
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 19 March 2015
Sec. Nutrigenomics
Volume 6 - 2015 | https://doi.org/10.3389/fgene.2015.00081
This article is part of the Research Topic The Human Gutome: Nutrigenomics of Host-Microbiome Interactions View all 8 articles
The availability of host and dietary carbohydrates in the gastrointestinal (GI) tract plays a key role in shaping the structure-function of the microbiota. In particular, some gut bacteria have the ability to forage on glycans provided by the mucus layer covering the GI tract. The O-glycan structures present in mucin are diverse and complex, consisting predominantly of core 1-4 mucin-type O-glycans containing α- and β- linked N-acetyl-galactosamine, galactose and N-acetyl-glucosamine. These core structures are further elongated and frequently modified by fucose and sialic acid sugar residues via α1,2/3/4 and α2,3/6 linkages, respectively. The ability to metabolize these mucin O-linked oligosaccharides is likely to be a key factor in determining which bacterial species colonize the mucosal surface. Due to their proximity to the immune system, mucin-degrading bacteria are in a prime location to influence the host response. However, despite the growing number of bacterial genome sequences available from mucin degraders, our knowledge on the structural requirements for mucin degradation by gut bacteria remains fragmented. This is largely due to the limited number of functionally characterized enzymes and the lack of studies correlating the specificity of these enzymes with the ability of the strain to degrade and utilize mucin and mucin glycans. This review focuses on recent findings unraveling the molecular strategies used by mucin-degrading bacteria to utilize host glycans, adapt to the mucosal environment, and influence human health.
The human gastrointestinal (GI) tract harbors a complex and dynamic population of microorganisms which contribute significantly to the maintenance of health and the onset and progression of disease (Sommer and Backhed, 2013). The intestinal epithelium surface is covered by a layer of mucus which differs in terms of composition, organization, and thickness along the GI tract (Pullan et al., 1994; Linden et al., 2008; Johansson et al., 2011; Juge, 2012; Ermund et al., 2013). In the colon, the mucus is divided into an outer layer which provides a nutrient-rich habitat for the microbiota and an inner layer firmly attached to the surface of the epithelium, and virtually free of bacteria (Johansson et al., 2008, 2011). There is an emerging paradigm that mucus is critical to the maintenance of a homeostatic relationship between the gut microbiota and their hosts, with subtle deviations from this dynamic interaction potentially resulting in major implications for health, among which are colitis, colorectal cancer, and susceptibility to infection, as extensively reviewed (McGuckin et al., 2011; Hansson, 2012; Sheng et al., 2012; Chen et al., 2014). Recent findings showed that alterations in mucosal carbohydrate availability impact on the composition of microbial species (Martens et al., 2008; Ng et al., 2013; Tong et al., 2014). The mucosal subpopulation is in a prime position to influence host immune responses. This review is focussed on the nutritional strategies used by gut bacteria to proliferate into the mucosal environment.
Mucins are the main structural components of mucus and play an integral and multifaceted role in the interaction between microbes and epithelial surfaces (Linden et al., 2008; Johansson et al., 2011). The expression profile of mucins varies among host tissues and particularly within the GI tract, which displays the highest and most diverse levels of mucin expression in the body (Linden et al., 2008). To date, more than 20 genes encoding mucins have been identified in humans, with their classification based on the arrangement of their monomeric polypeptide domains (Corfield, 2015). Mucins are broadly grouped as membrane-bound or secreted proteins (Moran et al., 2011; Corfield, 2015) (Table 1). Common to each mucin are an N-terminal signal peptide and a proline-threonine-serine (PTS) domain (Figure 1). The signal peptide is required for the targeting of the polypeptide to the endoplasmic reticulum (ER) and either extracellular secretion or insertion of the synthesized mucin into the cell membrane. The PTS domain is the site of extensive O-glycosylation with carbohydrates accounting for up to 80% of the total mucin mass (Gendler and Spicer, 1995). These PTS domains, comprised of variable number of tandem repeat (VNTR) domains, allow for a great degree of heterogeneity in mucins, due to variability in both, mucin length and extent of glycan attachment at these sites. This characteristic filamentous protein backbone decorated with outwardly protruding oligosaccharides results in the typical “bottle-brush”-like appearance of mucins (Bergstrom and Xia, 2013).
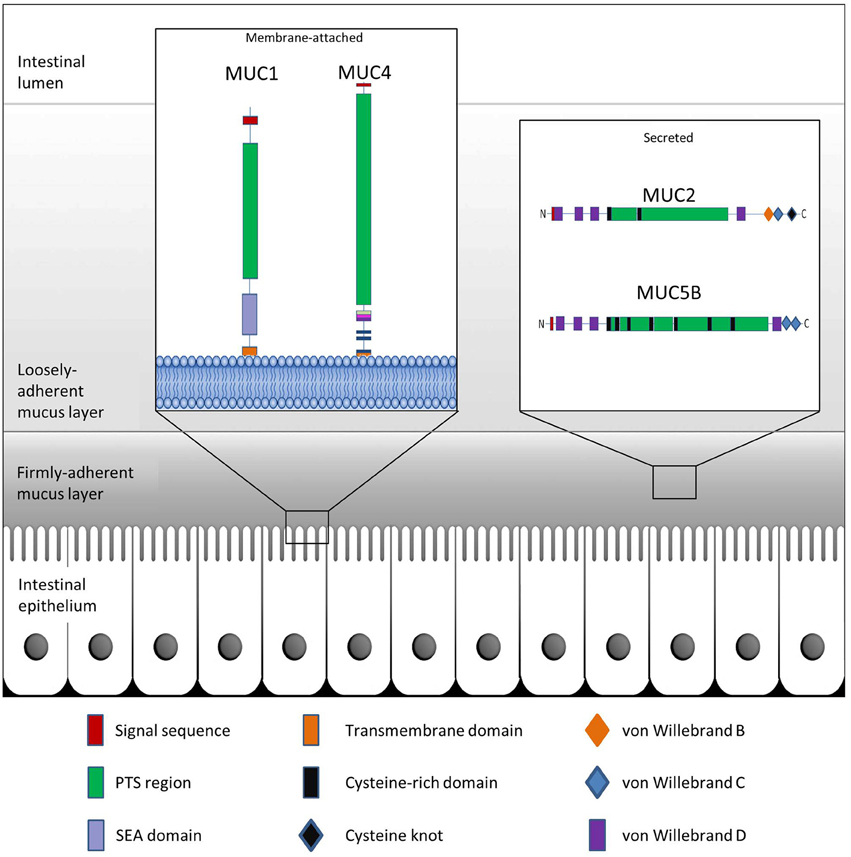
Figure 1. Schematic representation of GI mucus and mucins. The colonic epithelium is protected by mucin glycoproteins which are either membrane-attached (e.g., MUC1 and MUC4) or are secreted from goblet cells into the intestinal lumen (e.g., MUC2 and MUC5B). The secreted mucins further create a protective boundary in the form of a tightly-adherent mucus layer, which is devoid of bacteria, and a loosely-adherent mucus layer which provides a niche for intestinal bacteria.
Membrane-bound mucins are essential contributors of the glycocalyx of mucosal surfaces where they play important biological roles in cell-cell and cell-matrix interactions, and in cell signaling (Jonckheere et al., 2010). These mucins may be shed from the surface and integrate into the overlying mucus layer where they are able to influence the viscosity of the protective layer (Carrington et al., 2009). Secreted mucins are the main structural components of the mucus gel. Along the GI tract, synthesis and secretion of these polymeric glycoproteins take place in the goblet cells of the small intestine and colon, or the surface mucous cells of the stomach (Moncada et al., 2003). Characteristic properties of the secreted mucins are disulfide bridges which are formed among the cysteine-rich, cysteine knot, and von Willebrand C and D domains residing at the N- or C-termini of the glycoprotein monomers (Desseyn et al., 2008; Ambort et al., 2011) (Figure 1). MUC2 is the best characterized secreted mucin of the GI tract (Nilsson et al., 2014). Within the ER, newly synthesized MUC2 peptides immediately dimerize through the formation of disulfide bridges followed by transport to the Golgi apparatus (Asker et al., 1995, 1998; Ambort et al., 2012b). Here, the PTS domains of the mucin dimers are sites of elaborate glycosylation before further assembly into trimers in the trans-Golgi network and packaging into goblet cell vesicles in a pH- and Ca2+-dependent manner (Godl et al., 2002; Ambort et al., 2012a; Johansson and Hansson, 2012). As a monomer, fully glycosylated MUC2 exhibits a large size of approximately 2.5 MDa, while extensive polymerization may allow for sizes of greater than 100 MDa (Johansson et al., 2011). Following secretion of the mucin granules at the mucosal surface, the densely-packed mucin structures are hydrated and rapidly expand to a size approximately 3000-fold greater than in the granules, thus providing a dynamic barrier (Verdugo, 2012).
In addition to their protective and lubricating activities, mucins facilitate microbial tropism through the presentation of glycans which may impact colonization (e.g., Kobayashi et al., 2009; Gonzalez-Rodriguez et al., 2012; Etzold et al., 2014, for a review see Juge, 2012), and act as a nutritional source for microorganisms (e.g., Ruas-Madiedo et al., 2008; Crost et al., 2013, for a review see Marcobal et al., 2013). As such, mucin glycans have been proposed to play a key role in selecting microbial communities along and across the GI tract. Consistent with this hypothesis, recent studies in mouse models and humans showed an association between alteration in mucin glycosylation profile and deviations of overall community ecology along with altered abundances of specific microbes (Rausch et al., 2011; Wacklin et al., 2011, 2014; Pacheco et al., 2012; Kashyap et al., 2013; Sommer et al., 2014).
Mucins carry a vast array of oligosaccharide structures, with the glycosyltransferase profile expressed by the host determining the type of linkages and glycan structures present on the secreted mucins (Linden et al., 2004). The synthesis of mucin oligosaccharides starts with the transfer of N-acetylgalactosamine (GalNAc) to Ser and Thr residues of the mucin core (Bennett et al., 2012). Eight core structures of the mucin O-glycan chain have been identified (Brockhausen et al., 2009), with core 1-4 glycans most commonly found in intestinal mucins (Figure 2A). In humans, gastric and duodenal mucins generally contain the core 1 (Galβ1–3GalNAcα1-Ser/Thr) and the core 2 (Galβ1,3(GlcNAcβ1,6)GalNAcα1-Ser/Thr) structures (Robbe et al., 2004; Larsson et al., 2009), while the core 3 (GlcNAcβ1,3GalNAcαSer/Thr) structure is predominant in the small intestine (Robbe et al., 2004), and core 3 and 4 (GlcNAcβ1,6(GlcNAcβ1,3)GalNAcαSer/Thr) structures make up the majority of colonic mucin glycans (Robbe et al., 2004; Robbe-Masselot et al., 2009; Xia, 2010). Further studies revealed that MUC2 in the sigmoid colon mainly contains the core 3 structure (Robbe et al., 2004; Thomsson et al., 2012). These core structures can be further extended with galactose (Gal), N-acetylglucosamine (GlcNAc), GalNAc, fucose or sialic acid (Neu5Ac) sugar residues with the latter two frequently occupying terminal positions (Brockhausen et al., 2009). Alterations in mucin glycosylation have been associated with a number of diseases such as colitis, colonic cancer and inflammatory bowel diseases in humans (e.g., McGovern et al., 2010; Larsson et al., 2011; Rausch et al., 2011; Parmar et al., 2012; Forni et al., 2014) and mouse models (e.g., An et al., 2007; Stone et al., 2009; Fu et al., 2011). However, more work is needed to support the causal link between altered O-linked glycosylation and inflammation, as recently reviewed in Theodoratou and colleagues (Theodoratou et al., 2014).
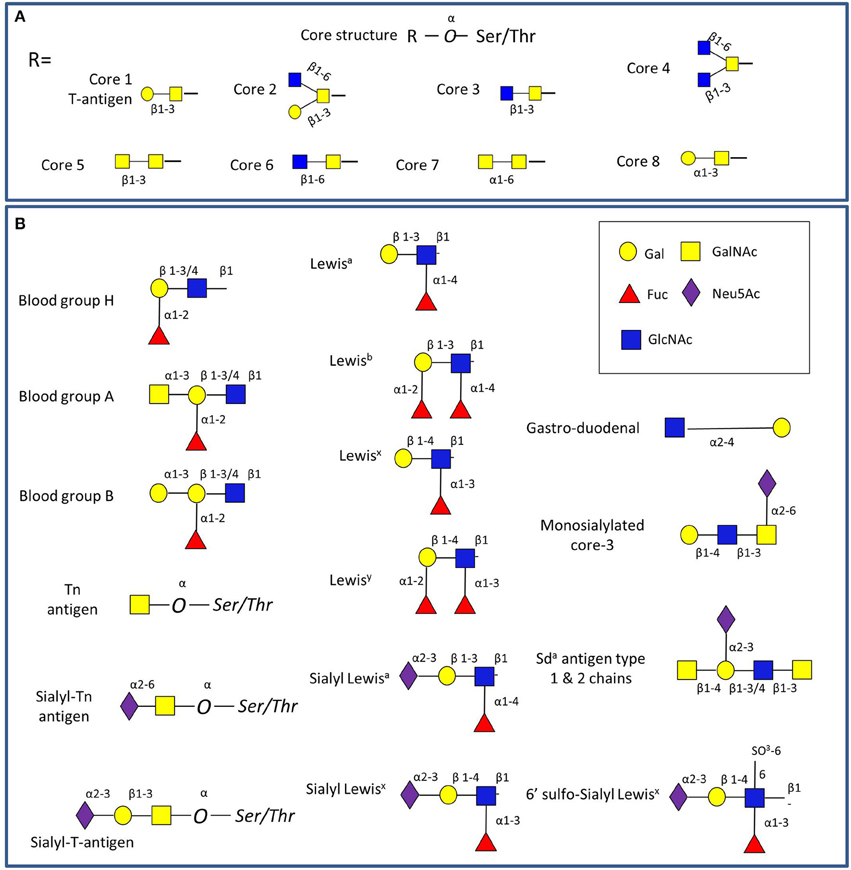
Figure 2. Schematic representation of GI mucin glycans. (A) The four common mucin type O-glycans (core 1-8) found in the GI tract. (B) Main glycan epitopes in GI mucins. The glycan sugars are represented using Glycan Builder (Ceronietal., 2007).
The main source of glycan diversity is provided by the peripheral terminal epitopes that show considerable variation (Figure 2B). The H1 structure (α1,2-fucose) is found in populations carrying the secretor gene (Mollicone et al., 1985), and individuals may also express the Lewis gene and the Leb histo-blood group antigen if they are secretors, while non-secretors express Lea (Kelly et al., 1995). Another phenotype (SeW—weak secretor) is characterized by the expression of both Lea and Leb antigens (Henry et al., 1995; Lindén et al., 2008). The presentation of the major mucin glycan epitopes, sialic acid and fucose, varies along the GI tract following opposing gradients with a decreasing gradient of fucose and ABH blood group expression and an increasing gradient of sialic acid from the ileum to the colon (Robbe et al., 2004). Interestingly, these gradients are reversed in mice, where the small intestine is dominated by sialylated structures and the colon with those terminating in fucose, potentially creating the need for additional considerations in the comparison of human and murine colonization studies (Holmen Larsson et al., 2013). These terminal mucin O-glycans have been proposed to serve as metabolic substrates, providing a nutritional advantage to bacteria which have adapted to the GI mucosal environment (Freitas et al., 2003; Severi et al., 2007; Pacheco et al., 2012; Vimr, 2013). Conversely, gut bacteria have the ability to affect the mucus barrier (Jakobsson et al., 2015) and mucin glycosylation (Hooper et al., 1999; Pickard et al., 2014).
The GI tract is heavily colonized by bacteria with most species belonging to the phyla Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Verrucomicrobia. The microbiota composition varies longitudinally along the GI tract but also transversally from the mucosa to the lumen (Zoetendal et al., 2002; Eckburg et al., 2005; Carroll et al., 2010). Defining the mucosa-associated bacteria composition is hampered by difficulties in (i) sampling, limiting the number and power of human studies, (ii) differences in the nature or definition of the mucosa samples (biopsies, mucus, or rectal swabs), and (iii) intrinsic inter-individual variability at the family, genera, or species level (see for example Hong et al., 2011). However, it is now clear that the composition of the mucosa-associated microbiota differs from that of the fecal microbiota in terms of relative abundance of the different phyla, although the results may vary between studies (Swidsinski et al., 2005; Frank et al., 2007; Carroll et al., 2010). For example, the percentage of Bacteroidetes phylum was shown to be higher in the colonic biopsies or rectal swabs of healthy human volunteers compared to the feces (Eckburg et al., 2005; Chen et al., 2012). In contrast, Firmicutes were enriched in the mucosa-associated bacteria of mice, especially members of the Lachnospiraceae and Ruminococcaceae families (Nava et al., 2011). Similarly, Van den Abbeele and collaborators showed that the Firmicutes phylum, especially members of Clostridium cluster XIVa, was significantly enriched in the mucus layer as opposed to the lumen, using an in vitro gut model inoculated with human fecal samples (Van Den Abbeele et al., 2013). These differences may be due to the different models used in these studies (e.g., in vivo vs. in vitro; human vs. mice) and sampling methods. In addition, a number of studies have focused on specific bacterial groups or species. For example, sulfate-reducing bacteria, acetogenic bacteria, and methanogenic archaea were shown to preferentially colonize the healthy human colon mucosa (Nava et al., 2012). An early study reported that Lactobacillus gasseri was a predominant Lactobacillus species in human biopsy samples (Zoetendal et al., 2002) whereas Bifidobacterium bifidum and Bifidobacterium longum were shown to be more abundant in the mucosa of germ-free rats inoculated with human fecal microbiota than in the lumen (Van Den Abbeele et al., 2011). Faecalibacterium prausnitzii, an abundant member of the microbiota with putative anti-inflammatory properties, has also been found in ileal, colonic, and rectal biopsies from healthy individuals (Lopez-Siles et al., 2014).
In the colon, the epithelium is covered by a thick gel of mucus, divided into two layers, an inner layer firmly attached to the epithelium and a loose outer layer (Atuma et al., 2001; Ermund et al., 2013). Johansson and collaborators demonstrated that the outer mucus layer is heavily colonized by bacteria, while the inner layer contains no or very few bacteria (Johansson et al., 2008). It is thus believed that in healthy conditions mucosa-associated bacteria are not in direct contact with the epithelium but are restricted to the outer mucus layer. Although the molecular mechanisms underpinning the adaptation of gut bacteria to mucus remain unclear, it is likely that their ability to utilize mucin glycans as a source of nutrients would confer a competitive advantage to those bacteria with the required repertoire of hydrolytic enzymes. The first mucin-degrading (or mucinolytic) bacteria studied were pathogens (e.g., Levy and Aminoff, 1980; Prizont, 1982; Slomiany et al., 1992), and thus for a long period mucin degradation had been associated with pathogenicity. However, it is now clear that mucin degradation is part of a normal turn-over process starting a few months after birth (Norin et al., 1985). To date, only a limited number of bacterial species/strains from the Bacteroidetes, Firmicutes, Actinobacteria, and Verrucomicrobia phyla have been studied for their ability to consume mucins (see below and Table 2).
The mucin-degrading ability of gut bacteria has been extensively studied in Bacteroidetes. An early study showed that all 22 strains of Bacteroides thetaiotaomicron tested were able to ferment glycosaminoglycans (GAG) but failed to ferment pig gastric mucin (PGM) or bovine submaxillary mucin (BSM) (Salyers et al., 1977a). However, later, B. thetaiotaomicron VPI-5482 was shown to be able to grow on different fractions of glycans purified from pig gastric mucosa, including an O-glycan rich fraction (Martens et al., 2008). Transcriptomic analyses highlighted specific polysaccharide-utilization loci (PULs) including genes coding for putative glycoside hydrolases (GHs) such as α-L-fucosidase, endo-β-N-acetylglucosaminidase, endo-β-galactosidase and α-mannosidase, which were up-regulated when B. thetaiotaomicron was grown on mucin O-glycans or in monoxenic mice as compared to in vitro glucose control. Interestingly, these PULs were not up-regulated when B. thetaiotaomicron was grown on GAG, as compared to glucose (Martens et al., 2008, 2011). Colonization competition experiments demonstrated that B. thetaiotaomicron mutants for O-glycan PULs were able to colonize germ-free mice in a similar way as the wild-type strain when mice were fed a plant glycan-rich diet, but were outcompeted by the wild-type on a simple-sugar diet (Martens et al., 2008). This indicates that B. thetaiotaomicron relies on mucin and other host-derived glycans for colonization. Genome analysis of Bacteroides fragilis confirmed that Bacteroides species contain a much larger number of genes encoding carbohydrate-active enzymes (CAZymes) compared to other sequenced gut bacteria (Kuwahara et al., 2004). In accordance with early studies demonstrating the ability of some B. fragilis strains to grow on mucin as sole carbon source (Salyers et al., 1977a; Roberton and Stanley, 1982), the B. fragilis genome contains a subset of PULs dedicated to host mucin O-glycan utilization; in particular, it has been shown that (i) loci involved in the binding, degradation, and transport of sialylated polysaccharides play an important role in the colonization of this bacterium in the gut (Nakayama-Imaohji et al., 2012) and (ii) the genes involved in sialic acid utilization are up-regulated when B. fragilis is grown in the presence of mucin O-glycans as compared to glucose (Marcobal et al., 2011). Some strains of Bacteroides vulgatus have also been shown to moderately degrade PGM but failed to utilize human MUC2 (Hoskins et al., 1992; Png et al., 2010) (Table 2).
In the Firmicutes phylum, Ruminococcus torques and Ruminococcus gnavus, both members of the Lachnospiraceae family (belonging to the C. coccoides group/cluster XIVa) have been shown to degrade mucins. In an early study, six R. torques strains out of nine tested, but none of the R. gnavus strains tested, were shown to have the capacity to ferment PGM (Salyers et al., 1977b). A few years later, R. gnavus ATCC 35913 and two R. torques strains (ATCC 35915 and VIII-239) were among the five strains isolated from human fecal samples for their ability to degrade mucins (Hoskins et al., 1985). Png and collaborators then confirmed that both R. gnavus and R. torques species were able to degrade and utilize human MUC2 as a sole carbon source (Png et al., 2010), providing further evidence of their adaptation to the human colonic mucosal environment. Several enzymatic activities were detected in the spent media of these strains grown with mucin that could explain their ability to degrade mucin (Hoskins et al., 1985, 1992; Corfield et al., 1992; Crost et al., 2013) (Table 2). Recently, the ability of R. gnavus strains to utilize mucin was shown to be strain-dependent, as also supported by comparative genomic and transcriptomic analyses (Crost et al., 2013), and in agreement with earlier findings (Salyers et al., 1977b).
B. bifidum ATCC 35914 and B. longum subsp. infantis VIII-240, from the Actinobacteria phylum, were also isolated by Hoskins and collaborators (Hoskins et al., 1985) as mucin degraders, and several enzymatic activities possibly involved in mucin degradation were detected in the spent media of these strains grown with mucin as sole carbon source (Hoskins et al., 1985, 1992; Corfield et al., 1992) (Table 2). Since then, B. longum subsp. infantis has been shown to grow on mucins, albeit moderately (Abe et al., 2010; Turroni et al., 2010; Kim et al., 2013), and the ability of B. bifidum to utilize mucins has been confirmed for several strains using different types of mucins, including human MUC2 (Ruas-Madiedo et al., 2008; Png et al., 2010; Turroni et al., 2010) (Table 2). Transcriptomic analyses of B. bifidum L22 and PLR2010 suggest the involvement of several enzymes in the process, e.g., α-L-fucosidase and endo-α-N-acetylgalactosaminidase (Ruas-Madiedo et al., 2008; Turroni et al., 2010) (Table 2). Few other Bifidobacteria species, including some strains of B. longum subsp. longum and Bifidobacterium breve, have also been shown to degrade mucins in vitro (Ruas-Madiedo et al., 2008; Turroni et al., 2010; Ruiz et al., 2011) (Table 2). β-N-acetylglucosaminidase and β-glucuronidase activities were increased in the spent medium of B. longum subsp. longum NCIMB8809 grown with human intestinal mucus, suggesting a role of these activities in mucin degradation. More recently, detailed genome analyses of Bifidobacteria have identified metabolic pathways for the degradation of mucin-type O-glycans and human milk oligosaccharides (HMOs) (Sela et al., 2008; Turroni et al., 2010) and several GHs have been functionally characterized supporting these findings (Yamamoto et al., 2010; Kiyohara et al., 2011) (see section below).
In the Verrucomicrobia phylum, Akkermansia muciniphila, a strictly anaerobic Gram-negative bacterial species, was recently identified as a key mucin degrader (Derrien et al., 2004). Initially isolated from a human fecal sample due to its ability to utilize mucins as sole carbon and nitrogen source, A. muciniphila has since been shown to be a common member of the human gut with a high prevalence and variable abundance, present both in feces and at the mucosal surface (Eckburg et al., 2005; Collado et al., 2007; Derrien et al., 2008). When A. muciniphila ATCC BAA-835 was grown with mucin as sole carbon source, several enzymatic activities potentially involved in mucin degradation were detected both in the spent medium and intracellularly (Derrien, 2007). However, although numerous genes encoding putative mucinolytic enzymes were found in the genome of the ATCC BAA-835 strain (Van Passel et al., 2011), the functional characterization of these proteins, and thus their role in mucin degradation, has not yet been reported.
Given the diversity and complexity of intestinal mucin glycan structures, strategies for deconstructing these molecules rely on the cooperative action of a number of proteases, sulfatases, and GHs encoded by the genome of mucin-degrading bacteria. These GHs include, but are not limited to, neuraminidases/sialidases (GH33), fucosidases (GH29 and GH95), exo- and endo-β-N-acetylglucosaminidases (GH84 and GH85), β-galactosidases (GH2, GH20, and GH42), α-N-acetylglucosaminidases (GH89), and α-N-acetylgalactosaminidases (GH101, GH129) (www.cazy.org/) (Lombard et al., 2014) (Figure 3 and Table S1).
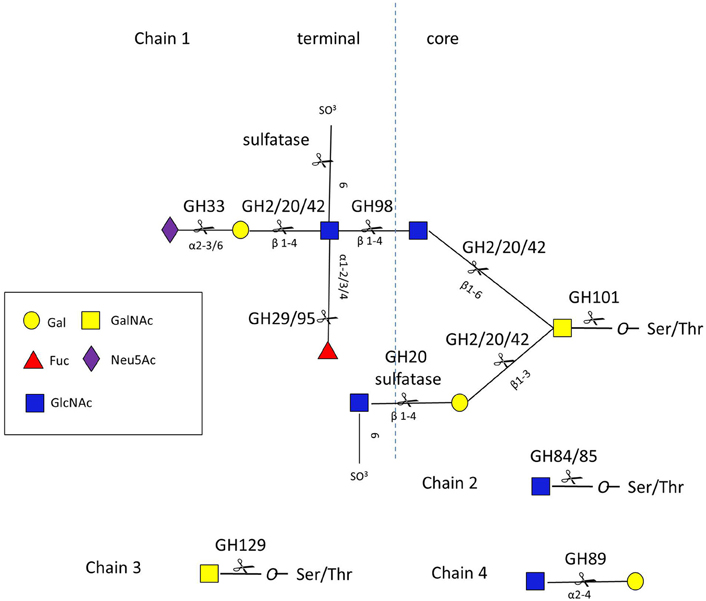
Figure 3. O-glycan chains showing sites of action of GHs and sulfatases. Chain 1 is a hypothetical mucin glycan chain, chain 2 is O-GlcNAc often found on other glycoproteins, chain 3 (Tn antigen) and chain 4 are found in gastro-duodenal mucin.
However, in sharp contrast to the number of genes predicted to be involved in mucin degradation (mainly based on transcriptomic analyses), only a few GHs have been biochemically characterized (see CAZy database; www.cazy.org). Detailed enzymatic characterization of putative mucin-degrading enzymes, and CAZymes in general, is essential for allowing accurate annotation of homologous gene sequences. Indeed, the process of reliable annotation of protein function from gene sequence is one of the main challenges for the effective use of metaproteomic data from the human microbiota (Kolmeder and De Vos, 2014). Currently, 46 bacterial GHs involved in mucin degradation have been functionally characterized, either by recombinant means, following identification of putative target genes by genomic analysis, or by purifying the bacterial enzyme following an activity screening (Table S1). Given the large diversity and complexity of mucin glycosylation, and the lack of amenable sources of mucins, functional characterization of mucin-degrading GHs often relies on the use of mucin-type oligosaccharides or synthetic substrates (such as para-Nitrophenol (pNP)- or 4-Methylumbelliferone (MU)-derivatized glycans) as surrogate substrates (Table S1). Owing to the paucity of purified proteins, only limited structural information is available on mucin-degrading enzymes from human gut bacteria, e.g., the absence of a GH129 crystal structure (www.cazy.org).
In addition to their catalytic domains, GHs may have one or more carbohydrate binding modules (CBMs) which mediate the adherence of CAZymes to their carbohydrate substrate. Currently, CBMs that recognize mucin glycans have been reported in families 32, 40, 47, and 51, as reviewed in Ficko-Blean and Boraston (2012a). These CBMs show specificity for terminal glycan motifs, such as Gal, GlcNAc, sialic acid, fucose, and histo-blood group antigens (Etzold and Juge, 2014). Other non-catalytic domains associated with these GHs include immunoglobulin domains, concanavalin A domains, or domains of unknown function (Table S1).
The genomic organization of mucin-degrading enzymes has been studied in few bacterial species. Bacteroidetes employ a highly organized system for glycan utilization whereby all genes involved in the degradation of dietary- or host-derived carbohydrate (GHs, sugar transport proteins, sugar sensors, regulatory proteins, etc.) are grouped together in a single PUL, as recently reviewed in Martens et al. (2009). Starch utilization systems (Sus) or Sus-like proteins have been particularly well studied in B. thetaiotaomicron (Martens et al., 2009). SusD proteins are cell envelope-associated proteins that mediate glycan binding, and a SusD like protein BT1043 of B. thetaiotaomicron has been implicated in O-glycan utilization of host mucins (Martens et al., 2008). Recently, the sialic acid utilizing protein NanU, a SusD family protein from B. fragilis, has been demonstrated to bind Neu5Ac with high affinity (Phansopa et al., 2014). Such organization in PULs is less apparent in Firmicutes. Generally the genes involved in the utilization of particular mucin glycans are clustered together in operons. For example, sialidases are often found in a cluster with other proteins involved in sialic acid catabolism (see below), and B. longum subsp. infantis has clusters dedicated to the metabolism of HMOs which share structural similarities with mucin core glycans (Sela et al., 2008).
The release of sialic acid from non-reducing ends is an initial step in the sequential degradation of mucins since the terminal location of sialic acid residues in the mucin oligosaccharide chains may prevent the action of other GHs. In bacteria, the genes involved in sialic acid metabolism are usually found clustered together forming what is denominated as a Nan cluster. Human gut bacteria that encode a Nan cluster include R. gnavus, Anaerotruncus colihominis, Dorea formicigenerans, Dorea longicatena, F. prausnitzii, Fusobacterium nucleatum, Lactobacillus sakei, Lactobacillus plantarum, and Lactobacillus salivarius (Almagro-Moreno and Boyd, 2009), B. fragilis (Brigham et al., 2009), and B. breve (Egan et al., 2014). Thus, the majority of the bacteria that harbor a Nan cluster colonize mucus regions of the human body, such as the gut, lung, bladder, or oral cavity, where sialic acid is highly abundant and can serve as a source of energy, carbon, and nitrogen (Almagro-Moreno and Boyd, 2009). However, some bacteria appear to have incomplete packages of enzymes for utilizing host sialic acids. For example, B. thetaiotaomicron encodes a sialidase and can release free sialic acid, but lacks the Nan operon required to consume the liberated monosaccharide and does not appear capable of consuming free sialic acid (Marcobal et al., 2011). On the other hand, Salmonella typhimurium and Clostridium difficile encode the Nan operon but each lacks the sialidase (Hoyer et al., 1992; Sebaihia et al., 2006), and thus rely on other sialidase-producing organisms to acquire this potential nutrient source from the mucosal environment (Vimr et al., 2004; Ng et al., 2013).
GH33 sialidases encoded by human gut bacteria vary in terms of their substrate specificity and enzymatic reaction. Although most of them are hydrolytic sialidases, releasing sialic acid from sialylated substrates, some display transglycosylation activities (see Table S1). For example the sialidase from B. bifidum JCM 1254 can transfer Neu5Ac to 1-alkanols, that of R. gnavus ATCC 29149 (RgNanH) is an intramolecular trans-sialidase (IT-sialidase) which releases 2,7 anhydro-Neu5Ac specifically from α2,3-linked sialyl conjugates (Crost et al., 2013), and NanI from Clostridium perfringens str 13 can hydrate the inhibitor 2-deoxy-2,3-dehydro-Neu5Ac to Neu5Ac (Newstead et al., 2008). Trans-sialidases show specificity for α2,3 linkages, whereas hydrolytic sialidases can often cleave α2,3, 2,6, or 2,8 linkages (e.g., B. thetaiotaomicron sialidase BTSA). However, the substrate specificity of many sialidases remains unknown as MU-Neu5Ac is often the only tested substrate (Table S1). Furthermore, few of these purified sialidases have been enzymatically characterized using mucin as a natural substrate (Table S1). Sialidases have been identified in the genomes of infant-derived Bifidobacteria, including two intracellular sialidases from B. longum subsp. infantis ATCC 15697 (Sela et al., 2008), two predicted extracellular exo-α-sialidases of B. bifidum PRL 2010 (Turroni et al., 2010), and a putative sialidase from B. breve (Egan et al., 2014). However, the only sialidase from this group of infant-associated bacteria to be functionally characterized is SiaBb2 from B. bifidum JCM 1254, a strain for which the genome sequence is not yet publicly available. SiaBb2 has a strong preference for α2,6 linkages and was shown to be sufficient to confer B. longum 105-A with the ability to degrade HMOs (Kiyohara et al., 2011).
In mucins, fucosyl residues can be found at the extremity of the O-glycosidic chain linked to galactose by α1,2 linkages or to GlcNAc by α1,3 linkages, whereas it is most commonly linked α1,6 to the reducing, terminal β-GlcNAc in human N-linked glycans. Fucosidase-encoding genes are widely distributed in the genome of gut bacteria and generally belong to GH29 and GH95 families, which differ in their enzymatic mechanisms; GH29 enzymes retain the anomeric conformation of the glycosidic bond (Katayama et al., 2005) whereas GH95 enzymes proceed via the inverting mechanism (Nagae et al., 2007). Transcriptional data suggest that GH29 and GH95 fucosidases play a key role in the ability of B. thetaiotaomicron VPI-5482 (Martens et al., 2008), B. longum subspecies infantis ATCC 15697 (Sela et al., 2012), B. bifidum JCM 1254 (Ashida et al., 2009), and R. gnavus ATCC 29149 (Crost et al., 2013) to utilize mucins as a source of carbon. However, the enzymatic characterization of members of the GH95 and GH29 family is often hampered by the fact that most of these enzymes are not active against synthetic fucosyl conjugates (Katayama et al., 2005), preventing high throughput activity screening. It is notable that among 495 bacterial GH95 enzymes being listed in the CAZy database to date (02 October 2014), only two have been biochemically characterized, one from B. longum subspecies infantis ATCC 15697 and one from B. bifidum JCM 1254 (Table S1), both from human gut commensal strains.
The substrate specificity of GH29 and GH95 fucosidases has been predominantly characterized in B. bifidum JCM 1254, where GH95 AfcA was shown to be specific for α1,2 linkages (Katayama et al., 2005) and GH29 AfcB for α1,3 and α1,4 linkages (Ashida et al., 2009); together these enzymes can remove fucose at the non-reducing termini except for those that are α1,6-linked (Ashida et al., 2009). AfcA and AfcB have been shown to be sufficient to confer B. longum 105-A with the ability to grow on 2-fucosyllactose (2′FL), 3-fucosyllactose (3′FL) and lacto-N-fucopentaose (LNFPII) (Ashida et al., 2009). The structural basis for the specificity of AfcA has been determined (Nagae et al., 2007).
B. thetaiotaomicron produces multiple fucosidases that cleave fucose from host glycans, resulting in high fucose availability in the gut lumen (Xu et al., 2003). The genome of B. thetaiotaomicron VPI-5482 encodes five GH95 and nine GH29 genes (www.cazy.org). Two of the GH29 genes have been expressed and shown to have α-fucosidase activity and have been classified in separate sub-families, i.e., GH29-A (BT_2970) has a relaxed specificity that can accommodate pNP-fucose (pNP-Fuc), whereas GH29-B (BT_2192) is specific for branched fuco-oligosaccharides found in Lewis blood groups (also present in mucin structures, see Figure 2B) (Sakurama et al., 2012) (Table S1). The structural basis for this specificity between the two B. thetaiotaomicron fucosidases was first expounded by Sakurama et al. (2012) using the structures from BT_2970 (GH29-A), (Lammerts Van Bueren et al., 2010) and BT_2192 (GH29-B) (PDB 3EYP, www.rcsb.org). Further structural analysis of BT_2192 elucidated the molecular mechanisms for the binding of the branched oligosaccharides and the unusual dual specificity of this enzyme, which also acts as a β-galactosidase (Guillotin et al., 2014). More detailed analysis of the substrate specificity of fucosidases is warranted to determine why B. thetaiotaomicron, and indeed other resident members of the human microbiota, have evolved to produce multiple α-fucosidases.
Both the blood group A antigen and B antigen can be cleaved from mucin by GH98 endo-β1,4-galactosidases, these have been characterized in Clostridium perfringens strains 10543 and 13 (Anderson et al., 2005; Table S1; Figure 3). The only structural information about this family of enzymes comes from Streptococcus pneumoniae str. and reveals a (α/β)8 barrel (Higgins et al., 2009). Once the terminal sugars and blood group antigens are removed, the mucin core glycans are exposed to further enzymatic degradation.
Mucin glycan core structures are cleaved from the Ser/Thr amino acids of the mucin protein backbone by endo-α-N-acetylgalactosaminidases, with that of B. bifidum (EndoBF) being the founding member of GH101 (Fujita et al., 2005). These enzymes differ in their specificity toward core glycan structure types (Table S1). For example, EndoBF is specific for the core 1 glycan (Galβ1,3GalNAc) (Katayama et al., 2005) while endo-α-N-acetylgalactosaminidases from Enterococcus faecalis and C. perfringens have a broader specificity (Ashida et al., 2008; Goda et al., 2008; Koutsioulis et al., 2008). The structural basis for this specificity has been elucidated for EndoBF (Suzuki et al., 2009). The α-N-acetylgalactosaminidase from B. bifidum JCM 1254 is the founding member of GH129 and differs from GH101 in that it targets the Tn antigen (GalNAcα1-Ser) found in gastroduodenal mucins (Kiyohara et al., 2012). GH129 is a small family with only 58 members to date (02 Oct 14), all of which are of bacterial origin. Many of the species encoding a GH129 are associated with the infant microbiota (Kiyohara et al., 2012), although Bacillus sp. also contain GH129 (www.cazy.org).
GH2, GH20, and GH42 β-galactosidases have been implicated in the degradation of type-1 and type-2 HMOs. Genome analysis of several Bifidobacteria species identified common metabolic pathways for the degradation of lacto-N-biose I (Galβ1,3GlcNAc, LNB) and galacto-N-biose (Galβ1,3GalNAc, GNB), a building block of the core 1 structure of mucin-type O-glycan, whereas the degradation of type-2 lacto-N-neo-tetraose (Galβ1,4GlcNAcβ1,3Galβ1,4Glc, LNnT), also present in the core 2 mucin-type O-glycans, involves a different pathway. In Bifidobacteria, the type-1 chain (Galβ1,3GlcNAcβ1-) is likely eliminated by GH20 lacto-N-biosidases (LnbB) (Wada et al., 2008), and the released LNB incorporated into the cytosol via a GNB/LNB transporter (Suzuki et al., 2009). Despite the quite rare occurrence in nature of β-galactosidases acting on type-1 chains, close lacto-N-biosidase homologs are present in the genomes of infant-gut associated Bifidobacteria that are known to consume type 1 Lacto-N-tetraose (Galβ1,3GlcNAcβ1,3Galβ1,4Glc, LNT), these are the GH42 LNT β-galactosidases (Yoshida et al., 2012). In Bifidobacteria, the type-2 chain (Galβ1,4GlcNAcβ1-) is sequentially degraded by GH2 β-galactosidase, BbgIII, acting on LacNAc and GH20 β-N-acetylhexosaminidases, BbhI and BbhII, specific for GlcNAcβ1,3Galβ1-R (Miwa et al., 2010). Although GH2 is a very common glycosidase present in intestinal bacteria, the presence of membrane bound β-galactosidases is limited to several bifidobacterial strains. In contrast to β-galactosidases, GH20 β-N-acetylhexosaminidases are relatively rare in the genome of enteric bacteria (Miwa et al., 2010). An unusual activity was reported for a GH20 enzyme from Prevotella strain RS2; the enzyme cleaved terminal 6-SO3-GlcNAc from sulfated mucin glycans, representing a novel activity within the GH20 family (Rho et al., 2005). Other sulfatases have also been characterized (Table S1), but since these do not recognize glycosidic bonds, they are not classified in the CAZy database.
Few GH84 and GH85 β-N-acetylglucosaminidases have been characterized so far and although these enzymes have been implicated in mucin metabolism [e.g., C. perfringens str 13 (Ficko-Blean and Boraston, 2005), and B. longum NCC2705 (Schell et al., 2002)], none have been enzymatically characterized using mucin glycans as substrates (Table S1). The crystal structure of the GH84 from B. thetaiotaomicron VPI-5482 has been elucidated (Dennis et al., 2006), but the activity of this enzyme has been studied with protein-O-GlcNAc and not mucin glycans.
The GH89, α-N-acetylglucosaminidase (AgnC), from C. perfringens (str 13124) has been studied structurally (Ficko-Blean et al., 2008; Ficko-Blean and Boraston, 2012b) and the highly similar enzyme from C. perfringens str 13 has been demonstrated to be active against PGM and cell surface mucin (from adenocarcinoma AGSα4GnT cells stably expressing GlcNAcα1,4Gal as O-glycans on the cell surface). These studies suggest a role for AgnC in the release of terminal GlcNAc (Fujita et al., 2011) from “Class III” gastroduodenal mucins (Nakajima et al., 2003), similar to GH129 (above).
Members of the human gut microbiota strains typically display a subset of glycan-degrading phenotypes that equip them to target just part of the overall glycan repertoire present at certain times or locations (Koropatkin et al., 2012). The ability of mucin-degrading bacteria to forage on the diversity and abundance of glycans present in GI mucus may have a role in early colonization by providing some bacteria with an endogenous source of nutrients before the introduction of dietary glycans. Due to the chemical similarity of HMOs and O-linked mucin glycans, bacteria have developed common strategies for degrading these complex carbohydrates, as reported in B. thetaiotaomicron and Bifibobacteria (Turroni et al., 2010; Marcobal et al., 2011). Bifidobacteria possess two distinct pathways for assimilation of O-glycans on gastroduodenal and intestinal mucins; GH homologs involved in mucin and HMO utilization are conserved in infant-associated bifidobacteria, suggesting a significant role for their adaptation within the infant gut (Turroni et al., 2010; Kiyohara et al., 2012). A B. thetaiotaomicron deletion mutant for O-glycan utilization used in a germ-free mouse colonization experiments was outcompeted >200-fold relative to the wild-type and the complemented-mutant bacteria and pre-weaned pups selectively retained the mucin-degrading wild-type and complemented strains (Martens et al., 2008), suggesting that mucin degradation may confer an ecological advantage to the bacteria inhabiting the mammalian GI tract. R. gnavus is an early infant colonizer of the human intestine (Favier et al., 2002) and in the top 15 species showing abundance in both adult and infant gut-enriched genes, in line with its adaptation to the intestinal habitat throughout life (Hattori and Taylor, 2009). A recent study showed that R. gnavus was predominant in breast milk-/goat milk-fed microbiotas compared to a more diverse collection of Lachnospiraceae in cow milk-fed babies (Tannock et al., 2013). Taken together, these findings suggest that the ability to forage on mucin glycans in the infant GI tract may contribute to the ability of gut bacteria species to establish early colonization (Koropatkin et al., 2012).
Microbial communities that are strongly associated with the mucosa are different from those that are frequently sampled from the feces (Ouwerkerk et al., 2013), with an overrepresentation of bacteria that degrade mucins (see above). A study using germ-free mice colonized with Escherichia coli and the mucin degrader B. fragilis, revealed that only B. fragilis penetrates the mucus layer in the mouse colon (Huang et al., 2011). Therefore, the ability to metabolize mucin O-linked glycans is likely to be a key factor in determining which bacteria species adapt to the mucosal environment for in vivo colonization of animals. Mucosa-associated bacteria, due to their intimate proximity to the host epithelium, are likely to play a significant role in human health and disease. Indeed the intestinal epithelium directly benefits from the products of microbial metabolism by absorbing short-chain fatty acids (SCFAs) such as butyrate, propionate, and acetate which have been demonstrated to reduce the risk of GI disorders. Nutritional strategies such as the administration of prebiotics can modulate the composition of the mucosal microbes, shifting mucin degradation to distal regions, where mucin-degrading bacteria may produce metabolites influencing the host (Van Den Abbeele et al., 2011).
During the past decade there has been increasing focus on gut microbiota as an influential factor on inflammatory disease development in both humans and animals. Inflammatory bowel disease (IBD) is characterized by an increase in total mucosa-associated bacteria (Schultsz et al., 1999). IBD patients have a disproportionate representation of mucin-degrading bacteria. A ~ 100-fold and >4-fold increase in R. torques and R. gnavus, respectively, was observed in macroscopically- and histologically-normal intestinal epithelia in cases of both Crohn's disease (CD) and ulcerative colitis (UC) (Png et al., 2010). In contrast, the most abundantly detected mucolytic bacterium in controls, A. muciniphila, was reduced many fold in CD and in UC (Png et al., 2010). Comparison between ileal mucosa samples of healthy individuals with patients suffering from ileal CD revealed an increased abundance of R. gnavus with a reduced abundance of F. prausnitzii in CD patients (Willing et al., 2010). The same findings were observed in fecal samples from CD patients compared to unaffected controls (Sokol et al., 2009; Joossens et al., 2011). A reduction of F. prausnitzii in mucosa-associated microbiota of CD patients is associated with a higher risk of postoperative recurrence of ileal CD (Sokol et al., 2008). In contrast, high prevalence of aggregative, adherent Escherichia coli strains has been reported in the mucosa-associated microbiota of CD patients (Darfeuille-Michaud et al., 2004; Thomazini et al., 2011). In CD and UC patients only the mucosa associated population of E. coli is augmented and the proliferation is prominent in the ileum of CD and rectum and sigmoid of both UC and CD patients which are sites where the lesions are usually observed (De Souza et al., 2012). A recent study examining CD, UC, and irritable bowel syndrome (IBS) patients showed that mucosa-associated F. prausnitzii and E. coli co-abundance can distinguish IBS and IBD phenotypes (Lopez-Siles et al., 2014). An earlier study reported that colonic biopsies from CD-afflicted patients compared with biopsies from normal control subjects had an increase in anaerobic bacteria; in the small bowel, CD patients had an increase in the R. gnavus subgroup with a decrease in the Clostridium leptum and Prevotella nigrescens subgroups (Prindiville et al., 2004). A different pattern was observed in patients with active UC, where R. gnavus was found abundantly present in the colonic mucosa of healthy subjects but lost during active UC (Nishikawa et al., 2009). It has been hypothesized that increased mucin-degrading bacteria in IBD provide increased substrate to sustain non-mucolytic mucosa-associated bacteria, which could explain the increased total mucosa-associated bacteria in IBD. The mucin degrader R. torques is also frequently associated with conditions such as IBS (Malinen et al., 2010). Altogether these studies point toward an important role of mucin-degrading bacteria in modulating gut inflammatory response at the mucosal surface. It has been proposed that excessive mucin degradation by intestinal bacteria may contribute to intestinal disorders, as access of luminal antigens to the intestinal immune system is facilitated (Ganesh et al., 2013). However, it is not known whether all mucin-degraders have the same effect. For example A. muciniphila may possess anti-inflammatory properties, as a high proportion of the bacteria has been correlated to protection against inflammation in diseases such as type 1 diabetes mellitus (Hansen et al., 2012), IBD (Png et al., 2010), atopic dermatitis (Candela et al., 2012), autism (Wang et al., 2011), type 2 diabetes mellitus (Ellekilde et al., 2014), and obesity (Everard et al., 2013; Le Chatelier et al., 2013). A. muciniphila treatment can reverse fat gain, serum lipopolysaccharide (LPS) levels, gut barrier function, and insulin resistance. With regards to mucus, an increase in A. muciniphila has been shown to correlate with an increase in the number of goblet cells, potentially underlying the improved glucose profiles seen after A. muciniphila administration (Shin et al., 2014). In light of these findings, future work is warranted to gain a better understanding of the role of mucin-degraders in metabolic syndromes.
The ability of enteric pathogens to thrive within the gut mucosal environment is intimately linked to the glycan metabolism of mucin-degrading bacteria. S. typhimurium accesses fucose and sialic acid within the lumen of the gut in a microbiota-dependent manner, and genetic ablation of the respective catabolic pathways reduces its competitiveness in vivo (Ng et al., 2013). Similarly, C. difficile expansion is aided by microbiota-induced elevation of sialic acid levels in vivo. Colonization of gnotobiotic mice with a sialidase-deficient mutant of B. thetaiotaomicron reduces free sialic acid levels resulting in C. difficile down-regulating its sialic acid catabolic pathway and exhibiting impaired expansion. These effects were reversed by exogenous dietary administration of free sialic acid (Ng et al., 2013). Furthermore, A. muciniphila was shown in gnotobiotic mice to exacerbate S. typhimurium-induced intestinal inflammation by its ability to disturb host mucus homeostasis (Ganesh et al., 2013). The GI pathogen, enterohaemorrhagic E. coli (EHEC), encodes a two-component system, termed FusKR, which responds to fucose and represses expression of virulence genes. During growth in mucin, B. thetaiotaomicron releases fucose from mucin, thereby activating the FusKR signaling cascade, modulating the virulence gene expression of EHEC (Pacheco et al., 2012). Taken together these studies indicate that mucin-derived monosaccharides made available by the microbiota profoundly influence the expansion of enteric pathogens within the gut.
Bacterial-mediated mucin glycan catabolism is an important component in gut colonization which impacts on microbiota ecology and gut health. Based on the studies reported in this review, gut bacteria strains appear to rarely produce the complete set of glycosidases necessary for the degradation of mucin glycans into their constituent monosaccharides and it is thus likely that in vivo complete degradation of mucins in the gut relies on the cooperative action of several microbial species. In recent years an alteration of the gut microbiota structure and function has been associated with an increasing number of diseases outside and inside the gut and diet has emerged as one of the most important factors believed to affect the composition and activity of the gut microbiome. However, the systematic contribution of mucin-degraders in gut homeostasis and dysbiosis has not yet been investigated. One of the reasons is that so far only a few bacteria species have been reported and characterized as “mucin-degraders.” Expanding our knowledge into the nature of different mucin-degrading bacteria and their differential roles in the GI tract is important to help develop new therapeutic approaches aimed at restoring eubiosis in inflammatory conditions and preventing infectious diseases caused by enteric pathogens.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC). This research was partly funded by the BBSRC Institute Strategic Programme BB/J004529/1: The Gut Health and Food Safety ISP and BBSRC responsive grants BB/K019554/1 and BB/L008602/1. We would like to thank Olivia Kober for her help with the preparation of some of the figures.
2′FL, 2-fucosyllactose; 3′FL, 3-fucosyllactose; AgnC, α-N-acetylglucosaminidase; BSM, Bovine submaxillary mucin; CAZyme, Carbohydrate-active enzyme; CBM, Carbohydrate-binding module; CD, Crohn's disease; ER, Endoplasmic reticulum; Fuc, Fucose; GAG, glycosaminoglycans; Gal, galactose; GalNAc, N-acetyl-galactosamine; GH, Glycoside hydrolase; GI, gastrointestinal; Glc, Glucose; GlcNAc, N-Acetyl-glucosamine; GNB, Galacto-N-biose; HMO, Human milk oligosaccharides; IBD, Inflammatory bowel disease; IBS, Irritable bowel syndrome; LNB, Lacto-N-biose; LnbB, Lacto-N-biosidases; LNFPII, lacto-N-fucopentaose; LNnT, Lacto-N-neo-tetraose; LNT, Lacto-N-tetraose; LPS, Lipopolysaccharide; MU, 4-Methylumbelliferone; Neu5Ac, N-Acetylneuraminic acid; PGM, Pig gastric mucin; pNP, para-Nitrophenol; pPGM, Purified pig gastric mucin; PUL, Polysaccharide-utilization loci; Pro, Proline; PTS, Proline-threonine-serine; Ser, Serine; Sus, Starch utilization system; Thr, Threonine; UC, Ulcerative colitis; VNTR, variable number of tandem repeat.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2015.00081/abstract
Abe, F., Muto, M., Yaeshima, T., Iwatsuki, K., Aihara, H., Ohashi, Y., et al. (2010). Safety evaluation of probiotic bifidobacteria by analysis of mucin degradation activity and translocation ability. Anaerobe 16, 131–136. doi: 10.1016/j.anaerobe.2009.07.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Almagro-Moreno, S., and Boyd, E. F. (2009). Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 9:118. doi: 10.1186/1471-2148-9-118
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ambort, D., Johansson, M. E., Gustafsson, J. K., Nilsson, H. E., Ermund, A., Johansson, B. R., et al. (2012a). Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc. Natl. Acad. Sci. U.S.A. 109, 5645–5650. doi: 10.1073/pnas.1120269109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ambort, D., Johansson, M. E. V., Gustafsson, J. K., Ermund, A., and Hansson, G. C. (2012b). Perspectives on mucus properties and formation-Lessons from the biochemical world. Cold Spring Harbor Perspect. Med. 2, 1–10. doi: 10.1101/cshperspect.a014159
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ambort, D., Van Der Post, S., Johansson, M. E., Mackenzie, J., Thomsson, E., Krengel, U., et al. (2011). Function of the CysD domain of the gel-forming MUC2 mucin. Biochem. J. 436, 61–70. doi: 10.1042/BJ20102066
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
An, G., Wei, B., Xia, B., McDaniel, J. M., Ju, T., Cummings, R. D., et al. (2007). Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 204, 1417–1429. doi: 10.1084/jem.20061929
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Anderson, K. M., Ashida, H., Maskos, K., Dell, A., Li, S. C., and Li, Y. T. (2005). A clostridial endo-beta-galactosidase that cleaves both blood group A and B glycotopes: the first member of a new glycoside hydrolase family, GH98. J. Biol. Chem. 280, 7720–7728. doi: 10.1074/jbc.M414099200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ashida, H., Maki, R., Ozawa, H., Tani, Y., Kiyohara, M., Fujita, M., et al. (2008). Characterization of two different endo-alpha-N-acetylgalactosaminidases from probiotic and pathogenic enterobacteria, Bifidobacterium longum and Clostridium perfringens. Glycobiology 18, 727–734. doi: 10.1093/glycob/cwn053
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ashida, H., Miyake, A., Kiyohara, M., Wada, J., Yoshida, E., Kumagai, H., et al. (2009). Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19, 1010–1017. doi: 10.1093/glycob/cwp082
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Asker, N., Axelsson, M. A., Olofsson, S. O., and Hansson, G. C. (1998). Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J. Biol. Chem. 273, 18857–18863. doi: 10.1074/jbc.273.30.18857
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Asker, N., Baeckstrom, D., Axelsson, M. A., Carlstedt, I., and Hansson, G. C. (1995). The human MUC2 mucin apoprotein appears to dimerize before O-glycosylation and shares epitopes with the ‘insoluble’ mucin of rat small intestine. Biochem. J. 308(Pt 3), 873–880.
Atuma, C., Strugala, V., Allen, A., and Holm, L. (2001). The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 280, G922–G929.
Audie, J. P., Janin, A., Porchet, N., Copin, M. C., Gosselin, B., and Aubert, J. P. (1993). Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J. Histochem. Cytochem. 41, 1479–1485. doi: 10.1177/41.10.8245407
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bennett, E. P., Mandel, U., Clausen, H., Gerken, T. A., Fritz, T. A., and Tabak, L. A. (2012). Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756. doi: 10.1093/glycob/cwr182
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bergstrom, K. S. B., and Xia, L. J. (2013). Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 23, 1026–1037. doi: 10.1093/glycob/cwt045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brigham, C., Caughlan, R., Gallegos, R., Dallas, M. B., Godoy, V. G., and Malamy, M. H. (2009). Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. J. Bacteriol. 191, 3629–3638. doi: 10.1128/JB.00811-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brockhausen, I., Schachter, H., and Stanley, P. (2009). “O-GalNAc Glycans,” in Essentials of Glycobiology, 2nd Edn., eds A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart, and M. E. Etzler (New York, NY: Cold Spring Harbor), 115–127.
Buisine, M. P., Devisme, L., Degand, P., Dieu, M. C., Gosselin, B., Copin, M. C., et al. (2000a). Developmental mucin gene expression in the gastroduodenal tract and accessory digestive glands. II. Duodenum and liver, gallbladder, and pancreas. J. Histochem. Cytochem. 48, 1667–1676. doi: 10.1177/002215540004801210
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Buisine, M. P., Devisme, L., Maunoury, V., Deschodt, E., Gosselin, B., Copin, M. C., et al. (2000b). Developmental mucin gene expression in the gastroduodenal tract and accessory digestive glands. I. Stomach. A relationship to gastric carcinoma. J. Histochem. Cytochem. 48, 1657–1666. doi: 10.1177/002215540004801209
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Candela, M., Biagi, E., Maccaferri, S., Turroni, S., and Brigidi, P. (2012). Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol. 20, 385–391. doi: 10.1016/j.tim.2012.05.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carrington, S., Clyne, M., Reid, C., Fitzpatrick, E., and Moran, A. (2009). “Microbial interaction with mucus and mucins,” in Microbial Glycobiology: Structures, Relevance and Applications, eds A. P. Moran, P. Brennan, O. Holst, and M. Von Itzstein (London: Elsevier), 655–672.
Carroll, I. M., Chang, Y. H., Park, J., Sartor, R. B., and Ringel, Y. (2010). Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2:19. doi: 10.1186/1757-4749-2-19
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ceroni, A., Dell, A., and Haslam, S. M. (2007). The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2:3. doi: 10.1186/1751-0473-2-3
Chang, S. K., Dohrman, A. F., Basbaum, C. B., Ho, S. B., Tsuda, T., Toribara, N. W., et al. (1994). Localization of mucin (Muc2 and Muc3) messenger-RNA and peptide expression in human normal intestine and colon-cancer. Gastroenterology 107, 28–36.
Chen, S. J., Liu, X. W., Liu, J. P., Yang, X. Y., and Lu, F. G. (2014). Ulcerative colitis as a polymicrobial infection characterized by sustained broken mucus barrier. World J. Gastroenterol. 20, 9468–9475. doi: 10.3748/wjg.v20.i28.9468
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, W., Liu, F., Ling, Z., Tong, X., and Xiang, C. (2012). Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 7:e39743. doi: 10.1371/journal.pone.0039743
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Collado, M. C., Derrien, M., Isolauri, E., De Vos, W. M., and Salminen, S. (2007). Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73, 7767–7770. doi: 10.1128/AEM.01477-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Corfield, A. P. (2015). Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta. 1850, 236–252. doi: 10.1016/j.bbagen.2014.05.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Corfield, A. P., Wagner, S. A., Clamp, J. R., Kriaris, M. S., and Hoskins, L. C. (1992). Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60, 3971–3978.
Crost, E. H., Tailford, L. E., Le Gall, G., Fons, M., Henrissat, B., and Juge, N. (2013). Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE 8:76341. doi: 10.1371/journal.pone.0076341
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C., Glasser, A. L., Barnich, N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421. doi: 10.1053/j.gastro.2004.04.061
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dennis, R. J., Taylor, E. J., Macauley, M. S., Stubbs, K. A., Turkenburg, J. P., Hart, S. J., et al. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 13, 365–371. doi: 10.1038/nsmb1079
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Derrien, M. (2007). Mucin Utilisation and Host Interactions of the Novel Intestinal Microbe Akkermansia Muciniphila. Ph.D. Thesis, Wageningen University.
Derrien, M., Collado, M. C., Ben-Amor, K., Salminen, S., and De Vos, W. M. (2008). The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74, 1646–1648. doi: 10.1128/AEM.01226-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Derrien, M., Vaughan, E. E., Plugge, C. M., and De Vos, W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476. doi: 10.1099/ijs.0.02873-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De Souza, H. L., De Carvalho, V. R., Romeiro, F. G., Sassaki, L. Y., Keller, R., and Rodrigues, J. (2012). Mucosa-associated but not luminal Escherichia coli is augmented in Crohn's disease and ulcerative colitis. Gut. Pathogens 4, 1–8. doi: 10.1186/1757-4749-4-21
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Desseyn, J. L., Tetaert, D., and Gouyer, V. (2008). Architecture of the large membrane-bound mucins. Gene 410, 215–222. doi: 10.1016/j.gene.2007.12.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Egan, M., O'connell Motherway, M., Ventura, M., and Van Sinderen, D. (2014). Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 80, 4414–4426. doi: 10.1128/AEM.01114-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellekilde, M., Krych, L., Hansen, C. H., Hufeldt, M. R., Dahl, K., Hansen, L. H., et al. (2014). Characterization of the gut microbiota in leptin deficient obese mice - Correlation to inflammatory and diabetic parameters. Res. Vet. Sci. 96, 241–250. doi: 10.1016/j.rvsc.2014.01.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ermund, A., Schutte, A., Johansson, M. E., Gustafsson, J. K., and Hansson, G. C. (2013). Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am. J. Physiol. Gastrointest Liver Physiol. 305, G341–G347. doi: 10.1152/ajpgi.00046.2013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Etzold, S., and Juge, N. (2014). Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 28C, 23–31. doi: 10.1016/j.sbi.2014.07.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Etzold, S., Kober, O. I., Mackenzie, D. A., Tailford, L. E., Gunning, A. P., Walshaw, J., et al. (2014). Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 16, 888–903. doi: 10.1111/1462-2920.12377
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Favier, C. F., Vaughan, E. E., De Vos, W. M., and Akkermans, A. D. (2002). Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68, 219–226. doi: 10.1128/AEM.68.1.219-226.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ficko-Blean, E., and Boraston, A. B. (2005). Cloning, recombinant production, crystallization and preliminary X-ray diffraction studies of a family 84 glycoside hydrolase from Clostridium perfringens. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 834–836. doi: 10.1107/S1744309105024012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ficko-Blean, E., and Boraston, A. B. (2012a). Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 22, 570–577. doi: 10.1016/j.sbi.2012.07.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ficko-Blean, E., and Boraston, A. B. (2012b). Structural analysis of a bacterial exo-alpha-D-N-acetylglucosaminidase in complex with an unusual disaccharide found in class III mucin. Glycobiology 22, 590–595. doi: 10.1093/glycob/cwr165
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ficko-Blean, E., Stubbs, K. A., Nemirovsky, O., Vocadlo, D. J., and Boraston, A. B. (2008). Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc. Natl. Acad. Sci. U.S.A. 105, 6560–6565. doi: 10.1073/pnas.0711491105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Forni, D., Cleynen, I., Ferrante, M., Cassinotti, A., Cagliani, R., Ardizzone, S., et al. (2014). ABO histo-blood group might modulate predisposition to Crohn's disease and affect disease behavior. J. Crohns. Colitis 8, 489–494. doi: 10.1016/j.crohns.2013.10.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Freitas, M., Tavan, E., Cayuela, C., Diop, L., Sapin, C., and Trugnan, G. (2003). Host-pathogens cross-talk. Indigenous bacteria and probiotics also play the game. Biol. Cell 95, 503–506. doi: 10.1016/j.biolcel.2003.08.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fu, J., Wei, B., Wen, T., Johansson, M. E., Liu, X., Bradford, E., et al. (2011). Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Invest. 121, 1657–1666. doi: 10.1172/JCI45538
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fujita, K., Oura, F., Nagamine, N., Katayama, T., Hiratake, J., Sakata, K., et al. (2005). Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 280, 37415–37422. doi: 10.1074/jbc.M506874200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fujita, M., Tsuchida, A., Hirata, A., Kobayashi, N., Goto, K., Osumi, K., et al. (2011). Glycoside hydrolase family 89 alpha-N-acetylglucosaminidase from Clostridium perfringens specifically acts on GlcNAc alpha1,4Gal beta1R at the non-reducing terminus of O-glycans in gastric mucin. J. Biol. Chem. 286, 6479–6489. doi: 10.1074/jbc.M110.206722
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ganesh, B. P., Klopfleisch, R., Loh, G., and Blaut, M. (2013). Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS ONE 8:e74963. doi: 10.1371/journal.pone.0074963
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gendler, S. J., and Spicer, A. P. (1995). Epithelial mucin genes. Annu. Rev. Physiol. 57, 607–634. doi: 10.1146/annurev.ph.57.030195.003135
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Goda, H. M., Ushigusa, K., Ito, H., Okino, N., Narimatsu, H., and Ito, M. (2008). Molecular cloning, expression, and characterization of a novel endo-alpha-N-acetylgalactosaminidase from Enterococcus faecalis. Biochem. Biophys. Res. Commun. 375, 441–446. doi: 10.1016/j.bbrc.2008.08.065
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Godl, K., Johansson, M. E., Lidell, M. E., Morgelin, M., Karlsson, H., Olson, F. J., et al. (2002). The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J. Biol. Chem. 277, 47248–47256. doi: 10.1074/jbc.M208483200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gonzalez-Rodriguez, I., Sanchez, B., Ruiz, L., Turroni, F., Ventura, M., Ruas-Madiedo, P., et al. (2012). Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl. Environ. Microbiol. 78, 3992–3998. doi: 10.1128/AEM.08024-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guillotin, L., Lafite, P., and Daniellou, R. (2014). Unraveling the substrate recognition mechanism and specificity of the unusual glycosyl hydrolase family 29 BT2192 from Bacteroides thetaiotaomicron. Biochemistry 53, 1447–1455. doi: 10.1021/bi400951q
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gum, J. R. Jr., Crawley, S. C., Hicks, J. W., Szymkowski, D. E., and Kim, Y. S. (2002). MUC17, a novel membrane-tethered mucin. Biochem. Biophys. Res. Commun. 291, 466–475. doi: 10.1006/bbrc.2002.6475
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hansen, C. H., Krych, L., Nielsen, D. S., Vogensen, F. K., Hansen, L. H., Sorensen, S. J., et al. (2012). Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55, 2285–2294. doi: 10.1007/s00125-012-2564-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hansson, G. C. (2012). Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 15, 57–62. doi: 10.1016/j.mib.2011.11.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hattori, M., and Taylor, T. D. (2009). The human intestinal microbiome: a new frontier of human biology. DNA Res. 16, 1–12. doi: 10.1093/dnares/dsn033
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Henry, S., Oriol, R., and Samuelsson, B. (1995). Lewis histo-blood group system and associated secretory phenotypes. Vox Sang. 69, 166–182. doi: 10.1111/j.1423-0410.1995.tb02591.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Higgins, M. A., Whitworth, G. E., El Warry, N., Randriantsoa, M., Samain, E., Burke, R. D., et al. (2009). Differential recognition and hydrolysis of host carbohydrate antigens by Streptococcus pneumoniae family 98 glycoside hydrolases. J. Biol. Chem. 284, 26161–26173. doi: 10.1074/jbc.M109.024067
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Higuchi, T., Orita, T., Nakanishi, S., Katsuya, K., Watanabe, H., Yamasaki, Y., et al. (2004). Molecular cloning, genomic structure, and expression analysis of MUC20, a novel mucin protein, up-regulated in injured kidney. J. Biol. Chem. 279, 1968–1979. doi: 10.1074/jbc.M304558200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ho, S. B., Niehans, G. A., Lyftogt, C., Yan, P. S., Cherwitz, D. L., Gum, E. T., et al. (1993). Heterogeneity of mucin gene-expression in normal and neoplastic tissues. Cancer Res. 53, 641–651.
Holmen Larsson, J. M., Thomsson, K. A., Rodriguez-Pineiro, A. M., Karlsson, H., and Hansson, G. C. (2013). Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am. J. Physiol. Gastrointest Liver Physiol. 305, G357–G363. doi: 10.1152/ajpgi.00048.2013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hong, P. Y., Croix, J. A., Greenberg, E., Gaskins, H. R., and Mackie, R. I. (2011). Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS ONE 6:e25042. doi: 10.1371/journal.pone.0025042
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T., and Gordon, J. I. (1999). A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. U.S.A. 96, 9833–9838. doi: 10.1073/pnas.96.17.9833
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hoskins, L. C., Agustines, M., McKee, W. B., Boulding, E. T., Kriaris, M., and Niedermeyer, G. (1985). Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J. Clin. Invest. 75, 944–953. doi: 10.1172/JCI111795
Hoskins, L. C., Boulding, E. T., Gerken, T. A., Harouny, V. R., and Kriaris, M. S. (1992). Mucin glycoprotein degradation by mucin oligosaccharide-degrading strains of human faecal bacteria. Characterisation of saccharide cleavage products and their potential role in nutritional support of larger faecal bacterial populations. Microb. Ecol. Health D 5, 193–207. doi: 10.3109/08910609209141586
Hoyer, L. L., Hamilton, A. C., Steenbergen, S. M., and Vimr, E. R. (1992). Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol. Microbiol. 6, 873–884. doi: 10.1111/j.1365-2958.1992.tb01538.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huang, J. Y., Lee, S. M., and Mazmanian, S. K. (2011). The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 17, 137–141. doi: 10.1016/j.anaerobe.2011.05.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Itoh, Y., Kamata-Sakurai, M., Denda-Nagai, K., Nagai, S., Tsuiji, M., Ishii-Schrade, K., et al. (2008). Identification and expression of human epiglycanin/MUC21: a novel transmembrane mucin. Glycobiology 18, 74–83. doi: 10.1093/glycob/cwm118
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jakobsson, H. E., Rodriguez-Pineiro, A. M., Schutte, A., Ermund, A., Boysen, P., Bemark, M., et al. (2015). The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177. doi: 10.15252/embr.201439263
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Johansson, M. E., and Hansson, G. C. (2012). Analysis of assembly of secreted mucins. Methods Mol. Biol. 842, 109–121. doi: 10.1007/978-1-61779-513-8_6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Johansson, M. E., Larsson, J. M., and Hansson, G. C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4659–4665. doi: 10.1073/pnas.1006451107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Johansson, M. E., Phillipson, M., Petersson, J., Velcich, A., Holm, L., and Hansson, G. C. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 15064–15069. doi: 10.1073/pnas.0803124105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jonckheere, N., Skrypek, N., and Van Seuningen, I. (2010). Mucins and pancreatic cancer. Cancers 2, 1794–1812. doi: 10.3390/cancers2041794
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Joossens, M., Huys, G., Cnockaert, M., De Preter, V., Verbeke, K., Rutgeerts, P., et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60, 631–637. doi: 10.1136/gut.2010.223263
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Juge, N. (2012). Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 20, 30–39. doi: 10.1016/j.tim.2011.10.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kashyap, P. C., Marcobal, A., Ursell, L. K., Smits, S. A., Sonnenburg, E. D., Costello, E. K., et al. (2013). Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 110, 17059–17064. doi: 10.1073/pnas.1306070110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Katayama, T., Fujita, K., and Yamamoto, K. (2005). Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J. Biosci. Bioeng. 99, 457–465. doi: 10.1263/jbb.99.457
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kelly, R. J., Rouquier, S., Giorgi, D., Lennon, G. G., and Lowe, J. B. (1995). Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem. 270, 4640–4649. doi: 10.1074/jbc.270.9.4640
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, J. H., An, H. J., Garrido, D., German, J. B., Lebrilla, C. B., and Mills, D. A. (2013). Proteomic analysis of Bifidobacterium longum subsp. infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLoS ONE 8:e57535. doi: 10.1371/journal.pone.0057535
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kiyohara, M., Nakatomi, T., Kurihara, S., Fushinobu, S., Suzuki, H., Tanaka, T., et al. (2012). alpha-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J. Biol. Chem. 287, 693–700. doi: 10.1074/jbc.M111.277384
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kiyohara, M., Tanigawa, K., Chaiwangsri, T., Katayama, T., Ashida, H., and Yamamoto, K. (2011). An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21, 437–447. doi: 10.1093/glycob/cwq175
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kobayashi, M., Lee, H., Nakayama, J., and Fukuda, M. (2009). Roles of gastric mucin-type O-glycans in the pathogenesis of Helicobacter pylori infection. Glycobiology 19, 453–461. doi: 10.1093/glycob/cwp004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kolmeder, C. A., and De Vos, W. M. (2014). Metaproteomics of our microbiome - developing insight in function and activity in man and model systems. J. Proteomics 97, 3–16. doi: 10.1016/j.jprot.2013.05.018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Koropatkin, N. M., Cameron, E. A., and Martens, E. C. (2012). How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335. doi: 10.1038/nrmicro2746
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Koutsioulis, D., Landry, D., and Guthrie, E. P. (2008). Novel endo-alpha-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology 18, 799–805. doi: 10.1093/glycob/cwn069
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kuwahara, T., Yamashita, A., Hirakawa, H., Nakayama, H., Toh, H., Okada, N., et al. (2004). Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. U.S.A. 101, 14919–14924. doi: 10.1073/pnas.0404172101
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lammerts Van Bueren, A., Ardevol, A., Fayers-Kerr, J., Luo, B., Zhang, Y., Sollogoub, M., et al. (2010). Analysis of the reaction coordinate of alpha-L-fucosidases: a combined structural and quantum mechanical approach. J. Am. Chem. Soc. 132, 1804–1806. doi: 10.1021/ja908908q
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Larsson, J. M., Karlsson, H., Crespo, J. G., Johansson, M. E., Eklund, L., Sjovall, H., et al. (2011). Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm. Bowel Dis. 17, 2299–2307. doi: 10.1002/ibd.21625
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Larsson, J. M. H., Karlsson, H., Sjövall, H., and Hansson, G. C. (2009). A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19, 756–766. doi: 10.1093/glycob/cwp048
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Levy, G. N., and Aminoff, D. (1980). Purification and properties of alpha-N-acetylgalactosaminidase from Clostridium perfringens. J. Biol. Chem. 255, 11737–11742.
Linden, S., Mahdavi, J., Hedenbro, J., Boren, T., and Carlstedt, I. (2004). Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem. J. 384, 263–270. doi: 10.1042/BJ20040402
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lindén, S., Mahdavi, J., Semino-Mora, C., Olsen, C., Carlstedt, I., Borén, T., et al. (2008). Role of ABO secretor status in mucosal innate immunity and Helicobacter pylori infection. PLoS Pathog. 4:e2. doi: 10.1371/journal.ppat.0040002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Linden, S. K., Sutton, P., Karlsson, N. G., Korolik, V., and McGuckin, M. A. (2008). Mucins in the mucosal barrier to infection. Mucosal Immunol. 1, 183–197. doi: 10.1038/mi.2008.5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lopez-Siles, M., Martinez-Medina, M., Busquets, D., Sabat-Mir, M., Duncan, S. H., Flint, H. J., et al. (2014). Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and Inflammatory Bowel Disease phenotypes. Int. J. Med. Microbiol. 304, 464–475. doi: 10.1016/j.ijmm.2014.02.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Malinen, E., Krogius-Kurikka, L., Lyra, A., Nikkila, J., Jaaskelainen, A., Rinttila, T., et al. (2010). Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 16, 4532–4540. doi: 10.3748/wjg.v16.i36.4532
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marcobal, A., Barboza, M., Sonnenburg, E. D., Pudlo, N., Martens, E. C., Desai, P., et al. (2011). Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10, 507–514. doi: 10.1016/j.chom.2011.10.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marcobal, A., Southwick, A. M., Earle, K. A., and Sonnenburg, J. L. (2013). A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23, 1038–1046. doi: 10.1093/glycob/cwt040
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martens, E. C., Chiang, H. C., and Gordon, J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. doi: 10.1016/j.chom.2008.09.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martens, E. C., Koropatkin, N. M., Smith, T. J., and Gordon, J. I. (2009). Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284, 24673–24677. doi: 10.1074/jbc.R109.022848
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martens, E. C., Lowe, E. C., Chiang, H., Pudlo, N. A., Wu, M., McNulty, N. P., et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. doi: 10.1371/journal.pbio.1001221
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McGovern, D. P., Jones, M. R., Taylor, K. D., Marciante, K., Yan, X., Dubinsky, M., et al. (2010). Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum. Mol. Genet. 19, 3468–3476. doi: 10.1093/hmg/ddq248
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McGuckin, M. A., Linden, S. K., Sutton, P., and Florin, T. H. (2011). Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278. doi: 10.1038/nrmicro2538
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Miller, R. S., and Hoskins, L. C. (1981). Mucin degradation in human colon ecosystems. Fecal population densities of mucin-degrading bacteria estimated by a “most probable number” method. Gastroenterology 81, 759–765.
Miwa, M., Horimoto, T., Kiyohara, M., Katayama, T., Kitaoka, M., Ashida, H., et al. (2010). Cooperation of beta-galactosidase and beta-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20, 1402–1409. doi: 10.1093/glycob/cwq101
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mollicone, R., Bara, J., Le Pendu, J., and Oriol, R. (1985). Immunohistologic pattern of type 1 (Lea, Leb) and type 2 (X, Y, H) blood group-related antigens in the human pyloric and duodenal mucosae. Lab. Invest. 53, 219–227.
Moncada, D. M., Kammanadiminti, S. J., and Chadee, K. (2003). Mucin and Toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 19, 305–311. doi: 10.1016/S1471-4922(03)00122-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Moran, A. P., Gupta, A., and Joshi, L. (2011). Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60, 1412–1425. doi: 10.1136/gut.2010.212704
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nagae, M., Tsuchiya, A., Katayama, T., Yamamoto, K., Wakatsuki, S., and Kato, R. (2007). Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum. J. Biol. Chem. 282, 18497–18509. doi: 10.1074/jbc.M702246200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nakajima, K., Ota, H., Zhang, M. X., Sano, K., Honda, T., Ishii, K., et al. (2003). Expression of gastric gland mucous cell-type mucin in normal and neoplastic human tissues. J. Histochem. Cytochem. 51, 1689–1698. doi: 10.1177/002215540305101213
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nakayama-Imaohji, H., Ichimura, M., Iwasa, T., Okada, N., Ohnishi, Y., and Kuwahara, T. (2012). Characterization of a gene cluster for sialoglycoconjugate utilization in Bacteroides fragilis. J. Med. Invest. 59, 79–94. doi: 10.2152/jmi.59.79
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nava, G. M., Carbonero, F., Croix, J. A., Greenberg, E., and Gaskins, H. R. (2012). Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 6, 57–70. doi: 10.1038/ismej.2011.90
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nava, G. M., Friedrichsen, H. J., and Stappenbeck, T. S. (2011). Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638. doi: 10.1038/ismej.2010.161
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Newstead, S. L., Potter, J. A., Wilson, J. C., Xu, G., Chien, C. H., Watts, A. G., et al. (2008). The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J. Biol. Chem. 283, 9080–9088. doi: 10.1074/jbc.M710247200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ng, K. M., Ferreyra, J. A., Higginbottom, S. K., Lynch, J. B., Kashyap, P. C., Gopinath, S., et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99. doi: 10.1038/nature12503
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nilsson, H. E., Ambort, D., Backstrom, M., Thomsson, E., Koeck, P. J., Hansson, G. C., et al. (2014). Intestinal MUC2 mucin supramolecular topology by packing and release resting on D3 domain assembly. J. Mol. Biol. 426, 2567–2579. doi: 10.1016/j.jmb.2014.04.027
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nishikawa, J., Kudo, T., Sakata, S., Benno, Y., and Sugiyama, T. (2009). Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand. J. Gastroenterol. 44, 180–186. doi: 10.1080/00365520802433231
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nordman, H., Davies, J. R., Lindell, G., De Bolos, C., Real, F., and Carlstedt, I. (2002). Gastric MUC5AC and MUC6 are large oligomeric mucins that differ in size, glycosylation and tissue distribution. Biochem. J. 364, 191–200.
Norin, K. E., Gustafsson, B. E., Lindblad, B. S., and Midtvedt, T. (1985). The establishment of some microflora associated biochemical characteristics in feces from children during the first years of life. Acta Paediatr. Scand. 74, 207–212. doi: 10.1111/j.1651-2227.1985.tb10951.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ouwerkerk, J. P., De Vos, W. M., and Belzer, C. (2013). Glycobiome: bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 27, 25–38. doi: 10.1016/j.bpg.2013.03.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pacheco, A. R., Curtis, M. M., Ritchie, J. M., Munera, D., Waldor, M. K., Moreira, C. G., et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–119. doi: 10.1038/nature11623
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Packer, L. M., Williams, S. J., Callaghan, S., Gotley, D. C., and McGuckin, M. A. (2004). Expression of the cell surface mucin gene family in adenocarcinomas. Int. J. Oncol. 25, 1119–1126.
Pallesen, L. T., Berglund, L., Rasmussen, L. K., Petersen, T. E., and Rasmussen, J. T. (2002). Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur. J. Biochem. 269, 2755–2763. doi: 10.1046/j.1432-1033.2002.02949.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Parmar, A. S., Alakulppi, N., Paavola-Sakki, P., Kurppa, K., Halme, L., Farkkila, M., et al. (2012). Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens 80, 488–493. doi: 10.1111/tan.12016
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Phansopa, C., Roy, S., Rafferty, J. B., Douglas, C. W., Pandhal, J., Wright, P. C., et al. (2014). Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem. J. 458, 499–511. doi: 10.1042/BJ20131415
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pickard, J. M., Maurice, C. F., Kinnebrew, M. A., Abt, M. C., Schenten, D., Golovkina, T. V., et al. (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. doi: 10.1038/nature13823
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Png, C. W., Linden, S. K., Gilshenan, K. S., Zoetendal, E. G., McSweeney, C. S., Sly, L. I., et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428. doi: 10.1038/ajg.2010.281
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Porchet, N., Pigny, P., Buisine, M. P., Debailleul, V., Degand, P., Laine, A., et al. (1995). Human mucin genes: genomic organization and expression of MUC4, MUC5AC and MUC5B. Biochem. Soc. Trans. 23, 800–805.
Prindiville, T., Cantrell, M., and Wilson, K. H. (2004). Ribosomal DNA sequence analysis of mucosa-associated bacteria in Crohn's disease. Inflamm. Bowel Dis. 10, 824–833. doi: 10.1097/00054725-200411000-00017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prizont, R. (1982). Degradation of intestinal glycoproteins by pathogenic Shigella flexneri. Infect. Immun. 36, 615–620.
Pullan, R. D., Thomas, G. A. O., Rhodes, M., Newcombe, R. G., Williams, G. T., Allen, A., et al. (1994). Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut 35, 353–359. doi: 10.1136/gut.35.3.353
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rausch, P., Rehman, A., Kunzel, S., Hasler, R., Ott, S. J., Schreiber, S., et al. (2011). Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. U.S.A. 108, 19030–19035. doi: 10.1073/pnas.1106408108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reis, C. A., David, L., Carvalho, F., Mandel, U., De Bolos, C., Mirgorodskaya, E., et al. (2000). Immunohistochemical study of the expression of MUC6 mucin and co-expression of other secreted mucins (MUC5AC and MUC2) in human gastric carcinomas. J. Histochem. Cytochem. 48, 377–388. doi: 10.1177/002215540004800307
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rho, J. H., Wright, D. P., Christie, D. L., Clinch, K., Furneaux, R. H., and Roberton, A. M. (2005). A novel mechanism for desulfation of mucin: identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J. Bacteriol. 187, 1543–1551. doi: 10.1128/JB.187.5.1543-1551.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Robbe, C., Capon, C., Coddeville, B., and Michalski, J. C. (2004). Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem. J. 384, 307–316. doi: 10.1042/BJ20040605
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Robbe-Masselot, C., Herrmann, A., Maes, E., Carlstedt, I., Michalski, J. C., and Capon, C. (2009). Expression of a core 3 disialyl-Le(x) hexasaccharide in human colorectal cancers: a potential marker of malignant transformation in colon. J. Proteome Res. 8, 702–711. doi: 10.1021/pr800740j
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Roberton, A. M., and Stanley, R. A. (1982). In vitro utilization of mucin by Bacteroides fragilis. Appl. Environ. Microbiol. 43, 325–330.
Ruas-Madiedo, P., Gueimonde, M., Fernandez-Garcia, M., De Los Reyes-Gavilan, C. G., and Margolles, A. (2008). Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 74, 1936–1940. doi: 10.1128/AEM.02509-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruiz, L., Gueimonde, M., Coute, Y., Salminen, S., Sanchez, J. C., De Los Reyes-Gavilan, C. G., et al. (2011). Evaluation of the ability of Bifidobacterium longum to metabolize human intestinal mucus. FEMS Microbiol. Lett. 314, 125–130. doi: 10.1111/j.1574-6968.2010.02159.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sakurama, H., Tsutsumi, E., Ashida, H., Katayama, T., Yamamoto, K., and Kumagai, H. (2012). Differences in the substrate specificities and active-site structures of two alpha-L-fucosidases (glycoside hydrolase family 29) from Bacteroides thetaiotaomicron. Biosci. Biotechnol. Biochem. 76, 1022–1024. doi: 10.1271/bbb.111004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Salyers, A. A., Vercellotti, J. R., West, S. E., and Wilkins, T. D. (1977a). Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33, 319–322.
Salyers, A. A., West, S. E., Vercellotti, J. R., and Wilkins, T. D. (1977b). Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34, 529–533.
Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., et al. (2002). The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U.S.A. 99, 14422–14427. doi: 10.1073/pnas.212527599
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schultsz, C., Van Den Berg, F. M., Ten Kate, F. W., Tytgat, G. N., and Dankert, J. (1999). The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 117, 1089–1097. doi: 10.1016/S0016-5085(99)70393-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sebaihia, M., Wren, B. W., Mullany, P., Fairweather, N. F., Minton, N., Stabler, R., et al. (2006). The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786. doi: 10.1038/ng1830
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sela, D. A., Chapman, J., Adeuya, A., Kim, J. H., Chen, F., Whitehead, T. R., et al. (2008). The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 18964–18969. doi: 10.1073/pnas.0809584105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sela, D. A., Garrido, D., Lerno, L., Wu, S., Tan, K., Eom, H. J., et al. (2012). Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 78, 795–803. doi: 10.1128/AEM.06762-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Severi, E., Hood, D. W., and Thomas, G. H. (2007). Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822. doi: 10.1099/mic.0.2007/009480-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sheng, Y. H., Hasnain, S. Z., Florin, T. H., and McGuckin, M. A. (2012). Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 27, 28–38. doi: 10.1111/j.1440-1746.2011.06909.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shin, N. R., Lee, J. C., Lee, H. Y., Kim, M. S., Whon, T. W., Lee, M. S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735. doi: 10.1136/gutjnl-2012-303839
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Slomiany, B. L., Murty, V. L., Piotrowski, J., Liau, Y. H., Sundaram, P., and Slomiany, A. (1992). Glycosulfatase activity of Helicobacter pylori toward gastric mucin. Biochem. Biophys. Res. Commun. 183, 506–513. doi: 10.1016/0006-291X(92)90511-I
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sokol, H., Seksik, P., Furet, J. P., Firmesse, O., Nion-Larmurier, L., Beaugerie, L., et al. (2009). Low Counts of Faecalibacterium prausnitzii in Colitis Microbiota. Inflamm. Bowel Dis. 15, 1183–1189. doi: 10.1002/ibd.20903
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sommer, F., Adam, N., Johansson, M. E., Xia, L., Hansson, G. C., and Backhed, F. (2014). Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS ONE 9:e85254. doi: 10.1371/journal.pone.0085254
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sommer, F., and Backhed, F. (2013). The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stone, E. L., Ismail, M. N., Lee, S. H., Luu, Y., Ramirez, K., Haslam, S. M., et al. (2009). Glycosyltransferase function in core 2-type protein O glycosylation. Mol. Cell. Biol. 29, 3770–3782. doi: 10.1128/MCB.00204-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Suzuki, R., Katayama, T., Kitaoka, M., Kumagai, H., Wakagi, T., Shoun, H., et al. (2009). Crystallographic and mutational analyses of substrate recognition of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biochem. 146, 389–398. doi: 10.1093/jb/mvp086
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P., and Lochs, H. (2005). Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tannock, G. W., Lawley, B., Munro, K., Gowri Pathmanathan, S., Zhou, S. J., Makrides, M., et al. (2013). Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 79, 3040–3048. doi: 10.1128/AEM.03910-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Theodoratou, E., Campbell, H., Ventham, N. T., Kolarich, D., Pucic-Bakovic, M., Zoldos, V., et al. (2014). The role of glycosylation in IBD. Nat. Rev. Gastroenterol. Hepatol. 11, 588–600. doi: 10.1038/nrgastro.2014.78
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thomazini, C. M., Samegima, D. A. G., Rodrigues, M. A. M., Victoria, C. R., and Rodrigues, J. (2011). High prevalence of aggregative adherent Escherichia coli strains in the mucosa-associated microbiota of patients with inflammatory bowel diseases. Int. J. Med. Microbiol. 301, 475–479. doi: 10.1016/j.ijmm.2011.04.015
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thomsson, K. A., Holmén-Larsson, J. M., Ångström, J., Johansson, M. E., Xia, L., and Hansson, G. C. (2012). Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1-and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology 22, 1128–1139. doi: 10.1093/glycob/cws083
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tong, M., McHardy, I., Ruegger, P., Goudarzi, M., Kashyap, P. C., Haritunians, T., et al. (2014). Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. 8, 2193–2206. doi: 10.1038/ismej.2014.64
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turroni, F., Bottacini, F., Foroni, E., Mulder, I., Kim, J. H., Zomer, A., et al. (2010). Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U.S.A. 107, 19514–19519. doi: 10.1073/pnas.1011100107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Den Abbeele, P., Belzer, C., Goossens, M., Kleerebezem, M., De Vos, W. M., Thas, O., et al. (2013). Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 7, 949–961. doi: 10.1038/ismej.2012.158
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Den Abbeele, P., Gerard, P., Rabot, S., Bruneau, A., El Aidy, S., Derrien, M., et al. (2011). Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ. Microbiol. 13, 2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Passel, M. W., Kant, R., Zoetendal, E. G., Plugge, C. M., Derrien, M., Malfatti, S. A., et al. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6:e16876. doi: 10.1371/journal.pone.0016876
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Verdugo, P. (2012). Supramolecular dynamics of mucus. Cold Spring Harbor Perspect. Med. 2, 1–15. doi: 10.1101/cshperspect.a009597
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vimr, E. R. (2013). Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013, 26. doi: 10.1155/2013/816713
Vimr, E. R., Kalivoda, K. A., Deszo, E. L., and Steenbergen, S. M. (2004). Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68, 132–153. doi: 10.1128/MMBR.68.1.132-153.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wacklin, P., Makivuokko, H., Alakulppi, N., Nikkila, J., Tenkanen, H., Rabina, J., et al. (2011). Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE 6:e20113. doi: 10.1371/journal.pone.0020113
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wacklin, P., Tuimala, J., Nikkila, J., Sebastian, T., Makivuokko, H., Alakulppi, N., et al. (2014). Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS ONE 9:e94863. doi: 10.1371/journal.pone.0094863
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wada, J., Ando, T., Kiyohara, M., Ashida, H., Kitaoka, M., Yamaguchi, M., et al. (2008). Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol. 74, 3996–4004. doi: 10.1128/AEM.00149-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, L., Christophersen, C. T., Sorich, M. J., Gerber, J. P., Angley, M. T., and Conlon, M. A. (2011). Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 77, 6718–6721. doi: 10.1128/AEM.05212-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Willing, B. P., Dicksved, J., Halfvarson, J., Andersson, A. F., Lucio, M., Zheng, Z., et al. (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xia, L. (2010). Core 3-derived O-glycans are essential for intestinal mucus barrier function. Meth. Enzymol. 479, 123–141. doi: 10.1016/S0076-6879(10)79007-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yamamoto, K., Katayama, T., Kitaoka, M., and Fushinobu, S. (2010). Analyses of bifidobacterial glycosidases involved in the metabolism of oligosaccharides. Biosci. Microfl. 29, 23–30. doi: 10.12938/bifidus.29.23
Yoshida, E., Sakurama, H., Kiyohara, M., Nakajima, M., Kitaoka, M., Ashida, H., et al. (2012). Bifidobacterium longum subsp. infantis uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22, 361–368. doi: 10.1093/glycob/cwr116
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zoetendal, E. G., Von Wright, A., Vilpponen-Salmela, T., Ben-Amor, K., Akkermans, A. D., and De Vos, W. M. (2002). Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68, 3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: gastrointestinal tract, gut bacteria, intestinal mucus, mucin degradation, O-glycosylation, glycoside hydrolase, carbohydrate, gut health and disease
Citation: Tailford LE, Crost EH, Kavanaugh D and Juge N (2015) Mucin glycan foraging in the human gut microbiome. Front. Genet. 6:81. doi: 10.3389/fgene.2015.00081
Received: 13 November 2014; Accepted: 16 February 2015;
Published: 19 March 2015.
Edited by:
Ines Thiele, University of Luxembourg, LuxembourgCopyright © 2015 Tailford, Crost, Kavanaugh and Juge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathalie Juge, The Gut Health and Food Safety Institute Strategic Programme, Institute of Food Research, Norwich Research Park, Norwich NR4 7UA, UKbmF0aGFsaWUuanVnZUBpZnIuYWMudWs=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.