- Department of Microbiology, The Ohio State Biochemistry Program, The Center for RNA Biology, The Ohio State University, Columbus, OH, United States
Among tRNA modification enzymes there is a correlation between specificity for multiple tRNA substrates and heteromultimerization. In general, enzymes that modify a conserved residue in different tRNA sequences adopt a heterodimeric structure. Presumably, such changes in the oligomeric state of enzymes, to gain multi-substrate recognition, are driven by the need to accommodate and catalyze a particular reaction in different substrates while maintaining high specificity. This review focuses on two classes of enzymes where the case for multimerization as a way to diversify molecular recognition can be made. We will highlight several new themes with tRNA methyltransferases and will also discuss recent findings with tRNA editing deaminases. These topics will be discussed in the context of several mechanisms by which heterodimerization may have been achieved during evolution and how these mechanisms might impact modifications in different systems.
Introduction
Critical to substrate binding specificity is the fact that enzymes need to achieve “high” affinity for their targets while ignoring non-targets. What makes this an especially difficult problem in cells is that substrates and non-substrates often look very similar, which tests the limits of enzyme-substrate recognition. This tenet is especially true of RNA binding proteins where high-affinity binding usually involves indirect readout of the phosphate backbone of RNA that must be combined with base- and shape-specific contacts to enable substrate discrimination. Many enzymes follow these rules to achieve effective target specificity, yet some face an additional obstacle, in that they must also maintain the ability to turn over during a catalytic cycle in order to yield a productive reaction. This issue is exacerbated when a single enzyme must target different substrates within a pool of nearly identical ones, as is the case faced by most tRNA modification enzymes.
A growing trend in the modification field is that many of the enzymes which recognize multiple substrates are heteromultimeric (Guy and Phizicky, 2014), where partnering may contribute to higher binding affinity and enable discrimination between nearly identical substrates. In a previous model based on observations with tRNA deaminases, it was suggested that homodimerization was necessary for the bacterial adenosine to inosine (A-to-I) deaminase to recognize a single tRNAArg substrate (Ragone et al., 2011; Spears et al., 2011). On the contrary to accommodate multiple tRNA substrates of different sequences, key recognition motifs in the eukaryotic tRNA deaminases are positioned further from the active site (Ragone et al., 2011). Critically, such evolutionary adaptations would not have been possible without a move toward heterodimerization. In general, one could imagine that many heterodimeric (heteromultimeric) enzymes arose by gene duplication, which then allowed the duplicated genes to accumulate mutations, aiding in the process of neo-functionalization. Such is the case of the eukaryotic tRNA deaminase ADAT2/ADAT3, for which the two subunits are very similar but not identical, strongly arguing for a gene duplication event that led to functional differentiation of each subunit. A similar explanation may be true of other heteromultimeric modification enzymes, especially of methyltransferases. In the following pages, we will discuss in greater detail both the nature and evolution of heteromultimeric enzymes, with a focus on methyltransferases and deaminases. A previous review touched on the topic of neofunctionalization in the context of pseudouridine synthases (Fitzek et al., 2018). Here neofunctionalization will be only discussed in passing. We will highlight current themes and concepts that have arisen from recently published structures of a number of methyltransferases and also introduce the new concept of enzyme co-activation, whereby seemingly unrelated enzymes inactive on a specific substrate become active after association; a new twist to the idea of neofunctionalization.
Interdependent Methyltransferases
Methyltransferases That Require Heterodimerization
Certain eukaryotic tRNA methyltransferases strictly function as two-subunit enzymes: where association between a structural and a catalytic subunit is required for tRNA methylation (Figure 1). While this review will detail these associations, many recent reviews more broadly cover the enzymology and biological role of tRNA methyltransferase enzymes (Guy and Phizicky, 2014; Swinehart and Jackman, 2015; McKenney and Alfonzo, 2016; Goto-Ito et al., 2017; Hori, 2017; Traube and Carell, 2017). Beyond improvement to overall enzyme efficiency, heteromultimeric methyltransferases show altered activity toward particular tRNA species, particularly in choice of nucleotides and/or sites that are modified in a specific tRNA. In general, and where they have been identified, the equivalent bacterial and archaeal enzymes for a particular methylation do not require a heterologous partner for activity. Many have speculated that as the non-coding RNA pool expanded, pairing a non-catalytic with a catalytic subunit evolved to increase tRNA substrate specificity and prevent rampant non-specific methylation. Heteromultimerization may also integrate environmental and metabolic cues needed for optimal translation profiles that promote homeostasis; these possibilities will be explored further in the following sections.
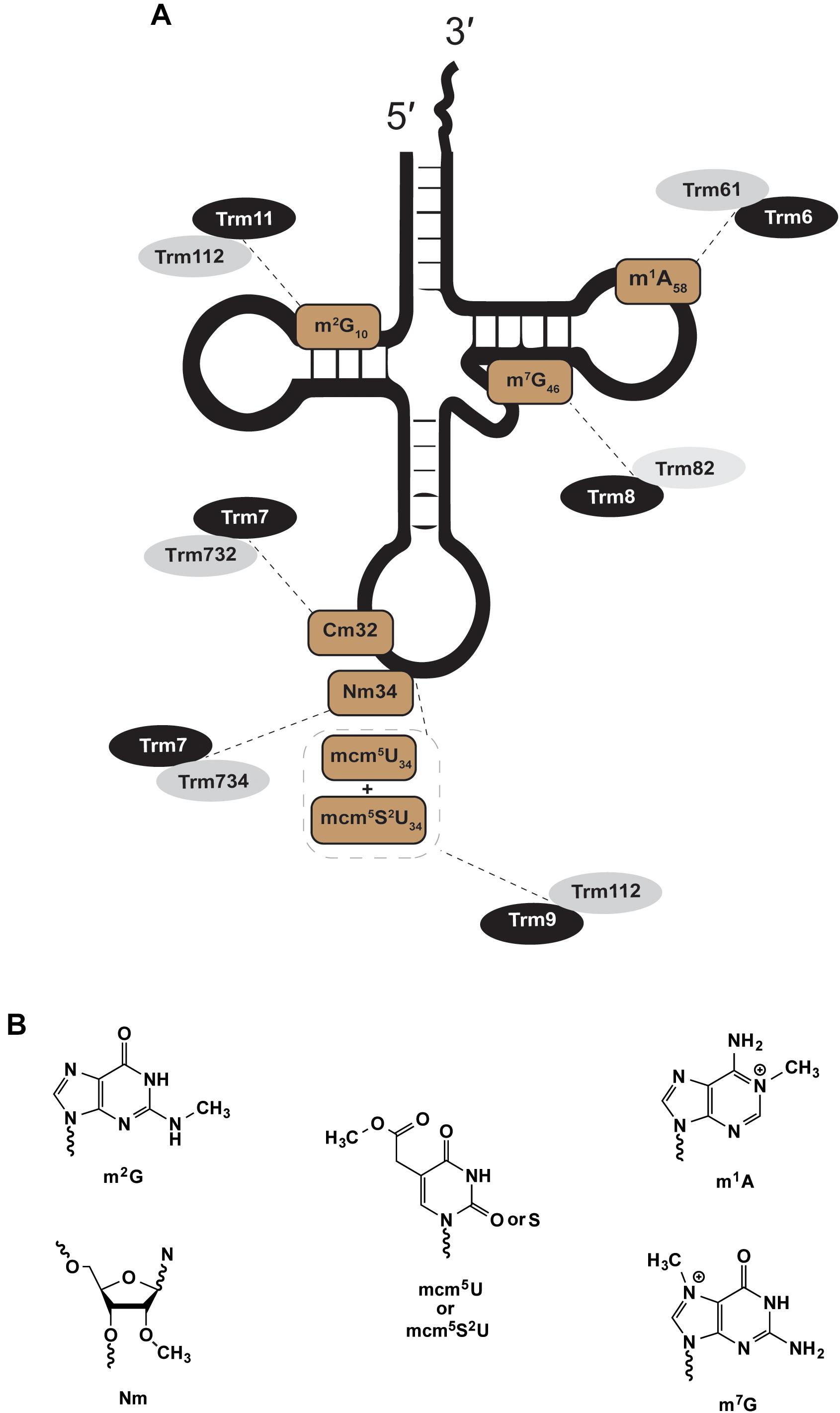
Figure 1. Eukaryotic two-subunit dependent methyltransferases. The figure shows examples of methylations catalyzed by heteromultimeric enzymes; a theme with eukaryotic modifications. (A) Shows the position and type of modification (brown) and the enzymes that catalyze such modifications (gray and black). (B) Chemical structures of modifications in (A).
Trm61/Trm6
The nuclear Trm61/Trm6 complex responsible for N-1 methyl adenosine at tRNA position 58 (Figure 1; m1A58) was first isolated from S. cerevisiae (Anderson et al., 1998, 2000). Trm61/Trm6 homologs exist in eukaryotic genomes of other yeast, protist, plant, and animal species (Bujnicki, 2001). The catalytic subunit Trm61 binds to co-substrate methyl donor S-adenosyl-L-methionine (SAM) through extensive hydrophobic and hydrogen bonding interactions (Wang M. et al., 2016). Trm61 association with Trm6 ensures high-affinity binding of Trm61/Trm6 to target tRNA substrates (Anderson et al., 2000; Ozanick et al., 2005, 2007; Finer-Moore et al., 2015; Wang M. et al., 2016). Disruption of Trm61 SAM binding eliminates m1A58 activity in vitro and in vivo, consistent with its assignment as the catalytic subunit (Anderson et al., 2000). Trm6 must associate with Trm61 for methyltransferase activity in vitro (Anderson et al., 2000), and in vivo S. cerevisiae trm6 or trm61 mutants lack m1A58-modified tRNA (Anderson et al., 1998). Human Trm61/Trm6 homologs can rescue the function of S. cerevisiae trm61 or trm6 mutants but only when expressed together, and maintain activity in vitro only when purified as a complex (Ozanick et al., 2005).
High-affinity binding of Trm61/Trm6 to tRNA substrates, such as tRNAiMet or tRNALys,3UUU, was originally thought to depend primarily on Trm6 as the RNA recognition subunit (Anderson et al., 2000; Ozanick et al., 2005). However, detailed mechanistic enzymology and crystal structures have refined this view, it is now clear that the Trm61/Trm6 holo-enzyme makes specific contacts with tRNA substrates (Ozanick et al., 2007; Finer-Moore et al., 2015; Wang M. et al., 2016). Together Trm61 and Trm6 create an L-shaped pocket to accommodate the tRNA substrate (Finer-Moore et al., 2015; Wang M. et al., 2016). The methylation site, nucleobase A58 is buried deep within the conventional L-shaped tRNA structure between D- and TΨC- stem-loop elements (Robertus et al., 1974). Crystallographic structure determination of the human Trm61/Trm6 complex bound to tRNALys,3UUU, revealed numerous protein–RNA interactions favoring separation of D- and TΨC- stem-loop elements, effectively allowing the enzyme to access the otherwise inaccessible N-1 of A58 (Finer-Moore et al., 2015).
Trm61/Trm6 has been suggested to arise through duplication and divergence from an ancestral TrmI-family enzyme, a family that catalyzes m1A58 formation in bacteria, and/or m1A57 in archaea (Grosjean et al., 1995; Bujnicki, 2001; Roovers et al., 2004; Ozanick et al., 2005, 2007; Guy and Phizicky, 2014; Finer-Moore et al., 2015; Wang M. et al., 2016). Purification of native TrmI from Mycobacterium tuberculosis and Thermus thermophilus yields stable TrmI homo-tetramers (Gupta et al., 2001; Droogmans et al., 2003; Barraud et al., 2008), a four subunit stoichiometry conserved in Trm61/Trm6, which forms a dimer of heterodimers (Ozanick et al., 2007; Finer-Moore et al., 2015). Many of the contact residues between TrmI subunits are maintained between Trm61/Trm6 subunits (Ozanick et al., 2007; Finer-Moore et al., 2015; Wang M. et al., 2016). The catalytic subunit Trm61 has high similarity to TrmI-family proteins, whereas Trm6 shows no apparent similarity to sequences deposited in current archaeal or bacterial databases. It is possible that Trm6 diverged from its ancestor, losing its ability to bind SAM, and maintaining only its RNA binding character. Interestingly, the tomato homolog of Trm6 (alias Gcd10) interacts with the dual methyltransferase/guanylyl transferase of the tobacco mosaic virus replicase complex (Osman and Buck, 1997; Taylor and Carr, 2000). This could indicate that other biologically relevant associations of Trm6 remain to be discovered. A recent report shows evidence that Trm61/Trm6 complexes catalyze low abundance m1A formation in regions of mRNA that loosely mimic tRNA TΨC-loops (Safra et al., 2017). A more detailed investigation of Trm61/Trm6 substrate specificity toward these mRNA structures is warranted.
Trm8/Trm82
The nuclear complex of Trm8/Trm82, responsible for N-7 methyl guanosine at position 46 (m7G46) in S. cerevisiae, shares many themes introduced in discussion of Trm61/Trm6 (Figure 1). Trm8 is a SAM-dependent methyltransferase that requires Trm82, a tryptophan-aspartic acid repeat (WD-repeat) homolog, for efficient activity (Alexandrov et al., 2002, 2005; Matsumoto et al., 2007; Muneyoshi et al., 2007; Leulliot et al., 2008). Deletion of either gene in yeast results in loss of m7G46 in tRNAs. Trm8/Trm82 can be purified as a stoichiometric complex, where co-expression is required for activity in cell-free wheat germ extracts (Matsumoto et al., 2008). Much like Trm61/Trm6, co-expression of the human homologs METTL1 (Trm8) and WDR4 (Trm82) restores m7G46 formation in Δtrm8 or Δtrm82 yeast strains, however individual expression of either human gene alone fails to complement these yeast mutants (Alexandrov et al., 2002). Disease mutants of either METTL1 or WDR4 that contribute to a rare form of primordial dwarfism, a prenatal growth deficiency that persists after birth, also result in human tRNAs that are deficient in m7G modification (Shaheen et al., 2015).
Trm8/Trm82 homologs are readily identifiable in yeast, protist, plant, and animal species. No apparent Trm8 or Trm82 homologs are currently found in archaeal genomes, which generally lack m7G46. Bacterial genomes do not contain an obvious Trm82 homolog, however, Trm8 shares sequence similarity with the bacterial TrmB-family (De Bie et al., 2003; Okamoto et al., 2004). Expression of TrmB homologs from Aquifex aeolicus or Escherichia coli in Δtrm82 or Δtrm8Δtrm82 yeast restores tRNA m7G46 formation without the need of an obvious cognate Trm82 (Alexandrov et al., 2005). This argues that Trm8 and TrmB may derive from a common ancestral protein, thematically consistent with the case of Trm61 and TrmI. However, unlike Trm61/Trm6, which may have arisen through duplication and drift, the partner subunit, Trm82, is a WD protein unrelated to any characterized methyltransferase.
Comparison of apo-Trm8/Trm82 to its tRNA bound form revealed that, unlike Trm61/Trm6, Trm82 makes no apparent RNA contacts in the context of a Trm8/Trm82 complex (Leulliot et al., 2008). This was corroborated by chemical crosslinking and small angle X-ray scattering experiments (Alexandrov et al., 2005; Leulliot et al., 2008). Although not formally tested, it is possible that Trm82 stabilizes conformations of Trm8 that promote substrate binding and/or productive methyl transfer. In turn, in vivo Trm8 levels are greatly reduced in Δtrm82 yeast (Alexandrov et al., 2005), suggesting that Trm82 stabilizes cellular pools of Trm8 by a hitherto unknown mechanism.
Trm9/Trm112 and Trm11/Trm112
Trm112 can partner with either Trm9 or Trm11 to form separate SAM-dependent tRNA methyltransferase complexes, which act on separate pools of cytoplasmic tRNA substrates, at different nucleotide sites, and produce distinct chemical products (Figure 1). In S. cerevisiae Trm11/Trm112 forms N-2 methylguanosine at position 10 (m2G10) in a broad pool of tRNA species (Purushothaman et al., 2005; Okada et al., 2009). Trm9/Trm112 catalyze a more specific terminal methylation in the multi-step biosynthesis of 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34) at the wobble position (position 34) in tRNAArgUCU and tRNAGluUUC in yeast (Kalhor and Clarke, 2003; Jablonowski et al., 2006; Begley et al., 2007), with additional tRNA isoacceptors in higher eukaryotes (Songe-Moller et al., 2010), including human tRNALysUUU and tRNASecUGA. Trm9/Trm112 can also methylate non-thiouridine substrates to form mcm5U. The biological significance of yeast mcm5U-wobble site modification was assayed in a Δtrm9 strain through comparative analyses of transcriptome, ribosomal footprinting and proteome data sets (Deng et al., 2015). Consistent with loss of their decoding function, hypomodified Trm9/Trm112 substrates tRNAArgUCU and tRNAGluUUC, resulted in significant repression of protein expression for transcripts enriched in AGA or GAA codons (Deng et al., 2015). Analogous to two-subunit complexes discussed already, single yeast mutants of trm112 or trm9 each lack mcm5U34-tRNAs (Kalhor and Clarke, 2003; Mazauric et al., 2010; Chen et al., 2011), while trm112 or trm11 mutants each lack m2G10-tRNAs (Purushothaman et al., 2005; Okada et al., 2009).
Work in E. coli showed that co-expression of S. cerevisiae Trm9 and Trm112 is necessary to produce an active enzyme, as singularly purified catalytic subunit Trm9 is inactive (Mazauric et al., 2010; Chen et al., 2011). A crystal structure of the Yarrowia lipolytica Trm9/Trm112 complex, which shares 50–60% identity to the S. cerevisiae enzyme, has been reported (Letoquart et al., 2015). Co-crystal structures with substrate SAM or tRNA have not been obtained, although putative SAM-binding residues of the catalytic subunit (Trm9) have been mapped (Letoquart et al., 2015). The yeast Trm9/Trm112 association is stabilized in part by a β-zipper formed between parallel β-sheets of Trm9 and Trm112. Additional inter-subunit contacts bury a large hydrophobic region of Trm9, likely improving its solubility, and thus the in vitro methyltransferase activity of Trm9/Trm112 (Letoquart et al., 2015).
The C-terminus of the multi-domain protein methyltransferase ALKBH8 contains a subdomain with high sequence similarity to Trm9, sufficient in formation of mcm5U34-style modifications at the tRNA wobble position. ALKBH8 contains additional functional domains: an N-terminal RNA recognition motif, an AlkB-related domain, and a zinc finger region upstream to the C-terminal Trm9 orthology region (Fu et al., 2010; Songe-Moller et al., 2010; Leihne et al., 2011). Additional proteins with similarity to ALKBH8 exist in plant and protozoans, but many lack the Trm9-like methyltransferase domain (Zdzalik et al., 2014). ALKBH8 homologs with Trm9-like domains in Mus musculus and Arabidopsis thaliana maintain strict requirement for Trm112 association to form mcm5U-tRNA (Songe-Moller et al., 2010; Leihne et al., 2011), whereas the requirement for human ALKBH8 partnering with a Trm112 ortholog for wobble site methyltransfer has not been formally tested. Binding experiments have been performed, where substrate mimetic anticodon stem loop sequences bind more tightly to ALKBH8 in the presence of Trm112 (Pastore et al., 2012). Curiously, full length in vitro transcribed tRNA sequences showed no enhancement of ALKBH8 binding in the presence of Trm112 (Pastore et al., 2012). Specific examination of what impact the RNA recognition motif has on human ALKBH8 tRNA substrate specificity versus just the Trm9-like domain with Trm112 may prove insightful, especially as the substrate tRNA pool appears to have expanded in higher order eukaryotes. More detailed descriptions of molecular interactions between Trm9/Trm112 complexes and mcm5U-tRNA substrates remain outstanding.
The requirement of the S. cerevisiae Trm11 catalytic subunit to associate with Trm112 for m2G10 methyltransferase activity has been shown in recombinant, purified protein mixtures and wheat germ cell-free assays (Purushothaman et al., 2005; Okada et al., 2009) Currently, no structure of an intact Trm11/Trm112 complex has been reported. Eukaryotes and archaea have readily identifiable Trm11 homologs, which are otherwise absent from bacteria. An archaeal version of the catalytic subunit Trm11 from Thermococcus kodakarensis with bound SAM has been crystallized (Hirata et al., 2016). Detailed substrate interaction studies of yeast Trm11/Trm112 complexes showed that Trm112 improves Trm11 binding affinity for SAM and tRNA substrate (Bourgeois et al., 2017b). Whether Trm112 directly interacts with tRNA in the context of a Trm11/Trm112 complex, similar to Trm61/Trm6, or solely provides allosteric support for Trm11 tRNA binding, as proposed for Trm8/Trm82, remains an open question.
As previously noted Trm11 homologs are absent from bacteria, while Trm9 mcm5U-type modifications are exclusive to eukaryotes, yet Trm112 is broadly conserved across every domain (van Tran et al., 2018). Trm112 pairs with additional protein partners beyond Trm9 and Trm11 to form two-subunit methyltransferases whose substrates include ribosomal RNA or translation release factors, a topic well-reviewed elsewhere (Guy and Phizicky, 2014; Bourgeois et al., 2017a). In formation of active tRNA methyltransferase complexes it is likely that cellular Trm112 levels are limiting (Ghaemmaghami et al., 2003; Studte et al., 2008; Sardana and Johnson, 2012). Trm112 co-purifies stoichiometrically with Trm11 (Bourgeois et al., 2017b), and overexpression of Trm11 in yeast decreases the amount of Trm112 that co-immunoprecipitates with Trm9 (Studte et al., 2008). Biologically relevant conditions that depend on the competition between these catalytic subunits for Trm112 await discovery.
Trm7/Trm732 and Trm7/Trm734
The catalytic subunit Trm7, a 2′-O ribose methyltransferase, acts at nucleotides C32 or N34 dependent upon cytoplasmic association with partner subunits Trm732 or Trm734, respectively (Figure 1). Nm32 and Nm34 modifications are observed in all three domains. Eukaryotic tRNAs that contain Nm32 and Nm34 likely rely on Trm7/Trm732 or Trm7/Trm734 homologs, which are mostly but not entirely conserved throughout deposited sequences of eukaryotic genomes. Nm32 and Nm34 in archaea and bacteria are catalyzed by methyltransferases not obviously related to Trm7, Trm732 or Trm734. Nm32 modification is catalyzed by TrmJ-family members in bacteria and archaea (Purta et al., 2006; Somme et al., 2014), while TrmL forms Nm34 in bacterial tRNAs (Benitez-Paez et al., 2010) and box C/D small nucleolar ribonucleoprotein complexes form Nm34 in archaeal tRNAs (Clouet d’Orval et al., 2001; Nolivos et al., 2005; Joardar et al., 2011). One of these archaeal Nm34 modification complexes, was shown to use the excised intron from a processed tRNATrp transcript as the guide RNA to direct Nm34 of pre-tRNATrp substrates (Clouet d’Orval et al., 2001).
In yeast and other eukaryotes, certain tRNAs contain both Cm32 and Nm34 modifications in tandem. The most broadly conserved tandem methylated substrates are tRNAPhe species. In yeast lack of Trm7-modified tRNAPhe activates the general amino acid control starvation response (Han et al., 2018), whereas specific mutant lesions of the human ortholog FTSJ1 are linked to non-syndromic X-linked intellectual disability (Guy et al., 2015). Yeast form tandem Cm32 and Nm34 on additional substrates tRNALeuUAA and tRNATrpCCA (Guy and Phizicky, 2014). Evidence for the formation of separate Trm7/Trm732 or Trm7/Trm734 two-subunit complexes initially came from immunoblot pull down assays (Guy et al., 2012). Additional genetic and biochemical evidence support the hypothesis that Trm7/Trm732 or Trm7/Trm734 act as separate two-subunit complexes in 2′-O ribose methylation of C32 or N34 on yeast tRNA substrates, respectively (Guy et al., 2012). The predicted Trm7 ortholog in humans, FTSJ1, requires the cognate human homolog THADA, for Cm32 activity. However, S. cerevisiae Trm732 is able to functionally complement FTSJ1 in the absence of THADA (Guy and Phizicky, 2015). The Trm732 partner protein contains a conserved domain of unknown function as well as multiple armadillo-like helical domains, a structural fold generally important for protein and nucleic acid interactions. The other Trm7 partner, Trm734, is a WD-repeat protein similar to Trm82 the partner subunit of Trm8 m7G46 methyltransferase. More precise descriptions of interactions between subunits of Trm7/Trm732 and Trm7/Trm734 complexes, and of formed hetero-dimer complexes with substrate tRNAs, have yet to be articulated.
Sequentially Ordered tRNA Methyltransferase Reactions
In most organisms, a tRNA sequence will contain well over a dozen post-transcriptional chemical modifications. An evergreen topic of discussion is whether modification enzymes act in a particular order, where modification at one site is informed by the modification status of other positions. In eukaryotes, compartmentalization of modification enzymes obviously results in the sequential order of some chemical transformations. For example, nascent tRNA transcripts are first modified in the nucleus; after export to the cytoplasm, tRNAs can be further modified. Because tRNAs can also be imported into organelles (chloroplast or mitochondria), these may receive other modifications in addition to those already obtained in the nuclear and cytoplasmic compartments. However, the sequential nature of certain tRNA methylation events cannot be explained by compartmentalization alone and may be an inherent property of enzymes that work in complexes or those who have achieved a heteromeric state. Alternatively, selection for increased specificity may be a leading factor in establishing sequentiallity, such may be the case of complex modification pathways such as those for wybutosine and threonylcarbamoyl synthesis.
With few exceptions, tRNA sequences encode for a purine at position 37 that is almost always post-transcriptionally modified. Guanosine at position 37 can be methylated, with more complex conversion to wybutosine (yW) in tRNAPhe (Thiebe and Poralla, 1973; Noma et al., 2006). When adenosine is located at position 37 it may also be modified. In all three domains, threonylcarbamoyl can be found on N-6 of adenosine at position 37 (t6A37), or similarly isopentenyl (i6A37) modification may occur. In eukaryotic tRNAPhe t6A37 and i6A37 are inhibitory of Trm7/Trm732 and Trm7/Trm734 Cm32 and Gm34 formation, and alternatively stimulate m3C32 formation by Trm140, or related enzymes in certain tRNA substrates (Guy et al., 2012; Arimbasseri et al., 2016; Han et al., 2017). In E. coli i6A37 similarly blocks the formation of Cm or Um at position 34, a reaction catalyzed by the TrmL family of enzymes (Han et al., 2017; Sokolowski et al., 2018). The extremophile Thermus thermophilus provides evidence for apparent temperature sensitive methylation circuits; at high temperatures the absence of m7G46 negatively impacts methylation at two other sites Gm18 and m1G37 (Tomikawa et al., 2010). A thematically similar report showed pseudouridylation at position 55 (Ψ55) impacts the formation of Gm18, m1A58, and m5s2U when cells are grown at lower temperatures (Ishida et al., 2011). Many more so-described sequential tRNA modification circuits exist beyond those covered here, and are reviewed elsewhere (Helm and Alfonzo, 2014; Maraia and Arimbasseri, 2017; Han and Phizicky, 2018; Sokolowski et al., 2018).
tRNA Editing by Deamination
Adenosine to Inosine (A-to-I) Editing
Chemical deamination of A-to-I in RNA sequences was observed 30 years prior to the discovery of any hydrolytic deaminase responsible for A-to-I activity that could act on polynucleotides (Bass and Weintraub, 1988). Since then, RNA adenosine deaminases have historically been divided into two broad classes based on their substrates: adenosine deaminases acting on mRNAs (ADARs) or adenosine deaminases acting on tRNAs (ADATs). Here we will focus exclusively on the tRNA deaminases (Figure 2). Inosine containing tRNAs are present in all domains of life; often tRNA A-to-I editing is essential. Unlike ADARs, which are generally promiscuous in A-to-I deamination of their targets (Bajad et al., 2017), ADATs show a more restricted A-to-I editing specificity, limited to three different tRNA sites: position 34 (wobble-position), 37 (3′ of the anticodon triplet) or 57 (in the TΨC-loop).
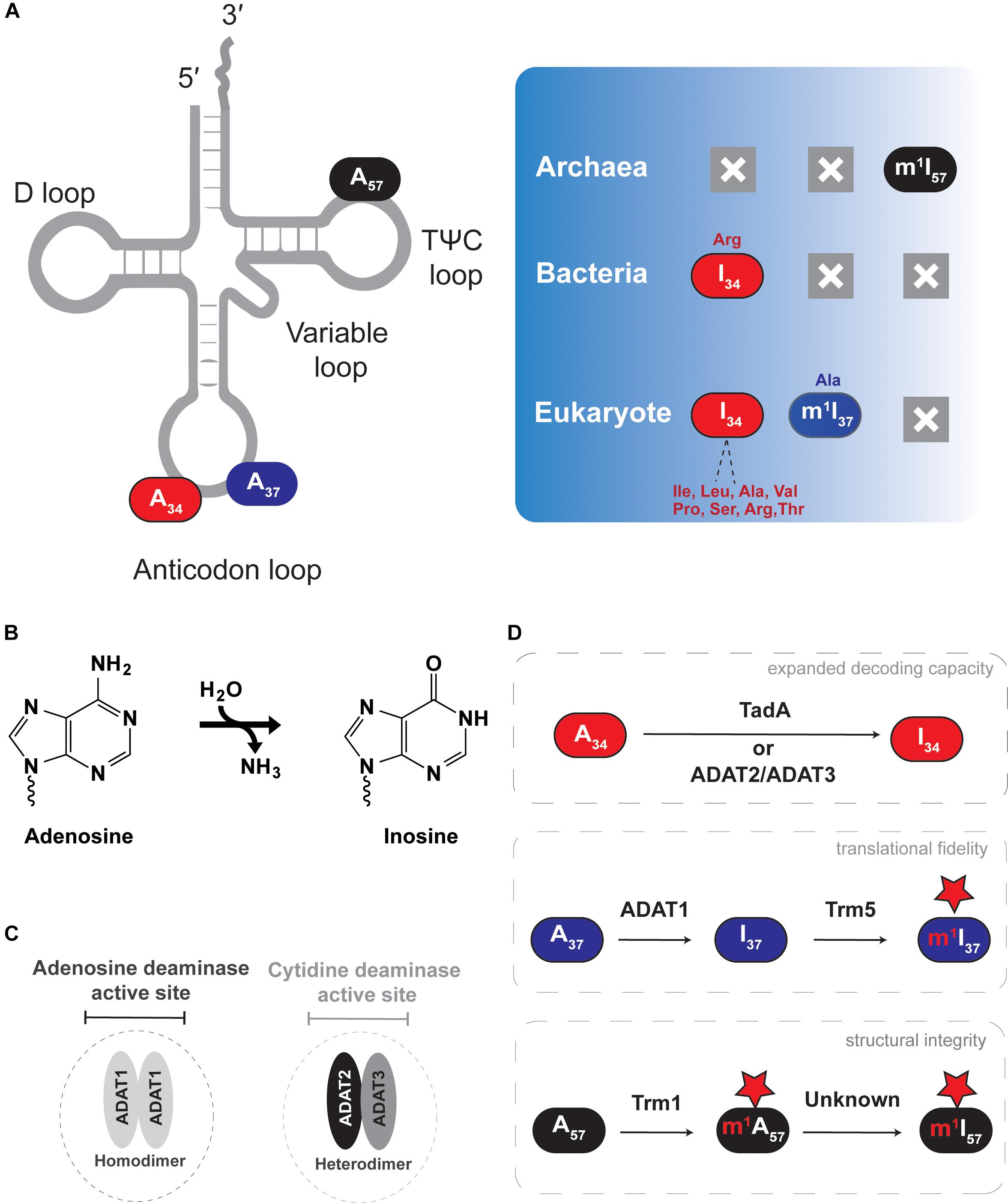
Figure 2. Adenosine-to-inosine (A-to-I) editing of tRNAs. The figure shows the different types of deamination reactions occurring in tRNAs from all domains of life. (A) Shows A-to-I edited nucleotide positions discussed in the text, with deamination end products and previous or subsequent methylation shown in the right panel. (B) Shows the general deamination reaction with ammonia as the leaving group and water as the key nucleophile. (C) Shows the characteristic active site of the different A-to-I deaminases. (D) Shows the effect each deamination has on tRNA function.
Inosine at the first position of the anticodon (I34) is essential in both eukaryotes and bacteria, but it does not occur in archaea (Figure 2). In eukaryotes, depending on the organism, roughly seven to eight cytoplasmic tRNAs have I34, while bacteria use I34 only in tRNAArg (Grosjean et al., 1996; Sprinzl et al., 1998). I34 increases the decoding capacity of tRNAs, allowing a single tRNA to decode three different codon (ending in U, C or A) and thus minimizing the number of necessary tRNA sequences that need to be genomically encoded. In addition, certain aminoacyl tRNA synthetases recognize and require the presence of I34 in tRNA substrates for productive aminoacylation (Droogmans and Grosjean, 1991; Senger et al., 1997; Gerber et al., 1998; Sprinzl et al., 1998; Losey et al., 2006). In bacteria, nearly all C-ending codons are read by tRNAs with an encoded G at position 34, except for tRNAArg which contains I34. Thus, despite a significantly more restricted pool of substrate tRNAs, I34 remains essential in bacteria.
A34-to-I34 deamination of eukaryotic tRNAs is catalyzed by the heterodimeric enzyme of (ADAT2/ADAT3 or Tad2/Tad3), which requires association between two paralogous sub-units for activity, while bacteria rely on the homodimeric enzyme ADATa (or TadA) (Auxilien et al., 1996; Wolf et al., 2002). In vitro, ADATa can efficiently recognize and deaminate a minimal substrate derived from the tRNAArg anticodon arm, while ADAT2/ADAT3 requires the entire tRNA for activity (Elias and Huang, 2005; Kuratani et al., 2005; Kim et al., 2006). Of special note, plant chloroplast also contains a single tRNAArg that undergoes A-to-I editing and relies on an ADATa-like enzyme, an observation consistent with the endosymbiotic theory of eukaryotic mitochondrial evolution (Delannoy et al., 2009; Karcher and Bock, 2009), but in general, mitochondria-encoded tRNAs do not contain inosine.
I37 and I57 are less widespread within organisms, where I37 is found in tRNAAla of certain eukaryotes (Gerber et al., 1998; Maas et al., 1999), and I57 has only been observed in tRNAs from archaea (Yamaizumi et al., 1982; Grosjean et al., 1995, 1996). Generally inosine at positions 37 and 57 can also be observed as methyl modified (m1I) as discussed later in Section “m1I Formation in Eukaryotes Versus Archaea.” I37 is formed by ADAT1, a homodimeric enzyme that shares key conserved residues with other deaminases: a conserved histidine and two cysteines that coordinate a catalytic Zn2+, as well as a conserved glutamate that participates in the final chemical step of inosine formation (Gerber et al., 1998; Gerber and Keller, 1999; Maas et al., 1999; Losey et al., 2006). I37 editing does not expand the decoding capacity of target tRNAs and its biological significance remains unclear. However, because of the importance of modifications at position 37 of the anticodon loop for reading-frame maintenance during translation, it is safe to assume a similar role for m1I37. The biological role of I57 in archaea is equally cryptic but its position in the backbone of the tRNA suggests a structural role.
All polynucleotide deaminases belong to the cytidine deaminase superfamily and require a Zn2+ for activity. However, phylogenetic analysis has revealed that ADAT1 closely aligns with mRNA specific ADARs, while ADAT2/ADAT3, responsible for inosine formation at position 34, strikingly resembles nucleic acid cytidine deaminases such as AID (activation-induced deaminases) or APOBEC (the apolipoprotein B mRNA editing enzyme) (Gerber and Keller, 2001). Recent investigation of the fungus Fusarium graminearum provides evidence that ADARs may have evolved from ADAT-like enzymes, as F. graminearum lack an obvious ADAR homolog, yet still contain A-to-I edited mRNAs (Wang C. et al., 2016; Bajad et al., 2017). These authors propose that ADATs may be responsible for A-to-I during the sexual life cycle of F. graminearum, which further suggests the double-stranded RNA binding domain found in ADARs was gained during the evolution from an ADAT-like ancestor, which lacks this domain (Gerber et al., 1998; Gerber and Keller, 1999; Maas et al., 1999; Losey et al., 2006). If true, it is equally possible that certain annotated ADATs may deaminate RNA substrates other than tRNAs, perhaps in complex with additional factors.
Cytidine to Uridine (C-to-U) tRNA Editing in Archaea and Eukarya
Certain archaea and eukaryotes contain C-to-U edited tRNAs, but in many instances the enzymes responsible remain to be identified (Figure 3) (Janke and Paabo, 1993; Marchfelder et al., 1996; Alfonzo et al., 1999; Fey et al., 2002; Randau et al., 2009; Grewe et al., 2011). In the archaeon Methanocryptus kandleri, CDAT8, a cytidine deaminase homolog, catalyzes C-to-U conversion at position 8 of tRNAs (Randau et al., 2009). For many tRNAs an encoded U at position 8 forms a Hoogsteen base pair with A14 to assist in the proper folding of mature L-shaped tRNAs (Romby et al., 1985). However, 30 out of 34 tRNAs that contain A14 in M. kandleri, are encoded with a cytidine at position 8 that must be deaminated by CDAT8 to form U8 (Randau et al., 2009) and ensure proper tRNA folding. The reason for having such a large number of tRNAs that require C-to-U editing in M. kandleri, rather than encoding for U8-containing transcripts remains obscure. One probable answer might be the extreme environment in which M. kandleri lives, which favors G:C pairing in the DNA for optimal genome stability, yet still requires C-to-U editing for proper tRNA folding (Randau et al., 2009).
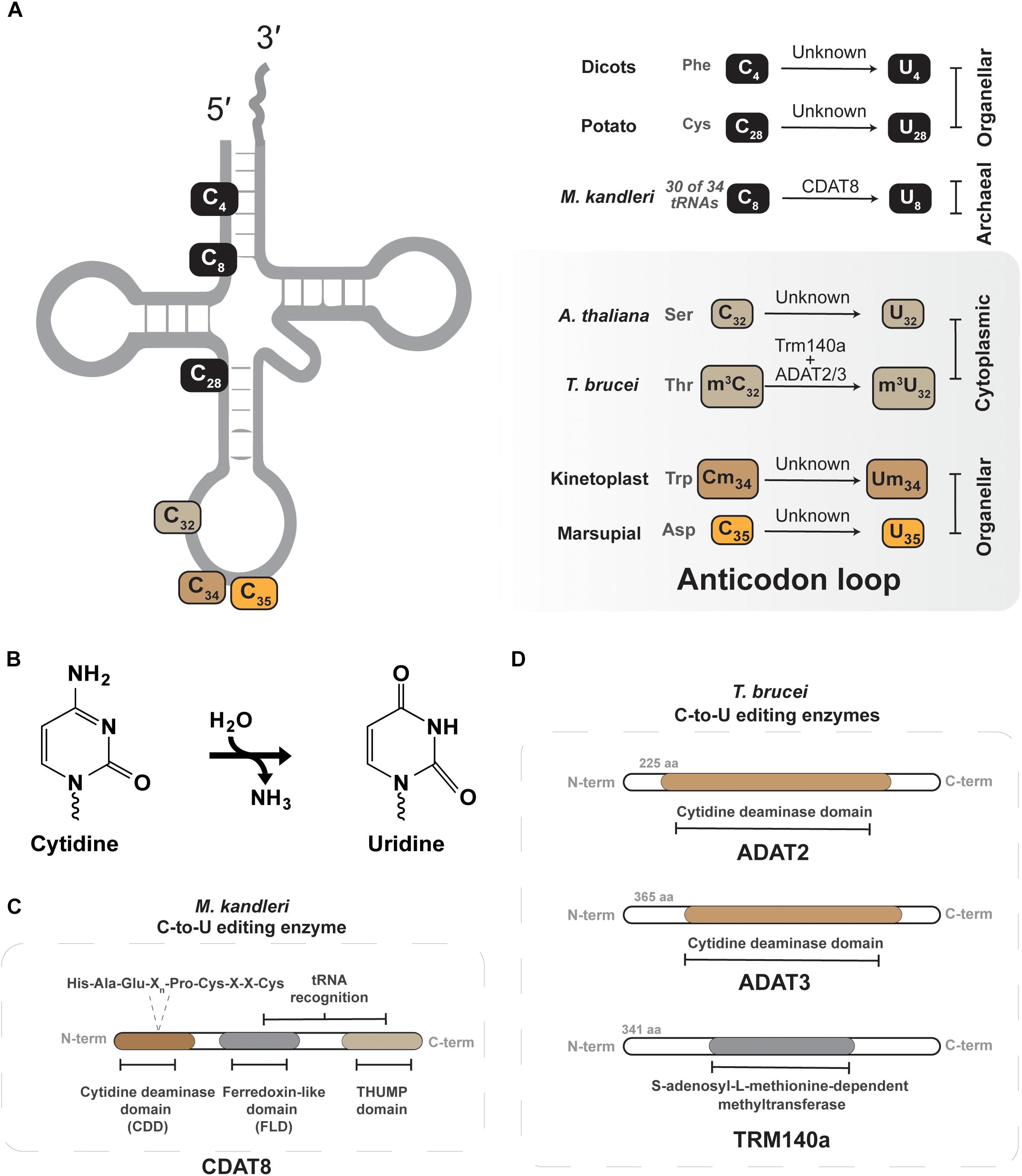
Figure 3. Cytidine-to-uridine (C-to-U) editing of tRNAs. (A) Shows the different C-to-U edited nucleotide positions that have been described in different tRNAs of different organisms. The organism and the nucleotide position along with tRNA identity are shown in the right panel. The enzyme identity is presented over the arrow. The gray panel denotes all known C-to-U editing events occurring in the anticodon loop. (B) Shows the C-to-U deamination reaction. (C) Depicts the conserved active-site residues in the tRNA C-to-U deaminase from archaea (CDAT8). (D) Depicts the active-site domain of both the ADAT2/3 and Trm140a of T. brucei, these enzymes interdependently.
C-to-U editing of tRNAs in eukarya was first reported in protists, plants and marsupial mitochondria (Janke and Paabo, 1993; Lonergan and Gray, 1993a,b; Marechal-Drouard et al., 1996). However, the marsupial system provided the first example of C to U editing at the anticodon nucleotides. These organisms do not encode a tRNA for decoding mitochondrial aspartate codons. To solve this issue, the anticodon of tRNAGlyGCC is C-to-U edited to tRNAAspGUC, which is recognized by the mitochondrial aspartyl tRNA synthetase to produce a functional ortholog to tRNAAsp (Janke and Paabo, 1993; Borner et al., 1996). This quirk of marsupial mitochondria paves the way for identification of similar mechanisms in nature, where an encoded tRNA gene can be edited to function as an alternative aminoacyl acceptor.
Leishmania tarentolae and Trypanosoma brucei, representative kinetoplastids, offer the only other example of anticodon C-to-U editing in tRNA (Alfonzo et al., 1999; Charriere et al., 2006). As in other organisms, the mitochondrial genome lacks certain tRNA genes that must be actively imported from the cytoplasm (Paris et al., 2009). In some instances, the mitochondrial translational code is not compatible with nuclear encoded tRNAs, as is the case for decoding UGA as tryptophan in mitochondria, not a stop codon (Barrell et al., 1979). In L. tarentolae and T. brucei UGA is used as a tryptophan codon in mitochondria, while the nucleus only contains a single-copy tRNATrpCCA to decode the canonical UGG codons. After import of tRNATrpCCA into the mitochondria, position 34 is C-to-U edited to create tRNATrpUCA as part of the mechanism that reassigned the UGA codons from stop to tryptophan. The enzyme responsible for this essential editing event is still unknown, but one would guess a deamination mechanism (Alfonzo et al., 1999; Charriere et al., 2006). Interestingly, only approximately 40–50% of the tRNATrp is edited after transport to the trypanosome mitochondria, raising questions as to how this balance is kept. The answer partly rests on the unusual thiolation at U33 in this tRNA, which negatively impacts C-to-U editing of C34 (Wohlgamuth-Benedum et al., 2009).
In the mitochondria of dicotyledon plants, C-to-U editing takes place outside the anticodon region and does not directly influence the decoding capacity of tRNAs (Marechal-Drouard et al., 1996; Fey et al., 2002; Ichinose and Sugita, 2016). In potato mitochondria, C-to-U editing corrects a mismatch-encoded pair C4:A69 in 5′ processed pre-tRNAPheGAA to U4:A69 (Binder et al., 1994; Marechal-Drouard et al., 1996). After editing 5′ processed pre-tRNAPheGAA can properly fold resulting in efficient removal of the 3′ trailer by mitochondrial RNaseZ (Kunzmann et al., 1998). In a modeling study of quillwort, Isoetes engelmannii, extensive C-to-U editing was predicted at 43 possible tRNA sites (Grewe et al., 2011; Ichinose and Sugita, 2016). The author′s obtained cDNA sequence data for 36 such sites, and among them 29 showed C-to-U editing (Grewe et al., 2011). Interestingly four sites showed U-to-C conversion, invoking the necessity of a likely transamination reaction (Grewe et al., 2011). There are other thematically similar C-to-U editing events that occur for tRNAs in plant mitochondria, and these are well-reviewed elsewhere (Fey et al., 2002; Paris et al., 2012).
Position 32 of the anti-codon stem loop is another site where tRNAs from multiple domains contain C-to-U conversions. It was first described in T. brucei that all three tRNAThr isoacceptors undergo C-to-U editing at position 32 (Rubio et al., 2006; Gaston et al., 2007). Trypanosoma brucei ADAT2/ADAT3, previously discussed in the context of A-to-I editing at position 34, has been shown to perform C-to-U editing of tRNAThrAGU at position 32 (Rubio et al., 2007; Rubio et al., 2017). However, this editing event first requires methylation at this site, discussed in more detail in Section “m3C-to-m3U.” Within the ADAT2/ADAT3 heterodimeric complex, the C-terminal region of ADAT2 assists in tRNA binding, while ADAT3 provides a structural role in formation of the catalytic deaminase core (Rubio et al., 2007), similar in arrangement as the two subunit methyltransferases discussed in Section “Interdependent Methyltransferases.” The biological significance of this editing is not exactly clear, but some evidence showed it is important for protein synthesis (Rubio et al., 2017), but C32 methylation and editing do not impact aminoacylation efficiency. Given its position in the anticodon loop, likely such effects in protein synthesis may be due to some function in translational efficiency or accuracy. Similar editing events have been recently described in Arabidopsis thaliana, where tRNASerAGA and tRNASerGCU are C-to-U edited at position 32 in the nucleocytoplasmic compartment by a presently unknown enzymatic mechanism (Zhou et al., 2014).
Interdependent Methylation and Editing and Its Biological Relevance
m1I Formation in Eukaryotes Versus Archaea
As discussed in Section “Adenosine to Inosine (A-to-I) Editing” A-to-I editing is found at three tRNA sites: position 34, 37, and 57. Intriguingly, inosines at position 37 and 57 can be methylated to form m1I37 and m1I57, by distinct chemical pathways (Yamaizumi et al., 1982; Grosjean et al., 1995, 1996). At position 37, after ADAT1 converts A-to-I, SAM-dependent Trm5 can act directly on N-1 of inosine (Gerber et al., 1998; Maas et al., 1999; Brule et al., 2004; Macbeth et al., 2005). As Trm5 is essential in most organisms catalyzing the generation of m1G37, the biological significance of its involvement in the modification of edited I37 remains unclear (Paris et al., 2013). The opposite order of events occurs at position 57 in archaea, where a SAM-dependent TrmI-family member must first methylate A57 before it becomes a substrate for deamination to inosine (Yamaizumi et al., 1982; Grosjean et al., 1995, 1996). The enzyme, or enzymatic complex, responsible for m1A57-to-m1I57 has not been identified, and the biological significance of this modification remains to be articulated. However, methylation followed editing also occurs in specific m3C-to-m3U conversions.
m3C-to-m3U
In trypanosomes, down regulation of ADAT2/ADAT3 expression reduces A34-to-I34 editing in tRNAThrAGU at position 34 and C-to-U editing at position 32 (Rubio et al., 2007). It was originally hypothesized that ADAT2/ADAT3, which has clear sequence similarity with cytidine deaminases, may be responsible, however, initial attempts to reconstitute C-to-U editing with T. brucei ADAT2/ADAT3 were not successful (Rubio et al., 2007). Since C32 of tRNAThrAGU is methylated to form m3C32, it suggested that C-to-U editing could require methylation prior to deamination (Figure 4) (Rubio et al., 2017), as discussed previously for archaeal m1A57-to-m1I57. The methyltransferase Trm140 was later identified as responsible for formation of m3C32 (D’Silva et al., 2011; Noma et al., 2011) and true to expectations, Trm140 and ADAT2/ADAT3 were shown to work interdependently to convert m3C32 to m3U32 in vitro (Rubio et al., 2017). Further experimentation also showed that Trm140 and ADAT2/ADAT3 are likely to work sequentially and as a complex (Rubio et al., 2017). How these two enzymes converge simultaneously on a single RNA substrate and act at the same nucleotide position remains an open question. However, recent studies demonstrated that the two enzymes bind their tRNA substrate synergistically, whereby binding affinity increases significantly if both proteins are present in the reaction (McKenney et al., 2018).
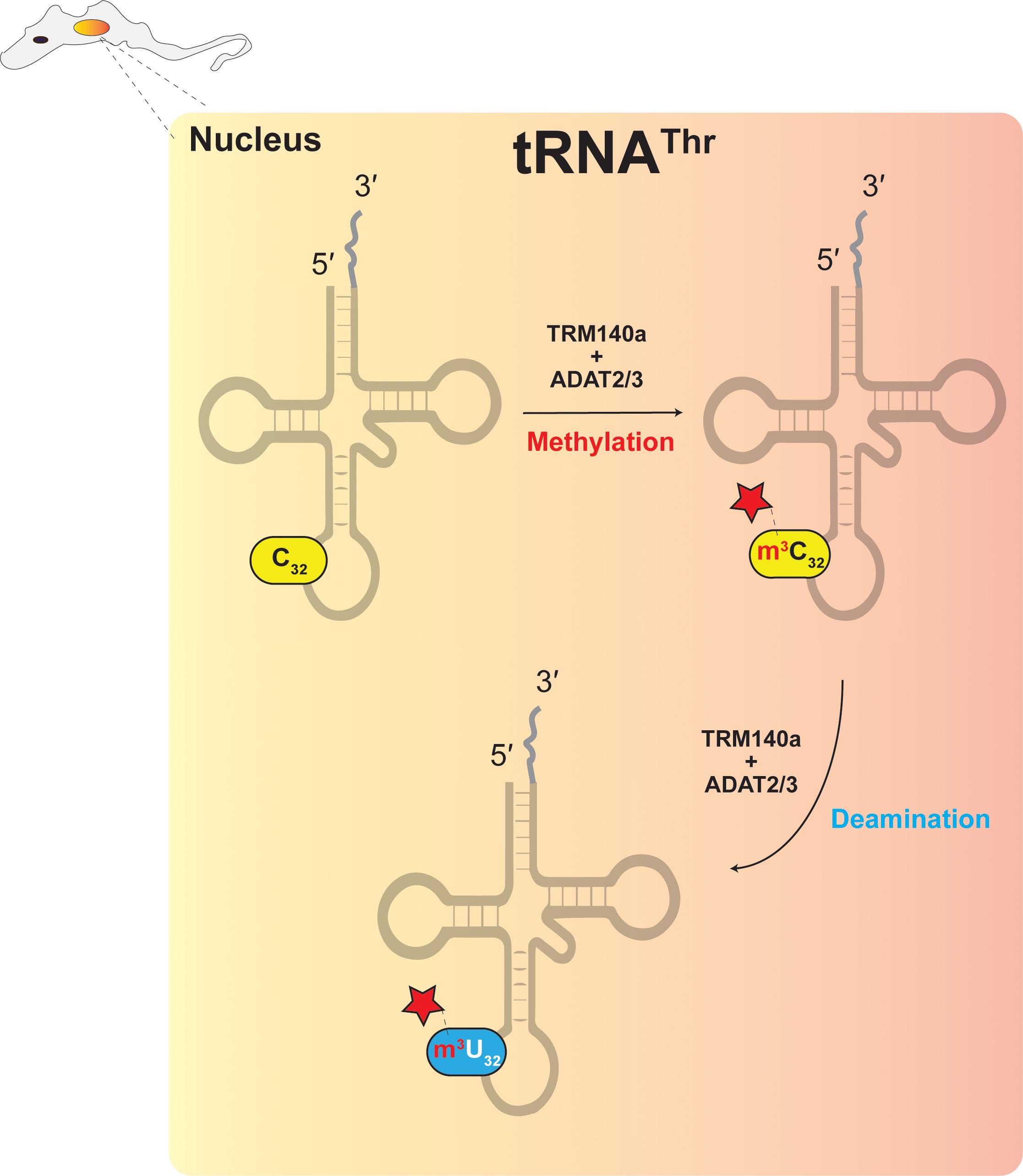
Figure 4. The interdependence model showing the necessary connection between RNA modification and RNA editing.
ADAT2/ADAT3 can deaminate DNA in vitro and in vivo, this mutagenic activity is dampened through association with Trm140 (Rubio et al., 2007, 2017). By extension a similar mechanism, may inform how the cytidine deaminase, AID, specifically targets genes that encode immunoglobulin receptors while leaving the rest of the genome unaffected during B cell somatic hypermutation (Teng and Papavasiliou, 2007). It is likely that additional protein factors influence specificity of AID toward its substrate genetic loci.
Concluding Remarks
Much has been written about the mechanisms that lead to, and determine the fate of, duplicated genes. It is clear that once gene duplication occurs, one copy is free to mutate via genetic drift, to accumulate mutations perhaps by a “neutral evolutionary ratchet” (Covello and Gray, 1993; Gray et al., 2010; Lukes et al., 2011). These mutations, in turn, can lead to total loss of function of the duplicated gene and the creation of pseudogenes. Alternatively, and more importantly, duplication may provide a powerful route to neofunctionalization or subfunctionalization of genes (Stoltzfus, 1999). The former makes the mutated duplicate gene acquire new functions different from that of the ancestral gene; the latter leads to a partitioning of labor so that each copy now carries a subset of the functions originally performed by the ancestral state. In this review, we have focused on various examples of modification enzymes, where evolution has pushed the system into one of the categories above. For example, in the case of the Trm61/Trm6 methyltransferase, sequence comparisons strongly suggest that these are paralogs. Here, the neutral acquisition of mutations without obvious gains in fitness led to a level of divergence that at some point may have become important in expanding substrate specificity, thus diversifying the number of targets the new enzyme could methylate.
In the case of Trm7/Trm732 and Trm7/Trm734, no evidence exists for gene duplication; neither Trm732 nor Trm734 have significant sequence conservation with known methyltransferases. Instead, they share similarity with other protein families, for example Trm734 with WD-domain family proteins. It may be that independent stochastic mutations accumulated in each gene for evolution of a dimerization interface that allowed each protein to interact with the Trm7 partner. In doing so, differential complex formation permitted a novel division of labor; one complex now targets position 32 of tRNAs and the other position 34. This is, of course, assuming that the catalytic subunit Trm7 derives from an ancestral gene that at some point was able to efficiently methylate both positions.
Similar arguments can be made with the tRNA A-to-I deaminase of trypanosomes, TbADAT2/TbADAT3, a heterodimeric enzyme comprised of two subunits encoded by clear paralogs (Rubio et al., 2007). Accumulation of mutations in one or both copies may have forced each paralog to strictly rely on the other for activity. Such cases echo themes observed in enzyme-prozyme complexes (Nguyen et al., 2013; Volkov et al., 2016), where in polyamine biosynthesis a catalytically dead paralog regulates the activity of an active paralogous enzyme. We (Gaston et al., 2007; Rubio et al., 2007), and others (Elias and Huang, 2005) have argued, that at least in the case of ADAT2/ADAT3 in eukaryotes, gene duplication led to the expansion in the specificity of the enzyme toward more molecular substrates. For example, ADATa, the homologous enzyme from bacteria is active as a homodimer, targets a single tRNAArg in vivo and a “minimalist” tRNAArg anti-codon stem loop is a sufficient substrate in vitro (Wolf et al., 2002). Not surprisingly the co-crystal structure of ADATa shows that residues near or at the active site are necessary for RNA binding (Losey et al., 2006). The eukaryotic counterpart ADAT2/ADAT3 recognizes seven to eight different tRNAs depending on the organism, but is only active on full length tRNAs (Auxilien et al., 1996). Years ago, we showed that one of the critical RNA binding domains of the T. brucei enzyme lies at the C-terminus of one subunit and that residues near the active site minimally contribute to substrate binding (Ragone et al., 2011). Thus, movement of critical binding residues away from the active site increased active site flexibility to allow for the observed expansion in substrate specificity. Again, such subtle, yet important, evolutionary changes in eukaryotic tRNA deaminases are only made possible by gene duplication and the previously proposed effects of “constructive neutral evolution” (Covello and Gray, 1993; Stoltzfus, 1999).
Finally let’s consider m3C/m3U editing and methylation at position 32 of several tRNAs in T. brucei (Figure 4) (also described in plants). The TbTrm140 m3C methyltransferase does not result from an obvious duplication of a deaminase gene and vice versa. While both enzymes come together to form a stable, active complex in the nucleus, the TbADAT2/TbADAT3 heterodimer is also active in the cytoplasm as a free enzyme catalyzing essential A-to-I deaminations (Rubio et al., 2017). In this particular case, both enzymes accumulated mutations in a neutral fashion, likely independent of each other. The question is how could TbTrm140 accumulate mutations without causing deleterious effects on the organism. The answer may involve TbMtase37, a paralog of Trm140 within the T. brucei genome, of currently unknown function (Fleming et al., 2016). This paralog might provide the necessary duplicate and essential function, which allowed TbTrm140 to mutationally drift and neofunctionalize with a seemingly unrelated enzyme like TbADAT2/TbADAT3 to modify and edit new substrates. What makes this case unusual is the fact that both enzymes have all the conserved residues required for activity and both may be active on other substrates, yet by themselves are totally inactive for methylation and deamination of tRNAThr position 32 of T. brucei. We thus introduce the concept of “enzyme co-activation,” whereby enzymes active with some substrates but inactive with others, gain new function upon their association and indeed co-activate each other.
Neo- or sub-functionalization includes a combination of non-adaptive and, later, adaptive mutations as originally suggested by Gray et al. (2010) Regardless of what factors or mechanisms are at play, the question then remains: Are there fitness gains to be made by organisms by the examples in this review? At least in trypanosomes, we have long appreciated the interdependent nature of RNA editing and modification, under the hypothesis that these events fine-tune translation to the ever-changing environmental conditions during the life cycle of these parasites (Paris et al., 2012). Interdependent modification and editing may serve to maintain levels of edited and unedited tRNAs in response to changes in environment or life stages. The same could be true of many other modifications and subsequent use of alternative substrates (Helm and Alfonzo, 2014); enzyme co-activation may provide an additional level of “tunability” to ensure fast responses to ever changing growth conditions, not only in response to stress, but also in maintenance of general cell homeostasis.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This article was funded in part by grants R56AI131248 and RO1GM132254 to JA and a Center of RNA Biology Postdoctoral Fellowship to JH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Alfonzo laboratory for useful discussions and comments.
References
Alexandrov, A., Grayhack, E. J., and Phizicky, E. M. (2005). tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11, 821–830. doi: 10.1261/rna.2030705
Alexandrov, A., Martzen, M. R., and Phizicky, E. M. (2002). Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8, 1253–1266. doi: 10.1017/S1355838202024019
Alfonzo, J. D., Blanc, V., Estevez, A. M., Rubio, M. A., and Simpson, L. (1999). C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 18, 7056–7062. doi: 10.1093/emboj/18.24.7056
Anderson, J., Phan, L., Cuesta, R., Carlson, B. A., Pak, M., Asano, K., et al. (1998). The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12, 3650–3662. doi: 10.1101/gad.12.23.3650
Anderson, J., Phan, L., and Hinnebusch, A. G. (2000). The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 5173–5178. doi: 10.1073/pnas.090102597
Arimbasseri, A. G., Iben, J., Wei, F. Y., Rijal, K., Tomizawa, K., Hafner, M., et al. (2016). Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA 22, 1400–1410. doi: 10.1261/rna.056259.116
Auxilien, S., Crain, P. F., Trewyn, R. W., and Grosjean, H. (1996). Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J. Mol. Biol. 262, 437–458. doi: 10.1006/jmbi.1996.0527
Bajad, P., Jantsch, M. F., Keegan, L., and O’Connell, M. (2017). A to I editing in disease is not fake news. RNA Biol. 14, 1223–1231. doi: 10.1080/15476286.2017.1306173
Barraud, P., Golinelli-Pimpaneau, B., Atmanene, C., Sanglier, S., Van Dorsselaer, A., Droogmans, L., et al. (2008). Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J. Mol. Biol. 377, 535–550. doi: 10.1016/j.jmb.2008.01.041
Barrell, B. G., Bankier, A. T., and Drouin, J. (1979). A different genetic code in human mitochondria. Nature 282, 189–194. doi: 10.1038/282189a0
Bass, B. L., and Weintraub, H. (1988). An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098. doi: 10.1016/0092-8674(88)90253-X
Begley, U., Dyavaiah, M., Patil, A., Rooney, J. P., DiRenzo, D., Young, C. M., et al. (2007). Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28, 860–870. doi: 10.1016/j.molcel.2007.09.021
Benitez-Paez, A., Villarroya, M., Douthwaite, S., Gabaldon, T., and Armengod, M. E. (2010). YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 16, 2131–2143. doi: 10.1261/rna.2245910
Binder, S., Marchfelder, A., and Brennicke, A. (1994). RNA editing of tRNA(Phe) and tRNA(Cys) in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet. 244, 67–74. doi: 10.1007/BF00280188
Borner, G. V., Morl, M., Janke, A., and Paabo, S. (1996). RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 15, 5949–5957. doi: 10.1002/j.1460-2075.1996.tb00981.x
Bourgeois, G., Letoquart, J., van Tran, N., and Graille, M. (2017a). Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 7:E7. doi: 10.3390/biom7010007
Bourgeois, G., Marcoux, J., Saliou, J. M., Cianferani, S., and Graille, M. (2017b). Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 45, 1971–1982. doi: 10.1093/nar/gkw1271
Brule, H., Elliott, M., Redlak, M., Zehner, Z. E., and Holmes, W. M. (2004). Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43, 9243–9255. doi: 10.1021/bi049671q
Bujnicki, J. M. (2001). In silico analysis of the tRNA:m1A58 methyltransferase family: homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett. 507, 123–127. doi: 10.1016/S0014-5793(01)02962-3
Charriere, F., Helgadottir, S., Horn, E. K., Soll, D., and Schneider, A. (2006). Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 103, 6847–6852. doi: 10.1073/pnas.0602362103
Chen, C., Huang, B., Anderson, J. T., and Bystrom, A. S. (2011). Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PLoS One 6:e20783. doi: 10.1371/journal.pone.0020783
Clouet d’Orval, B., Bortolin, M. L., Gaspin, C., and Bachellerie, J. P. (2001). Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 29, 4518–4529. doi: 10.1093/nar/29.22.4518
Covello, P. S., and Gray, M. W. (1993). On the evolution of RNA editing. Trends Genet. 9, 265–268. doi: 10.1016/0168-9525(93)90011-6
De Bie, L. G., Roovers, M., Oudjama, Y., Wattiez, R., Tricot, C., Stalon, V., et al. (2003). The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J. Bacteriol. 185, 3238–3243. doi: 10.1128/JB.185.10.3238-3243.2003
Delannoy, E., Le Ret, M., Faivre-Nitschke, E., Estavillo, G. M., Bergdoll, M., Taylor, N. L., et al. (2009). Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation. Plant Cell 21, 2058–2071. doi: 10.1105/tpc.109.066654
Deng, W., Babu, I. R., Su, D., Yin, S., Begley, T. J., and Dedon, P. C. (2015). Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLoS Genet. 11:e1005706. doi: 10.1371/journal.pgen.1005706
Droogmans, L., and Grosjean, H. (1991). 2′-O-methylation and inosine formation in the wobble position of anticodon-substituted tRNA-Phe in a homologous yeast in vitro system. Biochimie 73, 1021–1025. doi: 10.1016/0300-9084(91)90143-O
Droogmans, L., Roovers, M., Bujnicki, J. M., Tricot, C., Hartsch, T., Stalon, V., et al. (2003). Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 31, 2148–2156. doi: 10.1093/nar/gkg314
D’Silva, S., Haider, S. J., and Phizicky, E. M. (2011). A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA 17, 1100–1110. doi: 10.1261/rna.2652611
Elias, Y., and Huang, R. H. (2005). Biochemical and structural studies of A-to-I editing by tRNA:A34 deaminases at the wobble position of transfer RNA. Biochemistry 44, 12057–12065. doi: 10.1021/bi050499f
Fey, J., Weil, J. H., Tomita, K., Cosset, A., Dietrich, A., Small, I., et al. (2002). Role of editing in plant mitochondrial transfer RNAs. Gene 286, 21–24. doi: 10.1016/S0378-1119(01)00817-4
Finer-Moore, J., Czudnochowski, N., O’Connell, J. D. III, Wang, A. L., and Stroud, R. M. (2015). Crystal structure of the human tRNA m(1)A58 methyltransferase-tRNA(3)(Lys) complex: refolding of substrate tRNA allows access to the methylation target. J. Mol. Biol. 427, 3862–3876. doi: 10.1016/j.jmb.2015.10.005
Fitzek, E., Joardar, A., Gupta, R., and Geisler, M. (2018). Evolution of eukaryal and archaeal pseudouridine synthase Pus10. J. Mol. Evol. 86, 77–89. doi: 10.1007/s00239-018-9827-y
Fleming, I. M., Paris, Z., Gaston, K. W., Balakrishnan, R., Fredrick, K., Rubio, M. A., et al. (2016). A tRNA methyltransferase paralog is important for ribosome stability and cell division in Trypanosoma brucei. Sci. Rep. 6:21438. doi: 10.1038/srep21438
Fu, D., Brophy, J. A., Chan, C. T., Atmore, K. A., Begley, U., Paules, R. S., et al. (2010). Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell Biol. 30, 2449–2459. doi: 10.1128/MCB.01604-09
Gaston, K. W., Rubio, M. A., Spears, J. L., Pastar, I., Papavasiliou, F. N., and Alfonzo, J. D. (2007). C to U editing at position 32 of the anticodon loop precedes tRNA 5′ leader removal in trypanosomatids. Nucleic Acids Res. 35, 6740–6749. doi: 10.1093/nar/gkm745
Gerber, A., Grosjean, H., Melcher, T., and Keller, W. (1998). Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J. 17, 4780–4789. doi: 10.1093/emboj/17.16.4780
Gerber, A. P., and Keller, W. (1999). An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286, 1146–1149. doi: 10.1126/science.286.5442.1146
Gerber, A. P., and Keller, W. (2001). RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 26, 376–384. doi: 10.1016/S0968-0004(01)01827-8
Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., et al. (2003). Global analysis of protein expression in yeast. Nature 425, 737–741. doi: 10.1038/nature02046
Goto-Ito, S., Ito, T., and Yokoyama, S. (2017). Trm5 and TrmD: two enzymes from distinct origins catalyze the identical tRNA modification, m(1)G37. Biomolecules 7:32. doi: 10.3390/biom7010032
Gray, M. W., Lukes, J., Archibald, J. M., Keeling, P. J., and Doolittle, W. F. (2010). Cell biology. Irremediable complexity? Science 330, 920–921. doi: 10.1126/science.1198594
Grewe, F., Herres, S., Viehover, P., Polsakiewicz, M., Weisshaar, B., and Knoop, V. (2011). A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 39, 2890–2902. doi: 10.1093/nar/gkq1227
Grosjean, H., Auxilien, S., Constantinesco, F., Simon, C., Corda, Y., Becker, H. F., et al. (1996). Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie 78, 488–501. doi: 10.1016/0300-9084(96)84755-9
Grosjean, H., Constantinesco, F., Foiret, D., and Benachenhou, N. (1995). A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 23, 4312–4319. doi: 10.1093/nar/23.21.4312
Gupta, A., Kumar, P. H., Dineshkumar, T. K., Varshney, U., and Subramanya, H. S. (2001). Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol. 312, 381–391. doi: 10.1006/jmbi.2001.4935
Guy, M. P., and Phizicky, E. M. (2014). Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 11, 1608–1618. doi: 10.1080/15476286.2015.1008360
Guy, M. P., and Phizicky, E. M. (2015). Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 21, 61–74. doi: 10.1261/rna.047639.114
Guy, M. P., Podyma, B. M., Preston, M. A., Shaheen, H. H., Krivos, K. L., Limbach, P. A., et al. (2012). Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 18, 1921–1933. doi: 10.1261/rna.035287.112
Guy, M. P., Shaw, M., Weiner, C. L., Hobson, L., Stark, Z., Rose, K., et al. (2015). Defects in tRNA anticodon loop 2′-O-methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum. Mutat. 36, 1176–1187. doi: 10.1002/humu.22897
Han, L., Guy, M. P., Kon, Y., and Phizicky, E. M. (2018). Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 14:e1007288. doi: 10.1371/journal.pgen.1007288
Han, L., Marcus, E., D’Silva, S., and Phizicky, E. M. (2017). S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. RNA 23, 406–419. doi: 10.1261/rna.059667.116
Han, L., and Phizicky, E. M. (2018). A rationale for tRNA modification circuits in the anticodon loop. RNA 24, 1277–1284. doi: 10.1261/rna.067736.118
Helm, M., and Alfonzo, J. D. (2014). Posttranscriptional RNA modifications: playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 21, 174–185. doi: 10.1016/j.chembiol.2013.10.015
Hirata, A., Nishiyama, S., Tamura, T., Yamauchi, A., and Hori, H. (2016). Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 44, 6377–6390. doi: 10.1093/nar/gkw561
Hori, H. (2017). Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules 7:E23. doi: 10.3390/biom7010023
Ichinose, M., and Sugita, M. (2016). RNA editing and its molecular mechanism in plant organelles. Genes 8:E5. doi: 10.3390/genes8010005
Ishida, K., Kunibayashi, T., Tomikawa, C., Ochi, A., Kanai, T., Hirata, A., et al. (2011). Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39, 2304–2318. doi: 10.1093/nar/gkq1180
Jablonowski, D., Zink, S., Mehlgarten, C., Daum, G., and Schaffrath, R. (2006). tRNAGlu wobble uridine methylation by Trm9 identifies Elongator’s key role for zymocin-induced cell death in yeast. Mol. Microbiol. 59, 677–688. doi: 10.1111/j.1365-2958.2005.04972.x
Janke, A., and Paabo, S. (1993). Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 21, 1523–1525. doi: 10.1093/nar/21.7.1523
Joardar, A., Malliahgari, S. R., Skariah, G., and Gupta, R. (2011). 2′-O-methylation of the wobble residue of elongator pre-tRNA(Met) in Haloferax volcanii is guided by a box C/D RNA containing unique features. RNA Biol. 8, 782–791. doi: 10.4161/rna.8.5.16015
Kalhor, H. R., and Clarke, S. (2003). Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell Biol. 23, 9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003
Karcher, D., and Bock, R. (2009). Identification of the chloroplast adenosine-to-inosine tRNA editing enzyme. RNA 15, 1251–1257. doi: 10.1261/rna.1600609
Kim, J., Malashkevich, V., Roday, S., Lisbin, M., Schramm, V. L., and Almo, S. C. (2006). Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase. Biochemistry 45, 6407–6416. doi: 10.1021/bi0522394
Kunzmann, A., Brennicke, A., and Marchfelder, A. (1998). 5′ end maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 95, 108–113. doi: 10.1073/pnas.95.1.108
Kuratani, M., Ishii, R., Bessho, Y., Fukunaga, R., Sengoku, T., Shirouzu, M., et al. (2005). Crystal structure of tRNA adenosine deaminase (TadA) from Aquifex aeolicus. J. Biol. Chem. 280, 16002–16008. doi: 10.1074/jbc.M414541200
Leihne, V., Kirpekar, F., Vagbo, C. B., van den Born, E., Krokan, H. E., Grini, P. E., et al. (2011). Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. 39, 7688–7701. doi: 10.1093/nar/gkr406
Letoquart, J., van Tran, N., Caroline, V., Aleksandrov, A., Lazar, N., van Tilbeurgh, H., et al. (2015). Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 43, 10989–11002. doi: 10.1093/nar/gkv1009
Leulliot, N., Chaillet, M., Durand, D., Ulryck, N., Blondeau, K., and van Tilbeurgh, H. (2008). Structure of the yeast tRNA m7G methylation complex. Structure 16, 52–61. doi: 10.1016/j.str.2007.10.025
Lonergan, K. M., and Gray, M. W. (1993a). Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science 259, 812–816.
Lonergan, K. M., and Gray, M. W. (1993b). Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res. 21:4402. doi: 10.1093/nar/21.18.4402
Losey, H. C., Ruthenburg, A. J., and Verdine, G. L. (2006). Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159. doi: 10.1038/nsmb1047
Lukes, J., Archibald, J. M., Keeling, P. J., Doolittle, W. F., and Gray, M. W. (2011). How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 63, 528–537. doi: 10.1002/iub.489
Maas, S., Gerber, A. P., and Rich, A. (1999). Identification and characterization of a human tRNA-specific adenosine deaminase related to the ADAR family of pre-mRNA editing enzymes. Proc. Natl. Acad. Sci. U.S.A. 96, 8895–8900. doi: 10.1073/pnas.96.16.8895
Macbeth, M. R., Schubert, H. L., Vandemark, A. P., Lingam, A. T., Hill, C. P., and Bass, B. L. (2005). Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309, 1534–1539. doi: 10.1126/science.1113150
Maraia, R. J., and Arimbasseri, A. G. (2017). Factors that shape eukaryotic tRNAomes: processing, modification and anticodon-codon use. Biomolecules 7:E26. doi: 10.3390/biom7010026
Marchfelder, A., Brennicke, A., and Binder, S. (1996). RNA editing is required for efficient excision of tRNA(Phe) from precursors in plant mitochondria. J. Biol. Chem. 271, 1898–1903. doi: 10.1074/jbc.271.4.1898
Marechal-Drouard, L., Cosset, A., Remacle, C., Ramamonjisoa, D., and Dietrich, A. (1996). A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol. Cell Biol. 16, 3504–3510. doi: 10.1128/MCB.16.7.3504
Matsumoto, K., Tomikawa, C., Toyooka, T., Ochi, A., Takano, Y., Takayanagi, N., et al. (2008). Production of yeast tRNA (m(7)G46) methyltransferase (Trm8-Trm82 complex) in a wheat germ cell-free translation system. J. Biotechnol. 133, 453–460. doi: 10.1016/j.jbiotec.2007.11.009
Matsumoto, K., Toyooka, T., Tomikawa, C., Ochi, A., Takano, Y., Takayanagi, N., et al. (2007). RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett. 581, 1599–1604. doi: 10.1016/j.febslet.2007.03.023
Mazauric, M. H., Dirick, L., Purushothaman, S. K., Bjork, G. R., and Lapeyre, B. (2010). Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 285, 18505–18515. doi: 10.1074/jbc.M110.113100
McKenney, K. M., and Alfonzo, J. D. (2016). From prebiotics to probiotics: the evolution and functions of tRNA modifications. Life 6:E13. doi: 10.3390/life6010013
McKenney, K. M., Rubio, M. A. T., and Alfonzo, J. D. (2018). Binding synergy as an essential step for tRNA editing and modification enzyme codependence in Trypanosoma brucei. RNA 24, 56–66. doi: 10.1261/rna.062893.117
Muneyoshi, Y., Matsumoto, K., Tomikawa, C., Toyooka, T., Ochi, A., Masaoka, T., et al. (2007). Hetero subunit interaction and RNA recognition of yeast tRNA (m7G46) methyltransferase synthesized in a wheat germ cell-free translation system. Nucleic Acids Symp. Ser. 51, 359–360. doi: 10.1093/nass/nrm180
Nguyen, S., Jones, D. C., Wyllie, S., Fairlamb, A. H., and Phillips, M. A. (2013). Allosteric activation of trypanosomatid deoxyhypusine synthase by a catalytically dead paralog. J. Biol. Chem. 288, 15256–15267. doi: 10.1074/jbc.M113.461137
Nolivos, S., Carpousis, A. J., and Clouet-d’Orval, B. (2005). The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 33, 6507–6514. doi: 10.1093/nar/gki962
Noma, A., Kirino, Y., Ikeuchi, Y., and Suzuki, T. (2006). Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154. doi: 10.1038/sj.emboj.7601105
Noma, A., Yi, S., Katoh, T., Takai, Y., Suzuki, T., and Suzuki, T. (2011). Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA 17, 1111–1119. doi: 10.1261/rna.2653411
Okada, K., Muneyoshi, Y., Endo, Y., and Hori, H. (2009). Production of yeast (m2G10) methyltransferase (Trm11 and Trm112 complex) in a wheat germ cell-free translation system. Nucleic Acids Symp. Ser. 53, 303–304. doi: 10.1093/nass/nrp152
Okamoto, H., Watanabe, K., Ikeuchi, Y., Suzuki, T., Endo, Y., and Hori, H. (2004). Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 279, 49151–49159. doi: 10.1074/jbc.M408209200
Osman, T. A., and Buck, K. W. (1997). The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 71, 6075–6082.
Ozanick, S., Krecic, A., Andersland, J., and Anderson, J. T. (2005). The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290. doi: 10.1261/rna.5040605
Ozanick, S. G., Bujnicki, J. M., Sem, D. S., and Anderson, J. T. (2007). Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res. 35, 6808–6819. doi: 10.1093/nar/gkm574
Paris, Z., Fleming, I. M., and Alfonzo, J. D. (2012). Determinants of tRNA editing and modification: avoiding conundrums, affecting function. Semin. Cell Dev. Biol. 23, 269–274. doi: 10.1016/j.semcdb.2011.10.009
Paris, Z., Horakova, E., Rubio, M. A., Sample, P., Fleming, I. M., Armocida, S., et al. (2013). The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 19, 649–658. doi: 10.1261/rna.036665.112
Paris, Z., Rubio, M. A., Lukes, J., and Alfonzo, J. D. (2009). Mitochondrial tRNA import in Trypanosoma brucei is independent of thiolation and the Rieske protein. RNA 15, 1398–1406. doi: 10.1261/rna.1589109
Pastore, C., Topalidou, I., Forouhar, F., Yan, A. C., Levy, M., and Hunt, J. F. (2012). Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J. Biol. Chem. 287, 2130–2143. doi: 10.1074/jbc.M111.286187
Purta, E., van Vliet, F., Tkaczuk, K. L., Dunin-Horkawicz, S., Mori, H., Droogmans, L., et al. (2006). The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol. Biol. 7:23. doi: 10.1186/1471-2199-7-23
Purushothaman, S. K., Bujnicki, J. M., Grosjean, H., and Lapeyre, B. (2005). Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell Biol. 25, 4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005
Ragone, F. L., Spears, J. L., Wohlgamuth-Benedum, J. M., Kreel, N., Papavasiliou, F. N., and Alfonzo, J. D. (2011). The C-terminal end of the Trypanosoma brucei editing deaminase plays a critical role in tRNA binding. RNA 17, 1296–1306. doi: 10.1261/rna.2748211
Randau, L., Stanley, B. J., Kohlway, A., Mechta, S., Xiong, Y., and Soll, D. (2009). A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science 324, 657–659. doi: 10.1126/science.1170123
Robertus, J. D., Ladner, J. E., Finch, J. T., Rhodes, D., Brown, R. S., Clark, B. F., et al. (1974). Structure of yeast phenylalanine tRNA at 3 A resolution. Nature 250, 546–551. doi: 10.1038/250546a0
Romby, P., Moras, D., Bergdoll, M., Dumas, P., Vlassov, V. V., Westhof, E., et al. (1985). Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J. Mol. Biol. 184, 455–471. doi: 10.1016/0022-2836(85)90294-3
Roovers, M., Wouters, J., Bujnicki, J. M., Tricot, C., Stalon, V., Grosjean, H., et al. (2004). A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 32, 465–476. doi: 10.1093/nar/gkh191
Rubio, M. A., Gaston, K. W., McKenney, K. M., Fleming, I. M., Paris, Z., Limbach, P. A., et al. (2017). Editing and methylation at a single site by functionally interdependent activities. Nature 542, 494–497. doi: 10.1038/nature21396
Rubio, M. A., Pastar, I., Gaston, K. W., Ragone, F. L., Janzen, C. J., Cross, G. A., et al. (2007). An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc. Natl. Acad. Sci. U.S.A. 104, 7821–7826. doi: 10.1073/pnas.0702394104
Rubio, M. A., Ragone, F. L., Gaston, K. W., Ibba, M., and Alfonzo, J. D. (2006). C to U editing stimulates A to I editing in the anticodon loop of a cytoplasmic threonyl tRNA in Trypanosoma brucei. J. Biol. Chem. 281, 115–120. doi: 10.1074/jbc.M510136200
Safra, M., Sas-Chen, A., Nir, R., Winkler, R., Nachshon, A., Bar-Yaacov, D., et al. (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255. doi: 10.1038/nature24456
Sardana, R., and Johnson, A. W. (2012). The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol. Biol. Cell 23, 4313–4322. doi: 10.1091/mbc.E12-05-0370
Senger, B., Auxilien, S., Englisch, U., Cramer, F., and Fasiolo, F. (1997). The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry 36, 8269–8275. doi: 10.1021/bi970206l
Shaheen, R., Abdel-Salam, G. M., Guy, M. P., Alomar, R., Abdel-Hamid, M. S., Afifi, H. H., et al. (2015). Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 16:210. doi: 10.1186/s13059-015-0779-x
Sokolowski, M., Klassen, R., Bruch, A., Schaffrath, R., and Glatt, S. (2018). Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 409–418. doi: 10.1016/j.bbagrm.2017.12.003
Somme, J., Van Laer, B., Roovers, M., Steyaert, J., Versees, W., and Droogmans, L. (2014). Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA 20, 1257–1271. doi: 10.1261/rna.044503.114
Songe-Moller, L., van den Born, E., Leihne, V., Vagbo, C. B., Kristoffersen, T., Krokan, H. E., et al. (2010). Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell Biol. 30, 1814–1827. doi: 10.1128/MCB.01602-09
Spears, J. L., Gaston, K. W., and Alfonzo, J. D. (2011). Analysis of tRNA editing in native and synthetic substrates. Methods Mol. Biol. 718, 209–226. doi: 10.1007/978-1-61779-018-8_13
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. (1998). Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153. doi: 10.1093/nar/26.1.148
Stoltzfus, A. (1999). On the possibility of constructive neutral evolution. J. Mol. Evol. 49, 169–181. doi: 10.1007/PL00006540
Studte, P., Zink, S., Jablonowski, D., Bar, C., von der Haar, T., Tuite, M. F., et al. (2008). tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 69, 1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x
Swinehart, W. E., and Jackman, J. E. (2015). Diversity in mechanism and function of tRNA methyltransferases. RNA Biol. 12, 398–411. doi: 10.1080/15476286.2015.1008358
Taylor, D. N., and Carr, J. P. (2000). The GCD10 subunit of yeast eIF-3 binds the methyltransferase-like domain of the 126 and 183 kDa replicase proteins of tobacco mosaic virus in the yeast two-hybrid system. J. Gen. Virol. 81(Pt 6), 1587–1591. doi: 10.1099/0022-1317-81-6-1587
Teng, G., and Papavasiliou, F. N. (2007). Immunoglobulin somatic hypermutation. Annu. Rev. Genet. 41, 107–120. doi: 10.1146/annurev.genet.41.110306.130340
Thiebe, R., and Poralla, K. (1973). Origin of the nucleoside Y in yeast tRNAPhe. FEBS Lett. 38, 27–28. doi: 10.1016/0014-5793(73)80504-6
Tomikawa, C., Yokogawa, T., Kanai, T., and Hori, H. (2010). N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 38, 942–957. doi: 10.1093/nar/gkp1059
Traube, F. R., and Carell, T. (2017). The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 14, 1099–1107. doi: 10.1080/15476286.2017.1318241
van Tran, N., Muller, L., Ross, R. L., Lestini, R., Letoquart, J., Ulryck, N., et al. (2018). Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 46, 8483–8499. doi: 10.1093/nar/gky638
Volkov, O. A., Kinch, L., Ariagno, C., Deng, X., Zhong, S., Grishin, N., et al. (2016). Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog. eLife 5:e20198. doi: 10.7554/eLife.20198
Wang, C., Xu, J. R., and Liu, H. (2016). A-to-I RNA editing independent of ADARs in filamentous fungi. RNA Biol. 13, 940–945. doi: 10.1080/15476286.2016.1215796
Wang, M., Zhu, Y., Wang, C., Fan, X., Jiang, X., Ebrahimi, M., et al. (2016). Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae. Sci. Rep. 6:32562. doi: 10.1038/srep32562
Wohlgamuth-Benedum, J. M., Rubio, M. A., Paris, Z., Long, S., Poliak, P., Lukes, J., et al. (2009). Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J. Biol. Chem. 284, 23947–23953. doi: 10.1074/jbc.M109.029421
Wolf, J., Gerber, A. P., and Keller, W. (2002). tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 21, 3841–3851. doi: 10.1093/emboj/cdf362
Yamaizumi, Z., Ihara, M., Kuchino, Y., Gupta, R., Woese, C. R., and Nishimura, S. (1982). Archaebacterial tRNA contains 1-methylinosine at residue 57 in T psi C-loop. Nucleic Acids Symp. Ser. 11, 209–213.
Zdzalik, D., Vagbo, C. B., Kirpekar, F., Davydova, E., Puscian, A., Maciejewska, A. M., et al. (2014). Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PLoS One 9:e98729. doi: 10.1371/journal.pone.0098729
Keywords: translation, tRNA modification, mitochondria, inosine, deaminase, methylation
Citation: Dixit S, Henderson JC and Alfonzo JD (2019) Multi-Substrate Specificity and the Evolutionary Basis for Interdependence in tRNA Editing and Methylation Enzymes. Front. Genet. 10:104. doi: 10.3389/fgene.2019.00104
Received: 30 November 2018; Accepted: 30 January 2019;
Published: 14 February 2019.
Edited by:
Tohru Yoshihisa, University of Hyogo, JapanReviewed by:
Hiroyuki Hori, Ehime University, JapanYohei Kirino, Thomas Jefferson University, United States
Copyright © 2019 Dixit, Henderson and Alfonzo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan D. Alfonzo, YWxmb256by4xQG9zdS5lZHU=
†These authors have contributed equally to this work
 Sameer Dixit
Sameer Dixit Jeremy C. Henderson
Jeremy C. Henderson Juan D. Alfonzo
Juan D. Alfonzo