
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 07 June 2019
Sec. Genetics of Common and Rare Diseases
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00507
This article is part of the Research Topic Genetics of Kidney Diseases View all 11 articles
Chronic kidney disease is a worldwide health crisis, while diabetic kidney disease (DKD) has become the leading cause of end-stage renal disease (ESRD). DKD is a microvascular complication and occurs in 30–40% of diabetes patients. Epidemiological investigations and clinical observations on the familial clustering and heritability in DKD have highlighted an underlying genetic susceptibility. Furthermore, DKD is a progressive and long-term diabetic complication, in which epigenetic effects and environmental factors interact with an individual’s genetic background. In recent years, researchers have undertaken genetic and epigenetic studies of DKD in order to better understand its molecular mechanisms. In this review, clinical material, research approaches and experimental designs that have been used for genetic and epigenetic studies of DKD are described. Current information from genetic and epigenetic studies of DKD and ESRD in patients with diabetes, including the approaches of genome-wide association study (GWAS) or epigenome-wide association study (EWAS) and candidate gene association analyses, are summarized. Further investigation of molecular defects in DKD with new approaches such as next generation sequencing analysis and phenome-wide association study (PheWAS) is also discussed.
Diabetes is a major public health problem that is approaching epidemic proportions globally. According to the latest report from the IDF, the prevalence of diabetes will increase from 425 million persons in 2017 to 629 million by 2045 (IDF 20171). Diabetic kidney disease (DKD, previously termed diabetic nephropathy, DN) is a microvascular complication and progresses gradually over many years in approximately 30–40% of individuals with T1D and T2D mellitus (Harjutsalo and Groop, 2014; Thomas et al., 2015; Barrett et al., 2017). DKD is now the main cause of chronic kidney disease (CKD) worldwide and the leading cause of end-stage-renal disease (ESRD) requiring renal replacement therapy (dialysis or transplantation). The presence of CKD is the single strongest predictor of mortality for persons with diabetes (Dousdampanis et al., 2016; Papadopoulou-Marketou et al., 2017). Pathological findings in DKD include glomerular hypertrophy, mesangial matrix expansion, reduced podocyte number, glomerulosclerosis, tubular atrophy and tubulointerstitial fibrosis. Clinical criteria used to diagnose the subjects with DKD are urine ACR higher than 300 mg/g, while microalbuminuria is diagnosed when ACR is between 30–300 mg/g (Bouhairie and McGill, 2016). Accumulating evidence has indicated that podocyte loss and epithelial dysfunction play important roles in DKD pathogenesis with further progression associated with inflammation but the exact molecular mechanisms responsible for DKD are not fully known (Badal and Danesh, 2014; Reidy et al., 2014; Gnudi et al., 2016).
Both clinical and epidemiological studies have demonstrated that there is familial aggregation of DKD in different ethnic groups, indicating that genetic factors contribute to development of the disease. Furthermore, genetic risk factors in DKD interact with the environmental factors (for example, lifestyle, diet and medication) (Freedman et al., 2007a; Murea et al., 2012; Thomas et al., 2012; Kato and Natarajan, 2014). Figure 1 is a schematic diagram representing the relationship between genetic, epigenetic and environmental factors that are involved in the development and progression of DKD. Genetic studies of DKD are mainly focused on association analyses between genomic DNA variation (for example, single nucleotide polymorphisms, SNPs, copy number variants, CNVs, and microsatellites) and clinical phenotypes of the disease (Freedman et al., 2007a; Gu and Brismar, 2012; Thomas et al., 2012; Florez, 2016). Epigenetics studies of DKD examine potentially heritable changes in gene expression that occur without variation in the original DNA nucleotide sequence (Villeneuve and Natarajan, 2010; Kato and Natarajan, 2014; Thomas, 2016; Keating et al., 2018). Therefore, epigenetic studies of DKD may provide information to help understand how environmental factors modify the expression of genes that are involved in DKD progression. Combined genetic, epigenetic and phenotypic studies together may generate information to understand new pathogenic pathways and to search for new biomarkers for early diagnosis and prediction as part of prevention programs in DKD. The results may also be useful in finding novel targets for the treatment of DKD.
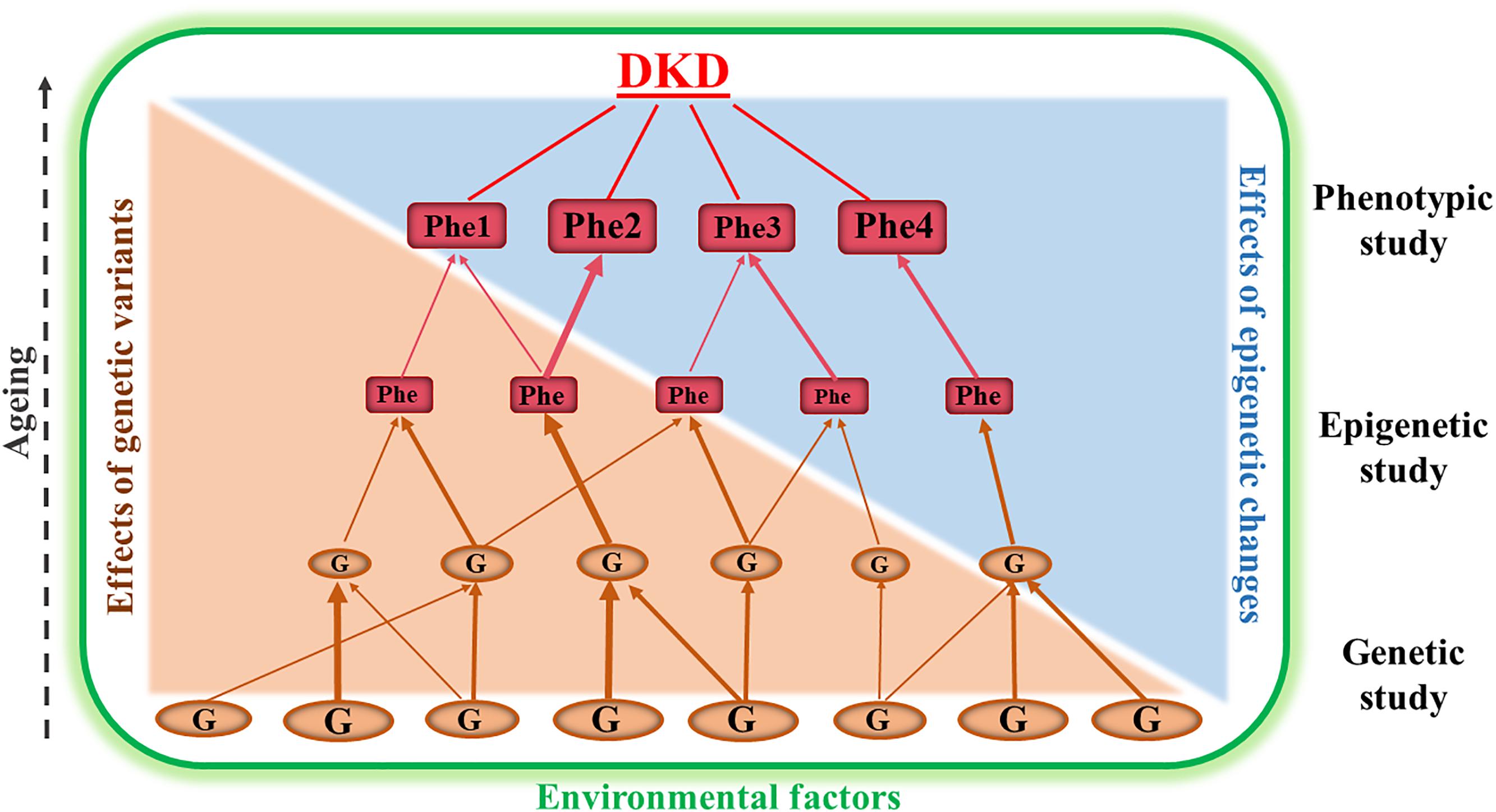
Figure 1. This is a schematic diagram representing the relationship between genetic, epigenetic and phenotypic studies in diabetic kidney disease (DKD). Genetic association studies are fundamentally important for identification of susceptibility or resistance genes (G). Epigenetic studies analyzing genomic DNA methylation changes, chromosome histone modification and ncRNA regulation are useful for dissecting the interaction of the genes with environmental factors. The combined data from genetic, epigenetic and phenotypic (Phe) studies may provide the opportunity for us to understand new pathways underlying the pathogenesis of DKD and to discover new biomarkers for early diagnosis and to find targets for prevention and treatment programs of this disease. The different sizes of the ‘G” and “Phe” represent the variation of genetic and phenotypic effects.
SNPs are the most common form of genomic DNA variation. The updated dbSNP database of more than 500 million reference SNPs (rs) with allele frequency data2 has provided fundamental information for genetic studies of complex diseases including, DKD. The genetic studies in DKD have implicated previously unsuspected biological pathways and subsequently improved our knowledge for understanding of the genetic basis of the disease. For most common traits studied in DKD, however, the identified genes and their SNPs only explain a fraction of associated risk, suggesting that human genomic DNA variations are only a part of underlying susceptibility to DKD. This has led to evolving interest in epigenetics to help explain some of the missing heritability of DKD. Epigenetic mechanisms mainly consist of DNA methylation, chromosome histone modification and non-coding RNA (ncRNA) regulation (Kato and Natarajan, 2014; Allis and Jenuwein, 2016). Epigenetic related ncRNAs include miRNA, siRNA, piRNA, and lncRNA (Holoch and Moazed, 2015). There are more than 30,000 identified CpG islands in the human genome. Detailed information for these CpG islands can be found in the public database3. The CpG islands are defined as stretches of DNA > 200 bp long with a GC percentage greater than 50% and an observed-to-expected CpG ratio of more than 60%. The CpG islands are often found at promoters and contain the 5′ end of the transcript, while DNA methylation occurs at 5′-cytosines of “CpG” dinucleotides4 (Cross and Bird, 1995). In DKD, the effects of DNA methylation have been studied in terms of transgenerational inheritance of the disease to explore environmental and other non-genetic factors that may influence epigenetic modifications in the genes involved in DKD (Deaton and Bird, 2011; Jones, 2012). Identification of differentially methylated CpG sites in promoters or other functional regions of genes and the analysis of the DNA methylation changes that are associated with DKD have become the most common approaches used in epigenetic studies of the disease (Villeneuve and Natarajan, 2010; Kato and Natarajan, 2014; Thomas, 2016). Furthermore, ncRNAs, particularly long ncRNAs are known to be involved in epigenetic processes. ncRNAs certainly play an important role in chromatin formation, histone modification, DNA methylation and consequently gene transcription silencing.
Genetic and epigenetic studies of DKD, initially using candidate gene approaches and more recently at genome-wide scale (known as GWAS and EWAS), have been undertaken to identify many genes conferring susceptibility or resistance to DKD. In this review, clinical phenotypes, research approaches and experimental designs that have been used for genetic and epigenetic studies of DKD are described. These research approaches and experimental designs can also be used for study of CKD. Current information from genetic and epigenetic studies of DKD is summarized. Further investigation of molecular defects in DKD with new generation sequencing analyses and phenome-wide association studies (PheWAS) are discussed.
Two major research approaches either at genome-wide scale or focused on candidate gene(s) have been widely used for comparative studies between cases (patients with DKD) and controls (diabetes patients without DKD). Case-control studies by recruiting large numbers of subjects can increase the statistical power of reported associations. The aim is to discover the genes presented differentially in genomic structure or genetic expression. Genome-wide or epigenome-wide association studies (GWAS or EWAS) are hypothesis-generating approaches (Rakyan et al., 2011; Do et al., 2017; Lappalainen and Greally, 2017). These study designs have benefited from rapid development of human genome research, including the creation of publicly available databases of SNPs, haplotypes and CpG islands and the rapid technical improvements in analyzing genomic variation using high-throughput techniques and high-density SNP or CpG arrays. Another approach is to focus on candidate genes and study a more limited number of genes potentially involved in the pathogenesis of DKD based upon our known knowledge or hypothesis. In genetic and epigenetic studies of DKD, DNA samples used are commonly extracted from peripheral blood samples because they are clinically accessible. Dick et al. (2014) have comparatively analyzed DNA methylation changes related to BMI by using both approaches of whole-blood DNA methylation profiling and adipose tissue specific methylation measurement. Data suggests that analysis of blood DNA methylation is worthwhile because the results can reflect the DNA methylation changes in relevant tissues for a particular phenotype. Nevertheless, there is still limited information concerning the correlation between whole blood DNA methylation profiles and kidney tissue specific DNA methylation changes in part due to the heterogeneity of cell types within the kidney. To improve the tissue specific DNA methylation analysis of kidney diseases, including DKD, it is necessary to construct biobanks of renal biopsies. Karolinska Institutet has established a biobank in KaroKidney with more than 750 renal biopsies5. The advantages and limitations of these two approaches, as well as the clinical materials and experimental design used in genetic and epigenetic studies of DKD are summarized in Table 1.
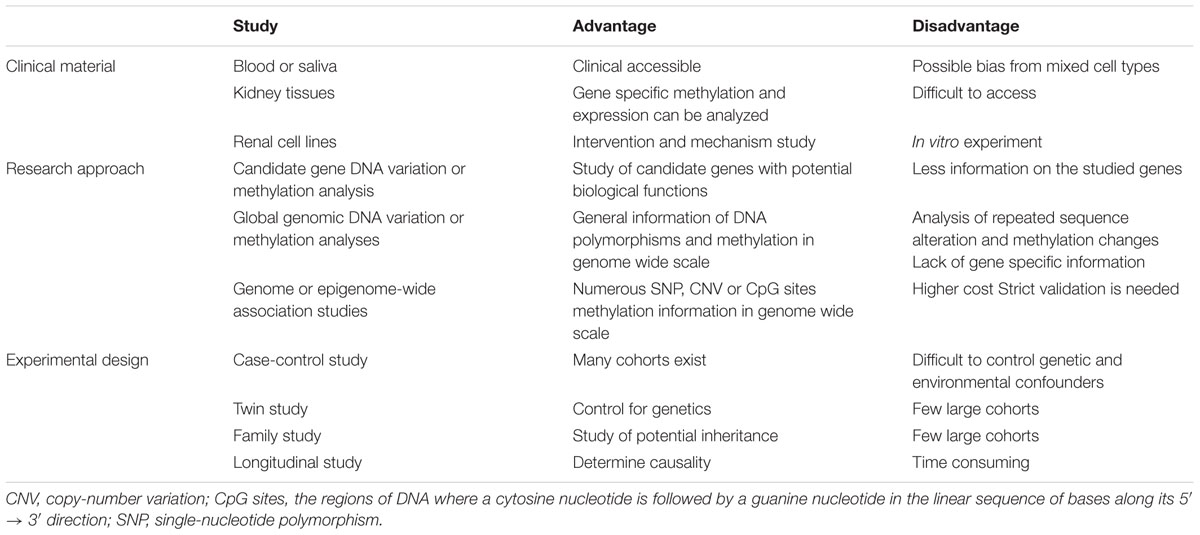
Table 1. Clinical material, research approaches and experimental designs used in genetic and epigenetic studies of diabetic kidney disease.
Considerable amounts of data from genetic studies in DKD have accumulated. A list of the genes that are reported to be associated with susceptibility or resistance to DKD are summarized in Table 2. The genes are listed in alphabetical order. Surprisingly, there are more than 150 genes. Most of them have been identified by genetic association studies employing candidate gene approaches over the past 20 years. Furthermore, a number of GWAS in DKD have been published in the last 10 years. By using GWAS approaches, approximately 33 genes have been found to be associated with the DKD, i.e., ABCG2, AFF3, AGER, APOL1, AUH, CARS, CERS2, CDCA7/SP3, CHN2, CNDP1, ELMO1, ERBB4, FRMD3, GCKR, GLRA3, KNG1, LIMK2, MMP9, NMUR2, MSRB3/HMGA2, MYH9, PVT1, RAET1L, RGMA/MCTP2, RPS12, SASH1, SCAF8/CNKSR3, SHROOM3, SLC12A3, SORBS1, TMPO, UMOD, and ZMIZ1 (Hanson et al., 2007; Sandholm et al., 2012, 2014; Maeda et al., 2013; Thameem et al., 2013; Bailey et al., 2014; Palmer et al., 2014; Guan et al., 2016; Teumer et al., 2016; Lim et al., 2017; Roden, 2017; Charmet et al., 2018; van Zuydam et al., 2018). However, most of these genes (∼80%) reportedly associated with DKD still need to be confirmed by further replication studies and detailed analysis of their functional role in DKD in experimental models. Polymorphisms in these candidate genes association with DKD studies are listed in Table 2A, while their potential biological relevance and genetic effects in DKD are briefly described. Of them, 34 genes are originally predicted by GWAS and the statistical association with DKD summarized in Table 2B.
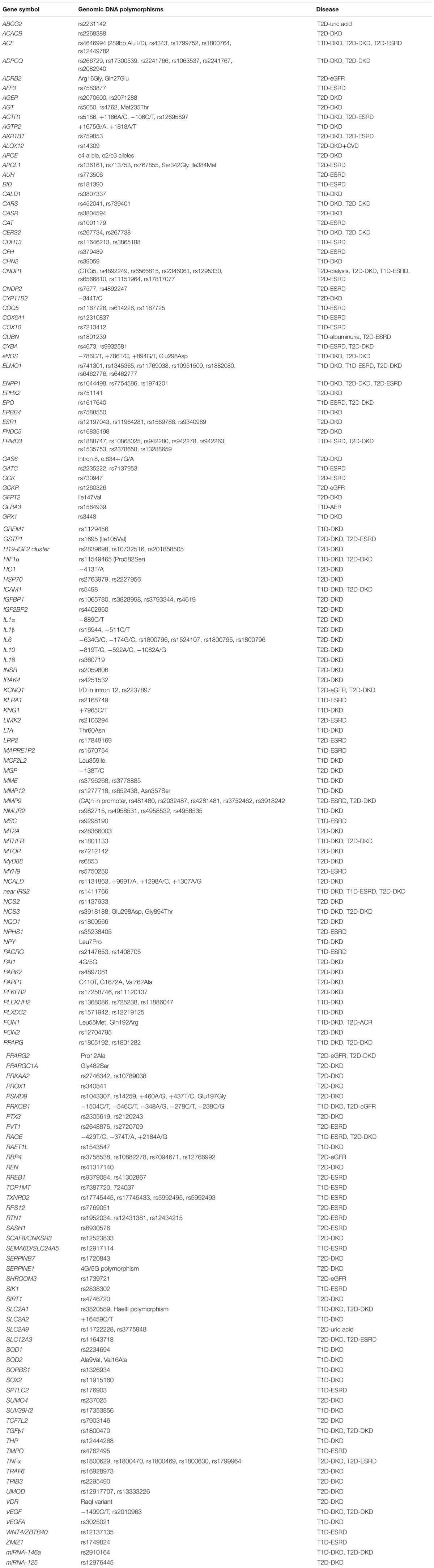
Table 2A. Current data from genetic association studies in diabetic kidney disease by using candidate gene approach.
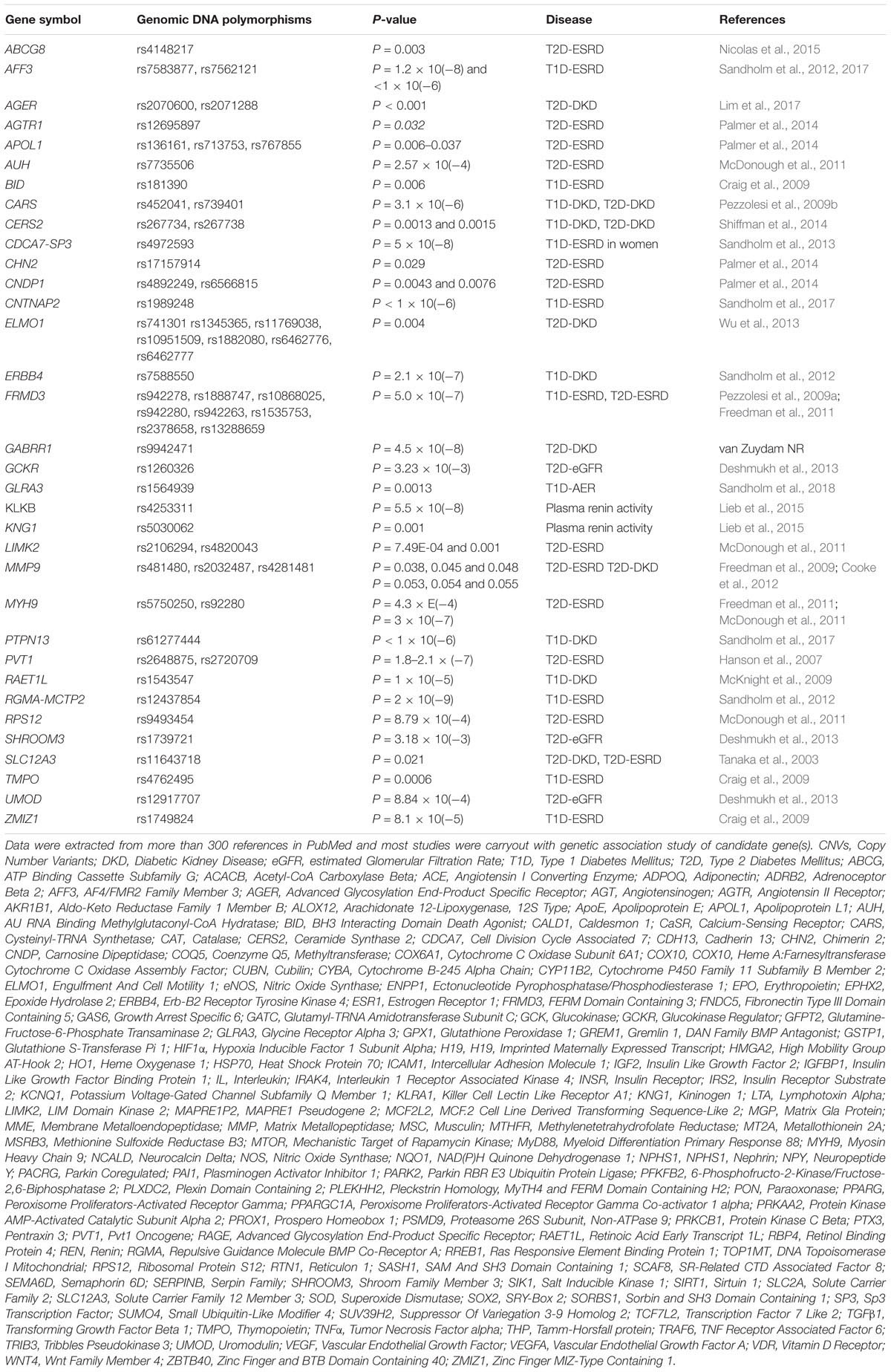
Table 2B. Current data from genetic association studies in diabetic kidney disease by using genome wide association approach.
The CNDP1 (carnosine dipeptidase 1) gene is located in chromosome 18q22.3 and contains 5-leucine (CTG) trinucleotide repeat length polymorphism (D18S880) in the coding region (Wanic et al., 2008). This trinucleotide repeat polymorphism is found to have gender specificity and to confer the susceptibility for DKD and ESRD in T2D (Albrecht et al., 2017b). Furthermore, serum carnosinase (CN-1) activity is negatively correlated with time on hemodialysis (Peters et al., 2016). In addition, several SNPs in this gene are also associated with DKD and ESRD (Janssen et al., 2005; Freedman et al., 2007b; McDonough et al., 2009; Alkhalaf et al., 2010; Mooyaart et al., 2010; Ahluwalia et al., 2011b; Chakkera et al., 2011; Kurashige et al., 2013). Interestingly, an experimental study in BTBR ob/ob mice has demonstrated that treatment with carnosine as the target of CNDP1 improves glucose metabolism and albuminuria, suggesting that carnosine may be a novel therapeutic strategy to treat patients with DKD (Albrecht et al., 2017a).
The ELMO1 (engulfment and cell motility 1) gene is located on chromosome p14.1 and encodes a member of the engulfment and cell motility protein family. The protein interacts with dedicator of cytokinesis proteins and subsequently promotes phagocytosis and cell migration. Increased expression of ELMO1 and dedicator of cytokinesis 1 may promote glioma cell invasion (Patel et al., 2010). Furthermore, several SNPs in this gene are found to be associated with DKD in both T1D and T2D (Shimazaki et al., 2005, 2006; Craig et al., 2009; Leak et al., 2009; Pezzolesi et al., 2009a; Hanson et al., 2010; Wu et al., 2013; Alberto Ramirez-Garcia et al., 2015; Bodhini et al., 2016; Hathaway et al., 2016; Mehrabzadeh et al., 2016; Sharma et al., 2016). The variants associated with DKD, however, are different in the several populations studied, suggesting the presence of allelic heterogeneity probably resulting from the diverse ancestral genetic backgrounds of the different racial groups.
The FRMD3 (FERM domain containing 3) gene is located in chromosome 9q21.32. The FRMD3 gene is expressed in adult brain, fetal skeletal muscle, thymus, ovaries, and podocytes (Ni et al., 2003). Pezzolesi et al. (2009b) have demonstrated that FRMD3 expression in kidneys of a DKD mouse model is decreased as compared with non-diabetic mice. Genetic polymorphisms in the FRMD3 gene are associated with DKD and ESRD in T1D and T2D (Freedman et al., 2011; Al-Waheeb et al., 2016). Furthermore, the members of the bone morphogenetic protein (BMP) interact with FRMD3, which implies that FRMD3 may influence the risk of DKD through regulation of the BMP pathway (Martini et al., 2013; Palmer and Freedman, 2013).
The MMP9 (matrix metallopeptidase 9) gene is located in chromosome 20q13.12. The MMP family members are involved in the breakdown of extracellular matrix (ECM) in physiological processes, such as tissue remodeling, reproduction and embryonic development, while MMP9 is the ninth member in the family. MMP9 may play an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. Moreover, MMPs, including MMP9, are zinc-dependent endopeptidases and the major proteases in ECM degradation. There are common variants such as rs3918242 (-1562C/T) and microsatellites (CA)n in the promoter region and several SNPs rs481480, rs2032487, rs4281481, rs3752462 and rs3918242 are found to be associated with the susceptibility to DKD (Hirakawa et al., 2003; Nair et al., 2008; Ahluwalia et al., 2009; Freedman et al., 2011; Cooke et al., 2012; Zhang et al., 2015; Feng et al., 2016).
Both UMOD (uromodulin) and SLC12A3 (solute carrier family 12 member 3) genes are located in the same chromosome but in short and long arms, respectively, i.e., 16p12.3 and 16q13. SLC12A3 is also known as thiazide-sensitive sodium-chloride cotransporter in kidney distal convoluted tubules, which is important for electrolyte homeostasis. Mutations in this gene are characterized by hypokalemic alkalosis combined with hypomagnesemia, low urinary calcium, but increased renin activity. Tanaka et al. (2003) performed a GWAS in Japanese T2D subjects and reported that the SLC12A3 Arg913Gln polymorphism was associated with reduced risk of DKD. Nishiyama et al. (2005) then conducted another 10-year longitudinal study in the same population. The results confirmed that the 913Gln allele of SLC12A3 Arg913Gln polymorphism conferred a protective effect in DKD (Nishiyama et al., 2005). More recently, Abu Seman et al. (2014) performed a further genetic study of SLC12A3 polymorphisms in a Malaysian population, including the meta-analysis of the association between the SLC12A3 Arg913Gln polymorphism and DKD from all the previous studies. SLC12A3 Arg913Gln polymorphism was found to be associated with T2D (P = 0.028, OR = 0.772, 95% CI = 0.612–0.973) and DKD (P = 0.038, OR = 0.547, 95% CI = 0.308–0.973) in the Malaysian cohort. The meta-analysis confirmed the protective effects of the SLC12A3 913Gln allele in DKD (Z-value = -1.992, P = 0.046, OR = 0.792). In addition, the authors investigated the role of slc12a3 expression in the progress of DKD with db/db mice and in kidney development with zebrafish embryos. With knockdown of zebrafish ortholog, slc12a3 led to structural abnormality of kidney pronephric distal duct at 1-cell stage. Slc12a3 mRNA and protein expression levels were upregulated in kidneys of db/db mice from 6, 12, and 26 weeks at the age. The authors thus concluded that SLC12A3 is a susceptibility gene in DKD, while allele 913Gln but not allele Arg913 has a preventive effect in the disease (Abu Seman et al., 2014). This association of the SLC12A3 Arg913Gln polymorphism with DKD has been very recently replicated in a Chinese population (Zhang et al., 2018). The UMOD gene encoded glycoprotein is synthesized exclusively in renal tubular cells and released into urine. Furthermore, UMOD may prevent urinary tract infection and inhibit formation of liquid containing supersaturated salts and subsequent formation of salt crystals. SNPs rs4293393 and rs1297707 in the UMOD gene are found to be associated with the susceptibility to DKD in T2D (Ahluwalia et al., 2011a; Prudente et al., 2017; van Zuydam et al., 2018).
The Human Genome Project has revealed that there are more than twenty thousand protein coding genes, and probably more than one million of RNA genes6. Genetic association studies of RNA gene polymorphisms with DKD are very limited. Up to date, only two SNPs, i.e., rs2910164 and rs12976445 in the genes for miRNA-146a and miRNA-125 have been found to be associated with DKD in T1D and T2D (Li et al., 2014; Kaidonis et al., 2016). Further investigation of RNA genetic variation conferring susceptibility to DKD needs to be undertaken.
Similar to genetic association studies, epigenome-wide (EWAS) and candidate gene DNA methylation analyses have been used for epigenetic studies of DKD. Current information from epigenetic studies in DKD are represented in Table 3. An EWAS suggested that several genes, including SLC22A12, TRPM6, AQP9, HP, AGTX, and HYAL2, may have epigenetic effects in DKD (VanderJagt et al., 2015). Interestingly, SLC22A12 encodes for urate anion transporter 1 (URAT1), which is a kidney-specific urate transporter that transports urate across the apical membrane of the proximal tubule in kidneys. Loss-of-function SLC22A12 mutations are associated with renal hypouricaemia and affected persons can develop exercise-induced acute kidney injury and are at increased risk of developing urate stones (Lee et al., 2008). TRPM6 is a member of transient receptor potential superfamily of cation channels. This gene is widely expressed in the body, including kidneys along the nephron. The TRPM6 channels are mainly located in the renal distal convoluted tubule, the site of active transcellular calcium and magnesium transport in the kidney (Felsenfeld et al., 2015). As described previously, several studies have implicated UMOD genetic polymorphisms in the susceptibility to DKD (Ahluwalia et al., 2011a; Prudente et al., 2017; van Zuydam et al., 2018). A recent study has demonstrated that UMOD regulates renal magnesium homeostasis through TRPM6 (Nie et al., 2018). Furthermore, analyses of the candidate genes such as IGFBP1 and MTHFR have also provided evidence that DNA methylation changes in these genes may be involved in the pathogenesis of DKD (Gu et al., 2013, 2014; Yang et al., 2016). Combining and analyzing data from genetic and epigenetic studies together may help understand some of the pathophysiology in DKD.
ncRNAs regulate gene expression at the post-transcriptional level and are involved in chromatin histone modification. Most of studies concerning histone modification and ncRNA dysregulation have been performed in diabetic animal models, while a few studies have been undertaken in subjects with DKD (Table 3). Reddy et al. (2014) have analyzed histone modification profiles in genes associated with DKD pathology and the modified regulation of these genes following treatment with the angiotensin II type 1 receptor (AT1R) blocker losartan. The data indicate that losartan attenuates key parameters of DKD and modifies gene expression, and reverses some epigenetic changes in db/db mice. Losartan also attenuates increased H3K9/14Ac at RAGE, PAI-1, and MCP-1 promoters in mesangial cells cultured under diabetic conditions (Reddy et al., 2014). In a recent study of subjects of T2D and diabetic complications (including DKD) (Dos Santos Nunes et al., 2018) the methylation profiles of miR gene were compared and related to the presence of diabetic complications. Results indicated that miRs can modulate the expression of a variety of genes and methylation changes of miR-9-3, miR-34a, and miR-137 were found to be associated with diabetic complications (Dos Santos Nunes et al., 2018). These two studies provide evidence suggesting that therapies targeting epigenetic regulators might be beneficial in the treatment of DKD.
Researchers have made major efforts to undertake well powered genetic and epigenetic studies in DKD to help understand its pathogenesis. The data, however, need to be confirmed by several strategies, for instance, replication studies could be performed with better selection of subjects with similar genetic background to limit influences from migration; intermarriage; cultural preferences; coupled with further investigation of DNA variation and methylation changes in RNA regulation genes and biological experiments to determine functional impact of these variants. Furthermore, new technologies for DNA and ncRNA sequencing analysis such as third generation sequencing and a PheWAS approach have recently been developed.
DNA sequencing analysis is used for determining the accurate order of nucleotides along chromosomes and genomes. Second-generation sequencing, commonly known as next-generation sequencing (NGS), has presently become popular in DNA sequencing analysis because NGS can enable a massively-paralleled approach capable of producing large numbers of reads at high coverages along the genome and therefore dramatically reduce the cost of DNA sequencing analysis (Treangen and Salzberg, 2011; Gu et al., 2018; Mone et al., 2018). Today, third-generation sequencing (often called as long-read sequencing) is a new generation sequencing method, which works by reading the nucleotide sequences at single molecule level in contrast to the first and second generations of DNA sequencing (van Dijk et al., 2018). Moreover, it is necessary to develop the molecular instruments for whole genome sequencing to make this new generation sequencing commercially available. The advanced sequencing technologies will improve genetic and epigenetic studies in DKD in the near future.
In the human genome, RNA genes are much more abundant than protein coding genes, while ncRNAs mainly include miRNAs and lncRNAs. Both forms of ncRNAs have been found to be involved in chromatin histone modifications, and subsequently can have epigenetic effects on the target genes. Therefore, identification of RNA genetic variation and investigation of biological alteration of these RNA genes should be included in research plans. Kato has very recently pointed out a hypothesis that transforming growth factor-β (TGF1β) may play an important role in early stage development of DKD, while some miRNAs and lncRNAs regulate the key molecules in the TGF1β pathway. These ncRNAs may be served as biomarkers for predicting the potential targets for prevention and treatment in DKD (Kato, 2018). Furthermore, Smyth et al. (2018) have compared Sanger sequencing and NGS to validate the five top ranked miRNAs that are predicted to be associated with DKD by EWAS. This study suggests that targeted NGS may offer a more cost-effective and sensitive approach and implied that the methylated miR-329-2, in which region SNP rs10132943 is located, and miR-429 where SNPs rs7521584 and rs112695918 exist, are associated with DKD (Smyth et al., 2018). Although these two studies are preliminary, they may be good examples to help direct further DKD research.
PheWAS is a new approach to analyze many phenotypes in comparison with a single genetic variant. This approach was originally described using electronic medical record (EMR) data from EMR-linked with a DNA biobank and also can be combined with GWAS and EWAS. Therefore, PheWAS has become a powerful tool to investigate the impact of genetic variation on drug response among many individuals and may expand our knowledge of new drug targets and effects (Pendergrass and Ritchie, 2015; Denny et al., 2016; Roden, 2017). Clearly, combined with GWAS and EWAS, PheWAS will provide us with the possibility to discover the associations with drug effects, including therapeutic response and side effect profiles in DKD (Hebbring, 2014).
Taken together, application of these advanced studies in DKD will be very useful not only for evaluating current data from genetic and epigenetic studies but also for generating new knowledge for dissecting the complexity of this disease.
The author confirms being the sole contributor of this work and has approved it for publication.
The study was supported by the Start Grant from China Pharmaceutical University.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACR, albumin-to-creatinine ratio; ADA, American Diabetes Association; BMI, body mass index; CNV, copy number variant; DKD, diabetic kidney disease; ESRD, end-stage renal disease; EWAS, epigenome-wide association study; GFR, glomerular filtration rate; GWAS, genome-wide association study; IDF, International Diabetes Federation; IHME, Institute for Health Metrics and Evaluation; LD, Linkage disequilibrium; PheWAS, phenome-wide association study; SNP, single nucleotide polymorphism; T1D, type 1 diabetes; T2D, type 2 diabetes; UAE, urinary albumin excretion.
Abu Seman, N., He, B., Ojala, J. R., Wan Mohamud, W. N., Östenson, C. G., Brismar, K., et al. (2014). Genetic and biological effects of sodium-chloride cotransporter (SLC12A3) in diabetic nephropathy. Am. J. Nephrol. 40, 408–416. doi: 10.1159/000368916
Ahluwalia, T. S., Khullar, M., Ahuja, M., Kohli, H. S., Bhansali, A., Mohan, V., et al. (2009). Common variants of inflammatory cytokine genes are associated with risk of nephropathy in type 2 diabetes among Asian Indians. PLoS One 4:e5168. doi: 10.1371/journal.pone.0005168
Ahluwalia, T. S., Lindholm, E., Groop, L., and Melander, O. (2011a). Uromodulin gene variant is associated with type 2 diabetic nephropathy. J. Hypertens. 29, 1731–1734. doi: 10.1097/HJH.0b013e328349de25
Ahluwalia, T. S., Lindholm, E., and Groop, L. C. (2011b). Common variants in CNDP1 and CNDP2, and risk of nephropathy in type 2 diabetes. Diabetologia 54, 2295–2302. doi: 10.1007/s00125-011-2178-5
Alberto Ramirez-Garcia, S., Charles-Niño, C., Mazariegos-Rubí, M., Rosalba Topete-González, L., Topete-González, R., Javier Flores-Alvarado, L., et al. (2015). Association of the ELMO1 gene (snp rs1345365) with development of type 2 diabetes mellitus in the Mexican mestizo population. Invest. Clin. 56, 341–355.
Albrecht, T., Schilperoort, M., Zhang, S., Braun, J. D., Qiu, J., Rodriguez, A., et al. (2017a). Carnosine attenuates the development of both type 2 diabetes and diabetic nephropathy in BTBR ob/ob mice. Sci. Rep. 7:44492. doi: 10.1038/srep44492
Albrecht, T., Zhang, S., Braun, J. D., Xia, L., Rodriquez, A., and Qiu, J. (2017b). The CNDP1 (CTG)(5) polymorphism is associated with biopsy-proven diabetic nephropathy, time on hemodialysis, and diabetes duration. J. Diabetes Res. 2017:9506730. doi: 10.1155/2017/9506730
Aldemir, O., Turgut, F., and Gokce, C. (2017). The association between methylation levels of targeted genes and albuminuria in patients with early diabetic kidney disease. Ren. Fail. 39, 597–601. doi: 10.1080/0886022X.2017.1358180
Alkhalaf, A., Bakker, S. J., Bilo, H. J., Gans, R. O., Navis, G. J., Postmus, D., et al. (2010). A polymorphism in the gene encoding carnosinase (CNDP1) as a predictor of mortality and progression from nephropathy to end-stage renal disease in type 1 diabetes mellitus. Diabetologia 53, 2562–2568. doi: 10.1007/s00125-010-1863-0
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Alvarez, M. L., Khosroheidari, M., Eddy, E., and Kiefer, J. (2013). Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS One 8:e77468. doi: 10.1371/journal.pone.0077468
Al-Waheeb, S., Alwohhaib, M., Abdelghani, A., Al-Sharrah, S., Al-Shafey, E., Al-Sahow, A., et al. (2016). Evaluation of associations between single nucleotide polymorphisms in the FRMD3 and CARS genes and diabetic nephropathy in a Kuwaiti population. Genet. Mol. Res. 15:gmr7619. doi: 10.4238/gmr.15017619
Badal, S. S., and Danesh, F. R. (2014). New insights into molecular mechanisms of diabetic kidney disease. Am. J. Kidney Dis. 63(2 Suppl. 2), S63–S83. doi: 10.1053/j.ajkd.2013.10.047
Bailey, J. N. C., Palmer, N. D., Ng, M. C. Y., Bonomo, J. A., Hicks, P. J., Hester, J. M., et al. (2014). Analysis of coding variants identified from exome sequencing resources for association with diabetic and non-diabetic nephropathy in African Americans. Hum. Genet. 133, 769–779. doi: 10.1007/s00439-013-1415-z
Barrett, E. J., Liu, Z., Khamaisi, M., King, G. L., Klein, R., Klein, B. E. K., et al. (2017). Diabetic microvascular disease: an endocrine society scientific statement. J. Clin. Endocrinol. Metab. 102, 4343–4410. doi: 10.1210/jc.2017-01922
Bell, C. G., Teschendorff, A. E., Rakyan, V. K., Maxwell, A. P., Beck, S., and Savage, D. A. (2010). Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med. Genomics 3:33. doi: 10.1186/1755-8794-3-33
Bock, F., Shahzad, K., Wang, H., Stoyanov, S., Wolter, J., Dong, W., et al. (2013). Activated protein C ameliorates diabetic nephropathy by epigenetically inhibiting the redox enzyme p66Shc. Proc. Natl. Acad. Sci. U.S.A. 110, 648–653. doi: 10.1073/pnas.1218667110
Bodhini, D., Chidambaram, M., Liju, S., Revathi, B., Laasya, D., Sathish, N., et al. (2016). Association of rs11643718 SLC12A3 and rs741301 ELMO1 variants with diabetic nephropathy in south Indian population. Ann. Hum. Genet. 80, 336–341. doi: 10.1111/ahg.12174
Cai, M., Bompada, P., Atac, D., Laakso, M., Groop, L., and De Marinis, Y. (2016). Epigenetic regulation of glucose-stimulated osteopontin (OPN) expression in diabetic kidney. Biochem. Biophys. Res. Commun. 469, 108–113. doi: 10.1016/j.bbrc.2015.11.079
Chakkera, H. A., Hanson, R. L., Kobes, S., Millis, M. P., Nelson, R. G., Knowler, W. C., et al. (2011). Association of variants in the carnosine peptidase 1 gene (CNDP1) with diabetic nephropathy in American Indians. Mol. Genet. Metab. 103, 185–190. doi: 10.1016/j.ymgme.2011.02.010
Charmet, R., Duffy, S., Keshavarzi, S., Gyorgy, B., Marre, M., Rossing, P., et al. (2018). Novel risk genes identified in a genome-wide association study for coronary artery disease in patients with type 1 diabetes. Cardiovasc. Diabetol. 17:61. doi: 10.1186/s12933-018-0705-0
Chen, J., Guo, Y., Zeng, W., Huang, L., Pang, Q., Nie, L., et al. (2014). ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. Am. J. Physiol. Renal Physiol. 306, F916–F925. doi: 10.1152/ajprenal.00697.2012
Cooke, J. N., Bostrom, M. A., Hicks, P. J., Ng, M. C., Hellwege, J. N., Comeau, M. E., et al. (2012). Polymorphisms in MYH9 are associated with diabetic nephropathy in European Americans. Nephrol. Dial. Transplant. 27, 1505–1511. doi: 10.1093/ndt/gfr522
Craig, D. W., Millis, M. P., and DiStefano, J. K. (2009). Genome-wide SNP genotyping study using pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to Type 1 diabetes. Diabet. Med. 26, 1090–1098. doi: 10.1111/j.1464-5491.2009.02846.x
De Marinis, Y., Cai, M., Bompada, P., Atac, D., Kotova, O., Johansson, M. E., et al. (2016). Epigenetic regulation of the thioredoxin-interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney Int. 89, 342–353. doi: 10.1016/j.kint.2015.12.018
Deaton, A. M., and Bird, A. (2011). CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022. doi: 10.1101/gad.2037511
Denny, J. C., Bastarache, L., and Roden, D. M. (2016). Phenome-wide association studies as a tool to advance precision medicine. Annu. Rev. Genomics Hum. Genet. 17, 353–373. doi: 10.1146/annurev-genom-090314-024956
Deshmukh, H. A., Palmer, C. N., Morris, A. D., and Colhoun, H. M. (2013). Investigation of known estimated glomerular filtration rate loci in patients with type 2 diabetes. Diabet. Med. 30, 1230–1235. doi: 10.1111/dme.12211
Dick, K. J., Nelson, C. P., Tsaprouni, L., Sandling, J. K., Aïssi, D., Wahl, S., et al. (2014). DNA methylation and body-mass index: a genome-wide analysis. Lancet 383, 1990–1998. doi: 10.1016/S0140-6736(13)62674-4
Do, C., Shearer, A., Suzuki, M., Terry, M. B., Gelernter, J., Greally, J. M., et al. (2017). Genetic-epigenetic interactions in cis: a major focus in the post-GWAS era. Genome Biol. 18:120. doi: 10.1186/s13059-017-1250-y
Dos Santos Nunes, M. K., Silva, A. S., Wanderley de Queiroga Evangelista, I., Modesto Filho, J., Alves Pegado Gomes, C. N., Ferreira do Nascimento, R. A., et al. (2018). Analysis of the DNA methylation profiles of miR-9-3, miR-34a, and miR-137 promoters in patients with diabetic retinopathy and nephropathy. J. Diabetes Complications 32, 593–601. doi: 10.1016/j.jdiacomp.2018.03.013
Dousdampanis, P., Trigka, K., and Mouzaki, A. (2016). Tregs and kidney: from diabetic nephropathy to renal transplantation. World J. Transplant. 6, 556–563. doi: 10.5500/wjt.v6.i3.556
Felsenfeld, A. J., Levine, B. S., and Rodriguez, M. (2015). Pathophysiology of calcium, phosphorus, and magnesium dysregulation in chronic kidney disease. Semin. Dial. 28, 564–577. doi: 10.1111/sdi.12411
Feng, S., Ye, G., Bai, S., Wei, H., Liao, X., and Li, L. (2016). Matrix metalloproteinase-9 -1562C/T gene polymorphism is associated with diabetic nephropathy. Biomed Res. Int. 2016:1627143. doi: 10.1155/2016/1627143
Freedman, B. I., Bostrom, M., Daeihagh, P., and Bowden, D. W. (2007a). Genetic factors in diabetic nephropathy. Clin. J. Am. Soc. Nephrol. 2, 1306–1316.
Freedman, B. I., Hicks, P. J., Bostrom, M. A., Comeau, M. E., Divers, J., Bleyer,A. J., et al. (2009). Non-muscle myosin heavy chain 9 gene MYH9 associations in African Americans with clinically diagnosed type 2 diabetes mellitus-associated ESRD. Nephrol. Dial. Transplant. 24, 3366–3371. doi: 10.1093/ndt/gfp316
Freedman, B. I., Hicks, P. J., Sale, M. M., Pierson, E. D., Langefeld, C. D., Rich, S. S., et al. (2007b). A leucine repeat in the carnosinase gene CNDP1 is associated with diabetic end-stage renal disease in European Americans. Nephrol. Dial. Transplant. 22, 1131–1135. doi: 10.1093/ndt/gfl717
Freedman, B. I., Langefeld, C. D., Lu, L., Divers, J., Comeau, M. E., Kopp, J. B., et al. (2011). Differential effects of MYH9 and APOL1 risk variants on FRMD3 association with diabetic ESRD in African Americans. PLoS Genet. 7:e1002150. doi: 10.1371/journal.pgen.1002150
Gilbert, R. E., Huang, Q., Thai, K., Advani, S. L., Lee, K., Yuen, D. A., et al. (2011). Histone deacetylase inhibition attenuates diabetes-associated kidney growth: potential role for epigenetic modification of the epidermal growth factor receptor. Kidney Int. 79, 1312–1321. doi: 10.1038/ki.2011.39
Gnudi, L., Coward, R. J. M., and Long, D. A. (2016). Diabetic nephropathy: perspective on novel molecular mechanisms. Trends Endocrinol. Metab. 27, 820–830. doi: 10.1016/j.tem.2016.07.002
Gu, H. F., and Brismar, K. (2012). Genetic association studies in diabetic nephropathy. Curr. Diabetes Rev. 8, 336–344. doi: 10.2174/157339912802083522
Gu, T., Falhammar, H., Gu, H. F., and Brismar, K. (2014). Epigenetic analyses of the insulin-like growth factor binding protein 1 gene in type 1 diabetes and diabetic nephropathy. Clin. Epigenetics 6:10. doi: 10.1186/1868-7083-6-10
Gu, T., Gu, H. F., Hilding, A., Sjöholm, L. K., Ostenson, C. G., Ekström, T. J., et al. (2013). Increased DNA methylation levels of the insulin-like growth factor binding protein 1 gene are associated with type 2 diabetes in Swedish men. Clin. Epigenetics 5:21. doi: 10.1186/1868-7083-5-21
Gu, W., Miller, S., and Chiu, C. Y. (2018). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. [Epub ahead of print].
Guan, M., Ma, J., Keaton, J. M., Dimitrov, L., Mudgal, P., Stromberg, M., et al. (2016). Association of kidney structure-related gene variants with type 2 diabetes-attributed end-stage kidney disease in African Americans. Hum. Genet. 135, 1251–1262. doi: 10.1007/s00439-016-1714-2
Hanson, R. L., Craig, D. W., Millis, M. P., Yeatts, K. A., Kobes, S., Pearson, J. V., et al. (2007). Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 56, 975–983. doi: 10.2337/db06-1072
Hanson, R. L., Millis, M. P., Young, N. J., Kobes, S., Nelson, R. G., Knowler, W. C., et al. (2010). ELMO1 variants and susceptibility to diabetic nephropathy in American Indians. Mol. Genet. Metab. 101, 383–390. doi: 10.1016/j.ymgme.2010.08.014
Harjutsalo, V., and Groop, P. H. (2014). Epidemiology and risk factors for diabetic kidney disease. Adv. Chronic Kidney Dis. 21, 260–266. doi: 10.1053/j.ackd.2014.03.009
Hathaway, C. K., Chang, A. S., Grant, R., Kim, H. S., Madden, V. J., Bagnell, C. R. Jr., et al. (2016). High Elmo1 expression aggravates and low Elmo1 expression prevents diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 113, 2218–2222. doi: 10.1073/pnas.1600511113
Hayashi, K., Sasamura, H., Nakamura, M., Azegami, T., Oguchi, H., Sakamaki, Y., et al. (2014). KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. J. Clin. Invest. 124, 2523–2537. doi: 10.1172/JCI69557
Hebbring, S. J. (2014). The challenges, advantages and future of phenome-wide association studies. Immunology 141, 157–165. doi: 10.1111/imm.12195
Hirakawa, S., Lange, E. M., Colicigno, C. J., Freedman, B. I., Rich, S. S., and Bowden, D. W. (2003). Evaluation of genetic variation and association in the matrix metalloproteinase 9 (MMP9) gene in ESRD patients. Am. J. Kidney Dis. 42, 133–142. doi: 10.1016/s0272-6386(03)00416-5
Holoch, D., and Moazed, D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84. doi: 10.1038/nrg3863
Janssen, B., Hohenadel, D., Brinkkoetter, P., Peters, V., Rind, N., Fischer, C., et al. (2005). Carnosine as a protective factor in diabetic nephropathy: association with a leucine repeat of the carnosinase gene CNDP1. Diabetes 54, 2320–2327. doi: 10.2337/diabetes.54.8.2320
Jones, P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi: 10.1038/nrg3230
Kaidonis, G., Gillies, M. C., Abhary, S., Liu, E., Essex, R. W., Chang, J. H., et al. (2016). A single-nucleotide polymorphism in the MicroRNA-146a gene is associated with diabetic nephropathy and sight-threatening diabetic retinopathy in Caucasian patients. Acta Diabetol. 53, 643–650. doi: 10.1007/s00592-016-0850-4
Kang, W. L., and Xu, G. S. (2016). Atrasentan increased the expression of klotho by mediating miR-199b-5p and prevented renal tubular injury in diabetic nephropathy. Sci. Rep. 6:19979. doi: 10.1038/srep19979
Kato, M. (2018). Noncoding RNAs as therapeutic targets in early stage diabetic kidney disease. Kidney Res. Clin. Pract. 37, 197–209. doi: 10.23876/j.krcp.2018.37.3.197
Kato, M., and Natarajan, R. (2014). Diabetic nephropathy–emerging epigenetic mechanisms. Nat. Rev. Nephrol. 10, 517–530. doi: 10.1038/nrneph.2014.116
Keating, S. T., van Diepen, J. A., Riksen, N. P., and El-Osta, A. (2018). Epigenetics in diabetic nephropathy, immunity and metabolism. Diabetologia 61, 6–20. doi: 10.1007/s00125-017-4490-1
Korabecna, M., Pazourkova, E., Horinek, A., Mokrejsova, M., and Tesar, V. (2013). Methylation status of immune response genes promoters in cell-free DNA differs in hemodialyzed patients with diabetic nephropathy according to the intensity of anemia therapy. Blood Purif. 36, 280–286. doi: 10.1159/000356094
Kurashige, M., Imamura, M., Araki, S., Suzuki, D., Babazono, T., Uzu, T., et al. (2013). The influence of a single nucleotide polymorphism within CNDP1 on susceptibility to diabetic nephropathy in Japanese women with type 2 diabetes. PLoS One 8:e54064. doi: 10.1371/journal.pone.0054064
Lappalainen, T., and Greally, J. M. (2017). Associating cellular epigenetic models with human phenotypes. Nat. Rev. Genet. 18, 441–451. doi: 10.1038/nrg.2017.32
Leak, T. S., Perlegas, P. S., Smith, S. G., Keene, K. L., Hicks, P. J., Langefeld, C. D., et al. (2009). Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann. Hum. Genet. 73, 152–159. doi: 10.1111/j.1469-1809.2008.00498.x
Lee, J. H., Choi, H. J., Lee, B. H., Kang, H. K., Chin, H. J., Yoon, H. J., et al. (2008). Prevalence of hypouricaemia and SLC22A12 mutations in healthy Korean subjects. Nephrology 13, 661–666. doi: 10.1111/j.1440-1797.2008.01029.x
Li, S. Y., Huang, P. H., Yang, A. H., Tarng, D. C., Yang, W. C., Lin, C. C., et al. (2014). Matrix metalloproteinase-9 deficiency attenuates diabetic nephropathy by modulation of podocyte functions and dedifferentiation. Kidney Int. 86, 358–369. doi: 10.1038/ki.2014.67
Lieb, W., Chen, M. H., Teumer, A., de Boer, R. A., Lin, H., and Fox, E. R. (2015). EchoGen consortium. Genome-wide meta-analyses of plasma renin activity and concentration reveal association with the kininogen 1 and prekallikrein genes. Circ. Cardiovasc. Genet. 8, 131–140. doi: 10.1093/hmg/ddr092
Lim, S. C., Dorajoo, R., Zhang, X., Wang, L., Ang, S. F., Tan, C. S. H., et al. (2017). Genetic variants in the receptor for advanced glycation end products (RAGE) gene were associated with circulating soluble RAGE level but not with renal function among Asians with type 2 diabetes: a genome-wide association study. Nephrol. Dial. Transplant. 32, 1697–1704. doi: 10.1093/ndt/gfw263
Maeda, S., Imamura, M., Kurashige, M., Araki, S., Suzuki, D., Babazono, T., et al. (2013). Replication study for the association of 3 SNP loci identified in a genome-wide association study for diabetic nephropathy in European type 1 diabetes with diabetic nephropathy in Japanese patients with type 2 diabetes. Clin. Exp. Nephrol. 17, 866–871. doi: 10.1007/s10157-013-0797-5
Martini, S., Nair, V., Patel, S. R., Eichinger, F., Nelson, R. G., Weil, E. J., et al. (2013). From single nucleotide polymorphism to transcriptional mechanism: a model for FRMD3 in diabetic nephropathy. Diabetes 62, 2605–2612. doi: 10.2337/db12-1416
Marumo, T., Yagi, S., Kawarazaki, W., Nishimoto, M., Ayuzawa, N., Watanabe, A., et al. (2015). Diabetes induces aberrant DNA methylation in the proximal tubules of the kidney. J. Am. Soc. Nephrol. 26, 2388–2397. doi: 10.1681/ASN.2014070665
McDonough, C. W., Hicks, P. J., Lu, L., Langefeld, C. D., Freedman, B. I., and Bowden, D. W. (2009). The influence of carnosinase gene polymorphisms on diabetic nephropathy risk in African-Americans. Hum. Genet. 126, 265–275. doi: 10.1007/s00439-009-0667-0
McDonough, C. W., Palmer, N. D., Hicks, P. J., Roh, B. H., An, S. S., Cooke, J. N., et al. (2011). A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 79, 563–572. doi: 10.1038/ki.2010.467
McKnight, A. J., Currie, D., Patterson, C. C., Maxwell, A. P., Fogarty, D. G., and Warren, 3/UK GoKinD Study Group. (2009). Targeted genome-wide investigation identifies novel SNPs associated with diabetic nephropathy. Hugo J. 3, 77–82. doi: 10.1007/s11568-010-9133-2
Mehrabzadeh, M., Pasalar, P., Karimi, M., Abdollahi, M., Daneshpour, M., Asadolahpour, E., et al. (2016). Association between ELMO1 gene polymorphisms and diabetic nephropathy in an Iranian population. J. Diabetes Metab. Disord. 15:43.
Mi, X., Tang, W., Chen, X., Liu, F., and Tang, X. (2016). Mitofusin 2 attenuates the histone acetylation at collagen IV promoter in diabetic nephropathy. J. Mol. Endocrinol. 57, 233–249. doi: 10.1530/JME-16-0031
Mone, F., Quinlan-Jones, E., and Kilby, M. D. (2018). Clinical utility of exome sequencing in the prenatal diagnosis of congenital anomalies: a review. Eur. J. Obstet. Gynecol. Reprod. Biol. 231, 19–24. doi: 10.1016/j.ejogrb.2018.10.016
Mooyaart, A. L., Zutinic, A., Bakker, S. J., Grootendorst, D. C., Kleefstra, N., van Valkengoed, I. G., et al. (2010). Association between CNDP1 genotype and diabetic nephropathy is sex specific. Diabetes 59, 1555–1559. doi: 10.2337/db09-1377
Murea, M., Ma, L., and Freedman, B. I. (2012). Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev. Diabet. Stud. 9, 6–22. doi: 10.1900/RDS.2012.9.6
Nair, S., Phillips, A. O., Norton, N., Spurlock, G., Williams, H. J., Craig, K. J., et al. (2008). Further evidence for the association of MMP9 with nephropathy in type 2 diabetes and application of DNA pooling technology to candidate gene screening. J. Nephrol. 21, 400–405.
Ni, X., Ji, C., Cao, G., Cheng, H., Guo, L., Gu, S., et al. (2003). Molecular cloning and characterization of the protein 4.1O gene, a novel member of the protein 4.1 family with focal expression in ovary. J. Hum. Genet. 48, 101–106. doi: 10.1007/s100380300015
Nicolas, A., Fatima, S., Lamri, A., Bellili-Muñoz, N., Halimi, J. M., Saulnier, P. J., et al. (2015). ABCG8 polymorphisms and renal disease in type 2 diabetic patients. Metabolism 64, 713–719. doi: 10.1016/j.metabol.2015.03.005
Nie, M., Bal, M. S., Liu, J., Yang, Z., Rivera, C., Wu, X. R., et al. (2018). Uromodulin regulates renal magnesium homeostasis through the ion channel transient receptor potential melastatin 6 (TRPM6). J. Biol. Chem. 293, 16488–16502. doi: 10.1074/jbc.RA118.003950
Nishiyama, K., Tanaka, Y., Nakajima, K., Mokubo, A., Atsumi, Y., Matsuoka, K., et al. (2005). Polymorphism of the solute carrier family 12 (sodium/chloride transporters) member 3, SLC12A3, gene at exon 23 (+78G/A: Arg913Gln) is associated with elevation of urinary albumin excretion in Japanese patients with type 2 diabetes: a 10-year longitudinal study. Diabetologia 48, 1335–1338. doi: 10.1007/s00125-005-1785-4
Palmer, N. D., and Freedman, B. I. (2013). Diabetic nephropathy: FRMD3 in diabetic nephropathy–guilt by association. Nat. Rev. Nephrol. 9, 313–314. doi: 10.1038/nrneph.2013.81
Palmer, N. D., Ng, M. C., Hicks, P. J., Mudgal, P., Langefeld, C. D., Freedman, B. I., et al. (2014). Evaluation of candidate nephropathy susceptibility genes in a genome-wide association study of African American diabetic kidney disease. PLoS One 9:e88273. doi: 10.1371/journal.pone.0088273
Papadopoulou-Marketou, N., Chrousos, G. P., and Kanaka-Gantenbein, C. (2017). Diabetic nephropathy in type 1 diabetes: a review of early natural history, pathogenesis, and diagnosis. Diabetes Metab. Res. Rev. 33:e2841. doi: 10.1002/dmrr.2841
Patel, M., Margaron, Y., Fradet, N., Yang, Q., Wilkes, B., Bouvier, M., et al. (2010). An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr. Biol. 20, 2021–2027. doi: 10.1016/j.cub.2010.10.028
Pendergrass, S. A., and Ritchie, M. D. (2015). Phenome-wide association studies: leveraging comprehensive phenotypic and genotypic data for discovery. Curr. Genet. Med. Rep. 3, 92–100. doi: 10.1007/s40142-015-0067-9
Peng, R., Liu, H., Peng, H., Zhou, J., Zha, H., Chen, X., et al. (2015). Promoter hypermethylation of let-7a-3 is relevant to its down-expression in diabetic nephropathy by targeting UHRF1. Gene 570, 57–63. doi: 10.1016/j.gene.2015.05.073
Peters, V., Kebbewar, M., Janssen, B., Hoffmann, G. F., Möller, K., and Wygoda, S. (2016). CNDP1 genotype and renal survival in pediatric nephropathies. J. Pediatr. Endocrinol. Metab. 29, 827–833. doi: 10.1515/jpem-2015-0262
Pezzolesi, M. G., Katavetin, P., Kure, M., Poznik, G. D., Skupien, J., Mychaleckyj, J. C., et al. (2009a). Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes 58, 2698–2702. doi: 10.2337/db09-0641
Pezzolesi, M. G., Poznik, G. D., Mychaleckyj, J. C., Paterson, A. D., Barati, M. T., and Klein, J. B. (2009b). Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes 58, 1403–1410.
Prudente, S., Di Paola, R., Copetti, M., Lucchesi, D., Lamacchia, O., and Pezzilli, S. (2017). The rs12917707 polymorphism at the UMOD locus and glomerular filtration rate in individuals with type 2 diabetes: evidence of heterogeneity across two different European populations. Nephrol. Dial. Transplant. 32, 1718–1722. doi: 10.1093/ndt/gfw262
Rakyan, V. K., Down, T. A., Balding, D. J., and Beck, S. (2011). Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541. doi: 10.1038/nrg3000
Reddy, M. A., Sumanth, P., Lanting, L., Yuan, H., Wang, M., Mar, D., et al. (2014). Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int. 85, 362–373. doi: 10.1038/ki.2013.387
Reidy, K., Kang, H. M., Hostetter, T., and Susztak, K. (2014). Molecular mechanisms of diabetic kidney disease. J. Clin. Invest. 124, 2333–2340. doi: 10.1172/JCI72271
Roden, D. M. (2017). Phenome-wide association studies: a new method for functional genomics in humans. J. Physiol. 595, 4109–4115. doi: 10.1113/JP273122
Sandholm, N., Forsblom, C., Mäkinen, V. P., McKnight, A. J., Osterholm, A. M., and He, B. (2014). Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia 57, 1143–1153. doi: 10.1007/s00125-014-3202-3
Sandholm, N., Haukka, J. K., Toppila, I., Valo, E., Harjutsalo, V., Forsblom, C., et al. (2018). Confirmation of GLRA3 as a susceptibility locus for albuminuria in Finnish patients with type 1 diabetes. Sci. Rep. 8:12408. doi: 10.1038/s41598-018-29211-1
Sandholm, N., McKnight, A. J., Salem, R. M., Brennan, E. P., Forsblom, C., and Harjutsalo, V. (2013). FinnDiane Study Group and the GENIE Consortium. Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J. Am. Soc. Nephrol. 24, 1537–1543. doi: 10.1681/ASN.2012111122
Sandholm, N., Salem, R. M., McKnight, A. J., Brennan, E. P., Forsblom, C., and Isakova, T. (2012). New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 8:e1002921. doi: 10.1371/journal.pgen.1002921
Sandholm, N., Van Zuydam, N., Ahlqvist, E., Juliusdottir, T., Deshmukh, H. A., and Rayner, N. W. (2017). The genetic landscape of renal complications in type 1 diabetes. J. Am. Soc. Nephrol. 28, 557–574. doi: 10.1681/ASN.2016020231
Sapienza, C., Lee, J., Powell, J., Erinle, O., Yafai, F., Reichert, J., et al. (2011). DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 6, 20–28. doi: 10.4161/epi.6.1.13362
Sayyed, S. G., Gaikwad, A. B., Lichtnekert, J., Kulkarni, O., Eulberg, D., Klussmann, S., et al. (2010). Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol. Dial. Transplant. 25, 1811–1817. doi: 10.1093/ndt/gfp730
Seman, N. A., Mohamud, W. N., Östenson, C. G., Brismar, K., and Gu, H. F. (2015). Increased DNA methylation of the SLC30A8 gene promoter is associated with type 2 diabetes in a Malay population. Clin. Epigenetics 7:30. doi: 10.1186/s13148-015-0049-5
Shan, Q., Zheng, G., Zhu, A., Cao, L., Lu, J., Wu, D., et al. (2016). Epigenetic modification of miR-10a regulates renal damage by targeting CREB1 in type 2 diabetes mellitus. Toxicol. Appl. Pharmacol. 306, 134–143. doi: 10.1016/j.taap.2016.06.010
Sharma, I., Dutta, R. K., Singh, N. K., and Kanwar, Y. S. (2017). High glucose-induced hypomethylation promotes binding of Sp-1 to myo-inositol oxygenase: implication in the pathobiology of diabetic tubulopathy. Am. J. Pathol. 187, 724–739. doi: 10.1016/j.ajpath.2016.12.011
Sharma, K. R., Heckler, K., Stoll, S. J., Hillebrands, J. L., Kynast, K., Herpel, E., et al. (2016). ELMO1 protects renal structure and ultrafiltration in kidney development and under diabetic conditions. Sci. Rep. 6:37172. doi: 10.1038/srep37172
Shiffman, D., Pare, G., Oberbauer, R., Louie, J. Z., Rowland, C. M., Devlin, J. J., et al. (2014). A gene variant in CERS2 is associated with rate of increase in albuminuria in patients with diabetes from ONTARGET and TRANSCEND. PLoS One 9:e106631. doi: 10.1371/journal.pone.0106631
Shimazaki, A., Kawamura, Y., Kanazawa, A., Sekine, A., Saito, S., Tsunoda, T., et al. (2005). Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes 54, 1171–1178. doi: 10.2337/diabetes.54.4.1171
Shimazaki, A., Tanaka, Y., Shinosaki, T., Ikeda, M., Watada, H., Hirose, T., et al. (2006). ELMO1 increases expression of extracellular matrix proteins and inhibits cell adhesion to ECMs. Kidney Int. 70, 1769–1776. doi: 10.1038/sj.ki.5001939
Smyth, L. J., Maxwell, A. P., Benson, K. A., Kilner, J., McKay, G. J., and McKnight, A. J. (2018). Validation of differentially methylated microRNAs identified from an epigenome-wide association study; Sanger and next generation sequencing approaches. BMC Res. Notes 11:767. doi: 10.1186/s13104-018-3872-x
Sun, G., Reddy, M. A., Yuan, H., Lanting, L., Kato, M., and Natarajan, R. (2010). Epigenetic histone methylation modulates fibrotic gene expression. J. Am. Soc. Nephrol. 21, 2069–2080. doi: 10.1681/ASN.2010060633
Sun, X. Y., Qin, H. J., Zhang, Z., Xu, Y., Yang, X. C., Zhao, D. M., et al. (2016). Valproate attenuates diabetic nephropathy through inhibition of endoplasmic reticulum stress-induced apoptosis. Mol. Med. Rep. 13, 661–668. doi: 10.3892/mmr.2015.4580.
Swan, E. J., Maxwell, A. P., and McKnight, A. J. (2015). Distinct methylation patterns in genes that affect mitochondrial function are associated with kidney disease in blood-derived DNA from individuals with Type 1 diabetes. Diabet. Med. 32, 1110–1115. doi: 10.1111/dme.12775
Tanaka, N., Babazono, T., Saito, S., Sekine, A., Tsunoda, T., Haneda, M., et al. (2003). Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes 52, 2848–2853. doi: 10.2337/diabetes.52.11.2848
Teumer, A., Tin, A., Sorice, R., Gorski, M., Yeo, N. C., and Chu, A. Y. (2016). Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes 65, 803–817. doi: 10.2337/db15-1313
Thameem, F., Igo, R. P. Jr., Freedman, B. I., Langefeld, C., Hanson, R. L., and Schelling, J. R. (2013). Family investigation of nephropathy and diabetes research group. A genome-wide search for linkage of estimated glomerular filtration rate (eGFR) in the family investigation of nephropathy and diabetes (FIND). PLoS One 8:e81888. doi: 10.1371/journal.pone.0081888
Thomas, M. C. (2016). Epigenetic mechanisms in diabetic kidney disease. Curr. Diab. Rep. 16:31. doi: 10.1007/s11892-016-0723-9
Thomas, M. C., Brownlee, M., Susztak, K., Sharma, K., Jandeleit-Dahm, K. A., Zoungas, S., et al. (2015). Diabetic kidney disease. Nat. Rev. Dis. Primers 1:15018. doi: 10.1038/nrdp.2015.18
Thomas, M. C., Groop, P. H., and Tryggvason, K. (2012). Towards understanding the inherited susceptibility for nephropathy in diabetes. Curr. Opin. Nephrol. Hypertens. 21, 195–202. doi: 10.1097/MNH.0b013e328350313e
Tikoo, K., Meena, R. L., Kabra, D. G., and Gaikwad, A. B. (2008). Change in post-translational modifications of histone H3, heat-shock protein-27 and MAP kinase p38 expression by curcumin in streptozotocin-induced type I diabetic nephropathy. Br. J. Pharmacol. 153, 1225–1231. doi: 10.1038/sj.bjp.0707666
Treangen, T. J., and Salzberg, S. L. (2011). Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13, 36–46. doi: 10.1038/nrg3117
van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi: 10.1016/j.tig.2018.05.008
van Zuydam, N. R., Ahlqvist, E., Sandholm, N., Deshmukh, H., Rayner, N. W., and Abdalla, M. S. (2018). A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 67, 1414–1427. doi: 10.2337/db17-0914
VanderJagt, T. A., Neugebauer, M. H., Morgan, M., Bowden, D. W., and Shah, V. O. (2015). Epigenetic profiles of pre-diabetes transitioning to type 2 diabetes and nephropathy. World J. Diabetes 6, 1113–1121. doi: 10.4239/wjd.v6.i9.1113
Villeneuve, L. M., and Natarajan, R. (2010). The role of epigenetics in the pathology of diabetic complications. Am. J. Physiol. Renal Physiol. 299, F14–F25. doi: 10.1152/ajprenal.00200.2010
Wang, T., Zhang, Y., Wang, N., Liu, Q., Wang, Z., Liu, B., et al. (2018). Synergistical action of the β2 adrenoceptor and fatty acid binding protein 2 polymorphisms on the loss of glomerular filtration rate in Chinese patients with type 2 diabetic nephropathy. Int. Urol. Nephrol. 50, 715–723. doi: 10.1007/s11255-018-1812-2
Wanic, K., Placha, G., Dunn, J., Smiles, A., Warram, J. H., and Krolewski, A. S. (2008). Exclusion of polymorphisms in carnosinase genes (CNDP1 and CNDP2) as a cause of diabetic nephropathy in type 1 diabetes: results of large case-control and follow-up studies. Diabetes 57, 2547–2551. doi: 10.2337/db07-1303
Watanabe, A., Marumo, T., Kawarazaki, W., Nishimoto, M., Ayuzawa, N., Ueda, K., et al. (2018). Aberrant DNA methylation of pregnane X receptor underlies metabolic gene alterations in the diabetic kidney. Am. J. Physiol. Renal Physiol. 314, F551–F560. doi: 10.1152/ajprenal.00390.2017
Wu, H. Y., Wang, Y., Chen, M., Zhang, X., Wang, D., Pan, Y., et al. (2013). Association of ELMO1 gene polymorphisms with diabetic nephropathy in Chinese population. J. Endocrinol. Invest. 36, 298–302. doi: 10.3275/8525
Yang, L., Zhang, Q., Wu, Q., Wei, Y., Yu, J., Mu, J., et al. (2018). Effect of TET2 on the pathogenesis of diabetic nephropathy through activation of transforming growth factor β1 expression via DNA demethylation. Life Sci. 207, 127–137. doi: 10.1016/j.lfs.2018.04.044
Yang, X. H., Cao, R. F., Yu, Y., Sui, M., Zhang, T., Xu, J. Y., et al. (2016). A study on the correlation between MTHFR promoter methylation and diabetic nephropathy. Am. J. Transl. Res. 8, 4960–4967.
Yuan, H., Reddy, M. A., Deshpande, S., Jia, Y., Park, J. T., Lanting, L. L., et al. (2016). Epigenetic histone modifications involved in profibrotic gene regulation by 12/15-lipoxygenase and its oxidized lipid products in diabetic nephropathy. Antioxid. Redox Signal. 24, 361–375. doi: 10.1089/ars.2015.6372
Zhang, H., Cai, X., Yi, B., Huang, J., Wang, J., and Sun, J. (2014). Correlation of CTGF gene promoter methylation with CTGF expression in type 2 diabetes mellitus with or without nephropathy. Mol. Med. Rep. 9, 2138–2144. doi: 10.3892/mmr.2014.2067
Zhang, L., Zhang, Q., Liu, S., Chen, Y., Li, R., Lin, T., et al. (2017). DNA methyltransferase 1 may be a therapy target for attenuating diabetic nephropathy and podocyte injury. Kidney Int. 92, 140–153. doi: 10.1016/j.kint.2017.01.010
Zhang, R., Zhuang, L., Li, M., Zhang, J., Zhao, W., Ge, X., et al. (2018). Arg913Gln of SLC12A3 gene promotes development and progression of end-stage renal disease in Chinese type 2 diabetes mellitus. Mol. Cell. Biochem. 437, 203–210. doi: 10.1007/s11010-017-3120-z
Zhang, Z., Wu, X., Cai, T., Gao, W., Zhou, X., Zhao, J., et al. (2015). Matrix metalloproteinase 9 gene promoter (rs3918242) mutation reduces the risk of diabetic microvascular complications. Int. J. Environ. Res. Public Health 12, 8023–8033. doi: 10.3390/ijerph120708023
Keywords: diabetic kidney disease, diabetes, end-stage renal disease, genetics, epigenetics, phenotypes
Citation: Gu HF (2019) Genetic and Epigenetic Studies in Diabetic Kidney Disease. Front. Genet. 10:507. doi: 10.3389/fgene.2019.00507
Received: 03 December 2018; Accepted: 08 May 2019;
Published: 07 June 2019.
Edited by:
Calli Dendrou, Wellcome Centre for Human Genetics (WT), United KingdomReviewed by:
Alexander Peter Maxwell, Queen’s University Belfast, United KingdomCopyright © 2019 Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harvest F. Gu, ZmVuZy5ndUBjcHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.