- Department of Biochemistry and Molecular Biology, College of Medicine, Howard University, Washington, DC, USA
RECQ1 is the most abundant member of the human RecQ family of DNA helicases genetically linked with cancer predisposition syndromes and well known for their functions in genome stability maintenance through DNA repair. Despite being the first discovered RecQ homolog in humans, biological functions of RECQ1 have remained largely underappreciated and its relevance to cellular transformation is yet unclear. RECQ1 is overexpressed and amplified in many clinical cancer samples. In silico evaluation of RECQ1 mRNA expression across the NCI-60 cancer cell lines predicts an association of RECQ1 with cancer cell migration, invasion, and metastasis. Consistent with this, latest work implicates RECQ1 in regulation of gene expression, especially of those associated with cancer progression. Functionally, silencing RECQ1 expression significantly reduces cell proliferation, migration, and invasion. Collectively, these results propose that discerning the role of RECQ1 in conferring proliferative and invasive phenotype to cancer cells could be useful in developing therapeutic strategies to block primary tumor progression and metastasis.
Introduction
The RecQ helicase family is a group of highly conserved DNA unwinding enzymes described as caretakers of the genome (Hickson, 2003; Brosh, 2013; Croteau et al., 2014). In humans, RECQ1, also known as RECQL or RECQL1, is the most abundant member of the RecQ helicase family represented by five homologs: RECQ1, WRN, BLM, RECQL4, and RECQL5 (Sharma and Brosh, 2008). Loss of function of three of the five members of the RecQ family are genetically linked with rare cancer susceptibility syndromes (Werner syndrome, Bloom syndrome, and Rothmund–Thomson syndrome) but the association of RECQ1 with cellular transformation is yet unclear. The five human RecQ helicases are expected to exhibit some functional redundancy, but the fact that the functional loss of one homolog cannot be substituted by others clearly indicates critical specialized functions.
RECQ1 is an integral component of replication complex in unperturbed cells and implicated in maintaining replication fork progression (Thangavel et al., 2010). Mechanistically, RECQ1 helicase binds and preferentially unwinds structural intermediates of DNA replication and repair, such as forked duplexes, D-loops, and Holliday junctions; and also exhibits an intrinsic ability to promote annealing of complementary single strand DNA (Sharma et al., 2005; Popuri et al., 2008). RECQ1-deficient cells are characterized by spontaneously elevated sister chromatid exchanges (Sharma and Brosh, 2008) that represent non-productive attempts to restart replication (Helleday, 2003). Furthermore, RECQ1-deficient cells accumulate DNA damage, display increased sensitivity to DNA damaging agents that induce stalled and collapsed replication forks, and exhibit chromosomal instability (Sharma and Brosh, 2007, 2008; Sharma et al., 2007). Thus it is believed that RECQ1 acts to restore productive DNA replication following stress and prevents subsequent genomic instability (Sharma and Brosh, 2008; Popuri et al., 2012; Berti et al., 2013; Sami and Sharma, 2013). RECQ1-deficiency is tolerated by cells to repair I-Sce induced double strand breaks by homologous recombination (Sharma et al., 2012), but a role of RECQ1 in replication stress induced recombination is possible given the demonstrated activity of RECQ1 protein to dissociate native Rad51-bound D-loops by branch migration (Branzei and Foiani, 2007; Bugreev et al., 2008).
Human RECQ1 is ubiquitously expressed in all cell types regardless of cell cycle stage (Kawabe et al., 2000). Previous studies have shown that RECQ1 is upregulated in rapidly dividing cells and its expression is higher in many cancer cell lines as compared to normal cells (Kawabe et al., 2000; Futami et al., 2008a). A meta-analyses of gene expression pattern included RECQ1 among the common signature genes for cancer as defined by the Gene Ontology Consortium (Xu et al., 2007). We noted that RECQ1 is overexpressed and amplified in many clinical cancer samples1. Forty three out of 447 differential expression analyses included RECQ1 in the top 10% upregulation list while only two did in the top 10% downregulation list in cancer versus normal2. Indeed, overexpression of RECQ1 has been experimentally demonstrated in human glioblastoma (Mendoza-Maldonado et al., 2011), ovarian cancer (Sanada et al., 2013), and head and neck squamous cell carcinoma (Arai et al., 2011). Furthermore, RECQ1 protein levels correlated with histological grade and Ki-67 labeling index in hepatocellular carcinoma (Futami et al., 2010) and high proliferative potential in ovarian cancer (Sanada et al., 2013). A polymorphism in RECQ1, A159C, has been associated with faster tumor progression and significantly reduced survival of pancreatic adenocarcinoma patients that received gemcitabine and radiotherapy (Li et al., 2006; Cotton et al., 2009). The A159C SNP is located in the 3′UTR and, thus, may functionally alter RECQ1 expression and affect clinical outcome. Remarkably, RECQ1 silencing significantly reduced proliferation of cancer cells as compared to normal cells in cell culture models and also suppressed tumor growth in mouse xenograft models (Sharma and Brosh, 2007; Futami et al., 2008a,b, 2010; Arai et al., 2011; Mendoza-Maldonado et al., 2011). Tumor cell growth was significantly inhibited in vitro by silencing RECQ1 in hypopharyngeal carcinoma cells and the combination treatment of RECQ1 siRNA and cis-platinum (II) diammine dichloride significantly augmented the in vivo anticancer effects of the drug (Arai et al., 2011). Dysfunctional checkpoint status, e.g., p53 in cancer cells may be linked to mitotic catastrophe following RECQ1 silencing (Futami et al., 2008a) but the ability of RECQ1 to facilitate recovery from replication stress could also be especially important for cancer cells (Popuri et al., 2012; Berti et al., 2013; Lu et al., 2013). Although these studies imply an association of RECQ1 with tumor growth, progression or differentiation, the molecular mechanisms through which RECQ1 might support malignant progression are not understood.
Recent analysis of genome-wide changes in gene expression has revealed a novel involvement of RECQ1 in regulation of gene expression in addition to its role in DNA damage repair (Li et al., 2014). Besides cell proliferation, the top 10 over-represented processes also included cellular movement and cell morphology suggesting that RECQ1 enhances the expression of multiple genes that play key roles in cell migration, invasion, and metastasis, including EZR, ITGA2, ITGA3, ITGB4, SMAD3, and TGFBR2. Functionally, silencing RECQ1 significantly reduced migration and invasion of the highly invasive MDA-MB-231 breast cancer cells indicating that RECQ1 plays a role in enhancing cell migration and invasion (Li et al., 2014). Consistent with the results from in vitro cell culture based study, high RECQ1 expression significantly associated (P = 4.2E-06) with poor overall survival in breast cancer in TCGA dataset (Gyorffy et al., 2010). High expression of RECQL4 was also associated with poor survival in breast cancer but the expression levels of WRN, BLM, and RECQL5 did not correlate with survival (Li et al., 2014).
A putative role in regulation of gene expression has been suggested by studies in Neurospora where a RECQ1 homolog mediates posttranscriptional gene silencing (Cogoni and Macino, 1999). Remarkably, rat RECQ1 was identified in a piRNA protein complex important for gene silencing (Lau et al., 2006), raising the possibility that human RECQ1 may function in a conserved mechanism. Although the mechanism of gene regulation by RECQ1 is yet unknown, the promoters of genes downregulated upon RECQ1 silencing were significantly enriched for a potential G4 sequence motif which are associated with potential to form G-quadruplex structures in DNA (G4 DNA; Li et al., 2014). Chromatin immunoprecipitation experiments demonstrated RECQ1 binding to G4 sequence motifs in the promoters of select downregulated genes indicating that RECQ1 may modulate gene expression by regulating the in vivo stability of G4 DNA structures. Elegant studies from the Maizels (Gray et al., 2014), Monnat and Harris (Nguyen et al., 2014) labs have recently identified that gene regulatory functions of XPB, XPD, and BLM helicases also involve G4 DNA suggesting that the binding to G4 motifs may be a common mechanism of transcriptional regulation by these distinct DNA helicases. Importantly, similar to XPB, but unlike BLM, RECQ1 binds but does not unwind G4 DNA in vitro (Popuri et al., 2008; Sharma, 2011). It is likely that binding to specific G4 sequence motifs by each of these helicases may characterize specific signaling and regulatory pathways associated with cancer. In addition to gene regulatory regions, G4 sequence motifs are also present at chromatin regions preferentially bound by RECQ1 such as replication origins, fragile sites, and telomeres indicating that RECQ1 functions are especially critical at genomic loci having potential to form secondary structures (Thangavel et al., 2010; Lu et al., 2013; Maizels and Gray, 2013; Popuri et al., 2014).
RECQ1 Expression Across the NCI-60 Human Tumor Cell Lines
By virtue of its overexpression in cancer and its association with cell proliferation, migration, and invasion, RECQ1 can potentially serve as a predictive biomarker and attractive target for cancer therapy. To assess gene expression pattern of RECQ1 in various tumors, we utilized a panel of 60 human tumor cell lines derived from nine different tissues of origin (NCI-60) that has been extensively characterized for gene expression and copy-number variations, and commonly used for genetic analysis and screening of potential chemotherapeutic agents (Shoemaker, 2006; Weinstein, 2006; Liu et al., 2010). We conducted in silico analysis of RECQ1 expression pattern across the NCI-60 employing a publicly accessible webtool CellMiner3 (Reinhold et al., 2012; Figure 1). RECQ1 mRNA expression in the NCI-60 panel is shown in Figure 1A both as average transcript intensity values and as the average z score that allows a better comparison of relative expression. Noticeably, the NCI-60 cell lines exhibit a wide spectrum of RECQ1 expression with transcript level values ranging from both lower and higher than the mean (Figure 1B). Cell lines melanoma LOXIMVI and central nervous system SF_268 showed the highest RECQ1 mRNA levels, whereas ovarian cancer cell lines OVCAR_4 and SK_OV_3 showed the lowest (Figure 1B). Nine out of the 10 melanoma cell lines and 4 out of 6 central nervous system cell lines expressed high RECQ1 mRNA (Figure 1B). SK_MEL_5 cells were the only melanoma cells within the NCI-60 panel with lower RECQ1 transcript levels. The lung and renal cell lines were most variable in RECQ1 expression and the ovarian cell lines expressed lowest levels of RECQ1 transcript. It remains to be tested whether RECQ1 transcript levels correlate with the protein level, but a statistically significant correlation with DNA copy number (Pearson’s correlation R = 0.38; P = 0.003) was observed with the estimated RECQ1 DNA copy number ranging from 3.67 for lung cancer cell line NCI_H322M to 1.35 in ovarian cancer cell line SK_OV_3. Expression profiles of five known RecQ family genes across the NCI-60 panel revealed no significant correlation of RECQ1 mRNA expression with WRN, BLM, or RECQL5; however, RECQ1 expression displayed significant inverse correlation with RECQL4 (Figure 1C).
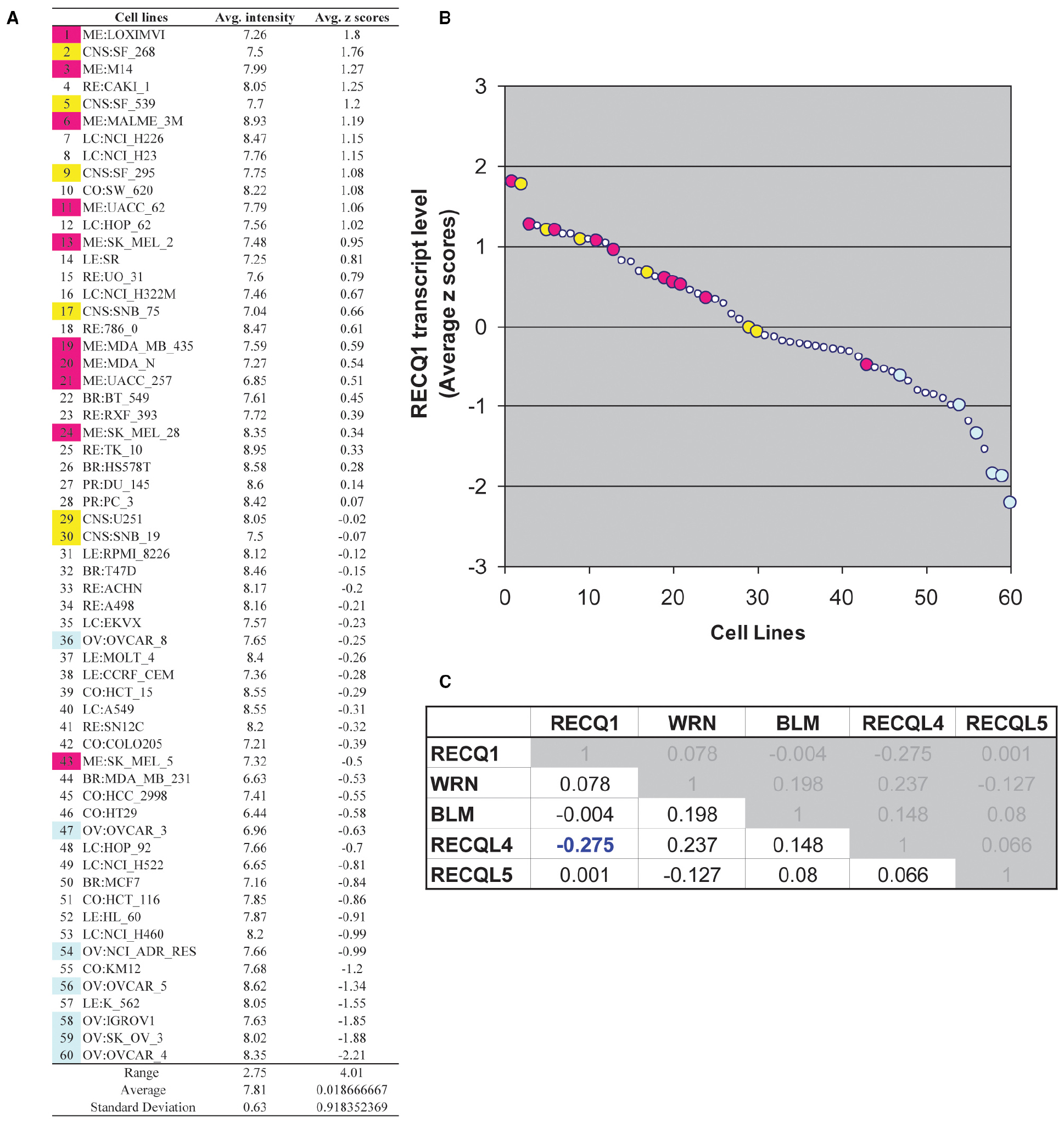
Figure 1. RECQ1 transcript levels in the NCI-60 human tumor cell lines. (A) Table of average intensity (determined from log 2 intensity values from Affymetrix microarrays) and their combined z-score means presented in descending order. (B) Scatter plot depicting the distribution of RECQ1 transcript levels across the NCI-60. The numerical series (1-to-60) for the cell lines from panel (A) correspond to the x-axis. Cell lines from the central nervous system (shown in yellow) and melanoma (shown in pink) displayed higher transcript levels of RECQ1, while ovarian cancer cell lines (shown in blue) displayed the least. (C) Cross-correlation among RecQ family. Pearson’s correlation coefficients between transcripts of the known five RecQ homologs in the NCI-60. Only significant correlation of RECQ1 in the NCI-60 is with the RECQL4 transcripts. Processing and normalization of transcript expression data from the NCI-60 has been described previously (Reinhold et al., 2012). Normalization of Affymetrix HG-U95, HG-U133, HG-U133 plus 2.0 and Affy HuEx 1.0 is done by GC robust multi-array average (GCRMA). Data is accessible at http://discover.nci.nih.gov by using RECQL (RECQ1) as input in Cellminer.
Recent studies using the NCI-60 panel have shown that the genes whose expression at the mRNA level is correlated over diverse cell lines are likely to function together in a molecular network regulating these processes (Kohn et al., 2012, 2014). We postulated that the ability of RECQ1 to promote tumor cell migration and invasion involves changes in expression of specific genes that are critical to these processes. Using the CellMiner NCI-60 analysis tools, we searched for genes that were significantly correlated with RECQ1-expression across the NCI-60 panel and had established functions in cell migration and invasion (Figure 2A). Notably, RECQ1 expression displayed significant positive correlation with mesenchymal differentiation markers vimentin, N-cadherin, and fibronectin (VIM, FN1, and CDH2), and transcription factors that promote these processes (TWIST, SNAI2, and ZEB2). Furthermore, RECQ1 expression correlated significantly and negatively with genes maintaining cellular structural integrity (KRT8) and epithelial marker E-cadherin (CDH1). Correlation of the mRNA transcript levels of RECQ1 with VIM and CDH1 across the NCI-60 tumor cell lines is depicted in Figure 2B. The NCI-60 gene expression profile for CDH1 is inversely correlated to that of the gene VIM which is considered as a hallmark of mesenchymal-like conversion of epithelial cells in carcinomas (Zeisberg and Neilson, 2009; Kohn et al., 2014).
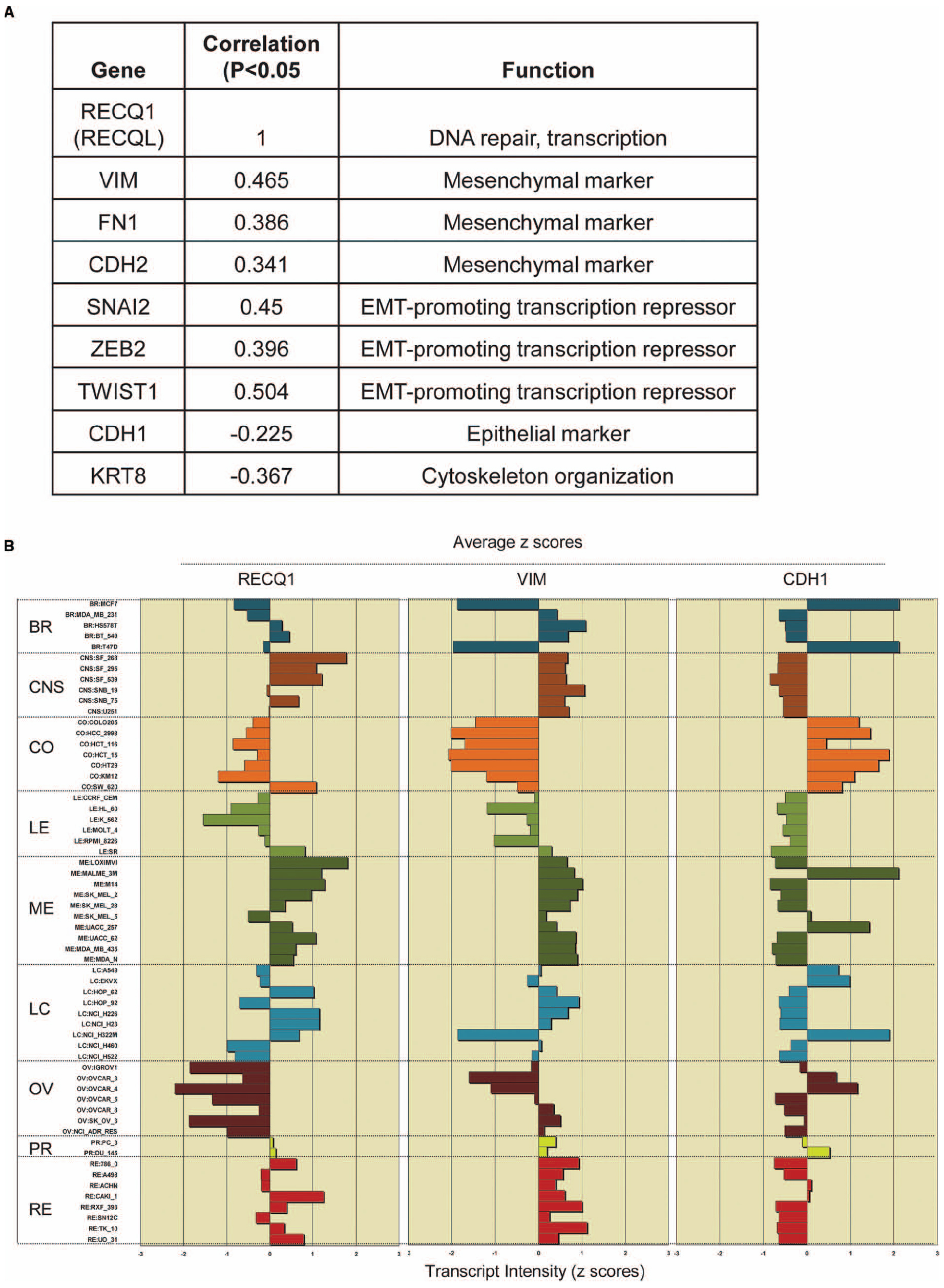
Figure 2. RECQ1 expression in the NCI-60 panel correlates with markers of tumor progression. (A) Significant correlations of RECQ1 with select genes involved the cell invasion, migration, and metastasis. The numbers in the table are expression profile correlations for expression of gene pairs in the NCI-60 panel. EMT, epithelial to mesenchymal transition; (B) RECQ1 expression is highly correlated with the mesenchymal marker vimentin. Comparison of expression profiles for RECQ1, VIM (vimentin), and CDH1 (E-cadherin) across the NCI-60 cell lines. Mean-centered transcript z-cores are plotted on the x-axis; bars to the right show increased expression, bars to the left show decreased expression relative to the expression mean. The cell lines on the y-axis are grouped by tissue of origin. BR, breast; CNS, central nervous system; CO, colon; LE, leukemia; ME, melanoma; LC, lung cancer; OV, ovarian; PR, prostrate; RE, renal. Data was generated querying RECQL (RECQ1) as input in Cellminer (http://discover.nci.nih.gov/cellminer/).
Predictive and Prognostic Potential of RECQ1 Expression in Cancer
Predictions from in silico analysis across the NCI-60 panel support previously reported overexpression of RECQ1 in various cancers and indicate RECQ1 expression to be especially significant in tumors of central nervous system origin and melanoma. Evidently, RECQ1 is highly expressed in human brain glioblastoma relative to control brain tissues and its depletion affects proliferation of glioblastoma cells and causes an increased level of DNA damages (Mendoza-Maldonado et al., 2011). Examining RECQ1 gene expression using the ONCOMINE mRNA microarray database4 revealed that RECQ1 is significantly upregulated in brain and central nervous system tumors, including glioblastoma, when compared to normal tissue. Overexpression of DNA repair genes is associated with metastasis in melanomas (Kauffmann et al., 2008), and the expression levels of RECQ1 significantly correlated with DNA repair genes displaying functional network which are commonly overexpressed in tumors with poorer prognosis in melanoma (Jewell et al., 2010). Given the demonstrated roles of RECQ1 in repairing DNA damage caused by chemotherapeutic agents such as ionizing radiation, camptothecin, and temozolomide, overexpression of RECQ1 may provide a survival advantage to melanoma cells by promoting the ability of cancer cells to tolerate genotoxic stress. In contrast, WRN and RECQL4 may have a tumor suppressor function in melanoma since melanoma has been reported in patients with loss of function mutations in these RecQ proteins (Howell and Bray, 2008; Monnat, 2010). Notably, within the RecQ family, RECQ1 expression correlated significantly and negatively with RECQL4 across the NCI-60 panel. Lao et al. (2013) have recently shown that the expression of BLM and RECQL4 is increased in primary colorectal cancer whereas expression levels of RECQ1 and RECQL5 are decreased. Consistent with the observation in primary colorectal cancer patient samples, low expression of RECQ1 and significantly high expression of RECQL4 were observed in colorectal cancer cell lines as compared to normal colonic mucosa indicating the feasibility of using cell lines to study the functional consequences of alterations in the expression levels of the RecQ helicases (Lao et al., 2013).
RecQ helicases contribute multiple biochemical activities to various DNA repair processes and loss of their functions leads to increased DNA damage, genomic instability, and enhanced sensitivity to certain chemotherapeutic agents (Brosh, 2013). Therefore, altered expression of RecQ helicases may be useful in predicting patient’s response to these DNA damaging drugs used for treatment of cancer. Indeed RECQ1 expression correlated with cisplatin resistance in oral squamous cell carcinoma (Zhang et al., 2006) and the depletion of RECQ1 significantly augmented the in vivo anticancer effects of the drug cis-platinum (II) diammine dichloride (Arai et al., 2011). This is consistent with the fact that cisplatin and related platinum drugs induce inter-strand cross links in DNA that impair progression of replication forks and RECQ1 helicase functions are important to restore productive DNA replication (Sami and Sharma, 2013). However, RECQ1 expression was found to be elevated in ovarian cancer cells that were sensitive to carboplatin as compared to those which were carboplatin-resistant (Peters et al., 2005) indicating complex mechanisms of platinum resistance (Stewart, 2007). Thus, a better correlation of RECQ1 expression with the molecular characteristics and heterogeneity of cancer needs to be established in order to test the use of RECQ1 as a potential biomarker.
In addition to unlimited proliferative potential, a tumor cell must acquire the ability to migrate and invade normal tissues to become fully malignant (Hanahan and Weinberg, 2011). Invasion of tumor cells and metastatic spread to distant organs relies on complex molecular interactions including diminished epithelial characteristics and enhanced mesenchymal attributes (Kalluri and Weinberg, 2009). RECQ1 expression across the NCI-60 panel significantly correlated with a loss of the epithelial marker CDH1 and acquisition of expression of the mesenchymal markers including VIM. Correlation with VIM in the NCI-60 panel raises the question whether RECQ1 expression is associated with the epithelial to mesenchymal transition (EMT) which is among the central mechanisms to induce invasiveness and metastasis of tumors. Indeed, RECQ1-depletion decreased cell migration and invasion in cervical adenocarcinoma HeLa and breast cancer MDA-MB-231 cell lines (Li et al., 2014). Similarly, RECQ1 silencing in oral squamous cell carcinoma SCC-9 cells downregulated the expression level of immunosuppressive factors that are necessary for regulating the migration of tumor cells (Tao et al., 2014). Collectively these results propose that RECQ1 may contribute to tumor progression by regulating key genes that promote cancer cell migration, invasion, and metastasis. It is plausible that similar to what has been shown for a few other proteins involved in DNA damage response and repair, RECQ1 participates in transcription regulation either by binding directly to DNA or through interaction with specific transcription factors (Featherstone and Jackson, 1999; Mullan et al., 2006; Kraus, 2008; Jaehnig et al., 2013; Broustas and Lieberman, 2014). A systematic investigation of RECQ1-regulated transcriptome may uncover the gene networks regulated by RECQ1 in the context of cancer progression.
Increased expression of several DNA repair proteins has been correlated with cellular invasiveness in cancer (Mitra et al., 2009; Barbano et al., 2011; Martinez-Marignac et al., 2011; Yuan et al., 2012); however, little effort has been made thus far to determine any role of RecQ helicase homologs in tumor progression and metastasis. Understanding the role of RECQ1 in conferring proliferative and invasive phenotype to cancer cells could be useful in developing therapeutic strategies to block primary tumor progression and metastases. Functional evidence from RECQ1 silencing, and the projected correlation with VIM expression in the NCI-60 panel implies an association of RECQ1 with more aggressive disease (Zeisberg and Neilson, 2009). Future studies should examine whether RECQ1 mRNA and/or protein levels can predict invasion and metastatic potential of a tumor. Predictions from large human cancer cell line panels that capture tumor heterogeneity, such as NCI-60, and the availability of clinically and molecularly annotated patient tumor datasets for multiple tumor types, such as TCGA, should provide additional opportunities to test the significance of RECQ1 in developing new in vitro preclinical models for cancer detection and treatment.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the NIGMS/NIH under award number SC1GM093999 and NSF under HU-ADVANCE-IT mini-grant. The content is solely the responsibility of the author and does not necessarily represent the official views of the funding agencies.
Footnotes
References
Arai, A., Chano, T., Futami, K., Furuichi, Y., Ikebuchi, K., Inui, T., et al. (2011). RECQL1 and WRN proteins are potential therapeutic targets in head and neck squamous cell carcinoma. Cancer Res. 71, 4598–4607. doi: 10.1158/0008-5472.CAN-11-0320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barbano, R., Copetti, M., Perrone, G., Pazienza, V., Muscarella, L. A., Balsamo, T., et al. (2011). High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient’s outcome. Int. J. Cancer 129, 536–545. doi: 10.1002/ijc.25736
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berti, M., Chaudhuri, A. R., Thangavel, S., Gomathinayagam, S., Kenig, S., Vujanovic, M., et al. (2013). Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20, 347–354. doi: 10.1038/nsmb.2501
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Branzei, D., and Foiani, M. (2007). RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 21, 3019–3026. doi: 10.1101/gad.1624707
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brosh, R. M. Jr. (2013). DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13, 542–558. doi: 10.1038/nrc3560
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Broustas, C. G., and Lieberman, H. B. (2014). DNA damage response genes and the development of cancer metastasis. Radiat. Res. 181, 111–130. doi: 10.1667/RR13515.1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bugreev, D. V., Brosh, R. M. Jr., and Mazin, A. V. (2008). RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 283, 20231–20242. doi: 10.1074/jbc.M801582200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cogoni, C., and Macino, G. (1999). Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286, 2342–2344. doi: 10.1126/science.286.5448.2342
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cotton, R. T., Li, D., Scherer, S. E., Muzny, D. M., Hodges, S. E., Catania, R. L., et al. (2009). Single nucleotide polymorphism in RECQL and survival in resectable pancreatic adenocarcinoma. HPB 11, 435–444. doi: 10.1111/j.1477-2574.2009.00089.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croteau, D. L., Popuri, V., Opresko, P. L., and Bohr, V. A. (2014). Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83, 519–552. doi: 10.1146/annurev-biochem-060713-035428
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Featherstone, C., and Jackson, S. P. (1999). Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 434, 3–15. doi: 10.1016/S0921-8777(99)00006-3
Futami, K., Kumagai, E., Makino, H., Goto, H., Takagi, M., Shimamoto, A., et al. (2008a). Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci. 99, 71–80. doi: 10.1111/j.1349-7006.2007.00647.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Futami, K., Kumagai, E., Makino, H., Sato, A., Takagi, M., Shimamoto, A., et al. (2008b). Anticancer activity of RecQL1 helicase siRNA in mouse xenograft models. Cancer Sci. 99, 1227–1236. doi: 10.1111/j.1349-7006.2008.00794.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Futami, K., Ogasawara, S., Goto, H., Yano, H., and Furuichi, Y. (2010). RecQL1 DNA repair helicase: a potential tumor marker and therapeutic target against hepatocellular carcinoma. Int. J. Mol. Med. 25, 537–545. doi: 10.3892/ijmm_00000375
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gray, L. T., Vallur, A. C., Eddy, J., and Maizels, N. (2014). G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 10, 313–318. doi: 10.1038/nchembio.1475
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gyorffy, B., Lanczky, A., Eklund, A. C., Denkert, C., Budczies, J., Li, Q., et al. (2010). An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731. doi: 10.1007/s10549-009-0674-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Helleday, T. (2003). Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 532, 103–115. doi: 10.1016/j.mrfmmm.2003.08.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hickson, I. D. (2003). RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3, 169–178. doi: 10.1038/nrc1012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Howell, S. M., and Bray, D. W. (2008). Amelanotic melanoma in a patient with Rothmund–Thomson syndrome. Arch. Dermatol. 144, 416–417. doi: 10.1001/archderm.144.3.416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jaehnig, E. J., Kuo, D., Hombauer, H., Ideker, T. G., and Kolodner, R. D. (2013). Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Rep. 4, 174–188. doi: 10.1016/j.celrep.2013.05.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jewell, R., Conway, C., Mitra, A., Randerson-Moor, J., Lobo, S., Nsengimana, J., et al. (2010). Patterns of expression of DNA repair genes and relapse from melanoma. Clin. Cancer Res. 16, 5211–5221. doi: 10.1158/1078-0432.ccr-10-1521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kalluri, R., and Weinberg, R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428. doi: 10.1172/jci39104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kauffmann, A., Rosselli, F., Lazar, V., Winnepenninckx, V., Mansuet-Lupo, A., Dessen, P., et al. (2008). High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene 27, 565–573. doi: 10.1038/sj.onc.1210700
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawabe, T., Tsuyama, N., Kitao, S., Nishikawa, K., Shimamoto, A., Shiratori, M., et al. (2000). Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene 19, 4764–4772. doi: 10.1038/sj.onc.1203841
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kohn, K. W., Zeeberg, B. M., Reinhold, W. C., and Pommier, Y. (2014). Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype. PLoS ONE 9:e99269. doi: 10.1371/journal.pone.0099269
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kohn, K. W., Zeeberg, B. R., Reinhold, W. C., Sunshine, M., Luna, A., and Pommier, Y. (2012). Gene expression profiles of the NCI-60 human tumor cell lines define molecular interaction networks governing cell migration processes. PLoS ONE 7:e35716. doi: 10.1371/journal.pone.0035716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kraus, W. L. (2008). Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302. doi: 10.1016/j.ceb.2008.03.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lao, V. V., Welcsh, P., Luo, Y., Carter, K. T., Dzieciatkowski, S., Dintzis, S., et al. (2013). Altered RECQ helicase expression in sporadic primary colorectal cancers. Transl. Oncol. 6, 458–469. doi: 10.1593/tlo.13238
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lau, N. C., Seto, A. G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D. P., et al. (2006). Characterization of the piRNA complex from rat testes. Science 313, 363–367. doi: 10.1126/science.1130164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, D., Frazier, M., Evans, D. B., Hess, K. R., Crane, C. H., Jiao, L., et al. (2006). Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J. Clin. Oncol. 24, 1720–1728. doi: 10.1200/JCO.2005.04.4206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X. L., Lu, X., Parvathaneni, S., Bilke, S., Zhang, H., Thangavel, S., et al. (2014). Identification of RECQ1-regulated transcriptome uncovers a role of RECQ1 in regulation of cancer cell migration and invasion. Cell Cycle 13. [Epub ahead of print].
Liu, H., D’Andrade, P., Fulmer-Smentek, S., Lorenzi, P., Kohn, K. W., Weinstein, J. N., et al. (2010). mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol. Cancer Ther. 9, 1080–1091. doi: 10.1158/1535-7163.MCT-09-0965
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lu, X., Parvathaneni, S., Hara, T., Lal, A., and Sharma, S. (2013). Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol. Cancer 12, 29. doi: 10.1186/1476-4598-12-29
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maizels, N., and Gray, L. T. (2013). The G4 genome. PLoS Genet. 9:e1003468. doi: 10.1371/journal.pgen.1003468
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martinez-Marignac, V. L., Rodrigue, A., Davidson, D., Couillard, M., Al-Moustafa, A. E., Abramovitz, M., et al. (2011). The effect of a DNA repair gene on cellular invasiveness: XRCC3 over-expression in breast cancer cells. PLoS ONE 6:e16394. doi: 10.1371/journal.pone.0016394
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mendoza-Maldonado, R., Faoro, V., Bajpai, S., Berti, M., Odreman, F., Vindigni, M., et al. (2011). The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol. Cancer 10, 83. doi: 10.1186/1476-4598-10-83
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitra, A., Jameson, C., Barbachano, Y., Sanchez, L., Kote-Jarai, Z., Peock, S., et al. (2009). Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology 55, 696–704. doi: 10.1111/j.1365-2559.2009.03448.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monnat, R. J. Jr. (2010). Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Semin. Cancer Biol. 20, 329–339. doi: 10.1016/j.semcancer.2010.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mullan, P. B., Quinn, J. E., and Harkin, D. P. (2006). The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25, 5854–5863. doi: 10.1038/sj.onc.1209872
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nguyen, G. H., Tang, W., Robles, A. I., Beyer, R. P., Gray, L. T., Welsh, J. A., et al. (2014). Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. U.S.A. 111, 9905–9910. doi: 10.1073/pnas.1404807111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peters, D., Freund, J., and Ochs, R. L. (2005). Genome-wide transcriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells. Mol. Cancer Ther. 4, 1605–1616. doi: 10.1158/1535-7163.mct-04-0311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Popuri, V., Bachrati, C. Z., Muzzolini, L., Mosedale, G., Costantini, S., Giacomini, E., et al. (2008). The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 283, 17766–17776. doi: 10.1074/jbc.M709749200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Popuri, V., Croteau, D. L., Brosh, R. M. Jr., and Bohr, V. A. (2012). RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle 11, 4252–4265. doi: 10.4161/cc.22581
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Popuri, V., Hsu, J., Khadka, P., Horvath, K., Liu, Y., Croteau, D. L., et al. (2014). Human RECQL1 participates in telomere maintenance. Nucleic Acids Res. 42, 5671–5688. doi: 10.1093/nar/gku200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reinhold, W. C., Sunshine, M., Liu, H., Varma, S., Kohn, K. W., Morris, J., et al. (2012). CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–3511. doi: 10.1158/0008-5472.CAN-12-1370
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sami, F., and Sharma, S. (2013). Probing genome maintenance functions of human RECQ1. Comput. Struct. Biotechnol. J. 6, e201303014. doi: 10.5936/csbj.201303014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sanada, S., Futami, K., Terada, A., Yonemoto, K., Ogasawara, S., Akiba, J., et al. (2013). RECQL1 DNA repair helicase: a potential therapeutic target and a proliferative marker against ovarian cancer. PLoS ONE 8:e72820. doi: 10.1371/journal.pone.0072820
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, S. (2011). Non-B DNA secondary structures and their resolution by RecQ helicases. J. Nucleic Acids 2011, 724215. doi: 10.4061/2011/724215
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, S., and Brosh, R. M. Jr. (2007). Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE 2:e1297. doi: 10.1371/journal.pone.0001297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, S., and Brosh, R. M. Jr. (2008). Unique and important consequences of RECQ1 deficiency in mammalian cells. Cell Cycle 7, 989–1000. doi: 10.4161/cc.7.8.5707
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, S., Phatak, P., Stortchevoi, A., Jasin, M., and Larocque, J. R. (2012). RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst.) 11, 537–549. doi: 10.1016/j.dnarep.2012.04.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, S., Sommers, J. A., Choudhary, S., Faulkner, J. K., Cui, S., Andreoli, L., et al. (2005). Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 280, 28072–28084. doi: 10.1074/jbc.M500264200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, S., Stumpo, D. J., Balajee, A. S., Bock, C. B., Lansdorp, P. M., Brosh, R. M., et al. (2007). RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol. Cell. Biol. 27, 1784–1794. doi: 10.1128/MCB.01620-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shoemaker, R. H. (2006). The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823. doi: 10.1038/nrc1951
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stewart, D. J. (2007). Mechanisms of resistance to cisplatin and carboplatin. Crit. Rev. Oncol. Hematol. 63, 12–31. doi: 10.1016/j.critrevonc.2007.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tao, J., Tao, S., Han, J., Zhou, Z., Zhang, X., Wang, H., et al. (2014). RECQL1 plays an important role in the development of tongue squamous cell carcinoma. Cell Physiol. Biochem. 33, 1579–1590. doi: 10.1159/000358721
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thangavel, S., Mendoza-Maldonado, R., Tissino, E., Sidorova, J. M., Yin, J., Wang, W., et al. (2010). Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol. 30, 1382–1396. doi: 10.1128/MCB.01290-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weinstein, J. N. (2006). Spotlight on molecular profiling: “integromic” analysis of the NCI-60 cancer cell lines. Mol. Cancer Ther. 5, 2601–2605. doi: 10.1158/1535-7163.MCT-06-0640
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, L., Geman, D., and Winslow, R. L. (2007). Large-scale integration of cancer microarray data identifies a robust common cancer signature. BMC Bioinformatics 8:275. doi: 10.1186/1471-2105-8-275
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yuan, S. S., Hou, M. F., Hsieh, Y. C., Huang, C. Y., Lee, Y. C., Chen, Y. J., et al. (2012). Role of MRE11 in cell proliferation, tumor invasion, and DNA repair in breast cancer. J. Natl. Cancer Inst. 104, 1485–1502. doi: 10.1093/jnci/djs355
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zeisberg, M., and Neilson, E. G. (2009). Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 119, 1429–1437. doi: 10.1172/JCI36183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: RecQ, helicase, gene expression, vimentin, metastasis, epithelial mesenchymal transition
Citation: Sharma S (2014) An appraisal of RECQ1 expression in cancer progression. Front. Genet. 5:426. doi: 10.3389/fgene.2014.00426
Received: 30 September 2014; Accepted: 19 November 2014;
Published online: 05 December 2014.
Edited by:
Fumiaki Uchiumi, Tokyo University of Science, JapanReviewed by:
Srinivas Ayyadevara, Central Arkansas Veterans Healthcare System, USADavid Lombard, University of Michigan, USA
Copyright © 2014 Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudha Sharma, Department of Biochemistry and Molecular Biology, College of Medicine, Howard University, 520 W Street, Northwest, Washington, DC 20059, USA e-mail:c3VkaGEuc2hhcm1hQGhvd2FyZC5lZHU=
 Sudha Sharma
Sudha Sharma