
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 02 August 2016
Sec. Pharmacogenetics and Pharmacogenomics
Volume 7 - 2016 | https://doi.org/10.3389/fgene.2016.00140
 Xiaoliang Wang1†
Xiaoliang Wang1† Zhipeng Liu2†
Zhipeng Liu2† Kai Wang3
Kai Wang3 Zhaowen Wang1
Zhaowen Wang1 Xing Sun1
Xing Sun1 Lin Zhong1
Lin Zhong1 Guilong Deng1
Guilong Deng1 Guohe Song1
Guohe Song1 Baining Sun4
Baining Sun4 Zhihai Peng1*
Zhihai Peng1* Wanqing Liu2*
Wanqing Liu2*Recent genome-wide association studies have identified that variants in or near PNPLA3, NCAN, GCKR, LYPLAL1, and TM6SF2 are significantly associated with non-alcoholic fatty liver disease (NAFLD) in multiple ethnic groups. Studies on their impact on NAFLD in Han Chinese are still limited. In this study, we examined the relevance of these variants to NAFLD in a community-based Han Chinese population and further explored their potential joint effect on NAFLD. Six single nucleotide polymorphisms (SNPs) (PNPLA3 rs738409, rs2294918, NCAN rs2228603, GCKR rs780094, LYPLAL1 rs12137855, and TM6SF2 rs58542926) previously identified in genome-wide analyses, to be associated with NAFLD were genotyped in 384 NAFLD patients and 384 age- and gender-matched healthy controls. We found two out of the six polymorphisms, PNPLA3 rs738409 (OR = 1.52, 95%CI: 1.19–1.96; P = 0.00087) and TM6SF2 rs58542926 (OR = 2.11, 95%CI: 1.34–3.39; P = 0.0016) are independently associated with NAFLD after adjustment for the effects of age, gender, and BMI. Our analysis further demonstrated the strong additive effects of the risk alleles of PNPLA3 and TM6SF2 with an overall significance between the number of risk alleles and NAFLD (OR = 1.64, 95%CI: 1.34–2.01; P = 1.4 × 10-6). The OR for NAFLD increased in an additive manner, with an average increase in OR of 1.52 per additional risk allele. Our results confirmed that the PNPLA3 and TM6SF2 variants were the most significant risk alleles for NAFLD in Chinese population. Therefore, genotyping these two genetic risk factors may help identify individuals with the highest risk of NAFLD.
Non-alcoholic fatty liver disease (NAFLD) is the most common liver disorder, whose prevalence is estimated to be 20–30% in Western industrialized countries (Bedogni et al., 2005). Although it is relatively less prevalent in China, the number of patients suffering NAFLD has approximately doubled in the past two decades (Fan, 2013).
The first genome-wide association study (GWAS) of NAFLD performed by Romeo et al. (2008) identified that rs738409 G allele of the patatin-like phospholipase domain-containing-3 (PNPLA3) is significantly associated with increased hepatic fat levels and susceptibility to NAFLD. Subsequent analyses demonstrated that PNPLA3 rs738409 is also associated with disease severity and progression (Sookoian and Pirola, 2011). A recent study demonstrated that the rs2294918 variant of the PNPLA3 gene is also significantly associated with hepatic steatosis in an Italian population, independent of rs738409 (Donati et al., 2016). Besides the variants associated with steatosis, Chalasani et al. (2010) identified several genetic variants associated with histologic features of NAFLD, including NAS score, degree of fibrosis, lobular inflammation, and serum levels of alanine aminotransferase. A subsequent study in a cohort of European ancestry led by the GOLD Consortium identified four additional NAFLD risk factors located in or near neurocan gene (NCAN), glucokinase regulatory protein gene (GCKR), lysophospholipase-like 1 gene (LYPLAL1), and protein phosphatase 1 regulatory subunit 3b (PPP1R3B).(Speliotes et al., 2011). Two subsequent studies aimed to verify these findings in multiethnic cohorts. Palmer et al. (2013) tested whether these European ancestry NAFLD-related variants were associated with computed tomography defined hepatic steatosis in African- and/or Hispanic-Americans. The results indicated that allele frequency and effect size varies in different ethnic groups. Another study led by Hernaez et al. (2013) investigated the association between these variants and ultrasound-measured hepatic steatosis in non-Hispanic white (NHW), non-Hispanic black (NHB), and Mexican American (MA) participants in National Health and Nutrition Examination Survey III (NHANES III). The results also showed the variance of allele frequency and effect size across ancestries (Hernaez et al., 2013). Most recently, a new variant rs58542926 in the transmembrane 6 superfamily member 2 (TM6SF2) gene was associated with higher liver fat through an exome-wide association study (Kozlitina et al., 2014). A multiethnic study consisting of 957 obese children and adolescents showed that the allele frequency and effect size of TM6SF2 rs58542926 in conferring NAFLD risk varies in Caucasians, African Americans, and Hispanics (Goffredo et al., 2016). Therefore, it is worthwhile examining the association of these variants with NAFLD in different ethnic settings. In particular, how these variants interact with each other and contribute together to the NAFLD susceptibility among each ethnic group remains to be explored.
East Asian populations account to the largest proportion of current total number of world living humans. However, they are among the least studied populations in terms of the genetic basis of NAFLD, which is incongruent with the fact that NAFLD is gaining prevalence in East Asian populations. In the present study, we aimed to evaluate the association of six previously identified polymorphisms, PNPLA3 rs738409, rs2294918, NCAN rs2228603, GCKR rs780094, LYPLAL1 rs12137855, and TM6SF2 rs58542926, with NAFLD in a community-based Han Chinese population. Additionally, we analyzed the joint effects of these genetic variants on the disease risk of NAFLD.
The characteristics of subjects enrolled in this study has been described before (Wang et al., 2015). Briefly, our study included 384 ultrasound (US) diagnosed NAFLD patients and 384 age- and gender-matched healthy controls from the same geographic areas as the patients. All the subjects were recruited from individuals conducting health examination in the Shanghai Jiao Tong University Affiliated First People’s Hospital (Shanghai, China). The mean age of both case and control groups was 45±13 years old, and both of them were consisted of 229 males and 155 females. Subjects with a history of heavy alcohol consumption (>20 g/day) and other specific diseases that could contribute to fatty liver, such as hepatitis B and C virus infection, drug-induced liver disease, were excluded from the study. US-based NAFLD diagnoses were made by one experienced operator without knowing clinical and biochemical evaluations of the participants. The guidelines used for diagnosis were adopted from the Chinese National Consensus Workshop on NAFLD (Zeng et al., 2008). Patients with moderate to severe fatty liver diseases were selected in order to increase the diagnostic accuracy and reliability (Hernaez et al., 2011). All subjects were unrelated and self-reported Han Chinese living in the Shanghai metropolitan area. The study conforms to the principles of the Declaration of Helsinki. All the participants have given their written informed consent, and the study method was approved by the ethical committee of the Shanghai Jiao Tong University.
Genomic DNA was extracted from whole blood of each participant according to the manufacturer’s instructions (Qiagen, CA, USA). Genotyping was conducted using a Taqman-based assay (Life Technologies, CA, USA) according to the manufacturer’s instructions. Note that the TM6SF2 rs58542926 polymorphism has been genotyped in these samples in our previous study (Wang et al., 2015). Hardy-Weinberg equilibrium (HWE) was tested for each SNP using the program PLINK (Version 1.07) (Purcell et al., 2007). The PPP1R3B rs4240624 was genotyped in our cohort. However, we found that the Taqman-based genotyping of this SNP does not follow the HWE given the deviated genotype distribution in our samples (P = 3.592×10-49). We further sequenced the DNA fragment containing this SNP in 20 randomly chosen samples. All these samples demonstrated monomorphic at this variant site, suggesting that this SNP may be a fake variant. We therefore removed this SNP from our analysis. The information of allele frequencies in East Asians and other populations was obtained from The 1000 Genomes Project (Genomes Project et al., 2015).
Chi-square test was used for the comparisons of allele and genotype frequencies between NAFLD patients and healthy controls. Multivariable analysis was used to assess the independent effect of each SNP on the prevalence of NAFLD by incorporating all six SNPs in a multiple logistic regression model with adjustment for age, gender, and BMI, under an assumption of an additive genetic model for each SNP, in which genotypes were coded as 0, 1, or 2 according to the number of minor allele for a specific individual. Odds ratio (OR) and 95% confidence interval (CI) were calculated in groups with different number of risk alleles, and Chi-square test was used for linear trend analysis. The group with 0 risk allele was set as reference group. The proportion of narrow sense heritability explained by susceptibility variants was estimated based on multifactorial liability threshold model (So et al., 2011). The median prevalence of NAFLD in China and narrow sense heritability used for estimation is 15% (Fan, 2013) and 38.6% (Schwimmer et al., 2009), respectively. All the statistical analyses were performed using statistical package R (version 3.2.1). P values less than 0.05 were considered statistically significant.
Genotyping was performed in 384 NAFLD patients and 384 age- and gender-matched healthy controls. The genotyping call rates for each SNP were 98.7% (PNPLA3 rs738409), 98.9% (PNPLA3 rs2294918), 97.9% (NCAN rs2228603), 99.3% (GCKR rs780094), 96.1% (LYPLAL1 rs12137855), and 95.4% (TM6SF2 rs58542926). The genotype distribution of all SNPs were in HWE in both case and control groups (all P > 0.05, data not shown). As shown in Table 1, the minor allele frequency (MAF) of SNPs genotyped in our study were comparable to the overall frequency in East Asians reported in The 1000 Genomes Project (Genomes Project et al., 2015).
The genotype distributions of these SNPs and their association with NAFLD are listed in Table 2. Note that the association between TM6SF2 rs58542926 polymorphism and NAFLD in these samples have been reported in our previous study (Wang et al., 2015). Two out of six variants, PNPLA3 rs738409 and TM6SF2 rs58542926, were significantly associated with NAFLD. The frequency of G allele of the PNPLA3 rs738409 variant was significantly higher in NAFLD patients compared to those in healthy controls (OR = 1.48, 95%CI: 1.20–1.82; P = 0.00023), and PNPLA3 GC and GG genotypes were significantly associated with increased risk of disease (GC: OR: 1.52, 95%CI: 1.11–2.07; P = 0.008; GG: OR: 2.24, 95%CI: 1.41–3.55; P = 0.00055). Similarly, T allele of TM6SF2 rs58542926 was more prevalent in the subjects with NAFLD (OR: 2.06, 95%CI: 1.34–3.17; P = 0.00086). The rest of SNPs, PNPLA3 rs2294918, GCKR rs780094, NCAN rs2228603, and LYPLAL1 rs12137855 did not show any independent significant association with NAFLD in the population.
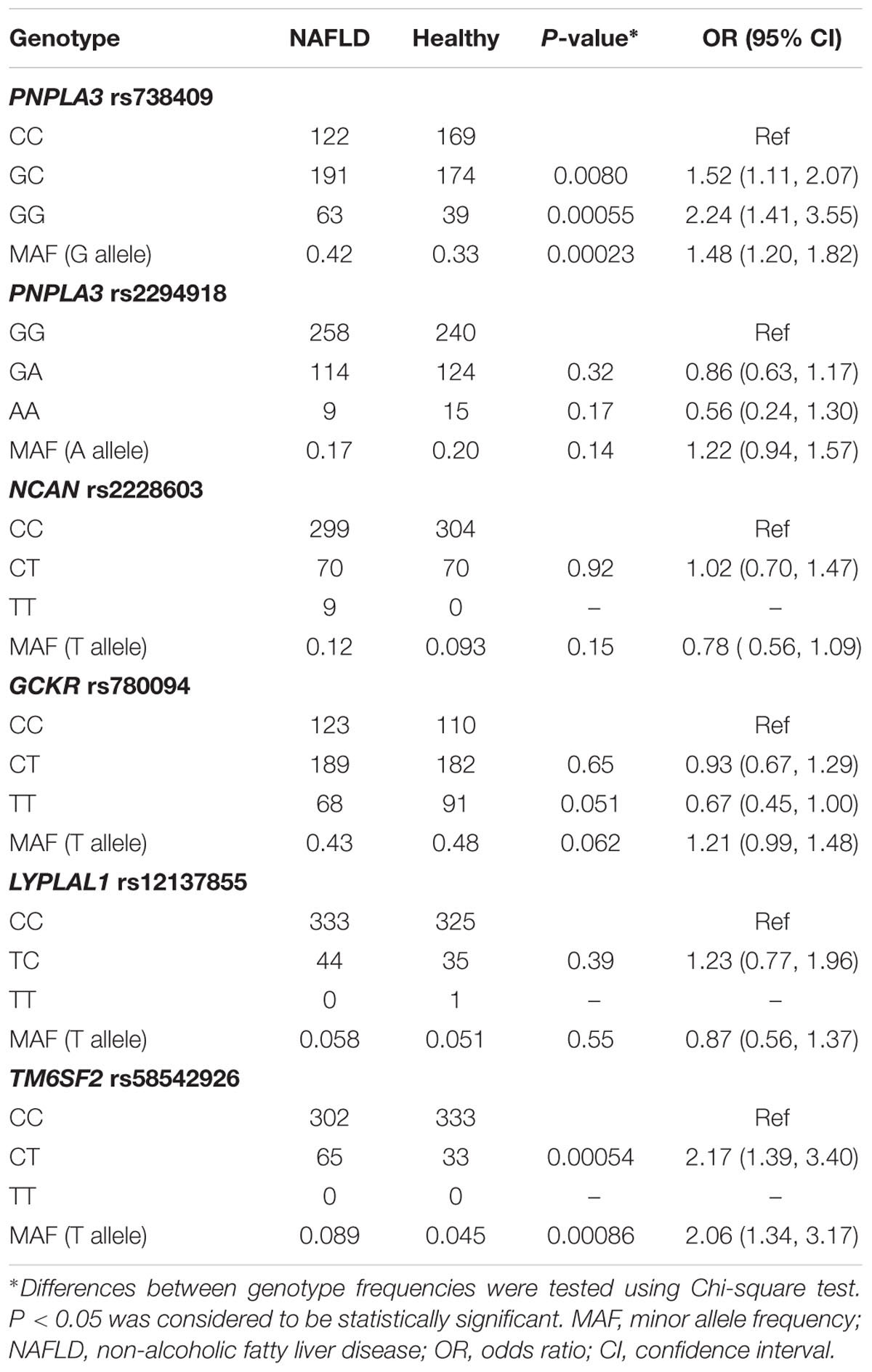
TABLE 2. Genotype distribution and MAF of PNPLA3 rs738409, rs2294918, NCAN rs2228603, GCKR rs780094, LYPLAL1 rs12137855, and TM6SF2 rs58542926.
In order to investigate the independent effect of each variant on the susceptibility to NAFLD, all the six SNPs were included in the multiple logistic regression model with age, gender, and BMI as covariates. As shown in Table 3, PNPLA3 rs738409 and TM6SF2 rs58542926 variants were significantly associated with NAFLD after adjustment for age, gender and BMI (P = 0.00087 and 0.0016, respectively).
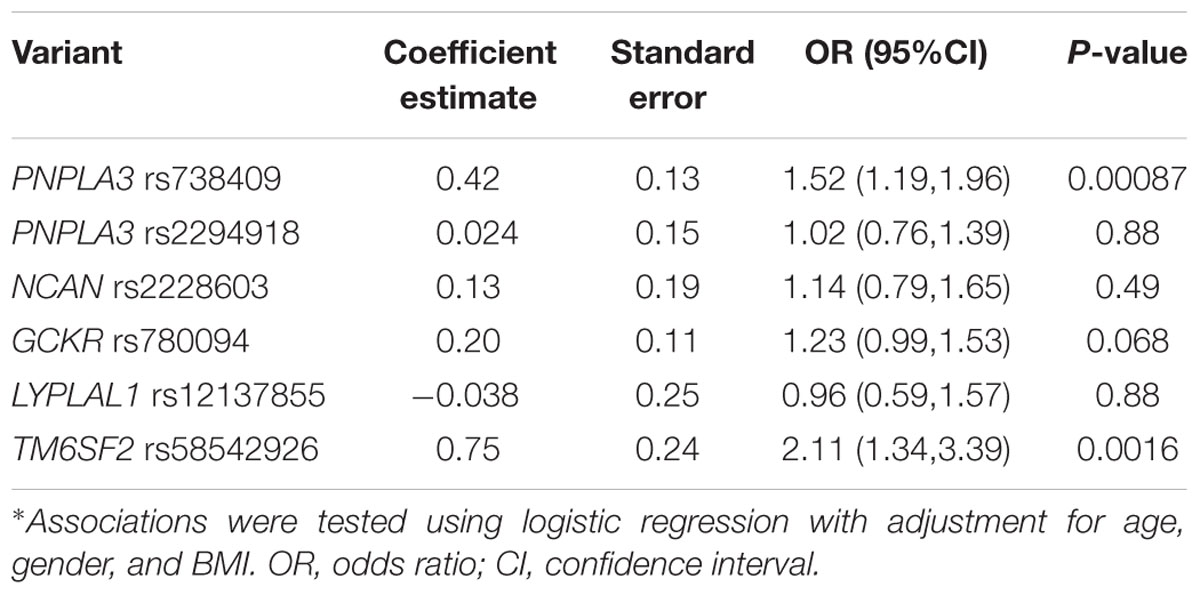
TABLE 3. Association between PNPLA3 rs738409, rs2294918, NCAN rs2228603, GCKR rs780094, LYPLAL1 rs12137855, and TM6SF2 rs58542926 variants and NAFLD*.
We further investigated the joint effect of the risk allele of PNPLA3 rs738409 and TM6SF2 rs58542926 on the risk of NAFLD. Figure 1 shows the proportion of NAFLD patients in the individuals harboring different number of risk alleles. Ninety-six NAFLD patients did not harbor any risk alleles, and we did not observe any individuals with four risk alleles in our study. The proportion of NAFLD patients increased significantly (P < 0.0001, Chi-square test for trend) along with the increase of the number of risk alleles. The overall significance between the number of risk alleles and NAFLD incidence is high, with a P value of 1.4×10-6, using a logistic regression model after adjustment for age, gender and BMI. We also plotted the proportion of each risk allele for healthy controls and NAFLD patients separately (Figure 2). Figure 3 shows the OR for NAFLD in the group of patients with different number of risk alleles, using the individuals with zero risk allele as the reference group. As shown in the Figure 3, the risk of NAFLD increases linearly with the number of risk alleles with an average of 1.52-fold increase in OR for each additional risk allele. The regression line fit excellently for the data with the R squared value of 0.992.
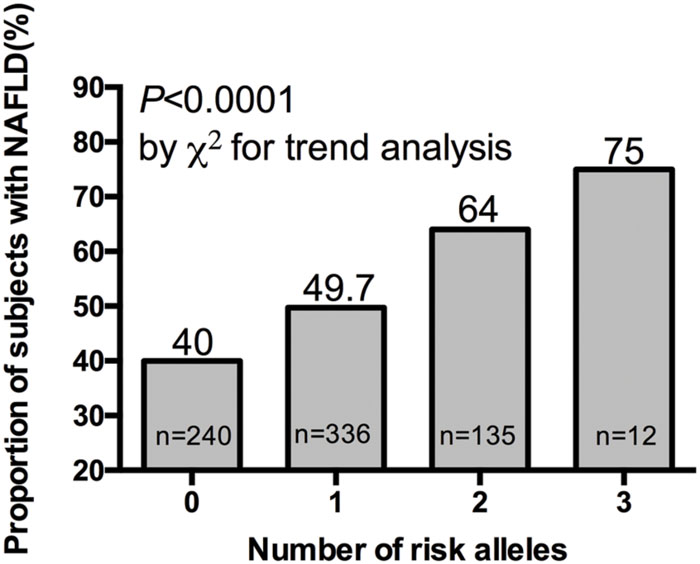
FIGURE 1. Proportion of NAFLD patient in 40, 336, 135, and 12 subjects harboring 0, 1, 2, and 3 risk alleles respectively.
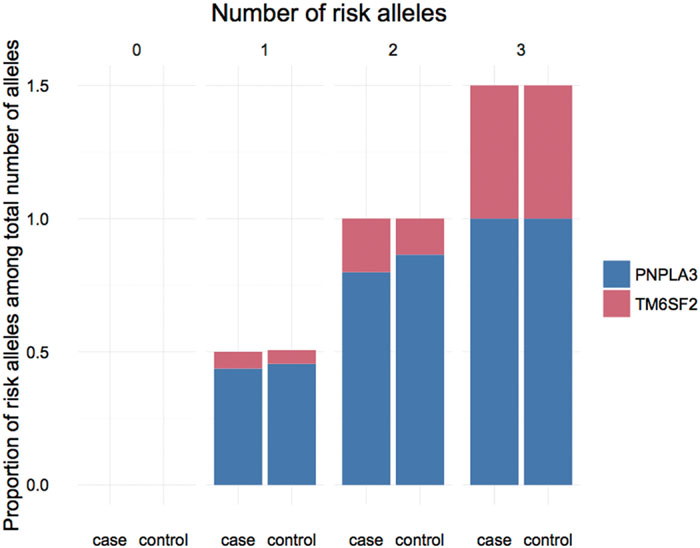
FIGURE 2. Proportion of PNPLA3 and TM6SF2 risk allele in NAFLD patients and healthy controls carrying different number of risk alleles.
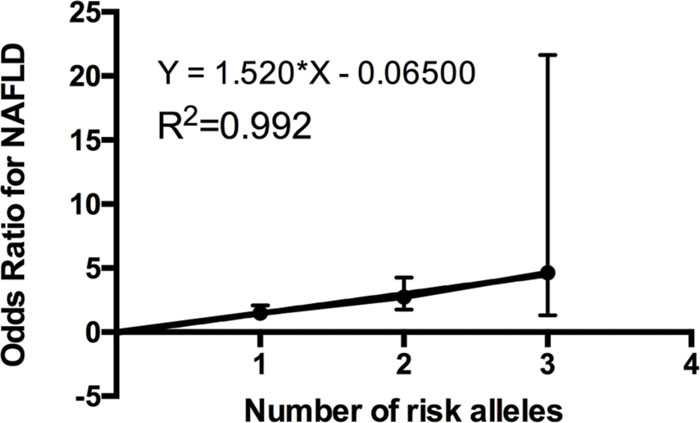
FIGURE 3. Odds ratio (ORs) (with 95% CI) for developing NAFLD is a linear function of the number of risk alleles. ORs obtained by multiple logistic regression after adjustment for age, gender and BMI. The actual odds ratio numbers are as follows: 0 (reference group OR = 1), 1 [OR = 1.48 (1.06–2.08)], 2 [OR = 2.73 (1.77–4.26)], and 3 [OR = 4.65 (1.32–21.65)].
Using a cohort of 384 NAFLD patients and 384 age- and gender-matched healthy controls, we confirmed that PNPLA3 rs738409 and TM6SF2 rs58542926 were independent risk factors conferred susceptibility to NAFLD in a Han Chinese population. Moreover, we examined the joint effects of PNPLA3 rs738409 and TM6SF2 rs58542926 polymorphisms. To our best of knowledge, this is the first study to report the additive effect of PNPLA3 rs738409 and TM6SF2 rs58542926 polymorphisms on NAFLD in a Chinese population.
PNPLA3 rs738409 was the first identified genetic variant that contributes to the differences in liver fat levels and the susceptibility to NAFLD (Romeo et al., 2008). Results in our study confirmed the association between PNPLA3 rs738409 and increased risk for NAFLD in a Han Chinese population, which emphasized its effects on NAFLD across multiple ethnic groups. TM6SF2 rs58542926 was the most recently identified genetic variant to be associated with NAFLD (Kozlitina et al., 2014). The subsequent studies demonstrated that this variant was also associated with the progression of NAFLD disease (Liu et al., 2014; Dongiovanni et al., 2015). TM6SF2 rs58542926 was initially identified in a multiethnic cohort consisting of African Americans, European Americans, and Hispanics. In our previous study, we first reported TM6SF2 rs58542926 polymorphism as an independent risk factor that contributes to NAFLD in a Chinese population (Wang et al., 2015). In the current study, we further investigated its joint effects with PNPLA3 rs738409 on the risk of developing NAFLD. Notably, the association between the number of risk alleles and NAFLD incidence is very significant, with a beta value of 1.52 and the mean OR of 4.65 in the individuals carrying three risk alleles, suggesting the value of early diagnosis of NAFLD by genotyping these two variants. An additive model of risk alleles was shown to fit very well in explaining the greater probability of NAFLD. Our findings in the Chinese population are consistent with previous study performed in other ethnic groups. Goffredo et al. (2016). have shown that the hepatic fat fraction increased as the sum of risk allele in TM6SF2 rs58542926, PNPLA3 rs738409, and GCKR rs1260326 in an obese children cohort including Caucasians, African Americans, and Hispanics. Our study confirmed this joint effect in a Chinese population, suggesting a new perspective in NAFLD classification and diagnosis using risk allele number counting. Moreover, the genetic variability in PNPLA3 rs738409, in TM6SF2 rs58542926, and in the combined effects explained 8.2, 9.6, and 19.2% of the heritability of NAFLD in Chinese population, respectively. This indicates additional possible unknown risk variants yet to be identified among Chinese NAFLD patients.
PNPLA3, which encodes a 481-residue protein, exhibits lipase activity against triglyceride (TG) in hepatocytes and retinyl esters in hepatic stellate cells (He et al., 2010; Huang et al., 2011; Pirazzi et al., 2014). Missense mutation (I148M) in PNPLA3 leads to a loss of function and thus promotes hepatic steatosis by limiting triglyceride hydrolysis (He et al., 2010; Smagris et al., 2015). Although the specific function of TM6SF2 is still unknown, Kozlitina et al. (2014) found that the Glu167Lys variant led to a lower levels of protein expression than wild-type TM6SF2 in cultured hepatocytes. A subsequent functional study indicated that the inhibition of TM6SF2 protein expression results in reduced secretion of TG-rich lipoproteins (TRLs), which increases cellular TG concentration (Mahdessian et al., 2014). Therefore, these two variants seem to affect the homeostasis processes of hepatic fat separately, whose functions are relatively independent but synergistically involved in the development of hepatic steatosis. Our results of the additive effects of the risk alleles of PNPLA3 and TM6SF2 on NAFLD were consistent with their biological functions, which highlights the possibility to identify high risk individuals by genotyping these two risk factors.
Inconsistent with the previous finding that GCKR rs780094 significantly increased the risk of NAFLD in obese Chinese children (Lin et al., 2014), we did not observe significant association between this variant and NAFLD. Given that the sample size and alleles frequencies of our study (n = 766, MAF = 0.45) were comparable to the previous study (n = 797, MAF = 0.49) (Lin et al., 2014), we speculated that GCKR rs780094 may only have a moderate effect on NAFLD incidence in Chinese population. On the other hand, participants enrolled in our study were adults with a mean age of 45 years old, which introduces more possible influence of environment in the study. The lack of consistency for GCKR variant suggests its potential interaction with environment. Similar to the results reported in the previous study (Lin et al., 2014), we did not find significant associations between either NCAN rs2228603 or LYPLAL1 rs12137855 and NAFLD in Han Chinese population. Recent studies have shown that conditioning on the TM6SF2 variant abolishes the association between NCAN variant and NAFLD while the reverse does not occur (Kozlitina et al., 2014), suggesting NCAN variant is not the causal variant associated with NAFLD. Our results of multivariable analysis confirmed this finding by exploring the independent effects of individual variant on NAFLD after conditioning on all the other SNPs. With regard to LYPLAL1 rs12137855 variant, as shown in the Table 1, the MAF of LYPLAL1 rs12137855 in East Asians is much lower than those in other populations, which reduces the power to detect the association given that our sample size is relatively small. Of course, this could be also due to the limited effect size of this variant in a Chinese population.
The NAFLD phenotype in our study was defined using an US-based diagnosis, which might cause concern about whether it is appropriate to detect NAFLD. However, according to a meta-analysis comparing the diagnostic accuracy and reliability of the US-based method to histology-based method (gold standard), the US-based method was fairly good in detecting moderate-severe fatty liver (Hernaez et al., 2011), and multiple genetic studies using the US-based diagnosis method have successfully demonstrated the associations between genetic polymorphisms and NAFLD (Hernaez et al., 2013; Sookoian et al., 2015).
We confirmed the associations between genetic variants in PNPLA3 and TM6SF2 genes and NAFLD risk in a Han Chinese population. We also demonstrated the additive effects of these two genetic risk factors on conferring risk of NAFLD in Han Chinese. The differences in allele frequencies of several other risk alleles that were identified in other populations but are not associated with NAFLD in Chinese indicates that additional unknown risk alleles are yet to be identified in Han Chinese populations.
XW sample collection, genotyping, data collection, manuscript drafting; ZL data analysis and manuscript drafting; KW sample collection, genotyping, and data collection; ZW, XS, LZ, GD, GS, and BS participated sample and data collection; ZP: coordinated and directed the study; WL conceived the study, data analysis, manuscript drafting.
This work was supported in part by the seed fund of the Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University (to WL), the Shanghai Pujiang Program (14PJD029, to WX), National Nature Science Foundation of China (81270557, 81472241, to WX).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer BL and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Project of Biomedical Engineering Research Foundation of Shanghai Jiao Tong University (YG2015MS33, to WX). We also would like to thank Ms. Shaminie Athinarayanan for her proofreading of this manuscript.
Bedogni, G., Miglioli, L., Masutti, F., Tiribelli, C., Marchesini, G., and Bellentani, S. (2005). Prevalence of and risk factors for nonalcoholic fatty liver disease: the Dionysos nutrition and liver study. Hepatology 42, 44–52. doi: 10.1002/hep.20734
Chalasani, N., Guo, X., Loomba, R., Goodarzi, M. O., Haritunians, T., Kwon, S., et al. (2010). Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology 139, 1567–1576, 1576.e1–1576.e6. doi: 10.1053/j.gastro.2010.07.057
Donati, B., Motta, B. M., Pingitore, P., Meroni, M., Pietrelli, A., Alisi, A., et al. (2016). The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology 63, 787–798. doi: 10.1002/hep.28370
Dongiovanni, P., Petta, S., Maglio, C., Fracanzani, A. L., Pipitone, R., Mozzi, E., et al. (2015). Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 61, 506–514. doi: 10.1002/hep.27490
Fan, J. G. (2013). Epidemiology of alcoholic and nonalcoholic fatty liver disease in China. J. Gastroenterol. Hepatol. 28(Suppl. 1), 11–17. doi: 10.1111/jgh.12036
Genomes Project, C., Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
Goffredo, M., Caprio, S., Feldstein, A. E., D’Adamo, E., Shaw, M. M., Pierpont, B., et al. (2016). Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: a multiethnic study. Hepatology 63, 117–125. doi: 10.1002/hep.28283
He, S., McPhaul, C., Li, J. Z., Garuti, R., Kinch, L., Grishin, N. V., et al. (2010). A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem. 285, 6706–6715. doi: 10.1074/jbc.M109.064501
Hernaez, R., Lazo, M., Bonekamp, S., Kamel, I., Brancati, F. L., Guallar, E., et al. (2011). Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology 54, 1082–1090. doi: 10.1002/hep.24452
Hernaez, R., McLean, J., Lazo, M., Brancati, F. L., Hirschhorn, J. N., Borecki, I. B., et al. (2013). Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin. Gastroenterol. Hepatol. 11, 1183–1190 e1182. doi: 10.1016/j.cgh.2013.02.011
Huang, Y., Cohen, J. C., and Hobbs, H. H. (2011). Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem. 286, 37085–37093. doi: 10.1074/jbc.M111.290114
Kozlitina, J., Smagris, E., Stender, S., Nordestgaard, B. G., Zhou, H. H., Tybjaerg-Hansen, A., et al. (2014). Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 46, 352–356. doi: 10.1038/ng.2901
Lin, Y. C., Chang, P. F., Chang, M. H., and Ni, Y. H. (2014). Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am. J. Clin. Nutr. 99, 869–874. doi: 10.3945/ajcn.113.079749
Liu, Y. L., Reeves, H. L., Burt, A. D., Tiniakos, D., McPherson, S., Leathart, J. B., et al. (2014). TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 5:4309. doi: 10.1038/ncomms5309
Mahdessian, H., Taxiarchis, A., Popov, S., Silveira, A., Franco-Cereceda, A., Hamsten, A., et al. (2014). TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. U.S.A. 111, 8913–8918. doi: 10.1073/pnas.1323785111
Palmer, N. D., Musani, S. K., Yerges-Armstrong, L. M., Feitosa, M. F., Bielak, L. F., Hernaez, R., et al. (2013). Characterization of European ancestry nonalcoholic fatty liver disease-associated variants in individuals of African and Hispanic descent. Hepatology 58, 966–975. doi: 10.1002/hep.26440
Pirazzi, C., Valenti, L., Motta, B. M., Pingitore, P., Hedfalk, K., Mancina, R. M., et al. (2014). PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum. Mol. Genet. 23, 4077–4085. doi: 10.1093/hmg/ddu121
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Romeo, S., Kozlitina, J., Xing, C., Pertsemlidis, A., Cox, D., Pennacchio, L. A., et al. (2008). Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40, 1461–1465. doi: 10.1038/ng.257
Schwimmer, J. B., Celedon, M. A., Lavine, J. E., Salem, R., Campbell, N., Schork, N. J., et al. (2009). Heritability of nonalcoholic fatty liver disease. Gastroenterology 136, 1585–1592. doi: 10.1053/j.gastro.2009.01.050
Smagris, E., BasuRay, S., Li, J., Huang, Y., Lai, K. M., Gromada, J., et al. (2015). Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61, 108–118. doi: 10.1002/hep.27242
So, H. C., Gui, A. H., Cherny, S. S., and Sham, P. C. (2011). Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 35, 310–317. doi: 10.1002/gepi.20579
Sookoian, S., Castano, G. O., Scian, R., Mallardi, P., Fernandez Gianotti, T., Burgueno, A. L., et al. (2015). Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology 61, 515–525. doi: 10.1002/hep.27556
Sookoian, S., and Pirola, C. J. (2011). Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology 53, 1883–1894. doi: 10.1002/hep.24283
Speliotes, E. K., Yerges-Armstrong, L. M., Wu, J., Hernaez, R., Kim, L. J., Palmer, C. D., et al. (2011). Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 7:e1001324. doi: 10.1371/journal.pgen.1001324
Wang, X., Liu, Z., Peng, Z., and Liu, W. (2015). The TM6SF2 rs58542926 T allele is significantly associated with non-alcoholic fatty liver disease in Chinese. J. Hepatol. 62, 1438–1439. doi: 10.1016/j.jhep.2015.01.040
Keywords: additive effects, PNPLA3, TM6SF2, NAFLD, Chinese
Citation: Wang X, Liu Z, Wang K, Wang Z, Sun X, Zhong L, Deng G, Song G, Sun B, Peng Z and Liu W (2016) Additive Effects of the Risk Alleles of PNPLA3 and TM6SF2 on Non-alcoholic Fatty Liver Disease (NAFLD) in a Chinese Population. Front. Genet. 7:140. doi: 10.3389/fgene.2016.00140
Received: 29 May 2016; Accepted: 19 July 2016;
Published: 02 August 2016.
Edited by:
Amit V. Pandey, University of Bern, SwitzerlandReviewed by:
Maria J. Prata, Instituto de Patologia e Imunologia Molecular da Universidade do Porto, PortugalCopyright © 2016 Wang, Liu, Wang, Wang, Sun, Zhong, Deng, Song, Sun, Peng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanqing Liu, bGl1NzgxQHB1cmR1ZS5lZHU= Zhihai Peng, cGVuZ3poaWhhaUBzanR1LmVkdS5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.