- Goodman Cancer Research Centre and Department of Biochemistry, McGill University, Montreal, QC, Canada
The sequences and structures of 3′-untranslated regions (3′UTRs) of messenger RNAs govern their stability, localization, and expression. 3′UTR regulatory elements are recognized by a wide variety of trans-acting factors that include microRNAs (miRNAs), their associated machinery, and RNA-binding proteins (RBPs). In turn, these factors instigate common mechanistic strategies to execute the regulatory programs encoded by 3′UTRs. Here, we review classes of factors that recognize 3′UTR regulatory elements and the effector machineries they guide toward mRNAs to dictate their expression and fate. We outline illustrative examples of competitive, cooperative, and coordinated interplay such as mRNA localization and localized translation. We further review the recent advances in the study of mRNP granules and phase transition, and their possible significance for the functions of 3′UTRs. Finally, we highlight some of the most recent strategies aimed at deciphering the complexity of the regulatory codes of 3′UTRs, and identify some of the important remaining challenges.
Introduction
Precise spatial and temporal regulation of gene expression is necessary for the proper development and homeostasis of organisms. Systems approaches indicate that post-transcriptional mechanisms, in particular translational repression is the most significant contributor to establishing a gene’s expression in mammalian cells (Schwanhausser et al., 2011). Post-transcriptional regulation is instated by mechanisms that control translation, stability, and localization of mRNAs. Such mechanisms converge on one or several distinctive features of mRNAs (Figure 1).
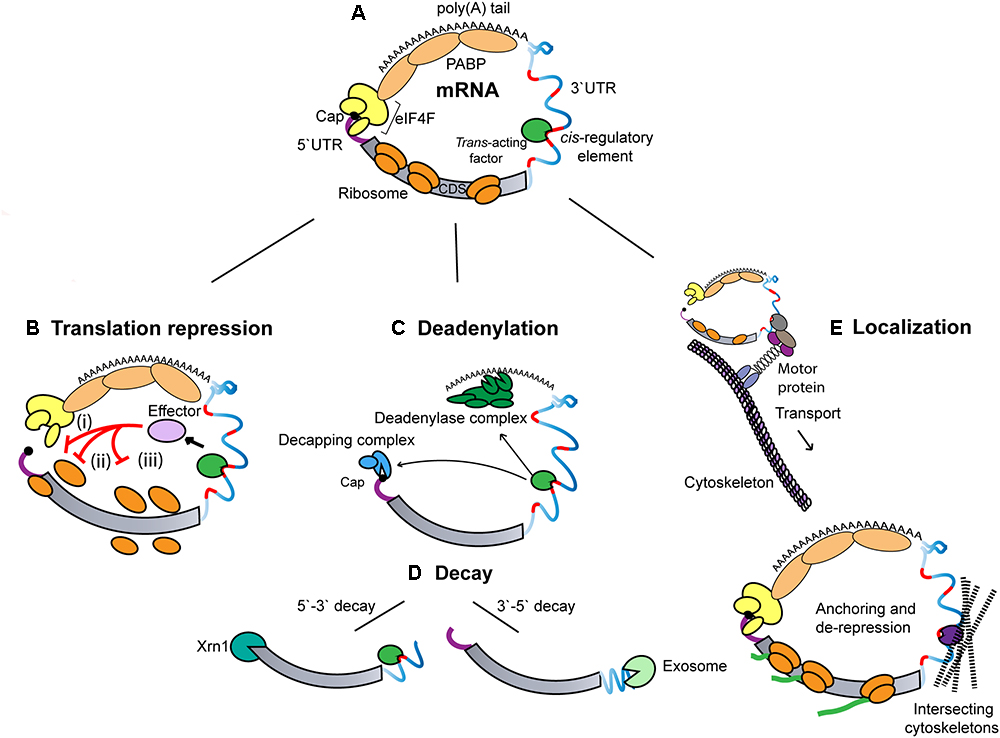
Figure 1. General modes and determinants of 3′UTR in post-transcriptional regulation. (A) Schematic illustration of the distinctive features of an eukaryotic mRNA. The 5′-terminal cap structure interacts with the 3′-terminal poly(A) tail of an mRNA through associated eIF4F and PABP. The coding sequence (CDS) is flanked by 5′- and 3′UTR, which harbors cis-regulatory sequences (marked in red) and provides a binding platform for trans-acting factors (green). (B) Translational repression mechanisms. (i) Competition/interference with cap-binding complex, eIF4F (ii) Inhibition of ribosomal subunit joining (iii) Inhibition of translation elongation. (C) Deadenylation and decapping. Recruitment of the CCR4-NOT deadenylase complex by trans-acting factors catalyzes the deadenylation of the mRNA target. This is often followed by the removal of the 5′-terminal cap structure by the decapping factors (DCP1-DCP2), and the associated co-factors. (D) mRNA decay. mRNAs that are deadenylated and decapped are rapidly degraded by either 5′- > 3′ exonuclease (XRN1) or 3′- > 5′ exonuclease (exosome). (E) RNA localization. Translationally repressed mRNAs are transported along the cytoskeleton to which it is tethered by RBPs and motor proteins. Upon reaching its destination, the mRNA is anchored, and its translation is de-repressed.
The coding sequence (CDS) of an mRNA is flanked by 5′- and 3′-untranslated regions (UTR). These sequences encode regulatory structures and sequences often referred to as cis-regulatory, or cis-acting elements. When unrepressed, interactions between the 5′-terminal cap, the eIF4F cap-binding complex (an assembly of eIF4E, eIF4A, and eIF4G), the 3′-terminal poly(A) tail and the associated poly(A) binding proteins (PABPs) lead to circularization of an mRNA (Gallie, 1991; Wells et al., 1998). mRNA circularization is thought to allow for synergy of the 5′-cap and poly(A) tail in potentiating translation initiation, and possibly also in stabilizing the mRNA (Sachs et al., 1997; Schwartz and Parker, 1999). Circularization brings 3′UTR cis-acting elements closer to the translation initiation machinery. Perhaps not surprisingly, 3′UTR-driven mechanisms determine the expression and fate of mRNAs by targeting the 5′-cap and 3′-poly(A) tail moieties and/or their associated cofactors.
The functional information encoded in the sequence and structure of 3′UTRs are decrypted and acted upon by an array of cellular regulatory factors (often referred to as trans-acting factors). Regulatory factors can be broken down into two distinct categories based on their direct molecular implication in (i) specific recognition of the 3′UTR sequence and structure, and (ii) execution of consequent activities. Factors involved in specific recognition include a variety of non-coding RNAs, such as microRNAs (miRNAs), and RNA-binding proteins (RBPs) to match the sequences and structural determinants encoded in 3′UTRs. A more limited diversity of effector machineries can be grouped in three effector activities: (i) translational control (Figure 1B), most often acting on translation initiation (Nelson et al., 2004; Humphreys et al., 2005; Chendrimada et al., 2007; Mathonnet et al., 2007; Zdanowicz et al., 2009), but also in some cases on translation elongation (Petersen et al., 2006; Gu et al., 2009), (ii) deadenylation and decay (Figures 1C,D), whereby deadenylation of an mRNA can be coupled to some degree to its decapping and decay, and (iii) localization (Figure 1E), which can be established through active RNA transport along the cytoskeleton and/or asymmetric anchoring of an mRNA in a cellular domain.
In many cases, including the examples presented below, more than one effector activity can be mobilized by a 3′UTR. Recognition and effector activities can involve synergistic, cooperative, or coordinated interactions dictated by the 3′UTR regulatory sequences themselves, but also by the cellular, sub-cellular, and biochemical context wherein the mRNA is found. mRNAs and the regulatory machineries are deeply affected by concentration, stoichiometry, affinities, RNA editing, protein post-translational modifications, and physical seclusion, all of which can change with cell identity or adaptation to environmental cues. Directly speaking to both cellular and biochemical contexts and re-emerging with the refining of different classes of RNA-protein condensates (referred to as mRNP granules) is the concept of phase transition. It remains less than clear how phase transition functionally intersects with 3′UTR regulatory mechanisms. Several hypotheses have recently been substantiated and will be discussed later in this review.
RNA-Binding Proteins (RBPs)
The human genome encodes more than 1,500 RBPs (reviewed in Hentze et al., 2018). Each one of these proteins is constituted of one or more RNA binding domains (RBD), which can be grouped in RBP families, and auxiliary domains that enable other interactions or carry out enzymatic activities (Gerstberger et al., 2014). Canonical RBDs that are often involved in 3′UTR recognition include RNA recognition motifs (RRM), K-Homology (KH) domain, several types of zinc finger domains, double-stranded RNA binding domain (dsRBD), Piwi/Argonaute/Zwille (PAZ) domain, Pumilio/FBF (PUF) domain, and Trim-NHL domain proteins (Lunde et al., 2007). Using intra-molecular or extra-molecular combinations of RBDs, RBPs can improve RNA recognition specificity, affinity, and avidity. Distinct surfaces of RBDs, specific motifs and auxiliary domains mediate the protein-protein interactions required to recruit and activate effector activities to mRNAs.
We will next review some well-characterized examples of how RBPs achieve these functions. Note that RBPs can also play a disruptive role on the activities guided by other regulatory elements in 3′UTRs. Those will be discussed later in this review.
PUF Proteins
Eukaryotic Pumilio and FEM-3 binding factor (PUF) proteins are part of a family of RBPs that can instigate translational repression, deadenylation and decay of targeted mRNAs. PUF proteins regulate a large number of mRNA targets involved in diverse biological functions. For example, Drosophila and Caenorhabditis elegans PUF proteins are important for the maintenance of stem cells (Wickens et al., 2002) and target mRNAs of central components of the Ras/MAPK, PI3K/Akt, NF-κB, and Notch signaling pathways (Kershner and Kimble, 2010). In mammalian cells, the precise dosage of PUF proteins is essential to fine-tune the expression of mRNAs encoding mitosis, DNA damage and DNA replication factors. Recently, PUF proteins were shown to be involved in a network of interactions with the NORAD lncRNA at its center, which prevents chromosomal instability (CIN) (Lee et al., 2016).
The PUF family of proteins binds RNAs bearing the 5′-UGUR (where R = purine) sequence (Quenault et al., 2011). The determinants of those interactions are understood to such an extent that a PUF protein’s specificity can actually be predicted (Hall, 2016). For example, the classical Drosophila Pumilio protein uses its eight α-helical Pumilio repeats to bind the eight-nucleotide sequence 5′-UGUANAUA. Furthermore, Pumilio proteins can be co-expressed. In Saccharomyces cerevisiae, co-expression of PUF proteins at different concentrations and with distinct binding affinities can result in competition for individual binding sites (Lapointe et al., 2015, 2017). Binding of PUF proteins to an mRNA typically leads to translational repression, deadenylation, and mRNA decapping. The yeast PUF-domain Mpt5p protein directly interacts with the ortholog of CAF1, one of the two catalytic subunits of the Carbon Catabolite Repressor-Negative on TATA (CCR4-NOT) deadenylase complex, through its RNA-binding domain (Goldstrohm et al., 2006). This interaction is conserved in metazoa, and C. elegans and human PUF homologs can also bind to the yeast CAF1 ortholog (Suh et al., 2009; Van Etten et al., 2012; Weidmann et al., 2014). PUF proteins can also repress mRNA expression by inducing their destabilization. Indeed, Mpt5p can recruit an eukaryotic translation initiation factor 4E (eIF4E)- binding protein to target mRNAs (Blewett and Goldstrohm, 2012). eIF4E-binding proteins block the interaction between eIF4E and eIF4G, and this typically prevents the recruitment of the 43S pre-initiation complex (PIC) to mRNAs (Haghighat et al., 1995). However, sometimes including this case, the interaction leads to the recruitment and activation of decapping and decay co-factors (Ferraiuolo et al., 2005; Nishimura et al., 2015).
Nanos and TRIM-NHL Proteins
The outcome of PUF protein binding to mRNA targets can be altered through interactions with other RBPs. This is the case for the prototypical Pumilio protein in the regulation of hunchback mRNA in Drosophila (Sonoda and Wharton, 2001), wherein its functions are highly dependent on Nanos and Brain Tumor (Brat) proteins. The RNA-binding specificity of Nanos is defined by its interactions with Pumilio, and Nanos directly interacts with the CCR4-NOT deadenylase complex to promote deadenylation of mRNAs (Curtis et al., 1997; Kraemer et al., 1999; Sonoda and Wharton, 1999; Kadyrova et al., 2007). Brat, a member of the broadly conserved TRIM-NHL family of proteins, forms a ternary complex with Pumilio and Nanos. This complex recruits the effector protein 4EHP to repress the translation of mRNAs (Cho et al., 2006). 4EHP is an eIF4E-like cap binding protein that does not interact with eIF4G and impairs ribosome recruitment to the mRNA (Rom et al., 1998). Unlike Nanos, Brat can stably bind RNA on its own through its NHL domain, and can also function independently of PUF proteins (Laver et al., 2015). Proteomic analysis of CCR4-NOT complex also suggests an interaction with Brat (Temme et al., 2010). It remains unknown whether this is a direct interaction and whether it contributes to and/or is necessary for mRNA repression. TRIM-NHL proteins exert a broader set of biological functions beyond their interplay with Pumilio in Drosophila embryo. They play critical roles in brain development, cell polarity, and sex determination (Tocchini and Ciosk, 2015). It is quite possible that this family drives different mechanisms in different cellular or physiological contexts, and that functional interactions with other RBP families may depend on the mRNA target and/or its genetic niche.
HuR and TTP Proteins
The presence of adenylate/uridylate (AU)-rich sequences in 3′UTRs has long been associated with regulation of mRNA stability (Barreau et al., 2005). Early computational analysis of human mRNA datasets estimated that 8% of mRNAs harbor AU-rich elements (Bakheet et al., 2006). While AU-rich sequences may be expected to contribute to the destabilization of 3′UTR folding structures, they are also directly recognized by a diversity of RBPs. Tristetraprolin (TTP) and its paralogs: butyrate response factors 1 and 2 (BRF-1/2), bind to AU-rich elements through their two zinc-finger domains and promote the decay of mRNAs (Lai et al., 2000). Here again, TTP or BRF direct mRNA destabilization by recruiting effectors of deadenylation, decapping, and 5′- and 3′-exonuclease activities (Lykke-Andersen and Wagner, 2005; Sandler et al., 2011). Interactions with effectors have been mapped to an auxiliary N-terminal domain, which is sufficient to trigger the decay of target mRNAs (Lykke-Andersen and Wagner, 2005). The XRN1 5′- > 3′ exonuclease is thought to be the enzyme effecting mRNA degradation instigated by TTP. It is recruited through the Enhancer of Decapping-4 (EDC4) scaffolding protein (Chang et al., 2014).
Not all AU-rich encoding mRNAs are subjected to degradation. In fact, closely similar sequences can instead lead to enhanced mRNA stability. Such a response often occurs when the HuR protein associates with AU-rich sequences (Brennan and Steitz, 2001). HuR is ubiquitously expressed and belongs to the Embryonic lethal abnormal vision (ELAV) family of proteins (Ma et al., 1996). The exact molecular mechanism used by HuR to confer mRNA stability is still being resolved (von Roretz et al., 2011). An early study showed that overexpression of HuR could slow the decay of mRNAs without impacting their deadenylation rates (Peng et al., 1998). The prevailing model proposes that HuR can stabilize AU-rich encoding mRNAs through competition for binding with factors such as TTP or a subset of miRNAs. Some of the keys to predicting whether an AU-rich sequence dictates degradation, stabilization or has no impact on an mRNA will likely lie in quantitative parameters such as stoichiometry of AU-rich elements and RBPs, and their binding affinities. Future studies may thus benefit from quantitative approaches in specific cell types.
microRNAs (miRNAs)
miRNAs are genome-encoded, ∼22-nucleotide (nt)-long RNA molecules which guide the associated proteins toward binding sites located in the 3′UTRs of mRNAs to repress their expression. miRNAs were first discovered in C. elegans where they regulate the heterochronic cascade of genes that pre-determines cell fate and developmental transitions (the lin-4 and let-7 miRNAs) (Lee et al., 1993; Wightman et al., 1993; Reinhart et al., 2000). A turning point for the fields of miRNAs and 3′UTRs was the identification of several let-7 homologs in other species including humans (Pasquinelli et al., 2000). This discovery coincided with important advances in sequencing technologies and sparked a concerted effort of miRNA sequencing and prediction, leading to the identification of thousands of new miRNAs (Lee and Ambros, 2001; Lau et al., 2001; Lagos-Quintana et al., 2001; Friedman et al., 2009). Currently, more than two thousand miRNAs have been identified in the human genome, and the miRbase database contains 48,885 mature miRNAs from a total of 271 species (Kozomara and Griffiths-Jones, 2014). Since their conservation across species has been shown, miRNAs have been implicated in a myriad of functional cascades across metazoans, including development, signaling, immune system, and metabolism (Ameres and Zamore, 2013). Conversely, their mis-expression or misregulation contributes to or plays instrumental roles in a variety of diseases ranging from heart disease to diabetes to cancer (Hesse and Arenz, 2014).
The base-pairing of miRNAs with 3′UTR sequences is quite distinct from what is to be expected from a ‘free’ single-stranded RNA of the same length. A miRNA’s target recognition kinetics and specificity are largely dictated by its interactions with the Argonaute protein within which it is bound in the cell (for a review, see Duchaine and Fabian, 2018). The miRNA strand is stretched across Argonaute’s croissant-shaped structure by interactions with its four domains. On its 5′ end, the miRNA interacts with the Mid and PIWI domains. Across a central cleft, the 3′ end of the miRNA is bound to the PAZ domain which closely interacts with the N-domain. Extensive interactions pre-orients the 5′-most bases of the miRNA (nts 2-8), a region called the seed, into a favorable conformation for pairing with target sequences. Target recognition through the seed is a two-step process wherein the rate limiting step is the pairing of nts 2–5 and the dissociation rate is largely determined by the pairing of nts 6–8 (Wee et al., 2012; Schirle et al., 2014; Chandradoss et al., 2015; Salomon et al., 2016). Multiple genomic studies and individual miRNA-binding sites have indicated that, alternative non-canonical routes of target recognition may be prevalent. For example, some miRNAs further use the 3′ end of the miRNA in target recognition (Broughton et al., 2016; Brancati and Großhans, 2018). Such alternative modes of target recognition likely involve dynamic interactions with the N-PAZ pair of Argonaute domains.
The importance of the interactions and molecular mechanics of the Argonaute scaffold in dictating miRNA targeting kinetics recently led the Zamore group to suggest that the miRNA/Argonaute (a minimal assembly referred to as RISC) behaves as a ‘programmable RNA-binding protein’ (Salomon et al., 2016). Incidentally, this analogy further extends to the effector activities that are mobilized by miRNAs, which largely overlap with effectors and mechanisms mobilized by RBPs. Metazoan Argonautes that are programmed by miRNAs also stably interact with the TNRC6 or GW182 family of proteins. This constitutes the core of a complex often referred to as miRNA Induced Silencing Complex or miRISC (Jonas and Izaurralde, 2015). In essence, GW182 proteins bridge interactions between Argonaute proteins and effector complexes including mRNA deadenylation, decapping and decay machineries. Here again, the CCR4-NOT complex plays a central and pivotal role (Fabian et al., 2011; Braun et al., 2013; Jonas and Izaurralde, 2015). We will thus next examine in more details the architecture, interactions and important functions of the CCR4-NOT complex in determining the fate of mRNAs.
The CCR4-NOT Complex: A Hub for 3′UTR Effector Activities
The CCR4-NOT complex plays a central role in the fate of an important diversity of mRNAs. Other deadenylases such as the PAN2/3 complex exert a regulatory function, but on a more limited subset of mRNAs and on population of longer poly(A) tails (Chen and Shyu, 2011). However, the CCR4-NOT complex seems to be responsible for most poly(A) tail controls in metazoan transcriptomes where it has been examined (Tucker et al., 2001; Temme et al., 2004; Yamashita et al., 2005; Schwede et al., 2008; Nousch et al., 2013). The CCR4-NOT complex integrates the effector functions in mechanisms initiated by a diversity of RNA-binding proteins and miRNAs (Figure 2). CCR4-NOT consists of two highly conserved modules: the CNOT1/2/3 proteins constitute a scaffolding module for all the subunits of the complex, while the catalytic module of the complex is formed by two deadenylases, EEP-type CCR4 and DEDD-type CAF1. Their functions partially overlap or compensate for each other in vivo, but CAF1 is believed to assume the bulk of the function in miRNA-directed deadenylation (Fabian et al., 2009). Beyond scaffolding the CCR4-NOT complex, the central CNOT1 subunit acts as a tether and directly interacts with GW182, TTP, Nanos, PUF, Smaug, and several other RNA-binding proteins in different cells and organisms (Wahle and Winkler, 2013).
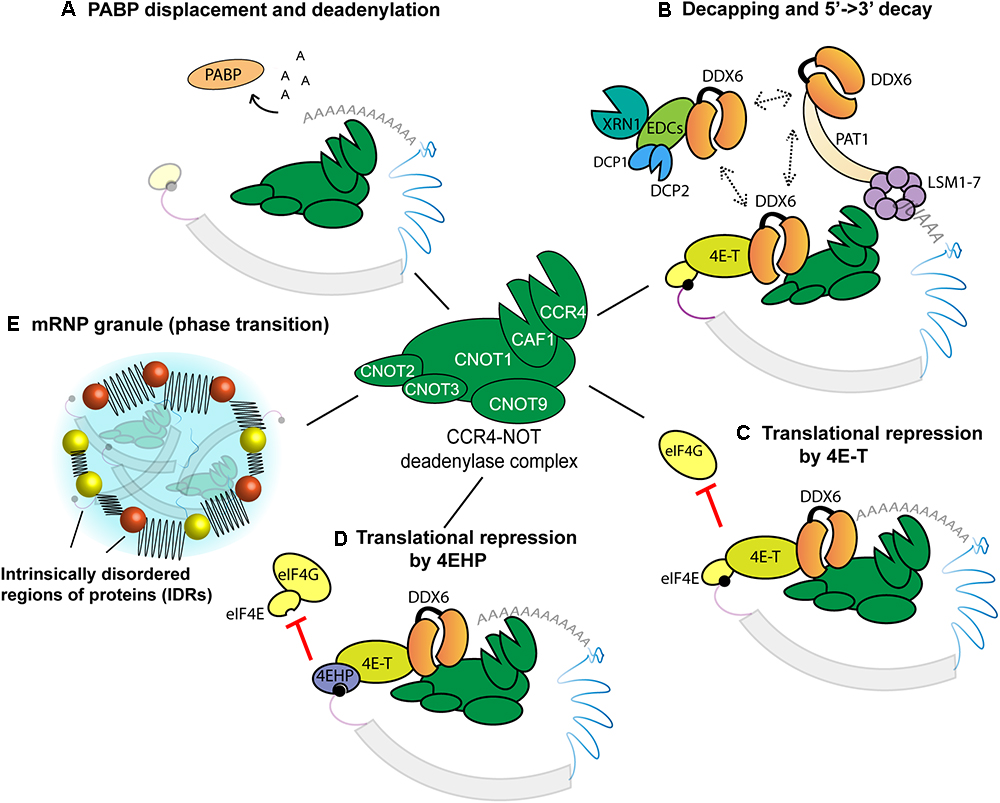
Figure 2. Roles of the CCR4-NOT complex and associated proteins in effecting 3′UTR-encoded gene regulation. The CCR4-NOT complex contains at least eight subunits, of which only six are shown here. (A) Inhibition of mRNA circularization by PABP displacement and deadenylation. (B) CCR4-NOT-directed mRNA decay. The CCR4-NOT complex deadenylates the mRNA and recruits DDX6. DDX6 promotes decay through three mutually exclusive interactions, with 4E-T, EDC-3, and PAT1 (dashed lines). (C,D) Inhibition of translation through CCR4-NOT. Note that all mechanisms depicted target initiation. (C) DDX6 recruits 4E-T to prevent the binding of eIF4G to eIF4E. (D) 4EHP is recruited to the cap through 4E-T/DDX6/CCR4-NOT complex. (E) Assembly of an mRNP granule. CCR4-NOT and intrinsically disordered regions (IDRs)-encoding proteins are sequentially recruited to target mRNAs to promote mRNP formation, possibly enabling or promoting phase transition.
Recruitment of the CCR4-NOT complex to mRNAs is associated with its deadenylation activities, but a different perspective on the function of this complex has recently emerged. The CCR4-NOT complex also recruits distinct activities such as decapping and exonucleases (Figure 2B) that are often coupled with deadenylation, but also with cap-binding and translation repression without mRNA deadenylation or decay (Figures 2C,D). Its interactions with intrinsically-disordered region (IDR)-encoding proteins that are components of the mRNP in the C. elegans embryo recently suggested a role in nucleating phase transition (Wu et al., 2017) (Figure 2E).
mRNA Deadenylation and Decay
In addition to its role in translation initiation, PABP is a cofactor of deadenylases, including the CCR4-NOT complex (Fabian et al., 2009; Huntzinger et al., 2013). In vitro, PABP accelerates the deadenylation of long 3′UTRs for which the poly(A) tail is distant to the regulatory sequences (Flamand et al., 2016). The first step in deadenylation of an mRNA is thought to be the displacement of PABP proteins from the poly(A) tail by cofactors recruited through the GW182 protein and CCR4-NOT complex (Moretti et al., 2012; Zekri et al., 2013). Removal of the poly(A) tail is then catalyzed by the CAF-1 and CCR4 deadenylases subunits (Figure 2A).
In metazoans, deadenylation is often tightly coupled with mRNA decapping and decay (Figure 2B). Earlier studies showed that following the shortening of a poly(A) tail below a certain threshold, an mRNA is subjected to first-order decay (Chen et al., 2008). mRNA deadenylation and decay are clearly coupled in early zebrafish embryo, where mRNA deadenylation instigated by the miR-430 family of miRNAs marks the initial step in the decay of an important fraction of maternal mRNAs in the Maternal-to-Zygotic Transition (MZT) (Giraldez et al., 2005, 2006). This is also obvious in Drosophila S2 cultured cells, where fully deadenylated mRNAs do not accumulate, and impairing the decapping enzymes Dcp1/2 is necessary to detect the deadenylated species (Eulalio et al., 2009). The LSM1-7 proteins are thought to form a ring-like complex around the remnants of the shortened poly(A) tail and to promote mRNA decapping and decay (Tharun, 2009).
A key protein, which physically couples the CCR4-NOT complex with decapping and decay, is the DEAD-box protein DDX6. DDX6 directly interacts with CNOT1 subunit and multiple decapping/decay factors, either simultaneously or through mutually exclusive interactions (Tritschler et al., 2009; Sharif et al., 2013; Chen et al., 2014; Mathys et al., 2014; Rouya et al., 2014; Nishimura et al., 2015; Ozgur et al., 2015). Interestingly, DDX6 also interacts with eIF4E-transporter (4E-T). This interaction is thought to increase the local concentration of decapping factors such as DCP2 around the 5′-cap, thus enabling competition with eIF4E (Nishimura et al., 2015). The removal of the 5′-cap structure by DCP2 seals the fate of the mRNA toward degradation via the 5′- > 3′ decay pathway mediated by XRN1 (Arribas-Layton et al., 2013). The activity of DCP2 is greatly enhanced by DCP1 and additional factors such as enhancers of decapping (EDC-3, EDC-4), PAT1, and the LSM1-7 complex (Jonas and Izaurralde, 2013) (Figure 2B). Alternative routes of mRNA decay have also been proposed, which would proceed from the 3′ end and through the cytoplasmic exosome complex (Chen and Shyu, 2011).
Translational Repression
mRNA deadenylation abolishes the physical and functional synergy between the 5′-cap and poly(A) tail, resulting in translational repression (Mishima et al., 2006; Wakiyama et al., 2007). However, strong evidence indicates that the CCR4-NOT complex can also participate in direct translational repression, through mechanisms that do not involve its deadenylase activities (Figures 2C,D). Using luciferase reporters engineered to block deadenylation, an early study showed that tethering of Xenopus or human CAF1 is sufficient to repress mRNAs (Cooke et al., 2010). Several other reports, using different experimental designs and systems, have since then confirmed the role of CCR4-NOT as a direct translational repressor (Braun et al., 2011; Chekulaeva et al., 2011; Flamand et al., 2016; Chapat et al., 2017). Models proposed to explain this activity have accumulated in recent years and were substantiated to different extents. Disruption of mRNA circularization by displacement of PABP through CCR4-NOT and its cofactors has been suggested as one mechanism (Zekri et al., 2013). Other mechanisms instead revolve around displacement of interactions with the 5′-cap of targeted mRNAs, and DDX6 is also central for these functions of CCR4-NOT.
DDX6 can recruit 4E-T whose interaction with eIF4E can displace eIF4G and thus mediate translational repression (Kamenska et al., 2016). Repression can also occur through the strong interaction between 4E-T and 4EHP (Joshi et al., 2004; Cho et al., 2005). Recruitment of this dimer to CCR4-NOT through DDX6 was recently involved in translation repression by miRNAs (Chapat et al., 2017). A subset of mRNAs is translationally regulated through this 4EHP-4E-T mechanism in mammalian cells, among which DUSP6 plays an important role in fine-tuning the ERK signaling cascade (Jafarnejad et al., 2018). This last study is unique in identifying a physiological purpose to one of the many CCR4-NOT ‘pure’ translational repression mechanisms. Indeed, the physiological importance has yet to be determined for most of those mechanisms, which were identified in cell culture and/or in vitro. It remains possible that distinct mechanisms will be predominant in different cellular contexts or on particular mRNA targets.
Cooperative and Competitive Interplay Among RBPs and miRISC
RBPs and miRISC can interact among themselves and with each other to alter the fate of mRNAs through either cooperation or competition. Considering the importance of 3′UTR sequences and the diversity and density of potential binding sites for RBPs and miRNAs, it is hard to expect otherwise. The median length of human 3′UTRs is 1,200 nt (Jan et al., 2011). On average each mRNA 3′UTR is bound by 14 RBPs (Plass et al., 2017), and ∼70% of vertebrate 3′UTRs encode multiple sites for different miRNA families (Friedman et al., 2009). Neither miRNA- nor RBP binding sites are distributed randomly in 3′UTR sequences. Early on, genomic studies have shown that miRNA-binding sites are more likely to be functional when they are located close to each other, or when located close to the ORF or the poly(A) tail (Grimson et al., 2007; Saetrom et al., 2007). Similarly, genomic analyses indicate that AU-rich sequences are associated with a greater functional output of nearby miRNA-binding sites, and computational analyses of the mammalian genomes indicate that recognition sites for PUF proteins and AU-rich sequences are enriched within 50 nt of binding sites for a subset of miRNAs (Jiang et al., 2013).
miRNA–miRNA Cooperativity
Signs that miRNA-mediated silencing acts through a cooperative mechanism were already visible in the seminal discovery papers in C. elegans. The 3′UTR of lin-14 encodes 7 potential base-pairing sites (Lee et al., 1993), while the lin-41 3′UTR harbors two let-7 miRNA-binding sites, separated by intervening sequences of 27 nt in length (Reinhart et al., 2000). If each of these individual sites were independently functional, some degree of redundancy could be expected, with their individual impairment having limited to no consequence. Instead, both let-7 sites in the lin-41 3′UTR are important in vivo (Vella et al., 2004). Likewise, binding sites for lin-4 and let-7, and multiple sites for lsy-6 functionally interact on the lin-28 and cog-1 mRNAs, respectively (Moss et al., 1997; Reinhart et al., 2000; Didiano and Hobert, 2008). In vitro and in vivo studies later demonstrated that miR-35 and miR-58 miRNAs cooperate in the deadenylation and the silencing of the C. elegans egl-1/BIM mRNA (Wu et al., 2010; Sherrard et al., 2017). In addition to the fore-mentioned early genomic studies, which support miRNA cooperativity, mammalian reporter assays clearly confirmed that a combination of sites exert a much more potent silencing output (Broderick et al., 2011). While some studies examined miRNA-binding site cooperativity on natural or fragments of 3′UTR sequences (Koscianska et al., 2015; Schouten et al., 2015), there are few detailed studies of miRNA-binding site interplay.
The mechanisms underlying miRNA cooperativity are still poorly resolved, but three models have been proposed and two have been substantiated experimentally. First, miRISC binding to nearby miRNA-binding sites can enhance their affinity for the 3′UTR (Broderick et al., 2011; Flamand et al., 2017). This type of cooperativity in target binding is in fact required for some non-seed miRNA-binding sites to be stably bound by miRISC and to be functional (Flamand et al., 2017). A second model involves the cooperative recruitment of effector machineries. In an embryonic cell-free system, a reporter mRNA bearing a single miRNA-binding site was not deadenylated, and could not recruit the CCR4-NOT complex, whereas a reporter encoding three adjacent miRNA-binding sites did so efficiently (Flamand et al., 2017). Whether this mode of cooperativity is especially important in the embryo and/or in C. elegans is not known at present. A third, mutually not exclusive, possibility could involve the cooperative activation of effector activities. CCR4-NOT recruitment by miRISC on 3′UTRs may not be sufficient on its own to trigger mRNA deadenylation and decay. A stoichiometric threshold, a specific configuration of target sites, post-translational modifications and/or conformation changes of miRISC may be required to trigger effector activation. These variations would be consistent with other protein/nucleic acid interaction paradigms, such as transcription factors.
RBP-miRISC 3′UTR Interactions
RBPs, miRNAs and the associated machineries can regulate their activities through cooperative or competitive interplay. It is likely that the mechanisms at work in cooperating miRNA-binding sites may also explain some of the RBP-miRNA cooperativity. Putative examples of direct interplay may include the cooperation of TTP with miR-16 in regulating TNF-alpha mRNA (Jing et al., 2005), and AU-rich sequences near the miR-16 binding site in the 3′UTR of COX-2 mRNA (Young et al., 2012). Positive interplay can also be indirect, through the modulation of global or local 3′UTR structures. Because they do not code, 3′UTRs can adopt complex folding structures, which can have positive or negative impacts on overlapping or nearby regulatory sequences. Structures can constitute determinants for the recognition of other RBPs, or limit binding to miRNA-binding sites. In turn, binding of miRISC or RBP to high-affinity sites can destabilize folding structures and facilitate access to nearby binding sites. This model explains the effect of Pumilio on the 3′UTR of the p27 tumor suppressor. Pumilio binding promotes a change in the local structure of the RNA that allows the binding of miR-221 and miR-222, leading to silencing of the p27 mRNA (Kedde et al., 2010). Similarly, a study showed that HuR could enhance the activity of let-7 on c-Myc mRNA. This is also likely through a change in the local structure of the RNA resulting in the unmasking of the let-7 binding site (Kim et al., 2009).
In the simplest form of antagonistic interaction, overlapping or nearby binding sites can lead to direct competition between RBPs and miRNAs/miRISC through steric hindrance. A survey by Keene and colleagues suggested that HuR prevents the function of abundant miRNAs on nearby and overlapping sites in a subset of mRNAs in HEK293 cells (Mukherjee et al., 2011). Similarly, Fillipowicz and colleagues showed that HuR could displace miRISC bound to a target mRNA thereby alleviating miRNA-mediated repression. This displacement occurs when HuR binds to AU-rich sequences 20–50 nt away from the miRNA-binding site (Kundu et al., 2012), again suggesting steric interference. The HuR example illustrates the fact that an RBP can have both positive or negative impacts on miRNA-binding site function, depending on 3′UTR structure and binding site positioning. It also highlights that interactions between 3′UTR structures, regulatory sequences and their trans-acting factors are precisely tuned through co-evolution.
Coordinated and Sequential 3′UTR Activities
Beyond simple positive or negative interplay, 3′UTR sequences can lead to the coordination of post-transcriptional mechanisms in both time and space. The mechanism underlying miRNA-mediated silencing is in itself a coordinated series of events wherein mRNA translation repression precedes deadenylation, which in turn precedes decapping and decay. Translation repression can be resolved in vitro in a mammalian cell-free system (Mathonnet et al., 2007), in vivo in cell culture (Djuranovic et al., 2012), and even occur at distinct but subsequent developmental stages during early zebrafish embryo development (Bazzini et al., 2012). The biological purpose of this series of events, however, remains to be fully elucidated. Some of these steps in the silencing mechanism may be expected to be at least partially redundant with regards to the impact on gene expression. However, one possibility is that translation inhibition enables faster repression, e.g., when a binary decision is promptly required. Another possibility is that this allows for reversible repression in the early steps, whereas decapping and decay may offer a more permanent decision.
RNA Localization
The coordination of 3′UTR-driven activities is clearly illustrated through examples of active mRNA transport and localization. A majority of mRNAs are localized to subcellular regions and most examples where the underlying mechanisms have been detailed involve 3′UTR regulatory elements (Jansen, 2001; Lécuyer et al., 2007). mRNA localization can be achieved through several mechanisms (reviewed in Martin and Ephrussi, 2009). In active mRNA transport, the mRNA is assembled in a ribonucleoprotein (mRNP) complex through the specific binding of a combination of RBPs to the 3′UTR of an mRNA (Figure 1E). Bound RBPs recruit effector proteins that repress translation and mediate interactions with motor proteins. The repressed mRNP is then transported via the cytoskeleton until it reaches its destination where it is anchored. The mRNA is then de-repressed at the appropriate time and place through a series of events involving displacement/competition by other RBPs, and/or post-translational modifications (reviewed in Besse and Ephrussi, 2008).
Oskar mRNA Localization
Localization of oskar mRNA in the Drosophila oocyte is the archetype, and remains one of the best-characterized examples of active mRNA transport (Ephrussi et al., 1991; Kim-Ha et al., 1991). oskar mRNA localization to the posterior pole of the oocyte occurs via microtubules through interactions with Staufen (Stau), tropomyosin and EJC components (Micklem et al., 2000; Zimyanin et al., 2008). Localized expression of oskar ensures proper patterning of the posterior body axis and germline fate (Kim-Ha et al., 1991). Mislocalization to the anterior pole leads to ectopic formation of abdomen and germ cells (Ephrussi and Lehmann, 1992), and absence of Oskar protein leads to loss of germ cells and aberrant abdominal segments (Lehmann and Nusslein-Volhard, 1986). Moreover, premature translation of localizing oskar mRNAs also results in patterning defects (Smith et al., 1992). Translational repression is achieved by Bruno RBP binding to multiple elements in the 3′UTR of oskar mRNA (Kim-Ha et al., 1995), which also recruits an eIF4E-binding protein, Cup (Wilhelm et al., 2003) to the mRNA. Similar to the 4E-T protein, Cup disrupts the interaction between eIF4E and eIF4G and prevents 43S pre-initiation complex binding to oskar mRNA (Nakamura et al., 2004). Bruno further represses oskar mRNA by promoting its oligomerization, a process which likely also contributes to rendering it inaccessible to the translation machinery (Chekulaeva et al., 2006). The Polypyrimidine Tract-Binding protein (PTB), which binds to multiple sites in oskar mRNA 3′UTR, is also essential for mRNA oligomerization and densely packed mRNP particles (Besse et al., 2009).
The fates of oskar and nanos mRNAs are closely linked in the Drosophila oocyte. nanos mRNA is also localized to the posterior pole, and is rapidly deadenylated and degraded elsewhere in the early embryo through the recruitment of CCR4-NOT complex by Smaug (Smibert et al., 1996, 1999; Bashirullah et al., 1999; Dahanukar et al., 1999; Zaessinger et al., 2006). Translation of nanos mRNA at the posterior pole is thought to be activated by the Oskar and Vasa proteins, but the exact underlying mechanism remains unclear (Ephrussi and Lehmann, 1992; Smith et al., 1992). Oskar could inhibit the function of Smaug, either by affecting the binding of Smaug to nanos mRNA or by interfering with the recruitment of the CCR4-NOT complex. It is also clear that some of the keys to solving the underlying mechanism will stem from the properties of phase transition in the posterior pole germ plasm (see below).
mRNA Routes in Mammalian Cells
An important variety of RNA localization events have been described in mammalian cells. Among them, the cascades dictated by the Zipcode and A2RE/RTS cis-acting elements provide well-delineated examples of how mammalian mRNAs can be sorted and locally translated in distinct cell types through information encoded in 3′UTRs. They also illustrate how localized cellular signaling can determine the precise site of translation of localized mRNAs.
Zipcode and the Zipcode-Binding Protein 1
β-actin mRNA localizes to the leading edge of the fibroblasts (Lawrence and Singer, 1986), and analogous mechanisms are thought to be at work in developing neurites and hippocampal dendrites (Bassell et al., 1998; Zhang et al., 1999, 2001; Eom et al., 2003; Shav-Tal and Singer, 2005). Localization of β-actin mRNA is instigated by the zipcode binding protein 1 (ZBP1) (Ross et al., 1997), which specifically binds a 54-nt long 3′UTR segment termed the ‘Zipcode’ (Kislauskis et al., 1994). The motor for β-actin mRNA localization in fibroblasts was only recently identified (Song et al., 2015). KIF11, a tubulin-associated motor, associates with the β-actin mRNPs wherein it directly interacts with ZBP. Disruption of this interaction in vivo leads to β-actin mRNA mis-localization and perturbs cell motility.
The exact nature of the mechanism responsible for the silencing of transported β-actin mRNAs remains unclear. Single-cell live imaging revealed an anti-correlation between the association of ZBP1 or ribosomes with β-actin mRNA (Wu et al., 2015). The authors thus proposed that the packaging of β-actin mRNA into mRNP granules may seclude mRNAs from ribosomes, and thus pre-empt translation. On one hand, the pervasive nature of mRNP granule formation in mRNA localization suggests that translation repression may be at least partly achieved through packaging of such mRNPs. On the other hand, the events leading to localized mRNA translational de-repression are rarely defined. For β-actin mRNA, this appears to result from signaling cascades locally converging on trans-acting factors. Upon reaching the endpoint of mRNA transport, phosphorylation of ZBP1 on a tyrosine residue by the protein kinase Src, which is closely associated with the cell membrane, disrupts RNA binding and relieves β-actin mRNA from translational repression (Hüttelmaier et al., 2005).
The A2RE/RTS Pathway
The A2 response element (A2RE) or RNA trafficking signal (RTS) is an 11-nt cis-acting element recognized by the heterogenous nuclear ribonucleoprotein A2 (hnRNP A2) and CArG-box binding factor A (CBF-A) proteins. The importance of this element was originally described in the transport of Myelin Basic Protein (MBP) mRNA in oligodendrocyte processes (Ainger et al., 1997; Carson et al., 1997; Hoek et al., 1998; Munro et al., 1999). A2RE/RTS-like sequences have since then been identified in a growing number of localized transcripts including BC1, αCaMKII, NG, ARC, BDNF, Prm2 mRNAs, and HIV RNAs (Mouland et al., 2001; Muslimov et al., 2006; Gao et al., 2008; Raju et al., 2011; Fukuda et al., 2013). Though the mechanism of translation inhibition remains unclear for most of these mRNAs, assembly of MBP mRNA molecules into granules somehow maintains the transcripts in a repressed state. Just like for β-actin mRNA, phosphorylation of a trans-acting factor is key to enable the translation of MBP mRNA, which is released at sites of glia-neuronal contacts through phosphorylation of hnRNP A2 and hnRNP F by the Fyn kinase (White et al., 2008, 2012).
Xenopus Oocyte mRNA Localization Pathways
The developing Xenopus laevis oocyte features mRNA localization examples that illustrate how elements in 3′UTRs direct toward distinct localization path in successive stages of development. During the six stages of oogenesis, RNAs localize along the animal/vegetal (A/V) axis of the oocyte (Kloc et al., 2001) through the early and late pathways. In the early pathway, germ plasm RNAs such as DEADSouth, Xpat, Xcat2, and Xdazl are transported by associating with a membrane-less structure termed the mitochondrial cloud (MC) or Balbiani body. This body contains germinal granules, endoplasmic reticulum, mitochondria, and is surrounded with bundles of intermediate filaments, which were suggested to play a role in maintaining its structure (Heasman et al., 1984; Forristall et al., 1995; Kloc and Etkin, 1995; Gard et al., 1997; King et al., 2005; Carotenuto and Tussellino, 2018). During stage II of oogenesis, the mitochondrial cloud expands between the nucleus and vegetal cortex. This expansion is thought to ‘push’ the germinal granules and RNAs toward the vegetal cortex where they are anchored on the cytoskeleton (Alarcon and Elinson, 2001; Wilk et al., 2005). Two distinct localization elements (LE) are encoded in the Xcat2 mRNA 3′UTR (Mosquera et al., 1993). A proximal 240 nt-long element is required for mitochondrial cloud localization (MCLE), whereas a distal ∼160 nt-long germinal granule localization element (GGLE) enables incorporation into germinal granules present inside the MC. Both localization signals are necessary for the proper localization of Xcat2 mRNA, which highlights the coordinated contributions of both 3′UTR elements (Kloc et al., 2000). Xcat2 mRNA is translationally repressed in the MC (MacArthur et al., 1999), and a few studies have implicated the RNA-binding protein Hermes in the repression of Xcat2 mRNP (King et al., 2005; Song et al., 2007; Nijjar and Woodland, 2013).
In the late localization pathway, mRNAs involved in somatic cell fates such as Vg1 and VegT are transported to the vegetal cortex in a microtubule-dependent mechanism (Kloc and Etkin, 1998). The 3′UTR of Vg1 mRNA encodes a 340-nt long LE, wherein clusters of short motifs are bound by the Vera and hnRNP I proteins (Deshler et al., 1997). The Vg1 LE is thought to be initially recognized by hnRNP I. This interaction remodels the Vg1 mRNP, which in turn allows Vera to bind Vg1 mRNA directly. Other factors are then recruited to the Vg1 mRNP including Staufen, Prrp and a kinesin motor to enact localization (Zhao et al., 2001; Yoon and Mowry, 2004; Lewis and Mowry, 2007; Lewis et al., 2008). Only after localizing to the vegetal cortex at the late stage IV of oogenesis is Vg1 mRNA translated (Dale et al., 1989; Tannahill and Melton, 1989). The spatiotemporal control of Vg1 mRNA translation is dictated by the 250-nt long translation-control element (TCE) encoded downstream of the Vg1 LE (Wilhelm et al., 2000; Otero et al., 2001). ElrB, a member of the ELAV family of RBPs, interacts with the TCE of Vg1 mRNA (Colegrove-Otero et al., 2005). This interaction correlates with the repression of the Vg1 mRNA, but how ElrB effects translational repression is not known.
mRNPs: Going Through Phases in the Lives of mRNAs
Mechanisms involving 3′UTR regulatory elements have long been associated with large mRNP granules. These granules can reach massive sizes by molecular standards (Brangwynne, 2013), often rivaling organelles. The list of large mRNP granules is rapidly expanding and includes P-bodies (originally named GW bodies), germ granules (also called polar granules and P granules, depending on species), stress granules, and the mRNA transport particles (Voronina et al., 2011), among others. Similarities and differences in the composition of large mRNPs have been documented (Eulalio et al., 2007a), mainly through comparison of associated markers by immunofluorescence. For example, stress granules are often distinguished from co-expressed P-bodies through exclusive colocalization of G3BP and DCP2, respectively (Ingelfinger et al., 2002; Tourriere et al., 2003; Kedersha and Anderson, 2007). In the early embryo, germ granules are distinguished from P-bodies through their association with germline markers such as PIE-1 in C. elegans (Strome, 2005). The absence of membranes in these organelle-sized particles and their scale led to their non-specific description as ‘large aggregates’ of RNA and proteins. A function in local mRNA concentration or storage for germ granules was naturally inferred from their scale and their concentration of maternal mRNAs in the oocyte (Noble et al., 2008; Voronina et al., 2011). Their importance in storage and protection of subsets of mRNAs from degradation was substantiated by well-defined examples, including the above-described nanos mRNA in Drosophila. The mRNA storage/protection model for mRNPs is also often associated with seclusion from the translational machinery. For example, in the developing oocytes of C. elegans, P granules help store translationally silent transcripts to prevent premature differentiation (Boag et al., 2008). Later in the embryo, P granules selectively repress somatic mRNAs in the P-lineage blastomeres, but not germline mRNAs to maintain germline fate and totipotency (Gallo et al., 2010; Updike et al., 2014).
While a role in mRNA storage makes sense and appears to be well supported, the biochemical nature of large mRNPs has remained elusive since the identification of the electron-dense ‘nuage’ structures in the early days of germline and developmental biology (Wilsch-Bräuninger et al., 1997). A breakthrough was recently made in the mechanisms of assembly and disassembly of mRNP granules. Hyman and colleagues showed that P granules in fact form by phase separation. Granules have liquid-like properties that permit dynamic fusing and exchange of components, but segregate from their surroundings like oil from water (Brangwynne et al., 2009). Similar properties were also described for P-bodies and stress granules in vitro (Lin et al., 2015; Molliex et al., 2015). Intrinsically disordered proteins (IDPs) or proteins with at least a portion of disordered regions (IDRs) are a critical component of phase transition and mRNPs (Brangwynne et al., 2015). It is suspected that most, if not all mRNP granules contain different IDPs/IDRs (Uversky, 2017), and the interactions and properties of these proteins can control mRNP contents. Another typical property is their propensity to scaffold multiple proteins through multivalent interaction networks (van der Lee et al., 2014). Alongside IDP/IDRs, mRNAs and their interactions contribute to mRNP dynamics, either in promoting (Lin et al., 2015), or modulating granule assembly (Hubstenberger et al., 2015; Seydoux, 2018). Thus, the nature of protein-protein and protein-RNA interactions which contribute to assembly and stability of mRNP granules are distinct from what is observed in stable complexes in aqueous phases. Phase separation instead is governed by weak multivalent interactions that segregate interacting macromolecules away from water at a critical concentration (Li et al., 2012; Hyman et al., 2014; Banani et al., 2017). Traditional protein-protein interaction studies based on co-immunoprecipitation and in vitro interaction assays may not be suitable to detect many, if not most of the interactions that occur in mRNPs. This, in turn, may be one of the reasons why proximity-based interaction mapping methods such as BioID were fruitful in mapping interactions in P-bodies and stress granules (Youn et al., 2018).
In light of the newly discovered properties of mRNPs, new and important questions have emerged. What are the folding and enzymatic differences that prevail in such phase-separated liquid droplets? How is the specific composition (if any) of an mRNP defined, and how are biochemical boundaries maintained or crossed between different types of mRNPs? Earlier work by the Seydoux group revealed that P granules and P-bodies closely interact, but do not merge in the C. elegans early embryo (Gallo et al., 2008). More recently, their work identified an important role for IDP MEG-3 in modulating the structural stability of P granules. Different enrichments in PGL-1 and MEG-3 proteins significantly altered mRNP properties and could limit access to RNA (Smith et al., 2016). In the Drosophila oocyte, nanos mRNPs progress along the cytoskeleton from smaller localization particles to the larger germ granules at the posterior pole. The Gavis group used quantitative single-molecule imaging to analyze the localization dynamics and assembly of mRNP germ granules in the Drosophila oocyte. Interestingly, single mRNP complexes that contain individual nanos transcripts merge into multi-mRNA granules at the posterior pole. This localized ‘growth’ appears to be exponential, rather than additive, which could be interpreted as mRNPs merging through phase transition into the germ plasm. In contrast, the oskar mRNA localizes as multi-copy mRNPs which are segregated from other mRNP granules once it reaches the posterior pole, and this exclusivity contributes to proper germline specification (Little et al., 2015). This suggests that single- or multi-mRNPs, can be differentially transported and locally stored. It further strengthens and refines the links between mRNPs and the transport and localization of mRNA granules.
The possible implications of this mechanism reach far beyond C. elegans and Drosophila oocytes and embryo. For example, analogous mRNP granules are likely common in mammalian neurons. A study took advantage of the preferential precipitation of IDPs by the chemical biotinylated isoxazole (b-isox) to fractionate mRNPs from mouse brain tissue (Han et al., 2012; Kato et al., 2012). mRNAs that precipitated with b-isox had on average 5-fold longer 3′UTRs compared to mRNAs recovered in the soluble fraction. Moreover, precipitated mRNAs encoded roughly 10-fold more binding sites for Pumilio proteins. This further suggests that 3′UTRs and their ability to bind multiple RBPs play an important role in mRNP assembly.
Originally named GW bodies because they contained an important fraction of the miRISC component GW182, P-bodies (for processing bodies) were later renamed because they also co-localized with decapping and decay proteins (Eystathioy et al., 2002, 2003). Because of this association, P-bodies have long been suspected to be sites of mRNA degradation (Sheth and Parker, 2003). They were also proposed as the site for RNAi, and several other mRNA decay activities (Unterholzner and Izaurralde, 2004; Jakymiw et al., 2005; Liu et al., 2005; Sheth and Parker, 2006). These functions, however, had been inferred and not directly demonstrated, and several studies challenged this role for P-bodies over the years (Chu and Rana, 2006; Eulalio et al., 2007b). Early on, a study by Izaurralde’s group revealed that while miRNA-mediated silencing promoted P-body formation, detectable P-bodies were not required for miRNA function (Eulalio et al., 2007b). More recently, the Weil group developed a FACS-based method to purify endogenous P-bodies and sequenced their RNA contents. With this method, they could not detect any mRNA decay intermediates (Hubstenberger et al., 2017). Interestingly, they also found that mRNAs in P-bodies were translationally repressed. They thus proposed that mRNP formation may increase the local concentration of translational repressors and thus maintain mRNA targets in a translationally repressed state. Similarly, another group monitored the dynamics of XRN1 (which mediates the 5′- > 3′ activity in many mRNA decay pathways) using an elegant dual fluorescent reporter design. Surprisingly, they noted that mRNA decay occurred throughout the cytoplasm, but not in P-bodies. This led them to also suggest that P-bodies are sites for mRNA storage, and not decay (Horvathova et al., 2017). This model nonetheless remains at striking odds with the localized concentration of decapping and decay enzymes in P-bodies.
Part of the solution to this conundrum may come from examining the composition and properties of P-bodies in different cellular lineages. The Seydoux group showed that the biochemical composition of P-bodies matured during early embryonic development, as it gained important decapping cofactors (Gallo et al., 2008). This stands to reason considering the dependence of mRNPs on the composition and concentration of proteins and mRNAs that are present in a particular context. P-bodies may have very different properties and functions in lineages as distinct as a neuron, an oocyte, an early blastomere, or an epithelial cell.
The properties of the proteins that are recruited to a 3′UTR target of miRISC or an RBP may also influence mRNP structure and activities. A recent study in C. elegans embryos suggested that recruitment of the CCR4-NOT complex and the associated IDR proteins by miRISC could nucleate mRNP assembly on target mRNAs. Recruitment of cell-lineage specified IDR proteins (such as PGL-1 or MEG-1/2) or co-factors of decapping and decay may enable progression into larger mRNP and toward context-dependent functions (Wu et al., 2017). In keeping with the importance of cellular context, a recent study by the Simard lab showed that miRISC has a distinct composition in C. elegans germline. While germline miRNA target reporters were silenced, single-molecule FISH methods revealed that targeting led to juxtaposition to P granules (germ granules) and also stabilized the targeted mRNA (Dallaire et al., 2018).
Lastly, a recent intriguing study showed that interactions between GW182 and the Argonaute could result in formation of miRISC droplets. This phase-separated condensate could in turn lead to sequestration of miRNA targets, and acceleration of their deadenylation in vitro (Sheu-Gruttadauria and MacRae, 2018). It thus seems likely that resolving the functions of P-bodies will be undissociable from the cellular expression and the sub-cellular concentration of mRNAs, IDRs, regulatory factors and effector machineries. Advances in quantitative methods to locally trace translation, mRNA deadenylation and decay in situ and in individual cell lineages may be important to resolve the apparent conflict that exists on the function of P-bodies.
Current Frontiers in 3′UTR Research
Great strides have been made in understanding the mechanisms underlying 3′UTR regulatory sequences and the factors that recognize and effect them. However, several dating problems remain unsolved and important new ones recently emerged. The above-mentioned resolution of the functions of phase transition mRNPs provides an example of an old problem that was recently visited with a new perspective. Other important problems came into focus with the emergence of next-generation sequencing, including alternative cleavage and polyadenylation (APA) (Edwalds-Gilbert et al., 1997), which generates significant diversity in 3′UTR isoforms. High-throughput sequencing identified multiple APA sites in at least 70% of known mammalian genes (Derti et al., 2012; Hoque et al., 2012). Most tissue-specific genes express single UTRs, but more than half of ubiquitously expressed genes are produced as multiple 3′UTR isoforms (Lianoglou et al., 2013). A different choice of polyadenylation sites in a 3′UTR has the potential to profoundly re-shape its structure and response elements, thus impacting mRNA stability, translation and localization. An interesting recent study even showed that an mRNA APA can alter the localization and expression of the membrane protein it encodes (Berkovits and Mayr, 2015). Not only is there an important diversity of 3′UTR isoforms, they are also dynamic in different cellular states. On average, proliferating cells (including several tumor-derived cell lines) express shorter 3′UTRs in mRNAs that are more stable and translated into more protein compared to the longer 3′UTR mRNAs expressed in differentiated cells (Sandberg et al., 2008; Ji et al., 2009; Mayr and Bartel, 2009). This led to the idea that shorter 3′UTR isoforms allowed mRNAs to avoid regulation by miRNAs and RBPs. This is likely an over-simplification and is not always the case, however, as shorter 3′UTRs can also mean more potent deadenylation (Flamand et al., 2016), and longer 3′UTRs can also mean regulatory sequences being buried in a more complex structure (Thivierge et al., 2018). Furthermore, some tissues like the brain (Ji et al., 2009; Hilgers et al., 2011; Ulitsky et al., 2012; Miura et al., 2013) have on average much longer 3′UTRs, potentially multiplying the folding structures and/or regulatory input, and thus the complexity of functional interplay.
The folding structures of 3′UTRs remain largely under-appreciated. This in itself is an important frontier, as structures can profoundly impact gene regulation (for reviews, see Jacobs et al., 2012; Kwok et al., 2015). Significant advances in chemical probes and next-generation sequencing now enable us to obtain genome-wide in vivo structures at single nucleotide resolution (Bevilacqua et al., 2016). Structures can be derived from in vivo transcripts, thus providing a perspective on the impact of developmental and cellular contexts, and the prevailing 3′UTR interactions (Spitale et al., 2015). Along those lines, a recent study analyzed changes in structures in zebrafish transcripts during MZT (Beaudoin et al., 2018), and revealed the interplay between ribosomes and the unwinding of mRNA secondary structures.
Improvements in throughput, library generation methods, and cost-effectiveness of next-generation sequencing now enable an integrated genomic perspective on multiple regulatory mechanisms. Massively parallel reporter assays (MPRA) have been used in the past to identify functional cis-regulatory elements in transcription and splicing (Melnikov et al., 2012; Rosenberg et al., 2015). Thousands of random sequences with unique tags are fused to reporters and introduced into cells, and their regulatory output is then quantified using high-throughput sequencing. A recent study used a similar technique to identify cis-regulatory elements in the 3′UTRs of maternal mRNAs in zebrafish that regulate mRNA decay (Rabani et al., 2017). The authors identified 2 stabilizing elements (polyU and UUAG sequences) and four destabilizing elements (GC- rich, AU-rich, Pumilio-binding sites, and miR-430-binding sequences).
Because so many mechanisms mobilize the deadenylase complex and its activities, sequencing libraries that allow the capture of poly(A) tail size, the end of the 3′UTR isoform, and the abundance of transcripts will provide insight on the impact of these key features on gene expression. Recent studies already identified distinct populations of poly(A) tail sizes in the transcriptome (Subtelny et al., 2014; Eichhorn et al., 2016; Lima et al., 2017).
Conclusion
We have reviewed how regulatory elements in 3′UTRs are recognized by miRNAs and RBPs, and some of the better-known mechanisms leading to the decisions on the fate of mRNAs. While genomic approaches are successful in unveiling the complexity and breadth of some of these mechanisms, each 3′UTR is also unique and has co-evolved closely in its genetic and cellular niche with its regulatory factors. Deciphering the 3′UTR code will also require detailing this uniqueness for each 3′UTR. Embracing genetics once more, this time through genome edition in model organisms, offers powerful new possibilities in linking the structures and sequences of 3′UTRs with mRNA fates in their physiological context.
Author Contributions
VM and TD contributed equally in writing the manuscript.
Funding
This work was supported by Canadian Institute of Health Research (CIHR) grants to TD (PJT-152900) and Fonds de Recherche du Québec-Santé (FRQS) Chercheur-Boursier Senior salary award to TD, The Charlotte and Leo Karassik Foundation Ph.D. fellowship award to VM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize for directly related work that we have not cited in this review. We would like to thank the members of the lab for their comments on the manuscript.
References
Ainger, K., Avossa, D., Diana, A. S., Barry, C., Barbarese, E., and Carson, J. H. (1997). Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 138, 1077–1087. doi: 10.1083/jcb.138.5.1077
Alarcon, V. B., and Elinson, R. P. (2001). RNA anchoring in the vegetal cortex of the Xenopus oocyte. J. Cell Sci. 114, 1731–1741.
Ameres, S. L., and Zamore, P. D. (2013). Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488. doi: 10.1038/nrm3611
Arribas-Layton, M., Wu, D., Lykke-Andersen, J., and Song, H. (2013). Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim. Biophys. Acta 1829, 580–589. doi: 10.1016/j.bbagrm.2012.12.006
Bakheet, T., Williams, B. R., and Khabar, K. S. (2006). ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 34, D111–D114.
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi: 10.1038/nrm.2017.7
Barreau, C., Paillard, L., and Osborne, H. B. (2005). AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150.
Bashirullah, A., Halsell, S. R., Cooperstock, R. L., Kloc, M., Karaiskakis, A., Fisher, W. W., et al. (1999). Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. Embo J. 18, 2610–2620. doi: 10.1093/emboj/18.9.2610
Bassell, G. J., Zhang, H., Byrd, A. L., Femino, A. M., Singer, R. H., Taneja, K. L., et al. (1998). Sorting of β-Actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 18, 251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998
Bazzini, A. A., Lee, M. T., and Giraldez, A. J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in Zebrafish. Science 336, 233–237. doi: 10.1126/science.1215704
Beaudoin, J. D., Novoa, E. M., Vejnar, C. E., Yartseva, V., Takacs, C. M., Kellis, M., et al. (2018). Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat. Struct. Mol. Biol. 25, 677–686. doi: 10.1038/s41594-018-0091-z
Berkovits, B. D., and Mayr, C. (2015). Alternative 3’ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367. doi: 10.1038/nature14321
Besse, F., and Ephrussi, A. (2008). Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9, 971–980. doi: 10.1038/nrm2548
Besse, F., López de Quinto, S., Marchand, V., Trucco, A., and Ephrussi, A. (2009). Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 23, 195–207. doi: 10.1101/gad.505709
Bevilacqua, P. C., Ritchey, L. E., Su, Z., and Assmann, S. M. (2016). Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 50, 235–266. doi: 10.1146/annurev-genet-120215-035034
Blewett, N. H., and Goldstrohm, A. C. (2012). A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol. Cell Biol. 32, 4181–4194. doi: 10.1128/MCB.00483-12
Boag, P. R., Atalay, A., Robida, S., Reinke, V., and Blackwell, T. K. (2008). Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182, 543–557. doi: 10.1083/jcb.200801183
Brancati, G., and Großhans, H. (2018). An interplay of miRNA abundance and target site architecture determines miRNA activity and specificity. Nucleic Acids Res. 46, 3259–3269. doi: 10.1093/nar/gky201
Brangwynne, C. P. (2013). Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203, 875–881. doi: 10.1083/jcb.201308087
Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. doi: 10.1126/science.1172046
Brangwynne, C. P., Tompa, P., and Pappu, R. V. (2015). Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904. doi: 10.1038/nphys3532
Braun, J. E., Huntzinger, E., Fauser, M., and Izaurralde, E. (2011). GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 44, 120–133. doi: 10.1016/j.molcel.2011.09.007
Braun, J. E., Huntzinger, E., and Izaurralde, E. (2013). “The role of GW182 proteins in miRNA-mediated gene silencing,” in Ten Years of Progress in GW/P Body Research, eds E. K. L. Chan, and M. J. Fritzler (New York, NY: Springer).
Brennan, C. M., and Steitz, J. A. (2001). HuR and mRNA stability. Cell Mol. Life Sci. 58, 266–277. doi: 10.1007/PL00000854
Broderick, J. A., Salomon, W. E., Ryder, S. P., Aronin, N., and Zamore, P. D. (2011). Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 17, 1858–1869. doi: 10.1261/rna.2778911
Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo, G. W., and Pasquinelli, A. E. (2016). Pairing beyond the seed supports MicroRNA targeting specificity. Mol. Cell 64, 320–333. doi: 10.1016/j.molcel.2016.09.004
Carotenuto, R., and Tussellino, M. (2018). Xenopus laevis oocyte as a model for the study of the cytoskeleton. C. R. Biol. 341, 219–227. doi: 10.1016/j.crvi.2018.04.001
Carson, J. H., Worboys, K., Ainger, K., and Barbarese, E. (1997). Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskeleton 38, 318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2
Chandradoss, S. D., Schirle, N. T., Szczepaniak, M., MacRae, I. J., and Joo, C. (2015). A dynamic search process underlies MicroRNA targeting. Cell 162, 96–107. doi: 10.1016/j.cell.2015.06.032
Chang, C. T., Bercovich, N., Loh, B., Jonas, S., and Izaurralde, E. (2014). The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res. 42, 5217–5233. doi: 10.1093/nar/gku129
Chapat, C., Jafarnejad, S. M., Matta-Camacho, E., Hesketh, G. G., Gelbart, I. A., Attig, J., et al. (2017). Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. U.S.A. 114, 5425–5430. doi: 10.1073/pnas.1701488114
Chekulaeva, M., Hentze, M. W., and Ephrussi, A. (2006). Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124, 521–533. doi: 10.1016/j.cell.2006.01.031
Chekulaeva, M., Mathys, H., Zipprich, J. T., Attig, J., Colic, M., Parker, R., et al. (2011). miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 18, 1218–1226. doi: 10.1038/nsmb.2166
Chen, C. Y., Ezzeddine, N., and Shyu, A. B. (2008). Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 448, 335–357. doi: 10.1016/S0076-6879(08)02617-7
Chen, C. Y., and Shyu, A. B. (2011). Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2, 167–183. doi: 10.1002/wrna.40
Chen, Y., Boland, A., Kuzuoğlu-Öztürk, D., Bawankar, P., Loh, B., Chang, C. T., et al. (2014). A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell 54, 737–750. doi: 10.1016/j.molcel.2014.03.034
Chendrimada, T. P., Finn, K. J., Ji, X., Baillat, D., Gregory, R. I., Liebhaber, S. A., et al. (2007). MicroRNA silencing through RISC recruitment of eIF6. Nature 447, 823–828. doi: 10.1038/nature05841
Cho, P. F., Gamberi, C., Cho-Park, Y. A., Cho-Park, I. B., Lasko, P., and Sonenberg, N. (2006). Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 16, 2035–2041. doi: 10.1016/j.cub.2006.08.093
Cho, P. F., Poulin, F., Cho-Park, Y. A., Cho-Park, I. B., Chicoine, J. D., Lasko, P., et al. (2005). A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by bicoid and the eIF4E Cognate 4EHP. Cell 121, 411–423. doi: 10.1016/j.cell.2005.02.024
Chu, C. Y., and Rana, T. M. (2006). Translation repression in human cells by MicroRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4:e210. doi: 10.1371/journal.pbio.0040210
Colegrove-Otero, L. J., Devaux, A., and Standart, N. (2005). The Xenopus ELAV Protein ElrB represses Vg1 mRNA Translation during Oogenesis. Mol. Cell Biol. 25, 9028–9039. doi: 10.1128/MCB.25.20.9028-9039.2005
Cooke, A., Prigge, A., and Wickens, M. (2010). Translational repression by deadenylases. J. Biol. Chem. 285, 28506–28513. doi: 10.1074/jbc.M110.150763
Curtis, D., Treiber, D. K., Tao, F., Zamore, P. D., Williamson, J. R., and Lehmann, R. (1997). A CCHC metal-binding domain in Nanos is essential for translational regulation. Embo J. 16, 834–843. doi: 10.1093/emboj/16.4.834
Dahanukar, A., Walker, J. A., and Wharton, R. P. (1999). Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4, 209–218. doi: 10.1016/S1097-2765(00)80368-8
Dale, L., Matthews, G., Tabe, L., and Colman, A. (1989). Developmental expression of the protein product of Vg1, a localized maternal mRNA in the frog Xenopus laevis. Embo J. 8, 1057–1065. doi: 10.1002/j.1460-2075.1989.tb03473.x
Dallaire, A., Frédérick, P. M., and Simard, M. J. (2018). Somatic and germline MicroRNAs form distinct silencing complexes to regulate their target mRNAs differently. Dev. Cell 47, 239–247.e4. doi: 10.1016/j.devcel.2018.08.022
Derti, A., Garrett-Engele, P., Macisaac, K. D., Stevens, R. C., Sriram, S., Chen, R., et al. (2012). A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183. doi: 10.1101/gr.132563.111
Deshler, J. O., Highett, M. I., and Schnapp, B. J. (1997). Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276, 1128–1131. doi: 10.1126/science.276.5315.1128
Didiano, D., and Hobert, O. (2008). Molecular architecture of a miRNA-regulated 3 UTR. RNA 14, 1297–1317. doi: 10.1261/rna.1082708
Djuranovic, S., Nahvi, A., and Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240. doi: 10.1126/science.1215691
Duchaine, T. F., and Fabian, M. R. (2018). Mechanistic insights into MicroRNA-mediated gene silencing. Cold Spring Harb. Perspect. Biol. doi: 10.1101/cshperspect.a032771 [Epub ahead of print].
Edwalds-Gilbert, G., Veraldi, K. L., and Milcarek, C. (1997). Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25, 2547–2561.
Eichhorn, S. W., Subtelny, A. O., Kronja, I., Kwasnieski, J. C., Orr-Weaver, T. L., and Bartel, D. P. (2016). mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife 5:e16955. doi: 10.7554/eLife.16955
Eom, T., Antar, L. N., Singer, R. H., and Bassell, G. J. (2003). Localization of a β-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 23, 10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003
Ephrussi, A., Dickinson, L. K., and Lehmann, R. (1991). Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66, 37–50. doi: 10.1016/0092-8674(91)90137-N
Ephrussi, A., and Lehmann, R. (1992). Induction of germ cell formation by oskar. Nature 358, 387–392. doi: 10.1038/358387a0
Eulalio, A., Behm-Ansmant, I., and Izaurralde, E. (2007a). P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8, 9–22.
Eulalio, A., Behm-Ansmant, I., Schweizer, D., and Izaurralde, E. (2007b). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 27, 3970–3981.
Eulalio, A., Huntzinger, E., Nishihara, T., Rehwinkel, J., Fauser, M., and Izaurralde, E. (2009). Deadenylation is a widespread effect of miRNA regulation. RNA 15, 21–32. doi: 10.1261/rna.1399509
Eystathioy, T., Chan, E. K., Tenenbaum, S. A., Keene, J. D., Griffith, K., and Fritzler, M. J. (2002). A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13, 1338–1351. doi: 10.1091/mbc.01-11-0544
Eystathioy, T., Jakymiw, A., Chan, E. K., Séraphin, B., Cougot, N., and Fritzler, M. J. (2003). The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9, 1171–1173. doi: 10.1261/rna.5810203
Fabian, M. R., Cieplak, M. K., Frank, F., Morita, M., Green, J., Srikumar, T., et al. (2011). miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 18, 1211–1217. doi: 10.1038/nsmb.2149
Fabian, M. R., Mathonnet, G., Sundermeier, T., Mathys, H., Zipprich, J. T., Svitkin, Y. V., et al. (2009). Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 35, 868–880. doi: 10.1016/j.molcel.2009.08.004
Ferraiuolo, M. A., Basak, S., Dostie, J., Murray, E. L., Schoenberg, D. R., and Sonenberg, N. (2005). A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170, 913–924. doi: 10.1083/jcb.200504039
Flamand, M. N., Gan, H. H., Mayya, V. K., Gunsalus, K. C., and Duchaine, T. F. (2017). A non-canonical site reveals the cooperative mechanisms of microRNA-mediated silencing. Nucleic Acids Res. 45, 7212–7225. doi: 10.1093/nar/gkx340
Flamand, M. N., Wu, E., Vashisht, A., Jannot, G., Keiper, B. D., Simard, M. J., et al. (2016). Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Res. 44, 5924–5935. doi: 10.1093/nar/gkw276
Forristall, C., Pondel, M., Chen, L., and King, M. L. (1995). Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development 121, 201–208.
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Fukuda, N., Fukuda, T., Sinnamon, J., Hernandez-Hernandez, A., Izadi, M., Raju, C. S., et al. (2013). The transacting factor CBF-A/Hnrnpab binds to the A2RE/RTS element of protamine 2 mRNA and contributes to its translational regulation during mouse spermatogenesis. PLoS Genet. 9:e1003858. doi: 10.1371/journal.pgen.1003858
Gallie, D. R. (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5, 2108–2116. doi: 10.1101/gad.5.11.2108
Gallo, C. M., Munro, E., Rasoloson, D., Merritt, C., and Seydoux, G. (2008). Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 323, 76–87. doi: 10.1016/j.ydbio.2008.07.008
Gallo, C. M., Wang, J. T., Motegi, F., and Seydoux, G. (2010). Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 330, 1685–1689. doi: 10.1126/science.1193697
Gao, Y., Tatavarty, V., Korza, G., Levin, M. K., and Carson, J. H. (2008). Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol. Biol. Cell 19, 2311–2327. doi: 10.1091/mbc.E07-09-0914
Gard, D. L., Cha, B. J., and King, E. (1997). The organization and animal–vegetal asymmetry of cytokeratin filaments in stage VI Xenopus oocytes is dependent upon F-actin and microtubules. Dev. Biol. 184, 95–114. doi: 10.1006/dbio.1997.8508
Gerstberger, S., Hafner, M., and Tuschl, T. (2014). A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845. doi: 10.1038/nrg3813
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., et al. (2005). MicroRNAs regulate brain morphogenesis in Zebrafish. Science 308, 833–838. doi: 10.1126/science.1109020
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. doi: 10.1126/science.1122689
Goldstrohm, A. C., Hook, B. A., Seay, D. J., and Wickens, M. (2006). PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13, 533–539. doi: 10.1038/nsmb1100
Grimson, A., Farh, K. K., Johnston, W. K., Garrett-Engele, P., Lim, L. P., and Bartel, D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105. doi: 10.1016/j.molcel.2007.06.017
Gu, S., Jin, L., Zhang, F., Sarnow, P., and Kay, M. A. (2009). Biological basis for restriction of microRNA targets to the 3 untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 16, 144–150. doi: 10.1038/nsmb.1552
Haghighat, A., Mader, S., Pause, A., and Sonenberg, N. (1995). Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. Embo J. 14, 5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x
Hall, T. M. (2016). De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 36, 116–121. doi: 10.1016/j.sbi.2016.01.010
Han, T. W., Kato, M., Xie, S., Wu, L. C., Mirzaei, H., Pei, J., et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. doi: 10.1016/j.cell.2012.04.016
Heasman, J., Quarmby, J., and Wylie, C. C. (1984). The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev. Biol. 105, 458–469. doi: 10.1016/0012-1606(84)90303-8
Hentze, M. W., Castello, A., Schwarzl, T., and Preiss, T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341. doi: 10.1038/nrm.2017.130
Hesse, M., and Arenz, C. (2014). MicroRNA maturation and human disease. Methods Mol. Biol. 1095, 11–25. doi: 10.1007/978-1-62703-703-7_2
Hilgers, V., Perry, M. W., Hendrix, D., Stark, A., Levine, M., and Haley, B. (2011). Neural-specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. U.S.A. 108, 15864–15869. doi: 10.1073/pnas.1112672108
Hoek, K. S., Kidd, G. J., Carson, J. H., and Smith, R. (1998). ‘hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA’. Biochemistry 37, 7021–7029. doi: 10.1021/bi9800247
Hoque, M., Ji, Z., Zheng, D., Luo, W., Li, W., You, B., et al. (2012). Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 10, 133. doi: 10.1038/nmeth.2288
Horvathova, I., Voigt, F., Kotrys, A. V., Zhan, Y., Artus-Revel, C. G., Eglinger, J., et al. (2017). The dynamics of mRNA turnover revealed by single-molecule imaging in single cells. Mol. Cell 68, 615–625.e9. doi: 10.1016/j.molcel.2017.09.030
Hubstenberger, A., Cameron, C., Noble, S. L., Keenan, S., and Evans, T. C. (2015). Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J. Cell Biol. 211, 703–716. doi: 10.1083/jcb.201504044
Hubstenberger, A., Courel, M., Bénard, M., Souquere, S., Ernoult-Lange, M., Chouaib, R., et al. (2017). P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e5. doi: 10.1016/j.molcel.2017.09.003
Humphreys, D. T., Westman, B. J., Martin, D. I., and Preiss, T. (2005). MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. U.S.A. 102, 16961–16966. doi: 10.1073/pnas.0506482102
Huntzinger, E., Kuzuoglu-Öztürk, D., Braun, J. E., Eulalio, A., Wohlbold, L., and Izaurralde, E. (2013). The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets’. Nucleic Acids Res. 41, 978–994. doi: 10.1093/nar/gks1078
Hüttelmaier, S., Zenklusen, D., Lederer, M., Dictenberg, J., Lorenz, M., Meng, X., et al. (2005). Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–525. doi: 10.1038/nature04115
Hyman, A. A., Weber, C. A., and Jülicher, F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58. doi: 10.1146/annurev-cellbio-100913-013325
Ingelfinger, D., Arndt-Jovin, D. J., Lührmann, R., and Achsel, T. (2002). ‘The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci’. RNA 8, 1489–1501.
Jacobs, E., Mills, J. D., and Janitz, M. (2012). The role of RNA structure in posttranscriptional regulation of gene expression. J. Genet. Genom. 39, 535–543. doi: 10.1016/j.jgg.2012.08.002
Jafarnejad, S. M., Chapat, C., Matta-Camacho, E., Gelbart, I. A., Hesketh, G. G., Arguello, M., et al. (2018). Translational control of ERK signaling through miRNA/4EHP-directed silencing. Elife 7:e35034. doi: 10.7554/eLife.35034
Jakymiw, A., Lian, S., Eystathioy, T., Li, S., Satoh, M., Hamel, J. C., et al. (2005). Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 7, 1267–1274. doi: 10.1038/ncb1334
Jan, C. H., Friedman, R. C., Ruby, J. G., and Bartel, D. P. (2011). Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469, 97–101. doi: 10.1038/nature09616
Jansen, R. P. (2001). mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2, 247–256. doi: 10.1038/35067016
Ji, Z., Lee, J. Y., Pan, Z., Jiang, B., and Tian, B. (2009). Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 7028–7033. doi: 10.1073/pnas.0900028106
Jiang, P., Singh, M., and Coller, H. A. (2013). Computational assessment of the cooperativity between RNA binding proteins and MicroRNAs in Transcript Decay. PLoS Comput. Biol. 9:e1003075. doi: 10.1371/journal.pcbi.1003075
Jing, Q., Huang, S., Guth, S., Zarubin, T., Motoyama, A., Chen, J., et al. (2005). Involvement of MicroRNA in AU-rich element-mediated mRNA instability. Cell 120, 623–634. doi: 10.1016/j.cell.2004.12.038
Jonas, S., and Izaurralde, E. (2013). The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 27, 2628–2641. doi: 10.1101/gad.227843.113
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi: 10.1038/nrg3965
Joshi, B., Cameron, A., and Jagus, R. (2004). Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 271, 2189–2203. doi: 10.1111/j.1432-1033.2004.04149.x
Kadyrova, L. Y., Habara, Y., Lee, T. H., and Wharton, R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519–1527. doi: 10.1242/dev.002212
Kamenska, A., Simpson, C., Vindry, C., Broomhead, H., Benard, M., Lee, B. P., et al. (2016). The DDX6-4E-T interaction mediates translational repression and P-body assembly. Nucleic Acids Res. 44, 6318–6334. doi: 10.1093/nar/gkw565
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. doi: 10.1016/j.cell.2012.04.017
Kedde, M., van Kouwenhove, M., Zwart, W., Oude, J. A., Vrielink, Elkon, R., et al. (2010). A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12, 1014–1020. doi: 10.1038/ncb2105
Kedersha, N., and Anderson, P. (2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61–81. doi: 10.1016/S0076-6879(07)31005-7
Kershner, A. M., and Kimble, J. (2010). Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. U.S.A. 107, 3936–3941. doi: 10.1073/pnas.1000495107
Kim, H. H., Kuwano, Y., Srikantan, S., Lee, E. K., Martindale, J. L., and Gorospe, M. (2009). HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748. doi: 10.1101/gad.1812509
Kim-Ha, J., Kerr, K., and Macdonald, P. M. (1995). Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell 81, 403–412. doi: 10.1016/0092-8674(95)90393-3
Kim-Ha, J., Smith, J. L., and Macdonald, P. M. (1991). Oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66, 23–35. doi: 10.1016/0092-8674(91)90136-M
King, M. L., Messitt, T. J., and Mowry, K. L. (2005). Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 97, 19–33. doi: 10.1042/BC20040067
Kislauskis, E. H., Zhu, X., and Singer, R. H. (1994). Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127, 441–451. doi: 10.1083/jcb.127.2.441
Kloc, M., Bilinski, S., Chan, A. P., Allen, L. H., Zearfoss, N. R., Etkin, L. D., et al. (2001). ‘RNA localization and germ cell determination in Xenopus’. Int. Rev. Cytol. 203, 63–91. doi: 10.1016/S0074-7696(01)03004-2
Kloc, M., Bilinski, S., Pui-Yee Chan, A., Etkin, L., and Etkin, D. (2000). The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3′UTR. Dev. Biol. 217, 221–229. doi: 10.1006/dbio.1999.9554
Kloc, M., and Etkin, L. D. (1995). Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development 121, 287–297.
Kloc, M., and Etkin, L. D. (1998). Apparent continuity between the messenger transport organizer and late RNA localization pathways during oogenesis in Xenopus. Mech. Dev. 73, 95–106. doi: 10.1016/S0925-4773(98)00041-0
Koscianska, E., Witkos, T. M., Kozlowska, E., Wojciechowska, M., and Krzyzosiak, W. J. (2015). ‘Cooperation meets competition in microRNA-mediated DMPK transcript regulation’. Nucleic Acids Res. 43, 9500–9518. doi: 10.1093/nar/gkv849
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Kraemer, B., Crittenden, S., Gallegos, M., Moulder, G., Barstead, R., Kimble, J., et al. (1999). NANOS-3 and FBF proteins physically interact to control the sperm–oocyte switch in Caenorhabditis elegans. Curr. Biol. 9, 1009–1018. doi: 10.1016/S0960-9822(99)80449-7
Kundu, P., Fabian, M. R., Sonenberg, N., Bhattacharyya, S. N., and Filipowicz, W. (2012). HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 40, 5088–5100. doi: 10.1093/nar/gks148
Kwok, C. K., Tang, Y., Assmann, S. M., and Bevilacqua, P. C. (2015). The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 40, 221–232. doi: 10.1016/j.tibs.2015.02.005
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi: 10.1126/science.1064921
Lai, W. S., Carballo, E., Thorn, J. M., Kennington, E. A., and Blackshear, P. J. (2000). Interactions of CCCH Zinc Finger Proteins with mRNA: binding of tristetraprolin-related zinc finger proteins to au-rich elements and destabilization OF mRNA’. J. Biol. Chem. 275, 17827–17837. doi: 10.1074/jbc.M001696200
Lapointe, C. P., Preston, M. A., Wilinski, D., Saunders, H. A. J., Campbell, Z. T., and Wickens, M. (2017). Architecture and dynamics of overlapped RNA regulatory networks. RNA 23, 1636–1647. doi: 10.1261/rna.062687.117
Lapointe, C. P., Wilinski, D., Saunders, H. A., and Wickens, M. (2015). Protein-RNA networks revealed through covalent RNA marks. Nat. Methods 12, 1163–1170. doi: 10.1038/nmeth.3651
Lau, N. C., Lim, L. P., Weinstein, E. G., and Bartel, D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. doi: 10.1126/science.1065062
Laver, J. D., Li, X., Ray, D., Cook, K. B., Hahn, N. A., Nabeel-Shah, S., et al. (2015). ‘Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition’. Genome Biol. 16:94. doi: 10.1186/s13059-015-0659-4
Lawrence, J. B., and Singer, R. H. (1986). Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415. doi: 10.1016/0092-8674(86)90326-0
Lécuyer, E., Yoshida, H., Parthasarathy, N., Alm, C., Babak, T., Cerovina, T., et al. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187. doi: 10.1016/j.cell.2007.08.003
Lee, R. C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864. doi: 10.1126/science.1065329
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The, C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Lee, S., Kopp, F., Chang, T. C., Sataluri, A., Chen, B., Sivakumar, S., et al. (2016). Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80. doi: 10.1016/j.cell.2015.12.017
Lehmann, R., and Nusslein-Volhard, C. (1986). Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell 47, 141–152. doi: 10.1016/0092-8674(86)90375-2
Lewis, R. A., Gagnon, J. A., and Mowry, K. L. (2008). PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus Oocytes. Mol. Cell Biol. 28, 678–686. doi: 10.1128/MCB.00999-07
Lewis, R. A., and Mowry, K. L. (2007). Ribonucleoprotein remodeling during RNA localization. Differentiation 75, 507–518. doi: 10.1111/j.1432-0436.2007.00159.x
Li, P., Banjade, S., Cheng, H. C., Kim, S., Chen, B., Guo, L., et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. doi: 10.1038/nature10879
Lianoglou, S., Garg, V., Yang, J. L., Leslie, C. S., and Mayr, C. (2013). Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396. doi: 10.1101/gad.229328.113
Lima, S. A., Chipman, L. B., Nicholson, A. L., Chen, Y. H., Yee, B. A., Yeo, G. W., et al. (2017). Short Poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 24, 1057–1063. doi: 10.1038/nsmb.3499
Lin, Y., Protter, D. S., Rosen, M. K., and Parker, R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219. doi: 10.1016/j.molcel.2015.08.018
Little, S. C., Sinsimer, K. S., Lee, J. J., Wieschaus, E. F., and Gavis, E. R. (2015). Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17, 558–568. doi: 10.1038/ncb3143
Liu, J., Rivas, F. V., Wohlschlegel, J., Yates, J. R. III, Parker, R., Hannon, G. J., et al. (2005). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7, 1261–1266. doi: 10.1038/ncb1333
Lunde, B. M., Moore, C., and Varani, G. (2007). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490. doi: 10.1038/nrm2178
Lykke-Andersen, J., and Wagner, E. (2005). Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19, 351–361. doi: 10.1101/gad.1282305
Ma, W. J., Cheng, S., Campbell, C., Wright, A., and Furneaux, H. (1996). Cloning and Characterization of HuR, a Ubiquitously Expressed Elav-like Protein. J. Biol. Chem. 271, 8144–8151. doi: 10.1074/jbc.271.14.8144
MacArthur, H., Bubunenko, M., Houston, D. W., and King, M. L. (1999). Xcat RNA is a translationally sequestered germ plasm component in Xenopus. Mech. Dev. 84, 75–88. doi: 10.1016/S0925-4773(99)00075-1
Martin, K. C., and Ephrussi, A. (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730. doi: 10.1016/j.cell.2009.01.044
Mathonnet, G., Fabian, M. R., Svitkin, Y. V., Parsyan, A., Huck, L., Murata, T., et al. (2007). MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317, 1764–1767. doi: 10.1126/science.1146067
Mathys, H., Basquin, J., Ozgur, S., Czarnocki-Cieciura, M., Bonneau, F., Aartse, A., et al. (2014). Structural and biochemical insights to the role of the CCR4-NOT Complex and DDX6 ATPase in MicroRNA Repression. Mol. Cell 54, 751–765. doi: 10.1016/j.molcel.2014.03.036
Mayr, C., and Bartel, D. P. (2009). Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684. doi: 10.1016/j.cell.2009.06.016
Melnikov, A., Murugan, A., Zhang, X., Tesileanu, T., Wang, L., Rogov, P., et al. (2012). Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277. doi: 10.1038/nbt.2137
Micklem, D. R., Adams, J., Grünert, S., and St Johnston, D. (2000). Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. Embo J. 19, 1366–1377. doi: 10.1093/emboj/19.6.1366
Mishima, Y., Giraldez, A. J., Takeda, Y., Fujiwara, T., Sakamoto, H., Schier, A. F., et al. (2006). Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 16, 2135–2142. doi: 10.1016/j.cub.2006.08.086
Miura, P., Shenker, S., Andreu-Agullo, C., Westholm, J. O., and Lai, E. C. (2013). Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 23, 812–825. doi: 10.1101/gr.146886.112
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. doi: 10.1016/j.cell.2015.09.015
Moretti, F., Kaiser, C., Zdanowicz-Specht, A., and Hentze, M. W. (2012). PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 19, 603–608. doi: 10.1038/nsmb.2309
Mosquera, L., Forristall, C., Zhou, Y., and King, M. L. (1993). ‘A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain’. Development 117, 377–386.
Moss, E. G., Lee, R. C., and Ambros, V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans, and Is Regulated by the lin-4 RNA. Cell 88, 637–646. doi: 10.1016/S0092-8674(00)81906-6
Mouland, A. J., Xu, H., Cui, H., Krueger, W., Munro, T. P., Prasol, M., et al. (2001). RNA trafficking signals in human immunodeficiency virus type 1. Mol. Cell Biol. 21, 2133–2143. doi: 10.1128/MCB.21.6.2133-2143.2001
Mukherjee, N., Corcoran, D. L., Nusbaum, J. D., Reid, D. W., Georgiev, S., Hafner, M., et al. (2011). Integrative regulatory mapping indicates that the RNA-binding protein HuR (ELAVL1) couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339. doi: 10.1016/j.molcel.2011.06.007
Munro, T. P., Magee, R. J., Kidd, G. J., Carson, J. H., Barbarese, E., Smith, L. M., et al. (1999). Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA Trafficking. J. Biol. Chem. 274, 34389–34395. doi: 10.1074/jbc.274.48.34389
Muslimov, I. A., Iacoangeli, A., Brosius, J., and Tiedge, H. (2006). Spatial codes in dendritic BC1 RNA. J. Cell Biol. 175, 427–439. doi: 10.1083/jcb.200607008
Nakamura, A., Sato, K., and Hanyu-Nakamura, K. (2004). Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell 6, 69–78. doi: 10.1016/S1534-5807(03)00400-3
Nelson, M. R., Leidal, A. M., and Smibert, C. A. (2004). Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. Embo J 23, 150–159. doi: 10.1038/sj.emboj.7600026
Nijjar, S., and Woodland, H. R. (2013). Localisation of RNAs into the germ plasm of vitellogenic Xenopus Oocytes’. PLoS One 8:e61847. doi: 10.1371/journal.pone.0061847
Nishimura, T., Padamsi, Z., Fakim, H., Milette, S., Dunham, W. H., Gingras, A. C., et al. (2015). The eIF4E-Binding Protein 4E-T Is a component of the mRNA decay machinery that bridges the 5′ and 3′ termini of target mRNAs. Cell Rep. 11, 1425–1436. doi: 10.1016/j.celrep.2015.04.065
Noble, S. L., Allen, B. L., Goh, L. K., Nordick, K., and Evans, T. C. (2008). Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559–572. doi: 10.1083/jcb.200802128
Nousch, M., Techritz, N., Hampel, D., Millonigg, S., and Eckmann, C. R. (2013). The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 126, 4274–4285. doi: 10.1242/jcs.132936
Otero, L. J., Devaux, A., and Standart, N. (2001). A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA 7, 1753–1767.
Ozgur, S., Basquin, J., Kamenska, A., Filipowicz, W., Standart, N., and Conti, E. (2015). Structure of a Human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep. 13, 703–711. doi: 10.1016/j.celrep.2015.09.033
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. doi: 10.1038/35040556
Peng, S. S., Chen, C. Y., Xu, N., and Shyu, A. B. (1998). RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. Embo J. 17, 3461–3470. doi: 10.1093/emboj/17.12.3461
Petersen, C. P., Bordeleau, M. E., Pelletier, J., and Sharp, P. A. (2006). Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21, 533–542. doi: 10.1016/j.molcel.2006.01.031
Plass, M., Rasmussen, S. H., and Krogh, A. (2017). Highly accessible AU-rich regions in 3′ untranslated regions are hotspots for binding of regulatory factors. PLoS Comput. Biol. 13:e1005460. doi: 10.1371/journal.pcbi.1005460
Quenault, T., Lithgow, T., and Traven, A. (2011). PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21, 104–112. doi: 10.1016/j.tcb.2010.09.013
Rabani, M., Pieper, L., Chew, G. L., and Schier, A. F. (2017). A massively parallel reporter assay of 3 UTR sequences identifies in vivo rules for mRNA Degradation. Mol. Cell 68, 1083–1094.e5. doi: 10.1016/j.molcel.2017.11.014
Raju, C. S., Fukuda, N., López-Iglesias, C., Göritz, C., Visa, N., and Percipalle, P. (2011). In neurons, activity-dependent association of dendritically transported mRNA transcripts with the transacting factor CBF-A is mediated by A2RE/RTS elements. Mol. Biol. Cell 22, 1864–1877. doi: 10.1091/mbc.e10-11-0904
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). ‘The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans’. Nature 403, 901–906. doi: 10.1038/35002607
Rom, E., Kim, H. C., Gingras, A. C., Marcotrigiano, J., Favre, D., Olsen, H., et al. (1998). Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem 273, 13104–13109. doi: 10.1074/jbc.273.21.13104
Rosenberg, A. B., Patwardhan, R. P., Shendure, J., and Seelig, G. (2015). Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163, 698–711. doi: 10.1016/j.cell.2015.09.054
Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L., and Singer, R. H. (1997). Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell Biol. 17, 2158–2165. doi: 10.1128/MCB.17.4.2158
Rouya, C., Siddiqui, N., Morita, M., Duchaine, T. F., Fabian, M. R., and Sonenberg, N. (2014). Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA 20, 1398–1409. doi: 10.1261/rna.045302.114
Sachs, A. B., Sarnow, P., and Hentze, M. W. (1997). Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89, 831–838. doi: 10.1016/S0092-8674(00)80268-8
Saetrom, P., Heale, B. S., Snøve, O. Jr., Aagaard, L., Alluin, J., and Rossi, J. J. (2007). Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res 35, 2333–2342. doi: 10.1093/nar/gkm133
Salomon, W. E., Jolly, S. M., Moore, M. J., Zamore, P. D., and Serebrov, V. (2016). Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell 166, 517–520. doi: 10.1016/j.cell.2016.06.048
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A., and Burge, C. B. (2008). Proliferating cells express mRNAs with shortened 3′ UTRs and fewer microRNA target sites. Science 320, 1643–1647. doi: 10.1126/science.1155390
Sandler, H., Kreth, J., Timmers, H. T., and Stoecklin, G. (2011). Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 39, 4373–4386. doi: 10.1093/nar/gkr011
Schirle, N. T., Sheu-Gruttadauria, J., and MacRae, I. J. (2014). Structural basis for microRNA targeting. Science 346, 608–613. doi: 10.1126/science.1258040
Schouten, M., Fratantoni, S. A., Hubens, C. J., Piersma, S. R., Pham, T. V., Bielefeld, P., et al. (2015). MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 5:12448. doi: 10.1038/srep12448
Schwanhausser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. doi: 10.1038/nature10098
Schwartz, D. C., and Parker, R. (1999). Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 5247–5256. doi: 10.1128/MCB.19.8.5247
Schwede, A., Ellis, L., Luther, J., Carrington, M., Stoecklin, G., and Clayton, C. (2008). A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 36, 3374–3388. doi: 10.1093/nar/gkn108
Seydoux, G. (2018). The P granules of C. elegans: a genetic model for the study of RNA–protein condensates. J. Mol. Biol. 430, 4702–4710. doi: 10.1016/j.jmb.2018.08.007
Sharif, H., Ozgur, S., Sharma, K., Basquin, C., Urlaub, H., and Conti, E. (2013). Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res. 41, 8377–8390. doi: 10.1093/nar/gkt600
Shav-Tal, Y., and Singer, R. H. (2005). RNA localization. J. Cell Sci. 118, 4077–4081. doi: 10.1242/jcs.02543
Sherrard, R., Luehr, S., Holzkamp, H., McJunkin, K., Memar, N., and Conradt, B. (2017). miRNAs cooperate in apoptosis regulation during C. elegans development’. Genes Dev. 31, 209–222. doi: 10.1101/gad.288555.116
Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808. doi: 10.1126/science.1082320
Sheth, U., and Parker, R. (2006). Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125, 1095–1109. doi: 10.1016/j.cell.2006.04.037
Sheu-Gruttadauria, J., and MacRae, I. J. (2018). Phase transitions in the assembly and function of human miRISC. Cell 173, 946–957.e16. doi: 10.1016/j.cell.2018.02.051
Smibert, C. A., Lie, Y. S., Shillinglaw, W., Henzel, W. J., and Macdonald, P. M. (1999). Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA 5, 1535–1547. doi: 10.1017/S1355838299991392
Smibert, C. A., Wilson, J. E., Kerr, K., and Macdonald, P. M. (1996). smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 10, 2600–2609. doi: 10.1101/gad.10.20.2600
Smith, J., Calidas, D., Schmidt, H., Lu, T., Rasoloson, D., and Seydoux, G. (2016). Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5:e21337. doi: 10.7554/eLife.21337
Smith, J. L., Wilson, J. E., and Macdonald, P. M. (1992). Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell 70, 849–859. doi: 10.1016/0092-8674(92)90318-7
Song, H. W., Cauffman, K., Chan, A. P., Zhou, Y., King, M. L., Etkin, L. D., et al. (2007). Hermes RNA-binding protein targets RNAs-encoding proteins involved in meiotic maturation, early cleavage, and germline development. Differentiation 75, 519–528. doi: 10.1111/j.1432-0436.2006.00155.x
Song, T., Zheng, Y., Wang, Y., Katz, Z., Liu, X., Chen, S., et al. (2015). Specific interaction of KIF11 with ZBP1 regulates the transport of beta-actin mRNA and cell motility. J. Cell Sci. 128, 1001–1010. doi: 10.1242/jcs.161679
Sonoda, J., and Wharton, R. P. (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712. doi: 10.1101/gad.13.20.2704
Sonoda, J., and Wharton, R. P. (2001). Drosophila brain tumor is a translational repressor. Genes Dev. 15, 762–773. doi: 10.1101/gad.870801
Spitale, R. C., Flynn, R. A., Zhang, Q. C., Crisalli, P., Lee, B., Jung, J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519:486. doi: 10.1038/nature14263
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H., and Bartel, D. P. (2014). Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71. doi: 10.1038/nature13007
Suh, N., Crittenden, S. L., Goldstrohm, A., Hook, B., Thompson, B., Wickens, M., et al. (2009). FBF and its dual control of gld-1 expression in the Caenorhabditis elegans Germline. Genetics 181, 1249–1260. doi: 10.1534/genetics.108.099440
Tannahill, D., and Melton, D. A. (1989). Localized synthesis of the Vg1 protein during early Xenopus development. Development 106, 775–785.
Temme, C., Zaessinger, S., Meyer, S., Simonelig, M., and Wahle, E. (2004). A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. Embo J. 23, 2862–2871. doi: 10.1038/sj.emboj.7600273
Temme, C., Zhang, L., Kremmer, E., Ihling, C., Chartier, A., Sinz, A., et al. (2010). Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 16, 1356–1370. doi: 10.1261/rna.2145110
Tharun, S. (2009). Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol. 6, 228–232. doi: 10.4161/rna.6.3.8282
Thivierge, C., Tseng, H. W., Mayya, V. K., Lussier, C., Gravel, S. P., and Duchaine, T. F. (2018). Alternative polyadenylation confers Pten mRNAs stability and resistance to microRNAs. Nucleic Acids Res. 46, 10340–10352. doi: 10.1093/nar/gky666
Tocchini, C., and Ciosk, R. (2015). TRIM-NHL proteins in development and disease. Semin. Cell Dev. Biol. 47-48, 52–59. doi: 10.1016/j.semcdb.2015.10.017
Tourriere, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J. M., Bertrand, E., et al. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831. doi: 10.1083/jcb.200212128
Tritschler, F., Braun, J. E., Eulalio, A., Truffault, V., Izaurralde, E., and Weichenrieder, O. (2009). Structural Basis for the Mutually Exclusive Anchoring of P Body Components EDC3 and Tral to the DEAD Box Protein DDX6/Me31B. Mol. Cell 33, 661–668. doi: 10.1016/j.molcel.2009.02.014
Tucker, M., Valencia-Sanchez, M. A., Staples, R. R., Chen, J., Denis, C. L., and Parker, R. (2001). ‘The Transcription Factor Associated Ccr4 and Caf1 Proteins Are Components of the Major Cytoplasmic mRNA Deadenylase in Saccharomyces cerevisiae’. Cell 104, 377–386. doi: 10.1016/S0092-8674(01)00225-2
Ulitsky, I., Shkumatava, A., Jan, C. H., Subtelny, A. O., Koppstein, D., Bell, G. W., et al. (2012). Extensive alternative polyadenylation during zebrafish development. Genome Res 22, 2054–2066. doi: 10.1101/gr.139733.112
Unterholzner, L., and Izaurralde, E. (2004). SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16, 587–596. doi: 10.1016/j.molcel.2004.10.013
Updike, D. L., Knutson, A. K., Egelhofer, T. A., Campbell, A. C., and Strome, S. (2014). Germ-Granule Components Prevent Somatic Development in the C. elegans Germline. Curr. Biol. 24, 970–975. doi: 10.1016/j.cub.2014.03.015
Uversky, V. N. (2017). Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18–30. doi: 10.1016/j.sbi.2016.10.015
van der Lee, R., Buljan, M., Lang, B., Weatheritt, R. J., Daughdrill, G. W., Dunker, A. K., et al. (2014). Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631. doi: 10.1021/cr400525m
Van Etten, J., Schagat, T. L., Hrit, J., Weidmann, C. A., Brumbaugh, J., Coon, J. J., et al. (2012). Human pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 287, 36370–36383. doi: 10.1074/jbc.M112.373522
Vella, M. C., Choi, E. Y., Lin, S. Y., Reinert, K., and Slack, F. J. (2004). The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 18, 132–137. doi: 10.1101/gad.1165404
von Roretz, C., Di Marco, S., Mazroui, R., and Gallouzi, I. E. (2011). Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip. Rev. RNA 2, 336–347. doi: 10.1002/wrna.55
Voronina, E., Seydoux, G., Sassone-Corsi, P., and Nagamori, I. (2011). RNA Granules in Germ Cells. Cold Spring Harb. Perspect. Biol. 3:a002774. doi: 10.1101/cshperspect.a002774
Wahle, E., and Winkler, G. S. (2013). RNA decay machines: deadenylation by the Ccr4–Not and Pan2–Pan3 complexes. Biochim. Biophys. Acta 1829, 561–570. doi: 10.1016/j.bbagrm.2013.01.003
Wakiyama, M., Takimoto, K., Ohara, O., and Yokoyama, S. (2007). Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 21, 1857–1862. doi: 10.1101/gad.1566707
Wee, L. M., Flores-Jasso, C. F., Salomon, W. E., and Zamore, P. D. (2012). Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151, 1055–1067. doi: 10.1016/j.cell.2012.10.036
Weidmann, C. A., Raynard, N. A., Blewett, N. H., Van Etten, J., and Goldstrohm, A. C. (2014). The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. RNA 20, 1298–1319. doi: 10.1261/rna.046029.114
Wells, S. E., Hillner, P. E., Vale, R. D., and Sachs, A. B. (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140. doi: 10.1016/S1097-2765(00)80122-7
White, R., Gonsior, C., Bauer, N. M., Krämer-Albers, E. M., Luhmann, H. J., and Trotter, J. (2012). Heterogeneous Nuclear Ribonucleoprotein (hnRNP) F is a novel component of oligodendroglial RNA transport granules contributing to regulation of myelin basic Protein (MBP) Synthesis. J. Biol. Chem. 287, 1742–1754. doi: 10.1074/jbc.M111.235010
White, R., Gonsior, C., Krämer-Albers, E. M., Stöhr, N., Hüttelmaier, S., and Trotter, J. (2008). Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J. Cell Biol. 181, 579–586. doi: 10.1083/jcb.200706164
Wickens, M., Bernstein, D. S., Kimble, J., and Parker, R. (2002). A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18, 150–157. doi: 10.1016/S0168-9525(01)02616-6
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. doi: 10.1016/0092-8674(93)90530-4
Wilhelm, J. E., Hilton, M., Amos, Q., and Henzel, W. J. (2003). Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 163, 1197–1204. doi: 10.1083/jcb.200309088
Wilhelm, J. E., Vale, R. D., and Hegde, R. S. (2000). Coordinate control of translation and localization of Vg1 mRNA in Xenopus oocytes. Proc. Natl. Acad. Sci. U.S.A. 97, 13132–13137. doi: 10.1073/pnas.97.24.13132
Wilk, K., Bilinski, S., Dougherty, M. T., and Kloc, M. (2005). Delivery of germinal granules and localized RNAs via the messenger transport organizer pathway to the vegetal cortex of Xenopus oocytes occurs through directional expansion of the mitochondrial cloud. Int. J. Dev. Biol. 49, 17–21. doi: 10.1387/ijdb.041906kw
Wilsch-Bräuninger, M., Schwarz, H., and Nüsslein-Volhard, C. (1997). A sponge-like structure involved in the association and transport of maternal products during Drosophila Oogenesis. J. Cell Biol. 139, 817–829. doi: 10.1083/jcb.139.3.817
Wu, B., Buxbaum, A. R., Katz, Z. B., Yoon, Y. J., and Singer, R. H. (2015). Quantifying protein-mRNA interactions in single live cells. Cell 162, 211–220. doi: 10.1016/j.cell.2015.05.054
Wu, E., Thivierge, C., Flamand, M., Mathonnet, G., Vashisht, A. A., Wohlschlegel, J., et al. (2010). Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell 40, 558–570. doi: 10.1016/j.molcel.2010.11.003
Wu, E., Vashisht, A. A., Chapat, C., Flamand, M. N., Cohen, E., Sarov, M., et al. (2017). A continuum of mRNP complexes in embryonic microRNA-mediated silencing. Nucleic Acids Res. 45, 2081–2098. doi: 10.1093/nar/gkw872
Yamashita, A., Chang, T. C., Yamashita, Y., Zhu, W., Zhong, Z., Chen, C. Y., et al. (2005). Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054. doi: 10.1038/nsmb1016
Yoon, Y. J., and Mowry, K. L. (2004). Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131, 3035–3045. doi: 10.1242/dev.01170
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69, 517–532.e11. doi: 10.1016/j.molcel.2017.12.020
Young, L. E., Moore, A. E., Sokol, L., Meisner-Kober, N., and Dixon, D. A. (2012). The mRNA stability factor HuR inhibits MicroRNA-16 Targeting of Cyclooxygenase-2. Mol. Cancer Res. 10, 167–180. doi: 10.1158/1541-7786.MCR-11-0337
Zaessinger, S., Busseau, I., and Simonelig, M. (2006). Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133:4573. doi: 10.1242/dev.02649
Zdanowicz, A., Thermann, R., Kowalska, J., Jemielity, J., Duncan, K., Preiss, T., et al. (2009). Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell 35, 881–888. doi: 10.1016/j.molcel.2009.09.009
Zekri, L., Kuzuoğlu-Öztürk, D., and Izaurralde, E. (2013). GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. Embo J. 32, 1052–1065. doi: 10.1038/emboj.2013.44
Zhang, H. L., Eom, T., Oleynikov, Y., Shenoy, S. M., Liebelt, D. A., Dictenberg, J. B., et al. (2001). Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 31, 261–275. doi: 10.1016/S0896-6273(01)00357-9
Zhang, H. L., Singer, R. H., and Bassell, G. J. (1999). Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J. Cell Biol. 147, 59–70. doi: 10.1083/jcb.147.1.59
Zhao, W. M., Jiang, C., Kroll, T. T., and Huber, P. W. (2001). A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. Embo J. 20, 2315–2325. doi: 10.1093/emboj/20.9.2315
Keywords: miRNAs, CCR4-NOT complex, RNA binding proteins (RBPs), phase transition, mRNP granules, translational repression, deadenylation, 3′untranslated region (UTR)
Citation: Mayya VK and Duchaine TF (2019) Ciphers and Executioners: How 3′-Untranslated Regions Determine the Fate of Messenger RNAs. Front. Genet. 10:6. doi: 10.3389/fgene.2019.00006
Received: 07 September 2018; Accepted: 07 January 2019;
Published: 24 January 2019.
Edited by:
Chiara Gamberi, Concordia University, CanadaReviewed by:
Michael Sheets, University of Wisconsin–Madison, United StatesPiergiorgio Percipalle, New York University Abu Dhabi, United Arab Emirates
Copyright © 2019 Mayya and Duchaine. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas F. Duchaine, dGhvbWFzLmR1Y2hhaW5lQG1jZ2lsbC5jYQ==
 Vinay K. Mayya
Vinay K. Mayya Thomas F. Duchaine
Thomas F. Duchaine