- 1Department of Biochemistry, School of Medicine, Ningbo University, Ningbo, China
- 2Zhejiang Key Laboratory of Pathophysiology, Medical School of Ningbo University, Ningbo, China
- 3Department of Chemical Pathology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong, China
- 4Laboratory for Genetics of Disease Susceptibility, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China
- 5Department of Cardiology, Ningbo No. 7 Hospital, Ningbo, China
- 6Department of Preventive Medicine, School of Medicine, Ningbo University, Ningbo, China
Introduction
Hypertension is among the first traits studied using genome-wide association study (GWAS). At first, GWAS by the Well-come Trust Case Control Consortium in 2007 did not identify any genome-wide significant single nucleotide polymorphism (SNP) (The Wellcome Trust Case Control Consortium, 2007), however, it was still asserted that GWAS would open the door to find the “missing heritability” of hypertension. In the following years, GWASs of hypertension were still unsuccessful in identifying robust loci until 2009. After shifting attention to the underlying quantitative traits (systolic blood pressure and diastolic blood pressure) and application of GWA meta-analyses of multiple cohorts to enlarge the sample size, the Global Blood Pressure Genetics (Global BPgen) Consortium and the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium identified 13 genome-wide significant signals (Levy et al., 2009; Newton-Cheh et al., 2009), although these SNPs had very small effect sizes and accounted for <0.2% of the overall blood pressure variation in the study populations.
Will GWAS Unlock the Genetic Basis of Hypertension?
There was a heated debate in HYPERTENSION on December 2010. Dominiczak and Munroe described the successes of aforementioned two studies, and predicted a bright future for GWAS in hypertension (Dominiczak and Munroe, 2010). On the contrary, Kurtz contended that GWAS had failed, and would continue to fail, to delineate the genetic basis of hypertension. He suggested that efforts and dollars should be shifted to other strategies and technologies that may hold greater chance for advancing our understanding of the genetic etiology of hypertension (Kurtz, 2010). Despite different opinions, both pro and con sides shared some common ground: low-frequency/rare variants are important, and the sample size should be further enlarged.
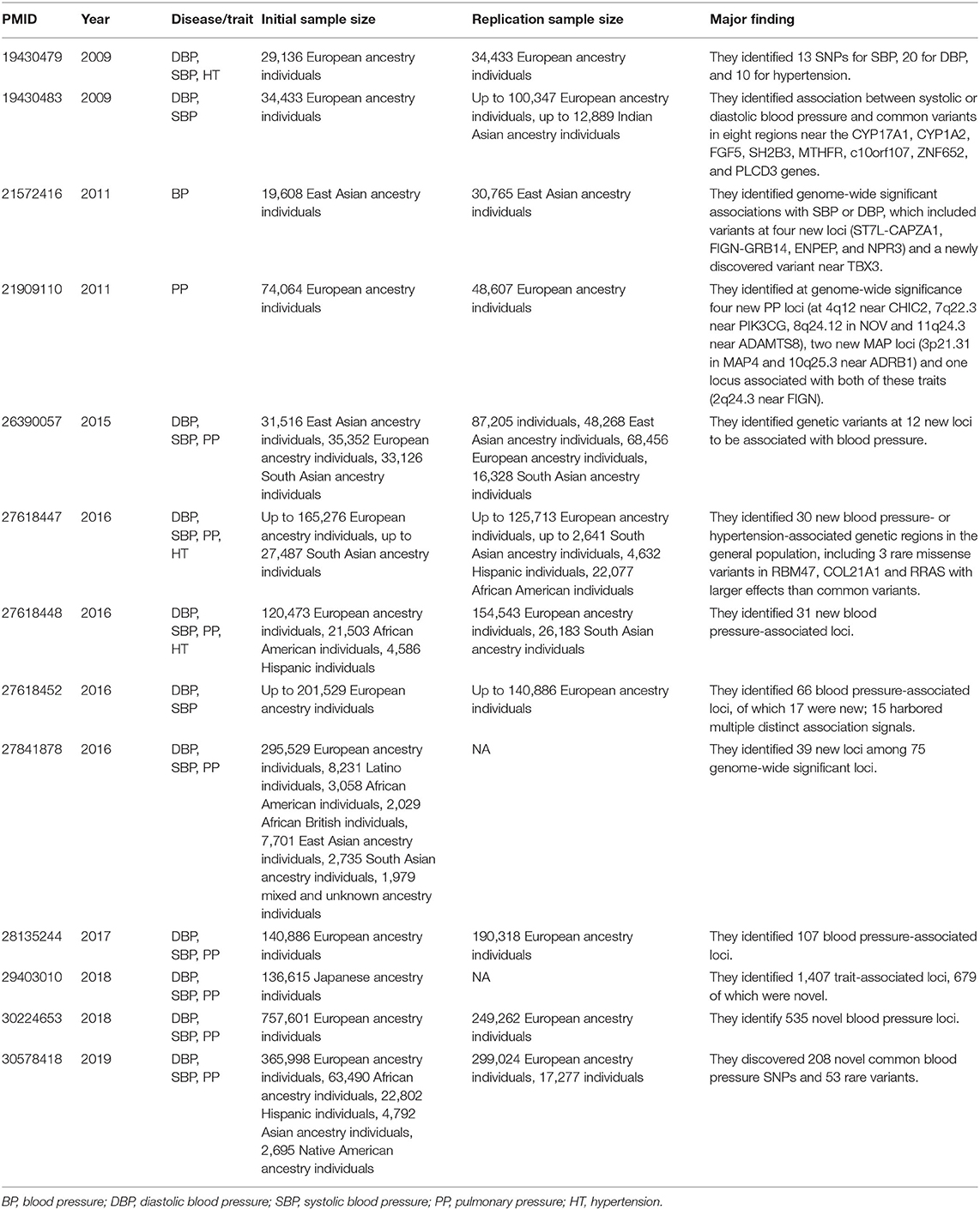
In 2016, three large-scale GWASs of hypertension were published in NATURE GENETICS (Ehret et al., 2016; Liu et al., 2016; Surendran et al., 2016). After applying newly designed microarray chips and meta-analyses of multiethnic populations with unprecedented large sample sizes (>300,000), these studies identified several new common loci of modest effects, and provided insights into the impact of low-frequency/rare variants. Subsequently, other genome-wide analyses of blood pressure traits (systolic, diastolic, and pulse pressure) were carried out in people of European ancestry drawn from UK Biobank (UKB), the International Consortium of Blood Pressure Genome Wide Association Studies (ICBP), the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, and other combined cohorts with very large sample size (Hoffmann et al., 2017; Warren et al., 2017; Evangelou et al., 2018). These studies identified hundreds of novel blood pressure loci that offer new biological insights into blood pressure regulation. Also, trans-ethnic genome-wide association study of blood pressure in up to 776,078 participants from the Million Veteran Program (MVP) and collaborating studies discovered 208 novel common blood pressure SNPs and 53 rare variants (Giri et al., 2019). Table 1 listed all high quality GWASs for hypertension or blood pressure published in NATURE GENETICS since 2007.
Gene–Environment Interplay
Thirteen years GWAS for hypertension, from the beginning “failure” to the recent “success,” can we say GWASs have deciphered the genetic architecture of hypertension? The answer is obviously NO. This is because of the fact that environmental factors and gene-environment interactions are likely major contributors to the development of hypertension, however, they were largely ignored in present GWASs (Cooper, 2018).
As widely accepted, hypertension is a consequence of significant interaction between genetic and environmental factors. Here, the meaning of “environment” is extensive, including intrauterine, postnatal and evolutionary environments. It has long been known that intrauterine environmental factors (e.g., maternal nutritional perturbation, toxin exposure, and stress during pregnancy) may result in hypertension in adult life. Recent epidemiological and experimental studies further indicated that paternal environmental factors, before conception and during sperm development, were also linked to the development of hypertension in later life (Li et al., 2016). On the other hand, classic epidemiological studies have addressed many postnatal environmental risk factors for hypertension, including living environments (cold temperature, air pollution, and toxins), life styles (lack of physical activity, psychological stress, smoking, alcohol abuse, drug use, improper nutrition, excessive salt intake, and obesity), and other demographic differences in age, gender, race, socioeconomic status, etc. From an evolutionary perspective, hypertension can be viewed as a maladaptation disease caused by the discrepancy between today's lifestyles and ancient adaptive genotypes. Physiologically, many risk factors for hypertension, such as enhanced salt and water avidity and vascular contractility, were adaptive traits associated with salt scarcity in the hot and humid climate of the ancestral African environment. As humans migrated out of Africa to cooler environments, the originally selected genes became maladaptive for new environments and turned into risk factors for hypertension (Young et al., 2005; Ji et al., 2016).
Discussion
Due to the complexity of essential hypertension, previous GWAS didn't find many robust variants. If more related factors are considered, and method like phenomics is used, future studies may help us to deeply understand this complex disease. As emphasized by Jeremy Berg, editor-in-chief of SCIENCE, genes alone do not determine our futures—environmental factors and chance also play important roles (Berg, 2016). Indeed, 13 years GWASs for hypertension really made a big step forward, however, if we leave behind another important factor, the environment, can also impede our progress. Incredibly, the environmental effects were ignored in almost all published GWASs of blood pressure traits or hypertension. Some studies were conducted in well-established cohorts like Global BPGen and CHARGE, and many environmental data should be available. Although analyzing the gene-environment interaction is still a great challenge, GWASs without consideration of environmental effects will definitely not go to address the key question regarding the genetic architecture of hypertension.
Author Contributions
L-dJ and JX conceived the opinion. L-dJ, NT, Z-fX, and JX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81402747), Funding of Science and Technology on Medicine and Health in Zhejiang Province (2019KY650), as well as the K.C. Wong Magna Fund in Ningbo University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Cooper, R. (2018). Hypertension, genes, and environment: challenges for prevention and risk prediction. Circulation 137, 662–664. doi: 10.1161/CIRCULATIONAHA.117.032196
Dominiczak, A. F., and Munroe, P. B. (2010). Genome-wide association studies will unlock the genetic basis of hypertension: pro side of the argument. Hypertension 56, 1017–1020. doi: 10.1161/HYPERTENSIONAHA.110.156208
Ehret, G. B., Ferreira, T., Chasman, D. I., Jackson, A. U., Schmidt, E. M., Johnson, T., et al. (2016). The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet. 48, 1171–1184. doi: 10.1038/ng.3667
Evangelou, E., Warren, H. R., Mosen-Ansorena, D., Mifsud, B., Pazoki, R., Gao, H., et al. (2018). Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425. doi: 10.1038/s41588-018-0205-x
Giri, A., Hellwege, J. N., Keaton, J. M., Park, J., Qiu, C., Warren, H. R., et al. (2019). Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat. Genet. 51, 51–62. doi: 10.1038/s41588-018-0303-9
Hoffmann, T. J., Ehret, G. B., Nandakumar, P., Ranatunga, D., Schaefer, C., Kwok, P. Y., et al. (2017). Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 49, 54–64. doi: 10.1038/ng.3715
Ji, L. D., Tang, N. L., and Xu, J. (2016). AGTR1 has undergone natural selection in Euro-Asian populations in relation to ambient temperature that predisposes Chinese populations to essential hypertension. Int. J. Cardiol. 209, 278–280. doi: 10.1016/j.ijcard.2016.02.031
Kurtz, T. W. (2010). Genome-wide association studies will unlock the genetic basis of hypertension.: con side of the argument. Hypertension 56, 1021–1025. doi: 10.1161/HYPERTENSIONAHA.110.156190
Levy, D., Ehret, G. B., Rice, K., Verwoert, G. C., Launer, L. J., Dehghan, A., et al. (2009). Genome-wide association study of blood pressure and hypertension. Nat. Genet. 41, 677–687. doi: 10.1038/ng.384
Li, J., Tsuprykov, O., Yang, X., and Hocher, B. (2016). Paternal programming of offspring cardiometabolic diseases in later life. J. Hypertens 34, 2111–2126. doi: 10.1097/HJH.0000000000001051
Liu, C., Kraja, A. T., Smith, J. A., Brody, J. A., Franceschini, N., Bis, J. C., et al. (2016). Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet. 48, 1162–1170. doi: 10.1038/ng.3660
Newton-Cheh, C., Johnson, T., Gateva, V., Tobin, M. D., Bochud, M., Coin, L., et al. (2009). Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 41, 666–676. doi: 10.1038/ng.361
Surendran, P., Drenos, F., Young, R., Warren, H., Cook, J. P., Manning, A. K., et al. (2016). Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 48, 1151–1161. doi: 10.1038/ng.3654
The Wellcome Trust Case Control Consortium (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. doi: 10.1038/nature05911
Warren, H. R., Evangelou, E., Cabrera, C. P., Gao, H., Ren, M., Mifsud, B., et al. (2017). Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 49, 403–415. doi: 10.1038/ng.3768
Keywords: hypertension, blood pressure, single nucleotide polymorphism, environment, gene
Citation: Ji L-d, Tang NLS, Xu Z-f and Xu J (2020) Genes Regulate Blood Pressure, but “Environments” Cause Hypertension. Front. Genet. 11:580443. doi: 10.3389/fgene.2020.580443
Received: 06 July 2020; Accepted: 23 October 2020;
Published: 09 November 2020.
Edited by:
Yue-miao Zhang, Peking University People's Hospital, ChinaReviewed by:
Haipeng Li, Partner Institute for Computational Biology, ChinaDong-Dong Wu, Kunming Institute of Zoology (CAS), China
Copyright © 2020 Ji, Tang, Xu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Xu, eHVqaW4xQG5idS5lZHUuY24=
 Lin-dan Ji1,2
Lin-dan Ji1,2 Nelson L. S. Tang
Nelson L. S. Tang Jin Xu
Jin Xu