
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 23 October 2020
Sec. Genomics of Plants and the Phytoecosystem
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.01001
This article is part of the Research TopicGenetics and Genomics to Enhance Crop Production, Towards Food SecurityView all 36 articles
Fusarium wilt (FW) disease is the key constraint to grain legume production worldwide. The projected climate change is likely to exacerbate the current scenario. Of the various plant protection measures, genetic improvement of the disease resistance of crop cultivars remains the most economic, straightforward and environmental-friendly option to mitigate the risk. We begin with a brief recap of the classical genetic efforts that provided first insights into the genetic determinants controlling plant response to different races of FW pathogen in grain legumes. Subsequent technological breakthroughs like sequencing technologies have enhanced our understanding of the genetic basis of both plant resistance and pathogenicity. We present noteworthy examples of targeted improvement of plant resistance using genomics-assisted approaches. In parallel, modern functional genomic tools like RNA-seq are playing a greater role in illuminating the various aspects of plant-pathogen interaction. Further, proteomics and metabolomics have also been leveraged in recent years to reveal molecular players and various signaling pathways and complex networks participating in host-pathogen interaction. Finally, we present a perspective on the challenges and limitations of high-throughput phenotyping and emerging breeding approaches to expeditiously develop FW-resistant cultivars under the changing climate.
Grain legumes being a rich source of dietary proteins and essential minerals serve as one of the vital components of human food (Graham and Vance, 2003; Bohra et al., 2015). Besides, grain legumes also supply essential micronutrients to the human population for combating increasing malnutrition related problems worldwide (Mudryj et al., 2014). Global legume production is severely challenged by a variety of fungal diseases (Kaiser et al., 2000), of which wilt caused by Fusarium oxysporum is one of the most destructive (Wade, 1929; Haware et al., 1978; Armstrong and Armstrong, 1981; Reddy et al., 1990; Kraft et al., 1998; Smith et al., 1999; Fall et al., 2001). It ranks fifth among top 10 fungal pathogens of research and economic importance (Dean et al., 2012). The fungus is soil-borne, and occurs as pathogenic (plant, animal, and human) as well as non-pathogenic strains (Leslie and Summerell, 2006). The pathogenic strains have been assigned to forma specialis (plural: formae speciales), abbreviated f. sp. (plural ff. spp.) based on host specificity (Baayen et al., 2000; Lievens et al., 2008). For instance, chickpea (Cicer arietinum L.) is affected by F. oxysporum f. sp. ciceris (Foc). Interestingly, Foc can also invade root tissues of other grain legumes, such as Vicia faba, Lens culinaris, Pisum sativum, and Cajanus cajan without causing external symptoms (Jiménez-Díaz et al., 2015). Presently Index Fungorum1 lists 124 special forms, whereas MycoBank2 lists 127 special forms of F. oxysporum. Following entry of Fusarium wilt (FW) pathogen through plant root, its colonization in the vascular system disrupts plant root-water continuum, leading to wilting symptoms and death of plant (Schäfer, 1994). Upon exposure to the wilt pathogen, plants recognize pathogen-associated molecular patterns (PAMPs) through their receptor protein pattern recognition receptors (PRRs) known as pattern triggered immunity (PTI) and pathogen effector triggered immunity (ETI), two important mechanisms for averting FW attacks (Zipfel and Robatzek, 2010; Langner et al., 2018). Among the various strategies devised to control FW disease, developing host plant resistance through breeding remains the most straightforward, economic, and sustainable approach. Therefore, physiological pathotypes or races, the subspecific ranks applied to formae speciales based on cultivar specificity, are extremely important to breeders for resistance breeding. However, race labels have been incoherent (Gerlagh and Blok, 1988; Correll, 1991; Kistler, 1997; Fourie et al., 2011) as numerous different race designation systems have been applied (Gabe, 1975; Risser et al., 1976; Armstrong and Armstrong, 1981) that created confusion (Kistler, 1997). Nevertheless, recent technological advances in molecular biology helped to overcome many of these hurdles. For instance, sequence-characterized amplified region (SCAR) markers facilitated race identification (Lievens et al., 2008; Epstein et al., 2017; Gilardi et al., 2017). However, laborious and prolonged pathogenicity tests are still required for the identification of new emerging races and resistance testing of the newly developed cultivars against the known races (Epstein et al., 2017; Gilardi et al., 2017). Understanding the genetic makeup of host plant resistance is crucial in this regard. In grain legumes, initial studies based on Mendelian genetics have elucidated a number of gene(s)/genetic determinants underlying resistance against FW (Wade, 1929; Hare et al., 1949; Upadhyaya et al., 1983a, b; Pandey et al., 1996; Coyne et al., 2000; Cross et al., 2000; Fall et al., 2001; Kotresh et al., 2006). Subsequent advances in genomics accelerated molecular mapping of FW resistance gene(s)/QTLs. To this end, availability of whole genome sequences of both plant and pathogen has shed deep insights into the host-pathogen relationship through leveraging comparative genomics approach (Varshney et al., 2012, 2013; Schmutz et al., 2014; Williams et al., 2016; Srivastava et al., 2018; Kreplak et al., 2019; Lonardi et al., 2019). In this review, we discuss the targeted improvement of plant resistance against FW using genomics-assisted approaches. The contributions of functional genomics toward delineating genomic regions/candidate gene(s) responsible for FW resistance in major grain legumes are highlighted. The potential of new omics approaches is discussed with respect to molecular players, signaling pathways, and complex networks underlying host-pathogen interactions. Finally, we present a perspective on high-throughput disease screening and emerging novel breeding techniques for developing FW-resistant cultivars under the changing climate.
Fusarium oxysporum is considered saprophytic because of its ability to survive on soil organic matter for several years (Alabouvette et al., 1993). The fungus survives in soil through producing chlamydospores that may serve as a reservoir of inoculums (Schippers and Van Eck, 1981). Subsequent to penetrating plant root epidermis, the pathogen invades the xylem vessels to cause wilting symptoms (Srinivas et al., 2019). The complete molecular mechanism of FW pathogenesis remains to be elucidated (Rep and Kistler, 2010). Genome-wide transcriptome profiling of conidial germination of one of the most virulent Indian races (race 1) of Foc has revealed germination-related genes and families of genes encoding secreted effectors, cell wall/pectin-degrading enzymes, metabolism related enzymes, transporters, and peptidases. Importantly, qRT-PCR confirmed the up-regulation of metabolism related enzymes during early infection, whereas up-regulation of most transporters and secondary metabolites important for tissue colonization and pathogenicity was confirmed during later stages (Sharma et al., 2016b).
Following establishment of the pathogen on plant roots, root penetration, and hyphal propagation of the FW pathogen causes a compromise in the host defense system (for detail see Rep and Kistler, 2010; Srinivas et al., 2019). At molecular level, the fungal pathogen recognizes a particular host and produces a range of cell wall-degrading enzymes including cellulases, pectinases, polygalacturonases, etc., in response to host plant derived hydrolases (viz., chitinase, glucanases) (Michielse et al., 2009; Swarupa et al., 2014; Husaini et al., 2018). Besides, the FW pathogen is also known to produce various mycotoxins/phytotoxins viz., fusaric acid (FSA), beauvericin, and enniatins in banana (López-Díaz et al., 2017; Liu et al., 2020; Shao et al., 2020). In parallel, the attacking pathogen integrates various signal transduction pathways mediated by mitogen activated protein (MAP) kinase cascades that transduce the signal downstream to the intracellular targets in response to the signal perceived by various receptors at cell surface during host infection (Widmann et al., 1999; Husaini et al., 2018). Thus, MAP plays a key role in regulating FW pathogenicity. Several genes are known to regulate host colonization and pathogenicity, which include FWO1 (Inoue et al., 2002), ClcI (Cañero and Roncero, 2008), chitin synthase V, DCW1, mannose- 6- phosphate isomerase gene, FOXG_11097 (Michielse et al., 2009), XlnR (Calero-Nieto et al., 2007), secreted in xylem (SIX) protein encoding genes (Thatcher et al., 2016), SIX1, SIX6, and FTF1 genes (NinÞo-Saìnchez et al., 2015) (for details see Husaini et al., 2018). Thus, successful disease occurrence by the FW pathogen demands a compromise in the plant defense system.
The PTI and the ETI activated by the PAMPs and pathogen effectors, respectively constitute the two tiers of the plant defense system. The PTI, presenting the first line of plant defense, ensues by the recognition of PAMPs via the host receptor protein PRRs (Zipfel and Robatzek, 2010; Beck et al., 2012; Lanubile et al., 2015). Following this, the plant evokes oxidative burst and ion influx that transduce the signals to different pathways by triggering down-stream signaling networks mediated by the protein kinases viz., mitogen-activated protein kinases (MAPK) and protein phosphorylation (Nakagami et al., 2005; Pedley and Martin, 2005; Boudsocq et al., 2010; Rodriguez et al., 2010; Bigeard et al., 2015; Lanubile et al., 2015). This is accompanied by the activation of multiple TFs that switch on various defense responsive genes including PR genes and the genes involved in hormone biosynthesis and signaling, protein and sugar metabolism (Castillejo et al., 2015; Lanubile et al., 2015; Kumar et al., 2016). As shown in Figure 1, once the PTI fails, the ETI forms the second line of immune response in which the plant defends itself against pathogen attacks through immune receptors encoded by the nucleotide binding leucine rich repeat (NB-LRR) class of R genes, thus enabling recognition of the effector molecules (Zipfel and Robatzek, 2010; Bigeard et al., 2015; Ma and Ma, 2016). This in turn triggers the plant innate immunity that inhibits the pathogen attack (Dodds and Rathjen, 2010). These resistance genes are overcome by the more virulent races of pathogen; hence, a broad-spectrum and durable host resistance is highly needed (Yin and Qiu, 2019; Li et al., 2020).
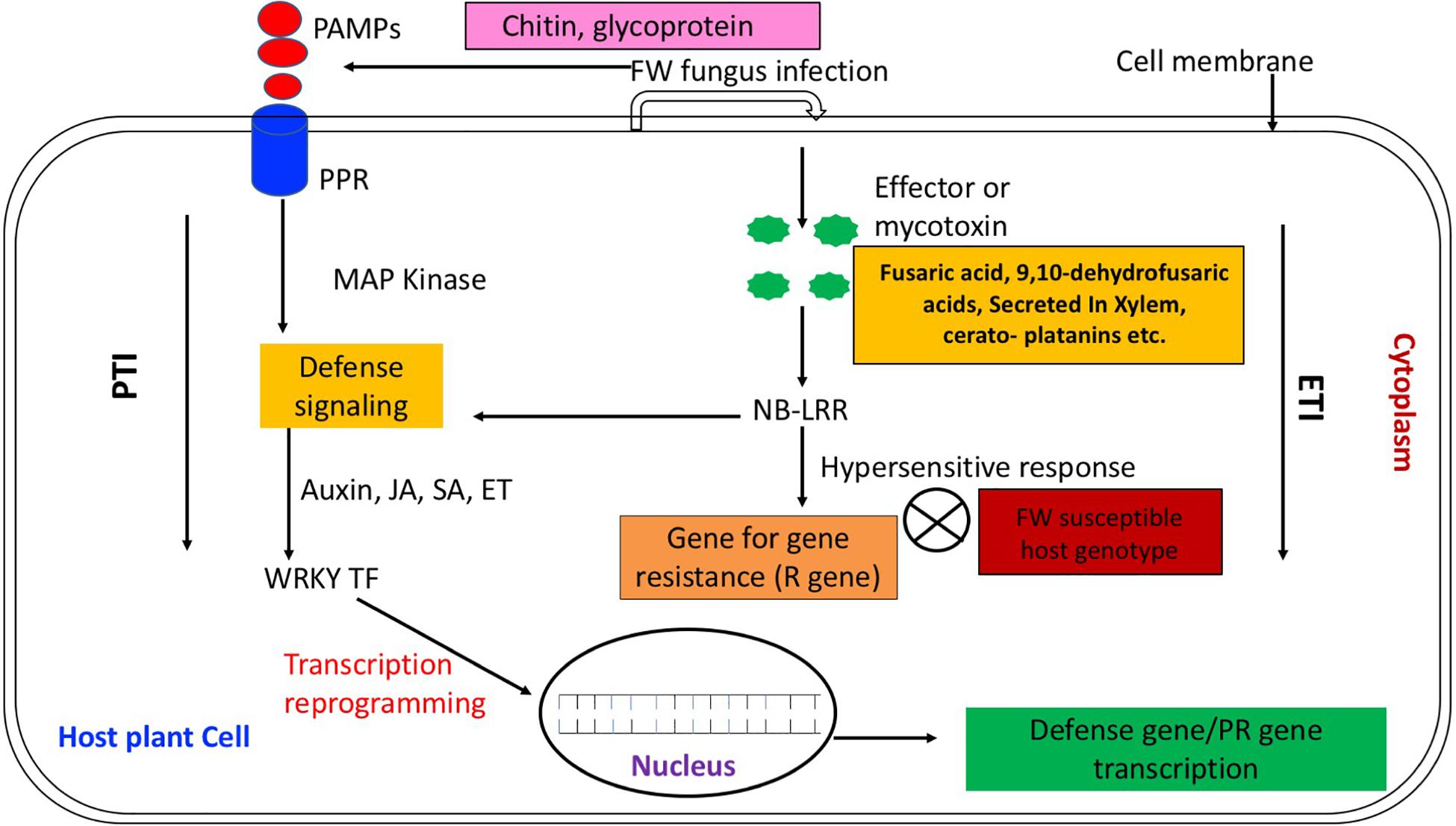
Figure 1. In PTI following the perception of PAMPs by PRRs, plant evokes signaling networks mediated by protein kinases viz., mitogen-activated protein kinases (MAPK) and protein phosphorylation (Nakagami et al., 2005; Boudsocq et al., 2010; Rodriguez et al., 2010; Lanubile et al., 2015). In turn, this is accompanied by activation of multiple TFs genes (via auxin, JA, ET) that ultimately switch on transcription of various defense responsive genes including PR genes (Castillejo et al., 2015). Once the PTI fails, the ETI forms the second line of immune response in which plant defends itself against pathogen attack through encoding immune receptors of the nucleotide binding leucine rich repeat (NB-LRR) class by R genes that enable recognizing the effector molecules (viz., fusaric acid) (Bani et al., 2014) produced by pathogen and initiate hypersensitive response (Zipfel and Robatzek, 2010; Ma and Ma, 2016). In case of susceptible host, the effector molecules remain unrecognized and there is no host pathogen hypersensitive response (Li et al., 2020).
Fusarium oxysporum f. sp. ciceris is prevalent in all major chickpea growing countries including India, Iran, Peru, Syria, Ethiopia, Mexico, and the United States (Nene et al., 1989; Halila and Strange, 1996). Up to 40% yield losses have been reported in chickpea due to FW, and the disease may lead to complete crop failure under congenial environment (Haware et al., 1978, 1986; Sharma et al., 2014). In recent times, FW has emerged as a severe threat to chickpea production because of a shift in chickpea area from long prevailing cool season of northern region to warm regions of southern and central India (Sharma and Pande, 2013).
Early wilting is manifested in the form of dull green discoloration within 25 days after sowing accounts for 77–94% yield loss (Jiménez-Díaz et al., 2015). In the case of “late wilt” dropping petioles and yellowing of leaf symptoms appear at podding stage, causing 24–65% yield loss (Jiménez-Díaz et al., 2015). Based on the symptoms produced on host plant, Foc races are grouped into two categories. Six races (1A, 2, 3, 4, 5, and 6) cause wilting symptoms (see Table 1), while two races (0 and 1B/C) cause yellowing symptoms in the plant during infection (Haware and Nene, 1982; Jimenez-Diaz et al., 1993; Kelly et al., 1994; Sharma et al., 2004). Among these Foc races, races 2, 3, and 4 are prevalent in India. Races 0, 1B/C, 5, and 6 are reported in Mediterranean and the United States, whereas the race 1A occurs in India, the United States, and the Mediterranean region (Jimenez-Diaz et al., 1993; Jiménez-Gasco et al., 2001; Landa et al., 2006). Gurjar et al. (2009) classified race 3 as Fusarium proliferatum based on phylogenetic analysis of translation elongation factor 1 alpha (TEF1-alpha) sequence data. The study demonstrated the reliable identification of Indian Foc races using a diverse DNA marker system. Earlier, Jiménez-Gasco and Jiménez-Díaz (2003) developed SCAR markers from race-specific random amplified polymorphic DNA (RAPD) markers for the identification of Foc and its pathogenic races. Change in the race scenario of Foc has been reported in India. Seventy Foc isolates, collected from 13 states and four crop cultivation zones of India, grouped into eight races based on their response to differential cultivars of chickpea. Further, characterization with four different molecular markers (RAPD, universal rice primers, SSR, and ISSR) grouped the isolates into eight clusters, which partially corresponded to the chickpea-growing zones and the racial distribution of the pathogen (Dubey et al., 2012). The presence of 47 isolates of different Foc races into one vegetative compatibility group (VCG) suggested its monophyletic origin (Nogales Moncada et al., 2009).
Fusarium oxysporum f. sp. tracheiphilum (Fot), a soil-borne fungus, severely challenges cowpea production (Armstrong and Armstrong, 1981). The disease is prevalent in all cowpea growing countries across the world including the United States, central valley of California (Hare, 1953; Smith et al., 1999), Australia (Summerell et al., 2011), Brazil and Nigeria (Armstrong and Armstrong, 1980; Assunção et al., 2003). Four Fot races (1, 2, 3, and 4) have been identified based on their pathogenic reactions to differential cowpea genotypes. Worldwide occurrence of race 3 has been reported among various Fot races (Hare, 1953; Smith et al., 1999). Broad patches of diseased cowpea plants with visible symptoms of chlorosis, wilting and stunting at the seedling stage or during flowering and early pod development result in high mortality and significant yield loss (Smith et al., 1999; Pottorff et al., 2012; see Table 1).
Wilt of common bean caused by F. oxysporum f. sp. phaseoli (FOP) results in substantial yield loss. FOP is commonly found in various common bean growing countries across the United States, China, Africa, and Latin America (Harter, 1929; Buruchara and Camacho, 2000). Elena and Papas (2002) reported that most of the isolates among 27 FOP isolates from Greece belonged to the same VCG. Earlier, VGC and molecular analysis of 128 FOP isolates collected from the field of El Barco de Avila in Spain suggested a difference between FOP isolates of Spain and America thus, supporting its pathogenic evolution (Alves-Santos et al., 1999).
The FOP invades plants through penetrating roots and colonizes the cortical cell. The hyphae move toward vascular parenchyma cells accompanied by a collapse of the xylem vessel and subsequent disruption of the water uptake from the roots (NinÞo-Saìnchez et al., 2015; Garces-Fiallos et al., 2017). Recent histological evidences support faster colonization of FOP in the susceptible genotypes in comparison to the resistant genotypes (Garces-Fiallos et al., 2017). A previous study by NinÞo-Saìnchez et al. (2015) explained the differential pattern of host plant colonization of highly virulent and weakly virulent strains of FOP based on expression analysis of different virulence genes viz., FTF1, SIX1, and SIX6. The infected plant displays wilting of older leaves followed by younger leaves and necrosis of the apex part (Fall et al., 2001; NinÞo-Saìnchez et al., 2015; Batista et al., 2017; Garces-Fiallos et al., 2017), internal stem discoloration (Batista et al., 2016), leading to death of the plant (Fall et al., 2001; Batista et al., 2017).
Fusarium oxysporum f. sp. lentis (Fol) is the causal agent of wilt disease in lentil (Erskine and Bayaa, 1996; Bayaa et al., 1998; Tosi and Cappelli, 2001). In India, 0.7–9.3% mortality has been reported at reproductive stage based on a survey of 116 districts of nine lentil growing states (Chaudhary et al., 2010). Similarly, severe disease incidence was recorded in 21 locations of Pakistan (Rubab et al., 2014). The pathogen survives in plant debris/soil and infects the host through the roots. The ability of the pathogen to survive in soil for long periods through chlamydospores further aggravates the situation. Evidence also suggests transmission of the pathogen through seeds (Erskine et al., 1990). In Algeria, based on the pathogenic variation of 32 isolates of Fol, Belabid et al. (2004) reported homogeneous behavior of Algerian Fol isolates with no variations in virulence and the existence of one Fol race. Although resistant or moderately resistant wilt varieties have been developed, variable responses of these varieties across agro-ecological niches imply the presence of high pathogen variability (Naimuddin and Chaudhary, 2009). No pathotypes within this formae specialis were identified until the study of Pouralibaba et al. (2016). Upon inoculation of 28 lentil resistant accessions with six Fol isolates with different geographical origins, a highly significant isolate × accession interaction resulted in the identification of four accessions as a putative differential set. The virulence pattern of 52 Fol isolates from Iran, Syria, and Algeria allowed the identification of seven pathotypes (1–7). Similarly, in India, Hiremani and Dubey (2018) identified eight races/pathotypes of the pathogen based on resistant and susceptible reactions on a set of differential cultivars. These studies pave the way for developing race specific wilt resistant lentil cultivars and help in the identification of new races from different lentil growing areas worldwide.
Wilt caused by F. oxysporum f. sp. pisi (Fop), is among the most significant yield reducers in pea growing areas (Kraft et al., 1981; Infantino et al., 2006; Rubiales et al., 2015). Among the four races (1, 2, 5, and 6) of Fop, races 1 and 5 are the most destructive, causing death of the plant (Jain et al., 2015). However, race 2 remains less pathogenic and wilting symptom appears in post podding stage. Based on VCG, isolates of Fop 1 and Fop 6 remained in a single VCG, whereas Fop5 belonged to a second VCG and Fop2 were present in another two VCGs (Kraft, 1994). Fop infects the root and interferes with the plant water movement, resulting in wilting symptoms (see Table 1). The other notable symptoms of wilt in pea include gray-green discoloration of foliage, chlorosis with unilateral wilting, orange discoloration in the vascular tissue (Jain et al., 2015).
Pigeonpea wilt is caused by Fusarium udum, a soil and seed borne pathogen, and the disease is reported to cause up to 100% yield loss in the susceptible cultivars (Reddy et al., 1990; Okiror, 2002). Occurrence of FW is reported predominantly in north and central parts of India, Kenya, and Malawi, and also in Ghana, Tanzania and Uganda (Nene, 1980). The annual losses caused by FW in pigeonpea have been estimated to be United States $71 million and 470,000 tons of grain in India and 30,000 tons of grain in Africa (Saxena et al., 2017).
Faba bean cultivation is severely affected by wilt [F. oxysporum f. fabae (FOF)] worldwide (Abdul Wahid et al., 1998; Dong et al., 2014). This disease was first reported by Abdel Rehim (1962) in Egypt.
Efforts to control FW disease with chemical agents have met with limited success. In view of the hazardous nature of fungicides to the environment, developing host plant resistance presents the most durable, economic and ecofriendly means to minimize the FW loss in grain legumes (Saxena, 2008; Jain et al., 2015). Therefore, harnessing the variation in plant traits that impart FW resistance could substantially improve the resistance level of various grain legumes. Considerable genetic variability has been observed in chickpea genotypes for FW response (Haware et al., 1992; Ali et al., 2002). Identification of resistant sources of FW in both kabuli (ICCV 2 and UC 15) and desi types (FLIP 85-20C, FLIP 85-29C, and FLIP 85-30C) by Ali et al. (2002) was consistent with the earlier report of Jimeìnez-Diìaz et al. (1991). More recently, Sharma et al. (2019) identified several chickpea FW resistant genotypes viz., ICCV 98505, ICCV 07105, ICCV 07111, ICCV 07305 based on GGE biplot analysis (see Table 2). Previously, Kumar et al. (1985) developed four kabuli resistant genotypes (ICCV 2, ICCV 3, ICCV 4, and ICCV 5) through a pedigree method. Recent advancements in genomic technologies in grain legumes have provided crop breeders with a set of more efficient tools for resistance breeding. Consequently, successful examples of genomics-assisted trait improvement for abiotic and biotic stresses are now available in legume crops (Bohra et al., 2014a, b; Varshney et al., 2015). In chickpea, a marker-assisted back crossing (MABC) scheme has allowed targeted transfer of genomic regions conferring FW resistance (foc4) from WR 315 to Annigeri 1 and JG 74, two elite yet FW-sensitive elite chickpea cultivars (Mannur et al., 2019). Thus, MABC derived products in chickpea such as Super Annigeri 1 and JG 74315-14 showed an 8% increase in yield and disease resistance over Annegiri and 53.5% increase in yield over JG74, respectively (Mannur et al., 2019). Likewise, genomic regions underlying foc1 and foc2 resistance were transferred from JG 315 to C 214 (Varshney et al., 2014) and from Vijay to Pusa 256 (Pratap et al., 2017) using MABC. The MABC-bred lines carrying favorable alleles such as ICCX-100175-349-2-2, ICCX-100175-382-4-6, and ICCX-100175-389-3-2 had high to moderate resistance against FW (foc1) under field conditions (Varshney et al., 2014). Marker-aided breeding schemes could enable efficient pyramiding of QTL into new cultivars, thus imparting on them durable resistance against multiple FW races. Apart from the cultivated pool, crop wild relatives (CWRs) of chickpea viz., Cicer reticulatum, Cicer echinospermum, Cicer bijugum, Cicer judaicum, Cicer pinnatifidum, and Cicer cuneatum have also been identified having traits that confer FW resistance (Nene and Haware, 1980; Kaiser et al., 1994; Singh et al., 1998).
Studies have shown significant genetic variability for Fol resistance in lentil (Bayaa et al., 1995; Pouralibaba et al., 2015).
Studies have shown significant genetic variability for Fol resistance in lentil (Bayaa et al., 1995; Pouralibaba et al., 2015). Screening in both controlled and field conditions led to the identification of three promising lines viz., 81S15, FLIP 2007-42 L, and FLIP 2009-18 L (Mohammadi et al., 2012) and BGE040548, BGE019708, BGE022526, BGE025720 (Pouralibaba et al., 2015) exhibiting Fol resistance. Recently, GGE biplot analysis has revealed two lentil genotypes PL101 and L4076 as promising sources for Fol resistance (Parihar et al., 2017). Similarly, resistant genotypes ILWL 79 and ILWL 113 (L. culinaris ssp. orientalis) and ILWL 138 (Lens nigricans ssp. ervoides) were screened against Fol from 219 wild lentils (Bayaa et al., 1995). Interspecific cross between ILL10829 (L. culinaris subsp. culinaris) and ILWL30 (Lens ervoides) has also revealed substantial genetic variability for Fol resistance (Singh et al., 2017).
Previously, based on glasshouse screening, Doling (1964) identified Delwiche Commando, New Era and New Season genotypes to be resistant to both Fop1 and Fop2 races of FW. Likewise, Kraft (1994) identified 74SN5 pea line to be an important source of resistance against all the four races Fop1, Fop2, Fop5, and Fop6. Subsequently, screening of a large set of 452 pea accessions collected from 24 countries resulted in the identification of 62 accessions to be resistant against Fop2 and 39 accessions out of these 62 also possessed resistant to Fop1 (McPhee et al., 1999). Additionally, PI 344012 a wild progenitor of pea displayed resistance against both Fop1 and Fop2 (McPhee et al., 1999). Given the screening of 117 pea genotypes against Fop1, Fop2, Fop5, and Fop6 under growth chamber, Neumann and Xue (2003) identified Radley and Princess cultivars exhibiting resistant reaction against Fop2, Fop5, and Fop6 races. Likewise, a thorough assessment of eighty accessions of Pisum spp. against Fop2 by recording detailed disease scoring based on the various parameters revealed significant genetic variation for Fop2 disease reaction (Bani et al., 2012). Further the authors reported eleven accessions namely JI 1412, JI 1559, JI 1760, P 23, P 42, P 614, P 627, P 633, P 639, P 650, and P 656 to be resistant against Fop2 (Bani et al., 2012). Evaluation of 34 pea genotypes against FW under wilt sick plot and artificially controlled conditions allowed for the identification of GP-6, GP-55, and GP-942 as highly resistant and GP-17, GP-48, GP-473, and GP-941 as resistant donors for Fop resistance (Shubha et al., 2016).
Considering the common bean earlier, Ribeiro and Hagedorn (1979) identified the Preto Uberabinha common bean cultivar to be resistant to FOP based on disease reaction of plant root inoculated with FOP microconidia. Subsequently, rigorous screening of 73 climbing and bush type accessions of common bean against FOP led to identification of 19 climbing type and 28 bush type accessions to be resistant against FOP. Further, two genotypes RWR 950 and G 685 displayed resistance even at higher inoculum density of FOP (Buruchara and Camacho, 2000). Based on the restriction of FOP xylem tissue colonization, Pastor-Corrales and Abawi (1987) reported Manteigão Fosco 11 genotype to be resistant against FOP. Likewise, UFSC-01 genotype was revealed to be resistant against FOP by checking the colonization of FOP inside the xylem vessel (Garces-Fiallos et al., 2017). Furthermore, availability of high throughput molecular maker based genome wide association study allowed the identification of 14 highly resistant common bean genotypes against FOP-SP1 race 6 (Leitão et al., 2020).
In pigeonpea, multi-location and multi-year testing of wider sets of germplasm and advanced breeding lines has revealed several promising lines for regular use in breeding programs. The resistant sources resulting from these evaluations include KPL 43, KPL 44, IPAs 16 F, 8 F, 9 F, and 12 F (Singh et al., 2011a) and ICPLs 20109, 20096, 20115, 20116, 20102, 20106, and 20094 (Sharma et al., 2016a; see Table 2). Notably, resistance to wilt is an essential prerequisite for variety identification and release in pigeonpea. Several pigeonpea varieties such as Asha (ICPL 87119), ICP 8863, BSMR 736, TS3R, IPA 203, BDN 708, BDN 711, etc., show considerable level of resistance to F. udum (Singh I. P. et al., 2016; Bohra et al., 2017).
In cowpea, genotypes CB46, CB3, 7964, and 8514 were identified as FW resistant based on three years evaluation at two different locations (Roberts et al., 1995). Similarly, genotypes CB46, CB27, and CB50 could serve as donors for developing FW resistant cowpea (Ehlers et al., 2000, 2009; Muchero et al., 2009). Since screening of large germplasm collections for FW resistance remains time consuming, thus availability of molecular markers linked with FW resistant gene(s) could circumvent the traditional screening methods (Ali et al., 2012; Jain et al., 2015).
In faba bean, variation was reported among 16 lines for FW resistance using inter-simple sequence repeat (ISSR), sequence related amplified polymorphism (SRAP) and SSR markers (Mahmoud and Abd El-Fatah, 2020). Based on the disease severity, Assiut-215, Roomy-3, Marut-2, and Giza-2 were found to be promising for FW resistance in faba bean.
Initial studies on genetic inheritance of FW resistance in grain legumes relied on Mendelian genetics. Literature in chickpea on FW inheritance suggests its control by major genes (Upadhyaya et al., 1983a, b; Kumar, 1998; Tullu et al., 1999). For example, genetic resistance against Foc1 results from the action of three independent loci h1, h2, and h3 (Upadhyaya et al., 1983a, b; Singh et al., 1987a, b). Kumar (1998) also advocated involvement of three separate loci controlling Foc2 resistance. Likewise, other researchers have reported digenic inheritance for other races (0 and 2) of Foc (Tullu et al., 1999; Rubio et al., 2003). Previously, the digenic (a, b) nature of Foc2 resistance was also established based on the disease reaction of F2 and F3 individuals derived from WR 315 × C 104 (Kumar, 1998). Whereas Tekeoglu et al. (2000) and Sharma et al. (2004) reported monogenic inheritance of Foc 3 and Foc 5 resistance. Concerning resistance against Foc4, Tullu et al. (1998, 1999) explained its monogenic and recessive nature in the genotype WR 315, however, its recessive and digenic nature was explained in Surutato 77. Likewise, Sharma et al. (2005) reported that the genetic resistance against each race 1A, 2, 3, 4, and 5 was controlled by a single gene. However, the genetic basis of resistance to races 1B/C and 6 is still to be studied.
Classical genetics studies in pea established that resistance to FW (races 1, 2, 5, and 6) was controlled by different genes of a dominant nature (Wade, 1929; Hare et al., 1949; Coyne et al., 2000; Haglund and Kraft, 2001). Resistance against Fop races 1, 5, and 6 was conferred by a single dominant gene, however, resistance to race 2 follows a quantitative pattern (Bani et al., 2012; McPhee et al., 2012). A study by Wade (1929) established that the Fop1 resistance was controlled by a single dominant gene (Fw), which was subsequently mapped on to LG III (Grajal-Martin and Muehlbauer, 2002).
Research on FOP resistance in common bean has shown presence of a single gene (Ribeiro and Hagedorn, 1979; Salgado et al., 1995; Cross et al., 2000; Fall et al., 2001), oligogene (Ribeiro and Hagedorn, 1979; Salgado et al., 1995; Cross et al., 2000; Fall et al., 2001; Batista et al., 2017) and polygenes (Salgado et al., 1995; Cross et al., 2000; Batista et al., 2016, 2017). Considering resistance against Fop4, occurrence of a single dominant gene (Salgado et al., 1995; Cross et al., 2000) as well as QTL was proposed (Fall et al., 2001).
Studies on the inheritance of FW resistance in pigeonpea indicate varying patterns such as two complementary genes (Pathak (1970), dominant monogenic (Pawar and Mayee, 1986; Pandey et al., 1996; Kotresh et al., 2006; Karimi et al., 2010), recessive monogenic (Jain and Reddy, 1995; Odeny et al., 2009), one dominant and one recessive gene with dominant suppressive epistatic (Saxena et al., 2012) as well as polygenic inheritance (Pal, 1934). Recent analysis of populations derived from four resistant and four susceptible parents suggested that the FW resistance was governed by one dominant gene each in BDN 2004-1 and BDN 2001-9 in comparison to two duplicate dominant genes in BWR 133 and two dominant complimentary genes in IPA 234 (Singh D. et al., 2016).
In lentil, limited research has been done on understanding the genetic basis of resistance to Fol. Kamboj et al. (1990) proposed the presence of five independently segregating genes for Fol resistance based on allelic tests of the crosses involving three Fol resistant lines (L 234, JL 446, and LP 286) and two susceptible lines (L 9-12 and JL 641). Subsequently, Eujayl et al. (1998) established monogenic dominant inheritance of Fol resistance based on F2:4 progenies [ILL 5588 × L 692–16−l(s)].
DNA marker technologies have facilitated locating/mapping of gene(s) controlling resistance against various races of Foc in chickpea (Mayer et al., 1997; Ratnaparkhe et al., 1998; Tullu et al., 1999; Tekeoglu et al., 2000; Winter et al., 2000; Rubio et al., 2003; Sharma et al., 2004; Cobos et al., 2005). Previously, Cobos et al. (2005) reported the Foc01/foc01 gene on LG 5 flanked by OPJ20600 and TR59 markers. Later, Halila et al. (2009) confirmed a second gene Foc02/foc02 (flanked by TS47 and TA59 markers) on LG2 following analysis of two mapping populations CA 2156 × JG 62 and CA 2139 × JG 62. Earlier, this gene was discovered by Rubio et al. (2003). Likewise, Jendoubi et al. (2016) fine mapped the Foc01/foc01 gene within an interval of 2 cM on LG5 using nearly isogenic lines (NILs). Of the 27 annotated genes, two candidate genes LOC101514038 and LOC101499491 are involved in disease resistance (Jendoubi et al., 2016). An SSR-based QTL analysis of F2:3 (C 214 × WR 315) elucidated two QTLs FW-Q-APR-6-1 and FW-Q-APR-6-2 on LG6 for Foc1 resistance (Sabbavarapu et al., 2013; see Table 3). The SSR marker TA103 was used for introgression of Foc1 from WR 315 to C 214 (Varshney et al., 2014). Previously, Gowda et al. (2009) located Foc1 flanked with SSRs TA110 and H3A12 on LG2. The authors also mapped the Foc2 (TA96-H3A12) and Foc3 (TA194- H1B06y) on LG2. However, Jingade and Ravikumar (2015) reported one major QTL GSSR 18-TC14801 on LG 1 for Foc1 resistance, which explained up to 71% phenotypic variation (PV). Subsequently, a major QTL FW-Q-APR-2-1 on CaLG02 and two other minor QTLs FW-Q-APR-4-1 and FW-Q-APR-6-1 on CaLG4 and CaLG6, respectively were identified for resistance against Foc1 and Foc3 (Garg et al., 2018). Considering Foc5, monogenic/oligogenic nature has been established for resistance loci on LG 2 (Tekeoglu et al., 2000; Winter et al., 2000; Castro et al., 2010). Recently, by using SNP in combination with SSR markers the candidate genomic region on LG2 was narrowed down to 820 kb (Caballo et al., 2019b). The authors also suggested involvement of a putative candidate gene LOC101511605 encoding CBL-interacting serine/threonine-protein kinase 8 in FW response.
In pea, genetic linkages of AFLP (McClendon et al., 2002), SSR (Loridon et al., 2005), SCAR (Okubara et al., 2005), and TRAP (Kwon et al., 2013) with FW was reported. Jain et al. (2015) identified a CAPS marker at 0.9 cM from the Fw locus on LG3, which could be effectively used for screening FW resistance in pea (see Table 3). Earlier, McPhee et al. (2012) reported two minor QTLs on LG 3 controlling resistance against Fop race 2.
In cowpea, SNP analysis of the population developed from CB27 × 24-125B-1 allowed identification of a 3.56 cM genomic region on LG1 for resistance to Fot3-1 (Pottorff et al., 2012). These marker-trait associations (MTAs) explained up to 27.8% PV for the resistance. Furthermore, a comparative analysis between cowpea and soybean genomes suggested four candidate genes from the Fot3-1 genomic region, which were related to leucine-rich repeat serine/threonine protein kinases (Pottorff et al., 2012). Likewise, two QTLs Fot4-1 and Fot4-2 imparting Fot4 resistance were identified on LG 5 and LG 3, respectively (Pottorff et al., 2014). Synteny analysis between soybean and cowpea suggested a role for candidate genes underlying the Fot4-1 and Fot4-2 QTLs that code for TIR–NBS–LRR proteins and leucine-rich repeat serine/threonine protein kinases (Pottorff et al., 2014; see Table 3).
In lentil, Eujayl et al. (1998) tagged Fw locus controlling resistance to Fol at 10.8 cM from RAPD marker (OPK−15900). Subsequently, Hamwieh et al. (2005) reported Fw locus on LG 6 flanked by SSR59-2B and AFLP marker p17m30710.
In pigeonpea, different research groups have found significant MTAs and candidate genes for FW by using SSR (Patil et al., 2017a) and SNP markers (Singh V. K. et al., 2016; Saxena et al., 2017).
Whole genome sequence information in combination with high-throughput DNA marker technologies has divulged massive amounts of genome-wide markers to analyze MTAs in large germplasm sets for traits including FW resistance.
In common bean, GWAS on a diverse collection of 162 Portuguese accessions showed nine significant SNPson chromosomes Pv04, Pv05, Pv07, and Pv08 for F. oxysporum f. sp. phaseoli strain FOP-SP1 race 6. Authors also reported that the resulting candidate genes are engaged in phytoalexin biosynthesis, hypersensitive response, and plant primary metabolism (Leitão et al., 2020). Similarly, GWAS of 96 genotypes in cowpea revealed 11 significant MTAs (on LG 1, 3, 4, 6, 8, 9, 10, and 11) explaining 4% PV related to leaf damage traits and seven significant MTAs (LG 3, 6, 10, 11) explaining 9.7% PV related to vascular discoloration (Wu et al., 2015). Among the significant MTAs, two SNPs 1_0691 and 1_1369 showed close proximity to the QTL Fot3-1 and Fot4-2, previously identified by Pottorff et al. (2012, 2014). A recent SSR-based association study of 89 pigeonpea lines phenotyped for 3 years in a wilt-sick field provided a set of six SSR markers, which were cross-validated in a biparental population segregating for FW (Patil et al., 2017a, b).
Availability of whole genome sequence information in chickpea (Varshney et al., 2013), common bean (Schmutz et al., 2014), cowpea (Lonardi et al., 2019), pea (Kreplak et al., 2019), and pigeonpea (Varshney et al., 2012) could allow identification of the candidate gene(s)/the genomic regions controlling disease resistance. Concurrently, draft genome assemblies of Fusarium udum F02845 (Srivastava et al., 2018) and Foc (Foc-38-1) and Fop (Fop-37622) (Williams et al., 2016) have shed new light onto virulence-related genes that enhance our current understanding of pathogenicity of FW and evolution of the host-pathogen interaction in the legume species. Further, comparative genomics of Fusarium species could also elucidate the host specific gene(s), effector gene(s) and the sequence conservation across legume-infecting isolates and other Fusarium spp. (Williams et al., 2016). Additionally, construction of fungal pangenome could offer deeper insights into novel pathogen effectors and how they defeat host plant resistance rapidly (Badet and Croll, 2020). Thus, growing refinements in deep sequencing chemistry have paved the way for whole genome re-sequencing (WGRS) of global legume germplasms for capturing the large structural variations (SVs) including the copy number variations and presence-absence variations controlling various traits of economic importance including disease resistance (Varshney et al., 2013; Thudi et al., 2016). In pigeonpea, a comparison of whole genome sequence information of FW-resistant genotypes (ICPL 87119, ICPL 20097, ICP 8863, and ICPL 99050) and FW-susceptible genotype (ICPB 2049) in combination with Seq-BSA of the resistant and susceptible bulks (ICPL 20096 × ICPL 332) revealed four candidate genes including C.cajan_03203 (Singh V. K. et al., 2016). These identified markers/candidate genes could be deployed for breeding FW resistance in pigeonpea. Genome-wide analysis using sequencing data of wilt responsive genotypes may help in pinpointing haplotypes responsible for resistance against multiple FW races, thus providing scope for gene pyramiding.
Prior to the discovery of digital transcriptome profiling, expressed sequenced tags (ESTs), cDNA-AFLP, and cDNA-RAPD were largely employed to find the gene(s) participating in the plant defense mechanisms and plant-pathogen interactions (Wise et al., 2007; Ashraf et al., 2009; Gupta et al., 2009; Gurjar et al., 2012; Xue et al., 2015).
In common bean, cDNA-AFLP analysis of FW-resistant and susceptible genotypes revealed differential expression of 423 transcript derived fragments (TDFs), of which 98 TDFs had annotated functions in signal transduction, protein synthesis and processing, RNA and energy metabolism, defense and stress responses (Xue et al., 2015). Furthermore, q-RT-PCR analysis confirmed FW-responsive expression of 19 candidate genes in CAAS 260205 (resistant) and BR 130 (susceptible) genotypes. Some important candidate genes viz., CBFi28, CBFi43 (ubiquitin protein), CBFi45 (poly-ubiquitin protein), CBFi76 (peroxidase), CBFi54, CBFi58 (calcium dependent protein kinase), CBFi83, and CBFi171 (NBS-LRR) had abundant expression in the resistant genotype (Xue et al., 2015).
In recent years, RNA-sequencing has enabled genome-wide surveys of transcriptomes to identify FW responsive candidate genes and their biological roles with greater precision and higher resolution (Li C. Y. et al., 2012; Kohli et al., 2014). Transcriptome analysis of four chickpea cultivars JG 62, ICCV 2, K 850, and WR 315 allowed several important “large effect” SNPs and Indels in the genomic regions controlling FW resistance (Jain et al., 2015). The underlying genomic region containing these SNPs and indels was predicted to be associated with defense related activity. Caballo et al. (2019a) functionally validated the genomic region controlling Foc (race 5) resistance in chickpea from resistant and sensitive NILs developed from the cross ILC 3279 × WR 315. Differential gene expression analysis at 24 h post inoculation (hpi) suggested two known candidate genes LOC101499873 (encoding chaperonin) and LOC101490851 and three novel candidate genes (LOC101509359, LOC101495941, and LOC101510206 encoding MADS-box transcription factor, MATE family protein and serine hydroxymethyl-transferase, respectively) to be related to defense activity against FW. Likewise, nine genes viz., LOC101503802, LOC101505941, LOC101506693 had significantly higher expression in FW sensitive NIL at 48 hpi than the FW resistant NIL (Caballo et al., 2019a). Transcriptome analysis of JG 62 and WR 315 in response to FW (race 1) infection uncovered abundance of differentially expressed transcripts related to various TFs, cellular transporters, sugar metabolism contributing to activate defense signaling against FW in chickpea (Gupta et al., 2013a; see Table 4). Further, network analysis also provided greater insights into the role of genes associated with defense components (MAP kinase, serine threonine kinase, etc.), reactive oxygen (superoxide dismutases, glutathione reductase, thioredoxin reductase, etc.), ATPase (myo-inositol phosphate, carboxylate synthase, etc.), significantly participating in the defense signaling against FW in chickpea (Gupta et al., 2013a). Similarly, differential expression was obtained by transcriptome profiling of chickpea genotypes JG 62 (Foc susceptible) and Digvijay (Foc resistant) for genes that are involved in lignification, hormonal homeostasis, plant defense signaling, reactive oxygen species (ROS) homeostasis, R-gene mediated defense in response to host-pathogen interaction (Upasani et al., 2017). An integrated analysis of transcriptome and metabolome data from the root samples of control and FOP infected seedlings in common bean demonstrated that pathogen establishment occurs after 24 h of infection, which is accompanied by timely induction of the defense mechanism. The study reinforced the proposition that the FOP defense system in common bean requires contributions from defense-related proteins such as glycosylphosphatidylinositol-anchored proteins (GPI-APs), signaling pathways mediated by hormones like salicylic acid, jasmonate and ethylene, and flavonoid biosynthesis pathway.
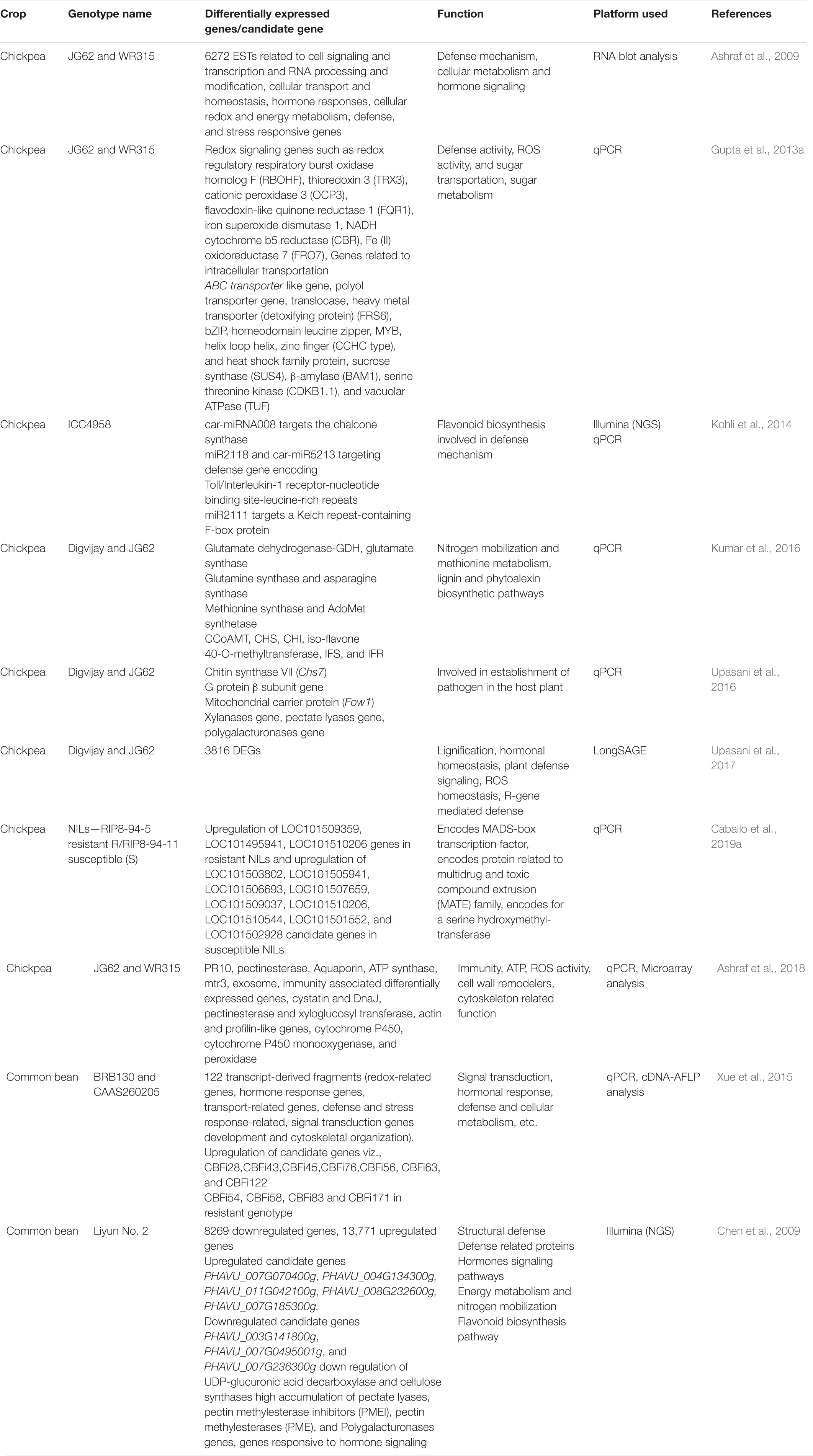
Table 4. Exhaustive list of various DEG and candidate genes contributing to FW resistance in grain legumes.
Further, to explore the role of micro RNAs (miRNAs) contributing to FW resistance, RNA-seq analysis of ICC 4958 uncovered known as well as novel miRNAs (car-miRNA008 targeting the chalcone synthase gene) involved in FW resistance in chickpea (Kohli et al., 2014). Among the identified miRNAs, miR530 showed 17-fold high expression, whereas miR156_1 and miR156_10 had slightly higher expression in response to FW infection (see Table 4).
A proteomics approach allows for the unraveling of the various proteins engaged in host-pathogen interaction and their role in defending the host plant against pathogen attacks (Castillejo et al., 2015; Kumar et al., 2016). Significant roles for myriads of proteins in host-pathogen interactions have been suggested either during the establishment of the pathogen in the susceptible host plant or protecting the host plant from pathogen invasion (Rep et al., 2002; Berrocal-Lobo and Molina, 2008; Mehta et al., 2008; Castillejo et al., 2010; Palomares-Rius et al., 2011; Gunnaiah et al., 2012). These proteins included chitinases, xylem proteinases, β-1,3-glucanases, proteinase inhibitors, pathogenesis-related (PR) proteins, leucine rich-repeat proteins, proline-rice glycoproteins, cellulose synthases, ankyrin repeat containing protein, thaumatin-like protein PR-5b, syntaxins to subtilin-like proteases in various plant species in response to FW infection (Yang et al., 1997; De Ascensao and Dubery, 2000; Li T. et al., 2012; Castillejo et al., 2015; Kumar et al., 2016; Silvia Sebastiani et al., 2017). In the host plant several enzymes viz., glutathione S-transferases, peroxidases, peroxiredoxin, uinone oxidoreductase, copper amine oxidase, caffeic acid O-methyltransferase, chalcone synthase, chalcone isomerase, isoflavone reductase, phenylalanine ammonia lyase, etc., show change in response to FW invasion (Klessig et al., 1998; Garcia-Limones et al., 2002; Gupta et al., 2013a; Kumar et al., 2016). However, limited information is available in grain legumes on the participation of anti-fungal proteins especially for FW (Palomares-Rius et al., 2011; Scarafoni et al., 2013; Kumar et al., 2016). To establish roles of proteins in disease development or prohibiting pathogen attack for disease progression in pea, Castillejo et al. (2015) performed proteomic analysis using two-dimensional electrophoresis (2-DE) and mass spectrometry (MALDI-TOF/TOF), and the study found 53 proteins engaged in the plant’s response to Fop race 2 infection. These proteins were found to affect carbohydrate and energy metabolism (viz., fructokinase-like protein, beta-amylase, phosphoglucomutase, cytoplasmic), nucleotide and amino acid metabolism (viz., apyrase S-type, adenosine kinase/copper ion binding), signal transduction and cellular process (viz., chalcone O-methyltransferase, 4-3-3-like protein), redox and homeostasis (viz., short-chain alcohol dehydrogenase SAD-C, short-chain alcohol dehydrogenase A), defense (endochitinase A2, beta-1,3-glucanase), and biosynthetic process (viz., NADPH: isoflavone oxidoreductase, glutamate decarboxylase) (Castillejo et al., 2015). In chickpea, the role of various defense-related proteins was observed in restricting FW infection in the genotypes Digvijay (FW resistant) and JG 62 (FW susceptible) (Kumar et al., 2016). Several ROS activating enzymes viz., glutathione peroxidase, glutaredoxin, glutathione S-transferase, ascorbate peroxidase, peroxiredoxin were abundant in Digvijay as compared to JG 62. Likewise, the genotype Digvijay was able to restrict FW pathogen attack than FW susceptible cultivar JG 62 due to the abundance of PR proteins (Kumar et al., 2016). Thus, proteomics could further illuminate our understanding of the unknown proteins involved in various signal transduction pathways for inducing host innate immunity against FW attack in grain legumes. Concurrently, this approach could also enable us to discover the novel pathogen effectors that drive the arms race between host plants and pathogens.
Like proteomics, a metabolomics approach has greatly advanced our understanding about various metabolites, hormonal crosstalks, and signaling molecules that participate in plant defense mechanisms against FW pathogenesis in crop plants including grain legumes (Gupta et al., 2010; Kumar et al., 2016). The various metabolites produced in response to FW include sugars like hexokinase, glucose-6-phosphate, sucrose synthase, trehalose, invertase, β-amylase, etc. (Morkunas and Ratajczak, 2014). These sugars play a key role in plant resistance against pathogen attacks by serving as substrate for supplying energy, causing oxidative burst and ROS generation, enhancing lignification of cell wall, and acting as signaling molecule in concert with various phytohormones to induce plant innate immunity (Morkunas et al., 2005, 2007; Nikraftar et al., 2013; Morkunas and Ratajczak, 2014).
Besides sugars, the other important metabolites that are implicated in FW pathogen attack and entry into host plant include various amino acids, organic acids (pyruvate, lactate, acetate, etc.), nucleotides and their derivatives, antioxidants, phytoalexins (lignans, pisatins), polyphenols, phenolic acids (monomers of lignin), calmodulins, flavonoids, lipids, and phenylpropanoids (Klessig et al., 1998; Rolland et al., 2002; Garcia-Limones et al., 2009; Kumar et al., 2015, 2016; Bani et al., 2018a, b). Several plant hormones that serve as the essential signaling molecules in regulation of host defense response against FW infection are ethylene, salicylic acid, and jasmonic acid (Kunkel and Brooks, 2002). The plant immunity or plant defense response mediated by these phytohormone is defeated or suppressed by the attacking pathogen through production of toxins or effectors (Ma and Ma, 2016).
Metabolite profiling of common bean in response to FOP demonstrated participation of UDP-glucuronic acid decarboxylase, cellulose synthases, and pectate lyases, amino acids, glycosyl phosphatidyl inositol-anchored proteins, and various phytohormones (salicylic acid, jasmonate, and ethylene), polyphenols, anthocyanins, flavanones, and flavones during host plant, and FOP interaction. Similarly, an abundance of various proteins contributing in glycolysis and TCA processes, defense related metabolites (endo beta-1,3-glucanase, chitinases, caffeic acid O-methyltransferase, and caffeoyl-CoA O-methyltransferase), phytoalexins (genistein, luteolin, quinone), flavonoids, isoflavonoids, and phenolic compounds was observed in Digvijay than in JG 62 (Kumar et al., 2016). However, significant decrease in various amino acids and sugars viz., sucrose and fructose in susceptible cultivar allows FW pathogens to invade and promote disease development (Kumar et al., 2016). Therefore, further advancements in metabolomics could enable elucidation of intricate network of various signaling molecules and hormonal crosstalk contributing to FW resistance in grain legumes.
A comparison of the different studies that analyzed changes in plant transcriptomes, proteomes and metabolomes in response to FW infection reinforces the role of chitinases, PR proteins, ROS activating enzymes, phenolic compounds, flavonoids, phytoalexins in imparting wilt resistance (Castillejo et al., 2015; Kumar et al., 2016; Upasani et al., 2017; Bani et al., 2018a, b). These studies also highlight the significance of molecules that participate in cellular metabolism including carbohydrate, protein, nucleotides (Castillejo et al., 2015; Xue et al., 2015; Kumar et al., 2016) and signaling pathways involving MAP kinase, serine threonine kinase and various phytohormones (Gupta et al., 2013a, b; Xue et al., 2015).
The declining cost of genotyping and accumulation of huge genotyping data have allowed pinpointing the targeted genomics regions and the underlying causative gene(s)/genomic regions for a variety of important traits including FW resistance. However, linking of this genomic information with the phenotype still remains a daunting task due to complexity of G × E interactions (Araus and Cairns, 2014). Current state-of-the-art high throughput phenotyping (HTP) approach has the potential to bridge the genotype-phenotype gap for various complex traits (Fiorani and Schurr, 2013; Araus and Cairns, 2014). Recent advances in high-resolution imaging platforms and sensor technologies have revolutionized our capacity to investigate plant disease interaction, screening of disease resistant lines and identifying plant disease at large scale and large field (Lowe et al., 2017; Shakoor et al., 2017; Singh et al., 2018). Several HTP approaches including field-based remote sensing, 3D scanning, unmanned aerial vehicles system in association with multispectral and thermal cameras, RGB based imaging, fluorescence imaging to hyperspectral image sensing are worth mentioning, and are routinely employed for precise understanding of plant-pathogen interaction, detecting early stage disease symptoms and assessing disease severity (Mahlein et al., 2012; Römer et al., 2012; Mahlein, 2016; Lowe et al., 2017; Zhang et al., 2019). Given the cumbersome techniques of identifying disease infected plants and monitoring of plant disease symptoms both manually and visually that may delay preventing disease progression, early detection of disease symptoms through various sophisticated imaging techniques could assist in taking early preventive measures for restricting disease progression and crop yield loss (Mahlein, 2016; Lowe et al., 2017; Ghosal et al., 2018). Among the various fluorescence-based imaging analysis techniques used, the chlorophyll fluorescence imaging technique estimating Fv/Fm remains a reliable phenotyping technique for monitoring plant-pathogen interaction and disease severity with greater precision (Berger et al., 2007; Bauriegel et al., 2011; Rousseau et al., 2013). Monitoring changes in leaf surface temperature of FW responsive genotypes through an infra-red system allowed identification of FW resistant and susceptible genotypes in pea (Rispail and Rubiales, 2015) and in Medicago truncatula (Rispail et al., 2015). Likewise, multi and hyperspectral imaging was used for early prediction of disease onset, differentiating healthy and diseased plants, quantifying disease infection and assessing disease severity in various plant species (for details see Lowe et al., 2017; Shakoor et al., 2017). Similarly, high resolution thermal and hyperspectral imaging approaches were used to detect early wilt of olive caused by Verticillium dahliae in a large acreage (Calderón et al., 2015). However, disease prediction accuracy completely depends on the curation and interpretation of acquired hyperspectral imaging data (Singh et al., 2018). Therefore, to improve prediction accuracy of various plant diseases, currently machine learning and deep learning (convolutional neural network and artificial neural networks) approaches have been combined with the hyperspectral imaging data (Mohanty et al., 2016; Sladojevic et al., 2016; Ghosal et al., 2018; Singh et al., 2018). Further interpretation of these images could allow us to identify disease with higher precision and accuracy and also assist in proper assessment of disease severity. Thus, in the era of “Crop breeding 4.0 driven by the big data,” HTP will be an integral component of genotype, phenotype, and environment-based decision-making models (Wallace et al., 2018; Jiang et al., 2019).
Grain legume cultivars with an enhanced level of FW resistance have been bred using conventional breeding approaches for many decades. However, this approach is time consuming, and demands (i) sufficient genetic variation in the breeding material and (ii) greater manpower for hybridization and handling of segregating population (Yin and Qiu, 2019). A continuous supply of FW-resistant varieties in response to evolving FW pathogens under changing climate demands adoption of efficient breeding technologies (Meuwissen et al., 2001; Gao, 2018; Ghosh et al., 2018).
Explosion of SNP data thanks to the high throughput genotyping platform has offered a great opportunity to adopt genomic selection (GS) in crop plants. GS is used to predict the genomic estimated breeding value of untested individuals using genome-wide marker data. The genotypic and phenotypic information of “training population” is used to train prediction models (Meuwissen et al., 2001; Jannink et al., 2010; Desta and Ortiz, 2014). This approach has been employed for developing disease resistant genotypes in wheat and its scope for breeding disease resistant genotypes has been discussed elsewhere (Poland and Rutkoski, 2016). Genome-wide predictions are yet to be employed for disease resistance in grain legumes.
Speed breeding (SB) or rapid generation advancement presents another promising means to reduce the crop generation time and accelerate the breeding program (Ghosh et al., 2018). This technology has allowed for the recovery of six generations per year in various crops including wheat, barley, chickpea, pea (Ghosh et al., 2018; Hickey et al., 2019). When combined with MAS, SB may dramatically accelerate the screening of the breeding populations against disease or targeted introgression of loci controlling resistance to susceptible genotypes.
To circumvent the cumbersome process of trait manipulation in plants, CRISPR/Cas9 based genome editing technology is revolutionizing plant biology and breeding by precise modification of target gene sequence using customized nucleases (Voytas, 2013; Sander and Joung, 2014; Wang et al., 2014; Langner et al., 2018). This technique has been successfully employed to improve plant resistance against various bacterial and fungal diseases (Li T. et al., 2012; Wang et al., 2014, 2016; Chandrasekaran et al., 2016; Nekrasov et al., 2017; Peng et al., 2017; Zhang et al., 2017; Langner et al., 2018). In future this technique could be harnessed for improving FW resistance in grain legumes. An integrated approach involving various omics technologies and novel breeding schemes for future designing of FW resistant grain legumes has been depicted in Figure 2.
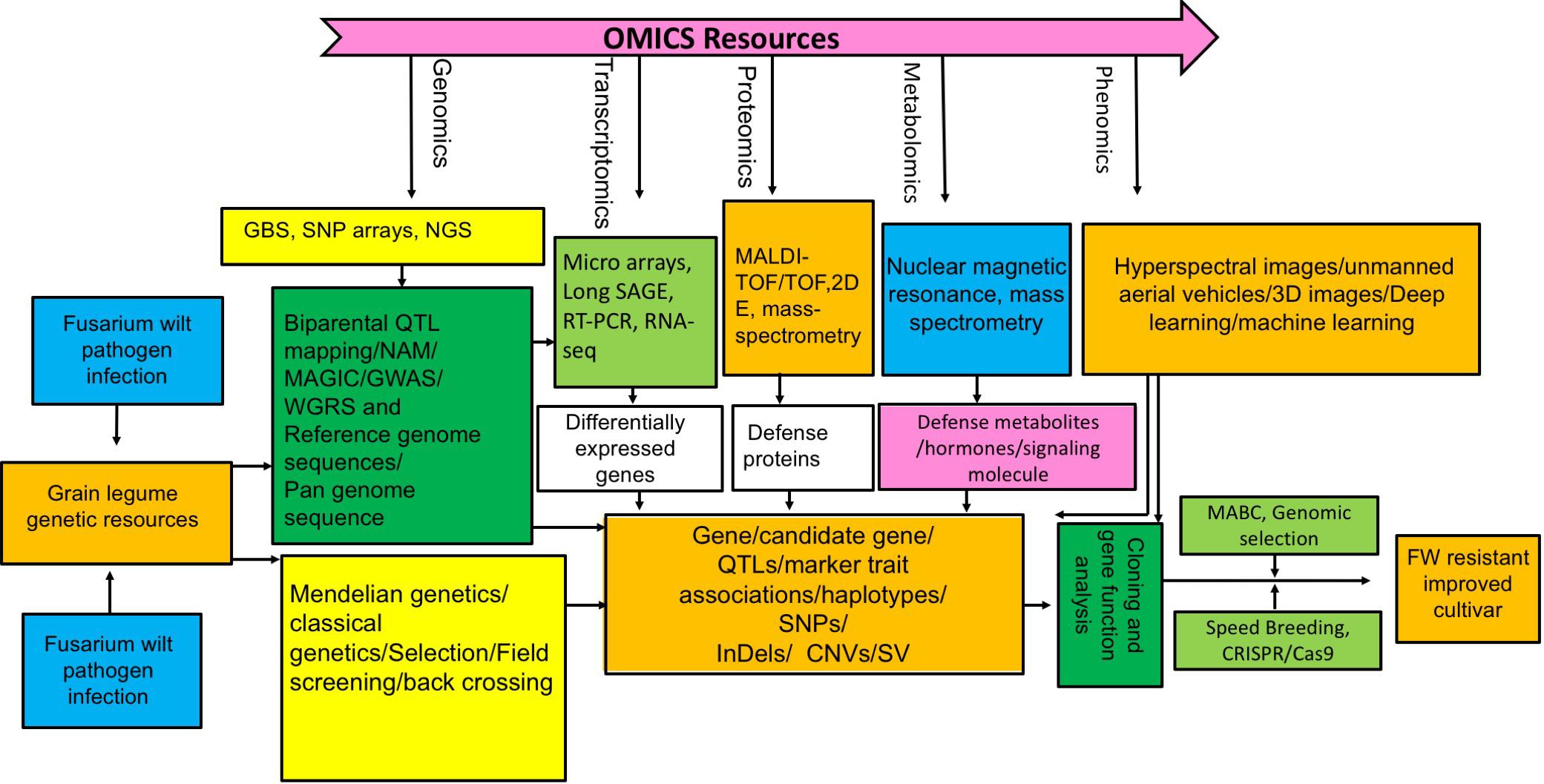
Figure 2. Integrated breeding, genetics, and “omics” scheme illustrating how to combat FW resistance in grain legume.
Severity and frequency of disease occurrence has seen a considerable rise in the wake of changing global climate, thus jeopardizing grain legume production worldwide. Breeding for FW resistance is a key breeding objective of crop improvement programs in grain legumes. Sourcing novel variations of FW resistance from unexploited CWRs and landraces needs greater attention to strengthen the genetic base. In parallel, pyramiding of different resistant gene(s) by adopting both standard backcrossing and DNA marker-aided approaches could expedite breeding of resistant cultivars. Advances in genomic technologies along with increasing genome sequence information could deepen knowledge about the resistant candidate genes/haplotypes to better breed FW-resistant grain legumes. Likewise, functional genomics could allow discovery of candidate loci, their biological functions and the molecular mechanisms underlying host-pathogen interactions. Importantly, emerging HTP phenotyping could illuminate the spatio-temporal aspects of host-pathogen interaction. Targeted and rapid manipulation of genomic loci responsible for FW resistance in grain legumes could be achieved with adoption of newer techniques like GS, SB, CRISPR/Cas9. An efficient combination of these new approaches paves the way for a steady stream of resistant legume cultivars that yield higher in increasing disease scenarios.
UCJ conceived the idea and wrote the manuscript with AB. UCJ developed the draft of the manuscript. AB and SKP contributed sections on omics technologies. SP contributed section on the FW infection and their effects on major grain legumes. AB edited the manuscript. All authors have read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors acknowledge support from Indian Council of Agricultural Research (ICAR), India.
Abawi, C. S., and Pastor-Corrales, M. A. (1990). Root Rots of Bean in Latin American and Africa: Diagnosis, Research Methodologies, and Management Strategies. Cali, CO: Centro Internacional de Agricultura Tropical (CIAT).
Abdel Rehim, M. A. (1962). Studies on the Organisms Causing Root Rot and Wilt of Horse Beans (Vicia faba var. equina) in U. A. R. Ph.D. thesis, Alexandria University, Alexandria.
Abdul Wahid, O. A., Ibrahim, M. E., and Omar, M. A. (1998). Oc- currence of soil suppressiveness to Fusarium wilt disease of broad bean in Ismailia Governorate. J. Phytopathol. 146, 431–435. doi: 10.1111/j.1439-0434.1998.tb04777.x
Agrawal, S. C., Singh, K., and Lal, S. S. (1991). “Plant protection of lentil in India,” in Lentil in South Asia ICAR-ICARDA Seminar, New Delhi, 147–167.
Alabouvette, C., Lemanceau, P., and Steinberg, C. (1993). Recent advances in the biological control of Fusarium wilts. Pestic. Sci. 37, 365–373. doi: 10.1002/ps.2780370409
Ali, H., ul Haq, M. A., Shah, T. M., Rahman, M., and Chen, W. (2012). Validation of molecular markers for resistance among Pakistani chickpea germplasm to races of Fusarium oxysporum f. sp. Ciceris. Eur. J. Plant Pathol. 132, 237–244. doi: 10.1007/s10658-011-9868-1
Ali, M. E. K., Inanga, S., and Sugimoto, Y. (2002). Soursce of resistance to Fusarium wilt of chickpea in Sudan. Phytopathol. Mediterr. 41, 163–169.
Alves-Santos, F. M., Benito, E. P., Eslava, A. P., and Diaz-Minguez, J. M. (1999). Genetic diversity of Fusarium oxysporum strains from common bean fields in Spain. Appl. Environ. Microbiol. 65, 3335–3340. doi: 10.1128/aem.65.8.3335-3340.1999
Araus, J. L., and Cairns, J. E. (2014). Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61. doi: 10.1016/j.tplants.2013.09.008
Armstrong, G. M., and Armstrong, J. K. (1980). Cowpea wilt Fusarium oxysporum f. sp. tracheiphilum race I from Nigeria. Plant Dis. 64, 954–955. doi: 10.1094/PD-64-954
Armstrong, G. M., and Armstrong, J. K. (1981). “Formae speciales and races of Fusarium oxysporum causing wilt disease,” in Fusarium: Diseases, Biology and Taxonomy, eds P. Nelson, T. Toussoun, and R. Cook (University Park, PA: Pennsylvania State University Press), 391–399.
Ashraf, N., Basu, S., Narula, K., Ghosh, S., Tayal, R., and Gangisetty, N. (2018). Integrative network analyses of wilt transcriptome in chickpea reveal genotype dependent regulatory hubs in immunity and susceptibility. Sci. Rep. 8:6528. doi: 10.1038/s41598-018-19919-5
Ashraf, N., Ghai, D., Barman, P., Basu, S., Gangisetty, N., Mandal, M. K., et al. (2009). Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genom. 10:415. doi: 10.1186/1471-2164-10-415
Assunção, I. P., Michereff, S. J., Mizubuti, E. S. G., and Brommonschenkel, S. H. (2003). Influence of Fusarium wilt intensity on cowpea yield. Fitopatol. Bras. 28, 615–619. doi: 10.1590/S0100-41582003000600006
Baayen, R. P., O’Donnell, K., Bonants, P. J., Cigelnik, E., Kroon, L. P., and Roebroeck, E. J. (2000). Gene genealogies and AFLP Analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 90, 891–900. doi: 10.1094/phyto.2000.90.8.891
Badet, T., and Croll, D. (2020). The rise and fall of genes: origins and functions of plant pathogen pangenomes. Curr. Opin. Plant Biol. 56, 65–73. doi: 10.1016/j.pbi.2020.04.009
Bani, M., Cimmino, A., Evidente, A., Rubiales, D., and Rispai, N. (2018a). Pisatin involvement in the variation of inhibition of Fusarium oxysporum f. sp. pisi spore germination by root exudates of Pisum spp. germplasm. Plant Pathol. 67, 1046–1054. doi: 10.1111/ppa.12813
Bani, M., Peìrez-De-Luque, A., Rubiales, D., and Rispail, N. (2018b). Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp. pisi in Pea. Front. Plant Sci. 9:199. doi: 10.3389/fpls.2018.00199
Bani, M., Rispail, N., Evidente, A., Rubiales, D., and Cimmino, A. (2014). Identification of the main toxins isolated from Fusarium oxysporum f. sp. pisi race 2 and their relation with isolates’ pathogenicity. J. Agric. Food Chem. 62, 2574–2580. doi: 10.1021/jf405530g
Bani, M., Rubiales, D., and Rispail, N. (2012). A detailed evaluation method to identify sources of quantitative resistance to Fusarium oxysporum f. sp. pisi race 2 within a Pisum spp. germplasm collection. Plant Pathol. 61, 532–542. doi: 10.1111/j.1365-3059.2011.02537.x
Batista, R. O., Oliveira, A. M. C., Silva, J. L. O., Nicoli, A., Carneiro, P. C. S., de Sousa Carneiro, J. E., et al. (2016). Resistance to Fusarium wilt in common bean. Crop Breed. Appl. Biotechnol. 16, 226–233. doi: 10.1590/1984-70332016v16n3a34
Batista, R. O., Silva, L. C., Moura, L. M., Souza, M. H., Carneiro, P. C. S., and Filho, J. L. S. (2017). Inheritance of resistance to Fusarium wilt in common bean. Euphytica 213:133. doi: 10.1007/s10681-017-1925-1
Bauriegel, E., Giebel, A., and Herppich, W. B. (2011). Hyperspectral and chlorophyll fluorescence imaging to analyse the impact of Fusarium culmorum on the photosynthetic integrity of infected wheat ears. Sensors 11, 3765–3779. doi: 10.3390/s110403765
Bayaa, B., Erskine, W., and Hamdi, A. (1995). Evaluation of a wild lentil collection for resistance to vascular wilt. Genet. Resour. Crop Evol. 42, 231–235. doi: 10.1007/bf02431257
Bayaa, B., Kumari, S. G., Akkaya, A., and Erskine, W. (1998). Survey of major biotic stresses of lentil in South-East Antalia. Turkey. Phytopathol. Mediterr. 37, 88–95.
Beck, M., Heard, W., Mbengue, M., and Robatzek, S. (2012). The INs and OUTs of pattern recognition receptors at the cell surface. Curr. Opin. Plant Biol. 15, 1–8. doi: 10.1016/j.pbi.2012.05.004
Belabid, L., Baum, M., Fortas, Z., Bouznad, Z., and Eujayl, I. (2004). Pathogenic and genetic characterisation of Algerian isolates of Fusarium oxysporum f. sp. lentis by RAPD and AFLP analysis. Afr. J. Biotechnol. 3, 25–31. doi: 10.5897/ajb2004.000-2005
Berger, S., Benediktyová, Z., Matouš, K., Bonfig, K. B., Mueller, M. J., Nedbal, L., et al. (2007). Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana. J. Expt. Bot. 58, 797–806. doi: 10.1093/jxb/erl208
Berrocal-Lobo, M., and Molina, A. (2008). Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci. 13, 145–150. doi: 10.1016/j.tplants.2007.12.004
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Bohra, A., Jha, R., Pandey, G., Patil, P. G., Saxena, R. K., Singh, I. P., et al. (2017). New hypervariable SSR markers for diversity analysis, hybrid purity testing and trait mapping in pigeonpea [Cajanus cajan (L.) Millspaugh]. Front. Plant Sci. 8:377. doi: 10.3389/fpls.2017.00377
Bohra, A., Jha, U. C., Kavi Kishor, P. B., Pandey, S., and Singh, N. P. (2014a). Genomics and molecular breeding in lesser explored pulse crops: current trends and future opportunities. Biotechnol. Adv. 32, 1410–1428. doi: 10.1016/j.biotechadv.2014.09.001
Bohra, A., Pandey, M. K., Jha, U. C., Singh, B., Singh, I. P., Datta, D., et al. (2014b). Genomics-assisted breeding in the four major pulse crops of developing countries: present status and prospects. Theor. Appl. Genet. 127, 1263–1291. doi: 10.1007/s00122-014-2301-3
Bohra, A., Sahrawat, K. L., Kumar, S., Joshi, R., Parihar, A. K., Singh, U., et al. (2015). Genetics- and genomics-based interventions for nutritional enhancement of grain legume crops: status and outlook. J. Appl. Genet. 56, 151–161. doi: 10.1007/s13353-014-0268-z
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., and He, P. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–423. doi: 10.1038/nature08794
Buddenhagen, I. W., Workneh, F., and Bosque-Pe rez, N. (1988). Chickpea improvement and chickpea diseases in California. Intern. Chickpea Newslett. 19, 9–10.
Buruchara, R. A., and Camacho, L. (2000). Common bean reaction to Fusarium oxysporum f. sp. phaseoli, the cause of severe vascular wilt in Central Africa. J. Phytopathol. 148, 39–45. doi: 10.1111/j.1439-0434.2000.tb04622.x
Caballo, C., Castro, P., Gil, J., Millan, T., Rubio, J., and Die, J. V. (2019a). Candidate genes expression profiling during wilting in chickpea caused by Fusarium oxysporum f. sp. ciceris race 5. PLoS One 14:e0224212. doi: 10.1371/journal.pone.0224212
Caballo, C., Madrid, E., Gil, J., Chen, W., Rubio, J., and Millan, T. (2019b). Saturation of genomic region implicated in resis- tance to Fusarium oxysporum f. sp. ciceris race 5 in chickpea. Mol. Breed. 39:16. doi: 10.1007/s11032-019-0932-4
Calderón, R., Navas-Cortés, J. A., and Zarco-Tejada, P. J. (2015). Early detection and quantification of Verticillium wilt in olive using hyperspectral and thermal imagery over large areas. Remote Sens. 7, 5584–5610. doi: 10.3390/rs70505584
Calero-Nieto, F., Di Pietro, A., Roncero, M. I., and Hera, C. (2007). Role of the transcriptional activator xlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant Microbe Interact. 20, 977–985. doi: 10.1094/mpmi-20-8-0977
Cañero, D. C., and Roncero, M. I. (2008). Influence of the chloride channel of Fusarium oxysporum on extracellular laccase activity and virulence on tomato plants. Microbiology. 154, 1474–1481. doi: 10.1099/mic.0.2007/015388-0
Castillejo, M. A., Curto, M., Fondevilla, S., Rubiales, D., and Jorriìn, J. V. (2010). Two- dimensional electrophoresis based proteomic analysis of the pea (Pisum sativum) in response to Mycosphaerella pinodes. J. Agric. Food Chem 58, 12822–12832. doi: 10.1021/jf1036917
Castillejo, M. Á, Bani, M., and Rubiales, D. (2015). Understanding pea resistance mechanisms in response to Fusarium oxysporum through proteomic analysis. Phytochemistry 115, 44–58. doi: 10.1016/j.phytochem.2015.01.009
Castillo, P., Navas-Cortés, J. A., Gomar-Tinoco, D., Di Vito, M., and Jiménez-Díaz, R. M. (2003). Interactions between Meloidogyne artiellia, the cereal and legume root-knot nematode, and Fusarium oxysporum f. sp. ciceris race 5 in chickpea. Phytopathology 93, 1513–1523. doi: 10.1094/phyto.2003.93.12.1513
Castro, P., Pistón, F., Madrid, E., Milla‘n, T., Gil, J., and Rubio, J. (2010). Development of chickpea near-isogenic lines for Fusarium wilt. Theor. Appl. Genet. 121, 1519–1526. doi: 10.1007/s00122-010-1407-5
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., and Pearlsman, M. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chaudhary, R. G., Saxena, D. R., Dhar, V., Singh, R. K., and Namdev, J. K. (2010). Prevalence of wilt-root rot and their associated pathogens at reproductive phase in lentil. Arch. Phytopathol. Plant Protect. 43, 996–1000. doi: 10.1080/03235400802214851
Chen, L., Wu, Q., He, W., He, T., Wu, Q., and Miao, Y. (2019). Combined De Novo transcriptome and metabolome analysis of common bean response to fusarium oxysporum f. sp. phaseoli infection. Int. J. Mol. Sci. 20:6278. doi: 10.3390/ijms20246278
Cobos, M., Fernandez, M., Rubio, J., Kharrat, M., Moreno, M., Gil, J., et al. (2005). A linkage map of chickpea (Cicer arietinum L.) based on populations from Kabuli × Desi crosses: location of genes for resistance to Fusarium wilt race 0. Theor. Appl. Genet. 110, 1347–1353. doi: 10.1007/s00122-005-1980-1
Cobos, M., Winter, P., Kharrat, M., Cubero, J., Gil, J., Millan, T., et al. (2009). Genetic analysis of agronomic traits in a wide cross of chickpea. Field Crops Res. 111, 130–145. doi: 10.1016/j.fcr.2008.11.006
Correll, J. C. (1991). The relationship between formae speciales, races, and vegetative compatibility groups in Fusarium oxysporum. Phytopathology 81, 1061–1064.
Coyne, C. J., Inglis, D. A., Whitehead, S. J., McClendon, M. T., and Muehlbauer, F. J. (2000). Chromosomal location of Fwf, the Fusarium wilt race 5 resistance gene in Pisum sativum. Pisum. Genet. 32, 20–22.
Cross, H., Brick, M. A., Schwartz, H. F., Panella, L. W., and Byrne, P. F. (2000). Inheritance of resistance to Fusarium wilt in two common bean races. Crop Sci. 40, 954–958. doi: 10.2135/cropsci2000.404954x
Davis, R. M., and Frate, C. A. (2007). UC IPM Pest Management Guide- Lines: Dry Beans Fusarium Wilt (Blackeyes/Cowpeas). Davis, CA: UC ANR Publication, 3446.
De Ascensao, A. R., and Dubery, I. A. (2000). Panama disease: Cell wall reinforcement in banana roots in response to elicitors from Fusarium oxysporum f. sp. cubense race four. Phytopathology 90, 1173–1180. doi: 10.1094/phyto.2000.90.10.1173
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., and Spanu, P. D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Desta, Z. A., and Ortiz, R. (2014). Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 19, 592–601. doi: 10.1016/j.tplants.2014.05.006
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Doling, D. A. (1964). Glasshouse testing of pea varieties for resistance to Fusarium Wilt. Plant Pathol. 13, 65–68. doi: 10.1111/j.1365-3059.1964.tb00265.x
Dong, Y., Dong, K., Zheng, Y., Tang, L., and Yang, Z. X. (2014). Faba bean Fusarium wilt (Fusarium oxysporum) control and its mechanism in different wheat varieties and faba bean in- tercropping system. J. Appl. Ecol. 25, 1979–1987.
Dubey, S. C., Priyanka, K., Singh, V., and Singh, B. (2012). Race Profiling and Molecular Diversity Analysis of Fusarium oxysporum f.sp. ciceris Causing Wilt in Chickpea. J. Phytopathol. 160, 576–587. doi: 10.1111/j.1439-0434.2012.01954.x
Dubey, S. C., Singh, B., and srinivasa, N. (2017). Evaluation of chickpea genotypes against Fusarium wilt for resistant sources. Indian Phytopath 70, 254–255. doi: 10.24838/ip.2017.v70.i2.70612
Ehlers, J. D., Hall, A. E., Patel, P. N., Roberts, P. A., and Matthews, W. C. (2000). Registration of ‘California Blackeye 27’ cowpea. Crop Sci. 40, 854–855.
Ehlers, J. D., Sanden, B. L., Frate, C. A., Hall, A. E., and Roberts, P. A. (2009). Registration of ‘California Blackeye 509cowpea. J. Plant Registrat. 3, 236–240. doi: 10.3198/jpr2009.01.0039crc
Elena, K., and Papas, A. C. (2002). Pathogenicity and vegetative compatibility of Fusarium oxysporum f.sp. phaseoli in Greece. J. Phytopathol. 150, 495–499. doi: 10.1046/j.1439-0434.2002.00786.x
Epstein, L., Kaur, S., Chang, P. L., Carrasquilla-Garcia, N., Lyu, G., Cook, D. R., et al. (2017). Races of the celery pathogen Fusarium oxysporum f. sp. apii are polyphyletic. Phytopathology 107, 463–473. doi: 10.1094/phyto-04-16-0174-r
Erskine, W., and Bayaa, B. (1996). Yield loss, incidence and inoculum density associated with vascular wilt of lentil. Phytopathol. Mediterr. 36, 24–32. doi: 10.2135/cropsci1996.0011183X0036000400055x
Erskine, W., Bayaa, B., and Dholli, M. (1990). The transmissibility of Fusarium oxysporum f. sp. lentis via seeds and the effect of some biotic and abiotic factors on its growth. Arab. J. Plant Protect. 8, 34–37. doi: 10.1111/j.1574-6941.2009.00777.x
Eujayl, I., Erskine, W., Bayaa, B., Baum, M., and Pehu, E. (1998). Fusarium vascular wilt in lentil: inheritance and identification of DNA markers for resistance. Plant Breed. 117, 497–499. doi: 10.1111/j.1439-0523.1998.tb01982.x
Eujayl, I., Erskine, W., Bayaa, B., Baum, M., and Pehu, E. (2006). Fusarium vascular wilt in lentil: inheritance and identification of DNA markers for resistance. Plant Breed. 117, 497–499. doi: 10.1111/j.1439-0523.1998.tb01982.x
Fall, A. L., Byrne, P. F., Jung, G., and Coyne, D. P. (2001). Detection and mapping of a major locus for Fusarium wilt resistance in common bean. Crop Sci. 41, 1494–1498. doi: 10.2135/cropsci2001.4151494x
Fiorani, F., and Schurr, U. (2013). Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 64, 267–291. doi: 10.1146/annurev-arplant-050312-120137
Fourie, G., Steenkamp, E. T., Ploetz, R. C., Gordon, T. R., and Viljoen, A. (2011). Current status of the taxonomic position of Fusarium oxysporum formae specialis cubense within the Fusarium oxysporum complex. Infec. Genet. Evol. 11, 533–542. doi: 10.1016/j.meegid.2011.01.012
Gabe, H. L. (1975). Standardization of nomenclature for pathogenic races of Fusarium oxysporum f. sp. lycopersici. Trans. Br. Mycol. Soc. 64, 156–159. doi: 10.1016/s0007-1536(75)80089-1
Gao, C. (2018). The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 39, 1–2. doi: 10.1007/978-1-0716-0616-2_1
Garces-Fiallos, F. R., de Borba, M. C., Schmidt, E. C., Bouzon, Z. L., and Stadnik, M. J. (2017). Delayed upward colonization of xylem vessels is associated with resistance of common bean to Fusarium oxysporum f. Sp phaseoli. Eur. J. Plant Pathol. 149, 477–489. doi: 10.1007/s10658-017-1197-6
Garcia-Limones, C., Dorado, G., Navas-Cortes, J. A., Jimenez-Diaz, R. M., and Tena, M. (2009). Changes in the redox status of chickpea roots in response to infection by Fusarium oxysporum f. sp. ciceris: apoplastic antioxidant enzyme activities and expression of oxi- dative stress-related genes. Plant Biol. 11, 194–203. doi: 10.1111/j.1438-8677.2008.00095.x
Garcia-Limones, C., Hervás, A., Navas-Cortés, J. A., Jiménez-Díaz, R. M., and Tena, M. (2002). Induction of an antioxidant enzyme system and other oxidative stress markers associated with compatible and incompatible interactions between chickpea (Cicer arietinum L.) and Fusarium oxysporum f. sp. ciceris. Physiol. Mol. Plant Pathol. 61, 325–337. doi: 10.1006/pmpp.2003.0445
Garg, T., Mallikarjuna, B. P., Thudi, M., Samineni, S., Singh, S., Sandhu, J. S., et al. (2018). Identification of QTLs for resistance to Fusarium wilt and Ascochyta blight in a recombinant inbred population of chickpea (Cicer arietinum L.). Euphytica 214:45. doi: 10.1007/s10681-018-2125-3
Gaur, P. M., Pande, S., Upadhyaya, H. D., and Rao, B. V. (2006). Extra-large Kabuli chickpea with high resistance to Fusarium wilt. SAT eJ 2, 1–2. doi: 10.9734/cjast/2019/v38i630409
Gerlagh, M., and Blok, W. J. (1988). Fusarium oxysporum f. sp. cucurbitacearum n.f. embracing all formae speciales of F. oxysporum attacking cucurbitaceous crops. Netherlands J Plant Pathol. 94, 17–31. doi: 10.1007/bf01999804
Ghosal, S., Blystone, D., Singh, A. K., Ganapathysubramanian, B., Singh, A., and Sarkar, S. (2018). An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci U.S.A. 115, 4613–4618. doi: 10.1073/pnas.1716999115
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., and Mendoza-Suárez, M. (2018). Speed breeding in growth cham- bers and glasshouses for crop breeding and model plant research. Nat. Protoc. 13, 2944–2963. doi: 10.1038/s41596-018-0072-z
Gilardi, G., Franco Ortega, S., Van Rijswick, P. C. J., Ortu, G., Gullino, M. L., and Garibaldi, A. (2017). A new race of Fusarium oxysporum f. sp. lactucae of lettuce. Plant Pathol. 66, 677–688. doi: 10.1111/ppa.12616
Gowda, S. J. M., Radhika, P., Kadoo, N., Mhase, L., and Gupta, V. (2009). Molecular mapping of wilt resistance genes in chickpea. Mol. Breed. 24, 177–184. doi: 10.1007/s11032-009-9282-y
Graham, P. H., and Vance, C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/pp.017004
Grajal-Martin, M. J., and Muehlbauer, F. J. (2002). Genomic location of the Fw gene for resistance to Fusarium wilt race 1 in peas. J. Hered. 93, 291–293. doi: 10.1093/jhered/93.4.291
Gunnaiah, R., Kushalappa, A. C., Duggavathi, R., Fox, S., and Somers, D. J. (2012). Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS One 7:e40695. doi: 10.1371/journal.pone.0040695
Gupta, S., Bhar, A., Chatterjee, M., and Das, S. (2013a). Fusarium oxysporum f. sp ciceri race 1 induced redox state alterations are coupled to downstream defense signaling in root tissues of chickpea (Cicer arietinum L.). PLoS One 8:e73163. doi: 10.1371/journal.pone.0073163
Gupta, S., Bhar, A., and Das, S. (2013b). Understanding the molecular defense responses of host during chickpea-Fusarium interplay: where do we stand? Funct. Plant Biol. 40, 1285–1297. doi: 10.1071/fp13063
Gupta, S., Chakraborti, D., Rangi, R. K., Basu, D., and Das, S. (2009). A molecular insight into the early events of chickpea (Cicer arietinum) and Fusarium oxysporum f. sp. ciceri (Race 1) interaction through cDNA-AFLP analysis. Phytopathology 99, 1245–1257. doi: 10.1094/phyto-99-11-1245
Gupta, S., Chakraborti, D., Sengupta, A., Basu, D., and Das, S. (2010). Primary metabolism of chickpea is the initial target of wound inducing early sensed Fusarium oxysporum f sp ciceri race 1. PLos One 5:e9030. doi: 10.1371/journal.pone.0009030
Gurjar, G., Barve, M., Giri, A., and Gupta, V. (2009). Identification of Indian pathogenic races of Fusarium oxysporum f. sp. ciceris with gene specific. ITS and random markers. Mycologia 101, 484–495. doi: 10.3852/08-085
Gurjar, G., Giri, A. P., and Gupta, V. S. (2012). Gene expression profiling during wilting in chickpea caused by Fusarium oxysporum f. sp. ciceri. Am. J. Plant Sci. 3, 190–201. doi: 10.4236/ajps.2012.32023
Haglund, W. A., and Kraft, J. M. (2001). “Fusarium wilt,” in Compendium of Pea Diseases, 2nd Edn, eds J. M. Kraft and F. L. Peger, (St. Paul, MN: APS Press), 14–16.
Halila, I., Cobos, M. J., Rubio, J., Millaìn, T., Kharrat, M., Marrakchi, M., et al. (2009). Tagging and mapping a second resistance gene for fusarium wilt race 0 in chickpea. Eur. J. Plant Pathol 124, 87–92. doi: 10.1007/s10658-008-9395-x
Halila, M. H., and Harrabi, M. M. (1990). Breeding for dual resistance to Ascochyta and wilt diseases in chickpea. Options Mediterr. Ser. Semin. 9, 163–170.
Halila, M. H., and Strange, R. N. (1996). Identification of the casual agent of wilt of chickpea in Tunisia as Fusarium oxysporum f. sp. ciceri race 0. Phytopathol. Mediterr. 35, 67–74.
Hamwieh, A., Udupa, S. M., Choumane, W., Sarker, A., Dreyer, F., Jung, C., et al. (2005). A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of fusarium vascular wilt resistance. Theor. Appl. Genet. 110, 669–677. doi: 10.1007/s00122-004-1892-5
Hare, W. W., Walker, J. C., and Delwiche, E. J. (1949). Inheritance of a gene for near-wilt resistance in the garden pea. J. Agric. Res. 78, 239–251.
Haware, M. P., Jimenez-Diaz, R. M., Amin, K. S., Phillips, J., and Halila, H. (1990). “Integrated management of wilt and root rots of chickpea,” in Chickpea in the Nineties: Proceedings of the Second International Work Shop on Chickpea Improvement, Patancheru, 129–137.
Haware, M. P., and Nene, Y. L. (1982). Races of Fusarium oxysporum f. sp. ciceris. Plant Dis. 66, 809–810. doi: 10.1094/PD-66-809
Haware, M. P., Nene, Y. L., and Natrajan, M. (1986). “Survival of Fusarium oxysporum f. sp. ciceri in soil in the absence of chickpea,” in National Seminar on Management of Soil-Borne Diseases of Crop Plants (abstract), Coimbatore, 1.
Haware, M. P., Nene, Y. L., Pundir, R. P. S., and Rao, J. N. (1992). Screening of world chickpea germplasm for resistance to Fusarium wilt. Field Crops Res. 30, 147–154. doi: 10.1016/0378-4290(92)90063-f
Haware, M. P., Nene, Y. L., and Rajeshwari, R. (1978). Eradication of Fusarium oxysporum f. sp. ciceri transmitted in chickpea seed. Phytopathology 68, 1364–1367. doi: 10.1094/phyto-68-1364
Helms, D., Pamella, L., Buddenhagen, I. W., Workneh, F., Tucker, C. L., Foster, K. W., et al. (1992). Registration of ‘UC 15’ chickpea. Crop Sci. 32:500. doi: 10.2135/cropsci1992.0011183X003200020051x
Hickey, L. T., Hafeez, A. N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotech. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hiremani, N., and Dubey, S. C. (2018). Race profiling of Fusarium oxysporum f. sp. lentis causing wilt in lentil. Crop Protect. 108, 23–30. doi: 10.1016/j.cropro.2018.02.010
Husaini, A. M., Sakina, A., and Cambay, S. R. (2018). Host–pathogen interaction in Fusarium oxysporum infections: where do we stand? Mol. Plant Microbe Int. 31, 889–898. doi: 10.1094/mpmi-12-17-0302-cr
Infantino, A., Kharrat, M., Riccioni, L., Coyne, C. J., McPhee, K. E., and Grunwald, N. J. (2006). Screening techniques and sources of resistance to root diseases in legumes. Euphytica 147, 201–221. doi: 10.1007/s10681-006-6963-z
Inoue, I., Namiki, F., and Tsuge, T. (2002). Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14, 1869–1883. doi: 10.1105/tpc.002576
Jain, K. C., and Reddy, M. V. (1995). Inheritance of resistance to Fusarium wilt in pigeonpea (Cajanus cajan (L) Millsp.). Indian J. Genet. 55, 434–437.
Jain, S., Weeden, N. F., Kumar, A., Chittem, K., and McPhee, K. (2015). Functional codominant marker for selecting the Fw gene conferring resistance to Fusarium wilt race 1 in pea. Crop Sci. 55, 2639–2646. doi: 10.2135/cropsci2015.02.0102
Jannink, J. L., Lorenz, A. J., and Iwata, H. (2010). Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9, 166–177. doi: 10.1093/bfgp/elq001
Jendoubi, W., Bouhadida, M., Millan, T., Kharrat, M., Gil, J., and Rubio, J. (2016). Identification of the target region including the Foc0 1/foc0 1 gene and development of near isogenic lines for resistance to Fusarium Wilt race 0 in chickpea. Euphytica 210, 119–133. doi: 10.1007/s10681-016-1712-4
Jiang, S., Cheng, Q., Yan, J., Fu, R., and Wang, X. (2019). Genome optimization for improvement of maize breeding. Theor. Appl. Genet. 133, 1491–1502. doi: 10.1007/s00122-019-03493-z
Jimenez-Diaz, R. M., Alcala-Jimenez, A. R., Hervas, A., and Traperocasas, J. L. (1993). “Pathogenic variability and hosts resistance in the Fusarium oxysporum f. sp. ciceris/Cicer arietinum pathosys- tem,”in Proceedings of the 3rd European Seminar on Fusarium Mycotoxins, Taxonomy, Pathogenicity and Host Resistance. Hodowsla Roslin Aklimatyazacja i Nasiennictwo. Poland: Plant Breed- ing and Acclimatization Institute, 87–94.
Jiménez-Díaz, R. M., Castillo, P., Jimeìnez-Gasco, M. M., Landa, B. B., and Navas-Corteìs, J. A. (2015). Fusarium wilt of chickpeas: biology, ecology and management. Crop Prot. 73, 16–27. doi: 10.1016/j.cropro.2015.02.023
Jiménez-Gasco, M. M., and Jiménez-Díaz, R. M. (2003). Development of a specific polymerase chain reaction-based assay for the identification of Fusarium oxysporum f. sp. ciceris and its pathogenic races 0, 1A, 5, and 6. Phytopathology 93, 200–209. doi: 10.1094/phyto.2003.93.2.200
Jiménez-Gasco, M. M., Pérez-Artés, E., and Jiménez-Díaz, R. M. (2001). Identification of pathogenic races 0, 1B/C, 5, and 6 of Fusarium oxysporum f. sp. ciceris with random amplified polymorphic DNA (RAPD). Eur. J. Plant Pathol. 107, 237–248.
Jimeìnez-Diìaz, R. M., Singh, K. B., Trapero-Casas, A., and Trapero-Casas, J. L. (1991). Resistance in kabuli chickpeas to Fusarium wilt. Plant Dis. 75, 914–918. doi: 10.1094/pd-75-0914
Jingade, P., and Ravikumar, R. L. (2015). Development of molecular map and identification of QTLs linked to Fusarium wilt resistance in chickpea. J. Genet. 94, 723–729. doi: 10.1007/s12041-015-0589-7
Kaiser, W. J., Alcalá-Jiménez, A. R., Hervás-Vargas, A., Trapero-Casas, J. L., and Jiménez-Díaz, R. M. (1994). Screening wild species for resistance to races 0 and 5 of Fusarium oxysporum f. sp. ciceris. Plant Dis. 78, 962–967.
Kaiser, W. J., Ramsay, M. D., Makkouk, K. M., Bretag, T. W., Acikgoz, N., Kumar, J., et al. (2000). “Foliar diseases of cool season food legumes and their control,” in Linking Research and Marketing Opportunities for Pulses in the 21st Century, ed. R. Knight (Dordrecht: Kluwer Academic Publishers), 437–455. doi: 10.1007/978-94-011-4385-1_41
Kamboj, R. K., Pandey, M. P., and Chaube, H. S. (1990). Inheritance of resistance to Fusarium wilt in Indian lentil germplasm (Lens culinaris Medik.). Euphytica 50, 113–117. doi: 10.1007/bf00023633
Kannaiyan, J., and Nene, Y. L. (1976). Reaction of lentil germplasm and cultivars against three root pathogens. Indian J Agric. Sci. 46, 165–167.
Karimi, R., Owuoche, J. O., and Silim, S. N. (2010). Inheritance of fusar- ium wilt resistance in pigeonpea (Cajanus cajan (L.) Millspugh. Indian J. Genet. 70, 271–276.
Kelly, A. G., Alcala-Jimenez, A. R., Bainbridge, B. W., Heale, J. B., Perez- Artes, E., and Jimenez-Diaz, R. M. (1994). Use of genetic fingerprinting and random amplified polymorphic DNA to characterize pathotypes of Fusarium oxysporum f. sp. ciceris infecting chickpea. Phytopathology 84, 1293–1298. doi: 10.1094/phyto-84-1293
Kistler, H. C. (1997). Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology 87, 474–479. doi: 10.1094/phyto.1997.87.4.474
Klessig, D. F., Durner, J., Shah, J., and Yang, Y. (1998). Salicylic acid-mediated signal transduction in plant disease resistance. Phytochem. Signal Plant Microbe Interact. 32, 119–137. doi: 10.1007/978-1-4615-5329-8_7
Kohli, D., Joshi, G., Deokar, A. A., Bhardwaj, A. R., Agarwal, M., Katiyar- Agarwal, S., et al. (2014). Identi cation and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS One 9:e108851. doi: 10.1371/journal.pone.0108851
Kotresh, H., Fakrudin, B., Punnuri, S. M., Rajkumar, B. K., Thudi, M., Paramesh, H., et al. (2006). Identification of two RAPD markers genetically linked to a recessive allele of a Fusarium wilt resistance gene in pigeonpea (Cajanus cajan L. Millsp.). Euphytica 149, 113–120. doi: 10.1007/s10681-005-9059-2
Kraft, J. M. (1994). Fusarium wilt of peas a review. Agronomie 14, 561–567. doi: 10.1051/agro:19940901
Kraft, J. M., Burke, D. W., and Haglund, W. A. (1981). “Fusarium diseases of beans, peas and lentils,” in Fusarium: Diseases, Biology and Taxonomy, eds P. E. Nelson, T. A. Toussoun, and R. J. Cook (University Park, PA: Pennsylvania State Univ. Press,), 142–156.
Kraft, J. M., Larsen, R. C., and Inglis, D. A. (1998). “Diseases of pea,” in The Pathology of Food and Pasture Legumes, eds D. J. Allen and J. M. Lenneì (WALLINGFORD:. CAB International), 325–370.
Kreplak, J., Madoui, M. A., Capal, P., Novak, P., Labadie, K., Aubert, G., et al. (2019). A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 51, 1411–1422.
Kumar, J., Haware, M. P., and Smithson, J. B. (1985). Registration of four short duration Fusarium wilt-resistant kabuli (garbanzo) chickpea germplasms. Crop Sci. 25, 576–577. doi: 10.2135/cropsci1985.0011183x002500030047x
Kumar, S. (1998). Inheritance of resistance to Fusarium wilt (race 2) in chickpea. Plant Breed. 117, 139–142. doi: 10.1111/j.1439-0523.1998.tb01467.x
Kumar, S. (2006). Inheritance of resistance to Fusarium wilt (race 2) in chickpea. Plant Breed. 117, 139–142. doi: 10.1111/j.1439-0523.1998.tb01467.x
Kumar, Y., Dholakia, B. B., Panigrahi, P., Kadoo, N. Y., Giri, A. P., and Gupta, V. S. (2015). Metabolic profiling of chickpea-Fusarium interaction identifies differential modulation of disease resistance pathways. Phytochemistry 116, 120–129. doi: 10.1016/j.phytochem.2015.04.001
Kumar, Y., Zhang, L., Panigrahi, P., Dholakia, B. B., Dewangan, V., Chavan, S. G., et al. (2016). Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnol. J. 14, 1589–1603. doi: 10.1111/pbi.12522
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/s1369-5266(02)00275-3
Kwon, S. J., Smykal, P., Hu, J., Kim, S. J., McGee, R. J., McPhee, K., et al. (2013). User-friendly markers linked to Fusarium wilt race 1 resistance Fw gene for marker-assisted selection in pea. Plant Breed. 132, 642–648. doi: 10.1111/pbr.12085
Landa, B. B., Navas-Cortes, J. A., Jimenez-Gasco, M. M., Katan, R. B., and Jimenez- Diìaz, R. M. (2006). Temperature response of chickpea cultivars to races of Fusa- rium oxysporum f. sp. ciceris, the causal agent of Fusarium wilt. Plant Dis. 90, 365–374. doi: 10.1094/pd-90-0365
Langner, T., Kamoun, S., and Belhaj, K. (2018). CRISPR crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. doi: 10.1146/annurev-phyto-080417-050158
Lanubile, A., Muppirala, U. K., Severin, A. J., Marocco, A., and Munkvold, G. P. (2015). Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genomics 16:1089. doi: 10.1186/s12864-015-2318-2
Leitão, S. T., Malosetti, M., Song, Q., van Eeuwijk, F., Rubiales, D., and Vaz Patto, M. C. (2020). natural variation in portuguese common bean germplasm reveals new sources of resistance Against Fusarium oxysporum f. sp. phaseoli and Resistance-Associated Candidate Genes. Phytopathology 110, 633–647. doi: 10.1094/phyto-06-19-0207-r
Leslie, J. F., and Summerell, B. A. (2006). The Fusarium Laboratory Manual. Ames, IA: Blackwell Publishing, doi: 10.1002/9780470278376
Li, C. Y., Deng, G. M., Yang, J., Viljoen, A., Jin, Y., Kuang, R. B., et al. (2012). Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 13:374. doi: 10.1186/1471-2164-13-374
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, W., Deng, Y. W., Ning, Y. S., Hu, Z. H., and Wang, G. L. (2020). Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 16, 1–25. doi: 10.1007/s11032-009-9323-6
Lievens, B., Rep, M., and Thomma, B. P. H. J. (2008). Recent development in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manage. Sci. 64, 781–788.
Liu, S., Li, J., Zhang, Y., Liu, N., Viljoen, A., Mostert, D., et al. (2020). Fusaric acid instigates the invasion of banana by Fusarium oxysporum f. sp. cubense TR4. New Phytol. 225, 913–929. doi: 10.1111/nph.16193
Lonardi, S., Muñoz-Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S. I., Lo, S., et al. (2019). The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98, 767–782.
López-Díaz, C., Rahjoo, V., Sulyok, M., Ghionna, V., Logrieco, A. F., Franzoni, M., et al. (2017). Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 19, 440–453. doi: 10.1111/mpp.12536
Loridon, K., McPhee, K., Morin, J., Dubreuil, P., Pilet-Nayel, M. L., Aubert, G., et al. (2005). Microsatellite marker polymor- phism and mapping in pea (Pisum sativum L.). Theor. Appl. Genet. 11, 1022–1031. doi: 10.1007/s00122-005-0014-3
Lowe, A., Harrison, N., and French, A. P. (2017). Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 13:80.
Ma, K. W., and Ma, W. (2016). Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol. Biol. 91, 713–725. doi: 10.1007/s11103-016-0452-0
Mahlein, A.-K. (2016). Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 100, 241–251. doi: 10.1094/pdis-03-15-0340-fe
Mahlein, A. K., Oerke, E. C., Steiner, U., and Dehne, H. W. (2012). Recent advances in sensing plant diseases for precision crop protection. Eur. J. Plant Pathol. 133, 197–209. doi: 10.1007/s10658-011-9878-z
Mahmoud, A. F., and Abd El-Fatah, B. E. (2020). Genetic diversity studies and identification of molecular and biochemical markers associated with Fusarium wilt resistance in cultivated faba bean (Vicia faba). Plant Pathol. J. 36, 11–28.
Mannur, D. M., Babbar, A., Thudi, M., Sabbavarapu, M. M., Roorkiwal, M., Sharanabasappa, B. Y., et al. (2019). Super Annigeri 1 and improved JG 74: two Fusarium wilt-resistant introgression lines developed using marker-assisted backcrossing approach in chickpea (Cicer arietinum L.). Mol. Breed. 39:2.
Mayer, M., Tullu, A., Simon, C., Kumar, J., Kaiser, W., Kraft, J., et al. (1997). Development of a DNA marker for Fusarium wilt resistance in chickpea. Crop Sci. 37, 1625–1637. doi: 10.2135/cropsci1997.0011183x003700050036x
McClendon, M. T., Inglis, D. A., McPhee, K. E., and Coyne, C. J. (2002). DNA markers linked to Fusarium wilt race 1 resistance in pea. J. Am. Soc. Hortic. Sci. 127, 602–607. doi: 10.21273/jashs.127.4.602
McPhee, K. E., Inglis, D. A., Gundersen, B., and Coyne, C. J. (2012). Mapping QTL for Fusarium wilt race 2 partial resistance in pea (Pisum sativum). Plant Breed. 131, 300–306. doi: 10.1111/j.1439-0523.2011.01938.x
McPhee, K. E., Tullu, A., Kraft, J. M., and Muehlbauer, F. J. (1999). Resistance to Fusarium wilt race 2 in the Pisum core collection. J. Amer. Soc. Hort. Sci. 124, 28–31. doi: 10.21273/jashs.124.1.28
Mehta, A., Brasileiro, A. C. M., Souza, D. S. L., Romano, E., Campos, M. A., Grossi- De-Sa, M. F., et al. (2008). Plant-pathogen interactions: what is proteomics telling us? FEBS J. 275, 3731–3746. doi: 10.1111/j.1742-4658.2008.06528.x
Meuwissen, T. H. E., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
Michielse, C. B., van Wijk, R., Reijnen, L., Cornelissen, B. J. C., and Rep, M. (2009). Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol. 10:R4.
Mohammadi, N., Puralibaba, H., Goltapeh, E. M., Ahari, A. B., and Sardrood, B. P. (2012). Advanced lentil lines screened for resistance to Fusarium oxysporum f. sp. lentis under greenhouse and field conditions. Phytoparasitica 40, 69–76. doi: 10.1007/s12600-011-0201-5
Mohanty, S. P., Hughes, D. P., and Salathé, M. (2016). Using deep learning for image-based plant disease detection. Front. Plant Sci. 7:1419. doi: 10.3389/fpls.2016.01419
Morales, G. J. A. (1986). Chickpea breeding programme in Sonora. Mexico. Int. Chickpea Newsl. 15, 11–12.
Morkunas, I., Kozłowska, M., Ratajczak, L., and Marczak, Ł (2007). Role of sucrose in the development of Fusarium wilt in lupine embryo axes. Physiol. Mol. Plant Pathol. 70, 25–37. doi: 10.1016/j.pmpp.2007.05.004
Morkunas, I., Marczak, Ł, Stachowiak, J., and Stobiecki, M. (2005). Sucrose- stimulated accumulation of isoflavonoids as a defense response of lupine to Fusarium oxysporum. Plant Physiol. Biochem. 43, 363–373. doi: 10.1016/j.plaphy.2005.02.011
Morkunas, I., and Ratajczak, L. (2014). The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant. 36, 1607–1619. doi: 10.1007/s11738-014-1559-z
Muchero, W., Diop, N. N., Bhat, P. R., Fenton, R. D., Wanamaker, S., Pottorff, M., et al. (2009). A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc. Natl. Acad. Sci. U.S.A. 106, 18159–18164. doi: 10.1073/pnas.0905886106
Mudryj, A. N., Yu, N., and Aukema, H. M. (2014). Nutritional and health benefits of pulses. Appl. Physiol. Nutr. Metab. 39, 1197–1204.
Naimuddin, and Chaudhary, R. G. (2009). Pathogenic variability in isolates of Fusarium oxysporum f. sp. lentis. Trends Biosci. 2, 50–52.
Nakagami, H., Pitzschke, A., and Hirt, H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10, 339–346. doi: 10.1016/j.tplants.2005.05.009
Navas-Cortes, J. A., Hau, B., and Jimenez-Diìaz, R. M. (1998). Effect of sowing date, host cultivar, and race of Fusarium oxysporum f. sp. ciceris on development of Fusarium wilt of chickpea. Phytopathology 88, 1338–1346. doi: 10.1094/phyto.1998.88.12.1338
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7:482.
Nene, Y. L. (1980). Screening chickpea for reìsistance to wilt. Plant Dis. 64, 379–380. doi: 10.1094/pd-64-379
Nene, Y. L., and Haware, M. P. (1980). screening chickpea for resistance to Wilt. Plant Dis. 64, 379–380. doi: 10.1094/pd-64-379
Nene, Y. L., Haware, M. P., Reddy, N. M. V., Philps, J. P., Castro, E. L., Kotasthane, S. R., et al. (1989). Identification of broad based and stable resistance to wilt and root-rots in chickpea. Indian Phytopathol. 42, 499–505.
Neumann, S., and Xue, A. G. (2003). Reactions of field pea cultivars to four races of Fusarium oxysporum f. sp pisi. Can. J. Plant Sci. 83, 377–379. doi: 10.4141/p02-048
Nikraftar, F., Taheri, P., Rastegar, M. F., and Tarighi, S. (2013). Tomato partial resistance to Rhizoctonia solani involves antioxidative defense mechanisms. Physiol. Mol. Plant Pathol. 81, 74–83. doi: 10.1016/j.pmpp.2012.11.004
NinÞo-Saìnchez, J., Tello, V., Casado-del Castillo, V., Thon, M. R., Benito, E. P., and Diìaz-Miìnguez, J. M. (2015). Gene expression patterns and dynamics of the colonization of common bean (Phaseolus vulgaris L.) by highly virulent and weakly virulent strains of Fusarium oxysporum. Front. Microbiol 6:234. doi: 10.3389/fmicb.2015.00234
Nogales Moncada, A. N., Jimenez-Diìaz, R. M., and Perez-Artes, E. (2009). Vegetative compatibility groups in Fusarium oxysporum f. sp. ciceris and F. oxysporum nonpathogenic to chickpea. J. Phytopathol. 157, 729–735. doi: 10.1111/j.1439-0434.2009.01562.x
Odeny, D. A., Githiri, S. M., and Kimani, P. M. (2009). Inheritance of resistance to Fusarium wilt in pigeonpea. J. Anim. Plant Sci. 2, 89–95.
Okiror, M. A. (2002). Genetics of resistance to Fusarium udum in pigeonpea [Cajanus cajan (L.) Millsp.]. Ind. J. Genet. Plant Breed. 62, 218–220.
Okubara, P. A., Keller, K. E., Mcclendon, M. T., Inglis, D. A., Mcphee, K. E., and Coyne, C. J. (2005). Y15_999fw, a dominant scar marker linked to the Fusarium wilt race 1 (fw) resistance gene in pea. Pisum Genet. 37, 32–35.
Omoigui, L. O., Danmaigona, C. C., Kamara, A. Y., Ekefan, E. J., and Timko, M. P. (2018). Genetic analysis of Fusarium wilt resistance in cowpea (Vigna unguiculata Walp.). Plant Breed. 137, 773–781. doi: 10.1111/pbr.12628
Palomares-Rius, J. E., Castillo, P., Navas-Corteìs, J. A., Jimeìnez-Diìaz, R. M., and Tena, M. (2011). A proteomic study of in-root interactions between chickpea pathogens: the root-knot nematode Meloidogyne artiellia and the soil-borne fungus Fusarium oxysporum f. sp. ciceris race 5. J. Proteom. 74, 2034–2051. doi: 10.1016/j.jprot.2011.05.026
Pandey, R. N., Pawar, S. E., and Bhatia, C. R. (1996). Inheritance of wilt resistance in pigeonpea. Indian J. Genet. 56, 305–308.
Parihar, A. K., Basandrai, A. K., Saxena, D. R., Kushwaha, K. P. S., Chandra, S., Sharma, K., et al. (2017). Biplot evaluation of test environments and identification of lentil genotypes with durable resistance to Fusarium wilt in India. Crop Pasture Sci. 68, 1024–1030. doi: 10.1071/cp17258
Pastor-Corrales, M. A., and Abawi, G. S. (1987). Reactions of selected bean germ plasms to infection by Fusarium oxysporumf. sp. phase- oli. Plant Dis. 71, 990–993. doi: 10.1094/pd-71-0990
Patel, P. N. (1985). “Fungal, bacterial and viral diseases of cowpeas in the USA,” in Cowpea Research, Production and Utilization, eds S. R. Singh and K. O. Rachie (Chichester: John Wiley and Sons), 205–213.
Pathak, G. N. (1970). “Red gram,” in Pulse Crops of India, (New Delhi: Indian Council of Agricultural Research), 14–53.
Patil, B. S., Ravikumar, R. L., Bhat, J. S., and Soregaon, D. (2014). Molecular mapping of QTLs for resistance to early and late Fusarium wilt in chickpea. Czech. J. Genet. Plant Breed. 50, 171–176. doi: 10.17221/188/2013-cjgpb
Patil, P. G., Bohra, A., Dubey, J., Saabale, P. R., Mishra, R. K., Pandey, G., et al. (2017a). Genetic analysis and molecular resistance to race 2 of Fusarium wilt in pigeonpea [Cajanus cajan (L.) Millsp.]. Crop Protect. 100, 117–123. doi: 10.1016/j.cropro.2017.06.016
Patil, P. G., Dubey, J., Bohra, A., Mishra, R. K., Saabale, P. R., Das, A., et al. (2017b). Association mapping to discover significant marker-trait associations for resistance against Fusarium wilt variant 2 in pigeonpea [Cajanus cajan (L.) Millspaugh] using SSR markers. J. Appl. Genet. 58, 307–319. doi: 10.1007/s13353-017-0400-y
Pawar, N. B., and Mayee, C. D. (1986). Reaction of pigeonpea genotypes and their crosses to Fusarium wilt. Indian Phytopathol. 39, 70–74.
Pedley, K. F., and Martin, G. B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8, 541–547. doi: 10.1016/j.pbi.2005.07.006
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., and Wu, L. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLob1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Poland, J., and Rutkoski, J. (2016). Advances and challenges in genomic selection for disease resistance. Annu. Rev. Phytopathol. 54, 79–98. doi: 10.1146/annurev-phyto-080615-100056
Pottorff, M., Wanamaker, S., Ma, Y. Q., Ehlers, J. D., Roberts, P. A., and Close, T. J. (2012). Genetic and physical mapping of candidate genes for resistance to Fusarium oxysporum f. sp. tracheiphilum race 3 in cowpea [Vigna unguiculata (L.) Walp]. PLoS One 7:e41600. doi: 10.1371/journal.pone.0041600
Pottorff, M. O., Li, G., Ehlers, J. D., Close, T. J., and Roberts, P. A. (2014). Genetic mapping, synteny, and physical location of two loci for Fusarium oxysporum f. sp. tracheiphilum race 4 resistance in cowpea [Vignaunguiculata (L.) Walp]. Mol. Breed. 33, 779–791. doi: 10.1007/s11032-013-9991-0
Pouralibaba, H. R., Rubiales, D., and Fondevilla, S. (2015). Identification of resistance to Fusarium oxysporum f. sp. lentis in Spanish lentil germplasm. Eur. J. Plant Pathol. 143, 399–405. doi: 10.1007/s10658-015-0692-x
Pouralibaba, H. R., Rubiales, D., and Fondevilla, S. (2016). Identification of pathotypes in Fusarium oxysporum f. sp. lentis. Eur. J. Plant Pathol. 144, 539–549. doi: 10.1007/s10658-015-0793-6
Pratap, A., Chaturvedi, S. K., Tomar, R., Rajan, N., Malviya, N., Thudi, M., et al. (2017). Marker-assisted introgression of resistance to Fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol. Gen. Genomics 293, 1237–1245. doi: 10.1007/s00438-017-1343-z
Ratnaparkhe, M., Santra, D., Tullu, A., and Muehlbauer, F. (1998). Inheritance of inter-simple-sequence-repeat polymorphisms and linkage with a Fusarium wilt resistance gene in chickpea. Theor. Appl. Genet. 96, 348–352. doi: 10.1007/s001220050747
Reddy, M. V., Nene, Y. L., Raju, T. N., Kannaiyan, J., Remanandan, P., Mengesha, M. H., et al. (1995). Registration of pigeonpea germplasm line ICP 9145 resistant to Fusarium wilt. Crop Sci. 35:1231. doi: 10.2135/cropsci1995.0011183x003500040092x
Reddy, M. V., Sharma, S. B., and Nene, Y. L. (1990). “Pigeonpea: disease management,” in The Pigeonpea, eds Y. L. Nene, S. D. Hall, and V. K. Sheila (Wallingford: CAB International), 303–348.
Rep, M., Dekker, H. L., Vossen, J. H., de Boer, A. D., Houterman, P. M., Speijer, D., et al. (2002). Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 130, 904–917. doi: 10.1104/pp.007427
Rep, M., and Kistler, H. C. (2010). The genomic organization of plant pathogenicity in Fusarium species. Curr. Opin. Plant Biol. 13, 420–426. doi: 10.1016/j.pbi.2010.04.004
Ribeiro, R. L. D., and Hagedorn, D. J. (1979). Inheritance and nature of resistance in beans to Fusarium oxysporum f. sp. phaseoli. Phytopathology 69, 859–861.
Rigert, K. S., and Foster, K. W. (1987). Inheritance of resistance to two races of Fusarium wilt in three cowpea cultivars. Crop Sci. 27, 220–224. doi: 10.2135/cropsci1987.0011183x002700020018x
Rispail, N., Bani, M., and Rubiales, D. (2015). Resistance reaction of Medicago truncatula genotypes to Fusarium oxysporum: effect of plant age, substrate and inoculation method. Crop Pasture Sci. 66, 506–515. doi: 10.1071/cp14216
Rispail, N., and Rubiales, D. (2015). Rapid and efficient estimation of pea resistance to the soil-borne pathogen Fusarium oxysporum by infrared imaging. Sensors 15, 3988–4000. doi: 10.3390/s150203988
Risser, G., Banihashemi, Z., and Davis, D. W. (1976). A proposed nomenclature of Fusarium oxysporum f. sp. melonis races and resistance genes in Cucumis melo. Phytopathology 66, 1105–1106. doi: 10.1094/phyto-66-1105
Roberts, P. A., Frate, C. A., Matthews, W. C., and Osterli, P. P. (1995). Interactions of virulent Meloigyne incognita and Fusarium wilt on resistant cowpea genotypes. Phytopathology 85, 1288–1295. doi: 10.1094/phyto-85-1288
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signalling in plants. Annu Rev Plant Biol. 61, 621–649.
Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14, S185–S205.
Römer, C., Wahabzada, M., Ballvora, A., Pinto, F., Rossini, M., Panigada, C., et al. (2012). Early drought stress detection in cereals: simplex volume maximisation for hyperspectral image analysis. Funct. Plant Biol. 39, 878–890. doi: 10.1071/fp12060
Rousseau, C., Belin, E., Bove, E., Rousseau, D., Fabre, F., and Berruyer, R. (2013). High throughput quantitative phenotyping of plant resistance using chlorophyll fluorescence image analysis. Plant Methods 9:17. doi: 10.1186/1746-4811-9-17
Rubab, A., Chaudhary, R. A., Farah, N., and Ghulam, S. (2014). Surveillance and morphological characterization of Fusarium isolates associated with lentil wilt. Pakistan J. Phytopathol. 26, 85–88.
Rubiales, D., Fondevilla, S., Chen, W., Gentzbittel, L., Higgins, T. J. V., and Castillejo, M. A. (2015). Achievements and challenges in legume breeding for pest and disease resistance. Crit. Rev. Plant Sci. 34, 195–236. doi: 10.1080/07352689.2014.898445
Rubio, J., Hajj-Moussa, E., Kharrat, M., Moreno, M. T., Millan, T., and Gil, J. (2003). Two genes and linked RAPD markers involved in resistance to Fusarium oxysporum f. sp. cicerisrace 0 in chickpea. Plant Breed. 122, 188–191. doi: 10.1046/j.1439-0523.2003.00814.x
Sabbavarapu, M. M., Sharma, M., Chamarthi, S. K., Swapna, N., Rathore, A., Thudi, M., et al. (2013). Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochytablight in chickpea (Cicer arietinum L.). Euphytica 193, 121–138. doi: 10.1007/s10681-013-0959-2
Salgado, M. O., Schwartz, H. F., and Brick, M. A. (1995). Inheritance of resistance to a Colorado race of Fusarium oxysporum f. sp. phaseoli in common beans. Plant Dis. 79, 279–281. doi: 10.1094/pd-79-0279
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355.
Sarker, A., Bayaa, B., and Erskine, W. (2001). Registration of six lentil germplasm lines with resistance to vascular wilt. Crop Sci. 41:1655. doi: 10.2135/cropsci2001.4151655x
Saxena, K. B., Kumar, R. V., Saxena, R. K., Sharma, M., Srivastava, R. K., Sultana, R., et al. (2012). Identification of dominant and recessive genes for resistance to Fusarium wilt in pigeonpea and their implication in breeding hybrids. Euphytica 188, 221–227. doi: 10.1007/s10681-012-0700-6
Saxena, R. K., Singh, V. K., Kale, S. M., Tathinen, S., Parupalli, S., Kumar, V., et al. (2017). Construction of genotyping-by-sequencing based high-density genetic maps and QTL mapping for Fusarium wilt resistance in pigeonpea. Sci. Rep. 7:1911.
Scarafoni, A., Ronchi, A., Prinsi, B., Espen, L., Assante, G., Venturini, G., et al. (2013). The proteome of exudates from germinating Lupinus albus seeds is secreted through a selective dual-step process and contains proteins involved in plant defence. FEBS J. 280, 1443–1459. doi: 10.1111/febs.12140
Schäfer, W. (1994). Molecular mechanisms of fungal pathogenicity to plants. Annu. Rev. Phytopathol. 32, 461–477. doi: 10.1146/annurev.py.32.090194.002333
Schippers, B., and Van Eck, W. H. (1981). “Formation and survival of chla- mydospores in Fusarium,” in Fusarium: Disease, Biology, and Taxonomy, eds P. Nelson, T. A. Tousson, and R. J. Cook (University Park, CO: Penn State University Press), 250–260.
Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713. doi: 10.1038/ng.3008
Shah, T. M., Atta, B. M., Mirza, J. I., and Haq, M. A. (2009). Screening of chickpea (Cicer arietinum) induced mutants against Fusarium wilt. Pak. J. Bot. 41, 1945–1955.
Shakoor, N., Lee, S., and Mockler, T. C. (2017). High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 38, 184–192. doi: 10.1016/j.pbi.2017.05.006
Shao, C., Xiang, D., Wei, H., Liu, S., Guo, L., Li, C., et al. (2020). Predicting Virulence of Fusarium Oxysporum F. Sp. cubense based on the production of mycotoxin using a linear regression model. Toxins 12:254. doi: 10.3390/toxins12040254
Sharma, K. D., Chen, W., and Muehlbauer, F. J. (2005). Genetics of chickpea resistance to five races of Fusarium wilt and a concise set of race differentials for Fusarium oxysporum f. sp. ciceris. Plant Dis. 89, 385–395. doi: 10.1094/pd-89-0385
Sharma, K. D., and Muehlbauer, F. J. (2007). Fusarium wilt of chickpea: physiological specialization, genetics of resistance and resistance gene tagging. Euphytica 157, 1–14. doi: 10.1007/s10681-007-9401-y
Sharma, K. D., Winter, P., Kahl, G., and Muehlbauer, F. J. (2004). Molecular mapping of Fusarium oxysporumf. sp. ciceris race 3 resistance gene in chickpea. Theor. Appl. Genet. 108, 1243–1256. doi: 10.1007/s00122-003-1561-0
Sharma, M., Ghosh, R., Tarafdar, A., Rathore, A., Kumar, A. V., Chobe, D. R., et al. (2019). Exploring the genetic cypher of chickpea (Cicer arietinum L.) through Identification and Multi-environment Validation of Resistant Sources against Fusarium wilt (Fusarium oxysporum f. sp. ciceris). Front. Sust. Food Syst. 3:78. doi: 10.3389/fsufs.2019.00078
Sharma, M., Ghosh, R., Telangre, R., Rathore, A., Saifulla, M., Mahalinga, D. M., et al. (2016a). Environmental influences on pigeon pea-Fusarium udum interactions and stability of genotypes to Fusarium wilt. Front. Plant Sci. 7:253. doi: 10.3389/fpls.2016.00253
Sharma, M., Sengupta, A., Ghosh, R., Agarwal, G., Tarafda, A., and Nagavardhini, N. (2016b). Genome wide transcriptome profiling of Fusarium oxysporum f sp. ciceris conidial germination reveals new insights into infection-related genes. Sci. Rep. 6:37353.
Sharma, M., Kiran Babu, T., Gaur, P. M., Ghosh, R., Rameshwar, T., Chaudhary, R. G., et al. (2012). Identification and multi-environment validation of resistance to Fusarium oxysporum f. spcicerisin chickpea. Field Crop Res. 135, 82–88. doi: 10.1016/j.fcr.2012.07.004
Sharma, M., Nagavardhini, A., Thudi, M., Ghosh, R., Pande, S., and Varshney, R. K. (2014). Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genomics 15:454. doi: 10.1186/1471-2164-15-454
Sharma, M., and Pande, S. (2013). Unravelling effects of temperature and soil moisture stress response on development of dry root rot [Rhizoctonia bataticola (Taub.) Butler] in chick- pea. Am. J. Plant Sci. 4, 584–589. doi: 10.4236/ajps.2013.43076
Sharma, M., Pande, S., Mangala, U. N., Tripathi, S., and Gaur, P. M. (2010). Genetic resistance in desi and kabuli chickpea lines to Fusarium wilt caused by Fusarium oxysporum f. sp. ciceris. Indian J. Plant Prot. 38, 57–62.
Shubha, K., Dhar, S., Choudhary, H., Dubey, S. C., and Sharma, R. K. (2016). Identification of resistant sources and inheritance of Fusarium wilt resistance in garden pea (Pisum sativum ssp. hortense). Indian J. Hort. 73, 356–361. doi: 10.5958/0974-0112.2016.00077.3
Silvia Sebastiani, M., Bagnaresi, P., Sestili, S., Biselli, C., Zechini, A., OrruÌ, L., et al. (2017). Transcriptome analysis of the melon-Fusarium oxysporum f. sp. melonis race 1.2 pathosystem in susceptible and resistant plants. Front. Plant Sci. 8:362. doi: 10.3389/fpls.2017.00362
Singh, A. K., Ganapathysubramanian, B., Sarkar, S., and Singh, A. (2018). Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 23, 883–898. doi: 10.1016/j.tplants.2018.07.004
Singh, D., Sinha, B., Rai, V. P., Singh, M. N., Singh, D. K., Kumar, R., et al. (2016). Genetics of Fusarium wilt resistance in pigeonpea (Cajanuscajan) and efficacy of associated SSR markers. Plant Pathol. J. 32, 95–101. doi: 10.5423/ppj.oa.09.2015.0182
Singh, D. V., Lal, S., and Singh, S. N. (1974). Breeding gram(Cicer arietinum L.) for resistance to wilt. Indian J. genet. Plant Breed. 34, 267–270.
Singh, F., Singh, I. P., and Mazumder, N. D. (2011a). Identification of Fusarium wilt resistant sources of long-duration pigeonpea (Cajanus cajan). Indian J. Agr. Sci. 81, 1046–1051.
Singh, F., Singh, I. P., Majumder, N. D., and Katiyar, P. K. (2011b). IPA 16 F (IC0574574; INGR10021), a Pigeonpea (Cajanuscajan) Germplasm with Resistance to Fusarium Wilt, Donor for Resistance to Sterility Mosaic Disease of Pigeon Pea. Indian J. Plant Genet. Resour. 24.
Singh, H., Kumar, J., and Haware, M. P. (1987a). “Genetics of resistance to Fusarium wilt in chickpeas,” in Genetics and Plant Pathogenesis, eds P. R. Day and G. J. Jellis (Oxford: Blackwell Scientific Publications), 339–342.
Singh, H., Kumar, J., Smithson, J. B., and Haware, M. P. (1987b). Complementation between genes for resistance to race 1 of Fusarium oxyspomm f. sp. ciceri in chickpea. Plant Pathol. 36, 539–543. doi: 10.1111/j.1365-3059.1987.tb02271.x
Singh, I. P., Bohra, A., and Singh, F. (2016). An overview of varietal development programme of pigeonpea in India. Legume Perspect. 11, 37–40.
Singh, K. B., Ocampo, B., and Robertson, L. D. (1998). Diversity for abiotic and biotic stress resistance in the wild annual Cicer species. Genet. Resour. Crop Evolut. 45, 9–17.
Singh, M., Rana, J. C., Singh, B., Kumar, S., Saxena, D. R., Saxena, A., et al. (2017). Comparative agronomic performance and reaction to Fusarium wilt of Lens culinaris × L. orientalis and L. culinaris × L. ervoides derivatives. Front. Plant Sci. 8:1162. doi: 10.3389/fpls.2017.01162
Singh, V. K., Khan, A. W., Saxena, R. K., Kumar, V., Kale, S. M., and Chitikineni, A. (2016). Next-generation sequencing for identification of candidate genes for Fusarium wilt and sterility mosaic disease in pigeonpea (Cajanus cajan). Plant Biotechnol. J. 14, 1183–1194. doi: 10.1111/pbi.12470
Sladojevic, S., Arsenovic, M., Anderla, A., Culibrk, D., and Stefanovic, D. (2016). Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 1–11. doi: 10.1155/2016/3289801
Smith, S. N., Helms, D. M., Temple, S. R., and Frate, C. (1999). The distribution of Fusarium wilt of blackeyed cowpeas within California caused by Fusarium oxysporum f. sp. tracheiphilum race 4. Plant Dis. 83, 694–694. doi: 10.1094/pdis.1999.83.7.694c
Srinivas, C., Devi, D. N., Murthy, K. N., Mohan, C. D., Lakshmeesha, T. R., and Singh, B. P. (2019). Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity– A review. Saud. J Biol. Sci. 26, 1315–1324. doi: 10.1016/j.sjbs.2019.06.002
Srivastava, A. K., Kashyap, P. L., Chakdar, H., Kumar, M., Srivastava, A. K., Yadav, J., et al. (2018). First de novo draft genome sequence of the pathogenic fungus Fusarium udum F02845, associated with pigeonpea (Cajanus cajan L. Millspaugh) wilt. Microbiol. Resour. Announc. 7, e1001–e1018.
Summerell, B. A., Leslie, J. F., Liew, E. C. Y., Laurence, M. H., and Bullock, S. (2011). Fusarium species associated with plants in Australia. Fungal Diversity 46, 1–27. doi: 10.1007/s13225-010-0075-8
Swarupa, V., Ravishankar, K. V., and Rekha, A. (2014). Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta 239, 735–751. doi: 10.1007/s00425-013-2024-8
Tekeoglu, M., Tullu, A., Kaiser, W. J., and Muehlbauer, F. J. (2000). Inheritance and linkage of two genes that confer resis- tance to Fusarium wilt in chickpea. Crop Sci. 40, 1247–1256. doi: 10.2135/cropsci2000.4051247x
Thatcher, L. F., Williams, A. H., Garg, G., Buck, S. G., and Singh, K. B. (2016). Transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. medicaginis during colonisation of resistant and susceptible Medicago truncatula hosts identifies differential pathogenicity profiles and novel candidate effectors. BMC Genomics 17:860. doi: 10.1186/s12864-016-3192-2
Thudi, M., Khan, A. W., Kumar, V., Gaur, P. M., Katta, K., Garg, V., et al. (2016). Whole genome resequencing reveals genome-wide variations among parental lines of 16 mapping populations in chickpea (Cicer arietinum). BMC. Plant Biol. 16:10. doi: 10.1186/s12870-015-0690-3
Tosi, L., and Cappelli, C. (2001). First report of Fusarium oxysporum F. Sp. lentis of lentil in Italy. Plant Dis. 85:562. doi: 10.1094/pdis.2001.85.5.562a
Tullu, A., Kaiser, W. J., Kraft, J. M., and Muehlbauer, F. J. (1999). A second gene for resistance to race 4 of Fusarium wilt in chickpea and linkage with a RAPD marker. Euphytica 109, 43–50.
Tullu, A., Muehlbauer, F. J., Simon, C. J., Mayer, M. S., Kumar, J., Kaiser, W. J., et al. (1998). Inheritance and linkage of a gene for resistance to race 4 of Fusarium wilt and RAPD markers in chickpea. Euphytica 102, 227–232.
Upadhyaya, H., Haware, M., Kumar, J., and Smithson, J. (1983a). Resistance to wilt in chickpea. I. Inheritance of late-wilting in response to race 1. Euphytica 32, 447–456. doi: 10.1007/bf00021454
Upadhyaya, H., Smithson, J., Haware, M., and Kumar, J. (1983b). Resistance to wilt in chickpea. Further evidence for two genes for resistance to race 1. Euphytica 32, 749–755. doi: 10.1007/bf00042155
Upasani, M. L., Gurjar, G. S., Kadoo, N. Y., and Gupta, V. S. (2016). Dynamics of colonization and expression of pathogenicity related genes in Fusarium oxysporum f.sp. ciceri during chickpea vascular wilt disease progression. PLoS One 11:e0156490. doi: 10.1371/journal.pone.0156490
Upasani, M. L., Limaye, B. M., Gurjar, G. S., Kasibhatla, S. M., Joshi, R. R., Kadoo, N. Y., et al. (2017). Chickpea-Fusarium oxysporum interaction transcriptome reveals di erential modulation of plant defense strategies. Sci. Rep. 7:7746.
Varshney, R., Mohan, S., Gaur, P., Chamarthi, S., Singh, V., Srinivasan, S., et al. (2014). Marker-assisted backcrossing to introgress resistance to Fusarium wilt race 1 and Ascochyta blight in C 214, an elite cultivar of chickpea. Plant Genome 7, 278–289.
Varshney, R. K., Chen, W., Li, Y., Bharati, A. K., Saxena, R. K., and Schlueter, J. A. (2012). Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30, 83–89. doi: 10.1038/nbt.2022
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., and Sharpe, A. G. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246.
Varshney, R. K., Kudapa, H., Pazhamala, L., Chitikineni, A., Thudi, M., Bohra, A., et al. (2015). Translational genomics in agriculture: some examples in grain legumes. Crit. Rev. Plant Sci. 34, 169–194. doi: 10.1080/07352689.2014.897909
Voytas, D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. doi: 10.1146/annurev-arplant-042811-105552
Wade, B. L. (1929). Inheritance of Fusarium Wilt Resistance in Canning Peas. Madison, WI: Agricultural Experiment Station of the University of Wisconsin, 1–32.
Wallace, J. G., Rodgers-Melnick, E., and Buckler, E. S. (2018). On the road to breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics. Annu. Rev. Genet. 52, 421–444. doi: 10.1146/annurev-genet-120116-024846
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., and Gao, Y. (2016). Enhanced rice blast resistance by CRISPR/Cas9- targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., and Goa, C. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Widmann, C., Gibson, S., Jarpe, M. B., and Johnson, G. L. (1999). Mitogen- activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180. doi: 10.1152/physrev.1999.79.1.143
Williams, A. H., Sharma, M., Thatcher, L. F., Azam, S., Hane, J. K., Sperschneider, J., et al. (2016). Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 17:191. doi: 10.1186/s12864-016-2486-8
Winter, P., Benko-Iseppon, A. M., Huttel, B., Ratnaparkhe, M., Tullu, A., Sonnante, G., et al. (2000). A linkage map of the chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum × C. reticulatumcross: localization of resistance genes for Fusarium wilt races 4 and 5. Theor. Appl. Genet. 101, 1155–1167. doi: 10.1007/s001220051592
Wise, R. P., Moscou, M. J., Bogdanove, A. J., and Whitham, S. A. (2007). Transcript profiling in host-pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369. doi: 10.1146/annurev.phyto.45.011107.143944
Wu, X., Wu, X., Xu, P., Wang, B., Lu, Z., and Li, G. (2015). Association mapping for Fusarium wilt resistance in Chinese asparagus bean germplasm. Plant Genome 8, 1–6. doi: 10.3835/plantgenome2014.11.008
Xue, R., Wu, J., Zhu, Z., Wang, L., Wang, X., Wang, S., et al. (2015). Differentially expressed genes in resistant and susceptible common bean (Phaseolus vulgaris L.) genotypes in response to Fusarium oxysporum f. sp. phaseoli. PLoS One 10:e0127698. doi: 10.1371/journal.pone.0127698
Yang, Y., Shah, J., and Klessig, D. F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. doi: 10.1101/gad.11.13.1621
Yin, K., and Qiu, J. L. (2019). Genome editing for plant disease resistance: applications and perspectives. Philos. Trans. R. Soc. B. 374:20180322. doi: 10.1098/rstb.2018.0322
Zhang, C., Chen, W., and Sankaran, S. (2019). High-throughput field phenotyping of Ascochyta blight disease severity in chickpea. Crop Protect. 125:104885. doi: 10.1016/j.cropro.2019.104885
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., and Gao, C. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Keywords: Fusarium wilt, genomics, molecular markers, genetic variability, grain legume
Citation: Jha UC, Bohra A, Pandey S and Parida SK (2020) Breeding, Genetics, and Genomics Approaches for Improving Fusarium Wilt Resistance in Major Grain Legumes. Front. Genet. 11:1001. doi: 10.3389/fgene.2020.01001
Received: 11 May 2020; Accepted: 06 August 2020;
Published: 23 October 2020.
Edited by:
Ajay Kumar, North Dakota State University, United StatesReviewed by:
Nicolas Rispail, Institute for Sustainable Agriculture, Spanish National Research Council, SpainCopyright © 2020 Jha, Bohra, Pandey and Parida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uday Chand Jha, dWRheV9nZW5lQHlhaG9vLmNvLmlu; dTk4MTE5ODFAZ21haWwuY29t; Abhishek Bohra, YWJoaS5vbWljc0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.