
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 19 June 2019
Sec. Evolutionary and Population Genetics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00583
This article is part of the Research Topic Genomic Advances and Challenges in Old and New World Camelids View all 20 articles
The alpaca (Vicugna pacos) is an economically important and cultural signature species in Peru. Thus, molecular genomic information about the genes underlying the traits of interest, such as fiber properties and color, is critical for improved breeding and management schemes. Current knowledge about the alpaca genome, particularly the chromosomal location of such genes of interest is limited and lags far behind other livestock species. The main objective of this work was to localize alpaca candidate genes for fiber growth and color using fluorescence in situ hybridization (FISH). We report the mapping of candidate genes for fiber growth COL1A1, CTNNB1, DAB2IP, KRT15, KRTAP13-1, and TNFSF12 to chromosomes 16, 17, 4, 16, 1, and 16, respectively. Likewise, we report the mapping of candidate genes for fiber color ALX3, NCOA6, SOX9, ZIC1, and ZIC5 to chromosomes 9, 19, 16, 1, and 14, respectively. In addition, since KRT15 clusters with five other keratin genes (KRT31, KRT13, KRT9, KRT14, and KRT16) in scaffold 450 (Vic.Pac 2.0.2), the entire gene cluster was assigned to chromosome 16. Similarly, mapping NCOA6 to chromosome 19, anchored scaffold 34 with 8 genes, viz., RALY, EIF2S2, XPOTP1, ASIP, AHCY, ITCH, PIGU, and GGT7 to chromosome 19. These results are concordant with known conserved synteny blocks between camelids and humans, cattle and pigs.
The alpaca (Vicugna pacos) is a domesticated South American camelid adapted to the Andean climate conditions. They are economically important in Peru as a fiber production species benefiting the small shareholders living in this geographical region (Quispe et al., 2009). Alpaca fiber is highly valued in the international market because of its softness and resistance (Crispín, 2008). Alpacas carry a cultural value because of their historical importance, millenary tradition, ancestral Peruvian identity and unique characteristics derived from their adaptation to the Andean geography and climate (Yucra, 2017). Alpaca meat is highly valued for its high protein and low cholesterol content (Hack, 2001), and continues serving rural population of Altiplano as an important source of protein (Cruz et al., 2017).
Management systems promoting the improvement of alpaca herd productivity have not yet been adopted widely (Quispe et al., 2009). Actual research is orientated to the application of genetic improvement technologies that would decrease fiber diameter, increase fleece weight, and establish uniform color herds (Morante et al., 2009). Genomic selection using single nucleotide polymorphisms (SNPs)-based genotype-phenotype associations, offers the best option presently available. To apply genomic selection in alpacas, it is necessary to identify and map SNPs throughout the genome and associate them with genes that control economic productive traits. In turn, mapping candidate genes already reported in association to color and fiber characteristics, as well as SNPs, will contribute to understanding the organization of the alpaca genome and genome-wide selection of appropriate markers to develop molecular marker microarrays.
Cytogenetic analysis has demonstrated that all camelids share the same chromosome number (2n = 74) with essentially similar chromosome morphology and banding patterns (Hsu and Benirschke, 1967; Taylor et al., 1968; Bianchi et al., 1986). The first camelid chromosome map was based on Zoo-FISH revealing evolutionarily conserved synteny segments across the dromedary, human, cattle and pig (Balmus et al., 2007). This information was instrumental for starting systematic gene mapping in these species and the first cytogenetics maps for the alpaca genome were developed only recently (Avila et al., 2014a,b, 2015). Because of difficulties to unambiguously identify camelid chromosomes (Di Berardino et al., 2006; Avila et al., 2014b), the 230 cytogenetically mapped markers in alpaca (Avila et al., 2014a) will serve as critical references for FISH-mapping new genes and markers.
The aim of this study was to cytogenetically map 11 alpaca candidate genes for fiber growth and coat color to progress the development of alpaca cytogenetic map and chromosomal anchoring the reference sequence.
Alpaca chromosome slides were prepared from peripheral blood lymphocytes of normal alpacas according to standard protocols (Raudsepp and Chowdhary, 2008). We used Concanavalin A (Con A from Canavalia ensiformis, 20 μg/ml; Sigma Aldrich) as the mitogen, instead of Pokeweed, because Con A stimulates better proliferation of alpaca blood lymphocytes (Avila et al., 2015).
Genes for cytogenetic mapping were retrieved from publications. Candidate genes regulating fiber growth characteristics, COL1A1, CTNNB1, DAB2IP, KRT15, and TNFSF12 (Fernandez, 2015), and KRTAP13-1 (Florez, 2016); candidate genes that regulate the expression of fiber color, NCOA6-agouti chimera (Chandramohan et al., 2013); ZIC1, ZIC5, and SOX9 which conform the neural crest gene regulatory network (Simoes-Costa and Bronner, 2013), and the ALX3 transcription factor that regulates melanocyte differentiation in striped rodents (Cuthill et al., 2017). Gene specific sequences were retrieved from VicPac 2.0.2 (GCA_000164845.3) at the NCBI (National Center for Biotechnology Information). Since each of the selected genes are members of gene super-families, sequences that characterized these super-families were identified using the BLASTp1 and Spling2 tools and manually removed from each gene FASTA sequence. This way unique sequences for each specific gene were obtained. The gene sequences were masked for repeats in RepeatMasker3. Gene-specific PCR primers were designed with Primer3 (Untergasser et al., 2012)4 and Primer-BLAST5 software packages. The primers were tested by in silico PCR6 and optimized on alpaca genomic DNA.
Overgo primers were designed manually from 36 to 52 bp size sequence within the PCR amplicon. We designed a 24 bp forward primer from the first nucleotide at the 5′ end position of the selected region. The reverse primer was designed starting at the 3′ end of the selected region, ending with 8 nucleotides overlapping the forward primer. The overlapping section and the single strand sections of the forward and reverse primers, contained 50–60 (±5) % GC (we used GC calculator7). PCR and overgo primers for each gene are presented in Table 1.
BAC clones containing sequences of the selected genes were identified as described by Avila et al. (2014b). Briefly, pools of radioactively labeled [(32P) dATP/dCTP] overgo primers were hybridized to CHORI-246 alpaca BAC library8 filters. Filters were exposed to autoradiography films and positive BAC clones were identified and picked from the library. BACs corresponding to individual genes were identified by PCR with gene-specific primers. BAC DNA was isolated with the Plasmid Midi Kit (Qiagen) and evaluated for quality by electrophoresis in 1% agarose gels.
BAC DNA labeling, hybridizations and signal detection were carried out according to standard protocols (Raudsepp and Chowdhary, 2008). The DNA of individual BACs was labeled with biotin or digoxigenin using DIG- or Biotin-Nick Translation Mix (Roche Diagnostics) and the manufacturer’s protocol. Because the known difficulties to unambiguously identify camelid chromosomes, we consulted Zoo-FISH data (Balmus et al., 2007) and the 230-marker cytogenetic map (Avila et al., 2014a) to infer the most probable chromosome location for each candidate gene. Based on these predictions, BACs containing new genes were co-hybridized with a differently labeled reference gene from the cytogenetic map (Table 2). Biotin- and dig-labeled probes were detected with avidin-FITC (Vector Laboratories) and anti-dig-rhodamine (Roche Applied Science), respectively. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and identified according to the nomenclature proposed by Balmus et al. (2007) and Avila et al. (2014b). Images were captured and analyzed using a Zeiss Axioplan 2 fluorescence microscope, equipped with the Isis Version 5.2 (MetaSystems GmbH) software. At least 10 images were captured and analyzed for each experiment.
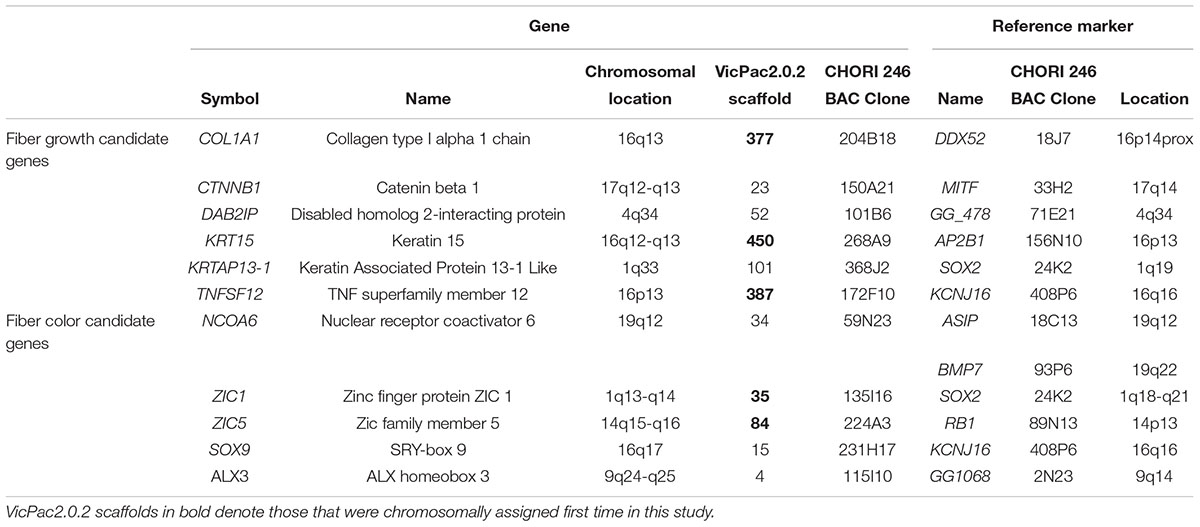
Table 2. Summary data of newly mapped genes and reference markers (Avila et al., 2014b).
Altogether, we identified 41 BAC clones that collectively contained the 11 genes of interest. Clones for individual genes were identified by PCR with gene-specific primers (Table 1), and one clone per each gene was selected for FISH mapping. In this manner, we assigned 11 BAC clones to eight different alpaca autosomes. Most of the candidate genes were mapped to a specific G-band or a range of G-bands (Table 2). Previously mapped reference markers (Avila et al., 2014b) confirmed chromosome identification and helped to position new genes in the centromere-telomere field (Figure 1). Four genes were located in chromosome 16 (chr16), and 2 genes in chr1, whereas the remaining five genes mapped to five different chromosomes (Figure 1 and Table 2). In chr19, the NCOA6 gene overlapped with ASIP in 19q12, and their relative order was resolved by interphase FISH using BMP7 as the second reference marker. The order of the three genes was revealed as cen-ASIP-NCOA6-BMP7-tel (Figure 1F, far right). Location of CTNNB1, DAB2IP, and SOX9 in chr17, chr4 and chr16, respectively, was confirmed by co-hybridized reference markers. No genes were assigned to chromosome arms that previously did not have a mapped marker. No discrepancies of the known conserved synteny blocks between camelids, cattle and human (Balmus et al., 2007) were observed.
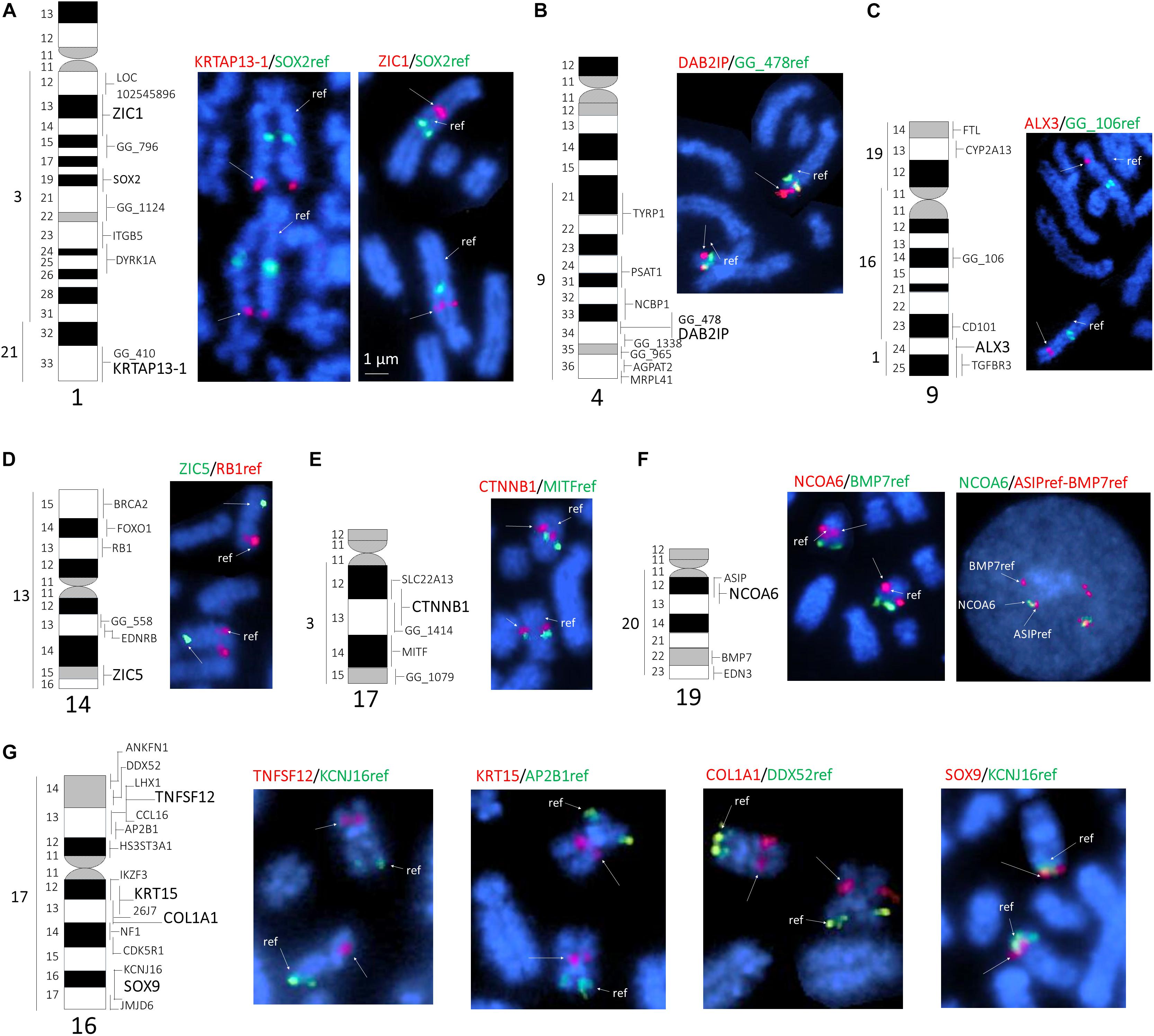
Figure 1. FISH mapping of selected genes to alpaca chromosomes. (A) ZIC1 and KRTAP13-1 to chr1; (B) DAB2IP to chr2; (C) ALX3 to chr9; (D) ZIC5 to chr14; (E) CTNNB1 to chr17; (F) NCOA6 to chr16, and (G) TNFSF12, KRT15, COL1A1, and SOX9 to chr16. The left side of each sub-figure includes a G-banded ideogram showing conserved synteny with human chromosomes (far left) and all cytogenetically mapped markers (right). The markers mapped in this study are in larger and bold font. The right side of each sub-figure shows partial metaphase spreads with dual-color FISH results. The newly mapped genes are denoted with arrows; reference genes for chromosome identification are denoted with arrows and “ref.” The symbols of newly mapped and reference genes are shown above each figure in green or red font colors, which match with the colors of FISH signals on chromosomes. In (F), we show refined ordering of NCOA6 in relation to ASIP in interphase chromosomes (far right) using BMP7 is an anchoring marker; the order is cen-ASIP-NCOA6-BMP7-tel.
We report the cytogenetic mapping of 11 new genes in the alpaca genome, which together with prior FISH maps (Avila et al., 2014a,b) takes the tally of all chromosomally mapped markers for this species to 241. This is not a high number but an important step forward for the development of chromosomally anchored reference genomes for the alpaca and other camelids. Furthermore, among the 11 markers mapped in this study, five belong to VicPac2.0.2 scaffolds that were not represented in previous maps (Avila et al., 2014b). This implies that the entire scaffold 35, anchored by ZIC1, maps to chr1; scaffold 84, anchored by ZIC5, maps to chr14, and scaffolds 377, 387, and 450, anchored by COL1A1, TNFSF12, and KRT15, respectively, map to chr16 (Table 2).
As the goal of this study was cytogenetic mapping of candidate genes related to fiber growth and color synthesis, we bioinformatically inspected all VicPac2.0.2 scaffolds containing the 11 mapped markers (Table 2) for additional genes of interest. In scaffold 450 (331,325 bp, NW_005883152.1), which was newly assigned to chr16q12-q13 by FISH mapping KRT15 (Figure 1G), there is a tandemly arranged cluster of five more keratin genes around KRT15, viz., 5′- KRT31- KRT15 -KRT13- KRT9- KRT14-KRT16 – 3′ (Fernández et al., 2019). Thus, our results allow the assignment of five more keratin genes to chr16q12-q13 (Table 3). This makes alpaca chr16 as a main target for identifying sequence variants potentially associated with hair texture and growth because three of the six candidate genes for fiber growth characteristics (Table 2), viz., KRT15 with the keratin cluster, COL1A1, and TNFSF12 (Fernandez, 2015), map to this chromosome. This also implies that phenotypic characteristics determined by these genes may show particular inheritance patterns due to close linkage. Among the many known molecular components of the mammalian hair follicle (Rompolas and Greco, 2013), keratins and collagens are perhaps most studied (Toivola et al., 2015) and associated with various hair texture characteristics in several mammalian species including humans (Shimomura et al., 2010), dogs (Cadieu et al., 2009), horses (Balmer et al., 2017; Morgenthaler et al., 2017) and alpacas (Fan et al., 2011). Here we considered as candidate genes for alpaca hair texture also genes related to apoptosis regulation and formation of the hair follicle, such as CTNNB1, TNFSF12, and DAB2IP. TNFSF12 and DAB2IP have roles in WNT/β-catenin signaling system (Xie et al., 2010), which controls hair follicle morphogenesis and stem cell differentiation in the skin (Huelsken et al., 2001). SNP variants in these genes have been associated with traits of interest (Farhadian et al., 2018) and used for genomic selection programs in sheep, goat (Rupp et al., 2016) and cattle (Wiggans et al., 2017).
Therefore, microsatellites that have been identified in the alpaca COL1A1, TNFSF12, and DAB2IP (Fernandez, 2015) are potential polymorphic markers for selection in this species.
Among the candidate genes for hair color, mapping NCOA6 to chr19q12 was of particular interest because it anchored a closely linked group of several other potential coat color genes from scaffold 34 (12,494,946 bp, NW_005882736.1) to this chromosome (Table 3). The closely linked gene cluster comprises RALY, EIF2S2, XPOTP1, ASIP, AHCY, ITCH, PIGU, NCOA6, and GGT7, of which only ASIP has been previously mapped (Avila et al., 2014b). In this study, we showed that NCOA6 is overlapping with ASIP in chr19q12 (Figure 1F) which is consistent with the known organization of the agouti locus in alpacas, where the 5′UTR of the ASIP gene contains 142 bp of the NCOA6 gene sequence (Chandramohan et al., 2013). The role of ASIP in regulation of pigment production in mammals is well established (Suzuki, 2013). Mutations in this gene have shown to cause the black coat color phenotype in different species, such as guinea pigs (Lai et al., 2019), black-bone chicken (Yu et al., 2019), sheep (Norris and Whan, 2008; Royo et al., 2008), Iranian Markhoz goats (Nazari-Ghadikolaei et al., 2018), donkeys (Abitbol et al., 2015), horses (Rieder et al., 2001), dogs (Kerns et al., 2004), cats (Eizirik et al., 2003), and impala antelope (Miller et al., 2016). In camelids the agouti signaling protein gene (ASIP) is involved in fiber color development in alpacas (Bathrachalam et al., 2011; Chandramohan et al., 2013), llamas (Daverio et al., 2016) and dromedaries (Almathen et al., 2018; Alshanbari et al., 2019). Sequence variants (SNPs) in other genes from this linkage group have been associated with color phenotypes in several mammalian species. For example, coat color of the Nanjiang Yellow goat has been associated with SNPs in the RALY-EIF2S2 locus (Guo et al., 2018), tandem duplication encompassing ASIP and AHCY coding regions and the ITCH promoter region have been reported as the genetic cause of the dominant white coat color of white/tan (AWt) agouti sheep (Norris and Whan, 2008), and RALY, ASIP, AHCY, and ITCH are associated with brown and black color coat in Iranian Markohz goat (Nazari-Ghadikolaei et al., 2018). Melanocytes, the cells that are responsible for skin pigmentation, are derived from neural crest cells from all axial levels (Betancur et al., 2010). Therefore, genes involved in neural crest generation, such as ZIC genes (Aruga, 2004), are potential candidates for fiber color development. Likewise, SOX9 is involved in the differentiation of neural crest cells into chondrocytes (Simões-Costa and Bronner, 2015) and cooperates with other cofactors in chondrocytes to regulate expression of COL2A1 in humans (Hattori et al., 2008). Furthermore, SOX9 is a key player in ultraviolet B radiation-induced melanocyte differentiation and pigmentation by directly regulating MITF (Passeron et al., 2007). MITF is involved in melanogenesis regulation in alpaca (Wang et al., 2017) and plays a role in the production of white coat color in the llama (Anello et al., 2019). Finally, ALX3 is involved in color differentiation in striped rodents (Cuthill et al., 2017), and proposed as a target melanoma gene fusion in humans (Berger et al., 2010). Also, Marín et al. (2018) used the genetic variation of MC1R and ASIP genes, that control coat color, to differentiate between wild and domestic South American camelids.
In summary, the findings of this study facilitate the improvement and chromosomal assignment of the alpaca genome reference sequence. This, in turn, is critical for correct assembly of newly sequenced individual animals and the discovery of sequence variants in candidate genes for fiber characteristics, coat color and other traits of interest. For instance, Alshanbari et al. (2019) have recently assign the MC1R gene to camelid chr21 that is not in line with the human-camelids Zoo-FISH synteny map. In addition, improving the alpaca cytogenetic map provides new molecular markers for clinical cytogenetics in alpacas and other camelids, thus facilitating chromosome identification in these complex karyotypes. Finally, cytogenetic mapping of specific genes refines the Zoo-FISH information (Balmus et al., 2007), reveals new evolutionary conserved synteny segments between camelids and other mammals, and adds to our knowledge about camelid chromosome evolution.
The cell cultures were prepared from alpaca blood samples obtained in accordance with the United States Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research and Training, approved by Animal Use Protocol AUP #2011-96, # 2018-0342 CA and CRRC #09-47 at Texas A&M University.
FPdL and GG conceived and supervised the study. MM conducted the experimental work. FA contributed to the BAC screening. MM and TR analyzed the data. MM wrote the manuscript in close consultation with FPdL, TR, and GG. All authors read and approved the final version of the manuscript.
The authors acknowledge the financial support from CONCYTEC through project 125-2015 FONDECYT, and VLIR-UOS through funding of the UNALM (IUC) programme.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Opinions of the author(s) do not automatically reflect those of either the Belgian Government or VLIR-UOS, and can neither bind the Belgian Government nor VLIR-UOS.
Abitbol, M., Legrand, R., and Tiret, L. (2015). A missense mutation in the agouti signaling protein gene (ASIP) is associated with the no light points coat phenotype in donkeys. Genet. Sel. Evol. 47:28. doi: 10.1186/s12711-015-0112-x
Almathen, F., Elbir, H., Bahbahani, H., Mwacharo, J., and Hanotte, J. (2018). Polymorphisms in MC1R and ASIP genes are associated with coat color variation in the arabian camel. J. Hered. 109, 700–706. doi: 10.1093/jhered/esy024
Alshanbari, F., Castaneda, C., Juras, R., Hillhouse, A., Mendoza, M. N., Gutiérrez, G. A., et al. (2019). Comparative FISH-Mapping of MC1R, ASIP, and TYRP1 in new and old world camelids and association analysis with coat color phenotypes in the dromedary (Camelus dromedarius). Front. Genet. 10:340. doi: 10.3389/fgene.2019.00340
Anello, M., Daverio, M. S., Silbestro, M. B., Vidal-Rioja, L., and Di Rocco, F. (2019). Characterization and expression analysis of KIT and MITF-M genes in llamas and their relation to white coat color. Anim. Genet. 50, 143–149. doi: 10.1111/age.12769
Avila, F., Baily, M., Perelman, P., Das, P., Pontius, J., Chowdhary, R., et al. (2014a). A comprehensive whole-genome integrated cytogenetic map for the alpaca (Lama pacos). Cytogenet. Genome. Res. 144, 196–207. doi: 10.1159/000370329
Avila, F., Das, P., Kutzler, M., Owens, E., Perelman, P., Rubes, J., et al. (2014b). Development and application of camelid molecular cytogenetic tools. J. Hered. 2014, 858–869. doi: 10.1093/jhered/ess067
Avila, F., Baily, M. P., Merriwether, D. A., Trifonov, V. A., Rubes, J., Kutzler, M. A., et al. (2015). A cytogenetic and comparative map of camelid chromosome 36 and the minute in alpacas. Chromo. Res. 23, 237–251. doi: 10.1007/s10577-014-9463-3
Balmer, P., Bauer, A., Pujar, S., McGarvey, K. M., Welle, M., Galichet, A., et al. (2017). A curated catalog of canine and equine keratin genes. PLoS One 12:e0180359. doi: 10.1371/journal.pone.0180359
Balmus, G., Trifonov, V. A., Biltueva, L. S., O’brien, P. C., Alkalaeva, E. S., Fu, B., et al. (2007). Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromo. Res. 15, 499–515.
Bathrachalam, C., La Manna, V., Renieri, C., and La Terza, A. (2011). “Asip and MC1R cDNA polymorphism in alpaca,” in Fibre Production in South American Camelids and Other Fibre Animals, Sprinter Book, eds M. Á Pérez, J. P. Gutiérrez, I. Cervantes, and M. J. Alcalde (Amsterdam: Wageningen Academic Publishers), 93–96.
Berger, M. F., Levin, J. Z., Vijayendran, K., Sivachenko, A., Adiconis, X., Maguire, J., et al. (2010). Integrative analysis of the melanoma transcriptome. Genome Res. 2010 Apr 20, 413–427. doi: 10.1101/gr.103697.109
Betancur, P., Bronner-Fraser, M., and Sauka-Spengler, T. (2010). Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 2010, 581–603. doi: 10.1146/annurev.cellbio.042308.113245
Bianchi, N. O., Larramendy, M. L., Bianchi, M. S., and Cortes, L. (1986). Karyological conservation in south american camelids. Experientia 42, 622–624.
Cadieu, E., Neff, M. W., Quignon, P., Walsh, K., Chase, K., Parker, H. G., et al. (2009). Coat variation in the domestic dog is governed by variants in three genes. Science 326, 150–153. doi: 10.1126/science.1177808
Chandramohan, B., Renieri, C., La Manna, V., and La Terza, A. (2013). The alpaca agouti gene: genomic locus, transcripts and causative mutations of eumelanic and pheomelanic coat color. Gene 521, 303–310. doi: 10.1016/j.gene.2013.03.060
Crispín, M. (2008). Productividad y distribución de fibra de alpaca en la región de Huancavelica: Un análisis comparativo entre Huancavelica y Puno. [dissertation/College degree]. Lima: Universidad Nacional Mayor de San Marcos.
Cruz, A., Cervantes, I., Burgos, A., Morante, R., and Gutierrez, J. P. (2017). Genetic parameters estimation for preweaning traits and their relationship with reproductive, productive and morphological traits in alpaca. Animal 11, 746–754. doi: 10.1017/S175173111600210X
Cuthill, I. C., Allen, W. L., Arbuckle, K., Caspers, B., Chaplin, G., Hauber, M. E., et al. (2017). The biology of color. Science 357:470.
Daverio, M. S., Rigalt, F., Romeroc, S., Vidal-Riojaa, L., and Di Rocco, F. (2016). Polymorphisms in MC1R and ASIP genes and their association with coat color phenotypes in llamas (Lama glama). Small Rumin. Res. 144, 83–89. doi: 10.1016/j.smallrumres.2016.08.003
Di Berardino, D., Nicodemo, D., Coppola, G., King, A. W., Ramunno, L., Cosenza, G. F., et al. (2006). Cytogenetic characterization of alpaca (Lama pacos, fam. Camelidae) prometaphase chromosomes. Cytogenet. Genome Res. 115, 138–144.
Eizirik, E., Yuhki, N., Johnson, W. E., Menotti-Raymond, M., Hannah, S. S., and O’Brien, S. J. (2003). Molecular genetics and evolution of melanism in the cat family. Curr. Biol. 13, 448–453. doi: 10.1016/S0960-9822(03)128-123
Fan, R., Dong, Y., Cao, J., Bai, R., Zhu, Z., Li, P., et al. (2011). Gene expression profile in white alpaca (Vicugna pacos) skin. Animal 5, 1157–1161. doi: 10.1017/S1751731111000280
Farhadian, M., Rafat, S. A., Hasanpur, K., Ebrahimi, M., and Ebrahimie, E. (2018). Cross-species meta-analysis of transcriptomic data in combination with supervised machine learning models identifies the common gene signature of lactation process. Front. Genet. 9:235. doi: 10.3389/fgene.2018.00235
Fernández, A. G., Gutiérrez, G. A., and Ponce de León, F. A. (2019). Bioinformatic identification of single nucleotide polymorphisms (SNPs) in candidate genes for fiber characteristics in alpacas (Vicugna pacos). Revista peruana de biología 26, 087–094. doi: 10.15381/rpb.v26i1.15911
Fernandez, D. (2015). Búsqueda de genes relacionados a la síntesis de la fibra y marcadores SSR en los ESTs de piel de alpaca Vicugna pacos. [dissertation/College degree]. Lima: Universidad Nacional Mayor de San Marcos.
Florez, F. (2016). Caracterización de marcadores genéticos en genes que codifican a proteínas asociadas a queratina y evaluación de la asociación del gen KRTAP11-1 al diámetro de fibra en alpaca (Vicugna pacos) siguiendo una aproximación de gen candidato. [dissertation/Master’s thesis]. Lima: Universidad Peruana Cayetano Heredia.
Guo, J., Tao, H., Li, P., Li, L., Zhong, T., Wang, L., et al. (2018). Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci Rep. 8:10405. doi: 10.1038/s41598-018-28719-w
Hack, W. (2001). The Peruvian Alpaca Meat and Hide Industries RIRDC Publication No. 01/19. Project No. TA001-18. https://www.agrifutures.com.au/wp-content/uploads/publications/01-019.pdf (accessed March 2011).
Hattori, T., Coustry, F., Stephens, S., Eberspaecher, H., Takigawa, M., Yasuda, H., et al. (2008). Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res. 36, 3011–3024. doi: 10.1093/nar/gkn150
Huelsken, J., Vogel, R., Erdmann, B., Cotsarelis, G., and Birchmeier, W. (2001). beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 18, 533–545.
Kerns, J. A., Newton, J., Berryere, T. G., Rubin, E. M., Cheng, J. F., Schmutz, S. M., et al. (2004). Characterization of the dog agouti gene and a non agouti mutation in German shepherd dogs. Mamm. Genome 15, 798–808. doi: 10.1007/s00335-004-2377-1
Lai, W., Hu, M., Zhu, W., Yu, F., Bai, C., Zhang, J., et al. (2019). A 4-bp deletion in the ASIP gene is associated with the recessive black coat colour in domestic guinea pigs (Cavia porcellus). Anim. Genet. 50, 190–191. doi: 10.1111/age.12766
Marín, J. C., Rivera, R., Varas, V., Cortés, J., Agapito, A., Chero, A., et al. (2018). Genetic variation in coat colour genes MC1R and ASIP provides insights into domestication and management of south american camelids. Front. Genet. 9:487. doi: 10.3389/fgene.2018.00487
Miller, S. M., Guthrie, A. J., and Harper, C. K. (2016). Single base-pair deletion in ASIP exon 3 associated with recessive black phenotype in impala (Aepyceros melampus). Anim. Genet. 47, 511–512. doi: 10.1111/age.12430
Morante, R., Goyache, F., Burgos, A., Cervantes, I., Pérez-Cabal, M. A., and Gutiérrez, J. P. (2009). Genetic improvement for alpaca fibre production in the peruvian altiplano: the pacomarca experience. Anim. Genetic Res. Infor. 2009, 37–43. doi: 10.1017/S1014233909990307
Morgenthaler, C., Diribarne, M., Capitan, A., Legendre, R., Saintilan, R., Gilles, M., et al. (2017). A missense variant in the coil1A domain of the keratin 25 gene is associated with the dominant curly hair coat trait (Crd) in horse. Genet. Sel. Evol. 49:85. doi: 10.1186/s12711-017-0359-355
Nazari-Ghadikolaei, A., Mehrabani-Yeganeh, H., Miarei-Aashtiani, S. R., Staiger, E. A., Rashidi, A., and Huson, H. J. (2018). Genome-wide association studies identify candidate genes for coat color and mohair traits in the Iranian markhoz goat. Front. Genet. 9:105. doi: 10.3389/fgene.2018.00105
Norris, B. J., and Whan, V. A. (2008). A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 18, 1282–1293. doi: 10.1101/gr.072090.107
Passeron, T., Valencia, J. C., Bertolotto, C., Hoashi, T., Le Pape, E., Takahashi, K., et al. (2007). SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc. Natl. Acad. Sci. U.S.A. 28, 13984–13989.
Quispe, E. C., Rodríguez, T. C., Iñiguez, L. R., and Mueller, J. P. (2009). Producción de fibra de alpaca, llama, vicuña y guanaco en Sudamérica. Anim. Genetic Res. Inform. 45, 1–14. doi: 10.1017/S1014233909990277
Raudsepp, T., and Chowdhary, B. P. (2008). FISH for mapping single copy genes. Methods Mol. Biol. 422, 31–49. doi: 10.1007/978-1-59745-581-7_3
Rieder, S., Taourit, S., Mariat, D., Langlois, B., and Guerin, G. (2001). Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm.Genome 12, 450–455. doi: 10.1007/s003350020017
Rompolas, P., and Greco, V. (2013). Stem cell dynamics in the hair follicle niche. Semin. Cell Dev. Biol. 2, 34–42.
Royo, L. J., Alvarez, I., Arranz, J. J., Fernandez, I., Rodriguez, A., Perez-Pardal, L., et al. (2008). Differences in the expression of the ASIP gene are involved in the recessive black coat colour pattern in sheep: evidence from the rare Xalda sheep breed. Anim. Genet. 39, 290–293. doi: 10.1111/j.1365-2052.2008.01712.x
Rupp, R., Mucha, S., Larroque, H., McEwan, J., and Conington, J. (2016). Genomic application in sheep and goat breeding. Anim Front. 2016, 39–44.
Shimomura, Y., Wajid, M., Petukhova, L., Kurban, M., and Christiano, A. M. (2010). Autosomal-dominant woolly hair resulting from disruption of keratin 74 (KRT74), a potential determinant of human hair texture. Am. J. Hum. Genet. 86, 632–638. doi: 10.1016/j.ajhg.2010.02.025
Simoes-Costa, M., and Bronner, M. E. (2013). Insights into neural crest development and evolution from genomic analysis. Genome Res. 2013, 1069–1080. doi: 10.1101/gr.157586.113
Simões-Costa, M., and Bronner, M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257. doi: 10.1242/dev.105445
Suzuki, H. (2013). Evolutionary and phylogeographic views on Mc1r and asip variation in mammals. Genes Genet. Syst. 2013, 155–164.
Taylor, K. M., Hungerford, D. A., Snyder, R. L., and Ulmer, F. A. Jr. (1968). Uniformity of karyotypes in the camelidae. Cytogenetics 7, 8–15.
Toivola, D. M., Boor, P., Alam, C., and Strnad, P. (2015). Keratins in health and disease. Curr. Opin. Cell Biol. 32, 73–81. doi: 10.1016/j.ceb.2014.12.008
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3 - new capabilities and interfaces. Nucleic Acids Res. 40:e115.
Wang, R., Chen, T., Zhao, B., Fan, R., Ji, K., Yu, X., et al. (2017). FGF21 regulates melanogenesis in alpaca melanocytes via ERK1/2-mediated MITF downregulation. Biochem. Biophys. Res. Commun. 490, 466–471. doi: 10.1016/j.bbrc.2017.06.064
Wiggans, G. R., Cole, J. B., Hubbard, S. M., and Sonstegard, T. S. (2017). Genomic selection in dairy cattle: the USDA Experience. Annu. Rev. Anim. Biosci. 5, 309–327. doi: 10.1146/annurev-animal-021815-111422
Xie, D., Gore, C., Liu, J., Pong, R. C., Mason, R., Hao, G., et al. (2010). Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 107, 2485–2490. doi: 10.1073/pnas.0908133107
Yu, S., Wang, G., and Liao, J. (2019). Association of a novel SNP in the ASIP gene with skin color in black-bone chicken. Anim. Genet. 50, 283–286. doi: 10.1111/age.12768
Keywords: alpaca, chromosomes, FISH, mapping, fiber, color, genes
Citation: Mendoza MN, Raudsepp T, Alshanbari F, Gutiérrez G and Ponce de León FA (2019) Chromosomal Localization of Candidate Genes for Fiber Growth and Color in Alpaca (Vicugna pacos). Front. Genet. 10:583. doi: 10.3389/fgene.2019.00583
Received: 29 October 2018; Accepted: 04 June 2019;
Published: 19 June 2019.
Edited by:
Pamela Burger, University of Veterinary Medicine Vienna, AustriaReviewed by:
Mohammed Piro, Agronomic and Veterinary Institute Hassan II, MoroccoCopyright © 2019 Mendoza, Raudsepp, Alshanbari, Gutiérrez and Ponce de León. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mayra N. Mendoza, TWF5cmEuTWVuZG96YS5jZXJuYUBvdXRsb29rLmNvbQ==; Gustavo Gutiérrez, Z3VzdGF2b2dyQGxhbW9saW5hLmVkdS5wZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.