
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 17 March 2020
Sec. Livestock Genomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00203
This article is part of the Research Topic Genetic Variability in Conservation and Selection Programs in the Post-Genomics Era View all 12 articles
Polyploidization often leads to “transcriptome shock,” and is considered an important factor in evolution of species. Analysis of the cell cycle, which is associated with survival in polyploidy, has proved useful in investigating polyploidization. Here, we used mRNA sequencing to investigate global expression in vitro (in cultured cells) and in vivo (in fin and liver tissues) in both the diploid and tetraploid Carassius auratus red var.. Differential expression (DE) of genes in diploid (7482, 36.0%) and tetraploid (3787, 18.2%) states suggested that in vitro and in vivo conditions dramatically change mRNA expression levels. However, of the 20,771 total shared expressed genes, 18,050 (87.0%), including 17,905 (86.2%) non-differentially expressed genes (DEGs) and 145 (0.7%) DEGs between diploids and tetraploids, showed the same expression trends in both cultured cells and liver tissues. Of the DEGs, four of seven genes in the cell cycle pathway had the same expression trends (upregulated in diploids and tetraploids) in both cultured cells and liver tissues. Quantitative PCR analysis confirmed the same expression trends in the nine DEGs associated with regulation of the cell cycle. This research on common characteristics between diploids and tetraploids provides insights into the potential molecular regulatory mechanisms of polyploidization. The steady changes that occur between diploids and tetraploids in vitro and in vivo show the potential value of studying polyploidy processes using cultured cell lines, especially with respect to cell cycle regulation.
Polyploidy occurs in plants, animals, and fungi (Comai, 2005; Blomme et al., 2006). It plays an important role in the evolutionary history of species by providing a large amount of genetic material, contributing to the genomic complexity, and further promoting speciation (Comai, 2005; Blomme et al., 2006; Otto, 2007). Polyploid breeding induced by artificial and natural mutagenesis is utilized to obtain cells and organisms with genome duplication, contributing to obtaining polyploid animals to achieve high genome plasticity, including allotetraploid hybrids of Carassius auratus red var. and Cyprinus carpio L. (Liu et al., 2001, 2016), polyploid channel catfish (Ictalurus punctatus) (Goudie et al., 1995), polyploid shellfish (Francesc et al., 2009), and autotetraploid C. auratus red var. × Megalobrama amblycephala (Qin et al., 2014).
Besides polyploid individuals, polyploidy has also been found in cells and tissues of diploid organisms, such as human muscle tissues, megakaryocytes, and hepatocytes (Parmacek and Epstein, 2009), as well as in some tissues under conditions of stress, such as aging seminal vesicle cells (Nguyen and Ravid, 2010). Additionally, polyploidy was shown to occur after administration of the drug cisplatin (Cantero et al., 2006) and the c-Jun N-terminal kinase inhibitor SP600123 (Zhou et al., 2016). Genetic instability in polyploid cells might lead to aneuploidy, thereby contributing to the formation of cancer (Storchova and Pellman, 2004). However, after self-breeding the allotetraploid progeny of C. auratus red var. and C. carpio L. for 26 generations, analysis of the chromosome number and reproductive fertility had revealed its genetic stability (Liu et al., 2001, 2016). To further study polyploid fish, the establishment of in vitro cell culture is necessary to analyze complex regulatory mechanisms including genome-wide additive and dominant expression in polyploid formation (Yoo et al., 2013).
Fibroblasts are the main cellular components of connective tissue, and can be easily obtained and cultured in vitro; they have been widely used to study the senescence of cells, cell damage, some congenital metabolic abnormalities and enzyme defects in basic medicine and clinical medicine research (Shima et al., 1980; Shima, 1988; Mahale et al., 2008; Swaminathan et al., 2016). Previously, cultured fibroblasts were obtained from the tail fin tissue of C. auratus red var. and their allotetraploid offspring (Huang et al., 2017). Here, we present an analysis of mRNA expression to investigate the cultured cells and tissues of diploid and tetraploid C. auratus red var.. We performed differential expression (DE) analysis between diploid and tetraploid samples in cultured fibroblasts and liver tissues. We also identified a number of mRNAs of differentially expressed genes (DEGs), and used quantitative (q) PCR to further confirm our findings in cultured cells and fin and liver tissues. Analysis of global expression in cultured cells and tissues should help to reveal whether in vitro cell lines can be used to research molecular expression and regulatory mechanisms in polyploid fish.
All experiments were approved by the Animal Care Committee of Hunan Normal University and followed guidelines of the Administration of Affairs Concerning Animal Experimentation of China. C. auratus red var. was distributed in natural waters of China, and tetraploid C. auratus red var. × C. carpio L. were obtained from self-crossing of the allodiploid hybrid F2 of C. auratus red var. (♀) × C. carpio L. (♂) (Liu et al., 2001, 2016). These individuals were bred and fed in pools under the same water temperature, dissolved oxygen content, and foraging conditions at the Engineering Research Center of Polyploid Fish Breeding and Reproduction of the State Education Ministry, China. Three individuals of each species were collected for further study.
Diploid cultured cells were obtained from the caudal fin of C. auratus red var., and tetraploid cultured cells were derived from the caudal fin of a tetraploid hybrid of C. auratus red var. (♀) × C. carpio L. (♂). Cells were cultured in complete growth medium composed of Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, United States), 10% fetal bovine serum (Invitrogen, Carlsbad, CA, United States), 0.1% 2-mercaptoethanol (Invitrogen, Carlsbad, CA, United States), 1 mM sodium pyruvate (Invitrogen, Carlsbad, CA, United States), and 1 mM non-essential amino acids (Invitrogen, Carlsbad, CA, United States). Cells were grown in 5% (v/v) CO2 at 28°C.
Before extracting total RNA, the ploidy level and DNA content of each sample were confirmed by flow cytometry. Diploid C. auratus red var. was used as a control group. Fish were anesthetized with 100 mg/L MS-222 (Sigma) before dissection. Fish tissues (∼0.2 cm2) were quickly rinsed with 70% alcohol and washed with phosphate-buffered saline. They were then digested with 0.25% trypsin (Invitrogen, Carlsbad, CA, United States) for 15–30 min.
Total RNA was extracted from cultured cells, fin and liver tissues in accordance with a standard TRIzol protocol (Invitrogen, Carlsbad, CA, United States) after RNALater removal (Hummon et al., 2007). The purified RNA was quantified using a 2100 Bioanalyzer system (Agilent). Then, the RNA was used to obtain first-strand cDNA synthesized using AMV reverse transcriptase (Fermentas), with an oligo (dT)12–18 primer at 42°C for 60 min and 70°C for 5 min.
For this study, we focused on the transcriptional regulation of C. auratus red var. in vitro and in vivo to investigate whether there is a difference in cell cycle regulation. Therefore, we obtained mRNA sequencing (seq) data of the liver tissue of diploid C. auratus red var. and tetraploid C. auratus red var. (♀) × C. carpio L. (♂) from the NCBI SRA database (SRR538839, SRR542431, SRR1535135, and SRR1536195) (Liu et al., 2016). Next, we submitted the mRNA-seq data of in vitro diploid C. auratus red var. and tetraploid C. auratus red var. (♀) × C. carpio L. cultured cells to the NCBI SRA (SRR7640867, SRR7640866, SRR7640869, and SRR7640868).
After removing read adapters and low-quality reads, quality of all clean reads of each library was assessed using the FastQC program1. Principal component analysis was used to examine the contribution of each gene to the separation of classes in the six liver transcriptomes based on Euclidean distances (Anders and Huber, 2010). mRNA-seq reads from each sample were mapped against the reference genome (C. auratus red var.2) using TopHat with default parameters (Trapnell et al., 2012). Negative effects of background noise were removed based on the read counts (≤2) of genes in all biological replicates. To compare DE between diploid and tetraploid C. auratus in vitro and in vivo, the values of fragments per kilobase of transcript per million mapped reads (Mortazavi et al., 2008) were calculated using Cufflinks (version 2.1.0) (Trapnell et al., 2012). The false discovery rate (FDR) was used to determine the threshold P-value in multiple tests and analysis. Genes with FDR ≤ 0.01 and fold change (FC) > 2 were defined as the DE threshold using the DEGseq package of the R program (version 2.13) (R Foundation for Statistical Computing, Vienna, Austria) (Wang et al., 2010). DEGs were annotated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
Quantitative (q) PCR primers to amplify 11 cell-cycle-regulated genes (lc3, smad6, p53, myc, gng10, id1, gng12, gadd45, jun, calm, and erg1) were designed using conserved regions of coding sequences in the reference genome (Supplementary Table 1). Primers were used to detect expression with the ABI Prism 7500 Sequence Detection System (Applied Biosystems) and the following amplification conditions: 50°C for 5 min then 95°C for 10 min, followed by 36 cycles of 95°C for 15 s and 60°C for 45 s. Each test was performed three times. Relative quantification was performed and melting curve analysis was used to verify the generation of a single product at the end of the assay. Triplicates of each sample were used both for standard curve generation and during experimental assays. The relative expression of each gene was calibrated with β-actin, and relative mRNA expression data were analyzed using the 2–Δ Δ Ct method (Livak and Schmittgen, 2001). The expression level of β-actin in induced tetraploid C. auratus red var. was estimated by the ratio of transcript abundance to the gene copy number using PCR and qPCR of DNA and RNA, respectively, extracted from cultured cells, caudal fin tissues, and liver tissues in diploid and tetraploid states. β-Actin expression was compared between diploid and tetraploid states.
To examine changes in the global transcriptomic profile between diploid and tetraploid C. auratus red var. in vitro and in vivo, 12 transcriptomes (from liver tissues and cultured cells; three individuals each from diploid and tetraploid) were obtained by paired-end sequencing. After initial adapter trimming and quality control, 535.9 million cleaned reads from the 12 libraries were obtained (Supplementary Table 2). Among these, 451.7 million cleaned reads were mapped against the reference genome of C. auratus red var.3 using TopHat (Supplementary Table 2). The heatmap based on Euclidean distances clustered the diploid liver and tetraploid cultured cell samples. These results indicated significant differences in expression between liver tissues and cultured cells (Figure 1). The analysis of expression levels between diploids and tetraploids revealed the presence of silent transcripts based on a threshold of >10 reads for each gene (Ren et al., 2016). Four shared silent genes were detected in both diploid cultured cells and diploid liver samples, while only one shared gene was found in both of the tetraploid samples (Supplementary Figure 1).
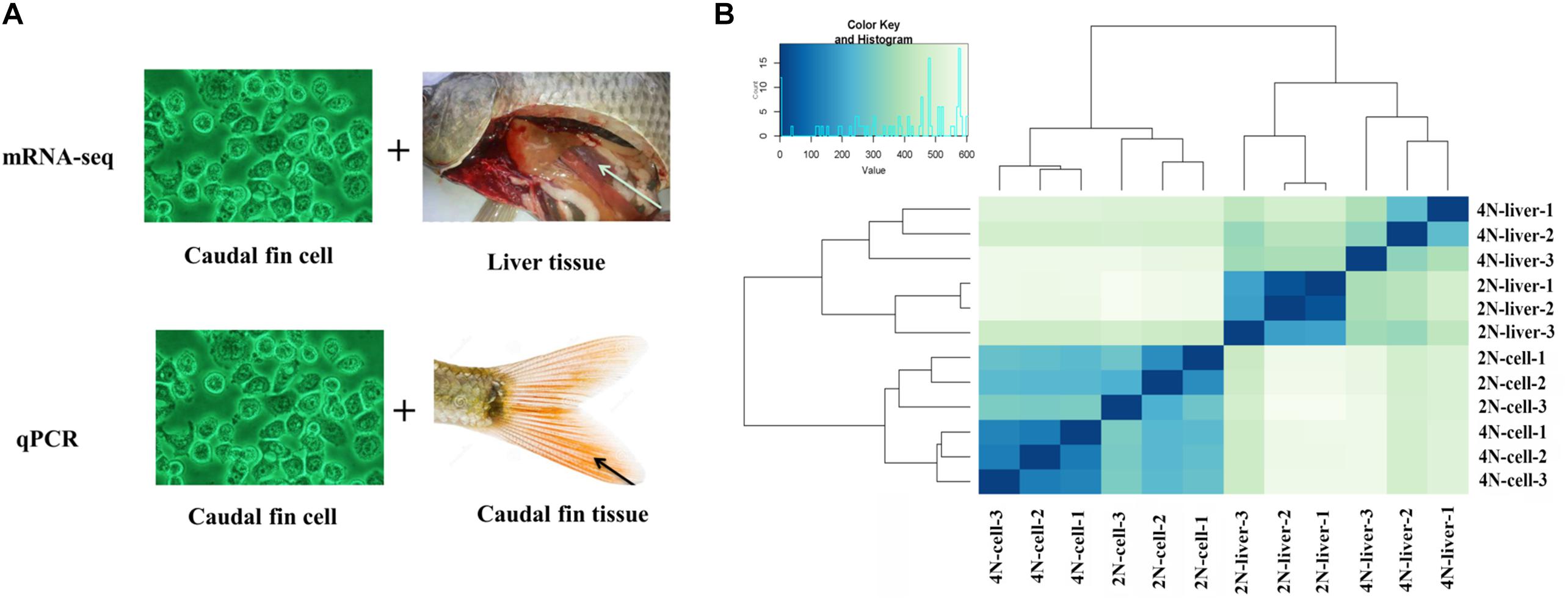
Figure 1. Strategy of the expression analysis and expression cluster in all samples. (A) mRNA-seq and qPCR methods were used to determine the expression levels in cultured cells, caudal fin tissues, and liver tissues. The comparison of “vs. 1” and “vs. 2” was used to assess the DE of in vivo and in vitro between diploids and tetraploids. The comparison of “vs. 3” and “vs. 4” was used to assess the DE of diploids and tetraploids between in vivo and in vitro. (B) Overall clustering of 12 samples including diploid and tetraploid liver tissues, and diploid (2N) and tetraploid (4N) cultured cells, using normalized count data calculated by Cufflinks. The heatmap drawn from all gene count data for the reference genome depicts the relationships of all transcriptomes.
After obtaining mapping information for all transcriptomes, we identified 20,771 shared expressed genes. To compare DE between in vitro and in vivo conditions, we performed DE analysis of diploid cultured cells and liver tissues (vs. 3 in Figure 1A) for all 20,771 expressed genes. A total of 3603 (17.3%) genes were found to be upregulated in diploid cultured cells, while 3879 (18.7%) were upregulated in diploid liver samples (Supplementary Figure 2). GO analysis of categories with the largest numbers of DEGs showed that 620 DEGs belonged to cell part (GO: 0044464) in the main category of cellular component, 1149 belonged to binding (GO: 0005488) in the main category of molecular function, and 1013 belonged to cellular process (GO: 0009987) in the main category of biological process (Supplementary Figure 3).
Next, we focused on differences in expression between tetraploid cultured cells and liver tissues (vs. 4 in Figure 1A), and identified 3787 (18.2%) DEGs (Supplementary Figure 2). Among these, 1258 were upregulated in cultured cells and 2529 were upregulated in the liver (Figure 2B). GO analysis of categories with the largest numbers of genes showed that 195 DEGs belonged to cell part (GO: 0044464) in the main category of cellular component, 557 belonged to binding (GO: 0005488) in the main category of molecular function, and 392 belonged to cellular process (GO: 0009987) in the main category of biological process (Supplementary Figure 3).
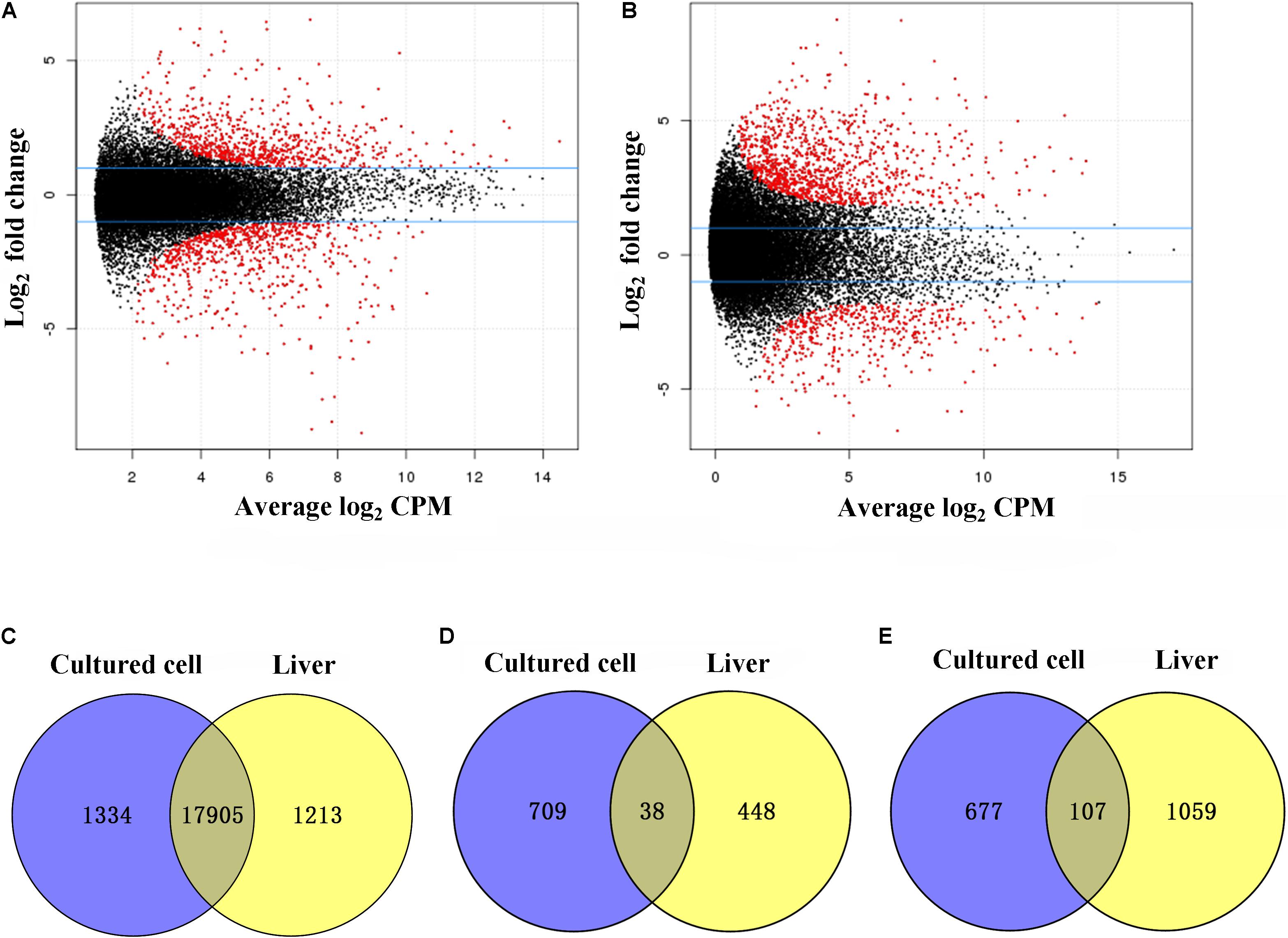
Figure 2. Differentially expressed genes (DEGs) between diploid and tetraploid states in cultured cells and liver tissues. (A) The distribution of DEGs in cultured cells. (B) The distribution of DEGs in liver tissues. (C) Shared genes with no DE in cultured cell and liver samples. Log2 counts per million (CPM). (D) Shared upregulated genes in diploid cultured cells and liver samples. (E) Shared upregulated genes in tetraploid cultured cells and liver samples.
A comparison of diploids and tetraploids can provide insights into the regulatory mechanisms associated with different ploidy levels. Therefore, we focused on DE analysis between diploid and tetraploid cultured cells of 20,771 genes (vs. 1 in Figure 1A), and found that 19,238 (92.6%) were not DEGs while 1,532 (7.4%) were DEGs; these included 747 (3.6%) that were upregulated in diploid cultured cells and 784 (3.8%) that were upregulated in tetraploid ones (Figure 2A). A comparison of diploid and tetraploid liver samples (vs. 2 in Figure 1A) showed that 486 (2.3%) genes were upregulated in diploid liver tissues, while 1166 (5.6%) were upregulated in tetraploid ones (Figure 2B). In total, 19,238 (92.6%) and 19,048 (92.1%) genes exhibited no significant DE in cultured cells (vs. 1 in Figure 1A) and liver tissues (vs. 2 in Figure 1A), respectively. Of the 20,771 total shared expressed genes, 18,050 (87.0%), including 17,905 (86.2%) non-DEGs and 145 (0.7%) DEGs, were found to have the same expression trend in the comparisons of “vs. 1” and “vs. 2” in Figure 1A (Figure 2C). Of these 145 DEGs, 38 (0.2%) showed upregulated expression in a diploid state, while 107 (0.5%) were upregulated in a tetraploid state (Figures 2D,E). Additionally, the 145 shared DEGs were displayed in a heatmap, in which diploid and tetraploid liver tissue and cultured cell samples were clustered together (Figure 3).
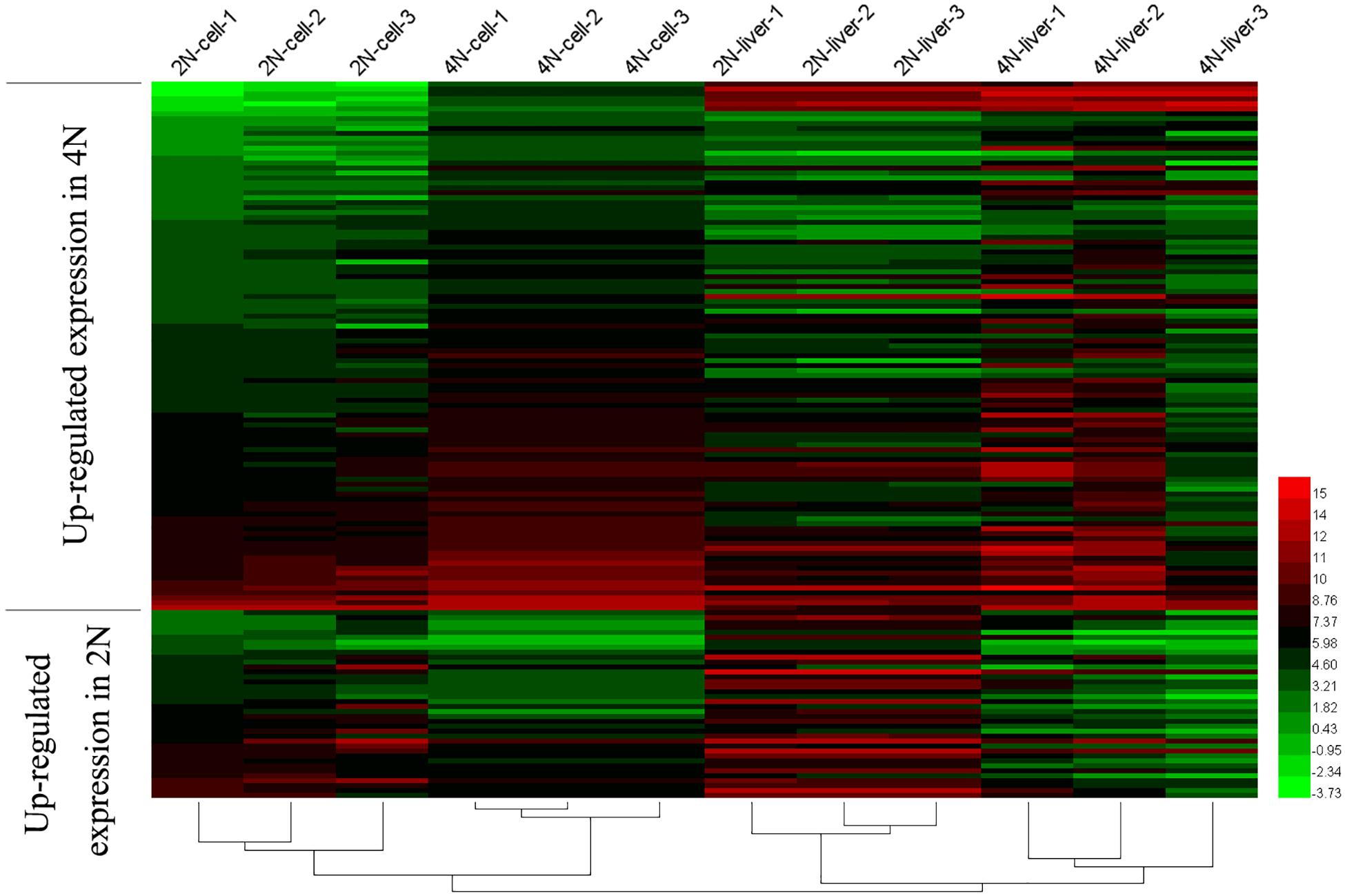
Figure 3. Shared DEGs between diploids and tetraploids in cultured cells and liver tissues. The 145 shared DEGs, including 38 that were upregulated in 2N and 107 that were upregulated in 4N, were detected from the comparison between diploid and tetraploid states.
To investigate changes in cell cycle regulation in vitro and in vivo, we next focused on KEGG pathways of the DEGs in our result (Supplementary Table 3). In comparison of diploid and tetraploid liver samples (vs. 1 and 2 in Figure 1A), the DEGs were shown to be mainly involved in the ribosome pathway (ko03010, 67 DEGs) and pathways associated with cancer (ko05200, 51 DEGs). Among these, 11 and 21 DEGs were associated with the cell cycle, respectively (Figure 4). Comparing of diploid and tetraploid cultured cells identified 15 DEGs in the cell cycle pathway. Of the seven DEGs shared between diploids and tetraploids in cultured cells and liver tissues, four showed the same expression trends as genes of the cell cycle pathway (Figure 4). Interestingly, three genes (ep300a, myc, and gadd45) exhibited the same DE trends between diploids and tetraploids, while three genes (smad4a, cul1a, and tp53) showed the opposite DE trends.
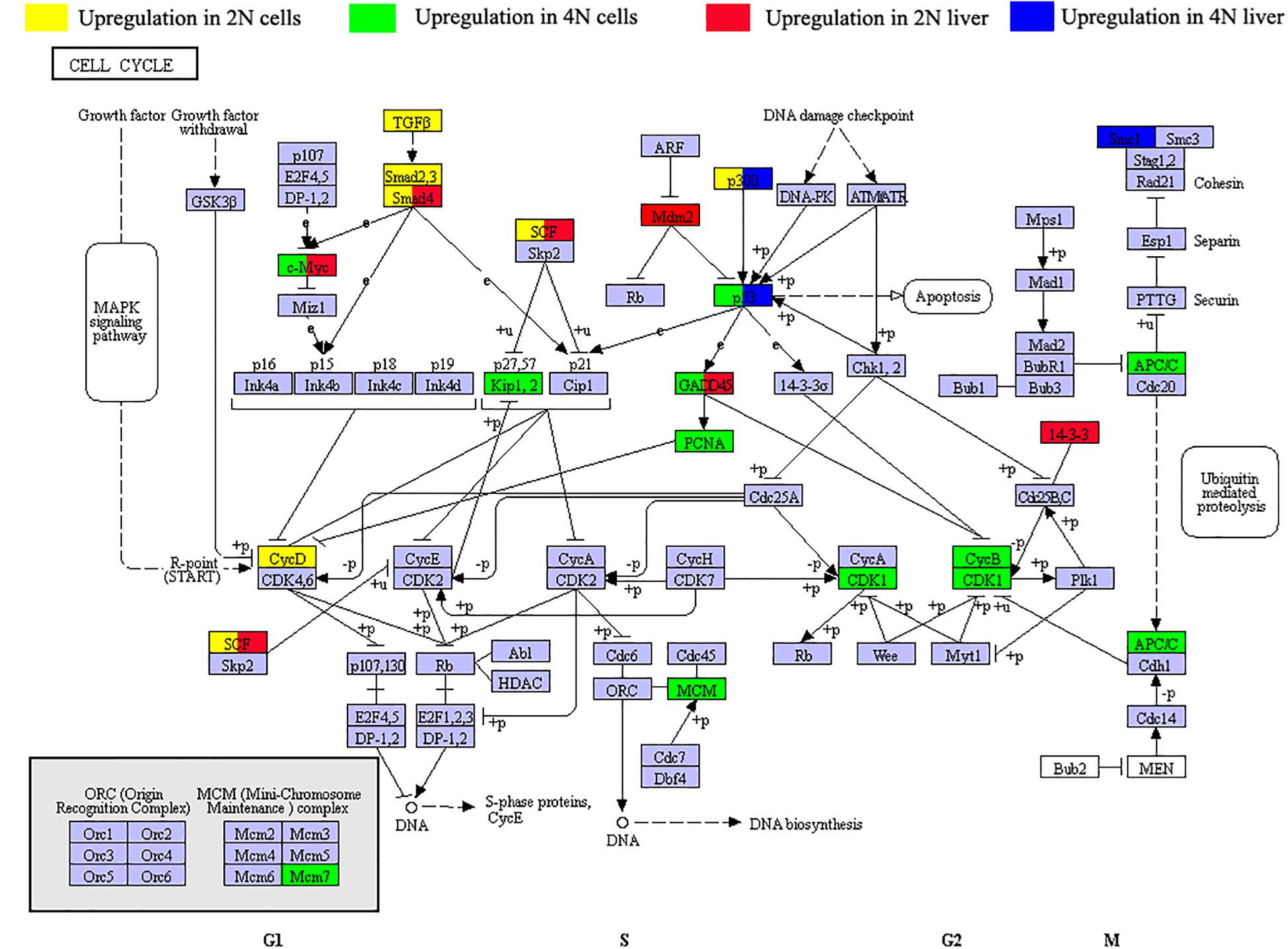
Figure 4. Distribution of differentially expressed genes (DEGs) of the cell cycle pathway (dre04110) in diploid and tetraploid states in vitro and in vivo. Green symbols represent upregulated expression in 2N cultured cells, yellow symbols represent upregulated expression in 4N cultured cells, red symbols represent upregulated expression in 2N liver, and blue symbols represent upregulated expression in 4N liver. Three genes (ep300a, myc, and gadd45) show the same DE trends between diploids and tetraploids. However, three other genes (smad4a, cul1a, and tp53) exhibited the opposite DE trends.
To better investigate expression differences in vitro and in vivo (Figure 5A), 11 DEGs including those in cell cycle pathways were analyzed with qPCR. This was performed in cultured cells and liver tissues, as well as in fin tissue from which the cultured cells had been generated. The different conditions between cultured cells and tissues resulted in major differences in expression profiles. To better describe gene regulation in the four samples, we established expression patterns based on relative levels in cultured cells and caudal fin tissue (Figures 5B–L). These patterns provided a clear perspective to assess differences between diploids and tetraploids in vitro and in vivo. The same relative expression patterns between diploids and tetraploids were detected in nine genes (smad6, p53, myc, id1, jun, gng10, gng12, gadd45, and calm), while different relative expression patterns were detected for the two other genes (lc3 and erg1) (Figure 5). These results of in vitro and in vivo exhibited the common trend of gene expression in cell-cycle-regulated genes accompanied with tetraploid formation.
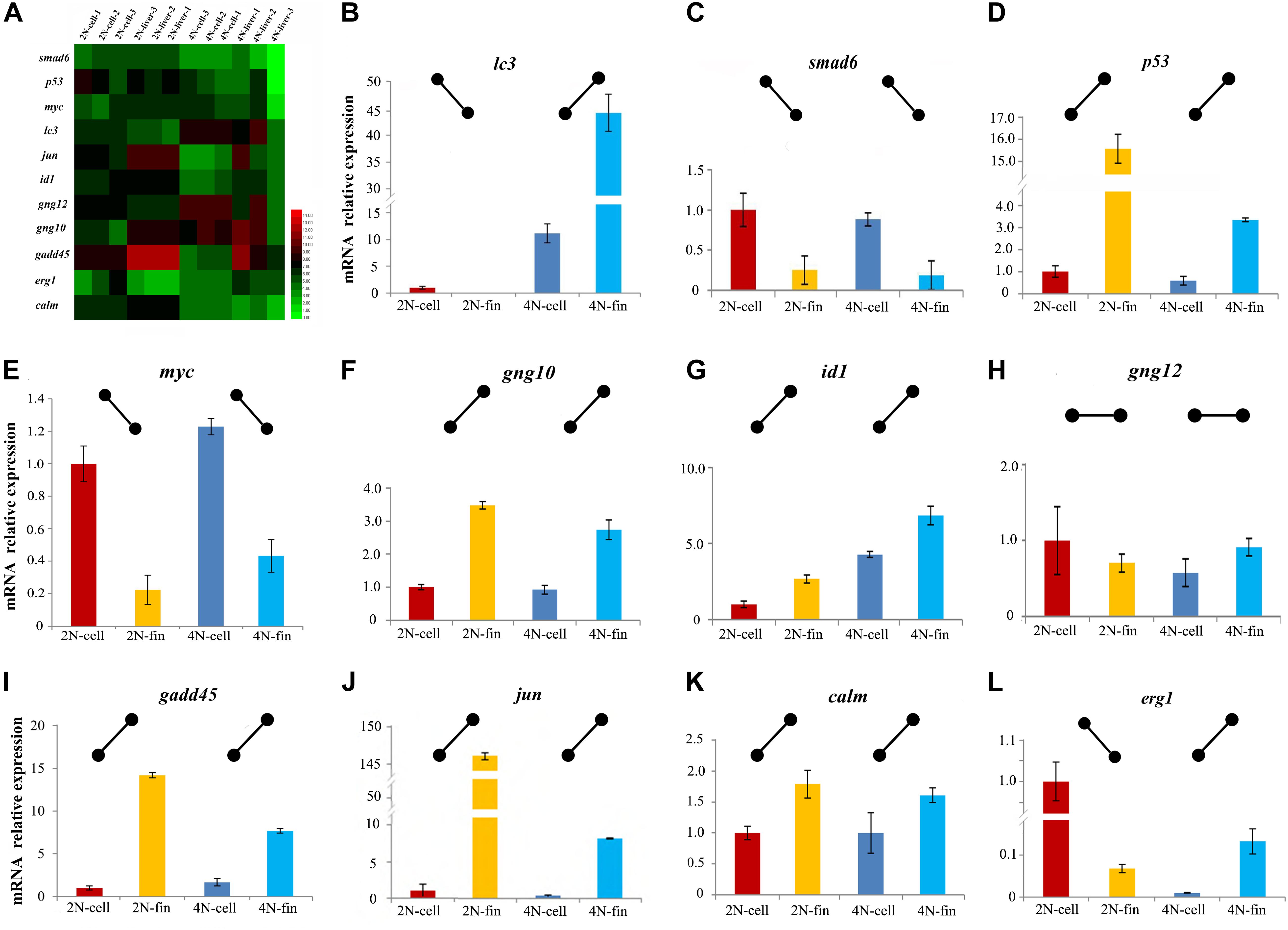
Figure 5. Expression levels of 11 genes detected by mRNA-seq and qPCR. (A) Heatmap of the expression distribution of 11 as detected by mRNA-seq in cultured cells and liver tissues. (B–L) The expression levels of 11 genes detected by qPCR in cultured cells and caudal fin.
Polyploidy were always observed in plant, but rarely in animals (Soltis et al., 2003). The formation of allotetraploid hybrids of C. auratus red var. and C. carpio L. provided an effective animal model to investigate mechanisms of polyploidy in animal (Liu et al., 2001, 2016). In comparison of diploid and tetraploid individuals, appropriate cell line were urgently needed to discover the different traits related to growth, fertility and disease resistance and various changes in molecular mechanisms for studying the potential mechanisms of these differences (Liu et al., 2001, 2009; Long et al., 2009; Ren et al., 2016). Here, we assessed the diploid and tetraploid cultured cell in gene expression level, and discussed them whether could be used to study polyploidy as comparison to in vitro.
Genome-wide expression profiles of polyploid culture cells and tissues in the present study provided a novel insight into the molecular mechanisms underlying the polyploidization effect in vitro and in vivo. To evaluate expression profile similarities between diploid and tetraploid states in vitro and in vivo, we performed DE analysis using mRNA-seq and qPCR. The analysis identified many DEGs between cells and liver tissues, not just in the diploid state but also in tetraploids (vs. 3 and 4 in Figure 1A) (Figure 2), indicating that marked changes in mRNA expression may be related to factors including changes in the cell microenvironment and the origin of the material (Arkhipchuk and Garanko, 2005). However, in the comparison between diploid and tetraploid samples (vs. 1 and 2 in Figure 1A), similar expression trends, including 38 shared upregulated genes in diploids, 107 shared upregulated genes in tetraploids, and 17,905 shared genes with no DE, were found in vitro and in vivo (Figures 2D,E). The results preliminary suggested that the relatively stable expression trends be maintained in most genes irrespective of in vivo and in vitro.
Dramatic mRNA expression changes often occurred with hybridization and polyploidization (Leggatt and Iwama, 2003; Osborn et al., 2003; Mallet, 2007). Some DEGs distributed were observed in some aquatic organisms, including oysters (Marie et al., 2006), protogynous wrasse (Jeong et al., 2009), rice field eel (Huang et al., 2005), rainbow trout (Cleveland and Weber, 2014), and gibel carp (Sun et al., 2010; Li et al., 2014). However, gene expression of polyploid cultured cell was rarely reported. Focused on cell-cycle-regulated genes, which play an important role in cell proliferation, ontogenesis and survival (Nishihara, 1997; Ashcroft and Vousden, 2001; Boxer and Dang, 2001; Ruzinova and Benezra, 2003; Wimmer et al., 2010; Wisdom et al., 2014; Valente et al., 2015), the 11 genes had been selected and performed with expression analysis using qPCR. The same expression trends were detected in nine genes between cultured cells from fin and caudal fin tissues (Figure 5), further suggesting that the common trends of gene expression were in cell-cycle-regulation irrespective of in vivo and in vitro. This research focused on common characteristics between diploids and tetraploids, providing us the gene expression changes of polyploidization in vitro and in vivo. Our findings indicate that the cultured cell line of this study appears to be an appropriate platform for polyploidy research, especially into the regulation of cell proliferation and adaptive regulation, although further comparisons of diploid and tetraploid material are necessary.
RNA-Seq data were submitted to NCBI SRA (SRR7640867, SRR7640866, SRR7640869, and SRR7640868).
All experiments were approved by the Animal Care Committee of Hunan Normal University and followed guidelines of the Administration of Affairs Concerning Animal Experimentation of China. All samples are raised in natural ponds, dissections are performed under sodium pentobarbital anesthesia, and all efforts are made to minimize suffering. This manuscript does not involve the use of any human data or tissue.
LR, SL, JiamL, and YX wrote and modified the manuscript. LR, JiahL, YF, and YH provided assistance extracting the raw material, and performing the qPCR experiment and bioinformatics analyses. YX and SL contributed to the conception and design of the study. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31772902, 31730098, 31430088, U19A2040, and 31702334), the National Key Research and Development Program of China (2018YFD0901202), the Hunan Provincial Natural Science and Technology Major Project (2017NK1031), the earmarked fund for China Agriculture Research System (CARS-45), the Key Research and Development Project of Hunan Province (2016NK2128), the Scientific Research Fund of the Hunan Provincial Education Department (16C0974), the Key Research and Development Program of Hunan Province (2018NK2072), and the Research Foundation of the Education Bureau of Hunan Province (17K058).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Zhanzhou Yao and Rurong Zhao for technical support with experimental equipment. We also thank Sarah Williams, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn), for editing the English text of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00203/full#supplementary-material
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Arkhipchuk, V. V., and Garanko, N. N. (2005). Using the nucleolar biomarker and the micronucleus test on in vivo fish fin cells. Ecotoxicol. Environ. Saf. 62, 42–52. doi: 10.1016/j.ecoenv.2005.01.001
Blomme, T., Vandepoele, K., De Bodt, S., Simillion, C., Maere, S., and Van de Peer, Y. (2006). The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7:R43. doi: 10.1186/gb-2006-7-5-r43
Boxer, L. M., and Dang, C. V. (2001). Translocations involving c-myc and c-myc function. Oncogene 20:5595. doi: 10.1038/sj.onc.1204595
Cantero, G., Pastor, N., Mateos, S., Campanella, C., and Cortes, F. (2006). Cisplatin-induced endoreduplication in CHO cells: DNA damage and inhibition of topoisomerase II. Mutat. Res. 599, 160–166. doi: 10.1016/j.mrfmmm.2006.02.006
Cleveland, B. M., and Weber, G. M. (2014). Ploidy effects on genes regulating growth mechanisms during fasting and refeeding in juvenile rainbow trout (Oncorhynchus mykiss). Mol. Cell. Endocrinol. 382, 139–149. doi: 10.1016/j.mce.2013.09.024
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Francesc, P., Andy, B., Jeanclaude, F. R., Martin, F. H., Pierrick, H., and Lorenzo, C. (2009). Polyploid fish and shellfish: production, biology and applications to aquaculture for performance improvement and genetic containment. Aquaculture 293, 125–156. doi: 10.1016/j.aquaculture.2009.04.036
Goudie, C. A., Simco, B. A., Davis, K. B., and Liu, Q. (1995). Production of gynogenetic and polyploid catfish by pressure-induced chromosome set manipulation. Aquaculture 133, 185–198. doi: 10.1016/0044-8486(94)00367-W
Huang, X., Guo, Y., Shui, Y., Gao, S., Yu, H., Cheng, H., et al. (2005). Multiple alternative splicing and differential expression of dmrt1 during gonad transformation of the rice field eel. Biol. Reprod. 73, 1017–1024. doi: 10.1095/biolreprod.105.041871
Huang, Y., Luo, Y., Liu, J., Gui, S., Wang, M., Liu, W., et al. (2017). A light-colored region of caudal fin: a niche of melanocyte progenitors in crucian carp (Cyprinus carpio L.). Cell Biol Int. 41:42. doi: 10.1002/cbin.10698
Hummon, A. B., Lim, S. R., Difilippantonio, M. J., and Ried, T. (2007). Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 42, 467–470. doi: 10.2144/000112401
Jeong, H. B., Park, J. G., Park, Y. J., Takemura, A., Hur, S. P., Lee, Y. D., et al. (2009). Isolation and characterization of DMRT1 and its putative regulatory region in the protogynous wrasse. Halichoeres tenuispinis. Gene 438, 8–16. doi: 10.1016/j.gene.2009.03.006
Leggatt, R. A., and Iwama, G. K. (2003). Occurrence of polyploidy in the fishes. Rev Fish Biol Fish. 13, 237–246. doi: 10.1023/B:RFBF.0000033049.00668.fe
Li, X. Y., Li, Z., Zhang, X. J., Zhou, L., and Gui, J. F. (2014). Expression characterization of testicular DMRT1 in both Sertoli cells and spermatogenic cells of polyploid gibel carp. Gene. 548, 119–125. doi: 10.1016/j.gene.2014.07.031
Liu, L., Cuiping, Y., Shaojun, L., Huan, Z., Dong, L., Zhen, L., et al. (2009). Cloning and evolutionary analysis of cyclin B gene introns in cyprinids with different ploidy levels. Prog. Nat. Sci. 19, 1103–1108. doi: 10.1016/j.pnsc.2009.03.001
Liu, S., Liu, Y., Zhou, G., Zhang, X., Luo, C., Feng, H., et al. (2001). The formation of tetraploid stocks of red crucian carp x common carp hybrids as an effect of interspecific hybridization. Aquaculture 192, 171–186.
Liu, S., Luo, J., Chai, J., Ren, L., Zhou, Y., Huang, F., et al. (2016). Genomic incompatibilities in the diploid and tetraploid offspring of the goldfish x common carp cross. Proc Natl Acad Sci U.S. A. 113, 1327–1332. doi: 10.1073/pnas.1512955113
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, Y., Zhong, H., Liu, S., Tao, M., Chen, L., Xiao, J., et al. (2009). Molecular characterization and genetic analysis of Gnrh2 and Gth in different ploidy level fishes. Prog. Nat. Sci. 19, 1569–1579. doi: 10.1016/j.pnsc.2009.06.002
Mahale, A. M., Khan, Z. A., Igarashi, M., Nanjangud, G. J., Qiao, R. F., Yao, S., et al. (2008). Clonal selection in malignant transformation of human fibroblasts transduced with defined cellular oncogenes. Cancer Res. 68, 1417–1426. doi: 10.1158/0008-5472.CAN-07-3021
Marie, V., Gonzalez, P., Baudrimont, M., Boutet, I., Moraga, D., Bourdineaud, J. P., et al. (2006). Metallothionein gene expression and protein levels in triploid and diploid oysters Crassostrea gigas after exposure to cadmium and zinc. Environ. Toxicol. Chem. 25, 412–418.
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nguyen, H. G., and Ravid, K. (2010). Polyploidy: Mechanisms and Cancer Promotion in Hematopoietic and Other Cells. New York, NY: Springer.
Nishihara, A. (1997). Smad6 inhibits signalling by the TGF-beta superfamily. Nature 389, 622–626. doi: 10.1038/39355
Osborn, T. C., Pires, J. C., Birchler, J. A., Auger, D. L., Chen, Z. J., Lee, H. S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19:141.
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. doi: 10.1016/j.cell.2007.10.022
Parmacek, M. S., and Epstein, J. A. (2009). Cardiomyocyte renewal. N. Engl. J. Med. 361, 86–88. doi: 10.1056/NEJMcibr0903347
Qin, Q., Wang, Y., Wang, J., Dai, J., Xiao, J., Hu, F., et al. (2014). The autotetraploid fish derived from hybridization of Carassius auratus red var. (female) x Megalobrama amblycephala (male). Biol. Reprod. 91, 93. doi: 10.1095/biolreprod.114.122283
Ren, L., Li, W., Tao, M., Qin, Q., Luo, J., Chai, J., et al. (2016). Homoeologue expression insights into the basis of growth heterosis at the intersection of ploidy and hybridity in Cyprinidae. Sci. Rep. 6:27040. doi: 10.1038/srep27040
Ruzinova, M. B., and Benezra, R. (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13, 410–418.
Shima, A. (1988). Fish cell culture: establishment of two fibroblast-like cell lines (OL-17 and OL-32) from fins of the medaka, oryzias latipes. In Vitro Cell Dev. Biol. 24, 294–298. doi: 10.1007/BF02628830
Shima, A., Nikaido, O., Shinohara, S., and Egami, N. (1980). Continued in vitro growth of fibroblast-like cells (RBCF-1) derived from the caudal fin of the fish, Carassius auratus. Exp. Gerontol. 15, 305–314.
Soltis, D. E., Soltis, P. S., and Tate, J. A. (2003). Advances in the study of polyploidy since Plant Speciation. New Phytol. 161, 173–191. doi: 10.1046/j.1469-8137.2003.00948.x
Storchova, Z., and Pellman, D. (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45–54. doi: 10.1038/nrm1276
Sun, M., Li, Z., and Gui, J. F. (2010). Dynamic distribution of spindlin in nucleoli, nucleoplasm and spindle from primary oocytes to mature eggs and its critical function for oocyte-to-embryo transition in gibel carp. J. Exp. Zool. A Ecol. Genet. Physiol. 313, 461–473. doi: 10.1002/jez.618
Swaminathan, T. R., Basheer, V. S., Gopalakrishnan, A., Sood, N., and Pradhan, P. K. (2016). A new epithelial cell line. Cytotechnology 68, 515–523. doi: 10.1007/s10616-014-9804-2
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Valente, G., Morani, F., Nicotra, G., Fusco, N., Peracchio, C., Titone, R., et al. (2015). Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in Ovarian Cancer. Biomed Res Int. 2014:462658. doi: 10.1155/2014/462658
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wimmer, P., Schreiner, S., Everett, R. D., Sirma, H., Groitl, P., and Dobner, T. (2010). SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29, 5511–5522. doi: 10.1038/onc.2010.284
Wisdom, R., Johnson, R. S., and Moore, C. (2014). c−Jun regulates cell cycle progression and apoptosis by distinct mechanisms. Embo J.. 18, 188–197. doi: 10.1093/emboj/18.1.188
Yoo, M. J., Szadkowski, E., and Wendel, J. F. (2013). Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110, 171–180. doi: 10.1038/hdy.2012.94
Keywords: polyploidy, in vitro, in vivo, cell cycle, mRNA expression
Citation: Ren L, Lu J, Fan Y, Hu Y, Li J, Xiao Y and Liu S (2020) Expression Profile Analysis of the Cell Cycle in Diploid and Tetraploid Carassius auratus red var.. Front. Genet. 11:203. doi: 10.3389/fgene.2020.00203
Received: 18 March 2019; Accepted: 21 February 2020;
Published: 17 March 2020.
Edited by:
Francesca Bertolini, Technical University of Denmark, DenmarkReviewed by:
Chuanju Dong, Henan Normal University, ChinaCopyright © 2020 Ren, Lu, Fan, Hu, Li, Xiao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yamei Xiao, eWFtZWl4QGh1bm51LmVkdS5jbg==; Shaojun Liu, bHNqQGh1bm51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.