
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet., 23 January 2020
Sec. Cancer Genetics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.01375
This article is part of the Research TopicThe Role of ncRNAs in Solid Tumors Prognosis: From Laboratory To Clinical UtilityView all 49 articles
 Yishu Li1†
Yishu Li1† Mengyu Du2†
Mengyu Du2† Shengsheng Wang1†
Shengsheng Wang1† Jin Zha2
Jin Zha2 Peijie Lei3
Peijie Lei3 Xueqi Wang4
Xueqi Wang4 Di Wu2
Di Wu2 Jianhua Zhang4
Jianhua Zhang4 Denggang Chen1
Denggang Chen1 Dong Huang1
Dong Huang1 Jing Lu5
Jing Lu5 Heng Li1
Heng Li1 Min Sun1,2,6*
Min Sun1,2,6*Background: SOX2 overlapping transcript (SOX2-OT) produces alternatively spliced long non-coding RNAs (lncRNA). Previous studies of the prognostic role of SOX2-OT expression met with conflicting results. The aim of this study was to properly consider the prognostic role of SOX2-OT expression in several cancers. In addition, the regulative mechanism of SOX2-OT is explored.
Methods: PubMed, EMBASE, and Cochrane Library and The Cancer Genome Atlas (TCGA) database were comprehensively explored to recover pertinent studies. We conducted an extensive inquiry to verify the implication of SOX2-OT expression in cancer patients by conducting a meta-analysis of 13 selected studies. Thirty-two TCGA databases were used to analyze the connection between SOX2-OT expression and both the overall survival (OS) and clinicopathological characteristics of cancer patients using R and STATA 13.0. Trial sequential analysis (TSA) was adopted in order to compute the studies’ power.
Results: Thirteen studies involving 1172 cancer patients and 32 TCGA cancer types involving 9676 cancer patients were eventually selected. Elevated SOX2-OT expression was significantly related to shorter OS (HR = 2.026, 95% CI: 1.691–2.428, P < 0.0001) and disease-free survival (DFS) (HR = 2.554, 95% CI: 1.261–5.174, P = 0.0092) in cancer patients. Meanwhile, TSA substantiated adequate power to demonstrate the relationship between SOX2-OT expression and OS. The cancer patients with elevated SOX2-OT expression were more likely to have advanced clinical stage (RR = 1.468, 95% CI: 1.106–1.949, P = 0.0079), earlier lymphatic metastasis (P = 0.0005), earlier distant metastasis (P < 0.0001), greater tumor size (P < 0.0001), and more extreme tumor invasion (P < 0.0001) compared to those with low SOX2-OT expression. Meta-regression and subgroup analysis revealed that follow-up time, sample type, and tumor type could significantly contribute to heterogeneity for survival outcomes. The follow-up time could significantly explain heterogeneity for tumor, node, metastasis (TNM) stage. Furthermore, up to 500 validated target genes were distinguished, and the gene oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses demonstrated that the validated targets of SOX2-OT were substantially enriched in cell adhesion, mRNA binding, and mRNA surveillance pathways.
Conclusions: Elevated expression of SOX2-OT predicted a poor OS and DFS. Overexpression of SOX2-OT was correlated with more advanced tumor stage, earlier lymphatic metastasis, earlier distant metastasis, larger tumor size, and deeper tumor invasion. SOX2-OT-mediated cell adhesion, mRNA binding, or mRNA surveillance could be intrinsic mechanisms for invasion and metastasis.
SOX2 overlapping transcript (SOX2-OT) is a long non-coding RNA located in 3q26.33 locus. Its third intron harbors SOX2 gene which encodes the transcription factor SOX2, an established pluripotency state modulator (Avilion et al., 2003; Fong et al., 2008; Han et al., 2018). Several studies revealed that SOX2-OT levels were consistently positively correlated with SOX2 levels. SOX2-OT plays a role in proliferation of cells and SOX2 regulation (Amaral et al., 2009; Hou et al., 2014; Shahryari et al., 2014; Shahryari et al., 2015).
It has been shown that lncRNA SOX2-OT is overexpressed in a number of human cancers as an oncogene promoting tumorigenesis and cancer progression, including ovarian cancer, breast cancer, pancreatic ductal adenocarcinoma, cholangiocarcinoma, hepatocellular carcinoma, esophageal squamous cell carcinoma, osteosarcoma, non-small cell lung cancer, and gastric cancer (Iranpour et al., 2016; Zhang et al., 2016; Zou et al., 2016; Wang et al., 2017a; Wang et al., 2017b; Han et al., 2018; Li et al., 2018a; Li et al., 2018b; Sun et al., 2018; Tian et al., 2018; Wei et al., 2018; Xie et al., 2018). SOX2-OT is co-upregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma and potentially involved in maintaining the pluripotent state of stem cells (Shahryari et al., 2014). Although these articles established the critical role of lncRNA SOX2-OT expression in some cancers, the prognostic value of SOX2-OT expression in numerous other cancers remained uncharacterized (Shahryari et al., 2015; Castro-Oropeza et al., 2018; Farhangian et al., 2018). In addition, inconsistent results were obtained in several studies on the association between SOX2-OT expression and clinical features such as tumor size, clinical stage, and tumor invasion (Shi and Teng, 2015; Zou et al., 2016; Wang et al., 2017a; Li et al., 2018b; Sun et al., 2018).
The evidence above showed that SOX2-OT is involved in tumor progression. Moreover, an earlier meta-analysis study published in 2018 had revealed that the overexpression of SOX2-OT was significantly correlated with the overall survival (OS), clinical stage, lymph node metastasis, distant metastasis, and tumor differentiation of cancers (Song et al., 2018). However, the sample size of the study was restricted, and the relationship between SOX2-OT and other clinicopathological characteristics was not explored (Song et al., 2018). As described below, we have conducted a more comprehensive trial sequential analysis (TSA) on the applicable literature and searched The Cancer Genome Atlas (TCGA) database to study the prognostic value of SOX2-OT in patients with several types of cancer. We additionally explored the potential target genes of SOX2-OT through gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, and the potential mechanisms of SOX2-OT in tumor progression are also discussed.
Studies on the prognostic roles of SOX2-OT in cancer patients that were published as of October 1st, 2019 were extracted from the electronic databases PubMed, EMBASE, and Cochrane Library using the terms (1) “SOX2-OT” OR “NCRNA00043” OR “SOX2OT” OR “SOX2 overlapping transcript” OR “SRY-box transcription factor 2 overlapping transcript” AND (2) “tumor OR cancer OR carcinoma OR neoplasm OR metastasis”. The search strategies are illustrated in Supplementary Table 1. The search and selection of articles for the study were conducted as described previously (Sun et al., 2019).
Studies entering this analysis met these requirements: (1) definitive diagnosis or histopathological confirmation for patients with cancer; (2) the expression of SOX2-OT must be measured by quantitative real-time polymerase chain reaction (qRT-PCR); (3) the hazard ratios (HRs) and their 95% confidence intervals (CIs) for survival parameters based on SOX2-OT expression levels were promptly available or could be calculated indirectly; and (4) the representative and accurate studies were selected to avoid unnecessary cohort overlapping. Studies that have satisfied the abovementioned inclusion requirements were further ruled out if they had any of the following features: (1) duplicated articles or data; (2) non-human studies; (3) review articles or letters; (4) articles in non-English languages.
The quality of the included studies was assessed using Newcastle-Ottawa Scale (NOS), with scores ≥ 6 considered high quality. A ‘‘star system’’ was applied for case-control studies (Supplementary Table 2).
The following information was extracted from each study: (1) first author; (2) publication year; (3) nationality, sample size, tumor type, and clinicopathological characteristics of involved patient population; (4) the assay method and cut-off value of SOX2-OT expression levels; (5) HRs of SOX2-OT expression for OS and disease-free survival (DFS). If the HRs for OS and DFS were calculated by both univariate and multivariate analyses, the latter were our first choice for these results and were adjusted for confounding factors. If a study did not report HRs, we estimated HRs and their corresponding 95% CIs using the procedure described by Parmar et al. (1998) and Tierney et al. (2007). The data of Kaplan-Meier curves were regained by Engauge Digitizer software (version 9.8, http://markummitchell.github.io/engauge-digitizer). This process was repeated three times to decrease variability. Discrepancies were resolved through discussion and review of extraction until consensus was reached on a final list of factors targeted by each study.
All the HRs and their 95% CIs were integrated to evaluate the association between SOX2-OT expression and prognosis. If the pooled HR < 1 and their 95% CI did not overlap the invalid line in the forest plot, the elevated expression of SOX2-OT predicted a good OS. The heterogeneity of the pooled results was examined via Cochrane’s Q test and Higgins’ I-squared. If P ≥ 0.1 and I2 ≤ 25%, we disregarded the influence of heterogeneity and pooled the overall result using a fixed effects model, otherwise employing the random effects model. Potential publication bias was assessed by a funnel plot and Egger’s test (Stuck et al., 1998) conducted using the “metafor” and “meta” packages of R (version 3.2.3). All of the abovementioned methods followed the Meta-analysis of Observational Studies in Epidemiology (MOOSE) Checklist.
Identification of eligible studies is summarized in Figure 1. We screened 122 articles for eligibility and identified 13 eligible studies. These eligible articles were published between 2014 and 2018 and included a total of 1172 participants who represented eight cancer types (Table 1). Most articles choose the mean and median as the cutoff value. Eight studies that used multivariate analysis of OS were included in the meta-analysis (Hou et al., 2014; Shi and Teng, 2015; Zhang et al., 2016; Zou et al., 2016; Wang et al., 2017a; Li et al., 2018a; Li et al., 2018b; Xie et al., 2018), the adjusted variables of the multivariate analysis were presented in Table 2. The other three studies provided survival curves (Zhang et al., 2017; Sun et al., 2018; Wei et al., 2018).
We carried out a meta-analysis of the association between SOX2-OT expression and OS and DFS. The results revealed that higher SOX2-OT expression predicted an unfavorable OS (n = 11, HR = 2.026, 95% CI: [1.691–2.428], P < 0.0001, I2 = 0%) (Figure 2A) and a poor DFS (n = 2, HR = 2.554, 95% CI: [1.261–5.174], P = 0.0092, I2 = 66.6%, Supplementary Figure 1, Table 3). No heterogeneity was identified according to a fixed effect model (I2 = 0%) (Figure 2A). The outcomes of publication bias analysis are listed in Table 3.
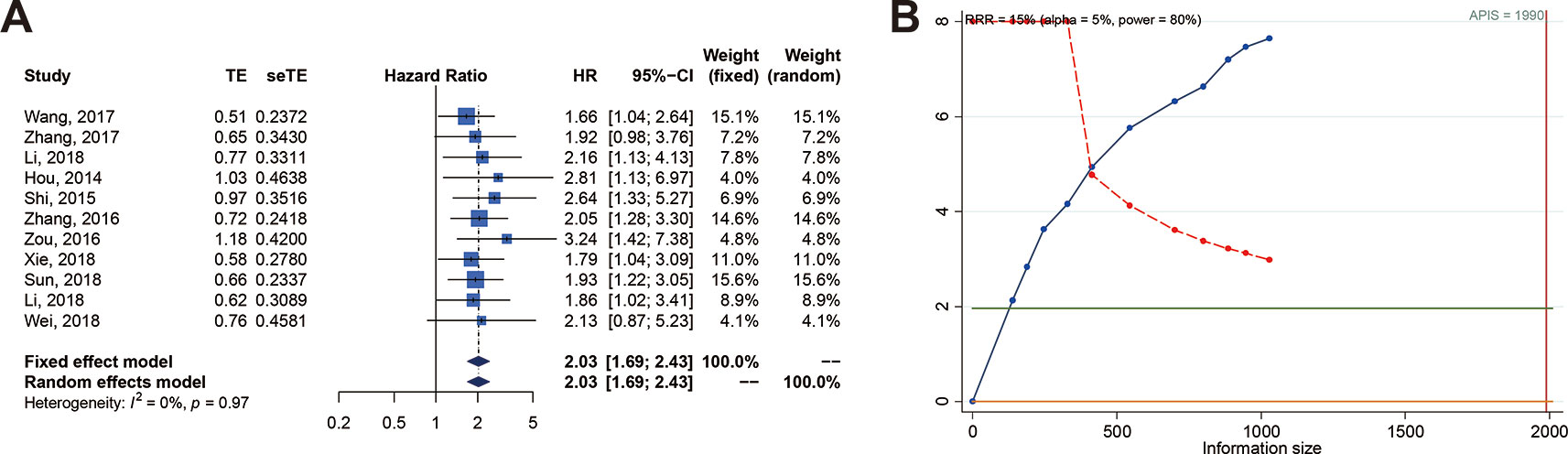
Figure 2 Relationship between SOX2 overlapping transcript (SOX2-OT) expression and overall survival (OS) in patients with various cancers. (A) Forest plot of SOX2-OT expression and OS. (B) trial sequential analysis (TSA) of 11 trials comparing OS of the high vs. low SOX2-OT expression. Heterogeneity adjustment required information size of 1990 participants calculated on basis of proportion of OS of 80%, RRR of 15%, α = 5%, β = 20%, power = 0.80, and I2 = 0%. Cumulative Z-curve crosses trial sequential monitoring boundary, showing sufficient evidence for a 15% increase in relative risk with high expression of SOX2-OT. Horizontal green lines illustrate the traditional level of statistical significance (P = 0.05).
We performed subgroup analyses of association between SOX2-OT expression and OS using 11 studies. The results showed the presence of a significant association between SOX2-OT expression and OS when the data were fully integrated from eight studies where OS was assessed with multivariate analysis (HR = 2.052, 95% CI: [1.661; 2.536], P < 0.0001, I2 = 0%) (Table 4). Furthermore, a significant relationship was revealed in the subgroup analyses for OS based on sample size (P < 0.0001), tumor type (P < 0.05), sample type (P < 0.05), and cut-off value (P < 0.01).
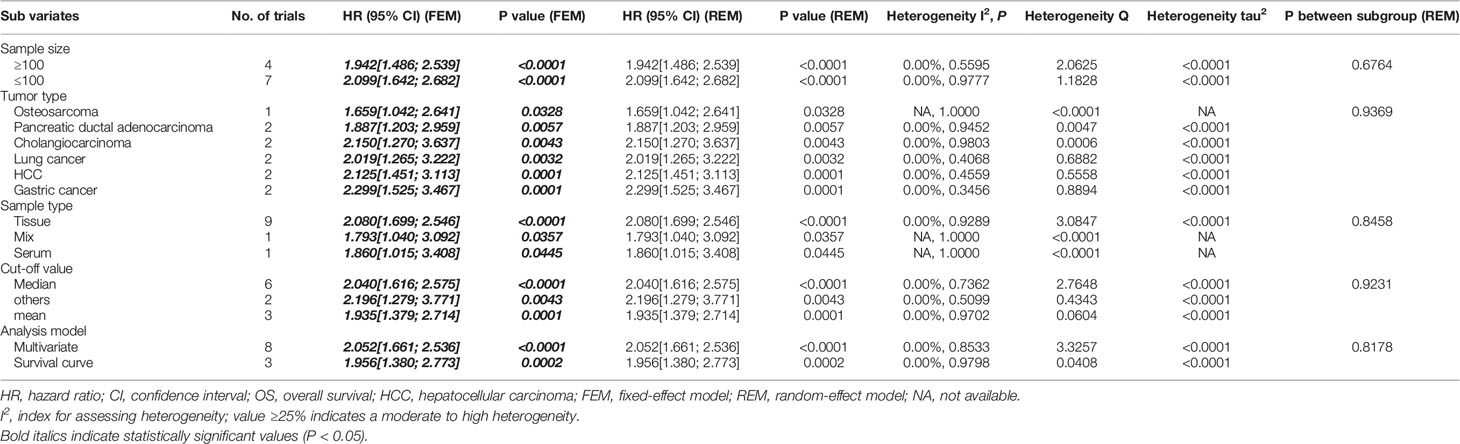
Table 4 Subgroup analysis of the association between SOX2-OT overexpression and OS in patients with different cancers.
Eight studies employed Cox multivariate analysis to survey the prognostic value of lncRNA SOX2-OT expression on the prognosis of cancer patients (Hou et al., 2014; Shi and Teng, 2015; Zhang et al., 2016; Zou et al., 2016; Wang et al., 2017a; Li et al., 2018a; Li et al., 2018b; Xie et al., 2018). An in-depth subgroup analysis is required to clearly define the values of the adjusted variables in multivariate analysis (Table 5). Subgroup analysis stratified by independent prognostic factors, such as clinical stage (P < 0.0001), lymph node metastasis (P < 0.0001), tumor differentiation (P < 0.0001), tumor size (P < 0.01), vascular invasion (P < 0.001), tumor depth (P < 0.001), distant metastasis (P < 0.0001), postoperative recurrence (P < 0.05), and smoking status (P < 0.05) (Table 5) demonstrated that a significant relationship existed between lncRNA SOX2-OT expression and OS.
We executed an analysis of the association between SOX2-OT expression and clinicopathological characteristics (Table 3). The results indicated that overexpression of SOX2-OT was significantly correlated with TNM stage. Higher SOX2-OT expression was associated with high TNM stage for several malignancies (n = 9, RR = 1.468; 95% CI: [1.106–1.949], P = 0.0079, I2 = 71.9%, Figure 3A). SOX2-OT expression was significantly correlated with lymphatic metastasis (n = 7, RR = 1.554, 95% CI: [1.211–1.994], P = 0.0005, I2 = 52.2%, Figure 3B), distant metastasis (n = 4, RR = 3.054, 95% CI: [1.866–4.999], P < 0.0001, I2 = 18.3%, Figure 3C), tumor size (n = 7, RR = 1.264, 95% CI: [1.019–1.566], P < 0.0329, I2 = 56.2%, Figure 3D), depth of tumor invasion (n = 3, RR = 1.552, 95% CI: [1.274–1.890], P < 0.0001, I2 = 0.0%, Figure 3E). However, SOX2-OT expression was not correlated with differentiation (n = 9, RR = 1.122, 95% CI: [0.800–1.573], P = 0.5062, I2 = 78.7%, Figure 3F), gender (n = 8, RR = 1.022, 95% CI: [0.921–1.134], P = 0.6798, I2 = 0.0%, Figure 3G), or age (n = 10, RR = 0.966, 95% CI: [0.821–1.138], P = 0.6812, I2 = 31.4%, Figure 3H).
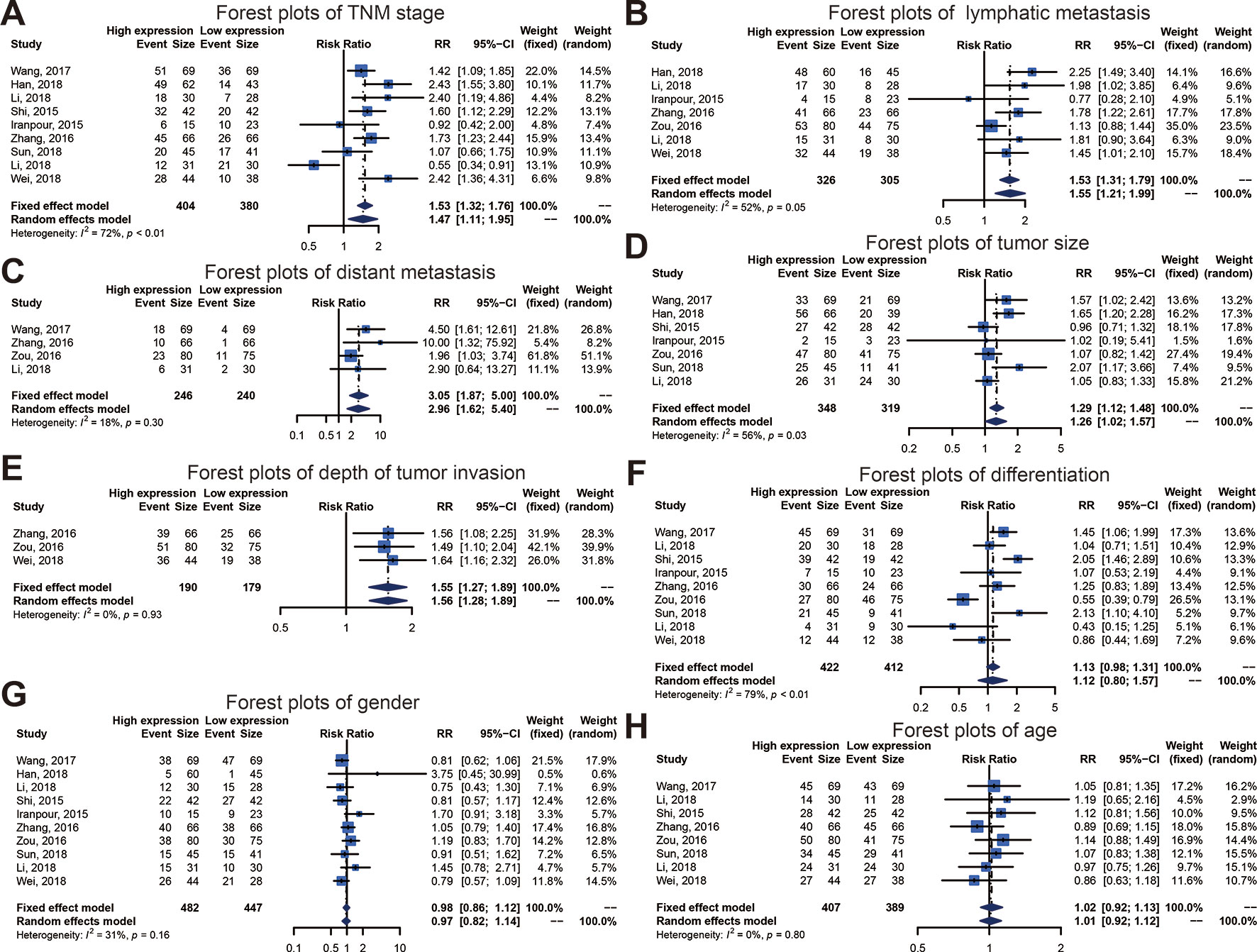
Figure 3 Forest plots of main clinical parameters under the upregulation or downregulation of SOX2 overlapping transcript (SOX2-OT). (A) tumor, node, metastasis (TNM) stage, (B) lymphatic metastasis, (C) distant metastasis, (D) tumor size, (E) depth of tumor invasion, (F) differentiation, (G) gender, and (H) age.
In order to examine the robustness of OS, the trial sequencing monitoring boundaries executed to the meta-analysis supposed a decrease in relative risk by 15%. The cumulative Z-curve crossed the trial sequential monitoring boundary for benefit, indicating that sufficient evidence exists for a 15% relative risk reduction (RRR) when SOX2-OT expression is low (Figure 2B).
Publication bias of the association between SOX2-OT expression and prognosis was inferred based on our Egger’s test (P < 0.05) (Figure 4A). No distinct biases of the correlation between SOX2-OT expression and clinicopathological characteristics were found across included studies on the basis of funnel plots and the P value of the Egger’s test (Figures 4B–I).
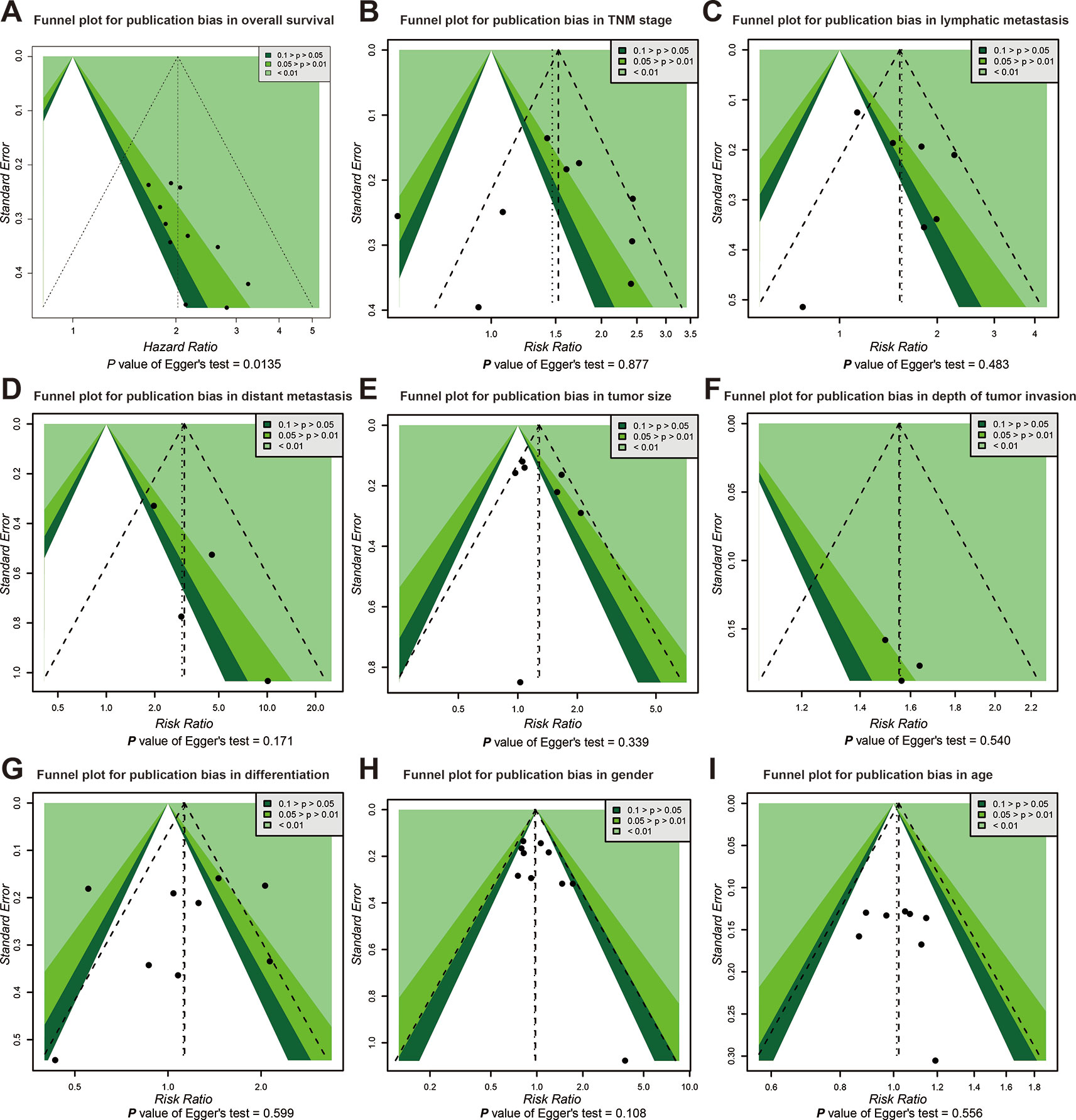
Figure 4 Funnel plot for publication bias in overall survival and clinicopathological characteristics. (A) overall survival (OS), (B) tumor, node, metastasis (TNM) stage, (C) lymphatic metastasis, (D) distant metastasis, (E) tumor size, (F) depth of tumor invasion, (G) differentiation, (H) gender, and (I) age.
To investigate the possible sources of heterogeneity, we gathered the original articles for subgroup analyses, based on various factors. Table 6 displays the outcomes of a meta-regression that examined the source of high heterogeneity for TNM stage. The follow-up time, sample type, and tumor type could significantly explain heterogeneity for survival outcomes in the post-hoc analysis (Table 6, Figure 5A). On the basis of the results of the meta-regression, we carried out a subgroup analysis on groups of patients with the follow-up time, sample type, and tumor type (Figures 5B–D). This subgroup analysis showed a significantly lower heterogeneity in the above 60 months follow-up group, the tissue group, or the Cholangiocarcinoma group, which suggested that the relationship between high SOX2-OT expression and TNM stage has stronger efficacy in these groups.
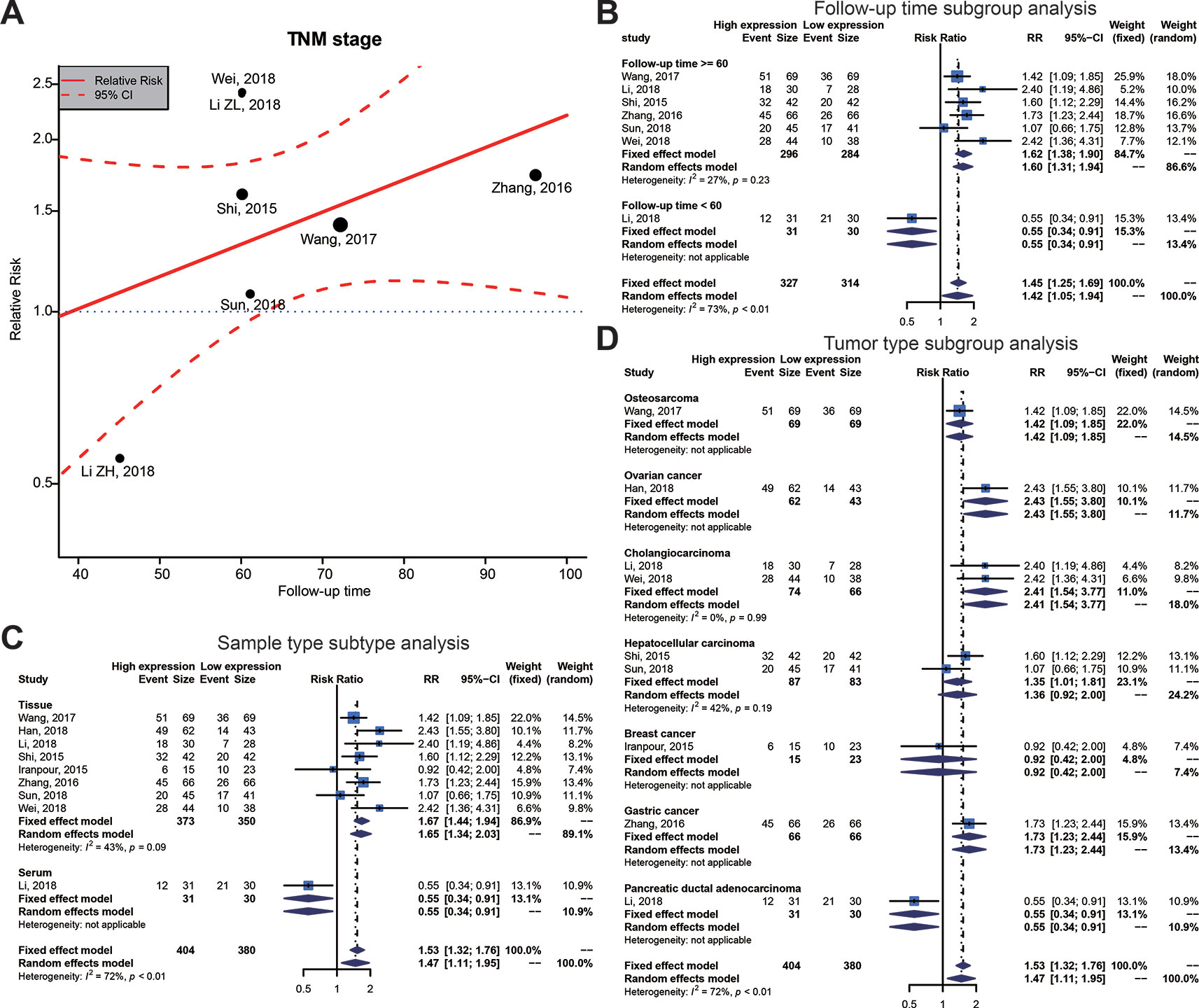
Figure 5 Meta-regression plot and subgroup analysis of tumor, node, metastasis (TNM) stage and follow-up time, sample type and tumor type. (A) Meta-regression plot correction of follow-up time and TNM stage. From the meta-regression plot correction, we determined that a follow-up time of more than 60 months correlated with higher TNM stage. The point of determination for differences in TNM stage is a follow-up time of about 60 months. (B) Follow-up time subgroup, (C) sample type subtype, and (D) tumor type subtype.
Meta-regression analysis (Supplementary Table 3) and stratified analysis (Supplementary Table 4) did not demonstrate heterogeneity between all potential factors and the other clinical parameters.
To validate the results of the meta-analysis, we employed tissue SOX2-OT expression data and the matching survival data from TCGA datasets. The results indicated that high SOX2-OT expression in tissues was not associated with worse OS in the pooled analysis of TCGA datasets for all the tumors (n = 32, HR = 1.078, 95% CI 0.922–1.262, P = 0.346, I2 = 66.3%) (Table 7, Supplementary Figure 2), which included 9676 patients with diversified types of cancer.
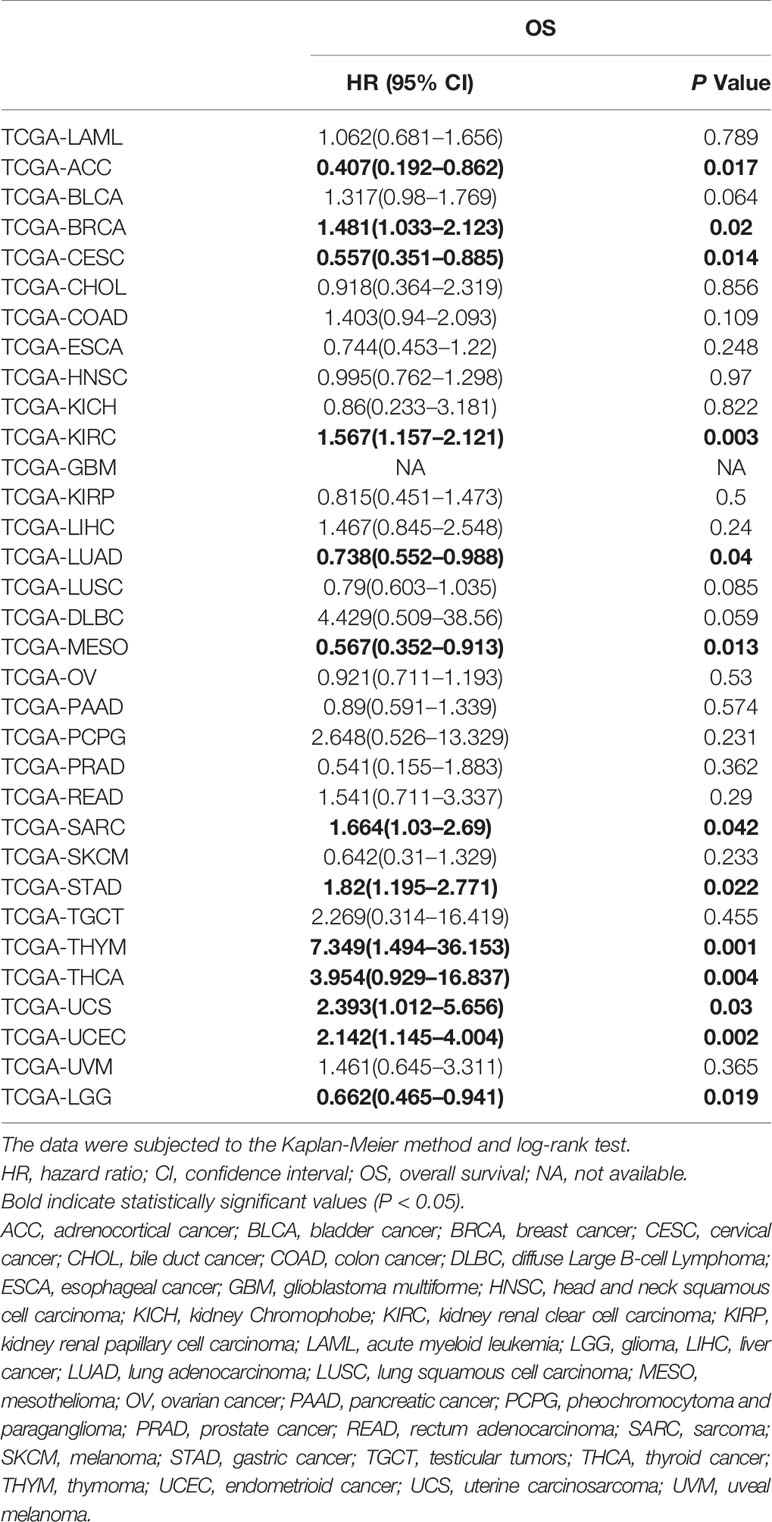
Table 7 HRs and corresponding 95% CIs of SOX2-OT overexpression in tumors based on The Cancer Genome Atlas (TCGA) datasets.
However, focusing on single tumor types combined with meta-analysis revealed that upregulation of SOX2-OT was significantly associated with worse OS in sarcoma (TCGA-SARC; HR = 1.664, 95% CI 1.03–2.69; P = 0.042, Figure 6A) and gastric cancer (TCGA-STAD; HR = 1.82, 95% CI 1.195–2.771; P = 0.022, Figure 6B), while the association was opposite in lung adenocarcinoma (TCGA-LUAD; HR = 0.738, 95% CI 0.552–0.988; P = 0.04) (Figure 6C). In the other tumor types, SOX2-OT expression was not associated with worse OS (Table 7, Supplementary Figures 3 and 4).
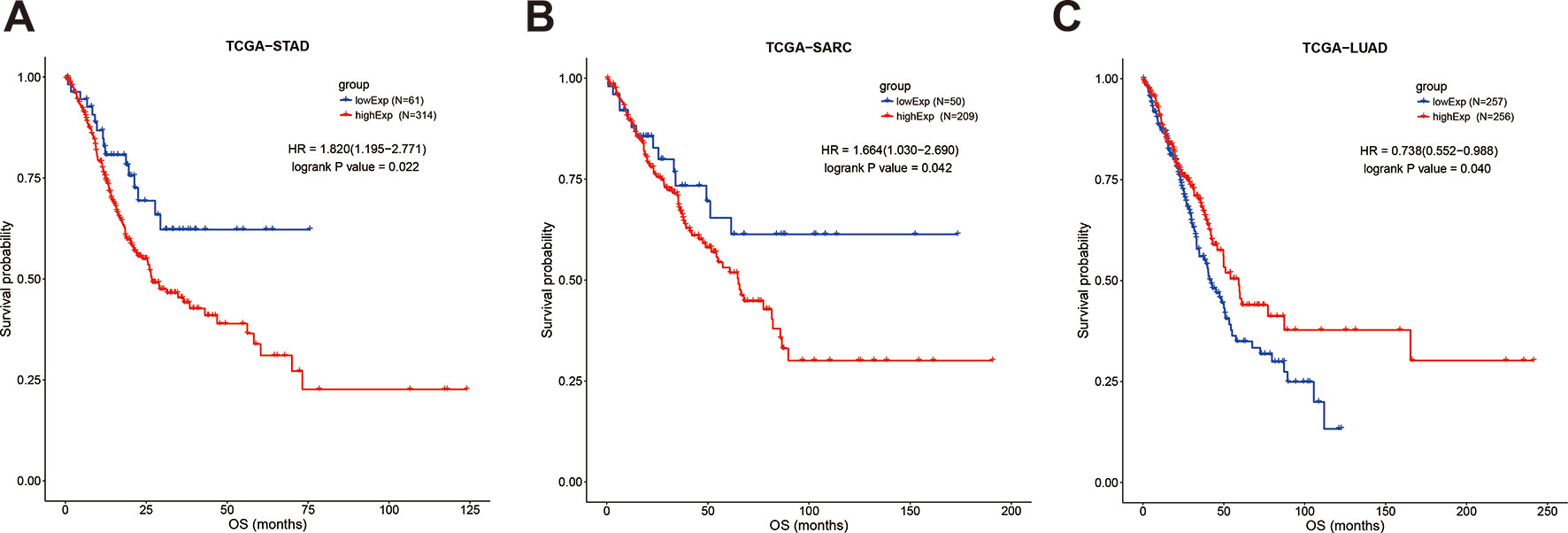
Figure 6 Kaplan-Meier survival curves for the overall survival of cancer patients, stratified by SOX2-OT expression levels. (A) TCGA-STAD, (B) TCGA−SARC, (C) TCGA−LUAD. STAD, gastric cancer; SARC, sarcoma; LUAD, lung adenocarcinoma.
To systematically analyze the underlying gene regulatory mechanisms of SOX2-OT, a total of 500 target genes were identified with Multi Experiment Matrix (MEM) (Supplementary Figure 5). GO and KEGG analyses were executed. Validated target genes of SOX2-OT enriched GO terms including cell adhesion, cell adhesion molecule (CAM) binding, mRNA binding, mRNA splicing via spliceosome, and MAPK cascade (Figure 7A). These relevant GO terms were considered as the most specific and useful for describing the concrete function of SOX2-OT. The visualization network is shown in Figure 7B. Furthermore, KEGG enrichment analysis indicated that SOX2-OT may play a critical role in cancers via several pathways including CAMs, retrograde endocannabinoid signaling, circadian entrainment, cAMP signaling pathway, and mRNA surveillance pathway (Figure 7C). These corresponding KEGG terms were considered as the most specific and useful for describing the concrete pathway of SOX2-OT. The visualization network is presented in Figure 7D.
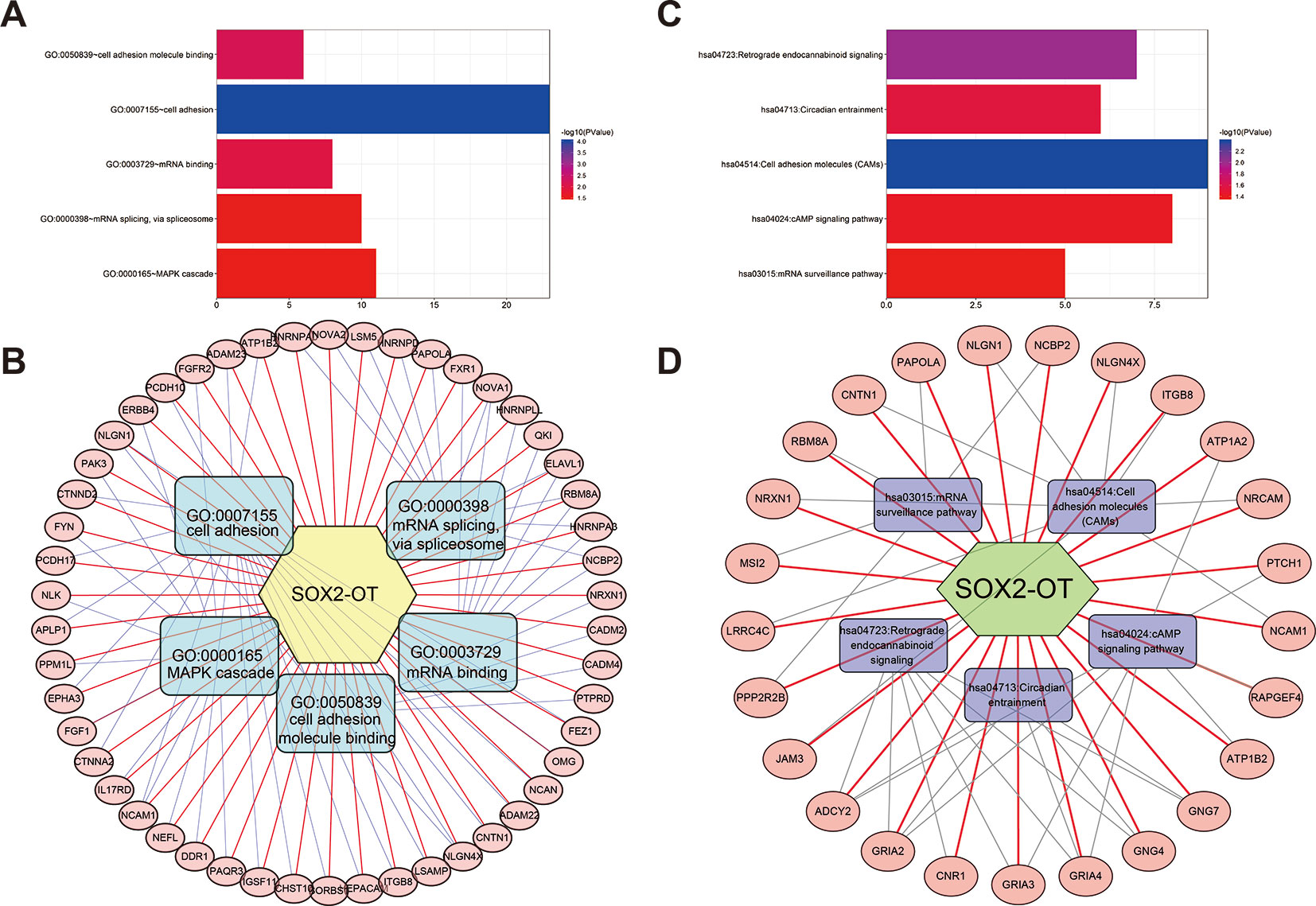
Figure 7 Significantly enriched gene ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of potential targets of long non-coding RNAs (lncRNA) SOX2 overlapping transcript (SOX2-OT) in cancer patients. (A) biological processes (BP), (B) the lncRNA SOX2-OT-GO-mRNA network was generated based on the Multi Experiment Matrix (MEM) and DAVID databases. (C) KEGG pathway. (D) the lncRNA SOX2-OT-KEGG-mRNA network was generated based on the MEM and DAVID databases.
Several studies have indicated that high expression of SOX2-OT is significantly related with the prognosis and clinicopathological outcomes in cancers (Hou et al., 2014; Shi and Teng, 2015; Iranpour et al., 2016; Zhang et al., 2016; Zou et al., 2016; Wang et al., 2017a; Wang et al., 2017b; Han et al., 2018; Li et al., 2018a; Li et al., 2018b; Sun et al., 2018; Wei et al., 2018; Xie et al., 2018). The crucial role that SOX2-OT may play in the progression of many cancers had been further outlined in reviews (Shahryari et al., 2015; Castro-Oropeza et al., 2018). A meta-analysis by Jing et al. proposed that the overexpression of SOX2-OT indicated higher TNM stage and a worse OS in cancer patients, but failed to predict distant metastasis and lymph node metastasis in Chinese cancer patients (Jing et al., 2017). Moreover, other studies since 2014 have investigated the relationship between SOX2-OT and the prognosis of cancer patients (Hou et al., 2014; Shi and Teng, 2015; Iranpour et al., 2016; Zhang et al., 2016; Zou et al., 2016; Wang et al., 2017a; Wang et al., 2017b; Han et al., 2018; Li et al., 2018a; Li et al., 2018b; Sun et al., 2018; Wei et al., 2018; Xie et al., 2018). The present study was performed to obtain a more definite conclusion and assess the potential mechanisms of SOX2-OT effects by integrating the outcomes of published studies and TCGA survival data and running GO and KEGG analyses.
The present meta-analysis of a combination of 1172 patients from 13 eligible studies with 9676 patients from TCGA investigated thoroughly the correlations between elevated expression of SOX2-OT and prognosis as well as clinicopathological outcomes in cancer patients. The NOS was applied to evaluate the quality of all the selected studies, and Egger’s test and Begg’s test were used to examine the publication bias. If the P value of the Egger’s test was less than 0.05, we also checked the reliability of the results by TSA.
Our results indicated that elevated expression of SOX2-OT was significantly related to worse prognosis indicators, with an OS of 2.026 (95% CI: 1.691–2.428), and a DFS of 2.554 (95% CI: 1.261–5.174). Regarding the clinicopathological characteristics of patients with cancers, our research suggested that high SOX2-OT expression was significantly associated with the invasion of cancers, as reveal by the tumor stage (RR = 1.468, 95% CI: 1.106–1.949), lymphatic metastasis (RR = 1.554, 95% CI: 1.211–1.994), distant metastasis (RR = 3.054, 95% CI: 1.866–4.999), tumor size (RR = 1.264, 95% CI: 1.019–1.566), and depth of tumor invasion (RR = 1.552, 95% CI: 1.274–1.890), but couldn’t predict histological differentiation, age, or gender.
According to our findings, SOX2-OT shows the potential to be used as a marker for progression and prognosis. A subgroup analysis indicated that elevated SOX2-OT expression was substantially associated with OS in sarcoma (SARC) and gastric cancer (STAD) patients, according to the publications and the TCGA datasets. As for pancreatic cancer (PAAD), bile duct cancer (CHOL), lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC), SOX2-OT overexpression was correlated with a bad prognosis in the publications. However, in the TCGA datasets, SOX2-OT was associated with a good prognosis although the results were not statistically significant; the corresponding HR values were 0.89 (95% CI: 0.591–1.339, P = 0.574), 0.918 (95% CI: 0.364–2.319, P = 0.856), 0.738 (95% CI: 0.552–0.988, P = 0.04), and 0.79 (95% CI: 0.603–1.035, P = 0.085), respectively. High expression of SOX2-OT in liver cancer (LIHC) in the TCGA datasets was correlated with an unfavorable prognosis (HR = 1.467, 95% CI: 0.845–2.548, P = 0.24) although the results were not statistically significant, which was consistent with the publications (Shi and Teng, 2015; Sun et al., 2018) (Tables 4 and 7). Kaplan-Meier analysis initially suggested that SOX2-OT overexpression was associated with a bad OS in adrenocortical cancer (ACC), cervical cancer (CESC), mesothelioma (MESO), and glioma (LGG), and associated with a worse OS in breast cancer (BRCA), kidney renal clear cell carcinoma (KIRC), thymoma (THYM), thyroid cancer (THCA), uterine carcinosarcoma (UCS), and endometrioid cancer (UCEC) according to the TCGA datasets (Table 7 and Supplementary Figures 3 and 4). Sampling error and publication bias may explain the inconsistent results between literature studies and studies on TCGA datasets.
Heterogeneity appeared in the clinicopathological aspects including tumor stage, lymphatic metastasis, and tumor size (P < 0.1). Since the presence of heterogeneity may affect the results of the meta-analysis, the heterogeneity has been dealt cautiously with a random effects model in order to reduce the effect of heterogeneity on the merged results. Publication bias was prominent in studies with OS data (P < 0.05) as showed by the Egger’s, Begg’s test, and funnel plots. Hence, the TSA data suggested the results of our study were statistically stable.
Recently, studies on the functioning of SOX2-OT in cancer have spread and cumulative evidence indicating that SOX2-OT could affect various biological behaviors of numerous tumors. Li et al. pointed out that SOX2-OT competitively binds to the miR-200 family to regulate the expression of SOX2, and SOX2-OT promotes epithelial-mesenchymal transition (EMT) and stem cell-like properties by regulating SOX2 expression, thereby promoting invasion and metastasis of pancreatic duct adenocarcinoma (Li et al., 2018a). Qu et al. proposed that SOX2-OT was highly expressed in gastric cancer cells, which promoted the expression of AKT2 by targeting miR-194-5p, thus elevating cell proliferation and metastasis (Qu and Cao, 2018). Finally, Wei et al. discovered that the upregulation of lncRNA SOX2-OT by transcription factor IRF4 promotes cell proliferation and metastasis in cholangiocarcinoma via upregulating SOX2, and activates PI3K/AKT signaling pathway via suppressing the nuclear transcription of PTEN (Wei et al., 2018).
The exact gene regulatory mechanisms of SOX2-OT remain poorly understood. Therefore, we uncovered the validated targeting genes of SOX2-OT through the MEM platform, and a comprehensive target gene network analysis was performed. The GO and KEGG pathway analysis together revealed that some CAMs and pathways may be regulated by SOX2-OT. SOX2-OT appears to play a critical role in the cancers via different pathways, including mRNA binding and mRNA splicing, similar to the post-transcriptional regulating functions of other lncRNAs. The above findings suggest that the elevation of SOX2-OT expression is associated with the processes of tumor invasion and metastasis, consistent with our findings.
Our study is consistent with the most recent study by Song et al. in which lncRNA SOX2-OT overexpression was significantly correlated with worse OS and more advanced clinical stages of solid tumors based on 943 cases from 10 studies, all of them being Asians (Song et al., 2018). Consistently, analysis of 481 patients from five studies by Jing et al. showed that high SOX2-OT expression predicted poor OS and more advanced tumor progression, but failed to predict distant metastasis and lymph node metastasis in Chinese cancer patients (Jing et al., 2017). Herein, we have performed a more comprehensive study on the clinicopathological significance of SOX2-OT expression in cancer patients. First, we included 13 eligible articles involving 1172 cancer patients and 32 TCGA cancer datasets involving 9676 cancer patients to investigate a total of 10,848 participants in our study. Second, we investigated both clinicopathological and prognostic significance of SOX2-OT expression based on comprehensive clinical data and performed a series of subgroup analyses based on prognostic types, adjusted variables in the multivariate analysis of OS, sample sizes, cancer types, sample types, cut-off values, analysis models, and clinicopathological characteristics. These stratifications increase our understanding of the clinicopathological significance of SOX2-OT expression in cancers. Third, TSA on the applicable literature was used to investigate reliability and conclusiveness of available evidence for the prognostic significance of SOX2-OT expression. Fourth, the prognostic value was validated using TCGA datasets and the potential functions were explored using GO and KEGG.
In this particular study, there were some limitations. As to this meta-analysis, different cut-off values and sample types of the selected articles contributed publication bias. Since direct results of survival analysis were unavailable, a divergence in HR values might significantly contribute to extract the survival data through the Kaplan-Meier curve. Consequently, in-depth study is required to investigate the clinical value and prognosis significance of SOX2-OT in cancers.
In order to increase the sample size, we used TCGA datasets for further analysis and validation, but only the results of gastric cancer and sarcoma were consistent with those based the publications. In order to clarify the mechanism by which SOX2-OT is involved in gastric cancer and sarcoma, further molecular biology experiment is warranted to explore other possible signaling pathways or target molecules.
In conclusion, our report shows that elevated SOX2-OT expression was significantly related with invasion and metastasis progress in cancers, implying shorter OS and DFS, a poorer TNM stage, higher rates of lymphatic and distant metastasis, larger tumor size, and deeper invasion. We also concluded that SOX2-OT plays a crucial role via a few pathways. Considering the limitations, further studies are necessary in order to better define the functions of SOX2-OT in cancers.
Participated in research design: YL, MD, MS, and DH. Performed data analysis: MS, SW, MD, JZ, PL, XW, DW, JZ, DC, and JL. Wrote or contributed to the writing of the manuscript: MS and HL.
This research was supported by the National Natural Science Foundation of China (81902498), Natural Science Foundation of Hubei Province of China (2019CFB177), Natural Science Foundation of Hubei Provincial Department of Education (Q20182105), Chen Xiao-ping Foundation for the development of science and technology of Hubei Provincial (CXPJJH11800001-2018333), Natural Science Foundation of Hubei Province of China (2016CFB530) and Faculty Development Foundation of Hubei University of Medicine (2014QDJZR01), and National Students’ platform for innovation and entrepreneurship training program (201810929005, 201810929009, 201810929068, and 201813249010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors gratefully acknowledge The Cancer Genome Atlas pilot project (established by NCI and NHGRI), which made the genomic data and clinical data available.
SOX2-OT, SOX2 overlapping transcript; lncRNA, Long non-coding RNAs; RNA-seq, RNA deep sequencing; TCGA, The Cancer Genome Atlas; OS, Overall survival; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MEM, Multi Experiment Matrix; HR, Hazard ratio; RR, Risk ratio; CI, Confidence interval; TSA, Trial sequential analysis; DFS, Disease free survival; TNM, Tumor, node, metastasis; q-RTPCR, Quantitative real-time polymerase chain reaction; NOS, Newcastle-Ottawa Scale; MOOSE, Meta-analysis of Observational Studies in Epidemiology; NA, Not available; ACC, Adrenocortical cancer; BLCA, Bladder cancer; BRCA, Breast cancer; CESC, Cervical cancer; CHOL, Bile duct cancer; COAD, Colon cancer; DLBC, Diffuse large B-cell lymphoma; ESCA, Esophageal cancer; GBM, Glioblastoma multiforme; HNSC, Head and neck squamous cell carcinoma; KICH, Kidney chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute myeloid leukemia; LGG, Glioma; LIHC, Liver cancer; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; MESO, Mesothelioma; OV, Ovarian cancer; PAAD, Pancreatic cancer; PCPG, Pheochromocytoma and paraganglioma; PRAD, Prostate cancer; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Melanoma; STAD, Gastric cancer; TGCT, Testicular tumors; THCA, Thyroid cancer; THYM, Thymoma; UCEC, Endometrioid cancer; UCS, Uterine carcinosarcoma; UVM, Uveal melanoma; CAMs, Cell adhesion molecules; EMT, Epithelial-mesenchymal transition; CP, Clinical parameters; BP, Biological processes.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01375/full#supplementary-material
Supplementary Figure 1 | Relationship of SOX2-OT expression and DFS in various cancers.
Supplementary Figure 2 | Meta-analysis of TCGA datasets estimating the association of SOX2-OT with the patients’ overall survival (OS).
Supplementary Figure 3 | Kaplan-Meier survival curves for overall survival of 16 cancers in TCGA, stratified by SOX2-OT expression levels. (A) ACC, (B) BLCA, (C) BRCA, (D) CESC, (E) CHOL, (F) COAD, (G) DLBC, (H) ESCA, (I) HNSC, (J) KICH, (K) KIRC, (L) KIRP, (M) LAML, (N) LGG, (O) LIHC, (P) LUAD.
Supplementary Figure 4 | Kaplan-Meier survival curves for overall survival of the other 16 cancers in TCGA, stratified by SOX2-OT expression levels. (A) LUSC, (B) MESO, (C) OV, (D) PAAD, (E) PCPG, (F) PRAD, (G) READ, (H) SARC, (I) SKCM, (J) STAD, (K) TGCT, (L) THCA, (M) THYM, (N) UCEC, (O) UCS, (P) UVM.
Supplementary Figure 5 | Totally 500 target genes of SOX2-OT were identified with Multi Experiment Matrix (MEM).
Supplementary Table 1 | Databases searching terms. (A) Search criterion of Medline (via PubMed, from inception to Jan 1st, 2019) (n=47), (B) Search criterion of Embase (from 1966 to Jan 20, 2019) (n=49), (C) Search criterion of Cochrane Library (Jan 1st, 2019) (n=1).
Supplementary Table 2 | Results of quality assessment using the Newcastle–Ottawa Scale for the included studies.
Supplementary Table 3 | Meta-regression analysis of heterogeneity for clinicopathological parameters in cancers.
Supplementary Table 4 | Subgroup analysis for SOX2-OT overexpression and clinicopathological parameters in cancers.
Amaral, P. P., Neyt, C., Wilkins, S. J., Askarian-Amiri, M. E., Sunkin, S. M., Perkins, A. C., et al. (2009). Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15 (11), 2013–2027. doi: 10.1261/rna.1705309
Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N., Lovell-Badge, R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17 (1), 126–140. doi: 10.1101/gad.224503
Castro-Oropeza, R., Melendez-Zajgla, J., Maldonado, V., Vazquez-Santillan, K. (2018). The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. (Dordr.) 41 (6), 585–603. doi: 10.1007/s13402-018-0406-4
Farhangian, P., Jahandoost, S., Mowla, S. J., Khalili, M. (2018). Differential expression of long non-coding RNA SOX2OT in gastric adenocarcinoma. Cancer Biomark. 23 (2), 221–225. doi: 10.3233/cbm-181325
Fong, H., Hohenstein, K. A., Donovan, P. J. (2008). Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26 (8), 1931–1938. doi: 10.1634/stemcells.2007-1002
Han, L., Zhang, W., Zhang, B., Zhan, L. (2018). Long non-coding RNA SOX2OT promotes cell proliferation and motility in human ovarian cancer. Exp. Ther. Med. 15 (2), 2182–2188. doi: 10.3892/etm.2017.5618
Hou, Z., Zhao, W., Zhou, J., Shen, L., Zhan, P., Xu, C., et al. (2014). A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int. J. Biochem. Cell Biol. 53, 380–388. doi: 10.1016/j.biocel.2014.06.004
Iranpour, M., Soudyab, M., Geranpayeh, L., Mirfakhraie, R., Azargashb, E., Movafagh, A., et al. (2016). Expression analysis of four long noncoding RNAs in breast cancer. Tumour Biol. 37 (3), 2933–2940. doi: 10.1007/s13277-015-4135-2
Jing, X., Lin, J., Wang, H., Tian, L., Tian, R., Zhang, Y., et al. (2017). Prognostic value of lncRNA SOX2OT for Chinese cancer patients: a meta-analysis. PloS One 12 (5), e0176889. doi: 10.1371/journal.pone.0176889
Li, Z., Jiang, P., Li, J., Peng, M., Zhao, X., Zhang, X., et al. (2018a). Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 37 (28), 3822–3838. doi: 10.1038/s41388-018-0237-9
Li, Z., Li, J., Ji, D., Leng, K., Xu, Y., Huang, L., et al. (2018b). Overexpressed long noncoding RNA Sox2ot predicts poor prognosis for cholangiocarcinoma and promotes cell proliferation and invasion. Gene 645, 131–136. doi: 10.1016/j.gene.2017.12.017
Parmar, M. K., Torri, V., Stewart, L. (1998). Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat. Med. 17 (24), 2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8
Qu, F., Cao, P. (2018). Long noncoding RNA SOX2OT contributes to gastric cancer progression by sponging miR-194-5p from AKT2. Exp. Cell Res. 369 (2), 187–196. doi: 10.1016/j.yexcr.2018.05.017
Shahryari, A., Rafiee, M. R., Fouani, Y., Oliae, N. A., Samaei, N. M., Shafiee, M., et al. (2014). Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells 32 (1), 126–134. doi: 10.1002/stem.1542
Shahryari, A., Jazi, M. S., Samaei, N. M., Mowla, S. J. (2015). Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front. Genet. 6, 196. doi: 10.3389/fgene.2015.00196
Shi, X. M., Teng, F. (2015). Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int. J. Clin. Exp. Pathol. 8 (4), 4008–4014.
Song, X., Yao, H., Liu, J., Wang, Q. (2018). The prognostic value of long noncoding RNA Sox2ot expression in various cancers: a systematic review and meta-analysis. Clin. Chim. Acta 484, 52–59. doi: 10.1016/j.cca.2018.05.038
Stuck, A. E., Rubenstein, L. Z., Wieland, D. (1998). Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ 316 (7129), 469; author reply 470–461.
Sun, J., Wei, X., Xu, L. (2018). Upregulation of lncRNA Sox2ot indicates a poor prognosis for patients with hepatocellular carcinoma and promotes cell invasion. Oncol. Lett. 16 (1), 1189–1195. doi: 10.3892/ol.2018.8725
Sun, M., Du, M., Zhang, W., Xiong, S., Gong, X., Lei, P., et al. (2019). Survival and clinicopathological significance of SIRT1 Expression in cancers: a meta-analysis. Front. Endocrinol. (Lausanne) 10, s. doi: 10.3389/fendo.2019.00121
Tian, T., Wang, M., Lin, S., Guo, Y., Dai, Z., Liu, K., et al. (2018). The impact of lncRNA dysregulation on clinicopathology and survival of breast cancer: a systematic review and meta-analysis. Mol. Ther. Nucleic Acids 12, 359–369. doi: 10.1016/j.omtn.2018.05.018
Tierney, J. F., Stewart, L. A., Ghersi, D., Burdett, S., Sydes, M. R. (2007). Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16. doi: 10.1186/1745-6215-8-16
Wang, P., Shan, L., Xue, L., Zheng, B., Ying, J., Lu, N. (2017a). Genome wide copy number analyses of superficial esophageal squamous cell carcinoma with and without metastasis. Oncotarget 8 (3), 5069–5080. doi: 10.18632/oncotarget.13847
Wang, Z., Tan, M., Chen, G., Li, Z., Lu, X. (2017b). LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2. IUBMB Life 69 (11), 867–876. doi: 10.1002/iub.1681
Wei, C. X., Wong, H., Xu, F., Liu, Z., Ran, L., Jiang, R. D. (2018). IRF4-induced upregulation of lncRNA SOX2-OT promotes cell proliferation and metastasis in cholangiocarcinoma by regulating SOX2 and PI3K/AKT signaling. Eur. Rev. Med. Pharmacol. Sci. 22 (23), 8169–8178. doi: 10.26355/eurrev_201812_16509
Xie, Y., Zhang, Y., Du, L., Jiang, X., Yan, S., Duan, W., et al. (2018). Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 12 (5), 648–658. doi: 10.1002/1878-0261.12188
Zhang, Y., Yang, R., Lian, J., Xu, H. (2016). LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am. J. Transl. Res. 8 (11), 5035–5043.
Zhang, J. J., Zhu, Y., Zhang, X. F., Liu, D. F., Wang, Y., Yang, C., et al. (2017). Yin Yang-1 suppresses pancreatic ductal adenocarcinoma cell proliferation and tumor growth by regulating SOX2OT-SOX2 axis. Cancer Lett. 408, 144–154. doi: 10.1016/j.canlet.2017.08.032
Keywords: SOX2-OT, cancer, prognosis, clinicopathological significance, meta-analysis
Citation: Li Y, Du M, Wang S, Zha J, Lei P, Wang X, Wu D, Zhang J, Chen D, Huang D, Lu J, Li H and Sun M (2020) Clinicopathological Implication of Long Non-Coding RNAs SOX2 Overlapping Transcript and Its Potential Target Gene Network in Various Cancers. 10:1375. doi: 10.3389/fgene.2019.01375
Received: 25 July 2019; Accepted: 17 December 2019;
Published: 23 January 2020.
Edited by:
Alfons Navarro, University of Barcelona, SpainReviewed by:
Zhijun Dai, Zhejiang University, ChinaCopyright © 2020 Li, Du, Wang, Zha, Lei, Wang, Wu, Zhang, Chen, Huang, Lu, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Sun, c3VubWluLTA3MTVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.