
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 27 February 2020
Sec. Nutrigenomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00049
This article is part of the Research Topic Development of Healthy and Nutritious Cereals: Recent Insights on Molecular Advances in Breeding View all 15 articles
With the ever-increasing world population, an extra 1.5 billion mouths need to be fed by 2050 with continuously dwindling arable land. Hence, it is imperative that extra food come from the marginal lands that are expected to be unsuitable for growing major staple crops under the adverse climate change scenario. Crop diversity provides right alternatives for marginal environments to improve food, feed, and nutritional security. Well-adapted and climate-resilient crops will be the best fit for such a scenario to produce seed and biomass. The minor millets are known for their high nutritional profile and better resilience for several abiotic stresses that make them the suitable crops for arid and salt-affected soils and poor-quality waters. Finger millet (Eleucine coracana) and foxtail millet (Setaria italica), also considered as orphan crops, are highly tolerant grass crop species that grow well in marginal and degraded lands of Africa and Asia with better nutritional profile. Another category of grains, called pseudo-cereals, is considered as rich foods because of their protein quality and content, high mineral content, and healthy and balance food quality. Quinoa (Chenopodium quinoa), amaranth (Amaranthus sp.), and buckwheat (Fagopyrum esculentum) fall under this category. Nevertheless, both minor millets and pseudo-cereals are morphologically different, although similar for micronutrient bioavailability, and their grains are gluten-free. The cultivation of these millets can make dry lands productive and ensure future food as well as nutritional security. Although the natural nutrient profile of these crop plant species is remarkably good, little development has occurred in advances in molecular genetics and breeding efforts to improve the bioavailability of nutrients. Recent advances in NGS have enabled the genome and transcriptome sequencing of these millets and pseudo-cereals for the faster development of molecular markers and application in molecular breeding. Genomic information on finger millet (1,196 Mb with 85,243 genes); S. italica, a model small millet (well-annotated draft genome of 420 Mb with 38,801 protein-coding genes); amaranth (466 Mb genome and 23,059 protein-coding genes); buckwheat (genome size of 1.12 Gb with 35,816 annotated genes); and quinoa (genome size of 1.5 Gb containing 54,438 protein-coding genes) could pave the way for the genetic improvement of these grains. These genomic resources are an important first step toward genetic improvement of these crops. This review highlights the current advances and available resources on genomics to improve nutrient bioavailability in these five suitable crops for the sustained healthy livelihood.
All foods have uniqueness in their composition, with a specific range of macro- and micronutrients in different combinations. Some foods are rich in carbohydrate (rice, wheat, maize, etc.), protein (pulses, spinach), fat (oilseeds, groundnut), or mineral (pearl millet, etc.), while some are nutrient-dense and have optimum combinations of nutrients with good digestibility (most of the minor millets, quinoa, etc.). These nutrient-dense foods with a proper mix of nutrients and high bioavailability are sometimes designated as superfoods, but the irony is that most of these are also considered as orphan crops because of lower cropped area, low demand, and low consumption. Minor millets and pseudo-cereals come under the category of underused and neglected crops.
Every day, arable non-stress areas are being converted into marginal lands at an alarming rate, which may further increase under the climate change scenario if business as usual (RCP8.5) continues without arresting the trend of global warming as per the IPCC AR5 (Porter et al., 2014). Recent studies based on compiled data from 53 countries with serious, alarming, or seriously alarming conditions clearly show that more than 50% (27 countries) had decreased consumed calories due to mean climate change (Ray et al., 2019). Although the productivity of major crops has been predicted to be lower under marginal environments, the good news is that the nutrient-rich underused neglected crops are very resilient to harsh environments (drought, salinity, and extreme temperature) and yield well with limited resources (Mabhaudhi et al., 2019). As per FAO estimates (FAO, 2017), most of the food needed for 2 billion more mouths by 2050 has to come from marginal environments. A “Next-Generation Green Revolution” is required to achieve food security, which is a much broader and systems-based approach, and this has to come from the areas that were left out of the first Green Revolution, to achieve future food security in a more sustainable way across all spectrums of society (Nusslein et al., 2016; Dhankher and Foyer, 2018). Huge scope exists for the genetic improvement of these crops but, unfortunately, only limited research programs are undertaking focused research on such crops worldwide. Small and complex flower shape has compounded the difficulty to handle these crops; hence, few genetic studies are being undertaken. This is one of the major factors for slowdown of the research on developing molecular markers for the agronomically important traits to be used in breeding programs. Most genomics studies focus on population structures, grouping, and evolution. The current paper is a review of the scientific work on neglected but nutrient-rich crops such as quinoa, amaranth, finger millet, foxtail millet, and buckwheat. In recent years, progress in addressing all forms of malnutrition has seen a declining trend, alarmingly slow, with >150 million children still stunted (Global Nutrition Report, 2018), but the movement has increased awareness among people and hence increased health-conscious people demanding healthy food relatively more often. This review will deal with five underused but nutritionally important and rich crops: finger millet (Eleucine coracana), foxtail millet (Setaria italica), quinoa (Chenopodium quinoa), amaranth (Amaranthus sp.), and buckwheat (Fagopyrum esculentum).
Bioavailability, in simple terms, refers to whatever is absorbed out of ingested food and goes into the bloodstream. Many factors could affect this process, but the prominent ones are the original profile of food, processing of food, and digestion efficiency. Nutritional composition is the mix of macronutrients (carbohydrate, fat, and protein) and micronutrients (minerals and vitamins) within a product, but their absorption efficiency in the body depends on the ability of food to be digested easily. Food that is absorbed easily maintains or improves health and energy status by providing appropriate macro- and micronutrients in balanced form and is thus called healthy food. Neglected or underused crops such as minor millets and pseudo-cereals were part of the common diet of ancient cultures but slowly, after the Green Revolution, the higher availability and accessibility of rice, wheat, and maize overtook these neglected crops and started providing >60% of the calorific intake through these three crops only, thus starting to create a nutrient-imbalanced diet. Not many studies exist on the bioavailability of the nutrients provided by nutrient-dense underused crops.
Millets serve as a good food source of carbohydrates, proteins, minerals, and vitamins. Although they are the basic food ingredient in the diets of millions of people living in the semi-arid and arid regions of the world, they are still sometimes referred to as orphan crops or even lost crops. These neglected crops are mostly cultivated in developing countries and their world production statistics show low volumes vis-à-vis other popular food crops. These neglected crops are important because of their contribution to biodiversity and climatic resilience, their rich nutrition profile, and their means of livelihood of the poor in various parts of the world (Belton and Taylor, 2004). Finger millet is one of the most efficient crops for nitrogen use efficiency (NUE) and it can grow well with less water requirement, hence well suited to semi-arid climates (Gupta et al., 2017). It is also responsive to nutrients but has the ability to do well under limited resources (Gull et al., 2014). The most important part is its excellent storing capacity without deterioration even with significant insect and pest attacks. This has earned it the popular name of “famine crop” as it can resist storage pests for as long as 10 years, ensuring a year-round food supply (Mgonja et al., 2007).
Among the millet crops, six crops are called minor millets due to their small size: finger millet (Eleusine coracana (L.) Gaertn.), foxtail millet (S. italica (L.) P. Beauv.), kodo millet (Paspalum scrobiculatum L.), proso millet (Panicum miliaceum L.), barnyard millet (Echinochloa spp.), and little millet (Panicum sumatrense Roth). All these small minor millets are known for their unique nutritional composition and resilience (Kumar et al., 2018). Only foxtail and finger millet from this group will be discussed here. Finger millet is relatively popular in India and many countries in Africa because of its resilience and nutrient-dense grain profile. It has been promoted in Africa to reduce anemic incidence in children (Tripathi and Patel, 2010; Udeh et al., 2017). The nutraceutical importance of finger millet lies in its high content of calcium (0.38%), protein (6–13%), dietary fiber (10–18%), carbohydrate (65–75%), and minerals (2.5–3.5%). Another quality of finger millet is that it is gluten free with low glycemic index (GI) hence suitable for the people suffering from gluten intolerance/coeliac disease as well as diabetes (Tables 1 and 2). It is rich in ergocalciferol (vitamin D) and essential amino acids (EAA) such as valine, phenyl-alanine, leucine, and histidine (Tables 3 and 4). Besides these important nutrients, it has phytates (0.48%), tannins (0.61%), phenolic compounds (0.3–3.0%), and trypsin inhibitory factors that affect the bioavailability of nutrients; that is why proper processing is important to exploit its positive nutritional qualities (Devi et al., 2014) (Tables 5 and 6). Chauhan and Sarita (2018) have reported increased bioavailability of minerals such as Fe and P and vitamins upon grain processing before consumption. Phytates and tannins have negative effects on the bioavailability of nutrients, but processing at germination and little fermentation of grains increase the availability of minerals, amino acids, and free sugars, along with digestibility (Sripriya et al., 1997). Finger millet is mostly consumed as flour, but its processing through germinating and fermenting makes its iron content much higher in grains (Tatala et al., 2007). Seed germination of finger millet can increase the bioavailability of iron from 0.75 to 1.25 mg/100 g and is a potential alternative to mitigate anemia (Tatala et al., 2007). The pre-processing of grains can drastically reduce the impact of anti-nutrients and can improve iron bioavailability and bioactive compounds, which are confirmed by several scientific studies (Tatala et al., 2007; Hithamani and Srinivasan, 2014; Udeh et al., 2017) (Tables 5 and 6).
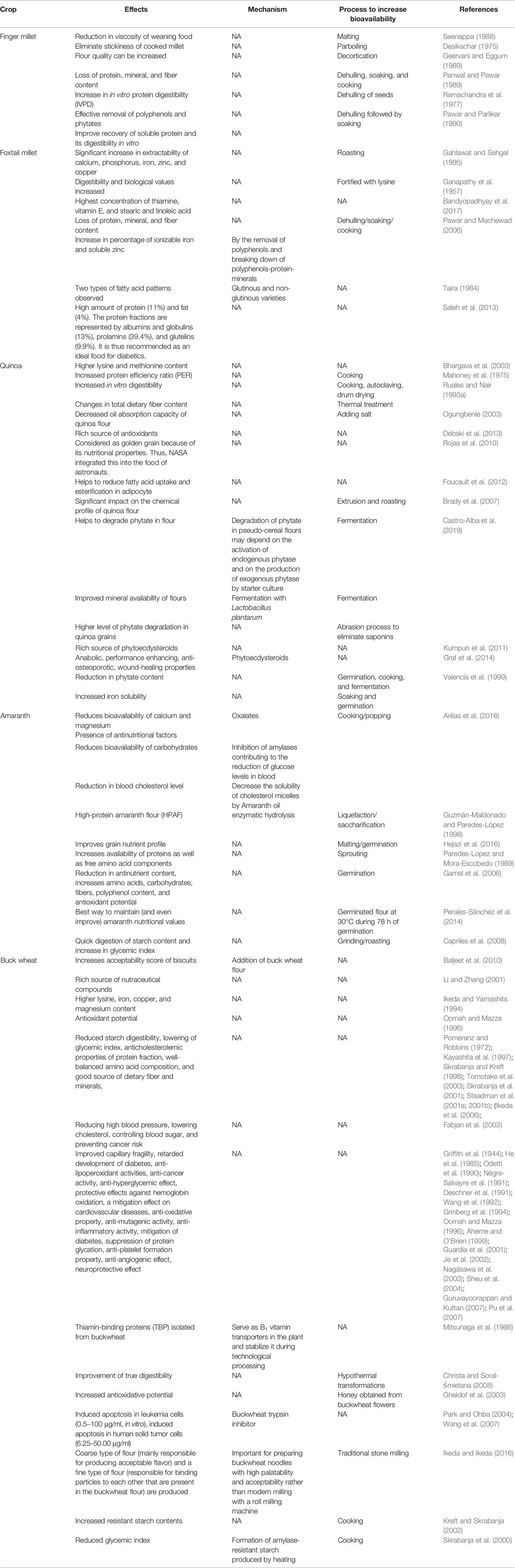
Table 5 Effects, mechanism, and process of increasing bioavailability of cereals and pseudo-cereal grains.

Table 6 Anti-nutrients and processes to decrease anti-nutritional activity in minor-millets and pseudo-cereals.
Singh et al. (2018) showed that traditional knowledge practiced by farmers to roast finger millet decreases phytochemical composition, moisture, protein, and antioxidant action, but increases fat, ash, and fiber and improves the bioavailability of iron and calcium. The improvement of iron bioavailability to reduce anemia happens via the biochemical changes in fortification with ferrous fumarate, ferric pyrophosphate (6 mg/kg), and zinc oxide (50 mg/kg) in finger millet flour (Tripathi and Patel, 2010). The diversity of finger millet offers a rich source of several antioxidants and calcium in the grains as polyphenols (0.3‒3.0%) that possess hypoglycemic, hypercholesterolemic, and anti-ulcerative properties (Chethan and Malleshi, 2007). Growing research interest exists in finger millet that could be attributed to several bioactive compounds such as ferulic acid-rich arabinoxylans, ferulic acid, caffeic acid, quercetin, and flavonoids, which are bio-accessible and have multiple therapeutic effects (Udeh et al., 2017). Hithamani and Srinivasan (2017) demonstrated that the bioavailability of phenolic compounds extracted from finger millet grain and co-administered to rats with piperine had a therapeutic benefit. The wide spectrum of phenolic compounds greatly enhances the nutraceutical potential of finger millet. Unprocessed and processed finger millet flour use in wafer and vermicelli (a fine noodle) have shown that bio-accessibility of Fe, Zn, and Ca through in vitro digestibility of starch (IVSD) and protein (IVPD) and bioactive polyphenols and flavonoids could be changed just by processing (Oghbaei and Prakash, 2012).
Finger millet [E. coracana (L.) Gaertn.] is a self-pollinated allotetraploid (2n = 4x = 36, AABB) species with a genome size of 1.593 Gb (Table 7). The 2C DNA amount in E. coracana is 3.36‒3.87 picogram (pg) (Mysore and Baird, 1997). It is an annual C4 herbaceous cereal crop belonging to family Poaceae and sub-family Chloridiodeae and exhibits morphological similarity to E. coracana subsp. africana and E. indica. Cytological studies, isozyme chloroplast DNA, and genomic in situ hybridization (GISH) have shown that the maternal diploid genome (AA) of E. coracana originated from E. indica whereas E. floccifolia is supposed to be the donor of the B genome to the polyploid species E. coracana (Bisht and Mukai, 2001). Because of its resilient nature, it is widely grown in arid and semi-arid areas of India and Africa.
Despite the nutritional benefits and climate-resilient nature of finger millet, the available genomic resources are limited, which has slowed the pace of genetic improvement of this crop (Saha et al., 2016). Immense morphological diversity is present in finger millet with a range of seed color correlated with protein and calcium content, time to maturity, and drought and salinity tolerance (Vadivoo et al., 1998; Tsehaye et al., 2006). Very few reports exist on the use of molecular markers for studying genetic diversity in finger millet, although the development and use of molecular markers in genomic studies of finger millet started a decade ago. Arya et al. (2009) developed 31 expressed sequence tag simple sequence repeats (EST-SSRs), out of which 17 were amplified and nine were found to be polymorphic between 11 elite germplasm accessions of finger millet of Indian and African origin. Reddy et al. (2012) identified 132 EST-based SSRs and developed 30 SSR primers for assessing genetic diversity in 15 finger millet accessions. Out of 30 EST-SSRs, 20 primers showed polymorphism and 13 primers were found to have polymorphism information content (PIC) value above 0.5. Using transcriptome data, Selvam et al. (2015) identified several SSRs and designed and validated 12 SSR primers on 23 finger millet accessions, where the primers showed an average PIC value of 0.67. Dida et al. (2007), using random HindIII, PstI, and SalI libraries, developed 82 genomic SSR markers and developed the first genetic map of finger millet using genomic SSRs, restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), and EST markers. The map covered 721 centiMorgan (cM) on genome A and 787 cM on genome B and consisted of 18 linkage groups. Phylogenetic studies using 45 genomic SSRs on 79 finger millet accessions showed that finger millet was domesticated in Africa first and was then introduced to India (Dida et al., 2008). Apart from work by Dida et al. (2007), 49 new polymorphic genomic SSR markers were developed by Musia (2013) using next-generation sequencing (NGS) data. Gimode et al. (2016) sequenced two genotypes of finger millet (KNE755 and KNE796) using Roche 454 and Illumina technologies and identified 10,327 SSRs and 23,285 single nucleotide polymorphism (SNP) and tested 101 of each across a diverse set of wild and cultivated finger millet accessions. Several other research groups identified EST-SSRs using sequences deposited in the NCBI database (Arya et al., 2009; Reddy et al., 2012; Babu et al., 2014b). The mean PIC value for 49 polymorphic SSRs tested was 0.42, whereas the mean PIC value for 80 polymorphic SNPs was 0.29. Genetic diversity analysis using molecular markers has reported low polymorphism showing a narrow genetic pool of cultivated finger millet genotypes (Muza et al., 1995). Using 14 polymorphic genomic SSRs and three genic SSRs, Arya et al. (2013) showed that African accessions have higher genetic diversity than Indian finger millet accessions. Apart from SSR markers, Kumar et al. (2016) identified 23,000 SNPs using genotyping by sequencing of 113 finger millet genotypes.
Finger millet has been reported to contain 5‒30 times higher calcium than other cereals (National Research Council, 1996) and 44.7% of the essential amino acids (Mbithi-Mwikya et al., 2000). Genetic diversity analysis of 103 finger millet genotypes using 36 EST-SSRs associated with opaque2 modifiers and 20 SSR primers associated with calcium transporters and calmodulin genes differentiated the finger millet genotypes based on protein and calcium content (Nirgude et al., 2014). Cereal endosperm proteins lack essential amino acids such as lysine and tryptophan and opaque 2 modifier (a bZIP transcription factor) is involved in regulating the accumulation of lysine and tryptophan in seed. A set of 67 functional SSR markers was developed and genetic diversity analysis in a global finger millet genotype collection for opaque2 modifier genes classified the genotypes into three clusters with high, medium, and low tryptophan content with few exceptions (Babu et al., 2014c). Association mapping studies identified markers associated with various agronomic traits such as days to flowering, tiller number, plant height, blast resistance, finger number, etc. (Bharathi, 2011; Babu et al., 2014a; Babu et al., 2014b). Association mapping studies for nutritional quality traits identified two QTLs associated with tryptophan content and one QTL associated with protein content, and the marker associated with tryptophan content showed an inverse relationship with protein content in finger millet (Babu et al., 2014c). Further, nine markers were identified to be associated with calcium content (Kumar et al., 2015b). Apart from SSR and SNP markers, the orthologous genes for amino acid composition and calcium content in grains of finger millet were identified and the SSR variations within these genes among the accessions differing in protein and calcium content were used for developing gene-specific functional SSR markers (Reddy et al., 2011; Nirgude et al., 2014). Kumar et al. (2015a) carried out transcriptome analysis in developing spikes of finger millet and identified SSR motifs in the genes encoding calcium transporters and seed storage proteins. Despite the markers and genetic materials identified, progress in marker-assisted selection for genetic improvement of finger millet has lagged because of poor understanding about the complex traits to be transferred to the genotypes of interest.
A few transcriptomics studies have been carried out to have a better understanding about the complexity of traits in finger millet. Salinity-responsive transcriptome profiling using a next-generation sequencing platform (Ion Proton) in contrasting finger millet genotypes led to the identification of the genes/pathways involved in an improved salt tolerance mechanism (Rahman et al., 2014). To understand the underlying mechanism of calcium accumulation in grains, Kumar et al. (2015a) carried out transcriptome analysis in the developing spikes of two finger millet genotypes (GPHCPB 45, a high-calcium genotype, and GPHCPB 1, a low-calcium genotype) using Illumina Hiseq-2000. It has been hypothesized that the accumulation of calcium in different tissues and genotypes of finger millet varies due to the differential expression of the genes involved in uptake, translocation, and accumulation of calcium in different tissues. Mirza et al. (2014) carried out expression analysis of the genes involved in calcium translocation and storage in two contrasting finger millet genotypes for calcium content and observed a two-pore channel (TPC1) and Ca(2+) ATPase that might be involved in calcium uptake and translocation, respectively, due to their strong expression in root, stem, and developing spikes; whereas Ca(2+)/H(+) antiporter (CAX1) might be involved in calcium accumulation in seeds due to its over-expression in developing spikes. The correlation between expression of these genes and calcium accumulation shows that these genes can be further used for a biofortification program (Table 8).
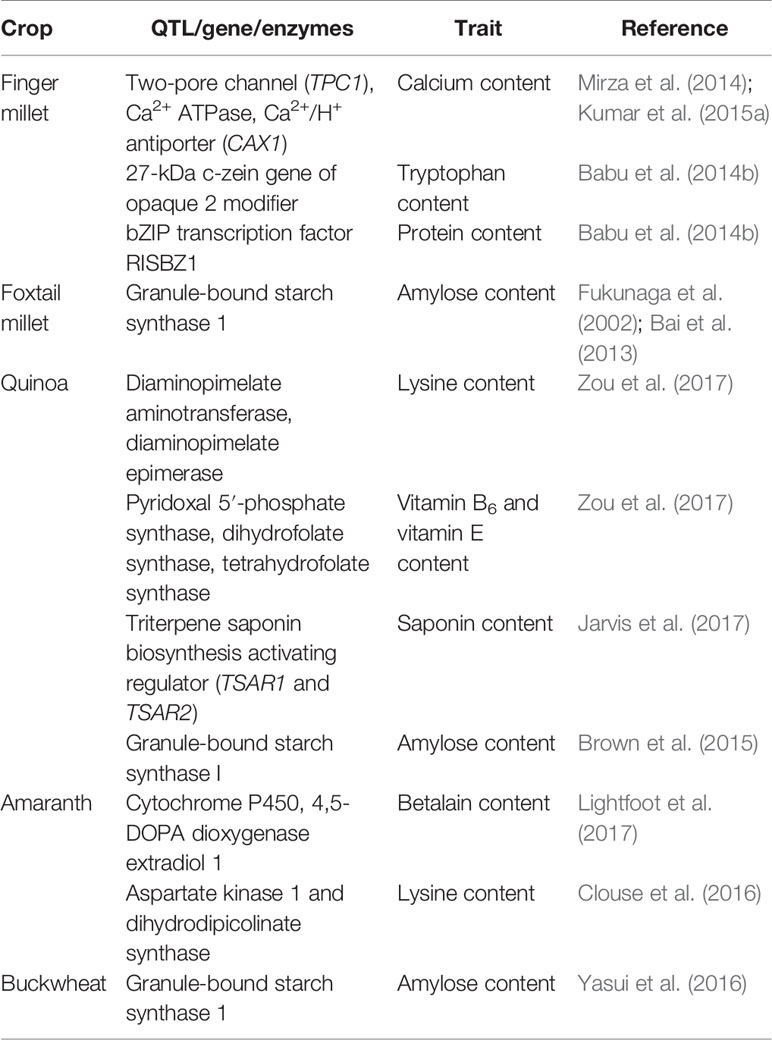
Table 8 Details of important QTLs/genes linked with accumulation of nutritional/anti-nutritional factors in underused crops.
To unravel and understand the complex genome of finger millet, two independent research programs were started for carrying out whole-genome sequencing of finger millet. These attempts were carried out by the Indo-Swiss collaborative program funded by the Department of Biotechnology, Ministry of Science and Technology, India, and coordinated by the University of Agricultural Sciences, Bangalore, India, in partnership with Functional Genomics Center Zurich (FGCZ), University of Zurich. The researchers sequenced the genome and transcriptome of a drought-tolerant and blast-resistant finger millet genotype (ML-365) using Illumina and SOLiD sequencing technologies (Hittalmani et al., 2017). The sequenced genome consisted of 1,196 Mb covering ~82% of the total genome size. Genome analysis revealed the presence of 85,243 genes and 49.92% of the genome was found to be consisting of repetitive DNA. Hatakeyama et al. (2018) carried out whole-genome sequencing and assembly of finger millet (cv. PR-202) using Illumina NextSeq500 and PacBio RS II systems. The assembled genome was found to be of 1,189 Mb, estimated to be covering 78.2% of the finger millet genome. Genome analysis resulted in the identification of 62,348 genes, of which 91% were functionally annotated. The whole-genome information on ML-365 and PR-202 can be used for candidate genes and marker identification, which can be further used in marker-assisted breeding programs for the genetic improvement of finger millet.
Foxtail millet (S. italica (L.) P. Beauv.), also called Italian, German, Hungarian, or Siberian millet, is a nutritional and natural staple used in many countries of East Asia for a long time. It has been cultivated in India, China, and many countries in Southeast Asia and Africa for many millennia and is quite popular in arid zones (Austin, 2006; Cheng and Dong, 2010; Mal et al., 2010). The grain of foxtail millet is quite rich in protein, fiber, and phosphorus compared with that of other minor millets (Muthamilarasan and Prasad, 2015). These grains are very rich in vitamin B1, B3, and B5 (Table 4), and also contain a much higher amount than other minor millets and common cereals such as rice, wheat, and maize (Cheng and Dong, 2010). Foxtail millet also contains all the EAA but has isoleucine, leucine, methionine, phenylalanine, threonine, tryptophan, and valine in significantly higher amounts (Table 3). This makes foxtail millet a highly important nutritive crop with rich genetic diversity for glutinous and non-glutinous grains with different lipid composition (Taira and Miyahara, 1983). Significant phenotypic variations provide ample opportunity for allele mining and the use of molecular markers to supplement breeding programs. This crop is also one of the most important C4 panicoid crops known for its small genome size (~490 Mb), short life cycle, and self-pollinating nature, which make it an excellent model crop for evolutionary studies within the panicoid grass system (Lata and Shivhare, 2017).
Foxtail millet, vis-à-vis other gramineous crops, is a naturally drought-tolerant crop (Goron and Raizada, 2015). It is a quite resilient crop and is better adapted to marginal environments, especially arid regions. It is an extremely suitable food for type 2 diabetics due to its low glycemic index (GI) as its starch digestibility is much lower than that of wheat (Table 1). However, its nutrient bioavailability could be further increased if it were processed by boiling or steaming (Pawar and Machewad, 2006; Ren et al., 2016) (Tables 5 and 6). Genotypic differences exist for antioxidant activities and some specific varieties such as SiA-2593 were identified as therapeutic and functional foods (Shejawale et al., 2016).
Foxtail millet starch is of great interest among entrepreneurs owing to its flexibility to make gels using foxtail millet flour and divalent cations such as CaCl2 and FeSO4 (Nagaprabha and Bhattacharya, 2016). S. italica is considered one of the best minor millet crops for anemic and diabetic people, thus reinforcing nutrition security besides food security (Bandyopadhyay et al., 2017). Processed protein from S. italica has been reported as a potential source of food additive (Mohamed et al., 2009). Ample diversity is available in India, China, France, Japan, Kenya, and Mexico gene banks for foxtail millet (Dwivedi et al., 2012), which can be exploited for various nutritional traits, including protein, as the protein content is higher than that of other selected small millets (Rao et al., 2011). Besides this, its rich composition of beta-glucans (42.6%) present in the fiber helps to enhance sugar and cholesterol metabolism, which ultimately prevents diabetes and cardiovascular diseases (Kumari and Thayumanavan, 1997; Itagi et al., 2012; Muthamilarasan and Prasad, 2015). Efforts on genomics are in progress to identify the underlying factors for the mechanisms that improve nutritional factors in foxtail millet (Zhang et al., 2012; Muthamilarasan and Prasad, 2015). Traditional simple as well as advanced food processing helps to improve the bioavailability of micronutrients (iron, zinc, and proteins) and renders them in a form that is easy to assimilate by the body, along with a significant decrease in anti-nutrients (polyphenols and phytate) (Pawar and Machewad, 2006; Saleh et al., 2013). Anti-nutrients exist but grain processing reduces them drastically; however, omics can contribute a lot to improving the bioavailability of micronutrients and reducing anti-nutrients such as phytic acid, polyphenols, and tannins, to decrease processing cost and time.
Foxtail millet (S. italica (L.) P. Beauv.) is one of the oldest domesticated cereals in the Old World. The genus Setaria belongs to the subtribe Cenchrinae and tribe Paniceae within the subfamily Panicoideae (Kellogg, 2015). Setaria is the largest genus in the Cenchrinae, consisting of ≈100 species and all possessing the C4 photosynthetic pathway. The small diploid genome (2n = 2x = 18; 513 Mb), self-pollination behavior, and short life cycle (6 weeks) have made Setaria an ideal model plant for functional genomics studies in millets as well as in cereals (Table 7).
Among millets, Setaria is the most deeply studied genus at both the genetic and molecular level. Various types of molecular markers used for genetic diversity and phylogenetic studies in S. italica, including RFLP (Wang et al., 1998; Fukunaga et al., 2002), random amplified polymorphic DNA (RAPD) (Schontz and Rether, 1999), AFLP (d'Ennequin et al., 2000), and SSRs (Lin, 2012; Trivedi et al., 2018), showed that foxtail millet genotypes differed genetically between different regions and Chinese landraces were found to be the most variable compared with landraces from other places. Wang et al. (1998) first reported the RFLP-based map consisting of 160 loci using an intervarietal cross of foxtail millet (Longgu 25 × Pagoda Flower Green), which was later used by Devos et al. (1998) to construct a comparative genetic map of foxtail millet and rice. Seeing the importance of SSR markers, Jia et al. (2007) developed 26 EST-SSRs. Later, Jia et al. (2009), using two genomic libraries enriched for (GA)n and (CA)n, identified 100 polymorphic SSR markers and developed a linkage map using 81 SSRs and 20 RFLP-anchored markers. Later, Lin et al. (2011) developed 45 polymorphic SSR markers from a RAPD-enriched library and used them for genetic diversity analysis as well as proving their cross-species transferability. Gupta et al. (2011; 2012) developed 98 intron-length polymorphic (ILP) and 147 genomic SSR markers and further showed the high-level cross-species transferability. Considering the importance and ease of microsatellite markers in MAS because of their high reproducibility, co-dominant nature, multiallelic variation, and abundance in genome work, Setaria was used to mine the genome wide SSRs by analyzing the genome sequence information. The genome-wide analysis of foxtail millet resulted in the identification of 28,342 microsatellite repeat-motifs spanning 405.3 Mb of the genome. Among the identified microsatellites, primers for 21,294 were designed and 15,573 markers were mapped on nine chromosomes to develop a high-density physical map of foxtail millet (Pandey et al., 2013). The validation of 159 developed markers in eight accessions of Setaria sp. showed 67% polymorphism and 89.3% cross-genera transferability across millet and non-millet species. Muthamilarasan et al. (2013) developed 5,123 ILP markers and proved their applicability in germplasm characterization, phylogenetic studies, transferability across species, and comparative mapping in millets and bioenergy grass species.
A milestone in the area of Setaria genomics was the release of the reference genome of foxtail millet cultivar “Zhang gu” using whole-genome shotgun sequencing combined with the Illumina second-generation sequencer covering ≈86% of the genome (Zhang et al., 2012). The sequencing resulted in the generation of 16,903 contigs and 439 scaffolds covering a total length of 423 Mb. Further sequence analysis identified 38,801 annotated genes. Apart from “Zhang gu,” a photo-thermo-sensitive male sterile line (A2) was sequenced and comparison of both genomes resulted in the identification of many SNPs (542,322), small insertions/deletions (33,587), and structural variants (10,839) between the two cultivars. A linkage map was constructed using an F2 population derived from “Zhang gu” and “A2” using 759 markers consisting of 118 SNPs and 641 structural variants (Zhang et al., 2012). S. italica accession “Yugu1” and Setaria viridis accession “A10” were sequenced using the ABI3730xl capillary sequencer (Bennetzen et al., 2012). The assembled genome was found to be 396.7 Mb covering 80% of the genome and genome analysis identified 24,000‒29,000 expressed genes. To date, foxtail millet is the only millet whose genome assembly is available to a chromosomal scale.
Because of the abiotic stress-tolerant nature of foxtail millet, several functional genomics tools have been applied to dissect its stress-tolerant nature (Zhang et al., 2007; Jayaraman et al., 2008; Lata et al., 2010; Lata and Prasad, 2011; Lata et al., 2011; Puranik et al., 2011; Puranik et al., 2013; Qi et al., 2013; Tang et al., 2017). Besides understanding the molecular basis of abiotic stress tolerance, GWAS studies have been carried out for mapping the QTLs underlying various agronomic traits in foxtail millet (Jia et al., 2013; Gupta et al., 2014; Jaiswal et al., 2019).
However, despite the enormous health benefits, few proper attempts have been made to understand the genetics and genomics of nutritional traits in foxtail millet. Resequencing of waxy landrace Shi-Li-Xiang (SLX) and fine mapping using an F2 population derived from SLX (waxy) × Yugu1 (non-waxy) identified a waxy locus harboring starch synthase-encoding GBSS 1 gene. Sequence analysis of GBSS 1 showed transposable elements confirming its waxy nature (Bai et al., 2013). To dissect the genetics and genomics of nutritional traits, genome and transcriptome data can be used to select genes associated with various pathways involved in biosynthesis and regulation of storage compounds and these can be exploited for understanding their role in the accumulation of various nutritional compounds.
Quinoa (C. quinoa Willd.), pronounced “keenwa,” is a dicotyledonous plant originated from South America, hence called an Andean grain. For seven millennia, Andean cultures from pre-Columbian time have been eating it and it has been part of their diet. It is actually a pseudo-cereal due to its morphological grain shape like grass crops. It is classified into five ecotypes, based on geographic adaptation in the center of diversity (Hinojosa et al., 2018):
● Valley = grown at 2,000 to 3,500 m.a.s.l. in Colombia, Ecuador, Peru, and Bolivia;
● Altiplano = grown at high altitudes of more than 3,500 m.a.s.l. around Titicaca Lake on the border of Bolivia and Peru;
● Salares = grown in the salt flats of Bolivia and Chile and having a high tolerance of salinity;
● Sea-level = grown in the low-altitude areas of southern and central Chile;
● Subtropical or yungas = grown in the low-altitude humid valleys of Bolivia and including late-flowering genotypes.
Quinoa is a highly resilient crop that not only has superior nutritional quality vis-à-vis common cereals but can also withstand environments where other crops have difficulty to grow (Choukr-Allah et al., 2016; Nanduri et al., 2019). Since 2013, when UN/FAO designated 2013 as the International Year of Quinoa, interest in this crop has increased markedly worldwide due to awareness, mainly because of its balanced nutritional profile and its potential as an alternative to feed the growing world population in a sustainable manner, especially when more food has to come from marginal environments (Jacobsen et al., 2013; Zurita-Silva et al., 2014). The natural selection process of quinoa cultivars took place under severe adverse conditions of the Andes, such as limited rainfall and extreme aridity (Martínez et al., 2009), and in salt-affected soils (Ruiz-Carrasco et al., 2011). That explains quinoa's built-in abiotic stress tolerance of aridity, salinity, highland, and frost; hence, it is well suited to marginal environments. Although quinoa is drought and salinity tolerant, it is sensitive to high-temperature stress. It can tolerate a wide range of temperatures (from −8°C to 35°C), but high temperature above 35°C during flowering results in a significant reduction in seed set and ultimately yield (Jacobsen et al., 2005). For example, studies in Italy (Pulvento et al., 2010), Morocco (Hirich et al., 2014), Germany (Präger et al., 2018), Portugal (Pires, 2017), India (Bhargava et al., 2006a), Egypt (Eisa et al., 2017), Mauritania (Bazile et al., 2016), and the United States (Peterson and Murphy, 2015; Walters et al., 2016) have reported that high temperatures reduce quinoa seed yield.
Quinoa has a unique balance between oil (4–9%), protein (averaging 16%, with high nutritional relevance due to the ideal balance of its essential amino acid content), and carbohydrate (64%) (Schlick and Bubenheim, 1996; Bhargava et al., 2006a; Vega-Gálvez et al., 2010) (Table 1). Lysine, one of the essential amino acids, which is usually much less in plant-based diets, is relatively high in quinoa, indeed very close to the standard set by FAO for human nutrition (Table 3). Because of its high starch content (51–61%), it can be used in the same way as cereals for flour production (Mastebroek et al., 2000; Ogungbenle, 2003; Repo-Carrasco et al., 2003; Bhargava et al., 2006a; Stikic et al., 2012). In addition, quinoa is a good source of vitamins, oil (high in omega 3, linoleic and linolenic acids, 55–66% of the lipid fraction), and natural antioxidants such as α and γ tocopherol, and it has more minerals such as Ca, Fe, K, Mg, Cu, and Mn than other cereals (Table 2) (Repo-Carrasco et al., 2003; Vega-Gálvez et al., 2010; Fuentes and Bhargava, 2011; Stikic et al., 2012). The International Center for Biosaline Agriculture (ICBA), based in Dubai, has been working on the suitability of quinoa for marginal environments since 2006 and has found high Fe content vis-à-vis major cereals such as wheat, rice, and maize. Five improved quinoa genotypes from ICBA (Q1 to Q5) have been analyzed for Fe content and it ranged from 49.55 to 133 ppm depending upon growing conditions, showing high genotype-by-environment-by-management (G × E × M) interaction. Quinoa is highly suitable for eating by diabetics due to its low glycemic index (GI) (Table 1). Quinoa seeds release important compounds such as phytoecdysteroids and 20-hydroxyecdysone (20HE) while germinating. These released bioactive phytochemicals from the seeds can be an excellent staple for developing anti-diabetic food products as these bioactive phytochemicals can decrease the glucose level in the blood and are a potential source to treat obesity and hyperglycemia (Graf et al., 2014). Quinoa grains are also gluten-free and are considered as complete protein as they contain all nine essential amino acids that the human body cannot produce itself and they have very high lysine content overall vis-à-vis other cereals (Jacobsen, 2003; Albugoch and Lilian, 2009; Alvarez-Jubete et al., 2010). Quinoa's exceptional nutritional qualities led NASA to include it as part of its astronauts' diet on long space missions. A NASA technical paper mentions that while no single food can supply all the essential life-sustaining nutrients, quinoa comes as close as any other in the plant or animal kingdom (Schlick and Bubenheim, 1996).
Despite these nutritional qualities, some antinutritional factors (triterpenoid glycoside) are present in quinoa. Saponins, when present in the seeds, confer bitterness. Natural occurrence of saponins in quinoa grain is usually higher but some native varieties have low saponin as well. Even though saponins can be removed by repeated washing or dehulling, this consumes additional resources on postharvest processing (Table 6). Enough genetic variation has been reported in saponin content in quinoa, varying from 0.2 g/kg in sweet genotypes to 11.3 g/kg in bitter genotypes based on dry matter (Mastebroek et al., 2000). Saponin is present in the seed coat and washed saponin solution from seeds could be used as a by-product in biopesticide and therapeutic compounds (Ruiz et al., 2017). Reducing saponin content could broaden quinoa production globally in a more economically sustainable manner. It is reportedly controlled by a recessive gene (TSARL1) and genotypes with low saponin could be developed using conventional breeding techniques with the help of MAS (Jarvis et al., 2017).
Additional mineral inhibitor can influence the bioavailability and bio-accessibility of minerals of quinoa. Plant-based diets usually have a low bioavailability of minerals (mainly zinc, iron, and calcium) due to the presence of inhibitors such as phytates and tannins that reduce absorption. Phytate (myo-inositol-6-phosphate) is the main inhibitor of zinc, iron, and calcium. Degradation of phytate is important to allow the bio-assimilation of minerals. Bioavailability of iron is reduced if the phytate/Fe molar ratio is higher than 1, and, for bioavailability of zinc, this can affect the relation of the phytate/Zn molar ratio when it is higher than 5 (Iglesias-Puig et al., 2015).
Quinoa is a rich source of minerals and it has much higher zinc (2.73–5.01 mg/100 g), iron (4.82–7.19 mg/100 g), and calcium (77.10–211.90 mg/100 g) than other cereals based on results from six varieties of quinoa from Chile (Miranda et al., 2012). Unfortunately, the levels of phytates were quite high in quinoa vis-à-vis other cereals. It is better if the phyate:zinc molar ratio (Phy: Zn) is <15 (Bindra et al., 1986), phytate:iron (Phy : Fe) ratio <1 (Hurrell, 2004), and phytate:calcium (Phy : Ca) ratio <0.17 (Umeta et al., 2005). Ratios above the desirable values indicate low bioavailability of the mineral in the grain. Therefore, phytate can affect the bio-assimilation of important minerals in food if the molar ratios are high above the critical values. However, germination, soaking, cooking, and fermentation decrease the phytate compound in grains of quinoa and allow the bio-assimilation of iron, although much washing to some extent reduces the vitamin and mineral content as well (Ruales and Nair, 1993a; Valencia et al., 1999; Lazarte et al., 2015) (Tables 5 and 6). Cooking does not affect the amount of soluble iron; rather, it increases 2–4 times after soaking and germination, 3‒5 times after fermentation, and 5‒8 times after fermentation of the germinated flour combined with reduced phytates (Valencia et al., 1999). Soluble iron thus could be available to anemic populations in processed products based on germinated quinoa and quinoa sprouts (Vega-Gálvez et al., 2010). Brady et al. (2007) compared quinoa flour when processed by steam pre-conditioning, extrusion, and roasting. Steam pre-conditioning had the least effect on the chemical profile of quinoa while extrusion and roasting changed the chemical profile a lot compared to raw quinoa. The extrusion and roasting techniques can reduce saponin and the bitter taste (Table 6). Bioactive polyphenols and flavonols in quinoa grains and sprouts can help to prevent oxidative stress (Paśko et al., 2009).
C. quinoa is an annual pseudo-cereal. It is an allotetraploid with 2n = 4x = 36 chromosomes having an estimated haploid genome size (C-value) of 1.005‒1.596 pg (Bennett and Smith, 1991; Stevens et al., 2006; Bhargava et al., 2007a; Palomino et al., 2008; Kolano et al., 2012) (Table 7). Quinoa has mostly smaller metacentric chromosomes of 0.94‒1.60 µm (Bhargava et al., 2006b; Palomino et al., 2008). Several studies on molecular understanding and genomics of quinoa have been carried out considering its nutritional importance. Most of the genetics and genomics studies have focused on either understanding its genetic diversity or evolutionary history whereas limited attempts have been made for genetic improvement of quinoa through molecular approaches. Most of the breeding has been carried out through mass selection for selecting high‐yielding, early maturing quinoa varieties with low saponin content as well as tolerance of biotic and abiotic stresses.
The genetic diversity and phylogenetic relationship studies have been carried out between cultivated species of quinoa and their wild relatives using various types of phenotypic, biochemical, and molecular markers. Wilson (1988a) used morphological and biochemical markers to study the genetic relationships between quinoa ecotypes and classified them into two broad groups: coastal types and Andean types (above 1,800 m.a.s.l.). The Andean ecotypes were further classified into northern and southern Andean quinoa. Further, phylogenetic study showed that the Altiplano was the center of origin of quinoa (Wilson, 1988b). Rojas et al. (2000) classified the 1,512 accessions of the Bolivian National Quinoa Collection into seven distinct groups using various morphological and agronomic traits. Bhargava et al. (2007b) studied genetic diversity in quinoa using morphological and quality traits and showed a high level of genetic variability among the accessions.
Random amplified polymorphic DNA (RAPD) markers were the first markers used to detect DNA polymorphisms among different quinoa accessions (Fairbanks et al., 1993; Ruas et al., 1999; del Castillo et al., 2007). Using RAPD markers, very low intraspecific variations were observed within C. quinoa (Ruas et al., 1999). del Castillo et al. (2007) studied the hierarchical structure of the genetic variation present in eight quinoa field populations from Bolivia and found that population structure was related to three major biogeographic zones: the northern and central Altiplano, the inter-Andean valley, and the southern Salar. Apart from RAPD, AFLP markers have been used to study genetic diversity in quinoa. Rodríguez and Isla (2009) used AFLP markers along with 20 phenotypic markers to characterize 14 accessions of quinoa and concluded that Chilean lowland germplasm might be genetically more diverse, and the germplasm clustered together into highland and lowland/coastal as earlier reported by Ruas et al. (1999). Using SSR-enriched library sequencing, 208 polymorphic SSR markers were identified for quinoa (Mason et al., 2005). Recently, because of their reproducibility and co-dominant nature, SSRs have been used to study genetic diversity in quinoa (Mason et al., 2005; Jarvis et al., 2008). Christensen et al. (2007) used 152 accessions of USDA and CIP-FAO quinoa collections for genetic diversity study using 35 SSR markers and found that the accessions from lowlands and highlands clustered together and identified the group of accessions that appears to be hybrid between lowland and highland. Later, the use of multiplex fluorescent SSR markers to understand the genetic diversity and phylogenetics of 59 quinoa accessions resulted in the separation of highland and lowland genotypes into two separate clusters (Fuentes et al., 2009). Recently, Zhang, T., et al (2017) carried out whole-genome resequencing of 11 quinoa accessions and identified various SSR, SNP, and Insertion/Deletion (InDel) markers. They further used the identified SSR and InDel markers to assess the genetic diversity of 129 quinoa accessions from the USDA collection. These studies using various types of markers showed that a strong population structure and huge genetic diversity exist among quinoa germplasm.
Seeing the importance of quinoa in food and nutritional security, several breeding programs for improving grain yield, earliness, disease resistance, and drought tolerance and reducing saponin content have begun. Molecular markers and linkage maps help in QTL mapping, which further helps in marker-assisted selection (MAS) for speeding up the breeding process. The first linkage map of quinoa was developed by Maughan et al. (2004) using 19 SSR, 6 RAPD, and 230 AFLP markers spanning 1,020 cM covering 60% of the genome, consisting of 35 linkage groups with an average marker density of 4.0 cM per marker. Later, Jarvis et al. (2008) developed 216 new SSR markers and a more enriched linkage map for quinoa by using new SSR and 75 AFLP markers consisting of 41 linkage groups covering 913 cM. Further, using two RIL populations, Maughan et al. (2012) mapped 511 SNPs across 29 linkage groups of quinoa spanning 1,404 cM with a marker density of 3.1 cM per marker. Recently, Jarvis et al. (2017) developed a high-density linkage map of quinoa consisting of 6,403 SNP markers through genotyping by sequencing (GBS) covering 2,034 cM on 18 linkage groups. Further, apart from molecular markers, several other genomic resources such as bacterial artificial chromosome (BAC) libraries and stress-responsive or tissue-specific transcriptome sequencing have been used, which has helped in gene discovery as well as identifying molecular markers. The first EST library of quinoa consisting of 424 ESTs was developed from seed and floral tissues (Coles et al., 2005). Stevens et al. (2006) developed a BAC library of quinoa using BamHI and EcoRI restriction enzymes consisting of 26,880 and 48,000 clones, respectively. Using the same BAC library, Balzotti et al. (2008) identified and characterized 11S globulin and 2S albumin seed storage proteins of quinoa, which they predicted were responsible for the relatively high protein content and ideal balance of amino acids in quinoa. Maughan et al. (2009) isolated and characterized the Salt Overly Sensitive 1 (SOS1) gene using a BAC library and reported that SOS1 is constitutively expressed in quinoa, unlike in other cereals in which mainly it is either stress-inducible or shows tissue-specific expression. Later, Walsh et al. (2015) carried out phylogenetic analysis in quinoa based on the sequence information of two introns of SOS1 and identified two distinct polyploid lineages.
The advances in next-generation sequencing technology and computational bioinformatics have accelerated genomics and transcriptomics research in quinoa. In the past few years, transcriptome studies have been carried out in quinoa to understand the molecular basis of drought and salinity tolerance. A drought-responsive transcriptome analysis carried out in two genotypes of quinoa using the Illumina HiSeq 2000 platform led to the identification of the genes involved in imparting drought tolerance to quinoa (Raney, 2012). Morales et al. (2017) performed transcriptome analysis in drought-tolerant Chilean quinoa genotype R49 and identified the drought-induced genes and pathways involved in providing drought stress tolerance in quinoa.
Using next-generation sequencing platforms, three research groups have independently completed quinoa genome sequencing (Yasui et al., 2016; Jarvis et al., 2017; Zou et al., 2017). Using two next-generation sequencing platforms, Illumina HiSeq 2500 (for short high-quality reads) and PacBio RSII (for longer reads and gap filling), Yasui et al. (2016) sequenced and assembled the draft genome of a quinoa inbred (Kd). The draft genome was found to be of 1.1 Gb size consisting of 24,847 scaffolds. The annotated genome of “Kd” was found to consist of 62,512 protein-coding genes around 535.5 Mb, leaving 49.2% of the genome as repetitive sequences. Further, a freely accessible Quinoa Genome DataBase (QGDB; http://quinoa.kazusa.or.jp/) was developed.
Zou et al. (2017) developed another draft genome sequence of an inbred line of quinoa (Real) using Illumina HiSeq 2500 and PacBio RSII platforms. The estimated genome size was 1.49 Gb covering 90.2% of the nuclear genome. Annotation of the draft genome sequence resulted in the identification of 54,438 protein-coding genes, of which 95.3% were functionally annotated. Approximately 65.5% of Real's genome consists of repeat sequences, 85.6% of which were found to be transposable elements comprising mostly retrotransposons. Further, to investigate protein quality, researchers analyzed comparative lysine, phenylalanine, and isoleucine content in three protein families [albumin, globulin, and late embryo abundant (LEA) proteins] and found that the lysine content of quinoa was significantly higher in all three protein families than in the other cereals such as wheat, rice, or maize. The high lysine content was not only at the free amino acid level but also for amino acid usage in seed protein sequences due to the presence of a high copy number of genes encoding the enzymes involved in converting aspartate into lysine. They also found that the high vitamin B and vitamin E content in quinoa is due to the presence of a high copy number of gene-encoding enzymes (pyridoxal 5′-phosphate synthase, dihydrofolate synthase, tetrahydrofolate synthase) involved in vitamin B6 and dihydrofolate biosynthesis (Table 8). Further, Zou et al. (2017) reported that the genes involved in ion sequestration, ABA homeostasis, and signaling are responsible for enhancing abiotic stress tolerance in quinoa. Based on transcriptome analysis, Zou et al. (2017) proposed a model for salt accumulation in salt bladders. Since few genes were found to be differentially expressed in epidermal salt bladders, this suggests that most of the transporter genes are constitutively active in salt sequestration in the bladders and that regulation of ion transport in bladder cells in response to salinity occurs at the protein level through protein phosphorylation (Zou et al., 2017).
Jarvis et al. (2017) published a more complete genome sequence of coastal Chilean quinoa accession PI 614886 (QQ74) using PacBio RSII and Illumina HiSeq 2500 sequencing platforms. The genome was assembled into 3,486 scaffolds spanning 1.39 Gb consisting of 44,776 genes and approximately 64% repetitive sequences. Along with quinoa accession PI 614886 (QQ74), the authors resequenced 15 other quinoa accessions along with five accessions of C. berlandieri and two of C. hircinum, which are supposed to be immediate tetraploid ancestors of quinoa. They further produced 18 pseudo-chromosomes of quinoa consisting of 1.18 Gb (i.e. ~80% of the predicted ~1.45 Gb haploid genome size of quinoa). Jarvis et al. (2017) identified two genes encoding basic Helix‐Loop‐Helix (bHLH) transcription factors involved in regulating the triterpenoid biosynthetic pathway associated with production in quinoa. They identified the triterpene saponin biosynthesis activating regulator like (TSARL1 and TSARL2) genes, of which TASRL2 was expressed in roots but not in flowers or immature seeds, whereas TSARL1 was over-expressed exclusively in seeds of bitter lines vis-à-vis sweet lines, suggesting that TSARL1 might be the functional TSAR ortholog involved in regulating biosynthesis of saponin in bitter quinoa genotypes (Table 8). Further, sequence analysis showed that alternative splicing on TSARL1 results in a premature stop codon, thereby translating a truncated protein with a compromised functional ability in forming homodimer to bind DNA for its activity.
Amaranth (Amaranthus sp.) is an ancient crop whose domestication and cultivation date back to around 8,000 years ago in Mayan civilization of South and Central America. There is no concrete evidence for the origin of Amaranthus. It was used as a staple food along with corn (maize) and beans in Mexico starting 1,400 years ago; however, its production declined after the collapse of Central American culture (Alvarez-Jubete et al., 2010). Amaranthus sp. is a highly nutritive pseudo-cereal, rich in proteins, vitamins, and minerals. Because of its nutraceutical value and climate resilience, amaranth has been relaunched and is being promoted as a suitable crop for food and nutritional security (Lakshmi and Vimala, 2000). Amaranth's leaves are consumed as vegetables and its grains as cereal. The amaranth family is divided into two sections: Amaranthus saucer and Blitopsis dumort (Allen, 1961). Based on its use, it has been grouped into grain amaranth, vegetable amaranth, and ornamental and weedy amaranth (Sauer, 1967). Grain amaranth has four species, A. hypochondriacus, A. cruentus, A caudatus, and A. edulis (Martínez-Cruz et al., 2014), whereas vegetable amaranth belongs to the section Blitopsis and has two major species, A. tricolor and A. lividis (Pal and Khoshoo, 1972; Madhusoodanan and Pal, 1981). As evident from recent past publications, amaranth has been attracting the attention of food technologists for exploring its functional aspects after the United States National Academy of Sciences showed its high nutritional value and agronomic potential (Monteros et al., 1994; Ulbricht et al., 2009).
Amaranth is a climate-resilient, fast-growing cereal-like plant. Despite its potential nutritional value, it has been underexploited. Protein content in the grain of amaranth species ranges from 13.1% to 21.0% with an average of 15%, which is comparatively higher than that of other cereals (Table 1) (Mlakar et al., 2009). Its amino acid profile makes it an attractive protein source as it contains significantly higher lysine content (4.9‒6.1%), which is an essential amino acid limiting in most cereals (Table 3). Amaranth protein is also richer in sulfur-containing amino acids (≈4.4%), which are limiting in pulse crops (Bressani, 1989). It is considered that if it is consumed along with other cereals, it will provide a “balanced” source of protein (Saunders and Becker, 1984). The balanced amino acid content in amaranth grain protein is close to the optimum protein reference pattern in the human diet as recommended by FAO/WHO (O' Brien and Price, 2008). The balanced nature of amino acid composition in amaranth grain protein is due to the presence of ≈65% of the protein in the embryo and only ≈35% in the perisperm, whereas in other grains ≈85% of the protein is present in endosperm, which is poorer in essential amino acids (Senft, 1979; Betschart et al., 1981). Mainly three amino acids (leucine, isoleucine, and valine) are limiting amino acid in amaranth grain but are present in excess in other grains. Amaranth flour has high glycemic index, therefore it is suitable for blending with other cereals (Table 1). Amaranth and wheat flour in 25:75 ratio bring down the GI quite low with good nutritional balance. Similarly amaranth and maize flour in a ratio of 1:1 nearly reaches the perfect score of 100 on the nutritionist's scale and also the combination of amaranth in wheat flour improves the nutritional value of baked products (Saunders and Becker, 1984; Bressani, 1989). Apart from the balanced profile of amino acid content, amaranth protein has high digestibility (≈90%), which further improves the bioavailability of amino acids when ingested. Apart from having good amino acid composition, amaranth protein is gluten-free, making it a choice of food for patients with coeliac disease incidence.
Besides protein, amaranth grain has higher oil content (5‒10%) than other cereals (Table 1). Amaranth oil contains 76‒77% unsaturated fatty acids consisting of primarily linoleic acid (25‒62%), oleic acid (19‒35%), palmitic acid (12‒25%), stearic acid (2.0‒8.6%), and linolenic acid (0.3‒2.2%). The ratio of saturated to unsaturated fatty acid ranged from 0.26 to 0.31 in oil of amaranth grain. Amaranth lipid is unique due to the presence of high biologically active compounds such as squalene (2‒7%), tocopherols (≈2%), phospholipids (up to 10%), and phytosterols (up to 2%) (Becker, 1994; León-Camacho et al., 2001; Berghofer and Schoenlechner, 2002). Squalene is an unsaturated hydrocarbon and is known as an obligatory precursor of sterols and has been reported to have antibacterial properties and anti-oxidative and anti-tumor effects in carcinogenesis.
Amaranth grain contains ≈60‒65% of carbohydrate, of which starch constitutes ≈57% and total dietary fiber constitutes ≈8‒16% (Table 1). Amaranth starch mostly consists of amylopectins (≈89.0‒99.9%) and is therefore known as “waxy starch” with unique characteristics of high viscosity and high gelatinization temperature. Despite high amylopectin content, the starch granules of amaranth are smaller (0.8‒2.5 μm) than those of other grains (3‒34 μm), providing them with high water-binding capacity, higher swelling power, lower gelatinization temperature, and high resistance to amylases, making amaranth a preferential source of starch in the food industry (Williams and Brenner, 1995; Pal et al., 2002) as well as providing high solubility and digestibility. The dietary fiber content in pale-colored amaranth seeds was 8%, whereas it was 16% in black-colored amaranth grains (Mlakar et al., 2009). In the leaves of vegetable amaranth, fiber content ranged from 6.95% to 9.65%, with an average of 8.39% ± 0.1% (Shukla et al., 2006).
Amaranth grains are rich sources of the minerals iron (72‒174 mg/kg), calcium (1,300‒2,850 mg/kg), phosphorus (455 mg/kg), sodium (160‒480 mg/kg), magnesium (2,300‒3,360 mg/kg), and zinc (36.2‒40.0 mg/kg) as well as the vitamin riboflavin (0.19‒0.23 mg/100 g of flour), ascorbic acid (4.5 mg/100 g), niacin (1.17‒1.45 mg/100 g), and thiamine (0.07‒0.10 mg/100 g) (Becker et al., 1981; Rastogi and Shukla, 2013) (Tables 2 and 4). Apart from the seeds, the leaves of vegetable amaranth (A. tricolor) are rich in minerals and have an average content of potassium of 3.7 ± 0.26 g/kg, calcium 1.7 ± 0.04 g/kg, magnesium 2.90 ± 0.01 g/kg, zinc 791.7 ± 28.98 mg/kg, iron 1,233.8 ± 50.02 mg/kg, manganese 108.1 ± 3.82 mg/kg, and nickel 222.6 ± 9.51 mg/kg (Shukla et al., 2006). Amaranth has all the necessary daily required vitamins up to a significant level and is an excellent source for reducing vitamin deficiency (Graebner et al., 2004). The leaves of A. tricolor contain 0.83 ± 0.02 mg/kg vitamin A (carotenoids) and 112.3 ± 5.0 mg/kg ascorbic acid (vitamin C) (Shukla et al., 2006). Despite these nutritional benefits, amaranth grain contains some antinutritional factors that can limit its food application. Amaranth grain contains growth inhibitors such as phytic acid (0.3‒0.6%), saponins (0.09–0.10%), and tannins, etc.
In the recent past, amaranth has become a crop of interest for its high nutritive value and great potential as a functional food given its cholesterol-lowering effect observed in animal models (Plate and Arêas, 2002; Mendonça et al., 2009). Despite the 75% in vitro digestibility of amaranth protein, net protein use ranges from 33.5% to 46% (Aguilar et al., 2015). Hejazi et al. (2016) showed that, after germination treatment at 28°C for 48 h, the nutritional quality and digestibility of amaranth resulted in not only improved protein digestibility (84%) but also decreased phytic acid and oxalate. Gamel et al. (2006) and Ferreira and Arêas (2010) demonstrated that amaranth extrusion increased calcium bioavailability in rats and suggested that amaranth can be a complementary source of dietary calcium once its bioavailability is favorably modified by the extrusion process (Tables 5 and 6). Whittaker and Ologunde (1990) demonstrated that iron supplementation through amaranth increases hemoglobin content because of the more absorbable form of iron in amaranth. Subramanian and Gupta (2016) reported that administration of amaranth extract through an oral dose increases the level of NO3‒ and NO2‒ in plasma as well as in saliva. The bio-accessibility of phenolic compounds was accessed in five wild (Amaranthus hybridus, Brachiaria brizantha, Panicum maximum, Rottboellia cochinchinensis, and Sorghum arundinaceum) and two domesticated cereal (Eleusine corocana and a red variety of Sorghum bicolor) grains found in Zimbabwe and showed that Amaranthus hybridus had the highest intestinal bio-accessibility (95.4 ± 0.01%) vis-à-vis the other cereals tested (Chitindingu et al., 2015). Serna-Saldivar et al. (2015) recommended the use of A. caudatus grain flour in maize tortillas to improve the bioavailability of nutrients and to reduce diabetes as well as an anthelmintic. Apart from these, no published studies have directly compared the relative bioavailability of other nutritional components of amaranth.
Amaranth is a C4 pseudo‐cereal and can be cultivated in a wide range of environments since it has good tolerance of drought and salinity (Kong et al., 2009; Teng et al., 2016).
The species of Amaranthus are difficult to characterize taxonomically because of the similarity existing among many of the species, small diagnostic parts, intermediate forms, and the broad geographic distribution (Mujica and Jacobsen, 2003). The taxonomic and phylogenetic position of the genus was investigated using various phenotypic and genotypic evaluations (Sauer, 1955; Stetter and Schmid, 2017). The most recent taxonomic classification divided it into three subgenera: A. albersia, A. acnida, and A. amaranthus (Mosyakin and Robertson, 1996; Costea and DeMason, 2001). Popa et al. (2010) used nuclear ribosomal DNA (rDNA) Internal Transcribed Spacer (ITS) regions for analyzing 92 accessions of Amaranthus and identified 12 out of 92 as weed. Xu and Sun (2001) reported low ITS divergence and poorly resolved phylogeny among the closely related taxa A. cruentus, A. caudatus, and A. hypochondriacus, and their putative wild progenitors A. hybridus, A. quitensis, and A. powellii. Chan and Sun (1997) used RAPD markers and reported that grain amaranths originated from a common ancestor, A. hybridus. Studies using isozyme markers have demonstrated a high degree of polymorphism between the populations, through which it may be possible to identify the intermediate stages of domestication.
Molecular markers are the essential tools in modern plant breeding research programs. The first attempt to develop and characterize the molecular markers for amaranth was carried out by Mallory et al. (2008), who sequenced 1,457 clones from microsatellite-enriched libraries and identified 353, out of which 179 were found to be polymorphic across the accessions of three grain amaranths. Maughan et al. (2009) applied a genomic reduction strategy with next-generation sequencing and identified 27,658 SNPs among four diverse amaranth accessions. Amaranthus hypochondriacus showed the maximum genetic diversity in terms of the number of polymorphic SNPs, whereas A. cruentus showed the lowest genetic diversity with 35 polymorphic SNPs only (Maughan et al., 2009). The reduced genetic diversity in A. cruentus is consistent with other studies carried out using different marker systems: SSRs, RFLPs, isozymes, and AFLPs (Chan and Sun, 1997; Xu and Sun, 2001; Mallory et al., 2008). Suresh et al. (2014) studied genetic diversity and population structure in 348 amaranth accessions belonging to 33 species using 11 SSR markers. The accessions were grouped into seven different clusters independent of species or geographic origin. The overall PIC value ranged from 0.436 to 0.898, with an average of 0.657, with observed heterozygosity from 0.056 to 0.876. The variation in 29 grain amaranth accessions was studied using 27 phenotypic and 16 RAPD markers, resulting in grouping of these accessions into five clusters at 87.5% similarity coefficient (Akin-Idowu et al., 2016). The molecular phylogeny of 94 accessions representing 35 Amaranthus species evaluated using SNP markers through genotyping by sequencing (GBS) showed that most accessions from the same species clustered together and they were classified into three subgenera with a few highly differentiated groups (Stetter and Schmid, 2017). It was also hypothesized that A. hybridus might be the ancestor of all three crop amaranth species and A. quitensis might be an intermediate between A. hybridus and A. caudatus.
Despite the great potentiality of amaranth as a source of nutritional food, few efforts have been made to explore its genomics. Stress-responsive transcriptome analysis of A. hypochondriacus was reported to understand the mechanism and identify the genes involved in adapting the species to survive against environmental stresses (Délano-Frier et al., 2011). The first attempt to sequence the draft genome of A. hypochondriacus used Illumina Genome Analyzer IIx with >100× coverage of estimated genome size of 466 Mb (Sunil et al., 2014). The annotation of A. hypochondriacus resulted in the identification of 24,829 protein-coding genes and 13.76% of the genome was found to consist of repeat elements. They further hypothesized that high lysine content in Amaranthus is due to the presence of only one ortholog of aspartate kinase 1 gene and high expression of the dihydrodipicolinate synthase (DHDPS) gene in seeds. Recently, a high-quality whole-genome sequencing of agronomically important amaranth cultivar “Plainsman” (A. hypochondriacus) was carried out using the Illumina HiSeq platform (Clouse et al., 2016). The assembled genome consisted of 377 Mb in 3,518 scaffolds covering 80.9% of the estimated genome size. Further, the genome consisted of 48% of the repeat sequences, of which Copia-like retrotransposons were predominantly present. Annotation of the genome led to the identification of 23,059 protein-coding genes. Clouse et al. (2016) further resequenced seven accessions of grain amaranths (A. hypochondriacus, A. caudatus, and A. cruentus) along with A. hybridus. SNP-based phylogenetic analysis confirmed that A. hybridus is the progenitor species of grain amaranths. Further, the researchers generated a physical map spanning 340 Mb of the whole genome of A. hypochondriacus. In addition, Lightfoot et al. (2017) performed single-molecule, real-time sequencing using the PacBio RSII system to close assembly gaps and used chromatin interaction mapping (Hi-C) to scaffold contigs and thereby improved their own previously reported Illumina-based assembly to a chromosome-scale assembly. The 16 largest scaffolds contain 98.0% of the assembly and represent the haploid chromosome number (n = 16). These researchers further produced physical and genetic maps and identified the betalain locus consisting of CYP76AD1 (cytochrome P450) and DODA1 (4,5-DOPA dioxygenase extradiol 1) involved in betalain biosynthesis, which controls stem color (Table 8).
Buckwheat (F. esculentum Monch) is a versatile dicotyledon annual crop and has been grown for centuries for its grains as well as greens to be used as food, feed, vegetable, and fodder, although it was neglected during the 20th century in western countries because of the increased yield of wheat during the Green Revolution (Cawoy et al., 2008). Nonetheless, it is recognized as a potential functional food source in China, Japan, and Taiwan. It is considered as a pseudo-cereal because of its usage as a conventional cereal and its chemical composition (Campbell, 1997). There are several species of buckwheat, out of which only nine species have agricultural and nutritional importance, among which only two are used for food purposes: common buckwheat (F. esculentum) and tartary buckwheat (F. tataricum). Common buckwheat is also known as “sweet buckwheat,” which is taller, has a thicker stem, and is a widely cultivated species in Asia, Europe, North America, and South Africa, whereas tartary buckwheat is mainly confined to the highlands of southwest China and the Himalayas (Ohnishi, 1993; Zhou et al., 2016).
In the recent past, buckwheat production has increased because of its nutraceutical properties and potential for use in the preparation of functional foods (Li and Zhang, 2001; Bonafaccia et al., 2003a; Bonafaccia et al., 2003b). Buckwheat is rich in nutrients and its seed contains 100‒125 mg/g of protein, 650‒750 mg/g of starch, 20‒25 mg/g of fat, and 20‒25 mg/g of mineral (Li and Zhang, 2001).
The protein content in buckwheat flour is higher than in commonly used cereals such as rice, wheat, millet, sorghum, maize, etc. Buckwheat protein primarily contains albumin (180 mg/g), globulin (430 mg/g), prolamin (8 mg/g), and glutelin (230 mg/g) (Javornik and Kreft, 1984; Ikeda et al., 1991; Ikeda and Asami, 2000). The amino acid profile of buckwheat protein is balanced compared with that of other cereals. Buckwheat protein is one of the richest in lysine (EAA) and arginine, which are generally limiting in other cereals (Table 3). The amino acid score for buckwheat protein is 100, which is highest among plant sources (Ikeda, 2002). Lys/Arg and Met/Gly ratios determine cholesterol-lowering effects and are lower in buckwheat than in most plants, suggesting that buckwheat protein should have cholesterol-lowering effects (Huff and Carroll, 1980; Sugiyama et al., 1985; Carroll and Kurowska, 1995). Consumption of food products derived from buckwheat could reduce the concentration of cholesterol in blood serum and glycemic and insulin indices (Skrabanja et al., 2001; Stokić et al., 2015). Buckwheat protein contains either no or very low gluten and is thus considered as gluten-free and is recommended to people suffering from coeliac disease. Despite these benefits, the major problem is the low digestibility of buckwheat protein in humans and animals due to the presence of anti-nutritional factors such as protease inhibitors (trypsin inhibitor) and tannins (Ikeda et al., 1986; Ikeda et al., 1991; Campbell, 1997). However, sprouting/germination of buckwheat seed reduces the protease inhibitors, thereby increasing protein digestibility (Kreft, 1983; Ookubo, 1992) (Table 6).
Starch is the major carbohydrate in buckwheat grain and it varies from 59% to 70% of the dry mass depending on climatic and cultivation conditions (Qian and Kuhn, 1999). Buckwheat seed starch contains 24% amylose and 76% amylopectin (Praznik et al., 1999) but it's digestibility is much slower (low GI) than other cereals like wheat, therefore good to be consumed by diabetic patients. Buckwheat flour is commonly used during fasting days in India.
The total dietary fiber in seeds varies from 5% to 11% (Izydorczyk et al., 2004). Buckwheat is the richest source of soluble carbohydrates such as fagopyrin and fagopyritols, which have been studied for their use in treating type II diabetes (Kawa et al., 2003). Buckwheat grains also contain from 1.5% to 4.0% lipids (Steadman et al., 2001a). Buckwheat is rich in minerals such as potassium, magnesium, and phosphorus (Table 2). Apart from this, buckwheat contains higher contents of zinc, copper, and manganese than other cereals (Ikeda et al., 1999; Steadman et al., 2001b). The bioavailability of zinc, copper, and potassium from buckwheat is high and 100 g of buckwheat flour can provide 13–89% of the recommended dietary allowance (RDA) for Zn, Cu, Mg, and Mn (Bonafaccia et al., 2003b). Buckwheat grains contain higher contents of vitamin B1 (thiamine), B2 (riboflavin), B3 (niacin and niacinamide), K (phylloquinones), and B6 and B6 (folates) than other cereals (Table 4). Tartary buckwheat contains higher vitamin B1, B2, and B3, whereas common buckwheat contains higher vitamin E (Ikeda et al., 2006). The contents of vitamin C, B1, and B6 can be further increased by germinating buckwheat. It has been reported that the content of vitamin C can be increased by up to 0.25 mg/g in buckwheat sprouts (Lintschinger et al., 1997; Kim et al., 2004). Buckwheat also contains trace elements such as selenium (0.0099–0.1208 mg/g), which provides resistance against cancer and AIDS (Shi et al., 2011). Besides being rich in high-quality protein and minerals, buckwheat is rich in many rare components such as flavones, flavonoids, phytosterols, fagopyrins, and thiamin-binding proteins, which are known to have healing effects against some chronic diseases. The presence of flavonoids such as rutin, quercetin, orientin, homoorientin, vitexin, and isovitexin in its leaves, flowers, seeds, and sprouts imparts buckwheat with nutraceutical properties (Zielińska et al., 2012; Raina and Gupta, 2015). Tartary buckwheat has higher flavonoid content (19.54 mg/g) than common buckwheat (0.28 mg/g) (Jiang et al., 2007). Buckwheat is the only grain crop with rutin content that is known to have antioxidant, anti-inflammatory, and anti-carcinogenic property and it reduces the fragility of blood vessels related to hemorrhagic diseases and hypertension in humans (Oomah and Mazza, 1996; Baumgertel et al., 2003). Whole buckwheat contains 2‒5 times more phenolic compounds, whereas buckwheat hull and bran contain 2‒7 times more antioxidant than oat and barley (Holasova et al., 2002; Zduńczyk et al., 2006). Because of its good nutritional profile and presence of various nutraceutical compounds with unique medicinal properties and adaptability toward harsh climatic conditions, buckwheat is a suitable alternative for food and nutritional security for marginal environments.
Buckwheat (Fagopyrum sp.), with its origin in China, is a pseudo-cereal belonging to family Polygonaceae. The genus has 19 species, of which F. esculentum (sweet buckwheat) and F. tataricum (bitter buckwheat) are the two species in the buckwheat genepool being cultivated predominantly in most parts of the world (Krkošková and Mrazova, 2005). Most of the species in the genus are diploid (2n = 2x = 16), except for F. cymosum and F. gracilipes being tetraploid (2n = 4x = 32) (Chrungoo et al., 2012; Farooq et al., 2016), with an estimated genome size of 540 Mb (Nagano et al., 2000) (Table 7). Despite having so many species, the genetic diversity in buckwheat has become depleted during the past few decades due to changing cropping patterns and food habits. Approximately 5,000 accessions of buckwheat have been collected in Southeast Asia, representing about 52% of the world's collection. China has the largest collection of buckwheat (2800), followed by Russia (2116), Ukraine (1600), and India (1050) (Zhou and Zhang, 1995; IPGRI-APO, 1999; Zhang et al., 2004; Zhou et al., 2018). The number of germplasm collections might be overrepresented due to duplications because of the exchange of germplasm between organizations within and between countries. It is an underused crop but holds tremendous potential due to its short life cycle (≈60 days), ability to grow and survive at higher altitude, and high-quality protein content.
A significant amount of research has been carried out to understand the properties of buckwheat proteins, flavonoids, flavones, phytosterols, thiamin-binding proteins, and other rare compounds (Tomotake et al., 2002; Kreft et al., 2006; Zielinski et al., 2009). However, little effort has been made in the development of molecular markers and genomic resources for buckwheat. RAPD has been used to study the relationship and estimate the genetic diversity between different accessions of Fagopyrum species (Sharma and Jana, 2002a; Sharma and Jana, 2002b) as well as to identify molecular markers linked with the homostylar (Ho) gene associated with self-compatibility (Aii et al., 1998). Nagano et al. (2001) used AFLP markers to identify markers tightly linked to the homostylar region and designed two primers to develop locus-specific markers for further use in marker-assisted breeding. Using the AFLP marker system, five markers linked to shattering habit in buckwheat were identified and a linkage map around the sht1 locus was constructed (Matsui et al., 2004). An interspecific linkage map was developed using F. esculentum and F. homotropicum covering 548.8 cM on eight linkage groups. Microsatellite markers showed comparatively higher polymorphism and expected heterozygosity than AFLP, RAPD, and RFLP markers (Powell et al., 1996). Konishi and Ohnishi (2006) developed 180 SSR markers by sequencing 2,785 clones, out of which 54 were found to be polymorphic. Ma et al. (2009) developed 136 new SSR markers for F. esculentum and used them for diversity analysis in related species of the genus Fagopyrum. However, out of the 136 SSRs, only 10 were found to be polymorphic on 41 accessions. Kishore et al. (2012) assessed genetic diversity and phylogenetic relationship in 75 accessions using 15 SSR markers and the estimated average gene diversity was 0.2098. Li et al. (2007) attempted to develop microsatellite markers for tartary buckwheat by constructing a genomic library enriched with (gT)n repeats. Apart from these markers, a couple of BAC libraries were constructed for F. homotropicum (Nagano et al., 2001) and F. esculentum (Yasui et al., 2008), which can be further used for identifying useful genes as well as developing markers. More efforts have been made to carry out research in molecular genetics and plant breeding in common buckwheat, but only fragmentary research efforts have been made in tartary buckwheat. Genetic maps have been developed for both F. esculentum (Yasui et al., 2004; Pan and Chen, 2010) and F. tataricum (Du et al., 2013) and QTLs governing photosensitivity (Hara et al., 2011) and stem length (Yabe et al., 2014) have been identified. However, these maps remain underused for mapping genes/QTLs due to the lack of enough informative markers distributed across all linkage groups. The whole genome de novo sequencing of “HeiFeng No. 1,” an elite tartary buckwheat cultivar, using Illumina HiSeq 2000 resulted in 204,340 contigs and 348 Mb of assembled sequences. Further, the sequence survey resulted in the identification of 24,505 SSR motifs. The researchers further carried out SSR fingerprinting of 64 accessions and predicted the population structure of tartary buckwheat (Hou et al., 2016).
Transcriptome analysis for floral structure (Logacheva et al., 2011), aluminum toxicity (Chen et al., 2017; Xu et al., 2017) and salt tolerance (Wu et al., 2017) has been carried out to understand the gene regulatory mechanism in buckwheat. Transcriptome analysis at 12 different developmental stages of seed development from fertilization to maturation led to the identification of 11,676 differentially expressed genes in tartary buckwheat (Huang et al., 2017). The candidate genes identified through various transcriptomic studies provide a rich genomic resource for further functional characterization for various traits in buckwheat through a reverse genetics approach.
The whole-genome sequencing of buckwheat (F. esculentum Moench) using Illumina HiSeq 2000 resulted in assembly of the genome in 387,594 scaffolds consisting of 1.17 Gb of data. Gene prediction analysis identified 35,816 annotated genes and developed the Buckwheat Genome DataBase (BGDB; http://buckwheat.kazusa.or.jp) (Yasui et al., 2016). Researchers further showed the applicability of genome sequence information in identifying genes involved in flavonoid, 2S albumin-type allergens biosynthesis, and granule bound starch synthases (GBSSs), and in controlling heteromorphic self-incompatibility. Zhang, L., et al (2017) sequenced the whole genome of tartary buckwheat (F. tataricum cv. Pinku1) using multiple Illumina NGS platforms and SMRT sequencing technology. A total 8,778 contigs assembled into 489.3 Mb (N50 = 550.7 kb), with maximum contig length of 6.64 Mb. The annotation of the genome resulted in the identification of 33,366 protein-coding genes. Further, the reference genome helped in identifying predicted genes involved in aluminum stress tolerance and abiotic stress responses as well as rutin biosynthesis and regulation. Rutin is a flavonoid with antioxidant property known for its ability to strengthen blood vessels, aiding in the usage of vitamin C and production of collagen. Identification of the genes involved in rutin biosynthesis will further help in increasing the rutin content in buckwheat by using modern genetic and genomics tools.
Improving the climate resilience of crops is crucial to the future food and nutritional security of marginal areas. Climate change is no longer a mirage but a reality that is evident from rising global temperature and more frequent drought, hot, cold, and rain spells worldwide. Every day, a large chunk of normal areas is being transformed into marginal lands, thus significantly decreasing productivity and endangering food security. More than 2 billion people that depend on major staple crops such as wheat, maize, and rice are suffering from “hidden hunger,” which has resulted in either malnutrition, due to deficiency of minerals, vitamins, and essential amino acids, or obesity, due to the surfeit of energy-rich carbohydrates. All of these major cereals are unable to tolerate climatic aberrations and marginality because of significant abiotic stresses. The need of the hour is to identify the crops and their varieties that can offer robust resistance against the harsh conditions of marginal environments and sustain food and nutritional security. Indeed, the targets for both globally healthy diets and sustained food production have to be met for 10 billion people on this planet by 2050. Several underused “neglected crops” are considered as “minor crops” and have received less importance globally in terms of both production and research. The five crops (finger millet, quinoa, foxtail millet, buckwheat, and amaranth) reviewed in this manuscript are nutrient-dense and are rich sources of macro- and micro-elements (e.g. protein with balanced amino acids, essential amino acids, vitamins, minerals, etc.). Small amounts of the minor millets and pseudo-cereals in the daily diet of people can ensure “no malnutrition.” Apart from being nutritionally rich, these underused crops are climate-resilient and suitable for cultivation in marginal environments. These neglected crops have huge potential for food and nutritional security through sustainable agriculture in marginal areas. This will help farmers to maintain productivity against a backdrop of rising temperatures, higher salinity, and increasing water scarcity and provide a practical and sustainable way of adapting to climate change. Unfortunately, the depth of scientific research on the yield and quality improvement of these minor millets and pseudo-cereals is extremely inadequate vis-à-vis that on the major staples; hence, international funding is extremely important to support research programs on genetic improvement for yield and nutritional traits for these crops.
JR and ST contributed on bioavailability part while HR contributed for genomics part. RS conceptualized, organized and finally polished the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved for publication.
The work was supported by the International Center for Biosaline Agriculture (ICBA) Dubai, UAE.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors acknowledge support from Dr. Ismahane Elouafi, Director General, and Ms. Seta Tutundjian, Director Programs, ICBA. We thank the reviewers for helpful comments and suggestions.
Aguilar, E. G., de Jesús Albarracín, G., Uñates, M. A., Piola, H. D., Camiña, J. M., Escudero, N. L. (2015). Evaluation of the nutritional quality of the grain protein of new amaranths varieties. Plant Foods Hum. Nutr. 70, 21–26. doi: 10.1007/s11130-014-0456-3
Aheme, S. A., O'Brien, N. M. (1999). Protection by the flavonoids myricetin, quercetin, and rutin against hydrogen peroxide-induced DNA damage in Caco-2 HepG2 cells. Nutr. Cancer 34, 160–166. doi: 10.1207/S15327914NC3402_6
Aii, J., Nagano, M., Penner, A. G., Campbell, G. C., Adachi, T. (1998). Identification of RAPD markers linked to the homostylar (Ho) gene in buckwheat. Japanese J. Breed. 48, 59–62. doi: 10.1270/jsbbs1951.48.59
Akin-Idowu, P., Gbadegesin, M., Orkpeh, U., Ibitoye, D., Odunola, O. (2016). Characterization of grain amaranth (Amaranthus spp.) germplasm in south west Nigeria using morphological, nutritional, and random amplified polymorphic DNA (RAPD) analysis. Resources 5, 6. doi: 10.3390/resources5010006
Albugoch, J., Lilian, E. (2009). “Chapter 1: Quinoa (Chenopodium quinoa Willd.): Composition, Chemistry, Nutritional, and Functional Properties,” in Advances in Food and Nutrition Research, vol. 58. (Academic Press), 1–31. doi: 10.1016/S1043-4526(09)58001-1
Alvarez-Jubete, L., Auty, M., Arendt, E. K., Gallagher, E. (2010). Baking properties and microstructure of pseudocereal flours in gluten-free bread formulations. Eur. Food Res. Technol. 230, 437. doi: 10.1007/s00217-009-1184-z
Amarowicz, R., Dykes, G. A., Pegg, R. B. (2008). Antibacterial activity of tannin constituents from Phaseolus vulgaris, Fagoypyrum esculentum, Corylus avellana and Juglans nigra. Fitoterapia 79 (3), 217–219. doi: 10.1016/j.fitote.2007.11.019
Arêas, J. A. G., Carlos-Menezes, A. C. C. C., Soares, R. A. M., Caballero, B., Finglas, P. M., Toldrá, F. (2016). Encyclopedia of Food and Health Vol. Amaranth (Oxford: Academic Press), 135–140. doi: 10.1016/B978-0-12-384947-2.00025-8
Arya, L., Verma, M., Gupta, V. K., Karihaloo, J. L. (2009). Development of EST SSRs in finger millet (Eleusine coracana ssp. coracana) and their transferability to pearl millet (Pennisetum glaucum). J. Plant Biochem. Biotechnol. 18, 97–100. doi: 10.1007/BF03263303
Arya, L., Verma, M., Gupta, V. K., Seetharam, A. (2013). Use of genomic and genic SSR markers for assessing genetic diversity and population structure in Indian and African finger millet (Eleusine coracana (L.) Gaertn.) germplasm. Plant Syst. Evol. 299, 1395–1401. doi: 10.1007/s00606-013-0822-x
Austin, D. F. (2006). Fox-tail millets (Setaria: Poaceae)—abandoned food in two hemispheres. Economic Bot. 60, 143–158. doi: 10.1663/0013-0001(2006)60[143:FMSPFI]2.0.CO;2
Babu, B. K., Agrawal, P. K., Pandey, D., Jaiswal, J. P., Kumar, A. (2014a). Association mapping of agro-morphological characters among the global collection of finger millet genotypes using genomic SSR markers. Mol. Biol. Rep. 41, 5287–5297. doi: 10.1007/s11033-014-3400-6
Babu, B. K., Agrawal, P. K., Pandey, D., Kumar, A. (2014b). Comparative genomics and association mapping approaches for opaque2 modifier genes in finger millet accessions using genic, genomic and candidate gene-based simple sequence repeat markers. Mol. Breed. 34, 1261–1279. doi: 10.1007/s11032-014-0115-2
Babu, B. K., Dinesh, P., Agrawal, P. K., Sood, S., Chandrashekara, C., Bhatt, J. C., et al. (2014c). Comparative genomics and association mapping approaches for blast resistant genes in finger millet using SSRs. PloS One 9, e99182. doi: 10.1371/journal.pone.0099182
Bai, H., Cao, Y., Quan, J., Dong, L., Li, Z., Zhu, Y., et al. (2013). Identifying the genome-wide sequence variations and developing new molecular markers for genetics research by re-sequencing a landrace cultivar of foxtail millet. PloS One 8, e73514. doi: 10.1371/journal.pone.0073514
Baljeet, S. Y., Ritika, B. Y., Roshan., L. Y. (2010). Studies on functional properties and incorporation of buckwheat flour for biscuit making. Int. Food Res. J. 17, 1067–1076.
Balzotti, M. R., Thornton, J. N., Maughan, P. J., McClellan, D. A., Stevens, M. R., Jellen, E. N., et al. (2008). Expression and evolutionary relationships of the Chenopodium quinoa 11S seed storage protein gene. Int. J. Plant Sci. 169, 281–291. doi: 10.1086/523874
Bandyopadhyay, T., Mehanathan, M., Prasad, M. (2017). Millets for next generation climate-smart agriculture. Front. Plant Sci. 8, 1266. doi: 10.3389/fpls.2017.01266
Baumgertel, A., Grimm, R., Eisenbeiß, W., Kreis, W. (2003). Purification and characterization of a flavonol 3-O-β-heterodisaccharidase from the dried herb of Fagopyrum esculentum Moench. Phytochemistry 64, 411–418. doi: 10.1016/S0031-9422(03)00418-7
Bazile, D., Jacobsen, S. E., Verniau, A. (2016). The global expansion of quinoa: trends and limits. Front. In Plant Sci. 7, 622. doi: 10.3389/fpls.2016.00622
Becker, R., Wheeler, E. L., Lorenz, K., Stafford, A. E., Grosjean, O. K., Betschart, A. A., et al. (1981). A compositional study of amaranth grain. J. Food Sci. 46, 1175–1180. doi: 10.1111/j.1365-2621.1981.tb03018.x
Becker, R. (1994). “Amaranth oil: composition, processing, and nutritional qualities,” in Amaranth biology, chemistry and technology. Ed. Paredes, L. O. (México: Instituto Politécnico Nacional), 133–139.
Belton, P. S., Taylor, J. R. (2004). Sorghum and millets: protein sources for Africa. Trends In Food Sci. Technol. 15, 94–98. doi: 10.1016/j.tifs.2003.09.002
Bennett, M. D., Smith, J. B. (1991). Nuclear DNA amounts in angiosperms. Philos. Trans.: Biol. Sci., 309–345. doi: 10.1098/rstb.1991.0120
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555. doi: 10.1038/nbt.2196
Berghofer, E., Schoenlechner, R. (2002). “Grain Amaranth,” in Pseudocereals and less common cereals, grain properties and utilization potential. Eds. Belton, P., Taylor, J. (Springer-Verlag), 219‒260.
Betschart, A. A., Irving, D. W., Shepherd, A. D., Saunders, R. M. (1981). Amaranthus cruentus: milling characteristics, distribution of nutrients within seed components, and the effects of temperature on nutritional quality. J. Food Sci. 46, 1181–1187. doi: 10.1111/j.1365-2621.1981.tb03019.x
Bharathi, A. (2011). Phenotypic and Genotypic Diversity of Global Finger Millet (Eleusine coracana (L.) Gaertn.) Composite Collection. (dissertation/Ph.D. thesis) (Tamil Nadu Agricultural University).
Bhargava, A., Shukla, S., Ohri, D. (2003). Genetic variability and heritability of selected traits during different cuttings of vegetable Chenopodium. Indian J. Genet. Plant Breed. 63 (4), 359‒360.
Bhargava, A., Shukla, S., Ohri, D.(2006a). Chenopodium quinoa—an Indian perspective. Ind. Crops Prod. 23, 73–87. doi: 10.1016/j.indcrop.2005.04.002
Bhargava, A., Shukla, S., Ohri, D. (2006b). Karyotypic studies on some cultivated and wild species of Chenopodium (Chenopodiaceae). Genet. Resour. Crop Evol. 53, 1309–1320. doi: 10.1007/s10722-005-3879-8
Bhargava, A., Shukla, S., Ohri, D. (2007a). Genome size variation in some cultivated and wild species of Chenopodium (Chenopodiaceae). Caryologia 60, 245–250. doi: 10.1080/00087114.2007.10797943
Bhargava, A., Shukla, S., Rajan, S., Ohri, D. (2007b). Genetic diversity for morphological and quality traits in quinoa (Chenopodium quinoa Willd.) germplasm. Genet. Resour. Crop Evol. 54, 167–173. doi: 10.1007/s10722-005-3011-0
Bindra, G. S., Gibson, R. S., Thompson, L. U. (1986). [Phytate][calcium]/[zinc] ratios in Asian immigrant lacto-ovo vegetarian diets and their relationship to zinc nutriture. Nutr. Res. 6, 475–483. doi: 10.1016/S0271-5317(86)80101-4
Bisht, M. S., Mukai, Y. (2001). Genomic in situ hybridization identifies genome donor of finger millet (Eleusine coracana). Theor. Appl. Genet. 102, 825–832. doi: 10.1007/s001220000497
Bonafaccia, G., Gambelli, L., Fabjan, N., Kreft, I. (2003a). Trace elements in flour and bran from common and tartary buckwheat. Food Chem. 83, 1–5. doi: 10.1016/S0308-8146(03)00228-0
Bonafaccia, G., Marocchini, M., Kreft, I. (2003b). Composition and technological properties of the flour and bran from common and tartary buckwheat. Food Chem. 80, 9–15. doi: 10.1016/S0308-8146(02)00228-5
Borges, J. T. S., Bonomo, R. C., Paula, C. D., Oliveira, L. C., Cesário, M. C. (2010). Physicochemical and nutritional characteristics and uses of Quinoa (Chenopodium quinoa Willd.). Temas Agrários 15 (1), 9–23.
Brady, K., Ho, C.-T., Rosen, R. T., Sang, S., Karwe, M. V. (2007). Effects of processing on the nutraceutical profile of quinoa. Food Chem. 100 (3), 1209–1216. doi: 10.1016/j.foodchem.2005.12.001
Bressani, R. (1989). The proteins of grain amaranth. Food Rev. Int. 5, 13–38. doi: 10.1080/87559128909540843
Brown, D. C., Cepeda-Cornejo, V., Maughan, P. J., Jellen, E. N. (2015). Characterization of the granule-bound starch synthase I gene in Chenopodium. Plant Genome 8, 1–12. doi: 10.3835/plantgenome2014.09.0051
Campbell, C. G. (1997). Buckwheat Fagopyrum esculentum Moench. promoting the conservation and use of underutilized and neglected crops 19 (Rome, Italy: Institute of Plant Genetics and Crop Plant Research; Gatersleben/International Plant Genetic Resources Institute).
Capriles, V. D., Lopes Almeida, E., Ferreira, E. R., Gomes Arêas, J. A., Steel, C. J., Chang, Y. K. (2008). Physical and sensory properties of regular and reduced-fat pound cakes with added amaranth flour. Cereal Chem. 85 (5), 614‒618. doi: 10.1094/CCHEM-85-5-0614
Carroll, K. K., Kurowska, E. M. (1995). Soy consumption and cholesterol reduction: review of animal and human studies. J. Nutr. 125, 594S‒597S. doi: 10.1093/jn/125.3_Suppl.594S
Castro-Alba, V., Lazarte, C. E., Perez-Rea, D., Carlsson, N.-G., Almgren, A., Bergenståhl, B., et al. (2019). Fermentation of pseudocereals quinoa, cañihua, and amaranth to improve mineral accessibility through degradation of phytate. J. Sci. Food Agric. 99, 5239–5248. doi: 10.1002/jsfa.9793
Cawoy, V., Kinet, J. M., Jacquemart, A. L. (2008). Morphology of nectaries and biology of nectar production in the distylous species Fagopyrum esculentum. Ann. Bot. 102, 675–684. doi: 10.1093/aob/mcn150
Chan, K. F., Sun, M. (1997). Genetic diversity and relationships detected by isozyme and RAPD analysis of crop and wild species of Amaranthus. Theor. Appl. Genet. 95, 865–873. doi: 10.1007/s001220050637
Chauhan, E., Sarita, S. (2018). Effects of processing (germination and popping) on the nutritional and anti-nutritional properties of finger millet (Eleusine coracana). Curr. Res. In Nutr. Food Sci. 6 (2), 566–572. doi: 10.12944/CRNFSJ.6.2.30
Chauhan, G. S., Eskin, N. A. M., Tkachuk, R. (1992). Nutrients and antinutrients in quinoa seed. Cereal Chem. 69 (1), 85–88.
Chen, W., Xu, J., Jin, J., Lou, H., Fan, W., Yang, J. (2017). Genome-wide transcriptome analysis reveals conserved and distinct molecular mechanisms of Al resistance in buckwheat (Fagopyrum esculentum Moench) leaves. Int. J. Mol. Sci. 18, 1859. doi: 10.3390/ijms18091859
Cheng, R., Dong, Z. (2010). Breeding and production of foxtail millet in China. In: Ed. Z. He APA Cereals in China. (CIMMYT, Mexico, pp 87–95.
Chethan, S., Malleshi, N. (2007). Finger millet polyphenols: characterization and their nutraceutical potential. Am. J. Food Technol. 2 (7), 582‒592. doi: 10.3923/ajft.2007.582.592
Chitindingu, K., Benhura, M. A., Muchuweti, M. (2015). In vitro bioaccessibility assessment of phenolic compounds from selected cereal grains: a prediction tool of nutritional efficiency. LWT-Food Sci. Technol. 63, 575–581. doi: 10.1016/j.lwt.2015.02.026
Choukr-Allah, R., Rao, N. K., Hirich, A., Shahid, M., Alshankiti, A., Toderich, K., et al. (2016). Quinoa for marginal environments: toward future food and nutritional security in MENA and Central Asia regions. Front. Plant Sci. 7, 346. doi: 10.3389/fpls.2016.00346
Christa, K., Soral-Śmietana, M. (2008). Buckwheat grains and buckwheat products: nutritional and prophylactic value of their components – a review. Czech J. Food Sci. 26, 153‒162. doi: 10.17221/1602-cjfs
Christensen, S. A., Pratt, D. B., Pratt, C., Nelson, P. T., Stevens, M. R., Jellen, E. N., et al. (2007). Assessment of genetic diversity in the USDA and CIP-FAO international nursery collections of quinoa (Chenopodium quinoa Willd.) using microsatellite markers. Plant Genet. Resour. 5, 82–95. doi: 10.1017/S1479262107672293
Chrungoo, N. K., Kreft, I., Sangma, S. C., Devadasan, N., Dohtdong, L., Chetri, U. (2012). “Genetic diversity in Himalayan buckwheats: a perspective for use in crop improvement programmes,” in Proceedings of the 12th International Symposium on Buckwheat (Pernica), 198–211.
Clouse, J. W., Adhikary, D., Page, J. T., Ramaraj, T., Deyholos, M. K., Udall, J. A., et al. (2016). The amaranth genome: genome, transcriptome, and physical map assembly. Plant Genome 9, 1–14. doi: 10.3835/plantgenome2015.07.0062
Coles, N. D., Coleman, C. E., Christensen, S. A., Jellen, E. N., Stevens, M. R., Bonifacio, A., et al. (2005). Development and use of an expressed sequenced tag library in quinoa (Chenopodium quinoa Willd.) for the discovery of single nucleotide polymorphisms. Plant Sci. 168, 439–447. doi: 10.1016/j.plantsci.2004.09.007
Comai, S., Bertazzo, A., Bailoni, L., Zancato, M., Costa, C. V. L., Allegri, G. (2007). The content of proteic and nonproteic (free and protein-bound) tryptophan in quinoa and cereal flours. Food Chem. 100 (4), 1350–1355. doi: 10.1016/j.foodchem.2005.10.072
Costea, M., DeMason, D. A. (2001). Stem morphology and anatomy in Amaranthus L. (Amaranthaceae), taxonomic significance. J. Torrey Bot. Soc. 128, 254–281. doi: 10.2307/3088717
Délano-Frier, J. P., Avilés-Arnaut, H., Casarrubias-Castillo, K., Casique-Arroyo, G., Castrillón-Arbeláez, P. A., Herrera-Estrella, L., et al. (2011). Transcriptomic analysis of grain amaranth (Amaranthus hypochondriacus) using 454 pyrosequencing: comparison with A. tuberculatus, expression profiling in stems and in response to biotic and abiotic stress. BMC Genomics 12, 363. doi: 10.1186/1471-2164-12-363
d'Ennequin, M. L. T., Panaud, O., Toupance, B., Sarr, A. (2000). Assessment of genetic relationships between Setaria italica and its wild relative S. viridis using AFLP markers. Theor. Appl. Genet. 100, 1061–1066. doi: 10.1007/s001220051387
Dayakar, R. B., Bhaskarachary, K., Arlene, C. G. D., Sudha, D. G., Vilas, A. T. (2017). Nutritional and Health Benefits of Millets (Rajendranagar, Hyderabad: ICAR_Indian Institute of Millets Research (IIMR)), 112.
Debski, B., Gralak, M. A., Bertrandt, J., Kłos, A. (2013). Minerals and polyphenols content of quinoa (Chenopodium quinoa Willd.) Plant. Probl. Hig. Epidemiol. 94 (2), 300‒304.
del Castillo, C., Winkel, T., Mahy, G., Bizoux, J.-P. (2007). Genetic structure of quinoa (Chenopodium quinoa Willd.) from the Bolivian altiplano as revealed by RAPD markers. Genet. Resour. Crop Evol. 54, 897–905. doi: 10.1007/s10722-006-9151-z
Deschner, E. E., Ruperto, J., Wong, G., Newmark, H. L. (1991). Quercetin and rutin as inhibitors of azoxymethanol-induced colonic neoplasia. Carcinogenesis 12, 1193–1196. doi: 10.1093/carcin/12.7.1193
Desikachar, H. S. R. (1975). Processing of maize, sorghum and millets for food uses. J. Sci. Ind. Res. 34, 231–237.
Devi, P. B., Vijayabharathi, R., Sathyabama, S., Malleshi, N. G., Priyadarisini, V. B. (2014). Health benefits of finger millet (Eleusine coracana L.) polyphenols and dietary fiber: a review. J. Food Sci. Technol. 51, 1021–1040. doi: 10.1007/s13197-011-0584-9
Devos, K. M., Wang, Z. M., Beales, J., Sasaki, T., Gale, M. D. (1998). Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theor. Appl. Genet. 96, 63–68. doi: 10.1007/s001220050709
Dhankher, O. P., Foyer, C. H. (2018). Climate resilient crops for improving global food security and safety. Plant Cell Environ. 41, 877–884. doi: 10.1111/pce.13207
Dida, M. M., Ramakrishnan, S., Bennetzen, J. L., Gale, M. D., Devos, K. M. (2007). The genetic map of finger millet, Eleusine coracana. Theor. Appl. Genet. 114, 321–332. doi: 10.1007/s00122-006-0435-7
Dida, M. M., Wanyera, N., Dunn, M. L. H., Bennetzen, J. L., Devos, K. M. (2008). Population structure and diversity in finger millet (Eleusine coracana) germplasm. Trop. Plant Biol. 1, 131–141. doi: 10.1007/s12042-008-9012-3
Dini, I., Schettino, O., Simioli, T., Dini, A. (2001). Studies on the constituents of Chenopodium quinoa seeds: Isolation and characterization of new triterpene saponins. J. Agric. Food Chem. 49 (2), 741–746. doi: 10.1021/jf000971y
Du, X., Zhang, Z., Wu, B., Li, Y., Yu, A. H. (2013). Construction of genetic linkage map in tartary buckwheat (Fagopyrum tataricum) using SSR. Chin. Agric. Sci. Bull. 29 (21), 61‒65.
Dwivedi, S. L., Upadhyaya, H. D., Senthilvel, S., Hash, C. T., Fukunaga, K., Diao, X., et al. (2012). Millets: genetic and genomic resources, in Plant Breeding Reviews, Ed. Janick, J.. Plant Breed. Rev. (Hoboken, NJ: John Wiley and Sons, Inc.), 247–374.
Eisa, S. S., Eid, M. A., Abd El-Samad, E. H., Hussin, S. A., Abdel-Ati, A. A., El-Bordeny, N. E., et al. (2017). Chenopodium quinoa Willd.: a new cash crop halophyte for saline regions of Egypt. Aust. J. Crop Sci. 11, 343. doi: 10.21475/ajcs.17.11.03.pne316
Fabjan, N., Rode, J., Kozir, I. J., Wang, Z., Zhang, Z., Kreft, I. (2003). Tartary buckwheat (Fagopyrum tartaricum Gaertn.) as a source of dietary rutin and quercitrin. J. Agric. Food Chem. 51 (22), 6452‒6455. doi: 10.1021/jf034543e
Fairbanks, D. J., Waldrigues, A., Ruas, C. F., Ruas, P. M., Maughan, P. J., Robison, L. R., et al. (1993). Efficient characterization of biological diversity using field DNA extraction and random amplified polymorphic DNA markers. Rev. Bras. Genet 16, 11–11.
FAO. (2017). The future of food and agriculture–Trends and challenges (Rome: Food and Agriculture Organisation).
Farooq, S., Rehman, R. U., Pirzadah, T. B., Malik, B., Dar, F. A., Tahir, I. (2016). “Cultivation, agronomic practices, and growth performance of buckwheat,” in Molecular breeding and nutritional aspects of buckwheat (Elsevier), 299–319.
Farro, P. C. A. (2008). Desenvolvimento de filmes biodegradáveis a partir de derivados do grão de quinoa (Chenopodium quinoa Willdenow) da variedade “Real.” Thesis, Faculdade de Engenharia de Alimentos (Campinas, Brazil: Universidade Estadual de Campinas). 303 pp.
Ferreira, T. A., Arêas, J. A. G. (2010). Calcium bioavailability of raw and extruded amaranth grains. Food Sci. Technol. 30, 532–538. doi: 10.1590/S0101-20612010000200037
Foucault, A.-S., Mathé, V., Lafont, R., Even, P., Dioh, W., Veillet, S., et al. (2012). Quinoa extract enriched in 20-hydroxyecdysone protects mice from diet-induced obesity and modulates adipokines expression. Obesity 20 (2), 270‒277. doi: 10.1038/oby.2011.257
Fuentes, F., Bhargava, A. (2011). Morphological analysis of quinoa germplasm grown under lowland desert conditions. J. Agron. Crop Sci. 197, 124–134. doi: 10.1111/j.1439-037X.2010.00445.x
Fuentes, F., Martinez, E. A., Hinrichsen, P. V., Jellen, E. N., Maughan, P. J. (2009). Assessment of genetic diversity patterns in Chilean quinoa (Chenopodium quinoa Willd.) germplasm using multiplex fluorescent microsatellite markers. Conserv. Genet. 10, 369–377. doi: 10.1007/s10592-008-9604-3
Fukunaga, K., Wang, Z., Kato, K., Kawase, M. (2002). Geographical variation of nuclear genome RFLPs and genetic differentiation in foxtail millet, Setaria italica (L.) P. Beauv. Genet. Resour. Crop Evol. 49, 95–101. doi: 10.1023/A:1013852007770
Gahlawat, P., Sehgal, S. (1994). Protein and starch digestibility and iron availability in developed weaning foods as affected by roasting. J. Hum. Nutr. Dietetics 7, 121–126. doi: 10.1111/j.1365-277X.1994.tb00419.x
Gahlawat, P., Sehgal, S. (1995). Improvement in HCl-extractability of minerals in home made weaning foods. Plant Foods Hum. Nutr., 47, 173–179. doi: 10.1007/BF01089267
Gamel, T. H., Jozef, P. L., Ahmed, S. M., Ahmed, A. D., Lila, A. S. (2006). Seed treatments affect functional and antinutritional properties of amaranth flours. J. Sci. Food Agric. 86 (7), 1095–1102. doi: 10.1002/jsfa.2463
Ganapathy, N. S., Chitra, R. G., Gokhale, S. K. (1957). The effect of the protein of Italian millet (Setaria italica) on nitrogen retention in albino rats. Indian J. Med. Res. 45, 395–399.
Geervani, P., Eggum, B. O. (1989). Nutrient composition and protein quality of minor millets. Plant Foods Hum. Nutr. (Netherlands) 39, 201. doi: 10.1007/BF01091900
Gheldof, N., Wang, X.-H., Engeseth, N. J. (2003). Buckwheat honey increases serum antioxidant capacity in humans. J. Agric. Food Chem. 51, 1500–1505. doi: 10.1021/jf025897t
Gimode, D., Odeny, D. A., de Villiers, E. P., Wanyonyi, S., Dida, M. M., Mneney, E. E., et al. (2016). Identification of SNP and SSR markers in finger millet using next generation sequencing technologies. PloS One 11, e0159437. doi: 10.1371/journal.pone.0159437
Global Nutrition Report(2018). https://globalnutritionreport.org/reports/global-nutrition-report-2018/burden-malnutrition/.
Gopalan, C., Rama Sastri, B. V., Balasubramanian, S. C. (1989). Nutritive Value of Indian Foods (NVIF), Revised & Updated by B G. Deosthala, and K. C. Pant (Reprinted 2007, 2011). National Institute of Nutrition, Indian Council of Medical Research (ICMR) Hyderabad-500007, India.
Goron, T. L., Raizada, M. N. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. In Plant Sci. 6, 157. doi: 10.3389/fpls.2015.00157
Graebner, I. T., Siqueira, E. M., Arruda, S. F., de Souza, E. M. (2004). Carotenoids from native Brazilian dark-green vegetables are bioavailable: a study in rats. Nutr. Res. 24, 671–679. doi: 10.1016/j.nutres.2003.10.012
Graf, B. L., Poulev, A., Kuhn, P., Grace, M. H., Lila, M. A., Raskin, I. (2014). Quinoa seeds leach phytoecdysteroids and other compounds with anti-diabetic properties. Food Chem. 163, 178–185. doi: 10.1016/j.foodchem.2014.04.088
Griffith, J. Q., Couch, J. F., Lindauer., M. A. (1944). Effect of rutin on increased capillary fragility in man. Proc. Soc. Exp. Biol. Med. 55, 228–229. doi: 10.3181/00379727-55-14532
Grinberg, L. N., Rachmilewitz, E. A., Newmark, H. (1994). Protective effects of rutin against haemoglobin oxidation. Biochem. Pharmacol. 48, 643‒649. doi: 10.1016/0006-2952(94)90040-x
Guardia, T., Rotelli, A. E., Juarez, A. O., Pelzer, L. E. (2001). Anti-inflammatory properties of plant flavonoids: effects of rutin, quercetin and hesperidin on adjuvant arthritis in rat. Farmaco 56, 683–687. doi: 10.1016/S0014-827X(01)01111-9
Gull, A., Jan, R., Nayik, G. A., Prasad, K., Kumar, P. (2014). Significance of finger millet in nutrition, health and value added products: a review. Magnesium (mg) 130, 120.
Gupta, S., Kumari, K., Das, J., Lata, C., Puranik, S., Prasad, M. (2011). Development and utilization of novel intron length polymorphic markers in foxtail millet (Setaria italica (L.) P. Beauv.). Genome 54, 586–602. doi: 10.1139/g11-020
Gupta, S., Kumari, K., Sahu, P. P., Vidapu, S., Prasad, M. (2012). Sequence-based novel genomic microsatellite markers for robust genotyping purposes in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Cell Rep. 31, 323–337. doi: 10.1007/s00299-011-1168-x
Gupta, S., Kumari, K., Muthamilarasan, M., Parida, S. K., Prasad, M. (2014). Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep. 33 (6), 881‒893. doi: 10.1007/s00299-014-1564-0
Gupta, S. M., Arora, S., Mirza, N., Pande, A., Lata, C., Puranik, S., et al. (2017). Finger millet: a “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front. In Plant Sci. 8, 643. doi: 10.3389/fpls.2017.00643
Guruvayoorappan, C., Kuttan, G. (2007). Antiangiogenic effect of rutin and its regulatory effect on the production of VEGF, IL-1 and TNF-a in tumor associated macrophages. J. Biol. Sci. 7, 1511‒1519. doi: 10.3923/jbs.2007.1511.1519
Guzmán-Maldonado, S. H., Paredes-López, O. (1998). Production of high-protein flours as milk substitutes. In. Funct. Prop. Proteins Lipids 708, 66–79. doi: 10.1021/bk-1998-0708.ch005. ACS Symposium Series 708. American Chemical Society.
Handoyo, T., Meda, T., Urisu, A., Adachi, T., Morita, M. (2006). Hypoallergenic buckwheat flour preparation by Rhizopus oligosporus and its application to soba noodle. Food Res. Int. 39, 598–605. doi: 10.1016/j.foodres.2005.12.003
Hara, T., Iwata, H., Okuno, K., Matsui, K., Ohsawa, R. (2011). QTL analysis of photoperiod sensitivity in common buckwheat by using markers for expressed sequence tags and photoperiod-sensitivity candidate genes. Breed. Sci. 61, 394–404. doi: 10.1270/jsbbs.61.394
Hatakeyama, M., Aluri, S., Balachadran, M. T., Sivarajan, S. R., Patrignani, A., Grüter, S., et al. (2018). Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 25, 39–47. doi: 10.1093/dnares/dsx036
He, J., Klag, M. J., Whelton, P. K., Mo, J. P., Chen, J. Y., Qian, M. C., et al. (1985). Oats and buckwheat intakes and cardiovascular disease risk factors in an ethnic minority of China. Am. J. Clin. Nutr. 61, 366–372. doi: 10.1093/ajcn/61.2.366
Hejazi, S. N., Orsat, V., Azadi, B., Kubow, S. (2016). Improvement of the in vitro protein digestibility of amaranth grain through optimization of the malting process. J. Cereal Sci. 68, 59–65. doi: 10.1016/j.jcs.2015.11.007
Hemalatha, S., Platel, K., Srinivasan, K. (2007a). Zinc and iron content and their bioaccessibility in cereals and pulses consumed in India. Food Chem. 102, 1328–1336. doi: 10.1016/j.foodchem.2006.07.015
Hemalatha, S., Platel, K., Srinivasan, K. (2007b). Influence of germination and fermentation on bioaccessibility of zinc and iron from food grains. Eur. J. Clin. Nutr. 61, 342–348. doi: 10.1038/sj.ejcn.1602524
Hinojosa, L., González, J. A., Barrios-Masias, F. H., Fuentes, F., Murphy, K. M. (2018). Quinoa abiotic stress responses: a review. Plants 7, 106. doi: 10.3390/plants7040106
Hirich, A., Choukr-Allah, R., Jacobsen, S. E. (2014). Quinoa in Morocco–effect of sowing dates on development and yield. J. Agron. Crop Sci. 200, 371–377. doi: 10.1111/jac.12071
Hithamani, G., Srinivasan, K. (2014). Effect of domestic processing on the polyphenol content and bio-accessibility in finger millet (Eleusine coracana) and pearl millet (Pennisetum glaucum). Food Chem. 164, 55–62. doi: 10.1016/j.foodchem.2014.04.107
Hittalmani, S., Mahesh, H. B., Shirke, M. D., Biradar, H., Uday, G., Aruna, Y. R., et al. (2017). Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18, 465. doi: 10.1186/s12864-017-3850-z
Holasova, M., Fiedlerova, V., Smrcinova, H., Orsak, M., Lachman, J., Vavreinova, S. (2002). Buckwheat—the source of antioxidant activity in functional foods. Food Res. Int. 35, 207–211. doi: 10.1016/S0963-9969(01)00185-5
Hotz, C., Gibson, R. S. (2007). Traditional Food-Processing and Preparation Practices to Enhance the Bioavailability of Micronutrients in Plant-Based Diets. J. Nutr. 137 (4), 1097–1100. doi: 10.1093/jn/137.4.1097
Hou, S., Sun, Z., Linghu, B., Xu, D., Wu, B., Zhang, B., et al. (2016). Genetic diversity of buckwheat cultivars (Fagopyrum tartaricum Gaertn.) assessed with SSR markers developed from genome survey sequences. Plant Mol. Biol. Rep. 34, 233–241. doi: 10.1007/s11105-015-0907-5
Huang, J., Deng, J., Shi, T., Chen, Q., Liang, C., Meng, Z., et al. (2017). Global transcriptome analysis and identification of genes involved in nutrients accumulation during seed development of rice tartary buckwheat (Fagopyrum tararicum). Sci. Rep. 7, 11792. doi: 10.1038/s41598-017-11929-z
Huff, M. W., Carroll, K. K. (1980). Effects of dietary protein on turnover, oxidation, and absorption of cholesterol, and on steroid excretion in rabbits. J. Lipid Res. 21, 546–548.
Hurrell. (2004). Phytic acid degradation as a means of improving iron absorption. Int. J. Vitam. Nutr. Res. 74, 445–452. doi: 10.1024/0300-9831.74.6.445
Iglesias-Puig, E., Monedero, V., Haros, M. (2015). Bread with whole quinoa flour and bifidobacterial phytases increases dietary mineral intake and bioavailability. LWT-Food Sci. Technol. 60, 71–77. doi: 10.1016/j.lwt.2014.09.045
Ikeda, K., Ikeda, S. (2016). “Chapter fifteen - Factors important for structural properties and quality of buckwheat products,” in Molecular Breeding and Nutritional Aspects of Buckwheat. Eds. Zhou, M., Kreft, I., Woo, S.-H., Chrungoo, N., Wieslander, G. (Academic Press), 193–202. doi: 10.1016/B978-0-12-803692-1.00015-8
Ikeda, S., Yamashita, Y. (1994). Buckwheat as a dietary source of zinc, copper and manganese. Fagopyrum 14, 29–34.
Ikeda, K., Oku, M., Kusano, T., Yasumoto, K. (1986). Inhibitory potency of plant antinutrients towards the in vitro digestibility of buckwheat protein. J. Food Sci. 511527 - (6), 1530. doi: 10.1111/j.1365-2621.1986.tb13851.x
Ikeda, K., Sakaguchi, T., Kusano, T., Yasumoto, K. (1991). Endogenous factors affecting protein digestibility in buckwheat. Cereal Chem. 68, 424–427.
Ikeda, S., Yamashita, Y., Kreft, I. (1999). Mineral composition of buckwheat by-products and its processing characteristics to konjak preparation. Fagopyrum 16, e94.
Ikeda, S., Yamashita, Y., Tomura, K., Kreft, I. (2006). Nutritional comparison in mineral characteristics between buckwheat and cereals. Fagopyrum 23, 5.
Ikeda, K. (2002). Buckwheat composition, chemistry, and processing. Adv. in Food Nutr. Res. 44, 395–434. doi: 10.1016/S1043-4526(02)44008-9
IPGRI-APO. (1999). Status reports on genetic resources of buckwheat. (Serdang, Malaysia; IPGRI regional office for Asia, the Pacific and Oceania). 73.
Itagi, S., Naik, R., Bharati, P., Sharma, P. (2012). Readymade foxtail millet mix for diabetics. Int. J. Sci. Nat. 3, 131–134.
Izydorczyk, M. S., You, S., Campbell, C. (2004). “Molecular characteristics of amylose and amylopectin in buckwheat starches,” in American Association of Cereal Chemists 2004 meeting. (St. Paul MN, USA. American Association of Cereal Chemists) Abstract available online: https://www.cerealsgrains.org/meetings/Documents/Pre2009Abstracts/2004Abstracts/a04ma17.htm.
Jacobsen, S. E., Monteros, C., Christiansen, J. L., Bravo, L. A., Corcuera, L. J., Mujica, A. (2005). Plant responses of quinoa (Chenopodium quinoa Willd.) to frost at various phenological stages. Eur. J. Agron. 22, 131–139. doi: 10.1016/j.eja.2004.01.003
Jacobsen, S.-E., Sørensen, M., Pedersen, S. M., Weiner, J. (2013). Feeding the world: genetically modified crops versus agricultural biodiversity. Agron. Sustain. Dev. 33, 651–662. doi: 10.1007/s13593-013-0138-9
Jacobsen, S.-E. (2003). The worldwide potential for quinoa (Chenopodium quinoa Willd.). Food Rev. Int. 19, 167–177. doi: 10.1081/FRI-120018883
Jaiswal, V., Gupta, S., Gahlaut, V., Muthamilarasan, M., Bandyopadhyay, T., Ramchiary, N., et al. (2019). Genome-wide association study of major agronomic traits in foxtail millet (Setaria italica L.) using ddRAD sequencing. Sci. Rep. 9, 5020. doi: 10.1038/s41598-019-41602-6
Jancurová, M., Minarovičová, L., Dandár, A. (2009). Quinoa: A review. Czech J. Food Sci. 27 (2), 71–79. doi: 10.17221/32/2008-CJFS
Jarvis, D. E., Kopp, O. R., Jellen, E. N., Mallory, M. A., Pattee, J., Bonifacio, A., et al. (2008). Simple sequence repeat marker development and genetic mapping in quinoa (Chenopodium quinoa Willd.). J. Genet. 87, 39–51. doi: 10.1007/s12041-008-0006-6
Jarvis, D. E., Ho, Y. S., Lightfoot, D. J., Schmöckel, S. M., Li, B., Borm, T. J. A., et al. (2017). The genome of Chenopodium quinoa. Nature 542, 307–312. doi: 10.1038/nature21370
Jayaraman, A., Puranik, S., Rai, N. K., Vidapu, S., Sahu, P. P., Lata, C., et al. (2008). cDNA-AFLP analysis reveals differential gene expression in response to salt stress in foxtail millet (Setaria italica L.). Mol. Biotechnol. 40, 241–251. doi: 10.1007/s12033-008-9081-4
Je, H. D., Shin, C. Y., Park, S. Y., Yim, S. H., Kum, C., Huh, I. H., et al. (2002). Combination of vitamin c and rutin on neuropathy and lung damage of diabetes mellitus rats. Arch. Pharm. Res. 25, 184–190. doi: 10.1007/BF02976561
Jia, X. P., Shi, Y. S., Song, Y. C., Wang, G. Y., Wang, T. Y., Li, Y. (2007). Development of EST-SSR in foxtail millet (Setaria italica). Genet. Resour. Crop Evol. 54, 233–236. doi: 10.1007/s10722-006-9139-8
Jia, X. P., Zhang, Z., Liu, Y., Zhang, C., Shi, Y., Song, Y., et al. (2009). Development and genetic mapping of SSR markers in foxtail millet [Setaria italica (L.) P. Beauv.]. Theor. Appl. Genet. 118, 821–829. doi: 10.1007/s00122-008-0942-9
Jia, G., Huang, X., Zhi, H., Zhao, Y., Zhao, Q., Li, W., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957. doi: 10.1038/ng.2673
Jiang, P., Burczynski, F., Campbell, C., Pierce, G., Austria, J. A., Briggs, C. J. (2007). Rutin and flavonoid contents in three buckwheat species Fagopyrum esculentum, F. tataricum, and F. homotropicum and their protective effects against lipid peroxidation. Food Res. Int. 40, 356–364. doi: 10.1016/j.foodres.2006.10.009
Johnson, D. L., Croissant, R. (1985). Quinoa production in Colorado. Service in action, no. 112 (Colorado, USA: Colorado State University, Cooperative Extension, Fort Collins).
Kaur, P., Purewal, S. S., Sandhu, K. S., Maninder, K., Salar, R. K. (2019). Millets: a cereal grain with potent antioxidants and health benefits. Food Meas. 13, 793–806. doi: 10.1007/s11694-018-9992-0
Kawa, J. M., Taylor, C. G., Przybylski, R. (2003). Buckwheat concentrate reduces serum glucose in streptozotocin-diabetic rats. J. Agric. Food Chem. 51, 7287–7291. doi: 10.1021/jf0302153
Kayashita, J., Shimaoka, I., Nakajoh, M., Yamazaki, M., Kato, N. (1997). Consumption of buckwheat protein lowers plasma cholesterol and raises fecal neutral sterols in cholesterol-fed rats because of its low digestibility. J. Nutr. 127, 1395–1400. doi: 10.1093/jn/127.7.1395
Kellogg, E. A. (2015). Flowering plants. Monocots. Poaceae (Springer International Publishing). Vol. 13, 416 pp. doi: 10.1007/978-3-319-15332-2
Khattab, R. Y., Arntfield, S. D. (2009). Nutritional quality of legume seeds as affected by some physical treatments. 2. Antinutritional factors. LWT Food Sci. Technol. 42 (6), 1113–1118. doi: 10.1016/j.lwt.2009.02.004
Kim, S. L., Kim, S. K., Park, C. H. (2004). Introduction and nutritional evaluation of buckwheat sprouts as a new vegetable. Food Res. Int. 37, 319–327. doi: 10.1016/j.foodres.2003.12.008
Kishore, G., Gupta, S., Pandey, A. (2012). Assessment of population genetic diversity of Fagopyrum tataricum using SSR molecular marker. Biochem. Syst. Ecol. 43, 32–41. doi: 10.1016/j.bse.2012.02.018
Kolano, B., Siwinska, D., Pando, L. G., Szymanowska-Pulka, J., Maluszynska, J. (2012). Genome size variation in Chenopodium quinoa (Chenopodiaceae). Plant Syst. Evol. 298, 251–255. doi: 10.1007/s00606-011-0534-z
Kong, X., Bao, J., Corke, H. (2009). Physical properties of Amaranthus starch. Food Chem. 113, 371–376. doi: 10.1016/j.foodchem.2008.06.028
Konishi, T., Ohnishi, O. (2006). A linkage map for common buckwheat based on microsatellite and AFLP markers. Fagopyrum 23, 1–6.
Koziol, M. J. (1990). “Composición química,” in Quinua Hacia su Cultivo Comercial, Chap. VIII. Eds. Wahli, C. (Brazil: Quito, Latinreco SA), 137–159.
Kreft, I., Skrabanja, V. (2002). Nutritional properties of starch in buckwheat noodles. J. Nutr. Sci. Vitaminol. 48, 47–50. doi: 10.3177/jnsv.48.47
Kreft, I., Fabjan, N., Yasumoto, K. (2006). Rutin content in buckwheat (Fagopyrum esculentum Moench) food materials and products. Food Chem. 98, 508–512. doi: 10.1016/j.foodchem.2005.05.081
Kreft, I. (1983). “Buckwheat breeding perspectives,” in Buckwheat Research 1983: Proceedings of the 2nd International Symposium on Buckwheat held in Miyazaki, Japan, Sept. 7–10, 1983. Eds. Nagatomo, T., Adachi, T. (Miyazaki, Japan: Kuroda-Toshado Printing Co., Japan), 3–12.
Krishnan, R., Dharmaraj, U., Malleshi, N. G. (2012). Influence of decortication, popping and malting on bioaccessibility of calcium, iron and zinc in finger millet. LWT - Food Sci. Technol. 48 (2), 169–174. doi: 10.1016/j.lwt.2012.03.003
Krkošková, B., Mrazova, Z. (2005). Prophylactic components of buckwheat. Food Res. Int. 38, 561–568. doi: 10.1016/j.foodres.2004.11.009
Kumar, A., Gaur, V. S., Goel, A., Gupta, A. K. (2015a). De novo assembly and characterization of developing spikes transcriptome of finger millet (Eleusine coracana): a minor crop having nutraceutical properties. Plant Mol. Biol. Rep. 33, 905–922. doi: 10.1007/s11105-014-0802-5
Kumar, A., Yadav, S., Panwar, P., Gaur, V. S., Sood, S. (2015b). Identification of anchored simple sequenceF repeat markers associated with calcium content in finger millet (Eleusine coracana). Proc. Natl. Acad. Sci. 85, 311–317. doi: 10.1007/s40011-013-0296-1
Kumar, A., Sharma, D., Tiwari, A., Jaiswal, J. P., Singh, N. K., Sood, S. (2016). Genotyping-by-sequencing analysis for determining population structure of finger millet germplasm of diverse origins. Plant Genome 9, 1–15. doi: 10.3835/plantgenome2015.07.0058
Kumar, A., Tomer, V., Kaur, A., Kumar, V., Gupta, K. (2018). Millets: a solution to agrarian and nutritional challenges. Agric. Food Secur. 7, 31. doi: 10.1186/s40066-018-0183-3
Kumari, S. K., Thayumanavan, B. (1997). Comparative study of resistant starch from minor millets on intestinal responses, blood glucose, serum cholesterol and triglycerides in rats. J. Sci. Food Agric. 75, 296–302. doi: 10.1002/(SICI)1097-0010(199711)75:3<296::AID-JSFA877>3.0.CO;2-X
Kumpun, S., Maria, A., Crouzet, S., Evrard-Todeschi, N., Girault, J.-P., Lafont, R. (2011). Ecdysteroids from Chenopodium quinoa Willd., an ancient Andean crop of high nutritional value. Food Chem. 125, 1226–1234. doi: 10.1016/j.foodchem.2010.10.039
Lakshmi, B., Vimala, V. (2000). Nutritive value of dehydrated green leafy vegetable powders. J. Food Sci. Technol. 37, 465–471.
Lata, C., Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lata, C., Shivhare, R. (2017). “Genetic Determinants of Abiotic Stress Tolerance in Foxtail Millet,” in The Foxtail Millet Genome (Springer), 85–104.
Lata, C., Sahu, P. P., Prasad, M. (2010). Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem. Biophys. Res. Commun. 393, 720–727. doi: 10.1016/j.bbrc.2010.02.068
Lata, C., Jha, S., Dixit, V., Sreenivasulu, N., Prasad, M. (2011). Differential antioxidative responses to dehydration-induced oxidative stress in core set of foxtail millet cultivars [Setaria italica (L.)]. Protoplasma 248, 817–828. doi: 10.1007/s00709-010-0257-y
Lazarte, C. E., Carlsson, N. G., Almgren, A., Sandberg, A.-S., Granfeldt, Y. (2015). Phytate, zinc, iron and calcium content of common Bolivian food, and implications for mineral bioavailability. J. Food Compos. Anal. 39, 111–119. doi: 10.1016/j.jfca.2014.11.015
León-Camacho, M., García-González, D. L., Aparicio, R. (2001). A detailed and comprehensive study of amaranth (Amaranthus cruentus L.) oil fatty profile. Eur. Food Res. Technol. 213, 349–355. doi: 10.1007/s002170100340
Li, S., Zhang, Q. H. (2001). Advances in the development of functional foods from buckwheat. Crit. Rev. In Food Sci. Nutr. 41, 451–464. doi: 10.1080/20014091091887
Li, Y. Q., Shi, T. L., Zhang, Z. W. (2007). Development of microsatellite markers from tartary buckwheat. Biotechnol. Lett. 29, 823–827. doi: 10.1007/s10529-006-9293-2
Lightfoot, D. J., Jarvis, D. E., Ramaraj, T., Lee, R., Jellen, E. N., Maughan, P. J. (2017). Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol. 15, 74. doi: 10.1186/s12915-017-0412-4
Lin, H. S., Chiang, C. Y., Chang, S. B., Kuoh, C. S. (2011). Development of simple sequence repeat (SSR) markers in Setaria italica (Poaceae) and cross-amplification in related species. Int. J. Mol. Sci. 12, 7835–7845. doi: 10.3390/ijms12117835
Lin, H. S. (2012). Genetic diversity in the foxtail millet (Setaria italica) germplasm as determined by agronomic traits and microsatellite markers. Aust. J. Crop Sci. 6, 342–349.
Lintschinger, J., Fuchs, N., Moser, H., Jäger, R., Hlebeina, T., Markolin, G., et al. (1997). Uptake of various trace elements during germination of wheat, buckwheat and quinoa. Plant Foods Hum. Nutr. 50, 223–237. doi: 10.1007/BF02436059
Logacheva, M. D., Kasianov, A. S., Vinogradov, D. V., Samigullin, T. H., Gelfand, M. S., Makeev, V. J., et al. (2011). De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genomics 12, 30. doi: 10.1186/1471-2164-12-30
Lopes, C. O., Dessimoni, G. V., Da Silva, M. C., Vieira, G., Pinto, N. A. V. D. (2009). Aproveitamento, composição nutricional e antinutricional da farinha de quinoa (Chenopodium quinoa). Alime. Nutr. 20 (4), 669–675.
Ma, K. H., Kim, N. S., Lee, G. A., Lee, S. Y., Lee, J. K., Yi, J. Y., et al. (2009). Development of SSR markers for studies of diversity in the genus Fagopyrum. Theor. Appl. Genet. 119, 1247–1254. doi: 10.1007/s00122-009-1129-8
Mabhaudhi, T., Chimonyo, V. G. P., Hlahla, S., Massawe, F., Mayes, S., Nhamo, L., et al. (2019). Prospects of orphan crops in climate change. Planta, 250, 695–708. doi: 10.1007/s00425-019-03129-y
Madhusoodanan, K. J., Pal, M. (1981). Cytology of vegetable amaranths. Bot. J. Linn. Soc. 82 (1), 61‒68. doi: 10.1111/j.1095-8339.1981.tb00950.x
Mahoney, A. W., Lopez, J. G., Hendricks, D. G. (1975). An evaluation of the protein quality of quinoa. J. Agric. Food Chem. 23, 90. doi: 10.1021/jf60198a035
Mal, B., Padulosi, S., Ravi, S. B. (2010). Minor millets in South Asia: learnings from IFAD-NUS Project in India and Nepal (Chennai, India: Bioversity International, Maccarese, Rome, Italy, and the M.S. Swaminathan Research Foundation), 1‒176.
Mallory, M. A., Hall, R. V., McNabb, A. R., Pratt, D. B., Jellen, E. N., Maughan, P. J. (2008). Development and characterization of microsatellite markers for the grain amaranths. Crop Sci. 48, 1098–1106. doi: 10.2135/cropsci2007.08.0457
Maradini, A. M., Ribeiro, M., Da Silva, J. T., Pinheiro, H. M., Paes, J. B., Dos Reis, J. S. (2017). Quinoa: Nutritional, Functional and Antinutritional Aspects. Crit. Rev. In Food Sci. Nutr. 57 (8), 1618–1630. doi: 10.1080/10408398.2014.1001811
Martínez, E. A., Jorquera-Jaramillo, C., Veas, E., Chia, E. (2009). El futuro de la quínoa en la región árida de Coquimbo: Lecciones y escenarios a partir de una investigación sobre su biodiversidad en Chile para la acción con agricultores locales. Rev. Geogr. Valparaíso, 42, 95–111.
Martínez-Cruz, O., Cabrera-Chávez, F., Paredes-López, O. (2014). Biochemical characteristics, and nutraceutical and technological uses of amaranth globulins. In: Eds. Milford, S. D., Globulins, S. D. Biochemistry, Production and Role in Immunity. (Nova Science Publishers, Inc), 41–70.
Mason, S. L., Stevens, M. R., Jellen, E. N., Bonifacio, A., Fairbanks, D. J., Coleman, C. E., et al. (2005). Development and use of microsatellite markers for germplasm characterization in quinoa (Chenopodium quinoa Willd.). Crop Sci. 45, 1618–1630. doi: 10.2135/cropsci2004.0295
Mastebroek, H. D., Limburg, H., Gilles, T., Marvin, H. J. P. (2000). Occurrence of sapogenins in leaves and seeds of quinoa (Chenopodium quinoa Willd). J. Sci. Food Agric. 80, 152–156. doi: 10.1002/(SICI)1097-0010(20000101)80:1<152::AID-JSFA503>3.0.CO;2-P
Matsui, K., Kiryu, Y., Komatsuda, T., Kurauchi, N., Ohtani, T., Tetsuka, T. (2004). Identification of AFLP makers linked to non-seed shattering locus (sht1) in buckwheat and conversion to STS markers for marker-assisted selection. Genome 47, 469–474. doi: 10.1139/g04-007
Maughan, P. J., Bonifacio, A., Jellen, E. N., Stevens, M. R., Coleman, C. E., Ricks, M., et al. (2004). A genetic linkage map of quinoa (Chenopodium quinoa) based on AFLP, RAPD, and SSR markers. Theor. Appl. Genet. 109, 1188–1195. doi: 10.1007/s00122-004-1730-9
Maughan, P. J., Turner, T. B., Coleman, C. E., Elzinga, D. B., Jellen, E. N., Morales, J. A., et al. (2009). Characterization of Salt Overly Sensitive 1 (SOS1) gene homoeologs in quinoa (Chenopodium quinoa Willd.). Genome 52, 647–657. doi: 10.1139/G09-041
Maughan, P. J., Smith, S. M., Rojas-Beltran, J. A., Elzinga, D., Raney, J. A., Jellen, E. N., et al. (2012). Single nucleotide polymorphism identification, characterization, and linkage mapping in quinoa. Plant Genome 5, 114–125. doi: 10.3835/plantgenome2012.06.0011
Mbithi-Mwikya, S., Ooghe, W., Van Camp, J., Ngundi, D., Huyghebaert, A. (2000). Amino acid profiles after sprouting, autoclaving, and lactic acid fermentation of finger millet (Eleusine coracana) and kidney beans (Phaseolus vulgaris L.). J. Agric. Food Chem. 48, 3081–3085. doi: 10.1021/jf0002140
Mendonça, S., Saldiva, P. H., Cruz, R. J., Arêas, J. A. (2009). Amaranth protein presents cholesterol-lowering effect. Food Chem. 116, 738–742. doi: 10.1016/j.foodchem.2009.03.021
Mgonja, M. A., Lenne, J. M., Manyasa, E., Sreenivasaprasad, S. (2007). Finger millet blast management in East Africa: creating opportunities for improving production and utilization of finger millet. Int. Crops Res. Inst. Semi-Arid Trop. 196.
Miranda, M., Vega-Gálvez, A., Quispe-Fuentes, I., Rodríguez, M. J., Maureira, H., Martínez, E. A. (2012). Nutritional aspects of six quinoa (Chenopodium quinoa Willd.) ecotypes from three geographical areas of Chile. Chil. J. Agric. Res. 72, 175.
Mirza, N., Taj, G., Arora, S., Kumar, A. (2014). Transcriptional expression analysis of genes involved in regulation of calcium translocation and storage in finger millet (Eleusine coracana L. Gartn.). Gene 550, 171–179. doi: 10.1016/j.gene.2014.08.005
Mishra, G., Joshi, D. C., Panda, B. K. (2014). Popping and puffing of cereal grains: a review. J. Grain Process. Storage 1 (2), 34–46.
Mitsunaga, T., Matsuda, M., Shimizu, M., Iwashima, A. (1986). Isolation and properties of thiamine-binding protein from bckwheat seed. Cereal Chem. 4, 332–335.
Mlakar, S. G., Turinek, M., Jakop, M., Bavec, M., Bavec, F. (2009). Nutrition value and use of grain amaranth: potential future application in bread making. Agricultura 6: 43–53.
Mohamed, T. K., Zhu, K., Issoufou, A., Fatmata, T., Zhou, H. (2009). Functionality, in vitro digestibility and physicochemical properties of two varieties of defatted foxtail millet protein concentrates. Int. J. Mol. Sci. 10, 5224–5238. doi: 10.3390/ijms10125224
Monteros, C. J., Nieto, C. C., Caicedo, C. V., Rivera, M. M., Vimos, C. N. (1994). INIAP-Alegría, primera variedad mejorada de amaranto para la sierra ecuatoriana. Boletin Divulgativo No. 245, Ecuador
Morales, A., Zurita-Silva, A., Maldonado, J., Silva, H. (2017). Transcriptional responses of Chilean quinoa (Chenopodium quinoa Willd.) under water deficit conditions uncovers ABA-independent expression patterns. Front. Plant Sci. 8, 216. doi: 10.3389/fpls.2017.00216
Mosyakin, S. L., Robertson, K. R. (1996). New infrageneric taxa and combinations in Amaranthus (Amaranthaceae). Ann. Botan. Fenn. (JSTOR), 33, 275–281.
Mounika, M., Devi, K. U., Devi, S. S. (2017). Bioavailability of iron from foxtail millet and sorghum millet recipes. Int. J. Agric. Sci. Res. (IJASR) 7 (4), 703–708. doi: 10.20546/ijcmas.2017.610.332
Mujica, A., Jacobsen, S. E. (2003). The genetic resources of Andean grain amaranths (Amaranthus caudatus L., A. cruentus L. and A. hypochondriacus L.) in America. Plant Genet. Resour. Newslett. 133, 41–44.
Musia, G. D. (2013). Identification of microsatellite markers for finger millet [Eleusine coracana (L.) Gaertn.] by analysis of Roche 454 GS-FLX Titanium sequence data. Dissertation/master's thesis (Nairobi, Kenya: Kenyatta University).
Muthamilarasan, M., Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi: 10.1007/s00122-014-2399-3
Muthamilarasan, M., Venkata Suresh, B., Pandey, G., Kumari, K., Parida, S. K., Prasad, M. (2013). Development of 5123 intron-length polymorphic markers for large-scale genotyping applications in foxtail millet. DNA Res. 21, 41–52. doi: 10.1093/dnares/dst039
Muza, F. R., Lee, D. J., Andrews, D. J., Gupta, S. C. (1995). Mitochondrial DNA variation in finger millet (Eleusine coracana L. Gaertn). Euphytica 81, 199–205. doi: 10.1007/BF00025434
Mysore, K. S., Baird, V. (1997). Nuclear DNA content in species of Eleusine (Gramineae): a critical re-evaluation using laser flow cytometry. Plant Syst. Evol. 207, 1–11. doi: 10.1007/BF00985206
Nègre-Salvayre, A., Affany, A., Hariton, C., Salvayre, R. (1991). Additional antilipoperoxidant activities of alpha-tocopherol and ascorbic acid on membrane-like systems are potentiated by rutin. Pharmacology 42, 262–272. doi: 10.1159/000138807
Nagano, M., Aii, J., Campbell, C., Kawasaki, S., Adachi, T. (2000). Genome size analysis of the genus Fagopyrum. Fagopyrum 17, 9.
Nagano, M., Aii, J., Kuroda, M., Campbell, C., Adachi, T. (2001). Conversion of AFLP markers linked to the Sh allele at the S locus in buckwheat to a simple PCR based marker form. Plant Biotechnol. 18, 191–196. doi: 10.5511/plantbiotechnology.18.191
Nagaprabha, P., Bhattacharya, S. (2016). Textural characterization of foxtail millet gels: effect of cations and hydrocolloids. J. Food Sci. Technol. 53 (1), 257–268. doi: 10.1007/s13197-015-2046-2
Nagasawa, T., Tabata, N., Ito, Y., Aiba, Y., Nishizawa, N., Kitts, D. D. (2003). Dietary G-Rutin suppresses glycation in tissue proteins of streptozotocin-induced diabetic rats. Mol. Cell. Biochem. 252, 141–147. doi: 10.1023/A:1025563519088
Nanduri, K. R., Hirich, A., Salehi, M., Saadat, S., Jacobsen, S. E. (2019). “Quinoa: a new crop for harsh environments,” in Sabkha Ecosystems (Springer), 301–333.
National Research Council. (1996). “Lost crops of Africa,” in Grains. Board on Science and Technology for International Development, vol. I. (Washington, D.C: National Academy Press).
Nirgude, M., Babu, B. K., Shambhavi, Y., Singh, U. M., Upadhyaya, H. D., Kumar, A. (2014). Development and molecular characterization of genic molecular markers for grain protein and calcium content in finger millet (Eleusine coracana (L.) Gaertn.). Mol. Biol. Rep. 41, 1189–1200. doi: 10.1007/s11033-013-2825-7
Nirmala, M., Subba Rao, M.V.S.S.T., Muralikrishna, G. (2000). Carbohydrates and their degrading enzymes from native and malted finger millet (Ragi, Eleusine coracana, Indaf-15). Food Chem. 69 (2), 175–180. doi: 10.1016/S0308-8146(99)00250-2
Nusslein, K., Parkash Dhankher, O., Xing, B., Smith-Doerr, L., Sacco, T., Maathuis, F., et al. (2016). Project management: food security needs social-science input. Nature 535, 37–37. doi: 10.1038/535037d
Odetti, P. R., Borgoglio, A., De Pascale, A., Rolandi, R., Adezati, L. (1990). Prevention of diabetes-increased aging effect on rat collagen-linked fluorescence by aminoguanidine and rutin. Diabetes 39, 796. doi: 10.2337/diab.39.7.796
Oghbaei, M., Prakash, J. (2012). Bioaccessible nutrients and bioactive components from fortified products prepared using finger millet (Eleusine coracana). J. Sci. Food Agric. 92, 2281–2290. doi: 10.1002/jsfa.5622
Ogungbenle, H. N. (2003). Nutritional evaluation and functional properties of quinoa (Chenopodium quinoa) flour. Int. J. Food Sci. Nutr. 54 (2), 153–158. doi: 10.1080/0963748031000084106
Ohnishi, O. (1993). Population genetics of cultivated common buckwheat, Fagopyrum esculentum Moench. VIII. Local differentiation of land races in Europe and the silk road. Japanese J. Genet. 68, 303–316. doi: 10.1266/jjg.68.303
Oliveira, A. C., Reis, S. M. P. M., Carvalho, E. M., Pimenta, F. M. V., Rios, K. R., Paiva, K. C., et al. (2003). Adições crescentes de ácido fitico à dieta não interferiram na digestibilidade da caseína e no ganho de peso em ratos. Rev. Nutr. 16 (2), 211–217. doi: 10.1590/S1415-52732003000200008
Ookubo, K. (1992). “Nutrition and functionality of soybean,” in Science of Soybean. Eds. Yamauchi, F., Ookubo, K. (Tokyo: Asakura-Shoten Press), 57–75.
Oomah, B. D., Mazza, G. (1996). Flavonoids and antioxidative activities in buckwheat. J. Agric. Food Chem. 44 (7), 1746–1750. doi: 10.1021/jf9508357
Paśko, P., Bartoń, H., Zagrodzki, P., Gorinstein, S., Fołta, M., Zachwieja, Z. (2009). Anthocyanins, total polyphenols and antioxidant activity in amaranth and quinoa seeds and sprouts during their growth. Food Chem. 115, 994–998. doi: 10.1016/j.foodchem.2009.01.037
Pal, M., Khoshoo, T. N. (1972). Evolution and improvement of cultivated amaranths: V. Inviability, weakness, and sterility in hybrids. J. Hered. 63 (2), 78‒82. doi: 10.1093/oxfordjournals.jhered.a108234
Pal, J., Singhal, R. S., Kulkarni, P. R. (2002). Physicochemical properties of hydroxypropyl derivative from corn and amaranth starch. Carbohydr. Polym. 48, 49–53. doi: 10.1016/S0144-8617(01)00209-0
Palomino, G., Hernández, L. T., de la Cruz Torres, E. (2008). Nuclear genome size and chromosome analysis in Chenopodium quinoa and C. berlandieri subsp. nuttalliae. Euphytica 164, 221. doi: 10.1007/s10681-008
Pan, S. J., Chen, Q. F. (2010). Genetic mapping of common buckwheat using DNA, protein and morphological markers. Hereditas 147, 27–33. doi: 10.1111/j.1601-5223.2009.02116.x
Pandey, G., Misra, G., Kumari, K., Gupta, S., Parida, S. K., Chattopadhyay, D., et al. (2013). Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. 20, 197–207. doi: 10.1093/dnares/dst002
Panwal, J. H., Pawar, V. D. (1989). Effect of soaking pearl millet in acid on the bleaching and functional properties of flours. J. Food Sci. Technol. 26 (2), 79–82.
Paredes-Lopez, O., Mora-Escobedo, R. (1989). Germination of amaranth seeds: effects on nutrient composition and color. J. Food Sci. 54, 761–762. doi: 10.1111/j.1365-2621.1989.tb04702.x
Park, S.-S., Ohba, H. (2004). Suppressive activity of protease inhibitors from buckwheat seeds against human T-acute lymphoblastic leukemia cell lines. Appl. Biochem. Biotechnol. 117, 65. doi: 10.1385/ABAB:117:2:065
Pawar, V. D., Machewad, G. M. (2006). Processing of foxtail millet for improved nutrient availability. J. Food Process. Preserv. 30 (3), 269‒279. doi: 10.1111/j.17454549.2006.00064.x
Pawar, V. D., Parlikar, G. S. (1990). Reducing the polyphenols and phytate and improving the protein quality of pearl millet by dehulling and soaking. J. Food Sci. Technol. 27 (3), 140–143. doi: 10.4236/fns.2011.28122
Perales-Sánchez, J. X., Reyes-Moreno, C., Gómez-Favela, M. A., Milán-Carrillo, J., Cuevas-Rodríguez, E. O., Valdez-Ortiz, A., et al. (2014). Increasing the antioxidant activity, total phenolic and flavonoid contents by optimizing the germination conditions of amaranth seeds. Plant Foods Hum. Nutr. 69, 196–202. doi: 10.1007/s11130-014-0430-0
Peterson, A., Murphy, K. (2015). Tolerance of lowland quinoa cultivars to sodium chloride and sodium sulfate salinity. Crop Sci. 55, 331–338. doi: 10.2135/cropsci2014.04.0271
Pires, J. L. (2017). Avaliação do Comportamento Agronómico da Quinoa (Chenopodium quinoa Willd.), em Diferentes Regimes Hídricos e Níveis de Fertilização Azotada, nas Condições Agroecológicas de Trás-os-Montes. Master"s Thesis (Bragança, Portugal: Instituto Politecnico de Bracanca Escola Superior Agraria).
Plate, A. Y., Arêas, J. A. (2002). Cholesterol-lowering effect of extruded amaranth (Amaranthus caudatus L.) in hypercholesterolemic rabbits. Food Chem. 76, 1–6. doi: 10.1016/S0308-8146(01)00238-2
Platel, K., Srinivasan, K. (2016). Bioavailability of Micronutrients from Plant Foods: An Update. Crit. Rev. in Food Sci. Nutr. 56 (10), 1608–1619. doi: 10.1080/10408398.2013.781011
Platel, K., Eipeson, S. W., Srinivasan, K. (2010). Bioaccessible Mineral Content of Malted Finger Millet (Eleusine coracana), Wheat (Triticum aestivum), and Barley (Hordeum vulgare). J. Agric. Food Chem. 58 (13), 8100–8103. doi: 10.1021/jf100846e
Pomeranz, Y., Robbins, G. S. (1972). Amino acid composition of buckwheat. J. Agric. Food Chem. 20 (2), 270–274. doi: 10.1021/jf60180a029
Popa, G., Cornea, C. P., Ciuca, M., Babeanu, N., Popa, O., Marin, D. (2010). Studies on genetic diversity in Amaranthus species using the RAPD markers. Tom 17 (2), 280–285.
Porter, J. R., Xie, L., Challinor, A. J., Cochrane, K., Howden, S. M., Iqbal, M. M., et al. (2014). Food security and food production systems in: Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. [Eds. Field, C. B., Barros, V. R., Dokken, D. J., Mach, K. J., Mastrandrea, M. D., Bilir, T. E., Chatterjee, M., Ebi, K. L., Estrada, Y. O., Genova, R. C., Girma, B., Kissel, E. S., Levy, A. N., MacCracken, S., Mastrandrea, P. R., White, L. L.]. (Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press), 485–533.
Powell, W., Morgante, M., Andre, C., Hanafey, M., Vogel, J., Tingey, S., et al. (1996). The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 2, 225–238. doi: 10.1007/BF00564200
Präger, A., Munz, S., Nkebiwe, P., Mast, B., Graeff-Hönninger, S. (2018). Yield and quality characteristics of different quinoa (Chenopodium quinoa Willd.) cultivars grown under field conditions in Southwestern Germany. Agronomy 8, 197. doi: 10.3390/agronomy8100197
Praznik, W., Mundigler, N., Kogler, A., Pelzl, B., Huber, A., Wollendorfer, M. (1999). Molecular background of technological properties of selected starches. Starch-Stärke 51, 197–211. doi: 10.1002/(SICI)1521-379X(199906)51:6<197::AID-STAR197>3.0.CO;2-K
Pu, F., Mishima, K., Irie, Motohashi, K., Tanaka, Y., Orito, K., et al. (2007). Neuroprotective effects of quercetin and rutin on spatial memory impairment in an 8-arm radial maze task and neuronal death induced by repeated cerebral ischemia in rats. J. Pharmacol. Sci. 104 (4), 329‒334. doi: 10.1254/jphs.FP0070247
Pulvento, C., Riccardi, M., Lavini, A., d'Andria, R., Iafelice, G., Marconi, E. (2010). Field trial evaluation of two Chenopodium quinoa genotypes grown under rain-fed conditions in a typical Mediterranean environment in South Italy. J. Agron. Crop Sci. 196, 407–411. doi: 10.1111/j.1439-037X.2010.00431.x
Puranik, S., Jha, S., Srivastava, P. S., Sreenivasulu, N., Prasad, M. (2011). Comparative transcriptome analysis of contrasting foxtail millet cultivars in response to short-term salinity stress. J. Plant Physiol. 168, 280–287. doi: 10.1016/j.jplph.2010.07.005
Puranik, S., Sahu, P. P., Mandal, S. N., Parida, S. K., Prasad, M. (2013). Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PloS One 8, e64594. doi: 10.1371/journal.pone.0064594
Qi, X., Xie, S., Liu, Y., Yi, F., Yu, J. (2013). Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 83, 459–473. doi: 10.1007/s11103-013-0104-6
Qian, J., Kuhn, M. (1999). Physical properties of buckwheat starches from various origins. Starch-Stärke 51, 81–85. doi: 10.1002/(SICI)1521-379X(199903)51:2<81::AID-STAR81>3.0.CO;2-I
Rahman, H., Jagadeeshselvam, N., Valarmathi, R., Sachin, B., Sasikala, R., Senthil, N., et al. (2014). Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol. Biol. 85, 485–503. doi: 10.1007/s11103-014-0199-4
Raina, A. P., Gupta, V. (2015). Evaluation of buckwheat (Fagopyrum species) germplasm for rutin content in seeds. Indian J. Plant Physiol. 20, 167–171. doi: 10.1007/s40502-015-0147-6
Ramachandra, G., Virupaksha, T. K., Shadaksharaswamy, M. (1977). Relation between tannin levels and in vitro protein digestibility in finger millet (Eleusine coracana Gaertn.). J. Agric. Food Chem. 25, 1101–1104. doi: 10.1021/jf60213a046
Raney, J. A. (2012). Transcriptome analysis of drought induced stress in Chenopodium quinoa. Am. J. Plant Sci. 05 (03), 338‒357. doi: 10.4236/ajps.2014.53047
Rao, B., Nagasampige, M. K., Ravikiran, M. (2011). Evaluation of nutraceutical properties of selected small millets. J. Pharm. BioAllied Sci. 3 (2), 277–279. doi: 10.4103/0975-7406.80775
Rastogi, A., Shukla, S. (2013). Amaranth: a new millennium crop of nutraceutical values. Crit. Rev. in Food Sci. Nutr. 53, 109–125. doi: 10.1080/10408398.2010.517876
Ray, D. K., West, P. C., Clark, M., Gerber, J. S., Prishchepov, A. V., Chatterjee, S. (2019). Climate change has likely already affected global food production. PloS One 14, e0217148. doi: 10.1371/journal.pone.0217148
Reddy, I. N. B. L., Reddy, D. S., Narasu, M. L., Sivaramakrishnan, S. (2011). Characterization of disease resistance gene homologues isolated from finger millet (Eleusine coracana L. Gaertn). Mol. Breed. 27, 315–328. doi: 10.1007/s11032-010-9433-1
Reddy, I. N. B. L., Narasu, M. L., Sivaramakrishnan, S. (2012). Identification and characterization of EST-SSRs in finger millet (Eleusine coracana (L.) Gaertn.). J. Crop Sci. Biotechnol. 15, 9–16. doi: 10.1007/s12892-011-0064-9
Ren, X., Chen, J., Molla, M. M., Wang, C., Diao, X., Shen, Q. (2016). In vitro starch digestibility and in vivo glycemic response of foxtail millet and its products. Food Funct. 7 (1), 372–379. doi: 10.1039/C5FO01074H
Repo-Carrasco, R., Espinoza, C., Jacobsen, S.-E. (2003). Nutritional value and use of the Andean crops quinoa (Chenopodium quinoa) and kañiwa (Chenopodium pallidicaule). Food Rev. Int. 19, 179–189. doi: 10.1081/FRI-120018884
Repo-Carrasco-Valencia, R. A., Encina, C. R., Binaghi, M. J., Greco, C. B., Ronayne de Ferrer, P. A. (2010). Effects of roasting and boiling of quinoa, kiwicha and kañiwa on composition and availability of minerals in vitro. J. Sci. Food Agric. 90, 2068–2073. doi: 10.1002/jsfa.4053
Rodríguez, L. A., Isla, M. T. (2009). Comparative analysis of genetic and morphologic diversity among quinoa accessions (Chenopodium quinoa Willd.) of the South of Chile and highland accessions. J. Plant Breed. Crop Sci. 1, 210–216.
Rojas, W., Barriga, P., Figueroa, H. (2000). Multivariate analysis of the genetic diversity of Bolivian quinua germplasm. Plant Genet. Resour. Newslett. 122, 16–23.
Rojas, W., Pinto, M., Soto, J. L., Alcocer, E. (2010). “Valor Nutricional, Agroindustrial y Funcional de Los Granos Andinos,” in Granos Andinos: Avances, Logros y Experiencias Desarrolladas en Quinua, Cañahua y Amaranto en Bolivia (Rome, Italy: Bioversity International), 151–164.
Ruales, J., Nair, B. M. (1993a). Content of fat, vitamins and minerals in quinoa (Chenopodium quinoa, Willd) seeds. Food Chem. 48, 131–136. doi: 10.1016/0308-8146(93)90047-J
Ruales, J., Nair, B. M. (1993b). Saponins, phytic acid, tannis and protease inhibitors in quinoa (Chenopodium quinoa Willd) seeds. Food Chem. 48 (2), 137–143. doi: 10.1016/0308-8146(93)90048-K
Ruas, P. M., Bonifacio, A., Ruas, C. F., Fairbanks, D. J., Andersen, W. R. (1999). Genetic relationship among 19 accessions of six species of Chenopodium L. by random amplified polymorphic DNA fragments (RAPD). Euphytica 105, 25–32. doi: 10.1023/A:1003480414735
Ruiz, K. B., Khakimov, B., Søren, B. E., Søren, B., Stefania, B., Sven-Erik, J. (2017). Quinoa seed coats as an expanding and sustainable source of bioactive compounds: an investigation of genotypic diversity in saponin profiles. Ind. Crops Prod. 104, 156–163. doi: 10.1016/j.indcrop.2017.04.007
Ruiz-Carrasco, K., Antognoni, F., Coulibaly, A. K., Lizardi, S., Covarrubias, A., Martínez, E. A., et al. (2011). Variation in salinity tolerance of four lowland genotypes of quinoa (Chenopodium quinoa Willd.) as assessed by growth, physiological traits, and sodium transporter gene expression. Plant Physiol. Biochem. 49, 1333–1341. doi: 10.1016/j.plaphy.2011.08.005
Saha, D., Gowda, M. C., Arya, L., Verma, M., Bansal, K. C. (2016). Genetic and genomic resources of small millets. Crit. Rev. In Plant Sci. 35, 56–79. doi: 10.1080/07352689.2016.1147907
Saleh, A. S. M., Zhang, Q., Chen, J., Shen, Q. (2013). Millet grains: nutritional quality, processing, and potential health benefits. Compr. Rev. In Food Sci. Food Saf. 12 (3), 281‒295. https://onlinelibrary.wiley.com/doi/abs/10.1111/1541-4337.12012.
Sauer, J. D. (1967). The grain amaranths and their relatives: a revised taxonomic and geographic survey. Ann. Missouri Bot. Garden 54, 103–137. doi: 10.2307/2394998
Saunders, R. M., Becker, R. (1984). Amaranthus: a potential food and feed resource. Adv. In Cereal Sci. Technol. (USA), 6, 357–396.
Schlick, G., Bubenheim, D. L. (1996). Quinoa: Candidate crop for NASA's Controlled Ecological Life Support Systems. 632–640 In: Ed. Janick, J., Janick Progress in new crops. (Arlington, VA: ASHS Press).
Schontz, D., Rether, B. (1999). Genetic variability in foxtail millet, Setaria italica (L.) P. Beauv.: identification and classification of lines with RAPD markers. Plant Breed. 118, 190–192. doi: 10.1046/j.1439-0523.1999.118002190.x
Seenappa, M. (1988). “Sorghum and Millets in East Africa with Reference to Their Use in Weaning Foods,” in Proceedings of a Workshop Held in Nairobi, Kenya, 12-16 October 1987, 39–54. Proceedings Series/IDRC (Nairobi, Kenya: International Development Research Centre, IDRC).
Selvam, J. N., Muthukumar, M., Rahman, H., Senthil, N., Raveendran, M. (2015). Development and validation of SSR markers in finger millet (Eleusine coracana Gaertn). Int. J. Trop. Agric. 33, 2055–2066.
Senft, J. P. (1979). “Protein quality of amaranth grain,” in Proceeding of second amaranth conference (Emmaus, PA: Rodale Press), 43‒47.
Serna-Saldivar, S. O., Gutiérrez-Uribe, J. A., García-Lara, S. (2015). “Phytochemical Profiles and Nutraceutical Properties of Corn and Wheat Tortillas,” in Tortillas. Eds. Rooney, L. W., Serna-Saldivar, S. O. (Elsevier Inc: (AACC) International Press), 65–96. doi: 10.1016/B978-1-891127-88-5.50003-7
Sharma, T., Jana, S. (2002a). Species relationships in Fagopyrum revealed by PCR-based DNA fingerprinting. Theor. Appl. Genet. 105, 306–312. doi: 10.1007/s00122-002-0938-9
Sharma, T. R., Jana, S. (2002b). Random amplified polymorphic DNA (RAPD) variation in Fagopyrum tataricum Gaertn. accessions from China and the Himalayan region. Euphytica 127, 327–333.
Shejawale, D. D., Hymavathi, T. V., Manorama, K., Zabeen, F. (2016). Effect of processing on nutraceutical properties of foxtail millet (Setaria italica) varieties grown in India. J. Food Meas. Charact. 10 (1), 16–23. doi: 10.1007/s11694-015-9271-2
Sheu, J. R., Hsiao, G., Chou, P. H., Shen, M. Y., Chou, D. S. (2004). Mechanisms involved in the antiplatelet activity of rutin, a glycoside of the flavonol quercetin, in human platelets. J. Agric. Food Chem. 52 (14), 4414–4418. doi: 10.1021/jf040059f
Shi, Z., Han, C., Huang, K. (2011). Research on Se content of different tartary buckwheat genotypes. Agric. Sci. Technol.-Hunan 12, 102–156.
Shibairo, S. I., Nyongesa, O., Onwonga, R., Ambuko, J. (2014). Variation of nutritional and anti-nutritional contents in finger millet (Eleusine coracana (L.) Gaertn) genotypes. J. Agric. Vet. Sci. 7 (11), 6–12. doi: 10.9790/2380-071110612
Shukla, S., Bhargava, A., Chatterjee, A., Srivastava, J., Singh, N., Singh, S. P. (2006). Mineral profile and variability in vegetable amaranth (Amaranthus tricolor). Plant Foods Hum. Nutr. 61, 21–26. doi: 10.1007/s11130-006-0004-x
Singh, N., David, J., Thompkinson, D. K., Seelam, B. S., Rajput, H., Morya, S. (2018). Effect of roasting on functional and phytochemical constituents of finger millet (Eleusine coracana L.). Pharma Innovation J. 7, 414–418.
Skrabanja, V., Kreft, I. (1998). Resistant starch formation following autoclaving of buckwheat (Fagopyrum esculentum Moench) groats: an in vitro study. J. Agric. Food Chem. 46 (5), 2020–2023. doi: 10.1021/jf970756q
Skrabanja, V., Helle, N. L., Kreft, I. (2000). Protein-polyphenol interactions and in vivo digestibility of buckwheat groat proteins. Pflügers Archiv - Eur. J. Physiol. 440, 129–131. doi: 10.1007/s004240000033
Skrabanja, V., Liljeberg Elmståhl, H. G., Kreft, I., Björck, I. M. (2001). Nutritional properties of starch in buckwheat products: studies in vitro and in vivo. J. Agric. Food Chem. 49, 490–496. doi: 10.1021/jf000779w
Spehar, C. R. (2007). Quinoa: Alternativa Para a Diversificação Agrícola e Alimentar. Eds. Planaltina, D. F. (Embrapa Cerrados, Brasil: Técnico). 103 pp.
Sripriya, G., Antony, U., Chandra, T. S. (1997). Changes in carbohydrate, free amino acids, organic acids, phytate and HCl extractability of minerals during germination and fermentation of finger millet (Eleusine coracana). Food Chem. 58 (4), 345–350. doi: 10.1016/S0308-8146(96)00206-3
Steadman, K. J., Burgoon, M. S., Lewis, B. A., Edwardson, S. E., Obendorf, R. L. (2001a). Buckwheat seed milling fractions: description, macronutrient composition and dietary fibre. J. Cereal Sci. 33, 271–278. doi: 10.1006/jcrs.2001.0366
Steadman, K. J., Burgoon, M. S., Lewis, B. A., Edwardson, S. E., Obendorf, R. L. (2001b). Minerals, phytic acid, tannin and rutin in buckwheat seed milling fractions. J. Sci. Food Agric. 81, 1094–1100. doi: 10.1002/jsfa.914
Stetter, M. G., Schmid, K. J. (2017). Analysis of phylogenetic relationships and genome size evolution of the Amaranthus genus using GBS indicates the ancestors of an ancient crop. Mol. Phylogenet. Evol. 109, 80–92. doi: 10.1016/j.ympev.2016.12.029
Stevens, M. R., Coleman, C. E., Parkinson, S. E., Maughan, P. J., Zhang, H. B., Balzotti, M. R., et al. (2006). Construction of a quinoa (Chenopodium quinoa Willd.) BAC library and its use in identifying genes encoding seed storage proteins. Theor. Appl. Genet. 112, 1593–1600. doi: 10.1007/s00122-006-0266-6
Stikic, R., Glamoclija, D., Demin, M., Vucelic-Radovic, B., Jovanovic, Z., Milojkovic-Opsenica, D., et al. (2012). Agronomical and nutritional evaluation of quinoa seeds (Chenopodium quinoa Willd.) as an ingredient in bread formulations. J. Cereal Sci. 55, 132–138. doi: 10.1016/j.jcs.2011.10.010
Stokić, E., Mandić, A., Sakač, M., Mišan, A., Pestorić, M., Šimurina, O., et al. (2015). Quality of buckwheat-enriched wheat bread and its antihyperlipidemic effect in statin treated patients. LWT-Food Sci. Technol. 63, 556–561. doi: 10.1016/j.lwt.2015.03.023
Subba Rao, M.V.S.S.T., Muralikrishna, G. (2002). Evaluation of the antioxidant properties of free and bound phenolic acids from native and malted finger millet (Ragi, Eleusine coracana Indaf-15). J. Agric. Food Chem. 50 (4), 889–892. doi: 10.1021/jf011210d
Subramanian, D., Gupta, S. (2016). Pharmacokinetic study of amaranth extract in healthy humans: a randomized trial. Nutrition 32, 748–753. doi: 10.1016/j.nut.2015.12.041
Sugiyama, K., Kushima, Y., Muramatsu, K. (1985). Effects of sulfur-containing amino acids and glycine on plasma cholesterol level in rats fed on a high cholesterol diet. Agric. Biol. Chem. 49, 3455–3461.
Sunil, M., Hariharan, A. K., Nayak, S., Gupta, S., Nambisan, S. R., Gupta, R. P., et al. (2014). The draft genome and transcriptome of Amaranthus hypochondriacus: a C4 dicot producing high-lysine edible pseudo-cereal. DNA Res. 21, 585–602. doi: 10.1093/dnares/dsu021
Suresh, S., Chung, J. W., Cho, G. T., Sung, J. S., Park, J. H., Gwag, J. G., et al. (2014). Analysis of molecular genetic diversity and population structure in Amaranthus germplasm using SSR markers. Plant Biosyst. Int. J. Dealing All Aspects Plant Biol. 148, 635–644. doi: 10.1080/11263504.2013.788095
Taira, H., Miyahara, T. (1983). Amylose content of foxtail millet [Setaria italica] starch (Japan: Report of National Food Research Institute).
Taira, H. (1984). Lipid content and fatty acid composition of nonglutinous and glutinous varieties of foxtail millet. J. Agric. Food Chem. 32, 369–371. doi: 10.1021/jf00122a047
Tang, S., Li, L., Wang, Y., Chen, Q., Zhang, W., Jia, G., et al. (2017). Genotype-specific physiological and transcriptomic responses to drought stress in Setaria italica (an emerging model for Panicoideae grasses). Sci. Rep. 7, 10009. doi: 10.1038/s41598-017-08854-6
Tatala, S., Ndossi, G., Ash, D., Mamiro, P. (2007). Effect of germination of finger millet on nutritional value of foods and effect of food supplement on nutrition and anaemia status in Tanzania children. Tanzania Health Res. Bull. 9, 77–86. doi: 10.4314/thrb.v9i2.14308
Teng, X. L., Chen, N., Xiao, X.-G. (2016). Identification of a catalase-phenol oxidase in betalain biosynthesis in red amaranth (Amaranthus cruentus). Front. Plant Sci. 6, 1228. doi: 10.3389/fpls.2015.01228
Tomotake, H., Shimaoka, I., Kayashita, J., Yokoyama, F., Nakajoh, M., Kato, N. (2000). A buckwheat protein product suppresses gallstone formation and plasma cholesterol more strongly than soy protein isolate in hamsters. J. Nutr. 130 (7), 1670‒1674. doi: 10.1093/jn/130.7.1670
Tomotake, H., Shimaoka, I., Kayashita, J., Nakajoh, M., Kato, N. (2002). Physicochemical and functional properties of buckwheat protein product. J. Agric. Food Chem. 50, 2125–2129. doi: 10.1021/jf011248q
Tripathi, B., Patel, K. (2010). Finger millet (Eleucine coracana) flour as a vehicle for fortification with zinc. J. Trace Elem. In Med. Biol. 24 (1), 46–51. doi: 10.1016/j.jtemb.2009.09.001
Trivedi, A. K., Arya, L., Verma, S. K., Tyagi, R. K., Hemantaranjan, A., Verma, M., et al. (2018). Molecular profiling of foxtail millet (Setaria italica (L.) P. Beauv) from Central Himalayan Region for genetic variability and nutritional quality. J. Agric. Sci. 156, 333–341. doi: 10.1017/S0021859618000382
Tsehaye, Y., Berg, T., Tsegaye, B., Tanto, T. (2006). Farmers' management of finger millet (Eleusine coracana L.) diversity in Tigray, Ethiopia and implications for on-farm conservation. Biodivers. Conserv. 15, 4289–4308. doi: 10.1007/s10531-005-3581-3
Udeh, H., Gyebi, D., Jideani, A. (2017). Finger millet bioactive compounds, bioaccessibility, and potential health effects: a review. Czech J. Food Sci. 35, 7–17. doi: 10.17221/206/2016-CJFS
Ulbricht, C., Abrams, T., Conquer, J., Costa, D., Grimes Serrano, J. M., Taylor, S., et al. (2009). An evidence-based systematic review of amaranth (Amaranthus spp.) by the natural standard research collaboration. J. Diet. Suppl. 6, 390–417. doi: 10.3109/19390210903280348
Umeta, M., West, C. E., Fufa, H. (2005). Content of zinc, iron, calcium and their absorption inhibitors in foods commonly consumed in Ethiopia. J. Food Compos. Anal. 18, 803–817. doi: 10.1016/j.jfca.2004.09.008
USDA National Nutrient Database for Standard Reference(2019). https://fdc.nal.usda.gov/. (accessed on 28 Oct 2019).
Vadivoo, A. S., Joseph, R., Ganesan, N. M. (1998). Genetic variability and diversity for protein and calcium contents in finger millet (Eleusine coracana (L.) Gaertn.) in relation to grain color. Plant Foods Hum. Nutr. 52, 353–364. doi: 10.1023/A:1008074002390
Valencia, U. S., Sandberg, A. S., Ruales, J. (1999). Processing of quinoa (Chenopodium quinoa, Willd): effects on in vitro iron availability and phytate hydrolysis. Int. J. Food Sci. Nutr. 50 (3), 203‒211. doi: 10.1080/096374899101247
Vega-Gálvez, A., Miranda, M., Vergara, J., Uribe, E., Puente, L., Martínez, E. A. (2010). Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: a review. J. Sci. Food Agric. 90, 2541–2547. doi: 10.1002/jsfa.4158
Walsh, B. M., Adhikary, D., Maughan, P. J., Emshwiller, E., Jellen, E. N. (2015). Chenopodium polyploidy inferences from Salt Overly Sensitive 1 (SOS1) data. Am. J. Bot. 102, 533–543. doi: 10.3732/ajb.1400344
Walters, H., Carpenter-Boggs, L., Desta, K., Yan, L., Matanguihan, J., Murphy, K. (2016). Effect of irrigation, intercrop, and cultivar on agronomic and nutritional characteristics of quinoa. Agroecol. Sustain. Food Syst. 40, 783–803. doi: 10.1080/21683565.2016.1177805
Wang, J., Liu, Z., Fu, X., Run, M. A. (1992). A clinical observation on the hypoglycaemic effect of Xinjiang buckwheat. In: Proceedings of the 5th International Symposium on Buckwheat. Eds. Lin, R., Zhou, M., Tao, Y., Li, J., Zhang, Z.. (Beijing, China: Agriculture Publishing House), 465–467
Wang, Z. M., Devos, K. M., Liu, C. J., Wang, R. Q., Gale, M. D. (1998). Construction of RFLP-based maps of foxtail millet, Setaria italica (L.) P. Beauv. Theor. Appl. Genet. 96, 31–36. doi: 10.1007/s001220050705
Wang, Z. H., Gao, L., Li, Y. Y., Zhang, Z., Yuan, J. M., Wang, H. W., et al. (2007). Induction of apoptosis by buckwheat trypsin inhibitor in chronic myeloid leukemia K562 cells. Biol. Pharm. Bull. 30 (4), 783–786. doi: 10.1248/bpb.30.783
Whittaker, P., Ologunde, M. O. (1990). Study on iron bioavailability in a native Nigerian grain amaranth cereal for young children, using a rat model. Cereal Chem. 67, 505–508.
Williams, J. T., Brenner, D. (1995). “Grain amaranth (Amaranthus species),” in Underutilized Crops: Cereals and Pseudocereals. Eds. Williams, J. T. (London, UK: Chapman & Hall), 129‒187.
Wilson, H. D. (1988a). Allozyme variation and morphological relationships of Chenopodium hircinum. Syst. Bot. 13, 215–228. doi: 10.2307/2419100
Wilson, H. D. (1988b). Quinua biosystematics I: domesticated populations. Economic Bot. 42, 461–477. doi: 10.1007/BF02862791
Wu, Q., Bai, X., Zhao, W., Xiang, D., Wan, Y., Yan, J., et al. (2017). De novo assembly and analysis of tartary buckwheat (Fagopyrum tataricum Gaertn.) transcriptome discloses key regulators involved in salt-stress response. Genes 8, 255. doi: 10.3390/genes8100255
Xu, F., Sun, M. (2001). Comparative analysis of phylogenetic relationships of grain amaranths and their wild relatives (Amaranthus; Amaranthaceae) using internal transcribed spacer, amplified fragment length polymorphism, and double-primer fluorescent intersimple sequence repeat markers. Mol. Phylogenet. Evol. 21, 372–387. doi: 10.1006/mpev.2001.1016
Xu, J. M., Fan, W., Jin, J. F., Lou, H. Q., Chen, W. W., Yang, J. L., et al. (2017). Transcriptome analysis of Al-induced genes in buckwheat (Fagopyrum esculentum Moench) root apex: new insight into Al toxicity and resistance mechanisms in an Al accumulating species. Front. Plant Sci. 8, 1141. doi: 10.3389/fpls.2017.01141
Yabe, S., Hara, T., Ueno, M., Enoki, H., Kimura, T., Nishimura, S., et al. (2014). Rapid genotyping with DNA micro-arrays for high-density linkage mapping and QTL mapping in common buckwheat (Fagopyrum esculentum Moench). Breed. Sci. 64, 291–299. doi: 10.1270/jsbbs.64.291
Yasui, Y., Wang, Y., Ohnishi, O., Campbell, C. G. (2004). Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self-pollinated relative Fagopyrum homotropicum. Genome 47, 345–351. doi: 10.1139/g03-126
Yasui, Y., Mori, M., Matsumoto, D., Ohnishi, O., Campbell, C. G., Ota, T. (2008). Construction of a BAC library for buckwheat genome research. Genes Genet. Syst. 83, 393–401. doi: 10.1266/ggs.83.393
Yasui, Y., Hirakawa, H., Oikawa, T., Toyoshima, M., Matsuzaki, C., Ueno, M., et al. (2016). Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid crop with great environmental adaptability and outstanding nutritional properties. DNA Res. 23, 535–546. doi: 10.1093/dnares/dsw037
Zduńczyk, Z., Flis, M., Zieliński, H., Wróblewska, M., Antoszkiewicz, Z., Juśkiewicz, J. (2006). In vitro antioxidant activities of barley, husked oat, naked oat, triticale, and buckwheat wastes and their influence on the growth and biomarkers of antioxidant status in rats. J. Agric. Food Chem. 54, 4168–4175. doi: 10.1021/jf060224m
Zhang, Z., Upadhayay, M. P., Zhao, Z., Lin, R., Zhou, M. D., Rao, V. R., et al. (2004). “Conservation and use of buckwheat biodiversity for the livelihood development,” in Proceedings of the 9th International Symposium on Buckwheat, Prague, Czech Republic, August 18-22, 2004, Czech Republic: Research Institute of Crop Production 442‒431.
Zhang, J., Liu, T., Fu, J., Zhu, Y., Jia, J., Zheng, J., et al. (2007). Construction and application of EST library from Setaria italica in response to dehydration stress. Genomics 90, 121–131. doi: 10.1016/j.ygeno.2007.03.016
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549. doi: 10.1038/nbt.2195
Zhang, G., Xu, Z., Gao, Y., Huang, X., Zou, Y., Yang, T. (2015). Effects of germination on the nutritional properties, phenolic profiles, and antioxidant activities of buckwheat. J. Food Sci. 80 (5), H1–H9. doi: 10.1111/1750-3841.12830
Zhang, L., Li, X., Ma, B., Gao, Q., Du, H., Han, Y., et al. (2017). The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol. Plant 10, 1224–1237. doi: 10.1016/j.molp.2017.08.013
Zhang, T., Gu, M., Liu, Y., Lv, Y., Zhou, L., Lu, H., et al. (2017). Development of novel InDel markers and genetic diversity in Chenopodium quinoa through whole-genome re-sequencing. BMC Genomics 18, 685. doi: 10.1186/s12864-017-4093-8
Zhou, M., Zhang, Z. (1995). Directory of germplasm collections – Buckwheat (Beijing, 26 pp: International Plant Genetic Resources Institute).
Zhou, M., Kreft, I., Suvorova, G., Tang, Y., Woo, S. H. (Eds.) (2018). Buckwheat germplasm in the world (Academic Press).
Zhou, M., Kreft, I., Woo, S. H., Chrungoo, N., Wieslander, G. (2016). Molecular breeding and nutritional aspects of buckwheat (Academic Press). 482 p.
Zielińska, D., Turemko, M., Kwiatkowski, J., Zieliński, H. (2012). Evaluation of flavonoid contents and antioxidant capacity of the aerial parts of common and tartary buckwheat plants. Molecules 17, 9668–9682. doi: 10.3390/molecules17089668
Zielinski, H., Michalska, A., Amigo-Benavent, M., del Castillo, M. D., Piskula, M. K. (2009). Changes in protein quality and antioxidant properties of buckwheat seeds and groats induced by roasting. J. Agric. Food Chem. 57, 4771–4776. doi: 10.1021/jf900313e
Zou, C., Chen, A., Xiao, L., Muller, H. M., Ache, P., Haberer, G., et al. (2017). A high-quality genome assembly of quinoa provides insights into the molecular basis of salt bladder-based salinity tolerance and the exceptional nutritional value. Cell Res. 27, 1327–1340. doi: 10.1038/cr.2017.124
Keywords: orphan crops, nutrient-rich crops, pseudo-cereals, molecular profiles, food and nutritional security, healthy crops, marginal environment
Citation: Rodríguez JP, Rahman H, Thushar S and Singh RK (2020) Healthy and Resilient Cereals and Pseudo-Cereals for Marginal Agriculture: Molecular Advances for Improving Nutrient Bioavailability. Front. Genet. 11:49. doi: 10.3389/fgene.2020.00049
Received: 07 November 2019; Accepted: 16 January 2020;
Published: 27 February 2020.
Edited by:
Felipe Klein Ricachenevsky, Universidade Federal de Santa Maria, BrazilReviewed by:
Joel B. Mason, Tufts University School of Medicine, United StatesCopyright © 2020 Rodríguez, Rahman, Thushar and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh K. Singh, ci5zaW5naEBiaW9zYWxpbmUub3JnLmFl; cmtzaW5naGlycmlAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.