
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 12 April 2019
Sec. Neurogenomics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00321
This article is part of the Research Topic Modulation of Central Nervous System Function by Mobile Genetic Elements in Health and Disease View all 6 articles
Social behavior and neuronal connectivity in rodents have been shown to be shaped by the prototypical T lymphocyte-derived pro-inflammatory cytokine Interferon-gamma (IFNγ). It has also been demonstrated that STAT1 (Signal Transducer And Activator Of Transcription 1), a transcription factor (TF) crucially involved in the IFNγ pathway, binds consensus sequences that, in humans, are located with a high frequency in the LTRs (Long Terminal Repeats) of the MER41 family of primate-specific HERVs (Human Endogenous Retroviruses). However, the putative role of an IFNγ/STAT1/MER41 pathway in human cognition and/or behavior is still poorly documented. Here, we present evidence that the promoter regions of intellectual disability-associated genes are uniquely enriched in LTR sequences of the MER41 HERVs. This observation is specific to MER41 among more than 130 HERVs examined. Moreover, we have not found such a significant enrichment in the promoter regions of genes that associate with autism spectrum disorder (ASD) or schizophrenia. Interestingly, ID-associated genes exhibit promoter-localized MER41 LTRs that harbor TF binding sites (TFBSs) for not only STAT1 but also other immune TFs such as, in particular, NFKB1 (Nuclear Factor Kappa B Subunit 1) and STAT3 (Signal Transducer And Activator Of Transcription 3). Moreover, IL-6 (Interleukin 6) rather than IFNγ, is identified as the main candidate cytokine regulating such an immune/MER41/cognition pathway. Of note, differences between humans and chimpanzees are observed regarding the insertion sites of MER41 LTRs in the promoter regions of ID-associated genes. Finally, a survey of the human proteome has allowed us to map a protein-protein network which links the identified immune/MER41/cognition pathway to FOXP2 (Forkhead Box P2), a key TF involved in the emergence of human speech. Our work suggests that together with the evolution of immune genes, the stepped self-domestication of MER41 in the genomes of primates could have contributed to cognitive evolution. We further propose that non-inherited forms of ID might result from the untimely or quantitatively inappropriate expression of immune signals, notably IL-6, that putatively regulate cognition-associated genes via promoter-localized MER41 LTRs.
Interferon gamma (IFNγ), the prototypical T Helper 1 (TH1) cytokine, is a T-cell derived pro-inflammatory molecule exerting several effects on innate immune cells and others on non-immune cells including neurons (Litteljohn et al., 2014). Specifically, IFNγ was recently shown to be a social behavior regulator and to shape neuronal connectivity in rodents (Filiano et al., 2016). Irrespective of cell type, binding of IFNγ to its receptor induces the transcriptional regulation of target genes via the recognition of promoter consensus sequences by the transcription factor STAT1 (Signal Transducer And Activator Of Transcription 1; Ramana et al., 2002; Green et al., 2017). In humans, an important share of the IFNγ/STAT1 pro-inflammatory pathway is mediated by the binding of STAT1 to consensus sequences localized in the Long Terminal Repeats (LTRs) of the MER41 family of Human Endogenous Retroviruses (HERVs; Chuong et al., 2016). Thus, in primates only, MER41 sequences located in the promoter regions of immune genes, serve as IFNγ-inducible enhancers that are indispensible for a full IFNγ-mediated immune response (Chuong et al., 2016). MER41 integrated into the genome of a primate ancestor 45–60 million years ago and a total of 7,190 LTR elements belonging to six subfamilies (MER41A–MER41G) are detectable in the modern human genome (Chuong et al., 2016). That being said, the hypothesis of an IFNγ/STAT1/MER41 pathway shaping social behavior and/or cognition in humans (Filiano et al., 2016) is not yet supported by experimental data. Addressing this issue is of importance as it could provide additional evidence on whether and how the immune system may translate environmental cues (including, possibly, cultural cues) into genomic regulatory pathways shaping behavior and/or cognition. Obviously, multiple research fields are concerned, from cognitive evolution to psychiatric disorders. As a first step, any human candidate gene(s) regulated by such a pathway need(s) to be identified. Additionally, it is important to determine if STAT1 is the sole immune TF that potentially regulate the transcription of behavior- and/or cognition-associated genes via MER41 LTRs in humans. Finally, it is worth considering whether the stepped integration of HERVs into the human genome, and more generally any evolutionary change dictated by infectious events, could be related to key cognitive specificities experienced by hominins. Indeed, it was recently hypothesized that the horizontal transfer of genetic material by viral and non-viral vectors might have prompted the emergence of language in the human species (Benítez-Burraco and Uriagereka, 2016).
To address these issues, we followed a bioinformatics workflow relying on the use of two recently generated web tools allowing a survey of HERV sequences and their associated transcription factor binding sites (TFBSs) in the entire human genome. Using this approach, we found that the promoter regions of genes causatively linked to genetically determined intellectual disability (ID) are highly significantly enriched in LTR sequences of the MER41 family. Such an enrichment was unique to both MER41, as compared to more than 130 explored HERVs, and to ID-associated genes, as compared to lists of genes associated with autism spectrum disorder (ASD) or schizophrenia. The MER41 LTRs that localize in the promoter regions of ID-associated genes harbor binding sites recognized by canonical immune TFs including STAT1, STAT3, and NFKB1. From these data, we performed phylogenetic comparisons between humans and chimpanzees regarding: (i) MER41 LTR insertion sites in the promoter regions of candidate ID-associated genes, (ii) protein sequences of the immune-related TFs binding MER41 LTRs in such promoter regions. This was so as to infer putative differences regarding the immune/MER41/cognition pathway, which might ultimately account for some of the cognitive differences between humans and chimpanzees. Finally, since FOXP2 is currently known as a relevant TF regulating aspects of brain development and functions which are important for the execution of speech-related motor programs (Spiteri et al., 2007; Vernes et al., 2007, 2011; Konopka et al., 2009; Oswald et al., 2017), we searched genomics and proteomics databases to map a putative functional interactome linking FOXP2 and the immune TFs binding MER41 LTRs. This way, we were able to unravel a HERV-driven evolutionary-determined connection between cognition and immunity with a potential impact on language evolution and the pathophysiology of ID.
A scheme summarizing the workflow followed in the present work is shown in Figure 1.
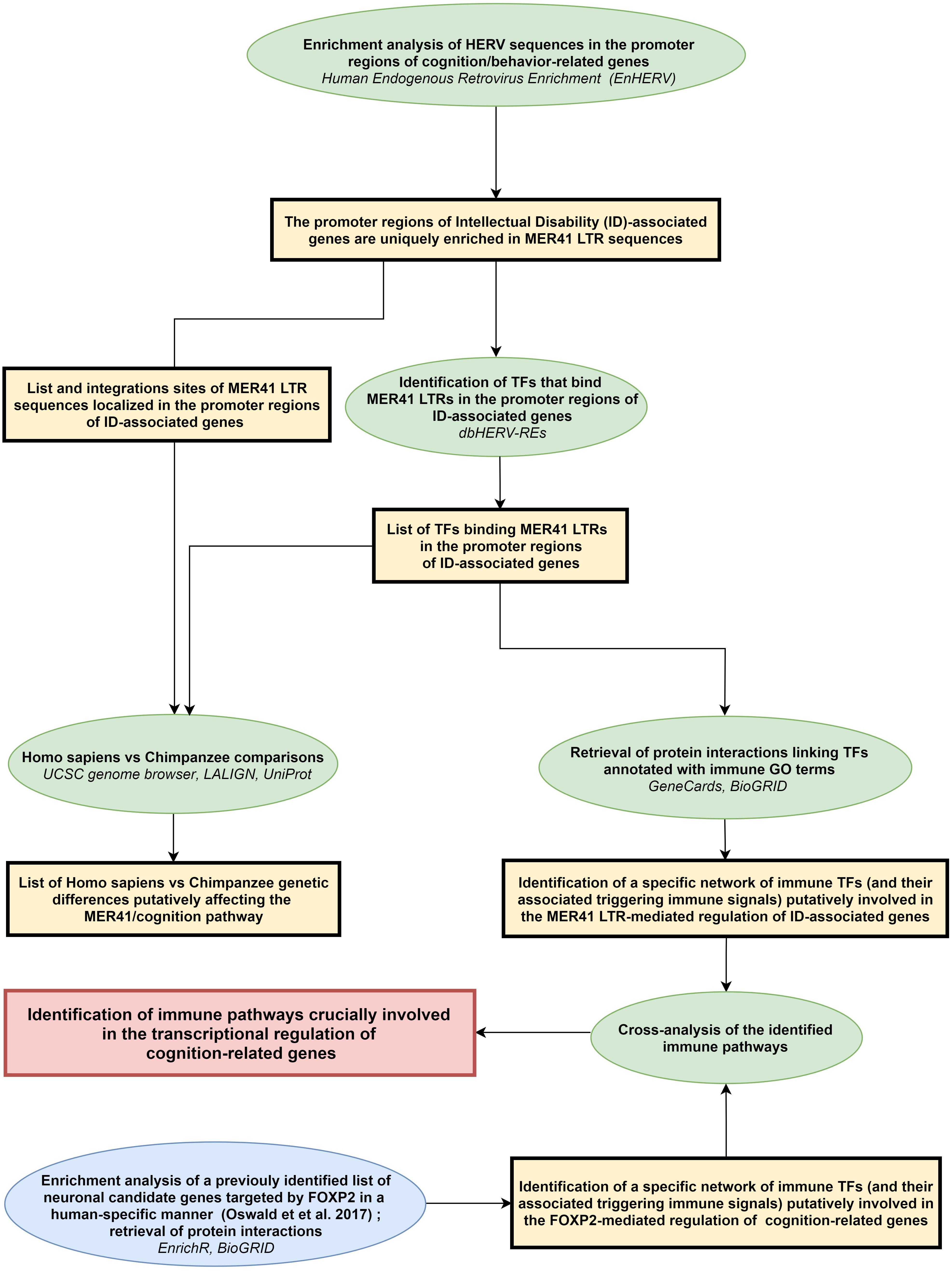
Figure 1. Workflow of the study. Rectangles (yellow or red) frame the main results obtained following each of the analytical steps briefly described in ellipse shapes (green or blue). Terms in italics correspond to the name of the bioinformatics tools used for each analytical step. LTR: long terminal repeat; ID: intellectual disability; TFs: transcription factors.
All the bioinformatics analyses were performed at least three times between December 2017 and February 2019. Bioinformatic tools and corresponding tasks performed in this study are described below.
1. The EnHERV database and web tool (Tongyoo et al., 2017): identifying human genes harboring MER41 LTR sequence(s) in the promoter region located 2 kb upstream the TSS; only solo LTRs oriented in the sense direction relative to the gene orientation were taken into account.
2. The db-HERV-REs database and web tool (Ito et al., 2017): identifying experimentally demonstrated TFBSs in HERV LTRs. The db-HERV-REs database has been generated by the re-analysis of 519 ChIP-Seq datasets provided by the ENCODE (ENCODE Project Consortium, 2004, 2012; Davis et al., 2018) and Roadmap (Roadmap Epigenomics Consortium et al., 2015) consortia.
3. The enrichment web platform Enrichr (Kuleshov et al., 2016): performing enrichments analyses on queried lists of genes. The Enrichr website allows surveying simultaneously 132 libraries gathering 245,575 terms and their associated lists of genes or proteins. Enrichment analysis tools provided by the Enrichr bioinformatics platform provides adjusted P-values computed from the Fisher’s exact test, Z-scores assessing deviation from an expected randomly obtained rank of P-values, and combined scores computed from the Z-scores and the adjusted P-values. We essentially focused our analysis on the well-recognized “GO term biological process” library (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) and on three ontology libraries based exclusively on text-mining: (i) the “Jensen TISSUES” library (Santos et al., 2015), to determine whether a list of genes is significantly associated with a specific tissue or cell type, (ii) the “Jensen COMPARTMENTS” library (Binder et al., 2014), to determine whether a list of genes is significantly associated with a specific cellular compartment or macromolecular complex, and (iii) the “Jensen DISEASES” library (Pletscher-Frankild et al., 2015), to determine whether a list of genes is significantly associated with a specific disease.
4. The UCSC genome browser (Rosenbloom et al., 2015): retrieving the sequences of MER41 LTRs and their precise localization in the promoter region of ID-associated genes in the human genome (Human genome assembly GRCh38/hg38) and in the Pan troglodytes genome (Chimpanzee genome assembly CSAC 2.1.4/panTro4).
5. The Swiss Institute of Bioinformatics (SIB) sequence alignment web tool LALIGN (SIB Swiss Institute of Bioinformatics Members, 2016): performing sequence comparisons between human and chimpanzee MER41 LTRs located in the promoter region of ID-associated genes. For each of these genes we checked the presence, nature and precise localization of MER41 LTR sequences in the promoter region.
6. The UniProt database of protein sequence and functional information (The UniProt Consortium, 2018): performing protein sequence alignments between Homo sapiens and Pan troglodytes for immune TFs binding MER 41 LTRs in the promoter regions of ID-associated genes.
7. The Brain RNA-Seq database (Zhang et al., 2016): exploring mRNA expression profiles obtained by RNA-Seq analyses in primary cultures of human neurons, astrocytes or macrophages/microglia.
8. The TISSUES database (Palasca et al., 2018): determining, for a given gene, which tissues harbor the highest levels of expression across a large range of normal human tissues. This database compiles results from four large expression atlases generated by pan-genomic and/or pan-proteomic analyses of normal human tissues (Su et al., 2004; Clark et al., 2007; Krupp et al., 2012; Fagerberg et al., 2014).
We queried the EnHERV database and web tool (Tongyoo et al., 2017) to determine whether candidate lists of cognition/behavior-related genes were enriched in genes harboring promoter-localized HERV LTRs (more precisely: sense-oriented solo HERV LTR sequence(s) localized in the promoter region located 2 kb upstream the TSS). We performed such an analysis successively for the 133 families of HERV that can be mined on the EnHERV website. Three lists of cognition/behavior-related genes were assessed (Supplementary Table 1): (i) a list of high confidence ASD susceptibility genes established by the SFARI consortium (Abrahams et al., 2013) and based on expert-operated manual curation of the literature, (ii) a recently established list of putative schizophrenia-causing genes inferred from the integrative analyses of genome wide association studies (Ma et al., 2018), and (iii) a list of genes for which mutations or deletions are considered as causative of intellectual disability based on a manual curation of the literature (Kochinke et al., 2016). As indicated in the original paper describing the EnHERV web tool (Tongyoo et al., 2017), results were considered as statistically significant when both following criteria were fulfilled: a Fisher exact test P-values <0.001 and an odds ratio >1. Using this approach, we found that the promoter regions of ID-associated genes were highly significantly enriched in MER41 LTRs (P-value = 0.0004; odds ratio = 4.28). Results were not significant for any of the other 132 HERV families that can be mined on the EnHERV website nor for the promoter regions of ASD- or schizophrenia-associated genes. Further supporting the specificity of our findings, when analyzing the 22 lists of non-CNS related genes provided as training lists by the EnHERV server, we did not find any significant enrichment in genes with promoter-localized MER41 LTR sequences. To confirm our findings, we retrieved from the EnHERV website the whole list of coding genes which, in humans, harbor a sense-oriented promoter-localized MER41 LTR sequence. On this list of 79 genes (Supplementary Table 2), we then performed enrichment analyses using the Enrichr website as described in the Materials and Methods section. We found no statistically significant enrichments with regard to “biological process” GO terms, tissue-specific expression or sub-cellular localization of gene products. However, text-mining enrichment analysis unraveled a significant enrichment in genes associated with the term “Intellectual disability” (Table 1).
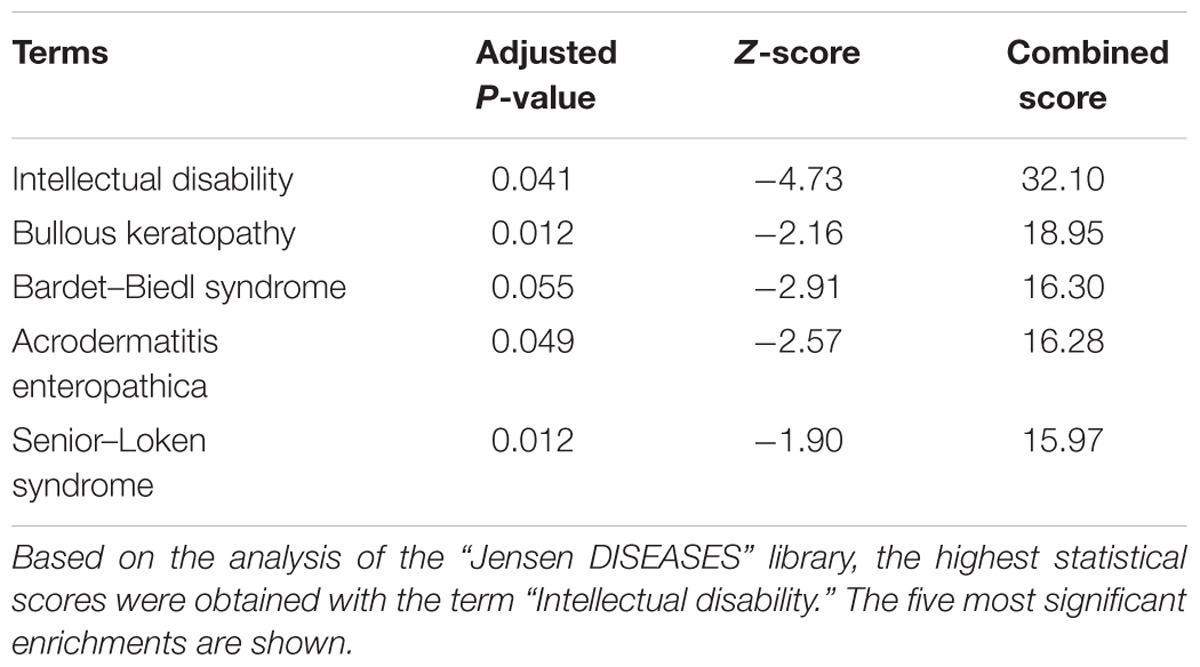
Table 1. Text mining-based enrichment analysis of the list of 79 coding genes harboring MER41 LTR sequences in their promoter region.
Since enrichment analysis based on text mining may be biased by the identification of a non-causative link between a given gene and the term “Intellectual disability”, we took into account only genes that had been identified as causative of ID (Kochinke et al., 2016). On this basis, out of 79 human genes harboring a MER41 LTR sequence in their promoter region, nine had an established causative link with ID. The genes and associated genetic conditions are summarized in Table 2.
1. BBS10 (Bardet-Biedl syndrome 10): Bardet-Biedl syndrome 10 (vision loss, obesity, polydactily, kidney abnormalities and intellectual disability).
2. DEAF1 (DEAF1 transcription factor): Mental retardation, autosomal dominant 24 (intellectual disability and impairments in adaptive behavior).
3. AP1S1 (Adaptor Related Protein Complex 1 Subunit Sigma 1): MEDNIK syndrome (Mental retardation, enteropathy, deafness, peripheral neuropathy, ichthyosis, and keratoderma).
4. ST3GAL5 (ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 5): Salt and pepper developmental regression syndrome (epilepsy, abnormal brain development and intellectual disability).
5. CDH15 (Cadherin 15): Mental retardation, autosomal dominant 3 (intellectual disability and impairments in adaptive behavior).
6. CEP290 (Centrosomal Protein 290): Bardet-Biedl syndrome 14 (vision loss, obesity, type 2 diabetes, hypercholesterolemia, polydactily, intellectual disability, impaired speech, delayed psychomotor development, and behavioral alterations).
7. GAMT (Guanidinoacetate N-Methyltransferase): Cerebral creatine deficiency syndrome 2 (epilepsy, intellectual disability, and altered speech development).
8. DDHD2 (DDHD Domain Containing 2): Spastic paraplegia 54, autosomal recessive (delayed psychomotor development, intellectual disability, and early-onset spasticity of the lower limbs).
9. GCSH (Glycine Cleavage System Protein H): Glycine encephalopathy (hypotonia, delayed psychomotor development, and epilepsy).
The “biological process” GO terms that annotate those nine genes are shown in Supplementary Table 3. Overall, these data point to a yet unrecognized potential link between promoter-localized MER41 LTRs and cognition.
TFBSs in LTRs from the MER41 family (MER41 A–E and MER41G) have been shown to vary depending of the MER41 member considered (Chuong et al., 2016). Using the EnHERV database and web tool, we have identified several MER41 members for which LTRs can be demonstrated in the promoter regions of ID-associated genes. As shown in Table 3, only three ID-associated genes harbored a MER41B LTR in their promoter region: CEP290, DDHD2, and GCSH. This indicates a potential transcriptional regulation of these three genes by the IFNγ/STAT1 pathway.
Interestingly, MER41 LTRs located in the promoter regions of ID-associated genes also include MER41A LTRs, which lack STAT1 binding sites (Chuong et al., 2016). This observation urged us to determine if other immune pathways (non-IFNγ/STAT1-mediated) may regulate the transcription of ID-associated genes via MER41 LTRs. To this aim we used the HERV database and web tool “db-HERV-RE” (Ito et al., 2017), which allow the identification of experimentally demonstrated TFBSs in HERV LTRs.
Using the approach described above, we have identified 32 TFs that bind MER41 LTRs in the promoter regions of ID-associated genes (Table 4).
As expected, MER41B LTR comprises a STAT1 consensus sequence while MER41A LTR does not. Interestingly, an YY1 consensus sequence is present in the LTRs of all the MER41A–E members. It is worth noting that mutations/deletions in YY1 are responsible for the Gabriele-De Vries syndrome, an autosomal dominant neurodevelopmental disorder characterized by intellectual disability, delayed psychomotor development and frequent autistic symptoms (Gabriele et al., 2017). Interestingly also, mutations in CTCF, another gene encoding a MER41 LTR-binding TF, are causally linked to “Mental retardation, autosomal dominant 21,” a developmental disorder characterized by significantly below-average general intellectual functioning associated with impairments in adaptive behavior (Gregor et al., 2013). Other inherited disorders associated to the above identified TF genes are summarized in Table 5 and notably include three groups of immune-related diseases induced by genetic alterations of the canonical immune TFs STAT1, STAT3, and NFKB1, respectively.
To summarize, besides STAT1, we have identified two canonical immune TFs, STAT3 and NFKB1, which bind specific MER41 LTRs in the promoter regions of ID-associated genes. Another TF, YY1, binds all MER41 LTRs in the promoter regions of ID-associated genes.
We then explored the BioGRID database of human protein interactions (Chatr-aryamontri et al., 2015) to determine whether STAT1, STAT3, and/or NFKB1 were reported to physically interact with each other and/or with YY1 and other TFs binding MER41 LTRs in the promoter regions of ID-associated genes. Interestingly, in the retrieved interaction network (Figure 2), we observed that YY1, via its interaction with NFKB1, is connected to a specific set of MER41 LTR-binding TFs that interact with STAT1, STAT3, and/or NFKB1. An analysis of the GO terms “Biological process” annotating each of these TFs indicates that besides STAT1, STAT3, and NFKB1, other members of this unique set of TFs exert immune functions (Supplementary Table 4). This is notably the case for YY1. Moreover, some of such immune functions are linked to specific cytokines among which IL-1 and IL-6 are the most commonly shared in the retrieved GO terms (Figure 3). This result indicates that the prototypical pro-inflammatory molecules IL-1 and IL-6 are possibly involved in the transcriptional regulation of ID-associated genes displaying promoter-localized MER41 LTRs.
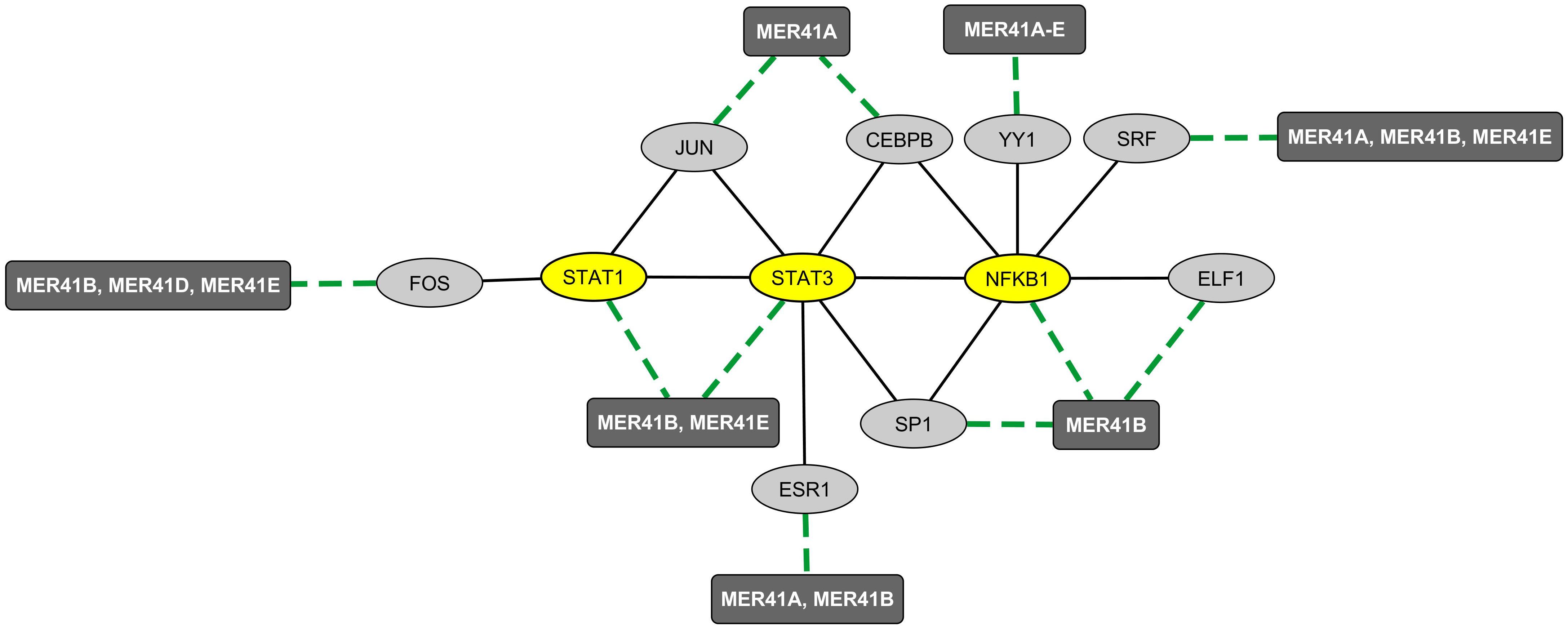
Figure 2. Mapping of the protein network formed by immune TFs that bind MER41 LTRs in the promoter regions of ID-associated genes. A survey of the human proteome was performed by querying the protein-protein interaction database BioGRID (Chatr-aryamontri et al., 2015). Ellipse shapes represent TFs and rectangles represent MER41 families displaying LTRs in the promoter regions of ID-associated genes. Black lines indicate protein-protein interactions. Green dashed lines indicate protein-DNA interactions between TFs and MER41 LTRs located in the promoter regions of ID-associated genes.
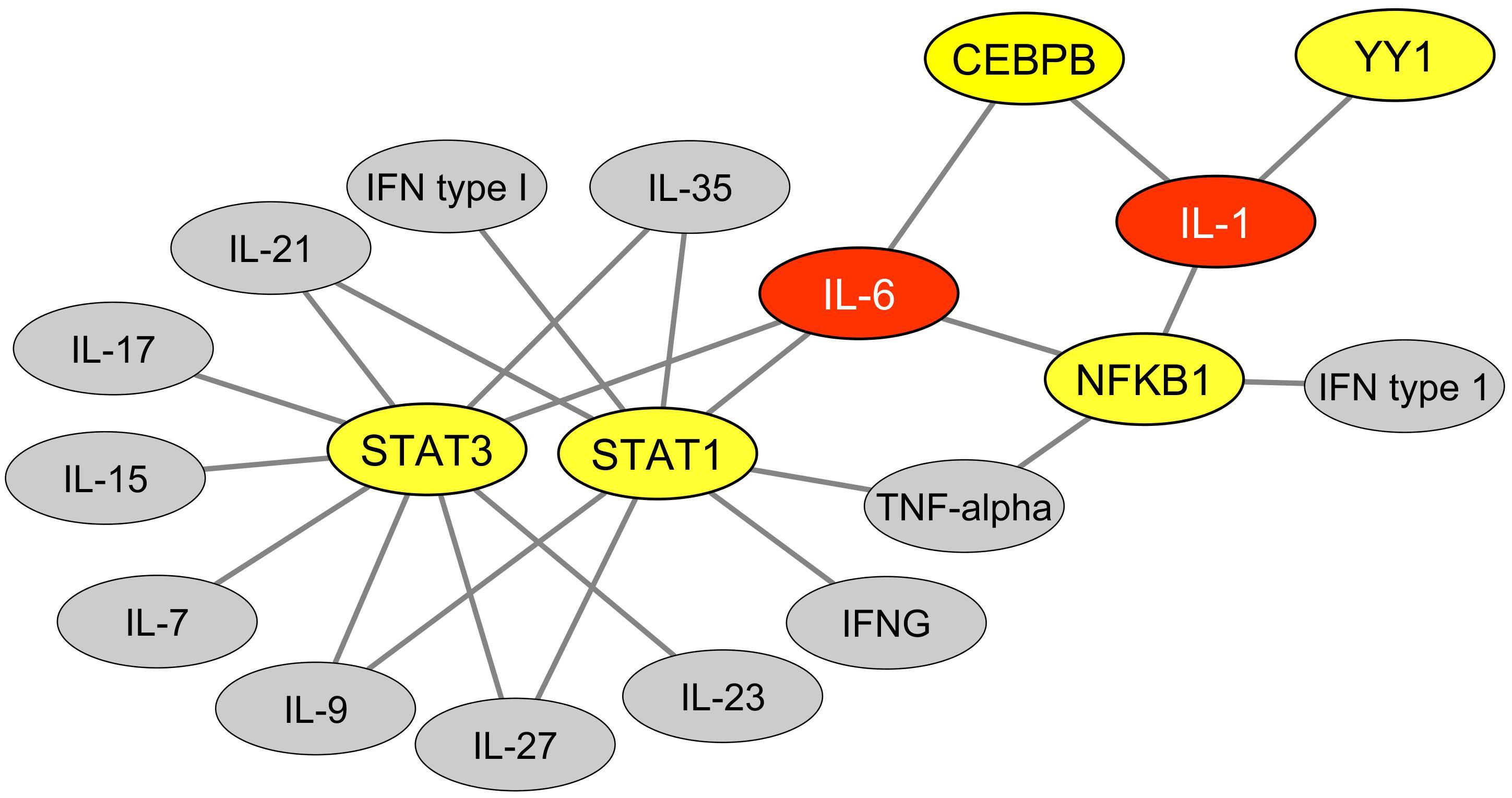
Figure 3. Cytokines functionally linked to TFs that bind MER41 LTRs in the promoter regions of ID-associated genes. An analysis was performed of the GO terms “biological process” that annotate each TFs which bind MER41 LTRs in the promoter regions of ID-associated genes. This allowed determining which cytokines are functionally linked with these TFs and potentially mediate an immune regulation of the MER41/cognition pathway. Such functional links are depicted as black lines. TFs are highlighted in yellow. Cytokines are highlighted in gray or, alternatively, in red for cytokines harboring three or more functional links with TFs. IFNG: interferon-gamma; IFN: interferon.
As mentioned, MER41 HERVs integrated the genome of a primate ancestor 45–60 million years ago. The process of so-called “ERV domestication” (Dewannieux and Heidmann, 2013) relies on mechanisms that are not only species-specific, but may have partly shaped speciation (Johnson, 2015). Accordingly, in primates, the species-specific domestication of MER41 HERVs translates into the existence of species-specific differences regarding the insertion sites and/or sequences of integrated (fixed) LTRs. On this basis, we investigated whether the promoters of ID-associated genes harbored the same MER41A-E LTRs in human and chimpanzees (Supplementary Table 5). Out of the nine candidate genes examined we found that five exhibited, in both species, MER41 LTR sequences belonging to the same family and displaying 95–100% homology (Supplementary Table 5). That being said, in two ID-associated genes MER41 LTR sequences were found at distances larger than 2 kb from the TSS in chimps and, for two other genes (CDH15 and GCSH), MER41 LTR sequences were absent, at least up to 10 kb from the TSS in chimps. These results are indicative of differences that may prove functionally relevant with regard to the MER41 LTR-mediated transcriptional regulation of specific ID-associated genes. This remains to be experimentally explored. It is of note that, according to the classification provided by the Gene ontology (GO) consortium (The Gene Ontology Consortium, 2017, 2019), three of the genes displaying such promoter-localized differences are annotated with “Biological process” GO terms that may possibly render an account of distinctive features between chimpanzees and human CNS (Supplementary Table 3). These include the terms “visual learning” and “locomotor behavior” for DDHD2, “hindbrain development” for CEP290 and “glycine catabolic process” for GCSH (glycine being a major inhibitory neurotransmitter). To complement these investigations, we also assessed whether key immune TFs putatively involved in the immune/MER41/cognition pathway exhibited humans vs. chimps differences regarding their amino acid sequences (Supplementary Table 6). Using the UniProt web tool “Align”, comparisons retrieved 100% homology between humans and chimps in the amino acid sequences of NFKB1, STAT1, STAT3, and CEBPB. A 99.7% homology was retrieved for YY1 and no functionally relevant amino-acid substitution in the compared YY1 sequences could be predicted according to the UniProt Align webtool.
FOXP2, a TF abundantly expressed in cortical neurons, is involved in the emergence of human speech (Vernes et al., 2007, 2011; Fisher and Scharff, 2009; Scharff and Petri, 2011; Xu et al., 2018). Until recently, the transcriptional activity FOXP2 was thought to rely on the recognition of specific TFBs by FOXP2/FOXP2 homodimers or by FOXP1/FOXP2 or FOXP4/FOXP2 heterodimers (Wang et al., 2003; Sin et al., 2015). However, a recent work established a short list of TFs that bind FOXP2 and are likely to form heterodimers that regulate FOXP2 availability and/or DNA binding properties in neurons (Estruch et al., 2018). Interestingly, YY1 was identified as one of the seven newly identified FOXP2-interacting TFs. We thus sought to determine whether YY1 could potentially represent a molecular link between FOXP2 and the immune/MER41/cognition pathway we identified. To this aim, we explored data obtained from a recent work attempting to identify a set of FOXP2 targets that are specific to human FOXP2 in neurons (Oswald et al., 2017). More precisely, data from this study were obtained by: (i) a meta-analysis of previous works reporting on FOXP2 neuronal targets (based notably on Chip-Seq analyses of the neuronal cell line SH-SY5Y; Spiteri et al., 2007; Vernes et al., 2007, 2011; Enard et al., 2009; Konopka et al., 2009; Hilliard et al., 2012) and (ii) a comparison of neuronal genes that are targeted by human FOXP2 vs. non-human primates orthologs of FOXP2 in the neuronal cell line SH-SY5Y (Oswald et al., 2017). A set of 40 candidate proteins encoded by FOXP2-targeted genes was identified. Protein interactors of these candidate targets were added in order to establish a final list of 80 proteins that are putatively regulated by FOXP2 in neurons in a human-specific manner. Interestingly, when performing enrichment analyses of the list of genes encoding such 80 proteins (Supplementary Table 7), we found a highly significant enrichment in genes that are either associated with immune-related terms, as identified by text mining (“immune System”, “NFKB complex”, “arthritis” and others), or linked to immune biological processes according to the GO term classification (“cellular response to IL-21,” “cellular response to IL-2” and others). Of note, the list of genes putatively regulated by FOXP2 in a human-specific manner is significantly enriched in genes involved in ”interleukin-6-mediated signaling pathway” (adjusted p-value: 0.0008) pointing again to IL-6 as a possible important player in the immune/MER41/cognition pathway. Finally, such a list of FOXP2 targets comprised STAT3 and several protein partners of STAT1, STAT3 and/or NFKB1. Integrating these data with the demonstrated interaction of FOXP2 with YY1 allows us to map a network of immune TFs that link FOXP2 to specific cognition-related (ID-associated) genes exhibiting promoter-localized MER41 LTRs (Figure 4).
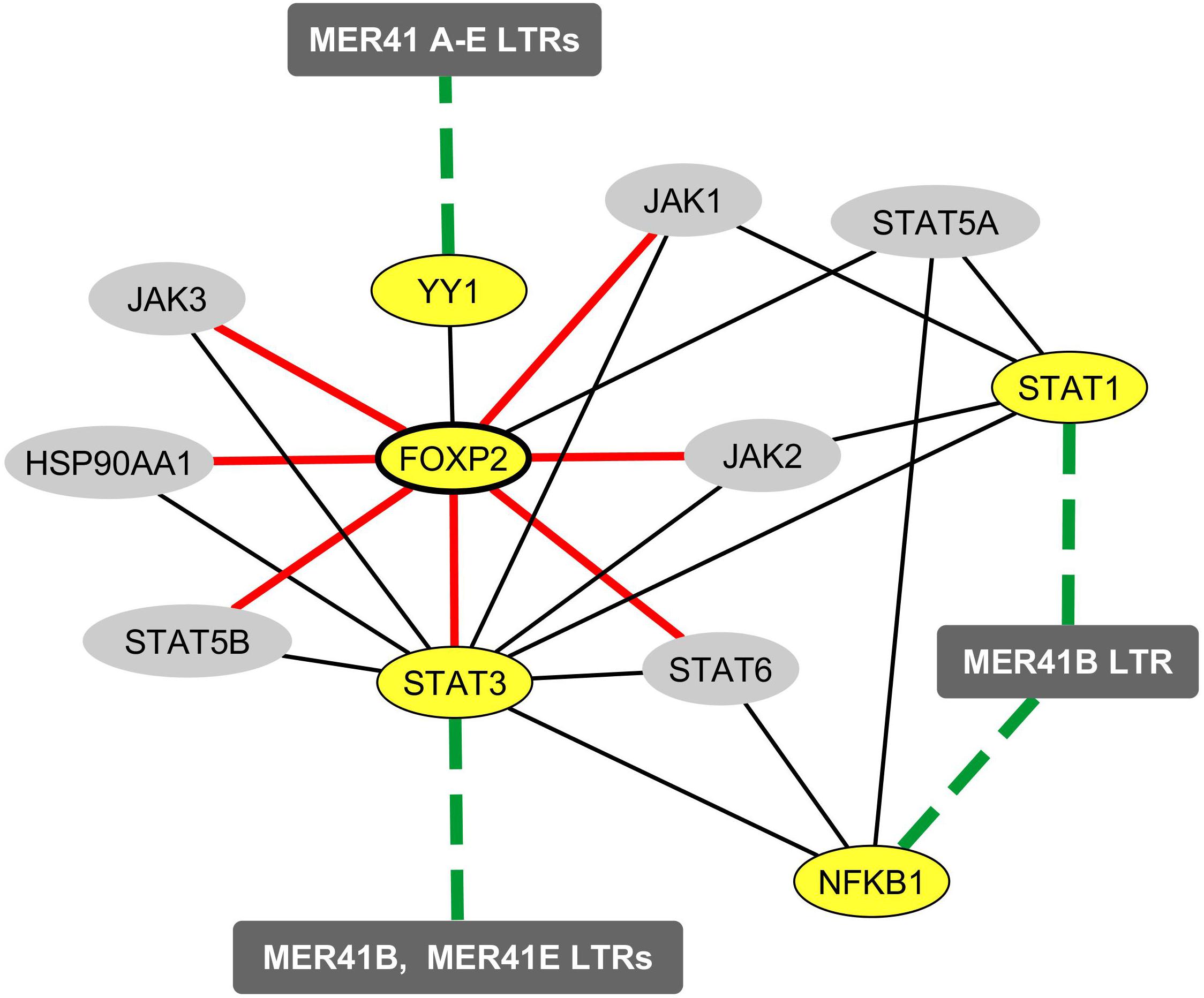
Figure 4. Mapping of the protein and functional network linking FOXP2 to immune TFs that bind MER41 LTRs in the promoter regions of cognition-related (ID-associated) genes. A survey of the human proteome was performed on the BioGRID database (Chatr-aryamontri et al., 2015) in order to map protein-protein interactions between FOXP2, STAT1, STAT3, NFKB1, YY1 and the molecules identified as being human-specifically regulated by FOXP2 in the neuronal cell line SH-SY-5Y (Oswald et al., 2017). Ellipse shapes highlighted in yellow represent FOXP2, its protein partner YY1 and the three immune TFs that bind MER41 LTRs in the promoter regions of cognition-related (ID-associated) genes (namely STAT1, STAT3 and NFKB1). Ellipse shapes highlighted in gray represent molecules that interact with these TFs and are putatively regulated by FOXP2 in a human-specific manner. Rectangles represent MER41 families displaying LTRs in the promoter regions of cognition-related (ID-associated) genes. Black lines indicate protein-protein interactions. Dashed green lines indicate protein-DNA interactions between TFs and MER41 LTRs located in the promoter regions of cognition-related (ID-associated) genes. Red lines indicate molecules that are targeted by FOXP2 in a human-specific manner.
To further assess the relevance of our findings, we surveyed two independent databases which allows determining the neural expression of key genes putatively involved in immune/MER41/cognition pathway. These databases comprise: (i) the “TISSUES” database (Palasca et al., 2018) which compiles manually curated expression results obtained in four distinct expression atlases (Su et al., 2004; Clark et al., 2007; Krupp et al., 2012; Fagerberg et al., 2014) covering a large range of normal human tissues and (ii) the recently launched “Brain RNA-Seq” database (Zhang et al., 2016) which allows exploring expression profiles observed in primary cultures of human neurons, astrocytes or macrophages/microglia. In our survey, the lymphocyte-specific gene markers CD3G and ZAP70 were used as negative controls. Data retrieved from the “Brain RNA-Seq database” showed that in cultured human neurons, CD3G and ZAP70 are expressed at levels considered as bellow the detection threshold (Supplementary Table 8). In contrast, all the immune genes examined which are putatively involved in the immune/MER41/cognition pathway (i.e., NFKB1, STAT1, STAT3, YY1, IL6R [Interleukin 6 Receptor], IL6ST [Interleukin 6 Signal Transducer] and IL6) were found to be expressed at detectable levels in cultured human neurons (Supplementary Table 8). The same results were retrieved when assessing mRNA levels in other neural cell types including fetal astrocytes, matures astrocytes, and macrophages/microglia (Supplementary Table 8). Interestingly, in this expression database, STAT1 mRNA levels reached higher levels in neurons than in macrophages/microglia (Supplementary Table 8). Of potential interest also, retrieved data showed that IL-6 is constitutively expressed by cultured macrophages/microglia derived from the brains of humans but not mice (data not shown). Regarding the expression pattern of candidate genes in normal human tissues, data retrieved from the “TISSUES” database showed that, as expected, CD3G and ZAP70 exhibited their higher levels of expression in lymphoid tissues (e.g., thymus, tonsils or lymph nodes; Supplementary Table 8). However, surprisingly, the prototypical immune-related genes STAT1, STAT3, NFKB1, IL6R and IL6ST were reported to display their highest (or second highest) levels of expression in the human brain (Supplementary Table 8). Similar results were obtained for YY1. Similarly, high levels IL-6 were reported in the spinal cord and pons (Supplementary Table 8). Altogether, these retrieved data indicate that all the immune players putatively involved in the immune/MER41/cognition pathway are actually expressed by human neural cells including neurons.
We have found that, in the human genome, the promoter regions of ID-associated genes are uniquely enriched in MER41 LTRs. More specifically, nine ID-associated genes that are putatively important in cognitive evolution exhibit MER41 LTRs in their promoter regions. As more than 100 families of HERV are integrated into our genome, it was important to determine whether our findings are specific to MER41 and to ID-associated genes, and if so to what extent. Among the 133 families of HERV explored here, MER41 is the only family whose LTRs were found with statistically high frequency in the promoter regions of ID-associated genes. It must be emphasized that, while many HERV families are inherited from ancestors common to all mammals, the MER41 family is detected exclusively in the genome of primates. Interestingly, we have observed substantial differences between humans and chimpanzees regarding the localization of MER41 LTRs in the promoter regions of ID-associated genes. These results suggest that the MER41 family of HERVs could have been involved in cognitive changes after our split from chimps. In this scheme, infection and horizontal transmission of the exogenous virus from which MER41 HERVs derive, would have occurred in a community of primate ancestors and would have led to germline infection, followed by vertical transmission and, in fine, endogenization. If so, genomic evolution from these primate ancestors would have been, at least in part, affected by the processes of HERV endogenization and domestication, which is itself mainly dictated by the host’s immune system (Dewannieux and Heidmann, 2013). Accordingly, differences regarding the insertion sites of MER41 LTRs in the promoter region of a large range of genes, including cognition-related (ID-associated) genes, might have played roles in cognitive speciation. It is worth noting that MER41 LTRs are not enriched in the promoter regions of ASD- or schizophrenia-related genes. This finding suggests that selected aspects of cognitive evolution in the primate genus are linked to MER41.
Our work also indicates that, in humans, immune regulation of the MER41/cognition pathway is not limited to IFNγ and its main downstream signaling molecule, STAT1. Indeed, the MER41 LTRs located in the promoter regions of cognition-related (ID-associated) genes harbor TFBSs for a group of five interacting immune-related TFs (STAT1, STAT3, NFKB1, YY1, and CEBPB) which are themselves functionally linked to multiple cytokines including IFNγ. Moreover, in this functional network, the prototypical pro-inflammatory cytokine IL-6 rather than IFNγ appears to be the main hub. Thus, cognitive evolution after our split from chimps might have been influenced by the process of endogenization and domestication of MER41 HERVs and by the parallel genomic evolution of immune genes. In this view, it is worth noting that, overall, immune genes harbor the highest levels of purifying selection in the human genome, which reflects the key functions of immunity in the defense against life-threatening infectious agents (Daub et al., 2013; Deschamps et al., 2016; Delgobo et al., 2019). This is notably the case for STAT1 (Deschamps et al., 2016) and for genes involved in the IL-6 pathway such as, in particular, IL-6, IL6ST, STAT3, and CEBPB (Daub et al., 2013; Delgobo et al., 2019).
Owing to its putatively important role in the immune/MER41/cognition pathway, YY1 deserves particular attention. Indeed, YY1 binding sites are observed in the LTRs from all MER41 subtypes (MER41 A to E) and YY1 is a direct protein partner of both NFKB1 and FOXP2, two TFs exerting major roles in immunity and language, respectively. Moreover, YY1 is not only recognized as being crucially involved in CNS development (as notably shown in the inherited brain disorder “Gabriel-de Vries syndrome”), but also as exerting major functions in the immune system. In particular, YY1 was demonstrated to inhibit differentiation and function of regulatory T cells by blocking Foxp3 expression (Hwang et al., 2016) and to regulate effector cytokine gene expression and T(H)2 immune responses (Guo et al., 2008). We previously proposed that the nervous and immune systems have somehow co-evolved to the benefits of both systems, particularly regarding cognitive evolution, including our language-readiness (Benítez-Burraco and Uriagereka, 2016; Nataf, 2017a,b). In this context, YY1 may represent a new molecular connection between immunity and cognition and, even more specifically, between immunity and speech or language more generally.
NFKB1 may also draw specific interest since its neuronal expression was reported to be essential to behavior and cognition in both invertebrates and mammals (Meffert and Baltimore, 2005; Mattson and Meffert, 2006; Kaltschmidt and Kaltschmidt, 2009; Dresselhaus et al., 2018). In the central nervous system of rodents, components of the NFKB complex are detectable in neuronal processes and in synapses under physiological conditions (Salles et al., 2014; Dresselhaus et al., 2018). Moreover, synaptic transmission as well as exposure to neurotrophins activate the NFKB pathway in neurons (Meffert and Baltimore, 2005; Mattson and Meffert, 2006; Kaltschmidt and Kaltschmidt, 2009). In turn, NFKB activation in neurons triggers the transcription of multiple neuronal genes that may favor cognition and shape behavior. This is notably the case for neuropeptide Y and BDNF (Snow and Albensi, 2016).
The immune-mediated retrotranscription of specific HERVs was shown possibly to negatively influence the outcome of CNS disorders (Douville et al., 2011; Kremer et al., 2013; Douville and Nath, 2017; Küry et al., 2018). While our work unravels the putative evolutionary-determined advantage conferred by the immune/MER41/cognition pathway in human, it also points to the potential weaknesses that are inherent to such a pathway. Indeed, we propose that alterations of the immune/MER41/cognition pathway might contribute to the development of non-inherited forms of ID. Such a dysfunction might be induced by the untimely or quantitatively inappropriate exposure of neurons to specific cytokines, notably IL-6 or IFNγ, which physiologically shape neurotransmission (Chourbaji et al., 2006; Baier et al., 2009; Victório et al., 2010; Litteljohn et al., 2014; Gruol, 2015) and are possibly involved in the immune/MER41/cognition pathway. Obviously, however, experiments are needed in order to start demonstrating that the immune/MER41/pathway actually operates in the human brain. In particular, in vitro experiments performed on human neural cells could allow determining whether or not IL-6 and/or IFNγ regulate the expression of ID-associated genes harboring promoter-localized MER41 LTRs and, if so, whether or not such a process occurs via the binding of STAT1, STAT3, NFKB1, YY1 and/or CEBPB to MER41 LTRs.
In any case, our work reinforces the notion of neuroimmune co-evolution that we previously put forward (Benítez-Burraco and Uriagereka, 2016; Nataf, 2017a,b). In this general frame, we would like to propose that, besides the potential role of endogenous immune cues (Nataf, 2017a,b), immune signals triggered by infectious agents, might have been important to cognitive evolution. In particular, depending on their pathogenicity, such infectious agents could have exerted a neuroimmune selection pressure over millions of years (e.g., via the self-domestication of HERVs) or during short periods of time (e.g., via the occurrence of life-threatening epidemics of viral or bacterial infections). In this view, our findings provide general support to the hypothesis previously enunciated by Piattelli-Palmarini and Uriagereka (Piattelli-Palmarini and Uriagereka, 2004), updated by Benítez-Burraco and Uriagereka (Benítez-Burraco and Uriagereka, 2016) which states that the recent emergence of linguistic skills would have been triggered by a fast propagating virus.
SN performed the bioinformatics analyses and wrote the manuscript. AB-B and JU wrote the manuscript.
This work benefited from private funds attributed to SN for bioinformatics analyses that do not relate with the content of this paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Marine Guillen from Histology Laboratory, UFR Lyon-Est, Université Claude Bernard Lyon 1 for the technical help she provided on internal quality control of bioinformatics analyses.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00321/full#supplementary-material
Abrahams, B. S., Arking, D. E., Campbell, D. B., Mefford, H. C., Morrow, E. M., Weiss, L. A., et al. (2013). SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4:36. doi: 10.1186/2040-2392-4-36
Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F., and Hamosh, A. (2015). OMIM.org: online Mendelian Inheritance in Man (OMIM)®, an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43, D789–D798. doi: 10.1093/nar/gku1205
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Baier, P. C., May, U., Scheller, J., Rose-John, S., and Schiffelholz, T. (2009). Impaired hippocampus-dependent and -independent learning in IL-6 deficient mice. Behav. Brain Res. 200, 192–196. doi: 10.1016/j.bbr.2009.01.013
Benítez-Burraco, A., and Uriagereka, J. (2016). The immune syntax revisited: opening new windows on language evolution. Front. Mol. Neurosci. 8:84. doi: 10.3389/fnmol.2015.00084
Binder, J. X., Pletscher-Frankild, S., Tsafou, K., Stolte, C., O’Donoghue, S. I., Schneider, R., et al. (2014). COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database 2014:bau012. doi: 10.1093/database/bau012
Chatr-aryamontri, A., Breitkreutz, B.-J., Oughtred, R., Boucher, L., Heinicke, S., Chen, D., et al. (2015). The BioGRID interaction database: 2015 update. Nucleic Acids Res. 43, D470–D478. doi: 10.1093/nar/gku1204
Chourbaji, S., Urani, A., Inta, I., Sanchis-Segura, C., Brandwein, C., Zink, M., et al. (2006). IL-6 knockout mice exhibit resistance to stress-induced development of depression-like behaviors. Neurobiol. Dis. 23, 587–594. doi: 10.1016/j.nbd.2006.05.001
Chuong, E. B., Elde, N. C., and Feschotte, C. (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. doi: 10.1126/science.aad5497
Clark, T. A., Schweitzer, A. C., Chen, T. X., Staples, M. K., Lu, G., Wang, H., et al. (2007). Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 8:R64. doi: 10.1186/gb-2007-8-4-r64
Daub, J. T., Hofer, T., Cutivet, E., Dupanloup, I., Quintana-Murci, L., Robinson-Rechavi, M., et al. (2013). Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 30, 1544–1558. doi: 10.1093/molbev/mst080
Davis, C. A., Hitz, B. C., Sloan, C. A., Chan, E. T., Davidson, J. M., Gabdank, I., et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801. doi: 10.1093/nar/gkx1081
Delgobo, M., Kozlova, E., Rocha, E. L., Rodrigues-Luis, G. F., Mendes, D. A., Mascarin, L., et al. (2019). Mycobacterium tuberculosis hijacks an evolutionary recent IFN-IL-6-CEBP axis linked to monocyte development and disease severity in humans. bioRxiv https://doi.org/10.1101/514943 doi: 10.1101/514943
Deschamps, M., Laval, G., Fagny, M., Itan, Y., Abel, L., Casanova, J.-L., et al. (2016). Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98, 5–21. doi: 10.1016/j.ajhg.2015.11.014
Dewannieux, M., and Heidmann, T. (2013). Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr. Opin. Virol. 3, 646–656. doi: 10.1016/j.coviro.2013.08.005
Douville, R., Liu, J., Rothstein, J., and Nath, A. (2011). Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 69, 141–151. doi: 10.1002/ana.22149
Douville, R. N., and Nath, A. (2017). Human Endogenous Retrovirus-K and TDP-43 Expression Bridges ALS and HIV Neuropathology. Front. Microbiol. 8:1986. doi: 10.3389/fmicb.2017.01986
Dresselhaus, E. C., Boersma, M. C. H., and Meffert, M. K. (2018). Targeting of NF-κB to dendritic spines is required for synaptic signaling and spine development. J. Neurosci. 38, 4093–4103. doi: 10.1523/JNEUROSCI.2663-16.2018
Enard, W., Gehre, S., Hammerschmidt, K., Hölter, S. M., Blass, T., Somel, M., et al. (2009). A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971. doi: 10.1016/j.cell.2009.03.041
ENCODE Project Consortium (2004). The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640. doi: 10.1126/science.1105136
ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
Estruch, S. B., Graham, S. A., Quevedo, M., Vino, A., Dekkers, D. H. W., Deriziotis, P., et al. (2018). Proteomic analysis of FOXP proteins reveals interactions between cortical transcription factors associated with neurodevelopmental disorders. Hum. Mol. Genet. 27, 1212–1227. doi: 10.1093/hmg/ddy035
Fagerberg, L., Hallström, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406. doi: 10.1074/mcp.M113.035600
Filiano, A. J., Xu, Y., Tustison, N. J., Marsh, R. L., Baker, W., Smirnov, I., et al. (2016). Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429. doi: 10.1038/nature18626
Fisher, S. E., and Scharff, C. (2009). FOXP2 as a molecular window into speech and language. Trends Genet. 25, 166–177. doi: 10.1016/j.tig.2009.03.002
Gabriele, M., Vulto-van Silfhout, A. T., Germain, P.-L., Vitriolo, A., Kumar, R., Douglas, E., et al. (2017). YY1 haploinsufficiency causes an intellectual disability syndrome featuring transcriptional and chromatin dysfunction. Am. J. Hum. Genet. 100, 907–925. doi: 10.1016/j.ajhg.2017.05.006
Green, D. S., Young, H. A., and Valencia, J. C. (2017). Current prospects of type II interferon γ signaling and autoimmunity. J. Biol. Chem. 292, 13925–13933. doi: 10.1074/jbc.R116.774745
Gregor, A., Oti, M., Kouwenhoven, E. N., Hoyer, J., Sticht, H., Ekici, A. B., et al. (2013). De novo mutations in the genome organizer CTCF cause intellectual disability. Am. J. Hum. Genet. 93, 124–131. doi: 10.1016/j.ajhg.2013.05.007
Gruol, D. L. (2015). IL-6 regulation of synaptic function in the CNS. Neuropharmacology 96, 42–54. doi: 10.1016/j.neuropharm.2014.10.023
Guo, J., Lin, X., Williams, M. A., Hamid, Q., and Georas, S. N. (2008). Yin-Yang 1 regulates effector cytokine gene expression and T(H)2 immune responses. J. Allergy Clin. Immunol. 122, 195–201.e5. doi: 10.1016/j.jaci.2008.03.012
Hilliard, A. T., Miller, J. E., Fraley, E. R., Horvath, S., and White, S. A. (2012). Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron 73, 537–552. doi: 10.1016/j.neuron.2012.01.005
Hwang, S. S., Jang, S. W., Kim, M. K., Kim, L. K., Kim, B.-S., Kim, H. S., et al. (2016). YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat. Commun. 7:10789. doi: 10.1038/ncomms10789
Ito, J., Sugimoto, R., Nakaoka, H., Yamada, S., Kimura, T., Hayano, T., et al. (2017). Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 13:e1006883. doi: 10.1371/journal.pgen.1006883
Johnson, W. E. (2015). Endogenous retroviruses in the genomics era. Annu. Rev. Virol. 2, 135–159. doi: 10.1146/annurev-virology-100114-054945
Kaltschmidt, B., and Kaltschmidt, C. (2009). NF-kappaB in the nervous system. Cold Spring Harb. Perspect. Biol. 1:a001271. doi: 10.1101/cshperspect.a001271
Kochinke, K., Zweier, C., Nijhof, B., Fenckova, M., Cizek, P., Honti, F., et al. (2016). Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am. J. Hum. Genet. 98, 149–164. doi: 10.1016/j.ajhg.2015.11.024
Konopka, G., Bomar, J. M., Winden, K., Coppola, G., Jonsson, Z. O., Gao, F., et al. (2009). Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature 462, 213–217. doi: 10.1038/nature08549
Kremer, D., Schichel, T., Förster, M., Tzekova, N., Bernard, C., van der Valk, P., et al. (2013). Human endogenous retrovirus type W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann. Neurol. 74, 721–732. doi: 10.1002/ana.23970
Krupp, M., Marquardt, J. U., Sahin, U., Galle, P. R., Castle, J., and Teufel, A. (2012). RNA-Seq Atlas—a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28, 1184–1185. doi: 10.1093/bioinformatics/bts084
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi: 10.1093/nar/gkw377
Küry, P., Nath, A., Créange, A., Dolei, A., Marche, P., Gold, J., et al. (2018). Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 24, 379–394. doi: 10.1016/j.molmed.2018.02.007
Litteljohn, D., Nelson, E., and Hayley, S. (2014). IFN-γ differentially modulates memory-related processes under basal and chronic stressor conditions. Front. Cell. Neurosci. 8:391. doi: 10.3389/fncel.2014.00391
Ma, C., Gu, C., Huo, Y., Li, X., and Luo, X.-J. (2018). The integrated landscape of causal genes and pathways in schizophrenia. Transl. Psychiatry 8:67. doi: 10.1038/s41398-018-0114-x
Mattson, M. P., and Meffert, M. K. (2006). Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death Differ. 13, 852–860. doi: 10.1038/sj.cdd.4401837
Meffert, M. K., and Baltimore, D. (2005). Physiological functions for brain NF-kappaB. Trends Neurosci. 28, 37–43. doi: 10.1016/j.tins.2004.11.002
Nataf, S. (2017a). Autoimmunity as a driving force of cognitive evolution. Front. Neurosci. 11:582. doi: 10.3389/fnins.2017.00582
Nataf, S. (2017b). Evolution, immunity and the emergence of brain superautoantigens. F1000Research 6:171. doi: 10.12688/f1000research.10950.1
Oswald, F., Klöble, P., Ruland, A., Rosenkranz, D., Hinz, B., Butter, F., et al. (2017). The FOXP2-Driven Network in Developmental Disorders and Neurodegeneration. Front. Cell Neurosci. 11:212. doi: 10.3389/fncel.2017.00212
Palasca, O., Santos, A., Stolte, C., Gorodkin, J., and Jensen, L. J. (2018). TISSUES 2.0: an integrative web resource on mammalian tissue expression. Database 2018:bay003. doi: 10.1093/database/bay003
Piattelli-Palmarini, M., and Uriagereka, J. (2004). “The immune syntax: the evolution of the language virus,” in Variation and Universals in Bioliguistics, ed. L. Jenkins (Oxford: Elsevier), 341–377.
Pletscher-Frankild, S., Pallejà, A., Tsafou, K., Binder, J. X., and Jensen, L. J. (2015). DISEASES: Text mining and data integration of disease–gene associations. Methods 74, 83–89. doi: 10.1016/j.ymeth.2014.11.020
Ramana, C. V., Gil, M. P., Schreiber, R. D., and Stark, G. R. (2002). Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 23, 96–101. doi: 10.1016/S1471-4906(01)02118-4
Roadmap Epigenomics Consortium, A., Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. doi: 10.1038/nature14248
Rosenbloom, K. R., Armstrong, J., Barber, G. P., Casper, J., Clawson, H., Diekhans, M., et al. (2015). The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681. doi: 10.1093/nar/gku1177
Salles, A., Romano, A., and Freudenthal, R. (2014). Synaptic NF-kappa B pathway in neuronal plasticity and memory. J. Physiol. Paris 108, 256–262. doi: 10.1016/j.jphysparis.2014.05.002
Santos, A., Tsafou, K., Stolte, C., Pletscher-Frankild, S., O’Donoghue, S. I., and Jensen, L. J. (2015). Comprehensive comparison of large-scale tissue expression datasets. PeerJ 3:e1054. doi: 10.7717/peerj.1054
Scharff, C., and Petri, J. (2011). Evo-devo, deep homology and FoxP2: implications for the evolution of speech and language. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 366, 2124–2140. doi: 10.1098/rstb.2011.0001
SIB Swiss Institute of Bioinformatics Members (2016). The SIB Swiss Institute of Bioinformatics’ resources: focus on curated databases. Nucleic Acids Res. 44, D27–D37. doi: 10.1093/nar/gkv1310
Sin, C., Li, H., and Crawford, D. A. (2015). Transcriptional Regulation by FOXP1, FOXP2, and FOXP4 Dimerization. J. Mol. Neurosci. 55, 437–448. doi: 10.1007/s12031-014-0359-7
Snow, W. M., and Albensi, B. C. (2016). Neuronal Gene Targets of NF-κB and Their Dysregulation in Alzheimer’s Disease. Front. Mol. Neurosci. 9:118. doi: 10.3389/fnmol.2016.00118
Spiteri, E., Konopka, G., Coppola, G., Bomar, J., Oldham, M., Ou, J., et al. (2007). Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am. J. Hum. Genet. 81, 1144–1157. doi: 10.1086/522237
Su, A. I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K. A., Block, D., et al. (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067. doi: 10.1073/pnas.0400782101
The Gene Ontology Consortium (2017). Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338. doi: 10.1093/nar/gkw1108
The Gene Ontology Consortium (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338. doi: 10.1093/nar/gky1055
The UniProt Consortium (2018). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46:2699. doi: 10.1093/nar/gky092
Tongyoo, P., Avihingsanon, Y., Prom-On, S., Mutirangura, A., Mhuantong, W., and Hirankarn, N. (2017). EnHERV: Enrichment analysis of specific human endogenous retrovirus patterns and their neighboring genes. PLoS One 12:e0177119. doi: 10.1371/journal.pone.0177119
Vernes, S. C., Oliver, P. L., Spiteri, E., Lockstone, H. E., Puliyadi, R., Taylor, J. M., et al. (2011). Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 7:e1002145. doi: 10.1371/journal.pgen.1002145
Vernes, S. C., Spiteri, E., Nicod, J., Groszer, M., Taylor, J. M., Davies, K. E., et al. (2007). High-Throughput Analysis of Promoter Occupancy Reveals Direct Neural Targets of FOXP2, a Gene Mutated in Speech and Language Disorders. Am. J. Hum. Genet. 81, 1232–1250. doi: 10.1086/522238
Victório, S. C. S., Havton, L. A., and Oliveira, A. L. R. (2010). Absence of IFNγ expression induces neuronal degeneration in the spinal cord of adult mice. J. Neuroinflammation 7, 7–77. doi: 10.1186/1742-2094-7-77
Wang, B., Lin, D., Li, C., and Tucker, P. (2003). Multiple domains define the expression and regulatory properties of foxp1 forkhead transcriptional repressors. J. Biol. Chem. 278, 24259–24268. doi: 10.1074/jbc.M207174200
Xu, S., Liu, P., Chen, Y., Chen, Y., Zhang, W., Zhao, H., et al. (2018). Foxp2 regulates anatomical features that may be relevant for vocal behaviors and bipedal locomotion. Proc. Natl. Acad. Sci. U.S.A. 115, 8799–8804. doi: 10.1073/pnas.1721820115
Keywords: intellectual disability, cognition, innate immunity, evolution, HERV
Citation: Nataf S, Uriagereka J and Benitez-Burraco A (2019) The Promoter Regions of Intellectual Disability-Associated Genes Are Uniquely Enriched in LTR Sequences of the MER41 Primate-Specific Endogenous Retrovirus: An Evolutionary Connection Between Immunity and Cognition. Front. Genet. 10:321. doi: 10.3389/fgene.2019.00321
Received: 05 October 2018; Accepted: 22 March 2019;
Published: 12 April 2019.
Edited by:
Avindra Nath, National Institute of Neurological Disorders and Stroke (NINDS), United StatesReviewed by:
Molly Hammell, Cold Spring Harbor Laboratory, United StatesCopyright © 2019 Nataf, Uriagereka and Benitez-Burraco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Serge Nataf, c2VyZ2UubmF0YWZAaW5zZXJtLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.