- 1Department of Biomedical Chemistry, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
- 2Department of Biotechnology, Okayama University, Okayama, Japan
Conventional tRNAs have highly conserved sequences, four-armed cloverleaf secondary structures, and L-shaped tertiary structures. However, metazoan mitochondrial tRNAs contain several exceptional structures. Almost all tRNAsSer for AGY/N codons lack the D-arm. Furthermore, in some nematodes, no four-armed cloverleaf-type tRNAs are present: two tRNAsSer without the D-arm and 20 tRNAs without the T-arm are found. Previously, we showed that in nematode mitochondria, an extra elongation factor Tu (EF-Tu) has evolved to support interaction with tRNAs lacking the T-arm, which interact with C-terminal domain 3 in conventional EF-Tu. Recent mitochondrial genome analyses have suggested that in metazoan lineages other than nematodes, tRNAs without the T-arm are present. Furthermore, even more simplified tRNAs are predicted in some lineages. In this review, we discuss mitochondrial tRNAs with divergent structures, as well as protein factors, including EF-Tu, that support the function of truncated metazoan mitochondrial tRNAs.
Introduction
As discussed in other articles in this special issue, conventional tRNAs are highly conserved: they have a four-armed cloverleaf secondary structure and L-shaped tertiary structure (Figures 1A,B; Jühling et al., 2009). However, some tRNAs encoded in mitochondrial DNA, particularly in metazoan (multi-cellular animal) mitochondria, have diverged from standard form tRNAs in a variety of ways. In this review, we focus on mitochondrial tRNAs (mt tRNAs) lacking either the dihydrouridine arm (D-arm) or the ribothimidine arm (T-arm; Figures 1C,D). The function of tRNA is to help decode mRNA into protein. tRNA collaborates with a variety of proteins from post-transcription to decoding in ribosomes. The unique characteristics of factors interacting with such shrunken tRNAs have been uncovered over the past several decades. In this review, these factors and their evolution will also be discussed.
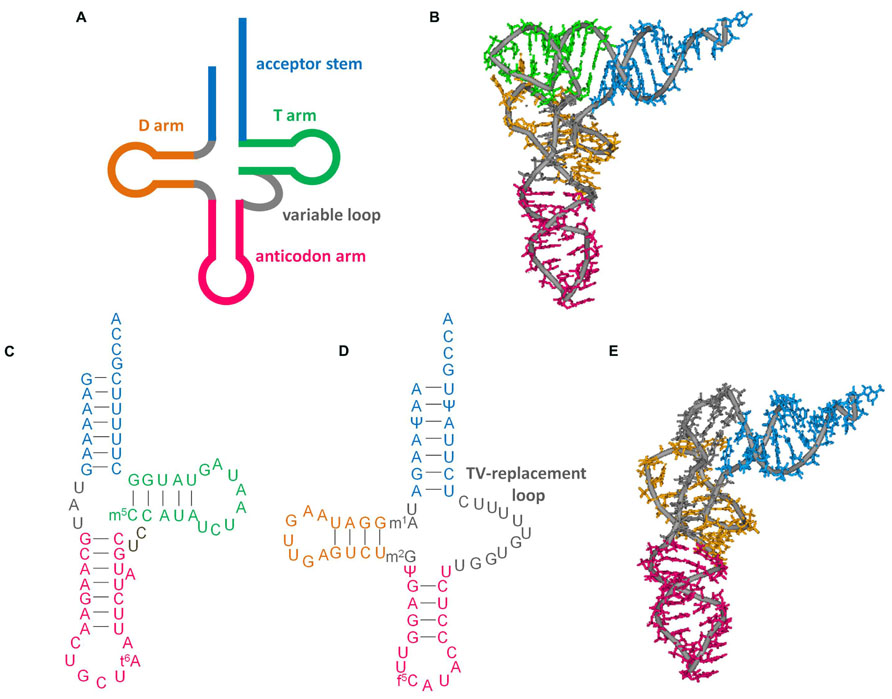
FIGURE 1. Secondary and tertiary structures of tRNAs. (A) Cloverleaf tRNA. (B) L-shape tertiary structure of cloverleaf tRNA (Saccharomyces cerevisiae tRNAPhe, PDB code 6TNA). (C) D-arm-lacking tRNA (bovine mt tRNASer(GCU), accession number: X15132. Modification data from tRNAdb (Jühling et al., 2009), tRNAdb ID: tdbR0000402). (D) T-arm-lacking tRNA (Ascaris suum mt tRNAMet, accession number: D28746; Watanabe et al., 1994; Sakurai et al., 2005b). (E) L-shape model of T-arm-lacking tRNA (Ohtsuki et al., 1998) built with reference to the crystal structure of yeast tRNAPhe. This model was built from computational analysis including molecular dynamics calculations to arrange the base locations according to tertiary interactions deduced from NMR observations (Ohtsuki et al., 1998), sequence alignment (Wolstenholme et al., 1994), and structural probing experiments (Watanabe et al., 1994).
Shrunken mt tRNAs
tRNAs Lacking the D-arm
In the 1970s, D-arm-lacking tRNAs were at first identified as non-tRNA molecules (a putative equivalent to cytoplasmic 5S ribosomal RNA) because of their short length (Dubin and Friend, 1972; Dubin et al., 1974; Baer and Dubin, 1980). Since the identification of genes in mitochondrial DNA, these truncated tRNAs have been proposed to be functional (Arcari and Brownlee, 1980; de Bruijn et al., 1980). Virtually all metazoan mitochondria have at least one of D-arm-lacking tRNA, namely tRNASer(GCU/UCU) for AGY or AGN codons (Figure 1C; Jühling et al., 2009). In addition, some animal mitochondria have additional D-arm-lacking tRNAs, such as tRNASer(UGA) in chromadorean nematodes (Okimoto et al., 1992), and tRNACys in some vertebrates (Seutin et al., 1994).
The secondary structures of D-arm-lacking tRNAs have been classified into several groups based on the base pairs in T and anticodon stems (Steinberg et al., 1994). Experimental verifications of the secondary and tertiary structures have been performed using chemical modification, limited enzymatic digestion, nuclear magnetic resonance (NMR) spectroscopy, and native gel electrophoresis (de Bruijn and Klug, 1983; Ueda et al., 1985; Hayashi et al., 1997; Frazer-Abel and Hagerman, 1999, 2004; Ohtsuki et al., 2002a). Although these results support the coaxial stacking of T and acceptor stems (Frazer-Abel and Hagerman, 2008), this idea is somewhat controversial, possibly because of the structural flexibility of the D-arm-lacking tRNAs themselves (Frazer-Abel and Hagerman, 2008). The shortest possible D-arm-lacking tRNA was suggested to be 54 nt long (Steinberg et al., 1994).
Aminoacylation and EF-Tu binding of D-arm-lacking tRNAs have been demonstrated (Ueda et al., 1985, 1992; Yokogawa et al., 1989, 2000; Kumazawa et al., 1991; Watanabe et al., 1994; Hanada et al., 2001; Shimada et al., 2001; Ohtsuki et al., 2002a; Chimnaronk et al., 2005; Suematsu et al., 2005). Translation with an unmodified D-arm-lacking mammalian mt tRNASer(GCU) derivative with a GAA anticodon has been investigated using a cell-free system (Hanada et al., 2001). The ability to form a ternary complex with EF-Tu/GTP of the tRNASer(GCU) derivative is similar to that of the tRNASer(UGA) derivative, which has a four-armed cloverleaf secondary structure (Hanada et al., 2001). However, the amount of peptides produced using tRNASer(GCU) derivative is lower than that produced using tRNASer(UGA) derivative (Hanada et al., 2001).
tRNAs Lacking the T-arm
T-arm-lacking tRNA genes were identified in nematode mitochondria in Wolstenholme et al. (1987). Since then, T-arm-lacking tRNA genes have also been found in mitochondrial DNA in other lineages of animals, such as Arthropoda (Masta, 2000). They have a TV replacement loop instead of a variable loop and T-arm (Figure 1D). Isolation of nematode mt T-arm-lacking tRNAs has been performed with preparative gel electrophoresis and/or solid-phase DNA affinity purification (Watanabe et al., 1994, 1997, Ohtsuki et al., 1998; Sakurai et al., 2005a,b).
Basically, intramolecular interactions in T-arm-lacking tRNAs are thought to be identical to those in conventional tRNAs, except for interactions between T- and D- arms, because of conservation around the D-arm and the similarity of the 5′ region in the TV replacement loop to the variable loop region (Watanabe et al., 1994; Wolstenholme et al., 1994). As an analog of cloverleaf-type tRNA, an L-shape-like structure of T-arm-lacking tRNAs has been proposed (Watanabe et al., 1994; Wolstenholme et al., 1994). Hypothesized interactions have been supported by chemical and enzymatic probing and NMR spectroscopy (Figure 1E; Watanabe et al., 1994; Ohtsuki et al., 1998).
The aminoacylation capacity of T-arm-lacking nematode mt tRNAs has been demonstrated with mt extract or recombinant enzymes (Watanabe et al., 1994; Ohtsuki et al., 1996; Chihade et al., 1998; Lovato et al., 2001; Sakurai et al., 2005b; Arita et al., 2006). Furthermore, the formation of a tertiary complex of a T-arm-lacking aminoacyl tRNA/EF-Tu/GTP has been detected (Ohtsuki et al., 2001; Arita et al., 2006).
In nematode mt T-arm-lacking tRNAs sequenced at the RNA level, the 1-methyladenosine at position 9 is strictly conserved (Watanabe et al., 1994, 1997; Sakurai et al., 2005a,b; see also Ohtsuki and Watanabe, 2007). This modification helps maintain the tertiary structure of the tRNA, and also aids in efficient aminoacylation and formation of the ternary complex with EF-Tu/GTP (Sakurai et al., 2005b).
tRNAs Potentially Lacking Both D- and T-arms
As mentioned above, the shortest biochemically characterized tRNA is a 54-nt long mt tRNASer(UCU) from the nematode Ascaris suum (Watanabe et al., 1994). A computational survey of mitochondrial tRNA genes predicted the presence of tRNA genes lacking both D- and T-arms (Jühling et al., 2012a,b). More recently, after the submission of the abstract of this review, RT-PCR analyses using 5′- and 3′-RACE showed that such putative tRNA genes are indeed transcribed, and the transcripts even have a 3′CCA sequence in the nematode Romanomermis culicivorax (Wende et al., 2014). Note that some tRNAs are imported from the cytoplasm into the mitochondria in some animals (Rubio et al., 2008), suggesting that imported tRNAs may function in place of mitochondrial-encoded putative tRNAs lacking both D- and T-arms. Thus, functional analysis of such extremely truncated putative tRNAs is critical.
Factors Interacting with tRNAs Lacking D- or T-arms
Aminoacyl tRNA Synthetases
Aminoacyl tRNA synthetases recognize a cognate tRNA and add an aminoacyl moiety to its 3′ end. The major recognition sites of the enzymes in tRNAs are the anticodon, a discriminator base at position 73, and the acceptor stem (Giegé et al., 1998). In fact, in the case of alanyl-tRNA synthetases, even the mitochondrial enzyme uses the acceptor stem as a major recognition site (Chihade et al., 1998). However, some of the enzymes also use the D-arm in tRNA [e.g., Escherichia coli isoleucyl-tRNA synthetase (Nureki et al., 1994)]. Whether the mitochondrial counterparts of such enzymes encoded in nuclear genome still use D-arms as recognition sites is an interesting issue yet to be investigated. If so, even if a tRNA lost its T-arm, the enzyme could still add the aminoacyl moiety to the shrunken tRNA.
On the other hand, seryl-tRNA synthetase (SerRS) uses recognition sites other than the anticodon and acceptor stem. Bacterial SerRS recognizes T- and characteristic long variable arms in bacterial tRNASer (Asahara et al., 1994) using the N-terminal coiled–coil region (Biou et al., 1994). However, metazoan mt tRNASer has lost its long variable arm, and even the D-arm is absent in tRNASer(GCU/UCU). Thus, how mt SerRS recognizes mt tRNASer without its long variable arm is an interesting question. Earlier studies suggested that mammalian mt SerRS can recognize not only mt tRNASer but also bacterial tRNASer; however, bacterial SerRS could not recognize mt tRNASer (Kumazawa et al., 1991). Also, mammalian mt SerRS recognizes the T-loop of both D-arm-lacking tRNASer and cloverleaf-type tRNASer(UGA) without the long variable arm, and further requires a T-loop/D-loop interaction with tRNASer(UGA) (dual-mode recognition; Ueda et al., 1992; Shimada et al., 2001). The crystal structure of a mammalian mt SerRS, a model of it complexed with tRNA, and mutational analyses suggest that the N-terminal coiled–coil region, the distal helix, and the C-tail interact with the T-arm of mt tRNASer (Chimnaronk et al., 2005, Figure 2). Mutational analysis of mammalian mt SerRS showed the substitution of some of the residues in N-terminal coiled–coil region (shown in stick model in Figure 2B) reduced the aminoacylation activities of either one of two mt tRNAsSer or both, suggesting that interaction of these residues with the tRNA in the enzyme-tRNA complex (Chimnaronk et al., 2005, Figure 2B). To maintain these interactions, the movement of N-terminal coiled–coil region (shown as a red arrow) is expected (Chimnaronk et al., 2005, Figure 2B). Furthermore, mutational analysis of the enzyme suggested that, for the recognition of tRNASer(UGA) which have T-loop/D-loop interaction, Arg24, Tyr28 and Arg32, in the distal helix and the Lys93 and Arg122 on the N-terminal coiled–coil region are important (Figure 2B). On the other hand, for the D-arm-lacking tRNASer(GCU), Arg24 and Arg32 flanking the distal helix, and Arg129 on the N-terminal helical region are crucial (Figure 2B). Thus, with the dual-mode recognition, mammalian mt SerRS recognizes two tRNASer isoacceptors which have different secondary structures using distinct sets of the residues.
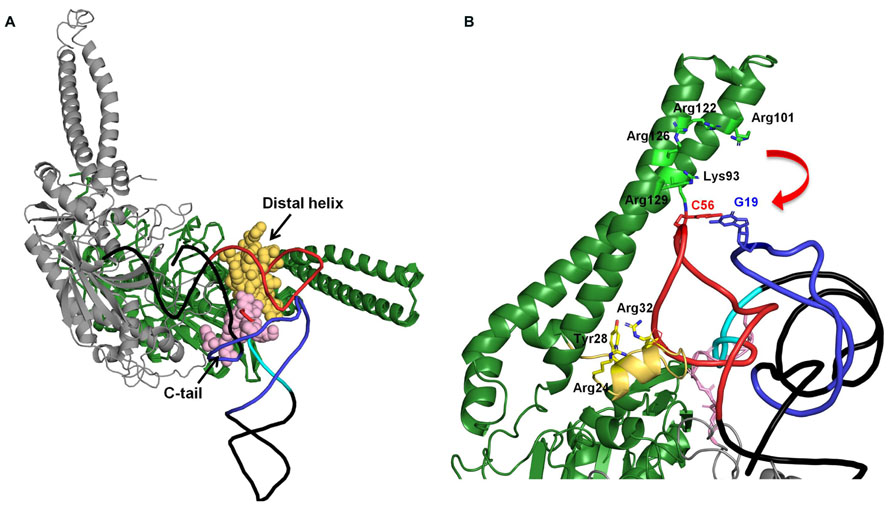
FIGURE 2. (A) Docking model of mammalian mt SerRS and yeast tRNAPhe (Chimnaronk et al., 2005). Two subunits of the enzyme are shown in green (monomer 1), and gray (monomer 2), respectively. Distal helix of monomer 1 and C-tail of monomer 2 which are mitochondrial-specific extensions and possibly interact with the tRNA, are shown in yellow and pink, respectively. In the tRNA, D-arm, variable loop, and T-arm are shown in purple, sky blue, and red, respectively. (B) Putative interactions between the N-terminal coiled–coil region and the C-tail distal helix with the T-arm of the tRNA. The interactions are inferred by mutational analysis of the enzyme (Chimnaronk et al., 2005). Residues in N-terminal coiled–coil region and distal helix involved in the interaction with tRNA are shown in stick model (Chimnaronk et al., 2005).
Interestingly, in chromadorean nematode mt, two tRNAsSer have lost their D-arms, but the remaining 20 tRNAs have lost their T-arms. It is of interest whether nematode mt SerRS also recognize the T-arm. If so, that may explain why only tRNAsSer have kept their T-arms in nematode mitochondria. However, it seems reasonable that the secondary structure of tRNA is governed by EF-Tu and ribosomes rather than aminoacyl-tRNA synthetase (ARS) during evolution, because the recognition mode of each ARS is constrained by only one or a few tRNA, while that of EF-Tu or the ribosome is constrained by over 20 tRNAs.
EF-Tu
The EF-Tu/GTP complex delivers aminoacyl-tRNAs to the A-site in ribosomes. Bacterial EF-Tu binds to the aminoacyl-moiety, a part of acceptor stem, and the T-arm (Nissen et al., 1995; Figure 3A), and it cannot bind to a tRNA analog missing the T-arm (Rudinger et al., 1994). In mitochondria, nuclear-encoded EF-Tu exists. Due to the presence of aminoacyl-tRNAs missing the T-arm in nematode mitochondria (Watanabe et al., 1994; Ohtsuki et al., 1996), the EF-Tu counterpart in nematode mitochondria should use an alternative binding mode for T-arm-lacking tRNAs. In fact, nematode mitochondria have two EF-Tu homologs, and only one of them (EF-Tu1) binds to T-arm-lacking tRNAs (Ohtsuki et al., 2001; Arita et al., 2006). Caenorhabditis elegans mt EF-Tu1 has an approximately 60 amino-acid extension at the C-terminus (domain 3′) that is essential for binding to T-arm-lacking tRNAs (Ohtsuki et al., 2001; Sakurai et al., 2006). The extension likely interacts with the D-arm region of T-arm-lacking tRNAs through positive charges in Lys residues (Sakurai et al., 2006; Figures 3B,C). Interestingly, C. elegans mt EF-Tu1 lacks binding ability to cloverleaf-type tRNAs (Ohtsuki et al., 2001), which are missing in C. elegans mitochondria (Okimoto et al., 1992). In another lineage of nematode, Trichinella, mitochondria have T-arm-lacking tRNAs, cloverleaf-type tRNAs, and D-arm-lacking tRNAsSer (Lavrov and Brown, 2001). Trichinella also have two EF-Tu homologs, and one of them (EF-Tu1) binds to both T-arm-lacking tRNAs and cloverleaf-type tRNAs (Arita et al., 2006). Interestingly, Trichinella mt EF-Tu1 has a 41-residue C-terminal extension shorter than that in C. elegans mitochondria (Arita et al., 2006). A mutant of C. elegans mt EF-Tu1 with a 13-residue deletion at the C-terminus (43-residue extension left) cannot bind to T-arm-lacking tRNAs (Sakurai et al., 2006). Although the detailed tRNA binding mode of Trichinella mt EF-Tu1 has not been elucidated, it could be similar but not identical to that of C. elegans mt EF-Tu1. Note that C. elegans EF-Tu1 binds to only T-arm-lacking tRNAs, while Trichinella EF-Tu1 binds to T-arm-lacking tRNA, D-arm-lacking tRNA, and cloverleaf tRNA (Ohtsuki et al., 2001; Arita et al., 2006).
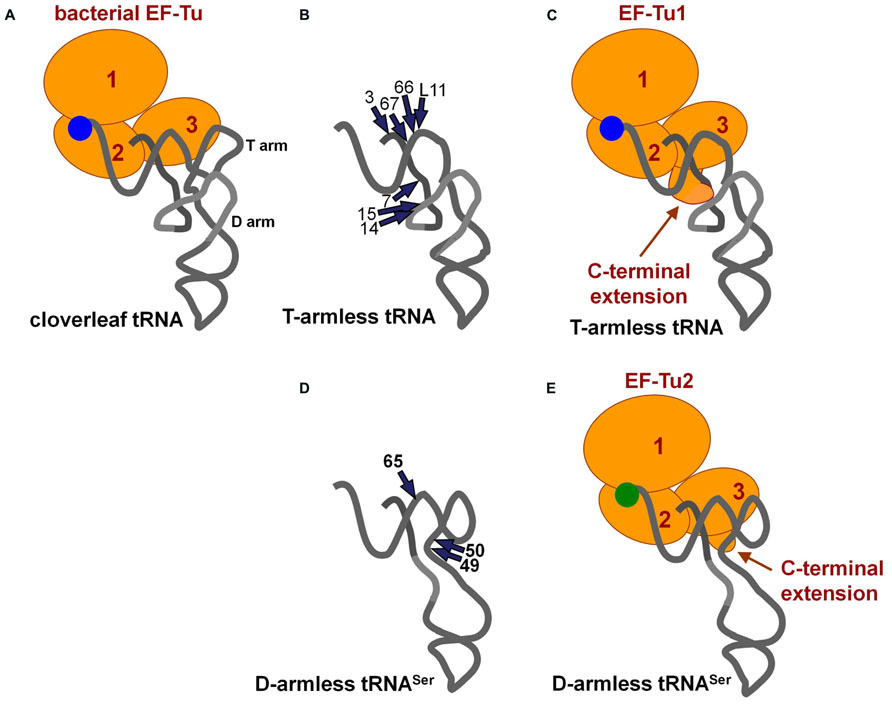
FIGURE 3. (A) Ternary complex of conventional tRNA/EF-Tu/GTP (Nissen et al., 1995). (B) Summary of ethylnitrosourea (ENU) modification interference studies of T-arm-lacking mt tRNA with C. elegans mt EF-Tu1 (Sakurai et al., 2006). The phosphate groups important for EF-Tu1 binding are shown with arrows. (C) A model of the ternary complex of T-arm-lacking tRNA/mt EF-Tu1/GTP (Sakurai et al., 2006). The model was based on the structure shown in (A) and in Figure 1E. A possible interaction between the C-terminal extension of EF-Tu1 and the region around position 9 in T-arm-lacking tRNA is suggested by modification interference as summarized in (B), cross-linking studies, and the tRNA binding activity of the C-terminal deletion mutants of EF-Tu1 (Ohtsuki et al., 2001; Sakurai et al., 2006). (D) Summary of ENU modification interference study of D-arm-lacking mt tRNA with C. elegans mt EF-Tu2 (Suematsu et al., 2005). The tRNA structure was inferred from the model proposed by Steinberg and his co-workers (Steinberg et al., 1994). The phosphate groups important for EF-Tu2 binding are shown by arrows. (E) A model of the ternary complex of D-arm-lacking mt tRNASer/C. elegans mt EF-Tu2/GTP (Suematsu et al., 2005). A possible interaction between the C-terminal extension of EF-Tu2 and acceptor-T helix in D-arm-lacking tRNA is suggested by modification interference study as summarized in (D), binding assays of mutant tRNAs, and the binding activity of EF-Tu mutants with C-terminal deletions (Suematsu et al., 2005).
Nematode mitochondria have another EF-Tu homolog, EF-Tu2. Nematode mt EF-Tu2 has a short (about 15-residue) C-terminal extension, and it binds to D-arm-lacking tRNASer, but not to T-arm-lacking or cloverleaf-type tRNAs (Ohtsuki et al., 2001; Suematsu et al., 2005; Arita et al., 2006). C. elegans mt EF-Tu2 binds to a region of the T-arm exposed due to the missing interaction between the T-arm and D-arm (Figures 3D,E; Suematsu et al., 2005). Interestingly, D-arm-lacking tRNAs in this species have anticodons for serine, and EF-Tu2 binds only to Ser-tRNA and accepts neither Ala-tRNA nor Val-tRNA with the same backbone (Ohtsuki et al., 2002b; Arita et al., 2006). This is likely due to the evolution of the aminoacyl-moiety binding pocket in EF-Tu2 to specialize in binding with the seryl moiety because of a unique adaptation in Ser-tRNA (Sato et al., 2006).
More recently, in some taxa other than the nematodes, such as Arthropoda, there have been mitochondrial T-arm-lacking tRNA genes discovered (Masta, 2000). Interestingly, there are two mt EF-Tu genes in arthropods (Ohtsuki and Watanabe, 2007). The functional differences between the two EF-Tu homologs in these species should be elucidated, and this project is in progress in our laboratory.
Future Perspectives
Besides ARS and EF-Tu, other factors such as tRNA terminal nucleotidyltransferases (CCA enzymes) and ribosomes could be interesting in terms of their interactions with shrunken mt tRNAs.
After the trimming of 3′ extra sequences, 3′CCA sequences are added to 3′ ends of pre-tRNAs by CCA enzymes (Deutscher, 1983). The bacterial CCA enzyme binds to the acceptor-T helix of pre-tRNA (Tomita et al., 2004), and thus a T-arm-lacking tRNA precursor is not a good substrate for the bacterial CCA enzyme (Tomari et al., 2002). On the other hand, the chromadorean nematode C. elegans has two genes for CCA enzymes, one of which encodes a putative mt CCA enzyme (Tomari et al., 2002). The recombinant (putative) mt CCA enzyme of C. elegans can recognize and efficiently add a CCA sequence, not only to conventional cloverleaf tRNAs, but also to T- or D-arm-lacking tRNAs (Tomari et al., 2002). It would be interesting to know how the nematode mt enzyme recognizes T-arm-lacking mt tRNAs efficiently.
During translation, conventional bacterial tRNA interacts with several sites in the ribosome (reviewed by Khade and Joseph, 2010). In bacterial ribosomes, the T-arm of P-site tRNA interacts with ribosomal protein L5 (Korostelev et al., 2006; Selmer et al., 2006). At the A-site, the T-arm of tRNA interacts with ribosomal protein L16 (Selmer et al., 2006; Voorhees et al., 2009). The residues in the A-site finger (helix 69) of 23S rRNA interact with the D-arm of tRNA at the A- and P-sites (Korostelev et al., 2006; Selmer et al., 2006; Voorhees et al., 2009). In a structural model of C. elegans mt rRNAs, the corresponding rRNA positions exist (Mears et al., 2002). At the E-site, residues in the T- and D-loops interact with ribosomal protein L1 and helices 76, 77, and 78 in 23S rRNA (e.g., the L1 stalk; Korostelev et al., 2006; Selmer et al., 2006). Interestingly, the corresponding regions in nematode mt rRNA are missing (Mears et al., 2002). In general, mitochondrial ribosomal proteins are enlarged compared to their counterparts in bacteria (Koc et al., 2000; Suzuki et al., 2001), suggesting that mitochondrial ribosomal proteins may have alternate binding modes for truncated tRNAs. Further structural analysis of metazoan mt ribosomes (Sharma et al., 2003; Greber et al., 2014) would be helpful to reveal the detailed interaction mode between mt ribosomes and shrunken mt tRNAs.
Structural alterations of metazoan mt tRNAs have been compensated for by several interacting factors. The mode of compensation by these factors may explain why metazoan tRNAs have undergone truncation during evolution. Further investigation into the detailed binding modes between shrunken tRNAs and the interacting factors that co-evolved with them will shed light on how truncated tRNAs evolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our studies described in this review were supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT), The Japan Society for the Promotion of Science (JSPS), the Kurata Memorial Hitachi Science and Technology Foundation and a grant from the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN), the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry. We thank Prof. K. Watanabe and Prof. K. Kita for their continued support, and Dr. Sarin Chimnaronk for the docking model coordinates of mammalian mt SerRS/ tRNA.
References
Arcari, P., and Brownlee, G. G. (1980). The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 8, 5207–5212. doi: 10.1093/nar/8.22.5207
Arita, M., Suematsu, T., Osanai, A., Inaba, T., Kamiya, H., Kita, K., et al. (2006). An evolutionary ‘intermediate state’ of mitochondrial translation systems found in Trichinella species of parasitic nematodes: co-evolution of tRNA and EF-Tu. Nucleic Acids Res. 34, 5291–5929. doi: 10.1093/nar/gkl526
Asahara, H., Himeno, H., Tamura, K., Nameki, N., Hasegawa, T., and Shimizu, M. (1994). Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristic tertiary structure. J. Mol. Biol. 236, 738–748. doi: 10.1006/jmbi.1994.1186
Baer, R. J., and Dubin, D. T. (1980). The sequence of a possible 5S RNA-equivalent in hamster mitochondria. Nucleic Acids Res. 8, 603–610. doi: 10.1093/nar/8.16.3603
Biou, V., Yaremchuk, A., Tukalo, M., and Cusack, S. (1994). The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263, 1404–1410. doi: 10.1126/science.8128220
Chihade, J. W., Hayashibara, K., Shiba, K., and Schimmel, P. (1998). Strong selective pressure to use G:U to mark an RNA acceptor stem for alanine. Biochemistry 37, 9193–9202. doi: 10.1021/bi9804636
Chimnaronk, S., Jeppesen, M. G., Suzuki, T., Nyborg, J., and Watanabe, K. (2005). Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 24, 3369–3379. doi: 10.1038/sj.emboj.7600811
de Bruijn, M. H., and Klug, A. (1983). A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire ‘dihydrouridine’ loop and stem. EMBO J. 2, 1309–1321.
de Bruijn, M. H., Schreier, P. H., Eperon, I. C., Barrell, B. G., Chen, E. Y., Armstrong, P. W., et al. (1980). A mammalian mitochondrial serine transfer RNA lacking the “dihydrouridine” loop and stem. Nucleic Acids Res. 8, 5213–5222. doi: 10.1093/nar/8.22.5213
Deutscher, M. P. (1983). “tRNA nucleotidyltransferase and the –C-C-A terminus of transfer RNA,” in Enzymes of Nucleic Acid Synthesis and Modification, Vol. 2, RNA Enzymes, ed. S.T. Jacob (Boca Raton, FL: CRC Press), 159–183.
Dubin, D. T., and Friend, D. A. (1972). Comparison of cytoplasmic and mitochondrial 4 S RNA from cultured hamster cells: physical and metabolic properties. J. Mol. Biol. 71, 163–175. doi: 10.1016/0022-2836(72)90344-0
Dubin, D. T., Jones, T. N., and Cleaves, G. R. (1974). An unmethylated “3 SE” RNA in hamster mitochondria: a 5 S RNA-equivalent? Biochem. Biophys. Res. Commun. 56, 401–406. doi: 10.1016/0006-291X(74)90856-0
Frazer-Abel, A. A., and Hagerman, P. J. (1999). Determination of the angle between the acceptor and anticodon stems of a truncated mitochondrial tRNA. J. Mol. Biol. 285, 581–593. doi: 10.1006/jmbi.1998.2320
Frazer-Abel, A. A., and Hagerman, P. J. (2004). Variation of the acceptor-anticodon interstem angles among mitochondrial and non-mitochondrial tRNAs. J. Mol. Biol. 343, 313–325. doi: 10.1016/j.jmb.2004.07.087
Frazer-Abel, A. A., and Hagerman, P. J. (2008). Core flexibility of a truncated metazoan mitochondrial tRNA. Nucleic Acids Res. 36, 5472–5481. doi: 10.1093/nar/gkn529
Giegé, R., Sissler, M., and Florentz, C. (1998). Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26, 5017–5035. doi: 10.1093/nar/26.22.5017
Greber, B. J., Boehringer, D., Leitner, A., Bieri, P., Voigts-Hoffmann, F., Erzberger, J. P., et al. (2014) Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519. doi: 10.1038/nature12890
Hanada, T., Suzuki, T., Yokogawa, T., Takemoto-Hori, C., Sprinzl, M., and Watanabe, K. (2001). Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells 6, 1019–1030. doi: 10.1046/j.1365-2443.2001.00491.x
Hayashi, I., Yokogawa, T., Kawai, G., Ueda, T., Nishikawa, K., and Watanabe, K. (1997). Assignment of imino proton signals of G-C base pairs and magnesium ion binding: an NMR study of bovine mitochondrial tRNASer(GCU) lacking the entire D arm. J. Biochem. 121, 1115–1122. doi: 10.1093/oxfordjournals.jbchem.a021703
Jühling, F., Mörl, M., Hartmann, R. K., Sprinzl, M., Stadler, P. F., and Pütz, J. (2009). tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162. doi: 10.1093/nar/gkn772
Jühling, F., Pütz, J., Bernt, M., Donath, A., Middendorf, M., Florentz, C., et al. (2012a). Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40, 2833–2845. doi: 10.1093/nar/gkr1131
Jühling, F., Pütz, J., Florentz, C., and Stadler, P. F. (2012b). Armless mitochondrial tRNAs in Enoplea (Nematoda). RNA Biol. 9, 1161–1166. doi: 10.4161/rna.21630
Khade, P., and Joseph, S. (2010). Functional interactions by transfer RNAs in the ribosome. FEBS Lett. 584, 420–426. doi: 10.1016/j.febslet.2009.11.034
Koc, E. C., Burkhart, W., Blackburn, K., Moseley, A., Koc, H., and Spremulli, L. L. (2000). A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J. Biol. Chem. 275, 32585–33291. doi: 10.1074/jbc.M003596200
Korostelev, A., Trakhanov, S., Laurberg, M., and Noller, H. F. (2006). Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077. doi: 10.1016/j.cell.2006.08.032
Kumazawa, Y., Himeno, H., Miura, K., and Watanabe, K. (1991). Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J. Biochem. 109, 421–427.
Lavrov, D. V., and Brown, W. M. (2001). Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics 157, 621–637.
Lovato, M. A., Chihade, J. W., and Schimmel, P. (2001). Translocation within the acceptor helix of a major tRNA identity determinant. EMBO J. 20, 4846–4853. doi: 10.1093/emboj/20.17.4846
Masta, S. E. (2000). Mitochondrial sequence evolution in spiders: intraspecific variation in tRNAs lacking the TΨC arm. Mol. Biol. Evol. 17, 1091–1100. doi: 10.1093/oxfordjournals.molbev.a026390
Mears, J. A., Cannone, J. J., Stagg, S. M., Gutell, R. R., Agrawal, R. K., and Harvey, S. C. (2002). Modeling a minimal ribosome based on comparative sequence analysis. J. Mol. Biol. 321, 215–234. doi: 10.1016/S0022-2836(02)00568-5
Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B. F., et al. (1995). Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472. doi: 10.1126/science.270.5241.1464
Nureki, O., Niimi, T., Muramatsu, T., Kanno, H., Kohno, T., Florentz, C., et al. (1994). Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J. Mol. Biol. 236, 710–724. doi: 10.1006/jmbi.1994.1184
Ohtsuki, T., Kawai, G., and Watanabe, K. (1998). Stable isotope-edited NMR analysis of Ascaris suum mitochondrial tRNAMet having a TV-replacement loop. J. Biochem. 124, 28–34. doi: 10.1093/oxfordjournals.jbchem.a022092
Ohtsuki, T., Kawai, G., Watanabe, Y., Kita, K., Nishikawa, K., and Watanabe, K. (1996). Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res. 24, 662–667. doi: 10.1093/nar/24.4.662
Ohtsuki, T., Kawai, G., and Watanabe, K. (2002a). The minimal tRNA: unique structure of Ascaris suum mitochondrial tRNASer(UCU) having a short T arm and lacking the entire D arm. FEBS Lett. 514, 37–43. doi: 10.1016/S0014-5793(02)02328-1
Ohtsuki, T., Sato, A., Watanabe, Y., and Watanabe, K. (2002b). A unique serine-specific elongation factor Tu found in nematode mitochondria. Nat. Struct. Biol. 9, 669–673. doi: 10.1038/nsb826
Ohtsuki, T., and Watanabe, Y. (2007). T-armless tRNAs and elongated elongation factor Tu. IUBMB Life 59, 68–75. doi: 10.1080/15216540701218722
Ohtsuki, T., Watanabe, Y., Takemoto, C., Kawai, G., Ueda, T., Kita, K., et al. (2001). An “elongated” translation elongation factor Tu for truncated tRNAs in nematode mitochondria. J. Biol. Chem. 276, 21571–21577. doi: 10.1074/jbc.M011118200
Okimoto, R., Macfarlane, J. L., Clary, D. O., and Wolstenholme, D. R. (1992). The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 130, 471–498.
Rubio, M. A., Rinehart, J. J., Krett, B., Duvezin-Caubet, S., Reichert, A. S., Söll, D., et al. (2008). Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl. Acad. Sci. U.S.A. 105, 9186–9191. doi: 10.1073/pnas.0804283105
Rudinger, J., Blechschmidt, B., Ribeiro, S., and Sprinzl, M. (1994). Minimalist aminoacylated RNAs as efficient substrates for elongation factor Tu. Biochemistry 33, 5682–5688. doi: 10.1021/bi00185a003
Sakurai, M., Ohtsuki, T., Suzuki, T., and Watanabe, K. (2005a). Unusual usage of wobble modifications in mitochondrial tRNAs of the nematode Ascaris suum. FEBS Lett. 579, 2767–2772. doi: 10.1016/j.febslet.2005.04.009
Sakurai, M., Ohtsuki, T., and Watanabe, K. (2005b). Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res. 33, 1653–1661. doi: 10.1093/nar/gki309
Sakurai, M., Watanabe, Y., Watanabe, K., and Ohtsuki, T. (2006). A protein extension to shorten RNA: elongated elongation factor-Tu recognizes the D-arm of T-armless tRNAs in nematode mitochondria. Biochem. J. 399, 249–256. doi: 10.1042/BJ20060781
Sato, A., Watanabe, Y., Suzuki, T., Komiyama, M., Watanabe, K., and Ohtsuki, T. (2006). Identification of the residues involved in the unique serine specificity of Caenorhabditis elegans mitochondrial EF-Tu2. Biochemistry 45, 10920–10927. doi: 10.1021/bi060536i
Selmer, M., Dunham, C. M., Murphy, F. V. IV, Weixlbaumer, A., Petry, S., Kelley, A. C., et al. (2006). Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942. doi: 10.1126/science.1131127
Sharma, M. R., Koc, E. C., Datta, P. P., Booth, T. M., Spremulli, L. L., and Agrawal, R. K. (2003). Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108. doi: 10.1016/S0092-8674(03)00762-1
Shimada, N., Suzuki, T., and Watanabe, K. (2001). Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J. Biol. Chem. 276, 46770–46778. doi: 10.1074/jbc.M105150200
Seutin, G., Lang, B. F., Mindell, D. P., and Morais, R. (1994). Evolution of the WANCY region in amniote mitochondrial DNA. Mol. Biol. Evol. 11, 329–340.
Steinberg, S., Gautheret, D., and Cedergren, R. (1994). Fitting the structurally diverse animal mitochondrial tRNAsSer to common three-dimensional constraints. J. Mol. Biol. 236, 982–989. doi: 10.1016/0022-2836(94)90004-3
Suematsu, T., Sato, A., Sakurai, M., Watanabe, K., and Ohtsuki, T. (2005). A unique tRNA recognition mechanism of Caenorhabditis elegans mitochondrial EF-Tu2. Nucleic Acids Res. 33, 4683–4691. doi: 10.1093/nar/gki784
Suzuki, T., Terasaki, M., Takemoto-Hori, C., Hanada, T., Ueda, T., Wada, A., et al. (2001). Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem. 276, 33181–33195. doi: 10.1074/jbc.M103236200
Tomari, Y., Suzuki, T., and Ueda, T. (2002). tRNA recognition by CCA-adding enzyme. Nucleic Acids Res. Suppl. 2002, 77–78. doi: 10.1093/nass/2.1.77
Tomita, K., Fukai, S., Ishitani, R., Ueda, T., Takeuchi, N., Vassylyev, D. G., et al. (2004). Structural basis for template-independent RNA polymerization. Nature 430, 700–704. doi: 10.1038/nature02712
Ueda, T., Ohta, T., and Watanabe, K. (1985). Large scale isolation and some properties of AGY-specific serine tRNA from bovine heart mitochondria. J. Biochem. 98, 1275–1284.
Ueda, T., Yotsumoto, Y., Ikeda, K., and Watanabe, K. (1992). The T-loop region of animal mitochondrial tRNASer(AGY) is a main recognition site for homologous seryl-tRNA synthetase. Nucleic Acids Res. 20, 2217–2222. doi: 10.1093/nar/20.9.2217
Voorhees, R. M., Weixlbaumer, A., Loakes, D., Kelley, A. C., and Ramakrishnan, V. (2009). Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 16, 528–533. doi: 10.1038/nsmb.1577
Watanabe, Y., Tsurui, H., Ueda, T., Furushima, R., Takamiya, S., Kita, K., et al. (1994). Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem. 269, 22902–22906.
Watanabe, Y., Tsurui, H., Ueda, T., Furushima-Shimogawara, R., Takamiya, S., Kita, K., et al. (1997). Primary sequence of mitochondrial tRNAArg of a nematode Ascaris suum: occurrence of unmodified adenosine at the first position of the anticodon. Biochim. Biophys. Acta 1350, 119–122. doi: 10.1016/S0167-4781(96)00211-4
Wende, S., Platzer, E. G., Jühling, F., Pütz, J., Florentz, C., Stadler, P. F., et al. (2014). Biological evidence for the world’s smallest tRNAs. Biochimie 100, 151–158. doi: 10.1016/j.biochi.2013.07.034
Wolstenholme, D. R., Macfarlane, J. L., Okimoto, R., Clary, D. O., and Wahleithner, J. A. (1987). Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl. Acad. Sci. U.S.A. 84, 1324–1328. doi: 10.1073/pnas.84.5.1324
Wolstenholme, D. R., Okimoto, R., and Macfarlane, J. L. (1994). Nucleotide correlations that suggest tertiary interactions in the TV-replacement loop-containing mitochondrial tRNAs of the nematodes, Caenorhabditis elegans and Ascaris suum. Nucleic Acids Res. 22, 4300–4306. doi: 10.1093/nar/22.20.4300
Yokogawa, T., Kumazawa, Y., Miura, K., and Watanabe, K. (1989). Purification and characterization of two serine isoacceptor tRNAs from bovine mitochondria by using a hybridization assay method. Nucleic Acids Res. 17, 2623–2638. doi: 10.1093/nar/17.7.2623
Keywords: mitochondrial tRNA, D-arm, T-arm, tRNA nucleotidyltransferase, aminoacyl-tRNA synthetase, elongation factor Tu, ribosome
Citation: Watanabe Y-i, Suematsu T and Ohtsuki T (2014) Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front. Genet. 5:109. doi: 10.3389/fgene.2014.00109
Received: 31 January 2014; Accepted: 12 April 2014;
Published online: 01 May 2014.
Edited by:
Akio Kanai, Keio University, JapanReviewed by:
Hyouta Himeno, Hirosaki University, JapanKozo Tomita, Biomedical Research Institute – National Institute of Advanced Industrial Science and Technology, Japan
Copyright © 2014 Watanabe, Suematsu and Ohtsuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoh-ichi Watanabe, Department of Biomedical Chemistry, Graduate School of Medicine, The University of Tokyo, Hongo, Bunkyo-ku, Tokyo 113-0033, Japan e-mail:eXdhdGFuYWJAbS51LXRva3lvLmFjLmpw;
Takashi Ohtsuki, Department of Biotechnology, Okayama University, 3-1-1 Tsushimanaka, Okayama 700-8530, Japan e-mail:b2h0c3VrQG9rYXlhbWEtdS5hYy5qcA==
†Present address: Takuma Suematsu, Department of Molecular and Cell Biology, Boston University, 72 East Concord Street, Boston, MA 02118, USA
 Yoh-ichi Watanabe
Yoh-ichi Watanabe Takuma Suematsu
Takuma Suematsu Takashi Ohtsuki
Takashi Ohtsuki