
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 19 February 2019
Sec. Livestock Genomics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00017
Camels have an important role in the lives of human beings, especially in arid regions, due to their multipurpose role and unique ability to adapt to harsh conditions. In spite of its enormous economic, cultural, and biological importance, the camel genome has not been widely studied. The size of camel genome is roughly 2.38 GB, containing over 20,000 genes. The unusual genetic makeup of the camel is the main reason behind its ability to survive under extreme environmental conditions. The camel genome harbors several unique variations which are being investigated for the treatment of several disorders. Various natural products from camels have also been tested and prescribed as adjunct therapy to control the progression of ailments. Interestingly, the camel employs unique immunological and molecular mechanisms against pathogenic agents and pathological conditions. Here, we broadly review camel classification, distribution and breed as well as recent progress in the determination of the camel genome, its size, genetic distribution, response to various physiological conditions, immunogenetics and the medicinal potential of camel gene products.
Camels contribute hugely to human survival in less agroecological parts of African, Asian and Arabian deserts. They have been used for transportation, as a source of food and for protection for a very long time. Nowadays, they are hugely important in many parts of the arid world as sustainable livestock species (Burger, 2016). Camels are members of the Camelidae family. Camelidae probably appeared in North America around 35 million years ago during the Eocene period (Reed, 1972). There are two major types – small and large camels – which are further subdivided into Camelus, Lama, and Vicugna genera. A clearly defined universal classification of camels does not exist. However, the most widely accepted classification is given in Figure 1 (Wu et al., 2014). They are generally differentiated on the basis of color, function and habitat. Camel breeds have roughly the same shape but diverge in body conformation, size and color (Al-Swailem et al., 2010). Large camelids include two domestic species: Camelus dromedarius, the single-humped camel, and Camelus bactrianus, the two-humped camel. The dromedary camels are also known as Arabian camels and are mostly prevalent in the hot and arid region between the east of Asia to the northern part of Africa. The dromedary camel was first domesticated around five thousand or six thousand years ago in the Arabian region (Trinks et al., 2012; Almathen et al., 2016). Bactrian camels are found in cold regions and desert of central Asia. Small camelids include the llama and the alpaca which are restricted to South America. A less widely present and reported form of large camelids is the tartary camel (C. bactrianus ferus) which are found in the remote areas of Mongolia and China. These wild bactrian camels are considered the lone survivors of Old World Camels (Bannikov, 1976; Ji et al., 2009). Their total number reported in 2012 was 730–880. This species is facing a population size reduction of at least 80% within the next 45–50 years (Hare, 2014).
Camels have several unique traits which enable them to survive and live in remote areas, high mountains and arid lands. They can withstand thirst and hunger for long periods in the most inhospitable ecological conditions. Genetic studies of camel have elucidated the role of several genes that enable them to adapt to desert conditions. Importantly, several studies have uncovered the beneficial property of camel products in the treatment of various diseases (Muyldermans et al., 2009; Agrawal et al., 2011; Jirimutu et al., 2012; Fitak et al., 2016).
This review covers several features of the Camelidae family, which distinguishes them even from closely related species. Some of the unique biological attributes and genomic variations make them fascinating not only for monetary purposes but also for the treatment of human ailment.
The camel number and distribution vary from region to region. While human population living in deserts has substantially decreased, the trend of nurturing camels in the desert is increasing at the global level. The camel population was estimated to be 27 million in 2014 according to the Food and Agriculture Organization (FAO). Interestingly, the annual growth rate of camel is higher than sheep, cattle, and horse (Faye and Bonnet, 2012). According to FAO statistics, the camel population is increasing with a yearly growth of 2.1%. The proportion of camels in the Domestic Herbivorous Biomass (DHB) was 1.1% in 1961 and this has increased to 1.5% in 2014 (Faye, 2016). Previously, camels were associated with nomadic communities. However, they are now an important part of modern life in arid regions (Breulmann et al., 2007). The distribution of camel population by region is given in Figure 2 (Faye, 2016). More than 80% of the world’s camel population is estimated to be in Africa. The camel population ranges from 120,000 to 998,000 in countries such as Turkmenistan, Egypt, Kazakhstan, Afghanistan, Tunisia, Oman, Saudi Arabia, Nigeria, China, Mongolia, Algeria, Eritrea, India, United Arab Emirates, Yemen, and Mali (Faye, 2016). Based on FAO statistics, the top 20 countries with camel population are listed in Table 1 (Food and Agriculture Organization [FAO], 2018). Faye and co-workers investigated the growth rate of camel in different countries and listed the countries with increasing and declining growth rates. They concluded that a decline in camel growth rate is due to a reduction in arid land devoted to camel rearing and a lack of universal selection programs for choosing the best breeds.
Several different breeds of camels exist and they are known locally under various names. Majority of camel breeds are differentiated based on phenotypic characteristics (Abdallah and Faye, 2012). The most common camel breeds are listed in Table 2.
Normally, each breed has a unique genetic makeup. However, traditional breeding is based on color phenotypes, which are correlated with certain economic and behavioral traits. With the increasing demand for sustainable food sources in desert regions, the interest in economic traits is also growing. Based on inherent local knowledge, economic traits (higher milk yield, drought resistance) are correlated with color phenotypes. With the advent of rapid genetic screening, beneficial traits in different breeds of camels are now being linked to mitochondrial and microsatellite markers.
It was observed that dromedary breeds of Nigerian origin are composed of a homogenous gene pool and no clear breed (Kurri and Kala) differentiation was noted by using molecular methods (Abdussamad et al., 2015). Ishag et al. (2010) studied single nucleotide polymorphism (SNP) in the growth hormone (GH) gene of six Sudanese camel breeds (Kenani, Lahwee, Rashaidi, Anafi, Bishari, and Kabbashi) using restriction fragment length polymorphism (RFLP). They identified a SNP 419C>T in intron 1 and determined that frequency of the C allele was high in pack camels (heavy weight) when compared to riding camel (light weight) (Bishari and Anafi). This substitution, associated with higher growth and body weight, has been used as a selective marker to assess genetic biodiversity in camel breeds (Abdel-Aziem et al., 2015). The association between GH polymorphism and body weight was also evaluated in four breeds of Arabian camels (Saheli, Majaheem, Waddah, and Homor) (Afifi et al., 2014). One SNP was identified in Waddah and Homor breeds at position 419C>T, two SNPs in Saheli breed at 419C>T and 450T>C, and thirteen SNPs in Majaheem breed. Together, these studies concluded that SNP 419C>T and 450T>C were correlated with greater body weight (Ishag et al., 2010; Afifi et al., 2014).
The camel possesses inimitable characteristics which enable them to survive in extreme desert conditions. They store energy in their humps in the form of fat which enables them to survive for longer period without food and water (Emmanuel and Nahapetian, 1980). The body temperature of camels can fluctuate from 34 to 41°C within the day (Schmidt-Nielsen et al., 1956). It has been reported that camels can easily lose water that is equivalent to more than 25% of its body weight (Macfarlane et al., 1963). Another study concluded that camels could lose water up to 30% of its body weight during dehydration while other mammals could die due to circulatory failure when the water loss exceeds 12% of their body weight (McKinley et al., 2001). Water balance in the body is controlled by several factors including tissue osmolality and most importantly blood osmolality (Andersson et al., 1980). Except camels, all mammalian erythrocytes are concave or spherical in shape. However, in camels it is highly ovaloid, flat, small and enucleated and circulates in large numbers (Goniakowska-Witalinska and Witalinski, 1976; Azwai et al., 2007). The unusual elliptical shape of camel red blood cells facilitates their flow in a dehydrated state and makes it possible to cross small capillaries (Eitan et al., 1976). Additionally, camel erythrocytes are very resistant to osmotic hemolysis and are able to swell up to 240% of their original volume without bursting (Oyewale et al., 2011). This might be due to the altered distribution of membrane phospholipids in its red blood cells (Warda and Zeisig, 2000). Kidneys of camels play a major role in the process of conservation of water through increasing the osmolarity of urine. It has a strong water reabsorption capacity and eliminates highly concentrated urine. The intestine of camels also reabsorbs water and, unlike other mammals, water loss through feces is low since the feces is very dry (Davidson, 2014). Interestingly, camels deal with high blood glucose level (twofold than other ruminants) without developing diabetes. They can also consume eight times more salt than sheep and cattle without showing any signs or symptoms of hypertension (Al-Ali et al., 1988; Ali, 1994; Jirimutu et al., 2012).
While camels have enormous cultural, economic and biological importance, very little is known about the camel genome and the evolutionary advantages it imparts. Domestic animals and livestock, including camels, hold unique features and genetic variations (Andersson and Georges, 2004). The first draft of both domestic and wild bactrian camel genomes were reported in 2012. The size of bactrian camel genome was reported as 2.38 Gb and contained 20,821 genes (Jirimutu et al., 2012). This was close to the camel genome size (2.02–2.40 Gb) estimated based on haploid DNA content (C-value). Based on phylogenetic analysis, the authors also revealed that the camel shared a common ancestor with even-toed ungulates around 55–60 million years ago. Immunoglobulin-like domains were the most common proteins found in the camel genome and the results were consistent with a previous report. After annotation, results also suggested that the rhodopsin-like G protein-coupled receptor family, which is widely involved in regulating signaling pathways of physiological and biological processes, was the most abundant protein family found in the genome. The authors also searched for heavy-chain antibodies (HCAbs) in the bactrian camel genome that are homologous to the human, alpaca and dromedary IgH gene. 17 VH (heavy-chain variable region), 7 DH (diversity region), 6 JH (joining region), and 10 CH (constant region) genes were identified. Their results showed that VH genes of bactrian camel contains mutations in the coding region of HCAbs which prevent them from binding to the light chains. Further, they predicted the presence of 20,821 bactrian camel genes. Additionally, each gene contained an average of 8 exons and 1,322 bp coding regions (CDS). Importantly, GC content of the CDS region was estimated to be 52% which was significantly higher than the whole genome. Wernersson et al. (2005) also reported similar differences in other mammals. A similar report, related to sequencing the genome of bactrian, dromedary and alpaca camels, was published in 2014. The genome size of bactrian camel reported in this study (2.45 Gb) was similar to the earlier report (Wu et al., 2014). Additionally, the authors reported that non-coding RNA genes of dromedary, bactrian and alpaca genomes shared similar copy numbers. Analysis of the bactrian camel genome showed that 34% of the DNA is repetitive which is lower than human (>50%) and other mammals (Lander et al., 2001; Elsik et al., 2009; Wade et al., 2009; Wu et al., 2014). However, this is close to dogs (34%) and mice (35%) (Waterston et al., 2002; Lindblad-Toh et al., 2005). Most of the repetitive DNA is present in the form of transposon derived repeats in bactrian camels. It was observed that long scattered elements are similar to other mammals whereas short interspersed elements have a lower frequency in bactrian camels when compared to other mammals. Based on these results it was suggested that this might be one of the reasons why the camel has a smaller genome size than other mammals (Jirimutu et al., 2012). Interestingly, primate genomes have multiple copies of Alu repeats. However, none exists in the bactrian camel genome. Additionally, 244,141 microsatellites or simple repeats loci were found in the camel genome which could be helpful in marker assisted selection among camels and quantitative trait locus mapping. A comparison of the features of camel genomes and other species are presented in Tables 3, 4.
Assessment of genetic variability among livestock species is critical to the regulation of genetic resources for their sustainable conservation and utilization. Characterization of genetic diversity within and among different breeds/populations is the prime strategy for management of animal biodiversity, mostly used in recognizing genetically unique structure (Excoffier and Lischer, 2010). Such steps are very important in the case of camels because their numbers have shown a rapid decline in the last decade. An innovative method of genome selection using dense marker maps covering all chromosomes suggested by Meuwissen and co-workers and the advent of next generation sequencing technologies have revolutionized the application of genetic information in livestock breeding programs (Hayes and Goddard, 2001). The proper implementation of genomic selection programs in different livestock species have produced remarkable results with regard to increasing the rate of genetic improvement (Hayes et al., 2009; Duchemin et al., 2012). Moreover, the estimation of large-scale genetic variation, particularly SNP markers, using whole genome sequencing could lead to the development of methods such as genome-wide association studies and genomic selection in camel breeding. Notwithstanding recent studies on camels at the genomic level, numerous genetic variants in different camel breeds remain to be identified and correctly annotated.
Heterozygosity is of significant interest in the study of genetic variations in populations. Jirimutu et al. (2012) conducted a study to determine heterozygous SNPs in wild and domestic camel genomes. The heterozygosity rate was calculated to be around 1.0 × 10-3 across the whole genome in both cases. Additionally, small indel number was also comparable between the two genomes. The authors further classified the SNPs in terms of gene annotations and measured the heterozygosity rates for coding and non-coding regions. Interestingly, domestic camel genome contained lower heterozygosity rate in the exonic region as compared to wild camel genome. This seems to support the idea of artificial selection of certain genes in the domestic species since strong artificial selection would reduce genetic diversity around specific loci (Doebley et al., 2006). The authors reported 2,816 regions containing 196 genes where the heterozygosity rate of the domestic camel was significantly lower than the wild one. Such genes were highly enriched in membrane receptors, particularly olfactory receptor activity, based on gene ontology analysis. Therefore, it was suggested that olfaction may be an important object of artificial selection during the domestication of bactrian camels. Further, Vijh et al. (2007) determined genetic variability and relationship among four camel breeds (Bikaneri, Jaisalmeri, Kutchi, and Mewari) using 23 microsatellite loci. A total of 252 alleles were detected across all the four populations. Interestingly, the calculated heterozygosity for the four breeds was 0.58 (Bikaneri), 0.57 (Jaisalmeri), 0.56 (Kutchi), and 0.60 (Mewari), which were lower than the expected heterozygosity value. Apart from this, mitochondrial DNA (mtDNA) has also been used to determine the genetic diversity and phylogeography in many animals (Chen et al., 2005; Lai et al., 2006). Researchers studied an 809 bp mtDNA fragment in 113 individuals from seven Chinese breeds, one Russian breed, three Mongolian breeds, two hybrid individuals and one wild breed to measure the genetic diversity and phylogeography of bactrian camel populations (Ming et al., 2017). They found 15 different haplotypes and the phylogenetic analysis proposed that domestic and wild bactrian camels belonged to two distinct lineages. Molecular variance analysis further suggested that wild and bactrian camels have different maternal origin. However, no significant genetic divergence was observed among domestic bactrian camels that belonged to different geographical locations. These findings implied that genetic admixture and gene flow might exist between these domesticated bactrian camels.
A recent study reported the first whole genome resequencing of individual camels from two distinct geographical regions (Khalkhali-Evrigh et al., 2018). In this study 692,908 indels and 4,727,238 SNPs were identified in the two camels. It was observed that most SNPs and indels were located in intergenic region based on in silico functional analysis. Additionally, the authors found 15,168 non-synonymous SNPs which were common to the three breeds (Yazd, Trod, and African dromedary) that could affect gene function and protein structure. In spite of this, much more needs to be done to improve our understanding of the camel genome and its role in breeding and genomic selection.
Besides physiological studies, genomic and transcriptomic analyses have recently unraveled the peculiarities of the unusual adaptations in camels (Jirimutu et al., 2012; Wu et al., 2014). Studies have investigated the role of ‘rapidly evolving genes’ in species differentiation and adaptation in camels (Kasahara et al., 2007; Muyldermans et al., 2009; Jirimutu et al., 2012; Wang et al., 2012). Rapid divergence of protein-coding genes are normally calculated by an increased ratio of non-synonymous-to-synonymous substitutions (dN/dS) (Jirimutu et al., 2012). Jirimutu et al. (2012) identified around 2,730 significantly faster evolving genes in camels than its closest cattle orthologs. These genes were enriched in metabolic pathways such as carbohydrate and lipid metabolism, insulin signaling pathways and adipocytokine signaling pathways. They hypothesized that these genes might have helped the camel to optimize their energy storage and production in the desert. Generally, monogastric animals have high blood glucose levels (3.5–5.0 mmol/l) than ruminants (2.5–3.5 mmol/l) (Elmahdi et al., 1997). The camel is a ruminant herbivores with an extensive forestomach. However, it has a high blood glucose level (6–8 mmol/l) when compared to other mammals. The results suggest that rapidly evolving genes like CYP2E and CYP2J could be involved in type II diabetes mellitus (Jirimutu et al., 2012). Two critical genes in the insulin signaling pathways – PI3K and AKT – have undergone rapid divergence in camels which could have changed their response to insulin (Wang et al., 2012). This finding strongly supports previously reported physiological experiments that demonstrated that high glucose level in camel blood is due to their strong insulin resistance (Kaske et al., 2001). The distribution of cytochrome P450 (CYP) genes, which are involved in the arachidonic acid metabolism were found to be quite different in camels when compared to other mammals. Genome sequence analysis of bactrain camels found a higher number of copies of the cytochrome P450 (CYP) genes such as CYP2J (11 copies) and CYP2E (2 copies) in camels when compared to closely related mammals and humans. But CYP4A (one copy) and CYP4F (two copies) genes were fewer than other mammals (Jirimutu et al., 2012). CYP2E and CYP2J help to transform arachidonic acid into 19(S)-hydroxy-eicosatetraenoic acid [19(S)-HETE], whereas CYP4F and CYP4A transform it into 20-HETE (Wang et al., 2012). 19(S)-HETE is a potent vasodilator of renal preglomerular vessels that stimulate water reabsorption and is potentially useful for the survival in deserts (Carroll et al., 1996). In addition, they also reported that multiple copies of CYP2J genes give them the ability to take large amount of salt without developing hypertension (Wang et al., 2012). The activity of CYP2J2 is regulated by high-salt diet and the suppression of this gene can lead to high blood pressure (Zhao et al., 2003). A study on the effects of dehydration on camel plasma levels of sodium and water-retaining hormone systems including the renin-angiotensin system, aldosterone and antidiuretic hormone, dehydration identified significant increase in serum sodium, creatinine, urea and plasma arginine vasopressin (AVP) levels while plasma aldosterone level was altered very little (Ali et al., 2012). These results agree with earlier reports that suggested that renin-angiotensin system is important for the maintenance of water balance during dehydration. Studies have also reported an increase of serum urea and creatinine levels during dehydration (Siebert and Macfarlane, 1971; Finberg et al., 1978). Camels are also able to withstand starvation while maintaining a persistent nitrogen level through urea-nitrogen recycling (Mousa et al., 1983).
Proteomic studies of dromedary organs have also been undertaken to explain a number of cellular mysteries related to surviving in arid conditions. The overexpression of α-actinin in the heart suggested an ability to adapt to fluctuations in blood concentration associated with alternative drought-rehydration periods. An increased expression of H+-ATPase in the brain is believed to provide a rapidly usable supply of energy (Warda et al., 2014).
Guanidinoacetate methyltransferase in the liver has a regulatory effect on high energy phosphate. It is a key enzyme involved in creatine phosphate synthesis, which is an important energy currency of the cell and has a protective role on Na+ and K+-ATPase. The hump fat tissue and kidney protein expression have been shown to favor cellular acidosis (Warda et al., 2014). Hump fat contained more proteins, well developed cytoskeleton and high levels of vimentin. The role of vimentin in the signal transduction pathway from a specific adrenoreceptor, β3 adrenoceptor (β3AR), to the activation of extra cellular signal regulated kinase (ERK) and its role in lipolysis make them an early marker of adipogenesis (Kumar et al., 2007). Additionally, the high level of glucagon in camel, with subsequent elevated basal blood glucose, is consistent with the role of vimentin in glucose transporter induced glucose adipocyte transport (Abdel-Fattah et al., 1999; Guilherme et al., 2000). These results strongly support the modulatory role of adipocyte vimentin as a reason for the tolerance of high blood glucose level in camels. Vimentin might operate as an inducer of a cellular trap for glucose and the abundance of vimentin in camel adipocytes could aid the morphology of the hump of the well-nourished camel and could be an adaptation for survival in arid conditions (Warda et al., 2014).
Camel hemoglobin is an interesting case study of adaptation to extremely high temperatures (Oyewale et al., 2011). Camel blood has a high concentration of glucose. However, hemoglobin (Hb) exhibits low glycation and exhibits higher electrophoretic mobility than hemoglobin from cattle or humans (Bazzi et al., 2013). Glycated hemoglobin A1C (HbA1C) is normally associated with diabetes. Glycosylated hemoglobin is formed by non-enzymatic attachment of glucose to N-terminal valine and internal lysine amino groups of hemoglobin (Rohlfing et al., 2002; Soranzo, 2011). The reaction between Hb and glucose is slow and irreversible (Higgins, 2012). The process of glycosylation in eukaryotes is based on the interaction between amino acid and glucose and could be categorized into five types: O-linked, C-linked, N-linked, P-linked, and G-linked (Chauhan et al., 2013). There is a direct relationship between HbA1C and blood glucose level in human and most other animals (Nathan et al., 2008; Bazzi et al., 2013). However, camels behave differently in the presence of high blood glucose. An in silico analysis on hemoglobin characteristics reported that hemoglobin beta chain (HBB) is resistant to N- and O-linked glycosylation especially in camels whereas, hemoglobin alpha chain (HBA) is susceptible to O-linked glycosylation. It has been reported that these factors, together with other post-translational modification, might be responsible for the protection of Hb from glycosylation (Thanka Christlet and Veluraja, 2001; Borai et al., 2011).
The unusual genetic architecture of camel is the reason behind its survival in harsh environmental conditions. The regulation of different genes in response to different stresses are summarized in Table 5.
Pastoralist survival in the desert relies heavily on camels. Recent advancement in genetic engineering, mainly in livestock and the incorporation of concepts of selective breeding with desired phenotypes, has made camel rearing a profitable profession in numerous countries (Beuzen et al., 2000; Olesen et al., 2000). SNPs in the camel genome play a vital role in the development of useful traits. Such genetic variations are listed in Table 6.
Camel sport is an integral part of communities in the Middle East region. Many individuals adopt rearing of racing camels as a profession and regular events are organized. The demand for special breeds of racing camels, with higher endurance and resilience, is quite high. Developing novel techniques to measure oxidative stress and endurance in racing camels has very high potential (Wilson, 1999). Several regions have declared camel as their national animal and organizes events at the national level to promote and highlight camel rearing among local residents.
The mitochondrial DNA (mtDNA) of dromedary camel is 16643 bp in size and encodes 23 subunits of the electron transfer chain associated with the production of cellular adenosine triphosphate (ATP) through oxidative phosphorylation (Cui et al., 2007). The cells that require a lot of energy in terms of ATP, such as neurons and muscles, maintain high copy number of mtDNA (Dickinson et al., 2013). Polyacrylamide gel electrophoresis was used to analyze malate dehydrogenase (Mdh) and malic (ME) isoenzymes in Arabian camel for energy production and racing. Results indicated the necessity of the mitochondrial Mdh-2 for energy production in racing breed and cytosolic Mdh-1 for lipogenesis and energy production in both breeds (Al-Harbi and Amer, 2012). Likewise, the ratio of mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) is also considered as a good predictor of the metabolic status of the tissue. This ratio was found to be high in racing camels when compared to dairy camels (Soman and Tinson, 2016).
Camels are relatively more resistant to major infectious diseases when compared to other livestock inhabiting the same geographical area. However, its immunogenome has not been well studied so far (Wernery and Kaaden, 2004; Muyldermans et al., 2009; Burger, 2016). The immune system of camels includes a unique heavy chain antibody homodimer. Camels are the only mammals that can produce heavy-chain antibodies (HCAbs) that lack the light chain (Hamers-Casterman et al., 1993; Tillib et al., 2014). However, little is known about the major histocompatibility complex (MHC) region of the camel genome (Antczak, 2013). The genomic MHC region contains immune response (IR) genes that play a crucial role in host–pathogen interactions (Janeway et al., 2001). Plasil and co-workers identified, mapped and characterized the MHC region of Old World camels.
The study investigated genomic localization, organization and sequence similarity of MHC genes and showed that camels possess MHC genes comparable to other mammals. Their results suggest that molecular diversity of MHC class II genes are significantly lower in Old World camels than in other mammals and the major part of the diversity resides in the DQB gene, which is not very well annotated in the camel genome (Plasil et al., 2016). MHC class I and class II genes contain antigen presenting molecules which are responsible for recognition of antigenic peptides expressed on the cell surface (Janeway et al., 2001). Other IR genes such as T cell receptors have evolved in camels due to mutations in the gamma (TRG) and delta (TRD) genes. Their genomic structure and organization have also been investigated (Ciccarese et al., 2014; Burger, 2016). New technology based on next generation sequencing could be used to probe and analyze the adaptive immune system. Millions of immunoglobulin sequences and T cell receptors from a single sample could be analyzed by amplifying in a single multiplex PCR reaction (Li et al., 2015). A study, which evaluated the sequence diversity of the constant and variable regions present in the 15 conventional heavy, 18 kappa and 35 lambda light chains of C. dromedarius and C. bactrianus by using standard Sanger sequencing, found that the functional sequences for these chains have been rearranged (Griffin et al., 2014).
The unique features of the camel genome and proteome not only enable them to survive and thrive under harsh environmental conditions but also make them less susceptible to various pathological conditions. Camels employ several intrinsic immunological and molecular mechanisms against pathogenic agents and pathological conditions (Fazil and Hofmann, 1981).
Therapeutic antibodies are an important treatment option against several auto-immune, infectious and malignant diseases. Monoclonal antibodies (mAb) are preferred in targeted drug delivery because they are relatively cheaper and have high specificity, stability and potency (Finlay and Almagro, 2012). Three types of immunoglobulins (IgA, IgG, and IgM) were discovered and separated in camels (Grover et al., 1983; Azwai et al., 1993). Conventional γ-immunoglobulins consist of two heavy chains, each containing three invariant (constant) domains (CH1–CH3), a single variable region (VH), and two light chains composed of a constant region (CL) and a variable region (VL). The VH and VL regions together form the paratope and it is the antigen-recognizing portion of the antibody (Jerne, 1955). Interestingly, camel antibodies are different from other animals. In addition to its conventional heterotetrameric structure (two heavy and two light chains), camels also possess non-conventional antibodies which are smaller (Hamers-Casterman et al., 1993). These immunoglobulins are now known as heavy-chain antibodies (HCAbs). Non-conventional 2-chain IgG structures are devoid of the light chain and heavy chain constant region CH1 (Harmsen and De Haard, 2007). Structural representations of the different antibodies are shown in Figure 3. The antigen binding site of HCAb is a single variable domain denoted as VHH (variable domain of camel heavy-chain-only antibody) (Muyldermans, 2001; De Genst et al., 2006; Deschacht et al., 2010). VHHs have the ability to penetrate and target small antigenic sites that conventional antibodies are unable to recognize (Paalanen et al., 2011). Moreover, HCAbs are usually absent in other mammals except some primitive fish (Nuttall et al., 2001). The new antigen receptor (NAR) and the Cos5 antibodies are some examples of antibodies obtained from sharks (Greenberg et al., 1995). Camels and some primitive fish possess natural antibodies composed only of heavy chains. The antigen binding site is formed by only one single domain, referred to as VHH (Zielonka et al., 2014). Antibody derived VHHs have gained importance because of its low size (15 kDa), stability, binding abilities and enhanced potency (Vincke et al., 2012). Additionally, the production of such antibodies in camels are easier than other animals (Alvarez-Rueda et al., 2007). The camel germline IGHV, IGLV, and IGKV genes show high degree of sequence homology with human counterparts (Achour et al., 2008; Klarenbeek et al., 2015). Therefore, active immunization of camels with human target proteins results in a better diversity of potential therapeutic antibodies due to the presence of more foreign epitopes (Tainer et al., 1985). This degree of foreignness is best exploited by immunizing camels against viral envelope proteins which results in a diverse panel of binders, allowing targeting of multiple epitopes on the same envelope protein (Hultberg et al., 2011; McCoy et al., 2012). The potential uses of camel derived antibodies are mentioned in Table 7.
Several studies have explored the structure and discovery of nanobodies, as well as their applications in immunity, oncology, neurodegenerative diseases, infections, and other ailments (Wesolowski et al., 2009; Altintas et al., 2012; Steeland et al., 2016; Beghein and Gettemans, 2017). VHHs are highly suitable for oral immunotherapy due to their resistance to extremes of pH and their ability to bind to the target at high concentrations of chaotropic agents (Harmsen and De Haard, 2007). Furthermore, due to their binding specificities, they are considered excellent modulators or inhibitors of receptors, enzymes or viruses (Desmyter et al., 2013; Hassaine et al., 2014). Camel nanobodies exhibit properties that are very useful in crystallizing mobile proteins (Ward et al., 2013). It can also be used to stabilize membrane protein structures. Nanobodies with high isoelectric point (pI) have the potential to cross the blood brain barrier (BBB) and, in future, could be adapted for brain imaging purposes (Li et al., 2012). The current limitation is that a large amount of nanobody needs to be injected to achieve the desirable effect. G protein-coupled receptors (GPCRs) are normally stabilized in the presence of G protein. Current crystal structures provide insights into the inactive states of several GPCRs. It is difficult to obtain an agonist-bound active-state GPCR structure due to the inherent instability of this state in the absence of a G protein. Nanobodies have the potential to stabilize GPCR in the absence of G protein (Steyaert and Kobilka, 2011). Studies have employed nanobodies as a stabilizing agent to achieve the stabilization of agonist-bound form of the beta-2 adrenoreceptor (b2Ar) and human M2 muscarinic acetylcholine receptor (Rasmussen et al., 2011; Kruse et al., 2013). Comparison with the inactive structure reveals subtle changes in the binding pocket. Therefore, such structures provide insights into the process of agonist binding and activation.
Hepatitis C virus (HCV) is a major health concern and is the main cause of chronic liver disease (Williams, 2006). It is estimated that over 180 million people suffer from this disease worldwide (Nguyen and Keeffe, 2005). The prevalence of HCV is likely to increase in the near future (Deuffic-Burban et al., 2007). Since no protective vaccine is available, the old treatment regimen of interferon alpha alone or in combination with ribavirin is currently widely adopted.
Lactoferrin (Lf) is a multifunctional glycoprotein that is usually present in milk. Its molecular weight is 80 kDa and it belongs to the transferrin family (Legrand et al., 2008). Lf also has a role as part of the innate immune system. Additionally, its antibacterial and antiviral potential have also been reported (Legrand et al., 2005). Lf has shown promising results against herpes simplex virus 1 and 2 (HSV-1 and HSV-2) (Marchetti et al., 1998). Lf also exhibits antiviral activity against HCV. The binding of human and bovine lactoferrin with envelope proteins of HCV inhibits the interaction of HCV virus with cellular receptors (Yi et al., 1997; Ikeda et al., 1998). These studies have shown that Lf shows its antiviral activity in the early stage of infection. Camel lactoferrin (cLf) inhibited the entry of HCV when cLf and HCV were preincubated (Redwan el and Tabll, 2007). More specifically, the C- and N-lobes of camel lactoferrin inhibits the activity of HCV (Redwan et al., 2014). Heparan sulfate (HS) is present on the cell surface of many animals. A host’s HS serves as a receptor for several pathogens. HS aids antiviral activity and binding of lactoferrin (Jenssen et al., 2004; Marchetti et al., 2004). It was demonstrated that camel lactoferrin has a higher antiviral activity against HCV than human, sheep and bovine lactoferrin (El-Fakharany et al., 2013). Apart from lactoferrin, camel milk casein has also shown promising activity against HCV. Furthermore, it has also been shown to possess apoptotic potential (Almahdy et al., 2011).
Cancer is a leading health problem worldwide. Chemotherapy is still considered as a fundamental treatment modality irrespective of its toxic side effects and high morbidity (Rood et al., 2004). Most of the drugs used for the management of cancers are derived from plant sources (Craig, 1997). Camel urine is a natural product used for the management of several diseases in the Arabian region. Cancer patients usually drink (100 ml/day) camel urine alone or mixed with milk. Camel urine is devoid of bad odor and toxicity due to low urea and lack of ammonia. Additionally, camel urine is basic (pH > 7.8), while human urine could be weakly acidic or weakly basic (Read, 1925). Research on camel urine has shown that it has antifungal and antibacterial activity and is able to protect the liver from CCL4 induced damage (Al-Bashan, 2011; Alzahrani and Alharbi, 2011). Gastroprotective and ulcer healing effects of camel urine have also been reported (Hu et al., 2017). Camel urine has potential activity as antiplatelet and anticancer agents as well (Alhaidar et al., 2011). Camel urine inhibits the induction of CYP1A1 gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). TCDD is a potent CYP1A1 inducer and a well-known carcinogen. This depicts the transcriptional regulation in which the binding of TCDD to a cytosolic transcription factor, the aryl hydrocarbon receptor (AhR), is the first step in a series of cellular events leading to carcinogenesis and mutagenesis. Research on camel urine has shown that camel urine, but not bovine, inhibits TCDD-mediated toxic effect by inhibiting the expression of CYP1A1 at both transcriptional and post-transcriptional levels (Alhaider et al., 2011). Good anticancer agents activate cell death or inhibit proliferation of tumor cells without affecting the growth of normal cells. Fortuitously, camel urine presents all these features (Al-Yousef et al., 2012). Anticancer activity of camel urine has also been demonstrated using GC-MS and ICP-MS methods (Ahamad et al., 2017). Studies using GC-MS and ICP-MS have shown a marked difference in urinary metabolites produced by camels. Camel urine metabolites like canavanine are also excreted by other mammals but the quantity is low when compared to camel. Canavanine, an arginine analog, is a by-product of amino acids and urea metabolism and it has been shown to possess potent activity against tumor cells. Camel urine has also displayed antimetastatic effect on breast cancer cells (Romli et al., 2017). Reported pharmacological activity of camel urine on different genes/proteins are listed in Table 8. A schematic diagram providing an overview of proteins and pathways affected by camel urine is given in Figure 4.
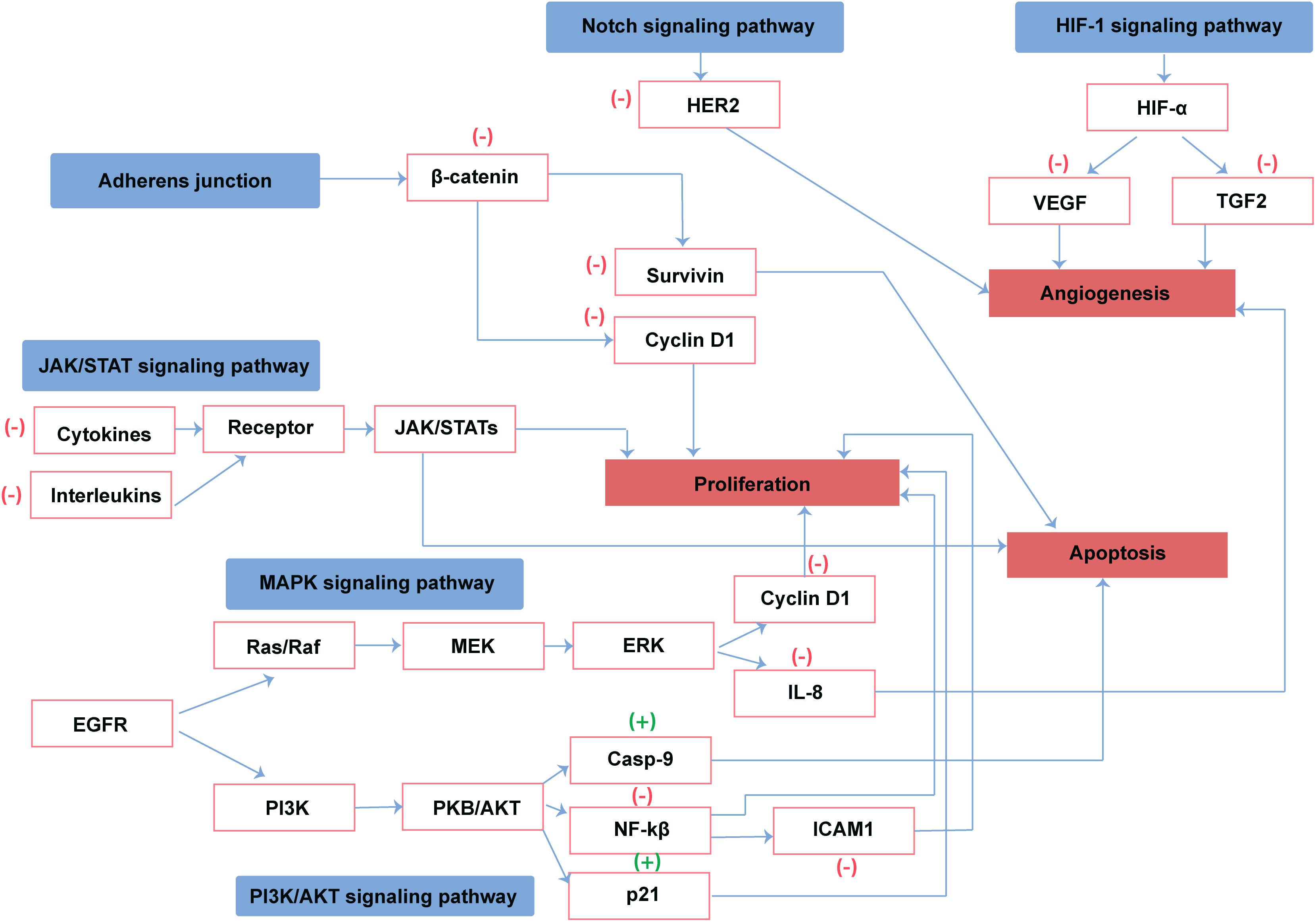
Figure 4. Reported direct and indirect effect of camel urine on proteins involved in cancer pathways. (+) represents activation and (–) represents the inhibition of the protein by camel urine.
Diabetes mellitus (DM) is a group of metabolic disorders characterized by an elevated level of blood glucose. It results from the inability of body to produce enough insulin (type 1 diabetes) or the failure to respond efficiently to the insulin produced by the pancreatic beta cells (type 2 diabetes) (Diamond, 2003). Worldwide prevalence of DM is expected to be 4.4% by 2030 (Wild et al., 2004). Metabolic control of hyperglycemia can be achieved through physical activity, diet with or without antidiabetic agents, which will help to prevent the likely risk of chronic complications (Lindstrom et al., 2006). Camel milk has exceptional biochemical properties (Wernery et al., 2006). The unique constituents of camel milk and their stability at a range of physiological conditions are different from other mammals (Shamsia, 2009). Camel milk has been used for a long time for a number of ailments (Yagil, 1982). One of its major roles is in controlling hyperglycemia in type 1 diabetes (Agrawal et al., 2005; Khan et al., 2013). Camel milk does not form coagulum in acidic environment and has a high buffering capacity. However, milk of other mammals readily forms coagulum in acid milieu (Wangoh, 1993). It was observed that camel milk protein has insulin-like properties and the lack of coagulum formation in the stomach allows it to pass rapidly and remains available for absorption in the intestine (Beg et al., 1986). Clinical studies showed that administration of camel milk as adjunct therapy with insulin to patients of type 1 diabetes reduced insulin requirement by up to 30% (Agrawal et al., 2007). Camel milk contains insulin like proteins which potentiates its interaction with insulin receptors (Mehaia et al., 1995; Malik et al., 2012). Studies have been conducted to determine the cellular and molecular mechanisms behind the insulin like properties of camel milk and its ability to control hyperglycemia (Agrawal et al., 2011). However, the mechanism has not been clearly elucidated yet. By using bioluminescence resonance energy transfer (BRET), it was observed that camel milk activated the human insulin receptor (hIR). Though it had no direct insulin like effect, it significantly potentiated the effect of insulin when pre-treated with camel milk (Abdulrahman et al., 2016). Camel milk also has a different content of casein, higher amount of vitamin B3 and lipid micelles that could enhance the body’s defense against free radicals (Al-Humaid et al., 2010).
The complications of DM, like neurological and vascular alterations, may worsen in the presence of high oxidative stress (Varvarovska et al., 2004). Normally, antioxidant enzymes such as catalases, superoxide dismutase (SOD), and glutathione peroxidase are responsible for scavenging reactive oxygen species (West, 2000). Camel milk has also shown antioxidant activity most probably by its chelating effects on toxicants (Shori, 2015). Another complication of DM is delayed wound healing. The presence of infectious agents prevents normal healing of wound. Camel milk contains whey protein and lacks beta-lactoglobulin. It exhibits a higher antioxidant activity than bovine milk and other whey proteins due to a high content of antioxidant amino acids (Cys, Met, Trp, Tyr, and Phe) (Marshall, 2004; Salami et al., 2010). Therefore, camel whey protein speeds up healing by increasing the immune response of wounded tissue cells (Badr, 2013). Camel milk also comprises of different proteins such as serum albumin, lactophorin, peptidoglycan recognition protein, and alpha lactalbumin protein (Kappeler et al., 2004). These proteins bind to lactic acid bacteria and other gram-positive bacteria with an affinity similar to that reported for the human and murine orthologs.
The camel has a special status among mammals domesticated by humans since they are highly adapted to the extreme desert ecosystem. It is a multipurpose animal used for dairy production, racing and transportation. The genetic makeup and variations in its genome allow them to survive these harsh conditions. The involvement and identification of key genes in the adaptation to desert environment may have applications in breeding programs. It may also provide insights into disease resistance in the future. Knowledge of the genetic differences between different types of camels may also assist in screening them for specific purposes like sports or dairy. Elucidation of the therapeutic role of camel products is ongoing. Clearly, further studies are required to decipher the precise molecular mechanisms that underpin their role in disease pathogenesis and the ability to stimulate the immune system.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by a United Arab Emirates University UPAR grant 31S243 to RV.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdallah, H. R., and Faye, B. (2012). Phenotypic classification of saudi arabian camel (Camelus dromedarius) by their body measurements. Emir. J. Food Agric. 24, 272–280.
Abdel-Aziem, S., Abdel-Kader, H., Alam, S. S., and Othman, O. E. (2015). Detection of MspI polymorphism and the single nucleotide polymorphism (SNP) of GH gene in camel breeds reared in Egypt. Afr. J. Biotechnol. 14, 752–757. doi: 10.5897/AJB2014.14374
Abdel-Fattah, M., Amer, H., Ghoneim, M. A., Warda, M., and Megahed, Y. (1999). Response of one-humped camel (Camelus dromedarius) to intravenous glucagon injection and to infusion of glucose and volatile fatty acids, and the kinetics of glucagon disappearance from the blood. Zentralbl. Veterinarmed. A 46, 473–481. doi: 10.1046/j.1439-0442.1999.00237.x
Abdelrahman, M. M., Aljumaah, R. S., and Ayadi, M. (2013). Selenium and iodine status of two camel breeds (Camelus dromedaries) raised under semi intensive system in Saudi Arabia. Ital. J. Anim. Sci. 12:e14. doi: 10.4081/ijas.2013.e14
Abdulrahman, A. O., Ismael, M. A., Al-Hosaini, K., Rame, C., Al-Senaidy, A. M., Dupont, J., et al. (2016). Differential Effects of camel milk on insulin receptor signaling - toward understanding the insulin-like properties of camel milk. Front. Endocrinol. 7:4. doi: 10.3389/fendo.2016.00004
Abdussamad, A., Charruau, P., Kalla, D., and Burger, P. (2015). Validating local knowledge on camels: colour phenotypes and genetic variation of dromedaries in the nigeria-niger corridor. Livest. Sci. 181, 131–136. doi: 10.1016/j.livsci.2015.07.008
Achour, I., Cavelier, P., Tichit, M., Bouchier, C., Lafaye, P., and Rougeon, F. (2008). Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J. Immunol. 181, 2001–2009. doi: 10.4049/jimmunol.181.3.2001
Af El-Kholy, M. Z., Shehata, M. F., Salem, M. A. I., El-Bahrawy, K. A., El-Halawany, N., and Hassanane, M. S. (2016). Association of single nucleotide polymorphisms for myogenic factor 5 and growth hormone genes with meat yield and quality traits in one humped camel (Camelus dromedarius). Asian J. Anim. Vet. Adv. 11, 263–271. doi: 10.3923/ajava.2016.263.271
Afifi, M., Metwali, E. M., and Brooks, P. H. (2014). Association between growth hormone single nucleotide polymorphism and body weight in four saudi camel (Camelus dromedarius) breeds. Pak. Vet. J. 34, 494–498.
Agrawal, R., Beniwal, R., Kochar, D., Tuteja, F., Ghorui, S., Sahani, M., et al. (2005). Camel milk as an adjunct to insulin therapy improves long-term glycemic control and reduction in doses of insulin in patients with type-1 diabetes: a 1 year randomized controlled trial. Diabetes. Res. Clin. Pract. 68, 176–177. doi: 10.1016/j.diabres.2004.12.007
Agrawal, R. P., Jain, S., Shah, S., Chopra, A., and Agarwal, V. (2011). Effect of camel milk on glycemic control and insulin requirement in patients with type 1 diabetes: 2-years randomized controlled trial. Eur. J. Clin. Nutr. 65, 1048–1052. doi: 10.1038/ejcn.2011.98
Agrawal, R. P., Saran, S., Sharma, P., Gupta, R. P., Kochar, D. K., and Sahani, M. S. (2007). Effect of camel milk on residual beta-cell function in recent onset type 1 diabetes. Diabetes Res. Clin. Pract. 77, 494–495. doi: 10.1016/j.diabres.2007.01.012
Ahamad, S. R., Alhaider, A. Q., Raish, M., and Shakeel, F. (2017). Metabolomic and elemental analysis of camel and bovine urine by GC-MS and ICP-MS. Saudi J. Biol. Sci. 24, 23–29. doi: 10.1016/j.sjbs.2015.09.001
Al-Ali, A. K., Husayni, H. A., and Power, D. M. (1988). A comprehensive biochemical analysis of the blood of the camel (Camelus dromedarius). Comp. Biochem. Physiol. B. 89, 35–37. doi: 10.1016/0305-0491(88)90257-X
Al-Atiyat, R. M., Suliman, G., AlSuhaibani, E., El-Waziry, A., Al-Owaimer, A., and Basmaeil, S. (2016). The differentiation of camel breeds based on meat measurements using discriminant analysis. Trop. Anim. Health Prod. 48, 871–878. doi: 10.1007/s11250-015-0990-5
Al-Bashan, M. M. (2011). In vitro assessment of the antimicrobial activity and biochemical properties of camel’s urine against some human pathogenic microbes. Middle East J. Sci. Res. 7, 947–958.
Alhaidar, A., Abdel Gader, A. G., and Mousa, S. A. (2011). The antiplatelet activity of camel urine. J. Altern. Complement. Med. 17, 803–808. doi: 10.1089/acm.2010.0473
Alhaider, A. A., Abdel Gader, A. G., Almeshaal, N., and Saraswati, S. (2014). Camel milk inhibits inflammatory angiogenesis via downregulation of proangiogenic and proinflammatory cytokines in mice. APMIS 122, 599–607. doi: 10.1111/apm.12199
Alhaider, A. A., El Gendy, M. A., Korashy, H. M., and El-Kadi, A. O. (2011). Camel urine inhibits the cytochrome P450 1a1 gene expression through an AhR-dependent mechanism in Hepa 1c1c7 cell line. J. Ethnopharmacol. 133, 184–190. doi: 10.1016/j.jep.2010.09.012
Al-Harbi, M. S., and Amer, S. A. (2012). Comparison of energy-related isoenzymes between production and racing Arabian camels. Adv. Biosci. Biotechnol. 3, 1124–1128. doi: 10.4236/abb.2012.38138
Al-Humaid, A., Mousa, H., El-Mergawi, R., and Abdel-Salam, A. (2010). Chemical composition and antioxidant activity of dates and dates-camel-milk mixtures as a protective meal against lipid peroxidation in rats. Am. J. Food Technol. 5, 22–30. doi: 10.3923/ajft.2010.22.30
Ali, M. A., Adem, A., Chandranath, I. S., Benedict, S., Pathan, J. Y., Nagelkerke, N., et al. (2012). Responses to dehydration in the one-humped camel and effects of blocking the renin-angiotensin system. PLoS One 7:e37299. doi: 10.1371/journal.pone.0037299
Ali, T. (1994). A Manual for the Primary Animal Health Care Worker. A Manual for the Primary Animal Health Care Worker. Rome: FAO.
Almahdy, O., El-Fakharany, E. M., El-Dabaa, E., Ng, T. B., and Redwan, E. M. (2011). Examination of the activity of camel milk casein against hepatitis C virus (genotype-4a) and its apoptotic potential in hepatoma and hela cell lines. Hepat. Mon. 11, 724–730. doi: 10.5812/kowsar.1735143X.722
Almathen, F., Charruau, P., Mohandesan, E., Mwacharo, J. M., Orozco-terWengel, P., Pitt, D., et al. (2016). Ancient and modern DNA reveal dynamics of domestication and cross-continental dispersal of the dromedary. Proc. Natl. Acad. Sci. U.S.A. 113, 6707–6712. doi: 10.1073/pnas.1519508113
Al-Swailem, A. M., Shehata, M. M., Abu-Duhier, F. M., Al-Yamani, E. J., Al-Busadah, K. A., Al-Arawi, M. S., et al. (2010). Sequencing, analysis, and annotation of expressed sequence tags for Camelus dromedarius. PLoS One 5:e10720. doi: 10.1371/journal.pone.0010720
Altintas, I., Kok, R. J., and Schiffelers, R. M. (2012). Targeting epidermal growth factor receptor in tumors: from conventional monoclonal antibodies via heavy chain-only antibodies to nanobodies. Eur. J. Pharm. Sci. 45, 399–407. doi: 10.1016/j.ejps.2011.10.015
Alvarez-Rueda, N., Behar, G., Ferré, V., Pugniere, M., Roquet, F., Gastinel, L., et al. (2007). Generation of llama single-domain antibodies against methotrexate, a prototypical hapten. Mol. Immunol. 44, 1680–1690. doi: 10.1016/j.molimm.2006.08.007
Al-Yousef, N., Gaafar, A., Al-Otaibi, B., Al-Jammaz, I., Al-Hussein, K., and Aboussekhra, A. (2012). Camel urine components display anti-cancer properties in vitro. J. Ethnopharmacol. 143, 819–825. doi: 10.1016/j.jep.2012.07.042
Alzahrani, S. H., and Alharbi, A. A. (2011). Antimicrobial activity of camel’s urine on methicillin-resistant staphylococcus aureus isolated from clinical specimens. Science 23, 251–268.
Amine, C. Y., Samir, G. S. B., Nasreddine, M., Nacera, T. A., and Nadhira, S.-M. (2013). Study of camelina biodiversity in southwestern of algeria. J. Life Sci. 7:416.
Andersson, B., Olsson, K., and Rundgren, M. (1980). ADH in regulation of blood osmolality and extracellular fluid volume. J. Parenter. Enteral Nutr. 4, 88–96. doi: 10.1177/014860718000400207
Andersson, L., and Georges, M. (2004). Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet. 5, 202–212. doi: 10.1038/nrg1294
Antczak, D. (2013). “Major histocompatibility complex genes of the dromedary camel,” in Proceedings of the Qatar Foundation Annual Research Conference, (Doha: BIOP), 15. doi: 10.5339/qfarf.2013.BIOP-015
Azwai, S. M., Abdouslam, O. E., Al-Bassam, L. S., Al Dawek, A. M., and Al-Izzi, S. A. L. (2007). Morphological characteristics of blood cells in clinically normal adult llamas (Lama glama). Vet. Arh. 77, 69–79.
Azwai, S. M., Carter, S. D., and Woldehiwet, Z. (1993). The isolation and characterization of camel (Camelus dromedarius) immunoglobulin classes and subclasses. J. Comp. Pathol. 109, 187–195. doi: 10.1016/S0021-9975(08)80262-9
Badr, G. (2013). Camel whey protein enhances diabetic wound healing in a streptozotocin-induced diabetic mouse model: the critical role of β-Defensin-1,-2 and-3. Lipids Health Dis. 12:46. doi: 10.1186/1476-511X-12-46
Balhuizen, A., Massa, S., Mathijs, I., Turatsinze, J. V., De Vos, J., Demine, S., et al. (2017). A nanobody-based tracer targeting DPP6 for non-invasive imaging of human pancreatic endocrine cells. Sci. Rep. 7:15130. doi: 10.1038/s41598-017-15417-2
Barnes, K., Ingram, J. C., Porras, O. H., Barros, L. F., Hudson, E. R., Fryer, L. G., et al. (2002). Activation of GLUT1 by metabolic and osmotic stress: potential involvement of AMP-activated protein kinase (AMPK). J. Cell Sci. 115(Pt 11), 2433–2442.
Bazzi, M. D., Quazani, M., Rabbani, N., Malik, A., Al, H. M., Elrobh, M. S., et al. (2013). Glycated hemoglobin in camel: minimal correlation with blood glucose level. Arch. Biol. Sci. 65, 911–917. doi: 10.2298/ABS1303911B
Beg, O. U., von Bahr-Lindström, H., Zaidi, Z. H., and Jörnvall, H. (1986). Characterization of a camel milk protein rich in proline identifies a new β-casein fragment. Regul. Pept. 15, 55–61. doi: 10.1016/0167-0115(86)90075-3
Beghein, E., and Gettemans, J. (2017). Nanobody technology: a versatile toolkit for microscopic imaging, protein-protein interaction analysis, and protein function exploration. Front. Immunol. 8:771. doi: 10.3389/fimmu.2017.00771
Behar, G., Sibéril, S., Groulet, A., Chames, P., Pugniere, M., Boix, C., et al. (2007). Isolation and characterization of anti-FcγRIII (CD16) llama single-domain antibodies that activate natural killer cells. Protein Eng. Des. Sel. 21, 1–10. doi: 10.1093/protein/gzm064
Behdani, M., Zeinali, S., Khanahmad, H., Karimipour, M., Asadzadeh, N., Azadmanesh, K., et al. (2012). Generation and characterization of a functional Nanobody against the vascular endothelial growth factor receptor-2; angiogenesis cell receptor. Mol. Immunol. 50, 35–41. doi: 10.1016/j.molimm.2011.11.013
Beuzen, N., Stear, M., and Chang, K. (2000). Molecular markers and their use in animal breeding. Vet. J. 160, 42–52. doi: 10.1053/tvjl.2000.0468
Borai, A., Livingstone, C., Abdelaal, F., Bawazeer, A., Keti, V., and Ferns, G. (2011). The relationship between glycosylated haemoglobin (HbA1c) and measures of insulin resistance across a range of glucose tolerance. Scand. J. Clin. Lab. Invest. 71, 168–172. doi: 10.3109/00365513.2010.547947
Breulmann, M., Böer, B., Wernery, U., Wernery, R., El Shaer, H., Alhadrami, G., et al. (2007). The Camel, from Tradition to Modern Times. Doha: The United Nations Educational.
Broisat, A., Hernot, S., Toczek, J., De Vos, J., Riou, L. M., Martin, S., et al. (2012). Nanobodies targeting mouse/human VCAM1 for the nuclear imaging of atherosclerotic lesions. Circ. Res. 110, 927–937. doi: 10.1161/CIRCRESAHA.112.265140
Burg, M. B., Ferraris, J. D., and Dmitrieva, N. I. (2007). Cellular response to hyperosmotic stresses. Physiol. Rev. 87, 1441–1474. doi: 10.1152/physrev.00056.2006
Burger, P. A. (2016). The history of Old World camelids in the light of molecular genetics. Trop. Anim. Health Prod. 48, 905–913. doi: 10.1007/s11250-016-1032-7
Callagy, G. M., Pharoah, P. D., Pinder, S. E., Hsu, F. D., Nielsen, T. O., Ragaz, J., et al. (2006). Bcl-2 is a prognostic marker in breast cancer independently of the nottingham prognostic index. Clin. Cancer Res. 12, 2468–2475. doi: 10.1158/1078-0432.CCR-05-2719
Carroll, M. A., Balazy, M., Margiotta, P., Huang, D., Falck, J., and McGiff, J. (1996). Cytochrome P-450-dependent HETEs: profile of biological activity and stimulation by vasoactive peptides. Am. J. Physiol. Regul. Integr. Comp. Physiol. 271, R863–R869. doi: 10.1152/ajpregu.1996.271.4.R863
Chan, P.-H., Pardon, E., Menzer, L., De Genst, E., Kumita, J. R., Christodoulou, J., et al. (2008). Engineering a camelid antibody fragment that binds to the active site of human lysozyme and inhibits its conversion into amyloid fibrils. Biochemistry 47, 11041–11054. doi: 10.1021/bi8005797
Chauhan, J. S., Rao, A., and Raghava, G. P. (2013). In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One 8:e67008. doi: 10.1371/journal.pone.0067008
Chen, S. Y., Su, Y. H., Wu, S. F., Sha, T., and Zhang, Y. P. (2005). Mitochondrial diversity and phylogeographic structure of Chinese domestic goats. Mol. Phylogenet. Evol. 37, 804–814. doi: 10.1016/j.ympev.2005.06.014
Cherifi, Y. A., Gaouar, S. B. S., Guastamacchia, R., El-Bahrawy, K. A., Abushady, A. M. A., Sharaf, A. A., et al. (2017). Weak genetic structure in northern african dromedary camels reflects their unique evolutionary history. PLoS One 12:e0168672. doi: 10.1371/journal.pone.0168672
Cheung, C. Y., and Ko, B. C. (2013). NFAT5 in cellular adaptation to hypertonic stress - regulations and functional significance. J. Mol. Signal. 8:5. doi: 10.1186/1750-2187-8-5
Chuluunbat, B., Charruau, P., Silbermayr, K., Khorloojav, T., and Burger, P. (2014). Genetic diversity and population structure of mongolian domestic bactrian camels (Camelus bactrianus). Anim. Genet. 45, 550–558. doi: 10.1111/age.12158
Ciccarese, S., Vaccarelli, G., Lefranc, M. P., Tasco, G., Consiglio, A., Casadio, R., et al. (2014). Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma (TRG) and delta (TRD) variable domains. Dev. Comp. Immunol. 46, 300–313. doi: 10.1016/j.dci.2014.05.001
Coppieters, K., Dreier, T., Silence, K., Haard, H. D., Lauwereys, M., Casteels, P., et al. (2006). Formatted anti–tumor necrosis factor α VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheumatol. 54, 1856–1866. doi: 10.1002/art.21827
Cortez-Retamozo, V., Lauwereys, M., Gobert, M., Conrath, K., Muyldermans, S., De Baetselier, P., et al. (2002). Efficient tumor targeting by single-domain antibody fragments of camels. Int. J. Cancer 98, 456–462. doi: 10.1002/ijc.10212
Cui, P., Ji, R., Ding, F., Qi, D., Gao, H., Meng, H., et al. (2007). A complete mitochondrial genome sequence of the wild two-humped camel (Camelus bactrianus ferus): an evolutionary history of camelidae. BMC Genomics 8:241. doi: 10.1186/1471-2164-8-241
De Genst, E., Saerens, D., Muyldermans, S., and Conrath, K. (2006). Antibody repertoire development in camelids. Dev. Comp. Immunol. 30, 187–198. doi: 10.1016/j.dci.2005.06.010
Deschacht, N., De Groeve, K., Vincke, C., Raes, G., De Baetselier, P., and Muyldermans, S. (2010). A novel promiscuous class of camelid single-domain antibody contributes to the antigen-binding repertoire. J. Immunol. 184, 5696–5704. doi: 10.4049/jimmunol.0903722
Desmyter, A., Farenc, C., Mahony, J., Spinelli, S., Bebeacua, C., Blangy, S., et al. (2013). Viral infection modulation and neutralization by camelid nanobodies. Proc. Natl. Acad. Sci. U.S.A. 110, E1371–E1379. doi: 10.1073/pnas.1301336110
Dettin, L., Rubinstein, N., Aoki, A., Rabinovich, G. A., and Maldonado, C. A. (2003). Regulated expression and ultrastructural localization of galectin-1, a proapoptotic beta-galactoside-binding lectin, during spermatogenesis in rat testis. Biol. Reprod. 68, 51–59. doi: 10.1095/biolreprod.102.006361
Deuffic-Burban, S., Poynard, T., Sulkowski, M. S., and Wong, J. B. (2007). Estimating the future health burden of chronic hepatitis C and human immunodeficiency virus infections in the United States. J. Viral Hepat. 14, 107–115. doi: 10.1111/j.1365-2893.2006.00785.x
Dickinson, A., Yeung, K. Y., Donoghue, J., Baker, M. J., Kelly, R. D., McKenzie, M., et al. (2013). The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 20, 1644–1653. doi: 10.1038/cdd.2013.115
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Duchemin, S. I., Colombani, C., Legarra, A., Baloche, G., Larroque, H., Astruc, J. M., et al. (2012). Genomic selection in the French Lacaune dairy sheep breed. J. Dairy Sci. 95, 2723–2733. doi: 10.3168/jds.2011-4980
Dumoulin, M., Last, A. M., Desmyter, A., Decanniere, K., Canet, D., Larsson, G., et al. (2003). A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme. Nature 424, 783–788. doi: 10.1038/nature01870
Eitan, A., Aloni, B., and Livne, A. (1976). Unique properties of the camel erythrocyte membrane, II. Organization of membrane proteins. Biochim. Biophys. Acta 426, 647–658. doi: 10.1016/0005-2736(76)90129-2
El-Fakharany, E. M., Sanchez, L., Al-Mehdar, H. A., and Redwan, E. M. (2013). Effectiveness of human, camel, bovine and sheep lactoferrin on the hepatitis C virus cellular infectivity: comparison study. Virol. J. 10:199. doi: 10.1186/1743-422X-10-199
Elmahdi, B., Sallmann, H. P., Fuhrmann, H., von Engelhardt, W., and Kaske, M. (1997). Comparative aspects of glucose tolerance in camels, sheep, and ponies. Comp. Biochem. Physiol. A Physiol. 118, 147–151. doi: 10.1016/S0300-9629(96)00449-5
Elsik, C. G., Tellam, R. L., Worley, K. C., Gibbs, R. A., Muzny, D. M., Weinstock, G. M., et al. (2009). The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324, 522–528. doi: 10.1126/science.1169588
Emmanuel, B., and Nahapetian, A. (1980). Fatty acid composition of depot fats, and rumen wall of the camel (Camelus dromedarius). Comp. Biochem. Physiol. B 67, 701–704. doi: 10.1016/0305-0491(80)90435-6
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Faye, B. (2016). The camel, new challenges for a sustainable development. Trop. Anim. Health Prod. 48, 689–692. doi: 10.1007/s11250-016-0995-8
Faye, B., and Bonnet, P. (2012). “Camel sciences and economy in the world: current situation and perspectives,” in Proceedings of the 3rd ISOCARD Conference, Muscat, 2–15.
Fazil, M., and Hofmann, R. (1981). Haltung und Krankheiten des Kamels. Tieraerztl. Prax. 9, 389–402.
Finberg, J., Yagil, R., and Berlyne, G. (1978). Response of the renin-aldosterone system in the camel to acute dehydration. J. Appl. Physiol. 44, 926–930. doi: 10.1152/jappl.1978.44.6.926
Finlay, W., and Almagro, J. (2012). Natural and man-made V-gene repertoires for antibody discovery. Front. Immunol. 2012:342. doi: 10.3389/fimmu.2012.00342
Fitak, R. R., Mohandesan, E., Corander, J., and Burger, P. A. (2016). The de novo genome assembly and annotation of a female domestic dromedary of North African origin. Mol. Ecol. Resour. 16, 314–324. doi: 10.1111/1755-0998.12443
Food and Agriculture Organization [FAO] (2018). FAOSTAT. Available at: http://www.fao.org/faostat/ [accessed November 25, 2018].
Gallazzini, M., and Burg, M. B. (2009). What’s new about osmotic regulation of glycerophosphocholine. Physiology 24, 245–249. doi: 10.1152/physiol.00009.2009
Gerondakis, S., Grumont, R., Gugasyan, R., Wong, L., Isomura, I., Ho, W., et al. (2006). Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene 25, 6781–6799. doi: 10.1038/sj.onc.1209944
Giambra, I. J., El Zubeir, I. E. Y. M., and Erhardt, G. (2013). Biochemical and molecular characterization of polymorphisms of αs1-casein in Sudanese camel (Camelus dromedarius) milk. Int. Dairy J. 28, 88–93. doi: 10.1016/j.idairyj.2012.09.002
Goniakowska-Witalinska, L., and Witalinski, W. (1976). Evidence for a correlation between the number of marginal band microtubules and the size of vertebrate erthrocytes. J. Cell Sci. 22, 397–401.
Greenberg, A. S., Avila, D., Hughes, M., Hughes, A., McKinney, E. C., and Flajnik, M. F. (1995). A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374, 168–173. doi: 10.1038/374168a0
Griffin, L. M., Snowden, J. R., Lawson, A. D., Wernery, U., Kinne, J., and Baker, T. S. (2014). Analysis of heavy and light chain sequences of conventional camelid antibodies from Camelus dromedarius and Camelus bactrianus species. J. Immunol. Methods 405, 35–46. doi: 10.1016/j.jim.2014.01.003
Grover, Y. P., Kaura, Y. K., Prasad, S., and Srivastava, R. N. (1983). Preliminary studies on camel serum immunoglobulins. Indian J. Biochem. Biophys. 20, 238–240.
Guilherme, A., Emoto, M., Buxton, J. M., Bose, S., Sabini, R., Theurkauf, W. E., et al. (2000). Perinuclear localization and insulin responsiveness of GLUT4 requires cytoskeletal integrity in 3T3-L1 adipocytes. J. Biol. Chem. 275, 38151–38159. doi: 10.1074/jbc.M003432200
Gutschalk, C. M., Yanamandra, A. K., Linde, N., Meides, A., Depner, S., and Mueller, M. M. (2013). GM-CSF enhances tumor invasion by elevated MMP-2, -9, and -26 expression. Cancer Med. 2, 117–129. doi: 10.1002/cam4.20
Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E. B., et al. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448. doi: 10.1038/363446a0
Harmsen, M., and De Haard, H. (2007). Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 77, 13–22. doi: 10.1007/s00253-007-1142-2
Hassaine, G., Deluz, C., Grasso, L., Wyss, R., Tol, M. B., Hovius, R., et al. (2014). X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281. doi: 10.1038/nature13552
Hayes, B., and Goddard, M. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
Hayes, B. J., Bowman, P. J., Chamberlain, A. J., and Goddard, M. E. (2009). Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92, 433–443. doi: 10.3168/jds.2008-1646
Herrera, M., León, G., Segura, A., Meneses, F., Lomonte, B., Chippaux, J. P., et al. (2005). Factors associated with adverse reactions induced by caprylic acid-fractionated whole IgG preparations: comparison between horse, sheep and camel IgGs. Toxicon 46, 775–781. doi: 10.1016/j.toxicon.2005.08.004
Higgins, T. (2012). HbA1c—An analyte of increasing importance. Clin. Biochem. 45, 1038–1045. doi: 10.1016/j.clinbiochem.2012.06.006
Hjort af Ornäs, A., and Dahl, G. (1991). Responsible Man: The Atmaan Beja of North-eastern Sudan. Stockholm: Stockholm University.
Hoover, H. E., Thuerauf, D. J., Martindale, J. J., and Glembotski, C. C. (2000). αB-crystallin gene induction and phosphorylation by mkk6-activated p38 a potential role for αb-crystallin as a target of the p38 branch of the cardiac stress response. J. Biol. Chem. 275, 23825–23833. doi: 10.1074/jbc.M003864200
Hu, Z., Chang, X., Pan, Q., Gu, K., and Okechukwu, P. N. (2017). Gastroprotective and ulcer healing effects of camel milk and urine in HCl/EtOH, non-steroidal anti-inflammatory drugs (Indomethacin), and water-restraint stress-induced ulcer in rats. Pharmacogn. Mag. 13, 559–565. doi: 10.4103/pm.pm_135_17
Huang, Y., Tracy, R., Walsberg, G. E., Makkinje, A., Fang, P., Brown, D., et al. (2001). Absence of aquaporin-4 water channels from kidneys of the desert rodent Dipodomys merriami merriami. Am. J. Physiol. Renal Physiol. 280, F794–F802. doi: 10.1152/ajprenal.2001.280.5.F794
Huet, H. A., Growney, J. D., Johnson, J. A., Li, J., Bilic, S., Ostrom, L., et al. (2014). Multivalent nanobodies targeting death receptor 5 elicit superior tumor cell killing through efficient caspase induction. mAbs 6, 1560–1570. doi: 10.4161/19420862.2014.975099
Hultberg, A., Temperton, N. J., Rosseels, V., Koenders, M., Gonzalez-Pajuelo, M., Schepens, B., et al. (2011). Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PLoS One 6:e17665. doi: 10.1371/journal.pone.0017665
Husein, M. A. (1987). Emic Notes on Camel breeds in Somalia (Camel Forum Working Paper No 17). Somalia: Somali Academy of Sciences.
Ikeda, M., Sugiyama, K., Tanaka, T., Tanaka, K., Sekihara, H., Shimotohno, K., et al. (1998). Lactoferrin markedly inhibits hepatitis C virus infection in cultured human hepatocytes. Biochem. Biophys. Res. Commun. 245, 549–553. doi: 10.1006/bbrc.1998.8481
Ishag, I., Reissmann, M., Peters, K., Musa, L.-A., and Ahmed, M. (2010). Phenotypic and molecular characterization of six Sudanese camel breeds. S. Afr. J. Anim. Sci. 40, 319–326.
Jahnichen, S., Blanchetot, C., Maussang, D., Gonzalez-Pajuelo, M., Chow, K. Y., Bosch, L., et al. (2010). CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20565–20570. doi: 10.1073/pnas.1012865107
Janeway, C. A., Travers, P., Walport, M., and Shlomchik, M. (2001). The Immune System in Health and Disease Immunobiology. New York, NY: Gerald Publishing.
Jenssen, H., Andersen, J. H., Uhlin-Hansen, L., Gutteberg, T. J., and Rekdal, Ø (2004). Anti-HSV activity of lactoferricin analogues is only partly related to their affinity for heparan sulfate. Antivir. Ther. 61, 101–109. doi: 10.1016/j.antiviral.2003.09.001
Jerne, N. K. (1955). The natural selection theory of antibody formation. Proc. Natl. Acad. Sci. U.S.A. 41, 849–857. doi: 10.1073/pnas.41.11.849
Ji, R., Cui, P., Ding, F., Geng, J., Gao, H., Zhang, H., et al. (2009). Monophyletic origin of domestic bactrian camel (Camelus bactrianus) and its evolutionary relationship with the extant wild camel (Camelus bactrianus ferus). Anim. Genet. 40, 377–382. doi: 10.1111/j.1365-2052.2008.01848.x
Jirimutu, Wang, Z., Ding, G., Chen, G., Sun, Y., Sun, Z., et al. (2012). Genome sequences of wild and domestic bactrian camels. Nat. Commun. 3:1202. doi: 10.1038/ncomms2192
Kappeler, S. R., Heuberger, C., Farah, Z., and Puhan, Z. (2004). Expression of the peptidoglycan recognition protein, PGRP, in the lactating mammary gland. J. Dairy Sci. 87, 2660–2668. doi: 10.3168/jds.S0022-0302(04)73392-5
Kasahara, M., Naruse, K., Sasaki, S., Nakatani, Y., Qu, W., Ahsan, B., et al. (2007). The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719. doi: 10.1038/nature05846
Kaske, M., Elmahdi, B., Engelhardt, W. V., and Sallmann, H.-P. (2001). Insulin responsiveness of sheep, ponies, miniature pigs and camels: results of hyperinsulinemic clamps using porcine insulin. J. Comp. Physiol. B 171, 549–556. doi: 10.1007/s003600100205
Khalkhali-Evrigh, R., Hafezian, S. H., Hedayat-Evrigh, N., Farhadi, A., and Bakhtiarizadeh, M. R. (2018). Genetic variants analysis of three dromedary camels using whole genome sequencing data. PLoS One 13:e0204028. doi: 10.1371/journal.pone.0204028
Khan, A. A., Alzohairy, M. A., and Mohieldein, A. H. (2013). Antidiabetic effects of camel milk in streptozotocin-induced diabetic rats. Am. J. Biochem. Mol. Biol. 3, 151–158. doi: 10.1186/1471-2172-14-31
Klarenbeek, A., Mazouari, K. E., Desmyter, A., Blanchetot, C., Hultberg, A., de Jonge, N., et al. (2015). Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. mAbs 7, 693–706. doi: 10.1080/19420862.2015.1046648
Kolling, J., and Wyse, A. T. (2010). Creatine prevents the inhibition of energy metabolism and lipid peroxidation in rats subjected to GAA administration. Metab. Brain Dis. 25, 331–338. doi: 10.1007/s11011-010-9215-9
Korashy, H. M., El Gendy, M. A., Alhaider, A. A., and El-Kadi, A. O. (2012). Camel milk modulates the expression of aryl hydrocarbon receptor-regulated genes, Cyp1a1, Nqo1, and Gsta1, in murine hepatoma Hepa 1c1c7 cells. J. Biomed. Biotechnol. 2012:782642. doi: 10.1155/2012/782642
Kruse, A. C., Ring, A. M., Manglik, A., Hu, J., Hu, K., Eitel, K., et al. (2013). Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106. doi: 10.1038/nature12735
Kumar, N., Robidoux, J., Daniel, K. W., Guzman, G., Floering, L. M., and Collins, S. (2007). Requirement of vimentin filament assembly for β3-adrenergic receptor activation of ERK MAP kinase and lipolysis. J. Biol. Chem. 282, 9244–9250. doi: 10.1074/jbc.M605571200
Lacroix, M., Toillon, R.-A., and Leclercq, G. (2006). p53 and breast cancer, an update. Endocr. Relat. Cancer 13, 293–325. doi: 10.1677/erc.1.01172
Lai, S. J., Liu, Y. P., Liu, Y. X., Li, X. W., and Yao, Y. G. (2006). Genetic diversity and origin of Chinese cattle revealed by mtDNA D-loop sequence variation. Mol. Phylogenet. Evol. 38, 146–154. doi: 10.1016/j.ympev.2005.06.013
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Legrand, D., Elass, E., Carpentier, M., and Mazurier, J. (2005). Lactoferrin: a modulator of immune and inflammatory responses. Cell. Mol. Life Sci. 62, 2549–2559. doi: 10.1007/s00018-005-5370-2
Legrand, D., Pierce, A., Elass, E., Carpentier, M., Mariller, C., and Mazurier, J. (2008). Lactoferrin structure and functions. Adv. Exp. Med. Biol. 606, 163–194. doi: 10.1007/978-0-387-74087-4_6
Leung, K. (2012). “99mTc(CO)3-Anti-carcinoembryonic antigen (CEA) humanized CEA5 graft nanobody,” in Molecular Imaging and Contrast Agent Database (MICAD) (Rockyville, MD: Bethesda).
Li, T., Bourgeois, J. P., Celli, S., Glacial, F., Le Sourd, A. M., Mecheri, S., et al. (2012). Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J. 26, 3969–3979. doi: 10.1096/fj.11-201384
Li, Z., Liu, G., Tong, Y., Zhang, M., Xu, Y., Qin, L., et al. (2015). Comprehensive analysis of the T-cell receptor beta chain gene in rhesus monkey by high throughput sequencing. Sci. Rep. 5:10092. doi: 10.1038/srep10092
Lindblad-Toh, K., Wade, C. M., Mikkelsen, T. S., Karlsson, E. K., Jaffe, D. B., Kamal, M., et al. (2005). Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819. doi: 10.1038/nature04338
Lindstrom, J., Ilanne-Parikka, P., Peltonen, M., Aunola, S., Eriksson, J. G., Hemio, K., et al. (2006). Sustained reduction in the incidence of type 2 diabetes by lifestyle intervention: follow-up of the finnish diabetes prevention study. Lancet 368, 1673–1679. doi: 10.1016/S0140-6736(06)69701-8
Lu, L., Timofeyev, V., Li, N., Rafizadeh, S., Singapuri, A., Harris, T. R., et al. (2009). α-Actinin2 cytoskeletal protein is required for the functional membrane localization of a Ca2+-activated K+ channel (SK2 channel). Proc. Natl. Acad. Sci. U.S.A. 106, 18402–18407. doi: 10.1073/pnas.0908207106
Macfarlane, W., Morris, R., and Howard, B. (1963). Turn-over and distribution of water in desert camels, sheep, cattle and kangaroos. Nature 197, 270–271. doi: 10.1038/197270a0
Malik, A., Al-Senaidy, A., Skrzypczak-Jankun, E., and Jankun, J. (2012). A study of the anti-diabetic agents of camel milk. Int. J. Mol. Med. 30, 585–592. doi: 10.3892/ijmm.2012.1051
Marchetti, M., Pisani, S., Antonini, G., Valenti, P., Seganti, L., and Orsi, N. (1998). Metal complexes of bovine lactoferrin inhibit in vitro replication of herpes simplex virus type 1 and 2. Biometals 11, 89–94. doi: 10.1023/A:1009217709851
Marchetti, M., Trybala, E., Superti, F., Johansson, M., and Bergström, T. (2004). Inhibition of herpes simplex virus infection by lactoferrin is dependent on interference with the virus binding to glycosaminoglycans. Virology 318, 405–413. doi: 10.1016/j.virol.2003.09.029
Maruo, Y., Gochi, A., Kaihara, A., Shimamura, H., Yamada, T., Tanaka, N., et al. (2002). ICAM-1 expression and the soluble ICAM-1 level for evaluating the metastatic potential of gastric cancer. Int. J. Cancer 100, 486–490. doi: 10.1002/ijc.10514
Massad, J. A. (2012). Colonial Effects: The Making of National Identity in Jordan. New York, NY: Columbia University Press.
Maussang, D., Mujic-Delic, A., Descamps, F. J., Stortelers, C., Vanlandschoot, P., Stigter-van Walsum, M., et al. (2013). Llama-derived single variable domains (nanobodies) directed against chemokine receptor CXCR7 reduce head and neck cancer cell growth in vivo. J. Biol. Chem. 288, 29562–29572. doi: 10.1074/jbc.M113.498436
McCoy, L. E., Quigley, A. F., Strokappe, N. M., Bulmer-Thomas, B., Seaman, M. S., Mortier, D., et al. (2012). Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J. Exp. Med. 209, 1091–1103. doi: 10.1084/jem.20112655
McKinley, M. J., McBurnie, M. I., and Mathai, M. L. (2001). Neural mechanisms subserving central angiotensinergic influences on plasma renin in sheep. Hypertension 37, 1375–1381. doi: 10.1161/01.HYP.37.6.1375
Meddeb-Mouelhi, F., Bouhaouala-Zahar, B., Benlasfar, Z., Hammadi, M., Mejri, T., Moslah, M., et al. (2003). Immunized camel sera and derived immunoglobulin subclasses neutralizing Androctonus australis hector scorpion toxins. Toxicon 42, 785–791. doi: 10.1016/j.toxicon.2003.10.021
Mehaia, M. A., Hablas, M. A., Abdel-Rahman, K. M., and El-Mougy, S. A. (1995). Milk composition of Majaheim, Wadah and Hamra camels in Saudi Arabia. Food Chem. 52, 115–122. doi: 10.1016/0308-8146(94)P4189-M
Mehta, S., Mishra, B., and Sahani, M. (2006). Genetic differentiation of Indian camel (Camelus dromedarius) breeds using random oligonucleotide primers. Anim. Genet. Resour. 39, 77–88. doi: 10.1017/S1014233900002157
Mehta, S. C. (2014). Genetic and demographic bottleneck analysis of Indian camel breeds by microsatellite markers. Trop. Anim. Health Prod. 46, 1397–1406. doi: 10.1007/s11250-014-0653-y
Mima, T., and Nishimoto, N. (2009). Clinical value of blocking IL-6 receptor. Curr. Opin. Rheumatol. 21, 224–230. doi: 10.1097/BOR.0b013e3283295fec
Minaeian, S., Rahbarizadeh, F., Zarkesh-Esfahani, S. H., Ahmadvand, D., and Broom, O. J. (2012). Neutralization of human papillomavirus by specific nanobodies against major capsid protein L1. J. Microbiol. Biotechnol. 22, 721–728. doi: 10.4014/jmb.1112.12001
Ming, L., Yi, L., Sa, R., Wang, Z., Wang, Z., and Ji, R. (2017). Genetic diversity and phylogeographic structure of Bactrian camels shown by mitochondrial sequence variations. Anim. Genet. 48, 217–220. doi: 10.1111/age.12511
Mocellin, S., Marincola, F. M., and Young, H. A. (2005). Interleukin-10 and the immune response against cancer: a counterpoint. J. Leukoc. Biol. 78, 1043–1051. doi: 10.1189/jlb.0705358
Mousa, H. M., Ali, K. E., and Hume, I. D. (1983). Effects of water deprivation on urea metabolism in camels, desert sheep and desert goats fed dry desert grass. Comp. Biochem. Physiol. A Comp. Physiol. 74, 715–720. doi: 10.1016/0300-9629(83)90574-1
Mukasa-Mugerwa, E. (1981). The Camel (Camelus dromedarius): A Bibliographical Review. Nairobi: ILRI.
Muyldermans, S. (2001). Single domain camel antibodies: current status. J. Biotechnol. 74, 277–302. doi: 10.1016/S1389-0352(01)00021-6
Muyldermans, S., Baral, T., Retamozzo, V. C., De Baetselier, P., De Genst, E., Kinne, J., et al. (2009). Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 128, 178–183. doi: 10.1016/j.vetimm.2008.10.299
Muyldermans, S., and Lauwereys, M. (1999). Unique single-domain antigen binding fragments derived from naturally occurring camel heavy-chain antibodies. J. Mol. Recognit. 12, 131–140. doi: 10.1002/(SICI)1099-1352(199903/04)12:2<131::AID-JMR454>3.0.CO;2-M
Nagai, S., and Toi, M. (2000). Interleukin-4 and breast cancer. Breast Cancer 7, 181–186. doi: 10.1007/BF02967457
Nathan, D. M., Kuenen, J., Borg, R., Zheng, H., Schoenfeld, D., and Heine, R. J. (2008). Translating the A1C assay into estimated average glucose values. Diabetes Care 31, 1473–1478. doi: 10.2337/dc08-0545
Nguyen, M. H., and Keeffe, E. B. (2005). Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin. Gastroenterol. Hepatol. 3(10 Suppl. 2), S97–S101. doi: 10.1016/S1542-3565(05)00711-1
Nuttall, S. D., Krishnan, U. V., Hattarki, M., De Gori, R., Irving, R. A., and Hudson, P. J. (2001). Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol. 38, 313–326. doi: 10.1016/S0161-5890(01)00057-8
Olesen, I., Groen, A. F., and Gjerde, B. (2000). Definition of animal breeding goals for sustainable production systems. J. Anim. Sci. 78, 570–582. doi: 10.2527/2000.783570x
Oyewale, J., Dzenda, T., Yaqub, L., Akanbi, D., Ayo, J., Owoyele, O., et al. (2011). Alterations in the osmotic fragility of camel and donkey erythrocytes caused by temperature, pH and blood storage. Vet. Arh. 81, 459–470.
Paalanen, M. M., Ekokoski, E., El Khattabi, M., Tuominen, R. K., Verrips, C. T., Boonstra, J., et al. (2011). The development of activating and inhibiting camelid VHH domains against human protein kinase C epsilon. Eur. J. Pharm. Sci. 42, 332–339. doi: 10.1016/j.ejps.2010.12.012
Paik, S. S., Jang, S. M., Jang, K. S., Lee, K. H., Choi, D., and Jang, S. J. (2009). Leptin expression correlates with favorable clinicopathologic phenotype and better prognosis in colorectal adenocarcinoma. Ann. Surg. Oncol. 16, 297–303. doi: 10.1245/s10434-008-0221-7
Pauciullo, A., Giambra, I. J., Iannuzzi, L., and Erhardt, G. (2014). The beta-casein in camels: molecular characterization of the CSN2 gene, promoter analysis and genetic variability. Gene 547, 159–168. doi: 10.1016/j.gene.2014.06.055
Plasil, M., Mohandesan, E., Fitak, R. R., Musilova, P., Kubickova, S., Burger, P. A., et al. (2016). The major histocompatibility complex in Old World camelids and low polymorphism of its class II genes. BMC Genomics 17:167. doi: 10.1186/s12864-016-2500-1
Prasad, C. P., Gupta, S. D., Rath, G., and Ralhan, R. (2007). Wnt signaling pathway in invasive ductal carcinoma of the breast: relationship between β-catenin, disheveled and cyclin D1 expression. Oncology 73, 112–117. doi: 10.1159/000120999
Put, S., Schoonooghe, S., Devoogdt, N., Schurgers, E., Avau, A., Mitera, T., et al. (2013). SPECT imaging of joint inflammation with Nanobodies targeting the macrophage mannose receptor in a mouse model for rheumatoid arthritis. J. Nucl. Med. 54, 807–814. doi: 10.2967/jnumed.112.111781
Qian, B. Z., Li, J., Zhang, H., Kitamura, T., Zhang, J., Campion, L. R., et al. (2011). CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225. doi: 10.1038/nature10138
Ramadan, S., and Inoue-Murayama, M. (2017). Advances in camel genomics and their applications: a review. J. Anim. Genet. 45, 49–58. doi: 10.5924/abgri.45.49
Rao, M., Gupta, R., and Dastur, N. (1970). Camels’ milk and milk products. Indian J. Dairy Sci. 23, 71–78.
Rasmussen, S. G., Choi, H. J., Fung, J. J., Pardon, E., Casarosa, P., Chae, P. S., et al. (2011). Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–180. doi: 10.1038/nature09648
Raziq, A., de Verdier, K., and Younas, M. (2010). Ethnoveterinary treatments by dromedary camel herders in the suleiman mountainous region in pakistan: an observation and questionnaire study. J. Ethnobiol. Ethnomed. 6:16. doi: 10.1186/1746-4269-6-16
Raziq, A., de Verdier, K., and Younas, M. (2011). Rapid change of strategy is necessary for development of dromedary camel pastoralism in the Cholistan desert of Pakistan. Pastoralism 1:3. doi: 10.1186/2041-7136-1-3
Redwan, E. M., El-Fakharany, E. M., Uversky, V. N., and Linjawi, M. H. (2014). Screening the anti infectivity potentials of native N- and C-lobes derived from the camel lactoferrin against hepatitis C virus. BMC Complement. Altern. Med. 14:219. doi: 10.1186/1472-6882-14-219
Redwan el, R. M., and Tabll, A. (2007). Camel lactoferrin markedly inhibits hepatitis C virus genotype 4 infection of human peripheral blood leukocytes. J. Immunoassay Immunochem. 28, 267–277. doi: 10.1080/15321810701454839
Reed, C. A. (1972). The origin of the domestic animals of africa. Science 176, 656–657. doi: 10.1126/science.176.4035.656
Rohlfing, C., Wiedmeyer, H. M., Little, R., Grotz, V. L., Tennill, A., England, J., et al. (2002). Biological variation of glycohemoglobin. Clin. Chem. 48, 1116–1118.
Romli, F., Abu, N., Khorshid, F. A., Syed Najmuddin, S. U. F., Keong, Y. S., Mohamad, N. E., et al. (2017). The growth inhibitory potential and antimetastatic effect of camel urine on breast cancer cells in vitro and in vivo. Integr. Cancer Ther. 16, 540–555. doi: 10.1177/1534735416656051
Rood, B. R., MacDonald, T. J., and Packer, R. J. (2004). Current treatment of medulloblastoma: recent advances and future challenges. Semin. Oncol. 31, 666–675. doi: 10.1053/j.seminoncol.2004.07.009
Roovers, R. C., Vosjan, M. J., Laeremans, T., el Khoulati, R., de Bruin, R. C., Ferguson, K. M., et al. (2011). A biparatopic anti-EGFR nanobody efficiently inhibits solid tumour growth. Int. J. Cancer 129, 2013–2024. doi: 10.1002/ijc.26145
Saerens, D., Kinne, J., Bosmans, E., Wernery, U., Muyldermans, S., and Conrath, K. (2004). Single domain antibodies derived from dromedary lymph node and peripheral blood lymphocytes sensing conformational variants of prostate-specific antigen. J. Biol. Chem. 279, 51965–51972. doi: 10.1074/jbc.M409292200
Salami, M., Moosavi-Movahedi, A. A., Ehsani, M. R., Yousefi, R., Haertlé, T., Chobert, J.-M., et al. (2010). Improvement of the antimicrobial and antioxidant activities of camel and bovine whey proteins by limited proteolysis. J. Agric. Food Chem. 58, 3297–3302. doi: 10.1021/jf9033283
Schmidt-Nielsen, K., Crawford, E., and Hammel, H. (1981). Respiratory water loss in camels. Proc. R. Soc. Lond. B 211, 291–303. doi: 10.1098/rspb.1981.0008
Schmidt-Nielsen, K., Schmidt-Nielsen, B., Jarnum, S., and Houpt, T. (1956). Body temperature of the camel and its relation to water economy. Am. J. Physiol. Cell. Physiol. Legacy Content 188, 103–112. doi: 10.1152/ajplegacy.1956.188.1.103
Shamsia, S. (2009). Nutritional and therapeutic properties of camel and human milks. Int. J. Genet. Mol. Biol. 1, 52–58. doi: 10.19082/1523
Shori, A. B. (2015). Camel milk as a potential therapy for controlling diabetes and its complications: a review of in vivo studies. J. Food Drug Anal. 23, 609–618. doi: 10.1016/j.jfda.2015.02.007
Siebert, B., and Macfarlane, W. (1971). Water turnover and renal function of dromedaries in the desert. Physiol. Zool. 44, 225–240. doi: 10.1086/physzool.44.4.30152494
Soman, S. S., and Tinson, A. (2016). Development and evaluation of a simple and effective real time PCR assay for mitochondrial quantification in racing camels. Mol. Cell. Probes 30, 326–330. doi: 10.1016/j.mcp.2016.07.006
Soranzo, N. (2011). Genetic determinants of variability in glycated hemoglobin (HbA 1c) in humans: review of recent progress and prospects for use in diabetes care. Curr. Diab. Rep. 11, 562–569. doi: 10.1007/s11892-011-0232-9
Steeland, S., Vandenbroucke, R. E., and Libert, C. (2016). Nanobodies as therapeutics: big opportunities for small antibodies. Drug Des. Discov. 21, 1076–1113. doi: 10.1016/j.drudis.2016.04.003
Steyaert, J., and Kobilka, B. K. (2011). Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. Biol. 21, 567–572. doi: 10.1016/j.sbi.2011.06.011
Szlosarek, P. W., and Balkwill, F. R. (2003). Tumour necrosis factor alpha: a potential target for the therapy of solid tumours. Lancet Oncol. 4, 565–573. doi: 10.1016/S1470-2045(03)01196-3
Tainer, J. A., Getzoff, E. D., Paterson, Y., Olson, A. J., and Lerner, R. A. (1985). The atomic mobility component of protein antigenicity. Annu. Rev. Immunol. 3, 501–539. doi: 10.1146/annurev.iy.03.040185.002441
Tanaka, K., Iwamoto, S., Gon, G., Nohara, T., Iwamoto, M., and Tanigawa, N. (2000). Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin. Cancer Res. 6, 127–134.
Thanka Christlet, T. H., and Veluraja, K. (2001). Database analysis of O-glycosylation sites in proteins. Biophys. J. 80, 952–960. doi: 10.1016/S0006-3495(01)76074-2
Thys, B., Schotte, L., Muyldermans, S., Wernery, U., Hassanzadeh-Ghassabeh, G., and Rombaut, B. (2010). In vitro antiviral activity of single domain antibody fragments against poliovirus. Antiviral Res. 87, 257–264. doi: 10.1016/j.antiviral.2010.05.012
Tillib, S. V., Vyatchanin, A. S., and Muyldermans, S. (2014). Molecular analysis of heavy chain-only antibodies of Camelus bactrianus. Biochemistry 79, 1382–1390. doi: 10.1134/S000629791412013X
Torimura, T., Sata, M., Ueno, T., Kin, M., Tsuji, R., Suzaku, K., et al. (1998). Increased expression of vascular endothelial growth factor is associated with tumor progression in hepatocellular carcinoma. Hum. Pathol. 29, 986–991. doi: 10.1016/S0046-8177(98)90205-2
Trinks, A., Burger, P., Beneke, N., and Burger, J. (2012). “Simulations of populations ancestry of the two-humped camel (Camelus bactrianus). Camels in Asia and North Africa,” in Interdisciplinary Perspectives on their Significance in Past and Present, eds E. Knoll and P. Burger (Vienna: Academy of Science Press), 79–86.
Ulmasov, H. A., Karaev, K. K., Lyashko, V. N., and Evgen’ev, M. B. (1993). Heat-shock response in camel (Camelus dromedarius) blood cells and adaptation to hyperthermia. Comp. Biochem. Physiol. B 106, 867–872. doi: 10.1016/0305-0491(93)90043-5
Vaneycken, I., Devoogdt, N., Van Gassen, N., Vincke, C., Xavier, C., Wernery, U., et al. (2011). Preclinical screening of anti-HER2 nanobodies for molecular imaging of breast cancer. FASEB J. 25, 2433–2446. doi: 10.1096/fj.10-180331
Varvarovska, J., Racek, J., Stetina, R., Sykora, J., Pomahacova, R., Rusavy, Z., et al. (2004). Aspects of oxidative stress in children with type 1 diabetes mellitus. Biomed. Pharmacother. 58, 539–545. doi: 10.1016/j.biopha.2004.09.011
Vijh, R. K., Tantia, M. S., Mishra, B., and Kumar, S. T. (2007). Genetic diversity and differentiation of dromedarian camel of India. Anim. Biotechnol. 18, 81–90. doi: 10.1080/10495390600648741
Vincke, C., Gutierrez, C., Wernery, U., Devoogdt, N., Hassanzadeh-Ghassabeh, G., and Muyldermans, S. (2012). Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol. Biol. 907, 145–176. doi: 10.1007/978-1-61779-974-7_8
Volpato, G., Dioli, M., and Di Nardo, A. (2017). Piebald camels. Pastoralism 7:3. doi: 10.1186/s13570-017-0075-3
Vosjan, M. J., Vercammen, J., Kolkman, J. A., Stigter-van Walsum, M., Revets, H., and van Dongen, G. A. (2012). Nanobodies targeting the hepatocyte growth factor: potential new drugs for molecular cancer therapy. Mol. Cancer Ther. 11, 1017–1025. doi: 10.1158/1535-7163.MCT-11-0891
Wade, C. M., Giulotto, E., Sigurdsson, S., Zoli, M., Gnerre, S., Imsland, F., et al. (2009). Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867. doi: 10.1126/science.1178158
Wang, R. C., Wei, Y., An, Z., Zou, Z., Xiao, G., Bhagat, G., et al. (2012). Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959. doi: 10.1126/science.1225967
Ward, A. B., Szewczyk, P., Grimard, V., Lee, C. W., Martinez, L., Doshi, R., et al. (2013). Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. U.S.A. 110, 13386–13391. doi: 10.1073/pnas.1309275110
Warda, M., Prince, A., Kim, H. K., Khafaga, N., Scholkamy, T., Linhardt, R. J., et al. (2014). Proteomics of old world camelid (Camelus dromedarius): Better understanding the interplay between homeostasis and desert environment. J. Adv. Res. 5, 219–242. doi: 10.1016/j.jare.2013.03.004
Warda, M., and Zeisig, R. (2000). Phospholipid-and fatty acid-composition in the erythrocyte membrane of the one-humped camel [Camelus dromedarius] and its influence on vesicle properties prepared from these lipids. Deut Tierarztl Woch 107, 368–373.
Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. doi: 10.1038/nature01262
Wendt, M. K., Tian, M., and Schiemann, W. P. (2012). Deconstructing the mechanisms and consequences of TGF-β-induced EMT during cancer progression. Cell Tissue Res. 347, 85–101. doi: 10.1007/s00441-011-1199-1
Wernersson, R., Schierup, M. H., Jorgensen, F. G., Gorodkin, J., Panitz, F., Staerfeldt, H. H., et al. (2005). Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genomics 6:70. doi: 10.1186/1471-2164-6-70
Wernery, U., and Kaaden, O. R. (2004). Foot-and-mouth disease in camelids: a review. Vet. J. 168, 134–142. doi: 10.1016/j.tvjl.2003.10.005
Wernery, U., Nagy, P., Bhai, I., Schiele, W., and Johnson, B. (2006). The effect of heat treatment, pasteurization and different storage temperatures on insulin concentrations in camel milk. Milchwissenschaft 61, 25–28.
Wesolowski, J., Alzogaray, V., Reyelt, J., Unger, M., Juarez, K., Urrutia, M., et al. (2009). Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med. Microbiol. Immunol. 198, 157–174. doi: 10.1007/s00430-009-0116-7
West, I. C. (2000). Radicals and oxidative stress in diabetes. Diabet. Med. 17, 171–180. doi: 10.1046/j.1464-5491.2000.00259.x
Wild, S., Roglic, G., Green, A., Sicree, R., and King, H. (2004). Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27, 1047–1053. doi: 10.2337/diacare.27.5.1047
Williams, R. (2006). Global challenges in liver disease. Hepatology 44, 521–526. doi: 10.1002/hep.21347
Wu, H., Guang, X., Al-Fageeh, M. B., Cao, J., Pan, S., Zhou, H., et al. (2014). Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 5:5188. doi: 10.1038/ncomms6188
Xavier, C., Vaneycken, I., D’Huyvetter, M., Heemskerk, J., Keyaerts, M., Vincke, C., et al. (2013). Synthesis, preclinical validation, dosimetry, and toxicity of 68Ga-NOTA-anti-HER2 Nanobodies for iPET imaging of HER2 receptor expression in cancer. J. Nucl. Med. 54, 776–784. doi: 10.2967/jnumed.112.111021
Yamin, G., Munishkina, L. A., Karymov, M. A., Lyubchenko, Y. L., Uversky, V. N., and Fink, A. L. (2005). Forcing nonamyloidogenic beta-synuclein to fibrillate. Biochemistry 44, 9096–9107. doi: 10.1021/bi048778a
Yancey, P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830. doi: 10.1242/jeb.01730
Yang, C., Trent, S., Ionescu-Tiba, V., Lan, L., Shioda, T., Sgroi, D., et al. (2006). Identification of cyclin D1- and estrogen-regulated genes contributing to breast carcinogenesis and progression. Cancer Res. 66, 11649–11658. doi: 10.1158/0008-5472.CAN-06-1645
Yi, M., Kaneko, S., Yu, D. Y., and Murakami, S. (1997). Hepatitis C virus envelope proteins bind lactoferrin. J. Virol. 71, 5997–6002.
Zhao, X., Pollock, D. M., Inscho, E. W., Zeldin, D. C., and Imig, J. D. (2003). Decreased renal cytochrome P450 2C enzymes and impaired vasodilation are associated with angiotensin salt-sensitive hypertension. Hypertension 41, 709–714. doi: 10.1161/01.HYP.0000047877.36743.FA
Keywords: camel genome, breeds, adaptations, immunogenomics, antibodies, anticancer
Citation: Ali A, Baby B and Vijayan R (2019) From Desert to Medicine: A Review of Camel Genomics and Therapeutic Products. Front. Genet. 10:17. doi: 10.3389/fgene.2019.00017
Received: 25 September 2018; Accepted: 14 January 2019;
Published: 19 February 2019.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
George E. Liu, Agricultural Research Service (USDA), United StatesCopyright © 2019 Ali, Baby and Vijayan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranjit Vijayan, cmFuaml0LnZAdWFldS5hYy5hZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.