
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 26 June 2020
Sec. Evolutionary and Genomic Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00661
This article is part of the Research Topic Genomics and Effectomics of Filamentous Plant Pathogens View all 20 articles
Filamentous pathogens, such as phytopathogenic oomycetes and fungi, secrete a remarkable diversity of apoplastic effector proteins to facilitate infection, many of which are able to induce cell death in plants. Over the past decades, over 177 apoplastic cell death-inducing proteins (CDIPs) have been identified in filamentous oomycetes and fungi. An emerging number of studies have demonstrated the role of many apoplastic CDIPs as essential virulence factors. At the same time, apoplastic CDIPs have been documented to be recognized by plant cells as pathogen-associated molecular patterns (PAMPs). The recent findings of extracellular recognition of apoplastic CDIPs by plant leucine-rich repeat-receptor-like proteins (LRR-RLPs) have greatly advanced our understanding of how plants detect them and mount a defense response. This review summarizes the latest advances in identifying apoplastic CDIPs of plant pathogenic oomycetes and fungi, and our current understanding of the dual roles of apoplastic CDIPs in plant-filamentous pathogen interactions.
Filamentous pathogens, such as oomycetes and fungi, are the causal agents of many of the world’s most serious plant diseases, causing extensive annual yield losses of crops worldwide (Giraldo and Valent, 2013; Sánchez-Vallet et al., 2018). Pathogenic oomycetes and fungi secrete a complex repertoire of effector proteins to establish successful interactions with host plants. These oomycete- or fungi-secreted effector proteins may function in the apoplast as well as within plant cells to interfere with host defense by a variety of mechanisms (Dou and Zhou, 2012; Lo Presti and Kahmann, 2017).
Studies from diverse filamentous pathogens have shown that many oomycete or fungal effector proteins possess the ability to induce cell death in plants (Gijzen and Nürnberger, 2006), including avirulence (AVR) proteins which trigger hypersensitive response (HR) upon recognition by cognate resistance (R) proteins (Kamoun, 2006; Ellis et al., 2009), nuclear-localized Crinkling and Necrosis proteins (CRNs) and nucleo-cytoplasmic RxLR proteins with capacity to induce plant cell death (Torto et al., 2003; Amaro et al., 2017), and extracellular cell death-inducing proteins (CDIPs) that function in the plant apoplast (Guo et al., 2019). The role of AVR proteins, CRN and RxLR effectors involved in the interactions of plants with filamentous pathogens have been reviewed extensively (Ellis et al., 2009; Giraldo and Valent, 2013; Amaro et al., 2017; Lo Presti and Kahmann, 2017). Here we focus this review on apoplastic CDIPs of filamentous plant pathogens and their roles in plant-pathogen interactions.
Because cell death plays an important role in the interactions of plants with pathogens, there has been a long-standing interest in the characterization of pathogenic molecules which are able to induce plant cell death (Gijzen and Nürnberger, 2006). Since the characterization of elicitins, a family of conserved small secreted proteins from oomycetes that induce necrosis in Nicotiana species, in the 1980s (Billard et al., 1988; Huet and Pernollet, 1989; Ricci et al., 1989), a large number of apoplastic CDIPs have been identified in oomycete and fungal plant pathogens (Tables 1, 2). These apoplastic CDIPs induce plant cell death in a non-race- or non-species-specific manner, and were initially defined or considered as “elicitors” or “toxins” (Gijzen and Nürnberger, 2006; Derevnina et al., 2016). The role of apoplastic CDIPs in the interactions of plants with filamentous pathogens has been controversial for a long time.
Over the past decades, tremendous progress has been made in understanding the biological functions of apoplastic CDIPs in filamentous oomycetes and fungi, such as their contribution to pathogen virulence and being recognized by plant cells. These findings have greatly enriched our knowledge on the roles of apoplastic CDIPs as virulence factors and how plants detect them and mount a defense response. In this review, we summarize the latest advances in identifying apoplastic CDIP effectors in plant pathogenic oomycetes and fungi, and our current understanding of the dual roles of apoplastic CDIPs in plant-filamentous pathogen interactions.
The purification and identification of extracellular proteins from phytopathogenic oomycetes that induce plant cell death began in the 1980s. Pernollet and colleagues purified three extracellular proteins, namely capsicein, cinnamomin and cryptogein, in culture filtrates of Phytophthora capsici, P. cinnamomi and P. cryptogea, respectively (Billard et al., 1988; Huet and Pernollet, 1989; Ricci et al., 1989). Determination of the amino acid sequences of these proteins led to definition of a novel protein family, called elicitins (Derevnina et al., 2016). Over the past decades, building on advances in molecular biology and genome sequencing, significant progress has been made in identifying extracellular proteins from phytopathogenic oomycetes that induce cell death in plants. To date, over 62 apoplastic CDIPs have been identified in 16 oomycete species (Table 1). While a few identified oomycete apoplastic CDIPs belong to PcF toxin, pectate lyase and glycoside hydrolase (GH) families or with no conserved domains, the majority are elicitins or Nep1 (necrosis- and ethylene-inducing peptide 1)-like proteins (NLPs) (Tables 1, 3).
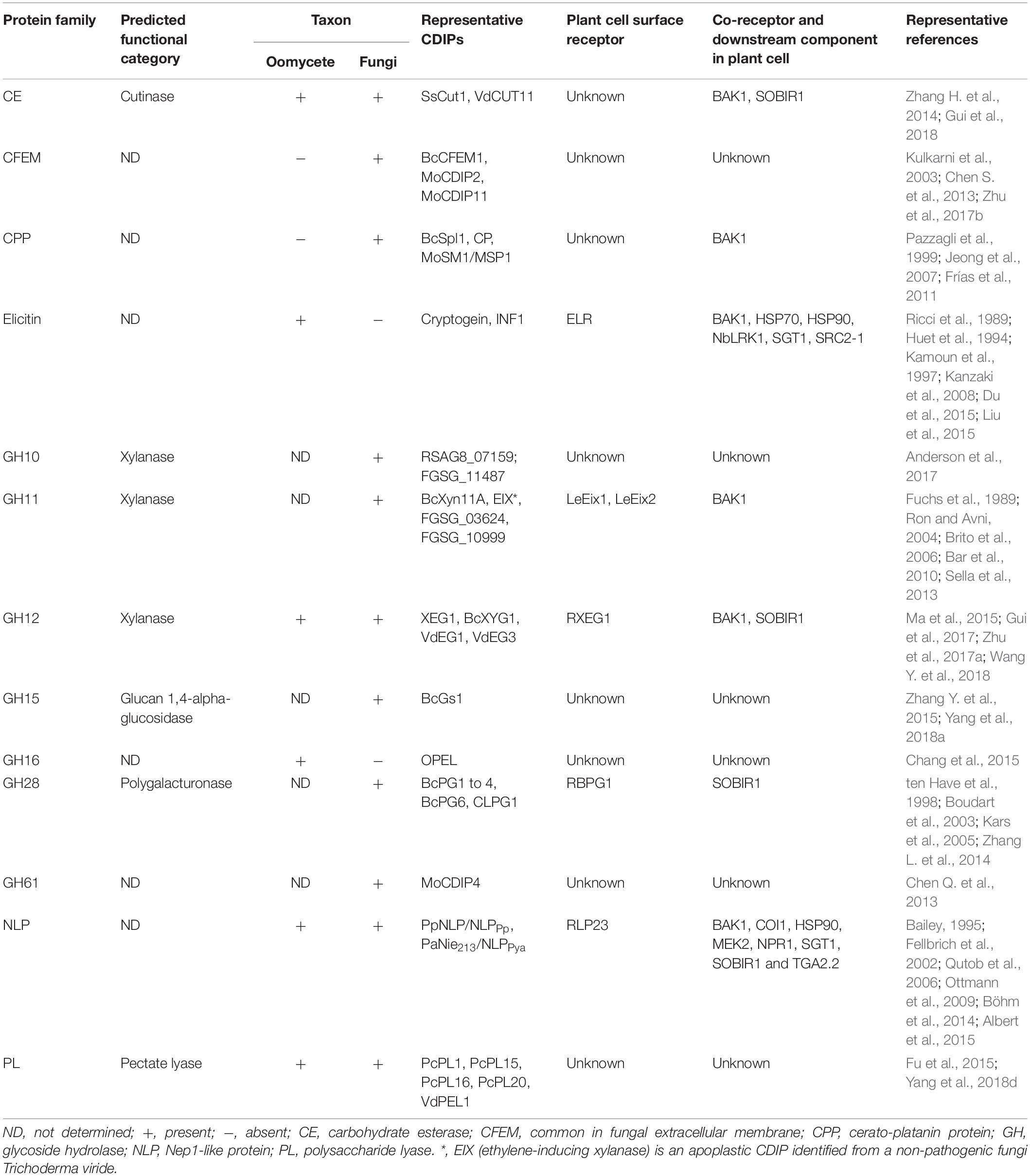
Table 3. Major protein families of apoplastic cell death-inducing proteins identified in oomycete and fungal plant pathogens.
Similarly, the availability of genome sequences of fungi has led to a rapid identification of apoplastic CIPDs from phytopathogenic fungi. To date, over 115 apoplastic CDIPs have been identified in 27 fungal species, including biotrophic, hemibiotrophic and necrotrophic pathogens (Table 2). The identified fungi apoplastic CDIPs include CFEM (common in fungal extracellular membrane)-containing proteins, cerato-platanin proteins (CPPs), GHs, NLPs, cutinases, and pectate lyases, and some proteins with other domains or with no conserved domains (Tables 2, 3). Among the identified oomycete and fungus apoplastic CDIPs, GHs, NLPs, Pectate lyases, and VmE02 homologues are widely distributed across oomycetes and fungi (Tables 1–3). On the other hand, many CDIPs are oomycete-specific or fungi-specific. For example, elicitins and elicitin-like proteins are unique to oomycetes, whereas CFEM-containing proteins and CPPs are unique to fungi (Table 3).
The CFEM domain, which contains eight conserved cysteines, is unique to fungi (Kulkarni et al., 2003; Zhang Z. N. et al., 2015). CFEM was first identified in a M. oryzae MAC1-interacting protein, ACI1 (Kulkarni et al., 2003). CFEM containing proteins are widely distributed in fungi (Zhang Z. N. et al., 2015). While some of CFEM containing proteins have been identified to have prominent roles in pathogenicity or virulence, such as MoPth11 and its ortholog FGRRES_16221 in Magnaporthe oryzae and Fusarium graminearum, respectively (Kou et al., 2017; Dilks et al., 2019), and CgCcw14, CgMam3 in Candida glabrata (Srivastava et al., 2014), several CFEM containing proteins have been identified to possess cell death-inducing activity.
The CFEM-containing protein BcCFEM1 from Botrytis cinerea, contains a CFEM domain at the N-terminus, and a glycosylphosphatidylinositol (GPI) anchored site at the C-terminus. BcCFEM1 was induced and expressed at high levels during early stages of infection on bean leaves. Targeted disruption of the BcCFEM1 gene reduced virulence, conidiation and stress tolerance in B. cinerea. Transient expression of BcCFEM1 in Nicotiana benthamiana leaves triggered obvious chlorosis (Zhu et al., 2017b), suggesting the involvement of BcCFEM1 in eliciting plant responses.
The M. oryzae genome harbors 19 proteins with the CFEM domain (Kou et al., 2017). Two CFEM-containing proteins MoCDIP2 and MoCDIP11 have been identified to induce cell in the non-host N. benthamiana and host rice cells (Chen S. et al., 2013; Guo et al., 2019). MoCDIP2 contains a CFEM domain at the N-terminus, and a GPI-anchored site at the C-terminus, whereas MoCDIP11 contains only a CFEM domain at the N-terminus. Transient expression assays in rice protoplasts or N. benthamiana leaves revealed that the signal peptides that led the secretion of proteins, were required for cell death inducing activity of MoCDIP2 and MoCDIP11, indicating that both two effectors function in the apoplast (Chen S. et al., 2013; Guo et al., 2019).
CPPs are small secreted cysteine-rich proteins that widely occur in filamentous fungi but not in bacteria, oomycetes, plants, or animals (Chen H. et al., 2013). CPPs have many aspects in common with expansins. Structural analysis revealed that CPPs have a double ψβ-barrel similar to the D1 domain of expansins (de Oliveira et al., 2011). Moreover, similar to expansins, CPPs possess properties to weaken cellulose (de O Barsottini et al., 2013; Baccelli et al., 2014b). Many CPPs have been shown to function as virulence factors in fungi, and on the other hand are able to induce cell death and elicit defense response in plants (Pazzagli et al., 2014).
The first CPP family CIDP, CP, was identified from the culture filtrates of Ceratocystis fimbriata f. sp. platani, the causal agent of the plane canker stain (Pazzagli et al., 1999). CP induced cell death in host and non-host plants, activated phytoalexin synthesis, pathogenesis-related (PR) gene expression, and mitogen-activated protein kinase (MAPK) phosphorylation, and triggered salicylic acid (SA) and ethylene (ET)-signaling pathways (Pazzagli et al., 1999; Scala et al., 2004; Baccelli et al., 2014a; Luti et al., 2016).
Till now, in addition to CP, 11 more CPP family CIDPs have been identified from different fungal pathogens (Table 2), such as BcSpl1 in B. cinerea (Frías et al., 2011, 2013), EPL1 in Colletotrichum falcatum (Ashwin et al., 2017), CgCP1 in Colletotrichum gloeosporioides (Wang W. et al., 2018), Pop1 in Ceratocystis populicola (Comparini et al., 2009), FgCPP2 in F. graminearum (Quarantin et al., 2016, 2019), FocCP1 in Fusarium oxysporum (Li et al., 2019a; Liu et al., 2019), HaCPL2 in Heterobasidion annosum sensu stricto (Chen et al., 2015), MoSM1/MSP1 (Jeong et al., 2007; Yang et al., 2009; Wang et al., 2016; Hong et al., 2017) in M. oryzae, MpCP1 in Moniliophthora perniciosa (Zaparoli et al., 2009), SsCP1 in Sclerotinia sclerotiorum (Yang et al., 2018b), and VdCP1 in Verticillium dahliae (Zhang et al., 2017a). Similar to CP, these 11 CPP family CIDPs induced strong necrosis and elicited defense responses in plants. For example, BcSpl1 induced strong necrosis, electrolyte leakage, cytoplasm shrinkage, and PR gene expression in plants including tomato, tobacco and Arabidopsis. In addition, BcSpl1 elicited systemic acquired resistance (SAR) in tobacco against Pseudomonas syringae and B. cinerea (Frías et al., 2011). The CPP family CIDPs possess similar biological properties as that of CP. For example, Pop1 and VdCP1 possessed chitin-binding properties (Baccelli et al., 2014b; Zhang et al., 2017a), and Pop1 and FgCPP2 exhibited the ability of loosening cellulose substrates and enhancing fungal cellulase activity in an expansin-like manner as well as CP (Baccelli et al., 2014b; Quarantin et al., 2016, 2019).
Elicitins are a family of small, highly conserved proteins secreted by Phytophthora and Pythium species (Derevnina et al., 2016). The first three elicitins, cryptogein, cinnamomin and capsicein were identified from the culture filtrates of P. cryptogea, P. cinnamomi and P. capsici, respectively (Billard et al., 1988; Huet and Pernollet, 1989; Ricci et al., 1989). Sequence analysis revealed that these elicitins share a conserved domain of 98 amino acids, which contains six cysteine residues at conserved positions forming three disulphide bridges (Boissy et al., 1996). Further studies revealed that elicitins are encoded by complex gene families (Kamoun et al., 1993; Panabieres et al., 1995; Jiang et al., 2006). In addition, Phytophthora and Pythium genomes contain a number of elicitin-like proteins possessing diverse, shorter or longer elicitin domains (Jiang et al., 2006).
Over 22 elicitin and elicitin-like CDIPs that induce cell death in certain plant species such as Nicotiana species and some Brassicaceae cultivars have been identified from different Phytophthora pathogens (Table 1). Among the elicitin and elicitin-like CDIPs, β-cryptogein and INF1 have been widely studied for their roles in the interactions of Phytophthora pathogens with plants. Cryptogein induced necrosis on tobacco leaves, triggered SAR and enhanced disease resistance in tobacco. Upon the SAR induction, the expression of the plant extracellular S-like RNase NE gene and its RNase activity were highly up-regulated, indicating that NE possibly associated with the cryptogein-induced SAR (Galiana et al., 1997). Cryptogein promoted the movement of plant respiratory burst oxidase homologues (RBOHs) from the Golgi cisternae to the plasma membrane, which may play a fundamental role for ROS production (Noirot et al., 2014). Cryptogein induced production of ROS, which was differentially regulated by the sphingolipid long-chain bases (LCBs) and their phosphorylated derivatives (LCB-Ps) in tobacco cells (Coursol et al., 2015). In addition, cryptogein induced production of NO, partly dependent on the ROS-dependent pathway, indicating that the defense responses induced by cryptogein involving interaction of the NO and ROS signaling pathways (Kulik et al., 2015). INF1 induced cell death on tobacco and potato leaves with the necrotic activity higher than that of other a-elicitins, such as cacto, parasiticein, capsicein and cryptogein, but less than that of β-cryptogein (Huet et al., 1994). INF1 induced the expression of chitinase, β-1,3-glucanase, phenylalanine ammonia-lyase (PAL), and PR1 genes, and rapid accumulation of H2O2 in tobacco (Sasabe et al., 2000; Huitema et al., 2005). INF1 induced the expression of NbrbohB in N. benthamiana, and silencing of NbrbohA or NbrbohB led to a reduction and delay of the necrotic reaction triggered by INF1 (Yoshioka et al., 2003), suggesting that INF1-induced cell death was dependent on the ROS burst.
Elicitins are able to bind sterols, phospholipids or fatty acids, and transport them between biological membranes (Mikes et al., 1997). However, the lipid-binding ability did not influence elicitin-induced response in tobacco. Investigations based on the mutant proteins of cryptogein with limited abilities to bind sterols revealed that induction of ROS synthesis, cytosol acidification and cell death in tobacco cells were not correlated with the sterol-binding abilities of the cryptogein proteins (Dokládal et al., 2012; Ptáčková et al., 2015).
Elicitins do not induce cell death in tomato plants. However, elicitins could induce immune responses in tomato, and enhance plant resistance against Phytophthora spp., bacterial wilt disease, and powdery mildew (Picard et al., 2000; Kawamura et al., 2009; Starý et al., 2019). INF1 induced the expression of jasmonic acid (JA)-responsive PR-6, LeATL6 and LOX-E, and ET-responsive PR-2b and ERF2, but not SA-responsive PR-1a and PR-2a in tomato leaves (Kawamura et al., 2009). Consistently, Starý et al. (2019) showed that cryptogein induced defense responses without cell death in tomato through JA- and ET-signaling pathways, but not SA-signaling pathway. These results indicated that elicitins triggered different signaling pathways between tobacco and tomato.
NLP family proteins are characterized by the presence of a common NPP1 (necrosis-inducing Phytophthora protein) domain. This family is widely distributed across taxa including oomycetes, fungi, and bacteria (Gijzen and Nürnberger, 2006). The first NLP protein, namely Nep1, was identified from culture filtrates of F. oxysporum (Bailey, 1995), and has been shown to induce necrosis in plants, such as Erythroxylum coca, Theobroma cacao and Arabidopsis, with activating PR gene expression, ROS production and other general defense response (Bailey et al., 2005; Bae et al., 2006). To date, over 39 NLP family CDIPs have been identified in phytopathogenic oomycetes and fungi. Among them, 20 NLP family CDIPs were from phytopathogenic oomycetes (Table 1), and the remaining 19 NLP CDIPs were from phytopathogenic fungi (Table 2).
NLP family CDIPs have been confirmed to induce cell death in dicot but not monocot plants. Among the identified NLP family CDIPs, PpNLP/NLPPp, PsojNIP and PaNie213/NLPPya have been extensively studied in the context of inducing cell death and immune responses in dicot plants. PpNLP/NLPPp induced necrosis in Arabidopsis, and activated PR genes expression, ROS production, callose apposition (Fellbrich et al., 2002), as well as the posttranslational expression of mitogen-activated protein kinase and production of nitric oxide and phytoalexin camalexin, suggesting dual roles of PpNLP/NLPPp as both toxin-like virulence factors and plant innate immunity triggers (Qutob et al., 2006). Determining and modeling of the structures of PaNie213/NLPPya, PpNLP/NLPPp, and a PccNLP protein from the phytopathogenic bacterium Pectobacterium carotovorum, revealed that NLPs displayed identical toxin folds which contributed to host infection and plant defense gene activation, suggesting that a common fold of the cytolytic toxin is required for both pathogen virulence and plant immunity activation (Ottmann et al., 2009). Böhm et al. (2014) further identified and characterized a pattern of 20 amino acid residues (nlp20) of cytotoxic NLPs that triggered immunity associated plant defenses in certain dicot plants.
Based on the finding that NLPs display a striking similarity to cytolytic sphingomyelin-binding actinoporins (Ottmann et al., 2009), glycosylinositol phosphorylceramide (GIPC) sphingolipids in eudicot plants were further shown to be bound by NLPs. The inositol phosphorylceramide in GIPCs was covalently bound to glucuronic acid and variable terminal hexoses which were different between eudicots and monocots. Eudicot GPICs typically carried two terminal sugars (series A), while monocots GIPCs bore three terminal sugars (series B) (Cacas et al., 2013). The absence of series A GIPCs lead to insensitivity to NLPs of monocot plants. Consistently, Arabidopsis mutants with altered GIPC composition suffered less cell death than the wild type upon NLP infiltration (Lenarčič et al., 2017). These results thus explained host specificity of cell death induced by NLPs in eudicot plants (Van den Ackerveken, 2017).
Phytopathogenic oomycetes and fungi secrete a large amount of effector proteins related to plant cell wall degradation, such as enzymes to degrade cellulose, xylan, pectin, etc. (Kubicek et al., 2014). Certain cell wall-degrading enzymes (CWDEs) have been proved to be associated with pathogen virulence, and some induced cell death and trigged defense responses in plants. CWDE family CDIPs have been identified in carbohydrate esterase (CE5), glycoside hydrolase (GH) and polysaccharide lyase (PL) families (Tables 1–3).
With functions to degrade plant cuticle or suberin polymers, the CE family cutinases has been associated with important roles in filamentous pathogen-plant interactions. Two cutinase family CDIPs, SsCut1 and VdCUT11 have been identified in S. sclerotiorum and V. dahliae (Zhang H. et al., 2014; Gui et al., 2018), respectively. SsCut1 induced cell death in both dicot and monocot species and activated plant resistance against S. sclerotiorum, P. nicotianae and Phytophthora sojae (Zhang H. et al., 2014). Further, SsCut1-induced cell death along with stomatal closure, ROS burst and NO production, was suppressed by silencing of a C2H2-type zinc finger gene NbCZF1 in N. benthamiana, showing that SsCut1-triggered defense could be mediated by NbCZF1-ROS-NO pathway (Zhang et al., 2016). VdCUT11 induced cell death and defense responses in N. benthamiana, cotton, and tomato plants, and the enzymatic activity was required for its cell death-inducing activity (Gui et al., 2018).
GHs hydrolyze the glycosidic bond between carbohydrates or between a carbohydrate and a noncarbohydrate moiety through acid catalysis (Zhao et al., 2013). About 18 GH domain-containing CDIPs have been identified (Table 3). The GH family CDIPs induced cell death as well as defense responses in host and nonhost plants. For example, BcXyn11A induced ROS burst, electrolyte leakage, cytoplasm shrinkage and PR gene expression, and these effects were dependent on its short 25-residue peptide (Frías et al., 2019). BcXYG1 triggered pattern-triggered immunity (PTI) response and systemic resistance in bean (Zhu et al., 2017a). PsXEG1 induced disease resistance in N. benthamiana and soybean (Ma et al., 2015). BcGs1 induced systemic resistance in tobacco and tomato against B. cinerea, Phytophthora syringae and tobacco mosaic virus (Zhang Y. et al., 2015). Furthermore, BcGs1 triggered ROS burst, PAL and peroxidase (POD) enzyme activity, and lignin accumulation in tomato (Yang et al., 2018a). Many of the GH family CDIPs have been shown to possess hydrolase activity. While hydrolase activity of BcPG2 was required for its cell death induction (Kars et al., 2005), the enzymatic activity of most identified GH family CDIPs was not necessary for cell death inducing activity. For example, the cell death inducing activity of Xyn11A, PsXEG1, BcXYG1, VdEG1 and VdEG3 was independent of their enzymatic activity (Brito et al., 2006; Ma et al., 2015; Gui et al., 2017; Zhu et al., 2017a).
The PL family pectate lyases play a critical role in pectin degradation. Four pectate lyases PcPL1, PcPL15, PcPL16 and PcPL20 in P. capsici, and one pectate lyase VdPEL1 in V. dahliae have been identified to have cell death inducing activity in plants (Fu et al., 2015; Yang et al., 2018d). PcPL1, PcPL15, PcPL16 and PcPL20 were highly expressed in P. capsici during infection of pepper, and transient expression of the four PcPLs induced severe cell death in pepper leaves (Fu et al., 2015). VdPEL1 induced cell death in N. benthamiana, tomato, soybean and cotton, and triggered defense responses and systemic resistance to B. cinerea and V. dahliae in N. benthamiana and cotton plants. Furthermore, the enzymatic activity was found to be necessary for cell death-inducing activity (Yang et al., 2018d).
Besides, a SGNH hydrolase subfamily protein BcXyl1 was identified to induce cell death in N. benthamiana, tomato, soybean and cotton (Yang et al., 2018c). BcXyl1 exhibited xylanase activity, but its cell death inducing activity was independent of the enzymatic activity. BcXyl1 triggered plant PTI responses with a pattern of 26-amino acid peptide.
Besides the above-mentioned CDIP families, a number of apoplastic CDIPs contain other conserved domains or no conserved domain (Tables 1, 2), indicating the incredible diversity of apoplastic CDIPs secreted by oomycetes and fungi. On the other hand, many of these apoplastic CDIPs are widely distributed across microbial taxa or different pathogen species and the homologs can induce cell death and defense responses in different plant species, indicating that recognition of these proteins is evolutionarily conserved. For example, the Valsa mali small cysteine-rich protein VmE02 induces cell death in N. benthamiana, tomato, pepper, Arabidopsis and apple and enhances resistance in N. benthamiana against S. sclerotiorum and P. capsic (Nie J. et al., 2019). VmE02 is widely conserved across oomycete and fungal species, and the homologs can induce cell death in N. benthamiana. Similarly, the Colletotrichum orbiculare NIS1 and its homologs from Colletotrichum higginsianum and Fusarium virguliforme (Yoshino et al., 2012; Chang et al., 2016), and the Rhynchosporium commune RcCDI1 and its homologues from M. oryzae, Neurospora crassa and Zymoseptoria tritici (Franco-Orozco et al., 2017), induced cell death in N. benthamiana. These results clearly supported that, although the physiological properties remain unknown, these apoplastic CDIPs are recognized as conserved patterns that induce defense responses in plants.
Phytopathogenic oomycetes and fungi initially colonize in the plant apoplast or extracellular space, and subsequently penetrate host cells. Therefore, the apoplastic effectors secreted by oomycetes and fungi are likely the primary weapons of filamentous plant pathogens. Despite the diversity of lifestyles as biotrophic, hemibiotrophic or necrotrophic, filamentous oomycetes and fungi secrete a high number of apoplastic CDIPs. Biotrophic pathogens feed on living plant cells (Giraldo and Valent, 2013). Whether apoplastic CDIPs of biotrophic pathogens actually cause plant cell death or function as important virulence factors during a natural infection remain to be determined (Tables 1, 2). In contrast, necrotrophic pathogens thrive on dead host tissues and take advantage of CDIP-triggered plant cell death. For example, NLP-triggered necrosis could aid infection by necrotrophic pathogens (Van den Ackerveken, 2017). Hemibiotrophic pathogens combine a biotrophic phase in early stages with a necrotrophic phage during later infection stages. Many apoplastic CDIPs of hemibiotrophic pathogens have been found to be highly expressed at late infection stages, suggesting that they contribute to the necrotrophic growth of hemibiotrophic pathogens (Qutob et al., 2002; Kelley et al., 2010).
Functional analysis based on overexpression, deletion or silencing of genes encoding apoplastic CDIPs have functionally proved many of them as important virulence factors in hemibiotrophic and necrotrophic pathogens (Tables 1, 2). Among the 62 identified oomycete apoplastic CDIPs, 12 have been proven to function as virulence factors that are required for pathogenicity or contribute to virulence. The P. cactorum SCR96 belongs to the PcF toxin family. Silencing of the scr96 gene in Phytophthora cactorum caused loss of pathogenicity on host plants, indicating that SCR96 is required for pathogenicity (Chen et al., 2016). In P. capsici, silencing of the genes encoding NLP-like proteins PcNLP2, PcNLP6 or PcNLP14, and the genes encoding pectate lyases PcPL1, PcPL15, PcPL16 or PcPL20 caused significantly reduced virulence on pepper (Feng et al., 2014; Fu et al., 2015), demonstrating the important roles of these apoplastic CDIPs in pathogen virulence. In Phytophthora palmivora, three fractions isolated from culture filtrates including high-molecular-weight glycoprotein, broad-molecular-weight glycoprotein and 42-kDa glycoprotein were observed to promote P. palmivora infection of rubber tree leaves (Pettongkhao and Churngchow, 2019). However, the virulence role of these three glycoproteins remains to be genetically confirmed. The P. sojae XEG1, a GH12 family CDIP, functions as a major virulence factor during infection. Both silencing and overexpression of the PsXEG1 gene in P. sojae severely impaired virulence (Ma et al., 2015). Interestingly, P. sojae also secretes a PsXLP1 (PsXEG1-like) apoplastic effector with a truncated GH12 domain functioning as a decoy to shield XEG1-mediated virulence (Ma et al., 2017).
Among the 115 identified fungi apoplastic CDIPs, 16 have been proven to be required for pathogenicity or contribute to virulence of both hemibiotrophic and necrotrophic pathogens. In B. cinerea, targeted deletion of the genes encoding BcCFEM1 (Zhu et al., 2017b), BcSpl1 (Frías et al., 2011), BcXyl1 (Yang et al., 2018c), or BcXyn11A (Brito et al., 2006) caused severely compromised virulence, suggesting that these CDIPs were important virulence factors of B. cinerea. Overexpression and deletion of BcXYG1 did not significantly affect B. cinerea infection on bean leaves. However, the BcXYG1 overexpression strains produced significantly earlier and more intense local necrosis, suggesting that BcXYG1 contributes to the establishment of infection in early stages (Zhu et al., 2017a). The C. gloeosporioides CgCP1 belongs to the CPP family. Knock-out of CgCP1 in C. gloeosporioides significantly reduced infection on rubber tree leaves (Wang W. et al., 2018). Similar to BcSpl1 and CgCP1, the CPP family CIDPs FocCP1, SsCP1 and VdCP1 function as important virulence factors. Deletion of FocCP1 in F. oxysporum (Liu et al., 2019), SsCP1 in S. sclerotiorum (Yang et al., 2018b), or VdCP1 in V. dahliae (Zhang et al., 2017a) caused significantly reduced virulence of pathogens on hosts banana, Arabidopsis or cotton, respectively. In M. oryzae, deletion of mohrip1, or mohrip2 remarkably compromised fungal virulence on rice (Nie H. et al., 2019; Nie H. Z. et al., 2019). MoHrip1 belongs to the Alt a 1 (AA1) family (Zhang et al., 2017b). Similarly, the V. dahliae PevD1, an AA1 family CDIP, is required for full virulence of V. dahliae on hosts (Zhang et al., 2019). In addition to VdCP1 and PevD1, VdCUT11, VdEG1, VdEG3, VdNLP1, VdNLP2, and VdPEL1 have been shown to play important roles in virulence of V. dahliae. Targeted deletion of VdCUT11 (Gui et al., 2018), VdEG1, VdEG3 (Gui et al., 2017), or VdPEL1 (Yang et al., 2018d) significantly compromised virulence of V. dahliae on cotton plants. Interestingly, while both VdNLP1 and VdNLP2 appear to be dispensable for V. dahliae infection in cotton plants (Zhou et al., 2012), VdNLP1 as well as VdNLP2 deletion strains were found to be significantly less pathogenic on tomato and Arabidopsis (Santhanam et al., 2013), demonstrating the functional diversification of the two NLP family CDIPs in virulence of V. dahliae.
Pathogenicity assays also revealed that many apoplastic CDIPs were not required for fungal pathogenicity or virulence. Targeted deletion of the genes encoding these proteins did not impair the virulence of pathogens (Table 2). One possibility why these apoplastic CDIPs were dispensable for virulence could be due to the functional redundancy with other apoplastic effectors (Yoshino et al., 2012; Guo et al., 2019).
In recent years, there have been breakthroughs in the identification of cell surface receptors recognizing apoplastic CDIPs. By screening T-DNA insertion mutants and natural accessions of Arabidopsis, Albert et al. (2015) identified T-DNA insertion alleles of RLP23 and Arabidopsis accessions carrying a frameshift mutation of RLP23 that were insensitive to the conserved nlp20 pattern found in most NLPs. RLP23 encodes a leucine-rich repeat-receptor-like protein (LRR-RLP), which binds extracellularly to nlp20, thereby mediating NLP-elicited immune response in Arabidopsis (Albert et al., 2015). Transgenic potato plants expressing RLP23 displayed enhanced resistance to oomycete and fungal pathogens, such as P. infestans and S. sclerotiorum (Albert et al., 2015), further supporting that RLP23 confers protection to oomycete and fungal pathogens. More recently, RXEG1 (Response to XEG1), an LRR-RLP that specifically recognizes the GH12 family CDIP XEG1, has been identified from N. benthamiana through a high-throughput virus-induced gene silencing (VIGS) screen (Wang Y. et al., 2018). RXEG1 interacts with XEG1 by the extracellular LRR domain in the apoplast, and regulates XEG1-induced plant cell death and immune responses.
Using a genetic mapping approach, an ELR (elicitin response) gene has been identified from a population derived from the cross of Solanum microdontum genotype mcd360-1 (responds to INF1 with a cell death response) with S. microdontum ssp. gigantophyllum gig714-1 (does not respond to INF1) (Du et al., 2015). ELR encodes an LRR-RLP, and ELR mediates extracellular recognition of a broad range of elicitins exhibiting relatively low sequence similarity, suggesting that ELR recognizes elicitins most likely based on domain similarity but not a small conserved peptide. Moreover, cultivated potato transformed with the ELR gene exhibited enhanced resistance to Phytophthora infestans. Overall, the results suggested ELR as a potential cell surface receptor to mediate response to elicitins. However, the physical association of ELR with INF proteins remains unclear (Du et al., 2015). Interestingly, two intracellular proteins, a lectin-like receptor kinase (NbLRK) and a pepper calcium-binding protein (SRC2-1), have been shown to interact with P. infestans INF1 (Kanzaki et al., 2008) and P. capsici PcINF1 (Liu et al., 2015), and mediate P. infestans INF1 or PcINF1-induced cell death, respectively. These results correspond with the speculation that elicitins could be possibly transported into plant cells through clathrin-mediated endocytosis (Leborgne-Castel et al., 2008). Hence, it would appear that plants possess multiple mechanisms to recognize elicitins.
The three cell surface receptors RLP23, ELR and RXEG1 have extracellular LRRs but lack a cytoplasmic signaling domain. Two LRR receptor-like kinases (LRR-RLKs) SUPPRESSOR OF BIR1-1 (SOBIR1) and/or BRI1-ASSOCIATED KINASE-1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE-3 (BAK1/SERK3) were shown to be essential for RLP23, ELR or RXEG1-induced cell death and immune responses that act as co-receptors to transduce signals to downstream elements. RLP23 forms a complex with SOBIR1 and recruits BAK1/SERK3 into a tripartite complex upon ligand binding (Albert et al., 2015); ELR associates with BAK1/SERK3 and mediates recognition of diverse elicitins from Phytophthora species (Du et al., 2015); RXEG1 associates with BAK1/SERK3 and SOBIR1 to transmit the XEG1-induced defense signal (Wang Y. et al., 2018).
BAK1/SERK3 and SOBIR1 appear to be general and central regulators of plant cell death and defense response induced by diverse apoplastic CDIPs besides nlp20, INF1 and XEG1. In Arabidopsis, bak1 mutants showed a significantly reduced sensitivity to the CPP family CIDP BcSpl1, indicating that BAK1 plays an important role in the perception of BcSpl1 (Frías et al., 2011). In N. benthamiana, VIGS-mediated silencing of BAK1 or SOBIR1, resulted in significant reductions of cell death induced by a number of apoplastic CDIPs, including BcXYG1 (Zhu et al., 2017a), BcXyl1 (Yang et al., 2018c), RcCD1 (Franco-Orozco et al., 2017), VdCUT11 (Gui et al., 2018), VdEG1, VdEG3 (Gui et al., 2017), VmE02 (Nie J. et al., 2019), MgNLP, Zt9, Zt11 or Zt12 (Kettles et al., 2017). These results indicate that BAK1/SERK3 and SOBIR1 are required for apoplastic CDIP-induced cell death.
Several cytosolic, or nucleo-cytoplasmic regulators have been shown to be important components in apoplastic CDIP-induced cell death and defense signal transduction pathways downstream of the cell surface receptor complexes. The ubiquitin ligase–associated protein SGT1 functions as a conserved component in both PTI and effector-triggered immunity (ETI) pathways. Silencing of SGT1 in N. benthamiana suppressed cell death induced by P. capsici PcINF1 (Liu Z. Q. et al., 2016), INF1, INF2A (Huitema et al., 2005), PiNPP1.1, PsojNIP (Kanneganti et al., 2006), BcSpl1 (Frías et al., 2011), NIS1 (Yoshino et al., 2012), NcCDI1 (Franco-Orozco et al., 2017), or VmE02 (Nie J. et al., 2019). In addition, VIGS-mediated silencing analysis revealed that HSP70, HSP90 (Kanzaki et al., 2003; Kanneganti et al., 2006; Yoshino et al., 2012; Nie J. et al., 2019), COI1, MEK2, NPR1, TGA2.2 (Kanneganti et al., 2006), and Avr9/Cf-9-INDUCED F-BOX1 (ACIF1) (Li et al., 2019c) were required for cell death induced by certain apoplastic CDIPs.
An emerging number of studies have shown that many apoplastic CDIPs are required for pathogenicity, or contribute to the virulence of hemibiotrophic and necrotrophic pathogens, demonstrating the important role of these apoplastic CDIPs as essential virulence factors. On the contrary, apoplastic CDIPs were shown to elicit plant defense responses. Over the past decades, studies have documented that many apoplastic CDIPs are recognized by plants as PAMPs, being able to trigger PR gene expression, phytoalexin synthesis, MAPK phosphorylation, SA-, JA/ET-signaling pathways, as well as resistance against pathogens. The recent findings of extracellular recognition of NLPs, elicitins and XEG1 by plant RLPs, RLP23, ELR and RXEG1 (Albert et al., 2015; Du et al., 2015; Wang Y. et al., 2018), respectively, have greatly advanced our understanding of the roles of apoplastic CDIPs in plant-pathogen interactions. More importantly, the identification of receptors for apoplastic CDIPs provides valuable gene resources for engineering crops with broad and durable disease resistance (Albert et al., 2015; Du et al., 2015).
Currently, the majority of apoplastic CDIPs have been characterized based on demonstrations in dicot plants, especially based on transient expression in N. benthamiana, even many apoplastic CDIPs were identified from monocot pathogens. Particularly, recognition receptors and downstream components that have been identified remain restricted to dicot plants. Given the paramount importance of monocot cereal plants, such as rice, wheat and maize, as staple crops, it would be important to determine recognition of apoplastic CDIPs in monocot hosts, which could help engineer monocot cereal crops with broad-spectrum resistance against filamentous pathogens.
YL, YH, and SC wrote the manuscript. MQ, JC, XC, and XG contributed with comments and inputs. ZW and SC revised the manuscript with comments and inputs from all authors. All authors commented on the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31171808 and 31901960) and funding from the State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops (SKL2018006 and SKL2019005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Albert, I., Böhm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Amaro, T. M. M. M., Thilliez, G. J. A., Motion, G. B., and Huitema, E. (2017). A perspective on CRN proteins in the genomics age: evolution, classification, delivery and function revisited. Front. Plant Sci. 8:99. doi: 10.3389/fpls.2017.00099
Anderson, J. P., Sperschneider, J., Win, J., Kidd, B., Yoshida, K., Hane, J., et al. (2017). Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci. Rep. 7:10410. doi: 10.1038/s41598-017-10405-y
Arenas, Y. C., Kalkman, E. R., Schouten, A., Dieho, M., Vredenbregt, P., Uwumukiza, B., et al. (2010). Functional analysis and mode of action of phytotoxic Nep1-like proteins of Botrytis cinerea. Physiol. Mol. Plant Pathol. 74, 376–386. doi: 10.1016/j.pmpp.2010.06.003
Ashwin, N. M. R., Barnabas, L., Ramesh Sundar, A., Malathi, P., Viswanathan, R., Masi, A., et al. (2017). Comparative secretome analysis of Colletotrichum falcatum identifies a cerato-platanin protein (EPL1) as a potential pathogen-associated molecular pattern (PAMP) inducing systemic resistance in sugarcane. J. Proteomics 169, 2–20. doi: 10.1016/j.jprot.2017.05.020
Ashwin, N. M. R., Barnabas, L., Ramesh Sundar, A., Malathi, P., Viswanathan, R., Masi, A., et al. (2018). CfPDIP1, a novel secreted protein of Colletotrichum falcatum, elicits defense responses in sugarcane and triggers hypersensitive response in tobacco. Appl. Microbiol. Biotechnol. 102, 6001–6021. doi: 10.1007/s00253-018-9009-2
Baccelli, I., Lombardi, L., Luti, S., Bernardi, R., Picciarelli, P., Scala, A., et al. (2014a). Cerato-platanin induces resistance in Arabidopsis leaves through stomatal perception, overexpression of salicylic acid- and ethylene-signalling genes and camalexin biosynthesis. PLoS One 9:e100959. doi: 10.1371/journal.pone.0100959
Baccelli, I., Luti, S., Bernardi, R., Scala, A., and Pazzagli, L. (2014b). Cerato-platanin shows expansin-like activity on cellulosic materials. Appl. Microbiol. Biotechnol. 98, 175–184. doi: 10.1007/s00253-013-4822-0
Bae, H., Kim, M. S., Sicher, R. C., Bae, H. J., and Bailey, B. A. (2006). Necrosis-and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiol. 141, 1056–1067. doi: 10.1104/pp.106.076869
Bailey, B. A. (1995). Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology 85, 1250–1255. doi: 10.1094/Phyto-85-1250
Bailey, B. A., Apel-Birkhold, P. C., Akingbe, O. O., Ryan, J. L., O’Neill, N. R., and Anderson, J. D. (2000). Nep1 Protein from Fusarium oxysporum Enhances Biological Control of Opium Poppy by Pleospora papaveracea. Phytopathology 90, 812–818. doi: 10.1094/PHYTO.2000.90.8.812
Bailey, B., Apel-Birkhold, P. C., and Luster, D. G. (2002). Expression of NEP1 by Fusarium oxysporum f. sp. erythroxyli after gene replacement and overexpression using polyethylene glycol-mediated transformation. Phytopathology 92, 833–841. doi: 10.1094/PHYTO.2002.92.8.833
Bailey, B. A., Bae, H., Strem, M. D., De Mayolo, G. A., Guiltinan, M. J., Verica, J. A., et al. (2005). Developmental expression of stress response genes in Theobroma cacao leaves and their response to Nep1 treatment and a compatible infection by Phytophthora megakarya. Plant Physiol. Biochem. 43, 611–622. doi: 10.1016/j.plaphy.2005.04.006
Baillieul, F., de Ruffray, P., and Kauffmann, S. (2003). Molecular cloning and biological activity of α-, β-, and γ-megaspermin, three elicitins secreted by Phytophthora megasperma H20. Plant Physiol. 131, 155–166. doi: 10.1104/pp.012658
Baillieul, F., Genetet, I., Kopp, M., Saindrenan, P., Fritig, B., and Kauffmann, S. (1995). A new elicitor of the hypersensitive response in tobacco: a fungal glycoprotein elicits cell death, expression of defence genes, production of salicylic acid, and induction of systemic acquired resistance. Plant J. 8, 551–560. doi: 10.1046/j.1365-313X.1995.8040551.x
Bar, M., Sharfman, M., Ron, M., and Avni, A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63, 791–800. doi: 10.1111/j.1365-313X.2010.04282.x
Billard, V., Bruneteau, M., Bonnet, P., Ricci, P., Pernollet, J., Huet, J., et al. (1988). Chromatographic purification and characterization of elicitors of necrosis on tobacco produced by incompatible Phytophthora species. J. Chromatogr. A 440, 87–94. doi: 10.1016/s0021-9673(00)94513-8
Böhm, H., Albert, I., Oome, S., Raaymakers, T. M., Van den Ackerveken, G., and Nürnberger, T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10:e1004491. doi: 10.1371/journal.ppat.1004491
Boissy, G., de La Fortelle, E., Kahn, R., Huet, J. C., Bricogne, G., Pernollet, J. C., et al. (1996). Crystal structure of a fungal elicitor secreted by Phytophthora cryptogea, a member of a novel class of plant necrotic proteins. Structure 4, 1429–1439. doi: 10.1016/s0969-2126(96)00150-5
Boudart, G., Charpentier, M., Lafitte, C., Martinez, Y., Jauneau, A., Gaulin, E., et al. (2003). Elicitor activity of a fungal endopolygalacturonase in tobacco requires a functional catalytic site and cell wall localization. Plant Physiol. 131, 93–101. doi: 10.1104/pp.011585
Brito, N., Espino, J. J., and González, C. (2006). The endo-β-1, 4-xylanase Xyn11A is required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 19, 25–32. doi: 10.1094/MPMI-19-0025
Bu, B., Qiu, D., Zeng, H., Guo, L., Yuan, J., and Yang, X. (2014). A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 33, 461–470. doi: 10.1007/s00299-013-1546-7
Cacas, J. L., Bure, C., Furt, F., Maalouf, J. P., Badoc, A., Cluzet, S., et al. (2013). Biochemical survey of the polar head of plant glycosylinositolphosphoceramides unravels broad diversity. Phytochemistry 96, 191–200. doi: 10.1016/j.phytochem.2013.08.002
Capasso, R., Cristinzio, G., Di Maro, A., Ferranti, P., and Parente, A. (2001). Syringicin, a new α-elicitin from an isolate of Phytophthora syringae, pathogenic to citrus fruit. Phytochemistry 58, 257–262. doi: 10.1016/s0031-9422(01)00201-1
Capasso, R., Cristinzio, G., Evidente, A., Visca, C., Ferranti, P., Blanco, F. D. V., et al. (1999). Elicitin 172 from an isolate of Phytophthora nicotianae pathogenic to tomato. Phytochemistry 50, 703–709. doi: 10.1016/s0031-9422(98)00539-1
Capasso, R., Di Maro, A., Cristinzio, G., De Martino, A., Chambery, A., Daniele, A., et al. (2008). Isolation, characterization and structure-elicitor activity relationships of hibernalin and its two oxidized forms from Phytophthora hibernalis Carne 1925. J. Biochem. 143, 131–141. doi: 10.1093/jb/mvm201
Chang, H. X., Domier, L. L., Radwan, O., Yendrek, C. R., Hudson, M. E., and Hartman, G. L. (2016). Identification of multiple phytotoxins produced by Fusarium virguliforme Including a phytotoxic effector (FvNIS1) associated with sudden death syndrome foliar symptoms. Mol. Plant Microbe Interact. 29, 96–108. doi: 10.1094/MPMI-09-15-0219-R
Chang, Y. H., Yan, H. Z., and Liou, R. F. (2015). A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol. Plant Pathol. 16, 123–136. doi: 10.1111/mpp.12166
Chen, H., Kovalchuk, A., Keriö, S., and Asiegbu, F. O. (2013). Distribution and bioinformatic analysis of the cerato-platanin protein family in Dikarya. Mycologia 105, 1479–1488.
Chen, H., Quintana, J., Kovalchuk, A., Ubhayasekera, W., and Asiegbu, F. O. (2015). A cerato-platanin-like protein HaCPL2 from Heterobasidion annosum sensu stricto induces cell death in Nicotiana tabacum and Pinus sylvestris. Fungal Genet. Biol. 84, 41–51. doi: 10.1016/j.fgb.2015.09.007
Chen, M., Zeng, H., Qiu, D., Guo, L., Yang, X., Shi, H., et al. (2012). Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS One 7:e37654. doi: 10.1371/journal.pone.0037654
Chen, M., Zhang, C., Zi, Q., Qiu, D., Liu, W., and Zeng, H. (2014). A novel elicitor identified from Magnaporthe oryzae triggers defense responses in tobacco and rice. Plant Cell Rep. 33, 1865–1879. doi: 10.1007/s00299-014-1663-y
Chen, Q., Chen, Z., Lu, L., Jin, H., Sun, L., Yu, Q., et al. (2013). Interaction between abscisic acid and nitric oxide in PB90-induced catharanthine biosynthesis of catharanthus roseus cell suspension cultures. Biotechnol. Prog. 29, 994–1001. doi: 10.1002/btpr.1738
Chen, S., Songkumarn, P., Venu, R., Gowda, M., Bellizzi, M., Hu, J., et al. (2013). Identification and characterization of in planta–expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol. Plant Microbe Interact 26, 191–202.
Chen, X. R., Huang, S. X., Zhang, Y., Sheng, G. L., Li, Y. P., and Zhu, F. (2018). Identification and functional analysis of the NLP-encoding genes from the phytopathogenic oomycete Phytophthora capsici. Mol. Genet. Genomics 293, 931–943. doi: 10.1007/s00438-018-1432-7
Chen, X. R., Huang, S. X., Zhang, Y., Sheng, G. L., Zhang, B. Y., Li, Q. Y., et al. (2017). Transcription profiling and identification of infection-related genes in Phytophthora cactorum. Mol. Genet. Genomics 293, 541–555. doi: 10.1007/s00438-017-1400-7
Chen, X. R., Li, Y. P., Li, Q. Y., Xing, Y. P., Liu, B. B., Tong, Y. H., et al. (2016). SCR96, a small cysteine-rich secretory protein of Phytophthora cactorum, can trigger cell death in the Solanaceae and is important for pathogenicity and oxidative stress tolerance. Mol. Plant Pathol. 17, 577–587. doi: 10.1111/mpp.12303
Chen, X. R., Zhang, B. Y., Xing, Y. P., Li, Q. Y., Li, Y. P., Tong, Y. H., et al. (2014). Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC Genomics 15:980. doi: 10.1186/1471-2164-15-980
Churngchow, N., and Rattarasarn, M. (2000). The elicitin secreted by Phytophthora palmivora, a rubber tree pathogen. Phytochemistry 54, 33–38. doi: 10.1016/S0031-9422(99)00530-0
Cobos, R., Calvo-Pena, C., Alvarez-Perez, J. M., Ibanez, A., Diez-Galan, A., Gonzalez-Garcia, S., et al. (2019). Necrotic and cytolytic activity on grapevine leaves produced by Nep1-like proteins of Diplodia seriata. Front. Plant Sci. 10:1282. doi: 10.3389/fpls.2019.01282
Comparini, C., Carresi, L., Pagni, E., Sbrana, F., Sebastiani, F., Luchi, N., et al. (2009). New proteins orthologous to cerato-platanin in various Ceratocystis species and the purification and characterization of cerato-populin from Ceratocystis populicola. Appl. Microbiol. Biotechnol. 84, 309–322. doi: 10.1007/s00253-009-1998-4
Coursol, S., Fromentin, J., Noirot, E., Bričre, C., Robert, F., Morel, J., et al. (2015). Long-chain bases and their phosphorylated derivatives differentially regulate cryptogein-induced production of reactive oxygen species in tobacco (Nicotiana tabacum) BY-2 cells. New Phytol. 205, 1239–1249. doi: 10.1111/nph.13094
Dagvadorj, B., Ozketen, A. C., Andac, A., Duggan, C., Bozkurt, T. O., and Akkaya, M. S. (2017). A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Sci. Rep. 7:1141. doi: 10.1038/s41598-017-01100-z
de O Barsottini, M. R., de Oliveira, J. F., Adamoski, D., Teixeira, P. J. P. L., do Prado, P. F. V., Tiezzi, H. O., et al. (2013). Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization. Mol. Plant Microbe Interact. 26, 1281–1293. doi: 10.1094/MPMI-05-13-0148-R
de Oliveira, A. L., Gallo, M., Pazzagli, L., Benedetti, C. E., Cappugi, G., Scala, A., et al. (2011). The structure of the elicitor cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double ψβ-barrel fold and carbohydrate binding. J. Biol. Chem. 286, 17560–17568. doi: 10.1074/jbc.M111.223644
Derevnina, L., Dagdas, Y. F., De la Concepcion, J. C., Bialas, A., Kellner, R., Petre, B., et al. (2016). Nine things to know about elicitins. New Phytol. 212, 888–895. doi: 10.1111/nph.14137
Dilks, T., Halsey, K., De Vos, R. P., Hammond-Kosack, K. E., and Brown, N. A. (2019). Non-canonical fungal G-protein coupled receptors promote Fusarium head blight on wheat. PLoS Pathog. 15:e1007666. doi: 10.1371/journal.ppat.1007666
Dokládal, L., Oboril, M., Stejskal, K., Zdráhal, Z., Ptácková, N., Chaloupková, R., et al. (2012). Physiological and proteomic approaches to evaluate the role of sterol binding in elicitin-induced resistance. J. Exp. Bot. 63, 2203–2215. doi: 10.1093/jxb/err427
Dou, D., and Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Du, J., Verzaux, E., Chaparro-Garcia, A., Bijsterbosch, G., Keizer, L. C., Zhou, J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1:15034. doi: 10.1038/nplants.2015.34
Ellis, J. G., Rafiqi, M., Gan, P., Chakrabarti, A., and Dodds, P. N. (2009). Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant. Biol. 12, 399–405. doi: 10.1016/j.pbi.2009.05.004
Fang, A., Han, Y., Zhang, N., Zhang, M., Liu, L., Li, S., et al. (2016). Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens. Mol. Plant Microbe Interact. 29, 405–416. doi: 10.1094/MPMI-09-15-0200-R
Fang, Y. L., Peng, Y. L., and Fan, J. (2017). The Nep1-like protein family of Magnaporthe oryzae is dispensable for the infection of rice plants. Sci. Rep. 7:4372. doi: 10.1038/s41598-017-04430-0
Fellbrich, G., Romanski, A., Varet, A., Blume, B., Brunner, F., Engelhardt, S., et al. (2002). NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390. doi: 10.1046/j.1365-313x.2002.01454.x
Feng, B. Z., Zhu, X. P., Fu, L., Lv, R. F., Storey, D., Tooley, P., et al. (2014). Characterization of necrosis-inducing NLP proteins in Phytophthora capsici. BMC Plant Biol. 14:126. doi: 10.1186/1471-2229-14-126
Franco-Orozco, B., Berepiki, A., Ruiz, O., Gamble, L., Griffe, L. L., Wang, S., et al. (2017). A new proteinaceous pathogen-associated molecular pattern (PAMP) identified in Ascomycete fungi induces cell death in Solanaceae. New Phytol. 214, 1657–1672. doi: 10.1111/nph.14542
Frías, M., Brito, N., and Gonzalez, C. (2013). The Botrytis cinereal cerato-platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 14, 191–196. doi: 10.1111/j.1364-3703.2012.00842.x
Frías, M., Gonzalez, C., and Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. doi: 10.1111/j.1469-8137.2011.03802.x
Frías, M., Gonzalez, M., Gonzalez, C., and Brito, N. (2016). BcIEB1, a Botrytis cinerea secreted protein, elicits a defense response in plants. Plant Sci. 250, 115–124. doi: 10.1016/j.plantsci.2016.06.009
Frías, M., González, M., González, C., and Brito, N. (2019). A 25-residue peptide from Botrytis cinerea xylanase BcXyn11A elicits plant defenses. Front. Plant Sci. 10:474. doi: 10.3389/fpls.2019.00474
Fu, L., Zhu, C., Ding, X., Yang, X., Morris, P. F., Tyler, B. M., et al. (2015). Characterization of cell-death-inducing members of the pectate lyase gene family in Phytophthora capsici and their contributions to infection of pepper. Mol. Plant Microbe Interact. 28, 766–775. doi: 10.1094/MPMI-11-14-0352-R
Fuchs, Y., Saxena, A., Gamble, H. R., and Anderson, J. D. (1989). Ethylene biosynthesis-inducing protein from cellulysin is an endoxylanase. Plant Physiol. 89, 138–143. doi: 10.1104/pp.89.1.138
Galiana, E., Bonnet, P., Conrod, S., Keller, H., Panabičres, F., Ponchet, M., et al. (1997). RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiol. 115, 1557–1567. doi: 10.1104/pp.115.4.1557
Garcia, O., Macedo, J. A., Tiburcio, R., Zaparoli, G., Rincones, J., Bittencourt, L. M., et al. (2007). Characterization of necrosis and ethylene-inducing proteins (NEP) in the basidiomycete Moniliophthora perniciosa, the causal agent of witches’ broom in Theobroma cacao. Mycol. Res. 111, 443–455. doi: 10.1016/j.mycres.2007.01.017
Gijzen, M., and Nürnberger, T. (2006). Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67, 1800–1807.
Giraldo, M. C., and Valent, B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. doi: 10.1038/nrmicro3119
González, M., Brito, N., and Gonzalez, C. (2017). The Botrytis cinerea elicitor protein BcIEB1 interacts with the tobacco PR5-family protein osmotin and protects the fungus against its antifungal activity. New Phytol. 215, 397–410. doi: 10.1111/nph.14588
Gui, Y. J., Chen, J. Y., Zhang, D. D., Li, N. Y., Li, T. G., Zhang, W. Q., et al. (2017). Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 19, 1914–1932. doi: 10.1111/1462-2920.13695
Gui, Y. J., Zhang, W. Q., Zhang, D. D., Zhou, L., Short, D. P. G., Wang, J., et al. (2018). A Verticillium dahliae extracellular cutinase modulates plant immune responses. Mol. Plant Microbe Interact. 31, 260–273. doi: 10.1094/MPMI-06-17-0136-R
Guo, X., Zhong, D., Xie, W., He, Y., Zheng, Y., Lin, Y., et al. (2019). Functional identification of novel cell death-inducing effector proteins from Magnaporthe oryzae. Rice 12:59. doi: 10.1186/s12284-019-0312-z
Hong, Y., Yang, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2017). Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. Rep. 7:41037. doi: 10.1038/srep41037
Huet, J., Sallé-Tourne, M., and Pernollet, J. (1994). Amino acid sequence and toxicity of the alpha elicitin secreted with ubiquitin by Phytophthora infestans. Mol. Plant Microbe Interact. 7, 302–304. doi: 10.1094/mpmi-7-0302
Huet, J. C., Mansion, M., and Pernollet, J. C. (1993). Amino acid sequence of the α-elicitin secreted by Phytophthora cactorum. Phytochemistry 34, 1261–1264. doi: 10.1016/0031-9422(91)80012-p
Huet, J. C., Nespoulous, C., and Pernollet, J. C. (1992). Structures of elicitin isoforms secreted by Phytophthora drechsleri. Phytochemistry 31, 1471–1476. doi: 10.1016/0031-9422(92)83089-h
Huet, J. C., and Pernollet, J. C. (1989). Amino acid sequence of cinnamomin, a new member of the elicitin family, and its comparison to cryptogein and capsicein. FEBS Lett. 257, 302–306. doi: 10.1016/0014-5793(89)81557-1
Huet, J. C., and Pernollet, J. C. (1993). Sequences of acidic and basic elicitin isoforms secreted by Phytophthora megasperma. Phytochemistry 33, 797–805. doi: 10.1016/0031-9422(93)85277-x
Huitema, E., Vleeshouwers, V. G., Cakir, C., Kamoun, S., and Govers, F. (2005). Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol. Plant Microbe Interact. 18, 183–193. doi: 10.1094/MPMI-18-0183
Jeong, J. S., Mitchell, T. K., and Dean, R. A. (2007). The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 273, 157–165. doi: 10.1111/j.1574-6968.2007.00796.x
Jiang, R. H., Tyler, B. M., Whisson, S. C., Hardham, A. R., and Govers, F. (2006). Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol. Biol. Evol. 23, 338–351. doi: 10.1093/molbev/msj039
Jin, H., Hartman, G. L., Nickell, C. D., and Widholm, J. M. (1996). Characterization and purification of a phytotoxin produced by Fusarium solani, the causal agent of soybean sudden death syndrome. Phytopathology 86, 277–282. doi: 10.1094/Phyto-86-277
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Kamoun, S., Klucher, K. M., Coffey, M. D., and Tyler, B. M. (1993). A gene encoding a host-specific elicitor protein of Phytophthora parasitica. Mol. Plant Microbe Interact. 6, 573–573. doi: 10.1094/MPMI-6-573
Kamoun, S., van West, P., de Jong, A. J., de Groot, K. E., Vleeshouwers, V. G., and Govers, F. (1997). A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol. Plant Microbe Interact. 10, 13–20. doi: 10.1094/MPMI.1997.10.1.13
Kamoun, S., Van West, P., Vleeshouwers, V. G., De Groot, K. E., and Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1425. doi: 10.2307/3870607
Kanneganti, T. D., Huitema, E., Cakir, C., and Kamoun, S. (2006). Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nep1-like protein PiNPP1. 1 and INF1 elicitin. Mol. Plant Microbe interact. 19, 854–863. doi: 10.1094/mpmi-19-0854
Kanzaki, H., Saitoh, H., Ito, A., Fujisawa, S., Kamoun, S., Katou, S., et al. (2003). Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 4, 383–391. doi: 10.1046/j.1364-3703.2003.00186.x
Kanzaki, H., Saitoh, H., Takahashi, Y., Berberich, T., Ito, A., Kamoun, S., et al. (2008). NbLRK1, a lectin-like receptor kinase protein of Nicotiana benthamiana, interacts with Phytophthora infestans INF1 elicitin and mediates INF1-induced cell death. Planta 228, 977–987. doi: 10.1007/s00425-008-0797-y
Kars, I., Krooshof, G. H., Wagemakers, L., Joosten, R., Benen, J. A., and van Kan, J. A. (2005). Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 43, 213–225. doi: 10.1111/j.1365-313X.2005.02436.x
Kawamura, Y., Takenaka, S., Hase, S., Kubota, M., Ichinose, Y., Kanayama, Y., et al. (2009). Enhanced defense responses in Arabidopsis induced by the cell wall protein fractions from Pythium oligandrum require SGT1, RAR1, NPR1 and JAR1. Plant Cell Physiol. 50, 924–934. doi: 10.1093/pcp/pcp044
Kelley, B. S., Lee, S. J., Damasceno, C. M., Chakravarthy, S., Kim, B. D., Martin, G. B., et al. (2010). A secreted effector protein (Sne1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J. 62, 357–366. doi: 10.1111/j.1365-313X.2010.04160.x
Kettles, G. J., Bayon, C., Canning, G., Rudd, J. J., and Kanyuka, K. (2017). Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana. New Phytol. 213, 338–350. doi: 10.1111/nph.14215
Khatib, M., Lafitte, C., Esquerré-Tugayé, M. T., Bottin, A., and Rickauer, M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162, 501–510. doi: 10.1111/j.1469-8137.2004.01043.x
Kleemann, J., Rincon-Rivera, L. J., Takahara, H., Neumann, U., van Themaat, E. V. L., van der Does, H. C., et al. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8:e1002643. doi: 10.1371/journal/ppat.1002643
Kou, Y. J., Tan, Y. H., Ramanujam, R., and Naqvi, N. I. (2017). Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 214, 330–342. doi: 10.1111/nph.14347
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Kulik, A., Noirot, E., Grandperret, V., Bourque, S., Fromentin, J., Salloignon, P., et al. (2015). Interplays between nitric oxide and reactive oxygen species in cryptogein signalling. Plant Cell Environ. 38, 331–348. doi: 10.1111/pce.12295
Kulkarni, R. D., Kelkar, H. S., and Dean, R. A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121. doi: 10.1016/S0968-0004(03)00034-3
Kulye, M., Liu, H., Zhang, Y. L., Zeng, H. M., Yang, X. F., and Qiu, D. W. (2012). Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defence-related genes and systemic acquired resistance in tobacco. Plant Cell Environ. 35, 2104–2120. doi: 10.1111/j.1365-3040.2012.02539.x
Leborgne-Castel, N., Lherminier, J., Der, C., Fromentin, J., Houot, V., and Simon-Plas, F. (2008). The plant defense elicitor cryptogein stimulates clathrin-mediated endocytosis correlated with reactive oxygen species production in bright yellow-2 tobacco cells. Plant Physiol. 146, 1255–1266. doi: 10.1104/pp.107.111716
Lenarčič, T., Albert, I., Böhm, H., Hodnik, V., Pirc, K., Zavec, A. B., et al. (2017). Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358, 1431–1434. doi: 10.1126/science.aan6874
Li, S., Dong, Y., Li, L., Zhang, Y., Yang, X., Zeng, H., et al. (2019a). The novel cerato-platanin-like protein FocCP1 from Fusarium oxysporum triggers an immune response in plants. Int. J. Mol. Sci. 20:2849. doi: 10.3390/ijms20112849
Li, S., Nie, H., Qiu, D., Shi, M., and Yuan, Q. (2019b). A novel protein elicitor PeFOC1 from Fusarium oxysporum triggers defense response and systemic resistance in tobacco. Biochem. Biophys Res. Commun. 514, 1074–1080. doi: 10.1016/j.bbrc.2019.05.018
Li, X. C., Sun, Y., Liu, N. N., Wang, P., Pei, Y. K., Liu, D., et al. (2019c). Enhanced resistance to Verticillium dahliae mediated by an F-box protein GhACIF1 from Gossypium hirsutum. Plant Sci. 284, 127–134. doi: 10.1016/j.plantsci.2019.04.013
Liu, M., Khan, N. U., Wang, N., Yang, X., and Qiu, D. (2016). The protein elicitor PevD1 enhances resistance to pathogens and promotes growth in Arabidopsis. Int. J. Biol. Sci. 12, 931–943. doi: 10.7150/ijbs.15447
Liu, S., Wu, B., Yang, J., Bi, F., Dong, T., Yang, Q., et al. (2019). A cerato-platanin family protein FocCP1 is essential for the penetration and virulence of Fusarium oxysporum f. sp. cubense tropical race 4. Int. J. Mol. Sci. 20:3785. doi: 10.3390/ijms20153785
Liu, Z. Q., Liu, Y. Y., Shi, L. P., Yang, S., Shen, L., Yu, H. X., et al. (2016). SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 6:21651. doi: 10.1038/srep21651
Liu, Z. Q., Qiu, A. L., Shi, L. P., Cai, J. S., Huang, X. Y., Yang, S., et al. (2015). SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 66, 3683–3698. doi: 10.1093/jxb/erv161
Lo Presti, L., and Kahmann, R. (2017). How filamentous plant pathogen effectors are translocated to host cells. Curr. Opin. Plant Biol. 38, 19–24. doi: 10.1016/j.pbi.2017.04.005
Luti, S., Caselli, A., Taiti, C., Bazihizina, N., Gonnelli, C., Mancuso, S., et al. (2016). PAMP activity of cerato-platanin during plant interaction: an-omic approach. Int. J. Mol. Sci. 17:866. doi: 10.3390/ijms17060866
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Ma, Z. H., Zhu, L., Song, T. Q., Wang, Y., Zhang, Q., Xia, Y. Q., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Mateos, F. V., Rickauer, M., and Esquerré-Tugayé, M. T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant Microbe Interact. 10, 1045–1053. doi: 10.1094/MPMI.1997.10.9.1045
M’Barek, S. B., Cordewener, J. H., Ghaffary, S. M. T., van der Lee, T. A., Liu, Z., Gohari, A. M., et al. (2015). FPLC and liquid-chromatography mass spectrometry identify candidate necrosis-inducing proteins from culture filtrates of the fungal wheat pathogen Zymoseptoria tritici. Fungal Genet. Biol. 79, 54–62. doi: 10.1016/j.fgb.2015.03.015
Mikes, V., Milat, M. L., Ponchet, M., Ricci, P., and Blein, J. P. (1997). The fungal elicitor cryptogein is a sterol carrier protein. FEBS Lett. 416, 190–192. doi: 10.1016/S0014-5793(97)01193-9
Mishra, A. K., Sharma, K., and Misra, R. S. (2009). Purification and characterization of elicitor protein from Phytophthora colocasiae and basic resistance in Colocasia esculenta. Microbiol. Res. 164, 688–693. doi: 10.1016/j.micres.2008.09.001
Motteram, J., Küfner, I., Deller, S., Brunner, F., Hammond-Kosack, K. E., Nürnberger, T., et al. (2009). Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain–containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 22, 790–799. doi: 10.1094/MPMI-22-7-0790
Mouton-Perronnet, F., Bruneteau, M., Denoroy, L., Bouliteau, P., Ricci, P., Bonnet, P., et al. (1995). Elicitin produced by an isolate of Phytophthora parasitica pathogenic to tobacco. Phytochemistry 38, 41–44. doi: 10.1016/0031-9422(94)0058X-A
Nespoulous, C., Huet, J. C., and Pernollet, J. C. (1992). Structure-function relationships of alpha and beta elicitins, signal proteins involved in the plant-Phytophthora interaction. Planta 186, 551–557. doi: 10.1007/bf00198035
Nie, H., Zhang, L., Zhuang, H., Yang, X., Qiu, D., and Zeng, H. (2019). Secreted protein MoHrip2 is required for full virulence of Magnaporthe oryzae and modulation of rice immunity. Appl. Microbiol. Biotechnol. 103, 6153–6167. doi: 10.1007/s00253-019-09937-2
Nie, H. Z., Zhang, L., Zhuang, H. Q., Shi, W. J., Yang, X. F., Qiu, D. W., et al. (2019). The secreted protein MoHrip1 is necessary for the virulence of Magnaporthe oryzae. Int. J. Mol. Sci. 20:1643. doi: 10.3390/ijms20071643
Nie, J., Yin, Z., Li, Z., Wu, Y., and Huang, L. (2019). A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 222, 995–1011. doi: 10.1111/nph.15631
Noda, J., Brito, N., and González, C. (2010). The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 10:38. doi: 10.1186/1471-2229-10-38
Noirot, E., Der, C., Lherminier, J., Robert, F., Moricova, P., Kiêu, K., et al. (2014). Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J. Exp. Bot. 65, 5011–5022. doi: 10.1093/jxb/eru265
Orsomando, G., Lorenzi, M., Raffaelli, N., Dalla Rizza, M., Mezzetti, B., and Ruggieri, S. (2001). Phytotoxic protein PcF, purification, characterization, and cDNA sequencing of a novel hydroxyproline-containing factor secreted by the strawberry pathogen Phytophthora cactorum. J. Biol. Chem. 276, 21578–21584. doi: 10.1074/jbc.M101377200
Ottmann, C., Luberacki, B., Küfner, I., Koch, W., Brunner, F., Weyand, M., et al. (2009). A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. U.S.A 106, 10359–10364.
Palmer, C. S., Saleeba, J. A., and Lyon, B. R. (2005). Phytotoxicity on cotton ex-plants of an 18.5 kDa protein from culture filtrates of Verticillium dahliae. Physiol. Mol. Plant Pathol. 67, 308–318. doi: 10.1016/j.pmpp.2006.05.003
Panabieres, F., Marais, A., Berre, J. Y., Penot, I., Fournier, D., Ricci, P., et al. (1995). Characterization of a gene cluster of Phytophthora cryptogea which codes for elicitins, proteins inducing a hypersensitive-like response in tobacco. Mol. Plant Microbe Interact. 8, 996–1003. doi: 10.1094/MPMI-8-0996
Pazzagli, L., Cappugi, G., Manao, G., Camici, G., Santini, A., and Scala, A. (1999). Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 274, 24959–24964. doi: 10.1074/jbc.274.35.24959
Pazzagli, L., Seidl-Seiboth, V., Barsottini, M., Vargas, W. A., Scala, A., and Mukherjee, P. K. (2014). Cerato-platanins: elicitors and effectors. Plant Sci. 228, 79–87. doi: 10.1016/j.plantsci.2014.02.009
Pettongkhao, S., and Churngchow, N. (2019). Novel cell death-inducing elicitors from Phytophthora palmivora promote infection on Hevea brasiliensis. Phytopathology 109, 1769–1778. doi: 10.1094/PHYTO-01-19-0002-R
Picard, K., Ponchet, M., Blein, J. P., Rey, P., Tirilly, Y., and Benhamou, N. (2000). Oligandrin. A proteinaceous molecule produced by the mycoparasite Pythium oligandrum induces resistance to Phytophthora parasitica infection in tomato plants. Plant Physiol. 124, 379–395. doi: 10.1104/pp.124.1.379
Ptáčková, N., Klempová, J., Obořil, M., Nedělová, S., Lochman, J., and Kašparovský, T. (2015). The effect of cryptogein with changed abilities to transfer sterols and altered charge distribution on extracellular alkalinization, ROS and NO generation, lipid peroxidation and LOX gene transcription in Nicotiana tabacum. Plant Physiol. Biochem. 97, 82–95. doi: 10.1016/j.plaphy.2015.09.009
Quarantin, A., Castiglioni, C., Schäfer, W., Favaron, F., and Sella, L. (2019). The Fusarium graminearum cerato-platanins loosen cellulose substrates enhancing fungal cellulase activity as expansin-like proteins. Plant Physiol. Biochem. 139, 229–238. doi: 10.1016/j.plaphy.2019.03.025
Quarantin, A., Glasenapp, A., Schafer, W., Favaron, F., and Sella, L. (2016). Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 109, 220–229. doi: 10.1016/j.plaphy.2016.10.001
Qutob, D., Kamoun, S., and Gijzen, M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. doi: 10.1046/j.1365-313x.2002.01439.x
Qutob, D., Kemmerling, B., Brunner, F., Kufner, I., Engelhardt, S., Gust, A. A., et al. (2006). Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18, 3721–3744. doi: 10.1105/tpc.106.044180
Ricci, P., Bonnet, P., Huet, J. C., Sallantin, M., Beauvais-Cante, F., Bruneteau, M., et al. (1989). Structure and activity of proteins from pathogenic fungi Phytophthora eliciting necrosis and acquired resistance in tobacco. Eur. J. Biochem. 183, 555–563. doi: 10.1111/j.1432-1033.1989.tb21084.x
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. doi: 10.1105/tpc.022475
Sánchez-Vallet, A., Fouché, S., Fuda, I., Hartmann, F. E., Soyer, J. L., Tellier, A., et al. (2018). The genome biology of effector gene evolution in filamentous plant pathogens. Annu. Rev. Phytopathol. 56, 21–40.
Santhanam, P., van Esse, H. P., Albert, I., Faino, L., Nürnberger, T., and Thomma, B. P. (2013). Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 26, 278–286. doi: 10.1094/MPMI-09-12-0222-R
Sasabe, M., Takeuchi, K., Kamoun, S., Ichinose, Y., Govers, F., Toyoda, K., et al. (2000). Independent pathways leading to apoptotic cell death, oxidative burst and defense gene expression in response to elicitin in tobacco cell suspension culture. Eur. J. Biochem. 267, 5005–5013. doi: 10.1046/j.1432-1327.2000.01553.x
Scala, A., Pazzagli, L., Comparini, C., Santini, A., Tegli, S., and Cappugi, G. (2004). Cerato-platanin, an early-produced protein by Ceratocystis fimbriata f. sp. platani, elicits phytoalexin synthesis in host and non-host plants. J. Plant Pathol. 86, 27–33. doi: 10.1111/j.1439-0329.2010.00668.x
Schouten, A., Van Baarlen, P., and Van Kan, J. A. (2008). Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associate with membranes and the nucleus of plant cells. New Phytol. 177, 493–505. doi: 10.1111/j.1469-8137.2007.02274.x
Sella, L., Gazzetti, K., Faoro, F., Odorizzi, S., D’Ovidio, R., Schäfer, W., et al. (2013). A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol. Biochem. 64, 1–10. doi: 10.1016/j.plaphy.2012.12.008
Sharathchandra, R. G., Geetha, N. P., Amruthesh, K. N., Kini, K. R., Sarosh, B. R., Shetty, N. P., et al. (2006). Isolation and characterisation of a protein elicitor from Sclerospora graminicola and elicitor-mediated induction of defence responses in cultured cells of Pennisetum glaucum. Funct. Plant Biol. 33, 267–278. doi: 10.1071/fp05197
Srivastava, V. K., Suneetha, K. J., and Kaur, R. (2014). A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem. J. 463, 103–114. doi: 10.1042/BJ20140598
Staats, M., van Baarlen, P., Schouten, A., and van Kan, J. A. (2007). Functional analysis of NLP genes from Botrytis elliptica. Mol. Plant Pathol. 8, 209–214. doi: 10.1111/j.1364-3703.2007.00382.x
Starý, T., Satková, P., Piterková, J., Mieslerová, B., Luhová, L., Mikulík, J., et al. (2019). The elicitin β-cryptogein’s activity in tomato is mediated by jasmonic acid and ethylene signalling pathways independently of elicitin-sterol interactions. Planta 249, 739–749. doi: 10.1007/s00425-018-3036-1
Teh, C. Y., Pang, C. L., Tor, X. Y., Ho, P. Y., Lim, Y. Y., Namasivayam, P., et al. (2018). Molecular cloning and functional analysis of a necrosis and ethylene inducing protein (NEP) from Ganoderma boninense. Physiol. Mol. Plant Pathol. 106, 42–48. doi: 10.1016/j.pmpp.2018.12.003
ten Have, A., Mulder, W., Visser, J., and van Kan, J. A. L. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 11, 1009–1016. doi: 10.1094/MPMI.1998.11.10.1009
Teng, W., Zhang, H., Wang, W., Li, D., Wang, M., Liu, J., et al. (2014). ALY proteins participate in multifaceted Nep1Mo-triggered responses in Nicotiana benthamiana and Arabidopsis thaliana. J. Exp. Bot. 65, 2483–2494. doi: 10.1093/jxb/eru136
Torto, T. A., Li, S., Styer, A., Huitema, E., Testa, A., Gow, N. A., et al. (2003). EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13, 1675–1685. doi: 10.1101/gr.910003
Tundo, S., Moscetti, I., Faoro, F., Lafond, M., Giardina, T., Favaron, F., et al. (2015). Fusarium graminearum produces different xylanases causing host cell death that is prevented by the xylanase inhibitors XIP-I and TAXI-III in wheat. Plant Sci. 240, 161–169. doi: 10.1016/j.plantsci.2015.09.002
Veit, S., Wörle, J. M., Nürnberger, T., Koch, W., and Seitz, H. U. (2001). A novel protein elicitor (PaNie) from Pythium aphanidermatum induces multiple defense responses in carrot, Arabidopsis, and tobacco. Plant Physiol. 127, 832–841. doi: 10.1104/pp.010350
Wang, B., Yang, X., Zeng, H., Liu, H., Zhou, T., Tan, B., et al. (2012). The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Appl. Microbiol. Biotechnol. 93, 191–201. doi: 10.1007/s00253-011-3405-1
Wang, J. Y., Cai, Y., Gou, J. Y., Mao, Y. B., Xu, Y. H., Jiang, W. H., et al. (2004). VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl. Environ. Microbiol. 70, 4989–4995. doi: 10.1128/AEM.70.8.4989-4995.200
Wang, W., An, B., Feng, L., He, C., and Luo, H. (2018). A Colletotrichum gloeosporioides cerato-platanin protein, CgCP1, contributes to conidiation and plays roles in the interaction with rubber tree. Can. J. Microbiol. 64, 826–834. doi: 10.1139/cjm-2018-0087
Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., et al. (2018). Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 9:594. doi: 10.1038/s41467-018-03010-8
Wang, Y., Hu, D., Zhang, Z., Ma, Z., Zheng, X., and Li, D. (2003). Purification and immunocytolocalization of a novel Phytophthora boehmeriae protein inducing the hypersensitive response and systemic acquired resistance in tobacco and Chinese cabbage. Physiol. Mol. Plant Pathol. 63, 223–232. doi: 10.1016/j.pmpp.2003.12.004
Wang, Y., Wu, J., Kim, S. G., Tsuda, K., Gupta, R., Park, S. Y., et al. (2016). Magnaporthe oryzae-secreted protein MSP1 induces cell death and elicits defense responses in rice. Mol. Plant Microbe Interact. 29, 299–312. doi: 10.1094/MPMI-12-15-0266-R
Yang, C., Liang, Y., Qiu, D., Zeng, H., Yuan, J., and Yang, X. (2018a). Lignin metabolism involves Botrytis cinerea BcGs1-induced defense response in tomato. BMC Plant Biol. 18:103. doi: 10.1186/s12870-018-1319-0
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018b). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Yang, Y., Yang, X., Dong, Y., and Qiu, D. (2018c). The Botrytis cinerea xylanase BcXyl1 modulates plant immunity. Front. Microbiol. 9:2535. doi: 10.3389/fmicb.2018.02535
Yang, Y., Zhang, Y., Li, B., Yang, X., Dong, Y., and Qiu, D. (2018d). A Verticillium dahliae pectate lyase induces plant immune responses and contributes to virulence. Front. Plant Sci. 9:1271. doi: 10.3389/fpls.2018.01271
Yang, Y., Zhang, H., Li, G., Li, W., Wang, X., and Song, F. (2009). Ectopic expression of MgSM1, a cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 7, 763–777. doi: 10.1111/j.1467-7652.2009.00442.x
Yao, Z., Rashid, K. Y., Adam, L. R., and Daayf, F. (2011). Verticillium dahliae’s VdNEP acts both as a plant defence elicitor and a pathogenicity factor in the interaction with Helianthus annuus. Can. J. Plant Pathol. 33, 375–388. doi: 10.1080/07060661.2011.579173
Yoshino, K., Irieda, H., Sugimoto, F., Yoshioka, H., Okuno, T., and Takano, Y. (2012). Cell death of Nicotiana benthamiana is induced by secreted protein NIS1 of Colletotrichum orbiculare and is suppressed by a homologue of CgDN3. Mol. Plant Microbe Interact. 25, 625–636. doi: 10.1094/MPMI-12-11-0316
Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., et al. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. doi: 10.1105/tpc.008680
Zaparoli, G., Cabrera, O. G., Medrano, F. J., Tiburcio, R., Lacerda, G., and Pereira, G. G. (2009). Identification of a second family of genes in Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao, encoding necrosis-inducing proteins similar to cerato-platanins. Mycol. Res. 113, 61–72. doi: 10.1016/j.mycres.2008.08.004
Zhang, H., Dong, S., Wang, M., Wang, W., Song, W., Dou, X., et al. (2010). The role of vacuolar processing enzyme (VPE) from Nicotiana benthamiana in the elicitor-triggered hypersensitive response and stomatal closure. J. Exp. Bot. 61, 3799–3812. doi: 10.1093/jxb/erq189
Zhang, H., Li, D., Wang, M., Liu, J., Teng, W., Cheng, B., et al. (2012). The Nicotiana benthamiana mitogen-activated protein kinase cascade and WRKY transcription factor participate in Nep1(Mo)-triggered plant responses. Mol. Plant Microbe Interact. 25, 1639–1653. doi: 10.1094/MPMI-11-11-0293
Zhang, H., Wu, Q., Cao, S., Zhao, T., Chen, L., Zhuang, P., et al. (2014). A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 86, 495–511. doi: 10.1007/s11103-014-0244-3
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164, 352–364. doi: 10.1104/pp.113.230698
Zhang, H., Zhao, T., Zhuang, P., Song, Z., Du, H., Tang, Z., et al. (2016). NbCZF1, a novel C2H2-Type zinc finger protein, as a new regulator of SsCut-induced plant immunity in Nicotiana benthamiana. Plant Cell Physiol. 57, 2472–2484. doi: 10.1093/pcp/pcw160
Zhang, Y., Gao, Y., Liang, Y., Dong, Y., Yang, X., and Qiu, D. (2019). Verticillium dahliae PevD1, an Alt a 1-like protein, targets cotton PR5-like protein and promotes fungal infection. J. Exp Bot. 70, 613–626. doi: 10.1093/jxb/ery351
Zhang, Y., Gao, Y., Liang, Y., Dong, Y., Yang, X., Yuan, J., et al. (2017a). The Verticillium dahliae snodprot1-like protein VdCP1 contributes to virulence and triggers the plant immune system. Front. Plant Sci. 8:1880. doi: 10.3389/fpls.2017.01880
Zhang, Y., Liang, Y., Dong, Y., Gao, Y., Yang, X., Yuan, J., et al. (2017b). The Magnaporthe oryzae Alt A 1-like protein MoHrip1 binds to the plant plasma membrane. Biochem. Biophys. Res. Commun. 492, 55–60. doi: 10.1016/j.bbrc.2017.08.039
Zhang, Y., Zhang, Y., Qiu, D., Zeng, H., Guo, L., and Yang, X. (2015). BcGs1, a glycoprotein from Botrytis cinerea, elicits defence response and improves disease resistance in host plants. Biochem. Biophys. Res. Commun. 457, 627–634. doi: 10.1016/j.bbrc.2015.01.038
Zhang, Z. G., Wang, Y. C., Li, J., Ji, R., Shen, G., Wang, S. C., et al. (2004). The role of SA in the hypersensitive response and systemic acquired resistance induced by elicitor PB90 from Phytophthora boehmeriae. Physiol. Mol. Plant Pathol. 65, 31–38. doi: 10.1016/j.pmpp.2004.11.001
Zhang, Z. N., Wu, Q. Y., Zhang, G. Z., Zhu, Y. Y., Murphy, R. W., Liu, Z., et al. (2015). Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 5:13032. doi: 10.1038/srep13032
Zhao, Z., Liu, H., Wang, C., and Xu, J. R. (2013). Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14:274. doi: 10.1186/1471-2164-14-274
Zheng, A., Lin, R., Zhang, D., Qin, P., Xu, L., Ai, P., et al. (2013). The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 4:1424. doi: 10.1038/ncomms2427
Zhou, B. J., Jia, P. S., Gao, F., and Guo, H. S. (2012). Molecular characterization and functional analysis of a necrosis-and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 25, 964–975. doi: 10.1094/MPMI-12-11-0319
Zhu, W., Ronen, M., Gur, Y., Minz-Dub, A., Masrati, G., Ben-Tal, N., et al. (2017a). BcXYG1, a secreted Xyloglucanase from Botrytis cinerea, triggers both cell death and plant immune responses. Plant Physiol. 175, 438–456. doi: 10.1104/pp.17.00375
Keywords: filamentous phytopathogen, apoplastic effector, cell death-inducing proteins, virulence factor, immune response
Citation: Li Y, Han Y, Qu M, Chen J, Chen X, Geng X, Wang Z and Chen S (2020) Apoplastic Cell Death-Inducing Proteins of Filamentous Plant Pathogens: Roles in Plant-Pathogen Interactions. Front. Genet. 11:661. doi: 10.3389/fgene.2020.00661
Received: 27 February 2020; Accepted: 01 June 2020;
Published: 26 June 2020.
Edited by:
Ramesh Raju Vetukuri, Swedish University of Agricultural Sciences, SwedenReviewed by:
Brett Merrick Tyler, Oregon State University, United StatesCopyright © 2020 Li, Han, Qu, Chen, Chen, Geng, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zonghua Wang, d2FuZ3poQGZhZnUuZWR1LmNu; Songbiao Chen, c29uZ2JpYW9fY2hlbkBob3RtYWlsLmNvbQ==; c2JjaGVuQGZqYWdlLm9yZw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.