
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 16 April 2018
Sec. Evolutionary and Population Genetics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00131
This article is part of the Research Topic Genetics, Evolution, and Conservation of Neotropical Fishes View all 34 articles
The study of patterns and evolutionary processes in neotropical fish is not always an easy task due the wide distribution of major fish groups in large and extensive river basins. Thus, it is not always possible to detect or correlate possible effects of chromosome rearrangements in the evolution of biodiversity. In the Astyanax genus, chromosome data obtained since the 1970s have shown evidence of cryptic species, karyotypic plasticity, supernumerary chromosomes, triploidies, and minor chromosomal rearrangements. In the present work, we map and discuss the main chromosomal events compatible with the molecular evolution of the genus Astyanax (Characiformes, Characidae) using mitochondrial DNA sequence data, in the search for major chromosome evolutionary trends within this taxon.
The role of chromosomal rearrangements in the evolution of organisms has been a matter of debate for many years. The initial observations that closely related species differ in their karyotypes was later supported by evidence that unbalanced rearrangements can interfere with gametogenesis, decrease gene flow, and reinforce reproductive isolation leading to speciation (Rieseberg, 2001; Navarro and Barton, 2003). On the other hand, some organisms tolerate a certain amount of chromosomal rearrangement, which is often referred to as karyotypic plasticity (Jónsson et al., 2014; Havelka et al., 2016). This complicates the task of explaining the possible role of rearrangements in the evolution of organisms, since the variation may result in speciation or stay as polymorphism within populations.
Since Moreira-Filho and Bertollo (1991), who proposed Astyanax scabripinnis as a species complex based on cytogenetic and morphometric data, chromosome variation has been regarded as part of speciation processes in Astyanax. Before that, however, it was already acknowledged that some populations could differ in their karyotype formulae and diploid numbers (Morelli et al., 1983). Thus, it is now known that populations currently assigned to A. scabripinnis and to the other nominal species are characterized by some degree of inter- or intra-population chromosome variation, with diploid numbers ranging from 46 to 50 chromosomes; such is the case of Astyanax fasciatus (Pazza et al., 2006). Molecular cytogenetics (i.e., satellite DNA and ribosomal genes localization) has also provided further taxonomically informative data (for a review, see Pazza and Kavalco, 2007).
Although little is known about the cause and effect of the rearrangements during the evolutionary process, the correlation between independent data sets such as molecular and chromosomal data suggest that some karyotypic signatures are associated with organismic evolution. Based on Cytochrome B (CytB) sequences, Mello et al. (2015) found three genetically distinct clades out of 17 Astyanax nominal species from the Iguaçu and adjacent river basins. Likewise, using Cytochrome Oxydase I (COI) sequences, Rossini et al. (2016) found five clades out of 64 nominal species plus 12 provisionally identified taxa. Based on DNA barcoding criteria, these authors identified only 21 morphological species. These studies also suggest the possibility of horizontal transfer that affects some species within otherwise cogent taxa. Indeed, hybridization may play a fundamental role in the genus speciation, especially when these events involve chromosomal characteristics. The matter is particularly difficult since the claims of possible natural hybrids in the specialized literature are rare (Artoni et al., 2006; Pazza et al., 2006; Sassi et al., 2018).
Thus, we analyzed mitochondrial sequences from individuals with known chromosome characteristics within the five clades proposed by Rossini et al. (2016) to describe the relations between DNA sequences and chromosomal rearrangements. We obtained a phylogenetic tree that depicts chromosomal characteristics that are strongly associated with the proposed clades.
In the present study we sequenced samples of 195 individuals from 16 nominal species of the genus Astyanax, deposited in the tissue collection of the Laboratory of Ecological and Evolutionary Genetics (LaGEEvo), of the Federal University of Viçosa, Rio Paranaíba campus. The species were chosen based on their chromosomal data as analyzed in previous works and because they were included in Rossini et al. (2016) clades. Geographic coordinates, Vouchers, and GenBank sequence access numbers are summarized in Supplementary Table S1. This study was carried out in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals by the Conselho Nacional de Controle de Experimentação Animal (CONCEA). Tissue samples of fish of the genus Astyanax deposited in the tissue bank of the Laboratory of Ecological and Evolutionary Genetics (LaGEEvo). All the specimens used had their chromosomal data analyzed in previous works. No additional animals were sacrificed to this study.
Our hypothesis on chromosome evolution was based on the following chromosome characters: diploid numbers, fundamental number (FN), location of 5S DNA ribosomal sites, presence/absence of a 5S rDNA site in a specific submetacentric, referred to as the “marker” chromosome by Almeida-Toledo et al. (2002) and Kavalco et al. (2005), and finally, the amount and distribution of the repetitive DNA As-51 probe (Mestriner et al., 2000).
Total DNA extraction from muscle or liver samples was carried out using commercial kits (PureLink Genomic DNA minikit, InvitrogenTM), according to the manufacturer’s instructions. Amplification of mitochondrial (mtDNA) subunits 6 and 8 of the ATP synthase enzyme gene (ATPase 6/8) was accomplished using the primers ATP8.2-L8331 (5′-AAAGCRTTRGCCTTTTAAAGC-3′) and CO3.2-H9236 (5′-GTTAGTGGTCAGGGCTTGGRTC-3′) (Sivasundar et al., 2001). PCR was performed in a final volume of 25 μL, with 2.5 μL of 10× Taq buffer, 1 μL of MgCl2, 1 μL of each primer, 0.2 μL Taq DNA polymerase, 12.8 μL ultrapure water, 1.5 μL of dNTP, and 5 μL of DNA. The amplification reactions were performed in a thermocycler at 95°C for initial denaturation (2 min) and 30 cycles of 94°C (30 s), 58°C (30 s), and 72°C (1 min). The PCR product was visualized on 1% agarose gel; purification and sequencing was performed by a third-party company (Macrogen, Korea).
Sequence editing was performed using Chromas Lite v2.01 and sequence identity was checked with BLASTn1. Sequences alignment was carried out with ClustalW v1.6 (Thompson et al., 1994) as implemented in MEGA v7 (Kumar et al., 2016). A Maximum Likelihood was obtained using the best model fit with MEGA v7 (Kumar et al., 2016), and chromosome characters were plotted on this ML phylogram. Phylogenetic signal was estimated using bootstrap (Felsenstein, 1985).
A total of 195 mitochondrial DNA sequences were obtained from individuals with known chromosomal characteristics, corresponding to 16 nominal species of Astyanax from the Neotropical region. The ATPase 8 gene yielded a 530 bp partial sequence without insertions, deletions, or stop codons, and the substitution model was Tn93 + G. The maximum likelihood phylogram indicated four main clades with strong bootstrap support. The main events of chromosome differentiation were plotted on a simplified phylogenetic tree (Figure 1) according to the trends observed in the analyzed specimens and data from the literature. At its root, and according to the chromosomal characteristics of closely related species, we propose a most recent common ancestor with a karyotype with 2n = 50 chromosomes, low FN, one or two pairs of chromosomes bearing 5S ribosomal rDNA sites at the terminal region, and multiple 18S rDNA sites.
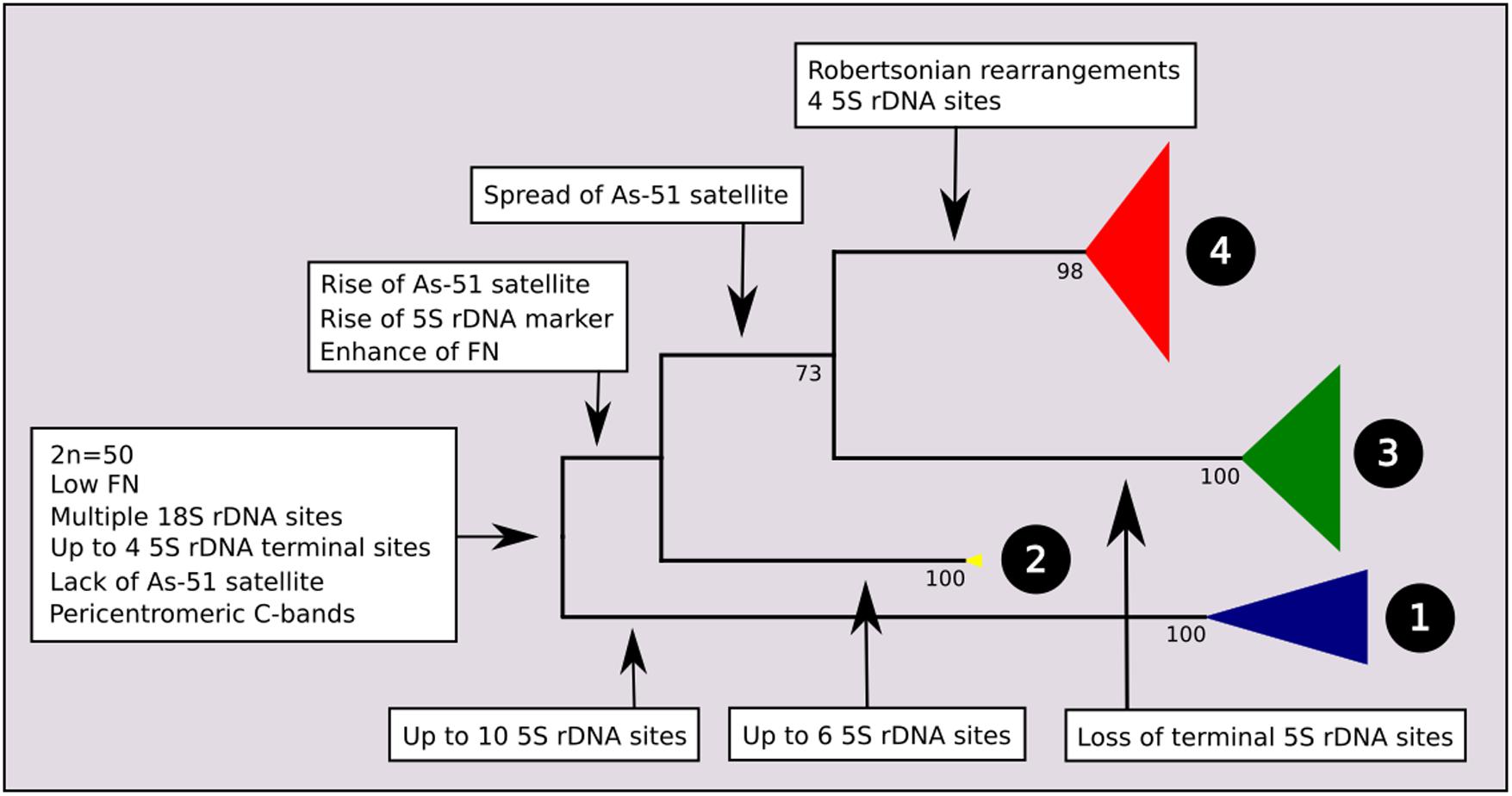
FIGURE 1. Molecular Phylogenetic analysis by Maximum Likelihood Method (MCL) with synapomorphies of each clade. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The tree with the highest log likelihood (–3974.2120) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the MCL approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.4001)]. The condensed tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 196 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 531 positions in the final dataset. Evolutionary analyses were conducted in MEGA7. Value of bootstrap related to 500 replicates is below of branches.
The first clade, composed of 36 individuals, corresponds to exclusively coastal species, here represented here by A. ribeirae, A. intermedius, A. giton, and A. hastatus (Figure 2), which have been recently proposed as members of the Probolodini (Silva, 2017). In this group, all individuals have 2n = 50 chromosomes; low FN; absence of repetitive DNA As51 and the 5S rDNA marker chromosome; little constitutive heterochromatin, distributed mainly in the pericentromeric region; a greater number of 5S rDNA sites (ranging from 6 to 10 sites) distributed in terminal regions; and a variable number of Ag-NORs and 18S rDNA sites.
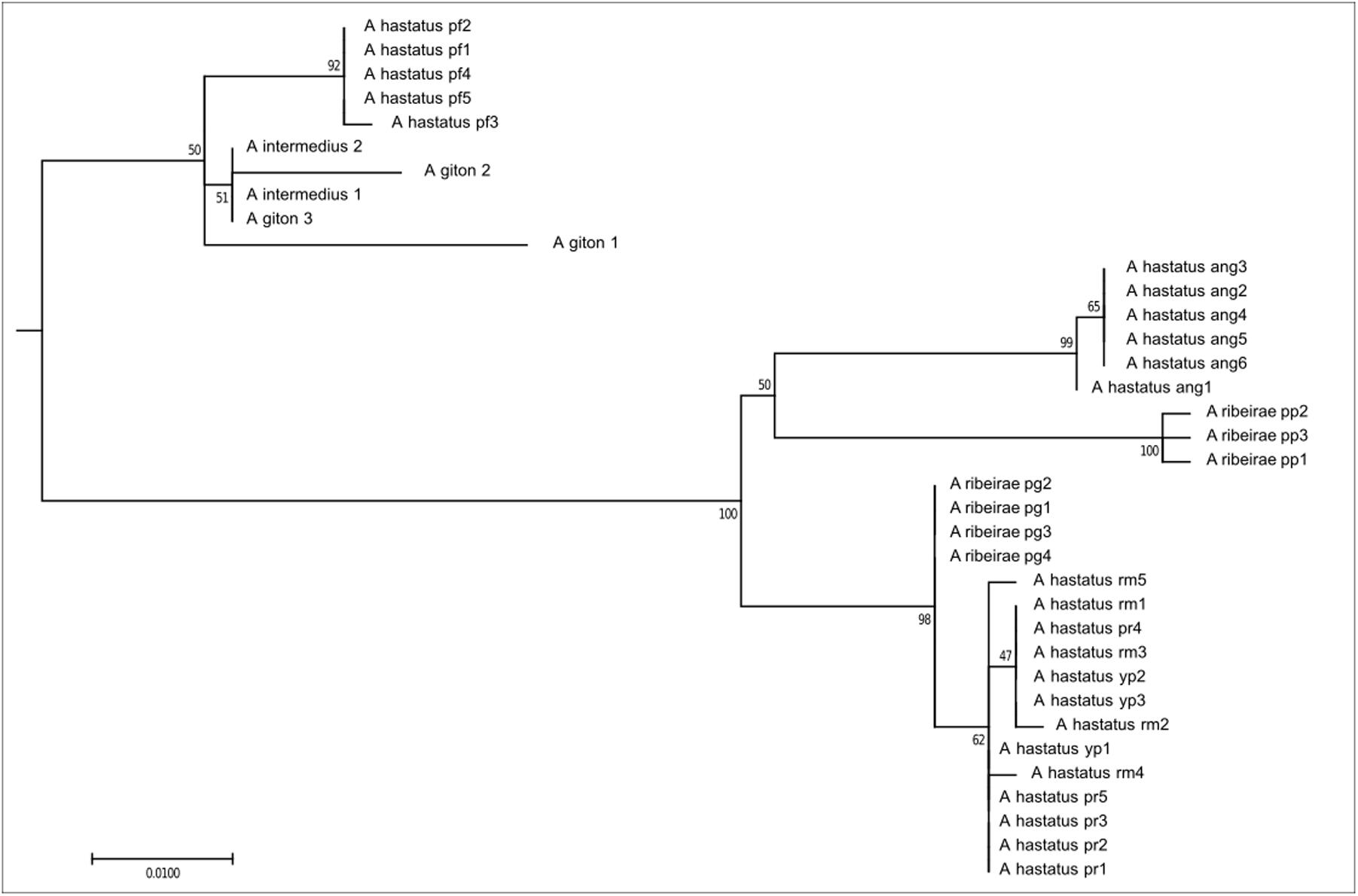
FIGURE 2. Molecular Phylogenetic of the Clade 1 analysis by Maximum Likelihood Method. Parameters described in Figure 1. Clade 1 is formed by species distributed only in coastal drainages as A. hastatus, A. ribeirae, A. intermedius, and A. giton.
This clade is composed of five individuals of Astyanax mexicanus (Figure 3). This species has a few repetitive As51 DNA sites and 5S rDNA is distributed on six sites, including the submetacentric, marker chromosome pair.

FIGURE 3. Molecular Phylogenetic of the Clade 2 analysis by Maximum Likelihood Method. Parameters described in Figure 1. Clade 2 is formed by A. mexicanus.
This clade comprises 71 individuals with oval humeral spots distributed in coastal, Upper Paraná, Paraguay, and São Francisco river basins, belonging to the Astyanax bimaculatus species complex, such as A. altiparanae, A. aff. bimaculatus, A. lacustris, A. assuncionensis, and A. abramis (Figure 4). They all have 50 chromosomes, with a predominance of submetacentric chromosomes, and consequently high FN numbers. The presence of As51 repetitive DNA in a few sites and the presence of 5S rDNA restricted to the submetacentric marker chromosome is remarkable. The distribution of heterochromatin and rDNA 18S/Ag-NORs is variable.
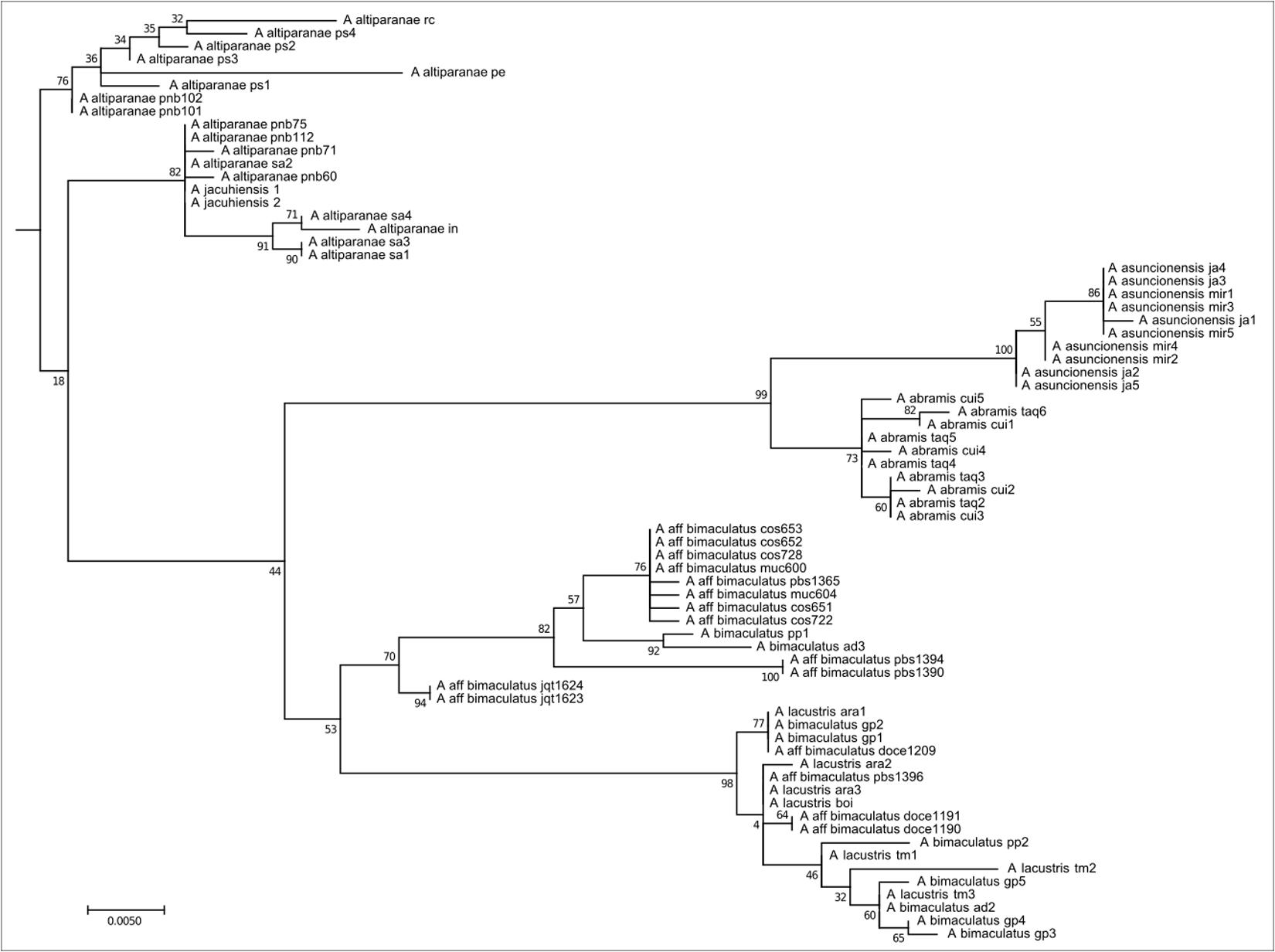
FIGURE 4. Molecular Phylogenetic of the Clade 3 analysis by Maximum Likelihood Method. Parameters described in Figure 1. Clade 3 is formed by species with humeral oval spot like the A. bimaculatus complex and the related species.
This clade encompasses species widely distributed in the Upper Paraná, São Francisco, and coastal river basins. Eighty four individuals of A. paranae, A. rivularis, A. bifasciatus, A. fasciatus, and A. bockmanni are represented in this clade (Figure 5). Species of this clade present high intra and interspecific chromosome variability, with 2n = 46 to 2n = 50 chromosomes. Chromosomes carrying As51 satellite DNA sites vary from none to 14 sites. Most species have four 5S rDNA sites, which may include the chromosomal marker pair. As in Clade 3, they have FN numbers, relatively few acrocentric chromosomes, 18S rDNA sites and variable Ag-NORs, and highly variable constitutive heterochromatin distribution.
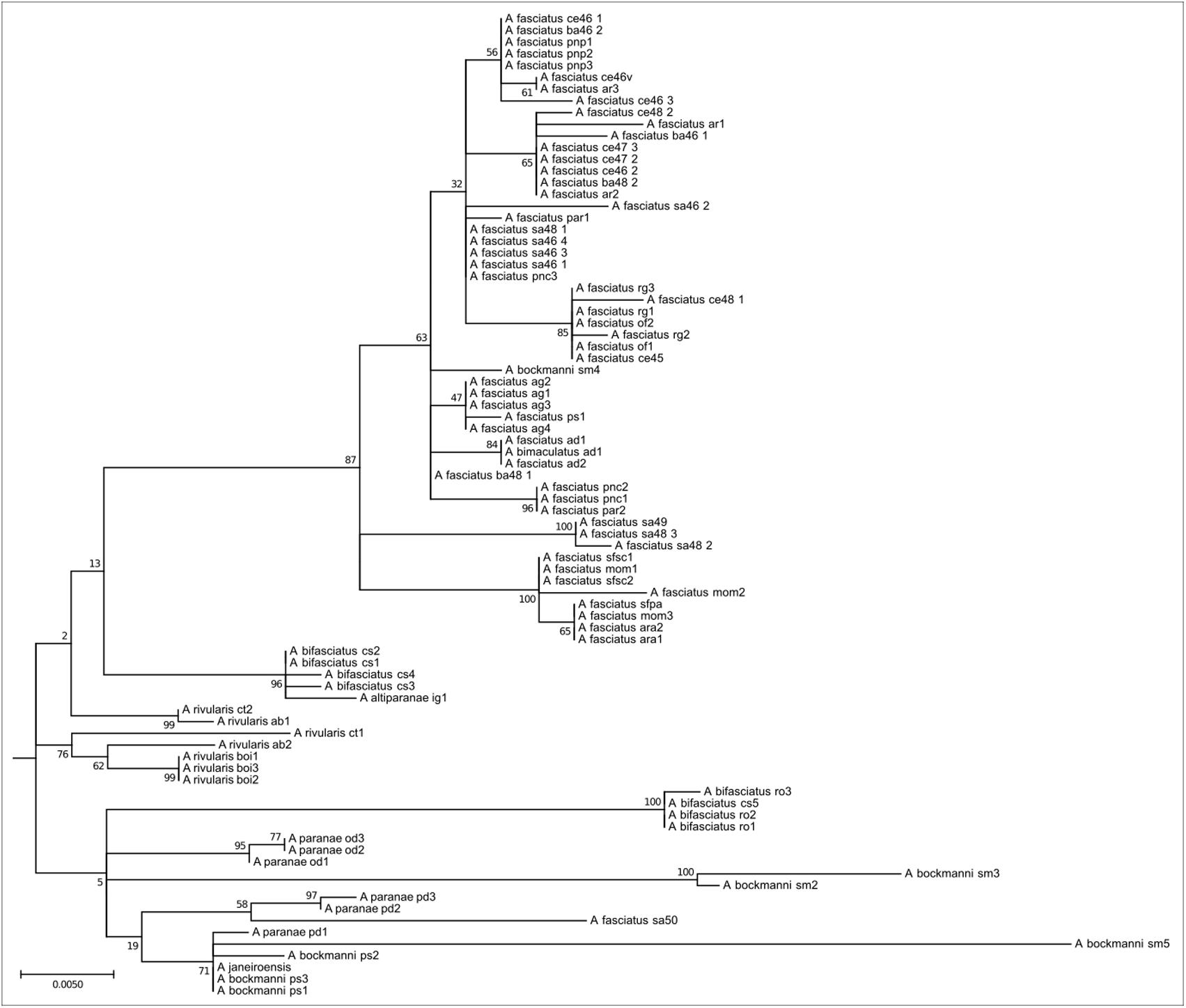
FIGURE 5. Molecular Phylogenetic of the Clade 4 analysis by Maximum Likelihood Method. Parameters described in Figure 1. Clade 4 is formed by red-tailed species and their related group as A. fasciatus, the A. scabripinnis group, A. bockmanni, and A. bifasciatus. This group is the most chromosome diverse in the genus and shows little mitochondrial divergence.
The phylogram obtained in the present work using the mitochondrial DNA sequence (ATPase subunit 6) is mostly congruent with that obtained by Mello et al. (2015) using the mitochondrial DNA sequence of the cytochrome b gene, as well as that obtained by Rossini et al. (2016) using the mitochondrial COI sequence. Karyotypical data allow to give further support to at least some monophyletic groupings within the current genus Astyanax, corresponding to the major clades, as it was proposed by Travenzoli et al. (2015) in Bryconidae. Despite this cohesion, some species seem to have haplotypes distributed in different clades, as demonstrated by Rossini et al. (2016). According to the authors, the COI sequence is not an appropriate tool to recover phylogenies, but rather to identify species (Hebert and Gregory, 2005). For the Astyanax genus, this sequence also seems to be unsuitable for species identification, since of the more than 70 nominal species analyzed by Rossini et al. (2016), only 21 were unequivocally identified by barcoding. The low genetic distances observed in the mitochondrial analyses among species of the Astyanax genus are often explained by the rapid divergence between them (Ornelas-García et al., 2008; Carvalho et al., 2011; Rossini et al., 2016). Therefore, multidisciplinary approaches may be more effective for reconstructing phylogenies within this genus. In lower systematic levels, molecular data associated with the chromosomal features were particularly effective to understand evolutionary patterns in A. aff. bimaculatus (Kavalco et al., 2011) and A. fasciatus (Pansonato-Alves et al., 2013; Kavalco et al., 2016).
Overlapping cytogenetic onto molecular phylograms offers an insight on large-scale chromosome evolutionary Astyanax. This analysis allows us to point out as the main specific chromosomal markers to be explored: the distribution patterns of 5S rDNA and As51 repetitive DNA, as they show signatures within the genus Astyanax. On the other hand, other characters such as location of the Nuclear Organizing Regions and C-banding shows patterns seem more informative at population level, as demonstrated by Jacobina et al. (2011) in Hoplias malabaricus (Erythrinidae). Although other markers have been used sporadically in chromosome studies of the genus Astyanax (Barbosa et al., 2015, 2017; Silva et al., 2015; Piscor and Parisi-Maltempi, 2016), their informative value will require intensive sampling in an array of species.
The genus Astyanax is currently incertae sedis in the Characidae family, and the evolutionary relations within the genus, as well as its phylogenetic relation to other genera of the family are quite challenging both from the morphological point of view (Mirande, 2009, 2010), and from the molecular systematic (Oliveira et al., 2011; Rossini et al., 2016). Despite the incomplete database, others genera related to Astyanax usually show more conservative cytogenetic characters, such as steady patterns of 50 chromosomes, multiple NORs and 5S ribosomal genes located in one or more pairs of chromosomes at the terminal region. These suite of characteristics have been reported in the genus Oligosarcus (Kavalco et al., 2005; Hattori et al., 2007; Barros et al., 2015), Deuterodon (Mendes et al., 2011; Coutinho-Sanches and Dergam, 2015), Hollandichthys and Ctenobrycon (Carvalho et al., 2002), plus the absence of the homology with the As51 repetitive DNA (Kavalco et al., 2005), suggesting that these characters are close to the evolutionary origins of Astyanax, being symplesiomorphies in clade 1.
The clade 1 obtained by our molecular analyses corresponds to clade 5 of Rossini et al. (2016). In the specimens analyzed we can observe the plesiomorphic chromosome characteristics (as a karyotype with 2n = 50 chromosomes and a low FN) and with good structure in A. ribeirae (Kavalco et al., 2010), A. intermedius, and A. giton (Kavalco and Moreira-Filho, 2003; Kavalco et al., 2004). In A. hastatus, different cytotypes with 2n = 50 chromosomes have been observed, constituting yet another species complex within the genus Astyanax (Kavalco et al., 2009), as corroborated by the present study (Figure 2). Also, a combination of molecular and chromosomal data suggests that the current A. hastatus may encompass more than one OTU (operational taxonomic unit) and so, more than one ESU (evolutionarily significant unit). The Astyanax species distributed in the coastal basins were added to the Probolodini group with species from the genus Probolodus, Deuterodon, and Myxiops and Hyphessobrycon luetkenii (Silva, 2017). These other species are less studied by cytogenetic methods, but until now, the chromosome characteristics seems to be shared (Mendes et al., 2011; Coutinho-Sanches and Dergam, 2015), bringing new evidence supporting the group. Unfortunately, As51 repetitive DNA in these coastal distribution species has not been confirmed so far (Kavalco et al., 2007, 2009).
Smaller variations in the number of 5S rDNA sites can be observed among these species, although they are always located in the proximal or distal region of acrocentric chromosomes, reaching up to 10 markings in A. intermedius and A. giton, which differ by a pericentric inversion (Kavalco et al., 2004). The hypothesis that these characteristics are a symplesiomorphy can be corroborated in independent chromosome data, such as those obtained for A. taeniatus (Cunha et al., 2016), which is also included in the same clade 5 by Rossini et al. (2016). The evolutionary dynamics of this gene are related not only to variations in non-transcribed spacers, but also to syntenia with long and short interspersed nuclear elements, non-long terminal repeat retrotransposons, U-snRNA families, and microsatellite polymorphisms (Rebordinos et al., 2013). According to these authors, polymorphisms in non-transcribed regions are observed in fish. Polymorphisms in transcribed regions do not appear to interfere with the cellular activity of 5S rDNA, and the molecular diversity of the 5S rDNA gene families is greater than the chromosome diversity (Rebordinos et al., 2013).
At the root of clades 2, 3, and 4, it is possible to hypothesize two main chromosomal events: the occurrence one of the 5S rDNA sites to the proximal position of a specific submetacentric chromosome pair, which has been considered as a marker (Almeida-Toledo et al., 2002) and the presence of As51 repetitive DNA in the Astyanax genome.
The 5S rDNA in clade 2 appears on a pair of two-arm chromosomes in the proximal region of the centromere (Kavalco and Almeida-Toledo, 2007). This marker may have arisen by pericentric inversion from an acrocentric chromosome bearing the 5S rDNA site, which should represent the most basal character state.
In addition to this site, A. mexicanus also presents four more 5S rDNA sites on acrocentric chromosomes, being distal markings on one pair and proximal markings on another (Kavalco and Almeida-Toledo, 2007). The isolation of the group represented by the A. mexicanus is congruent with some molecular data that relate ichthyofauna invasions in Central America with the genesis of the Panamá Isthmus (around 3.3 Mya; Ornelas-García et al., 2008), as well as with clustering with DNA barcoding (Rossini et al., 2016). Unfortunately, cytogenetic data for the species of this group are scarce, but it would not be surprising to have independent autapomorphies of the karyotype evolution of the cisandine species of Astyanax.
As51 satellite DNA is partly repetitive tandem DNA and holds similarities with transposable elements that were isolated from a population of A. scabripinnis carrier of chromosome B (Mestriner et al., 2000). In this population, this DNA was located mainly in the B chromosome, besides two to four sites in the distal region of acrocentric chromosomes. Considering the possible origin from transposable elements, it is appropriate to assume that their distribution was initially restricted to a few sites and subsequently spread in the genome by intrinsic mechanisms of the repetitive sequence amplification. In fact, the individuals of A. mexicanus that form clade 2 in the present work have few sites that carry this satellite DNA, and that it is shown more diffusely than in other species, which suggests a smaller number of copies or even only partial homology (Kavalco and Almeida-Toledo, 2007). This divergence might be related to independent evolution of A. mexicanus relative to the A. scabripinnis strain that donated this probe.
The increase of As51 satellite DNA sites characterizes the common ancestral root of clades 3 and 4, in contrast to the loss of 5S rDNA sites on acrocentric chromosomes observed in members of Clade 3. Clade 3 is composed of the species complex “A. bimaculatus,” represented herein by A. altiparanae, A. aff. bimaculatus, A. lacustris, A. asuncionensis, and A. abramis. Specimens from this clade present 2n = 50 chromosomes, with a higher FN, multiple Nucleolar Organizing Regions, and consequently multiple sites of 18S rDNA, according to the samples analyzed (Kavalco et al., 2011) and others independent studies (Almeida-Toledo et al., 2002; Fernandes and Martins-Santos, 2004, 2006; Peres et al., 2008; Hashimoto et al., 2011; Giongo et al., 2013).
Clade 3 is characterized by a synapomorphy: only one pair of chromosomes carrying a rDNA 5S site on its pericentromeric region (Kavalco et al., 2011) and in the literature (Almeida-Toledo et al., 2002; Fernandes and Martins-Santos, 2006; Peres et al., 2008, 2012; Hashimoto et al., 2011; Paiz et al., 2015). One exception is A. abramis, which presents in addition to the site of the chromosomal marker pair, a pair of extra-acrocentric chromosomes with proximal marking (Paiz et al., 2015; Piscor et al., 2015). This differentiated pattern may be an autapomorphy of A. abramis, whose molecular distinctiveness within the group was also evident in our analysis (Figure 5). Additionally, the pattern of occurrence of a 5S rDNA carrier pair can be seen in other species related to the A. bimaculatus group, as in A. janeiroensis (Vicari et al., 2008), A. goyacensis (Santos et al., 2013), and A. elachylepis (Santos et al., 2016).
In turn, there is little data on the distribution of As51 satellite DNA in this group, besides previous reports (Kavalco et al., 2011). In these species, small sites are observed, suggesting relatively lower number of in tandem copies and more restricted distribution through the karyotype, except for A. bimaculatus from coastal basins, in which As51 satDNA is absent (Kavalco et al., 2011). On the other hand, in Astyanax janeiroensis, a considerable number of very conspicuous sites are observed (Vicari et al., 2008; Kantek et al., 2009b). Although the Catalog of Fishes (Eschmeyer et al., 2017) characterizes the distribution of A. janeiroensis as being “in Brazil”, Melo (2001) suggests that its distribution is restricted to the basins of the Paraíba do Sul and other coastal drainages. These discrepant and independent occurrences in A. janeiroensis and A. bimaculatus can be a result of the historical biogeography of coastal drainages, affected by stream capture from continental basins and complex dispersal due to marine regressions and transgressions (Pereira et al., 2013). Fish faunas in these regions are characterized by a high degree of endemism and low species richness (Albert and Reis, 2011).
Clade 4 comprises a group of species with the highest chromosomal variability in the genus, with 2n ranging from 46 to 50 chromosomes and including the species complexes A. scabripinnis (encompassing A. rivularis and A. paranae) and A. fasciatus, as well the species A. bockmanni and A. bifasciatus (formerly referred to as Astyanax sp. B). Equally variable among the species of this clade is the number of NOR/rDNA 18S sites and the constitutive heterochromatin distribution patterns. The molecular divergence among these species is apparently quite recent, with low genetic distance indices as observed in the clade 1 and by Rossini et al. (2016) using the COI gene. Despite some level of structuring, low bootstrap indexes preclude further hypotheses on the genetic diversification of the group. There is no detailed chromosome information on the other species analyzed by Rossini et al. (2016) belonging to this clade, except for A. parahybae (Kavalco and Moreira-Filho, 2003; Kavalco et al., 2004) presenting 2n = 48 chromosomes, and A. schubarti (Morelli et al., 1983) presenting 2n = 36 chromosomes. Unfortunately, we were unable to obtain the mitochondrial DNA sequence from A. parahybae in the present work.
Among the specimens analyzed, this clade presents some species with a conserved chromosome number, always with 2n = 50 chromosomes, such as A. bockmanni and A. bifasciatus (Fazoli et al., 2003; Kavalco et al., 2009; Hashimoto and Porto-Foresti, 2010). On the other hand, the others species present high levels of numerical chromosome variation.
This diploid number (2n = 50) was also observed in the A. paranae specimens from the Paranaíba and some A. rivularis specimens from the São Francisco river basin, although other specimens of the Paranaíba river basin also had 2n = 46 chromosomes (data not shown, in preparation). Both species belong to the historical A. scabripinnis species complex, characterized by broad sympatric and allopatric karyotype variation (Moreira-Filho and Bertollo, 1991, among others).
Finally, a diploid number that varies from 2n = 46 or 2n = 48 was observed among the analyzed specimens of A. fasciatus (Pazza et al., 2006; Kavalco et al., 2016) that was also observed in other populations (Artoni et al., 2006; Medrado et al., 2008; Pansonato-Alves et al., 2013). This variation is considered common in the species, although 2n = 50 chromosomes have already been reported (Artoni et al., 2006).
In relation to rDNA 5S, most of the species/populations already analyzed have the same following pattern: one site on the marker chromosome plus a pair of acrocentric chromosomes with a rDNA 5S site in the proximal region; this was also observed in the specimens analyzed in the present work (Kavalco et al., 2004, 2007, 2009, 2016; Pazza et al., 2006) and in the literature available for A. bockmanni, A. fasciatus, and A. parahybae (Almeida-Toledo et al., 2002; Hashimoto et al., 2011; Silva et al., 2013; Daniel et al., 2015). Despite this relatively conserved pattern, there are two autapomorphies in the 5S rDNA phenotype in A. fasciatus. Medrado et al. (2015) reported the occurrence of a small variation in the distribution of 5S rDNA sites from a population of Astyanax aff. fasciatus where the marker site of the metacentric chromosome is absent, an evident populational autapomorphy. We also considered the additional occurrence of two 5S rDNA sites of A. aff. fasciatus from the Paraíba do Sul river basin an autapomorphy. In total, six 5S rDNA-bearing chromosomes are detected, represented always by the marker chromosome pair plus two st/a chromosomes pairs in this sample (Kavalco et al., 2016).
In relation to the A. scabripinnis group, the most studied from the cytogenetic point of view (Pazza and Kavalco, 2007), several populations of the Coastal, São Francisco, and Upper Paraná rivers show the same mentioned pattern (Mantovani et al., 2005; Fernandes and Martins-Santos, 2006; Peres et al., 2008; Vicari et al., 2008). However, other studies have already demonstrated, in addition to these standard sites, more distal or proximal sites in other pairs of acrocentric chromosomes in populations of coastal rivers and the Upper Paraná basin (Kavalco et al., 2004).
Although absent in our analyses, A. schubarti is a species that has available chromosome data and is found in clade 1 of Rossini et al. (2016), the clade analogous to clade 4 of this work. Its low chromosomal number (2n = 36) and high FN suggest the occurrence of Robertsonian events of chromosome fusion at its origin (Morelli et al., 1983). In fact, this species shows the 5S rDNA sites located in the proximal region of two pairs of metacentric chromosomes (Almeida-Toledo et al., 2002). Interestingly, A. currentinus, a species recently described (Mirande et al., 2015), presents 2n = 36 chromosomes and the same distribution pattern of A. schubarti 5S rDNA, with an additional odd site (Paiz et al., 2015). The authors suggest that these species may belong to the same morphological group and are phylogenetically related. Unfortunately, there are no available phylogenetic data to support this hypothesis.
Among the specimens used in the present study, populations of the A. fasciatus and A. scabripinnis species complexes presented sites with homology with the As51 satellite DNA (Kavalco et al., 2007, 2013; Pazza et al., 2008). On the other hand, this probe showed no homology in the chromosomes of A. bockmanni (Kavalco et al., 2009). Among the species included in this group and in the clade 1 of Rossini et al. (2016), in only one population of A. rivularis (cited as A. scabripinnis) this repetitive sequence is absent (Abel et al., 2006). On the other hand, other species belonging to clade 1 of Rossini et al. (2016) have already presented homology with As51 satellite DNA, such as A. parahybae (Kavalco et al., 2007) and A. serratus (quoted as Astyanax sp.; Kantek et al., 2009a). Among these species, the available data are mainly concentrated in the group A. scabripinnis/paranae/rivularis (Abel et al., 2006; Kantek et al., 2009b; Barbosa et al., 2015, 2017), and in A. fasciatus (Abel et al., 2006; Kantek et al., 2009b; Medrado et al., 2015). The distribution of this satellite DNA in A. fasciatus seems to follow a biogeographic pattern, with an increase in the number of sites in drainage populations in the interior of the continent, and a decrease in coastal populations (Kavalco et al., 2013; Medrado et al., 2015). This pattern does not appear to be the same in the A. scabripinnis group and its cryptic species.
The absence of a clear biogeographic pattern of satellite DNA distribution in the A. scabripinnis populations should be related to the fact that these fish inhabit isolated headwaters. In addition to facilitating the occurrence of vicariance processes, this ecology keeps low levels of gene flow among, with consequent reduction of population sizes and possible strong effect of genetic drift, resulting in random of As51 satDNA. This could easily generate the biogeographic gaps that occur in the genomic distribution and the existence of populations in which this repetitive DNA is absent, even being a basal characteristic for the group. In fact, it is not yet proven if there is any adaptive role for this DNA, even though we have evidence that it is present in a larger amount in the species with the greatest chromosome diversity, with extensive polymorphisms such as A. fasciatus. The occurrence of the As51 satDNA predominantly in supernumerary chromosomes (Mestriner et al., 2000) may imply exogenous origin.
The data of the literature obtained in the present study still cannot answer definitively the questions about the origin and evolution of these sequences within the lineages of Astyanax, only to indicate trends within specific clades. Can As51 satDNA promote the chromosomal rearrangements that generate the diversity in diploid numbers and karyotype formulas observed in Astyanax; or can the genome of species with chromosomal plasticity, such as those of the genus, facilitate the dispersion of repetitive DNAs? To answer this question, we need more information about the role of this DNA in the karyotype and organismic evolution of the group. Fortunately, the genus Astyanax, due to the great concentration of studies, is an excellent model to answer these questions of comparative genomics.
RP and KK designed the research and performed the analyses. All the authors collected data and contributed to the writing and review of the manuscript.
This study was financed by the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG; APQ-01247-08, APQ-00756-09, and APQ-01510-11).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the Manejo e Conservação de Ecossistemas Naturais e Agrários (MCENA) post-graduate program, Federal University of Viçosa, Campus Florestal.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00131/full#supplementary-material
Abel, L. D. S., Mantovani, M., and Moreira-Filho, O. (2006). Chromosomal distribution of the As-51 satellite DNA in two species complexes of the genus Astyanax (Pisces, Characidae). Genet. Mol. Biol. 29, 448–452. doi: 10.1590/S1415-47572006000300008
Albert, J. S., and Reis, R. E. (eds). (2011). Historical Biogeography of Neotropical Freshwater Fishes. Los Angeles: University of California Press, 408.
Almeida-Toledo, L. F., Ozouf-Costaz, C., Foresti, F., Bonillo, C., Porto-Foresti, F., and Daniel-Silva, M. F. Z. (2002). Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae). Cytogenet. Genome Res. 97, 229–233. doi: 10.1159/000066609
Artoni, R. F., Shibatta, O. A., Gross, M. C., Schneider, C. H., Almeida, M. C., Vicari, M. R., et al. (2006). Astyanax aff. fasciatus Cuvier, 1819 (Teleostei; Characidae): evidences of a species complex in the Upper rio Tibagi basin (Paraná, Brazil). Neotrop. Ichtyol. 4, 197–202. doi: 10.1590/S1679-62252006000200005
Barbosa, P., Leal, E. V., Silva, M., Almeida, M. C., Moreira-Filho, O., and Artoni, R. F. (2017). Variability and evolutionary implications of repetitive DNA dynamics in genome of Astyanax scabripinnis (Teleostei. Characidae). Comp. Cytogenet. 11, 143–162. doi: 10.3897/CompCytogen.v11i1.11149
Barbosa, P., Oliveira, L. A., Pucci, M. B., Santos, M. H., Moreira-Filho, O., Vicari, M. R., et al. (2015). Identification and chromosome mapping of repetitive elements in the Astyanax scabripinnis (Teleostei: Characidae) species complex. Genetica 143, 55–62. doi: 10.1007/s10709-014-9813-2
Barros, L. C., Santos, U., Cioffi, M. B., and Dergam, J. A. (2015). Evolutionary divergence among Oligosarcus spp. (Ostariophysi, Characidae) from the São Francisco and Doce river basins: Oligosarcus solitarius Menezes, 1987 shows the highest rates of chromosomal evolution in the Neotropical region. Zebrafish 12, 102–110. doi: 10.1089/zeb.2014.1030
Carvalho, D. C., Oliveira, D. A. A., Pompeu, P. S., Leal, C. G., Oliveira, C., and Hanner, R. (2011). Deep barcode divergence in Brazilian freshwater fishes: the case of the São Francisco River basin. Mitochondrial DNA 22, 80–86. doi: 10.3109/19401736.2011.588214
Carvalho, M. L., Oliveira, C., and Foresti, F. (2002). Cytogenetic analysis of five species of the subfamily Tetragonopterinae (Teleostei, Characiformes, Characidae). Caryologia 55, 181–188. doi: 10.1080/00087114.2002.10589275
Coutinho-Sanches, N., and Dergam, J. A. (2015). Cytogenetic and molecular data suggest Deuterodon pedri Eigenmann, 1907 (Teleostei: Characidae) is a member of an ancient coastal group. Zebrafish 12, 357–365. doi: 10.1089/zeb.2014.1068
Cunha, M. S., Reis, V. J. C., and Dergam, J. A. (2016). Closely related syntopic cytotypes of Astyanax taeniatus (Jenyns, 1842) from the Upper Piranga River, Upper Doce basin in Southeastern Brazil. Zebrafish 13, 112–117. doi: 10.1089/zeb.2015.1163
Daniel, S. N., Penitente, M., Hashimoto, D. T., Silva, D. M. Z. A., Foresti, F., and Porto-Foresti, F. (2015). New insights into karyotypic relationships among populations of Astyanax bockmanni (Teleostei, Characiformes) of different watersheds. Zebrafish 12, 181–188. doi: 10.1089/zeb.2014.1053
Eschmeyer, W. N., Fricke, R., and van der Laan, R. (2017). Catalog of Fishes: Genera, Species, References. Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp [accessed October 10, 2017].
Fazoli, L. C., Silva, V. A. B., Portela-Castro, A. L., and Júlio, H. F. Jr. (2003). Chromosome characterization of Astyanax sp B (Characidae, Tetragonopterinae), and endemic species of the Iguaçu river, Paraná, Brazil. Cytologia 68, 389–394. doi: 10.1508/cytologia.68.389
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fernandes, C. A., and Martins-Santos, I. C. (2004). Cytogenetic studies in two populations of the Astyanax altiparanae (Pisces, Characiformes). Hereditas 141, 328–332. doi: 10.1111/j.1601-5223.2004.01832.x
Fernandes, C. A., and Martins-Santos, I. C. (2006). Chromosomal location of 5S and 18S rRNA genes in three sympatric cytotypes of Astyanax scabripinnis (Characiformes, Characidae) from the Ivaí river basin, state of Paraná, Brazil. Caryologia 59, 253–259. doi: 10.1080/00087114.2006.10797923
Giongo, P., Sampaio, W. M. S., Fernandes, A., Dergam, J. A., Kavalco, K. F., and Pazza, R. (2013). Diversidade cariotípica de Astyanax asuncionensis (Teleostei, Characiformes) na microbacia do rio Sepotuba (Alto Paraguai). Evol. Cons. Biodivers. 3, 53–58. doi: 10.7902/ecb.v3i2.9
Hashimoto, D. T., Ferguson-Smith, M. A., Rens, W., Foresti, F., and Porto-Foresti, F. (2011). Chromosome mapping of H1 histone and 5S rRNA gene clusters in three species of Astyanax (Teleostei, Characiformes). Cytogenet. Genome Res. 134, 64–71. doi: 10.1159/000323512
Hashimoto, D. T., and Porto-Foresti, F. (2010). Chromosome polymorphism of heterochromatin and nucleolar regions in two populations of the fish Astyanax bockmanni (Teleostei: Characiformes). Neotrop. Ichthyol. 8, 861–866. doi: 10.1590/S1679-62252010000400016
Hattori, R. S., Daniel-Silva, M. F. Z., and Almeida-Toledo, L. F. (2007). Karyotype characterization and gene mapping of 5S and 18S rDNA in three species of Oligosarcus (Teleostei: Characidae). Caryologia 60, 372–378. doi: 10.1080/00087114.2007.10797961
Havelka, M., Bytyutskyy, D., Symonová, R., Ráb, P., and Flajshans, M. (2016). The second highest chromosome count among vertebrates is observed in cultured sturgeon and is associated with genome plasticity. Genet. Sel. Evol. 48:12. doi: 10.1186/s12711-016-0194-0
Hebert, P. D. N., and Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Mol. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Jacobina, U. P., Paiva, E., and Dergam, J. A. (2011). Pleistocene karyotypic divergence in Hoplias malabaricus (Bloch, 1794) (Teleostei: Erythrinidae) populations in southeastern Brazil. Neotrop. Ichthyol. 9, 325–333. doi: 10.1590/S1679-62252011005000023
Jónsson, H., Schubert, M., Seguin-Orlando, A., Ginolhac, A., Petersen, L., Fumagalli, M., et al. (2014). Speciation with gene flow in equids despite extensive chromosomal plasticity. Porc. Natl. Acad. Sci. U.S.A. 111, 18655–18660. doi: 10.1073/pnas.1412627111
Kantek, D. L. Z., Cestari, M. M., Bertollo, L. A. C., and Moreira-Filho, O. (2009a). Characterization of the constitutive heterochromatin of Astyanax sp. D (Characiformes: Characidae) from the Upper Iguaçu river (PR). Folia Biol. 57, 1–2.
Kantek, D. L. Z., Vicari, M. R., Peres, W. Z. M., Cestari, M. M., Artoni, R. F., Bertollo, L. A. C., et al. (2009b). Chromosomal location and distribution of As51 satellite DNA in five species of the genus Astyanax (Teleostei, Characidae, Incertae sedis). J. Fish Biol. 75, 408–421. doi: 10.1111/j.1095-8649.2009.02333.x
Kavalco, K. F., and Almeida-Toledo, L. F. (2007). Molecular cytogenetics of blind mexican tetra and comments on the karyotypic characteristics of genus Astyanax (Teleostei, Characidae). Zebrafish 4, 103–111. doi: 10.1089/zeb.2007.0504
Kavalco, K. F., and Moreira-Filho, O. (2003). Cytogenetical analyses in four species of the genus Astyanax (Pisces, Characidae) from Paraíba do Sul river basin. Caryologia 56, 453–461. doi: 10.1080/00087114.2003.10589358
Kavalco, K. F., Pazza, R., and Almeida-Toledo, L. F. (2009). Astyanax bockmanni Vari and Castro, 2007: an ambiguous karyotype in the Astyanax genus. Genetica 136, 135–139. doi: 10.1007/s10709-008-9325-z
Kavalco, K. F., Pazza, R., and Almeida-Toledo, L. F. (2010). Molecular cytogenetics of Astyanax ribeirae (Teleostei, Characidae), an endemic characin of the Atlantic rainforest. Nucleus 53, 51–54. doi: 10.1007/s13237-010-0006-9
Kavalco, K. F., Pazza, R., Brandão, K. d. O., and Almeida-Toledo, L. F. (2013). Biogeographic patterns in the chromosomal distribution of a satellite DNA in the banded tetra Astyanax fasciatus (Teleostei: Characiformes). Org. Divers. Evol. 13, 67–76. doi: 10.1007/s13127-012-0100-8
Kavalco, K. F., Pazza, R., Brandão, K. d. O., Garcia, C., and Almeida-Toledo, L. F. (2011). Comparative cytogenetics and molecular phylogeography in the group Astyanax altiparanae – Astyanax aff. bimaculatus (Teleostei, Characidae). Cytogenet. Genome Res. 134, 108–119. doi: 10.1159/000325539
Kavalco, K. F., Pazza, R., Bertollo, L. A. C., and Moreira-Filho, O. (2004). Gene mapping of 5S rDNA sites in eight fish species from the Paraíba do Sul river basin, Brazil. Cytogenet. Genome Res. 106, 107–110. doi: 10.1159/000078567
Kavalco, K. F., Pazza, R., Bertollo, L. A. C., and Moreira-Filho, O. (2005). Molecular cytogenetics of Oligosarcus hepsetus (Teleostei, Characiformes) from two Brazilian locations. Genetica 124, 85–91. doi: 10.1007/s10709-005-0176-6
Kavalco, K. F., Pazza, R., Bertollo, L. A. C., and Moreira-Filho, O. (2007). Satellite DNA sites in four species of the genus Astyanax (Teleostei, Characiformes). Genet. Mol. Biol. 30, 529–535. doi: 10.1590/S1415-47572007000400005
Kavalco, K. F., Pazza, R., Brandão, K. O., Garcia, C., Bertollo, L. A. C., and Almeida-Toledo, L. F. (2016). Chromosomal diversification higher than molecular variation in Astyanax aff. fasciatus (Teleostei, Characidae). Zebrafish 13, 345–353. doi: 10.1089/zeb.2016.1272
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Mantovani, M., Abel, L. D. S., and Moreira-Filho, O. (2005). Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 123, 211–216. doi: 10.1007/s10709-004-2281-3
Medrado, A. S., Affonso, P. R. A. M., Carneiro, P. L. S., Vicari, M. R., Artoni, R. F., and Costa, M. A. (2015). Allopatric divergence in Astyanax aff. fasciatus Cuvier, 1819 (Characidae, Incertae sedis) inferred from DNA mapping and chromosomes. Zool. Anz. 257, 119–129. doi: 10.4238/2011.November.22.5
Medrado, A. S., Figueiredo, A. V. A., Waldschmidt, A. M., Affonso, P. R. A. M., and Carneiro, P. L. S. (2008). Cytogenetic and morphological diversity in populations of Astyanax fasciatus (Teleostei, Characidae) from Brazilian northeastern river basins. Genet. Mol. Biol. 31, 208–214. doi: 10.1590/S1415-47572008000200007
Mello, R., Maniglia, T. C., Prioli, S. M. A. P., and Prioli, A. J. (2015). Genetic and biogeographical relationships among species of Astyanax (Teleostei, Characidae) in Brazilian river basins. Genet. Mol. Res. 14, 15356–15364. doi: 10.4238/2015.November.30.13
Melo, F. A. G. (2001). Revisão taxonômica das espécies do gênero Astyanax Baird e Girard, 1854 (Teleostei: Characiformes: Characidae) da região da Serra dos Órgãos. Arq. Mus. Nac. 59, 1–46.
Mendes, M. M., Rosa, R., Giuliano-Caetano, L., and Dias, A. L. (2011). Karyotype diversity of four species of the incertae sedis group (Characidae) from different hydrographic basins: analysis of AgNORs, CMA3 and 18S rDNA. Genet. Mol. Res. 10, 3596–3608. doi: 10.4238/2011.November.22.5
Mestriner, C. A., Galetti, P. M. Jr., Valentini, S. R., Ruiz, I. R. G., Abel, L. D. S., Moreira-Filho, O., et al. (2000). Structural and functional evidence that a B chromosome in the characid fish Astyanax scabripinnis is an isochromosome. Heredity 85, 1–9. doi: 10.1046/j.1365-2540.2000.00702.x
Mirande, J. M. (2009). Weighted parsimony phylogeny of the family Characidae (Teleostei: Characiformes). Cladistics 25, 574–613. doi: 10.1111/j.1096-0031.2009.00262.x
Mirande, J. M. (2010). Phylogeny of the family Characidae (Teleostei: Characiformes): from characters to taxonomy. Neotrop. Ichthyol. 8, 385–568. doi: 10.1590/S1679-62252010000300001
Mirande, J. M., Azpelicueta, M. M., and Aguilera, G. (2015). Redescription of Astyanax correntinus (Holmberg, 1891) (Teleostei: Characiformes: Characidae), more than one hundred years after original description. Zool. Abh. 55, 9–15.
Moreira-Filho, O., and Bertollo, L. A. C. (1991). Astyanax scabripinnis (Pisces, Characidae): a species complex. Braz. J. Genet. 14, 331–357.
Morelli, S., Bertollo, L. A. C., Foresti, F., Moreira-Filho, O., and Toledo Filho, S. A. (1983). Cytogenetic considerations on the genus Astyanax (Pisces, Characidae). I. Karyotypic variability. Caryologia 36, 235–244. doi: 10.1080/00087114.1983.10797664
Navarro, A., and Barton, N. H. (2003). Accumulation postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57, 447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x
Oliveira, O., Avelino, G. S., Abe, K. T., Mariguela, T. C., Benine, R. C., Ortí, G., et al. (2011). Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 11:275. doi: 10.1186/1471-2148-11-275
Ornelas-García, C. P., Domínguez-Domínguez, O., and Doadrio, I. (2008). Evolutionary history of the fish genus Astyanax Baird & Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol. Biol. 8:340. doi: 10.1186/1471-2148-8-340
Paiz, L. M., Baumgärtner, L., Graça, W. J., and Margarido, V. P. (2015). Basic cytogenetics and physical mapping of ribosomal genes in four Astyanax species (Characiformes, Characidae) collected in Middle Paraná river, Iguassu National Park: considerations on taxonomy and systematics of the genus. Comp. Cytogenet. 9, 51–65. doi: 10.3897/CompCytogen.v9i1.9002
Pansonato-Alves, J. C., Hilsdorf, A. W. S., Utsunomia, R., Silva, D. M. Z. A., Oliveira, C., and Foresti, F. (2013). Chromosomal mapping of repetitive DNA and Cytochrome c Oxidase I sequence analysis reveal differentiation among sympatric samples of Astyanax fasciatus (Characiformes, Characidae). Cytogenet. Genome Res. 141, 133–142. doi: 10.1159/000354885
Pazza, R., and Kavalco, K. F. (2007). Chromosomal evolution in the Neotropical characin Astyanax (Teleostei, Characidae). Nucleus 50, 519–543.
Pazza, R., Kavalco, K. F., and Bertollo, L. A. C. (2006). Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). 1. Karyotype analysis, Ag-NORs and mapping of the 18S and 5S ribosomal genes in sympatric karyotypes and their possible hybrid forms. Cytogenet. Genome Res. 112, 313–319. doi: 10.1159/000089886
Pazza, R., Kavalco, K. F., and Bertollo, L. A. C. (2008). Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). 2 – Chromosomal location of a satellite DNA. Cytogenet. Genome Res. 122, 61–66. doi: 10.1159/000151317
Pereira, L. H. G., Hanner, R., Foresti, F., and Oliveira, C. (2013). Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genet. 14:20. doi: 10.1186/1471-2156-14-20
Peres, W. A. M., Bertollo, L. A. C., Buckup, P. A., Blanco, D. R., Kantek, D. L. Z., and Moreira-Filho, O. (2012). Invasion, dispersion and hybridization of fish associated to river transposition: karyotypic evidence in “bimaculatus group” (Characiformes: Characidae). Rev. Fish Biol. Fish. 22, 519–526. doi: 10.1007/s11160-011-9246-2
Peres, W. A. M., Bertollo, L. A. C., and Moreira-Filho, O. (2008). Physical mapping of the 18S and 5S ribosomal genes in nine Characidae species (Teleostei, Characiformes). Genet. Mol. Biol. 31, 222–226. doi: 10.1590/S1415-47572008000200009
Piscor, D., Alves, A. L., and Parisi-Maltempi, P. P. (2015). Chromosomal microstructure diversity in three Astyanax (Characiformes, Characidae) species: comparative analysis of ghe chromosomal locations of the 18S and 5S rDNAs. Zebrafish 12, 81–90. doi: 10.1089/zeb.2014.1036
Piscor, D., and Parisi-Maltempi, P. P. (2016). Microsatellite organization in the B chromosome and A chromosome complement in Astyanax (Characiformes, Characidae) species. Cytogenet. Genome Res. 148, 45–51. doi: 10.1159/000444728
Rebordinos, L., Cross, I., and Merlo, A. (2013). High evolutionary dynamism in 5S rDNA of fish: state of the art. Cytogenet. Genome Res. 141, 103–113. doi: 10.1159/000354871
Rieseberg, L. H. (2001). Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358. doi: 10.1016/S0169-5347(01)02187-5
Rossini, B. C., Oliveira, C. A. M., Melo, F. A. G., Bertaco, V. A., Astarloa, R. N. D., Rosso, J. J., et al. (2016). Highlighting Astyanax species diversity through DNA Barcoding. PLoS One 11:e0167203. doi: 10.1371/journal.pone.0167203
Santos, L. P., Castro, J. P., Francisco, C. M., Vicari, M. R., Almeida, M. C., Goll, L. G., et al. (2013). Cytogenetic analysis in the neotropical fish Astyanax goyacensis Eigenmann, 1908 (Characidae, incertae sedis): karyotype description and occurrence of B microchromosomes. Mol. Cytogenet. 6:48. doi: 10.1186/1755-8166-6-48
Santos, L. P., Francisco, C. M., Castro, J. P., Barbosa, P., Morelli, S., Oliveira, L. A., et al. (2016). Karyoevolutionary inferences in enigmatic taxon Astyanax (Teleostei: Characidae). Zebrafish 13, 578–583. doi: 10.1089/zeb.2016.1291
Sassi, F. M. C., Resende, S. V., Pazza, R., and Kavalco, K. F. (2018). “Natural Hybridization between chromosomal discrepant species and the role of hybrid speciation in the Genus Astyanax,” in An Essential Guide to Cytogenetics, eds N. Norris and C. Miller (Hauppauge, NY: Nova Science Publisher).
Silva, D. M. Z. A., Pansonato-Alves, J. C., Utsonomia, R., Daniel, S. N., Hashimoto, D. T., Oliveira, C., et al. (2013). Chromosomal organization of repetitive DNA sequences in Astyanax bockmanni (Teleostei, Characiformes): dispersive location, association and co-localization in the genome. Genetica 141, 329–336. doi: 10.1007/s10709-013-9732-7
Silva, D. M. Z. A., Utsonomia, R., Pansonato-Alves, J. C., Oliveira, C., and Foresti, F. (2015). Chromosomal mapping of repetitive DNA sequences in five species of Astyanax (Characiformes, Characidae) reveals independent location of U1 and U2 snRNA sites and association of U1 snRNA and 5S rDNA. Cytogenet. Genome Res. 146, 144–152. doi: 10.1159/000438813
Silva, P. C. (2017). Sistemática Integrativa - Diversidade e Relações de Deuterodon e Gêneros Afins. Doctoral thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, 188.
Sivasundar, A., Bermingham, E., and Ortí, G. (2001). Population structure and biogeography of migratory freshwater fishes (Prochilodus: Characiformes) in major South American Rivers. Mol. Ecol. 10, 407–417. doi: 10.1046/j.1365-294X.2001.01194.x
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Travenzoli, N. M., Silva, P. C., Santos, U., Zanuncio, J. C., Oliveira, C., and Dergam, J. A. (2015). Cytogenetic and molecular data demonstrate that the Bryconinae (Ostariophysi, Bryconidae) species from Southeastern Brazil form a phylogenetic and Phylogeographic unit. PLoS One 10:e0137843. doi: 10.1371/journal.pone.0137843
Keywords: cytotaxonomy, molecular evolution, chromosomal rearrangements, mtDNA, chromosomal symplesiomorphy, chromosomal synapomorphy, chromosomal autapomorphy
Citation: Pazza R, Dergam JA and Kavalco KF (2018) Trends in Karyotype Evolution in Astyanax (Teleostei, Characiformes, Characidae): Insights From Molecular Data. Front. Genet. 9:131. doi: 10.3389/fgene.2018.00131
Received: 01 November 2017; Accepted: 03 April 2018;
Published: 16 April 2018.
Edited by:
Roberto Ferreira Artoni, State University of Ponta Grossa, BrazilReviewed by:
Wenyan Du, Alforex Seeds LLC, United StatesCopyright © 2018 Pazza, Dergam and Kavalco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rubens Pazza, cnBhenphQHVmdi5icg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.