
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 22 March 2016
Sec. Pharmacogenetics and Pharmacogenomics
Volume 7 - 2016 | https://doi.org/10.3389/fgene.2016.00037
 Brooke L. Fridley1*
Brooke L. Fridley1* Taraswi M. Ghosh2
Taraswi M. Ghosh2 Alice Wang1
Alice Wang1 Rama Raghavan1
Rama Raghavan1 Junqiang Dai1
Junqiang Dai1 Ellen L. Goode3
Ellen L. Goode3 Jatinder K. Lamba4
Jatinder K. Lamba4Background: The standard treatment for epithelial ovarian cancer (EOC) patients with advanced disease is carboplatin-paclitaxel combination therapy following initial debulking surgery, yet there is wide inter-patient variation in clinical response. We sought to identify pharmacogenomic markers related to carboplatin-paclitaxel therapy.
Methods: The lymphoblastoid cell lines, derived from 74 invasive EOC patients seen at the Mayo Clinic, were treated with increasing concentrations of carboplatin and/or paclitaxel and assessed for in vitro drug response using MTT viability and caspase3/7 apoptosis assays. Drug response phenotypes IC50 (effective dose at which 50% of cells are viable) and EC50 (dose resulting in 50% induction of caspase 3/7 activity) were estimated for each patient to paclitaxel and carboplatin (alone and in combination). For each of the six drug response phenotypes, a genome-wide association study was conducted.
Results: Statistical analysis found paclitaxel in vitro drug response phenotypes to be moderately associated with time to EOC recurrence (p = 0.008 IC50; p = 0.058 EC50). Although no pharmacogenomic associations were significant at p < 5 × 10−8, seven genomic loci were associated with drug response at p < 10−6, including at 4q21.21 for carboplatin, 4p16.1 and 5q23.2 for paclitaxel, and 3q24, 10q, 1q44, and 13q21 for combination therapy. Nearby genes of interest include FRAS1, MGC32805, SNCAIP, SLC9A9, TIAL1, ZNF731P, and PCDH20.
Conclusions: These results suggest the existence of genetic loci associated with response to platinum-taxane therapies. Further research is needed to understand the mechanism by which these loci may impact EOC clinical response to this commonly used regimen.
Epithelial ovarian cancer (EOC) is the fifth leading cause of cancer death among women in the United States (6% of cancer deaths); in 2015, it is estimated that 14,180 women will die from the disease (Siegel et al., 2015). The standard treatment for patients with advanced disease is initial debulking surgery followed by carboplatin-paclitaxel combination chemotherapy (Marsh, 2009). Five-year overall survival remains around 45% (Marsh, 2009), yet there is a wide inter-patient variation in response. Currently there are few reliable prognostic biomarkers for the classification of patients and treatment response.
Platinating agents, such as carboplatin, interfere with DNA via inter-strand, intra-strand, and DNA-protein crosslinks, thereby causing DNA damage and prevention of cell division and growth, resulting in cell-cycle arrest and apoptosis (Dekou et al., 2001). Although platinum-based drugs are widely used in cancer treatment, many tumors are completely resistant to these drugs, and no clinical response is attained. Major molecular mechanisms underlying this resistance might involve alteration in platinum inactivation or reduced intracellular accumulation by uptake/efflux transporters, increased repair of adducts, increased adduct tolerance or failure of apoptotic pathway. Taxane agents, such as paclitaxel, are commonly used chemotherapeutic drugs often in combination with platinating agents. Taxanes block cell division by binding to α-tubulin, stabilizing the microtubules, thus resulting in cell death (Huizing et al., 1995; Jordan and Wilson, 2004). The development of taxane resistance is common, where response has been linked to metabolism and disposition molecules, such as, cytochrome P450s and drug transporters (e.g., ABCB1, ABCG2, ABCC1, ABCC2, and SLC01B3; Hewett et al., 2002; Rodriguez-Antona, 2010). In particular, studies in colorectal cell lines and tumor tissue have shown that CYP2C8 may play a role in paclitaxel resistance and the CYP3A may be involved in local inactivation of paclitaxel (Martinez et al., 2002; Garcia-Martin et al., 2006). Additionally, CYP2C8 and CYP3A4 are high involved in the metabolism of paclitaxel in patients with ovarian cancer (Bergmann et al., 2011), while breast cancer patients carrying the *3 variant of CYP2C8 have better response to paclitaxel, but at an increase in peripheral neurotoxicity (Hertz et al., 2012).
However, these molecules do not explain all the variation in taxane response or resistance. Inherited variation in many of the genes encoding these molecules have been assessed for association with clinical outcome with inconsistent results (Peethambaram et al., 2011; Johnatty et al., 2013; White et al., 2013); genome-wide searches to date have also failed to identify variants associated with outcome after correction for genome-wide testing (p < 5 × 10−8). Patient-derived cell line based model systems represent a novel way to identify genomic predictors of drug response. Although lymphoblastoid cell lines (LCLs) derived from participants in the international HapMap project have been used to identify genomic predictors of cytotoxic effects of various chemotherapeutic agents (Li et al., 2008, 2010; Niu et al., 2010; Huang et al., 2011; Wu et al., 2011), they are limited as they are not derived from the EOC population but from healthy individuals.
In this study, we generated LCLs derived from Mayo Clinic EOC patients, conducted in vitro cytotoxic studies, and associated in vitro drug response phenotypes with germline genotype. Utilizing patient-derived LCLs, as opposed to commercially available LCLs, allows us to screen and directly correlate in vitro phenotypes and clinical responses. These genome-wide association scans (GWAS) should contribute to the identification of predictive markers of treatment responses and ultimately improve clinicians' ability to tailor therapy decisions for EOC patients.
Prior to initiation of chemotherapy, ovarian cancer patients diagnosed at the Mayo Clinic between 2000 and 2003 provided blood for immediate germline DNA extraction and for the creation of Epstein Barr Virus (EBV)-transformed LCLs. Samples from 74 patients were successfully transformed and subjected to in vitro drug testing. All patients provided informed written consent, including for passive and active follow-up, using protocols approved by the appropriate Institutional Review Board at the Mayo Clinic in Rochester, MN.
In vitro cellular chemo-sensitivity studies of LCLs were performed in two batches (N = 33, N = 41) using identical procedures and assays. Cells were maintained in RPMI1640 media supplemented with 2 mM L-glutamine, and 15% fetal bovine serum at 37°C under 5% CO2. Following 24 h incubation, LCLs were treated with increasing concentrations of carboplatin and/or paclitaxel (in duplicate). The concentrations of carboplatin were 0, 5, 10, 20, 40, 80, and 128 μM, while for paclitaxel were 0, 4.5, 7.5, 10, 20, 40, and 80 nM when used as single agents. In drug combination experiments, we used half of the doses for each drug in increasing doses (i.e., 2.5 μM carboplatin + 2.25 nM paclitaxel for “dose level 1” and so on). Cell viability 48 h post-treatment was determined using standard MTT assay (Li et al., 2008; Gamazon et al., 2010). Caspase3/7 (Promega) apoptosis assays were performed at the same time in parallel plates. A Synergy 3 plate reader (BioTek Instruments) was used to read absorbance (cell viability using MTT) or fluorescence (for caspase3/7 activity) intensities.
Four parameter logistic dose response curves were fit to the in vitro drug response measurements (cell survival and caspase3/7 activity assays) for each LCL and treatment (paclitaxel, carboplatin, and combination), where Yi is the measurement at dose i (Di), α α is the estimated bottom of the curve (i.e., measurement as Di → ∞), β → ∞), β is the estimated top of the curve (i.e., measurement as Di → 0), ϕ is the slope of the curve, and θ is the inflection point of the curve (i.e., concentration giving a response half way between the top and bottom of the curve). The estimated inflection point of the curve was used as the drug response phenotype. That is, using these dose-response curves, we estimated the relative IC50 (effective dose that kills 50% of the cells) for MTT cell viability and the relative EC50 (concentration required to induce caspase 3/7 activity by 50%) for caspase3/7 activity. For simplicity of presentation, we will refer to these quantitative values as the IC50 for the MTT cytotoxicity assays and EC50 for the caspase 3/7 assays. We then applied rank-based inverse Gaussian transformation (i.e., Van de Waerden rank transformation). Summaries of the IC50 and EC50 values for both experimental batches are presented in Supplemental Table 1. We observed a high degree of correlation between many of the drug response phenotypes, as expected (Supplemental Figure 1).
Germline DNA was genotyped on the Illumina Infinium 610K array, as previously described (Pharoah et al., 2013). All samples had genotype call rate >95% and were predicted by STRUCTURE (Pritchard et al., 2000) analysis to have greater than 80% European ancestry. SNPs were excluded with call rate < 95%, Hardy-Weinberg Equilibrium p < 10−4, or no variation in this set. Using the 1000 Genomes Project (Durbin et al., 2010) as reference, imputation was completed with mach and minimac in a two-step process (Howie et al., 2012), resulting in data on more than 30 million SNPs. Assessment of imputation quality was completed and high quality imputed markers (r2 ≥ 0.30 and MAF ≥ 0.01) were retained (6,243,550 SNPs).
The association of each SNP with in vitro drug response phenotypes IC50 or EC50 was evaluated with linear models using the expected genotype or “dosage” (i.e., additive or dose-response/trend model). Thus, a negative effect estimate indicates that the carriers of the minor/rare allele have lower IC50 (EC50) values (i.e., were more “sensitive” to treatment). The two in vitro experimental batches were analyzed separately followed by meta-analysis was conducted using metal (Willer et al., 2010), with weights applied for the number of samples in each group. We completed the GWAS analyses for each individual in vitro drug response phenotype, as opposed to a combined analysis with all phenotypes model together, due to the difference in mechanism of action between the drugs (i.e., not in the same drug class; Fridley et al., 2012). For annotation of results across gene regions, SNPs were mapped to genes within 2 KB using Biofilter (assembly CRCh37.p10, genome build 104.0; Bush et al., 2009). Pathway analysis used Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com).
We examined the relationships of in vitro phenotypes with time to recurrence of EOC (40 of 74 patients had recurred or died). Of the 74 patients with LCLs included in this study, 51 had available information on the first two treatments used: 43 patients were treated with paclitaxel / carboplatin, 2 treated with paclitaxel/cisplatin, 3 treated with carboplatin/topotecan, and 3 treated with carboplatin/other less common agent. Paclitaxel in vitro drug response phenotypes were moderately associated with time to EOC recurrence (HR = 1.90 per unit increase in MTT IC50, p = 0.008; HR = 1.84 per unit increase in caspase 3/7 EC50, p = 0.058; Figure 1; Supplemental Table 2). This suggests that patients whose LCLs demonstrated greater sensitivity to the chemotherapeutics tested had improved outcome; as Figure 1 illustrates, LCLs that were sensitive to paclitaxel (as reflected by having low IC50 and low EC50 values) were from patients with longer time to progression as compared to patients with LCLs with high values. Although based on a small sample size, this provides, for the first time, a link between in vitro chemo-sensitivity testing and clinical outcome in EOC.
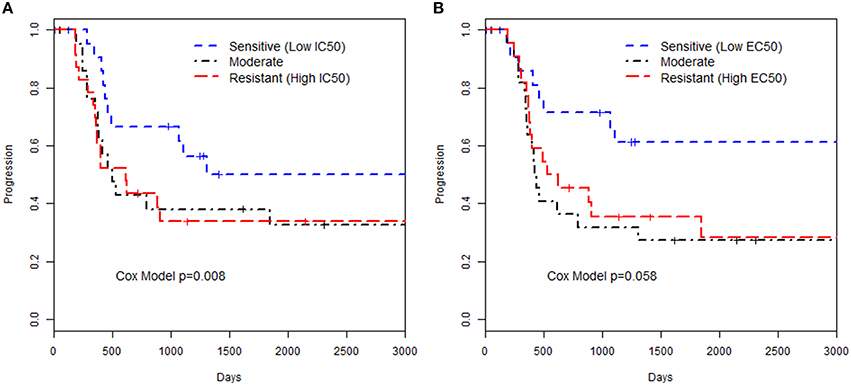
Figure 1. Kaplan-Meier curves for time to progression and paclitaxel in vitro phenotypes. The groupings were defined by 3 quartiles representing low, medium or high values for the phenotype based (A) MTT assay IC50 or (B) Caspase 3/7 EC50 assay. The p-value presented is from a Cox proportional hazards model with the in vitro phenotype modeled as a continuous measurement on the log-scale.
Results of genome-wide association analyses for each drug response phenotype are presented in Figure 2. Regions with p < 10−6 are highlighted and are further displayed in Figure 3. Table 1 presents the SNPs associated with the drug response phenotype with p < 10−6. Overall, we found a greater proportion of significant results (e.g., at p < 10−6) for the combination therapy as compared to the single agent therapies. In particular, we found strong SNP associations with combination therapy in the following gene regions: SLC9A9 (MAF = 0.41, p = 6 × 10−7), TIAL1 (MAF = 0.23, p = 7.3 × 10−7), ZNF731P (MAF = 0.39, p = 6.6 × 10−7), and PCDH20 (MAF = 0.42, p = 8.2 × 10−7). None of these regions were found to be moderately associated with single agent carboplatin or paclitaxel IC50 in other pharmacogenomic studies involving commercially available LCLs (Huang et al., 2011; Niu et al., 2012).
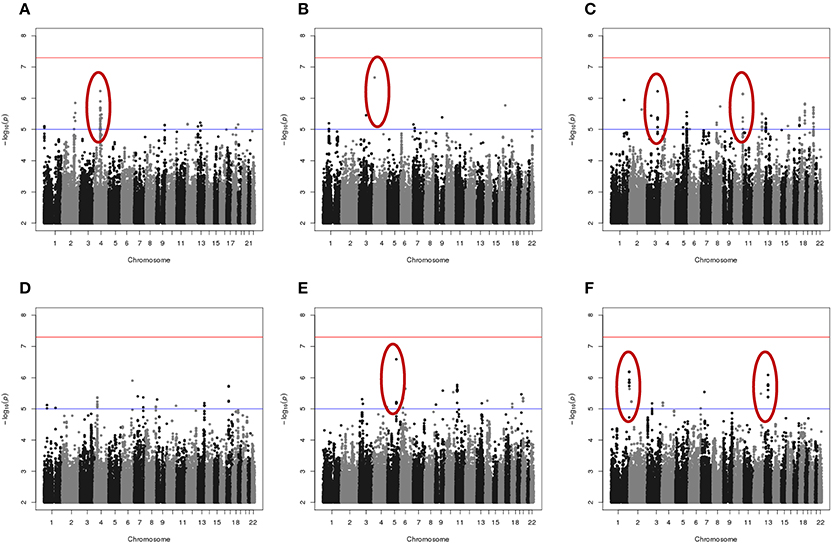
Figure 2. Manhattan plots of the single SNP meta-analysis genome-wide association results based the six in vitro drug response measures among ovarian cancer patient LCLs. (A) Carboplatin MTT; (B) Paclitaxel MTT; (C) Combination of carboplatin and paclitaxel MTT; (D) Carboplatin Caspase3/7; (E) Paclitaxel Caspase3/7; and (F) Combination of carboplatin and paclitaxel Caspase3/7. Blue line indicates p = 0.00001; Red line indicates p = 5 × 10−8. Highlighted regions (circled) are displayed in Figure 3.
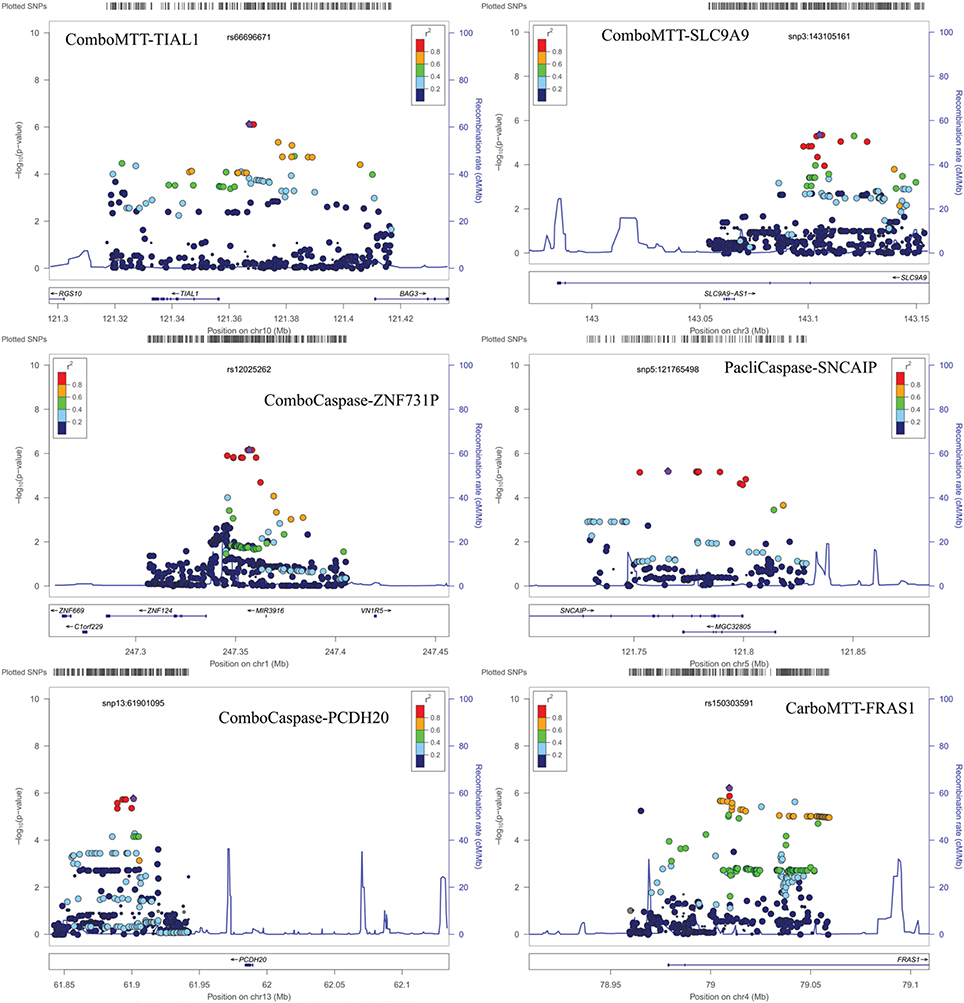
Figure 3. Locus Zoom plots for regions with p < 10−6. Note: BOD1L1 region not presented as only one SNP in region with p < 0.001.
We evaluated potential overlap of loci associated with both phenotypes for a given drug. Only one SNP was found to be associated with a p < 10−4 with same direction of the effect for MTT IC50 and caspase3/7 EC50 values for paclitaxel, carboplatin, or combination treatment. An intronic SNP rs35067965 in COLEC12 (chromosome 18, bp 455396) was associated with response to paclitaxel (MTT IC50 p = 2.2 × 10−5, caspase 3/7 EC50 p = 3.8 × 10−5; Table 2). We also examined overlap of associations at the level of genes, considering SNPs within 20 kb. This showed consistency of IC50 and EC50 results for paclitaxel response and COLEC12 and revealed similar IC50 and EC50 associations for carboplatin response in the gene regions of CTIF and CDH4. As presented in Table 3, additional regions showed joint associations with response to multiple drugs, including variants in protein coding regions of BRE, EML6, CTNNA2, LRP1B, EYS, NKAIN2, ANTXRL, COL13A1, and MTCL1 (SNPs in gene regions with p < 0.0001).
Because of the suggested association between in vitro paclitaxel MTT IC50 response and time to EOC recurrence (Supplemental Table 2), we also examined recurrence association with SNPs rs185229225 (intronic BOD1L1) rs35067965 (intronic COLEC12) and rs1525599 (intronic LRP1B) which were associated with paclitaxel MTT IC50 (Tables 1–3, respectively). However, none of these SNPs were associated with time to recurrence with a nominal p < 0.05 (data not shown).
In this proof of concept study, we explored use of LCLs derived from EOC patients followed for clinical response as a model for discovery of pharmacogenomics markers. LCLs were treated with varying concentration of the chemotherapeutics agents (carboplatin and paclitaxel and, uniquely, their combination) that were used for the treatment and cellular chemo-sensitivity was determined by measuring cell viability and activation of caspase activity (as a marker of apoptosis) post drug treatment. Genome-wide association studies were performed to identify inherited markers associated with these measures of in vitro chemo-sensitivity (i.e., MTT IC50 and caspase 3/7 EC50 values) and the relationships between in vitro measures and clinical outcome was explored.
Although the sample size was small limiting the power of the study, some of the implicated biologically interesting genes are worthy of discussion. Pathway analysis of genes with SNPs showing association with one of the drug response phenotypes (at p < 10−6), both phenotypes for a given drug (at p < 10−4), or multiple drugs for any phenotype (at p < 10−4) found enrichment in genes related to malignant solid tumor and epithelial cancers (Supplemental Figure 2A). Among the top canonical pathways represented by these genes were “Epithelial Adherens Junction Signaling,” “Sertoli Cell Junction Signaling,” and “Endometrial Cancer Signaling” (Supplemental Figure 2B).
In addition, genes such as CTNNA2 and CDH4, both tumor suppressor genes with role in cell adhesion were found to be associated with chemo-sensitivity in carboplatin alone or combination treatments. SNPs in CTNNA2- catenin (cadherin-associated protein) alpha2, a structural constituent of cytoskeleton and cadherin binding was associated with in vitro cytotoxicity to carboplatin alone as well as in combination with paclitaxel. CTNNA2 has been shown to be frequently mutated in laryngeal carcinomas with mutations predictive of poor prognosis (Fanjul-Fernandez et al., 2013). Additionally, SNPs within CTNNA2 have recently been implicated in breast cancer (Haryono et al., 2015) and its role in tumor progression and metastasis has been suggested for multiple cancers (Mcgranahan et al., 2015). Variants in CTNNA2 have also been implicated in schizophrenia (Mexal et al., 2008) and alcohol addiction (Song and Zhang, 2014). CTNNA2 SNPs associated with carboplatin and paclitaxel MTT IC50 were both intronic and present functional relevance of these is not known. CDH4, codes for cadherin, and has been implicated in nasopharyngeal carcinoma (Du et al., 2011) and aberrant methylation of CDH4 promoter has been colorectal and gastric cancer (Miotto et al., 2004). Our results identified two intronic SNPs (rs2748151 and rs113594423) that were associated with carboplatin resistant as measured by cell death (IC50) and apoptosis (caspase 3/7 EC50). Variants in PCDH20, another member of cadherin family, were also found to be associated in vitro drug response. PCHD20 codes for protocadherin20 and functions as a tumor suppressor by interacting with Wnt/b-catenin signaling (Chen et al., 2015; Lv et al., 2015).
Another gene with role in cell adhesion identified in our study was FRAS1, which encodes for an extracellular matrix protein and is involved in the regulation of epidermal-basement membrane adhesion and organogenesis during development. Inherited mutations in FRAS1, and FREM2, have been associated with development of Fraser syndrome. FRAS1 has also been implicated in ERK signaling and influence migration and invasion of lung cancer cell line by influencing FAK signaling (Zhan et al., 2014), suggesting its role in tumorigenesis and metastasis of lung cancer. Although the genes described above are involved cell adhesion/cell migration, the functional significance of the intronic SNPs identified in this study is not known and would require further investigation.
Two intronic variants within BRE were found to be associated with caspase 3/7 levels for carboplatin and combination therapy (indel rs5830067 and rs7572664). BRE encodes for Brain and reproductive Organ-Expressed (TNFRSF1A modulator) and is a component of BRCA1-A DNA damage repair complex that recognizes Lys 62linked ubiquitinated H2A and H2Ax at DNA lesions, resulting in recruitment of BRCA1-BARD1 to double strand DNA breaks (Li et al., 1995). BRE expression has been shown to be predictive of disease free survival in non-familial breast cancer patients (Noordermeer et al., 2012) and recent studies show its involvement in both intrinsic and extrinsic apoptotic pathways by influencing XIAP (Chui et al., 2014). Variation within EML6, which is involved assembly dynamics of microtubules, was found to be associated with platinum-sensitivity which was of interest since paclitaxel's mechanism of action involved disruption of microtubules; however no evidence exists in the literature on functional relevance of these particular SNPs within EML6. No other genes involved in microtubule protein were identified with respect to paclitaxel chemo-sensitivity.
Lastly, two intronic variants within LRP1B (low density lipoprotein related protein 1B) were associated with paclitaxel and combination therapy drug response phenotypes. LRP1B is a tumor suppressor with decreased expression in several primary cancers and is among 10 most significantly deleted genes across 3312 cancer samples (Langbein et al., 2002; Sonoda et al., 2004; Nakagawa et al., 2006; Prazeres et al., 2011). In renal cell cancer, down-regulation of LRP1B has been shown to regulate cell motility and actin cytoskeleton reorganization (Lu et al., 2013). Germline SNPs/ haplotype in LRP1B have been associated with aging without cognitive decline (Poduslo et al., 2010); however, associations of germline SNPs with incidence/progression of cancer and pharmacogenomics have yet to be reported.
In summary, using a patient-derived cell-based model system to generate several in vitro drug response phenotypes on a clinically followed set of EOC cases we have identified genetic loci associated with response to platinum-taxane therapies. Overall our results identified germ-line SNPs in multiple cell adhesion molecules and several tumor suppressor genes (PCDH20, LRP1B, CDH4, and CTNNA2). However, none of the most associated SNPs were reported by Huang et al. (2011) or associated with mRNA gene expression in lymphoblastoid cell lines (http://www.ncbi.nlm.nih.gov/projects/gap/eqtl/index.cgi). Further studies are needed to determine if these SNPs are truly associated with drug response or if they represent false-positive findings. Similar to other studies comparing in vitro chemo-sensitivity with clinical outcomes (Huang et al., 2011; Gamazon et al., 2013), our findings suggest that in vitro response to paclitaxel correlates with time to disease recurrence indicating that this model may have utility in several types of future studies. On possible explanation for the observation that paclitaxel correlates with recurrence and not carboplatin may be the fact that the majority of EOC patients eventually develop platinum resistant tumors and the main factor related to future response maybe attributed to response to taxane therapy. Further research is needed to understand the mechanism by which genomic loci impact clinical response in ovarian cancer patients to the most common regimen used in the treatment of ovarian cancer following surgery.
Conceived and designed the study: BF, JL, EG. Collected cell lines and clinical information: EG. Genotyping data: EG, BF. Performed drug assays: TG, JL. Statistical Analyses: RR, JD, AW, BF. Wrote the paper: BF, JL, EG. Reviewed Manuscript: ALL.
Funding for this research was provided by National Institute of Health (R01 CA122443, P50 CA136393, P30 CA168524, P30 CA15083, P20 GM103418, R21 CA182715), the Minnesota Partnership, the Fraternal Order of Eagles Cancer Research Fund, and the Minnesota Ovarian Cancer Alliance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer (EG-M) and Handling Editor declared their shared affiliation, and the Handling Editor states that the process nevertheless met the standards of a fair and objective review.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2016.00037
Bergmann, T. K., Brasch-Andersen, C., Green, H., Mirza, M., Pedersen, R. S., Nielsen, F., et al. (2011). Impact of CYP2C8*3 on paclitaxel clearance: a population pharmacokinetic and pharmacogenomic study in 93 patients with ovarian cancer. Pharmacogenomics J. 11, 113–120. doi: 10.1038/tpj.2010.19
Bush, W. S., Dudek, S. M., and Ritchie, M. D. (2009). Biofilter: a knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pac. Symp. Biocomput. 14, 368–379. doi: 10.1142/9789812836939_0035
Chen, T., Long, B., Ren, G., Xiang, T., Li, L., Wang, Z., et al. (2015). Protocadherin20 acts as a tumor suppressor gene: epigenetic inactivation in nasopharyngeal carcinoma. J. Cell. Biochem. 116, 1766–1775. doi: 10.1002/jcb.25135
Chui, Y. L., Ma, C. H., Li, W., Xu, Z., Yao, Y., Lin, F. K., et al. (2014). Anti-apoptotic protein BRE/BRCC45 attenuates apoptosis through maintaining the expression of caspase inhibitor XIAP in mouse Lewis lung carcinoma D122 cells. Apoptosis 19, 829–840. doi: 10.1007/s10495-013-0963-y
Dekou, V., Whincup, P., Papacosta, O., Ebrahim, S., Lennon, L., Ueland, P. M., et al. (2001). The effect of the C677T and A1298C polymorphisms in the methylenetetrahydrofolate reductase gene on homocysteine levels in elderly men and women from the British regional heart study. Atherosclerosis 154, 659–666. doi: 10.1016/S0021-9150(00)00522-0
Du, C., Huang, T., Sun, D., Mo, Y., Feng, H., Zhou, X., et al. (2011). CDH4 as a novel putative tumor suppressor gene epigenetically silenced by promoter hypermethylation in nasopharyngeal carcinoma. Cancer Lett. 309, 54–61. doi: 10.1016/j.canlet.2011.05.016
Durbin, R. M., Abecasis, G. R., Altshuler, D. L., Auton, A., Brooks, L. D., Durbin, R. M., et al. (2010). A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073. doi: 10.1038/nature09534
Fanjul-Fernandez, M., Quesada, V., Cabanillas, R., Cadinanos, J., Fontanil, T., Obaya, A., et al. (2013). Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun. 4, 2531. doi: 10.1038/ncomms3531
Fridley, B. L., Jenkins, G. D., Batzler, A., Wang, L., Ji, Y., Li, F., et al. (2012). Multivariate models to detect genomic signatures for a class of drugs: application to thiopurines pharmacogenomics. Pharmacogenomics J. 12, 105–110. doi: 10.1038/tpj.2010.83
Gamazon, E. R., Huang, R. S., Cox, N. J., and Dolan, M. E. (2010). Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl. Acad. Sci. U.S.A. 107, 9287–9292. doi: 10.1073/pnas.1001827107
Gamazon, E. R., Lamba, J. K., Pounds, S., Stark, A. L., Wheeler, H. E., Cao, X., et al. (2013). Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood 121, 4366–4376. doi: 10.1182/blood-2012-10-464149
Garcia-Martin, E., Pizarro, R. M., Martinez, C., Gutierrez-Martin, Y., Perez, G., Jover, R., et al. (2006). Acquired resistance to the anticancer drug paclitaxel is associated with induction of cytochrome P450 2C8. Pharmacogenomics 7, 575–585. doi: 10.2217/14622416.7.4.575
Haryono, S. J., Datasena, I. G., Santosa, W. B., Mulyarahardja, R., and Sari, K. (2015). A pilot genome-wide association study of breast cancer susceptibility loci in Indonesia. Asian Pac. J. Cancer Prev. 16, 2231–2235. doi: 10.7314/APJCP.2015.16.6.2231
Hertz, D. L., Motsinger-Reif, A. A., Drobish, A., Winham, S. J., Mcleod, H. L., Carey, L. A., et al. (2012). CYP2C8*3 predicts benefit/risk profile in breast cancer patients receiving neoadjuvant paclitaxel. Breast Cancer Res. Treat. 134, 401–410. doi: 10.1007/s10549-012-2054-0
Hewett, M., Oliver, D. E., Rubin, D. L., Easton, K. L., Stuart, J. M., Altman, R. B., et al. (2002). PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 30, 163–165. doi: 10.1093/nar/30.1.163
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J., and Abecasis, G. R. (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959. doi: 10.1038/ng.2354
Huang, R. S., Johnatty, S. E., Gamazon, E. R., Im, H. K., Ziliak, D., Duan, S., et al. (2011). Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin. Cancer Res. 17, 5490–5500. doi: 10.1158/1078-0432.CCR-11-0724
Huizing, M. T., Misser, V. H., Pieters, R. C., Ten Bokkel Huinink, W. W., Veenhof, C. H., Vermorken, J. B., et al. (1995). Taxanes: a new class of antitumor agents. Cancer Invest. 13, 381–404. doi: 10.3109/07357909509031919
Johnatty, S. E., Beesley, J., Gao, B., Chen, X., Lu, Y., Law, M. H., et al. (2013). ABCB1 (MDR1) polymorphisms and ovarian cancer progression and survival: a comprehensive analysis from the Ovarian Cancer Association Consortium and The Cancer Genome Atlas. Gynecol. Oncol. 131, 8–14. doi: 10.1016/j.ygyno.2013.07.107
Jordan, M. A., and Wilson, L. (2004). Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265. doi: 10.1038/nrc1317
Langbein, S., Szakacs, O., Wilhelm, M., Sukosd, F., Weber, S., Jauch, A., et al. (2002). Alteration of the LRP1B gene region is associated with high grade of urothelial cancer. Lab. Invest. 82, 639–643. doi: 10.1038/labinvest.3780458
Li, F., Fridley, B. L., Matimba, A., Kalari, K. R., Pelleymounter, L., Moon, I., et al. (2010). Ecto-5'-nucleotidase and thiopurine cellular circulation: association with cytotoxicity. Drug Metab. Dispos. 38, 2329–2338. doi: 10.1124/dmd.110.035220
Li, L., Fridley, B., Kalari, K., Jenkins, G., Batzler, A., Safgren, S., et al. (2008). Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 68, 7050–7058. doi: 10.1158/0008-5472.CAN-08-0405
Li, L., Yoo, H., Becker, F. F., Ali-Osman, F., and Chan, J. Y. (1995). Identification of a brain- and reproductive-organs-specific gene responsive to DNA damage and retinoic acid. Biochem. Biophys. Res. Commun. 206, 764–774. doi: 10.1006/bbrc.1995.1108
Lu, G., Zhang, G., Zhang, C., Chen, C., and Liu, R. (2013). A study of 131iodine-labeling of histamine-indomethacin: its in vivo therapeutic effect and anti-tumor mechanisms in Lewis-bearing lung cancer. Radiat. Oncol. 8:74. doi: 10.1186/1748-717X-8-74
Lv, J., Zhu, P., Yang, Z., Li, M., Zhang, X., Cheng, J., et al. (2015). PCDH20 functions as a tumour-suppressor gene through antagonizing the Wnt/beta-catenin signalling pathway in hepatocellular carcinoma. J. Viral Hepat. 22, 201–211. doi: 10.1111/jvh.12265
Marsh, S. (2009). Pharmacogenomics of taxane/platinum therapy in ovarian cancer. Intern. J. Gynecol. Cancer 19 (Suppl. 2), S30–S34. doi: 10.1111/igc.0b013e3181c10513
Martinez, C., Garcia-Martin, E., Pizarro, R. M., Garcia-Gamito, F. J., and Agundez, J. A. (2002). Expression of paclitaxel-inactivating CYP3A activity in human colorectal cancer: implications for drug therapy. Br. J. Cancer 87, 681–686. doi: 10.1038/sj.bjc.6600494
Mcgranahan, N., Favero, F., De Bruin, E. C., Birkbak, N. J., Szallasi, Z., and Swanton, C. (2015). Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra254. doi: 10.1126/scitranslmed.aaa1408
Mexal, S., Berger, R., Pearce, L., Barton, A., Logel, J., Adams, C. E., et al. (2008). Regulation of a novel alphaN-catenin splice variant in schizophrenic smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 759–768. doi: 10.1002/ajmg.b.30679
Miotto, E., Sabbioni, S., Veronese, A., Calin, G. A., Gullini, S., Liboni, A., et al. (2004). Frequent aberrant methylation of the CDH4 gene promoter in human colorectal and gastric cancer. Cancer Res. 64, 8156–8159. doi: 10.1158/0008-5472.CAN-04-3000
Nakagawa, T., Pimkhaokham, A., Suzuki, E., Omura, K., Inazawa, J., and Imoto, I. (2006). Genetic or epigenetic silencing of low density lipoprotein receptor-related protein 1B expression in oral squamous cell carcinoma. Cancer Sci. 97, 1070–1074. doi: 10.1111/j.1349-7006.2006.00283.x
Niu, N., Qin, Y., Fridley, B. L., Hou, J., Kalari, K. R., Zhu, M., et al. (2010). Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 20, 1482–1492. doi: 10.1101/gr.107672.110
Niu, N., Schaid, D. J., Abo, R. P., Kalari, K., Fridley, B. L., Feng, Q., et al. (2012). Genetic association with overall survival of taxane-treated lung cancer patients - a genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study. BMC Cancer 12:422. doi: 10.1186/1471-2407-12-422
Noordermeer, S. M., Wennemers, M., Bergevoet, S. M., Van Der Heijden, A., Tonnissen, E., Sweep, F. C., et al. (2012). Expression of the BRCA1 complex member BRE predicts disease free survival in breast cancer. Breast Cancer Res. Treat. 135, 125–133. doi: 10.1007/s10549-012-2122-5
Peethambaram, P., Fridley, B. L., Vierkant, R. A., Larson, M. C., Kalli, K. R., Elliott, E. A., et al. (2011). Polymorphisms in ABCB1 and ERCC2 associated with ovarian cancer outcome. Int. J. Mol. Epidemiol. Genet. 2, 185–195.
Pharoah, P. D., Tsai, Y. Y., Ramus, S. J., Phelan, C. M., Goode, E. L., Lawrenson, K., et al. (2013). GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45, 362–370, 370e361-362. doi: 10.1038/ng.2564
Poduslo, S. E., Huang, R., and Spiro, A. III. (2010). A genome screen of successful aging without cognitive decline identifies LRP1B by haplotype analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B, 114–119. doi: 10.1002/ajmg.b.30963
Prazeres, H., Torres, J., Rodrigues, F., Pinto, M., Pastoriza, M. C., Gomes, D., et al. (2011). Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene 30, 1302–1317. doi: 10.1038/onc.2010.512
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Rodriguez-Antona, C. (2010). Pharmacogenomics of paclitaxel. Pharmacogenomics 11, 621–623. doi: 10.2217/pgs.10.32
Siegel, R. L., Miller, K. D., and Jemal, A. (2015). Cancer statistics, 2015. CA Cancer J. Clin. 65, 5–29. doi: 10.3322/caac.21254
Song, C., and Zhang, H. (2014). TARV: tree-based analysis of rare variants identifying risk modifying variants in CTNNA2 and CNTNAP2 for alcohol addiction. Genet. Epidemiol. 38, 552–559. doi: 10.1002/gepi.21843
Sonoda, I., Imoto, I., Inoue, J., Shibata, T., Shimada, Y., Chin, K., et al. (2004). Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma. Cancer Res. 64, 3741–3747. doi: 10.1158/0008-5472.CAN-04-0172
White, K. L., Vierkant, R. A., Fogarty, Z. C., Charbonneau, B., Block, M. S., Pharoah, P. D. Goode, E.L., et al. (2013). Analysis of over 10,000 Cases finds no association between previously reported candidate polymorphisms and ovarian cancer outcome. Cancer Epidemiol. Biomarkers Prev. 22, 987–992. doi: 10.1158/1055-9965.EPI-13-0028
Willer, C. J., Li, Y., and Abecasis, G. R. (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. doi: 10.1093/bioinformatics/btq340
Wu, T. Y., Fridley, B. L., Jenkins, G. D., Batzler, A., Wang, L., and Weinshilboum, R. M. (2011). Mycophenolic acid response biomarkers: a cell line model system-based genome-wide screen. Int. Immunopharmacol. 11, 1057–1064. doi: 10.1016/j.intimp.2011.02.027
Keywords: pharmacogenomics, genome-wide association, carboplatin, paclitaxel, ovarian cancer, cell viability, apoptosis, lymphoblastoid cell lines
Citation: Fridley BL, Ghosh TM, Wang A, Raghavan R, Dai J, Goode EL and Lamba JK (2016) Genome-Wide Study of Response to Platinum, Taxane, and Combination Therapy in Ovarian Cancer: In vitro Phenotypes, Inherited Variation, and Disease Recurrence. Front. Genet. 7:37. doi: 10.3389/fgene.2016.00037
Received: 07 January 2016; Accepted: 04 March 2016;
Published: 22 March 2016.
Edited by:
José A. G. Agúndez, University of Extremadura, SpainReviewed by:
Elena García-Martín, Universidad de Extremadura, SpainCopyright © 2016 Fridley, Ghosh, Wang, Raghavan, Dai, Goode and Lamba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brooke L. Fridley, YmZyaWRsZXlAa3VtYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.