- State Key Laboratory of Developmental Biology of Freshwater Fish, Engineering Research Center of Polyploid Fish Reproduction and Breeding of the State Education Ministry, College of Life Sciences, Hunan Normal University, Changsha, China
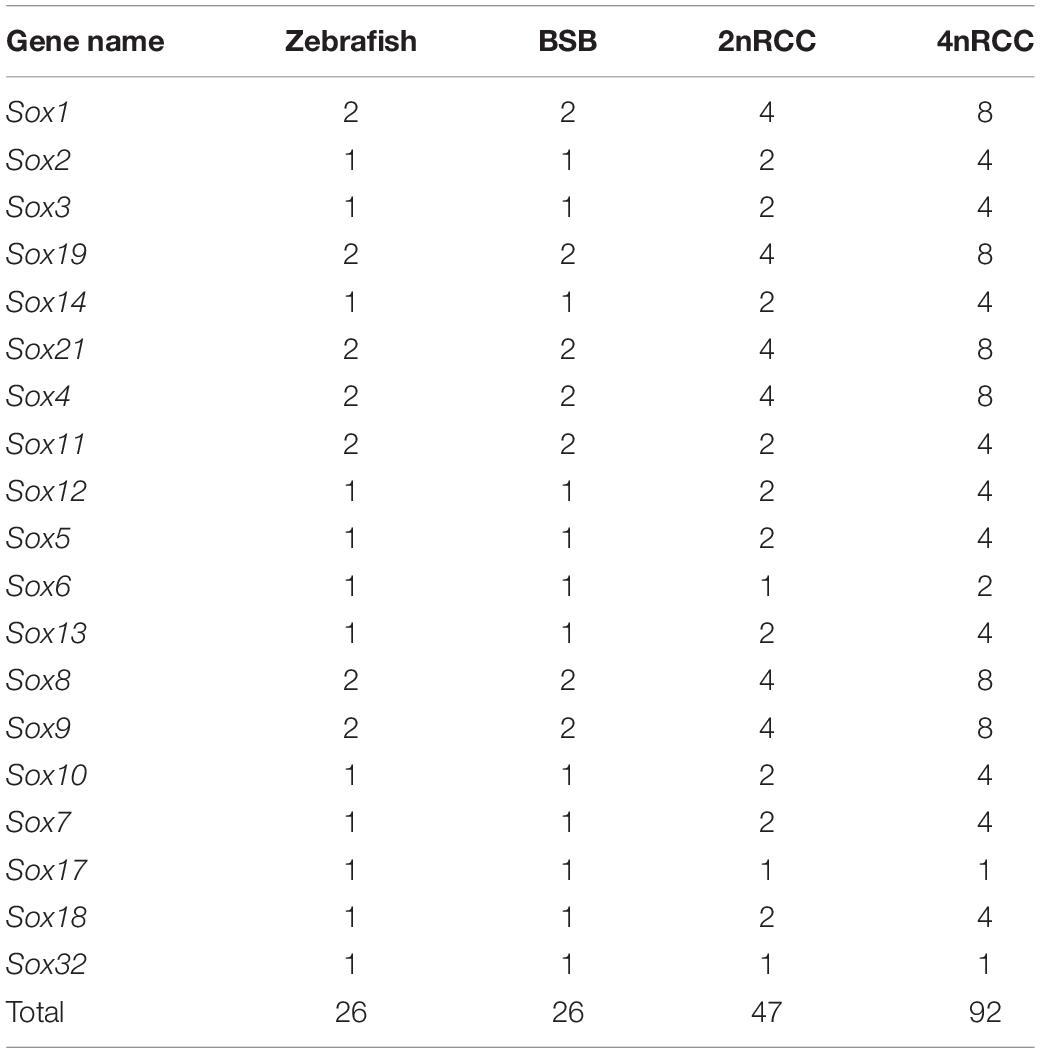
The Sox gene family encoded transcription factors that played key roles in developmental processes in vertebrates. To further understand the evolutionary fate of the Sox gene family in teleosts, the Sox genes were comprehensively characterized in fish of different ploidy levels, including blunt snout bream (2n = 48, Megalobrama amblycephala, BSB), goldfish (2n = 100, Carassius auratus red var., 2nRCC), and autotetraploid C. auratus (4n = 200, 4nRCC). The 4nRCC, which derived from the whole genome duplication (WGD) of 2nRCC, were obtained through the distant hybridization of 2nRCC (♀) × BSB (♂). Compared with the 26 Sox genes in zebrafish (2n = 50, Danio rerio), 26, 47, and 92 putative Sox genes were identified in the BSB, 2nRCC, and 4nRCC genomes, respectively, and classified into seven subfamilies (B1, B2, C, D, E, F, and K). Comparative analyses showed that 89.36% (42/47) of Sox genes were duplicated in 2nRCC compared with those in BSB, while 97.83% (90/92) of Sox genes were duplicated in 4nRCC compared with those in 2nRCC, meaning the Sox gene family had undergone an expansion in BSB, 2nRCC, and 4nRCC, respectively, following polyploidization events. In addition, potential gene loss, genetic variations, and paternal parent SNP locus insertion occurred during the polyploidization events. Our data provided new insights into the evolution of the Sox gene family in polyploid vertebrates after several rounds of WGD events.
Introduction
Several rounds of whole genome duplication (WGD) events were known to have taken place during the evolution of vertebrates (Voldoire et al., 2017). The two rounds (2R) of WGD events in vertebrates might be traced back to 500–600 million years ago (Mya), and teleosts were widely believed to have undergone teleost-specific WGD (TSGD or third round, 3R) events approximately 320–350 Mya (Venkatesh, 2003; Guo et al., 2009; Van de Peer et al., 2009; Li et al., 2014; Zhang et al., 2018). The TSGD might have resulted in the wide variety of the teleosts. More than 32,500 species of fish existed in nature, making them the most diverse group of vertebrates (Cossins and Crawford, 2005; Liu, 2014; Wang et al., 2018). Moreover, the common carp (Cyprinus carpio) and the goldfish (Carassius auratus), which represented the “tetraploid state” in relation to other members of the Cyprinidae family (Danio rerio), were thought to have undergone an additional round (fourth round, 4R) of genome duplication (Ohno et al., 1967; Larhammar Dan, 1994; David et al., 2003; Wang et al., 2012; Chen et al., 2019; Xu et al., 2019). Hence, the natural polyploids formed by genome duplication would have subsequently become new diploids or paleopolyploids through diploidization and differentiation (Jenczewski and Alix, 2004; Parisod et al., 2010; Song et al., 2012; Qin et al., 2016; Zhou and Gui, 2017). Genome duplication could thus be considered an important process in species origination and evolution.
The goldfish (2nRCC, 2n = 100) is raised over all Asia for food and as an ornamental pet (Liu et al., 2016; Chen et al., 2019), while Megalobrama amblycephala (blunt snout bream, 2n = 48, BSB) is an economically important freshwater fish (Ren et al., 2019). In previous studies, we artificially obtained fertile allotetraploids (4nRB, 4n = 148, F1) from the distant hybridization of 2nRCC (♀) × BSB (♂) (Liu et al., 2007). The fertile 4nRCC (4n = 200) was obtained through the fertilization of diploid sperm and eggs caused by the abnormal chromosome behavior during meiosis of 4nRB, and the 4nRCC lineage (F2–F14) was established through continuous self-crossing (Qin et al., 2014; Hu et al., 2019). The newly synthesized 4nRCC possessed four sets of chromosomes from 2nRCC and produced diploid gametes, and there were obvious phenotypic differences between 2nRCC and 4nRCC (Qin et al., 2014; Qin Q. et al., 2018; Qin Q. B. et al., 2018). It has been stated that studies of recently polyploid fishes and their closely related species might be essential to investigate the evolutionary processes occurring in polyploid genomes (gene families) after polyploidization (Moghadam et al., 2005; Guo et al., 2009). Thus, the newly synthesized 4nRCC might be one of the best model species to study polyploidization and genome duplication.
The Sox gene family shared a high mobility group (HMG) box, which contained a DNA-binding domain of 79 amino acids (aa) (Chiang et al., 2001; Yokoi et al., 2002; Hett and Ludwig, 2005). Based on conserved protein domains and nucleic acid sequences, the Sox gene family was divided into 11 groups from SoxA to SoxK (Yu et al., 2018). The Sox genes were involved in the regulation of diverse growth-related and developmental processes, such as chondrogenesis, neurogenesis, and sex determination and differentiation (Jiang et al., 2013). For example, the Sox9a gene presented significant sex correlations in both 4nRCC and 2nRCC (Huang et al., 2020). Currently, extensive studies had identified the Sox gene family in various species, and the number of Sox genes had been found to vary greatly (Wei et al., 2016; Yu et al., 2017). In teleosts, 19 Sox genes were found in Japanese medaka (Oryzias latipes, 3R), 24 in pufferfish (Fugu rubripes, 3R), 25 in catfish (Ictalurus punctatus, 3R) and Japanese flounder (Paralichthys olivaceus, 3R), 26 in zebrafish (D. rerio, 3R), and 27 in tilapia (Oreochromis niloticus, 3R), while 49 putative Sox genes were present in common carp (C. carpio, 4R) (Cui et al., 2011; Wei et al., 2016; Yu et al., 2018; Zhang et al., 2018). Obviously, from 3R to 4R, more Sox genes existed in common carp than in other species, suggesting that most Sox genes also underwent expansion. Natural and synthetic polyploid fish might provide ideal models for examining the evolutionary behavior of Sox genes in duplicated genomes after polyploidization events (Guo et al., 2009). For the important role of Sox gene family in developmental processes in Cyprinidae fishes with different ploidy levels, studies of this family may provide a better understanding the effect of genome duplication.
In this study, we analyzed the Sox gene sequences of BSB (2n = 48), 2nRCC (2n = 100), and 4nRCC (4n = 200) compared with those of zebrafish (2n = 50). Based on all the available Sox genes of zebrafish, the putative Sox gene sequences of BSB and 2nRCC were obtained using the available genome databases of BSB and goldfish, respectively. Subsequently, PCR surveys were performed for Sox gene sequences in 4nRCC. This study aimed to perform a comprehensive investigation of the diversity and phylogenetic relationships of the Sox gene family in BSB, 2nRCC, and 4nRCC, thus elucidating the genomic effects of polyploidization events.
Materials and Methods
Materials
All the individuals of BSB, 2nRCC, and 4nRCC in this paper were cultured in similar pond with area of 666.7 m2 in the State Key Laboratory of Developmental Biology of Freshwater Fish, Hunan Normal University, Changsha, Hunan, China. The 4nRCC was produced from the same fish lineage of BSB in this study. The individuals used as samples in this study weighed about 100 g and were anesthetized with 100 mg/L MS-222 (Sigma–Aldrich, St. Louis, MO, United States) prior to dissection.
Preparation of Chromosome Spreads
In this study, to better understand the changes in chromosome number doubling after genome replication in BSB, 2nRCC, and 4nRCC, chromosome counts were performed using kidney tissue from 10 individuals each of BSB, 2nRCC, and 4nRCC. A detailed method can be found in Liu et al. (2007). Twenty metaphase spreads of chromosomes for each individual were photographed under a microscope.
Identification of the Sox Genes in the BSB and 2nRCC Genomes
All available Sox genes were identified in the zebrafish genome1. These Sox genes were used to search for their respective BSB and 2nRCC counterparts in the available BSB and 2nRCC genomic resources (project ID: PRJNA481500) using Blast (V2.5.0) software (Chen et al., 2019; Ren et al., 2019). The CDs lengths and aa numbers of the putative Sox genes were identified from the BSB and 2nRCC genomes.
RNA Isolation and RT-PCR
According to the spatial expression profiles of the tilapia and Japanese flounder Sox genes, most Sox genes show high expression in five adult tissues, including brains, testes, ovaries, hearts, and kidneys (Wei et al., 2016; Yu et al., 2018). Hence, total RNA was isolated from the brains, testes, ovaries, hearts, and kidneys tissues of 10 4nRCC individuals using RNAiso reagent (TaKaRa, Japan) following the manufacturer’s protocol. Total RNA integrity was detected by 1.5% agarose gel electrophoresis and a Synergy 2 Multi-Mode Microplate Reader (BioTek2), and the samples were stored at −80°C. The first-strand cDNA was synthesized using the Maxima H Minus First Strand cDNA Synthesis Kit with dsDNase (Thermo Scientific, United States) in a 20 μL reaction volume.
PCR Amplification, Cloning, and Sequencing of 4nRCC Sox Genes
In the genetic composition, the 4nRCC were derived from the WGD of 2nRCC (Qin et al., 2014). Based on the CDs of the 2nRCC putative Sox genes, primers (Supplementary Table S1) were designed by Primer 5.0 software to amplify 4nRCC Sox genes with cDNA as templates. The primers were synthesized by Tsingke (Beijing, China). PCR was performed in a final volume of 50 μL using LA Taq (TaKaRa). The amplification conditions were as follows: 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 50–63°C, and 2 min at 72°C; and 5 min at 72°C for final extension (Wang et al., 2017). All PCR products were cloned into the pMD18-T vector (TaKaRa) and transferred into Escherichia coli DH5α (Sangon, China). To avoid PCR errors, at least 60 positive clones for each gene were obtained after screening by PCR amplification and sequenced by Tsingke. All sequences were analyzed using Blast3.
Phylogenetic Analysis, Gene Nomenclature, and Identification of Conserved Motifs
Multiple sequence alignments of the CDs and derived aa sequences of the Sox genes in zebrafish, BSB, 2nRCC, and 4nRCC were performed using BioEdit software. Phylogenetic analysis was used to better understand the evolution of Sox genes in these species using MrBayes version 3.1.2. The mixed model was chosen for the Sox genes as no substitution model could be decided according the aa sequence. MrBayes was run for 10 million generations with two runs and four chains in parallel and a burn-in of 25%, and the analysis was terminated after the average standard deviation of the split frequencies fell under 0.01 (Luo et al., 2019). The final trees were visualized in FIGTREE (V1.4.4) software. Phylogenetic analysis is considered to be one of the most important pieces of evidence for the gene annotation and nomenclature of non-model species (Jiang et al., 2016). All the Sox genes from BSB, 2nRCC, and 4nRCC were named after their closely related zebrafish Sox genes. When more than one Sox gene from 2nRCC or 4nRCC was clustered with certain zebrafish Sox genes, Roman-letter suffixes were added to each Sox gene (for instance, 2nRCC_Sox1a-1, 2nRCC_Sox1a-2, 4nRCC_Sox1a-1, 4nRCC_Sox1a-2, 4nRCC_Sox1a-3, 4nRCC_Sox1a-4). Generally, Sequence motif is defined as a sequence pattern of DNA, RNA, or aa sequence. Conserved motifs were checked using the online Multiple Expectation Maximization for Motif Elicitation (MEME4) program (Zhao et al., 2018).
Statistical Analysis
Analyses of variance and pairwise comparisons of the data were performed with SPSS 17.0 software.
Results
Examination of Chromosome Number
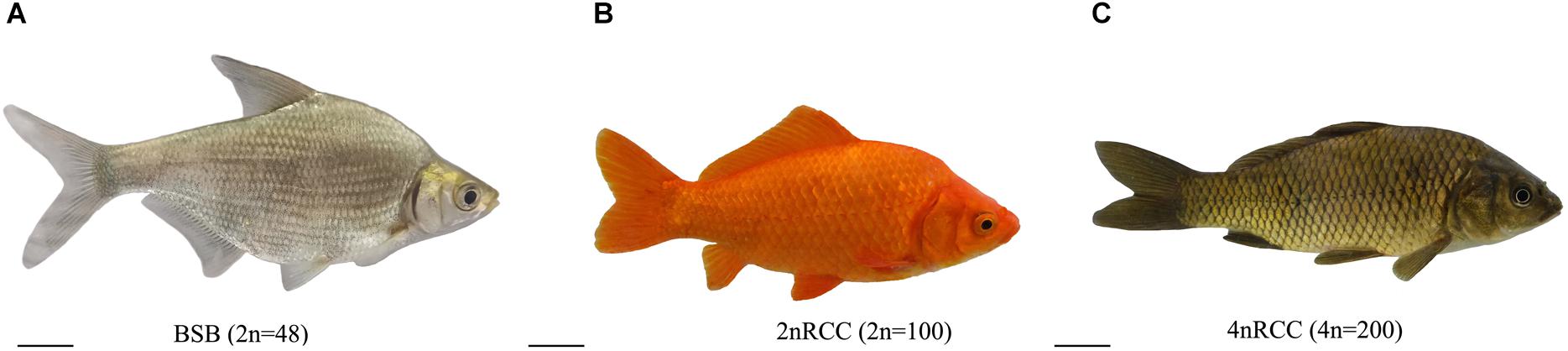
The appearances of BSB, 2nRCC, and 4nRCC were presented in Figure 1. The chromosome number distributions in BSB, 2nRCC, and 4nRCC are shown in Table 1 and Figure 2. In the BSB samples (Figure 1A), 92.5% of the chromosomal metaphases possessed 48 chromosomes (Table 1 and Figure 2A). Among the 2nRCC samples (Figure 1B), 90.5% of the chromosomal metaphases possessed 100 chromosomes (Table 1 and Figure 2B). Among the 4nRCC samples (Figure 1C), 85% of the chromosomal metaphases had 200 chromosomes (Table 1 and Figure 2C), indicating that distant hybridization could result in polyploidization events accompanied by chromosome number doubling.
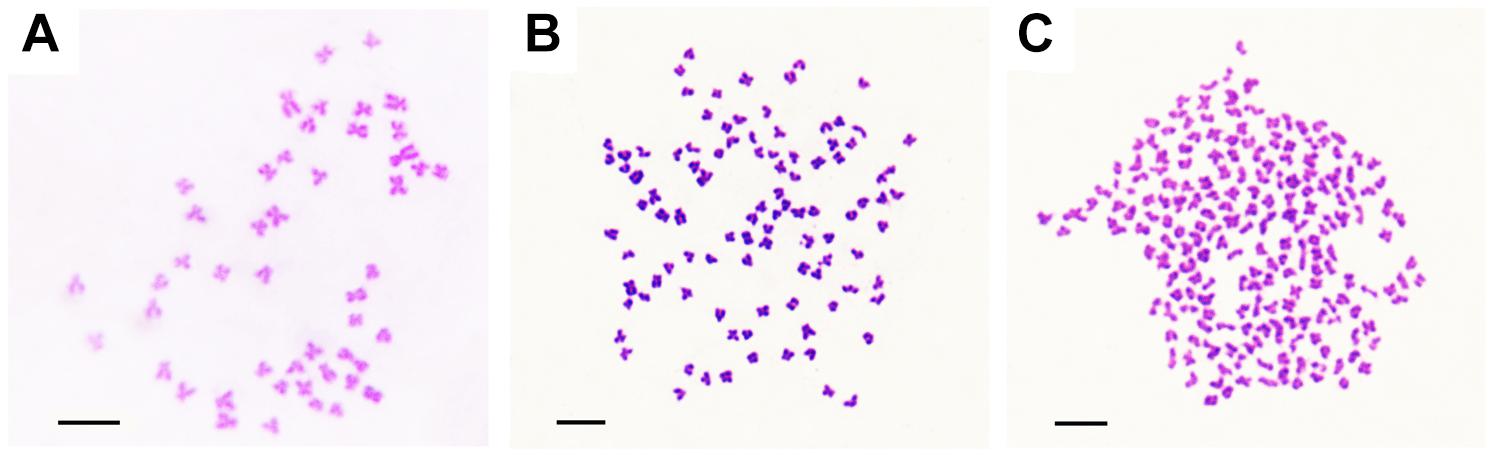
Figure 2. The chromosomes at metaphase in BSB, 2nRCC, and 4nRCC. (A) The 48 chromosomes of BSB. (B) The 100 chromosomes of 2nRCC. (C) The 200 chromosomes of 4nRCC. Bar = 20 μm.
Identification of Sox Genes in BSB, 2nRCC, and 4nRCC
A total of 26 Sox genes were identified in the zebrafish genome1 (Yu et al., 2018). By searching for these sequences in the BSB and 2nRCC genomes, we identified 26 and 47 putative Sox genes in BSB and 2nRCC, respectively (Supplementary Tables S2, S3). In addition, a total of 92 Sox genes were identified in 4nRCC by PCR survey (Supplementary Table S4). All the coding sequences (CDs) from 4nRCC were confirmed to be Sox gene sequences via the NCBI website database5. Through a comprehensive comparison, we noticed that the number of Sox genes between BSB and zebrafish were the same, indicating that BSB had also undergone a TSGD event; we also noticed an important expansion of the Sox gene family following genome duplication (Table 2): compared with BSB, 89.36% (42/47) of Sox genes were duplicated in 2nRCC due to the 4R WGD event; from 2nRCC to 4nRCC, 97.83% (90/92) of Sox genes were duplicated due to a specific WGD event. These results indicated that the number of most Sox genes in each species were generally associated with ploidy level.
SNP Loci From the Paternal Parent in 4nRCC and Sequence Analyses
We examined the sequence divergence of all Sox genes in BSB, 2nRCC, and 4nRCC. The identity for most Sox genes between 4nRCC and BSB was lower than that between 4nRCC and 2nRCC. However, as shown in Figure 3, SNP loci were found in the 4nRCC sequencing results that were identical to the BSB sequence but different from that of 2nRCC. According to the multiple alignment, synonymous mutations and several base insertions/deletions were observed in the coding regions of most Sox genes among BSB, 2nRCC, and 4nRCC. For instance, some SNP mutations were observed in all Sox14 genes (Figure 4A), and all Sox14 genes had higher identities in the aa sequence than in the CDs (Figure 4B), suggesting that most mutations were synonymous. As shown in Figure 5, two six-base insertions/deletions and a 15-base insertion/deletion were found in Sox1b; two two-amino-acid insertions/deletions and a five-amino-acid insertion/deletion were found in the corresponding putative aa sequence.
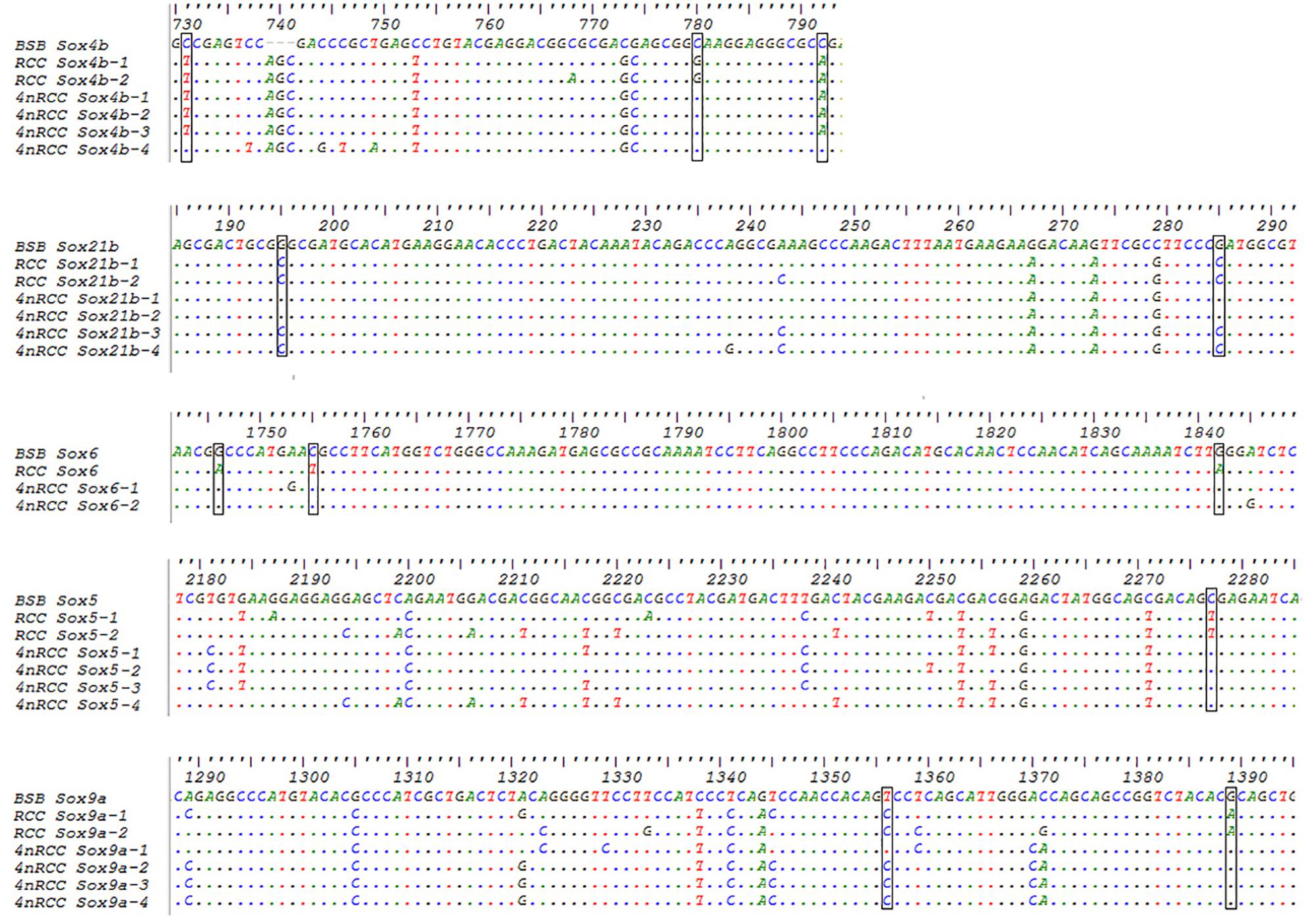
Figure 3. The SNP loci from BSB found in the 4nRCC sequencing results. The dots represented sequence identity, and the hyphens represented insertions/deletions. The black boxes showed the SNP loci.
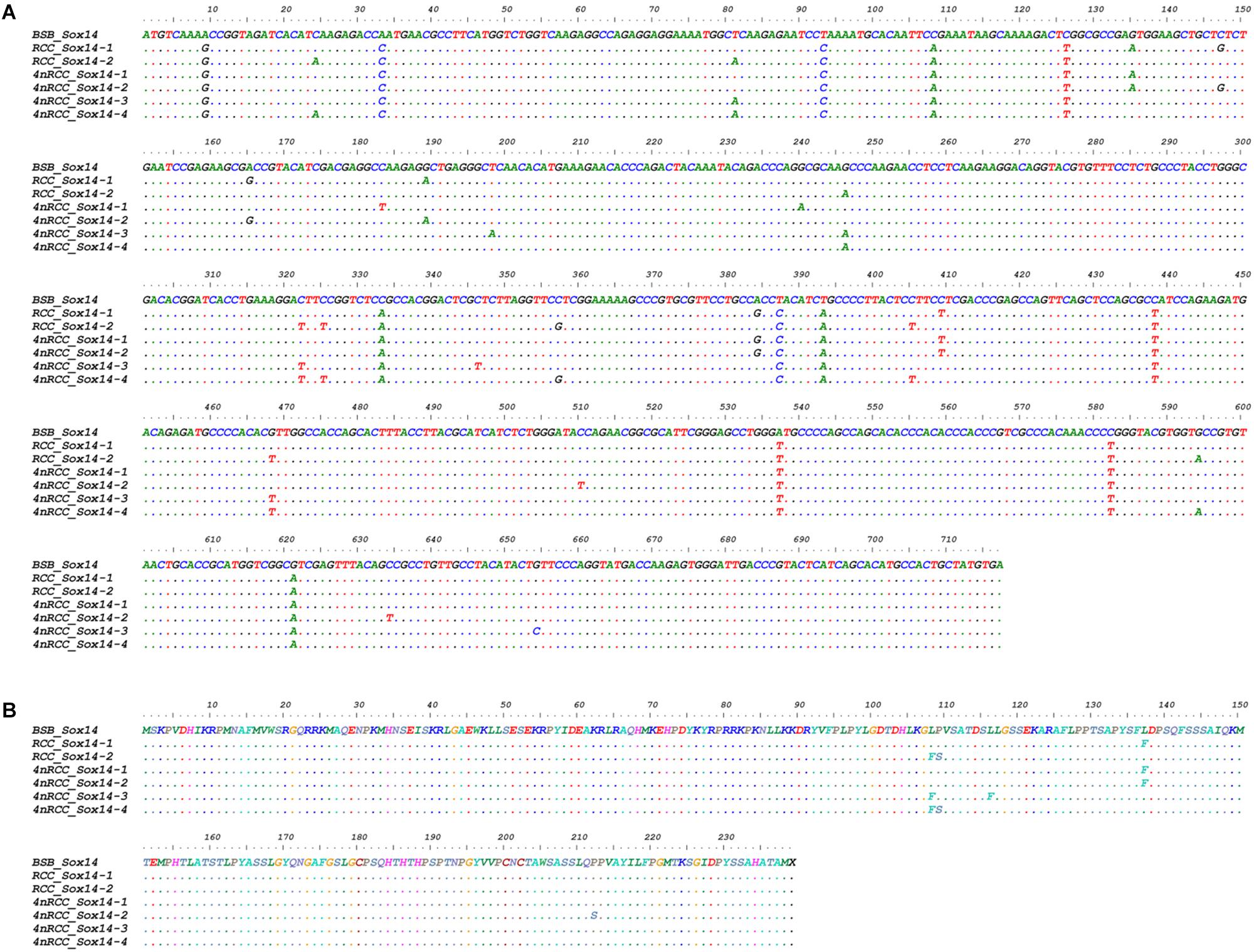
Figure 4. Alignment of the Sox14 CDs (A) and derived aa sequences (B) of BSB, 2nRCC, and 4nRCC. The dots represented sequence identity.

Figure 5. Alignment of the Sox1b CDs (A) and derived aa sequences (B) of BSB, 2nRCC, and 4nRCC. The dots represented sequence identity, and the hyphens represented insertions/deletions. Base insertions/deletions and aa insertions/deletions were framed by red boxes.
Phylogenetic Analysis
We further used the putative aa sequences of the Sox genes to build phylogenetic trees for zebrafish, BSB, 2nRCC, and 4nRCC (Figure 6). The phylogenetic analysis showed that the Sox genes of BSB, 2nRCC, and 4nRCC were clustered with their respective counterparts from zebrafish, providing strong evidence for correctly naming and grouping the Sox genes of BSB, 2nRCC, and 4nRCC and indicating that all the genes in the Sox gene family were highly conserved. All the putative Sox genes could be divided into seven subfamilies. In BSB, six putative genes were clustered in subfamily B1, three in subfamily B2, five in subfamily C, three in subfamily D, five in subfamily E, three in subfamily F, and one in subfamily K. In 2nRCC, 12 putative genes were clustered in subfamily B1, six in subfamily B2, eight in subfamily C, five in subfamily D, 10 in subfamily E, five in subfamily F, and one in subfamily K. In 4nRCC, the PCR survey showed 24 putative genes in subfamily B1, 12 in subfamily B2, 16 in subfamily C, 10 in subfamily D, 20 in subfamily E, nine in subfamily F, and one in subfamily K. The names and numbers of as for each Sox gene in BSB, 2nRCC, and 4nRCC are listed in Supplementary Tables S1–S3, respectively. However, some Sox genes, such as Sox17 and Sox32, retained only one copy in these four fishes. Some other Sox genes, including Sox6 and Sox11, retained their original number and did not undergo duplication (Table 2) after the 4R WGD event.
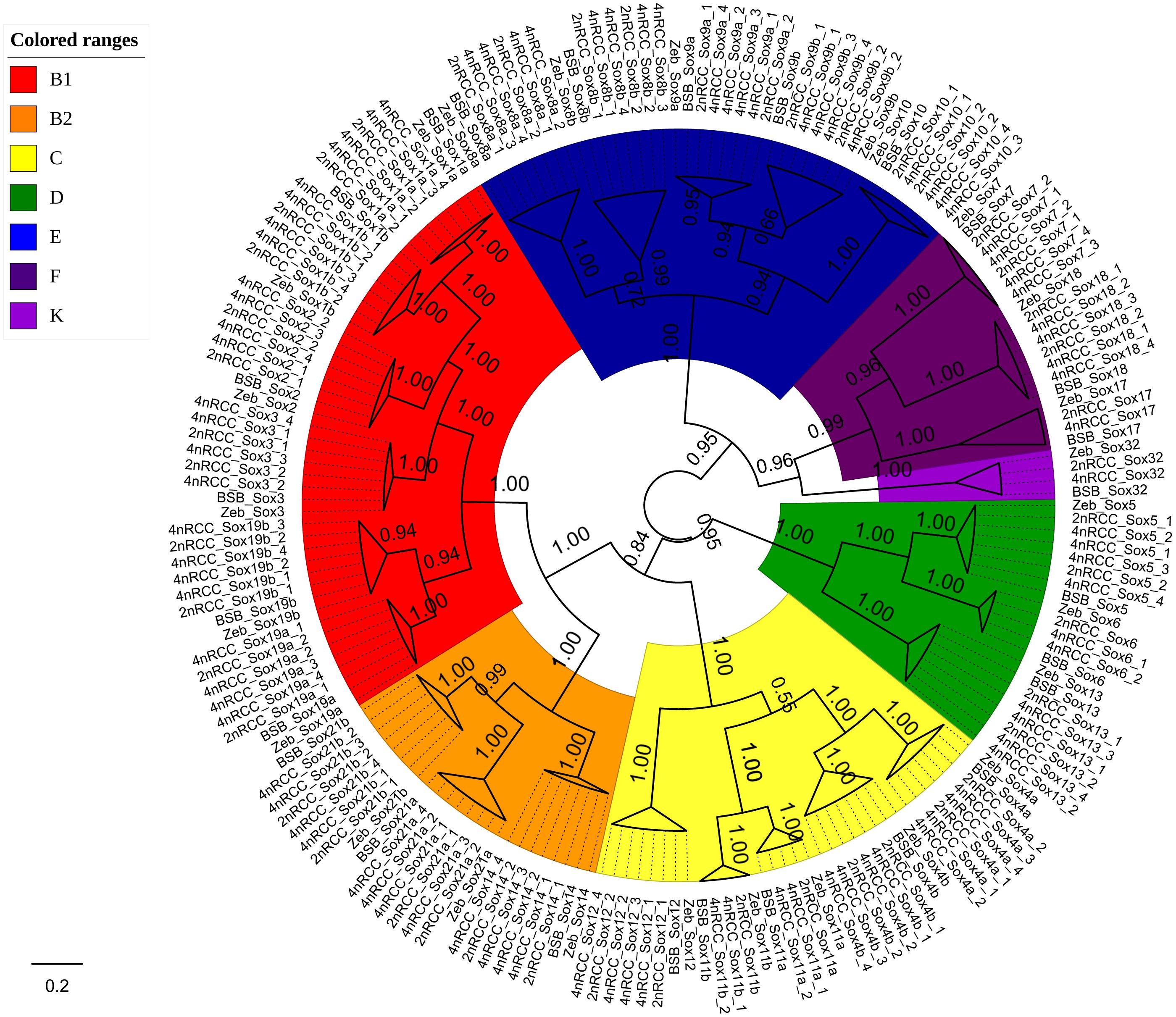
Figure 6. Phylogenetic analyses of the amino acid sequences of all the Sox genes in zebrafish (Zeb), BSB, 2nRCC, and 4nRCC. The phylogenetic tree constructed using MrBayes with mixed model; MCMC = 10 million generations. Different branch colors denoted different Sox subfamilies.
Identification of Conserved Motifs
A total of 10 conserved motifs (motifs 1–10) are identified in Figure 7. In general, subfamily B1, subfamily B2, subfamily C, subfamily D, and subfamily E had ten motifs (1, 2, 3, 4, 5, 6, 7, 8, 9, 10); subfamily F had nine motifs (1, 2, 3, 4, 6, 7, 8, 9, 10); and subfamily K had seven motifs (1, 2, 3, 4, 5, 7, 8). Comparing these motifs with the conserved HMG box sequence of vertebrate Sox proteins (DHVKRPMNAFMVWSRGERRKIAQQNPDMHNSEISKRLG KRWKLLSESEKRPFIEEAERLRAQHMKDYPDYKYRPRRKKK) (Yu et al., 2018), we found that complete motifs 1, 2, 3, 4, and 7 and partial motif 5 constituted the conserved HMG domain. Sox genes in the same subfamily shared similar conserved motifs that were clearly distinguishable from those in other subfamilies.
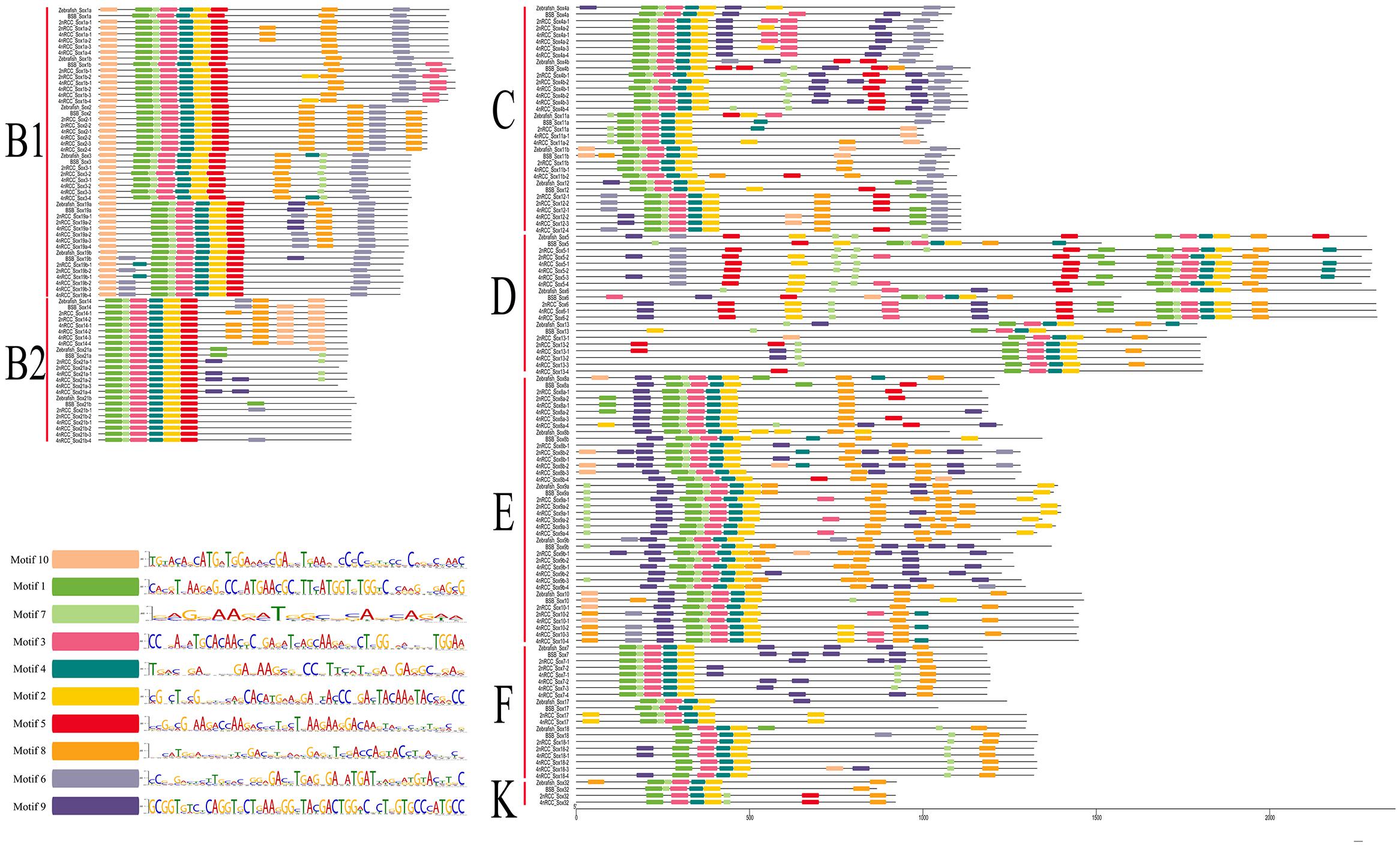
Figure 7. Conserved motifs of seven subfamilies (B1, B2, C, D, E, F, and K) of all Sox genes in zebrafish, BSB, 2nRCC, and 4nRCC. Conserved motifs were checked using the online Multiple Expectation Maximization for Motif Elicitation (MEME, http://alternate.meme-suite.org/) program. Different colors denoted different motifs in Sox genes, and the sequence of motif 1–10 in Sox genes was presented.
Discussion
Autopolyploids were traditionally considered to arise by the WGD of a species, whereas allopolyploids were produced by hybridization and the merging of genomes from different species (Julca et al., 2018). Evidence has accumulated that an additional round (4R) of genome duplication in some Cyprinidae species occurred approximately 8–12 Mya, suggesting that those Cyprinidae species, which include goldfish and common carp, are natural allotetraploids (Ma et al., 2014; Wei et al., 2016; Chen et al., 2019; Omori and Kon, 2019; Xu et al., 2019). In previous study, we successfully obtained the synthetic line 4nRCC, which was derived from a specific WGD event of 2nRCC induced by distant hybridization (Qin et al., 2014). In this study, we noticed an important expansion of the Sox gene family in 4nRCC (4n = 200): 97.83% (90/92) of Sox genes were duplicated compared with 2nRCC (2n = 100). Similarly, the duplication rate of Sox genes in 2nRCC reached 89.36% (42/47) compared with those in BSB (3R, 2n = 48), while the number of chromosomes in 2nRCC was approximately twice that in BSB. Hence, our findings also provided evidence that 2nRCC was a tetraploid and had undergone 4R WGD.
Lineage-specific gene duplication and gene loss events were frequently observed in fish genomes during evolution; these events represented one of the primary driving forces for genome evolution and provided useful information for gene evolution studies (Fortna et al., 2004; Jiang et al., 2016; Zhang et al., 2018; Han et al., 2019). Gene duplication was the key mechanism for generating new genes and biological processes, which would be conducive to the evolution of species (Fortna et al., 2004). Moreover, a “1-2-4 (-8 in teleosts)” rule had been proposed for the evolution of vertebrate gene and genome duplications (Axel Meyer, 1999; Meyer and Van de Peer, 2005). In this study, all duplicated genes, except for Sox6, Sox11, Sox17, and Sox32, were consistent with this proposal, creating new homologous Sox genes following the genome duplication. For instance, there were four Sox19a genes in 4nRCC (artificial tetraploid, 4n = 200), two Sox19a genes in 2nRCC (diploidized tetraploid, 2n = 100), and only one in BSB (diploid, 2n = 48). In addition, phylogenetic analyses and identification of conserved motifs demonstrated that all Sox genes were conserved. Following the emergence of new genes, the regulatory networks during the development of vertebrates might become more complicated to cope with the complex and ever-changing natural environment (Kondrashov, 2012). The specific functions of Sox genes in 4nRCC and 2nRCC remain unclear, which need to be further investigated. Thus, our findings also served to support the “1-2-4 (-8 in teleost fishes)” rule in natural and synthetic polyploid fishes. According to this rule and the numbers of most of the duplicated Sox genes, we speculated that the newly synthesized 4nRCC might in fact be octoploid. Of course, further experiments are required to verify this hypothesis.
Similar duplication events were also observed in other gene families. For example, there were at least 16 FGFs in common carp (4R) with duplicated copies comparable to the eight FGFs in zebrafish (3R) (Jiang et al., 2016). A special result was also found in sturgeons that seven Hox clusters including 68 Hox genes were identified (Cheng et al., 2019). Although the sturgeons did not undergo the teleost-specific genome duplication (TSGD) event, they have experienced their own lineage-specific polyploidizations with one or more rounds of genome duplication (Jaillon et al., 2004; Crow et al., 2012). We speculated that the similar duplication events might also occur in other gene families in 4nRCC.
Some Sox genes (Sox6, Sox11, Sox17, and Sox32) retained their original number and did not undergo duplication in the 4R WGD or specific WGD. Potential gene loss were also observed in other gene families, such as the Fibroblast growth factors (FGFs) in common carp (4R) and the ATP-binding cassette (ABC) transporter gene superfamily in catfish (3R) (Liu et al., 2013; Jiang et al., 2016). Although a genome duplication should be accompanied by the doubling of all genes, gene losses might occur in the state of natural or synthetic polyploidy following WGD events for reasons of critical selective pressure; organisms may need to overcome genomic incompatibility through genomic changes. It is worth noting that imperfect genome assembly and annotation may also be one of the reasons for “gene loss,” especially on polyploids that have recently experienced WGD events (Jiang et al., 2016).
Interestingly, deletions and mutations were also observed in the coding regions of most Sox genes, indicating that WGD events could lead to genomic sequence diversity and that genomic alterations were part of adapting to polyploidization. Although 4nRCC was derived from a specific WGD event of 2nRCC induced by distant hybridization, SNP loci derived from the paternal parent were also detected in 4nRCC. In previous studies, chimeric SNP loci (or chimeric genes) were also detected in other hybrid lineage, which might produce structural changes by affecting normal transcriptional processing (Liu et al., 2016; Wu et al., 2020). Hence, we speculated that the expansion of diversity in the Sox gene family further revealed that distant hybridization as a catalyst accelerated the process of polyploidization and speciation, and these processes in polyploid formation provided more insight into the origins of natural and synthetic polyploid vertebrates.
Conclusion
This study revealed an important expansion of the Sox gene family in Cyprinidae fishes with different ploidy levels and represented a comparative analysis of natural and synthetic polyploids. In this study, a total of 26, 47, and 92 putative Sox genes were identified in the BSB, 2nRCC, and 4nRCC genomes, respectively. Comparative analyses revealed that the Sox gene family had undergone duplication in Cyprinidae fishes following WGD events. Multiple alignment, phylogenetic, and conserved motif analyses revealed that most of the Sox genes were well conserved during evolution. Our data provided valuable information about genome duplication and new insights into the expansion and evolution of Sox genes in polyploid vertebrates after several rounds of WGD events; these findings also suggested that distant hybridization was an effective method to accelerate rates of speciation or evolution.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Fish treatments were carried out according to the regulations for protected wildlife and the Administration of Affairs Concerning Animal Experimentation and approved by the Science and Technology Bureau of China. Approval from the Department of Wildlife Administration was not required for the experiments conducted in this manuscript.
Author Contributions
SL, QQ, and XH designed the study. XH provided the preliminary data that supported this study and wrote the manuscript. CW and KG performed the bioinformatics analysis. XH, CW, and KG participated in phylogenetic analysis and discussions. QC, HQ, CZ, and TY participated in the sequence alignment. LY, WF, and YW prepared the chromosome spreads. SL, QQ, and CW provided expert comments. QG also participated in phylogenetic analysis and reconstructed the phylogenetic tree. All authors read and approved the final manuscript.
Funding
This research was financially supported by grants from the National Natural Science Foundation of China (Grant Nos. 31430088, 31730098, and U19A2040), the earmarked fund for China Agriculture Research System (Grant No. CARS-45), the Key Research and Development Program of Hunan Province (Grant No. 2018NK2072), Hunan Provincial Natural Science and Technology Major Project (Grant No. 2017NK1031), and the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (Grant No. 20134486).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shi Wang, Yuwei Zhou, and Liu Cao for their advice on bioinformatics analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00804/full#supplementary-material
TABLE S1 | PCR primers designed based on the putative CDs of the goldfish Sox genes.
TABLE S2 | Inventory of the Sox genes in BSB.
TABLE S3 | Inventory of the Sox genes in 2nRCC.
TABLE S4 | Inventory of the Sox genes in 4nRCC.
Abbreviations
2nRCC, Carassius auratus red var.; 4nRCC, autotetraploid Carassius auratus; BSB, Megalobrama amblycephala; CDs, coding sequences; SNP, single nucleotide polymorphism.
Footnotes
- ^ http://asia.ensembl.org/index.html
- ^ www.biotek.com
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://alternate.meme-suite.org/
- ^ http://www.ncbi.nlm.nih.gov
References
Axel Meyer, M. S. (1999). Gene and genome duplications in vertebrates_ the one-tofour (to eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11, 699–704. doi: 10.1016/s0955-0674(99)00039-3
Chen, Z., Omori, Y., Koren, S., Shirokiya, T., Kuroda, T., and Miyamoto, A. (2019). De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci Adv. 5:eaav0547. doi: 10.1126/sciadv.aav0547
Cheng, P., Huang, Y., Du, H., Li, C., Lv, Y., Ruan, R., et al. (2019). Draft genome and complete hox-cluster characterization of the sterlet (Acipenser ruthenus). Front. Genet. 10:776. doi: 10.3389/fgene.2019.00776
Chiang, E. F., Pai, C. I., Wyatt, M., Yan, Y. L., Postlethwait, J., and Chung, B. (2001). Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 231, 149–163. doi: 10.1006/dbio.2000.0129
Cossins, A. R., and Crawford, D. L. (2005). Fish as models for environmental genomics. Nat. Rev. Genet. 6, 324–333. doi: 10.1038/nrg1590
Crow, K. D., Smith, C. D., Cheng, J. F., Wagner, G. P., and Amemiya, C. T. (2012). An independent genome duplication inferred from Hox paralogs in the American paddlefish–a representative basal ray-finned fish and important comparative reference. Genome Biol. Evol. 4, 937–953. doi: 10.1093/gbe/evs067
Cui, J., Shen, X., Zhao, H., and Nagahama, Y. (2011). Genome-wide analysis of Sox genes in Medaka (Oryzias latipes) and their expression pattern in embryonic development. Cytogenet. Genome Res. 134, 283–294. doi: 10.1159/000329480
David, L., Blum, S., Feldman, M. W., Lavi, U., and Hillel, J. (2003). Recent duplication of the common carp (Cyprinus carpio L.) genome as revealed by analyses of microsatellite loci. Mol. Biol. Evol. 20, 1425–1434. doi: 10.1093/molbev/msg173
Fortna, A., Kim, Y., MacLaren, E., Marshall, K., Hahn, G., Meltesen, L., et al. (2004). Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2:E207. doi: 10.1371/journal.pbio.0020207
Guo, B., Tong, C., and He, S. (2009). Sox genes evolution in closely related young tetraploid cyprinid fishes and their diploid relative. Gene 439, 102–112. doi: 10.1016/j.gene.2009.02.016
Han, Z., Liu, Y., Deng, X., Liu, D., Liu, Y., Hu, Y., et al. (2019). Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 20:101. doi: 10.1186/s12864-019-5455-1
Hett, A. K., and Ludwig, A. (2005). SRY-related (Sox) genes in the genome of European Atlantic sturgeon (Acipenser sturio). Genome 48, 181–186. doi: 10.1139/g04-112
Hu, F., Fan, J., Qin, Q., Huo, Y., Wang, Y., and Wu, C. (2019). The sterility of allotriploid fish and fertility of female autotriploid fish. Front. Genet. 10:377. doi: 10.3389/fgene.2019.00377
Huang, X., Qin, Q., Gong, K., Wu, C., Zhou, Y., Chen, Q., et al. (2020). Comparative analyses of the Sox9a-Amh-Cyp19a1a regulatory Cascade in Autotetraploid fish and its diploid parent. BMC Genet. 21:35. doi: 10.1186/s12863-020-00840-8
Jaillon, O., Aury, J., Brunet, F., Petit, J. R., Stangethomann, N., Mauceli, E., et al. (2004). Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957. doi: 10.1038/nature03025
Jenczewski, E., and Alix, K. (2004). From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23, 21–45. doi: 10.1080/07352680490273239
Jiang, L., Zhang, S., Dong, C., Chen, B., Feng, J., Peng, W., et al. (2016). Genome-wide identification, phylogeny, and expression of fibroblast growth genes in common carp. Gene 578, 225–231. doi: 10.1016/j.gene.2015.12.027
Jiang, T., Hou, C. C., She, Z. Y., and Yang, W. X. (2013). The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol. Biol. Rep. 40, 2187–2194. doi: 10.1007/s11033-012-2279-3
Julca, I., Marcethouben, M., Vargas, P., and Gabaldón, T. (2018). Phylogenomics of the olive tree (Olea europaea) reveals the relative contribution of ancient allo- and autopolyploidization events. BMC Biol. 16:15. doi: 10.1186/s12915-018-0482-y
Kondrashov, F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279, 5048–5057. doi: 10.1098/rspb.2012.1108
Larhammar Dan, R. C. (1994). Molecular genetic aspects of tetraploidy in the common carp Cyprinus carpio.pdf. Mol. Phylogenet. Evol. 3, 59–68. doi: 10.1006/mpev.1994.1007
Li, X. Y., Zhang, X. J., Li, Z., Hong, W., Liu, W., Zhang, J., et al. (2014). Evolutionary history of two divergent Dmrt1 genes reveals two rounds of polyploidy origins in gibel carp. Mol. Phylogenet. Evol. 78, 96–104. doi: 10.1016/j.ympev.2014.05.005
Liu, S., Li, Q., and Liu, Z. (2013). Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PLoS One 8:e63895. doi: 10.1371/journal.pone.0063895
Liu, S., Luo, J., Chai, J., Ren, L., Zhou, Y., and Huang, F. (2016). Genomic incompatibilities in the diploid and tetraploid offspring of the goldfish x common carp cross. Proc. Natl. Acad. Sci. U.S.A. 113, 1327–1332. doi: 10.1073/pnas.1512955113
Liu, S., Qin, Q., Xiao, J., Lu, W., Shen, J., and Li, W. (2007). The formation of the polyploid hybrids from different subfamily fish crossings and its evolutionary significance. Genetics 176, 1023–1034. doi: 10.1534/genetics.107.071373
Luo, K., Wang, S., Fu, Y., Zhou, P., Huang, X., Gu, Q., et al. (2019). Rapid genomic DNA variation in newly hybridized carp lineages derived from Cyprinus carpio (female symbol) x Megalobrama amblycephala (male symbol). BMC Genet. 20:87. doi: 10.1186/s12863-019-0784-2
Ma, W., Zhu, Z. H., Bi, X. Y., Murphy, R. W., Wang, S. Y., Gao, Y., et al. (2014). Allopolyploidization is not so simple: evidence from the origin of the tribe Cyprinini (Teleostei: Cypriniformes). Curr. Mol. Med. 14, 1331–1338. doi: 10.2174/1566524014666141203101543
Meyer, A., and Van de Peer, Y. (2005). From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27, 937–945. doi: 10.1002/bies.20293
Moghadam, H. K., Ferguson, M. M., and Danzmann, R. G. (2005). Evidence for Hox gene duplication in rainbow trout (Oncorhynchus mykiss): a tetraploid model species. J. Mol. Evol. 61, 804–818. doi: 10.1007/s00239-004-0230-5
Ohno, S., Muramoto, J., Christian, L., and Atkin, N. B. (1967). Diploid-tetraploid relationship among old-world members of the fish family Cyprinidae. Chromosoma 23, 1–9. doi: 10.1007/bf00293307
Omori, Y., and Kon, T. (2019). Goldfish: an old and new model system to study vertebrate development, evolution and human disease. J. Biochem. 165, 209–218. doi: 10.1093/jb/mvy076
Parisod, C., Holderegger, R., and Brochmann, C. (2010). Evolutionary consequences of autopolyploidy. New Phytol. 186, 5–17. doi: 10.1111/j.1469-8137.2009.03142.x
Qin, Q., Huo, Y., Liu, Q., Wang, C., Zhou, Y., and Liu, S. (2018). Induced gynogenesis in autotetraploids derived from Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). Aquaculture 495, 710–714. doi: 10.1016/j.aquaculture.2018.06.028
Qin, Q., Lai, Z., Cao, L., Xiao, Q., Wang, Y., and Liu, S. (2016). Rapid genomic changes in allopolyploids of Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). Sci. Rep. 6:34417.
Qin, Q., Wang, Y., Wang, J., Dai, J., Xiao, J., Hu, F., et al. (2014). The autotetraploid fish derived from hybridization of Carassius auratus red var. (female) x Megalobrama amblycephala (male). Biol. Reprod. 91:93.
Qin, Q. B., Liu, Q. W., Zhou, Y. W., Wang, C. Q., Qin, H., Zhao, C., et al. (2018). Differential expression of HPG-axis genes in autotetraploids derived from red crucian carp Carassius auratus red var., female symbol x blunt snout bream Megalobrama amblycephala, male symbol. J. Fish Biol. 93, 1082–1089. doi: 10.1111/jfb.13818
Ren, L., Li, W., Qin, Q., Dai, H., Han, F., and Xiao, J. (2019). The subgenomes show asymmetric expression of alleles in hybrid lineages of Megalobrama amblycephala x Culter alburnus. Genome Res. 29, 1805–1815. doi: 10.1101/gr.249805.119
Song, C., Liu, S., Xiao, J., He, W., Zhou, Y., Qin, Q., et al. (2012). Polyploid organisms. Sci. China Life Sci. 55, 301–311.
Van de Peer, Y., Maere, S., and Meyer, A. (2009). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732. doi: 10.1038/nrg2600
Venkatesh, B. (2003). Evolution and diversity of fish genomes. Curr. Opin. Genet. Dev. 13, 588–592. doi: 10.1016/j.gde.2003.09.001
Voldoire, E., Brunet, F., Naville, M., Volff, J. N., and Galiana, D. (2017). Expansion by whole genome duplication and evolution of the sox gene family in teleost fish. PLoS One 12:e0180936. doi: 10.1371/journal.pone.0180936
Wang, J. T., Li, J. T., Zhang, X. F., and Sun, X. W. (2012). Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genomics 13:96. doi: 10.1186/1471-2164-13-96
Wang, S., Tang, C., Tao, M., Qin, Q., Zhang, C., and Luo, K. (2018). Establishment and application of distant hybridization technology in fish. Sci. China Life Sci. 62, 22–45. doi: 10.1007/s11427-018-9408-x
Wang, Y. D., Qin, Q. B., Yang, R., Sun, W. Z., Liu, Q. W., Huo, Y. Y., et al. (2017). Hox genes reveal genomic DNA variation in tetraploid hybrids derived from Carassius auratus red var. (female) x Megalobrama amblycephala (male). BMC Genet. 18:86. doi: 10.1186/s12863-017-0550-2
Wei, L., Yang, C., Tao, W., and Wang, D. (2016). Genome-wide identification and transcriptome-based expression profiling of the sox gene family in the nile tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 17:270. doi: 10.3390/ijms17030270
Wu, C., Huang, X., Chen, Q., Hu, F., Zhou, L., Gong, K., et al. (2020). The formation of a new type of hybrid culter derived from a hybrid lineage of Megalobrama amblycephala (♀) × Culter alburnus (♂). Aquaculture 525:735328. doi: 10.1016/j.aquaculture.2020.735328
Xu, P., Xu, J., Liu, G., Chen, L., Zhou, Z., and Peng, W. (2019). The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat. Commun. 10:4625.
Yokoi, H., Kobayashi, T., Tanaka, M., Nagahama, Y., Wakamatsu, Y., Takeda, H., et al. (2002). Sox9 in a teleost fish, medaka (Oryzias latipes): evidence for diversified function of Sox9 in gonad differentiation. Mol. Reprod. Dev. 63, 5–16. doi: 10.1002/mrd.10169
Yu, H., Du, X., Li, X., Qu, J., Zhu, H., Zhang, Q., et al. (2018). Genome-wide identification and transcriptome-based expression analysis of sox gene family in the Japanese flounder Paralichthys olivaceus. J. Oceanol. Limnol. 36, 1731–1745. doi: 10.1007/s00343-018-7216-4
Yu, J., Zhang, L., Li, Y., Li, R., Zhang, M., Li, W., et al. (2017). Genome-wide identification and expression profiling of the SOX gene family in a bivalve mollusc Patinopecten yessoensis. Gene 627, 530–537. doi: 10.1016/j.gene.2017.07.013
Zhang, S., Chen, X., Wang, M., Zhang, W., Pan, J., and Qin, Q. (2018). Genome-wide identification, phylogeny and expressional profile of the Sox gene family in channel catfish (Ictalurus punctatus). Comp. Biochem. Physiol. D Genomics Proteomics 28, 17–26. doi: 10.1016/j.cbd.2018.03.001
Zhao, J., Zhai, Z., Li, Y., Geng, S., Song, G., Guan, J., et al. (2018). Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. Plant Sci. 9:1282. doi: 10.3389/fpls.2018.01282
Keywords: Sox gene family, whole genome duplication, autotetraploid, polyploidization, hybridization
Citation: Huang X, Wu C, Gong K, Chen Q, Gu Q, Qin H, Zhao C, Yu T, Yang L, Fu W, Wang Y, Qin Q and Liu S (2020) Sox Gene Family Revealed Genetic Variations in Autotetraploid Carassius auratus. Front. Genet. 11:804. doi: 10.3389/fgene.2020.00804
Received: 22 April 2020; Accepted: 06 July 2020;
Published: 28 July 2020.
Edited by:
Shu-Hong Zhao, Huazhong Agricultural University, ChinaReviewed by:
Chuanju Dong, Henan Normal University, ChinaJie Mei, Huazhong Agricultural University, China
Copyright © 2020 Huang, Wu, Gong, Chen, Gu, Qin, Zhao, Yu, Yang, Fu, Wang, Qin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinbo Qin, MzAwNTU3NzZAcXEuY29t; Shaojun Liu, bHNqQGh1bm51LmVkdS5jbg==
†These authors have contributed equally to this work
 Xu Huang
Xu Huang Chang Wu†
Chang Wu† Wen Fu
Wen Fu Yude Wang
Yude Wang Shaojun Liu
Shaojun Liu