- 1The First School of Clinical Medicine, Lanzhou University, Lanzhou, China
- 2The First Hospital of Lanzhou University, Lanzhou, China
- 3Key Laboratory for Resources Utilization Technology of Unconventional Water of Gansu Province, Gansu Membrane Science and Technology Research Institute Co., Ltd., Lanzhou, China
- 4The Second Hospital of Lanzhou University, Lanzhou, China
- 5Changjiang Scholar’s Laboratory/Guangdong Provincial Key Laboratory for Diagnosis and Treatment of Breast Cancer, Shantou University Medical College, Shantou, China
In recent years, the prevalence of obesity and cancer have been rising. Since this poses a serious threat to human health, the relationship between the two has attracted much attention. This study examined whether fat mass and obesity-associated (FTO) genes are linked, taking into account a Genome-wide Association Study (GWAS) that revealed multiple single nucleotide polymorphism sites (SNPs) of the FTO gene, indicating an association between obesity and cancer in different populations. FTO proteins have been proved to participate in adipogenesis and tumorigenesis with post-transcriptional regulation of downstream molecular expression or through the target of the mammalian target protein rapamycin (mTOR). FTO inhibitors have also been found to share anti-obesity and anti-cancer effects in vivo. In this review, we comprehensively discuss the correlation between obesity and cancer by measuring FTO gene polymorphism, as well as the molecular mechanism involved in these diseases, emphasizing FTO as the common genetic basis of obesity and cancer.
Introduction
The morbidity of obesity and cancer is increasing year by year in most countries around the world and represents a threat to human health (Ng et al., 2014; Siegel et al., 2018). Obesity causes changes in the body’s physiological and hormonal environments that promote many diseases, including diabetes and cardiovascular diseases. Obesity has been proven to increase the risks of at least 13 different types of cancers, such as esophageal adenocarcinoma, colon cancer, endometrial cancer, postmenopausal breast cancer, kidney cancer, and hematopoietic cancers (Calle and Kaaks, 2004; Goodwin and Stambolic, 2015). Of all Americans diagnosed with cancer in 2014, the overweight and obese population account for 40% (Steele et al., 2017). Furthermore, another prospective study of large samples of Americans confirmed that 14% of cancer deaths in males and 20% of females are due to them being overweight or having obesity (Calle et al., 2003). The biological mechanism of obesity and cancer are complex, including obesity-related hormones, growth factors, multiple signaling pathways, and chronic inflammation (Chen, 2011; Vucenik and Stains, 2012). In recent years, FTO SNPs have been firmly associated with increased body mass index (BMI) and higher risks of various types of cancers in people of multiple races, and the role of FTO SNPs in the development of obesity and cancer has been gradually revealed (Loos and Yeo, 2014; Hernández-Caballero and Sierra-Ramírez, 2015; Deng et al., 2018a; Chen and Du, 2019). This review details this role and the molecular mechanisms of FTO in obesity and cancer, as well as its potential clinical applications as a therapeutic target.
FTO Gene and Functions
In 1999, FTO was first cloned by exon trapping analysis in Fused toes (Ft) mutation mice (Peters et al., 1999). Initially, FTO was expected to be associated with programmed cell death because scientists observed that heterozygous mice with Ft mutation developed syndactyly in the forelimb part and thymus hyperplasia (Van Der Hoeven et al., 1994). In 2007, the GWAS study identified FTO as an obesity sensitivity gene, and multiple SNPs in the intron 1 region were strongly associated with BMI, body fat rate, waist circumference, hip circumference, and energy intake (Dina et al., 2007; Frayling et al., 2007; Scuteri et al., 2007). As a result, the gene was named as the fat mass and obesity-associated (FTO) gene and has received extensive attention.
According to current genomics research, the FTO gene only exists in vertebrates and a few kinds of marine algae with highly conserved nucleotide and amino acid sequences (Robbens et al., 2008). The human FTO gene is located on chromosome 16q12.2, encoding a 2-oxoglutarate (2-OG) Fe(II)-dependent AlkB family dioxygenase, with a total length of 410.50 kb including 9 exons and 8 introns. About 3.4 kb upstream of FTO gene was Merkel’s diverticulum syndrome-associated gene (RPGRIP1L), and its downstream was close to Iroquois gene family (including IRX3, IRX5, IRX6) (Supplementary Figure). FTO is extensively expressed in adipose tissues and the skeletal muscles of human tissues, with the highest expression in the hypothalamus in the region that controls energy balance, namely the arcuate nucleus, which indicates that it may play a critical role in regulating appetite and energy metabolism (Frayling et al., 2007).
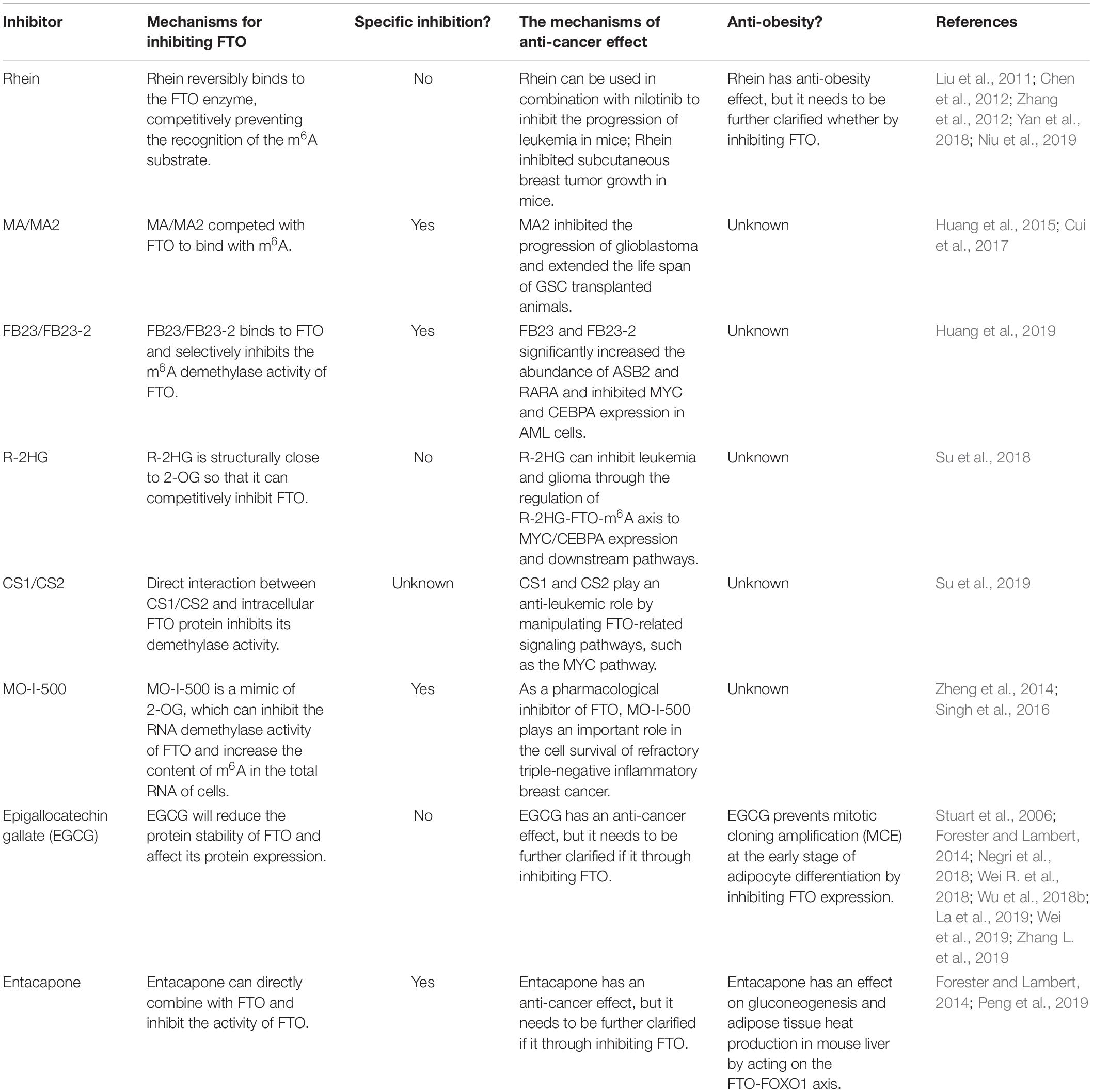
In 2007, Thomas et al. revealed that the FTO gene encodes Fe(II)/2-OG dependent demethylase, which is the ninth AlkB family protein found in mammals (also called ALKBH9) (Gerken et al., 2007). They also used purified FTO protein from recombinant mice or humans that can catalyze the demethylation of 3-methylthymine(3-meT) and 3-methyluracil (3-meU) with the help of Fe(II)/2-OG (Gerken et al., 2007; Jia et al., 2008). Later, He et al. found that N6-methyladenosine (m6A) in nuclear RNA was a main substrate of the FTO (Jia et al., 2011). Therefore, the FTO was identified as the first RNA demethylase, thus initiating a wave of research on epigenetic modifications of RNA. Since then, the complex and diverse functions of FTO proteins have been gradually revealed. FTO can bind to multiple types of RNAs, including mRNA, snRNA, and tRNA, and can demethylate m6A and N6,2′-O-dimethyladenosine (m6Am) in mRNA, m6A in U6RNA, m6Am in snRNAs, and N1-methyladenosine (m1A) in tRNA (Wei J. et al., 2018; Figure 1A). However, m6A is the most favorable nucleobase substrate of FTO (Zhang X. et al., 2019).
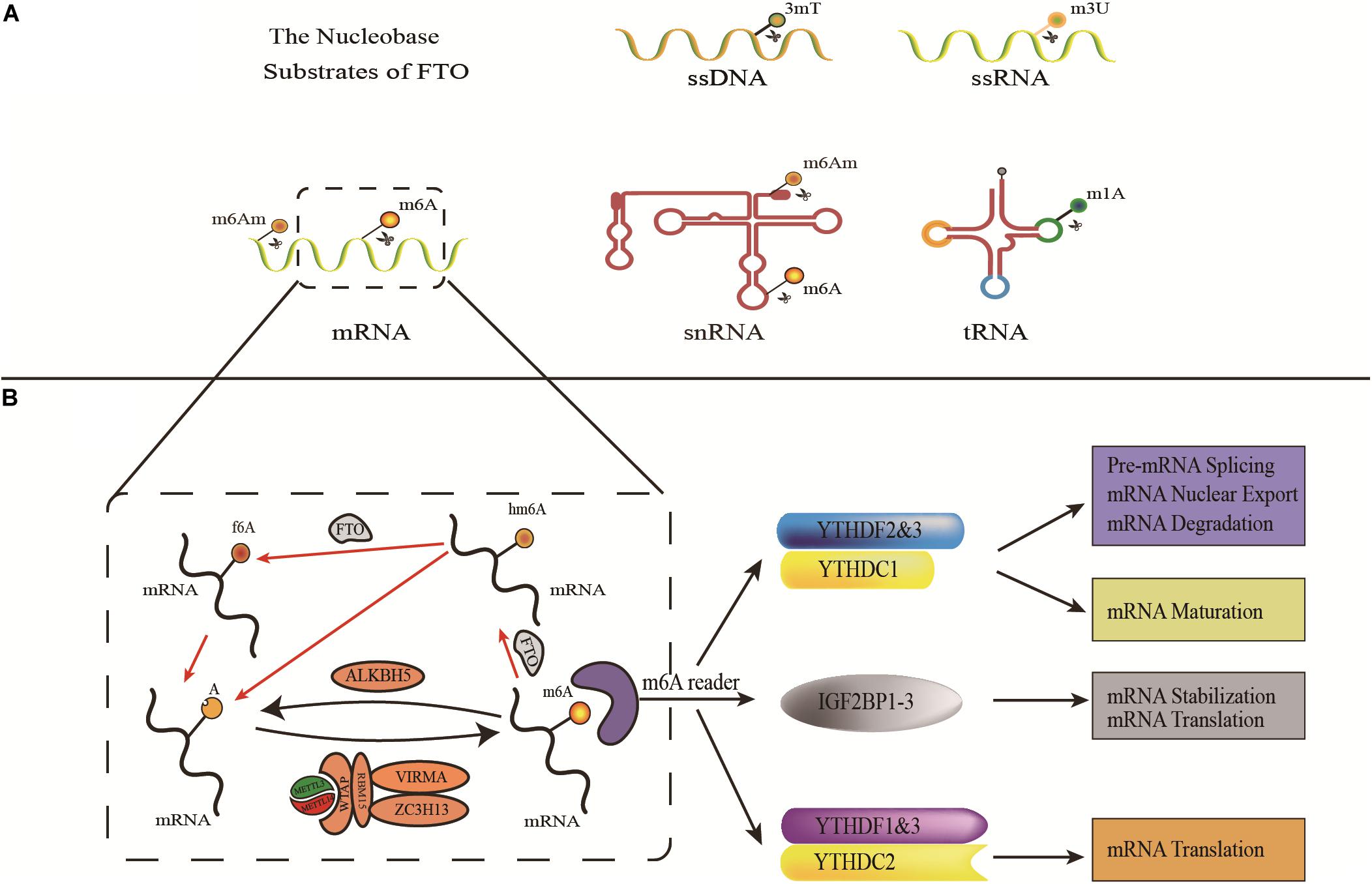
Figure 1. The demethylases activity of FTO. (A) The nucleobase substrates of FTO. (B) m6A modification is induced by METTL3, METTL14, and their cofactors(writers), reversed by FTO and ALKBH5 (erasers), and functionally facilitated by m6A binding proteins including YTHDF1-3, YTHDC1-2, and IGF2BP1-3(readers).
M6A, methylation modification on the sixth nitrogen atom of adenine (Wei et al., 1975) is the most common mRNA methylation enriched in the 3′-untranslated region (3′-UTRs), between the stop codon and the start codon (Roundtree et al., 2017). M6A modifications were subjected to reversible and dynamic regulations including writers (METTL3, METTL14, and WTAP), erasers (FTO and ALKBH5), and readers (YTH domain family and IGF2BPs) (Aik et al., 2014; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014; Wang et al., 2014, 2015, 2016; Huang et al., 2018; Liao et al., 2018; Figure 1B). Based on this extensive existence and dynamic regulation, m6A plays an important role in post-transcriptional regulatory processes such as RNA splicing, nuclear production, degradation, and translation (Fustin et al., 2013; Wang et al., 2014, 2015; Bartosovic et al., 2017; Huang et al., 2018).
Association of FTO SNPs With Obesity or Cancer
Since FTO has been identified as the first obesity-related gene. By conducting GWAS analysis, researchers have found that FTO SNPs are associated with obesity and higher risks of various cancers in multiracial populations (Supplementary Figure).
Association of FTO SNPs With Obesity
The connection between FTO SNPs and BMI was first found in European people with diabetes. The classic BMI-related FTO SNPs were rs9939609 (T/A), and compared with those who did not carry the risk allele, 16% of adults who carried the homozygous risk allele gained nearly 3 kg in weight, and the risk of obesity increased by 1.67 times (Frayling et al., 2007). Serial GWAS studies on obesity-related traits in people of European descent have confirmed the important role of the FTO locus, and many other FTO SNPs in the intron 1 region have been reported, such as rs9930506, rs1421085, rs8050136, rs1121980, and so forth (Dina et al., 2007; Scuteri et al., 2007; Haupt et al., 2008; Cauchi et al., 2009). The obesity-associated FTO SNPs in East Asian populations are comparable to that of people of European descent. The risk allele A of FTO SNP rs9939609 was closely related to obesity and BMI in Chinese, ethnic Chinese, Malaysian, Singaporean, East, and South Asian people (Chang et al., 2008; Tan et al., 2008; Li et al., 2012). A large-scale meta-analysis targeting GWAS analysis of East Asian populations found that FTO SNPs rs17817449 have the most significant correlation with BMI in people of Chinese descent (Wen et al., 2012). In recent years, global studies have associated the rs9939609 variant with higher obesity risks in other populations (including Brazilian people, early adolescence in China, and adults in Shiraz, Iran) (Fonseca et al., 2019; Jiang et al., 2019; Mehrdad et al., 2020b). It is also associated with increased BMI and waist circumference (in Brazilian youths) (Reuter et al., 2016), adipose tissue distribution (in Italian people), and increased metabolic syndrome susceptibility (in Chinese populations) (Merra et al., 2020; Wang et al., 2020). In terms of the potential mechanisms between mutations in FTO and increased risks of obesity, studies have proven the role of FTO in the influence of food intake. People carrying FTO risk alleles and are inclined to higher energy intake foods like fat or proteins, reduced satiety, resulting in overeating, and many even lose control when eating (Cecil et al., 2008; Sonestedt et al., 2009; Tanofsky-Kraff et al., 2009; Ahmad et al., 2011). Another large-scale meta-analysis showed that the homozygous FTO risk allele was associated with a 27% lower risk of obesity in physically active adults (Kilpeläinen et al., 2011).
Association of FTO SNPs With Cancer
To date, it has been studied that variants of FTO rs9939609, rs8050136, rs1477196, rs6499640, rs1121980, rs17817449, rs11075995, rs8047395, and rs7206790 have an association with a higher risk of cancers (Hernández-Caballero and Sierra-Ramírez, 2015). The most typical FTO SNP rs9939609 was associated with lung cancer, renal cancer, breast cancer, prostate cancer, pancreatic cancer, endometrial cancer (Delahanty et al., 2011; Kaklamani et al., 2011; Lin et al., 2013; Huang et al., 2017). Multiple SNPs in the intron 1 region of the FTO (including rs9939609, rs1477196, rs7206790, rs8047395) have been correlated with the risk of breast cancer, with rs1477196 strongly associated (Kaklamani et al., 2011). Interestingly, Da et al. observed that the interaction of FTO and MC4R polymorphisms showed a strong association with breast cancer: there was a 4.59-fold increased risks for women who have the allele combination C/T/C (FTO rs1121980/FTO rs9939609/MC4R rs17782313) (Da Cunha et al., 2013). In addition, in 2013, Iles et al. found an association between FTO rs16953002 and rs12596638 and melanoma susceptibility. However, these two SNPs are located in intron 8 of the FTO gene rather than intron 1 (the BMI-related region). This suggests that the association between the FTO variant and a wide range of diseases may play a role beyond BMI (Iles et al., 2013).
FTO Is Involved in the Pathogenesis of Obesity and Cancer
FTO, as the RNA m6A Demethylase, Is Involved in the Development of Obesity and Cancer
FTO proteins are widely involved in both adipogenesis and tumorigenesis by m6A-dependent demethylase activity which influences several mRNA processing events (Table 1).

Table 1. FTO proteins are widely involved in both adipogenesis and tumorigenesis by m6A-dependent demethylase activity.
FTO, as the RNA m6A Demethylase, Is Involved in the Development of Obesity
FTO proteins are involved in the development of obesity by affecting the m6A level of hormones related to eating or molecules related to adipogenesis (Figure 2A). In 2013, Efthimia et al. found that FTO over-expression limited the m6A modification of ghrelin mRNA in cell models and increased ghrelin mRNA and peptide levels concomitantly. This article provided insights into how FTO predisposes to stimulated energy intake and obesity in humans (Karra et al., 2013). Simultaneously, substantial evidence has proved that FTO participates in the process of adipogenesis (Ben-Haim et al., 2015). Zhao et al. found that FTO regulates the exonic splicing of the adipogenic regulator RUNX1T1 by influencing the level of m6A around the splice site, thereby modulating cell differentiation (Zhao et al., 2014). Moreover, FTO affects adipogenesis by regulating the process of mitotic clonal expansion (MCE), which is a prerequisite for adipocyte differentiation that occurs within 48 h of adipogenic stimulation (Merkestein et al., 2015). The overexpression of FTO can induce MCE and regulate the differentiation of preadipocytes by influencing the expression of m6A-dependent transcription factors (Tang et al., 2003; Merkestein et al., 2015; Zhang et al., 2015). Furthermore, Wu et al. found that FTO regulated adipogenesis by dominating cell cycle proteins such as CCNA2 and CDK2 by m6A-YTHDF2 dependent pathway, revealing a new mechanism about anti-obesity and anti-adipogenesis activity of Epigallocatechin gallate (EGCG) (Wu et al., 2018a,b).
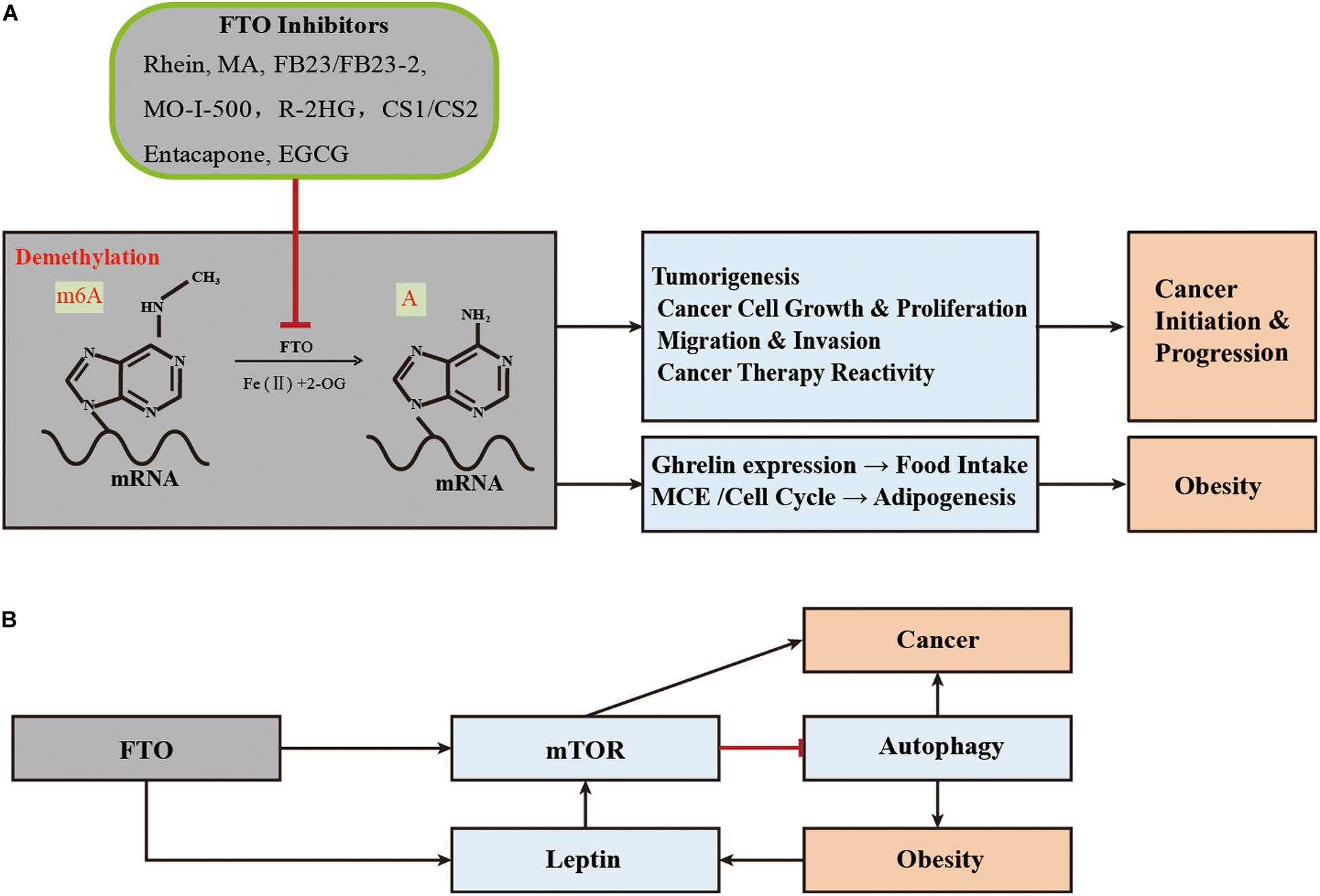
Figure 2. Schematic illustration of the roles of FTO in obesity and tumorigenesis/drug response. (A) As the m6A demethylase, FTO post-transcriptionally regulates expression of its critical target genes and thereby contributes to obesity (through affecting adipogenesis and food intake) and cancers (including tumorigenesis, cancer cell growth, migration, and drug response). MA, meclofenamic acid; R-2HG, 2-hydroxyglutarate; EGCG, Epigallocatechin gallate; (B) FTO regulates the development and progression of obesity and cancer through the mTOR or adipofactor-mTOR pathway.
FTO, as the RNA m6A Demethylase, Regulates the Malignant Phenotype and Therapeutic Response of Cancer Cells
FTO is highly expressed in many cancer tissues, which plays the role of an oncogene in an m6A-dependent way and participates in the regulation of the malignant phenotype of cancer cells (Figure 2A). In 2017, Li et al. found that FTO enhanced leukemia oncogene-mediated cell transformation and leukemogenesis and inhibited all-trans-retinoic acid (ATRA)-induced AML cell differentiation by affecting the expression of targets mRNAs such as ASB2 and RARA, through reducing the level of m6A (Li et al., 2017). Moreover, the elevated level of m6A was found to promote the growth, self-renewal, and tumorigenesis of Glioblastoma Stem Cells (GSC) as well as prolong the lifespan of GSC-grafted mice (Cui et al., 2017). In 2018, Niu et al. showed that FTO can promote breast cancer cell malignant phenotype through epigenetically demethylated m6A in BNIP3 mRNA 3′UTR (Niu et al., 2019). Similarly, the overexpression level of FTO was also found in gastric cancer, advanced non-small cell lung cancer, and hepatocellular carcinoma, which can regulate cell proliferation and/or migration/invasion through targeting demethylation for the m6A of Wnt/PI3K-Akt, USP7, MZF1, or PKM2, respectively (Liu et al., 2018; Li et al., 2019a,b; Zhang C. et al., 2019).
FTO may also have an impact on the therapeutic response of cancer (Figure 2A). In 2018, Zhou et al. found that FTO enhanced the chemo-radiotherapy resistance of cervical squamous cell carcinoma both in vitro and in vivo through influencing the expression of ß-catenin by reducing m6A levels (Zhou et al., 2018). The overexpression of FTO in leukemia cells can promote its expression by catalyzing the demethylation of cell proliferation-related genes such as m6A of MERTL and BCL-2 mRNA and affecting the generation of resistance phenotypes in the treatment with tyrosine kinase inhibitors (TKI) (Yan et al., 2018). Recently, He et al. found that the down-regulation of FTO made melanoma cells sensitive to interferon-gamma (IFN-γ) and anti-PD-1. FTO played a crucial role by promoting melanoma tumorigenesis and anti-PD-1 resistance (Yang et al., 2019).
FTO-mTOR Axis Affects Obesity and Cancer
The mammalian target protein rapamycin (mTOR) is an atypical serine/threonine kinase, which is the core component of regulating mRNA translation and can promote cell growth according to environmental signals (Laplante and David, 2012). mTOR binds to a variety of chaperone proteins to form two different kinase complexes, i.e., mTOR complex 1 (mTORC1) and mTORC2.
In 2013, in vivo experiments demonstrated that FTO played a role in the coupling of amino acid level and mTORC1 signaling pathway. FTO deficient cells showed reduced activation of the mTORC1 pathway, decreased mRNA translation rate, and increased autophagy (Cheung et al., 2013; Gulati et al., 2013). mTORC1 is a negative regulator of autophagy, which is a major cellular digestion process. In response to nutrition and environmental stress, autophagy plays a critical role in the occurrence and progression of obesity and cancer (White and Dipaola, 2009; Kim and Guan, 2015; Zhang Y. et al., 2018). Furthermore, mTOR activates the Warburg effect by inducing PKM2 and other glycolytic enzymes under normoxic conditions (Sun et al., 2011). In summary, mTORC1 can regulate the development of obesity and cancer through autophagy or direct activation of downstream signaling pathways (Laplante and David, 2012).
Mutations in the FTO gene raise blood levels of leptin, a known mediator or growth factor between obesity and colon cancer, which activates a variety of pathways associated with colon cancer (Drew, 2012; Mehrdad et al., 2020a). In addition, leptin has been suggested as an intermediate link between obesity and breast or prostate cancer (Stattin et al., 2001; Barone et al., 2020). Intriguingly, mTOR is also one of the signal mediators of obesity related factors, such as leptin, adiponectin, and inflammatory cytokines, through the Akt/PI3K or AMPK pathways (Maya-Monteiro and Bozza, 2008; Wang et al., 2012; Mauro et al., 2018). This seems to coincide with the FTO-mTOR pathway, discussed in the previous paragraph. In summary, FTO can directly or indirectly target mTOR, thus regulating the occurrence and progress of obesity and cancer in a broad manner (Figure 2B).
Effects of FTO Inhibitors in Obesity and Cancer
With the gradual disclosure of the important functions of FTO as mRNA demethylase in many diseases, the crystal structure of FTO has been resolved since 2010, and the development and applications of its specific inhibitors have attracted extensive attention (Han et al., 2010). FTO inhibitors that have been shown to have anti-obesity or anti-cancer effects in vitro or in vivo are summarized in Table 2.
Effects of FTO Inhibitors in Obesity
In 2012, Yang et al. reported natural product Rhein as an inhibitor of human FTO demethylase, which can competitively bind to the FTO catalytic site (Chen et al., 2012). Before this study, Rhein was thought to prevent or even reverse weight gain and obesity caused by high-fat diets (Liu et al., 2011; Zhang et al., 2012). The catechin EGCG, another natural compound rich in green tea, was found to play anti-obesity and anti-adipogenesis roles through the FTO-m6A-YTHDF2 axis (Wu et al., 2018b). Recently, Peng et al. identified Entacapone as a potential FTO inhibitor, which has the effect of reducing weight and lowering blood glucose (Peng et al., 2019). It was initially approved as an adjunctive therapy combined with levodopa and carbidopa for the treatment of Parkinson’s disease. Entacapone had an effect on gluconeogenesis and adipogenesis in the liver of mice by acting on an FTO-FOXO1 regulatory axis (Peng et al., 2019).
Effects of FTO Inhibitors in Cancer
As for cancers, there are more studies on the application of FTO inhibitors, especially in the treatment of leukemia, glioblastoma, and breast cancer. Compared with single drug therapy, Rhein combined with nilotinib is a more effective treatment for leukemia in mice (Yan et al., 2018). Recently, Yang et al. identified meclofenamic acid (MA) as a highly selective inhibitor of FTO, which can compete with FTO binding for the m6A-containing nucleic acids (Huang et al., 2015). The inhibitor FB23 was designed and synthesized from the chemical scaffold of MA, which exhibited a more potent inhibition for FTO demethylation in vitro (Huang et al., 2019). Its bioisostere FB23-2 can inhibit the leukemogenesis in cells and in the patient-derived xenografted (PDX) mouse model (Huang et al., 2019). Su et al. found that R-2HG (oncometabolite produced by mutant isocitrate dehydrogenase 1/2 (IDH1/2) enzymes), compounds CS1 and CS2 were also targeted inhibitors of FTO. By inhibiting its demethylation function, they affected related signaling pathways (such as the MYC pathway) and played an active role in inhibiting the proliferation of AML cells in vivo and in vitro (Su et al., 2018, 2019). Compounds CS1 and CS2 extended the overall survival of transplanted mice with primary MLL-AF16 cells and made AML cells sensitive to other curative drugs, such as decitabine, a tyrosine kinase inhibitor, and IDH2mut inhibitor (Su et al., 2019). Comparing the anti-leukemic activities of the four FTO inhibitors, CS1 and CS2 showed higher activity in inhibiting cell viability, and their IC50 values were 10–30 times lower than FB23-2 or MO-I-500 (Su et al., 2019). MO-I-500 is another FTO inhibitor, which selectively inhibits the demethylation of FTO and increases the m6A levels in cells (Zheng et al., 2014). In addition, among the above-mentioned FTO inhibitors, Rhein and MO-I-500 have been reported to significantly inhibit the growth ability of breast cancer cells in vivo and in vitro (Singh et al., 2016; Niu et al., 2019). MA2 (the ethyl ester form of MA) and R-2HG had inhibitory effects on glioma (Cui et al., 2017; Su et al., 2018; Deng et al., 2018b). Compared with MA, MA2 has a better cell penetration, significantly increased m6A methylation in cells, suppresses glioblastoma progression, and prolongs the lifespan of GSC-grafted animals (Huang et al., 2015; Cui et al., 2017). Interestingly, the previous anti-obesity EGCG, Entacapone, also had an inhibitory effect on cancer. For example, EGCG had an inhibitory effect on lung cancer, breast cancer, colon cancer, metastatic pancreatic cancer, and prostate cancer or had a sensitivity to chemotherapy. It was noted that it can be used as an adjuvant drug in cancer treatments (Negri et al., 2018; Wei R. et al., 2018; La et al., 2019; Wei et al., 2019). The combination of Entacapone and EGCG can synergistically enhance the growth inhibitor of lung cancer cell lines (Forester and Lambert, 2014).
In summary, more FTO inhibitors are displaying positive therapeutic effects in animal disease models, and represent promising therapeutic targets for obesity and cancer (Figure 2A).
Controversy on the Mechanism of Association Between FTO Risk Alleles and Diseases
Single nucleotide polymorphism sites are the main form of human genome DNA sequence variation and can regulate gene expression. From FTO polymorphisms which have a risk for obesity and cancer to the specific mechanisms regulating these diseases through nucleic acid demethylation of FTO proteins, FTO SNPs seem to regulate the expression level of FTO and affect its enzymatic function, playing an important role in obesity and cancer. Some studies support this hypothesis. For example, In 2008, a study of a Mexican population revealed that in obese patients, the FTO risk allele was significantly correlated with high FTO expression (Villalobos-Comparán et al., 2008). Subsequently, Tea et al. and Efthimia et al. revealed that FTO mRNA caused by the risk allele was more abundant than non-risk alleles at least in blood cells (Berulava and Horsthemke, 2010; Karra et al., 2013). Unfortunately, the mechanism for the correlation between FTO SNPs and obesity or cancer has been elusive. So far, there are no studies that provide indisputable evidence for these associations.
Moreover, some studies have suggested that FTO SNPs may be associated with obesity by regulating the expression of adjacent genes (Tung et al., 2014; Figure 3A). The Leibel group found that the rs8050136 of the intron 1 region of FTO overlapped with the binding site of transcription factor Cut Like Homeobox 1 (CUX1). This SNP nucleotide type can affect the transcriptional activation of FTO and retinitis pigmentosa GTPase regulator interacting protein 1 like (RPGRIP1L) by CUX1 P110 (Stratigopoulos et al., 2008, 2011). For individuals with the obesity risk allele at rs8050136, the expression of RPGRIP1L and FTO in the hypothalamus were decreased due to the low binding affinity of CUX1 P110 to DNA, RPGRIP1L can affect the location of leptin receptors and leptin signaling in neurons and lead to increased food intake and obesity (Stratigopoulos et al., 2011). In addition, Jowett et al. found that in combination with gene variation and expression data from the human cohort, the A allele of rs8050136 was positively associated with the expression level of RBL2, and an increase in RBL2 level might help to restrict the clonal expansion of A population of precursor adipose cells during development (Jowett et al., 2010). Moreover, Smemo et al. found that these sites also contained an enhancer sequence that can bind to the promoter of IRX3. Using expression quantitative trait loci (eQTL), they found that obesity-related SNPs such as rs9930506 were correlated with the expression of IRX3 in human brain samples. Mice lacking IRX3 lost 25–30% of their body weight through increasing basal metabolic rate and browning of white adipose tissues (Smemo et al., 2014). The study of Claussnitzer et al. also supported the regulatory relationships between FTO SNPs and IRX3 expression. They suggested that changes in the rs1421085 risk allele led to a double expression of IRX3 and IRX5 through disruption of the conservative motif of the ARID5B repressor in the early stage of adipocyte differentiation. In this case, brown fat cells transform automatically into white fat cells and lower down the mitochondrial thermogenesis by five times (Claussnitzer et al., 2015). This study explained the correlation between FTO SNPs and obesity by using the effect of the autonomous transformation of fat cells on thermogenesis. These studies provided a plausible mechanism for the correlation between SNP variation of FTO intron 1 and obesity (Figure 3B).
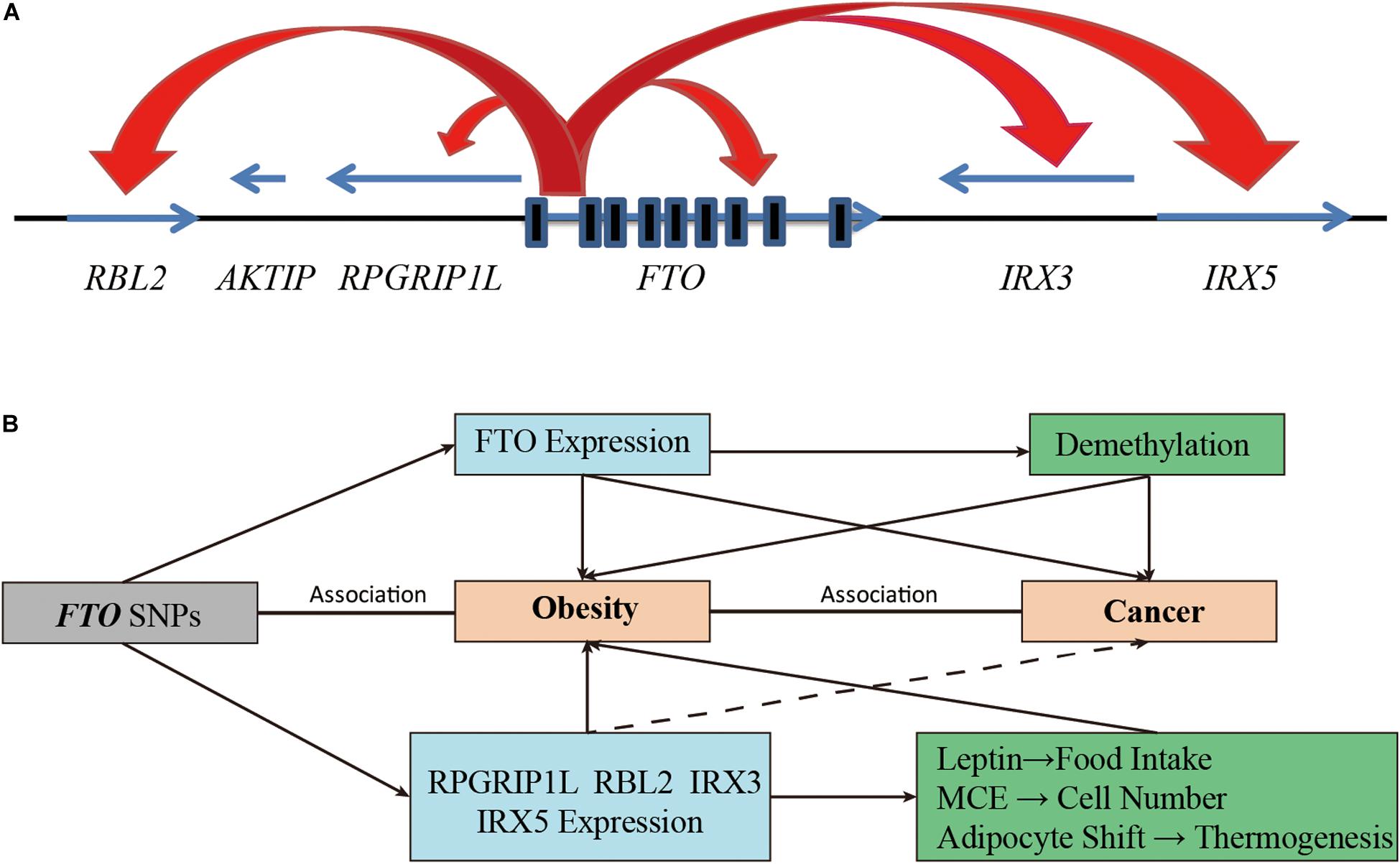
Figure 3. Increasing functional complexity around the FTO Locus. (A) SNPs in the intron 1 of FTO can regulate the expression of FTO itself and adjacent genes (including RBL2, RPGRIP1L, IRX3, and IRX5). (B) The FTO SNPs are involved in the development of obesity and cancer by affecting the demethylation function of FTO or by regulating the expression of adjacent genes.
Adjacent genes (RPGRIP1L, RBL2, IRX3, IRX5) regulated by FTO SNPs are also involved in the occurrence and progression of cancer in various ways (Figure 3B). For example, studies have shown that RPGRIP1L was one of the tumor suppressor genes of human hepatocellular carcinoma (Lin et al., 2009). RBL2, a member of the retinoblastoma (Rb) family, was inactivated by cell cycle kinases and was the basis of various cancer types (Pentimalli et al., 2015). The emerging role of RBL2 in aging and apoptosis also appeared to play an active part in tumor inhibition (Pentimalli et al., 2015). IRX3 was highly expressed in approximately 30% of patients with AML, and high expression of IRX3 alone can perpetuate hematopoietic stem cells and progenitor cells (HSPC) in bone marrow cultures and induce lymphoid leukemia in vivo (Somerville et al., 2018). In addition, IRX3 and IRX5 have been reported to participate in the transition from intestinal adenoma to colorectal cancer by negatively regulating the Dpp/TGF-ß pathway (Martorell et al., 2014). IRX5 alone has also been reported to be an oncogenic gene in hepatocellular carcinoma, colorectal cancer, prostate cancer, and non-small cell lung cancer by regulating cancer cell cycle and apoptosis (Myrthue et al., 2008; Zhang D.L. et al., 2018; Zhu et al., 2019, 2020). Although there is no direct evidence that FTO SNPs are associated with cancer by affecting the expression of adjacent genes, we cannot rule it out as a possibility.
Based on these findings, the correlation between FTO SNPs and obesity and cancer may be due to the regulation of FTO enzyme activity or expression of adjacent genes. However, more convincing and systematic research studies are needed to decipher the causal mechanism between FTO non-coding variants and obesity or cancer. A healthy lifestyle such as proper diet and moderate exercise is recommended to minimize the negative effects of obesity susceptibility genes before we can fully understand the underlying mechanisms.
Conclusion
Although the specific mechanisms for FTO polymorphism and high risk of obesity and cancer are elusive, the correlation is definite. As for FTO, it can participate in the disease progression of obesity and cancer in m6A-dependent post-transcriptional regulation, or by targeting mTOR. More importantly, some drugs have been shown to inhibit obesity, and some cancers such as leukemia, glioblastoma, and breast cancer by targeting FTO. In particular, FB23, R-2HG, and CS1/CS2 have shown anti-leukemia effects through in vivo experiments, and MA2 can also inhibit the activities of glioblastoma cells in vivo. This evidence suggests FTO as the common genetic basis of obesity and cancer and a potential target for obesity and some cancers.
Author Contributions
WY oversaw and guided the process of writing this manuscript. NL wrote and edited the manuscript. YL, YZ, SP, XN, HX, and JL put forward suggestions for the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Gansu Natural Science Foundation (18JR3RA343) and the Science and Technology Bureau 2018 Fund of the Chengguan District (2018KJGG0037).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to the First School of Clinical Medicine of Lanzhou University and the Key Laboratory for Resources Utilization Technology of Unconventional Water of Gansu Province.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.559138/full#supplementary-material
References
Ahmad, T., Lee, I., Paré, G., Chasman, D., Rose, L., Ridker, P. M., et al. (2011). Lifestyle interaction with fat mass and obesity-associated (fto) genotype and risk of obesity in apparently healthy U.S. Women. Diabetes Care 34, 675–680. doi: 10.2337/dc10-0948
Aik, W., Scotti, J. S., Choi, H., Gong, L., Demetriades, M., Schofield, C. J., et al. (2014). Structure of human rna n 6-methyladenine demethylase alkbh5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42, 4741–4754. doi: 10.1093/nar/gku085
Barone, I., Giordano, C., Bonofiglio, D., Andò, S., and Catalano, S. (2020). The weight of obesity in breast cancer progression and metastasis: clinical and molecular perspectives. Semin. Cancer Biol. 60, 274–284. doi: 10.1016/j.semcancer.2019.09.001
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase fto targets pre-mrnas and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Ben-Haim, M. S., Moshitch-Moshkovitz, S., and Rechavi, G. (2015). Fto: linking m 6 a demethylation to adipogenesis. Cell Res. 25, 3–4. doi: 10.1038/cr.2014.162
Berulava, T., and Horsthemke, B. (2010). The obesity-associated snps in intron 1 of the fto gene affect primary transcript levels. Eur. J. Hum. Genet. 18, 1054–1056.
Calle, E. E., and Kaaks, R. (2004). Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature Rev. Cancer 4, 579–591. doi: 10.1038/nrc1408
Calle, E. E., Rodriguez, C., Walker-Thurmond, K., and Thun, M. J. (2003). Overweight, obesity, and mortality from cancer in a prospectively studied cohort of us adults. N. Engl. J. Med. 348, 1625–1638.
Cauchi, S., Stutzmann, F., Cavalcanti-Proença, C., Durand, E., Pouta, A., Hartikainen, A. L., et al. (2009). Combined effects of mc4r and fto common genetic variants on obesity in european general populations. J. Mol. Med. 87, 537–546. doi: 10.1007/s00109-009-0451-6
Cecil, J. E., Tavendale, R., Watt, P., Hetherington, M. M., and Palmer, C. N. (2008). An obesity-associated fto gene variant and increased energy intake in children. N. Engl. J. Med. 359, 2558–2566.
Chang, Y., Liu, P., Lee, W., Chang, T., Jiang, Y., Li, H., et al. (2008). Common variation in the fat mass and obesity-associated (fto) gene confers risk of obesity and modulates bmi in the chinese population. Diabetes 57, 2245–2252. doi: 10.2337/db08-0377
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of cell-active n 6-methyladenosine rna demethylase fto inhibitor. J. Am. Chem. Soc. 134, 17963–17971.
Chen, J. (2011). Multiple signal pathways in obesity-associated cancer. Obes. Rev. 12, 1063–1070. doi: 10.1111/j.1467-789X.2011.00917.x
Chen, J., and Du, B. (2019). Novel positioning from obesity to cancer: fto, an m6a rna demethylase, regulates tumour progression. J. Cancer Res. Clin. Oncol. 145, 19–29. doi: 10.1007/s00432-018-2796-0
Cheung, M. K., Gulati, P., O’rahilly, S., and Yeo, G. S. (2013). Fto expression is regulated by availability of essential amino acids. Int. J. Obes. 37, 744–747. doi: 10.1038/ijo.2012.77
Claussnitzer, M., Dankel, S., Kim, K., Quon, G., Meuleman, W., Haugen, C., et al. (2015). Fto obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 373, 895–907. doi: 10.1056/NEJMoa1502214
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). M6a rna methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi: 10.1016/j.celrep.2017.02.059
Da Cunha, P. A., De Carlos, B. L. K., Sereia, A. F., Kubelka, C., Ribeiro, M. C., Fernandes, B. L., et al. (2013). Interaction between obesity-related genes, fto and mc4r, associated to an increase of breast cancer risk. Mol. Biol. Rep. 40, 6657–6664. doi: 10.1007/s11033-013-2780-3
Delahanty, R. J., Beeghly-Fadiel, A., Xiang, Y.-B., Long, J., Cai, Q., Wen, W., et al. (2011). Association of obesity-related genetic variants with endometrial cancer risk: a report from the shanghai endometrial cancer genetics study. Am. J. Epidemiol. 174, 1115–1126.
Deng, X., Su, R., Stanford, S., and Chen, J. (2018a). Critical enzymatic functions of fto in obesity and cancer. Front. Endocrinol. 9:396. doi: 10.3389/fendo.2018.00396
Deng, X., Su, R., Weng, H., Huang, H., Li, Z., and Chen, J. (2018b). Rna n-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28, 507–517. doi: 10.1038/s41422-018-0034-6
Dina, C., Meyre, D., Gallina, S., Durand, E., Körner, A., Jacobson, P., et al. (2007). Variation in fto contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726. doi: 10.1038/ng2048
Drew, J. (2012). Molecular mechanisms linking adipokines to obesity-related colon cancer: focus on leptin. Proc. Nutr. Soc. 71, 175–180. doi: 10.1017/s0029665111003259
Fonseca, A. C. P. D., Abreu, G. D. M., Zembrzuski, V. M., Junior, M. C., Carneiro, J. R. I., Neto, J. F. N., et al. (2019). The association of the fat mass and obesity-associated gene (fto) rs9939609 polymorphism and the severe obesity in a brazilian population. Diabetes Metab. Syndr. Obes. Targets Ther. 12, 667–684. doi: 10.2147/DMSO.S199542
Forester, S. C., and Lambert, J. D. (2014). Synergistic inhibition of lung cancer cell lines by (-)-epigallocatechin-3-gallate in combination with clinically used nitrocatechol inhibitors of catechol-o-methyltransferase. Carcinogenesis 35, 365–372. doi: 10.1093/carcin/bgt347
Frayling, T. M., Timpson, N. J., Weedon, M. N., Zeggini, E., Freathy, R. M., Lindgren, C. M., et al. (2007). A common variant in the fto gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894.
Fustin, J. M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). Rna-methylation-dependent rna processing controls the speed of the circadian clock. Cell 155, 793–806. doi: 10.1016/j.cell.2013.10.026
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated fto gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. doi: 10.1126/science.1151710
Goodwin, P. J., and Stambolic, V. (2015). Impact of the obesity epidemic on cancer. Annu. Rev. Med. 66, 281–296. doi: 10.1146/annurev-med-051613-012328
Gulati, P., Cheung, M. K., Antrobus, R., Church, C. D., Harding, H. P., Tung, Y. C., et al. (2013). Role for the obesity-related fto gene in the cellular sensing of amino acids. Proc. Natl. Acad. Sci. U.S.A. 110, 2557–2562. doi: 10.1073/pnas.1222796110
Han, Z., Niu, T., Chang, J., Lei, X., Zhao, M., Wang, Q., et al. (2010). Crystal structure of the fto protein reveals basis for its substrate specificity. Nature 464, 1205–1209. doi: 10.1038/nature08921
Haupt, A., Thamer, C., Machann, J., Kirchhoff, K., Stefan, N., Tschritter, O., et al. (2008). Impact of variation in the fto gene on whole body fat distribution, ectopic fat, and weight loss. Obesity 16, 1969–1972. doi: 10.1038/oby.2008.283
Hernández-Caballero, M. E., and Sierra-Ramírez, J. A. (2015). Single nucleotide polymorphisms of the fto gene and cancer risk: an overview. Mol. Biol. Rep. 42, 699–704. doi: 10.1007/s11033-014-3817-y
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of rna n-methyladenosine by igf2bp proteins enhances mrna stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Huang, X., Zhao, J., Yang, M., Li, M., and Zheng, J. (2017). Association between fto gene polymorphism (rs9939609 t/a) and cancer risk: a meta-analysis. Eur. J Cancer Care 26:e12464. doi: 10.1111/ecc.12464
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-molecule targeting of oncogenic fto demethylase in acute myeloid leukemia. Cancer Cell 35, 677.e10–691.e10. doi: 10.1016/j.ccell.2019.03.006
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic acid selectively inhibits fto demethylation of m6a over alkbh5. Nucleic Acids Res. 43, 373–384. doi: 10.1093/nar/gku1276
Iles, M. M., Law, M. H., Stacey, S. N., Han, J., Fang, S., Pfeiffer, R., et al. (2013). A variant in fto shows association with melanoma risk not due to bmi. Nat. Genet. 45:428.
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear rna is a major substrate of the obesity-associated fto. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jia, G., Yang, C.-G., Yang, S., Jian, X., Yi, C., Zhou, Z., et al. (2008). Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and rna by mouse and human fto. FEBS Lett. 582, 3313–3319. doi: 10.1016/j.febslet.2008.08.019
Jiang, Y., Mei, H., Lin, Q., Wang, J., Liu, S., Wang, G., et al. (2019). Interaction effects of fto rs9939609 polymorphism and lifestyle factors on obesity indices in early adolescence. Obes. Res. Clin. Pract. 13, 352–357. doi: 10.1016/j.orcp.2019.06.004
Jowett, J. B., Curran, J. E., Johnson, M. P., Carless, M. A., Göring, H. H., Dyer, T. D., et al. (2010). Genetic variation at the fto locus influences rbl2 gene expression. Diabetes 59, 726–732. doi: 10.2337/db09-1277
Kaklamani, V., Yi, N., Sadim, M., Siziopikou, K., Zhang, K., Xu, Y., et al. (2011). The role of the fat mass and obesity associated gene (fto) in breast cancer risk. BMC Med. Genet. 12:52. doi: 10.1186/1471-2350-12-52
Karra, E., O’daly, O. G., Choudhury, A. I., Yousseif, A., Millership, S., Neary, M. T., et al. (2013). A link between fto, ghrelin, and impaired brain food-cue responsivity. J. Clin. Investig. 123, 3539–3551.
Kilpeläinen, T. O., Qi, L., Brage, S., Sharp, S. J., Sonestedt, E., Demerath, E., et al. (2011). Physical activity attenuates the influence of fto variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 8:e1001116. doi: 10.1371/journal.pmed.1001116
Kim, Y. C., and Guan, K. L. (2015). Mtor: a pharmacologic target for autophagy regulation. J. Clin. Investig. 125, 25–32. doi: 10.1172/jci73939
La, X., Zhang, L., Li, Z., Li, H., and Yang, Y. (2019). (-)-epigallocatechin gallate (egcg) enhances the sensitivity of colorectal cancer cells to 5-fu by inhibiting grp78/nf-κb/mir-155-5p/mdr1 pathway. J. Agric. Food Chem. 67, 2510–2518. doi: 10.1021/acs.jafc.8b06665
Laplante, M., and David, M. S. (2012). Mtor signaling in growth control and disease. Cell 149, 285–293.
Li, H., Kilpeläinen, T., Liu, C., Zhu, J., Liu, Y., Hu, C., et al. (2012). Association of genetic variation in fto with risk of obesity and type 2 diabetes with data from 96,551 east and south asians. Diabetologia 55, 981–995. doi: 10.1007/s00125-011-2370-7
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019a). The m6a demethylase fto promotes the growth of lung cancer cells by regulating the m6a level of usp7 mrna. Biochem. Biophys. Res. Commun. 512, 479–485. doi: 10.1016/j.bbrc.2019.03.093
Li, J., Zhu, L., Shi, Y., Liu, J., Lin, L., and Chen, X. (2019b). M6a demethylase fto promotes hepatocellular carcinoma tumorigenesis via mediating pkm2 demethylation. Am. J. Transl. Res. 11:6084.
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). Fto plays an oncogenic role in acute myeloid leukemia as a n6-methyladenosine rna demethylase. Cancer Cell 31, 127–141. doi: 10.1016/j.ccell.2016.11.017
Liao, S., Sun, H., and Xu, C. (2018). Yth domain: a family of n-methyladenosine (ma) readers. Genomics Proteomics Bioinformatics 16, 99–107. doi: 10.1016/j.gpb.2018.04.002
Lin, Y., Ueda, J., Yagyu, K., Ishii, H., Ueno, M., Egawa, N., et al. (2013). Association between variations in the fat mass and obesity-associated gene and pancreatic cancer risk: a case–control study in japan. BMC Cancer 13:337. doi: 10.1186/1471-2407-13-337
Lin, Y. W., Yan, M. D., Shih, Y. L., and Hsieh, C. B. (2009). The basal body gene, rpgrip1l, is a candidate tumour suppressor gene in human hepatocellular carcinoma. Eur. J. Cancer 45, 2041–2049. doi: 10.1016/j.ejca.2009.04.012
Liu, J., Ren, D., Du, Z., Wang, H., Zhang, H., and Jin, Y. (2018). M6a demethylase fto facilitates tumor progression in lung squamous cell carcinoma by regulating mzf1 expression. Biochem. Biophys. Res. Commun. 502, 456–464. doi: 10.1016/j.bbrc.2018.05.175
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A mettl3-mettl14 complex mediates mammalian nuclear rna n6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, Q., Zhang, X.-L., Tao, R.-Y., Niu, Y.-J., Chen, X.-G., Tian, J.-Y., et al. (2011). Rhein, an inhibitor of adipocyte differentiation and adipogenesis. J. Asian Nat. Prod. Res. 13, 714–723.
Loos, R. J. F., and Yeo, G. S. H. (2014). The bigger picture of fto—the first gwas-identified obesity gene. Nat. Rev. Endocrinol. 10, 51–61. doi: 10.1038/nrendo.2013.227
Martorell, Ò, Barriga, F. M., Merlos-Suárez, A., Stephan-Otto Attolini, C., Casanova, J., Batlle, E., et al. (2014). Iro/irx transcription factors negatively regulate dpp/tgf-β pathway activity during intestinal tumorigenesis. EMBO Rep. 15, 1210–1218. doi: 10.15252/embr.201438622
Mauro, L., Naimo, G. D., Gelsomino, L., Malivindi, R., Bruno, L., Pellegrino, M., et al. (2018). Uncoupling effects of estrogen receptor α on lkb1/ampk interaction upon adiponectin exposure in breast cancer. FASEB J. 32, 4343–4355. doi: 10.1096/fj.201701315R
Maya-Monteiro, C., and Bozza, P. (2008). Leptin and mtor: partners in metabolism and inflammation. Cell Cycle 7, 1713–1717.
Mehrdad, M., Doaei, S., Gholamalizadeh, M., Fardaei, M., Fararouei, M., and Eftekhari, M. H. (2020a). Ftoassociation of rs9939609 polymorphism with serum leptin, insulin, adiponectin, and lipid profile in overweight adults. Adipocyte 9, 51–56. doi: 10.1080/21623945.2020.1722550
Mehrdad, M., Fardaei, M., Fararouei, M., and Eftekhari, M. H. (2020b). The association between fto rs9939609 gene polymorphism and anthropometric indices in adults. J. Physiol. Anthropol. 39:14. doi: 10.1186/s40101-020-00224-y
Merkestein, M., Laber, S., Mcmurray, F., Andrew, D., Sachse, G., Sanderson, J., et al. (2015). Fto influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 6, 1–9. doi: 10.1038/ncomms7792
Merra, G., Gualtieri, P., Cioccoloni, G., Falco, S., Bigioni, G., Tarsitano, M., et al. (2020). Fto rs9939609 influence on adipose tissue localization in the italian population. Eur. Rev. Med. Pharmacol. Sci. 24, 3223–3235. doi: 10.26355/eurrev_202003_20689
Myrthue, A., Rademacher, B. L., Pittsenbarger, J., Kutyba-Brooks, B., Gantner, M., Qian, D., et al. (2008). The iroquois homeobox gene 5 is regulated by 1,25-dihydroxyvitamin d3 in human prostate cancer and regulates apoptosis and the cell cycle in lncap prostate cancer cells. Clin. Cancer Res. 14, 3562–3570. doi: 10.1158/1078-0432.Ccr-07-4649
Negri, A., Naponelli, V., Rizzi, F., and Bettuzzi, S. (2018). Molecular targets of epigallocatechin-gallate (egcg): a special focus on signal transduction and cancer. Nutrients 10:1936. doi: 10.3390/nu10121936
Ng, M., Fleming, T., Robinson, M., Thomson, B., Graetz, N., Margono, C., et al. (2014). Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the global burden of disease study 2013. Lancet 384, 766–781. doi: 10.1016/s0140-6736(14)60460-8
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). Rna n6-methyladenosine demethylase fto promotes breast tumor progression through inhibiting bnip3. Mol. Cancer 18, 46. doi: 10.1186/s12943-019-1004-4
Peng, S., Xiao, W., Ju, D., Sun, B., Hou, N., Liu, Q., et al. (2019). Identification of entacapone as a chemical inhibitor of fto mediating metabolic regulation through foxo1. Sci. Transl. Med. 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
Pentimalli, F., Esposito, L., Forte, I. M., Iannuzzi, C. A., and Giordano, A. (2015). Abstract lb-080: reactivating rbl2/p130 oncosuppressive function as a new, possible antitumoral strategy. Cancer Res. 75, 18–22.
Peters, T., Ausmeier, K., and Rüther, U. (1999). Cloning of fatso (fto), a novel gene deleted by the fused toes (ft) mouse mutation. Mamm. Genome 10, 983–986. doi: 10.1007/s003359901144
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian wtap is a regulatory subunit of the rna n6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Reuter, C. P., Rosane De Moura Valim, A., Gaya, A. R., Borges, T. S., Klinger, E. I., Possuelo, L. G., et al. (2016). Fto polymorphism, cardiorespiratory fitness, and obesity in brazilian youth. Am. J. Hum. Biol. 28, 381–386. doi: 10.1002/ajhb.22798
Robbens, S., Rouzé, P., Cock, J., Spring, J., Worden, A., and Van De Peer, Y. (2008). The fto gene, implicated in human obesity, is found only in vertebrates and marine algae. J. Mol. Evol. 66, 80–84. doi: 10.1007/s00239-007-9059-z
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic rna modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6a writers reveals two distinct classes of mrna methylation at internal and 5′ sites. Cell Rep. 8, 284–296. doi: 10.1016/j.celrep.2014.05.048
Scuteri, A., Sanna, S., Chen, W. M., Uda, M., Albai, G., Strait, J., et al. (2007). Genome-wide association scan shows genetic variants in the fto gene are associated with obesity-related traits. PLoS Genet. 3:e115. doi: 10.1371/journal.pgen.0030115
Siegel, R. L., Jemal, A., Wender, R. C., Gansler, T., Ma, J., and Brawley, O. W. (2018). An assessment of progress in cancer control. CA Cancer J. Clin. 68, 329–339. doi: 10.3322/caac.21460
Singh, B., Kinne, H. E., Milligan, R. D., Washburn, L. J., Olsen, M., and Lucci, A. (2016). Important role of fto in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS One 11:e0159072. doi: 10.1371/journal.pone.0159072
Smemo, S., Tena, J. J., Kim, K.-H., Gamazon, E. R., Sakabe, N. J., Gómez-Marín, C., et al. (2014). Obesity-associated variants within fto form long-range functional connections with irx3. Nature 507, 371–375. doi: 10.1038/nature13138
Somerville, T. D. D., Simeoni, F., Chadwick, J. A., Williams, E. L., Spencer, G. J., Boros, K., et al. (2018). Derepression of the iroquois homeodomain transcription factor gene irx3 confers differentiation block in acute leukemia. Cell Rep. 22, 638–652. doi: 10.1016/j.celrep.2017.12.063
Sonestedt, E., Roos, C., Gullberg, B., Ericson, U., Wirfält, E., and Orho-Melander, M. (2009). Fat and carbohydrate intake modify the association between genetic variation in the fto genotype and obesity. Am. J. Clin. Nutr. 90, 1418–1425. doi: 10.3945/ajcn.2009.27958
Stattin, P., Söderberg, S., Hallmans, G., Bylund, A., Kaaks, R., Stenman, U. H., et al. (2001). Leptin is associated with increased prostate cancer risk: a nested case-referent study. J. Clin. Endocrinol. Metab. 86, 1341–1345. doi: 10.1210/jcem.86.3.7328
Steele, C. B., Thomas, C. C., Henley, S. J., Massetti, G. M., Galuska, D. A., Agurs-Collins, T., et al. (2017). Vital signs: trends in incidence of cancers associated with overweight and obesity – United States, 2005–2014. MMWR 66, 1052–1058. doi: 10.15585/mmwr.mm6639e1
Stratigopoulos, G., Leduc, C. A., Cremona, M. L., Chung, W. K., and Leibel, R. L. (2011). Cut-like homeobox 1 (cux1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa gtpase regulator-interacting protein-1-like (rpgrip1l) genes and coordinates leptin receptor signaling. J. Biol. Chem. 286, 2155–2170.
Stratigopoulos, G., Padilla, S. L., Leduc, C. A., Watson, E., Hattersley, A. T., Mccarthy, M. I., et al. (2008). Regulation of fto/ftm gene expression in mice and humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1185–R1196.
Stuart, E. C., Scandlyn, M. J., and Rosengren, R. J. (2006). Role of epigallocatechin gallate (egcg) in the treatment of breast and prostate cancer. Life Sci. 79, 2329–2336. doi: 10.1016/j.lfs.2006.07.036
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2hg exhibits anti-tumor activity by targeting fto/ma/myc/cebpa signaling. Cell 172, 90.e23–105.e23. doi: 10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Han, L., Gao, M., Wunderlich, M., et al. (2019). Effective novel fto inhibitors show potent anti-cancer efficacy and suppress drug resistance. Blood 134, 233–233. doi: 10.1182/blood-2019-124535
Sun, Q., Chen, X., Ma, J., Peng, H., Wang, F., Zha, X., et al. (2011). Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type m2 is critical for aerobic glycolysis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 108, 4129–4134. doi: 10.1073/pnas.1014769108
Tan, J., Dorajoo, R., Seielstad, M., Sim, X., Ong, R. T., Chia, K., et al. (2008). Fto variants are associated with obesity in the chinese and malay populations in singapore. Diabetes 57, 2851–2857. doi: 10.2337/db08-0214
Tang, Q.-Q., Otto, T. C., and Lane, M. D. (2003). Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 44–49.
Tanofsky-Kraff, M., Han, J. C., Anandalingam, K., Shomaker, L. B., Columbo, K. M., Wolkoff, L. E., et al. (2009). The fto gene rs9939609 obesity-risk allele and loss of control over eating. Am. J. Clin. Nutr. 90, 1483–1488.
Tung, Y. C. L., Yeo, G. S. H., O’rahilly, S., and Coll, A. P. (2014). Obesity and fto: changing focus at a complex locus. Cell Metab. 20, 710–718.
Van Der Hoeven, F., Schimmang, T., Volkmann, A., Mattei, M., Kyewski, B., and Rüther, U. (1994). Programmed cell death is affected in the novel mouse mutant fused toes (ft). Development 120, 2601–2607.
Villalobos-Comparán, M., Flores-Dorantes, M. T., Villarreal-Molina, M. T., Rodríguez-Cruz, M., García-Ulloa, A. C., Robles, L., et al. (2008). The fto gene is associated with adulthood obesity in the mexican population. Obesity 16, 2296–2301.
Vucenik, I., and Stains, J. (2012). Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann. N.Y. Acad. Sci. 1271, 37–43. doi: 10.1111/j.1749-6632.2012.06750.x
Wang, D., Chen, J., Chen, H., Duan, Z., Xu, Q., Wei, M., et al. (2012). Leptin regulates proliferation and apoptosis of colorectal carcinoma through pi3k/akt/mtor signalling pathway. J. Biosci. 37, 91–101. doi: 10.1007/s12038-011-9172-4
Wang, D., Wu, Z., Zhou, J., and Zhang, X. (2020). Rs9939609 polymorphism of the fat mass and obesity-associated (fto) gene and metabolic syndrome susceptibility in the chinese population: a meta-analysis. Endocrine 69, 278–285. doi: 10.1007/s12020-020-02280-x
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol. Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger rna stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger rna translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wei, C. M., Gershowitz, A., and Moss, B. (1975). Methylated nucleotides block 5′ terminus of hela cell messenger rna. Cell 4, 379–386. doi: 10.1016/0092-8674(75)90158-0
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P., et al. (2018). Differential ma, ma, and ma demethylation mediated by fto in the cell nucleus and cytoplasm. Mol. Cell 71, 973.e5–985.e5. doi: 10.1016/j.molcel.2018.08.011
Wei, R., Mao, L., Xu, P., Zheng, X., Hackman, R., Mackenzie, G., et al. (2018). Suppressing glucose metabolism with epigallocatechin-3-gallate (egcg) reduces breast cancer cell growth in preclinical models. Food Funct. 9, 5682–5696. doi: 10.1039/c8fo01397g
Wei, R., Penso, N. E. C., Hackman, R. M., Wang, Y., and Mackenzie, G. G. (2019). Epigallocatechin-3-gallate (egcg) suppresses pancreatic cancer cell growth, invasion, and migration partly through the inhibition of akt pathway and epithelial-mesenchymal transition: enhanced efficacy when combined with gemcitabine. Nutrients 11:1856. doi: 10.3390/nu11081856
Wen, W., Cho, Y. S., Zheng, W., Dorajoo, R., Kato, N., Qi, L., et al. (2012). Meta-analysis identifies common variants associated with body mass index in east asians. Nat. Genet. 44, 307–311. doi: 10.1038/ng.1087
White, E., and Dipaola, R. S. (2009). The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316. doi: 10.1158/1078-0432.Ccr-07-5023
Wu, R., Liu, Y., Yao, Y., Zhao, Y., Bi, Z., Jiang, Q., et al. (2018a). Fto regulates adipogenesis by controlling cell cycle progression via m6a-ythdf2 dependent mechanism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1323–1330. doi: 10.1016/j.bbalip.2018.08.008
Wu, R., Yao, Y., Jiang, Q., Cai, M., Liu, Q., Wang, Y., et al. (2018b). Epigallocatechin gallate targets fto and inhibits adipogenesis in an mrna m 6 a-ythdf2-dependent manner. Int. J. Obes. 42, 1378–1388. doi: 10.1038/s41366-018-0082-5
Yan, F., Al-Kali, A., Zhang, Z., Liu, J., Pang, J., Zhao, N., et al. (2018). A dynamic n 6-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 28, 1062–1076. doi: 10.1038/s41422-018-0097-4
Yang, S., Wei, J., Cui, Y.-H., Park, G., Shah, P., Deng, Y., et al. (2019). M 6 a mrna demethylase fto regulates melanoma tumorigenicity and response to anti-pd-1 blockade. Nat. Commun. 10, 1–14. doi: 10.1038/s41467-019-10669-0
Zhang, C., Zhang, M., Ge, S., Huang, W., Lin, X., Gao, J., et al. (2019). Reduced m6a modification predicts malignant phenotypes and augmented wnt/pi3k-akt signaling in gastric cancer. Cancer Med. 8, 4766–4781.
Zhang, D. L., Qu, L. W., Ma, L., Zhou, Y. C., Wang, G. Z., Zhao, X. C., et al. (2018). Genome-wide identification of transcription factors that are critical to non-small cell lung cancer. Cancer Lett. 434, 132–143. doi: 10.1016/j.canlet.2018.07.020
Zhang, L., Xie, J., Gan, R., Wu, Z., Luo, H., Chen, X., et al. (2019). Synergistic inhibition of lung cancer cells by egcg and nf-κb inhibitor bay11-7082. J. Cancer 10, 6543–6556. doi: 10.7150/jca.34285
Zhang, M., Zhang, Y., Ma, J., Guo, F., Cao, Q., Zhang, Y., et al. (2015). The demethylase activity of fto (fat mass and obesity associated protein) is required for preadipocyte differentiation. PLoS One 10:e0133788. doi: 10.1371/journal.pone.0133788
Zhang, X., Wei, L.-H., Wang, Y., Xiao, Y., Liu, J., Zhang, W., et al. (2019). Structural insights into fto’s catalytic mechanism for the demethylation of multiple rna substrates. Proc. Natl. Acad. Sci. U.S.A. 116, 2919–2924. doi: 10.1073/pnas.1820574116
Zhang, Y., Fan, S., Hu, N., Gu, M., Chu, C., Li, Y., et al. (2012). Rhein reduces fat weight in db/db mouse and prevents diet-induced obesity in c57bl/6 mouse through the inhibition of pparγ signaling. PPAR Res. 2012: 374936.
Zhang, Y., Sowers, J. R., and Ren, J. (2018). Targeting autophagy in obesity: from pathophysiology to management. Nat. Rev. Endocrinol. 14, 356–376. doi: 10.1038/s41574-018-0009-1
Zhao, X., Yang, Y., Sun, B.-F., Shi, Y., Yang, X., Xiao, W., et al. (2014). Fto-dependent demethylation of n6-methyladenosine regulates mrna splicing and is required for adipogenesis. Cell Res. 24, 1403–1419. doi: 10.1038/cr.2014.151
Zheng, G., Cox, T., Tribbey, L., Wang, G. Z., Iacoban, P., Booher, M. E., et al. (2014). Synthesis of a fto inhibitor with anticonvulsant activity. ACS Chem. Neurosci. 5, 658–665. doi: 10.1021/cn500042t
Zhou, S., Bai, Z. L., Xia, D., Zhao, Z. J., Zhao, R., Wang, Y. Y., et al. (2018). Fto regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (cscc) by targeting β-catenin through mrna demethylation. Mol. Carcinogenesis 57, 590–597.
Zhu, L., Dai, L., Yang, N., Liu, M., Ma, S., Li, C., et al. (2020). Transcription factorirx5 promotes hepatocellular carcinoma proliferation and inhibits apoptosis by regulating the p53 signalling pathway. Cell Biochem. Funct. 38, 621-629. doi: 10.1002/cbf.3517
Keywords: obesity, cancer, FTO, SNP, M6A modification, mTOR, FTO inhibitors
Citation: Lan N, Lu Y, Zhang Y, Pu S, Xi H, Nie X, Liu J and Yuan W (2020) FTO – A Common Genetic Basis for Obesity and Cancer. Front. Genet. 11:559138. doi: 10.3389/fgene.2020.559138
Received: 05 May 2020; Accepted: 02 September 2020;
Published: 16 November 2020.
Edited by:
Xiang Shu, Vanderbilt University Medical Center, United StatesReviewed by:
Jie Tan, Huazhong University of Science and Technology, ChinaMinghua Wang, Soochow University Medical College, China
Copyright © 2020 Lan, Lu, Zhang, Pu, Xi, Nie, Liu and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzhen Yuan, eXVhbnd6aEBsenUuZWR1LmNu
 Ning Lan
Ning Lan Ying Lu1,2,3
Ying Lu1,2,3 Shuangshuang Pu
Shuangshuang Pu Huaze Xi
Huaze Xi Xin Nie
Xin Nie Jing Liu
Jing Liu