- 1State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control, Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2College of Forestry, Henan University of Science and Technology, Luoyang, China
As a conserved protein family, WRKY has been shown to be involved in multiple biological processes in plants. However, the mechanism of functional diversity for WRKYs in pepper has not been well elucidated. Here, a total of 223 WRKY members from solanaceae crops including pepper, tomato and potato, were analyzed using comparative genomics. A tremendous genetic variation among WRKY members of different solanaceous plants or groups was demonstrated by the comparison of some WRKY features, including number/size, group constitution, gene structure, and domain composition. The phylogenetic analysis showed that except for the known WRKY groups (I, IIa/b/c/d/e and III), two extra WRKY subgroups specifically existed in solanaceous plants, which were named group IIf and group IIg in this study, and their genetic variations were also revealed by the characteristics of some group IIf and IIg WRKYs. Except for the extensive genetic variations, certain degrees of conservatism for solanaceae WRKYs were also revealed. Moreover, the variant zinc-finger structure (CX4,7CX22-24HXC) in group III of solanaceae WRKYs was identified. Expression profiles of CaWRKY genes suggested their potential roles in pepper development and stress responses, and demonstrated a functional division pattern for pepper CaWRKYs. Furthermore, functional analysis using virus induced gene silencing (VIGS) revealed critical roles of two CaWRKYs (CaWRKY45 and CaWRKY58) in plant responses to disease and drought, respectively. This study provides a solid foundation for further dissection of the evolutionary and functional diversity of solanaceae WRKYs in crop plants.
Introduction
The WRKY transcription factor (TF) family is one of the largest TF families in higher plants, which has been extensively studied for its important regulatory roles in multiple biological processes relating to plant growth, development and responses to stress (Pandey and Somssich, 2009; Li et al., 2013a; Dang et al., 2014; Cai et al., 2015). As a sequence specific DNA-binding TFs, the WRKYproteins contain at least one WRKY domain, which is responsible for the binding of special cis-element (w-box: TTGACC/T) (Rushton et al., 2010). As the common features of WRKY TFs, the WRKY domain is typically composed of a highly conserved peptide (WRKYGQK motif) and a zinc-finger structure (C2H2:CX4-5CX22-23HXH or C2HC:CX7CX23HXC) (Rushton et al., 1996, 2010; Eulgem and Somssich, 2007), which form a four-stranded β-sheet and a zinc-binding pocket formed by the conserved Cys/His residues located at one end of the β-sheet (Eulgem et al., 2000; Xie et al., 2005; Yamasaki et al., 2005).
Based on the number of WRKY domains and the corresponding zinc-finger structure, the WRKY proteins were initially classified into three groups, namely group I, II, and III (Eulgem et al., 2000). The group I WRKY members contain two WRKY domains with two CX4CX22-23HXH zinc-finger motifs, the group II WRKYs have one WRKY domain with one CX4CX22-23HXH zinc-finger motif, and the group III WRKYs contain one WRKY domain with one CX7CX23HXC zinc-finger motif (Rushton et al., 1996, 2010; Eulgem and Somssich, 2007). The group II WRKYs could be further divided into five subgroups, such as subgroup IIa, IIb, IIc, IId, and IIe (Eulgem et al., 2000; Xie et al., 2005). However, later studies on phylogenetic relationship reclassified the group II into IIa+IIb, II c, and IId+IIe (Wu et al., 2005; Zhang and Wang, 2005; Huang et al., 2012). It is well recognized that WRKY proteins originated from the lower photosynthetic and non-photosynthetic eukaryotes. As the group I WRKY members only exist in lower plants, they are now believed to be the evolutionary ancestors of the other WRKYs (Wu et al., 2005; Zhang and Wang, 2005; Cheng et al., 2012).
Since the first identification of plant WRKY protein in sweet potato (Ishiguro and Nakamura, 1994), the whole genome level WRKY identification has been accomplished in a wide variety of plant species (Wu et al., 2005; Zhang and Wang, 2005; Mangelsen et al., 2008; Rushton et al., 2008; Ling et al., 2011; Tripathi et al., 2012), including Arabidopsis (Rushton et al., 2010), rice (Ross et al., 2007), tomato (Huang et al., 2012) and potato (Huang and Liu, 2013). Many identified WRKYs have been shown to be functional in a broad range of biological processes, including biotic or abiotic stress responses (Li et al., 2013b; Dang et al., 2014; Zhou et al., 2015), hormone responses (Antoni et al., 2011; Bakshi and Oelmuller, 2014; Zhang et al., 2015) and other developmental processes (Robatzek and Somssich, 2002; Miao et al., 2004; Jiang and Yu, 2009; Zentgraf et al., 2010; Suttipanta et al., 2011; Besseau et al., 2012). SlWRKY39, a group IIa member in tomato, was reported to play important role in disease resistance against Pseudomonas syringae pv. tomato DC3000 (PstDC3000), as well as in osmotic stress and drought tolerance (Sun et al., 2015). Another group I member defined as SlDRW1, is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress in tomato (Liu et al., 2014). In pepper, CaWRKY40 from group IIa, plays an important role in the modulation of both high temperature tolerance and Ralstonia solanacearum resistance, and this regulation seems to be dependent on the transcriptional activity of CaWRKY06 (group IIb member) (Dang et al., 2013; Cai et al., 2015). In potato, the function of WRKY TF was barely reported, except for the StWRKY01 (group II member), which might play certain roles in the defense against Erwinia carotovora subsp. atroseptica and Phytophthora infestans (Dellagi et al., 2000). Despite being a highly conserved protein family characterized by the conserved WRKY-domain and specific w-box (TTGACC/T) recognition pattern, the functional diversity of WRKY TFs has not been systematically elucidated yet. So far, several hypotheses, including the specificity determinant of the nucleotides sequence adjacent to the W-box core sequence (Ciolkowski et al., 2008), the temporal and spatial expression differences of WRKY genes decided by their own structural variations, and interacting partners (co-activators, chromatin remodelers) modulating patterns (Andreasson et al., 2005; Qiu et al., 2008; Wang et al., 2010; Cheng et al., 2012), were proposed.
Although many WRKYs have been analyzed using bioinformatics tools in some solanaceous species, the genetic or functional diversity of solanaceae WRKYs has barely been studied. Thus, based on the genome databases of the three representative solanaceous plants (pepper, potato and tomato) (The Potato Genome Sequencing Consortium, 2011; The Tomato Genome Consortium, 2012; Cheng et al., 2016), totally 223 solanaceae WRKY members were collected (Huang et al., 2012; Huang and Liu, 2013; Cheng et al., 2016), and their extensive genetic variations were comparatively analyzed in the current study. Moreover, the functional diversity of CaWRKYs in pepper was studied through the analysis of tissue-specific expression pattern and stress responses.
Materials and Methods
Collection of WRKY Proteins From Solanaceous Plant Genomes
The bioinformation of WRKYs (accession ID, group classification etc.) in pepper, tomato and potato were collected from previous publications (Huang et al., 2012; Huang and Liu, 2013; Cheng et al., 2016). The detail information of the WRKYs (gene/protein sequences) was further collected from Pepper Informatics Hub (PIH1), Sol Genomics Network (SGN2) and Potato Genome Project (PGP3) according to their corresponding accession IDs, respectively (Supplementary Table S1). The HMM profile of the conserved WRKY domain (Pfam: PF03106) from Pfam 27.0 database4 was used for the validation of all collected WRKY sequences.
Phylogenetic Analysis and Intron-Exon Configuration of Solanaceae WRKY Genes
The alignments of amino acid sequences of 72 complete WRKY domains from pepper, 94 complete WRKY domains from tomato, 92 complete WRKY domains from potato, 84 complete WRKY domains from Arabidopsis and 102 complete WRKY domains from rice (both N- and C-terminal domains included), were performed using ClustalX 1.83 with default settings (Thompson et al., 1997)5. An unrooted phylogenetic tree was conducted based on the alignment data using MEGA 5.0 with neighbor joining method and maximum-likelihood method, respectively (Tamura et al., 2011). Relative branch support was evaluated using bootstraps (1000 replicates), branch lengths were calculated by pairwise comparison of genetic distances, and missing data were treated by pairwise deletions of gaps. The intron-exon structure was visualized using GSDS 2.06 based on genomic DNA sequences of the 223 solanaceae (pepper, tomato and potato) WRKYs. The coefficient of variation (CV) on intron numbers was calculated by the formula of STDEV/AVERAGE in Excel 2007 (Supplementary Table S1).
WRKY Domain Structure and Motif Analyses
The specific WRKY domain sequences of all WRKYs from pepper, tomato and potato were collected using SMART7. The WRKY domain sequences were aligned using ClustalX 1.83 with default settings (see Footnote 5), converted to “CLUSTAL” format and visualized by Bioedit 7.0 (Supplementary Table S2). The sequence logos of WRKY domains were generated online using weblogo8. All retrieved WRKY sequences were subjected to domain analysis by using the Conserved Domain Database (CDD) programs9 (Supplementary Table S3).
Chromosome Mapping and Comparative Analysis of WRKYs Among Solanaceous Plants
Chromosome mapping of 223 solanaceae WRKY genes was performed with MapDraw V2.1 (Zheng et al., 2006) based on their gene chromosomal localization information derived from SGN, PGR and PGD, respectively. Online platform PGDD10 was used for comparative genomics analysis of WRKYs from pepper, tomato and potato. Based on the constructed chromosome mappings, correlative tomato-potato-pepper WRKY genes were labeled and connected with black lines.
Expression Analysis of CaWRKY Genes and Virus-Induced Gene Silencing (VIGS) of CaWRKYs
Total RNA was isolated from collected pepper samples (Root, stem, Leaf, flower, bud, green fruit, red fruit, and seed). Total RNA kit according to the manufacturer’s protocol (Tiangen, Beijing, China), and reverse transcribed into cDNA using the FastQuant RT Kit (Tiangen, Beijing, China). Genscript online tool11 was used for the total 61 pairs of CaWRKY-specific primers design (Supplementary Table S4). The real-time PCR reactions were carried out in 20 μl reaction mixture containing 10 μl SuperMix, 0.4 μl of each primer (20 μM), 1 μl diluted (10×) sample cDNA, and 8.2 μl sterile distilled water. RT-qPCR assay was performed using the following program: 30 s at 94°C, followed by 40 cycles of 5 s at 94°C, 15 s at 55°C and 10 s at 72°C. The pepper gene UBI-1 (Capana04g000407) (F: AAGGAAATGTGTGTCTCAAC; R: TCCAAATGCCAAACTTCTAG) was used as an internal control for the normalization of expression levels of the target genes (Livak and Schmittgen, 2001). The relative gene expression was calculated according to Livak and Schmittgen (2001). Three independent biological replicates were performed.
Due to highly conservation among pepper CaWRKY members, SGN VIGS online tool12 was used to ensure the specificity of target CaWRKY genes. The virus-induced gene silencing (VIGS) target fragments of CaWRKY22, CaWRKY45, and CaWRKY58 were amplified with gene-specific primers (Supplementary Table S5) and ligated into the VIGS vector pTRV, which was subsequently transformed into Agrobacterium tumefaciens strain GV3101 for VIGS analyses. VIGS analyses were performed as described previously (Wang et al., 2015). The empty pTRV vector was used as control. Four weeks after agro-infiltration, pepper plants were infected with PstDC3000 and B. cinerea. The bacterial strain PstDC3000 were grown overnight in King’s B medium containing rifampicin (50 mg/mL) and kanamycin (25 mg/mL). The bacterial cells were harvested and suspended in 10 mM MgCl2. The cells were then diluted to 1 × 106 cfu/ml and infiltrated into the abaxial surface of the leaves (Cheng et al., 2012). Growth of B. cinerea were performed as previously described (Zheng et al., 2006), and the inoculation was conducted by spot infection of 2 μl bacterial fluids (2 × 105 spor/ml). For abiotic stress treatments, 4-week-old pepper seedlings were exposed to osmotic stress by irrigation of 20% PEG6000 for 12 h, or exposed to drought by withholding water for 9 d, or to heat by elevating the temperature to 42°C for 3 h, and leaves were collected at the end of each treatment. The relative water content (%) in leaves was measured following the drying method (Wang et al., 2018). All materials were frozen in liquid nitrogen and used for the following expression analyses. For each treatment, three replicates (four plants in one replicate) were examined. DMRT (Duncan’s multiple range test) was used for statistical analysis.
Results
Comprehensive Evolutionary Analysis of WRKY Proteins in Solanaceous Plants
Here, totally 223 solanaceae WRKYs, as well as the WRKYs of two model plants (100 OsWRKYs from rice, 75 AtWRKYs from Arabidopsis), were collected for comparative analysis together. According to Table 1, the number of WRKY proteins is not linearly correlated with the genome size of the corresponding plant species (Table 1). For example, with the very large genome size of 3500-magabase, only 61 CaWRKYs were detected from pepper. On the contrary, totally 100 WRKYs were identified from rice (only 390-magabase genome) (Table 1).
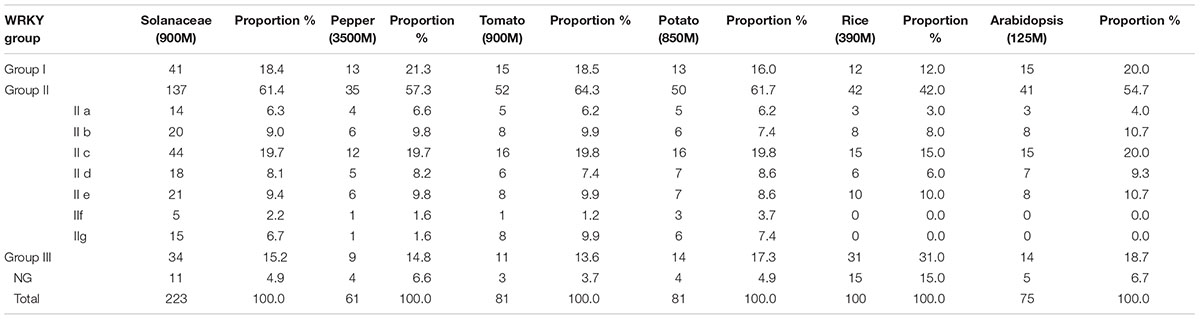
Table 1. Group constitution of WRKYs in Solanaceae (pepper, tomato and potato) and other plants (rice and Arabidopsis).
Although pepper, tomato and potato all belong to the solanaceae family, they could be further differentiated into genus levels, such as the Capsicum genus (pepper) and Solanum genus (tomato, potato), respectively. According to Table 1, some genetic variations on WRKY numbers were reflected on genus level, as the number of WRKYs in tomato and potato (the Solanum genus) are both 81, but only 61 WRKY members in pepper (the Capsicum genus) were identified, indicating the occurrence of WRKY gene loss/gain events during the evolution of solanaceous plants. Further analysis in group-wise manner showed that the events of gene loss/gain were mainly occurred in group II (especially IIc and IIg), as there are 35 group II CaWRKYs in pepper (including 2 IIc, 1 IIg), 52 group II SlWRKYs in tomato (including 16 IIc, 8 IIg), and 50 group II StWRKYs in potato (including 16 IIc, 6 IIg), respectively (Table 1).
As shown in Table 2, the average size [encoding nucleotide residues (bp) and amino acids (aa)] of CaWRKYs, SlWRKYs, and StWRKYs are 1075.0bp/355.7aa, 1060.0bp/352.3aa, and 1017.4bp/388.1aa, respectively, suggesting the presence of size variations in WRKY families of different solanaceous plants. Moreover, the distinctive differences on the average WRKY size of most groups between pepper (the Capsicum genus) and tomato/potato (the Solanum genus) were observed (Table 2). The average size of different WRKY groups were obviously more similar between tomato and potato, the average size of pepper WRKY groups were either larger (group I, IIa, IIf, and IIg) or smaller (for IIb, IIc, IId, IIe, and III) than their counterparts in tomato and potato.
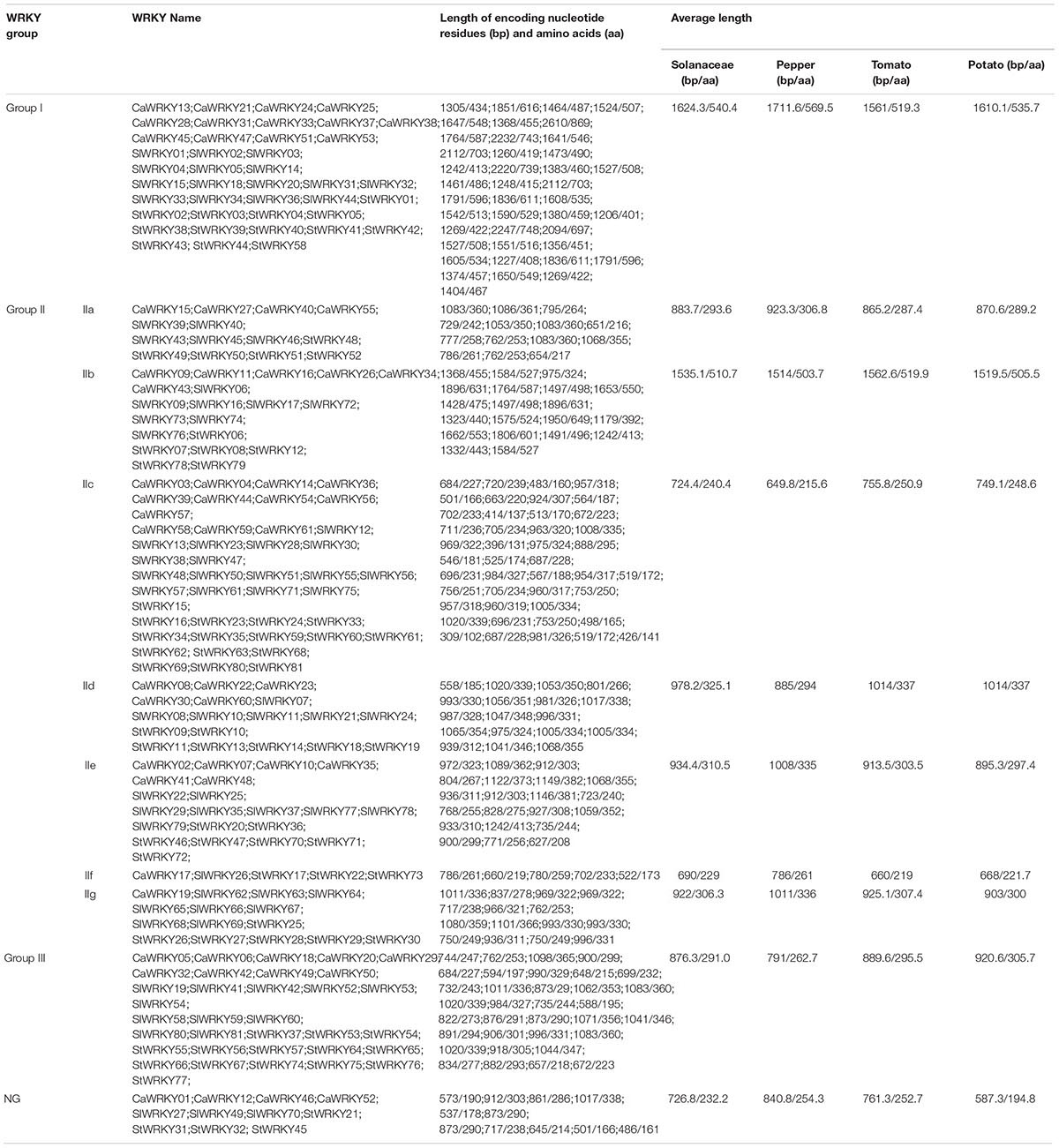
Table 2. Group classification and length of encoding nucleotide residues (bp)/amino acids (aa) of WRKYs in pepper, tomato and potato.
It is noteworthy that although both tomato and potato belong to the same genus, as well as owning the same number of WRKY members, some unconspicuous variations on WRKY group features, including WRKY group constitution and average size of group members, were also detected (Tables 1, 2).
Phylogenetic Analysis of WRKY Gene Family in Solanaceous Plants
To better study the phylogenetic relationship of the 223 WRKYs in solanaceous plants, we constructed an unrooted phylogenetic tree based on the alignment of protein sequences of 444 complete WRKY domains (including N-terminal (I-N) and C-terminal (I-C) domains of group I) from pepper, tomato, potato, rice and Arabidopsis (Figure 1). Incomplete WRKY domains, including WRKY domains of CaWRKY01/CaWRKY12 in pepper, SlWRKY27/SlWRKY70 in tomato, and StWRKY21/StWRKY31/StWRKY32 in potato (Supplementary Table S1), were excluded from the phylogenetic analysis. Three main clusters including different WRKY groups were observed, which are cluster A (I-C+I-N+IIc), cluster B (IIa+IIb), and cluster C (IId+IIe+III+two outstanding expansions in group II) (Figure 1), respectively. In cluster A, the C-terminal WRKY domains of group I (I-C) and WRKY domains of group IIc seem to be more closely related on evolutionary level, and a unique expansion including five WRKY members (AtWRKY49, AtWRKY59, OsWRKY17, CaWRKY46, and SlWRKY49) was observed (Figure 1). In cluster C, two distinct gene expansion events adjacent to group IId+IIe and group III were identified, respectively. These two expansions were only composed of solanaceae WRKY members, and were tentatively defined as group IIf (CaWRKY17, SlWRKY26, StWRKY17, StWRKY22, and StWRKY73) and group IIg (CaWRKY19, SlWRKY62, SlWRKY63, SlWRKY64, SlWRKY65, SlWRKY66, SlWRKY67, SlWRKY68, StWRKY25, StWRKY26, StWRKY27, StWRKY28, StWRKY29, and StWRKY30) in this study (Figure 1 and Tables 1, 2). Eleven WRKYs were defined as None Group (NG) due to their incomplete WRKY domains (CaWRKY01, CaWRKY12, SlWRKY27, SlWRKY70, StWRKY21, StWRKY31, and StWRKY32) or phylogenetic uniqueness (CaWRKY46, CaWRKY52, SlWRKY49, and StWRKY45) (Figure 1 and Supplementary Table S1) (Huang et al., 2012; Huang and Liu, 2013).
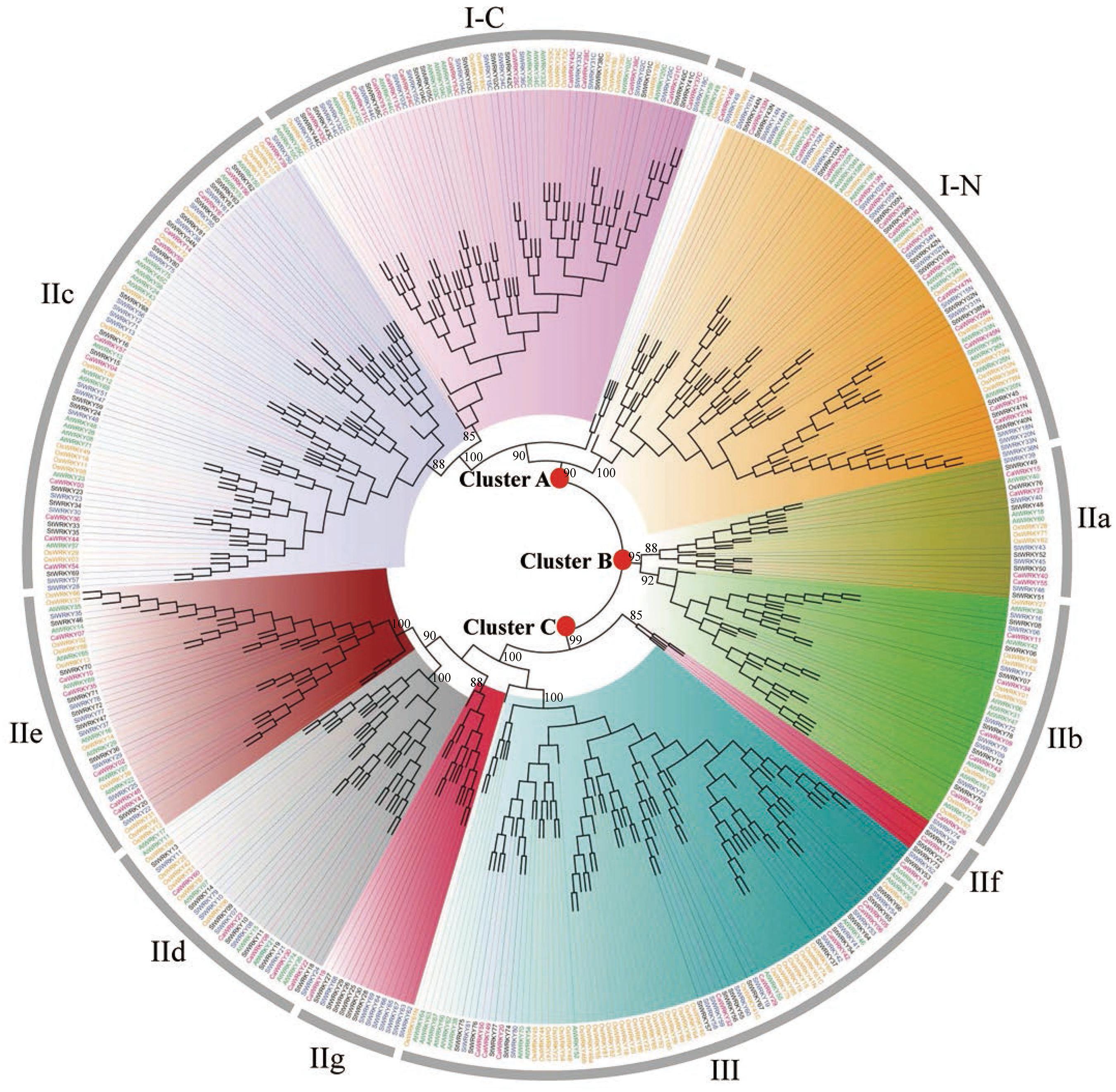
Figure 1. Phylogenetic tree of WRKY proteins from pepper, tomato, potato, Arabidopsis and rice using the neighbor joining method by MEGA 5.0. The WRKYs of each group were clustered together and labeled with different colors. The root nodes of Cluster A, Cluster B, and Cluster C were marked with red solid circles. Group IIf and IIg were two newly identified subgroups that only comprised WRKYs from Solanaceous plants. WRKYs from pepper (CaWRKY), tomato (SlWRKY), potato (StWRKY), Arabidopsis (AtWRKY), and rice (OsWRKY) are labeled in red, blue, black, green and yellow, respectively.
Group-Wise Structural Analysis of Solanaceae WRKY Genes
The structural analysis on 223 WRKY genes from pepper, tomato and potato were visualized using GSDS 2.0 in group-wise manner (Figure 2). Intron number of the WRKY genes demonstrated extensive variations, such as 0 (e.g., StWRKY45), 1 (e.g., CaWRKY14), 2 (e.g., SlWRKY36), 3 (e.g., CaWRKY25), 4 (e.g., StWRKY38), 5 (e.g., CaWRKY51), and 9 (CaWRKY33) (Supplementary Table S1). Nevertheless, a certain degree of conservatism was demonstrated by the dominated intron number of 2, which accounts for more than 50% (114/223) of the intron numbers in solanaceae WRKY genes. The intron number of different WRKY group members also displayed certain variations, as the group I (ranged from 2 to 5), group IIa (ranged from 2 to 4), and group IIb (ranged from 2 to 5) seemed to own relatively more introns than the other group members (ranged from 0 to 3) (Supplementary Table S1). The intron numbers are most variable among group I WRKY members (CV = 0.31), and relatively conserved among group IIa (CV = 0.17) and group IIe (CV = 0.11) WRKY members (Supplementary Table S1). Notably, WRKYs of group IIf (CaWRKY17, SlWRKY26, StWRKY17, StWRKY22, and StWRKY73) had distinct gene structure compared to the counterparts of other groups, due to their zero intron structure (Figure 2). A group-wise intron phase distribution was reflected by the intron phase profile analysis of the solanaceae WRKY genes, as all three different intron phases (0, 1, 2) could be detected in the WRKY genes of group I, IIc, IIg, III and NG, two intron phases (1 and 2) were found in group IId and IIe WRKYs, while only one intron phase (0) was detected in the WRKYs of group IIa and IIb. As for the intron phase pattern (pattern of intron phase constitution), three dominated (frequency > 70%) patterns were identified in some WRKY groups, including 0-0-0 pattern in group IIa (frequency of 71.4%), 2-2 pattern in group IId (frequency of 77.8%) and group IIe (frequency of 76.2%) (Supplementary Table S1). Thus, both the intron phase profile and intron phase pattern analysis suggest high genetic variations occurred in group I, IIc, IIg, III, and on the contrary, relatively genetic conservatism in group IIa, IId, and IIe (Figure 2 and Supplementary Table S1).

Figure 2. The group-wise analysis of WRKY gene structures from pepper (Capsicum annuum; CaWRKY), tomato (Solanum lycopersicum; SlWRKY), and potato (Solanum tuberosum; StWRKY). The upstream/downstream, exon and intron are shown with blue rectangle, yellow cylinder and black line, respectively. Intron phase numbers (0, 1, 2) are labeled on the top of each WRKY gene sketch map.
Domain Composition and Conserved Amino Acids/Motif at the WRKY Domain Region of WRKY Proteins in Solanaceous Plants
As the most conserved structures of WRKY domain, the heptapeptide WRKYGQK and C2H2/C2HC zinc-finger structure are critical for DNA-binding and subsequent transcriptional regulation (Rushton et al., 1996, 2010). According to Figure 3, the WRKY domains of different solanaceous plants were relatively conservative as we expected. Nevertheless, some variations of the conserved heptapeptide WRKYGQK 7 structure, such as KKKGEK (CaWRKY58), WRKYGKK (SlWRKY60), and WHKYGQR (StWRKY55), were still detected. Except for the WRKYGQK polypeptide and Cys (C)/His (H) of zinc-finger, more highly conserved (occurrence rate ≥ 90%) amino acids (aa) were identified, including seven aa [Asp (D), Pro (P), Tyr (Y), Tyr (Y), Lys (K), Val (V) and Tyr (Y)] in pepper, five aa (D, P, Y, K and Y) in tomato, and four aa (D, Y, K and Y) in potato, respectively (Figure 3). Thus, the four common amino acids, D (4 amino acids pre-WRKYGQK), Y (3 amino acids pre C1), K (4 amino acids after C2) and Y (4 amino acids pre H1), are the most conserved amino acids of solanaceae WRKY domains, which may play conservative and important roles in the WRKY proteins of solanaceous plants.
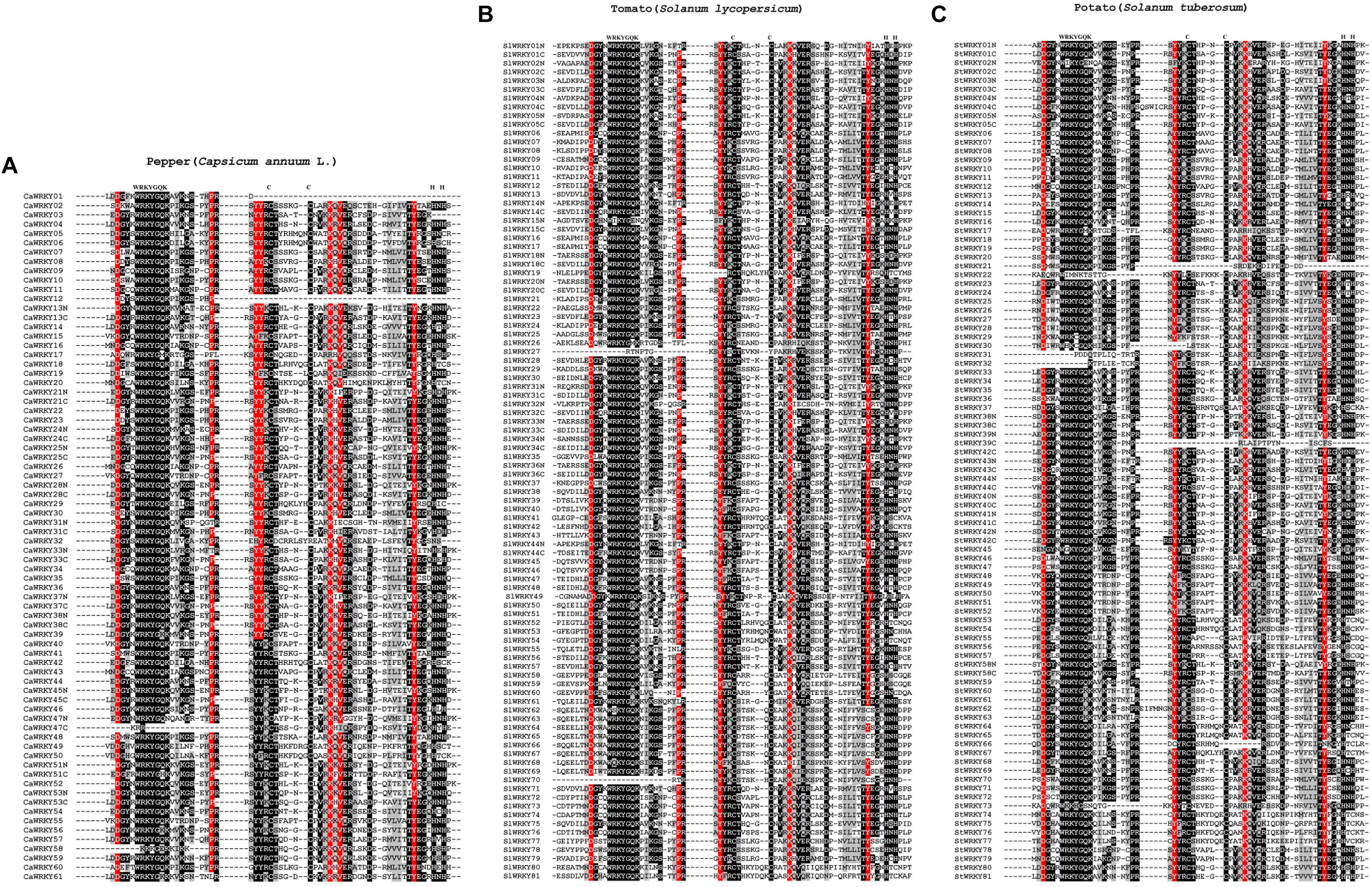
Figure 3. The sequence alignment of WRKY domains from solanaceous plants. (A) Pepper, (B) Tomato, and (C) Potato. The conserved amino acids were emphasized with black background. WRKYs from pepper, tomato and pepper were clustered together, respectively. The position of conserved heptapeptide (WRKYGQK) and zinc-finger consisting amino acids (C-C-H-H/C) were labeled on top. Amino acids with over 90% frequency were emphasized with red background.
The domain composition was further analyzed in a group-wise manner (Figure 4). The corresponding WRKY domain frame that recognizes WRKY domain sequences of each group was generated. For example, motif [DGY]-X(1)-WRKYGQK-[VTRDNP]-X(1)-[PRAY]-X(2)-C-[SFAP]-X(1)-C-[PVKKKVQRS]-X(2)-[D]-X(1)-[SI]-X(1)-[VA]-X(1)-[YEGE]-H-[N]-H was conducted to recognize all group IIa members (Supplementary Table S2). According to Figure 4, the heptapeptide WRKYGQK is extremely conserved among WRKY domains of group IIa, IIb, IId and IIe. On the contrary, the variations (WRKYGEN, WRKYGHK, WRKYGKK, WRKYGMK, etc.) were widely distributed in the WRKY domains of group IIc, IIf, IIg, and III. The WRKY domain frames exhibited different degrees of variations among different groups, as WRKY domains of group IIa, IIb, and IId own over 40 highly conserved amino acids (including WRKYGQK and C/H of zinc-finger), while the N-terminal of group I, group IIf and group III only have conserved amino acids of less than 30 (Supplementary Table S2). As for the detailed sequences of zinc-finger structures, the results showed that the variation of C2H2/C2HC zinc-finger structure was mainly occurred on the amino acid number between the first Cys (C1) and the second Cys (C2) or C2 and the first/only His (H1/H). The number of amino acids (4) between C1 and C2 of group I, as well as between C2 and H1 of group II (23) WRKY domains, were both consistent. On the contrary, the number of amino acids between C1 and C2, as well as between C2 and H1 of group III WRKY domains seemed to be genetically inconstant (4 or 7 between C1 and C2; 22, 23, or 24 between C2 and H1) (Supplementary Table S2).
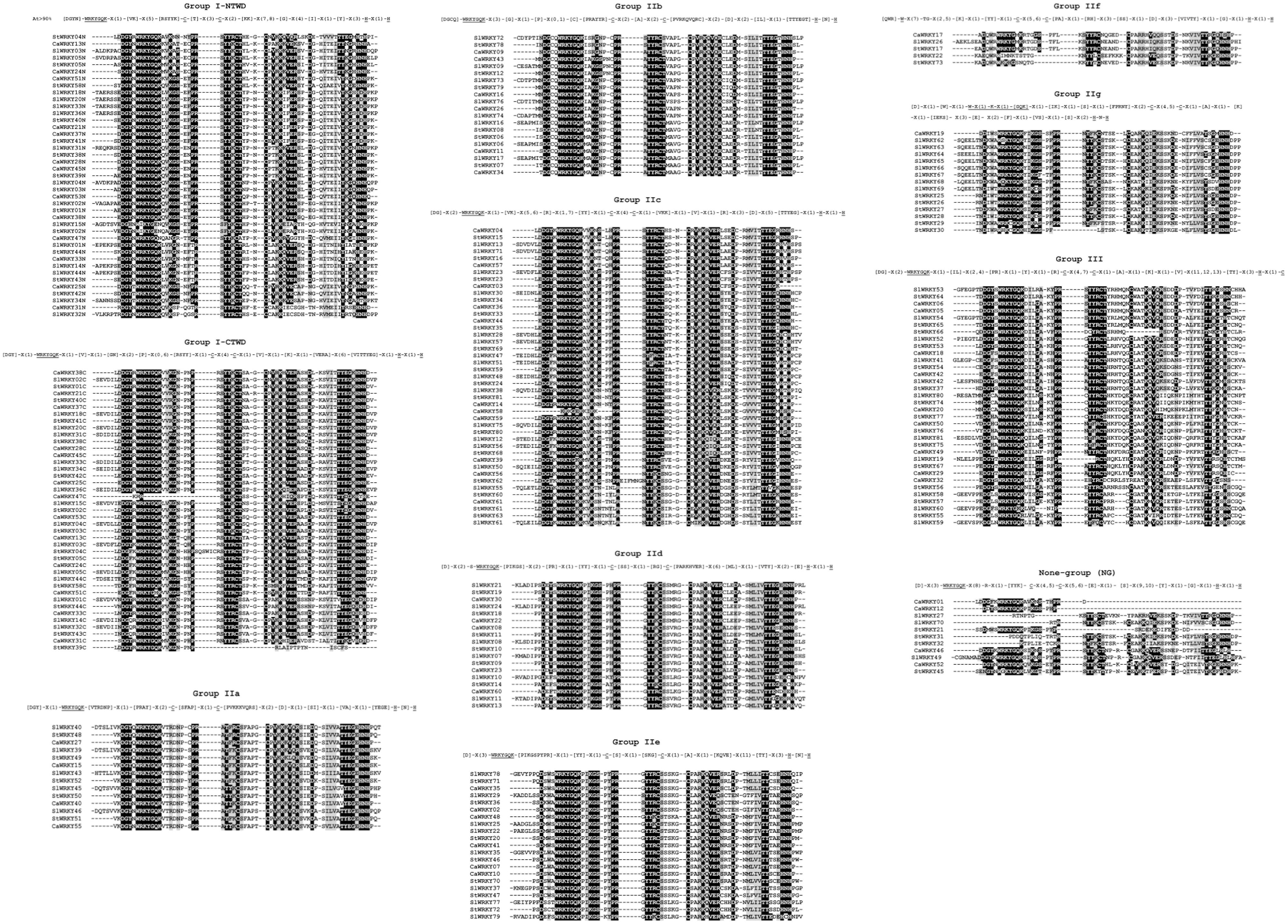
Figure 4. The group-wise sequence alignment of WRKY domains from solanaceous plants (pepper, tomato and potato). The group frames based on the conservatism of corresponding amino acids of WRKY groups were presented on top of each sequence alignment. The amino acids with occurrence rate over 90% were labeled in []; “X” indicate the random amino acids; The symbolic heptapeptide WRKYGQK and C2H2 of zinc-finger structure were annoted with underline.
The group-wise sequence logo motif analysis further revealed the genetic variation situation of motifs in WRKY domain regions of each group (Supplementary Figure S1). As the motif of over five successive amino acids was defined as conserved motif in this study, group IIb has the most conserved motifs (WRKYGQK, CPRAYYR, PVRKQVQRC, and TTYEGT), while group III has no conserved motif at all (Supplementary Figure S1), indicating the occurrence of more intense variation in group III WRKY domains.
Domain Composition Analysis in WRKY Proteins of Solanaceous Plants
Some functional domains except for the WRKY domain in solanaceae WRKYs were also detected, which were mainly existed in the WRKY members of group II, especially in the group IId members of pepper and group IIa/IIb/IId members of tomato and potato (Supplementary Table S3). Plant-ZN-ClUST (pfam10533) was the dominated domain of group IId WRKYs in pepper, tomato and potato, the function of the Plant-ZN-ClUST domain is still unclear. Some other common domains were also detected in the group IIa and group IIb WRKYs of tomato and potato. For instance, SlWRKY40 and StWRKY48 (group IIa members) both contain PHA03255 (BDLF3: PHA03255) and SSP160 (Special lobe-specific silk protein: pfam06933) domain. SlWRKY06, SlWRKY17, StWRKY06, and SlWRKY07 (group IIb members) all carried the common bZIP-Maf-small domain (Basic leucine zipper (bZIP) domain of small musculoaponeurotic fibrosarcoma (Maf) proteins: cd14717), which might be involved in various cell functions including proliferation, apoptosis, survival, and morphogenesis (Hang and Stein, 2011; Vanderford, 2011; Kannan et al., 2012). The larger average size of group IIb and IId WRKY proteins in tomato and potato might be caused by the extensively existence of extra domains (Table 1). No other domain except for WRKY was detected in group IIe, IIf, IIg, or III WRKY members of solanaceous plants (Supplementary Table S3). In summary, the domain composition of WRKYs that belong to different groups was also a reflection of genetic conservatism in group IIa, IIb, and IId, and genetic variation in group IIe, IIf, IIg, and III, as the WRKY groups with more common domains tend to be more genetic and functionally conserved.
Orthologue Analysis of WRKY Genes in Solanaceous Plants
A Best-BLAST approach was employed to identify candidate StWRKY/SlWRKY/CaWRKY orthologies among potato, tomato and pepper. In defining probable orthologous pairs (two WRKYs) or combinations (three WRKYs), we required that orthologous proteins belong to the same structural class (based on phylogenetic tree) (Figure 1) and have a pairwise identity of over 90% (based on sequence blast). Corresponding candidate orthologous pairs or combinations were also filtered by chromosome localization (Grube et al., 2000; Xu et al., 2016). Totally 119 WRKY protein pairs were detected as bona fide orthologs, labeled on the corresponding chromosomes, and linked with lines to highlight the relative positions in their genomes (Figure 5). Of the 119 WRKY orthologous pairs identified, nearly 60% (71) of which were SlWRKY/StWRKY pairs, compared to only 23 SlWRKY/CaWRKY and 25 StWRKY/CaWRKY pairs, respectively (Figure 5), which confirmed the notable variation between the Capsicum genus (pepper) and the Solanum genus (tomato and potato). The 19 orthologous combinations were identified from seven different chromosomes, as chromosome 02 (2), chromosome 03 (5), chromosome 04 (1), chromosome 06 (3), chromosome 07 (6), chromosome 08 (1), and chromosome 10 (1), indicating the relatively lower degree of genetic variation of WRKYs in these chromosomes, especially in chromosome 03 and chromosome 07 (Figure 5).
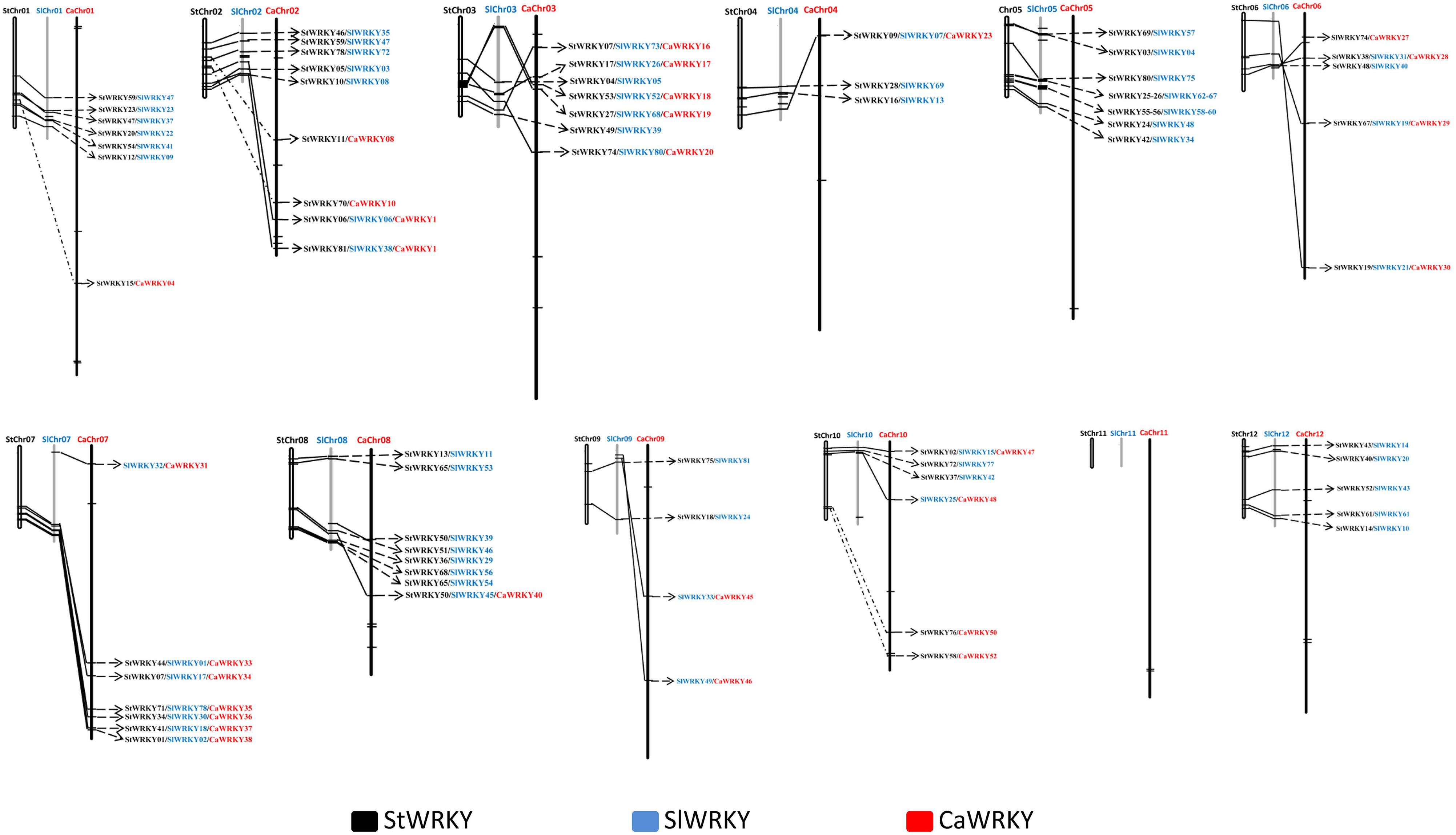
Figure 5. Orthologue analysis of WRKY genes in potato (Solanum tuberosum), tomato (Solanum lycopersicum) and pepper (C. annuum). The white, gray and black columns represent the chromosomes that belong to potato, tomato, and pepper, respectively. The StWRKYs, SlWRKYs, and CaWRKYs were labeled in black, blue and red, respectively. The orthologures of StWRKY, SlWRKY, and CaWRKY were connected with black lines.
Expression Analysis of CaWRKY Genes in Various Pepper Tissues or Under Different Biotic/Abiotic Stresses
To explore the functional diversity of solanaceae WRKY proteins, we analyzed the tissue expression pattern of the 61 CaWRKY genes by RT-qPCR (Figure 6A). Three representative tissue expression patterns of the CaWRKYs were demonstrated, including constitutive expression (expressed in all tested tissues), low expression (barely expressed in any of the tissue tested), specific/preferential expression (significantly high expression in a specific tissue). According to Figure 6A, half of the constitutively expressed CaWRKY genes [CaWRKY30 (IId), CaWRKY31 (I), CaWRKY33 (I), CaWRKY35 (IIe), CaWRKY38 (I), CaWRKY41 (IIe), CaWRKY45 (I), CaWRKY46 (NG), CaWRKY47 (I), and CaWRKY55 (IIa)] belong to group I, and none belongs to group III CaWRKY in this expression pattern. On the contrary, thirteen CaWRKY genes including CaWRKY03 (IIc), CaWRKY04 (IIc), CaWRKY06 (III), CaWRKY07 (IIe), CaWRKY17 (IIf), CaWRKY18 (III), CaWRKY48 (IIe), CaWRKY54 (IIc), CaWRKY56 (IIc), CaWRKY57 (IIc), CaWRKY58 (IIc), CaWRKY60 (IId), and CaWRKY61 (IIc), were found to be barely expressed in any of the tissue tested (Figure 6A). Group IIc CaWRKYs was the dominated (7/13) members of the low expression pattern, and none of group I CaWRKY was detected in this pattern. As for the specific/preferential expression pattern, relatively more CaWRKY genes including seed specifically/preferentially expressed CaWRKs [CaWRKY19 (IIf), CaWRKY51 (I), CaWRKY52 (NG)], leaf specifically/preferentially expressed CaWRKYs [CaWRKY12 (NG), CaWRKY39 (IIc)], root specifically/preferentially expressed CaWRKYs [CaWRKY15 (IIa), CaWRKY26 (IIb)], stem specifically/preferentially expressed CaWRKY04 (IIc), and full ripening fruit specifically/preferentially expressed CaWRKYs [CaWRKY01 (NG), CaWRKY02 (IIe), CaWRKY09 (IIb), CaWRKY10 (IIe), CaWRKY11 (IIb), CaWRKY36 (IIc), CaWRKY37 (I), CaWRKY40 (IIa), CaWRKY42 (III), CaWRKY49 (III), and CaWRKY59 (IIc)], were respectively identified (Figure 6A). The remaining CaWRKYs were expressed in two or more different tissues with diversified expression patterns (Figure 6A).
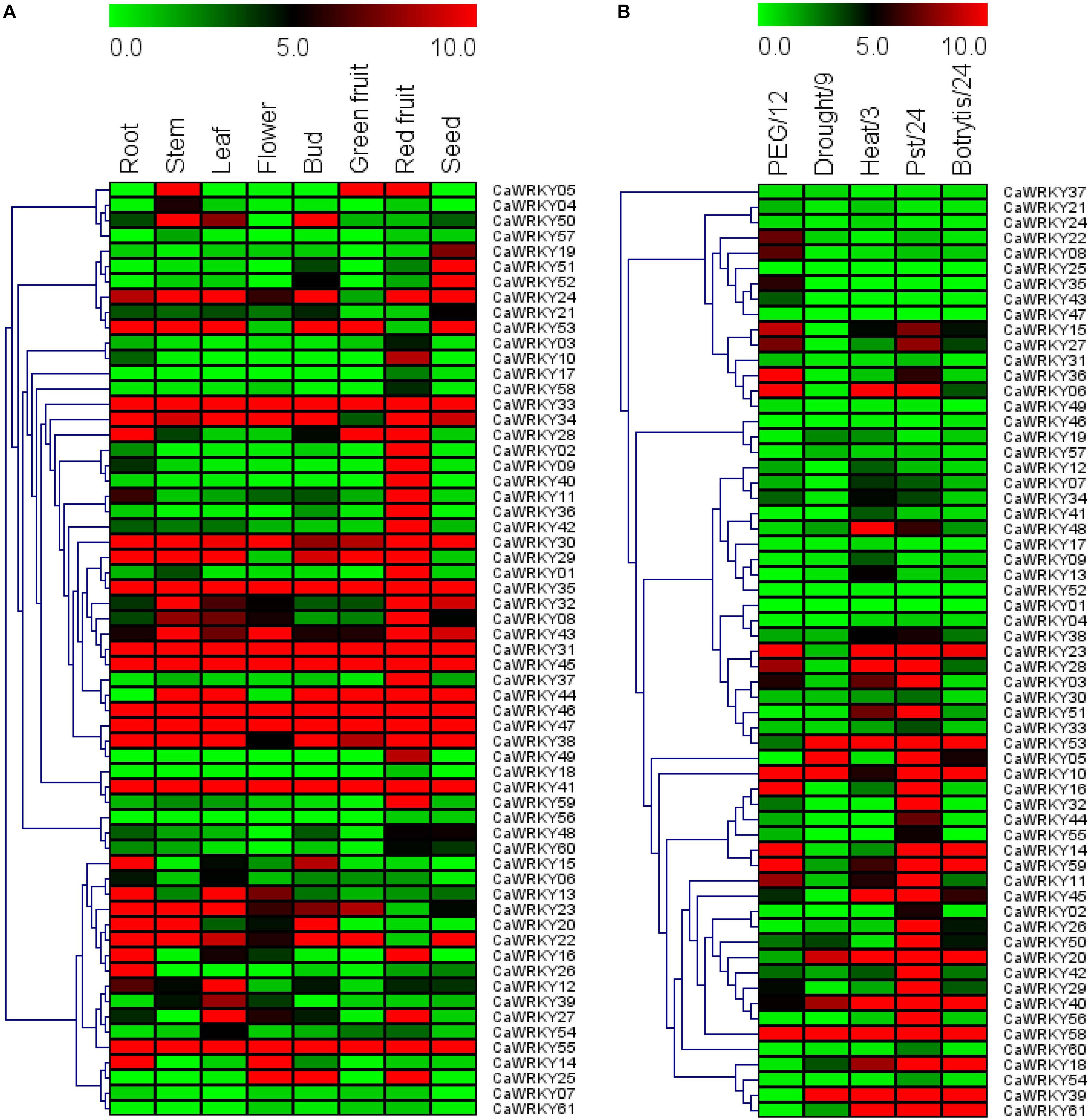
Figure 6. Expression profiles of 61 CaWRKY genes in various pepper tissues (A) and under different biotic/abiotic stresses (B). (A) CaWRKY gene expression was determined by RT-qPCR. CaWRKYs are indicated as rows and tissues as columns. Green, black and red elements represented down-regulated, no change and up-regulated, respectively. (B) CaWRKY gene expression was determined by RT-qPCR. CaWRKYs are represented as rows and stress treatment/time point as columns (PEG/12: 20% PEG6000 for 12 h; Drought/9: Water withheld for 9 days; Heat: 42°C for 3 h; Pst/24: PstDC3000 infection for 24 h; Botrytis/24: Botrytis cinerea infection for 24 h. The vertical dendrograms represent the similarity degree of tissue expression profile (A) and stress response profile (B) among the 61 CaWRKY genes.
In addition to biotic stress, abiotic stresses, such as drought, salinity and extreme temperatures are important factors that constrain agricultural productivity worldwide (Chen et al., 2013; Wang et al., 2016; Li et al., 2018). According to the stress response pattern of CaWRKYs reflected in Figure 6B, more than half (32/61) of CaWRKY genes were significantly induced by one or more biotic/abiotic stresses tested (osmotic stress, drought, heat, PstDC3000 and B. cinerea). Overall, compared to abiotic stresses (PEG, drought and heat), CaWRKYs were more sensitive to pathogen (PstDC3000 and B. cinerea) infections. For example, 25 CaWRKY genes were significantly up-regulated under PstDC3000 infection, and eleven CaWRKYs were induced by B. cinerea. Meanwhile, only eight PEG-induced, five drought-induced and eleven heat-induced CaWRKY genes were detected. It is worth mentioning that nearly 70% (7/13) of the group I CaWRKYs seemed to be insensitive to any of the stress imposed, while nearly 90% (8/9) of the group III CaWRKYs (CaWRKY49) was induced by at least one of the stresses tested.
Functional Analyses Suggested Differential Roles of CaWRKYs in Disease Resistance and Abiotic Stress Responses in Pepper
To directly analyze the biological functions of CaWRKY genes from pepper, we first screened for the 32 CaWRKY genes that were induced by at least one of the stresses tested (Figure 7B), and obtained the corresponding gene silencing lines through Virus induced gene silence (VIGS) technology. The resistance of the 32 CaWRKY gene silenced plants to PstDC3000, B. cinerea and drought were tested, and several representative phenotypes determined by the silencing of some CaWRKY genes were detected in this study (Figure 7).
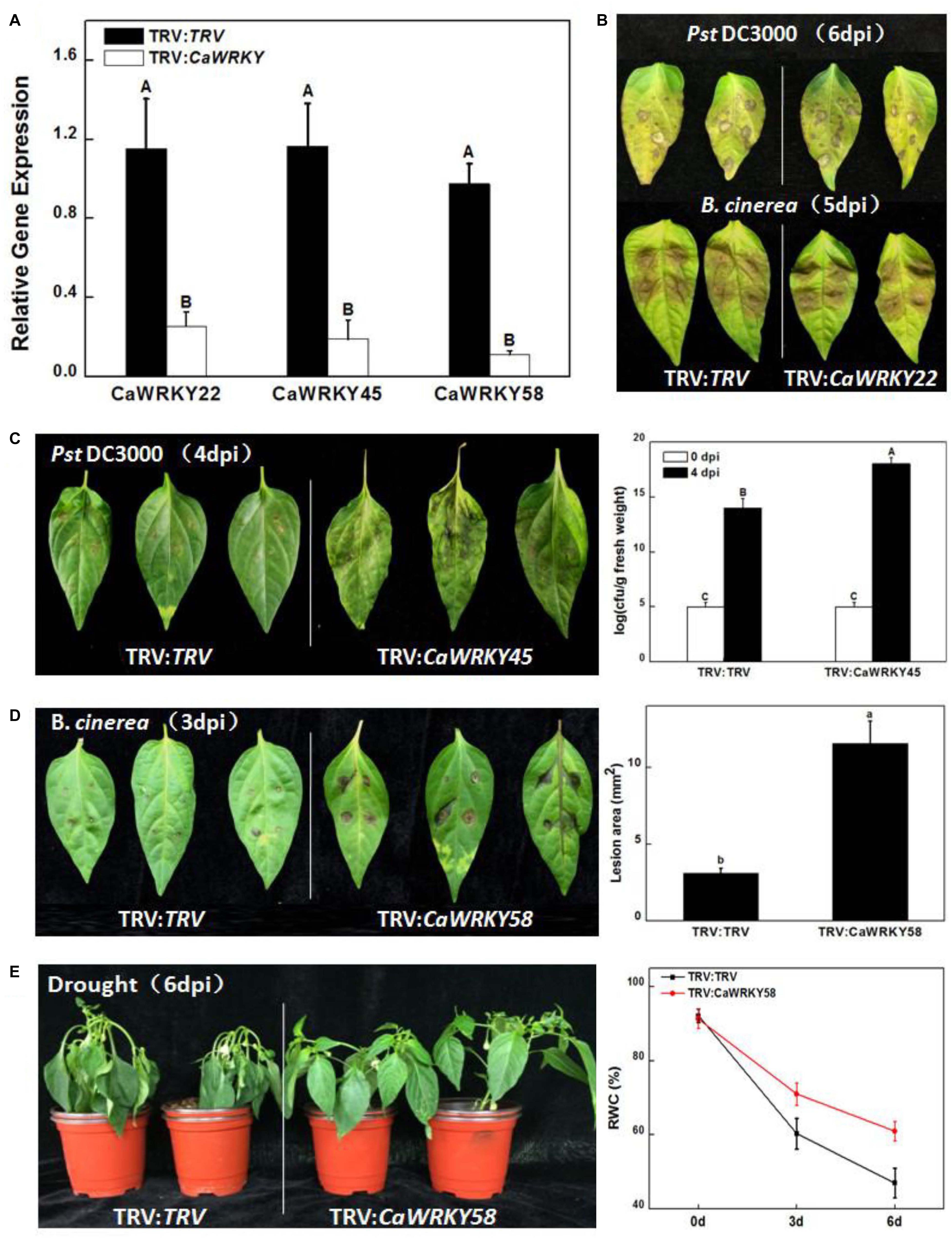
Figure 7. Functions of the CaWRKY genes in disease resistance and abiotic stress responses indicated by Virus induced gene silencing (VIGS). (A) Silencing efficiency. Pepper plants infiltrated with Agrobacterium suspensions carrying an empty pTRV vector served as control plants. Expression levels of CaWRKYs in control and VIGS-treated plants were detected by RT-qPCR assays. (B) PstDC3000 (Up) and B. cinerea (down) infections. Photographs were taken at 6 dpi days past inoculation) and 5 dpi, respectively. (C) Enhanced susceptibility to PstDC3000. pTRV-control plants (TRV:TRV) and CaWRKY45-silenced plants (TRV:CaWRKY45) were inoculated with PstDC3000 and the picture was taken at 4 dpi (Left). According to DMRT (Duncan’s multiple range test), means of the colony-forming units (cfu) in leaves of TRV:CaWRKY45 is significantly higher than control at 4 dpi (P < 0.01) (Right). (D) Enhanced susceptibility to B. cinerea. TRV:TRV and TRV:CaWRKY58 plants were inoculated with B. cinerea and the photograph was taken at 3 dpi (Left). Lesion diameter was measured and statistically calculated for all plants. Significant difference between lesion diameters of the silenced plants and that of the control plants is indicated at P < 0.05 according to DMRT (Right). (E) Drought stress response. Phenotypes of the CaWRKY58-silenced and control plants after withholding water for 6 days (Left). Comparisons of TRV:CaWRKY58 and TRV:TRV control plants at 6th day after drought treatment. Twelve plants were used for each treatment. The experiments were conducted three times independently. Values represent mean ± SD (n = 3).
The silencing effects were partially demonstrated in Figure 7A, from which the reduced expression patterns of CaWRKY22, CaWRKY45, and CaWRKY58 were evident. Figure 7B was intended as negative control, showing that most silencing plants had no phenotype, just like CaWRKY22 silenced plants. CaWRKY45 silenced plants showed compromised resistance to PstDC3000 (Figure 7C). Disease lesions developed quicker and stronger in CaWRKY45 silenced plants and the bacterial growth was about 5-log higher. Consistently, the expression of CaWRKY45 was highly induced by PstDC3000, but not B. cinerea. CaWRKY45 was consistently expressed in all pepper tissues detected (Figure 6A), which may point to a life-keeping as well as defense-related role of this important WRKY protein. Another gene, CaWRKY58, showed reduced resistance to B. cinerea. As shown in Figure 7D, both images and measurement demonstrated significantly increased lesion area caused by B. cinerea at 3 days post-inoculation in CaWRKY58-silenced plants. In contrast to CaWRKY45, CaWRKY58 was expressed at very low level in most tissues (except in red fruit), and was induced by all stresses (both biotic and abiotic). Thus, CaWRKY58 may be purely stress-responsive or stress-related protein. Interestingly, the CaWRKY58-silenced plants were also more tolerant to drought stress (Figure 7E).
Discussion
Extensive Genetic Variations Occur in Solanaceae WRKYs
With the advances in genome sequencing technology, the whole genome sequences of tomato, potato, and pepper were subsequently accomplished (The Tomato Genome Consortium, 2012; Qin et al., 2014), and the corresponding WRKY TF families of these three plants were unveiled in recent years (Huang et al., 2012; Huang and Liu, 2013; Cheng et al., 2016). Here, a comparative analysis was conducted on the WRKY families of pepper, tomato and potato, and extensive genetic variations of the solanaceae WRKYs were demonstrated by the variations of WRKY number/size, group constitution, phylogenetic relationship, gene structure, and domain composition. Moreover, significant genetic variations were observed between the Capsicum genus (pepper) and the Solanum genus (tomato, potato) (Figure 5 and Tables 1, 2), and some minor variations on WRKY group constitution and average WRKY size of each group were also detected between tomato and potato that both belong to the Solanum genus (Tables 1, 2).
Many genetic variations were revealed in group-wise manner. For example, WRKY genes of group I were most variable on the gene structure characteristics (intron number, intron phase profile, intron phase pattern) among all solanaceae WRKY groups (Figure 2 and Supplementary Table S1). The group II (especially group IIc and IIg) was the main group in which the WRKY gene loss/gain variations occurred in solanaceous plants (Table 1). The group III WRKYs experienced the most intense genetic variation during evolutionary process, revealed by the least number of conserved amino acids (only 24), zero number of motif in WRKY domain, and highly variant zinc-finger structure of this group members (Supplementary Figure S1 and Supplementary Table S2). Moreover, the only existence of WRKY domain (no other common functional domain detected) in group III WRKY members was also an indication of high variability of this group (Supplementary Table S3). As for the solanaceae specific WRKY groups including group IIf and IIg (Figure 1), group IIf had the unique gene structure of no intron and relatively smaller size (average size of 690 bp in solanaceae, average size of 786 bp in pepper, average size of 660 bp in tomato, average size of 668 bp in potato) (Figure 2 and Table 2), and group IIg was the main group in which variation in WRKY number occurred between the Capsicum genus and Solanum genus (Table 1). Considering these distinctive characteristics, further analysis is required to elucidate the evolutionary specificity of these two solanaceae-specific WRKY groups in the future.
It is well accepted that the existence of almost invariable WRKYGQK motif and C2H2/C2HC zinc-finger is the symbolic structure of WRKY domains (Eulgem et al., 2000; Xie et al., 2005), which are necessary but not sufficient to judge a WRKY. Previous studies have shown that WRKYGQK is not completely invariable (Xie et al., 2005; Huang et al., 2012; Xu et al., 2016), and some TFs other than WRKY also contain zinc-finger structures (Babu et al., 2006; Duan et al., 2007; Shi et al., 2014). Here, various WRKYGQK variants in WRKYs of solanaceous plants were identified, more than half of them were from potato (Figure 3), and the remaining variants were mainly distributed across the solanaceae WRKY domains of group IIc, IIf, IIg and III (Figure 4). As for zinc-finger, our group-wise frame analysis (Supplementary Table S2) showed a variant zinc-finger structure (CX4,7CX23HXC) of group III in solanaceae WRKYs, which differs from the well recognized CX7CX23HXC zinc-finger structure (Rushton et al., 2010; Cheng et al., 2012).
Certain Degrees of Genetic Conservatism Present in Solanaceae WRKYs
Although this study was mainly focused on the genetic variation analysis, certain extent of conservatism in the solanaceae WRKYs was also demonstrated in this study. The most direct evidence was the existence of four common conserved amino acids in the solanaceae WRKY domains, which are the D (4 amino acids pre-WRKYGQK), Y (3 amino acids pre the first C of zinc finger), K (4 amino acids after the second C of zinc finger), and Y (4 amino acids pre the first/only H of zinc finger) (Figure 3). Moreover, the analysis of WRKY number, group constitution and average WRKY size, domain composition and ortholog pairs showed certain degree of genetic conservatism between the WRKYs of tomato and potato (Figures 1–5 and Tables 1, 2). The group-wise analysis of WRKY gene structure features (intron number, intron phase, intron phase pattern) showed that group IIa and IIe were relatively more conserved compared to WRKY genes of other groups, determined by their lower CV of intron number, simpler composition of intron phase (0, 2) and corresponding intron phase pattern (0-0-0, 2-2) (Supplementary Table S1).
Gene Expression Profile Analysis of Pepper CaWRKYs
Although the full genome sequence of pepper was recently completed (Qin et al., 2014), no systematic research on CaWRKYs of pepper had been conducted so far. Herein, we concluded that the constitutively expressed CaWRKYs (10 members) and low expressed CaWRKYs (13 members) both possessed relatively low percentage of the whole CaWRKY gene family, which revealed the potential functional diversity of CaWRKYs in pepper. The group-wise analysis showed that the CaWRKYs of group I were the main part of constitutive expression pattern members (50%), while no group I CaWRKY was detected in the low expression pattern members. Furthermore, CaWRKYs of group IIc seemed to be the dominant members in low expression pattern, and no group III CaWRKY was found to be constitutively expressed in all the tested tissues. As for the specifical/preferential expressed pattern, CaWRKYs that belong to various groups were included, and no special distribution role was detected. Nevertheless, these specific/preferentially expressed CaWRKYs could be related to the particular character or functions of the tissues where they expressed. To date, most studies on WRKY TFs were focused on their resistance functions, and plant leaves were always collected for relative research. Our study showed the possibility that CaWRKYs may play corresponding roles in the development of various pepper organs, especially fruit maturation, as a large number of CaWRKYs were specifically expressed in the red fruits in general. According to the responsiveness of CaWRKYs to various biotic and abiotic stressors, more than 50% of CaWRKY genes were sensitive to one or more biotic/abiotic stresses tested (osmotic stress, drought, heat, PstDC3000 and B. cinerea). Nevertheless, nearly 70% of group I CaWRKYs seemed to be insensitive to any of the stress used, on the contrary, nearly 90% of group III CaWRKYs was induced by at least one of the stresses imposed, which indicated a high activity of stress response of group III. As both the tissue expression and stress response pattern of CaWRKYs were somehow correlated with their group classification, we propose that WRKY TFs may play their specific biological roles in group-wise manner, which is worthy a lot of subsequent study.
Some Pepper CaWRKYs Play Critical Roles in Disease Resistance and Abiotic Stress Responses
Before the release of PGD (Release 2.01), eight WRKY protein encoding genes from pepper had been reported to play critical roles in biological processes, including pathogen resistance and high-temperature tolerance (Dang et al., 2013, 2014; Cai et al., 2015; Xu et al., 2016). Consistent with earlier studies, CaWRKY10 (previously named CaWRKY27) was induced by most stresses except heat, while CaWRKY15 was induced by PEG and PstDC3000 and CaWRKY34 (previously named CaWRKY6) was induced by heat stress. CaWRKY45 (named CaWRKY-a) was induced by both heat and PstDC3000. Interestingly, CaWRKY58 (previously named as CaWRKY-b) was “most stress responsive,” as its tissue-specific expressions were low in general, but its stress-responsive expressions were high in all stresses tested. Nevertheless, the function study of CaWRKY proteins in pepper was still very limited, compared to Arabidopsis, rice and tomato. Just like its counterparts in other plants, CaWRKYs seem to be closely involved in various stress responsive processes, revealed by the drastic expression changes of most CaWRKY genes under two disease infections (PstDC3000 and B. cinerea) or three abiotic stresses (osmotic stress, drought and high temperature). From a functional point of view, our VIGS experiments demonstrated a positive role of CaWRKY58 in disease resistance (B. cinerea) and a negative role in drought tolerance. This WRKY protein can be multi-functional in stress responses, and its role in other stresses and the correlation and connection of all those stress responsive pathways are the appealing research directions for the future. What’s more, CaWRKY58 might have multiple interacting partners to carry out all its functions in various stresses. CaWKRY45 was shown by VIGS to positively regulate PstDC3000 defense pathways. This phenotype fitted with the PstDC3000 responsive expression of CaWRKY45. In support to our findings, earlier publication (using the name CaWRKY-a) suggested that TMV, Xcv and salicylic acid (SA)-induced expression of CaWRKY45. It will be interesting to further identify its interacting partners and find out the affected pathways (either PTI or ETI) of CaWRKY45 in pepper. It is surprising that many defense responsive CaWRKY genes showed no phenotype in response to the stresses we tested. Perhaps, the limitations of VIGS or the gene redundancy may hide the phenotype in silenced plants in our study. However, the currently discovered phenotypes shed some light on the functionality of the CaWRKY family, and further researches will be continued to unravel the underlining mechanisms.
Conclusion
The WRKY gene family has been demonstrated to be involved in various biological processes including plant development and responses to biotic/abiotic stresses (Pandey and Somssich, 2009; Li et al., 2013a; Dang et al., 2014; Cai et al., 2015). The comprehensive and systematic comparative analysis of 223 WRKY members from solanaceae crops (pepper, tomato and potato) demonstrated tremendous genetic variations among WRKY members of different solanaceous plants or groups, as well as certain degrees of conservatism for some solanaceae WRKYs. Moreover, the expression analysis and functional exploration of CaWRKYs in pepper provide insight into the functional divergence of the WRKY gene family from pepper. The results of bioinformatics and functional analysis might provide basic resources for further dissection of the evolutionary clues of WRKYs in solanaceae crop plants, and also elucidation of the functional diversity of CaWRKYs in pepper.
Author Contributions
HW and GZ supervised and designed the experiments. YC, QY, ZY, and MR performed the experiments. YC and GA conceived the project and wrote the manuscript with contributions from ZL and RW.
Funding
This research was partially supported by National Key Research and Development Program of China (2018YFD1000800), State Key Laboratory Breeding Base for the Zhejiang Sustainable Pest and Disease Control (2010DS700124-ZZ1903 and 2010DS700124-ZZ1807), National Natural Science Foundation of China (31772294), National Key Research and Development Program (2018YCGC005), Zhejiang Provincial major Agricultural Science and Technology Projects of New Varieties Breeding (2016C02051), Zhejiang Provincial Natural Science Foundation of China (LY18C150008), General Program from the National key research and development program (2017YFD0101902), The earmarked fund for China Agriculture Research System (CARS-23-G-44).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00492/full#supplementary-material
FIGURE S1 | The group-wise logo motif analysis of WRKY domains from solanaceous plants (pepper, tomato and potato). The bit score for each position in the sequence and the group names are indicated to the left. The blue solid circle and brown solid square label the conserved heptapeptide and zinc-ginger structure, respectively. The green triangle below the logo indicates the distinct motif uniquely existing in the corresponding group. The red asterisks below indicate the highly conserved amino acids in each group.
TABLE S1 | The WRKY intron features of solanaceous plants.
TABLE S2 | The group-wise analysis of WRKY domain frame, conserved amino acids number and zinc-finger structure.
TABLE S3 | Domain composition analysis in WRKY proteins of solanaceous plants.
TABLE S4 | Primers of CaWRKY genes for RT-qPCR analysis.
TABLE S5 | Primers of CaWRKY genes used for VIGS analysis.
Footnotes
- ^http://pepperhub.hzau.edu.cn
- ^https://solgenomics.net/
- ^http://potatogenome.berkeley.edu/nsf5
- ^http://pfam.xfam.org/
- ^http://www.clustal.org/
- ^http://gsds.cbi.pku.edu.cn
- ^http://smart.emblheidelberg.de/
- ^http://weblogo.berkeley.edu/
- ^http://www.ncbi.nlm.nih.gov/cdd
- ^http://chibba.agtec.uga.edu/duplication/index/locus
- ^www.genscript.com
- ^http://vigs.solgenomics.net/
References
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. doi: 10.1038/sj.emboj.7600737
Antoni, R., Rodriguez, L., Gonzalez-Guzman, M., Pizzio, G. A., and Rodriguez, P. L. (2011). News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr. Opin. Plant Biol. 14, 547–553. doi: 10.1016/j.pbi.2011.06.004
Babu, M. M., Lyer, L. M., Balaji, S., and Aravind, L. (2006). The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 34, 6505–6520. doi: 10.1093/nar/gkl888
Bakshi, M., and Oelmuller, R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav. 9:e27700. doi: 10.4161/psb.27700
Besseau, S., Li, J., and Palva, E. T. (2012). WRKY54 and WRKY70 cooperate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679. doi: 10.1093/jxb/err450
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Chen, S., Jin, W., Liu, A., Zhang, S., Liu, D., Wang, F., et al. (2013). Arbuscular mycorrhizal fungi (AMF) increase growth and secondary metabolism in cucumber subjected to low temperature stress. Sci. Hortic. 160, 222–229. doi: 10.1016/j.scienta.2013.05.039
Cheng, Y., Yao, Z. P., Ruan, M. Y., Ye, Q., Wang, Q., Zhou, G. Z., et al. (2016). In silico identification and characterization of the WRKY gene superfamily in pepper (Capsicum annuum L.). Genetics Mol. Res. 15:gmr15038675. doi: 10.4238/gmr.15038675
Cheng, Y., Zhou, Y., Yang, Y., Chi, Y. J., Zhou, J., Chen, J. Y., et al. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. doi: 10.1104/pp.112.196816
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Dang, F., Wang, Y., She, J., Lei, Y., Liu, Z., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup II e WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150, 397–411. doi: 10.1111/ppl.12093
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Dellagi, A., Helibronn, J., Avrova, A. O., Montesano, M., Palva, E. T., Stewart, H. E., et al. (2000). A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol. Plant Microbe Interact. 13, 1092–1101. doi: 10.1094/mpmi.2000.13.10.1092
Duan, M. R., Nan, J., Liang, Y. H., Mao, P., Lu, L., Li, L., et al. (2007). DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 35, 1145–1154. doi: 10.1093/nar/gkm001
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Grube, R. C., Radwanski, E. R., and Jahn, M. (2000). Comparative genetics of disease resistance within the solanaceae. Genet 155, 873–887.
Hang, Y., and Stein, R. (2011). MafA and MafB activity in pancreatic β cells. Trends Endocrin. Met. 22, 364–373. doi: 10.1016/j.tem.2011.05.003
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Gen. Genomics 287, 495–513. doi: 10.1007/s00438-012-0696-6
Huang, S. X., and Liu, Y. S. (2013). The bioinformatics analysis of WRKY transcription factors in potato. Chin. J. App. Environ. Biol. 19, 205–214. doi: 10.1186/s12864-015-1575-4
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA-encoding a novel DNA-binding protein, Spf1, that recognizes Sp8 sequences in the 5′ upstream regions of genes-coding for sporamin and β-amylase from sweet-potato. Mol. Gen. Genet. 244, 563–571.
Jiang, W., and Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. doi: 10.1186/1471-2229-9-96
Kannan, M. B., Solovieva, V., and Blank, V. (2012). The small MAF transcription factors MAFF. MAFG and MAFK: current knowledge and perspectives. Biochim. Biophys. Acta. 1823, 1841–1846. 10.1016/j.bbamcr.2012.06.012
Li, C., Li, Y., Li, Y., and Fu, G. (2018). Cultivation techniques and nutrient management strategies to improve productivity of rain-fed maize in semi-arid regions. Agric. Water Manage. 210, 149–157. doi: 10.1016/j.agwat.2018.08.014
Li, J., Besseau, S., Törönen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013a). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, J., Nie, X., Tan, J. L., and Berger, F. (2013b). Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Nati. Acad. Sci. U.S.A. 110, 15479–15484. doi: 10.1073/pnas.1305175110
Ling, J., Jiang, W., Zhang, Y., Yu, H., Mao, Z., Gu, X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12:471. doi: 10.1186/1471-2164-12-471
Liu, B., Hong, Y. B., Zhang, Y. F., Li, X. H., Huang, L., Zhang, H. J., et al. (2014). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227, 145–156. doi: 10.1016/j.plantsci.2014.08.001
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mangelsen, E., Kilian, J., Berendzen, K. W., Kolukisaoglu, U. H., Harter, K., Jansson, C., et al. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9:194. doi: 10.1186/1471-2164-9-194
Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-004-2142-6
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5125–5140. doi: 10.1073/pnas.1400975111
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Robatzek, S., and Somssich, I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. doi: 10.1101/gad.222702
Ross, C. A., Liu, Y., and Shen, Q. X. J. (2007). The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1744-7909.2007.00504.x
Rushton, P. J., Bokowiec, M. T., Han, S., Zhang, H., Brannock, J. F., Chen, X., et al. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147, 280–295. doi: 10.1104/pp.107.114041
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends. Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, P. J., Torres, J. T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I. E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700. doi: 10.1002/j.1460-2075.1996.tb00953.x
Shi, H., Wang, X., and Cheng, F. (2014). The Cys2/His2-type zinc finger transcription factor ZAT6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and CBFs in Arabidopsis. Plant Physiol. 165, 1367–1379. doi: 10.1104/pp.114.242404
Sun, X. C., Gao, Y. F., Li, H. R., Yang, S. Z., and Liu, Y. S. (2015). Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 58, 52–60. doi: 10.1007/s12374-014-0407-4
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the ter-penoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
The Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tripathi, P., Rabara, R. C., Langum, T. J., Boken, A. K., Rushton, D. L., Boomsma, D. D., et al. (2012). The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics 13:270. doi: 10.1186/1471-2164-13-270
Vanderford, N. L. (2011). Regulation of β-cell-specific and glucose-dependent MafA expression. Islets 3, 35–37. doi: 10.4161/isl.3.1.14032
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271.x
Wang, X.-L., Qin, R.-R., Sun, R.-H., Wang, J.-J., Hou, X.-G., Qi, L., et al. (2018). No post-drought compensatory growth of corns with root cutting based on cytokinin induced by roots. Agric. Water Manage. 205, 9–20. doi: 10.1016/j.agwat.2018.04.035
Wang, X.-L., Wang, J.-J., Sun, R.-H., Hou, X.-G., Zhao, W., Shi, J., et al. (2016). Correlation of the corn compensatory growth mechanism after post-drought rewatering with cytokinin induced by root nitrate absorption. Agric. Water Manage. 166, 77–85. doi: 10.1016/j.agwat.2015.12.007
Wang, Y., Cai, S., Yin, L., Shi, K., Xia, X., Zhou, Y., et al. (2015). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Auto 11, 2033–2047. doi: 10.1080/15548627.2015.1098798
Wu, K. L., Guo, Z. J., Wang, H. H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Xie, Z., Zhang, Z. L., Zou, X. L., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xu, H., Watanabe, K. A., Zhang, L., and Shen, Q. J. (2016). WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 0, 1–13. doi: 10.1093/dnares/dsw025
Yamasaki, K., Kigawa, T., and Inoue, M. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17, 944–956. doi: 10.1105/tpc.104.026435
Zentgraf, U., Laun, T., and Miao, Y. (2010). The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 89, 133–137. doi: 10.1016/j.ejcb.2009.10.014
Zhang, L., Gu, L., Ringler, P., Smith, S., Rushton, P. J., and Shen, Q. J. (2015). Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 236, 214–222. doi: 10.1016/j.plantsci.2015.04.014
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1.
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. doi: 10.1111/j.1365-313x.2006.02901.x
Keywords: WRKY, solanaceae, genetic variation, genetic conservatism, functional diversity
Citation: Cheng Y, Ahammed GJ, Yao Z, Ye Q, Ruan M, Wang R, Li Z, Zhou G and Wan H (2019) Comparative Genomic Analysis Reveals Extensive Genetic Variations of WRKYs in Solanaceae and Functional Variations of CaWRKYs in Pepper. Front. Genet. 10:492. doi: 10.3389/fgene.2019.00492
Received: 20 January 2019; Accepted: 06 May 2019;
Published: 28 May 2019.
Edited by:
Genlou Sun, Saint Mary’s University, CanadaReviewed by:
Jingyin Yu, University of Georgia, United StatesBayram Cevik, Isparta University of Applied Sciences, Turkey
Copyright © 2019 Cheng, Ahammed, Yao, Ye, Ruan, Wang, Li, Zhou and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guozhi Zhou, aHVvaHUxMDA1QDEyNi5jb20=; Q2hpbmF6aG91Z3pAMTYzLmNvbQ==; Hongjian Wan, d2FuaG9uZ2ppYW5Ac2luYS5jb20=
 Yuan Cheng
Yuan Cheng Golam Jalal Ahammed
Golam Jalal Ahammed Zhuping Yao1
Zhuping Yao1 Zhimiao Li
Zhimiao Li Hongjian Wan
Hongjian Wan