- 1Key Laboratory of Germplasm Enhancement, Physiology and Ecology of Food Crops in Cold Region, Ministry of Education, Northeast Agricultural University, Harbin, China
- 2College of Life Science, Northeast Agricultural University, Harbin, China
Heavy metal exposure is a serious environmental stress in plants. However, plants have evolved several strategies to improve their heavy metal tolerance. Heavy metal-associated proteins (HMPs) participate in heavy metal detoxification. Here, we identified 46 and 55 HMPs in rice and Arabidopsis, respectively, and named them OsHMP 1–46 and AtHMP 1–55 according to their chromosomal locations. The HMPs from both plants were divided into six clades based on the characteristics of their heavy metal-associated domains (HMA). The HMP gene structures and motifs varied greatly among the different classifications. The HMPs had high collinearity and were segmentally duplicated. A cis-element analysis revealed that the HMPs may be regulated by different transcription factors. An expression profile analysis disclosed that only eight OsHMPs were constitutive in rice tissues. Of these, the expression of OsHMP37 was far higher than that of the other seven genes while OsHMP28 was expressed exclusively in the roots. For Arabidopsis, nine AtHMPs presented with very high transcript levels in all organs. Most of the selected OsHMPs were differentially expressed in various tissues under different heavy metal stresses. Only OsHMP09, OsHMP18, and OsHMP22 showed higher expression levels in all tissues under different heavy metal stresses. In contrast, most of the selected AtHMPs had nearly constant expression levels in different tissues under various heavy metal stresses. The AtHMP20, AtHMP23, AtHMP25, AtHMP31, AtHMP35, AtHMP46 expression levels under different heavy metal stresses were higher in the leaves and roots. The foregoing discoveries elucidated HMP evolution in monocotyledonous and dicotyledonous plants and may helpful functionally characterize HMPs in the future.
Introduction
Heavy metal pollution is a serious environmental problem associated with agricultural development and industrialization. It exerts negative effects on plants and threatens human health by causing heavy metal accumulating in food crops (Boyd, 2010). Certain heavy metals such as zinc (Zn) and copper (Cu) are microelements essential for plant growth and metabolism (Yuan et al., 2012). At micro-level concentrations, these heavy metals function as cofactors for enzymes in photosynthesis, biomacromolecule synthesis, electron transport, and other metabolic processes (Ricachenevsky et al., 2013). However, essential heavy metal overaccumulation is toxic to plant cells and inhibits their growth (Thomine and Vert, 2013; Cambrollé et al., 2015). Certain non-essential heavy metals such as cadmium (Cd) and lead (Pb) are phytotoxic at very low concentrations and adversely affect plant growth and development (Hayat et al., 2012; Gill et al., 2013).
Plants have evolved homeostatic mechanisms such as preventing root metal ion uptake and reducing long-distance metal ion transport to increase their tolerance for these substances (Clemens et al., 2002). In plant cells, metal ions can be rendered less harmful by transport, chelation, traffic, and vacuolar sequestration (Hall, 2002). Plants activate various signaling pathways and defense mechanisms that synthesize stress-related proteins in response to heavy metal exposure (Mourato et al., 2015). Heavy metal-associated proteins (HMPs) play key roles in heavy metal transport and detoxification in plant cells. HMPs are metalloproteins or metallochaperone-like proteins containing heavy metal-associated (HMA) domains (Tehseen et al., 2010; Zhang X. D. et al., 2018). The HMA domain is conserved and comprises ~30 amino acid residues. It occurs in several proteins that transport or detoxify heavy metals (Bull and Cox, 1994) and contains two cysteine residues that bind and transfer copper, cadmium, cobalt, zinc, and other heavy metal ions (Gitschier et al., 1998).
As a rule, plant proteins containing HMA domains fall into one of the following groups: HPPs (heavy metal-associated plant proteins), HIPPs (heavy metal-associated isoprenylated plant proteins) (de Abreu-Neto et al., 2013), ATX1-like (Puig et al., 2007), and P1B-ATPase (Pedersen et al., 2012). Previous studies focused mainly on the functions of P1B-type ATPase HMPs. Nine and eight P1B-type ATPase HMPs were identified in rice and Arabidopsis, respectively (Pedersen et al., 2012; Zhiguo et al., 2018). Of these, OsHMA3 was found to control the root-to-shoot Cd translocation rates (Miyadate et al., 2011). OsHMA4 sequesters Cu in root cell vacuoles and limits Cu transport to the grain and its accumulation there (Huang et al., 2016). In Arabidopsis, the Cu-translocating ATPase AtHMA5 is induced by high Cu levels and causes the efflux of excess Cu from the cytosol to the plasma membrane (Andrés-Colás et al., 2006; Kobayashi et al., 2008). Several ATX1-like metallochaperones have been functionally identified in Arabidopsis and rice (Zhang X. D. et al., 2018). A previous study reported that the ATX1-like Cu chaperones ATX1 and CCH in rice, Arabidopsis, and soybean transferred Cu to yeast Ccc2 P1B-type ATPase which, in turn, enhanced its antioxidant mechanism (Puig et al., 2007). The HPP and HIPP clades contain the largest number of HMPs but only a few genes in them have been functionally investigated. AtHIPP3 (AtHMP52 in the present study) was identified as an upstream controller of stress- and development-related regulatory networks. It is also involved in the salicylate-dependent pathogen response pathway and in flower and seed development (Zschiesche et al., 2015).
Rice (Oryza sativa L.) and Arabidopsis thaliana L. are research models for monocotyledonous and dicotyledonous plants, respectively. In earlier research, different heavy metal gene families in various species were studied or classified separately (Li et al., 2015; Fang et al., 2016; Zhiguo et al., 2018; Khan et al., 2019). Here, we identified all heavy metal-associated proteins in rice and Arabidopsis, including HPP, HIPP, ATX, CCH, CCS, and P1B-ATPase HMPs, by repeated HMM searches in silico. We analyzed HMP chromosomal distributions, gene synteny, phylogeny, gene structures, motif compositions, cis-elements, expression patterns, and heavy metal stress responses in different tissues of rice and Arabidopsis. The aims of this study were to clarify the evolutionary and taxonomic relationships among the heavy metal-associated proteins and identify their expression patterns in different tissues and under various types of metal ion stress. This information may serve as a theoretical basis for the elucidation of the mechanisms of heavy metal tolerance and plant-metal interactions.
Materials and Methods
Identification and Sequence Analysis of HMP Family Genes in Rice and Arabidopsis
The HMPs of various plant species were identified according to a previously described method (Li et al., 2019). The Hidden Markov Model of the HMA domain (PF00403) was downloaded from the Pfam database (http://pfam.xfam.org/) (El-Gebali et al., 2019). The amino acid, genome, and CDS sequence assemblies were downloaded from the EnsemblPlants database (http://plants.ensembl.org/index.html) (Kersey et al., 2018). Candidate proteins were sought with the HMMSEARCH program (https://www.ebi.ac.uk/Tools/hmmer/search/hmmsearch) based on the Bio-Linux system (Dr. Tracey Timms-Wilson, Centre for Ecology & Hydrology (CEH), Oxfordshire, UK). Only proteins with E-value < 0.01 were selected and they were verified against the Pfam and InterPro databases (http://www.ebi.ac.uk/interpro/) (Mitchell et al., 2019).
The MEME program (http://meme-suite.org/) identified conserved HMP family protein motifs. The HMP family gene structures were displayed using Gene Structure Display Server tools (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). The chromosomal locations of the HMP family genes were mapped according to their TIGR numbers onto a rice or Arabidopsis linkage map using online tools (Kurata and Yamazaki, 2006; Lamesch et al., 2012). The isoelectric points and molecular weights of the HMP family proteins were estimated with ExPASy (http://expasy.org/) (Artimo et al., 2012). Subcellular localizations of the HMP family proteins were predicted by Cell-PLoc v. 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) (Chou and Shen, 2010).
Phylogenetic Analysis
A multiple alignment was performed on the HMA domain sequences of various plant species using MEGA v. 7.0 (https://www.megasoftware.net/) (Kumar et al., 2016). Unrooted trees were constructed by the maximum likelihood (ML) method with the following parameters: Poisson correction; pairwise deletion; 1,000 bootstrap replicates.
Gene Duplication Analysis
Synteny blocks of various plant genomes were downloaded from the Plant Genome Duplication Database (PGDD, http://chibba.agtec.uga.edu/duplication/) (Lee et al., 2013). Duplicated HMP gene pairs were connected by solid lines.
Cis-Element Analysis of HMP Family
HMP family gene promoters were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html#) (Goodstein et al., 2011). The PLACE database (https://sogo.dna.affrc.go.jp/) was used to analyze the cis-regulatory elements on the HMP family gene promoters (Higo et al., 1999).
Plant Growth Conditions and Treatments
Nipponbare rice seeds (O. sativa L. ssp. japonica) were surface-sterilized with 10% (w/v) sodium hypochlorite solution for 30 min, sown onto 1/2 MS (Murashige & Skoog) solid medium, and grown in a light incubator. After 2 weeks, seedlings at the two-true-leaf stage were transplanted into Hoagland's nutrient solution and cultured under 14 h light at 28°C, 10 h dark at 22°C, and RH = 70%. Arabidopsis thaliana L. (Heyn) cv. Columbia plants were grown on germination medium (GM) agar plates for 2 weeks as described previously (Qin et al., 2008), transferred to vermiculite, and grown under a 16 h light/8 h dark photoperiod.
For the heavy metal treatments, the plants were exposed to 100 μM CdCl2, 100 μM CuSO4, 500 μM Pb (NO3)2, and 500 μM ZnSO4 (Feng, 2011; Li et al., 2015; Fu et al., 2017). Three biological replicates were prepared per treatment. The control treatment consisted of normal nutrient solution or medium. All other culture conditions were the same as those described above. The samples were harvested after treatment for 1, 3, 12, and 24 h and immediately placed in liquid nitrogen and stored at −80°C until use. The experimental procedure was repeated at least thrice.
Expression Analysis of HMP Gene Family
To analyze the HMP gene expression profiles in different tissues, RNA-seq data were downloaded from the Expression Atlas database (https://www.ebi.ac.uk/gxa/home) (Papatheodorou et al., 2017). RNA-seq data for Arabidopsis used in the present research are available in the Sequence Read Archive database under accession number SRP013631 and in the GEO database under accession number GSE38612 and GSE108751. RNA-seq data for rice are available in the DNA Data Bank of Japan Sequence Read Archive under accession numbers DRR001024–DRR001051 and in the Sequence Read Archive database under accession numbers SRP008505, SRP008469, and SRP008821 and in the GEO database under accession number GSE34895. Heatmaps were generated with HemI from the normalized value by row for the signatures in transcripts per million (TPM).
Total RNA used for quantitative real-time PCR analysis was extracted from the plant tissues with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and treated with DNase I to eliminate any DNA contamination. RNA quality was assessed by gel electrophoresis and the RNA was stored at −80°C until use. First-strand cDNA (10 μL) was synthesized according to the instructions for the PrimeScript™ RT Master Mix (Takara Biomedical Technology (Beijing) Co., Ltd., Beijing, China). Quantitative real-time PCR was performed as described previously (Li et al., 2019). The gene-specific primers used in the quantitative real-time PCR are listed in Table S1.
Statistical Analyses
Statistical analyses were completed using the Statistical Program for Social Sciences (release 19.0, SPSS Inc., IBM, www.ibm.com) and Microsoft Excel 2016.
Results
Genome-Wide Identification, Chromosomal Distributions and Synteny Analysis of HMP in Rice and Arabidopsis
Forty-six and 55 candidate rice and Arabidopsis HMP genes, respectively, were identified by a Hidden Markov Model for the HMA domain. All HMPs were mapped onto chromosomes and named OsHMP01-OsHMP46 and AtHMP01-AtHMP55 according to the gene orders on their respective chromosomes. OsHMP26, OsHMP36, AtHMP01, AtHMP03, AtHMP14, AtHMP21, AtHMP25, AtHMP30, AtHMP37, AtHMP38, AtHMP42, AtHMP43, AtHMP53, and AtHMP54 had two alternative splicings, OsHMP45, OsHMP46, AtHM46, and AtHMP47 had three alternative splicings, and AtHMP52 had four alternative splicings. Characteristics of the rice and Arabidopsis HMPs are summarized in Table S2.
OsHMP07 was the smallest protein (69 amino acids) while OsHMP11 was the largest (1,012 amino acids). Their molecular weights range from 7.65 to 108.48 kDa and their predicted isoelectric points varied from 5.01 (OsHMP35) to 11.07 (OsHMP40). Thirty-one OsHMPs were in the nuclei, 12 in the chloroplasts, and nine in the cell membranes. AtHMP44 was the smallest protein (77 amino acids) while AtHMP51 was the largest (1,001 amino acids). The molecular weights range from 8.87 to 107.39 kDa and their predicted isoelectric points varied from 4.88 (AtHMP47) to 10.14 (AtHMP34). Thirty-nine HMPs were in the nuclei, ten in the chloroplasts, five in the cell membranes, four in the cytoplasms, and four in the mitochondria. AtHMP13 and AtHMP40 were in the cytoplasms, AtHMP24 was in the extracellular spaces, AtHMP29 was in the vacuoles, AtHMP29 and AtHMP32 were in the cell walls, and AtHMP40 was in the peroxisomes. However, the subcellular localization of HMPs in rice and Arabidopsis in the present study is only based on purely computational prediction, the validation in situ should be performed in the future research.
The chromosomal locations of the HMPs were identified by extracting chromosomal data. Figure 1 shows that the HMPs were unevenly and non-randomly distributed on the chromosomes. Chr1 (chromosomal 1) contained the largest number of OsHMPs (10) while Chr9, Chr11, and Chr12 contained only one each. Chr1–4 contain 29 HMPs whereas Chr5–12 had only 2–3 OsHMPs. Thus, the OsHMPs were distributed mainly on Chr1–4. In contrast, the AtHMPs were more evenly distributed on the chromosomes. As shown in Figure 1B, the longer Chr1 and Chr5 contained more AtHMPs (15 and 14, respectively) while the shorter Chr2 and Chr4 contained fewer AtHMPs (6 and 9, respectively).
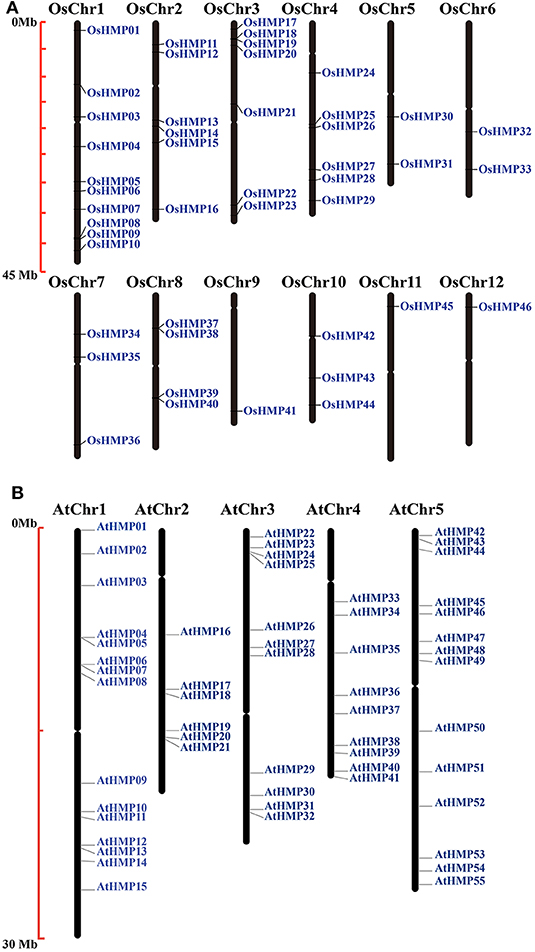
Figure 1. Chromosomal locations of heavy-metal-associated genes in rice (A) and Arabidopsis (B). Black bars represent the chromosomes. Chromosome numbers are shown at the tops of the bar. Heavy-metal-associated genes are labeled at the right of the chromosomes. Scale bar on the left indicates the chromosome lengths (Mb).
The OsHMPs had five pairs of clustered genes (OsHMP08 and OsHMP09, OsHMP18 and OsHMP19, OsHMP25 and OsHMP26, OsHMP37 and OsHMP38, and OsHMP39 and OsHMP40). Twelve AtHMPs were clustered into six tandem duplication event regions on the chromosomes (AtHMP04 and AtHMP05, AtHMP06 and AtHMP07, AtHMP12 and AtHMP13, AtHMP20 and AtHMP21, AtHMP24 and AtHMP25, and AtHMP31 and AtHMP32). Figure 2A shows that seven segmental duplication events with 14 OsHMPs were identified and localized to duplicated segments on chromosomes 1, 2, 3, 4, 5, 7, and 10. The Arabidopsis HMPs exhibited eight segmental duplication events distributed on all chromosomes (Figure 2B). Therefore, synteny of the HMPs was highly conserved and non-diverse.
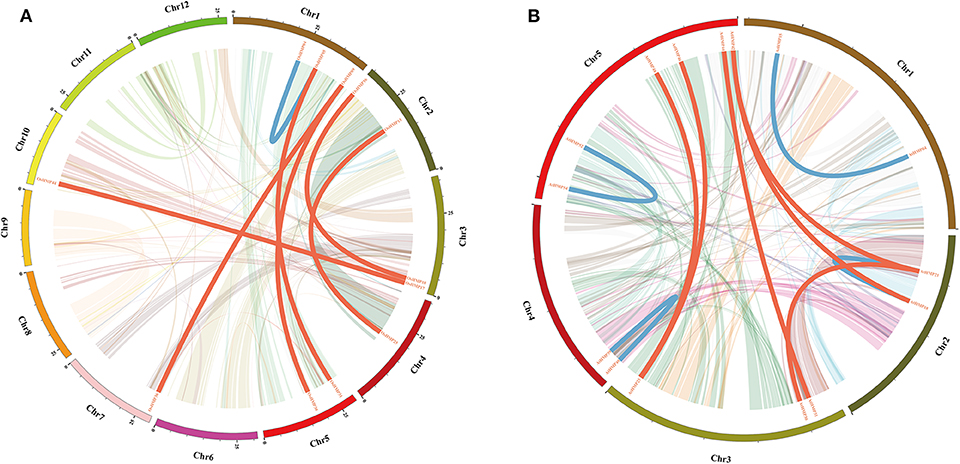
Figure 2. Schematic representations of segmental duplications of rice (A) and Arabidopsis (B) heavy-metal-associated genes. Different color lines indicate all synteny blocks in rice or Arabidopsis genome between each chromosome. Thick red lines indicate duplicated heavy-metal-associated gene pair on different chromosome, and thick blue lines indicate duplicated gene pair on same chromosome. The chromosome number is indicated at the bottom of each chromosome. Scale bar marked on the chromosome indicating chromosome lengths (Mb).
To expand our investigation of orthologous HMP family genes between rice and other species, we constructed 10 syntenic rice maps associated with the monocots Brachypodium distachyon, Oryza brachyantha, Triticum aestivum, Setaria italica, and Zea mays (Figure 3) and the dicots Brassica rapa, Cucumis sativus, Glycine max, Gossypium raimondii, and Solanum tuberosum (Figure S1). According to their relationships, Oryza brachyantha had the greatest syntenic conservation (39 HMP orthologous gene pairs distributed on all chromosomes) followed by Zea mays (37), Brachypodium distachyon (34), Triticum aestivum (30), and Setaria italica (29). Therefore, the syntenic relationships among the HMP family genes in various species are relatively conservative. Ten homologous genes between rice and soybean were detected followed by Cucumis sativus (2). Only one OsHMPs showed a syntenic relationship with those in Brassica rapa, Gossypium raimondii, and Solanum tuberosum. OsHMP01, OsHMP05, OsHMP07, OsHMP09, OsHMP11, OsHMP12, OsHMP17, OsHMP18, OsHMP22, OsHMP25, OsHMP27, OsHMP28, OsHMP29, OsHMP30, OsHMP35, OsHMP43, and OsHMP46 were associated with all homologous gene pairs between rice and other monocots species. Therefore, these genes may have been implicated in the OsHMP family during gene duplication.
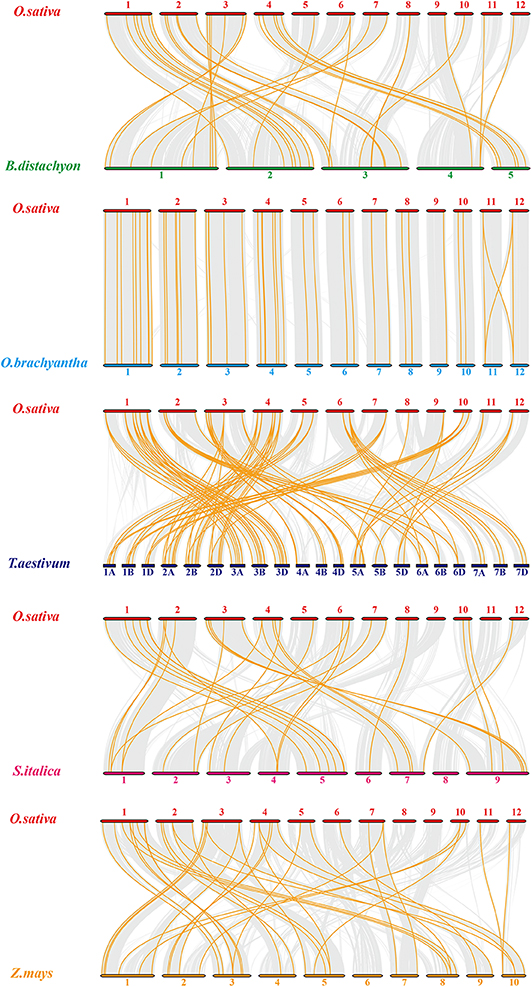
Figure 3. Synteny analysis of heavy-metal-associated genes between rice and Brachypodium distachyon, Oryza brachyantha, Triticum aestivum, Setaria italica, and Zea mays. Gray lines in the background indicate the collinear blocks within rice and other plant genomes, while the orange lines highlight the syntenic OsHMP gene pairs. The species names with the prefixes “O. sativa,” “B. distachyon,” “O. brachyantha,” “T. aestivum,” “Setaria italica,” and “Z. mays” indicate Oryza sativa, Brachypodium distachyon, Oryza brachyantha, Triticum aestivum, Setaria italica, and Zea mays, respectively. Different color bars represent the chromosomes of different species. The chromosome number is labeled at the top or bottom of each chromosome.
For Arabidopsis orthologous HMP family genes between Arabidopsis and other species, fifty-four AtHMPs showed synteny with those in Brassica rapa followed by Glycine max (49), Cucumis sativus (28), Gossypium raimondii (20), and Solanum tuberosum (10) (Figure 4). In addition, only three of the AtHMPs showed syntenic relationships with those in Brachypodium distachyon, Setaria italica, and Triticum aestivum, followed by Oryza brachyantha (1). No HMP gene duplication event was detected between Arabidopsis and Zea mays (Figure S2). Furthermore, only one orthologous HMP gene was detected between rice and Arabidopsis (Figure S3). Syntenic analysis of the HMPs of Arabidopsis and the other five dicotyledonous species disclosed that AtHMP20, AtHMP42, and AtHMP43 were associated with ≥ 1 syntenic gene. This association may have played a central role in the gene duplication of the HMP gene family. The foregoing results show that numerous HMPs could be produced by gene replication. We calculated the Ka/Ks ratios of the HMP syntenic gene pairs to clarify the selective pressure on the HMP gene family (Tables S3, S4). Most of gene pairs had Ka/Ks < 1. However, some syntenic gene pairs showed Ka/Ks > 1, indicating that these genes might have undergone positive selective pressure.
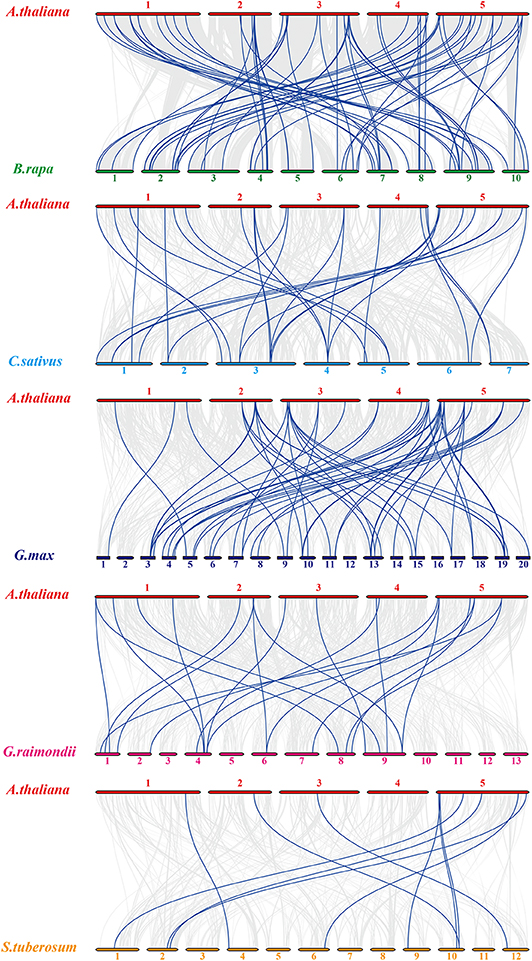
Figure 4. Synteny analysis of heavy-metal-associated genes between Arabidopsis thaliana and Brassica rapa, Cucumis sativus, Glycine max, Gossypium raimondii, and Solanum tuberosum. Gray lines in the background indicate the collinear blocks within rice and other plant genomes, while the blue lines highlight the syntenic AtHMP gene pairs. The species names with the prefixes “A. thaliana,” “B. rapa,” “C. sativus,” “G. max,” “G. raimondii,” and “S. tuberosum” indicate Arabidopsis thaliana and Brassica rapa, Cucumis sativus, Glycine max, Gossypium raimondii, and Solanum tuberosum, respectively. Different color bars represent the chromosomes of different species. The chromosome number is labeled at the top or bottom of each chromosome.
Multiple Sequence Alignment, Phylogenetic Analysis, and Classification of HMPs
We examined the phylogenetic relationships of the HMPs by multiple sequence alignment of their HMA domains. Figure 5 shows that the core sequences for each classification in the HMA domain were highly conserved. Fifty-two OsHMPs and 73 AtHMPs had the highly conserved sequence “CXXC” (where “X” denotes different amino acids) while the AtHMP11 in the H3 category varied by one amino acid (“C” to “F”). For each classification, the H2 and H4 groups exhibit relatively more conservative domains. The “XX”s of the “CXXC” in their HMA domains were nearly always “DG,” “EG,” or “VG” except for “TG” in OsHMP23 in the H4 group and “DK” in OsHMP29 of the H2 group. The P1B-ATPase group presented with divergence of its HMA domains. Thus, the functions of the proteins in the P1B group may differ from those in the other groups.
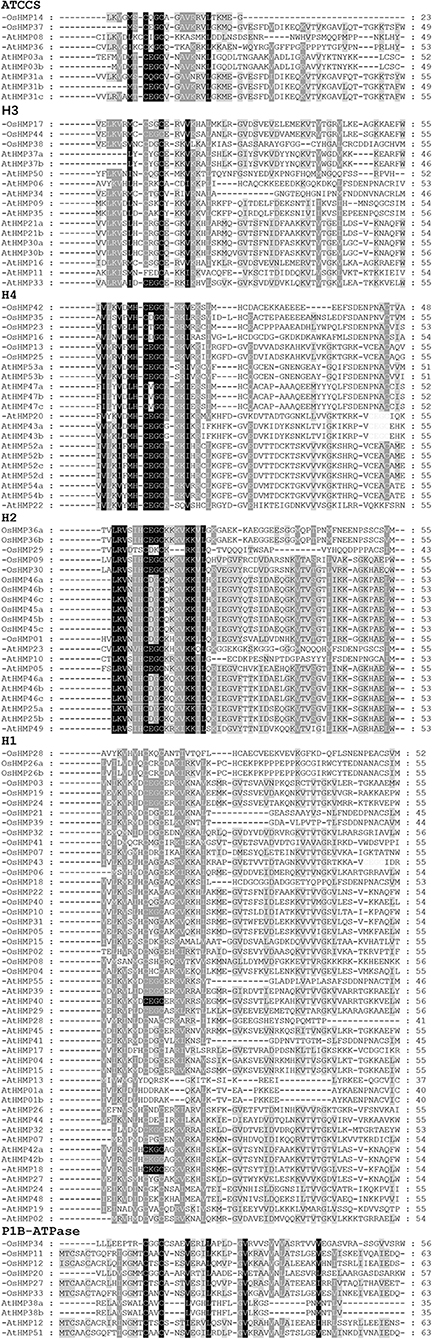
Figure 5. Multiple Alignment of rice and Arabidopsis HMP and selected heavy-metal-associated domain amino acid sequences. “ATCCS,” “H1,” “H2,” “H3,” “H4,” and “P1B-ATPase” represent different HMP proteins classification.
To expand our investigation of the phylogenetic relationships among HMPs, we built a maximum likelihood phylogenetic tree according to the results of the multiple sequence alignment of the HMA domains. Figure 6 shows that all HMPs could be divided into the subfamilies H1, H2, ATCCS, P1B-ATPase, H3, and H4 according to their structure or function. Certain genes previously identified as P1B-ATPase members including GmHMA1, GmHMA12, GmHMA17, and GmHMA20 in soybean, ZmHMA4 and ZmHMA9 in maize, PtHMA1 in Populus trichocarpa, and SbHMA4 in sorghum were classified into the P1B-ATPase clade. The credibility of this classification was confirmed by the phylogenetic tree.
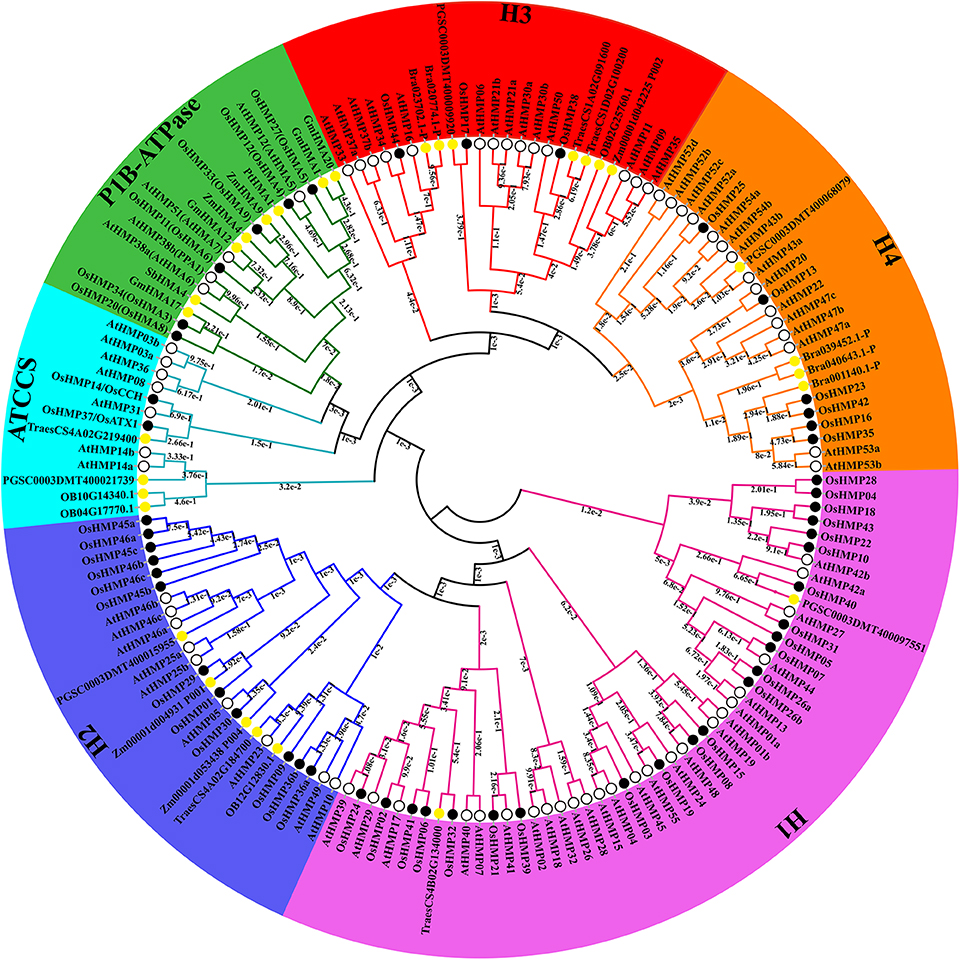
Figure 6. Phylogenetic relationships among 156 heavy-metal-associated proteins in rice, Arabidopsis, soybean, maize, potato, wheat, sorghum, wild rice, Brachypodium distachyon, and Populus trichocarpa. The maximum likelihood tree was created using MEGA v. 7.0 (bootstrap value = 1,000) and the bootstrap value of each branch is displayed. The black solid circles, hollow circles, and yellow solid circles represent heavy-metal-associated proteins from rice, Arabidopsis, and other species, respectively.
The HMA domains of the proteins in the H1 and H2 groups were located in the N termination. The amino acid sequences of the HMPs in the H1 clade were shorter than those in the H2 clade. The amino acid sequences in the H3 subfamily were also shorter than those in the H2 family. The HMA domains of certain proteins in the H3 subfamily were located in the C termination. Certain chloroplast-targeted copper chaperone proteins were detected in both the H1 and H3 subfamilies. Based on the structural differences among the HMA domains (Figure 5), the proteins in the H1 and H2 could not cluster into one group. The ATCCS clade may be implicated in metal cation transport for various peroxidases such as copper/zinc superoxide dismutase. The main function of P1B-ATPase is to help transport metal ions across biological membranes (Axelsen and Palmgren, 1998; Argüello, 2003). The biological functions of the proteins in the H4 clade, which contained two HMA domains, have not yet been established. However, it is known that the HMPs in this subfamily include isoprenylated FARNESYLATED PROTEIN 3-RELATED proteins which are involved in heavy metal detoxification and are responsive to Cd2+, Hg2+, Fe2+, and Cu2+ (Suzuki et al., 2002; Crowell and Huizinga, 2009).
HMP Structure and Motif Composition
We analyzed the HMP gene structure to identify the differences between the HMPs from various subfamilies in rice and Arabidopsis. Figure 7 shows that the number of HMP exons was discontinuously distributed from 1 to 16. By combining the gene structure (Figure 7B) with the phylogenetic tree (Figure 7A), we found that the HMP exon numbers in various subgroups were related to their classification. All genes from the H1, H3, H4, ATCCS, and H2 clades contained 2–4 exons whereas OsHMP46a and OsHMP46c from the H2 clade contained eight exons, AtHMP11 from the H3 clade contained five exons, and AtHMP03a from the ATCCS clade contained six exons. The P1B-ATPase genes in these groups contained more exons than those in other branches. Moreover, the HMPs nucleotide sequence lengths varied among different classifications. The genes of the P1B-ATPase clade had longer nucleotide sequences than those of the other groups. The nucleotide sequences of the genes in the H2 and H4 groups were longer than those in the H1, H3, and ATCCS groups but shorter than those in the P1B-ATPase subgroup except for OsHMP14 from ATCCS whose structure may naturally vary. Homologous genes usually have similar characteristics. For this reason, the numbers of exons and the lengths of the genes are similar within the same subfamily.
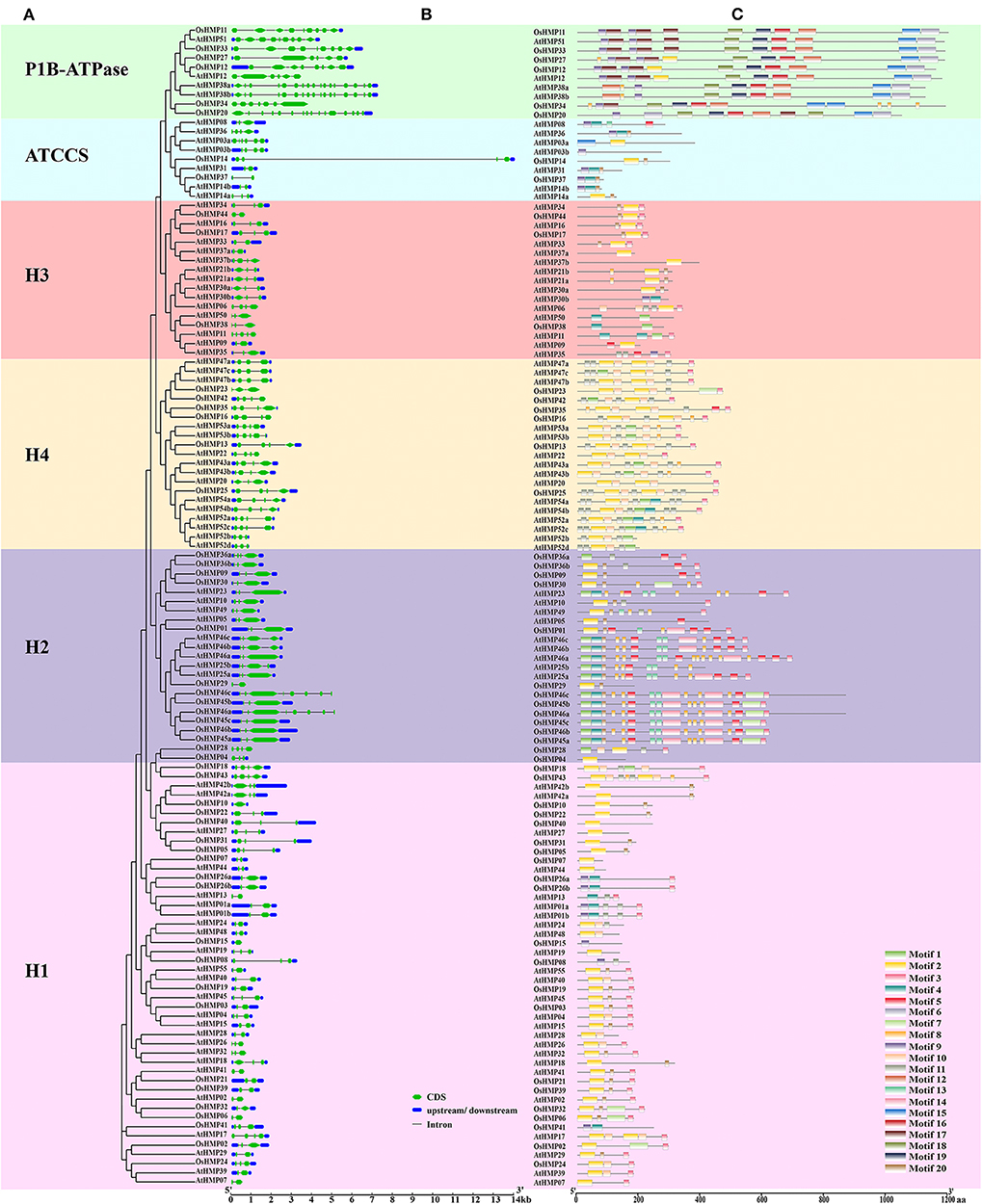
Figure 7. Phylogenetic analysis, gene structure and motif analysis of heavy-metal-associated genes in rice and Arabidopsis. (A) Phylogenetic tree of HMP proteins between rice and Arabidopsis. The maximum likelihood tree was created using MEGA v. 7.0. P1B-ATPase, ATCCS, H1, H2, H3, and H4 are marked with different colors. (B) Gene structure of HMP genes in rice and Arabidopsis. A schematic diagram was constructed by the Gene Structure Display Server 2.0. Exons, introns, and untranslated regions are marked by green double-sided wedge, black lines, and blue round-corner rectangles, respectively. The scale bar at the bottom estimates the lengths of the exons, introns, and untranslated regions. (C) Motif composition of HMP proteins in rice and Arabidopsis. Motif analysis was performed using the MEME program. Boxes of different colors represent the various motifs. Their location in each sequence is marked. Motif sequence logo is shown in Figure S4. The scale bar at the bottom indicates the lengths of the HMP protein sequences.
We analyzed the HMP motif compositions to establish the functions of the HMP protein. We identified 20 motifs with E < 1.8 × 10−45. The motif sequence logo is presented in Figure S4. Motifs 1, 2, and 9 were identified as HMA domains with several altered amino acid residues. Figure 7C shows that 30 HMP proteins contained motif 1, 87 proteins contained motif 2, and 26 proteins contained motif 9. There were significant differences in the HMP protein motifs among the different classifications (Figures 7A,C). P1B-ATPase had a longer amino acid sequence than the other subgroups. It also contained specific motifs such as 6, 15, 16, 17, 18, and 19. Thus, the HMP proteins in the P1B-ATPase must have specialized physiological functions. The H4 clade had numerous short motifs and was densely distributed on the amino acid sequences. The H2 subgroup also contained several short motifs but its distribution was not as dense as that of the H4 clade. The H1, H3, and ATCCS groups all had similar motif distributions. The H1, H3, and ATCCS groups had fewer motifs and shorter amino acid sequences than the other three categories. Therefore, these proteins may have auxiliary physiological functions such as enzyme subunits or molecular chaperones.
HMP Cis-Element Analysis
To clarify upstream HMP regulation, the promoter sequences (−1,500 bp upstream of the HMP genomic sequence) of 101 rice and Arabidopsis HMPs were submitted to New PLACE and the cis-elements were investigated. In the present study, only the cis-elements common to all rice and Arabidopsis HMPs were displayed. As shown in Figure 8, seven cis-elements common to the OsHMPs were scanned out. DOFCOREZM and WRKY71OS were core sequences for transcription factor (TFs) binding (by Dof TFs and WRKY TFs) (Yanagisawa, 2000; Xie et al., 2005), ARR1AT was a type of response regulator (Ross et al., 2004), CACTFTPPCA1 was in the distal region of phosphoenolpyruvate carboxylase (Gowik et al., 2004), GTGANTG10 and POLLEN1LELAT52 were pollen-specific expression elements indicating that HMPs may participate in pollen biosynthesis (Rogers et al., 2001; Filichkin et al., 2004), and MYCCONSENSUSAT was abscisic acid (ABA) and a cold- and dehydration-responsive element. Thus, HMPs may be involved in environmental stress tolerance (Lee et al., 2005; Agarwal et al., 2006).
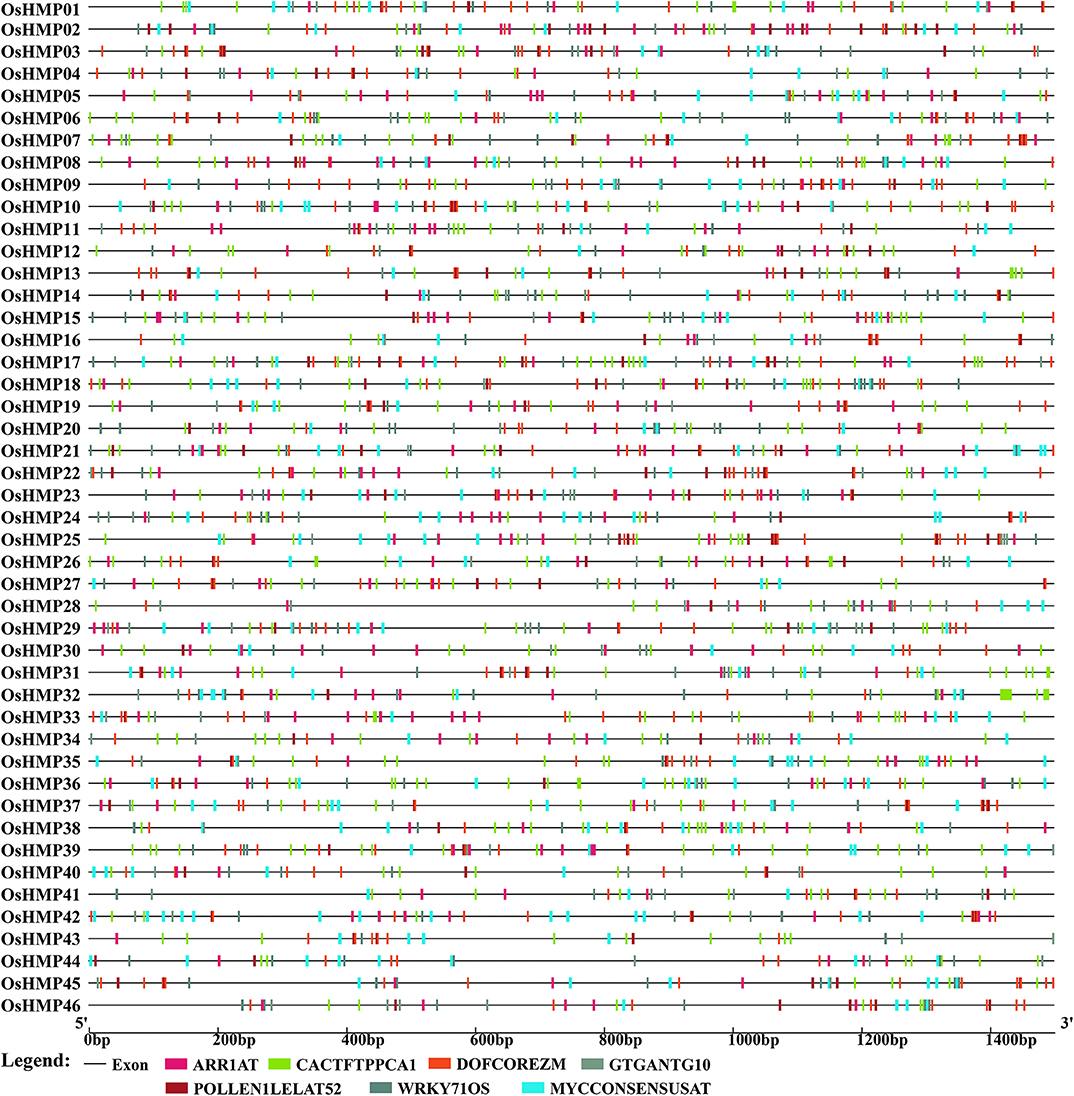
Figure 8. Predicted cis-elements in the promoter regions of the rice heavy-metal-associated genes. All promoter sequences (−1,500 bp upstream genomic sequence) were analyzed. The heavy-metal-associated genes are shown on the left side of the figure. The scale bar at the bottom indicates the length of promoter sequence. Different cis-elements were labeled by rectangle of different color.
The AtHMPs contained all of the cis-elements detected in the OsHMPs (Figure 9 and Table S5). For this reason, the regulatory mechanism for HMP may not significantly differ between monocotyledons and dicotyledons. The AtHMPs also contained a specific cis-element not found in rice, namely, TAAAGSTKST1, which may be a target site for a trans-acting Dof protein controlling guard cell-specific gene expression (Plesch et al., 2001). The cis-acting element analysis indicated that the HMPs may be regulated by numerous transcription factors.

Figure 9. Predicted cis-elements in the promoter regions of the Arabidopsis heavy-metal-associated genes. All promoter sequences (−1,500 bp upstream genomic sequence) were analyzed. The heavy-metal-associated genes are shown on the left side of the figure. The scale bar at the bottom indicates the length of promoter sequence. Different cis-elements were labeled by rectangle of different color.
HMP Expression Profiles in Various Rice and Arabidopsis Tissues
We evaluated the OsHMPs expression profiles in different tissues via the RNA-seq data (Figure 10 and Table S6). Figure 10A shows that the gene expression levels varied greatly among different tissues. Several genes were more strongly upregulated in the carpels, emerging inflorescences, leaves, pistils, embryos, and shoots than they were in the other organs. In contrast, no highly expressed genes were detected in the pollen sperm cells. Most of the OsHMPs in the pollen sperm cells, endosperms, and microgametophyte vegetative cells had low transcript levels.
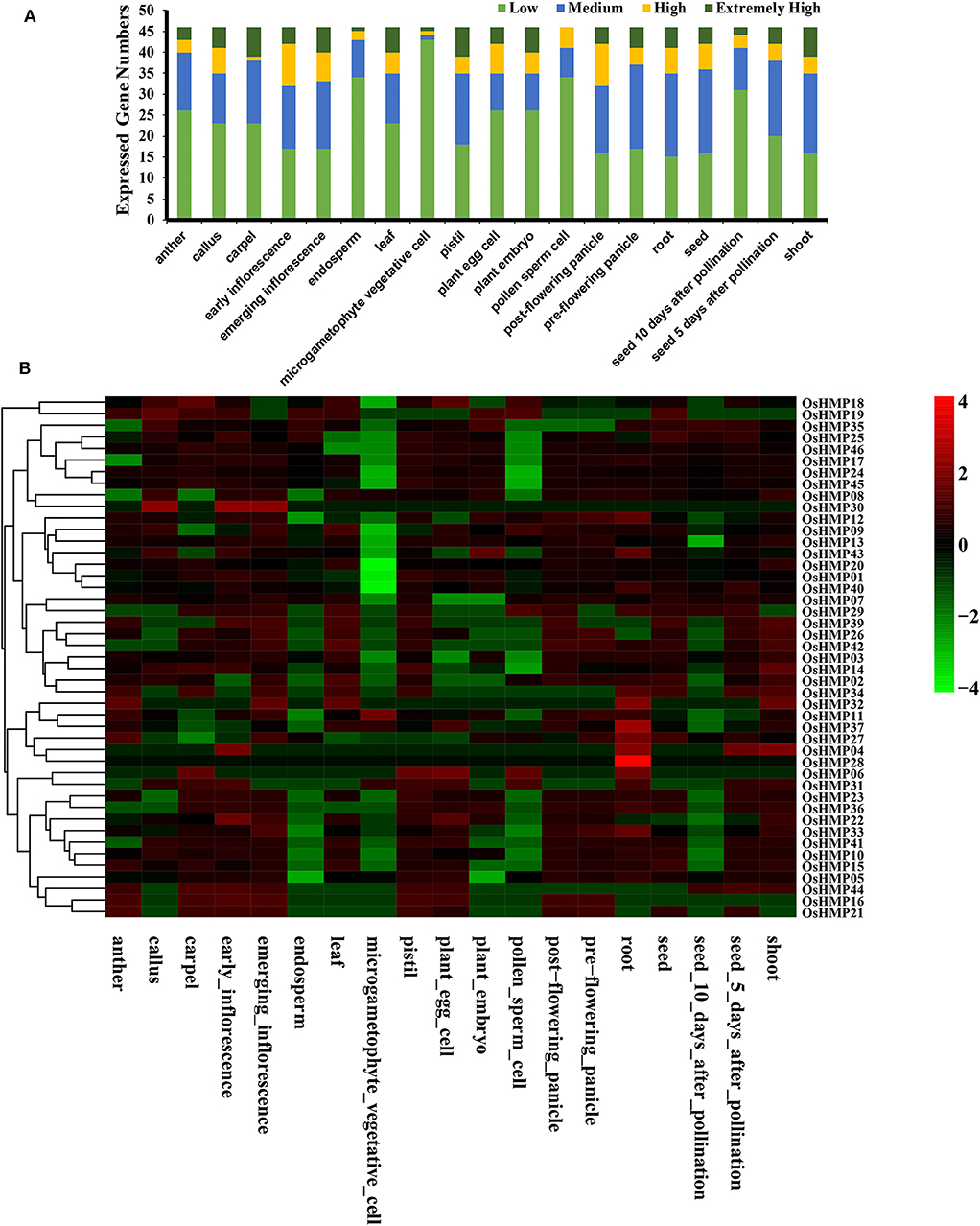
Figure 10. Expression pattern of the rice heavy-metal-associated gene family in various tissues or stages. (A) Numbers of expressed genes in each organ. Expression data of the rice HMP genes were downloaded from the Expression Atlas database. Extremely high: TPM > 100, high: 100 ≥TPM >50, medium: 50 ≥ TPM > 5, low: 5 ≥ TPM > 0; (B) Expression patterns of the rice HMP genes in various tissues. Heatmaps were generated using HemI from the normalized value by row for the signatures in transcripts per million (TPM). Transcript levels are depicted by different colors on the scale. Green and red represent low and high expression levels, respectively.
All OsHMPs were expressed in ≥ 1 tissue. No pseudogenes were found in the present study (Figure 10B). Only OsHMP28 was exclusively expressed in the root. Twelve OsHMPs were expressed in all 19 samples tested (TPM > 0) while eight OsHMPs were constitutive (TPM > 1 in all samples). OsHMP37 (OsATX1) clustered into the ATCCS group had far higher transcript levels than the other seven genes in all tissues. Conversely, OsHMP06, OsHMP16, OsHMP30, OsHMP31, and OsHMP41 were expressed either at low levels or not at all in every tissue. Certain OsHMPs preferentially expressed in specific tissues. OsHMP04 expressed only in the roots and shoots, OsHMP19 expressed only in the calli, OsHMP29 expressed only in pollen sperm cells, OsHMP32, OsHMP34 and OsHMP36 expressed only in the roots, and OsHMP44 expressed only in the carpels. The early inflorescences presented with higher transcript abundances than all other organs.
In general, very few AtHMPs had extremely high transcript levels in any tissue (Figure 11A and Table S7). The AtHMPs exhibited tissue-specific expression while most of the AtHMPs maintained very low transcript levels in all tissues. Of all 55 AtHMPs, AtHMP07, AtHMP08, AtHMP13, AtHMP17, AtHMP18, AtHMP22, AtHMP26, AtHMP34, AtHMP36, and AtHMP55 presented with low transcript levels in all tissues whereas those for AtHMP03, AtHMP14, AtHMP20, AtHMP23, AtHMP25, AtHMP31, AtHMP35, AtHMP46, and AtHMP51 remained high. Certain AtHMPs expressed in only one tissue. AtHMP02, AtHMP06, AtHMP24, and AtHMP32 expressed exclusively in the roots whereas AtHMP11 expressed exclusively in the fruits and AtHMP50 expressed exclusively in the flowers. AtHMP01, AtHMP04, AtHMP05, AtHMP21, and AtHMP49 in the fruits, AtHMP04, AtHMP27, AtHMP39, and AtHMP47 in the leaves, and AtHMP15, AtHMP30, and AtHMP54 in the roots displayed low expression levels but were highly upregulated in all other tissues. In contrast, AtHMP10 and AtHMP37 in the flowers, AtHMP29 in the fruits, and AtHMP12, AtHMP16, AtHMP19, AtHMP33, AtHMP41, and AtHMP43 in roots exhibited high transcript levels compared to those in the other organs (Figure 11B). HMPs displayed tissue-specific expression which could indicate their particular roles in various mechanisms.
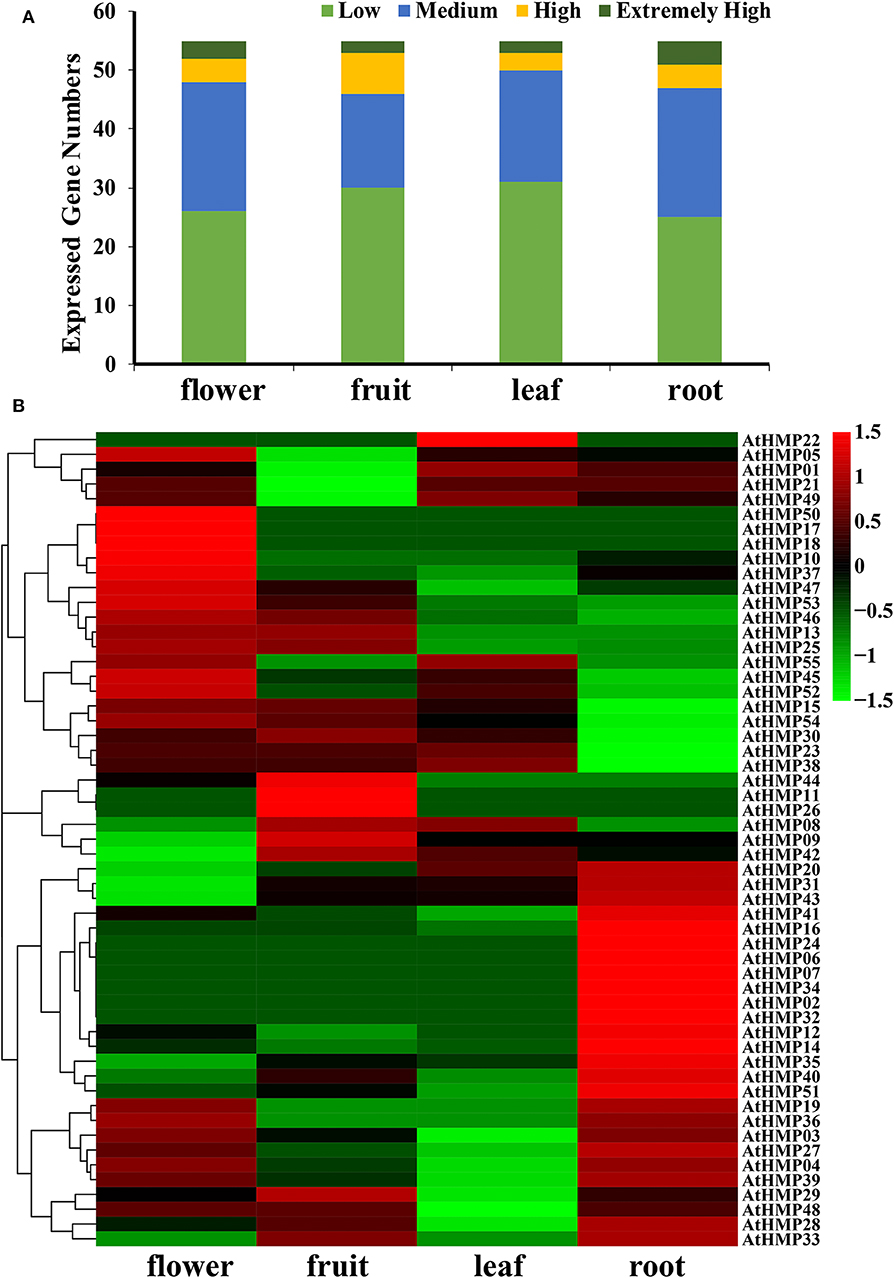
Figure 11. Expression pattern of the Arabidopsis heavy-metal-associated gene family in various tissues or stages. (A) Numbers of expressed genes in each organ. Expression data of the Arabidopsis HMP genes were downloaded from the Expression Atlas database. Extremely high: TPM > 100, high: 100 ≥ TPM > 50, medium: 50 ≥ TPM > 5, low: 5 ≥ TPM > 0; (B) Expression patterns of the Arabidopsis HMP genes in various tissues. Heatmaps were generated using HemI from the normalized value by row for the signatures in transcripts per million (TPM). Transcript levels are depicted by different colors on the scale. Green and red represent low and high expression levels, respectively.
OsHMP and AtHMP Expression Pattern Analysis in Responses to Heavy Metal Ion Stress
We subjected rice and Arabidopsis seedlings to Cu2+, Cd2+, Zn2+, and Pb2+, selected 12 genes in rice and nine genes in Arabidopsis that were positively expressed in various organs (de Abreu-Neto et al., 2013; Xie et al., 2018), and performed qRT-PCR (Figures 12, 13). The OsHMP expression levels generally varied greatly under different heavy metal ion treatments in different tissues. The OsHMP18 and OsHMP22 expression levels under Cu2+ stress, OsHMP09 and OsHMP22 under Cd2+ stress, OsHMP09 under Zn2+ stress, and OsHMP22 under Pb2+ stress showed extremely significant difference in all tissues between the control at least one time point (Figure 12). Certain genes had stronger transcript levels in one particular tissue under specific types of heavy metal ion stress. OsHMP09 and OsHMP27 maintained extremely high expression levels both in the leaves and shoots under Cu2+ stress. Furthermore, in all tissues, OsHMP12 was significantly and highly upregulated at all time points under Cu2+ stress relative to the control (Figure 12A). Moreover, OsHMP25 in the leaves and OsHMP27 in the shoots positively responded to Cd2+ stress at least one time point (Figure 12B). Under the Zn2+ treatment, the OsHMP11 expression level in the leaves, OsHMP22 in the roots, OsHMP18 in the shoots and roots extremely significantly differed from that of the control. Although OsHMP14 exhibited extremely significant difference between the control in the leaves and roots under Zn2+ stress, its transcript levels did not significantly differ from that for the control at all time points in the shoots (Figure 12C). Under the Pb2+ treatment, the OsHMP11 expression level in the leaves and OsHMP18 in the shoot and roots were extremely significantly upregulated. In contrast, neither OsHMP14 in the shoots nor OsHMP09, OsHMP12, and OsHMP27 in the roots were significantly upregulated in response to the Pb2+ treatment at all time points (Figure 12D).
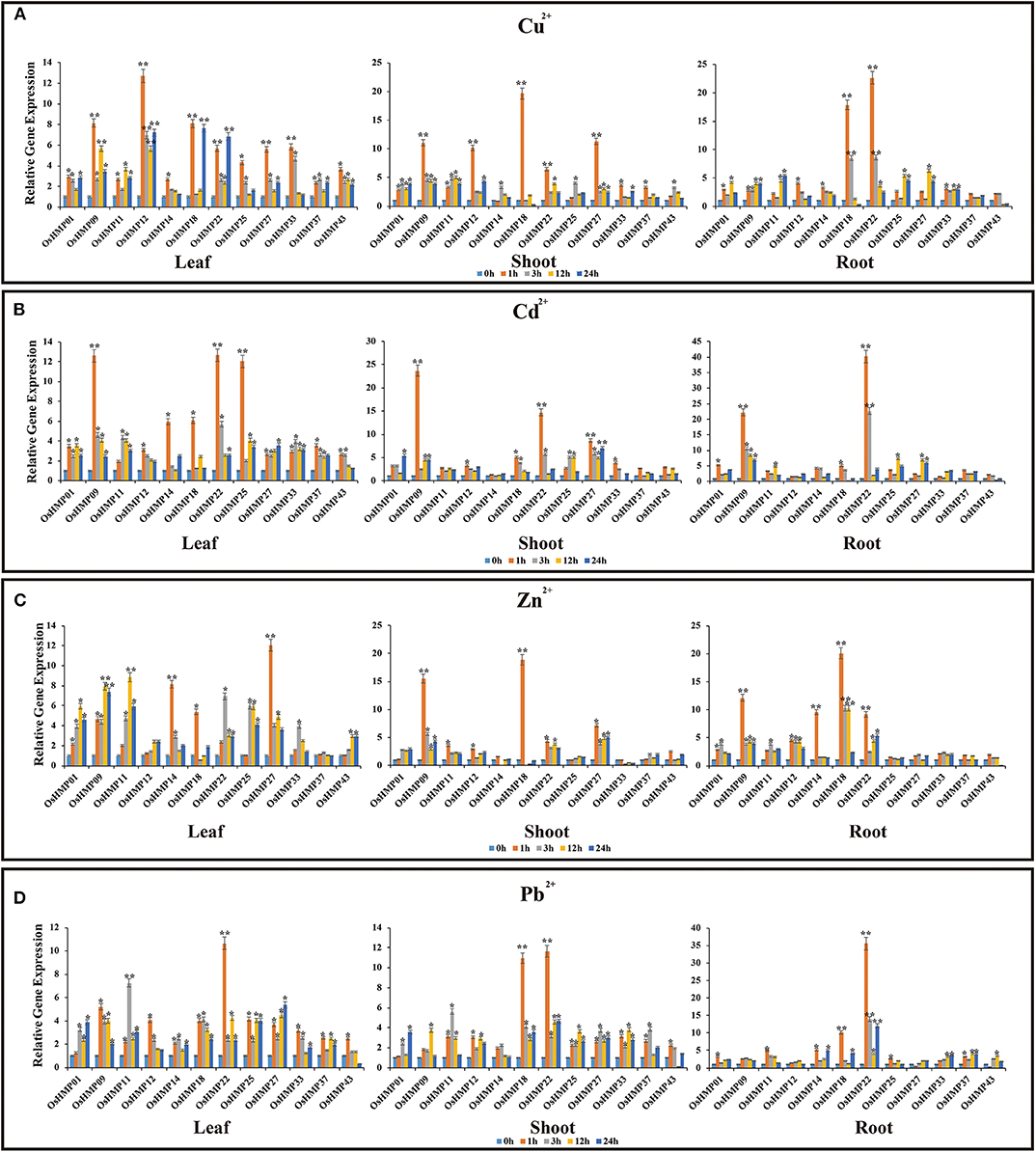
Figure 12. Expression profiles of 12 selected OsHMPs in response to Cu2+(A), Cd2+(B), Zn2+(C), and Pb2+(D) stresses in 2-weeks old rice seedlings after treatment for 1, 3, 12, and 24 h. Data represent means (±SD) of three biological replicates. Vertical bars indicate standard deviations. Asterisks indicate corresponding genes significantly upregulated or downregulated between the treatment and control (n = 12, *p < 0.05; **p < 0.01; Student's t-test).
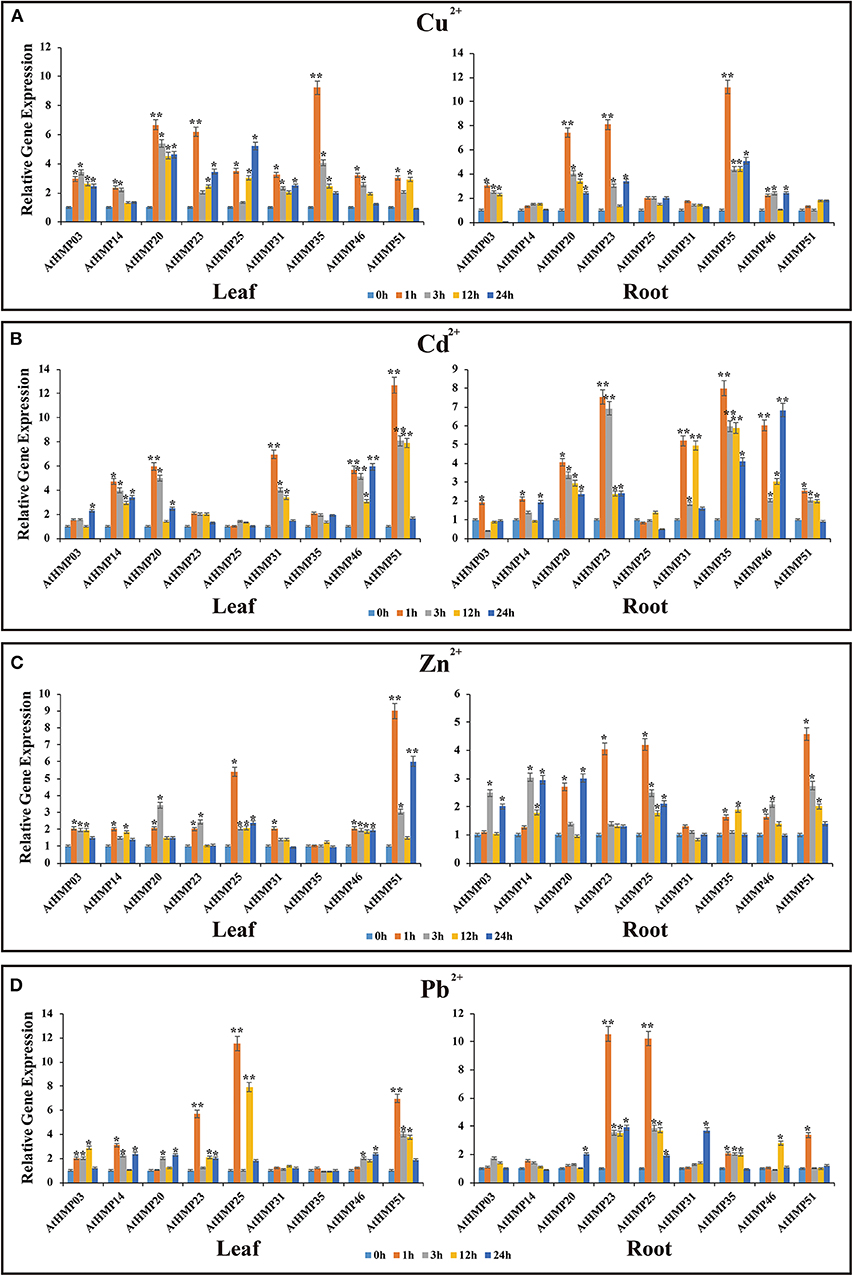
Figure 13. Expression profiles of 9 selected AtHMPs in response to Cu2+(A), Cd2+(B), Zn2+(C), and Pb2+(D) stresses in 2-weeks old Arabidopsis seedlings after treatment for 1, 3, 12, and 24 h. Data represent means (±SD) of three biological replicates. Vertical bars indicate standard deviations. Asterisks indicate corresponding genes significantly upregulated or downregulated between the treatment and control (n = 12, *p < 0.05; **p < 0.01; Student's t-test).
Under heavy metal stress, the AtHMP transcript levels were dramatically lower than those of the OsHMPs (Figure 13). Overall, the relative AtHMP expression levels varied greatly among different heavy metal treatments. Under Cu2+ stress, the expression levels of AtHMP20, AtHMP23, and AtHMP35 in both leaves and roots markedly differed from those for the control at least one time point (Figure 13A). Under Cd2+ stress, the AtHMP31 and AtHMP46 transcript levels in the leaves and roots, AtHMP20 and AtHMP51 in the leaves, AtHMP23 and AtHMP35 in the roots extremely significantly differed from those of the control (Figure 13B). Conversely, under the Cd2+ treatment, the AtHMP25 transcript level did not significantly differ from that for the control at all time points in all tissues. Under Zn2+ stress, only the expression level of AtHMP51 in the leaves showed extremely significant difference from the control at least one time point. Moreover, in the roots, nine AtHMPs were significantly upregulated under Zn2+ stress (Figure 13C). Under the Pb2+ treatment, the AtHMP23 and AtHMP25 transcript levels in both leaves and roots, AtHMP51 in the leaves extremely significantly differed from those for the control (Figure 13D). In conclusion, these selected HMP genes are induced by at least one type of heavy metal ion, but the expression levels are different in various tissues.
In order to investigate the expression patterns of all HMPs under various heavy metal stresses, we further analyzed the expression levels in the roots under Cu2+ and Cd2+ stresses by using the transcript data from GEO database (Figures S5, S6). Sixteen of the 46 OsHMPs were differentially expressed (|Log2 fold change| ≥ 1) both at treatment for 1 and 3 h under Cu2+ stress. Fifteen of 46 OsHMPs were only differentially expressed at treatment for 1 h under Cu2+ stress. Consistent with the Cu2+ treatment, 14 of the 46 OsHMPs were differentially expressed (|Log2 fold change| ≥ 1) both at treatment for 1 and 3 h under Cd2+ stress, and 13 OsHMPs were only differentially expressed at treatment for 1 h under Cd2+ stress. Although OsHMP04, OsHMP07, OsHMP13, OsHMP15, OsHMP29, OsHMP32, and OsHMP44 showed low expression levels under normal growth condition, these genes were upregulated under heavy metal stress (Figure 10B and Figure S5). For AtHMPs, 37 of the 55 AtHMPs were differentially expressed (|Log2 fold change| ≥ 1) under 1.3 or 1.6 μM Cu2+ stress, and 33 AtHMPs were differentially expressed under 3 or 15 μM Cd2+ stress. Compared with the expression profiles in various Arabidopsis tissues under normal growth condition, AtHMP05, AtHMP06, AtHMP07, AtHMP09, AtHMP10, AtHMP11, AtHMP30, AtHMP32, AtHMP38, AtHMP41, AtHMP43, AtHMP50, AtHMP52, AtHMP54, and AtHMP55 were upregulated under heavy metal stress (Figure 11B and Figure S6). These results indicated that some HMPs expression only upregulate under heavy metal stress instead of normal growth condition, implied heavy metal-associated proteins have specific expression patterns under various heavy metal stresses.
Discussion
Collinearity of the HMPs in rice and Arabidopsis has not been systematically studied. As a rule, gene synteny consists of tandem and segmental duplications (Cannon et al., 2004). Here, we detected 12 pairs of syntenic OsHMPs and 14 pairs of syntenic AtHMPs (Figure 2). Both tandem duplication (five pairs in rice and six pairs in Arabidopsis) and segmental duplication (seven pairs in rice and eight pairs in Arabidopsis) were involved in HMP collinearity. A previous study demonstrated that syntenic conservatism increased with the number of duplicated genes in the same gene family (Willis et al., 2017). The present study revealed that the synteny of HMPs are highly conservative. By comparing Arabidopsis and rice collinearity, we identified 17 homologous gene pairs among various rice species but only three homologous gene pairs among different Arabidopsis species. The collinearity differences observed between monocots and dicots is that generally synteny is maintained at a much higher level in the same species (Eckardt, 2001).
Protein structure determines its function (Aebersold and Mann, 2016). Here, alignment of the HMA domain sequences showed that the core sequences of the HMA domain in each group, namely, “CXXC,” were highly conserved (Figure 5). However, AtHMP11, OsHMP23, and OsHMP29 may have been affected by environment or genetic recombination during evolution and could have undergone natural variation. For these reasons, AtHMP11, OsHMP23, and OsHMP29 may have unique functions among the HMPs and could be applied toward population genetics or gene function research in the future.
All HMPs were divided into six clades according to the characteristics of the HMA domain (Figure 6). In a previous study, HPP and HIPP were divided into five groups and the proteins in the ATCCS group were also included (Khan et al., 2019). The latter were named in the present study. To elucidate the evolutionary relationships among the HMPs, we placed those involved in peroxidase in a separate group and divided HPP and HIPP into four groups based on their HMA domain characteristics. The highly important P1B-ATPase HMPs were also displayed in the evolutionary tree along with HPP and HIPP. Previously, they had only been investigated separately. Although certain HMPs belong to the P1B-ATPase classification, there are P1B-ATPase proteins such as LOC_Os06g47550, LOC_Os06g48720, At4g37270, At4g30110, At4g30120, and At2g19110 that do not contain HMA domains (Pedersen et al., 2012; Zhiguo et al., 2018).
Subsequent gene structure and motif analyses established that genes within the same subfamilies were relatively conserved (Figure 7). However, the properties of the HMPs and their proteins varied widely under the different classifications. Thus, HMPs may be functionally diverse in plants. Comparative analysis of the HMPs in rice and Arabidopsis disclosed that the HMPs in Arabidopsis had more alternative splicing than those in rice. An earlier study revealed that the differences in the exons/introns among gene family members were indicative of their vital roles in the evolution of these genes under environmental stress (Laloum et al., 2018). Thus, Arabidopsis HMPs could have been subjected to more environmental stress than rice during evolution.
Determination of the cis-element distributions on gene promoters clarifies the signaling pathways in which HMPs are implicated. Here, it was found that the cis-elements common to all HMP promoters participated in transcription factor regulation and pollen-specific expression. Distribution of the HMP cis-acting elements resembled that for the RMP gene class, which disclosed several cis-elements distributed on the RMP gene promoters including ARR1AT, DOFCOREZM, GTGANTG10, POLLEN1LELAT52, and CACTFTPPCA1 (Nguyen et al., 2016). All of these were detected in the rice and Arabidopsis HMPs. The RMP genes are preferentially expressed in pollen. GTGANTG10 and POLLEN1LELAT52 are pollen-specific cis-acting elements (Nguyen et al., 2016). Therefore, the HMPs genes may play critical roles in pollen and could be focal points for future gene function studies.
Previous functional analyses of plant HMPs focused mainly focus on the mechanisms of their responses to various heavy metal ions (Hasan et al., 2017). Here, we identified ARR1AT, DOFCOREZM, and WRKY71OS as cis-acting elements in the HMP genes participating in transcriptional regulation. The Dof and WRKY transcription factors play roles in several abiotic and biotic stresses (Lindemose et al., 2013) and present with the attributes of zinc finger-like motifs. Several members of the WRKY and Dof families respond to heavy metal stress. AtWRKY22, AtWRKY25, and AtWRKY29 in the leaves and roots of 3-wk Arabidopsis plants were induced by exposure to 2 μM Cu2+ (Opdenakker et al., 2012). An earlier study showed that Dof transcription factors were substantially upregulated under CdCl2 stress (Xu et al., 2019). However, another investigation reported that transcription factors are probably upregulated in response to the peroxide stress caused by heavy metal exposure (Dubey et al., 2014). On the other hand, the findings of this work suggested that the expression levels of these transcription factors also increase in direct response to heavy metal stress by binding the HMP cis-elements.
Gene expression specificity analyses of different plant tissues have demonstrated that these genes are implicated in heavy metal ion transport and detoxification in various organs (Liu et al., 2019). In previous studies, HIPP and HPP expression in rice and Arabidopsis and P1B-ATPase HMP expression in Populus trichocarpa were tissue-specific (de Abreu-Neto et al., 2013; Li et al., 2015). In the present study, the HMP expression levels varied widely among plant tissues. Only eight HMPs in rice and nine HMPs in Arabidopsis were constitutive in different tissues (Figures 10, 11). Therefore, these HMPs also participate in biological functions not related to stress response. OsHMP37 (OsATX1) clustered into the ATCCS group and showed far higher transcript levels than the other genes in all tissues. Previous research indicated that ATX1 participates in peroxide disposition and serves as a chaperone for Cu transport in plant cells (Shin and Yeh, 2012; Zhang Y. Y. et al., 2018). Nevertheless, certain genes remained at extremely low transcript level in all organs examined. OsHMP06, OsHMP16, OsHMP30, OsHMP31, OsHMP41, AtHMP18, AtHMP22, AtHMP26, AtHMP34, AtHMP36, and AtHMP55 were only slightly expressed or not expressed at all in every tissue evaluated. An earlier study disclosed that the downregulation of certain genes helps conserve their ancestral functions (Qian et al., 2010). Consequently, these HMPs may have been conserved from their ancestors and are only inducible under special conditions.
Earlier research on heavy metal-associated proteins revealed that they play critical roles in metal ion distribution in plants (Li et al., 2015). In Arabidopsis, AtHIPP06 was induced by Cd2+, Hg2+, Fe2+, and Cu2+ (de Abreu-Neto et al., 2013) while AtHIPP26 participated in Cd2+ and Zn2+ traffic (Barth et al., 2009). AtHIPP06 and AtHIPP26 overexpression increased plant Cd2+ tolerance whereas triple knockout of AtHIPP20/21/22 caused Cd2+ hypersensitivity in Arabidopsis (Tehseen et al., 2010). The present study investigated the expression patterns of HMPs with relatively higher expression levels in various organs under different heavy metal ion stresses. In this way, the ions transported by these HMPs in rice and Arabidopsis were identified. Most of the selected OsHMPs were differentially expressed in various tissues under metal ion stress. In contrast, most of the selected AtHMPs were expressed at the same levels in different tissues under heavy metal ions stress. The HMPs were involved in ion transport and responded to ≥ 1 type of ion stress. Nevertheless, few HMPs responded equally to all metal ions. Only OsHMP11, OsHMP14, OsHMP18, and AtHMP23 positively responded to all four types of heavy metal cations (Figures 12, 13). However, these four genes belong to different clades (Figure 6). In Populus, the PtHMA1–PtHMA4 phylogenetic cluster with the Zn/Cd/Co/Pb subclass of HMAs was induced by Cu and Ag (Li et al., 2015). These results suggested there may be no significant correlation between HMP classification and the types of heavy metal cations. However, the expression levels OsHMP and AtHMP are different and there was no significant correlation between gene expression and its classification. OsHMP37 (OsATX1) showed higher expression levels in the roots than the leaves or shoots. OsATX1 overexpression lowered the Cu2+ concentrations in the roots but raised them in the shoots (Zhang Y. Y. et al., 2018). Thus, OsATX1 is expressed mainly in the roots and is, therefore, a root-specific stress response gene. Under normal conditions, it is constitutively expressed. Our research has preliminary analyzed the property, evolution, classification and function of HMP genes in dicotyledonous or monocotyledonous plants, which has laid a theoretical foundation for the future cultivation and breeding of those crops that are suffered from heavy metal pollution. In the future study, more molecular experiments, such as overexpression or CRISPR, can be performed on HMP genes to further verify their function.
Conclusions
HMPs participate in numerous biological processes in plants. Here, 46 OsHMPs in rice and 55 AtHMPs in Arabidopsis were identified in silico. A gene duplication analysis showed that HMPs are conserved among various plant species and that some of them may have originated from a common ancestor. A phylogenetic analysis divided the HMPs into six subfamilies named H1–H4, ATCCS, and P1B-ATPase. The HMP gene structures and conserved domains varied greatly among clades. Nevertheless, they shared common cis-elements involved in regulatory transcription factors and pollen-specific expression. A gene expression profile analysis indicated that HMP expression varied substantially among different plant tissues. Only eight OsHMPs and nine AtHMPs were constitutive in various tissues. qRT-PCR analysis revealed that HMPs were induced in response to exposure to various heavy metal ions. The present study helped elucidate the biological functions of the HMPs in rice and Arabidopsis.
Data Availability Statement
RNA-seq data used in the present research are available in the Sequence Read Archive database under accession number SRP013631, SRP008505, SRP008469, and SRP008821 and in the GEO database under accession number GSE38612, GSE34895, and GSE108751 and in the DNA Data Bank of Japan Sequence Read Archive under accession numbers DRR001024-DRR001051.
Author Contributions
Conceptualization: JL. Data curation: XM. Formal analysis: JS and XL. Funding acquisition: DZ. Investigation: JW. Project administration: HZha. Resources: HL. Software: HZhe. Supervision: DZ. Writing—original draft: JL. Writing—review and editing: MZ. All authors have read, edited, and approved the current version of the manuscript.
Funding
This work was supported by grants from National Natural Science Foundation (31701507) and National Key Research and Development Plan (2017YFD0300501).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage for English language editing. We extend our thanks to the reviewers and editors for their careful review and helpful comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00477/full#supplementary-material
Figure S1. Synteny analysis of heavy-metal-associated genes between rice and Brassica rapa, Cucumis sativus, Glycine max, Gossypium raimondii, and Solanum tuberosum. Gray lines in the background indicate the collinear blocks within rice and other plant genomes, while the orange lines highlight the syntenic OsHMP gene pairs. The species names with the prefixes “O. sativa,” “B. rapa,” “C. sativus,” “G. max,” “G. raimondii,” and “S. tuberosum” indicate Oryza sativa and Brassica rapa, Cucumis sativus, Glycine max, Gossypium raimondii, and Solanum tuberosum, respectively. Different color bars represent the chromosomes of different species. The chromosome number is labeled at the top or bottom of each chromosome.
Figure S2. Synteny analysis of heavy-metal-associated genes between Arabidopsis and Brachypodium distachyon, Oryza brachyantha, Triticum aestivum, Setaria italica, and Zea mays. Gray lines in the background indicate the collinear blocks within Arabidopsis and other plant genomes, while the blue lines highlight the syntenic AtHMP gene pairs. The species names with the prefixes “A. thaliana,” “B. distachyon,” “O. brachyantha,” “T. aestivum,” “Setaria italica,” and “Z. mays” indicate Arabidopsis thaliana, Brachypodium distachyon, Oryza brachyantha, Triticum aestivum, Setaria italica, and Zea mays, respectively. Different color bars represent the chromosomes of different species. The chromosome number is labeled at the top or bottom of each chromosome.
Figure S3. Synteny analysis of heavy-metal-associated genes between Arabidopsis and rice. Gray lines in the background indicate the collinear blocks within Arabidopsis and rice genomes, while the red lines highlight the syntenic HMP gene pairs between Arabidopsis and rice. The species names with the prefixes “A. thaliana” and “O. sativa,” indicate Arabidopsis thaliana, and Oryza sativa, respectively. Different color bars represent the chromosomes of different species. The chromosome number is labeled at the top or bottom of each chromosome.
Figure S4. Motif sequences of heavy-metal associated proteins in Arabidopsis and rice.
Figure S5. Expression patterns of the rice HMP genes in various heavy metal stresses. Heatmaps were generated using HemI from the normalized value by row for the signatures in transcripts per million (TPM). Transcript levels are depicted by different colors on the scale. Blue and red represent low and high expression levels, respectively.
Figure S6. Expression patterns of the Arabidopsis HMP genes in various heavy metal stresses. Heatmaps were generated using HemI from the normalized value by row for the signatures in transcripts per million (TPM). Transcript levels are depicted by different colors on the scale. Blue and red represent low and high expression levels, respectively.
Table S1. Primers for quantitative real-time PCR.
Table S2. Detailed information of all HMP genes identified in the rice and Arabidopsis genome.
Table S3. Synteny rice HMP gene pairs and Ka/Ks ratios.
Table S4. Synteny Arabidopsis HMP gene pairs and Ka/Ks ratios.
Table S5. Cis-elements of HMPs in rice and Arabidopsis.
Table S6. RNA-seq data of rice HMP genes in different tissues.
Table S7. RNA-seq data of Arabidopsis HMP genes in different tissues.
References
Aebersold, R., and Mann, M. (2016). Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355. doi: 10.1038/nature19949
Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Andrés-Colás, N., Sancenon, V., Rodriguez-Navarro, S., Mayo, S., Thiele, D. J., Ecker, J. R., et al. (2006). The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. 45, 225–236. doi: 10.1111/j.1365-313X.2005.02601.x
Argüello, J. M. (2003). Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J. Membr. Biol. 195, 93–108. doi: 10.1007/s00232-003-2048-2
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Axelsen, K. B., and Palmgren, M. G. (1998). Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101. doi: 10.1007/PL00006286
Barth, O., Vogt, S., Uhlemann, R., Zschiesche, W., and Humbeck, K. (2009). Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol. Biol. 69, 213–226. doi: 10.1007/s11103-008-9419-0
Boyd, R. S. (2010). Heavy metal pollutants and chemical ecology: exploring new frontiers. J Chem. Ecol. 36, 46–58. doi: 10.1007/s10886-009-9730-5
Bull, P. C., and Cox, D. W. (1994). Wilson disease and menkes disease: new handles on heavy-metal transport. Trends Genet. 10, 246–252. doi: 10.1016/0168-9525(94)90172-4
Cambrollé, J., Garcia, J. L., Figueroa, M. E., and Cantos, M. (2015). Evaluating wild grapevine tolerance to copper toxicity. Chemosphere 120, 171–178. doi: 10.1016/j.chemosphere.2014.06.044
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chou, K.-C., and Shen, H.-B. (2010). Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Development 109:1091. doi: 10.1038/nprot.2007.494
Clemens, S., Palmgren, M. G., and Krämer, U. (2002). A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci. 7, 309–315. doi: 10.1016/S1360-1385(02)02295-1
Crowell, D. N., and Huizinga, D. H. (2009). Protein isoprenylation: the fat of the matter. Trends Plant Sci. 14:170. doi: 10.1016/j.tplants.2008.12.001
de Abreu-Neto, J. B., Turchetto-Zolet, A. C., de Oliveira, L. F. V., Zanettini, M. H. B., and Margis-Pinheiro, M. (2013). Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280, 1604–1616. doi: 10.1111/febs.12159
Dubey, S., Shri, M., Misra, P., Lakhwani, D., Bag, S. K., Asif, M. H., et al. (2014). Heavy metals induce oxidative stress and genome-wide modulation in transcriptome of rice root. Funct. Integr. Genom. 14, 401–417. doi: 10.1007/s10142-014-0361-8
Eckardt, N. A. (2001). Everything in its place: conservation of gene order among distantly related plant species. Plant Cell 13, 723–725. doi: 10.2307/3871334
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Fang, X. L., Wang, L., Deng, X. J., Wang, P., Ma, Q. B., Nian, H., et al. (2016). Genome-wide characterization of soybean P-1B-ATPases gene family provides functional implications in cadmium responses. BMC Genomics 17:376. doi: 10.1186/S12864-016-2730-2
Feng, L. (2011). Effects of lead stress on seed germination and physiological traits in rice. Modern Agric. Sci. Tech. 17, 58–59. doi: 10.3969/j.issn.1007-5739.2011.17.026
Filichkin, S. A., Leonard, J. M., Monteros, A., Liu, P.-P., and Nonogaki, H. (2004). A novel endo-beta-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol. 134, 1080–1087. doi: 10.1104/pp.103.035998
Fu, X.-Z., Tong, Y.-H., Zhou, X., Ling, L.-L., Chun, C.-P., Cao, L., et al. (2017). Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 629, 1–8. doi: 10.1016/j.gene.2017.07.072
Gill, S. S., Hasanuzzaman, M., Nahar, K., Macovei, A., and Tuteja, N. (2013). Importance of nitric oxide in cadmium stress tolerance in crop plants. Plant Physiol. Biochem. 63, 254–261. doi: 10.1016/j.plaphy.2012.12.001
Gitschier, J., Moffat, B., Reilly, D., Wood, W. I., and Fairbrother, W. J. (1998). Solution structure of the fourth metal-binding domain from the Menkes copper-transporting ATPase. Nat. Struct. Biol. 5, 47–54. doi: 10.1038/nsb0198-47
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gowik, U., Burscheidt, J., Akyildiz, M., Schlue, U., Koczor, M., Streubel, M., et al. (2004). cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16, 1077–1090. doi: 10.1105/tpc.019729
Hall, J. L. (2002). Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 53, 1–11. doi: 10.1093/jexbot/53.366.1
Hasan, M. K., Cheng, Y., Kanwar, M. K., Chu, X. Y., Ahammed, G. J., and Qi, Z. Y. (2017). Responses of plant proteins to heavy metal stress-a review. Front. Plant Sci. 8:1492. doi: 10.3389/Fpls.2017.01492
Hayat, S., Khalique, G., Irfan, M., Wani, A. S., Tripathi, B. N., and Ahmad, A. (2012). Physiological changes induced by chromium stress in plants: an overview. Protoplasma 249, 599–611. doi: 10.1007/s00709-011-0331-0
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, X. Y., Deng, F., Yamaji, N., Pinson, S. R., Fujii-Kashino, M., Danku, J., et al. (2016). A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 7:12138. doi: 10.1038/ncomms12138
Kersey, P. J., Allen, J. E., Allot, A., Barba, M., Boddu, S., Bolt, B. J., et al. (2018). Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 46, D802–D808. doi: 10.1093/nar/gkx1011
Khan, I. U., Rono, J. K., Zhang, B. Q., Liu, X. S., Wang, M. Q., Wang, L. L., et al. (2019). Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotox. Environ. Safe. 175, 8–18. doi: 10.1016/j.ecoenv.2019.03.040
Kobayashi, Y., Kuroda, K., Kimura, K., Southron-Francis, J. L., Furuzawa, A., Kimura, K., et al. (2008). Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 148, 969–980. doi: 10.1104/pp.108.119933
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurata, N., and Yamazaki, Y. (2006). Oryzabase. an integrated biological and genome information database for rice. Plant Physiol. 140, 12–17. doi: 10.1104/pp.105.063008
Laloum, T., Martin, G., and Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23, 140–150. doi: 10.1016/j.tplants.2017.09.019
Lamesch, P., Berardini, T. Z., Li, D. H., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lee, B. H., Henderson, D. A., and Zhu, J. K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175. doi: 10.1105/tpc.105.035568
Lee, T. H., Tang, H., Wang, X., and Paterson, A. H. (2013). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, 1152–1158. doi: 10.1093/nar/gks1104
Li, D. D., Xu, X. M., Hu, X. Q., Liu, Q. G., Wang, Z. C., Zhang, H. Z., et al. (2015). Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa. Front. Plant Sci. 6:1149. doi: 10.3389/fpls.2015.01149
Li, J. M., Zhang, M. H., Sun, J., Mao, X. R., Wang, J., Wang, J. G., et al. (2019). Genome-wide characterization and identification of trihelix transcription factor and expression profiling in response to abiotic stresses in rice (Oryza sativa L.). Int. J. Mol. Sci. 20:251. doi: 10.3390/Ijms20020251
Lindemose, S., O'Shea, C., Jensen, M. K., and Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Liu, J. K., Gao, Y. F., Tang, Y. L., Wang, D., Chen, X. M., Yao, Y. N., et al. (2019). Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front. Genet. 10:345. doi: 10.3389/Fgene.2019.00345
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi: 10.1093/nar/gky1100
Miyadate, H., Adachi, S., Hiraizumi, A., Tezuka, K., Nakazawa, N., Kawamoto, T., et al. (2011). OsHMA3, a P-1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189, 190–199. doi: 10.1111/j.1469-8137.2010.03459.x
Mourato, M. P., Moreira, I. N., Leitao, I., Pinto, F. R., Sales, J. R., and Martins, L. L. (2015). Effect of heavy metals in plants of the Genus Brassica. Int. J. Mol. Sci. 16, 17975–17998. doi: 10.3390/ijms160817975
Nguyen, T. D., Moon, S., Nguyen, V. N. T., Gho, Y., Chandran, A. K. N., Soh, M. S., et al. (2016). Genome-wide identification and analysis of rice genes preferentially expressed in pollen at an early developmental stage. Plant Mol. Biol. 92, 71–88. doi: 10.1007/s11103-016-0496-1
Opdenakker, K., Remans, T., Keunen, E., Vangronsveld, J., and Cuypers, A. (2012). Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environ. Exp. Bot. 83, 53–61. doi: 10.1016/j.envexpbot.2012.04.003
Papatheodorou, I., Fonseca, N. A., Keays, M., Tang, Y. A., Barrera, E., Bazant, W., et al. (2017). Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 46, D246–D251. doi: 10.1093/nar/gkx1158
Pedersen, C. N. S., Axelsen, K. B., Harper, J. F., and Palmgren, M. G. (2012). Evolution of plant P-type ATPases. Front. Plant Sci. 3:31. doi: 10.3389/Fpls.2012.00031
Plesch, G., Ehrhardt, T., and Mueller-Roeber, B. (2001). Involvement of TAAAG elements suggests a role of Dof transcription factors in guard cell-specific gene expression. Plant J. 28, 455–464. doi: 10.1046/j.1365-313X.2001.01166.x
Puig, S., Mira, H., Dorcey, E., Sancenon, V., Andres-Colas, N., Garcia-Molina, A., et al. (2007). Higher plants possess two different types of ATX1-like copper chaperones. Biochem. Biophys. Res. Commun. 354, 385–390. doi: 10.1016/j.bbrc.2006.12.215
Qian, W. F., Liao, B. Y., Chang, A. Y. F., and Zhang, J. Z. (2010). Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 26, 425–430. doi: 10.1016/j.tig.2010.07.002
Qin, F., Sakuma, Y., Tran, L. S. P., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Ricachenevsky, F. K., Menguer, P. K., Sperotto, R. A., Williams, L. E., and Fett, J. P. (2013). Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 4:144. doi: 10.3389/Fpls.2013.00144
Rogers, H. J., Bate, N., Combe, J., Sullivan, J., Sweetman, J., Swan, C., et al. (2001). Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol. Biol. 45, 577–585. doi: 10.1023/a:1010695226241
Ross, E. J., Stone, J. M., Elowsky, C. G., Arredondo-Peter, R., Klucas, R. V., and Sarath, G. (2004). Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55, 1721–1731. doi: 10.1093/jxb/erh211
Shin, L. J., and Yeh, K.-C. (2012). Overexpression of Arabidopsis ATX1 retards plant growth under severe copper deficiency. Plant Signal. Behav. 7, 1082–1083. doi: 10.4161/psb.21147
Suzuki, N., Yamaguchi, Y., Koizumi, N., and Sano, H. (2002). Functional characterization of a heavy metal binding protein CdI19 from Arabidopsis. Plant J. 32, 165–173. doi: 10.1046/j.1365-313X.2002.01412.x
Tehseen, M., Cairns, N., Sherson, S., and Cobbett, C. S. (2010). Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2, 556–564. doi: 10.1039/c003484c
Thomine, S., and Vert, G. (2013). Iron transport in plants: better be safe than sorry. Curr. Opin. Plant Biol. 16, 322–327. doi: 10.1016/j.pbi.2013.01.003
Willis, N. A., Frock, R. L., Menghi, F., Duffey, E. E., Panday, A., Camacho, V., et al. (2017). Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature 551, 590–595. doi: 10.1038/nature24477
Xie, T., Chen, C. J., Li, C. H., Liu, J. R., Liu, C. Y., and He, Y. H. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 19:490. doi: 10.1186/S12864-018-4880-X
Xie, Z., Zhang, Z.-L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.2307/4356804
Xu, Z. G., Dong, M., Peng, X. Y., Ku, W. Z., Zhao, Y. L., and Yang, G. Y. (2019). New insight into the molecular basis of cadmium stress responses of wild paper mulberry plant by transcriptome analysis. Ecotox. Environ. Safe. 171, 301–312. doi: 10.1016/j.ecoenv.2018.12.084
Yanagisawa, S. (2000). Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 21, 281–288. doi: 10.1046/j.1365-313x.2000.00685.x
Yuan, L. Y., Yang, S. G., Liu, B. X., Zhang, M., and Wu, K. Q. (2012). Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 31, 67–79. doi: 10.1007/s00299-011-1140-9
Zhang, X. D., Sun, J. Y., You, Y. Y., Song, J. B., and Yang, Z. M. (2018). Identification of Cd-responsive RNA helicase genes and expression of a putative BnRH 24 mediated by miR158 in canola (Brassica napus). Ecotox. Environ. Saf. 157, 159–168. doi: 10.1016/j.ecoenv.2018.03.081
Zhang, Y. Y., Chen, K., Zhao, F. J., Sun, C. J., Jin, C., Shi, Y. H., et al. (2018). OsATX1 interacts with heavy metal P1B-type ATPases and affects copper transport and distribution. Plant Physiol. 178, 329–344. doi: 10.1104/pp.18.00425
Zhiguo, E., Li, T. T., Chen, C., and Wang, L. (2018). Genome-wide survey and expression analysis of P-1B-ATPases in rice, maize and sorghum. Rice Sci. 25, 208–217. doi: 10.1016/j.rsci.2018.06.004
Zschiesche, W., Barth, O., Daniel, K., Bohme, S., Rausche, J., and Humbeck, K. (2015). The zinc-binding nuclear protein HIPP3 acts as an upstream regulator of the salicylate-dependent plant immunity pathway and of flowering time in Arabidopsis thaliana. New Phytol. 207, 1084–1096. doi: 10.1111/nph.13419
Keywords: Arabidopsis, gene duplication, heavy-metal stress, phylogenetic analysis, rice
Citation: Li J, Zhang M, Sun J, Mao X, Wang J, Liu H, Zheng H, Li X, Zhao H and Zou D (2020) Heavy Metal Stress-Associated Proteins in Rice and Arabidopsis: Genome-Wide Identification, Phylogenetics, Duplication, and Expression Profiles Analysis. Front. Genet. 11:477. doi: 10.3389/fgene.2020.00477
Received: 18 January 2020; Accepted: 17 April 2020;
Published: 08 May 2020.
Edited by:
Million Tadege, Oklahoma State University, United StatesReviewed by:
Cheng Qin, Zunyi Vocational and Technical College, ChinaKranthi Varala, Purdue University, United States
Copyright © 2020 Li, Zhang, Sun, Mao, Wang, Liu, Zheng, Li, Zhao and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Detang Zou, em91ZHRuZWF1QDEyNi5jb20=
 Jiaming Li
Jiaming Li Minghui Zhang
Minghui Zhang Jian Sun
Jian Sun Xinrui Mao1
Xinrui Mao1