mimics NC) were co-transfected into HEK 293T cells. Dual luciferase reporter assay showed that miR-497-5p dramatically suppressed the luciferase activity of WT Smurf2 than that of the negative control, and there was no significant alteration in the luciferase activity of mutated Smurf2 (Figure 5E). The above demonstrated that miR-497-5p negatively regulated Smurf2 by directly binding to the 3′UTR of its mRNA. In other words, Smurf2 is a target gene of miR-497-5p.
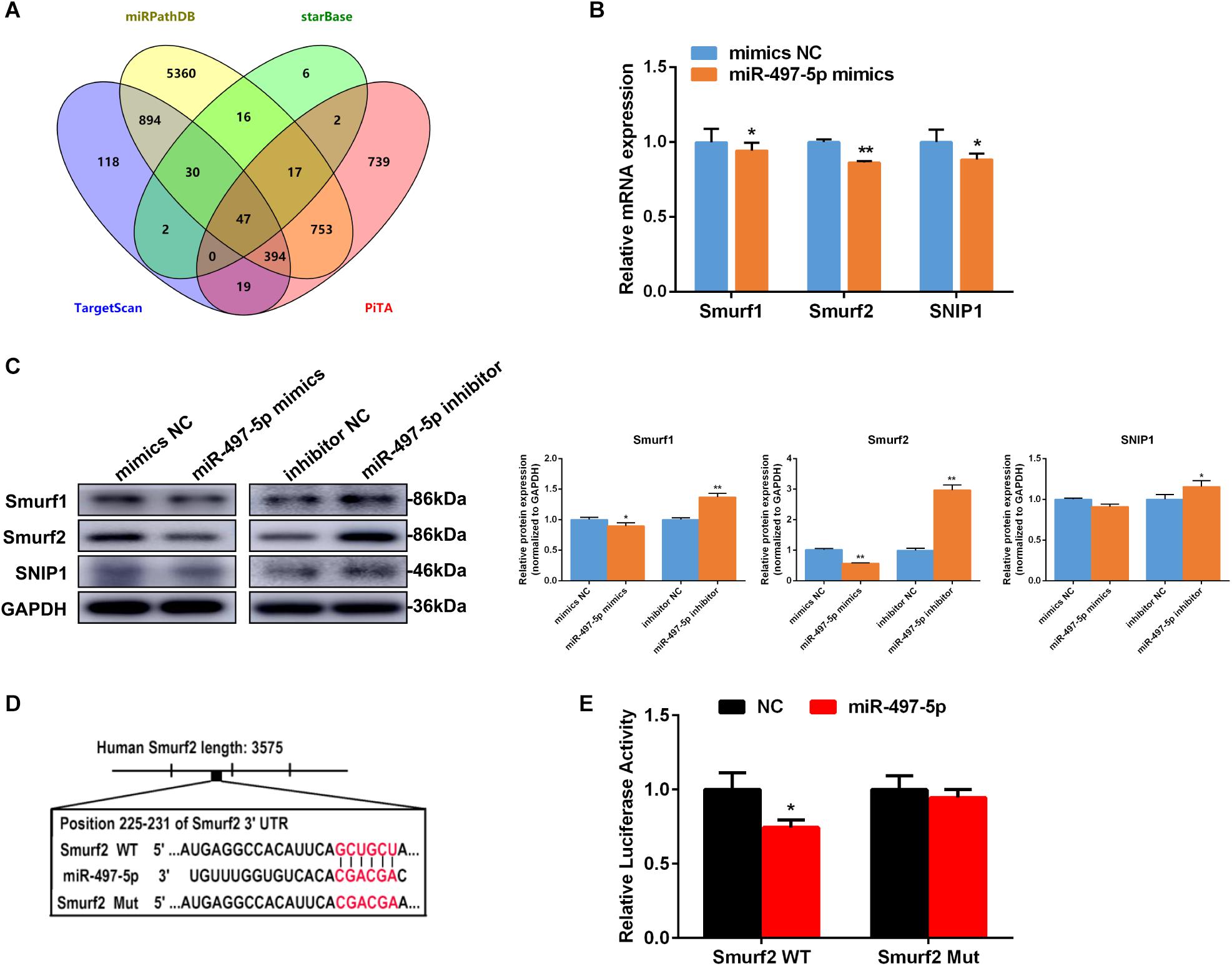
Figure 5. miR-497-5p directly targets Smurf2. (A) Venn diagram showed number of miR-497-5p target genes predicted by performing TargetScan, miRPathDB, starBase and PiTA. (B,C) Candidate gene expressions in SCAP detected by qRT-PCR and western blot after transfected with the miR-497-5p mimics/inhibitor. (D) Schematic diagrams indicating the Smurf2 wild-type (WT) contains miR-497-5p binding sites. (E) Luciferase activities of Smurf2 3′UTR WT or Mut reporter plasmids. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
Smurf2 Is Involved in Osteo/Odontogenic Differentiation of SCAP
In this study, Smurf2 mRNA expression decreased on day 3 and remained low level to day 21 during osteo/odontogenic differentiation of SCAP (Figure 6A). To confirm the effect of Smurf2 on osteo/odontogenic differentiation of SCAP, Smurf2 expression was interfered by transfecting siSmurf2 in SCAP. The knockdown efficiency was estimated by qRT-PCR and western blot. As our expected, the siSmurf2 could markedly decreased Smurf2 expression (Figures 6B,C). Moreover, Smurf2 knockdown significantly promoted the ALP activity and osteo/odontogenic genes expression (Figures 6D–F). The Alizarin red staining also confirmed that knockdown of Smurf2 accelerated the osteogenic differentiation of SCAP (Figure 6G). These results demonstrated that Smurf2 was involved in osteo/odontogenesis of SCAP.
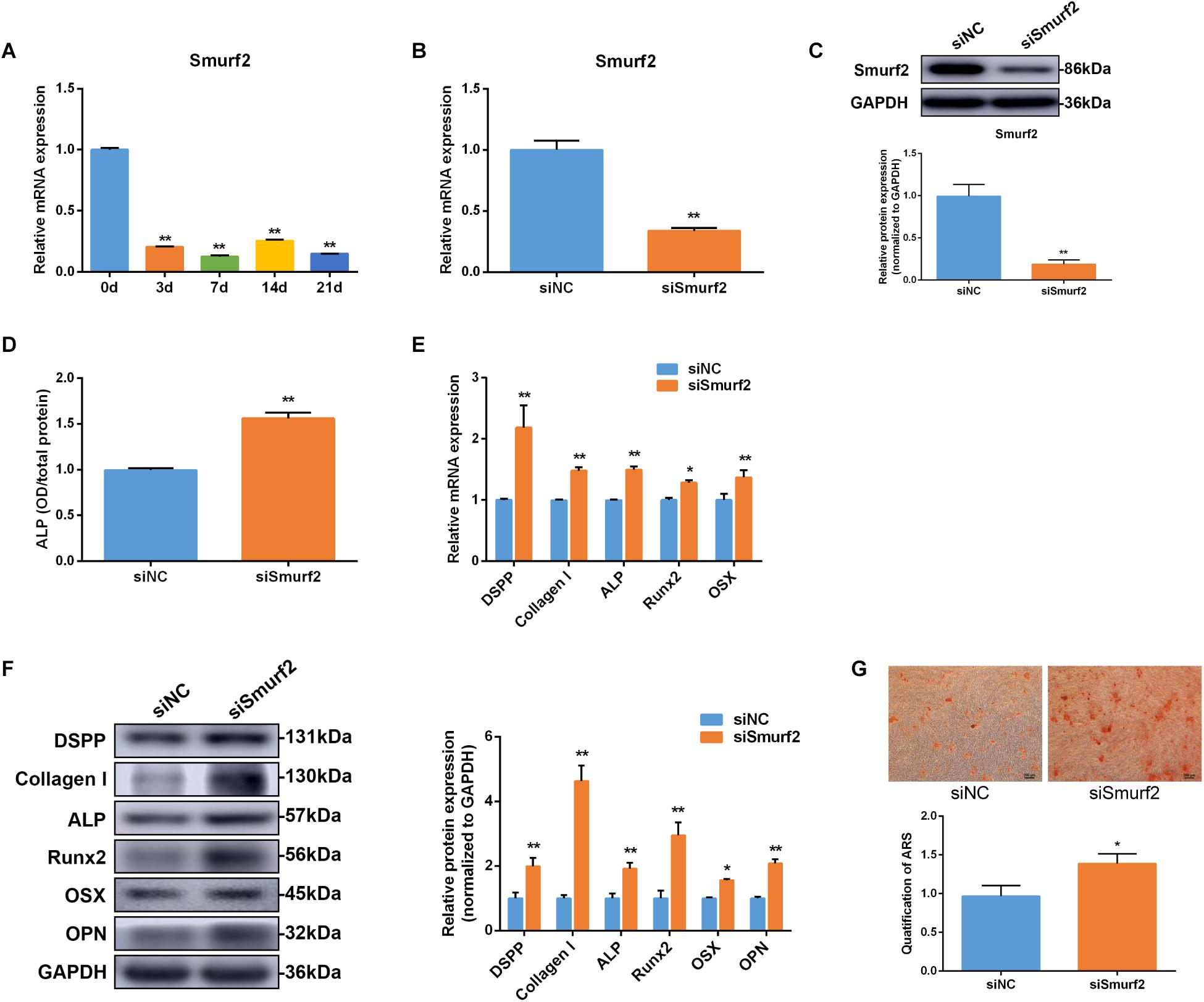
Figure 6. Smurf2 was involved in osteo/odontogenic differentiation of SCAP. (A) Smurf2 mRNA expression was examined via qRT-PCR at different time points during osteo/odontogenic differentiation of SCAP. (B,C) The knockdown efficiency of Smurf2 siRNA was confirmed by qRT-PCR and western blot. (D) ALP activity was detected on day 7 after Smurf2 knockdown. (E,F) mRNA and protein expression of osteo/odontogenic markers were examined by qRT-PCR and western blot after Smurf2 knockdown. (G) Alizarin red staining and quantitative assay after Smurf2 knockdown. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
Smurf2 Knockdown Blocked the Inhibitory Effect of miR-497-5p Inhibitor
To further confirm whether the effect of miR-497-5p on osteo/odontogenic differentiation depended on Smurf2 in SCAP, we co-transfected the miR-497-5p inhibitor and siSmurf2 or their respective negative controls into SCAP. As expected, the inhibitory effects of miR-497-5p inhibitor on SCAP were suppressed after Smurf2 knockdown (Figure 7A). The result indicated that miR-497-5p regulated osteo/odontogenic differentiation of SCAP through targeting Smurf2.
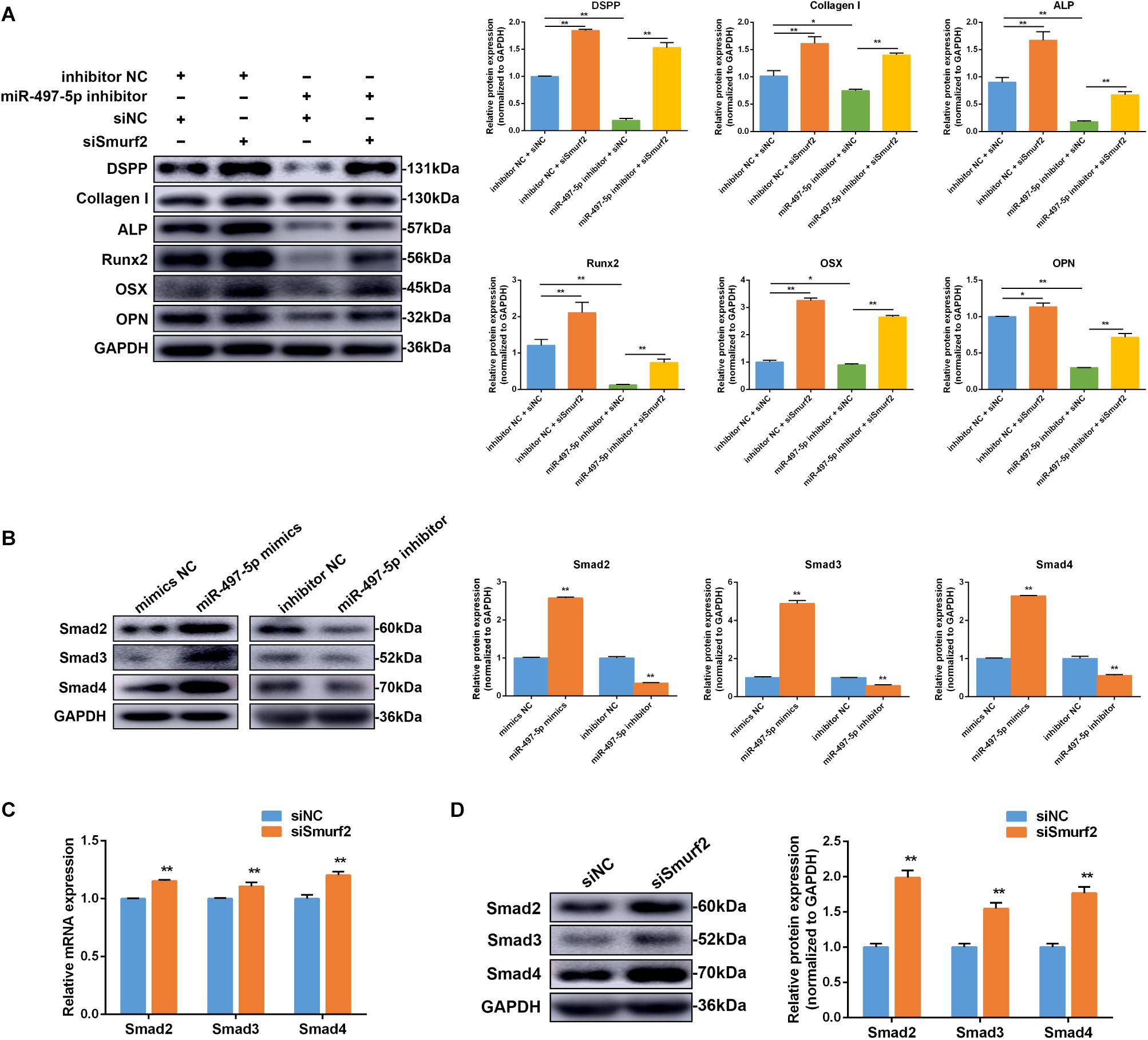
Figure 7. miR-497-5p modulated osteo/odontogenic differentiation of SCAP via Smad signaling pathway by targeting Smurf2. (A) The protein expression of osteo/odontogenic markers after co-transfected with miR-497-5p inhibitor and siSmurf2 or their respective negative controls. (B) Smad2, Smad3 and Smad4 protein expression level following miR-497-5p mimics and inhibitor transfection in SCAP. (C,D) Smad2, Smad3, and Smad4 mRNA and protein levels examined via qRT-PCR and western blot following Smurf2 knockdown in SCAP. Data are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with control group.
MiR-497-5p Regulated Osteo/Odontogenic Differentiation in SCAP via the Smad Signaling Pathway
To determine if miR-497-5p increased SCAP osteo/odontogenic differentiation through the Smad signaling pathway, miR-497-5p mimics or inhibitor were transfected into cells. Western blot revealed Smad2, Smad3 and Smad4 expression levels were upregulated after transfection with miR-497-5p mimics and downregulated after transfection with miR-497-5p inhibitors (Figure 7B). The results indicated that Smad signaling pathway was involved in miR-497-5p-mediated osteo/odontogenic differentiation. Moreover, we further confirmed that mRNA and protein levels of Smad2, Smad3 and Smad4 were highly upregulated following transfection of siSmurf2 into SCAP (Figures 7C,D). All these results demonstrated that miR-497-5p promote osteo/odontogenic differentiation of SCAP via Smad signaling pathway by targeting Smurf2.
Discussion
Recent studies indicated that miRNAs were involved in diverse biological processes including cell proliferation, differentiation, apoptosis, and carcinogenesis. MiR-497 was reported to be upregulated or downregulated in various cancers, thereby suggesting its diverse roles in different tissues (Yan et al., 2008; Lan et al., 2014; Maura et al., 2015). However, its role in osteogenic/odontogenic differentiation and related mechanisms require further investigation. In the present study, we first discovered miR-497-5p was upregulated during osteo/odontogenic differentiation in human SCAP and overexpression of miR-497-5p increased its osteo/odontogenic differentiation of SCAP. This is consistent with a previous study, which reported that overexpression of miR-497∼195 promotes CD31hiEMCNhi endothelial cells angiogenesis and osteogenesis (Yang et al., 2017). Contrary to our findings, miR-497 suppressed proliferation and osteogenic differentiation in human primary mesenchymal stromal/stem cells (MSC) (Almeida et al., 2016). These results might be attributed to distinct origin of the cells utilized in several differentiation studies. Human MSC and SCAP, as postnatal mesenchymal stem cells, share some similarity, however, they have their own characteristics. The tissue-specific expression of miRNAs corresponded with the diversity of their functions. The SCAP of developing tooth root have their distinct biological characteristics (Sonoyama et al., 2006, 2008).
To investigate the molecular mechanism by which miR-497-5p promoted human SCAP osteo/odontogenic differentiation, we searched for potential target genes of miR-497-5p. We identified Smurf2 was a direct target gene of miR-497-5p by using bioinformatics analysis and dual luciferase reporter assay. Smurf2 is a member of E3 ubiquitin ligases (Lin et al., 2000; Zhang et al., 2001). It interacted with Smad proteins (important proteins of TGF-β/Smad signaling pathway) and other osteogenic-related genes, leading to their degradation and thereby the negative regulation of the TGF-β/Smad signaling pathway and osteogenic differentiation process (Zhang et al., 2001; Izzi and Attisano, 2004; Kaneki et al., 2006; Lönn et al., 2008). Kaneki et al. (2006) found that tumor necrosis factor inhibited bone formation by promoting Runx2 degradation via upregulation of Smurf1 and Smurf2 in vitro and in vivo. The latest study showed that TRAF4 up-regulated the osteogenic differentiation of MSCs by acting as an E3 ubiquitin ligase to mediate the ubiquitination of Smurf2 and cause Smurf2 degradation (Li et al., 2019). In our study, overexpression of miR-497-5p significantly suppressed the expression of Smurf2. In contrast, inhibition of miR-497-5p increased Smurf2 expression. More importantly, dual luciferase reporter gene assay confirmed that Smurf2 was a direct target of miR-497-5p.
Despite it has been reported that Smurf2 is the target gene of miR-497 in lung cancer cells (Dong-Kyu et al., 2019), miRNA profiles are tissue-specific. Specific cell type differentiation is a process involving complex network transcription factors. However, the regulation of miR-497-5p and Smurf2 on osteo/odontogenic differentiation in SCAP is largely unknown. Therefore, we further investigated the effects of Smurf2 on osteo/odontogenic differentiation in SCAP. The finding showed that Smurf2 was downregulated during the odontogenic differentiation of SCAP and Smurf2 knockdown significantly elevated expression of osteo/odontogenic markers, thus resembling the effect of miR-497-5p overexpression. A rescue effect was observed, wherein Smurf2 knockdown could block the effect of miR-497-5p inhibition in SCAP osteo/odontogenic differentiation. In conclusion, our study demonstrated that miR-497-5p enhanced osteo/odontogenic differentiation in SCAP through suppressing Smurf2 expression.
Smad signaling pathway plays a crucial role in osteogenic differentiation. The ubiquitin-proteasome degradation is a vital mechanism regulating TGF-β/Smad pathway. Smurf2, an important component of ubiquitin-proteasome system, participates in its regulation (Kavsak et al., 2000; Zhang et al., 2001; Ganji et al., 2015). The signal transduction of the TGF-β/Smad pathway mainly depends on the Smad proteins, which act as critical intracellular receptors, including Smad1-8 (Massagué and Wotton, 2000). For example, Smurf2 interacted with Smad7 to compose a complex in the nucleus, then entered the cytoplasm to degrade TβRI, and inhibits the signal transduction of TGF-β (Ganji et al., 2015). It also has been reported that Smurf2 combining Smad7-induced output and collection activated TGF-β receptors. Then the receptor and Smad7 were degraded via the proteasome and lysosomal pathways (Kavsak et al., 2000). In this study, we observed Smad2, Smad3 and Smad4 protein levels were upregulated by miR-497-5p mimics and decreased by miR-497-5p inhibitors, indicating Smad signaling pathway was activated by miR-497-5p. Furthermore, when Smurf2 was knocked down by siRNA, Smad2, Smad3 and Smad4 mRNA and protein levels were upregulated. Our results suggested that miR-497-5p could directly target Smurf2 and regulate the osteo/odontogenic differentiation through Smad signaling pathway.
In the future study, additional strategies might investigate the effect of miR-497-5p on the osteo/odontogenic differentiation under the undifferentiated condition and more studies could be performed to confirm these factors by well-designed in vivo experiments.
In summary, our study was the first to demonstrate that miR-497-5p acted as a positive regulator of osteo/odontogenic differentiation in SCAP. Importantly, miR-497-5p promoted osteo/odontogenic differentiation by targeting Smurf2 and modulating the Smad signaling pathway as depicted in Figure 8. Our findings reveal a novel function of miR-497-5p in the osteo/odontogenic differentiation, and suggest the application of miR-497-5p as a potential target to enhance dental pulp/dentine regeneration and to develop new therapeutic approaches in dental tissue regeneration.
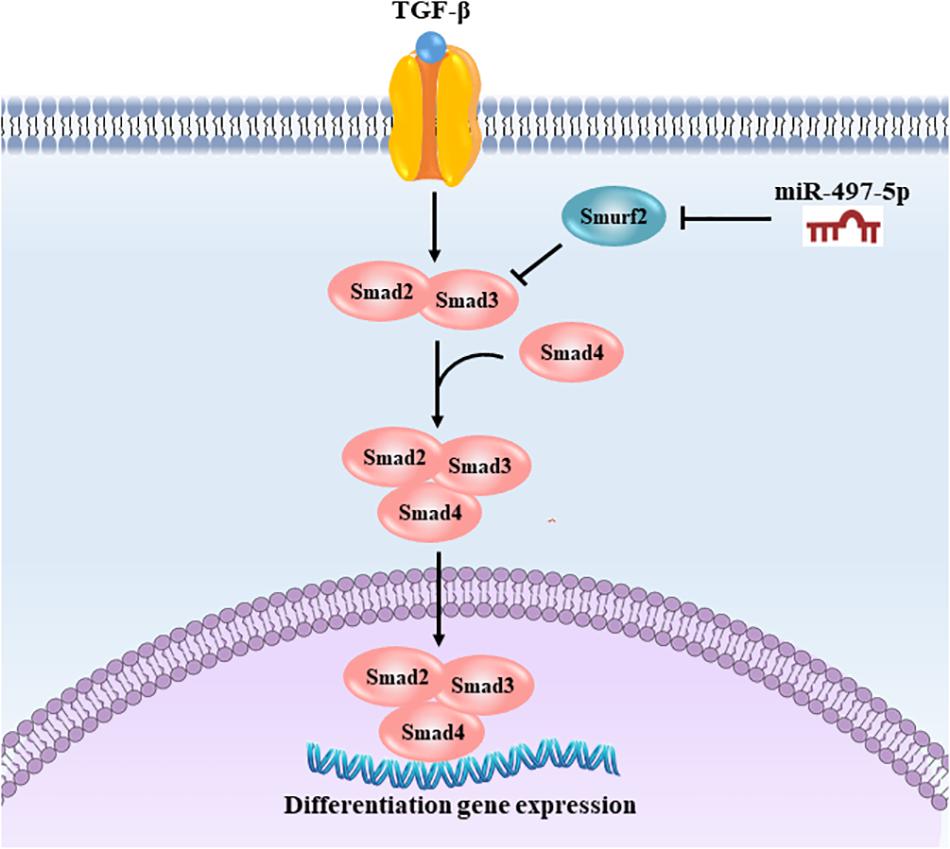
Figure 8. Schematic illustration of miR-497-5p regulating osteo/odontogenic differentiation of SCAP. miR-497-5p suppresses Smurf2 expression, resulting in upregulating Smad signaling pathway, thereby promoting osteo/odontogenic differentiation.
Data Availability Statement
The microarray data has been deposited in GEO: GSE154466.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the School and Hospital of Stomatology, Shandong University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
JL, XW, CZ, and YW contributed in conception and design. JL, XW, MS, and JD contributed in performing experiments. JL, JY, and WZ contributed in data collection, interpretation and statistical analysis. JL and XW contributed in drafting manuscript. CZ and YW contributed in critically revise manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the Natural Science Foundation of Shandong Province (ZR2017MH104), the Natural Science Foundation of Shandong Province (ZR2017BH050) and the General Research Fund (GRF) grants from the Research Grants Council of Hong Kong (grant no. HKU17111619).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Permission to Reuse and Copyright
All authors approved the reuse and copyright licenses of this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.582366/full#supplementary-material
References
Abe, S., Yamaguchi, S., Watanabe, A., Hamada, K., and Amagasa, T. (2008). Hard tissue regeneration capacity of apical pulp derived cells (APDCs) from human tooth with immature apex. Biochem. Biophys. Res. Commun. 371, 90–93. doi: 10.1016/j.bbrc.2008.04.016
Almeida, M. I., Silva, A. M., Vasconcelos, D. M., Almeida, C. R., Caires, H., Pinto, M. T., et al. (2016). miR-195 in human primary mesenchymal stromal/stem cells regulates proliferation, osteogenesis and paracrine effect on angiogenesis. Oncotarget 7, 7–22. doi: 10.18632/oncotarget.6589
Bakopoulou, A., Leyhausen, G., Volk, J., Tsiftsoglou, A., Garefis, P., Koidis, P., et al. (2011). Comparative analysis of in vitro osteo/odontogenic differentiation potential of human dental pulp stem cells (DPSCs) and stem cells from the apical papilla (SCAP). Arch. Oral Biol. 56, 709–721. doi: 10.1016/j.archoralbio.2010.12.008
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Chen, S., Zheng, Y., Zhang, S., Jia, L., and Zhou, Y. (2017). Promotion effects of miR-375 on the osteogenic differentiation of human adipose-derived mesenchymal stem cells. Stem Cell Rep. 8, 773–786. doi: 10.1016/j.stemcr.2017.01.028
Chrepa, V., Austah, O., and Diogenes, A. (2017). Evaluation of a commercially available hyaluronic acid Hydrogel (Restylane) as injectable scaffold for dental pulp regeneration: an in vitro evaluation. J. Endod. 43, 257–262. doi: 10.1016/j.joen.2016.10.026
Dong-Kyu, C., Jinyoung, P., Moonsoo, C., Eunmi, B., Mihue, J., Young, S. Y., et al. (2019). MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-β receptor I ubiquitination. Mol. Oncol. 13, 2663–2678. doi: 10.1002/1878-0261.12581
Ganji, A., Roshan, H. M., Varasteh, A., Moghadam, M., and Sankian, M. (2015). The effects of WW2/WW3 domains of Smurf2 molecule on TGF-beta signaling and arginase I gene expression. Cell Biol. Int. 39, 690–695. doi: 10.1002/cbin.10446
Hammond, S. M., Caudy, A. A., and Hannon, G. J. (2001). Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2, 110–119. doi: 10.1038/35052556
Huang, G. T., Yamaza, T., Shea, L. D., Djouad, F., Kuhn, N. Z., Tuan, R. S., et al. (2010). Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng. Part A 16, 605–615. doi: 10.1089/ten.TEA.2009.0518
Izzi, L., and Attisano, L. (2004). Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene 23, 2071–2078. doi: 10.1038/sj.onc.1207412
Julia, W., Stephanie, J., Sarina, K., Richard, G. I, and Sven, D. (2009). Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234. doi: 10.1038/ncb0309-228
Kaneki, H., Guo, R., Chen, D., Yao, Z., Schwarz, E. M., Zhang, Y. E., et al. (2006). Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J. Biol. Chem. 281, 4326–4333. doi: 10.1074/jbc.M509430200
Kavsak, P., Rasmussen, R. K., Causing, C. G., Bonni, S., Zhu, H., Thomsen, G. H., et al. (2000). Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6, 1365–1375. doi: 10.1016/S1097-2765(00)00134-9
Koutsoumparis, A., Vassili, A., Bakopoulou, A., Ziouta, A., and Tsiftsoglou, A. S. (2018). Erythropoietin (rhEPOa) promotes endothelial transdifferentiation of stem cells of the apical papilla (SCAP). Arch. Oral Biol. 96, 96–103. doi: 10.1016/j.archoralbio.2018.09.001
Lan, J., Xue, Y., Chen, H., Zhao, S., Wu, Z., Fang, J., et al. (2014). Hypoxia-induced miR-497 decreases glioma cell sensitivity to TMZ by inhibiting apoptosis. FEBS Lett. 588, 3333–3339. doi: 10.1016/j.febslet.2014.07.021
Li, J., Wang, P., Xie, Z., Wang, S., Cen, S., Li, M., et al. (2019). TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 26, 2652–2666. doi: 10.1038/s41418-019-0328-3
Lin, X., Liang, M., and Feng, X. H. (2000). Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 275, 36818–36822. doi: 10.1074/jbc.C000580200
Lindsay, M. A. (2008). microRNAs and the immune response. Trends Immunol. 29, 343–351. doi: 10.1016/j.it.2008.04.004
Liu, F., Wang, X., Yang, Y., Hu, R., Wang, W., and Wang, Y. (2019a). The suppressive effects of miR-508-5p on the odontogenic differentiation of human dental pulp stem cells by targeting glycoprotein non-metastatic melanomal protein B. Stem Cell Res. Ther. 10:35. doi: 10.1186/s13287-019-1146-8
Liu, J., Du, J., Chen, X., Yang, L., Zhao, W., Song, M., et al. (2019b). The effects of mitogen-activated protein kinase signaling pathways on lipopolysaccharide-mediated osteo/odontogenic differentiation of stem cells from the apical papilla. J. Endod. 45, 161–167. doi: 10.1016/j.joen.2018.10.009
Lönn, P., Morén, A., Raja, E., Dahl, M., and Moustakas, A. (2008). Regulating the stability of TGFβ receptors and Smads. Cell Res. 19, 21–35. doi: 10.1038/cr.2008.308
Massagué, J., and Wotton, D. (2000). Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19, 1745–1754. doi: 10.1093/emboj/19.8.1745
Maura, F., Cutrona, G., Mosca, L., Matis, S., Lionetti, M., Fabris, S., et al. (2015). Association between gene and miRNA expression profiles and stereotyped subset #4 B-cell receptor in chronic lymphocytic leukemia. Leuk. Lymphoma 56, 3150–3158. doi: 10.3109/10428194.2015.1028051
Na, S., Zhang, H., Huang, F., Wang, W., Ding, Y., Li, D., et al. (2016). Regeneration of dental pulp/dentine complex with a three-dimensional and scaffold-free stem-cell sheet-derived pellet. J. Tissue Eng. Regen. Med. 10, 261–270. doi: 10.1002/term.1686
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Sonoyama, W., Liu, Y., Fang, D., Yamaza, T., Seo, B.-M., Zhang, C., et al. (2006). Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS One 1:e79. doi: 10.1371/journal.pone.0000079
Sonoyama, W., Liu, Y., Yamaza, T., Tuan, R. S., Wang, S., Shi, S., et al. (2008). Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J. Endod. 34, 166–171. doi: 10.1016/j.joen.2007.11.021
Sun, F., Wan, M., Xu, X., Gao, B., Zhou, Y., Sun, J., et al. (2014). Crosstalk between miR-34a and Notch signaling promotes differentiation in apical papilla stem cells (SCAPs). J. Dent. Res. 93, 589–595. doi: 10.1177/0022034514531146
Tang, J., Zhang, Z., Jin, X., and Shi, H. (2018). miR-383 negatively regulates osteoblastic differentiation of bone marrow mesenchymal stem cells in rats by targeting Satb2. Bone 114, 137–143. doi: 10.1016/j.bone.2018.06.010
Wan, F., Gao, L., Lu, Y., Ma, H., Wang, H., Liang, X., et al. (2016). Proliferation and osteo/odontogenic differentiation of stem cells from apical papilla regulated by Zinc fingers and homeoboxes 2: an in vitro study. Biochem. Biophys. Res. Commun. 469, 599–605. doi: 10.1016/j.bbrc.2015.11.135
Wang, W., Wang, X., Li, C., Chen, T., Zhang, N., Liang, Y., et al. (2019). Huaier suppresses breast cancer progression via linc00339/miR-4656/CSNK2B signaling pathway. Front. Oncol. 9:1195. doi: 10.3389/fonc.2019.01195
Yan, L. X., Huang, X. F., Shao, Q., Huang, M. Y., Deng, L., Wu, Q. L., et al. (2008). MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348–2360. doi: 10.1261/rna.1034808
Yang, H., Ma, L., Han, X., Yang, L., Tian, C., and Wang, Y. (2012). The effects of tumor necrosis factor-alpha on mineralization of human dental apical papilla cells. J. Endod. 38, 960–964. doi: 10.1016/j.joen.2012.04.005
Yang, M., Li, C. J., Sun, X., Guo, Q., Xiao, Y., Su, T., et al. (2017). MiR-497 approximately 195 cluster regulates angiogenesis during coupling with osteogenesis by maintaining endothelial Notch and HIF-1alpha activity. Nat. Commun. 8:16003. doi: 10.1038/ncomms16003
Yao, S., Zhao, W., Ou, Q., Liang, L., Lin, X., and Wang, Y. (2017). MicroRNA-214 suppresses osteogenic differentiation of human periodontal ligament stem cells by targeting ATF4. Stem Cells Int. 2017:3028647. doi: 10.1155/2017/3028647
Keywords: miR-497-5p, stem cells from apical papilla, Smurf2, Smad signaling pathway, osteo/odontogenic differentiation
Citation: Liu J, Wang X, Song M, Du J, Yu J, Zheng W, Zhang C and Wang Y (2020) MiR-497-5p Regulates Osteo/Odontogenic Differentiation of Stem Cells From Apical Papilla via the Smad Signaling Pathway by Targeting Smurf2. Front. Genet. 11:582366. doi: 10.3389/fgene.2020.582366
Received: 11 July 2020; Accepted: 12 October 2020;
Published: 30 October 2020.
Edited by:
Kai Kretzschmar, University Hospital Würzburg, GermanyReviewed by:
Naoki Iwamoto, Nagasaki University Hospital, JapanPaul Sharpe, King’s College London, United Kingdom
Copyright © 2020 Liu, Wang, Song, Du, Yu, Zheng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Wang, d2FuZ3lhbjE5NjVAc2R1LmVkdS5jbg==; Chengfei Zhang, emhhbmdjZkBoa3UuaGs=
†These authors have contributed equally to this work
 Junqing Liu
Junqing Liu Xiaolong Wang
Xiaolong Wang Mengxiao Song
Mengxiao Song Jing Du
Jing Du Jiali Yu1
Jiali Yu1 Wenzhou Zheng
Wenzhou Zheng Chengfei Zhang
Chengfei Zhang Yan Wang
Yan Wang