- 1Department of Epidemiology, University of Alabama at Birmingham School of Public Health, Birmingham, AL, USA
- 2Department of Biostatistics, University of Alabama at Birmingham School of Public Health, Birmingham, AL, USA
We evaluated the association of 56 candidate SNPs identified in two published genome-wide association studies (GWAS) of uterine leiomyoma (UL), or fibroids, with the risk and tumor size in the multi-ethnic uterine fibroid study (NIEHS-UFS). The selected SNPs were genotyped in 916 premenopausal women of African American (AA) and European American (EA) descents and their association with the outcomes was evaluated in race-stratified models and in meta-analysis of risk in NIEHS-UFS and discovery and replication GWAS in the Japanese population. We report moderate associations of variant rs4954368 in THSD7B (thrombospondin, type I, domain containing 7B) with tumor size in pooled analysis of AA and EA samples (P = 0.004), and at TNRC6B (trinucleotide repeat containing 6B) variants rs138039 and rs139909 in EA (P = 0.001 and 0.008, respectively). The most significant associations with risk in meta-analysis were observed at TNRC6B variants rs739182 (P = 3.7 × 10−10) and rs2072858 (P = 1.1 × 10−9) and were stronger than those reported in the discovery GWAS (P = 2.01 × 10−8 and 2.58 × 10−8, respectively). The present study failed to replicate the associations reported for CCDC57 and FASN in a discovery GWAS in populations of European descent. Consistent with previous replication studies in the Right From the Start Study (RFTS) and the BioVU DNA repository, we provide independent evidence for association of TNRC6B with both risk and size of UL. The present study is the first to report a replicated association of THSD7B with UL, albeit with tumor size and not with risk.
Introduction
Uterine leiomyoma (UL), or fibroids, are common hormone-dependent tumors that arise in the smooth muscle cells of the uterus. These tumors develop in >75% of North American women over the reproductive age and become clinically apparent in about one-third (Cramer and Patel, 1990). Although benign, UL cause significant gynecologic morbidities including menorrhagia, pelvic pain, urinary incontinence, infertility, and pregnancy complications (Stewart, 2001; Walker and Stewart, 2005). Uterine fibroids are the primary indication for hysterectomy, with an incidence rate of 5.5 per 1000 women in the United States and accounting for about $9.4 billion of public health burden (Farquhar and Steiner, 2002).
Estrogen and progesterone are believed to promote the growth of these tumors and long-term unopposed estrogens are associated with risk (Andersen, 1996). Factors influencing gonadal steroid metabolisms, such as obesity, diet, or exercise show apparent association with risk (Schwartz et al., 2000). Age (increasing risk with increasing premenopausal age), menopause (risk decreases with menopause), and African American (AA) ethnicity (higher risk compared with that of non-Hispanic Whites), which was originally tested in NIEHS-UFS (National Institute of Environmental Health Sciences-Uterine Fibroid Study) (Baird et al., 2003) are established risk factors. The basis for the higher prevalence among African Americans is not well-understood. The genetic contribution to this ethnic disparity was investigated in admixture-based genome-wide scans of the AA population in BWHS (Black Women's Health Study) (Wise et al., 2012) and recently in NIEHS-UFS (Zhang et al., 2015). Both the BWHS (Wise et al., 2012) and NIEHS-UFS found a decreased mean percentage of European ancestry in UL cases compared to controls. The negative correlation between European ancestry and UL prevalence suggests that genetic factors may explain the different prevalence between AA and European American (EA) women.
Genetic predisposition to UL was evidenced in early familial aggregation studies (Vikhlyaeva et al., 1995; Sato et al., 2002). Nonetheless, no convergent views on a potential mechanism of pathogenesis have emerged from the genome linkage scans and genome-wide association studies (GWAS) reported to date (Gross et al., 2004; Cha et al., 2011; Eggert et al., 2012; Wise et al., 2012). These GWA studies pointed to TNRC6B (trinucleotide repeat containing 6B) and BET1L (Bet1 golgi vesicular membrane trafficking protein-like) as the most significant candidate genes in the Japanese population (Cha et al., 2011) and to FASN (fatty acid synthase) and the nearby CCDC57 (coiled-coil domain containing 57) gene in the European American population of the WGHS (Women Genome Health study) (Eggert et al., 2012) and Australian populations. Furthermore, genetic association studies of UL in European American women enrolled in the Right From The Start (RFTS) cohort and the Biorepository at Vanderbilt University (BioVU) study reported replications of the associations with BET1L and TNRC6B (Edwards et al., 2013a,b). Recently, in an independent follow-up study to published linkage (Eggert et al., 2012) and admixture (Wise et al., 2012) signals for UL, we reported association data in NIEHS-UFS implicating variants in genes encoding components of the extracellular matrix (ECM) (Aissani et al., 2014), consistent with the current paradigm for altered ECM in UL pathogenesis.
Whole genome sequencing of tumors have identified several genes involved in UL pathology (Makinen et al., 2011; Mehine et al., 2013) and cytogenetic analyses detected recurrent chromosomal aberrations like those affecting the high mobility group AT-hook 2 (HMGA2) gene on chromosome 12 in band q14.3 in tumorigenesis (Heim et al., 1988; Turc-Carel et al., 1988). In contrast to these somatic mutations, germline mutations affecting highly conserved amino acids have been reported for the fumarate hydratase-encoding gene (FH) on chromosome 1q43 in tumor syndromes (Alam et al., 2001; Tomlinson et al., 2002) but only in rare cases of non-syndromic UL (Barker et al., 2002; Kiuru et al., 2002).
Replicating GWAS candidate SNPs in independent studies of populations with different ancestry, especially in the high-risk African American population, is an important undertaking toward validation of the association results reported in discovery GWAS. Another motivation for conducting the present study is the lack of shared candidate SNPs between the two available GWAS. In the present study, we sampled 916 clinically ascertained premenopausal women participants in the multi-ethnic NIEHS-UFS to evaluate the association of candidate SNPs (single nucleotide polymorphisms) from GWA studies conducted in the Japanese (JPN), EA, and Australian (AUS) populations, as well as a few SNPs in or nearby candidate genes, with the risk of UL and tumor size. We report double-sided p-values uncorrected for multiple testing from meta-analyses and from race-stratified analyses for the two predominant AA (393 cases and 132 controls) and EA (195 cases and 196 controls) ethnic groups from logistic regression models with or without adjustment for covariates.
Materials and Methods
Study Population
The characteristics of the study population were reported in previous studies (Aissani et al., 2013, 2014). Briefly, women aged 35–51 years, were randomly selected from a computerized list of members of a prepaid urban health plan for enrollment in the NIEHS-UFS (Baird et al., 2003). Self-administered questionnaires were used to collect demographic data. Reproductive and gynecologic history data were collected during a telephone interview. About 92% of the study population was self-identified as AA or non-Hispanic EA. Among the premenopausal women enrolled in the study and who had ultrasound examinations (n = 1119), 1045 (93%) had available DNA specimens and were of African American (n = 574), non-Hispanic European American (394) or other (n = 77) ethnicity. The NIEHS-UFS and the present sub-study were approved by the Human Subject's Review boards at the NIEHS, George Washington University and University of Alabama at Birmingham, respectively. Participants to NIEHS-UFS were fully informed and were enrolled only after full written consents were obtained.
For meta-analysis, model estimates were available only from the GWAS in the Japanese population (Cha et al., 2011). This GWAS sampled 1607 individuals clinically diagnosed with UL and 1328 individual controls for the discovery study, and 3466 clinically diagnosed UL cases 3245 individual controls for the replication study. The GWAS obtained written patient consents in accordance with the Ethical Committee at each of The Institute of Medical Science of the University of Tokyo and the Center for Genomic Medicine of RIKEN.
Covariates
Covariates included age, age at menarche, parity (pregnancies after age 25; earlier births were not significantly related to fibroid development in the NIEHS-UFS cohort (Dunson and Baird, 2002; Baird and Dunson, 2003), body mass index (BMI), and physical activity.
Ascertainment
Fibroid status was assessed by ultrasonography at baseline or by medical record review in 80 and 90% of AA and EA participants, respectively. For the study participants who had recent pelvic ultrasound examinations, the radiology records were examined for the presence of fibroids. The remaining participants were asked to have a pelvic ultrasound examination at a primary care site. If neither ultrasounds nor medical record reviews could be conducted, the concerned participants were excluded from the present study. Clinical examinations consisted of both transabdominal and transvaginal ultrasonographies. The abdominal portion evaluated fibroid changes arising from the upper uterus that could be missed by the transvaginal approach alone. Tumor size was classified in three categories (Small, Medium, and Large) measured by the diameter of the tumors (S ≤ 2 cm, 2 cm < M < 4 cm, L ≥ 4 cm). For participants diagnosed with multiple tumors, the largest tumor determined the size category.
No clinical information on the size of fibroids was available in the GWAS sample used in meta-analysis. Compared with the reported family history among cases (21.03 and 16.56% for the discovery and replication samples, respectively), only 5.11 and 1.94% of the controls in the discovery and replication sets, respectively, reported family history of UL (Cha et al., 2011).
SNP Selection and Typing
Illumina iSelect assay (Illumina Inc., San Diego, CA) was used to type a total of 63 SNPs selected among the most significantly associated SNPs in GWAS. We also included additional SNPs in associated genes or genes adjacent to them, as well as a small number of SNPs in the candidate HMGA2. Reliability in typing data was assessed by inclusion of blind intra- and inter-plate duplicates representative of each of the study population groups. For accurate assessment of African descent, we included a set of 25 unrelated Yoruban (YRI) samples. A total of 2682 SNPs (from an extended set of 4363 SNPs typed in a previous study and also that included 147 ancestry informative markers) that were common to our data and to the majority of the HapMap III reference populations was used to assess ethnic membership of quality control samples. About 35% of the SNPs were from chromosome 1q43; the remaining SNPs have genome-wide distribution (Aissani et al., 2014). All SNPs in the extended set were selected to have a minor allele frequency (MAF) of at least 5% and most of them are inter-population tagging SNPs relevant to the ethnic backgrounds of the NIEHS-UFS study populations (Aissani et al., 2013).
Principal component analysis was used to assess the Japanese ethnicity of the case and control individuals sampled for the GWAS (Cha et al., 2011). The control individuals studied in the discovery and replication GWAS were older than the cases (mean ages 42.5 vs. 48.9 years and 45.2 vs. 62.3 years in discovery and replication sets, respectively). This study design is important to keep downward the rate of misclassification, which can be substantial in case and control studies where controls are not clinically ascertained. The participants to NIEHS-UFS were all ≥35 years of age and about two-thirds were ≥40 years of age.
Statistical Analysis
Quality Control
SNP calls were checked for adherence to Hardy–Weinberg equilibrium (HWE) in each of the affection status and population stratum, and SNPs deviating (p < 0.01) from HWE in UL-free controls were excluded.
Analytical Design
Association of SNPs with risk of UL and tumor size was evaluated separately in each population stratum using dichotomous and polytomous logistic regression modeling, respectively, in SAS (SAS, Cary, NC), and in meta-analyses by fitting random effect models to the data in the metafor R package (http://www.jstatsoft.org/v36/i03/) (Viechtbauer, 2010). Heterogeneity between studies was assessed by the Cochran's Q-test that is part of DerSimonian-Laird estimator in the pooling method (DerSimonian and Laird, 1986). Because the discovery GWAS data were derived from models with no adjustments for the covariates, for comparisons, we report data from both adjusted and unadjusted models in NIEHS-UFS and from unadjusted models in the meta-analysis of NIEHS-UFS and Japanese GWAS.
Previous studies in NIEHS-UFS have assessed the levels of the ordinal covariates that influenced or modified the risk of UL (Baird et al., 1998, 2007; Baird and Dunson, 2003); we therefore modeled these covariates accordingly. Specifically, we modeled age as a continuous variable and BMI as an ordinal variable (based on categories <25, 25–29.99, 30–34.99, 35+). The other covariates were age of menarche modeled as a dichotomous variable (age ≤11 years vs. other ages), exercise as a 4-level ordinal variable (lower third, middle third, next sixth, top sixth as described (Baird et al., 2007) and parity defined as having or not having given birth at age 25 and older (binary variable).
The association of SNP genotypes with UL outcomes was tested in principal component analysis (PCA)-defined groups assuming additive genetic effects as described (Aissani et al., 2013). For the polytomous models, the assumption for proportional odds in both or in either AA or EA study population was checked and reported if not met. UL was modeled as a dichotomous outcome (case-control design comparing UL-free category to the combination of tumor size categories S, M, and L) or as a polytomous outcome comparing UL size categories (S vs. M and L and S and M vs. L).
SNP tests that passed Bonferroni-corrected significance thresholds, which were p = 0.05/56= 9 × 10−4 for multiple testing of 56 quality control-filtered SNPs in NIEHS-UFS and genome-wide significance p = 10−7 in meta-analysis of NIEHS-UFS and discovery GWAS, were deemed to be statistically significant.
Results
Characteristics of the participants available for testing the genetic association with the risk of UL (case-control design) and tumor size (case-only design) were reported in a previous study (Aissani et al., 2013, 2014). Except for a unique individual of European descent, typing was achieved in the entire study population (n = 1045). The call rate was >98% in about 92% of sampled individuals. Among the 1045 typed samples, 42 (4.0%) with call rates <90% were excluded from the analysis; this yielded a total of 1003 samples for analysis. The overall concordance rate between duplicates was 99.6%, implying that a false discovery due to typing errors is unlikely. Discriminant analysis corrected ethnic group inference for several individuals who self-identified as “other populations” to either EA or AA ancestry (Supplementary Figure 1). Sixteen individuals who self-identified as African American (AA) and one individual as a non-Hispanic European American (EA) clustered more closely with the Yoruban African population (YRI). This observation prompted the exclusion of this subset of 17 individuals, resulting in a total of 986 individuals (525 AA, 391 EA, and 70 other) available for analysis.
Of the 63 assayed SNPs, 56 passed the QC filters including adherence to HWE in the control groups. Tables 1, 2 display the results of association with the risk and tumor size outcomes, respectively, for the most significant (Bonferroni-uncorrected P ≤ 0.05) SNPs in any of the tested models. Race-stratified analyses and meta-analyses were restricted to the two largest ethnic groups AA and EA. To allow comparisons with the discovery GWAS, which were not adjusted for the covariates modeled in the present study, we report results from models with or without adjustment for covariates (Supplementary Tables S1, S2). The results showed no significant differences between the adjusted and unadjusted models.
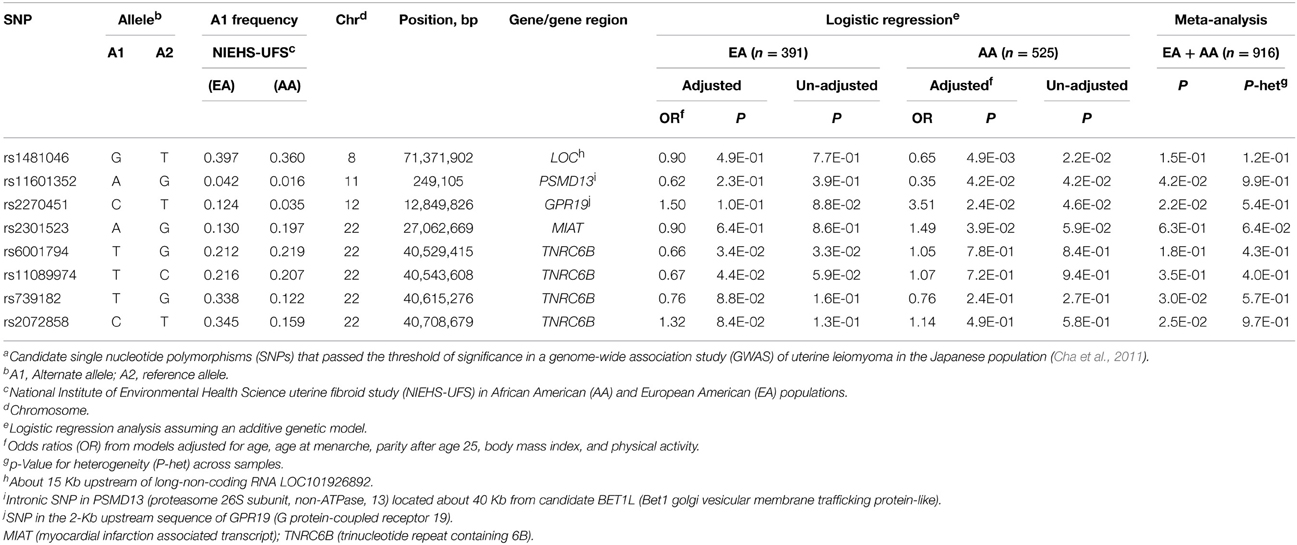
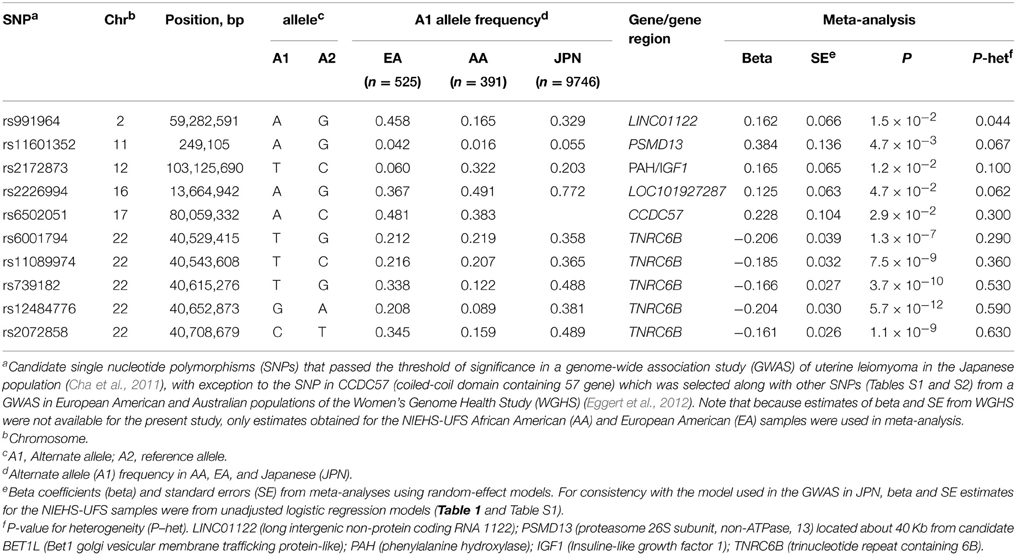
Table 1. Association of candidate SNPs awith risk of uterine fibroids in the NIEHS uterine fibroid study.
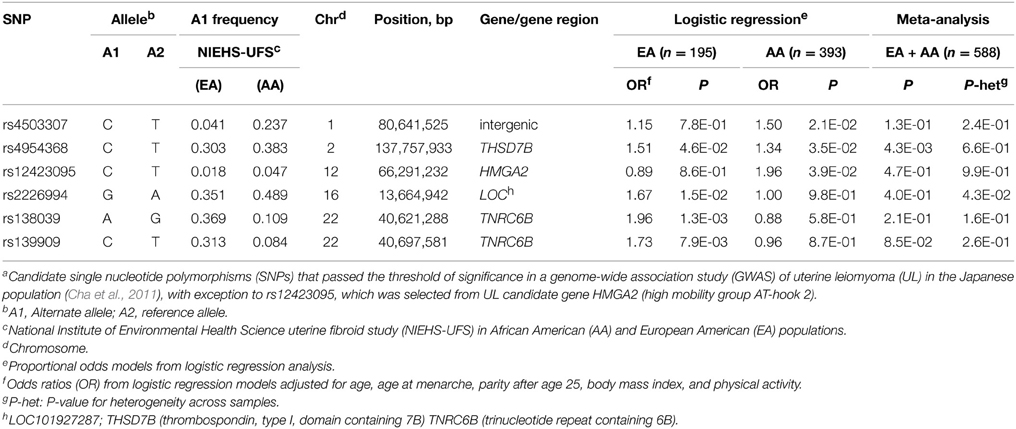
Table 2. Association of candidate SNPs a with the size of uterine fibroids in NIEHS uterine fibroid study.
Race-stratified analyses of risk showed moderate associations (P < 0.05) with 6 SNPs, one in each of chromosomes 8, 11, and 12, and 3 on chromosome 22 (Table 1). The most significant association (P = 0.005) occurred in the AA group at rs1481046 located about 15 Kb upstream of a long-non-coding RNA (LOC101926892) of uncharacterized function. Among the associated SNPs, three locate to or near previously replicated BETL1 and TNRC6B genes (Edwards et al., 2013b). The intronic SNP rs11601352 in PSMD13 (proteasome 26S subunit, non-ATPase, 13) is located about 40 Kb from the replicated candidate BET1L. The moderate association with this SNP was observed only in the AA group and in meta-analysis (P = 0.042), whereas the association with the two other SNPs, rs6001794, and rs11089974, in TNRC6B was observed in the EA group but not in the meta-analysis. The two other associated SNPs in the AA group, rs2270451 (P = 0.024) and rs2301523 (P = 0.039), are located in the 2-Kb upstream sequence of GPR19 (G protein-coupled receptor 19) and within the non-protein coding RNA MIAT (myocardial infarction associated transcript), respectively. Two TNRC6B intronic SNPs, rs739182, and rs2072858, showed moderate associations (P = 0.030 and P = 0.025, respectively) but only in meta-analyses; these SNPs are in strong linkage disequilibrium (LD) in EA (r2 = 0.95; Supplementary Table S4) and in moderate LD in AA (r2 = 0.58; Supplementary Table S5).
Association with tumor size was tested in case-only designs using proportional odds models and assuming an additive genetic model with adjustments for covariates. No variants that associated with risk also associate with tumor size; however, SNPs in TNRC6B other than those affecting risk influenced significantly (P = 0.001 at rs138039 and P = 0.008 at rs139909) tumor size in EA (Table 2). Examination of the LD pattern in TNRC6B showed that these two SNPs are in LD with each other both in EA (r2 = 0.74) and AA (r2 = 0.76) but not with any other tested TNRC6B SNP (r2 < 0.05) in AA (Supplementary Table S5) or in EA (r2 < 0.35) (Supplementary Table S4).
Variant rs12423095 in HMGA2 showed a moderate association with tumor size in AA only although no significant heterogeneity was observed between AA and EA (Supplementary Table S2), possibly indicating a spurious association due to the low minor allele frequency at this SNP. Consistent with these data, rs12423095 showed weak or no LD (r2 < 0.15) with the other tested HMGA2 SNPs in both study groups. The sole association with tumor size observed separately in AA (P = 0.035) and EA (P = 0.046), and in meta-analysis (P = 0.004) was with the intronic variant rs4954368 in THSD7B (thrombospondin, type I, domain containing 7B gene) (Table 2 and Table S2). Of note, given that none of the significantly associated SNPs failed to meet the assumption for proportional odds, these results imply single OR for the contrasted ordinal outcomes.
We re-evaluated the association of the full set of 56 SNPs with risk in meta-analyses of NIEHS-UFS (n = 916) and the discovery (n = 3035) and replication (n = 6711) GWAS in the Japanese population (Cha et al., 2011). The results of the meta-analyses showed significant associations (P < 1.3 × 10−7) with 5 of the 8 tested SNPs in TNRC6B and a moderate association at the candidate PSMD13/BET1L region (rs11601352, P = 0.0047) (Table 3). In particular, TNRC6B variants rs739182 (P = 3.7 × 10−10) and rs2072858 (P = 1.1 × 10−9) showed stronger associations than in the discovery GWAS (P = 2.01 × 10−8 and 2.58 × 10−8, respectively). No association was observed at the TNRC6B variants rs138039 and rs139909 that associated moderately with tumor size in the EA group (Table 2). Examination of the minor allele frequency (MAF) at these two SNPs showed higher MAFs in EA compared to AA, and opposite minor and major alleles in the Japanese population (Table S3).
Moderate heterogeneities between the studies were observed at three gene loci including PSMD13/BET1L but not at the significantly associated TNRC6B or at the moderately associated CCDC57. Of note, the variant rs6502051 in CCDC57, which maps 3 Kb away from the top candidate FASN in WGHS (Eggert et al., 2012) was not reported in the Japanese GWAS. This SNP showed weak LD with our tested SNPs in FASN in EA and AA (Supplementary Tables S4, S5). The slightly different results obtained for this SNP (P = 0.13 in Table S1 and P = 0.029 in Table 3) stem from the use of models with (Table 1 and Table S1) and without (Table 3) adjustment for covariates.
Strong heterogeneity (P = 3.2 × 10−4) across the studies was also observed at the THSD7B variant rs4954368 in the test of association with the risk outcome (Table S3).
Discussion
In the present NIEHS-UFS, we attempted replications of GWAS findings for uterine fibroids in the Japanese population and reported moderate and significant associations in race-stratified analyses and in meta-analyses depending on the SNP and outcome (risk or tumor size) under consideration. The most striking results were replicated associations of several variants in TNRC6B with either risk or tumor size. Based on the observed LD patterns across the associated SNPs, our data suggest the presence of two common TNRC6B haplotypes associated independently with risk or size of UL. Our data in NIEHS-UFS are consistent with the previous replication of TNRC6B SNP associations with risk and tumor size among European Americans in the BioVu and RFTS studies (Edwards et al., 2013a,b); however, differences between the association results from these replication studies were also observed. First, only few SNP associations could be compared because RFST/BioVU studies were focused on the three top GWAS genes (BET1L, TNRC6B, and near SLK genes) whereas the NIEHS-UFS examined all genome-wide significant SNPs. Among the rare overlapping SNPs in these replication studies TNRC6B rs12484776 was associated moderately with risk in RFTS and BioVu, and with increasing fibroid volume in RFTS whereas in NIEHS-UFS no association was observed with this SNP. The association observed in the meta-analysis of NIEHS-UFS and GWAS samples (Table 3) is solely explained by the latter samples. Second, while none of these studies reported evidence for TNRC6B rs139909 association with risk, association with tumor size (OR = 1.73; p = 0.008) was observed in the EA group in our study, potentially suggesting decreased power in the AA group that can be explained by the low MAF at this SNP.
We also reported moderate associations of intronic variant rs4954368 in THSD7B (thrombospondin, type I, domain containing 7B) with tumor size in race-stratified analyses and combined analyses of AA and EA in NIEHS-UFS but not in meta-analysis of risk in NIEHS-UFS and prior GWAS samples. The negative association with risk in the meta-analysis of the two studies is not inconsistent with the results from the separate studies, which showed no association in NIEHS-UFS and strong heterogeneity (P = 1.8 × 10−5) in the reported meta-analysis of the discovery and replication GWAS in the Japanese study (Cha et al., 2011). The reported association of THSD7B with tumor size may not be a spurious finding because (i) it was observed in a case-only design, (ii) the strength and direction of the association are similar in each study population, and (iii) the EA and AA groups share the allele “C” associated with tumor size.
A potential explanation to the observed association of rs4954368 with risk in the discovery GWAS but only with tumor size in NIEHS-UFS is the non-clinical ascertainment of the controls in the discovery GWAS. Non-clinical ascertainment of controls in cross-sectional studies of UL may be prone to high misclassification rates because women with small tumors are likely to be asymptomatic and consequently might not draw clinical attention. In the setting of high misclassification rates (large proportions of cases misclassified as controls), case and control designs would also test an ordinal outcome (small tumors vs. tumors with varying sizes). Thus, despite an appropriate and robust design of the reported GWAS (controls older than cases), misclassifications cannot be excluded. Alternatively, the negative association of THSD7B with risk and positive association with tumor size in NIEHS-UFS may reflect a pathogenic mechanism whereby the tagged causal variant in THSD7B is a secondary mutation rather than a driver mutation. Another possible explanation is the level of circulating estrogen, which affects the production of thrombospondins and may differ in NIEHS-UFS and the GWAS.
THSD7B encodes a member of the thrombospondins (TSP) which are potent inhibitors of angiogenesis and therefore may be important in controlling tumor growth (Vallbo and Damber, 2005). TSPs are secreted by various kinds of cells including endothelial and smooth muscle cells. These secreted molecules affect cell functions by modulating cell-matrix interactions. This finding converges with those from our recent study in NIEHS-UFS that implicated variants of encoded components of the extra-cellular matrix (Aissani et al., 2014).
We also reported that the intronic variant rs138039 in TNRC6B is associated with tumor size in EA with a significance level close to the Bonferroni-corrected threshold of P = 0.0009 and that the lower MAF in AA likely precluded detection of the association. The association of tumor size with a different TNRC6B variant (rs12484776) in RFTS (Edwards et al., 2013a), albeit moderately (P = 0.024), supports a candidate status for this gene because the observed association with a different variant in the same gene in RFTS reduces the possibility of biased results for rs138039 and rs139909 in NIEHS-UFS (not tested in RFTS). Given that these associated variants most likely tag rather than being the actual causal variant(s), cryptic difference in the pattern of linkage disequilibrium between the EA samples in NIEHS-UFS and RFTS or genetic heterogeneity (different causal variants) may explain the observed results.
We were not able to assess the replicated association with the BET1L 3′UTR variant rs2280543 that was reported in RFTS and BioVU; this variant did not pass the QC filter in our study. Nonetheless, the GWAS discovery variant rs11601352 in the adjacent PSMD13 gene was shown to associate moderately with the risk of UL in AA and in meta-analysis in NIEHS-UFS (P = 0.042). The low frequency (MAF = 0.02) of the associated allele “A” at this SNP site, as well as that at rs2280543 in EA in RFTS and BioVU, precludes conclusive statements. Thus, validation of this association in other study populations with higher MAF is needed. Variant rs11601352 is not represented in the 1000 Genomes database and the highest MAF reported for this SNP in the HapMap III database is in populations of Mexican ancestry (MEX; MAF = 0.15), followed by Asian ancestries (China and Japan) with a MAF of 5%.
The function of TNRC6B is not known; however, this gene was highlighted as a candidate in GWAS for height (Estrada et al., 2009) and for age-at-menarche (Guo et al., 2006), which is a risk factor for UL (Marshall et al., 1997; Baird et al., 2003; Wise et al., 2004). Although no association was observed between TNRC6B variants and age-at-menarche in NIEHS-UFS (unpublished data), validation of the reported association in independent GWAS could imply that the encoded product of TNRC6B functions in the causal pathway. Of potential relevance to these independent replications of TNRC6B association with UL, the expression of the TNRC6A paralog was reported to be markedly dysregulated in large fibroids compared to small fibroids (log2 fold change = −1.00, p = 0.0008) (Guo et al., 2014). The TNRC6 paralogs encode members of the GW182 protein family that are essential components of the miRNA pathway (Yao et al., 2012), which has been implicated in the pathogenesis of uterine fibroids (Marsh et al., 2008; Wei and Soteropoulos, 2008).
We also reported a moderate association of the common CCDC57 variant rs6502051 with protection against UL in the AA group; however, the decreased level of significance for this SNP after controlling for the covariates suggested potential confounding effects. Furthermore, the negative association of the other common SNPs in this gene as well as in the nearby FASN with either outcome or study group remains unclear and requires confirmation in independent study populations. Finally, several SNP associations including those reported for GPR19, HMGA2, MIAT, PSMD13/BET1L and for other annotated or non-annotated gene loci had either low minor allele frequencies, were observed only in unadjusted models or that the association was lost or dropped significantly in meta-analysis of EA and AA or of EA, AA, and JPN samples.
The relevance of the encoded product of THSD7B, the sole gene that consistently associated with tumor size in EA and AA populations, to UL pathogenesis is not known. Very little is known about the function of this gene; it encodes a member of the thrombospondin family which are matricellular modulators of cell function (Bornstein, 2001). Of note, another member of the thrombospondin-encoding genes, THBS1 (alias TSP-1), was reported to be down-regulated in uterine fibroids (Behera et al., 2007) and TSP-1 production has been shown to be directly controlled by estrogens in estrogen receptor-positive breast cancer cells (Hyder et al., 2009), suggesting that TSP-1 may function in the estrogen receptor-positive UL cells as well. Thus, the possibility that the product of THSD7B functions in the same pathway as TSP-1 cannot be excluded.
In conclusion, the present work replicated the association of TNRC6B with risk and size of UL consistent with the first reported replication of these associations in the RFTS and BioVU studies. Of potential relevance to the pathogenesis of UL, we reported an association of the thrombospondin-encoding THSD7B with tumor size in both European and African American populations.
Author Contributions
BA supervised the study design, data analyses and writing of the manuscript. KZ provided analytic support for the study design and data analyses. HW performed quality control of the data and statistical analyses. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grant R01-HD064398 (to BA) from the National Institute of Child Health and Human Development (NIH-NICHD) and the National Institute for Environmental Health Science (NIH-NIEHS). The authors are grateful to Dr. Donna Day Baird, the principal investigator of the NIEHS Uterine Fibroid Study, for her support. The authors thank the women who participated to the NIEHS-UFS for their valuable contribution to this research.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2015.00241
References
Aissani, B., Wiener, H., and Zhang, K. (2013). Multiple hits for the association of uterine fibroids on human chromosome 1q43. PLoS ONE 8:e58399. doi: 10.1371/journal.pone.0058399
Aissani, B., Zhang, K., and Wiener, H. (2014). Follow-up to genome-wide linkage and admixture mapping studies implicates components of the extracellular matrix in susceptibility to and size of uterine fibroids. Fertil. Steril. 103, 528–534.e513. doi: 10.1016/j.fertnstert.2014.10.025
Alam, N. A., Bevan, S., Churchman, M., Barclay, E., Barker, K., Jaeger, E. E., et al. (2001). Localization of a gene (MCUL1) for multiple cutaneous leiomyomata and uterine fibroids to chromosome 1q42.3-q43. Am. J. Hum. Genet. 68, 1264–1269. doi: 10.1086/320124
Andersen, J. (1996). Growth factors and cytokines in uterine leiomyomas. Semin. Reprod. Endocrinol. 14, 269–282. doi: 10.1055/s-2007-1016336
Baird, D. D., and Dunson, D. B. (2003). Why is parity protective for uterine fibroids? Epidemiology 14, 247–250. doi: 10.1097/01.EDE.0000054360.61254.27
Baird, D. D., Dunson, D. B., Hill, M. C., Cousins, D., and Schectman, J. M. (2003). High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am. J. Obstet. Gynecol. 188, 100–107. doi: 10.1067/mob.2003.99
Baird, D. D., Dunson, D. B., Hill, M. C., Cousins, D., and Schectman, J. M. (2007). Association of physical activity with development of uterine leiomyoma. Am. J. Epidemiol. 165, 157–163. doi: 10.1093/aje/kwj363
Baird, D. D., Dixon, D., and Sandler, D. P. (1998). African Americans at higher risk than whites for uterine fibroids: ultrasound evidence Am. J. Epidemiol. 147, S90.
Barker, K. T., Bevan, S., Wang, R., Lu, Y. J., Flanagan, A. M., Bridge, J. A., et al. (2002). Low frequency of somatic mutations in the FH/multiple cutaneous leiomyomatosis gene in sporadic leiomyosarcomas and uterine leiomyomas. Br. J. Cancer 87, 446–448. doi: 10.1038/sj.bjc.6600502
Behera, M. A., Feng, L., Yonish, B., Catherino, W., Jung, S. H., and Leppert, P. (2007). Thrombospondin-1 and thrombospondin-2 mRNA and TSP-1 and TSP-2 protein expression in uterine fibroids and correlation to the genes COL1A1 and COL3A1 and to the collagen cross-link hydroxyproline. Reprod. Sci. 14, 63–76. doi: 10.1177/1933719107309591
Bornstein, P. (2001). Thrombospondins as matricellular modulators of cell function. J. Clin. Invest. 107, 929–934. doi: 10.1172/JCI12749
Cha, P. C., Takahashi, A., Hosono, N., Low, S. K., Kamatani, N., Kubo, M., et al. (2011). A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat. Genet. 43, 447–450. doi: 10.1038/ng.805
Cramer, S. F., and Patel, A. (1990). The frequency of uterine leiomyomas. Am. J. Clin. Pathol. 94, 435–438.
DerSimonian, R., and Laird, N. (1986). Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188. doi: 10.1016/0197-2456(86)90046-2
Dunson, D. B., and Baird, D. D. (2002). A proportional hazards model for incidence and induced remission of disease. Biometrics 58, 71–78. doi: 10.1111/j.0006-341X.2002.00071.x
Edwards, T. L., Hartmann, K. E., and Velez Edwards, D. R. (2013a). Variants in BET1L and TNRC6B associate with increasing fibroid volume and fibroid type among European Americans. Hum. Genet. 132, 1361–1369. doi: 10.1007/s00439-013-1340-1
Edwards, T. L., Michels, K. A., Hartmann, K. E., and Velez Edwards, D. R. (2013b). BET1L and TNRC6B associate with uterine fibroid risk among European Americans. Hum. Genet. 132, 943–953. doi: 10.1007/s00439-013-1306-3
Eggert, S. L., Huyck, K. L., Somasundaram, P., Kavalla, R., Stewart, E. A., Lu, A. T., et al. (2012). Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata. Am. J. Hum. Genet. 91, 621–628. doi: 10.1016/j.ajhg.2012.08.009
Estrada, K., Krawczak, M., Schreiber, S., van Duijn, K., Stolk, L., van Meurs, J. B., et al. (2009). A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum. Mol. Genet. 18, 3516–3524. doi: 10.1093/hmg/ddp296
Farquhar, C. M., and Steiner, C. A. (2002). Hysterectomy rates in the United States 1990-1997. Obstet. Gynecol. 99, 229–234. doi: 10.1016/S0029-7844(01)01723-9
Gross, K. L., Panhuysen, C. I., Kleinman, M. S., Goldhammer, H., Jones, E. S., Nassery, N., et al. (2004). Involvement of fumarate hydratase in nonsyndromic uterine leiomyomas: genetic linkage analysis and FISH studies. Genes Chromosomes Cancer 41, 183–190. doi: 10.1002/gcc.20079
Guo, H., Zhang, X., Dong, R., Liu, X., Li, Y., Lu, S., et al. (2014). Integrated analysis of long noncoding RNAs and mRNAs reveals their potential roles in the pathogenesis of uterine leiomyomas. Oncotarget 5, 8625–8636.
Guo, Y., Shen, H., Xiao, P., Xiong, D. H., Yang, T. L., Guo, Y. F., et al. (2006). Genomewide linkage scan for quantitative trait loci underlying variation in age at menarche. J. Clin. Endocrinol. Metab. 91, 1009–1014. doi: 10.1210/jc.2005-2179
Heim, S., Nilbert, M., Vanni, R., Floderus, U. M., Mandahl, N., Liedgren, S., et al. (1988). A specific translocation, t(12;14)(q14-15;q23-24), characterizes a subgroup of uterine leiomyomas. Cancer Genet. Cytogenet. 32, 13–17. doi: 10.1016/0165-4608(88)90305-6
Hyder, S. M., Liang, Y., and Wu, J. (2009). Estrogen regulation of thrombospondin-1 in human breast cancer cells. Int. J. Cancer 125, 1045–1053. doi: 10.1002/ijc.24373
Kiuru, M., Lehtonen, R., Arola, J., Salovaara, R., Jarvinen, H., Aittomaki, K., et al. (2002). Few FH mutations in sporadic counterparts of tumor types observed in hereditary leiomyomatosis and renal cell cancer families. Cancer Res. 62, 4554–4557.
Makinen, N., Mehine, M., Tolvanen, J., Kaasinen, E., Li, Y., Lehtonen, H. J., et al. (2011). MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 334 252–255. doi: 10.1126/science.1208930
Marsh, E. E., Lin, Z., Yin, P., Milad, M., Chakravarti, D., and Bulun, S. E. (2008). Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil. Steril. 89, 1771–1776. doi: 10.1016/j.fertnstert.2007.05.074
Marshall, L. M., Spiegelman, D., Barbieri, R. L., Goldman, M. B., Manson, J. E., Colditz, G. A., et al. (1997). Variation in the incidence of uterine leiomyoma among premenopausal women by age and race. Obstet. Gynecol. 90, 967–973. doi: 10.1016/S0029-7844(97)00534-6
Mehine, M., Kaasinen, E., Makinen, N., Katainen, R., Kampjarvi, K., Pitkanen, E., et al. (2013). Characterization of uterine leiomyomas by whole-genome sequencing. N. Engl. J. Med. 369, 43–53. doi: 10.1056/NEJMoa1302736
Sato, F., Mori, M., Nishi, M., Kudo, R., and Miyake, H. (2002). Familial aggregation of uterine myomas in Japanese women. J. Epidemiol. 12, 249–253. doi: 10.2188/jea.12.249
Schwartz, S. M., Marshall, L. M., and Baird, D. D. (2000). Epidemiologic contributions to understanding the etiology of uterine leiomyomata. Environ Health Perspect 108(Suppl. 5), 821–827. doi: 10.1289/ehp.00108s5821
Tomlinson, I. P., Alam, N. A., Rowan, A. J., Barclay, E., Jaeger, E. E., Kelsell, D., et al. (2002). Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30, 406–410. doi: 10.1038/ng849
Turc-Carel, C., Dal Cin, P., Boghosian, L., Terk-Zakarian, J., and Sandberg, A. A. (1988). Consistent breakpoints in region 14q22-q24 in uterine leiomyoma. Cancer Genet. Cytogenet. 32, 25–31. doi: 10.1016/0165-4608(88)90307-X
Vallbo, C., and Damber, J. E. (2005). Thrombospondins, metallo proteases and thrombospondin receptors messenger RNA and protein expression in different tumour sublines of the Dunning prostate cancer model. Acta Oncol. 44, 293–298. doi: 10.1080/02841860410002806
Viechtbauer, W. (2010). Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48.
Vikhlyaeva, E. M., Khodzhaeva, Z. S., and Fantschenko, N. D. (1995). Familial predisposition to uterine leiomyomas. Int. J. Gynaecol. Obstet. 51, 127–131. doi: 10.1016/0020-7292(95)02533-I
Walker, C. L., and Stewart, E. A. (2005). Uterine fibroids: the elephant in the room. Science 308, 1589–1592. doi: 10.1126/science.1112063
Wei, J. J., and Soteropoulos, P. (2008). MicroRNA: a new tool for biomedical risk assessment and target identification in human uterine leiomyomas. Semin. Reprod. Med. 26, 515–521. doi: 10.1055/s-0028-1096131
Wise, L. A., Palmer, J. R., Harlow, B. L., Spiegelman, D., Stewart, E. A., Adams-Campbell, L. L., et al. (2004). Reproductive factors, hormonal contraception, and risk of uterine leiomyomata in African-American women: a prospective study. Am. J. Epidemiol. 159, 113–123. doi: 10.1093/aje/kwh016
Wise, L. A., Ruiz-Narvaez, E. A., Palmer, J. R., Cozier, Y. C., Tandon, A., Patterson, N., et al. (2012). African ancestry and genetic risk for uterine leiomyomata. Am. J. Epidemiol. 176, 1159–1168. doi: 10.1093/aje/kws276
Yao, B., La, L. B., Chen, Y. C., Chang, L. J., and Chan, E. K. (2012). Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep. 13, 1102–1108. doi: 10.1038/embor.2012.160
Keywords: uterine fibroids, tumor size, thrombospondin, extracellular matrix, meta-analysis, genetic association, African American, reproductive medicine
Citation: Aissani B, Zhang K and Wiener H (2015) Evaluation of GWAS candidate susceptibility loci for uterine leiomyoma in the multi-ethnic NIEHS uterine fibroid study. Front. Genet. 6:241. doi: 10.3389/fgene.2015.00241
Received: 06 March 2015; Accepted: 29 June 2015;
Published: 14 July 2015.
Edited by:
Kari North, University of North Carolina at Chapel Hill, USAReviewed by:
Inga Reynisdóttir, Landspitali - The National University Hospital of Iceland, IcelandGeetha Chittoor, University of North Carolina at Chapel Hill, USA
Jennelle Hodge, Cedars-Sinai Medical Center, USA
Copyright © 2015 Aissani, Zhang and Wiener. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brahim Aissani, Department of Epidemiology, R217J, University of Alabama at Birmingham School of Public Health, 1665 University Blvd, Birmingham, AL, USA,YmFpc3NhbmlAdWFiLmVkdQ==
 Brahim Aissani
Brahim Aissani Kui Zhang
Kui Zhang Howard Wiener
Howard Wiener