- 1Department of Biological Sciences, Faculty of Science, Kuwait University, Kuwait, Kuwait
- 2Human Genetics Unit, Department of Pathology, Faculty of Medicine, Kuwait University, Kuwait, Kuwait
Common variants of Apolipoprotein A5 (APOA5) have been associated with lipid levels yet very few studies have reported full sequence data from various ethnic groups. The purpose of this study was to analyse the full APOA5 gene sequence to identify variants in 100 healthy Kuwaitis of Arab ethnicities and assess their association with variation in lipid levels in a cohort of 733 samples. Sanger method was used in the direct sequencing of the full 3.7 Kb APOA5 and multiple sequence alignment was used to identify variants. The complete APOA5 sequence in Kuwaiti Arabs has been deposited in GenBank (KJ401315). A total of 20 reported single nucleotide polymorphisms (SNPs) were identified. Two novel SNPs were also identified: a synonymous 2197G>A polymorphism at genomic position 116661525 and a 3′ UTR 3222 C>T polymorphism at genomic position 116660500 based on human genome assembly GRCh37/hg:19. Five SNPs along with the two novel SNPs were selected for validation in the cohort. Association of those SNPs with lipid levels was tested and minor alleles of three SNPs (rs2072560, rs2266788, and rs662799) were found significantly associated with TG and VLDL levels. This is the first study to report the full APOA5 sequence and SNPs in an Arab ethnic group. Analysis of the variants identified and comparison to other populations suggests a distinctive genetic component in Arabs. The positive association observed for rs2072560 and rs2266788 with TG and VLDL levels confirms their role in lipid metabolism.
Introduction
Apolipoprotein gene (APOA5) was first described in 2001 as a result of comparative genome sequencing between mouse and man (Pennacchio et al., 2001). It is located on chromosome 11q23 in the APOA1/C3/A4 gene cluster. The human APOA5 gene usually gives a 366 amino acid long apoA5 protein (Pennacchio et al., 2001) which is predominantly produced in the liver. ApoA5 protein can be found both intracellulary and associated with cell membrane structures (Nilsson et al., 2011). In plasma, apoA5 is present as a monomer on chylomicrons, VLDL and frequently on HDL at extremely low concentration (Nilsson et al., 2011). The specific role of apoA5 has not been yet clearly defined, however, since its discovery to date many studies have been conducted to investigate its effect on lipid metabolism (Weinberg et al., 2003; Olofsson, 2005; Beckstead et al., 2007; Forte et al., 2009; Guardiola et al., 2012).
Several mechanisms have been proposed to explain apoA5 mode of action. Based on such findings, it was concluded that in plasma, apoA5 induced both an increase in lipolysis, through the increase in lipoprotein lipase LPL activity, and an elevated removal of lipoprotein particles, particularly, the triglycerides-rich lipoproteins (TG-RL) (Grosskopf et al., 2005). Also, apoA5 serves as a ligand for an endothelial cell surface protein called glycosylphosphatidylinositol high-density lipoprotein binding protein 1 (GPIHBP1) (Sharma et al., 2012). It was found that GPIHBP1 plays a particular and critical role in the lipolysis processing of chylomicrons promoting LPL lipolytic processing of the TG content of these particles (Beckstead et al., 2003; Beigneux et al., 2007; Sharma et al., 2012). In addition to its important role in modulating plasma TG levels, some experiments have indicated apoa5 effect on reducing HDL-C levels (Beckstead et al., 2003; Grosskopf et al., 2005; Cha et al., 2014). Beside its extracellular effect, previous investigation of apoA5 impact has pointed an action within hepatocytes to modulate hepatic TG metabolism and secretion (Weinberg et al., 2003; Olofsson, 2005). In hepatocytes, apoA5 was found responsible for the attenuation of the second-step of VLDL particle maturation, essential for the formation of TG-rich VLDL and therefore impairing liver VLDL assembly and secretion (Walther and Farese, 2012).
However, recent advances in genome-wide association studies (GWAS) have provided a fairly good view of the contribution of APOA5 variants to dyslipidemia in several populations (Talmud, 2007; Li et al., 2014; Zubair et al., 2016; Kefi et al., 2017; Sumegi et al., 2017). The minor allele of several common APOA5 SNPs such as rs662799 (-1131T>C) and rs3135506 (S19W) have been reported to be consistently and significantly associated with plasma triglyceride (TG) levels (Baum et al., 2003; Chaaba et al., 2005) and the risk of cardiovascular disease (Talmud et al., 2002; Lai et al., 2004) in several populations. In addition to the well characterized common APOA5 variants, rare variants in the APOA5 gene have been documented and implicated in the development of dyslipidemia and associated diseases (Priore Oliva et al., 2006; Ouatou et al., 2014). But while that disease etiology is common between populations, risk variants can often be population-specific. Therefore, these variants and others have been identified and genotyped in several populations by various molecular methods and the results have not been always been consistent among different ethnicities (Pullinger et al., 2008; Cha et al., 2014; Li et al., 2014; Ouatou et al., 2014; Sumegi et al., 2017).
Few studies have provided full sequence analysis of the APOA5 gene locus and limited studies have been conducted on APOA5 variants and their possible associations among Arab countries (Chaaba et al., 2005; Ouatou et al., 2014; Kefi et al., 2017).
Kuwait is a small country that is situated on the north-east of the Arabian Peninsula and at the northern end of the Arabian Gulf. Historically, the population of Kuwait comprises early settlers originating from the tribes of neighboring Arabian and Persian countries (particularly from Saudi Arabia, Iraq and Iran) and from the nomadic Arabs of the desert, living on the fringes of the Arabian Peninsula, called Bedouins (Teebi, 1994; Al-Bustan et al., 2005; Casey, 2007). The prevalence of disorders such as type 2 diabetes, obesity, hypertension and dyslipidemia has increased (Alwan and King, 1995; AlMajed et al., 2011; Alhyas et al., 2012; Channanath et al., 2013). In 2013, (WHO) estimates that dyslipidemia is showing alarmingly high prevalence and emerging as a risk factor for coronary disease in Kuwait (World Health Organization, 2013). While the settlements and the subsequent admixtures have shaped the genetics of Kuwait, understanding the genetic structure of Kuwaiti population remains a big challenge but necessary to aid studies in defining genetic and molecular mechanisms underlying causes of dyslipidemia and other complex diseases.
Therefore, the main objective of this study was to re-sequence the full APOA5 gene loci in Kuwaiti Arabs to identify potential variants that could be associated with variation in serum lipid levels and to validate a selection of the variants in a cohort. This study is the first to report full sequence data and analysis at the APOA5 gene loci in an Arabic ethnic group.
Materials and Methods
Studied Samples
This study was undertaken within the guidelines set by the Declaration of Helsinki, and the protocol was approved by the Local Ethical Committee at Kuwait University. One hundred apparently healthy Kuwaiti Arabs (50 males and 50 females) were recruited from volunteers who attend the regional polyclinics or the major hospitals in Kuwait. Written informed consent was obtained from all subjects. The age range of the subjects was 18–69 years, and the body mass index (BMI; in kg/m2) range was 19–30. Medical and family medical history of hypertension, hypercholesterolemia, hyperglyceridemia, diabetes and cardiovascular diseases were documented. Ethnicity was verified by tracing both maternal and paternal lineages at least four generations using pedigree analysis. The selected subjects were divided into three groups according to their Cholesterol (TC) levels. There were 20 subjects with high TC levels (5.2–6.77 mmol/L), 20 with low TC levels (2.48–3.9 mmol/L) and a remaining 60 with normal TC levels (TC: 4–5.16 mmol/L). Association of identified and selected variants were analyzed in a cohort of 733 samples of the Kuwaiti General population. The cohort included randomized samples of the Kuwaiti general population with documented lipid profiles recruited at various hospitals during routine check-up whose informed consent was obtained individually following the ethical guidelines stated earlier. The sample consisted of 425 females and 279 males with ages ranging between 18 to 76. For each sample, phenotypic variables including BMI and family history of hypercholesterolemia, hypertension, diabetes and heart diseases were recorded. Relevant phonetic data was documented for each sample. A summary of the sample description is provided in Table 1.
Biochemical Measurements
Blood samples were taken intravenously after an overnight fast. Serum lipid levels in each sample were determined by enzymatic methods with commercially available kits and performed on a UniCel DxC 800 Synchron Clinical System from Beckman Coulter (USA) in the Clinical Chemistry facility at Al-Amiri Hospital in Kuwait.
DNA Analysis
Total genomic DNA was extracted from whole blood using salting-out procedure described by Miller et al. (1988). A region of 3.7 Kb of APOA5 gene was amplified from each individual in eight overlapping segments using newly designed primer sets (n = 8 pairs) using Primer3 Input software version 0.4.0 (//Frodo.wi.mit.edu/) (Supplementary Table 1). Amplification reactions were performed by PCR in an Applied Biosystems Fast thermal cycler (Version 1.01, Life Technologies, USA). PCR products were purified using Nucleospin® extract II column Kit (Clontech Laboratories, Inc., Version No. PR48598) following the suggested protocol (Macherey-Nagel, Germany). Cycle sequencing was then performed according to the manufacturer's instructions using the BigDye Terminator v.3.1 in a Fast Thermal Cycler (Life Technologies, Applied Biosystems, USA). The extension products were purified using BigDye® XTerminator™ Kit (Life Technologies, Applied Biosystems, USA). Capillary electrophoresis was then performed in ABI-3130xl Genetic Analyzer (Life Technologies, Applied Biosystems, USA).
Sequence Analysis and Polymorphisms Identification
The obtained DNA sequences for each sample were analyzed using the AB DNA Sequencing Analysis Software version 5.3.1 (Life Technologies, Applied Biosystems, USA). The sequences from the two reactions were compared for quality assurance and were aligned using ClustalW software (Multiple Alignment Tool) [37] to screen for all novel and/or mutations. Multiple sequence alignment among all samples and in comparison, to the reference sequence (NT_033899.8) was performed in order to confirm the genotype of each sample and to determine the presence of “common,” “rare,” and “novel” SNPs. Information about reported SNPs were obtained from NCBI and Ensembl databases (http://www.ncbi.nlm.nih.gov/, http://www.ensembl.org/).
Validation of Novel SNPs and Association of Common SNPs Identified
A total of 7 SNPs, including the two novels were validated a cohort of 736, with the exception of rs662799 which was only validated in 549 samples of the cohort due to technical difficulties and limited resources. The variants were analyzed using real-time PCR [ABI 7800HT Realtime PCR (GS01/02)] with customized primer and probe sets for the two novel SNP's and commercially available primer and probe sets for the four remaining SNPs (Supplementary Table 2, Applied Biosystems # 4351379). The genotyping assay and protocol was followed based on the manufacturer's recommendations for Taqman™ Genotyping Master Mix (Applied Biosystems # 4371355).
Haplotype Analysis
Linkage disequilibrium (r2) (D′) between the selected SNPs was analyzed using Haploview (version 4.2.) (Barrett et al., 2005). Pairwise r2 and D′ value between each two of SNPs was calculated. An r2 value of 1 means that the two loci are in complete linkage disequilibrium, in contrast an r2 value of 0 means that the two loci are in linkage equilibrium. In this study, r2 values more than 0.7 was considered to signify that two loci are in linkage disequilibrium. Haploview (version 4.2.) was also used to make graphical representations of LD. Haplotypes blocks were defined according to Gabriel et al. definition (Gabriel et al., 2002).
Statistical Analysis
Allele and genotype frequencies for each variant (novel, rare and common) were estimated by simple gene counting method for both the sequenced samples (n = 100: 22 variants) and for the cohort (n = 733: 6 variants). Hardy-Weinberg equilibrium (HWE) was also tested using the GENEPOP software (Version 4.2) (Rousset, 2008). Statistical analyses on the differences in the genotypic and allele frequency distribution were determined with regards to age, gender and BMI using Kruskal-Wallis ANOVA test and were reported as mean ± standard error. A two-tailed p-value of 0.05 was considered statistically significant. TG, HDL-C, LDL-C, VLDL were natural log-transformed to achieve approximate normal distribution before further analysis. In addition, Logistic regression analyses was preformed to investigate the association of the studied SNPs with lipid level variations. Genetic modeling of the significant variants was also performed. All these tests were performed using the R software (version 3.3.1) utilizing the following packages SNPassoc, psych, genetics, and MASS (R Core Team, 2016). Person's Chi-square test was also used to investigate the significant of the different distribution of haplotypes frequencies between the high and normal TG patient's groups. For all tests, the statistical significance level was set at p < 0.05.
Results
Genetic Variants
The full APOA5 gene including the promoter region, exons and introns, downstream and upstream regions was successfully sequenced for all 100 Kuwaiti Arab samples. The revealed APOA5 sequence with all of the detected SNPs was deposited in the NCBI GenBank under accession number KJ401315. Twenty-two genetic variants, two of which are novel, were observed by scanning the resulted chromatograms (Supplementary Figure 1) as well as by aligning the sequences with a reference APOA5 sequence (NT_033899.8). The majority of the identified SNPs were in the non-coding region in which; five were intragenic, seven in the 3′ UTR, one in the 5′ UTR, two in the downstream region and two in the upstream region. Further, three missense mutations (rs3135506, rs3135507, rs369952307) plus a previously reported synonymous variant (rs12287066) and a novel synonymous variant in exon 4 (2197G>A) were reported (Table 2). Each novel variant was identified in a heterozygous state in two different individuals. The novel synonymous SNP 2197G>A (at genomic position 116661525; GRCh37/hg:19) causes a G to A transition at position. The individual carrying this variant is a 27 years old female that has a high (28 kg/m2), low (0.89 mmol/L) and high (3.52 mmol/L). The second novel variant found in the 3′UTR (at genomic position 116660500; GRCh37/hg:19) results from a C to T transition at position 3,222 belongs to a 20 years old female that has a normal BMI (20.7 kg/m2) as well as a normal lipid profile. Functional prediction analysis employing SIFT, TargetScan, SNPNexus tools revealed that the two novel SNP's are simple sequence variants.
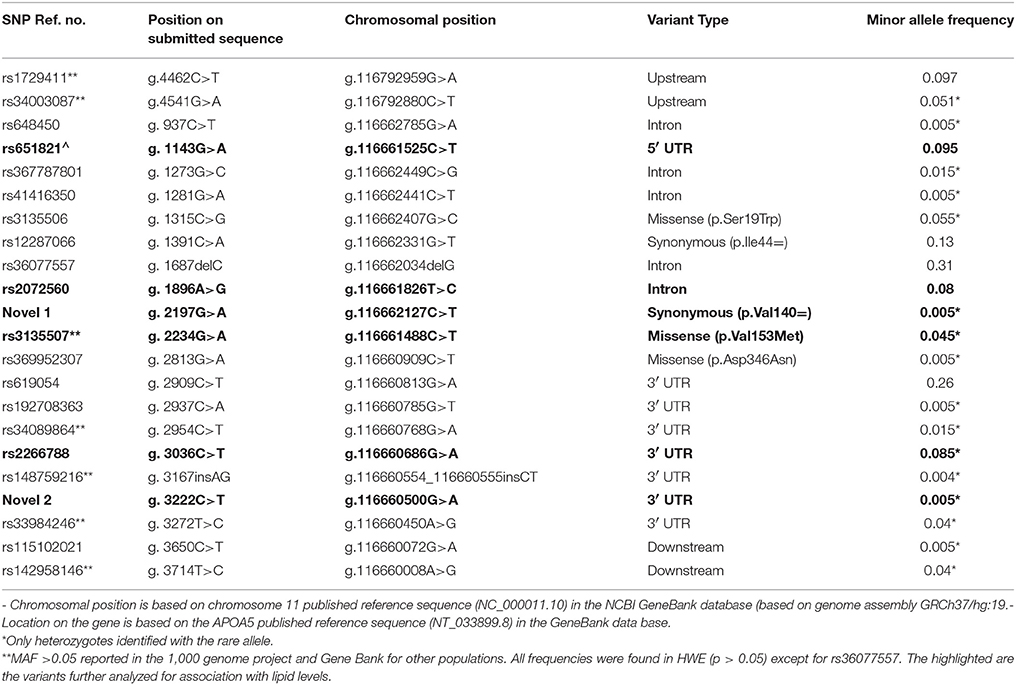
Table 2. Characteristics of the 22 identified SNPs by re-sequencing the APOA5 gene locus in Kuwaiti Arabs (n = 100).
Genotype and Allele Frequencies
All variants were in HWE. Nine variants were common between Kuwaiti Arabs with a minor allele frequency (MAF) above 0.05. The other remaining variants (n = 13) were identified as rare with a minor allele frequency < 0.05 including the two novel SNPs (Table 2).
Four of the detected variants (rs2266788, rs651821, rs2072560, and rs3135506) plus the commonly studied rs662799 (−1,131 T>C) SNP that had previously shown to be associated with lipid levels were selected for further analysis to determine their association with variation in serum lipid levels among a Kuwait cohort. The two novel variants were also selected for the validation and further statistical analysis.
Validation and Association of Selected Variants With Serum Lipid Levels
Real-time PCR was used to validate and genotype a cohort of 736 samples of the general Kuwaiti population. A sample of the real-time PCR allelic discrimination plot for the novel variants using the newly designed primers and probes sets are provided in the Supplementary Figures 2A–F. A summary of the genotype and allele frequencies determined in the 736 samples of the Kuwaiti cohort are provided in Table 3. Both novel variants were only identified in a heterozygous state in < 1 % of the cohort (Table 3) in which Novel 2 was only detected in the original sequenced sample while Novel 1 was further identified in an additional three samples implicating them as “private” variants.
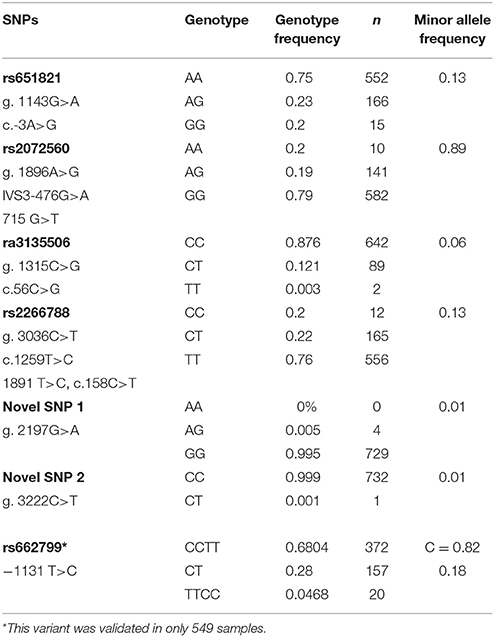
Table 3. Genotypic and allelic frequencies of the selected 7 APOA5 variants in the cohort of Kuwaiti Arabs (n = 733).
The association between the genotypes of the seven variants with variation in serum lipid levels are summarized in Table 4. SNPs rs2072560 and rs2266788 showed a significant association with TG reporting p-values of 0.04 and 0.02 respectively. Individuals carrying the minor A allele of rs2072560 (n = 107) displayed higher TG levels (>0.98 mmol/L) than individuals (0.92 mmol/L ± 0.04) with the GG genotype (n = 404). Carriers of the C allele of rs2266788 (n = 125) also displayed higher TG (>0.99 mmol/L) than individuals (0.92 mmol/L ± 0.04) with the TT genotype (n = 386). SNPs rs2072560 and rs2266788 also showed significant association with VLDL in which carriers of the A allele of rs2072560 (n = 106) showed increased VLDL levels (>0.41 mmol/L) than those (0.39 mmol/L ± 0.02) having the GG genotype (n = 397). Also, individuals with the C allele of rs2266788 (n = 123) showed higher levels of VLDL (>0.42 mmol/L) than the TT genotype (0.39 mmol/L ± 0.02; n = 380). Significant association (p = 0.01) of the minor C allele of rs662799 was observed with lower levels of TG (−0.46 mmol/l) as well as lower VLDL levels (−1.29 mmol/L, p = 0.03) in the 459 samples analyzed of the cohort (n = 733). On the other hand, the novel SNPs did not reveal any significant association with variation in lipid levels.
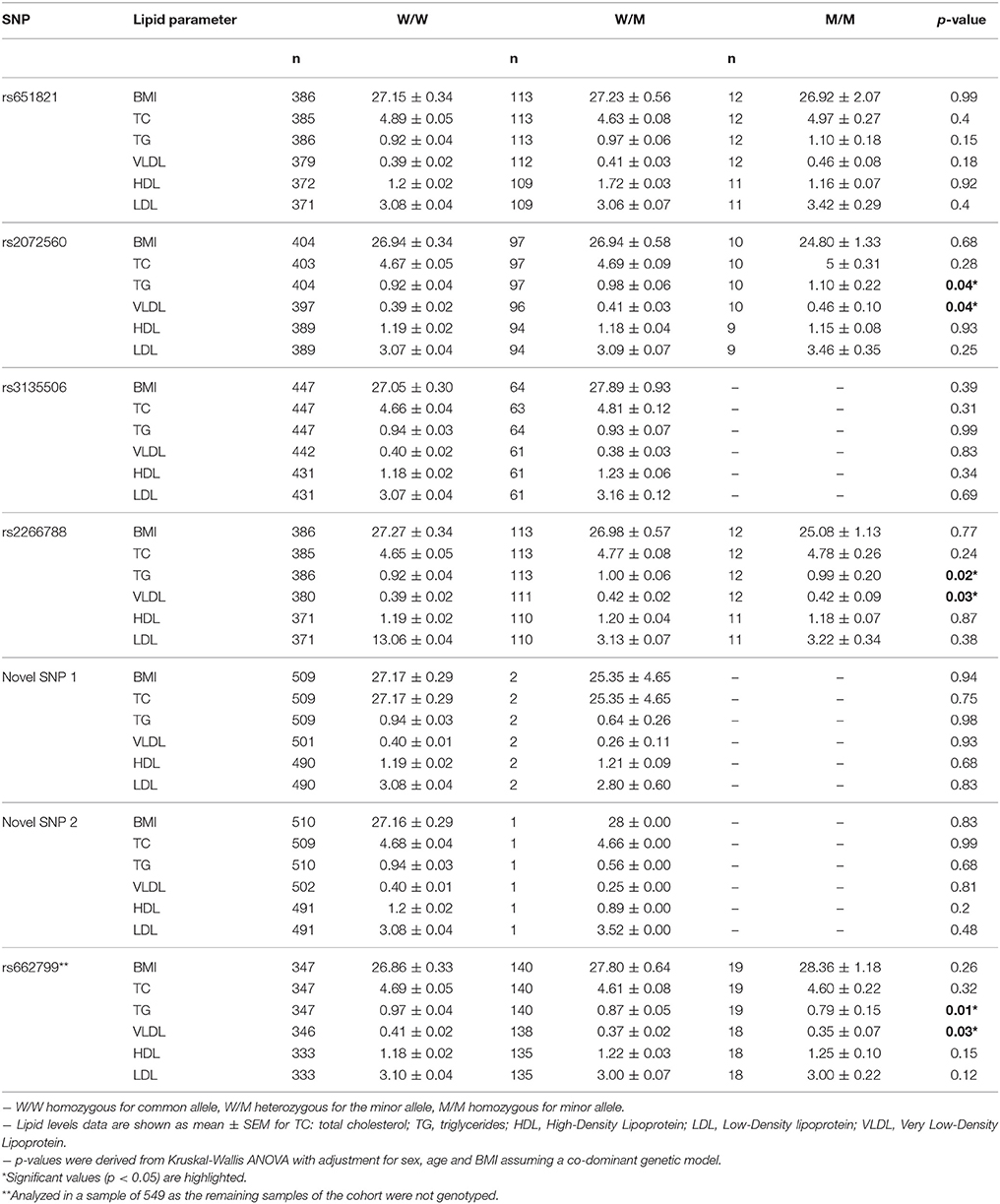
Table 4. Association of the 7 variants at the APOA5 gene locus with lipid profiles in the Kuwaiti Cohort (n = 733).
Multivariate Analysis and Genetic Modeling of the Associated Variants
The rs2072560 SNP was found to be an independent predictive factor for both TG and VLVD when controlling for age, sex and BMI with a similar odds ratio of 1.42 (95% CI: 1.03 –1.95, p = 0.03). In addition, rs2266788 SNP was also found to be an independent predictive factor for both TG and VLVD when controlling for age, sex and BMI with an odds ratio of 1.15 (95% CI: 1.03–1.27, p = 0.01) and an odds ratio of 1.25 (95% CI: 0.92–1.68, p = 0.02) respectively. A significant (p = 0.01) decrease of 0.73 OR was also observed for homozygous genotypes of the minor C allele for rs662799 concerning TG serum levels. These results are demonstrated in Table 5.
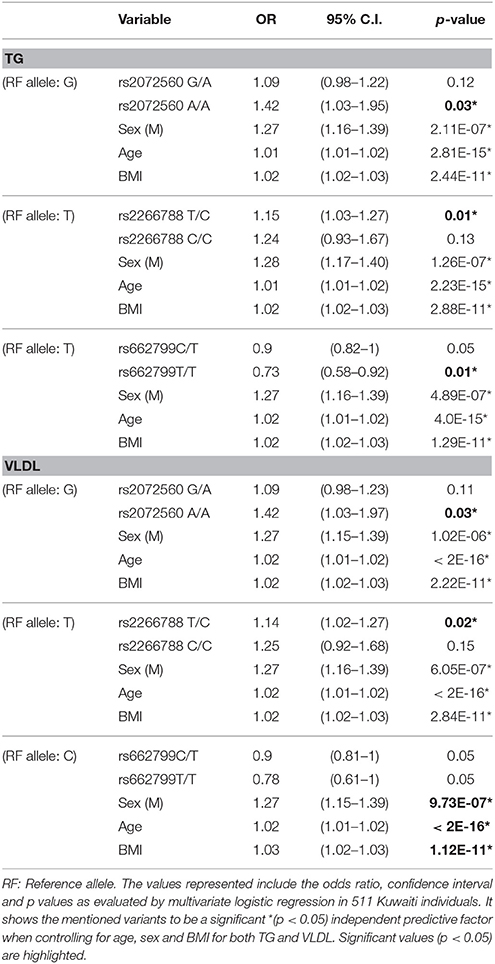
Table 5. Results of multivariate analysis of the APOA5 rs2072560, rs2266788 and rs662799 with triglycerides (TG) and Very Low-Density Lipoprotein (VLDL) levels in the cohort (n = 733).
The genetic modeling of the significant variants was preformed, and results are summarized in Table 6. A significant association of SNP rs2072560 with TG and VLDL was confirmed in which AA genotype shows a 42% increase of having higher TG and VLDL levels regardless of age, sex and BMI as compared to the wildtype GG with an odds ratio of 1.42 (95% CI 1.03–1.95, p-value of 0.04) and odds ratio of 1.42 (95% CI 1.03–1.97, p-value of 0.04) respectively. Moreover, a significant p-value for all modes of inheritance was observed with regards to variant to rs2072560 in association to TG, VLDL except for the over dominant model. Similarly, rs2266788 also revealed genetic association with TG and VLDL regardless of age, sex and BMI. The CC genotype for the this variant showed a 25% increase chance of having high TG and VLDL in comparison with the wild type genotype TT with an odds ratio of 1.25 (95% CI 0.93–1.68, p-value of 0.02) and odds ratio of 1.25 (95% CI 0.92–1.68, p-value of 0.03) respectively. A significant genetic association for all modes of inheritance was also observed for rs662799 with -TG, VLDL except for the over dominant model.
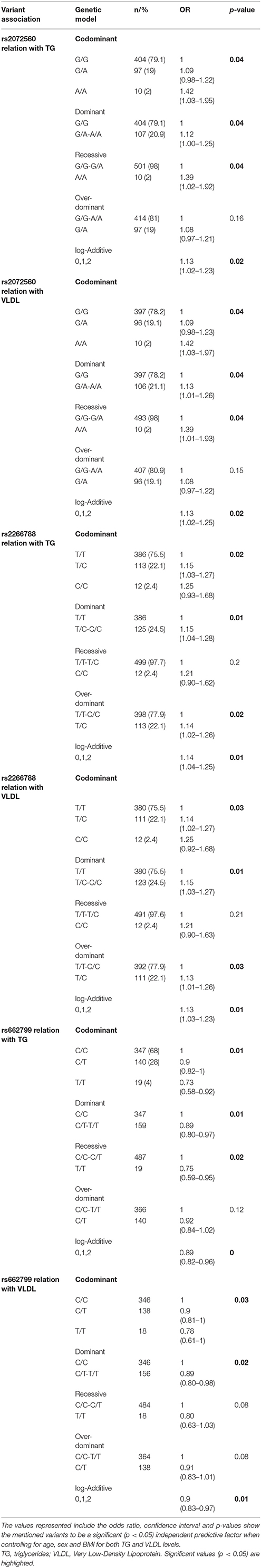
Table 6. Genetic modeling of APOA5 rs2072560, rs2266788 and rs662799 with TG and VLDL levels in the cohort (n = 733).
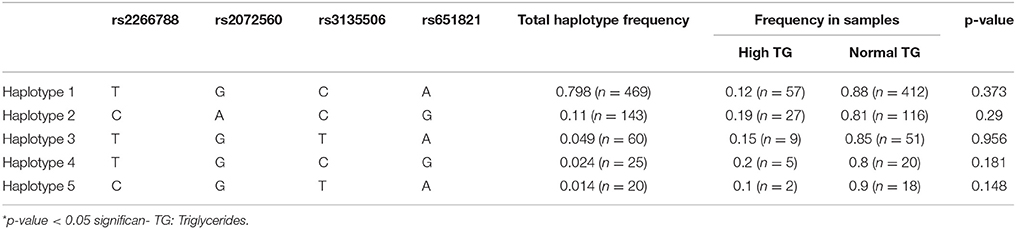
Haplotype Analysis
Linkage disequilibrium (LD) and haplotype analysis between the selected SNPs was expressed in D′ and r2 as presented in Figure 1. Both rs2266788 and rs2072560 SNPs showed disequilibrium (D′ = 1, r2 = 0.834). In addition, SNPs rs2072560 and rs651821 were also found significantly linked (D′ = 1, r2 = 0.799). Another strong LD was observed between SNPs re2266788 and rs651821 (D′ = 0.831, r2 = 0.662). However, rs3135506 SNP showed a perfect D′ value of 1 with SNPs rs2072560 and rs651821 but low r2 of 0.008 and 0.01, respectively. On the other hand, five major haplotypes of rs2266788, rs2072560, rs3135506, and rs651821 alleles were identified with TGCA as the common haplotype followed by CACG haplotype in which they represented 79.8 and 11 %, respectively (Table 7). The remaining haplotypes, TGTA, TGCG and CGTA accounted for 4.9, 2.4, and 1.4%, respectively. In total, those five haplotypes accounted for 99.5% of all of the haplotypes in this cohort. Testing the distribution of haplotypes frequencies between high and normal TG level groups indicated no significant association of the resulted haplotypes and TG levels (P > 0.05) (Table 7).
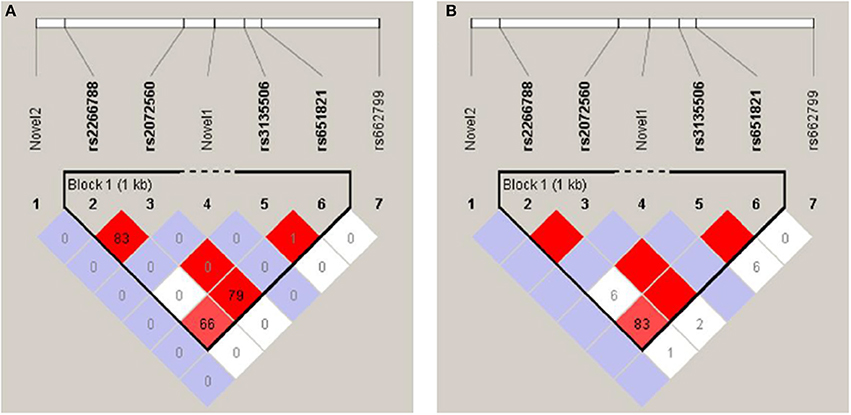
Figure 1. Linkage disequilibrium plots of the studied variants: rs2266788, rs651821, rs2072560, SNP rs3135506, rs662799, novel 1, and novel 2 constructed by Haploview (version 4.2.) (A) LD plot representing r2 values between studied APOA5 SNPs (B) LD plot representing D′ values between studied APOA5 SNPs.
Discussion
In several studies, apoA5 displayed substantial impact on lipid levels especially TG leading to the consensus that one of its core functions is TG modulation. Variations in the genetic region encoding this protein has therefore been associated with lipid imbalances among different ethnicities. The present study has re-sequenced the full APOA5 gene to screen for reported and novel variants and investigate their association with levels of plasma lipids. Re-sequencing the full APOA5 region in the sampled Kuwaiti Arabs resulted in the identification of 22 genetic variants; 2 novels and 20 previously reported SNPs. Distributions of SNPs were found to be different across the functional component of the APOA5 gene locus. As expected, the majority of the detected SNPs were found to be in non-coding regions (n = 17) than in coding exons (n = 5). In 2008, Luis Barreiro and his colleagues hypothesized that non-synonymous mutations and variations within 5′UTR and 3′UTR experience increased selective pressure over intronic and synonymous mutations (Quach et al., 2009). Therefore, results from this study suggest that the Kuwaiti Arabs population is under certain selection pressure. This selection could be toward fixing beneficial APOA5 alleles that fits better either with their genetic and environmental makeup or toward removing disadvantageous APOA5 alleles. Most of the detected SNPs in the Kuwaiti Arabs samples were in the 3′UTR (n = 7), including a novel SNP. A growing body of evidence has revealed the important role of the 3′UTR in epigenetics as well as regulation of gene expression and that its dysfunction is linked to the pathophysiology of many diseases (Scheper et al., 2007).
SNPs identified in the exonic region revealed three missense variants. One is rs369952307 (c. 1036G>A) SNP that has been recently identified in an African American individual by the group of researchers working on the NHLBI GO ESP (NHLBI Exome Sequencing Project (ESP) 2011). No information about the sample medical condition was provided in that study. In this study, a 22 years old male (BMI = 20.7 Kg/m2) with a normal lipid profile was identified with SNP rs369952307. This sample and the ESP reported sample were identified with a heterozygote genotype. The variant is an outcome of a G to an A transition in exon 4 resulting in an Aspartic (Asp) to Asparagine (Asn) substitution at residue 346 at the C-terminal region of the precursor apoA5 protein. Aspartic acid is acidic, polar and charged amino acid that plays important roles in maintaining the solubility and ionic character of proteins (Patel et al., 2011) unlike asparagine that is neutral, polar and uncharged amino acid. This substitution may therefore affect apoA5 lipid binding ability since this region-terminal has been reported to modulate protein lipid binding activity. To characterize and understand the effect of this variant, a large-scale validation of rs369952307 (c. 1036G>A) SNP among African Americans, Kuwaiti Arabs and other populations is warranted.
The most common identified SNPs were rs36077557 and rs619054 (c.*31C>T) with MAF of 31 and 26%, respectively. Those SNPs showed approximately similar allele frequencies like Caucasians and African Americans but diverse from East Asian population (†The International HapMap, 2003; The Genomes Project, 2015). However, both SNPs where not selected for association study as they did not show strong implication in lipid levels in previous studies. Moreover, the rare SNPs identified in the sampled Kuwaiti Arabs are rare among all studied populations in the 1,000 genome project with slightly higher frequencies among Africans. These results may be consistent with the out of Africa theory which states that humans originated in Africa and expanded to Eurasia about one million years ago (Excoffier and Ray, 2008). The bottleneck associated with the expansion may result in those rare SNPs being rarer or lost in non-African populations. These observed interethnic differences in the allelic frequencies between different populations could be due to the distinctive processes of natural selection and adaptation to variable environmental conditions and demographic changes. Consequently, such processes may exert high influence on disease development like dyslipidemia.
Moreover, testing the effect of the commonly studied APOA5 variants on lipid profile of Kuwaiti Arabs showed controversial results. The functional rs3135506 (S19W) SNP that alters apoA5 signal peptide causing reduced production of the mature thereby affecting apoA5 levels (Olivier et al., 2004; Talmud et al., 2005) have been confirmed to be associated with high TG levels in different populations such as Caucasians (Talmud et al., 2002), African Americans (Klos et al., 2005), and Europeans (De Andrade et al., 2011). In this study however, no association between rs3135506 SNP with TG levels nor with other lipid levels was observed. Yet these findings are consistent with those reported in East Asians (Lai et al., 2003; Li et al., 2011; Yin et al., 2011). Both Kuwaiti Arabs and East Asians showed lower MAF of SNP rs3135506 (0.003, 0.01, respectively) as compared to the populations that reflected the pathological effect of the SNP. This lack of association may be attributed to the ethnicity-specific background and/or the differences in other genetic and environmental factors that may modify the gene effect.
In contrast, the usual association of SNPs rs662799, rs2072560, and rs2266788 minor alleles with elevated TG levels was detected amongst Kuwaiti Arabs (p = 0.01, p = 0.04, p = 0.02, respectively). These findings are comparable with results found in other populations such as Mongolians (Chuluun-Erdene et al., 2017), Caucasians (Talmud et al., 2002), Uyghur population (Li et al., 2014) African Americans (Talmud et al., 2002; Lai et al., 2004), East Asians (Endo et al., 2002; Nabika et al., 2002), and Pakistani population (Shahid et al., 2017). The MAFs of both rs2072560 and rs2266788 in Kuwaiti Arab population was estimated to be 0.8, which resemble frequencies observed between African Americans and Caucasians (Pennacchio et al., 2002). In contrast, rs662799 frequency was extremely low (MAF = 0.04) when compared to other populations such as Chinese (0.3) (Liu et al., 2005), Singaporeans (0.29) (Lai et al., 2003) and Japanese (0.34) (Endo et al., 2002; Nabika et al., 2002). However, it is closer to Caucasians (0.08) (Pennacchio et al., 2001), Hispanic Americans (0.13) (Pennacchio et al., 2002) (Aouizerat et al., 2003) and Tunisians (0.13) (Chaaba et al., 2005). Again, divergence in allelic frequencies between different populations is imputed to different gene-gene or gene-environment interactions among ethnicities.
Moreover, minor alleles of these three SNPs were also found significantly associated with increased VLDL levels in Kuwaiti Arabs. These findings may explain the effect of those variants on apoA5 mechanism of action. Previous studies has explained two mechanisms were apoA5 modulate lipid levels especially TG which are enhancing the catabolism of TG-rich lipoproteins through LPL (Schaap et al., 2004; Merkel et al., 2005) and inhibiting hepatic VLDL, major carrier of TG, assembly and secretion (Weinberg et al., 2003; Schaap et al., 2004). Therefore, the observed elevated TG and VLDL levels associated with SNPs rs662799, rs2072560, and rs2266788 at the APOA5 locus may reflect the impact of those SNPs on apoA5 mechanism of action. Moreover, a newly discussed evidence on the functionality of the three variants proposed that their rare alleles particularly of rs2266788 create an illegitimate miR-binding site that causes liver posttranscriptional downregulation of APOA5 by miR-485-4p, a noncoding RNA (miRNA) expressed in the human liver (Caussy et al., 2014). The study suggests that base pairing occurred between APOA5 mRNA carrying the rare alleles and the miR-485-4p leading to mRNA degradation or translation repression (Caussy et al., 2014).
Other studies suggest that the effect of rs2072560 and rs2266788 SNPs is due to their linkage with rs662799 as a leading SNP located in the promoter region. However, linkage analysis results in this study revealed that both SNPs are not linked with rs662799 SNP. The five different haplotypes observed in the Kuwaiti Arab population were very similar to the commonly studied APOA*1, APOA*2, and APOA*3 haplotypes except that haplotypes observed between Kuwaiti Arabs did not involve alleles of rs662799 SNP. In addition, two more haplotypes were observed. The first comprises a mutant allele of rs651821 SNP and the least common includes mutant alleles of both rs2266788 and rs3135506 SNPs. Despite the significant association of some of the detected SNPs with TG levels, the resulted haplotypes frequencies between high and normal TG level patient groups was not significant. These results are different from most of the studies conducted on other populations, but again they may be attributed to the differences in haplotypes frequencies between the different ethnicities as well as other ethnic-specific variations (Li et al., 2014; Sumegi et al., 2017). These findings also indicate the independent role of each variant exerting effect on APOA5 function or expression rather than in concert.
Ethics Statement
This study has been approved by the Local Ethical Committee at Kuwait University. The sample and phenotypic data collection protocol and informed consents used were in accordance to the year 2000 revised version of the 1975 Helsinki guidelines. Informed consent from each participant in this study was obtained.
Author Contributions
AJ performed all the experiments, analyzed all the data and prepared the manuscript. SA-B prepared the project proposal and study design, supervised the molecular genetic analysis and sample collection and documentation assisted with the data interpretation and preparation of the manuscript. WA-K supervised and assisted with the sequence alignment, its annotations and submission of SNPS and revised the manuscript. AA-S participated in the study design, supervised and assisted with the statistical analysis and revised the manuscript. HA conducted the statistical analysis for the validation and association of the selected variants among the cohort samples. All the authors have read and approved the final manuscript.
Funding
The data in the manuscript was generated from two projects funded by the Research Sector of Kuwait University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported and funded by Kuwait University Research Administration, Project Grant No. YS02/12 and SL09/07. The authors would also like to acknowledge the General Facility Project (GS01/02) for the use of the ABI 3130xl Gene Analyzer. The authors extend their deepest appreciation and gratitude to all the participants in this study and to the technical assistance provided by Mrs. Babitha G. Annice and Mrs. Sheela Thankakkon.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00112/full#supplementary-material
Supplementary Table 1. Characteristics and sequences of each primer set (forward and reverse primers) obtained from Primer3 program (version 0.4.0). The length (in base pair) of the overlapping APOA5 gene regions that each set amplifies is also shown.
Supplementary Table 2. Primer sequence and probes for the 7 APOA5 variants genotyped by Taqman Allelic Discrimnation Assay using Real-time PCR.
Supplementary Figure 1. (A,B) A chromatograph generated by the sequence at the APOA5 gene locus showing the novel variants at genomic positions 11:116661525 (A) and 11:116660500 (B). The arrow indicates the position of the novel variant which appears as a single peak that was produced by sequencing the target region with the forward primer.
Supplementary Figure 2. (A–F) A sample plot from the real-time PCR allelic discrimination assay showing the major allele on the x-axis against the minor allele on the y-axis. Three clusters are shown on each plot representing the three possible genotypes represented by the blue dots for the homozygous wildtype, red dots for the homozygous mutant allele and the green dots for the heterozygous. while near the origin the no template control (NTC) is shown. The genotype assay for each APOA5 polymorphism is demonstrated as follows: rs2266788 (A), rs651821 (B), rs2072560 (C), SNP rs3135506 (D), rs662799 (E), novel 1 (F).
References
Al-Bustan, S. A., Alnaqeeb, M. A., Annice, B. G., Ibrhim, G., Al-Rubaian, J., Ahmed, A. H., et al. (2005). Apolipoprotein E genotyping among the healthy Kuwaiti population. Hum. Biol. 77, 487–498. doi: 10.1353/hub.2005.0058
Alhyas, L., McKay, A., and Majeed, A. (2012). Prevalence of Type 2 diabetes in the states of The co-operation council for the Arab states of the Gulf: a systematic review. PLoS ONE 7:e40948. doi: 10.1371/journal.pone.0040948
AlMajed, H. T., AlAttar, A. T., Sadek, A. A., AlMuaili, T. A., AlMutairi, O. A., Shaghouli, A. S., et al. (2011). Prevalence of dyslipidemia and obesity among college students in Kuwait. Alex. J. Med. 47, 67–71. doi: 10.1016/j.ajme.2010.12.003
Alwan, A., and King, H. (1995). Diabetes in the Eastern Mediterranean (Middle East) region: the world health organization responds to a major public health challenge. Diabet. Med. 12, 1057–1058. doi: 10.1111/j.1464-5491.1995.tb00421.x
Aouizerat, B. E., Kulkarni, M., Heilbron, D., Drown, D., Raskin, S., Pullinger, C. R., and Kane, J. P. (2003). Genetic analysis of a polymorphism in the human apoA-V gene: effect on plasma lipids. J. Lipid Res. 44, 1167–1173. doi: 10.1194/jlr.M200480-JLR200
Barrett, J. C., Fry, B., Maller, J., and Daly, M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. doi: 10.1093/bioinformatics/bth457
Baum, L., Tomlinson, B., and Thomas, G. N. (2003). APOA5-1131T>C polymorphism is associated with triglyceride levels in Chinese men. Clin. Genet. 63, 377–379. doi: 10.1034/j.1399-0004.2003.00063.x
Beckstead, J. A., Oda, M. N., Martin, D. D., Forte, T. M., Bielicki, J. K., Berger, T., and Ryan, R. O. (2003). Structure-function studies of human apolipoprotein A-V: a regulator of plasma lipid homeostasis. Biochemistry 42, 9416–9423. doi: 10.1021/bi034509t
Beckstead, J. A., Wong, K., Gupta, V., Wan, C. P., Cook, V. R., Weinberg, R. B., and Ryan, R. O. (2007). The C terminus of apolipoprotein A-V modulates lipid-binding activity. J. Biol. Chem. 282, 15484–15489. doi: 10.1074/jbc.M611797200
Beigneux, A. P., Davies, B. S., Gin, P., Weinstein, M. M., Farber, E., Qiao, X., and Young, S. G. (2007). Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5, 279–291. doi: 10.1016/j.cmet.2007.02.002
Casey, M. S. (2007). “History of Kuwait,” in Desert Origins and Settlement, M. S. Casey (Westport, CT: Greenwood Press), 15–28.
Caussy, C., Charrière, S., Marçais, C., Di Filippo, M., Sassolas, A., Delay, M., and Moulin, P. (2014). An APOA5 3′ UTR variant associated with plasma triglycerides triggers APOA5 downregulation by creating a functional miR-485-5p binding site. Am. J. Hum. Genet. 94, 129–134. doi: 10.1016/j.ajhg.2013.12.001
Cha, S., Yu, H., Park, A. Y., and Song, K. H. (2014). Effects of apolipoprotein A5 haplotypes on the ratio of triglyceride to high-density lipoprotein cholesterol and the risk for metabolic syndrome in Koreans. Lipids Health Dis. 13:45. doi: 10.1186/1476-511X-13-45
Chaaba, R., Attia, N., Hammami, S., Smaoui, M., Mahjoub, S., Hammami, M., et al. (2005). Association of SNP3 polymorphism in the apolipoprotein A-V gene with plasma triglyceride level in Tunisian type 2 diabetes. Lipids Health Dis. 4:1. doi: 10.1186/1476-511X-4-1
Channanath, A. M., Farran, B., Behbehani, K., and Thanaraj, T. A. (2013). State of diabetes, hypertension, and comorbidity in kuwait: showcasing the trends as seen in native versus expatriate populations. Diabetes Care 36, e75–e75. doi: 10.2337/dc12-2451
Chuluun-Erdene, A., Dagdan, B., Sengeragchaa, O., Sanjmyatav, P., Janlav, M. (2017). Association of apolipoprotein A-V gene polymorphisms and lipid metabolism in mongolian patients with metabolic syndrome. Cent. Asian J. Med. Sci. 3, 140–147. doi: 10.24079/CAJMS.2017.06.006
De Andrade, F. M., Maluf, S. W., Schuch, J. B., Voigt, F., Barros, A. C., Lucatelli, J. F., et al. (2011). The influence of the S19W SNP of the APOA5 gene on triglyceride levels in southern Brazil: interactions with the APOE gene, sex and menopause status. Nutr. Metab. Cardiovasc. Dis. 21, 584–590. doi: 10.1016/j.numecd.2009.12.013
Endo, K., Yanagi, H., Araki, J., Hirano, C., Yamakawa-Kobayashi, K., and Tomura, S. (2002). Association found between the promoter region polymorphism in the apolipoprotein A-V gene and the serum triglyceride level in Japanese schoolchildren. Hum. Genet. 111, 570–572. doi: 10.1007/s00439-002-0825-0
Excoffier, L., and Ray, N. (2008). Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. (Amst) 23, 347–351. doi: 10.1016/j.tree.2008.04.004
Forte, T. M., Shu, X., and Ryan, R. O. (2009). The ins (cell) and outs (plasma) of apolipoprotein A-V. J. Lipid Res. 50(Suppl.), S150–S155. doi: 10.1194/jlr.R800050-JLR200
Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstiel, B., and Altshuler, D. (2002). The structure of haplotype blocks in the human genome. Science 296, 2225–2229. doi: 10.1126/science.1069424
Grosskopf, I., Baroukh, N., Lee, S. J., Kamari, Y., Harats, D., Rubin, E. M., et al. (2005). Apolipoprotein A-V deficiency results in marked hypertriglyceridemia attributable to decreased lipolysis of triglyceride-rich lipoproteins and removal of their remnants. Arterioscler. Thromb. Vasc. Biol. 25, 2573–2579. doi: 10.1161/01.ATV.0000186189.26141.12
Guardiola, M., Alvaro, A., Vallve, J. C., Rosales, R., Sola, R., Girona, J., et al. (2012). APOA5 gene expression in the human intestinal tissue and its response to in vitro exposure to fatty acid and fibrate. Nutr. Metab. Cardiovasc. Dis. 22, 756–762. doi: 10.1016/j.numecd.2010.12.003
Kefi, R., Hechmi, M., Dallali, H., Elouej, S., Jmel, H., Halima, Y. B., et al. (2017). Association of apolipoprotein A5 gene variants with metabolic syndrome in Tunisian population. Ann. Endocrinol. (Paris) 78, 146–155. doi: 10.1016/j.ando.2017.01.005
Klos, K. L., Hamon, S., Clark, A. G., Boerwinkle, E., Liu, K., and Sing, C. F. (2005). APOA5 polymorphisms influence plasma triglycerides in young, healthy African Americans and whites of the CARDIA Study. J. Lipid Res. 46, 564–571. doi: 10.1194/jlr.M400437-JLR200
Lai, C. Q., Demissie, S., Cupples, L. A., Zhu, Y., Adiconis, X., Parnell, L. D., and Ordovas, J. M. (2004). Influence of the APOA5 locus on plasma triglyceride, lipoprotein subclasses, and CVD risk in the Framingham Heart Study. J. Lipid Res. 45, 2096–2105. doi: 10.1194/jlr.M400192-JLR200
Lai, C. Q., Tai, E. S., Tan, C. E., Cutter, J., Chew, S. K., Zhu, Y. P. et al. (2003). The APOA5 locus is a strong determinant of plasma triglyceride concentrations across ethnic groups in Singapore. J. Lipid Res. 44, 2365–2373. doi: 10.1194/jlr.M300251-JLR200
Li, S., Hu, B., Wang, Y., Wu, D., Jin, L., and Wang, X. (2014). Influences of APOA5 variants on plasma triglyceride levels in Uyghur population. PLoS ONE 9:e110258. doi: 10.1371/journal.pone.0110258
Li, Y. Y., Yin, R. X., Lai, C. Q., Li, M., Long, X. J., Li, K. L., et al. (2011). Association of apolipoprotein A5 gene polymorphisms and serum lipid levels. Nutr. Metab. Cardiovasc. Dis. 21, 947–956. doi: 10.1016/j.numecd.2010.04.004
Liu, H., Zhang, S., Lin, J., Li, H., Huang, A., Xiao, C., et al. (2005). Association between DNA variant sites in the apolipoprotein A5 gene and coronary heart disease in Chinese. Metab. Clin. Exp. 54, 568–572. doi: 10.1016/j.metabol.2004.11.009
Merkel, M., Loeffler, B., Kluger, M., Fabig, N., Geppert, G., Pennacchio, L. A., et al. (2005). Apolipoprotein AV accelerates plasma hydrolysis of triglyceriderich lipoproteins by interaction with proteoglycan-bound lipoprotein lipase. J. Biol. Chem. 280, 21553–21560. doi: 10.1074/jbc.M411412200
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. doi: 10.1093/nar/16.3.1215
Nabika, T., Nasreen, S., Kobayashi, S., and Masuda, J. (2002). The genetic effect of the apoprotein AV gene on the serum triglyceride level in Japanese. Atherosclerosis 165, 201–204. doi: 10.1016/S0021-9150(02)00252-6
Nilsson, S. K., Heeren, J., Olivecrona, G., and Merkel, M. (2011). Apolipoprotein A-V; a potent triglyceride reducer. Atherosclerosis 219, 15–21. doi: 10.1016/j.atherosclerosis.2011.07.019
Olivier, M., Wang, X., Cole, R., Gau, B., Kim, J., Rubin, E. M., et al. (2004). Haplotype analysis of the apolipoprotein gene cluster on human chromosome 11. Genomics 83, 912–923. doi: 10.1016/j.ygeno.2003.11.016
Olofsson, S. O. (2005). ApoA-V: the regulation of a regulator of plasma triglycerides. Arterioscler. Thromb. Vasc. Biol. 25, 1097–1099. doi: 10.1161/01.ATV.0000167518.27125.b1
Ouatou, S., Ajjemami, M., Charoute, H., Sefri, H., Ghalim, N., Rhaissi, H., et al. (2014). Association of APOA5 rs662799 and rs3135506 polymorphisms with arterial hypertension in Moroccan patients. Lipids Health Dis. 13:60. doi: 10.1186/1476-511X-13-60
Patel, R. V., Kumari, P., Rajani, D. P., and Chikhalia, K. H. (2011). Synthesis and antimycobacterial activity of various 1-(8-Quinolinyloxy)-3-piperazinyl(piperidinyl)-5-(4-cyano-3-trifluoromethylphenyl amino)-s-triazines. Acta Chim. Slov. 58, 802–810.
Pennacchio, L. A., Olivier, M., Hubacek, J. A., Cohen, J. C., Cox, D. R., Fruchart, J. C., and Rubin, E. M. (2001). An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294, 169–173. doi: 10.1126/science.1064852
Pennacchio, L. A., Olivier, M., Hubacek, J. A., Krauss, R. M., Rubin, E. M., and Cohen, J. C. (2002). Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum. Mol. Genet. 11, 3031–3038. doi: 10.1093/hmg/11.24.3031
Priore Oliva, C., Tarugi, P., Calandra, S., Pisciotta, L., Bellocchio, A., Bertolini, S., and Schaap, F. G. (2006). A novel sequence variant in APOA5 gene found in patients with severe hypertriglyceridemia. Atherosclerosis 188, 215–217. doi: 10.1016/j.atherosclerosis.2006.04.010
Pullinger, C. R., Aouizerat, B. E., Movsesyan, I., Durlach, V., Sijbrands, E. J., Nakajima, K., and Kane, J. P. (2008). An apolipoprotein A-V gene SNP is associated with marked hypertriglyceridemia among Asian-American patients. J. Lipid Res. 49, 1846–1854. doi: 10.1194/jlr.P800011-JLR200
Quach, H., Barreiro, L. B., Laval, G., Zidane, N., Patin, E., Kidd, K. K., and Quintana-Murci, L. (2009). Signatures of purifying and local positive selection in human miRNAs. Am. J. Hum. Genet. 84, 316–327. doi: 10.1016/j.ajhg.2009.01.022
R Core Team (2016). R: A Language and Environment for Statistical Computing (Version 3.3.1). Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/
Rousset, F. (2008). genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Schaap, F. G., Rensen, P. C. N., Voshol, P. J., Vrins, C., van der Vliet, H. N., Chamuleau, R. A. F. M., et al. (2004). ApoAV reduces plasma triglycerides by inhibiting very low density lipoprotein-triglyceride (VLDL-TG) production and stimulating lipoprotein lipase-mediated VLDL-TG hydrolysis. J. Biol. Chem. 279, 27941–27947. doi: 10.1074/jbc.M403240200
Scheper, G. C., van der Knaap, M. S., and Proud, C. G. (2007). Translation matters: protein synthesis defects in inherited disease. Nat. Rev. Genet. 8, 711–723. doi: 10.1038/nrg2142
Shahid, S. U., Shabana, N. A., Cooper, J. A., Rehman, A., and Humphries, S. E. (2017). Common variants in the genes of triglyceride and HDL-C metabolism lack association with coronary artery disease in the Pakistani subjects. Lipids Health Dis. 16:24. doi: 10.1186/s12944-017-0419-4
Sharma, V., Ryan, R. O., and Forte, T. M. (2012). Apolipoprotein A-V dependent modulation of plasma triacylglycerol: a puzzlement. Biochim. Biophys. Acta 1821, 795–799. doi: 10.1016/j.bbalip.2011.12.002
Sumegi, K., Duga, B., Melegh, B. I., Banfai, Z., Kovesdi, E., Maasz, A., et al. (2017). Marked Differences of Haplotype Tagging SNP Distribution, Linkage, and Haplotype Profile of APOA5 Gene in Roma Population Samples. Pathol. Oncol. Res. 2, 853–861. doi: 10.1007/s12253-017-0197-3
Talmud, P. J. (2007). Rare APOA5 mutations–clinical consequences, metabolic and functional effects: an ENID review. Atherosclerosis 194, 287–292. doi: 10.1016/j.atherosclerosis.2006.12.010
Talmud, P. J., Hawe, E., Martin, S., Olivier, M., Miller, G. J., Rubin, E. M., and Humphries, S. E. (2002). Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum. Mol. Genet. 11, 3039–3046. doi: 10.1093/hmg/11.24.3039
Talmud, P. J., Palmen, J., Putt, W., Lins, L., and Humphries, S. E. (2005). Determination of the functionality of common APOA5 polymorphisms. J. Biol. Chem. 280, 28215–28220. doi: 10.1074/jbc.M502144200
Teebi, A. S. (1994). Autosomal recessive disorders among Arabs: an overview from Kuwait. J. Med. Genet. 31, 224–233. doi: 10.1136/jmg.31.3.224
The Genomes Project, C. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
†The International HapMap, C. (2003). The International HapMap project. Nature 426:789. doi: 10.1038/nature02168
Walther, T. C., and Farese, R. V. Jr. (2012). Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714. doi: 10.1146/annurev-biochem-061009-102430
Weinberg, R. B., Cook, V. R., Beckstead, J. A., Martin, D. D., Gallagher, J. W., Shelness, G. S., et al. (2003). Structure and interfacial properties of human apolipoprotein A-V. J. Biol. Chem. 278, 34438–34444. doi: 10.1074/jbc.M303784200
Yin, R. X., Li, Y. Y., and Lai, C. Q. (2011). Apolipoprotein A1/C3/A5 haplotypes and serum lipid levels. Lipids Health Dis. 10:140. doi: 10.1186/1476-511X-10-140
Keywords: APOA5, re-sequencing, SNPs, genetic association, Kuwait, Arabs
Citation: Jasim AA, Al-Bustan SA, Al-Kandari W, Al-Serri A and AlAskar H (2018) Sequence Analysis of APOA5 Among the Kuwaiti Population Identifies Association of rs2072560, rs2266788, and rs662799 With TG and VLDL Levels. Front. Genet. 9:112. doi: 10.3389/fgene.2018.00112
Received: 06 December 2017; Accepted: 21 March 2018;
Published: 09 April 2018.
Edited by:
Noor Ahmad Shaik, King Abdulaziz University, Saudi ArabiaReviewed by:
Xusheng Wang, St. Jude Children′s Research Hospital, United StatesMuhammad Tariq, University of Tabuk, Saudi Arabia
Narasimha Reddy Parine, King Saud University, Saudi Arabia
Copyright © 2018 Jasim, Al-Bustan, Al-Kandari, Al-Serri and AlAskar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzanne A. Al-Bustan, cy5hbGJ1c3RhbkBrdS5lZHUua3c=
 Anfal A. Jasim
Anfal A. Jasim Suzanne A. Al-Bustan1*
Suzanne A. Al-Bustan1* Huda AlAskar
Huda AlAskar