
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 10 June 2020
Sec. Epigenomics and Epigenetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00609
This article is part of the Research Topic Genetic and Epigenetic Regulation of Insect Development, Reproduction, and Phenotypic Plasticity View all 24 articles
Besides the function of preventing metamorphosis in insects, the juvenile hormone (JH) plays a role in female reproduction; however, the underlying mechanism is largely unknown. The methoprene-tolerant (Met) protein belongs to a family of basic helix-loop-helix–Per-Arnt-Sim (bHLH-PAS) transcription factors and functions as the JH intracellular receptor. In this study, two full length cDNAs encoding Met (CsMet1 and CsMet2) were isolated from the rice stem borer, Chilo suppressalis. Structural analysis revealed that both CsMet1 and CsMet2 exhibited typical bHLH, PAS-A, PAS-B, and PAC (PAS C terminal motif) domains. Comparative analysis of transcript level using reverse transcription-quantitative PCR (RT-qPCR) revealed that CsMet1 was predominant in almost all examined developmental stages and tissues. Treatment with methoprene in vivo induces the transcription of both CsMet1 and CsMet2. Notably, injection of dsCsMet1 and dsCsMet2 suppressed the expression levels of vitellogenin (CsVg) and Vg receptor (CsVgR). These findings revealed the potential JH signaling mechanism regulating C. suppressalis reproduction, and provided evidence that RNAi-mediated knockdown of Met holds great potential as a control strategy of C. suppressalis.
The rice stem borer, Chilo suppressalis (Walker) (Lepidoptera: Crambidae), is one of the most serious rice pests in Asia, Middle East, and southern Europe, and causes large crop losses through feeding on the stems of rice. In China, the change of cultivation patterns has led to the frequent outbreaks of C. suppressalis in recent years (Mao et al., 2019). To date, spraying chemical insecticides remains the primary strategy for controlling C. suppressalis. However, intensive use of insecticides has driven C. suppressalis to develop resistance to a wide range of insecticides (Su et al., 2014; Lu et al., 2017; Yao et al., 2017). Hence, the development of RNA interference (RNAi)-mediated disruption of reproduction represents an alternative control strategy of C. suppressalis (Miao et al., 2020).
Insect reproduction is intricately regulated by three hormones, including the juvenile hormones (JHs), ecdysteroids, and insulin (Smykal and Raikhel, 2015; Roy et al., 2018; Al Baki et al., 2019). JH exerts its function through its intracellular receptor methoprene-tolerant (Met), a member of the family of the basic helix-loop-helix (bHLH)-Per-Arnt-Sim (PAS) transcription factors (Konopova and Jindra, 2007; Jindra et al., 2013, 2015). Since the first characterization of Met in Drosophila melanogaster (Wilson and Fabian, 1986), Met has been identified from a broad range of insect species, such as Aedes aegypti (Wang et al., 2007), Tribolium castaneum (Konopova and Jindra, 2007), Nilaparvata lugens (Lin et al., 2015), and Helicoverpa armigera (Ma et al., 2018). Interestingly, two Met paralogs, Met and Germ cell-expressed (Gce), are present across 12 Drosophila species (Baumann et al., 2010a, b). While mosquitoes and beetles possess only a single gene, two Met genes have also been identified in several Lepidopteran insects, including Danaus plexippus (Zhan et al., 2011), Operophtera brumata (Derks et al., 2015), and Bombyx mori (Guo et al., 2012; Suetsugu et al., 2013; Kayukawa and Shinoda, 2015). However, the role of two Met genes in the reproduction of Lepidopteran insects remains largely unknown.
The reproductive success of insects depends on vitellogenin (Vg) biosynthesis and uptake of Vg into developing oocytes mediated by vitellogenin receptor (VgR). Recently, we have characterized the C. suppressalis CsVg and CsVgR at the molecular levels (Huang et al., 2016; Miao et al., 2020). In this study, full-length cDNAs of two paralogous Met genes (named as CsMet1 and CsMet2) were isolated from C. suppressalis. We report the structural features and temporal–spatial expression patterns of CsMet1 and CsMet2. Furthermore, RNAi was employed to reveal the role of CsMet1 and CsMet2 in the regulation of CsVg and CsVgR.
Chilo suppressalis larvae was collected from rice stubbles of Yangzhou (32.39°N, 119.42°E) in 2013, and reared on an artificial diet in an incubator at 28 ± 1°C, 70 ± 5% RH, and a 16-h light/8-h dark photoperiod.
To examine the developmental expression profiles of CsMet, individuals were collected from larvae (third, fourth, fifth, and sixth instar), pupae at intervals of 2 days from pupation, and adults at intervals of 12 h from eclosion. For tissue expression analysis, tissues (including head, epidermis, midgut, hemolymph, and fat body) were dissected from the fifth-instar larva. All the samples were frozen immediately in liquid nitrogen and stored at -80°C until RNA isolation. Each experiment was performed with three biological replicates containing three to 10 individuals.
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, United States). First-strand cDNA was synthesized from total RNA using the PrimescriptTM First-Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturers’ instruction.
The amino acid (aa) sequences of B. mori BmMet1 (GenBank: NP001108458) and BmMet2 (GenBank: BAJ05086) were searched against the transcriptome database of C. suppressalis in Insect Base1, and specific primer pairs were designed based on the sequences of putative transcripts (Table 1). PCR reactions were performed with LA TaqTM DNA polymerase (TaKaRa, Dalian, China). To obtain full length cDNA sequences of CsMet1 and CsMet2, 5′-RACE and 3′-RACE were conducted using the SMART RACE cDNA Amplification Kit (Clontech, Mountain View, CA, United States), following the manufacturers’ instructions. Gene specific primers (GSPs) used for RACE are listed in Table 1.
RT-PCR and RACE products were subcloned into the pMD18−T vector (TaKaRa) and sequenced. The molecular mass and isoelectric point (pI) of the deduced protein sequences were predicted by using the online ExPASy proteomics server2. Conserved domains were predicted by NCBI conserved domain search tool3 or by alignment to other published insect Mets. A phylogenetic tree was constructed with MEGA 7.0 using the neighbor-joining method with a p-distance model and a pairwise deletion of gaps (Kumar et al., 2016). The reliability of the NJ tree topology was statistically evaluated by bootstrap analysis with 1000 replicates.
Reverse transcription-quantitative qPCR reactions were performed on the Bio-Rad CFX-96TM Real-time PCR system using TB GreenTM Premix Ex TaqTM (Takara, Dalian, China) following the manufacturers’ instructions. GSPs used are listed in Table 1. The stably expressed gene encoding EF1-α was used as a reference gene (Hui et al., 2011; Meng et al., 2019). The mRNA levels were normalized to reference gene with the 2–ΔΔCT method (Livak and Schmittgen, 2001). The means and standard errors for each time point were obtained from the average of three biologically independent samples.
To study effects of JH on expression of CsMet, newly emerged adult females were injected with 1 μL JH analog methoprene (3 μg/μL, Sigma–Aldrich, St. Louis, MO, United States) or same volume of acetone as control. Insects were collected for RT-qPCR analysis at 1, 6, 12, and 24 h after treatment, respectively, and experiments contained three biological replications.
Double-strand RNAs (dsRNAs) against CsMet1 or CsMet2 were synthesized using TranscriptAidTM T7 High Yield Transcription Kit (Thermo Fisher Scientific, Waltham, MA, United States). GSPs for dsRNA synthesis are listed in Table 1. Using a Nanoliter 2010 injector system (WPI, Sarasota, FL, United States), 1 μL solution of dsRNA (3 μg/μL) was injected into the abdomen of 6-day-old female pupae under a stereomicroscope. Equal dose of dsRNA for enhanced green fluorescent protein (EGFP) was injected as a control. A total of 40 pupae were injected per treatment, and each treatment was performed in triplicate. The treatment was repeated in newly emerged female adults (within 24 h), and insects were collected at 24 and 48 h after injection, respectively, for expression analysis of CsMet1, CsMet2, CsVg, and CsVgR by RT-qPCR.
The statistical analysis was done using graphpad.prism.6 by one-way analysis of variance, followed by student’s t-test. All data are presented as the mean ± SE.
The full length CsMet1 cDNA (GenBank accession number MN906993) was 2673 bp, with a 460 bp 5’-terminal untranslated region (UTR), a 1587 bp open reading frame (ORF), and a 626 bp 3′-UTR. The deduced CsMet1 protein contained 528 aas with a predicted mass and pI of 60.34 kDa and 8.35, respectively. The 2824 bp CsMet2 cDNA (GenBank accession number MN906994) contained an 85 bp 5′-UTR, a 2598 bp ORF encoding an 865 aa residue protein with a molecular mass of 97.37 kDa and a pI of 6.68, and a 141 bp 3′-UTR.
Structural analysis showed that the deduced protein of CsMet1 and CsMet2 had conserved domains of bHLH-PAS protein family including bHLH (DNA binding and dimerization regions), PAS-A (dimerization region), PAS-B (ligand binding and dimerization region), and PAC (PAS C terminal motif) domains (dimerization region) (Figure 1). Alignment of aa sequences demonstrated that CsMet1 shared identity with other insect Met orthologs including B. mori BmMet1 (60.98%) and BmMet2 (21.88%), D. melanogaster DmMet (27.49%), and T. castaneum TcMet (28.30%). CsMet2 shared 20.55, 44.19, 17.19, and 22.90% identity with BmMet1, BmMet2, DmMet, and TcMet, respectively. The aa identity between CsMet1 and CsMet2 was 21.71% (Figure 1). Phylogenetic analysis revealed that CsMet1 and CsMet2 were clustered into Lepidopteran group 1 and group 2 Met, respectively (Figure 2).
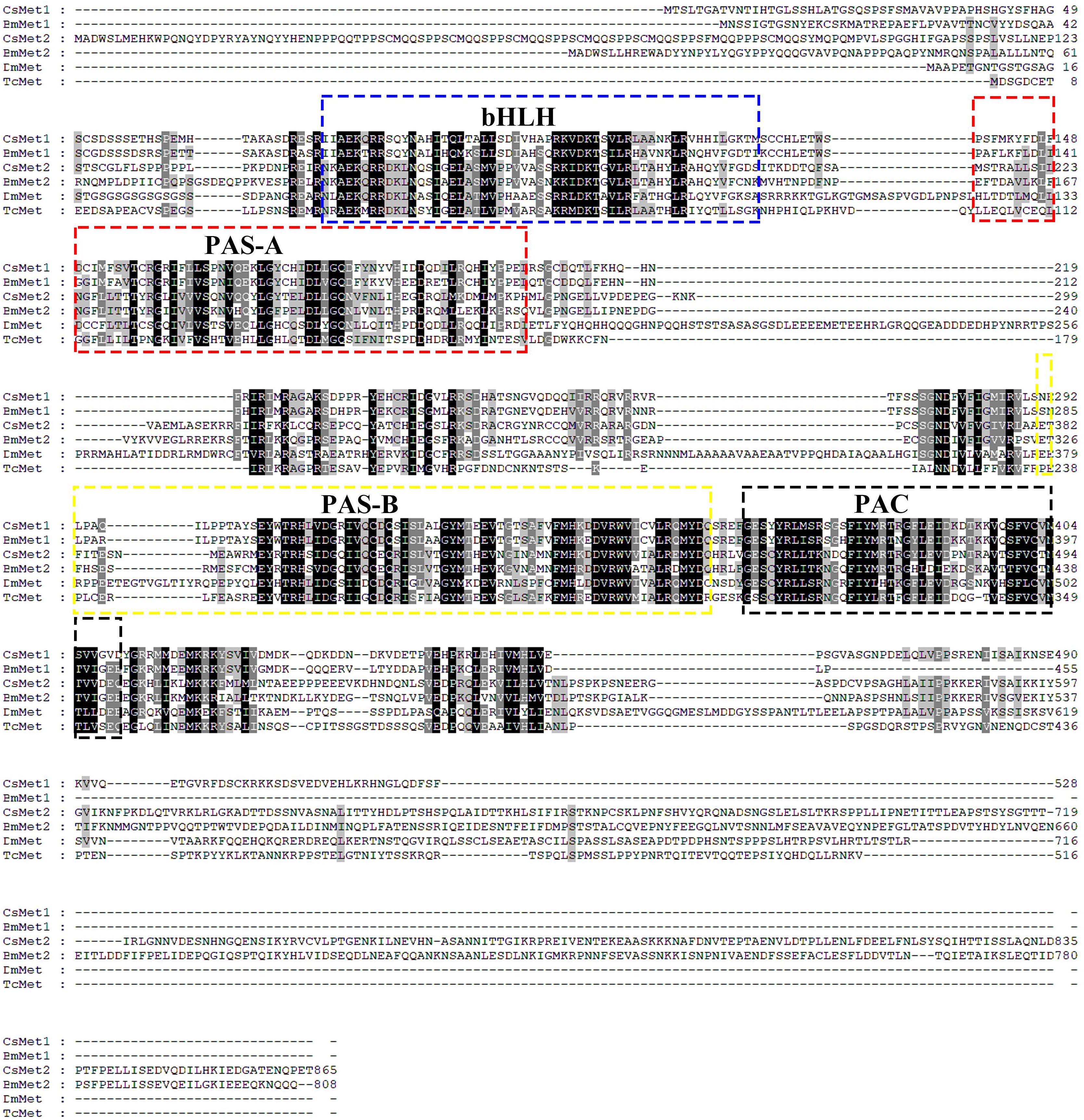
Figure 1. Amino acid sequence alignments of CsMet1, CsMet2 and other representative insect Mets. Met sequences were obtained from the following GenBank entries: NP001108458 for B. mori BmMet1, BAJ05086 for B. mori BmMet2, AAC14350 for D. melanogaster DmMet and NP 001092812 for T. castaneum TcMet.

Figure 2. Phylogenetic relationships of CsMet1, CsMet2 and other insect Mets. The neighbor-joining tree was constructed by MEGA 7.0. Numbers indicated bootstrap support values (%) based on 1,000 replicates. Sequences were retrieved from GenBank protein database.
The temporal expression patterns of CsMet1 and CsMet2 were detected from third instar to 3-day-old female adults and determined by RT-qPCR analysis. The results showed that the transcription level of CsMet1 and CsMet2 was detectable during all selected developmental stages. Specifically, the expression of CsMet1 was relatively stable in the larval stage, sharply decreased in the prepupal stage, maintained at a low level during the pupal stage, and reached a peak in 12-h-old female adults (Figure 3A). The expression of CsMet2 gradually increased from fourth instar to 5-day-old pupae. In the adult stage, the highest and lowest expression levels of CsMet2 were observed in 48-h-old and 72-h-old female adults, respectively (Figure 3B).
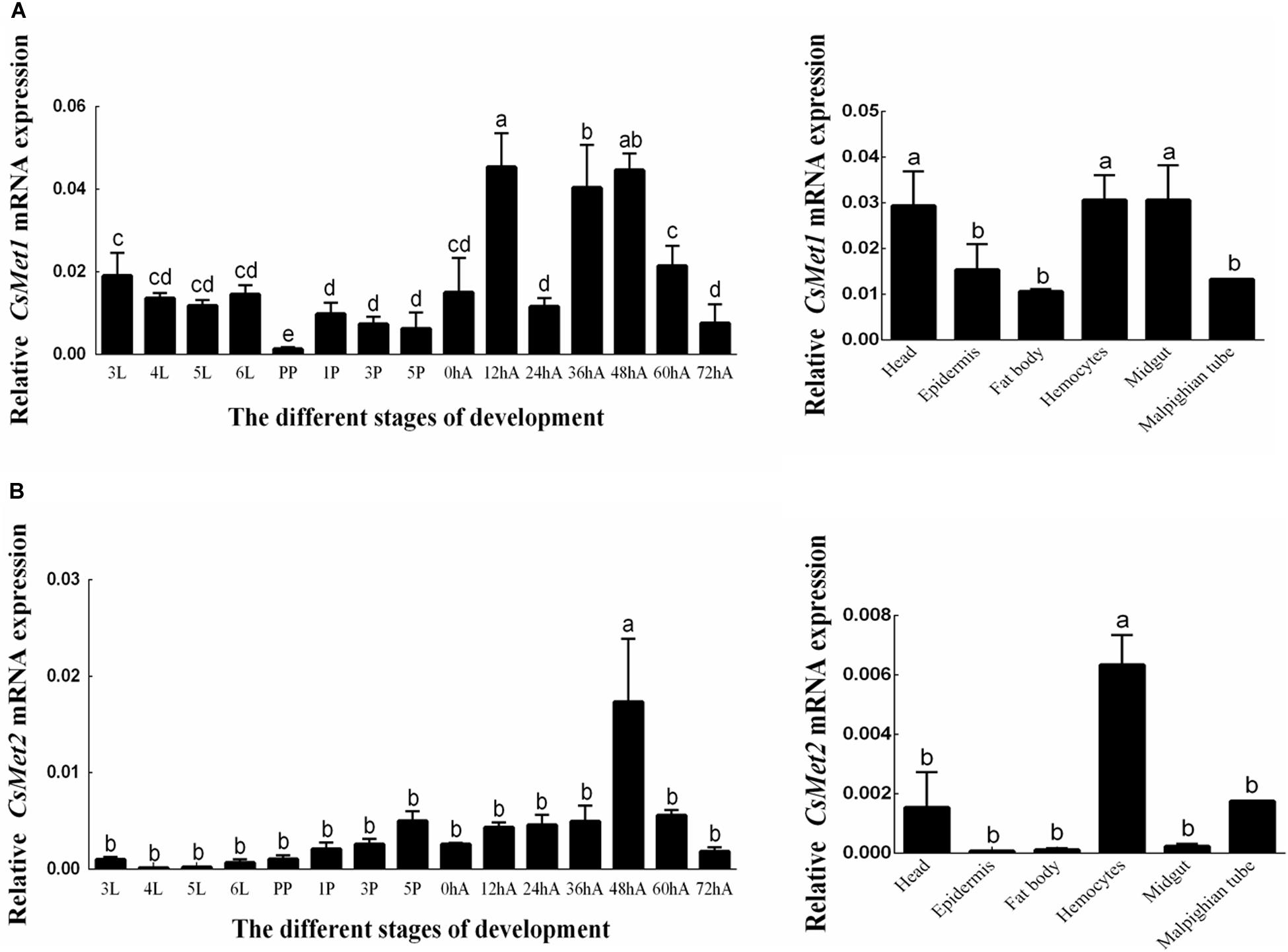
Figure 3. Expression profiles of CsMet1 (A) and CsMet2 (B) in different developmental stages and various tissues from 5th instar larva. 3L: 33rd instar larvae; 4L: 4th instar larvae; 5L: 5th instar larvae; 6L: 6th instar larvae; PP: prepupae; 1P: 1-day-old pupae; 3P: 3-day-old pupae; 5P: 5-day-old pupae; 0hA: newly emerged female adults; 12hA: 12-h-old female adults; 24hA: 24-h-old female adults; 36hA: 36-h-old female adults; 48hA: 48-h-old female adults; 60hA: 60-h-old female adults; 72hA: 72-h-old female adults. Bars not sharing a common letter are significantly different.
Analysis of the spatial expression profiles of CsMet1 and CsMet2 in fifth-instar larva showed that CsMet1 was highly expressed in head, hemocytes, and midgut, while relatively low expression was observed in epidermis, fat body, and malpighian tube (Figure 3A). The highest expression level of CsMet2 was observed in hemocytes, followed by head and Malpighian tube, whereas low expression was observed in epidermis, fat body, and midgut (Figure 3B).
To examine whether CsMet1 and CsMet2 were regulated by JH, JHA methoprene was injected into the newly emerged female adults. Compared with control, the results showed that CsMet1 expression was upregulated by 2.74, 2.41, and 1.63 times at 1, 6, and 12 h after injection, respectively (Figure 4A). Similarly, transcription level of CsMet2 was increased by 1.95, 1.45, and 3.68 times at 1, 6, and 1 2 h post-treatment, respectively (Figure 4B).
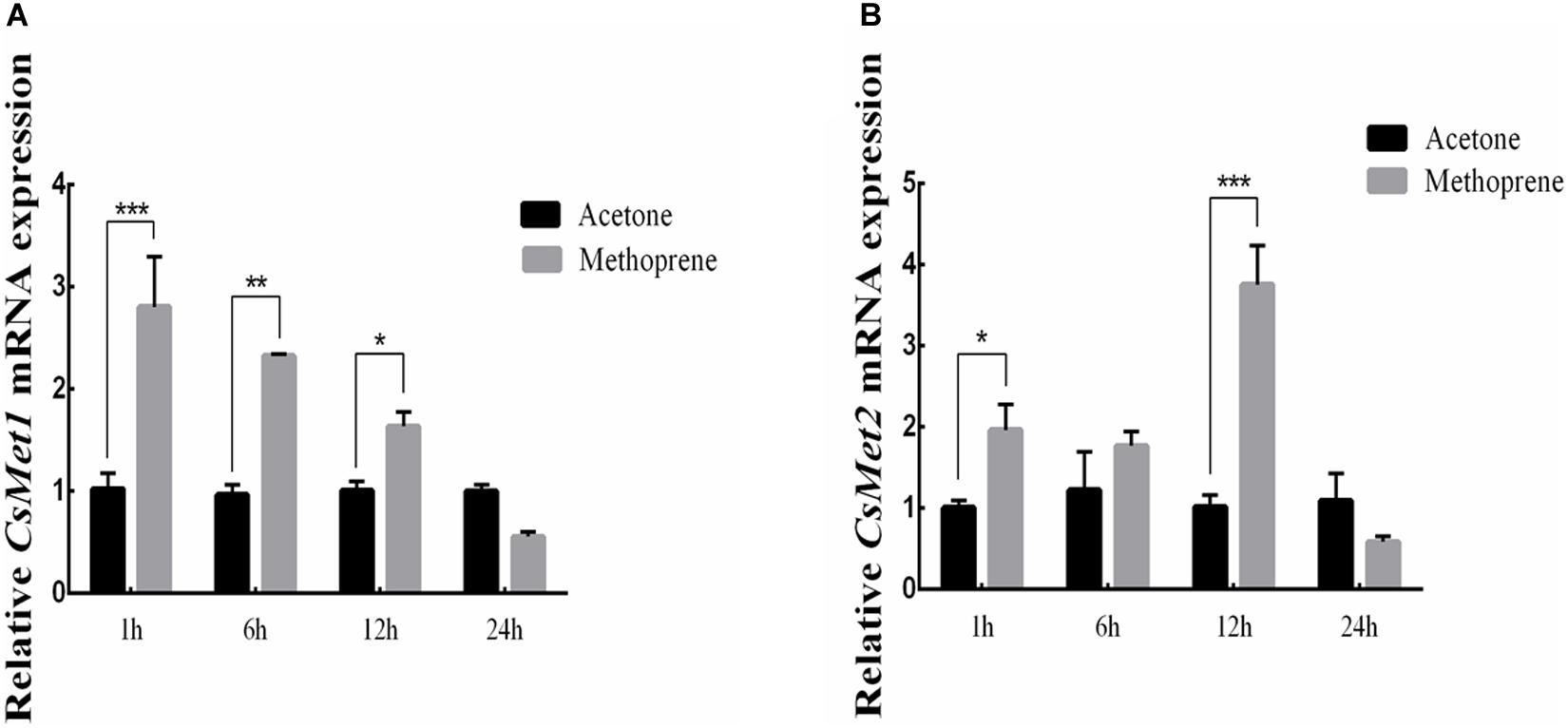
Figure 4. Relative mRNA levels of CsMet1 (A) and CsMet2 (B) in the whole body insects after methoprene treatment in newly emerged female adults. The bars represent mean ± SE of three biological replicates. Asterisks indicate the significant differences between the groups (*P < 0.05, **P < 0.01, and ***P < 0.001).
To explore the role of CsMet1 and CsMet2 in the regulation of CsVg and CsVgR, female adults were collected at 24 and 48 h after the second dsRNA injection, respectively. The transcript level of CsMet1 in dsCsMet1-injected female adults was suppressed by 78.74% (24 h) and 70.32% (48 h) compared with dsEGFP-injected insects, whereas the expression of CsMet2 was significantly reduced by 94.69% (24 h) and 92.79% (48 h) (Figure 5A). Meanwhile, RNAi-mediated suppression of CsMet1 and CsMet2 decreased CsVg expression by 70.85 and 44.37% at 48 h post-treatment, respectively (Figure 5B). CsVgR expression in the dsCsMet1-treated and dsCsMet2-treated group decreased significantly by 52.91 and 62.98% at 24 h post-injection, respectively (Figure 5C). Furthermore, phenotypic observation revealed that silencing of both CsMet1 and CsMet2 resulted in suppressed ovarian development with decreased number of mature follicles (Figure 6).
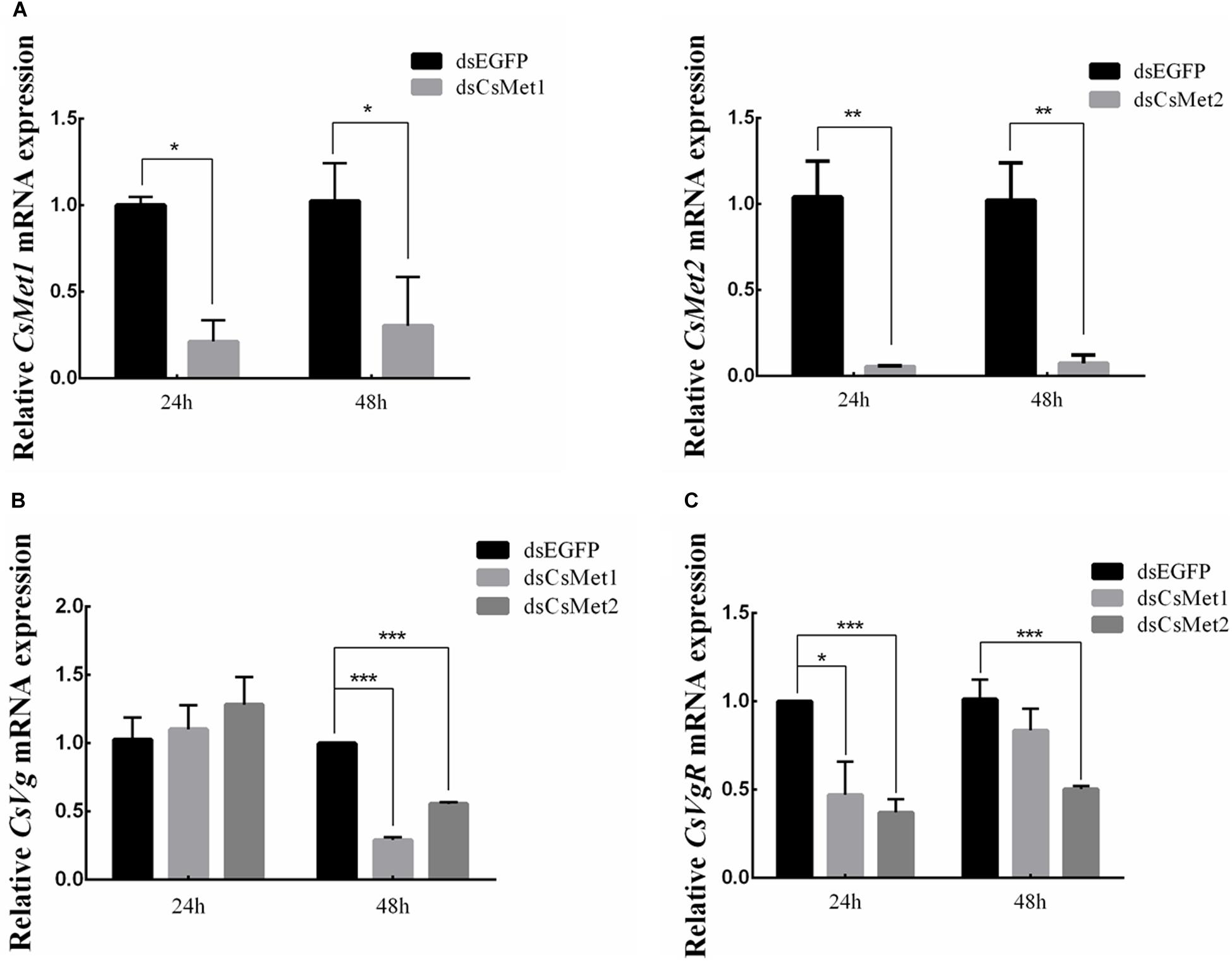
Figure 5. Knockdown of CsMet1 and CsMet2. dsRNAs were injected into 6-day-old female pupae followed by the second injection into the newly emerged female moths. The expression levels of CsMet1, CsMet2 (A), CsVg (B), and CsVgR (C) in whole body insects were examined at 24 h and 48 h after treatment. The bars represent mean ± SE of three biological replicates. Asterisks indicate the significant differences between the groups (*P < 0.05, **P < 0.01, and ***P < 0.001).
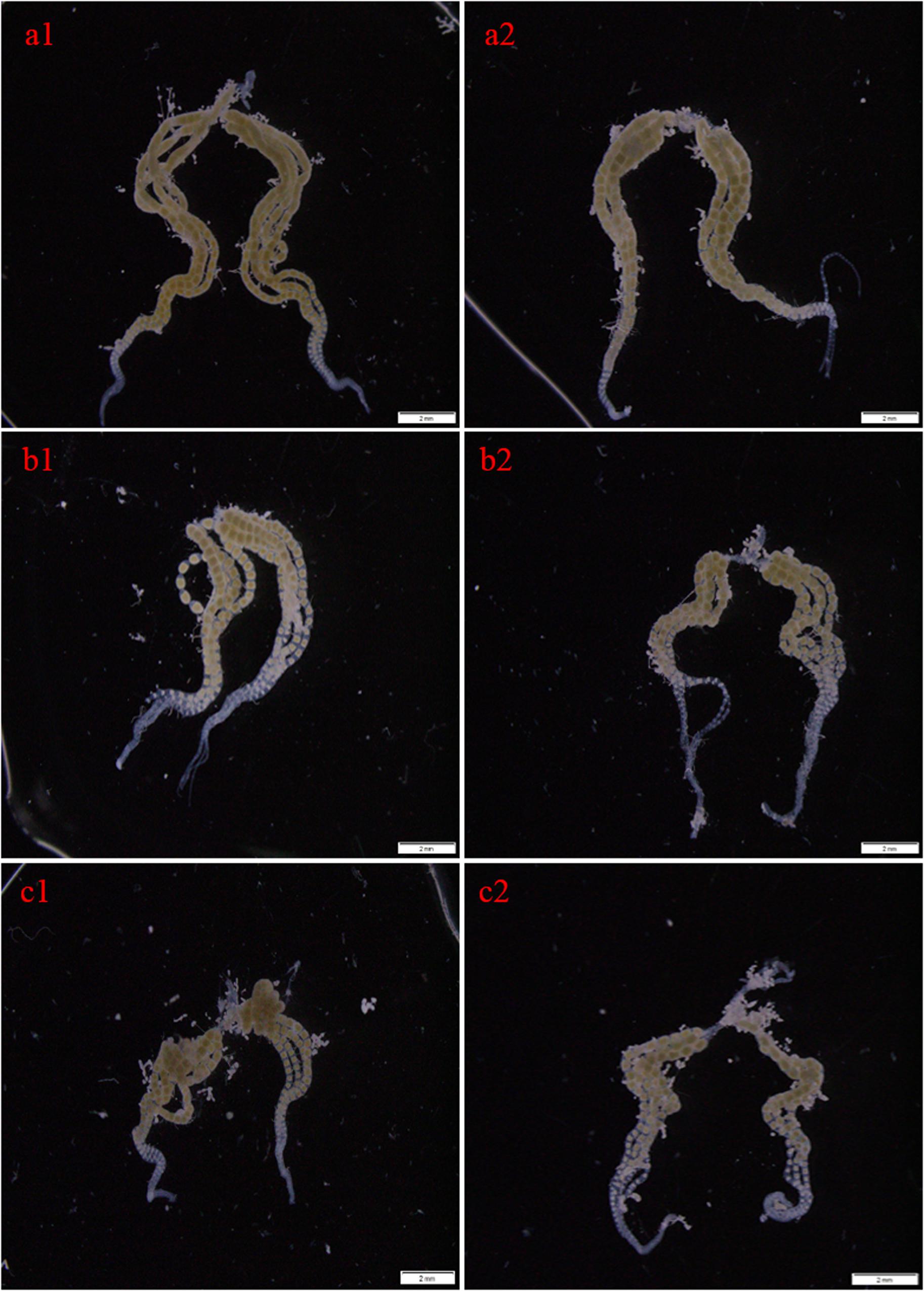
Figure 6. Effects of RNAi-mediated knockdown of CsMet1 and CsMet2 on ovary development with dsEGFP as a control. The ovaries were dissected from female adults at 24 h after the second injection of dsEGFP (a1, a2), dsCsMet1 (b1, b2) and dsCsMet2 (c1, c2).
The important roles played by JH in insect development and reproduction prompted the study on JH signaling, and the breakthrough had been the identification of transcription factor Met as an intracellular receptor for JH (Charles et al., 2011). Binding of JH to Met triggers dimerization of Met with another bHLH-PAS protein Taiman (Tai) to form a functional complex, which interacts with JH response elements (JHREs) of target genes (Smykal and Raikhel, 2015). While the Met null allele was expected to result in a lethal phenotype, the paralogous Met gene in D. melanogaster, Gce, ensured survival of the met null mutants (Baumann et al., 2010a, b). Similarly, two Met genes were also recently identified in several Lepidopteran insects (Zhan et al., 2011; Guo et al., 2012; Suetsugu et al., 2013; Derks et al., 2015; Kayukawa and Shinoda, 2015). However, only a single Met gene was found in those from other insect orders including A. aegypti (Dipteran, Wang et al., 2007), T. castaneum (Coleopteran, Konopova and Jindra, 2007), Blattella germanica (Blattarian, Lozano and Belles, 2014), N. lugens (Hemipteran, Lin et al., 2015), Sitodiplosis mosellana (Dipteran, Cheng et al., 2018), and so on. In this study, we isolated two paralogous Met genes, CsMet1 and CsMet2, from C. suppressalis. Both proteins showed conserved domain organization of bHLH-PAS protein family (Ashok et al., 1998; Kayukawa and Shinoda, 2015; Ma et al., 2018). Phylogenetic analysis revealed that CsMet1 and CsMet2 were clustered into Lepidopteran group 1 and group 2 Met, respectively, providing further evidence of the occurrence of Met gene duplication in Lepidopteran insects.
The temporal and spatial expression patterns of a gene may provide hints as to its function. In this study, we found a marked reduction in expression levels of CsMet1 in the prepupae stage, supporting its involvement in larval–pupal metamorphosis. Similar results were also observed in HaMet1 of H. armigera (Ma et al., 2018) and SmMet of S. mosellana (Cheng et al., 2018), and was in agreement with the fact that JH exerts its anti-metamorphic effect through its receptor Met (Parthasarathy et al., 2008; Li et al., 2019). CsMet2 transcript was hardly detected during the larval stage, and its expression reached the peak in 48-h-old female adults, suggesting that CsMet2 was involved in JH signaling in adults, as had been reported for BmMet2 in B. mori (Kayukawa and Shinoda, 2015). For tissue expression, CsMet1 was highly expressed in larval head, midgut, and hemocytes, whereas CsMet2 showed the highest expression in larval hemocytes. Interestingly, the temporal and spatial expression analysis revealed that CsMet1 was dominant at almost all developmental stages and in all tissues examined. This result was somewhat different from the observation in B. mori that BmMet2 transcript was more abundant than BmMet1 transcript in the adult stage (Kayukawa and Shinoda, 2015). Whether CsMet1 played a more important role in C. suppressalis needed further study in the future.
Juvenile hormone plays a crucial role in insect reproduction, but its molecular mode of action only became clear recently. It has been reported in H. armigera, Diploptera punctata, N. lugens, and S. mosellana that Met is significantly upregulated by JH III or JHA methoprene (Marchal et al., 2014; Zhao et al., 2014; Lin et al., 2015; Cheng et al., 2018). Our hormone treatment results also indicated that CsMet1 and CsMet2 expression was upregulated after JHA methoprene treatment. Further RNAi analysis revealed that knockdown of CsMet1 and CsMet2 resulted in significant decline of both CsVg and CsVgR expression in female adults, leading to decreased number of mature follicles in ovaries. Similarly, expression of Vg is regulated by JH via Met in A. aegypti, H. armigera, D. punctata, N. lugens, and Bactrocera dorsalis (Zou et al., 2013; Marchal et al., 2014; Lin et al., 2015; Ma et al., 2018; Yue et al., 2018). Downregulation of VgR transcription as well as suppressed ovarian development were also observed in dsMet-treated H. armigera and Colaphellus bowringi (Ma et al., 2018; Liu et al., 2019). Whether expression of Vg and VgR were directly regulated by Met was not well known. It had been recently reported that knockdown of Krüppel-homolog 1 (Kr-h1), a direct target of JH-Met signaling, also decreased VgR expression in C. bowringi (Liu et al., 2019). On the other hand, Met is also a repressor of 20E pathway gene expression. It is likely that Met regulated the relative genes of 20E pathway (Zhao et al., 2014), which contributes to vitellogenis to some extent. Further study needs to be carried out to confirm these presumptions.
All datasets generated for this study are included in the article.
JW conceived and designed the study. LM, NZ, HJ, FD, XY, and XX performed the experiments. LM, JW, KQ, and XM wrote the manuscript. All authors reviewed and approved this submission.
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest under Grant No. 201303017 and National Natural Science Foundation of China (31701807).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Al Baki, M. A., Lee, D. W., Jung, J. K., and Kim, Y. (2019). Insulin signaling mediates previtellogenic development and enhances juvenile hormone-mediated vitellogenesis in a lepidopteran insect, Maruca vitrata. BMC Dev. Biol. 19:14. doi: 10.1186/s12861-019-0194-8
Ashok, M., Turner, C., and Wilson, T. G. (1998). Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 95, 2761–2766. doi: 10.1073/pnas.95.6.2761
Baumann, A., Barry, J., Wang, S., Fujiwara, Y., and Wilson, T. G. (2010a). Paralogous genes involved in juvenile hormone action in Drosophila melanogaster. Genetics 185, 1327–1336. doi: 10.1534/genetics.110.116962
Baumann, A., Fujiwara, Y., and Wilson, T. G. (2010b). Evolutionary divergence of the paralogs methoprene tolerant (Met) and germ cell expressed (gce) within the genus Drosophila. J. Insect. Physiol. 56, 1445–1455. doi: 10.1016/j.jinsphys
Charles, J. P., Iwema, T., Epa, V. C., Takaki, K., Rynes, J., and Jindra, M. (2011). Ligand-binding properties of a juvenile hormone receptor, methoprene-tolerant. Proc. Natl. Acad. Sci. U.S.A. 108, 21128–21133. doi: 10.1073/pnas.1116123109
Cheng, W. N., Li, X. J., Zhao, J. J., and Salzman, K. Z. (2018). Cloning and characterization of Methoprene-tolerant (Met) and Krüppel homolog 1 (Kr-h1) genes in the wheat blossom midge, Sitodiplosis mosellana. Insect. Sci. 27, 292–303. doi: 10.1111/1744-7917
Derks, M. F., Smit, S., Salis, L., Schijlen, E., Bossers, A., Mateman, C., et al. (2015). The genome of winter moth (Operophtera brumata) providesa genomic perspective on sexual dimorphism and phenology. Genome. Biol. Evol. 7, 2321–2332. doi: 10.1093/gbe/evv145
Guo, E., He, Q., Liu, S., Tian, L., Sheng, Z., Peng, Q., et al. (2012). MET is required for the maximal action of 20-hydroxyecdysone during Bombyx metamorphosis. PLoS One 7:e53256. doi: 10.1371/journal.pone.0053256
Huang, L., Lu, M. X., Han, G. J., Du, Y. Z., and Wang, J. J. (2016). Sublethal effects of chlorantraniliprole on development, reproduction and vitellogenin gene (CsVg) expression in the rice stem borer, Chilo suppressalis. Pest. Manag. Sci. 72, 2280–2286. doi: 10.1002/ps.4271
Hui, X. M., Yang, L. W., He, G. L., Yang, Q. P., Han, Z. J., and Li, F. (2011). RNA interference of ace1 and ace2 in Chilo suppressalis reveals their different contributions to motor ability and larval growth. Insect Mol. Biol. 20, 507–518. doi: 10.1111/j.1365-2583
Jindra, M., Bellés, X., and Shinoda, T. (2015). Molecular basis of juvenile hormone signaling. Curr. Opin. Insect. Sci. 11, 39–46. doi: 10.1016/j.cois.2015.08.004
Jindra, M., Palli, S. R., and Riddiford, L. M. (2013). The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204. doi: 10.1146/annurev-ento-120811-153700
Kayukawa, T., and Shinoda, T. (2015). Functional characterization of two paralogous JH receptors, methoprene tolerant 1 and 2, in the silkworm, Bombyx mori (Lepidoptera: Bombycidae). Jpn. Soc. Appl. Entomol. Zool. 50, 383–391. doi: 10.1007/s13355-015-0345-8
Konopova, B., and Jindra, M. (2007). Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc. Natl. Acad. Sci. U.S.A. 104, 10488–10493. doi: 10.1073/pnas.0703719104
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, K., Jia, Q. Q., and Li, S. (2019). Juvenile hormone signaling- a mini review. Insect. Sci. 26, 600–606. doi: 10.1111/1744-7917.12614
Lin, X., Yao, Y., and Wang, B. (2015). Methoprene-tolerant (Met) and Krüpple-homologue 1 (Kr-h1) are required for ovariole development and egg maturation in the brown plant hopper. Sci. Rep. 5:18064. doi: 10.1038/srep18064
Liu, W., Guo, S., Sun, D., Zhu, L., Zhu, F., Lei, C. L., et al. (2019). Molecular characterization and juvenile hormone-regulated transcription of the vitellogenin receptor in the cabbage beetle Colaphellus bowringi. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 229, 69–75. doi: 10.1016/j.cbpa.2018.12.004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lozano, J., and Belles, X. (2014). Role of methoprene-tolerant (met) in adult morphogenesis and in adult ecdysis of Blattella germanica. PLoS One 9:e103614. doi: 10.1371/journal.pone.0103614
Lu, Y. H., Wang, G. R., Zhong, L. Q., Zhang, F. C., Bai, Q., Zheng, X. S., et al. (2017). Resistance monitoring of Chilo suppressalis (Walker) (Lepidoptera: Crambidae) to chlorantraniliprole in eight field populations from east and central china. Crop. Prot. 100, 196–202. doi: 10.1016/j.cropro.2017.07.006
Ma, L., Zhang, W., Liu, C., Chen, L., Xu, Y., Xiao, H., et al. (2018). Methoprene-tolerant (Met) is indispensable for larval metamorphosis and female reproduction in the cotton bollworm Helicoverpa armigera. Front. Physiol. 9:1601. doi: 10.3389/fphys.2018.01601
Mao, K., Li, W., Liao, X., Liu, C., Qin, Y., Ren, Z., et al. (2019). Dynamics of insecticide resistance in different geographical populations of Chilo suppressalis (Lepidoptera: Crambidae) in China 2016-2018. J. Econ. Entomol. 112, 1866–1874. doi: 10.1093/jee/toz109
Marchal, E., Hult, E. F., Huang, J., Pang, Z., Stay, B., and Tobe, S. S. (2014). Methoprene-tolerant (met) knockdown in the adult female cockroach, Diploptera punctata completely inhibits ovarian development. PLoS One 9:e106737. doi: 10.1371/journal.pone.0106737
Meng, X. K., Miao, L. J., Ge, H. C., Yang, X. M., Dong, F., Xu, X., et al. (2019). Molecular characterization of glutamate-gated chloride channel and its possible roles in development and abamectin susceptibility in the rice stem borer, Chilo suppressalis. Pestic. Biochem. Physiol. 155, 72–80. doi: 10.1016/j.pestbp
Miao, L. J., Zhang, N., Jiang, H., Dong, F., Yang, X. M., Xu, X., et al. (2020). Molecular characterization and functional analysis of the vitellogenin receptor in the rice stem borer, Chilo suppressalis. Arch. Insect Biochem. Physiol. 103:e21636. doi: 10.1002/arch.21636
Parthasarathy, R., Tan, A. J., and Palli, S. R. (2008). bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech. Dev. 125, 601–616. doi: 10.1016/j.mod.2008.03.004
Roy, S., Saha, T. T., Zou, Z., and Raikhel, A. S. (2018). Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 63, 489–511. doi: 10.1146/annurev-ento-020117-043258
Smykal, V., and Raikhel, A. S. (2015). Nutritional control of insect reproduction. Curr. Opin. Insect. Sci. 11, 31–38. doi: 10.1016/j.cois.2015.08.003
Su, J., Zhang, Z., Wu, M., and Gao, C. (2014). Changes in insecticide resistance of the rice striped stem borer (Lepidoptera: Crambidae). J. Econ. Entomol. 107, 333–341. doi: 10.1603/ec13285
Suetsugu, Y., Futahashi, R., Kanamori, H., Keiko, K. O., Sasanuma, S., Narukawa, J., et al. (2013). Large scale full-length cDNA sequencing reveals a unique genomic landscape in a Lepidopteran model insect, Bombyx mori. G3 3, 1481–1492. doi: 10.1534/g3.113.006239
Wang, S. L., Baumann, A., and Wilson, T. G. (2007). Drosophila melanogaster Methoprene-tolerant (Met) gene homologs from three mosquito species: members of PAS transcriptional factor family. J. Insect. Physiol. 53, 246–253. doi: 10.1016/j.jinsphys.2006.07.011
Wilson, T. G., and Fabian, J. A. (1986). A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev. Biol. 118, 190–201. doi: 10.1016/0012-1606(86)90087-4
Yao, R., Zhao, D. D., Zhang, S., Zhou, L. Q., Wang, X., Gao, C. F., et al. (2017). Monitoring and mechanisms of insecticide resistance in Chilo suppressalis (Lepidoptera: Crambidae), with special reference to diamides. Pest. Manag. Sci. 73, 1169–1178. doi: 10.1002/ps.4439
Yue, Y., Yang, R. L., Wang, W. P., Zhou, Q. H., Chen, E. H., Yuan, G. R., et al. (2018). Involvement of Met and Kr-h1 in JH-mediated reproduction of female Bactrocera dorsalis (Hendel). Front Physiol. 9:482. doi: 10.3389/fphys.2018.00482
Zhan, S., Merlin, C., Boore, J. L., and Reppert, S. M. (2011). The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185. doi: 10.1016/j.cell.2011.09.052
Zhao, W. L., Liu, C. Y., Liu, W., Wang, D., Wang, J. X., and Zhao, X. F. (2014). Methoprene-tolerant 1 regulates gene transcription to maintain insect larval status. J. Mol. Endocrinol. 53, 93–104. doi: 10.1530/JME-14-0019
Keywords: Chilo suppressalis, juvenile hormone, methoprene-tolerant, vitellogenin, vitellogenin receptor, RNAi
Citation: Miao L, Zhang N, Jiang H, Dong F, Yang X, Xu X, Qian K, Meng X and Wang J (2020) Involvement of Two Paralogous Methoprene-Tolerant Genes in the Regulation of Vitellogenin and Vitellogenin Receptor Expression in the Rice Stem Borer, Chilo suppressalis. Front. Genet. 11:609. doi: 10.3389/fgene.2020.00609
Received: 20 February 2020; Accepted: 19 May 2020;
Published: 10 June 2020.
Edited by:
Fei Li, Zhejiang University, ChinaReviewed by:
Weihua Ma, Huazhong Agricultural University, ChinaCopyright © 2020 Miao, Zhang, Jiang, Dong, Yang, Xu, Qian, Meng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianjun Wang, d2FuZ2pqQHl6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.