- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL, USA
Mobile DNA in the genome is subject to RNA-targeted epigenetic control. This control regulates the activity of transposons, retrotransposons and genomic proviruses. Many different life history experiences alter the activities of mobile DNA and the expression of genetic loci regulated by nearby insertions. The same experiences induce alterations in epigenetic formatting and lead to trans-generational modifications of genome expression and stability. These observations lead to the hypothesis that epigenetic formatting directed by non-coding RNA provides a molecular interface between life history events and genome alteration.
Introduction
Understanding the functional organization of the genome and its evolutionary history are key goals of modern molecular biology. The task has become more interesting and complex as we learn more the details of cell processes involving the genome. Recent applications of high resolution technologies to genome expression in animals reveal a dynamic four-dimensional interactive control architecture incompatible with prior notions that genomes contain discrete functional segments of DNA (“genes”) (Mercer and Mattick, 2013). This review will focus on the role of epigenetic regulation of viruses and mobile genetic elements as a key interface between the activities of these agents of evolutionary change and inputs from cell and organism life histories. The hypothesis developed as a result of the review is that disruption of epigenetic silencing constitutes a major target for life history activation of cellular functions for genome change. This likely occurs after genome replication, possibly by changes in small non-coding (snc) RNAs, typically on the order of 20–50 nucleotides long.
Mobile DNA is a Major and Functionally Significant Component of Genomes
One of the major surprises to come from the initial sequencing of the human genome was the high abundance of dispersed mobile repeat elements (Consortium, 2001). Today, we estimate that at least two-thirds of our genomes is composed of mobile DNA (De Koning et al., 2011). The human genome is not exceptional in its high content of mobile DNA (http://shapiro.bsd.uchicago.edu/TableII.1.shtml).
We increasingly recognize that viruses contribute to cell genomes (Kokosar and Kordis, 2013). They provide sequences for non-coding ncRNAs (Frias-Lasserre, 2012), sites for transcriptional control (Peaston et al., 2004; Dunn et al., 2005; Maksakova et al., 2006; Conley et al., 2008), and elements important in epigenetic regulation (Brunmeir et al., 2010; Conley and Jordan, 2012). Similar transcriptional and epigenetic regulatory contributions are made by mobile genetic elements (http://shapiro.bsd.uchicago.edu/Table5C-1.MobileElementsFoundtobeExaptedascis-RegulatoryControlSitesinAnimals.html) (Youngson et al., 2005; Kinoshita et al., 2007; Suzuki et al., 2007; Fujimoto et al., 2008; Gehring et al., 2009; Pask et al., 2009; Nakayashiki, 2011).
Mobile DNA is a major source of novel coding information. One mechanism is the process known as “exonization,” when splice signals are utilized in newly inserted DNA segments (http://shapiro.bsd.uchicago.edu/Origin_of_New_Protein_Domains.html). New coding sequences also form by reverse transcription of processed RNAs and genome insertion of the cDNAs, sometimes producing chimeric fusions with existing exons (http://shapiro.bsd.uchicago.edu/Table 5B. Reports of retrogenes in plant and animal genomes.html) (Long, 2001; Betrán et al., 2002; Fu et al., 2010).
It is now clear that mobile genetic elements play a key role in establishing and rewiring genomic networks (http://shapiro.bsd.uchicago.edu/Table5C-1.MobileElementsFoundtobeExaptedascis-RegulatoryControlSitesinAnimals.html) (Feschotte, 2008; Lindblad-Toh et al., 2011; Lowe et al., 2011; Testori et al., 2012; Kokosar and Kordis, 2013). Moreover, mobile element proliferation is a key factor in the formation of very large genomes (http://shapiro.bsd.uchicago.edu/Genome_Size.html).
The potential functional importance of distributed mobile DNA in genomes grows rapidly as evidence accumulates for pervasive genome transcription (http://shapiro.bsd.uchicago.edu/PervasiveGenomeTranscription.html) and for the regulatory role of non-coding RNAs (ncRNAs) in genome expression, including the functional juxtaposition of distant genome regions to activate transcription (http://shapiro.bsd.uchicago.edu/NonCodingRNAinGenomeExpression.html). Mobile elements participate in this long-range genomic communication and provide the sequences of many ncRNAs (Kapusta et al., 2013).
Cells use RNA-Targeted Epigenetic Control to Inhibit the Activity of Mobile DNA
Given the high content of mobile DNA in many genomes, an important question is: what prevents all the mobility systems from destroying genome integrity? In eukaryotic cells, a major control mechanism is sncRNA-directed epigenetic formatting into silent chromatin (Law and Jacobsen, 2010; Castel and Martienssen, 2013).
Both prokaryotes and eukaryotes have systems for capturing fragments from invading DNA molecules and placing the fragments into special loci encoding sncRNAs (Dumesic and Madhani, 2014). In prokaryotes, these loci are called CRISPRs (clustered regular interspersed palindromic repeats) (http://shapiro.bsd.uchicago.edu/CRISPRs.html) (Marraffini and Sontheimer, 2010; Garrett et al., 2011; Bikard and Marraffini, 2013; Watanabe et al., 2013). The RNA transcripts from CRISPRs are processed into sncRNAs that target cleavage of homologous invading DNA and also inactivation of complementary mRNA (Djordjevic et al., 2012). The details of the RNA processing and interference activities are well-characterized, but the acquisition of DNA fragments is poorly understood. The process must be very rapid, because viral infection yields cells that survive the initial infection with appropriate fragments added to their CRISPR repertoire (Barrangou et al., 2007).
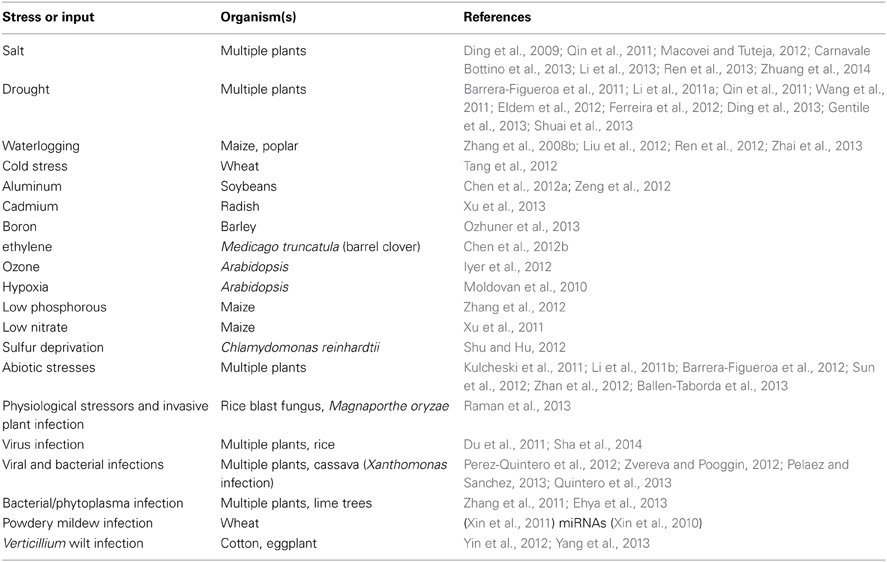
Virtually all eukaryotes investigated, with the notable exception of budding yeast, have mechanisms for sncRNA-directed chromatin silencing. They are based on members of the Argonaute family of sncRNA-processing proteins (http://shapiro.bsd.uchicago.edu/microRNA-directedchromatinsilencing.html). Plants and animals have independently evolved distinct mechanisms of processing the sncRNAs for the Argonaute family systems, but both groups use targeted epigenetic regulatory processes to defend against virus infection (Ding and Voinnet, 2007; Csorba et al., 2009) and prevent genome instability (Table 1). Like prokaryotes, Drosophila has specific genomic loci where it acquires fragments of invading DNA to encode the targeting sncRNAs (Brennecke et al., 2007, 2008; Handler et al., 2013).
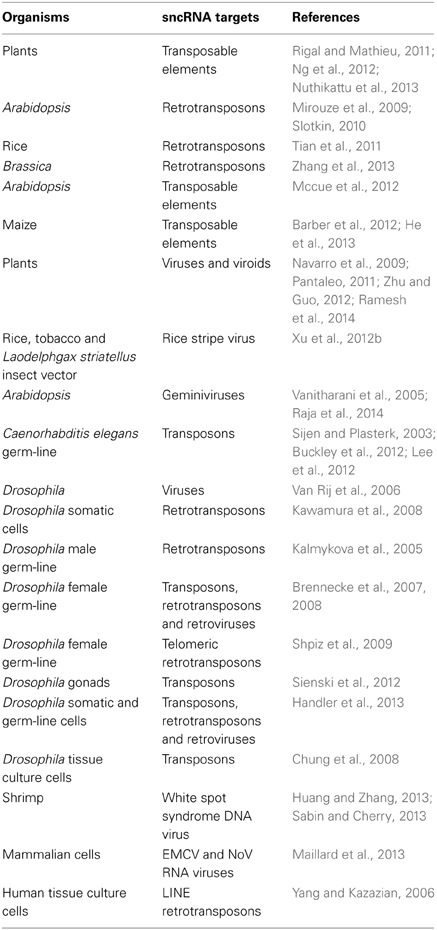
Table 1. Genome immunity by sncRNA targeting of mobile DNA (see also http://shapiro.bsd.uchicago.edu/TableII.9.shtml for earlier references).
Life History Events Destabilize Genomes and Activate Mobile DNA
Anyone who has studied real-time genome changes quantitatively knows that mutation frequencies depend upon the treatment of the experimental organism prior to measurement. A wide variety of life history events influence the natural genetic engineering (NGE) functions that generate mutations, especially mobile elements (Table 2; Shapiro, 2011). In some cases, the genome instabilities are large scale and last multiple cell or organismal generations.
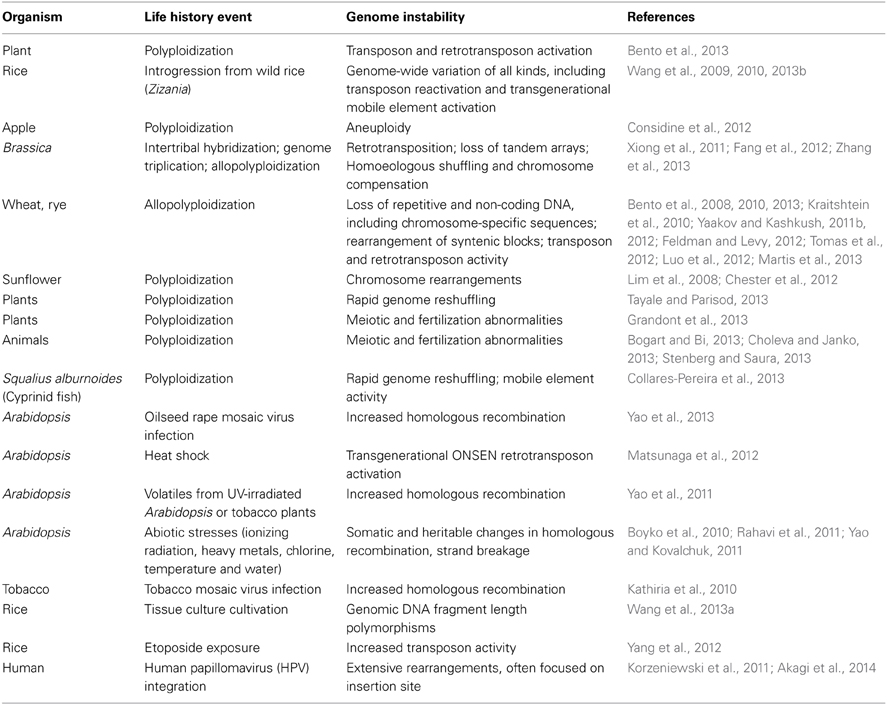
Table 2. Life history events that lead to genome destabilization (see also http://shapiro.bsd.uchicago.edu/TableII.8.shtml for earlier references).
Many observations demonstrate responses of the circuits controlling NGE functions to biological and abiotic inputs. It is particularly significant that many such responses occur following exceptional cell interactions with viruses or with other cells, either by infection or by hybridization (Table 2). As we might expect, the introduction of alien DNA or chromatin into a cell often has disruptive effects on genome homeostasis (Shapiro, 2014).
Epigenetic Changes in Response to Life History Events
One of the most active research areas in the second decade of the 21st century is analyzing the impact of life history events on the epigenetic layers of cell regulatory architecture (Table 3) (Chinnusamy and Zhu, 2009a; Vandegehuchte and Janssen, 2013). The observed epigenetic responses include alterations to cytosine methylation in DNA (Chinnusamy and Zhu, 2009b), histone modifications in nucleosomes, and sncRNAs (Ruiz-Ferrer and Voinnet, 2009; Ng et al., 2012) as well as transgenerational inheritance of complex novel phenotypes (Zucchi et al., 2012), frequently induced by stress (Boyko and Kovalchuk, 2010). The phenomenon of hybrid vigor, or heterosis, increasingly is viewed as an alteration in sncRNA-targeted epigenetic formatting stimulated by the encounter of two distinct genome control regimes (Groszmann et al., 2011; Miller et al., 2012; Shivaprasad et al., 2012).
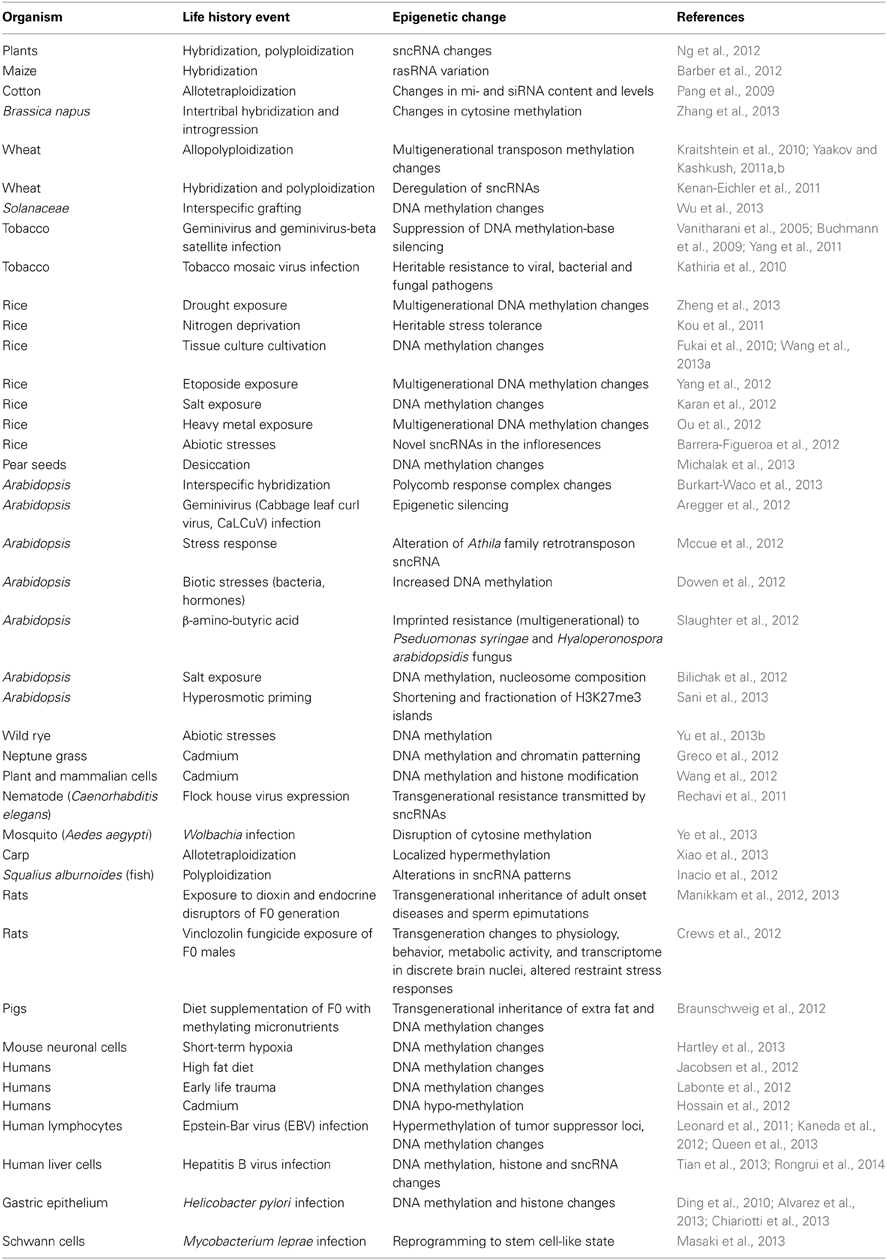
Table 3. Life history events that induce epigenetic changes (see also http://shapiro.bsd.uchicago.edu/TableII.10.shtml for earlier references).
Many of the studies demonstrating induced epigenetic modifications also document accompanying genome instabilities and emphasize their evolutionary potential (Madlung and Wendel, 2013). It is noteworthy that many of the same stimuli are involved in both genomic and epigenomic responses in plants (Hegarty et al., 2013) and animals (Arkhipova and Rodriguez, 2013). The common stimuli include infection and symbiosis (Hamon and Cossart, 2008; Bierne et al., 2012; Takahashi, 2014), hybridization and changes in ploidy.
Direct Interactions between NGE Activities and Epigenetic Regulatory Functions
In addition to disruption of sncRNA-targeted inhibition, there is limited but growing evidence that NGE functions acting on DNA molecules interact directly with epigenetic control factors. There is convincing evidence of the connection between NGE and the epigenome in DNA damage repair and retroviral or retrotransposon insertions into chromosomes.
Epigenetic Involvement in DNA Proofreading and Repair
There are recent reports that a specific histone modification (H3K36me3) primes DNA mismatch repair (Schmidt and Jackson, 2013), that H3K56 acetylation affects mismatch repair (Kadyrova et al., 2013), that hypoacetylation of H3K56 by HDACs 1 and 2 facilitates recruitment of non-homologous end-joining (NHEJ) proteins (Miller et al., 2010; Munoz-Galvan et al., 2013), and that nucleosome remodeling is integral to DS break repair (Seeber et al., 2013). Longstanding observations document the involvement of a specific histone, gamma-H2AX, in DS break repair and NHEJ (Kinner et al., 2008; Altaf et al., 2009; Dickey et al., 2009b; Redon et al., 2009; Firsanov et al., 2011; Chen et al., 2013). A direct role in chromatin remodeling for DNA repair has been claimed for another H2 analog, H2A.Z (Xu et al., 2012a).
Published reports indicate that H2AX incorporation into chromatin suppresses conversion of single-strand nicks to DS breaks (Franco et al., 2006) and affects the processing of the ends of broken DNA molecules (Helmink et al., 2011). H2AX operates in phosphorylated form (Rogakou et al., 1998; Kinner et al., 2008).
Beyond the role of H2AX, chromatin dynamics play an essential role in DNA repair and genome homeostasis (Lahue and Frizzell, 2012; Shi and Oberdoerffer, 2012). Many reports claim repair roles for chromatin regulators, remodeling complexes and nucleosome exchange factors (Ryan and Owen-Hughes, 2011):
– in DNA damage tolerance (Conaway and Conaway, 2009; Falbo et al., 2009);
– after exposure to ionizing radiation (Hunt et al., 2013);
– in UV damage responses (Palomera-Sanchez and Zurita, 2011; Yu et al., 2011);
– in DS break repair by NHEJ and HR (Van Attikum and Gasser, 2005; Williams et al., 2007; Robert et al., 2011; Xu and Price, 2011; Price and D'andrea, 2013);
– in PolyADP-dependent DNA repair (Ahel et al., 2009);
– in NER as well as DS break repair (Osley et al., 2007; Czaja et al., 2012; Yu et al., 2013a).
Nucleosome disassembly is probably necessary for certain repair processes (Linger and Tyler, 2007; Amouroux et al., 2010; Gospodinov and Herceg, 2013), and histone modifications affect damage-induced checkpoint signaling (Chen and Tyler, 2008). Once repair is complete, nucleosome modifications are reversed, and H2AX~P is eliminated from chromatin (Svetlova et al., 2010). So-called “bystander” cells, which are not subjected to DNA damage but are in the same culture as irradiated cells, also display H2AX phosphorylation (Sokolov et al., 2007; Dickey et al., 2009a, 2011).
A key feature of genome repair is that H2AX-marked damaged DNA mobilizes to subnuclear “repair centers” where homologous recombination and NHEJ proteins also localize (Lisby and Rothstein, 2005; Plate et al., 2008; Bekker-Jensen and Mailand, 2010). A role for chromatin in mobilization of damaged DNA has been proposed (Seeber et al., 2013), but multiple sources of evidence are lacking.
Retroviral and Retrotransposon Integrases
A more extensive case for NGE-chromatin interactions comes from analysis of retroviral and retrotransposon insertion specificities (Zhang and Mager, 2012). Each type of retrovirus displays a characteristic insertion specificity for its provirus (Lewinski et al., 2006). A number of targeting mechanisms involve epigenetic formatting molecules.
In budding yeast, Ty1 retrotransposon integrase contacts an H2A/H2B interface upstream of RNA polymerase III initiation sites (Baller et al., 2012; Bridier-Nahmias and Lesage, 2012; Mularoni et al., 2012). Histone deacetylase Hos2 and Trithorax group protein Set3 stimulate this nucleosome-targeted integration (Mou et al., 2006), and chromatin remodeling factor Isw2p is also implicated (Bachman et al., 2005). In contrast, the Ty5 retrotransposon inserts in silent chromatin, targeted by binding of its integrase to the Sir4 heterochromatin nucleating factor (Xie et al., 2001; Dai et al., 2007; Brady et al., 2008; Baller et al., 2011).
HIV and other lentiviral targeted integration into actively transcribed regions of the genome is associated with transcription-associated histone modifications, including H3 acetylation, H4 acetylation, and H3 K4 methylation, but is disfavored in regions rich in transcription-inhibiting modifications, which include H3K27me3 and DNA CpG methylation (Wang et al., 2007). The specificity results from integrase tethering by the LEDGF/p75 chromatin-binding growth factor (Vanegas et al., 2005; Llano et al., 2006; Ciuffi, 2008; Meehan and Poeschla, 2010; Zheng et al., 2010; Christ and Debyser, 2013). Replacing the LEDGF/p75 domain that interacts with expressed chromatin by the CBX1 domain, which binds histones H3K9me2 or H3K9me3 found in pericentric heterochromatin, targets HIV insertions to silent chromatin regions (Gijsbers et al., 2010).
Murine leukemia virus (MuLV) insertion targeting to initiation sites upstream of actively transcribed regions involves integrase interactions with bromodomain proteins BRD2, BRD3, and BRD4 (De Rijck et al., 2013; Gupta et al., 2013; Sharma et al., 2013a). Interestingly, chromatin recognition bromodomain protein BRD4 antagonizes HIV provirus reactivation (Zhu et al., 2012).
Certain retrotransposons are specifically targeted to centromeres (Wolfgruber et al., 2009; Birchler and Presting, 2012; Tsukahara et al., 2012; Sharma et al., 2013b), which have a special chromatin configuration characterized by centromeric versions of H3 (Henikoff and Dalal, 2005; Vos et al., 2006; Partridge, 2008; Zhang et al., 2008a). Centromeric retrotransposons in rice are highly associated with H3K9me2, a hallmark for heterochromatin (Neumann et al., 2007). Some centromeric retrotransposons encode integrase proteins with histone-binding chromodomains at their carboxy-termini (Neumann et al., 2011). Chromodomains recognize lysine methylation (Blus et al., 2011; Yap and Zhou, 2011; Eissenberg, 2012).
It is probably not coincidental that the most widely distributed group of retrotransposons among all eukaryotic clades are the “chromoviruses,” which are so named because they have chromodomains in their integrase proteins (Gorinsek et al., 2004; Kordis, 2005; Novikov et al., 2012; Weber et al., 2013). A chromodomain has been reported to target fungal chromovirus MAGGY insertions to heterochromatin marked by H3K9me2/me3 (Gao et al., 2008). An integrase chromodomain also participates in activator protein-targeted insertion of fission yeast retrotransposon Tf1 upstream of RNA polymerase II transcription start sites (Hizi and Levin, 2005; Chatterjee et al., 2009).
DNA Transposons
In contrast with many retrotransposons that interact with nucleosomes, the DNA transposon Hermes inserts preferentially in budding yeast into nucleosome-free regions of the genome (Gangadharan et al., 2010). The widely used P element DNA transposons in Drosophila show targeting (called “P element homing”) by incorporating binding sites for various regulatory factors, including chromatin insulators (Bender and Hudson, 2000; Fujioka et al., 2009) and Polycomb group response elements (Kassis, 2002; Cheng et al., 2012).
Epigenetic Reformatting After DNA Replication and ncRNAs as Potential Agents for Transmitting Experience to the Genome
While the evidence is increasingly abundant for effects of different life history events on epigenetic regulation in general, and on genome homeostasis in particular, it is far from clear how those effects occur (Lim and Brunet, 2013). We know very little about the connections between cell sensors and epigenetic (re)formatting complexes (Erdel et al., 2011; Narlikar et al., 2013). Deciphering those connections is currently an important research goal.
DNA replication provides a key decision point for maintaining or changing chromatin configurations (Poot et al., 2005; Liu and Gong, 2011; Mermoud et al., 2011). The replication apparatus must disassemble chromatin for polymerization and then reassemble chromatin once replication is complete. Replication takes place only in dividing cells, and transgenerational inheritance of epigenetic states must involve the proliferating cells that give rise to gametes. Transfer of outside information from somatic tissues to the germline has been reported in mammals (Sharma, 2013; Skinner et al., 2013). And epigenetic windows of susceptibility to environmental insults have been suggested during sperm development (Soubry et al., 2014). Since there is no segregated germ line in plants and eukaryotic microbes, the same cells that experience environmental inputs can also be the progenitors of gametes.
A number of different factors have been found or hypothesized to participate in post-replication chromatin restoration: histone chaperones (Budhavarapu et al., 2013), RNA editing and sncRNAs (Savva et al., 2013), chromatin remodeler SMARCAD1 (Mermoud et al., 2011), chromatin assembly factor 1 (Huang and Jiao, 2012), histone chaperon FACT (Winkler and Luger, 2011) and Swi/Snf complexes (Neves-Costa and Varga-Weisz, 2006; Ryan and Owen-Hughes, 2011; Zhu et al., 2013), and ISW1 complexes (Erdel and Rippe, 2011).
One frequently overlooked feature of post-replication reestablishment of epigenetic formatting is where in the nucleus it might occur. Replication takes place in specialized “replication factories” (Vago et al., 2009; Guillou et al., 2010). Does chromatin reestablishment occur in the same location or does it involve migration of newly replicated DNA segments to distinct subnuclear “chromatin factories,” like the ones that exist in the nucleolus for heterochromatin formation on rRNA-encoding DNA (Guetg and Santoro, 2012)? If so, such post-replication relocalization would be guided by the nucleoskeleton and lncRNAs (Mercer and Mattick, 2013; Mercer et al., 2013) and might present an attractive target for stress response and sensory input signaling (Weiner et al., 2012).
It is notable that changes to ncRNAs are frequently cited with regard to the impact of life history events on the genome (Sunkar et al., 2007; Khraiwesh et al., 2012; Lelandais-Briere et al., 2012; Nakaminami et al., 2012; Amaral et al., 2013). In the plant literature, there is documentation of numerous ncRNA changes in response to particular biotic and abiotic stress regimes (Table 4).
A number of observations about resistance to biotic and abiotic stresses are consistent with a key role for ncRNA changes in life history responses. Several viruses encode siRNA suppressors to overcome host defenses (Jiang et al., 2012; Omarov and Scholthof, 2012; Guo and Lu, 2013). Transgenic constructs encoding constitutive miRNA expression can lead to salt and drought tolerance in creeping bentgrass (Zhou et al., 2013), to immunity against blast fungus in rice (Li et al., 2014), and in Arabidopsis to greater salt and alkalinity sensitivity (Gao et al., 2011). Acquired aphid resistance in Arabidopsis involves sncRNA changes (Kettles et al., 2013), and most acquired stress resistances in plants display transgenerational epigenetic inheritance (Holeski et al., 2012; Luna and Ton, 2012; Slaughter et al., 2012).
Speculative Conclusions about an Epigenetic Interface between Experience and Genome Change
Mobile DNA and other NGE functions are the key agents for adaptively significant changes in genome organization and DNA sequences. The data reviewed and tabulated above establish the importance of RNA-directed chromatin formatting in the regulation and operation of mobile elements, viruses and DNA repair functions. In addition, there is a remarkable correlation between the life history events that activate NGE functions to destabilize genomes and those that lead to alteration of chromatin states and DNA methylation patterns.
The preceding observations lead to the plausible hypothesis that epigenetic regulation serves as a key interface between organismal life history and the agents that restructure genomic DNA. This hypothesis is supported by the limited number of cases where empirical observations have established direct molecular connections between NGE functions and components of the epigenetic control system: histones, nucleosomes, and chromatin reformatting complexes.
If, as I expect, further research bolsters the epigenome-NGE correlations and connections documented above, then we need to ask: what components(s) of the epigenetic control apparatus communicate information about experience to NGE operators? We do not know the answer to this fundamental question. However, the data reported in Table 4 indicate that ncRNAs are good candidates for key intermediates in the experience-genome signal transduction process. If this is so, then ncRNAs are logical molecular targets for modulating genome change toward potentially adaptive outcomes. Let us hope that research aimed at examining this proposal deepens our understanding of how life history impacts both epigenetic and genome change operations (Tables 2–4), whether or not my speculation ultimately proves to be correct.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author is grateful to the editors for the invitation to contribute to this special issue and for the opportunity to comment on the relationship between life history and genome change.
References
Ahel, D., Horejsi, Z., Wiechens, N., Polo, S. E., Garcia-Wilson, E., Ahel, I., et al. (2009). Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243. doi: 10.1126/science.1177321
Akagi, K., Li, J., Broutian, T. R., Padilla-Nash, H., Xiao, W., Jiang, B., et al. (2014). Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 24, 185–199. doi: 10.1101/gr.164806.113
Altaf, M., Auger, A., Covic, M., and Cote, J. (2009). Connection between histone H2A variants and chromatin remodeling complexes. Biochem. Cell Biol. 87, 35–50. doi: 10.1139/O08-140
Alvarez, M. C., Santos, J. C., Maniezzo, N., Ladeira, M. S., Da Silva, A. L., Scaletsky, I. C., et al. (2013). MGMT and MLH1 methylation in Helicobacter pylori-infected children and adults. World J. Gastroenterol. 19, 3043–3051. doi: 10.3748/wjg.v19.i20.3043
Amaral, P. P., Dinger, M. E., and Mattick, J. S. (2013). Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief. Funct. Genomics 12, 254–278. doi: 10.1093/bfgp/elt016
Amouroux, R., Campalans, A., Epe, B., and Radicella, J. P. (2010). Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 38, 2878–2890. doi: 10.1093/nar/gkp1247
Aregger, M., Borah, B. K., Seguin, J., Rajeswaran, R., Gubaeva, E. G., Zvereva, A. S., et al. (2012). Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 8:e1002941. doi: 10.1371/journal.ppat.1002941
Arkhipova, I. R., and Rodriguez, F. (2013). Genetic and epigenetic changes involving (retro)transposons in animal hybrids and polyploids. Cytogenet. Genome Res. 140, 295–311. doi: 10.1159/000352069
Bachman, N., Gelbart, M. E., Tsukiyama, T., and Boeke, J. D. (2005). TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 19, 955–964. doi: 10.1101/gad.1299105
Ballen-Taborda, C., Plata, G., Ayling, S., Rodriguez-Zapata, F., Becerra Lopez-Lavalle, L. A., Duitama, J., et al. (2013). Identification of cassava MicroRNAs under abiotic stress. Int. J. Genomics 2013, 857986. doi: 10.1155/2013/857986
Baller, J. A., Gao, J., Stamenova, R., Curcio, M. J., and Voytas, D. F. (2012). A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res. 22, 704–713. doi: 10.1101/gr.129585.111
Baller, J. A., Gao, J., and Voytas, D. F. (2011). Access to DNA establishes a secondary target site bias for the yeast retrotransposon Ty5. Proc. Natl. Acad. Sci. U.S.A. 108, 20351–20356. doi: 10.1073/pnas.1103665108
Barber, W. T., Zhang, W., Win, H., Varala, K. K., Dorweiler, J. E., Hudson, M. E., et al. (2012). Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. U.S.A. 109, 10444–10449. doi: 10.1073/pnas.1202073109
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Barrera-Figueroa, B. E., Gao, L., Diop, N. N., Wu, Z., Ehlers, J. D., Roberts, P. A., et al. (2011). Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 11:127. doi: 10.1186/1471-2229-11-127
Barrera-Figueroa, B. E., Gao, L., Wu, Z., Zhou, X., Zhu, J., Jin, H., et al. (2012). High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 12:132. doi: 10.1186/1471-2229-12-132
Bekker-Jensen, S., and Mailand, N. (2010). Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst.) 9, 1219–1228. doi: 10.1016/j.dnarep.2010.09.010
Bender, W., and Hudson, A. (2000). P element homing to the Drosophila bithorax complex. Development 127, 3981–3992. Available online at: http://dev.biologists.org/content/127/18/3981.long
Bento, M., Gustafson, P., Viegas, W., and Silva, M. (2010). Genome merger: from sequence rearrangements in triticale to their elimination in wheat-rye addition lines. Theor. Appl. Genet. 121, 489–497. doi: 10.1007/s00122-010-1325-6
Bento, M., Pereira, H. S., Rocheta, M., Gustafson, P., Viegas, W., and Silva, M. (2008). Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in triticale. PLoS ONE 3:e1402. doi: 10.1371/journal.pone.0001402
Bento, M., Tomas, D., Viegas, W., and Silva, M. (2013). Retrotransposons represent the most labile fraction for genomic rearrangements in polyploid plant species. Cytogenet. Genome Res. 140, 286–294. doi: 10.1159/000353308
Betrán, E., Thornton, K., and Long, M. (2002). Retroposed new genes out of the X in Drosophila. Genome Res. 12, 1854–1859. doi: 10.1101/gr.6049
Bierne, H., Hamon, M., and Cossart, P. (2012). Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2:a010272. doi: 10.1101/cshperspect.a010272
Bikard, D., and Marraffini, L. A. (2013). Control of gene expression by CRISPR-Cas systems. F1000Prime Rep. 5:47. doi: 10.12703/p5-47
Bilichak, A., Ilnystkyy, Y., Hollunder, J., and Kovalchuk, I. (2012). The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 7:e30515. doi: 10.1371/journal.pone.0030515
Birchler, J. A., and Presting, G. G. (2012). Retrotransposon insertion targeting: a mechanism for homogenization of centromere sequences on nonhomologous chromosomes. Genes Dev. 26, 638–640. doi: 10.1101/gad.191049.112
Blus, B. J., Wiggins, K., and Khorasanizadeh, S. (2011). Epigenetic virtues of chromodomains. Crit. Rev. Biochem. Mol. Biol. 46, 507–526. doi: 10.3109/10409238.2011.619164
Bogart, J. P., and Bi, K. (2013). Genetic and genomic interactions of animals with different ploidy levels. Cytogenet. Genome Res. 140, 117–136. doi: 10.1159/000351593
Boyko, A., Golubov, A., Bilichak, A., and Kovalchuk, I. (2010). Chlorine ions but not sodium ions alter genome stability of Arabidopsis thaliana. Plant Cell Physiol. 51, 1066–1078. doi: 10.1093/pcp/pcq048
Boyko, A., and Kovalchuk, I. (2010). Transgenerational response to stress in Arabidopsis thaliana. Plant Signal. Behav. 5, 995–998. doi: 10.1371/journal.pone.0009514
Brady, T. L., Schmidt, C. L., and Voytas, D. F. (2008). Targeting integration of the Saccharomyces Ty5 retrotransposon. Methods Mol. Biol. 435, 153–163. doi: 10.1007/978-1-59745-232-8_11
Braunschweig, M., Jagannathan, V., Gutzwiller, A., and Bee, G. (2012). Investigations on transgenerational epigenetic response down the male line in F2 pigs. PLoS ONE 7:e30583. doi: 10.1371/journal.pone.0030583
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi: 10.1016/j.cell.2007.01.043
Brennecke, J., Malone, C. D., Aravin, A. A., Sachidanandam, R., Stark, A., and Hannon, G. J. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392. doi: 10.1126/science.1165171
Bridier-Nahmias, A., and Lesage, P. (2012). Two large-scale analyses of Ty1 LTR-retrotransposon de novo insertion events indicate that Ty1 targets nucleosomal DNA near the H2A/H2B interface. Mob. DNA 3:22. doi: 10.1186/1759-8753-3-22
Brunmeir, R., Lagger, S., Simboeck, E., Sawicka, A., Egger, G., Hagelkruys, A., et al. (2010). Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 6:e1000927. doi: 10.1371/journal.pgen.1000927
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., and Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/jvi.01771-08
Buckley, B. A., Burkhart, K. B., Gu, S. G., Spracklin, G., Kershner, A., Fritz, H., et al. (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. doi: 10.1038/nature11352
Budhavarapu, V. N., Chavez, M., and Tyler, J. K. (2013). How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 6:32. doi: 10.1186/1756-8935-6-32
Burkart-Waco, D., Ngo, K., Dilkes, B., Josefsson, C., and Comai, L. (2013). Early disruption of maternal-zygotic interaction and activation of defense-like responses in Arabidopsis interspecific crosses. Plant Cell 25, 2037–2055. doi: 10.1105/tpc.112.108258
Carnavale Bottino, M., Rosario, S., Grativol, C., Thiebaut, F., Rojas, C. A., Farrineli, L., et al. (2013). High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS ONE 8:e59423. doi: 10.1371/journal.pone.0059423
Castel, S. E., and Martienssen, R. A. (2013). RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112. doi: 10.1038/nrg3355
Chatterjee, A. G., Leem, Y. E., Kelly, F. D., and Levin, H. L. (2009). The chromodomain of Tf1 integrase promotes binding to cDNA and mediates target site selection. J. Virol. 83, 2675–2685. doi: 10.1128/JVI.01588-08
Chen, C. C., and Tyler, J. (2008). Chromatin reassembly signals the end of DNA repair. Cell Cycle 7, 3792–3797. doi: 10.4161/cc.7.24.7188
Chen, L., Wang, T., Zhao, M., Tian, Q., and Zhang, W. H. (2012a). Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235, 375–386. doi: 10.1007/s00425-011-1514-9
Chen, L., Wang, T., Zhao, M., and Zhang, W. (2012b). Ethylene-responsive miRNAs in roots of Medicago truncatula identified by high-throughput sequencing at whole genome level. Plant Sci. 184, 14–19. doi: 10.1016/j.plantsci.2011.11.007
Chen, W. T., Alpert, A., Leiter, C., Gong, F., Jackson, S. P., and Miller, K. M. (2013). Systematic identification of functional residues in mammalian histone H2AX. Mol. Cell. Biol. 33, 111–126. doi: 10.1128/mcb.01024-12
Cheng, Y., Kwon, D. Y., Arai, A. L., Mucci, D., and Kassis, J. A. (2012). P-element homing is facilitated by engrailed polycomb-group response elements in Drosophila melanogaster. PLoS ONE 7:e30437. doi: 10.1371/journal.pone.0030437
Chester, M., Gallagher, J. P., Symonds, V. V., Cruz Da Silva, A. V., Mavrodiev, E. V., Leitch, A. R., et al. (2012). Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. U.S.A. 109, 1176–1181. doi: 10.1073/pnas.1112041109
Chiariotti, L., Angrisano, T., Keller, S., Florio, E., Affinito, O., Pallante, P., et al. (2013). Epigenetic modifications induced by Helicobacter pylori infection through a direct microbe-gastric epithelial cells cross-talk. Med. Microbiol. Immunol. 202, 327–337. doi: 10.1007/s00430-013-0301-6
Chinnusamy, V., and Zhu, J. K. (2009a). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Chinnusamy, V., and Zhu, J. K. (2009b). RNA-directed DNA methylation and demethylation in plants. Sci. China C Life Sci. 52, 331–343. doi: 10.1007/s11427-009-0052-1
Choleva, L., and Janko, K. (2013). Rise and persistence of animal polyploidy: evolutionary constraints and potential. Cytogenet. Genome Res. 140, 151–170. doi: 10.1159/000353464
Christ, F., and Debyser, Z. (2013). The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology 435, 102–109. doi: 10.1016/j.virol.2012.09.033
Chung, W. J., Okamura, K., Martin, R., and Lai, E. C. (2008). Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18, 795–802. doi: 10.1016/j.cub.2008.05.006
Ciuffi, A. (2008). Mechanisms governing lentivirus integration site selection. Curr. Gene Ther. 8, 419–429. doi: 10.2174/156652308786848021
Collares-Pereira, M. J., Matos, I., Morgado-Santos, M., and Coelho, M. M. (2013). Natural pathways towards polyploidy in animals: the Squalius alburnoides fish complex as a model system to study genome size and genome reorganization in polyploids. Cytogenet. Genome Res. 140, 97–116. doi: 10.1159/000351729
Conaway, R. C., and Conaway, J. W. (2009). The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci. 34, 71–77. doi: 10.1016/j.tibs.2008.10.010
Conley, A. B., and Jordan, I. K. (2012). “Endogenous retroviruses and the epigenome,” in Viruses: Essential Agents of Life, ed G. Witzany (Dordrecht: Spriinger), 309–323.
Conley, A. B., Piriyapongsa, J., and Jordan, I. K. (2008). Retroviral promoters in the human genome. Bioinformatics 24, 1563–1567. doi: 10.1093/bioinformatics/btn243
Considine, M. J., Wan, Y., D'antuono, M. F., Zhou, Q., Han, M., Gao, H., et al. (2012). Molecular genetic features of polyploidization and aneuploidization reveal unique patterns for genome duplication in diploid Malus. PLoS ONE 7:e29449. doi: 10.1371/journal.pone.0029449
Consortium, I. H. G. S. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Crews, D., Gillette, R., Scarpino, S. V., Manikkam, M., Savenkova, M. I., and Skinner, M. K. (2012). Epigenetic transgenerational inheritance of altered stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 9143–9148. doi: 10.1073/pnas.1118514109
Csorba, T., Pantaleo, V., and Burgyan, J. (2009). RNA silencing: an antiviral mechanism. Adv. Virus Res. 75, 35–71. doi: 10.1016/s0065-3527(09)07502-2
Czaja, W., Mao, P., and Smerdon, M. J. (2012). The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int. J. Mol. Sci. 13, 11954–11973. doi: 10.3390/ijms130911954
Dai, J., Xie, W., Brady, T. L., Gao, J., and Voytas, D. F. (2007). Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol. Cell 27, 289–299. doi: 10.1016/j.molcel.2007.06.010
De Koning, A. P., Gu, W., Castoe, T. A., Batzer, M. A., and Pollock, D. D. (2011). Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384. doi: 10.1371/journal.pgen.1002384
De Rijck, J., De Kogel, C., Demeulemeester, J., Vets, S., El Ashkar, S., Malani, N., et al. (2013). The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 5, 886–894. doi: 10.1016/j.celrep.2013.09.040
Dickey, J. S., Baird, B. J., Redon, C. E., Sokolov, M. V., Sedelnikova, O. A., and Bonner, W. M. (2009a). Intercellular communication of cellular stress monitored by gamma-H2AX induction. Carcinogenesis 30, 1686–1695. doi: 10.1093/carcin/bgp192
Dickey, J. S., Redon, C. E., Nakamura, A. J., Baird, B. J., Sedelnikova, O. A., and Bonner, W. M. (2009b). H2AX: functional roles and potential applications. Chromosoma 118, 683–692. doi: 10.1007/s00412-009-0234-4
Dickey, J. S., Zemp, F. J., Altamirano, A., Sedelnikova, O. A., Bonner, W. M., and Kovalchuk, O. (2011). H2AX phosphorylation in response to DNA double-strand break formation during bystander signalling: effect of microRNA knockdown. Radiat. Prot. Dosimetry 143, 264–269. doi: 10.1093/rpd/ncq470
Ding, D., Zhang, L., Wang, H., Liu, Z., Zhang, Z., and Zheng, Y. (2009). Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 103, 29–38. doi: 10.1093/aob/mcn205
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Ding, S. Z., Goldberg, J. B., and Hatakeyama, M. (2010). Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 6, 851–862. doi: 10.2217/fon.10.37
Ding, Y., Tao, Y., and Zhu, C. (2013). Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 64, 3077–3086. doi: 10.1093/jxb/ert164
Djordjevic, M., Djordjevic, M., and Severinov, K. (2012). CRISPR transcript processing: a mechanism for generating a large number of small interfering RNAs. Biol. Direct 7:24. doi: 10.1186/1745-6150-7-24
Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U.S.A. 109, E2183–E2191. doi: 10.1073/pnas.1209329109
Du, P., Wu, J., Zhang, J., Zhao, S., Zheng, H., Gao, G., et al. (2011). Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 7:e1002176. doi: 10.1371/journal.ppat.1002176
Dumesic, P. A., and Madhani, H. D. (2014). Recognizing the enemy within: licensing RNA-guided genome defense. Trends Biochem. Sci. 39, 25–34. doi: 10.1016/j.tibs.2013.10.003
Dunn, C. A., Van De Lagemaat, L. N., Baillie, G. J., and Mager, D. L. (2005). Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene 364, 2–12. doi: 10.1016/j.gene.2005.05.045
Ehya, F., Monavarfeshani, A., Mohseni Fard, E., Karimi Farsad, L., Khayam Nekouei, M., Mardi, M., et al. (2013). Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS ONE 8:e66372. doi: 10.1371/journal.pone.0066372
Eissenberg, J. C. (2012). Structural biology of the chromodomain: form and function. Gene 496, 69–78. doi: 10.1016/j.gene.2012.01.003
Eldem, V., Celikkol Akcay, U., Ozhuner, E., Bakir, Y., Uranbey, S., and Unver, T. (2012). Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS ONE 7:e50298. doi: 10.1371/journal.pone.0050298
Erdel, F., Krug, J., Langst, G., and Rippe, K. (2011). Targeting chromatin remodelers: signals and search mechanisms. Biochim. Biophys. Acta 1809, 497–508. doi: 10.1016/j.bbagrm.2011.06.005
Erdel, F., and Rippe, K. (2011). Chromatin remodelling in mammalian cells by ISWI-type complexes–where, when and why? FEBS J. 278, 3608–3618. doi: 10.1111/j.1742-4658.2011.08282.x
Falbo, K. B., Alabert, C., Katou, Y., Wu, S., Han, J., Wehr, T., et al. (2009). Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat. Struct. Mol. Biol. 16, 1167–1172. doi: 10.1038/nsmb.1686
Fang, L., Cheng, F., Wu, J., and Wang, X. (2012). The impact of genome triplication on tandem gene evolution in Brassica rapa. Front. Plant Sci. 3:261. doi: 10.3389/fpls.2012.00261
Feldman, M., and Levy, A. A. (2012). Genome evolution due to allopolyploidization in wheat. Genetics 192, 763–774. doi: 10.1534/genetics.112.146316
Ferreira, T. H., Gentile, A., Vilela, R. D., Costa, G. G., Dias, L. I., Endres, L., et al. (2012). microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 7:e46703. doi: 10.1371/journal.pone.0046703
Feschotte, C. (2008). Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397–405. doi: 10.1038/nrg2337
Firsanov, D. V., Solovjeva, L. V., and Svetlova, M. P. (2011). H2AX phosphorylation at the sites of DNA double-strand breaks in cultivated mammalian cells and tissues. Clin. Epigenetics 2, 283–297. doi: 10.1007/s13148-011-0044-4
Franco, S., Gostissa, M., Zha, S., Lombard, D. B., Murphy, M. M., Zarrin, A. A., et al. (2006). H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell 21, 201–214. doi: 10.1016/j.molcel.2006.01.005
Frias-Lasserre, D. (2012). Non coding RNAs and viruses in the framework of the phylogeny of the genes, epigenesis and heredity. Int. J. Mol. Sci. 13, 477–490. doi: 10.3390/ijms13010477
Fu, B., Chen, M., Zou, M., Long, M., and He, S. (2010). The rapid generation of chimerical genes expanding protein diversity in zebrafish. BMC Genomics 11:657. doi: 10.1186/1471-2164-11-657
Fujimoto, R., Kinoshita, Y., Kawabe, A., Kinoshita, T., Takashima, K., Nordborg, M., et al. (2008). Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 4:e1000048. doi: 10.1371/journal.pgen.1000048
Fujioka, M., Wu, X., and Jaynes, J. B. (2009). A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development 136, 3077–3087. doi: 10.1242/dev.036467
Fukai, E., Umehara, Y., Sato, S., Endo, M., Kouchi, H., Hayashi, M., et al. (2010). Derepression of the plant Chromovirus LORE1 induces germline transposition in regenerated plants. PLoS Genet. 6:e1000868. doi: 10.1371/journal.pgen.1000868
Gangadharan, S., Mularoni, L., Fain-Thornton, J., Wheelan, S. J., and Craig, N. L. (2010). DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 21966–21972. doi: 10.1073/pnas.1016382107
Gao, P., Bai, X., Yang, L., Lv, D., Pan, X., Li, Y., et al. (2011). osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 38, 237–242. doi: 10.1007/s11033-010-0100-8
Gao, X., Hou, Y., Ebina, H., Levin, H. L., and Voytas, D. F. (2008). Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 18, 359–369. doi: 10.1101/gr.7146408
Garrett, R. A., Vestergaard, G., and Shah, S. A. (2011). Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 19, 549–556. doi: 10.1016/j.tim.2011.08.002
Gehring, M., Bubb, K. L., and Henikoff, S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324, 1447–1451. doi: 10.1126/science.1171609
Gentile, A., Ferreira, T. H., Mattos, R. S., Dias, L. I., Hoshino, A. A., Carneiro, M. S., et al. (2013). Effects of drought on the microtranscriptome of field-grown sugarcane plants. Planta 237, 783–798. doi: 10.1007/s00425-012-1795-7
Gijsbers, R., Ronen, K., Vets, S., Malani, N., De Rijck, J., Mcneely, M., et al. (2010). LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 18, 552–560. doi: 10.1038/mt.2010.36
Gorinsek, B., Gubensek, F., and Kordis, D. (2004). Evolutionary genomics of chromoviruses in eukaryotes. Mol. Biol. Evol. 21, 781–798. doi: 10.1093/molbev/msh057
Gospodinov, A., and Herceg, Z. (2013). Shaping chromatin for repair. Mutat. Res. 752, 45–60. doi: 10.1016/j.mrrev.2012.10.001
Grandont, L., Jenczewski, E., and Lloyd, A. (2013). Meiosis and its deviations in polyploid plants. Cytogenet. Genome Res. 140, 171–184. doi: 10.1159/000351730
Greco, M., Chiappetta, A., Bruno, L., and Bitonti, M. B. (2012). In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/err313
Groszmann, M., Greaves, I. K., Albertyn, Z. I., Scofield, G. N., Peacock, W. J., and Dennis, E. S. (2011). Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. U.S.A. 108, 2617–2622. doi: 10.1073/pnas.1019217108
Guetg, C., and Santoro, R. (2012). Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics 7, 811–814. doi: 10.4161/epi.21072
Guillou, E., Ibarra, A., Coulon, V., Casado-Vela, J., Rico, D., Casal, I., et al. (2010). Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 24, 2812–2822. doi: 10.1101/gad.608210
Guo, X., and Lu, R. (2013). Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J. Virol. 87, 5414–5423. doi: 10.1128/jvi.00148-13
Gupta, S. S., Maetzig, T., Maertens, G. N., Sharif, A., Rothe, M., Weidner-Glunde, M., et al. (2013). Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J. Virol. 87, 12721–12736. doi: 10.1128/jvi.01942-13
Hamon, M. A., and Cossart, P. (2008). Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4, 100–109. doi: 10.1016/j.chom.2008.07.009
Handler, D., Meixner, K., Pizka, M., Lauss, K., Schmied, C., Gruber, F. S., et al. (2013). The genetic makeup of the Drosophila piRNA pathway. Mol. Cell 50, 762–777. doi: 10.1016/j.molcel.2013.04.031
Hartley, I., Elkhoury, F. F., Heon Shin, J., Xie, B., Gu, X., Gao, Y., et al. (2013). Long-lasting changes in DNA methylation following short-term hypoxic exposure in primary hippocampal neuronal cultures. PLoS ONE 8:e77859. doi: 10.1371/journal.pone.0077859
He, G., Chen, B., Wang, X., Li, X., Li, J., He, H., et al. (2013). Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol. 14:R57. doi: 10.1186/gb-2013-14-6-r57
Hegarty, M., Coate, J., Sherman-Broyles, S., Abbott, R., Hiscock, S., and Doyle, J. (2013). Lessons from natural and artificial polyploids in higher plants. Cytogenet. Genome Res. 140, 204–225. doi: 10.1159/000353361
Helmink, B. A., Tubbs, A. T., Dorsett, Y., Bednarski, J. J., Walker, L. M., Feng, Z., et al. (2011). H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature 469, 245–249. doi: 10.1038/nature09585
Henikoff, S., and Dalal, Y. (2005). Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15, 177–184. doi: 10.1016/j.gde.2005.01.004
Hizi, A., and Levin, H. L. (2005). The integrase of the long terminal repeat-retrotransposon tf1 has a chromodomain that modulates integrase activities. J. Biol. Chem. 280, 39086–39094. doi: 10.1074/jbc.M506363200
Holeski, L. M., Jander, G., and Agrawal, A. A. (2012). Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. 27, 618–626. doi: 10.1016/j.tree.2012.07.011
Hossain, M. B., Vahter, M., Concha, G., and Broberg, K. (2012). Low-level environmental cadmium exposure is associated with DNA hypomethylation in Argentinean women. Environ. Health Perspect. 120, 879–884. doi: 10.1289/ehp.1104600
Huang, H., and Jiao, R. (2012). Roles of chromatin assembly factor 1 in the epigenetic control of chromatin plasticity. Sci. China Life Sci. 55, 15–19. doi: 10.1007/s11427-012-4269-z
Huang, T., and Zhang, X. (2013). Host defense against DNA virus infection in shrimp is mediated by the siRNA pathway. Eur. J. Immunol. 43, 137–146. doi: 10.1002/eji.201242806
Hunt, C. R., Ramnarain, D., Horikoshi, N., Iyengar, P., Pandita, R. K., Shay, J. W., et al. (2013). Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 179, 383–392. doi: 10.1667/rr3308.2
Inacio, A., Pinho, J., Pereira, P. M., Comai, L., and Coelho, M. M. (2012). Global analysis of the small RNA transcriptome in different ploidies and genomic combinations of a vertebrate complex–the Squalius alburnoides. PLoS ONE 7:e41158. doi: 10.1371/journal.pone.0041158
Iyer, N. J., Jia, X., Sunkar, R., Tang, G., and Mahalingam, R. (2012). microRNAs responsive to ozone-induced oxidative stress in Arabidopsis thaliana. Plant Signal. Behav. 7, 484–491. doi: 10.4161/psb.19337
Jacobsen, S. C., Brons, C., Bork-Jensen, J., Ribel-Madsen, R., Yang, B., Lara, E., et al. (2012). Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia 55, 3341–3349. doi: 10.1007/s00125-012-2717-8
Jiang, L., Wei, C., and Li, Y. (2012). Viral suppression of RNA silencing. Sci. China Life Sci. 55, 109–118. doi: 10.1007/s11427-012-4279-x
Kadyrova, L. Y., Mertz, T. M., Zhang, Y., Northam, M. R., Sheng, Z., Lobachev, K. S., et al. (2013). A reversible histone h3 acetylation cooperates with mismatch repair and replicative polymerases in maintaining genome stability. PLoS Genet. 9:e1003899. doi: 10.1371/journal.pgen.1003899
Kalmykova, A. I., Klenov, M. S., and Gvozdev, V. A. (2005). Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 33, 2052–2059. doi: 10.1093/nar/gki323
Kaneda, A., Matsusaka, K., Aburatani, H., and Fukayama, M. (2012). Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 72, 3445–3450. doi: 10.1158/0008-5472.can-11-3919
Kapusta, A., Kronenberg, Z., Lynch, V. J., Zhuo, X., Ramsay, L., Bourque, G., et al. (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9:e1003470. doi: 10.1371/journal.pgen.1003470
Karan, R., Deleon, T., Biradar, H., and Subudhi, P. K. (2012). Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 7:e40203. doi: 10.1371/journal.pone.0040203
Kassis, J. A. (2002). Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46, 421–438. doi: 10.1016/S0065-2660(02)46015-4
Kathiria, P., Sidler, C., Golubov, A., Kalischuk, M., Kawchuk, L. M., and Kovalchuk, I. (2010). Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 153, 1859–1870. doi: 10.1104/pp.110.157263
Kawamura, Y., Saito, K., Kin, T., Ono, Y., Asai, K., Sunohara, T., et al. (2008). Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453, 793–797. doi: 10.1038/nature06938
Kenan-Eichler, M., Leshkowitz, D., Tal, L., Noor, E., Melamed-Bessudo, C., Feldman, M., et al. (2011). Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188, 263–272. doi: 10.1534/genetics.111.128348
Kettles, G. J., Drurey, C., Schoonbeek, H. J., Maule, A. J., and Hogenhout, S. A. (2013). Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 198, 1178–1190. doi: 10.1111/nph.12218
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kinner, A., Wu, W., Staudt, C., and Iliakis, G. (2008). Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694. doi: 10.1093/nar/gkn550
Kinoshita, Y., Saze, H., Kinoshita, T., Miura, A., Soppe, W. J., Koornneef, M., et al. (2007). Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49, 38–45. doi: 10.1111/j.1365-313X.2006.02936.x
Kokosar, J., and Kordis, D. (2013). Genesis and regulatory wiring of retroelement-derived domesticated genes: a phylogenomic perspective. Mol. Biol. Evol. 30, 1015–1031. doi: 10.1093/molbev/mst014
Kordis, D. (2005). A genomic perspective on the chromodomain-containing retrotransposons: chromoviruses. Gene 347, 161–173. doi: 10.1016/j.gene.2004.12.017
Korzeniewski, N., Spardy, N., Duensing, A., and Duensing, S. (2011). Genomic instability and cancer: lessons learned from human papillomaviruses. Cancer Lett. 305, 113–122. doi: 10.1016/j.canlet.2010.10.013
Kou, H. P., Li, Y., Song, X. X., Ou, X. F., Xing, S. C., Ma, J., et al. (2011). Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.). J. Plant Physiol. 168, 1685–1693. doi: 10.1016/j.jplph.2011.03.017
Kraitshtein, Z., Yaakov, B., Khasdan, V., and Kashkush, K. (2010). Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186, 801–812. doi: 10.1534/genetics.110.120790
Kulcheski, F. R., De Oliveira, L. F., Molina, L. G., Almerao, M. P., Rodrigues, F. A., Marcolino, J., et al. (2011). Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12:307. doi: 10.1186/1471-2164-12-307
Labonte, B., Suderman, M., Maussion, G., Navaro, L., Yerko, V., Mahar, I., et al. (2012). Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatry 69, 722–731. doi: 10.1001/archgenpsychiatry.2011.2287
Lahue, R. S., and Frizzell, A. (2012). Histone deacetylase complexes as caretakers of genome stability. Epigenetics 7, 806–810. doi: 10.4161/epi.20922
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Lee, H. C., Gu, W., Shirayama, M., Youngman, E., Conte, D. Jr., and Mello, C. C. (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. doi: 10.1016/j.cell.2012.06.016
Lelandais-Briere, C., Sorin, C., Crespi, M., and Hartmann, C. (2012). [Non-coding RNAs involved in plant responses to environmental constraints]. Biol. Aujourdhui. 206, 313–322. doi: 10.1051/jbio/2012032
Leonard, S., Wei, W., Anderton, J., Vockerodt, M., Rowe, M., Murray, P. G., et al. (2011). Epigenetic and transcriptional changes which follow Epstein-Barr virus infection of germinal center B cells and their relevance to the pathogenesis of Hodgkin's lymphoma. J. Virol. 85, 9568–9577. doi: 10.1128/jvi.00468-11
Lewinski, M. K., Yamashita, M., Emerman, M., Ciuffi, A., Marshall, H., Crawford, G., et al. (2006). Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2:e60. doi: 10.1371/journal.ppat.0020060
Li, B., Duan, H., Li, J., Deng, X. W., Yin, W., and Xia, X. (2013). Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol. Biol. 81, 525–539. doi: 10.1007/s11103-013-0010-y
Li, B., Qin, Y., Duan, H., Yin, W., and Xia, X. (2011a). Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 62, 3765–3779. doi: 10.1093/jxb/err051
Li, H., Dong, Y., Yin, H., Wang, N., Yang, J., Liu, X., et al. (2011b). Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 11:170. doi: 10.1186/1471-2229-11-170
Li, Y., Lu, Y. G., Shi, Y., Wu, L., Xu, Y. J., Huang, F., et al. (2014). Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 164, 1077–1092. doi: 10.1104/pp.113.230052
Lim, J. P., and Brunet, A. (2013). Bridging the transgenerational gap with epigenetic memory. Trends Genet. 29, 176–186. doi: 10.1016/j.tig.2012.12.008
Lim, K. Y., Soltis, D. E., Soltis, P. S., Tate, J., Matyasek, R., Srubarova, H., et al. (2008). Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS ONE 3:e3353. doi: 10.1371/journal.pone.0003353
Lindblad-Toh, K., Garber, M., Zuk, O., Lin, M. F., Parker, B. J., Washietl, S., et al. (2011). A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482. doi: 10.1038/nature10530
Linger, J. G., and Tyler, J. K. (2007). Chromatin disassembly and reassembly during DNA repair. Mutat. Res. 618, 52–64. doi: 10.1016/j.mrfmmm.2006.05.039
Lisby, M., and Rothstein, R. (2005). Localization of checkpoint and repair proteins in eukaryotes. Biochimie 87, 579–589. doi: 10.1016/j.biochi.2004.10.023
Liu, Q., and Gong, Z. (2011). The coupling of epigenome replication with DNA replication. Curr. Opin. Plant Biol. 14, 187–194. doi: 10.1016/j.pbi.2010.12.001
Liu, Z., Kumari, S., Zhang, L., Zheng, Y., and Ware, D. (2012). Characterization of miRNAs in response to short-term waterlogging in three inbred lines of Zea mays. PLoS ONE 7:e39786. doi: 10.1371/journal.pone.0039786
Llano, M., Saenz, D. T., Meehan, A., Wongthida, P., Peretz, M., Walker, W. H., et al. (2006). An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464. doi: 10.1126/science.1132319
Long, M. (2001). Evolution of novel genes. Curr. Opin. Genet. Dev. 11, 673–680. doi: 10.1016/S0959-437X(00)00252-5
Lowe, C. B., Kellis, M., Siepel, A., Raney, B. J., Clamp, M., Salama, S. R., et al. (2011). Three periods of regulatory innovation during vertebrate evolution. Science 333, 1019–1024. doi: 10.1126/science.1202702
Luna, E., and Ton, J. (2012). The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal. Behav. 7, 615–618. doi: 10.4161/psb.20155
Luo, J., Hao, M., Zhang, L., Chen, J., Zhang, L., Yuan, Z., et al. (2012). Microsatellite mutation rate during allohexaploidization of newly resynthesized wheat. Int. J. Mol. Sci. 13, 12533–12543. doi: 10.3390/ijms131012533
Macovei, A., and Tuteja, N. (2012). microRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 12:183. doi: 10.1186/1471-2229-12-183
Madlung, A., and Wendel, J. F. (2013). Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140, 270–285. doi: 10.1159/000351430
Maillard, P. V., Ciaudo, C., Marchais, A., Li, Y., Jay, F., Ding, S. W., et al. (2013). Antiviral RNA interference in mammalian cells. Science 342, 235–238. doi: 10.1126/science.1241930
Maksakova, I. A., Romanish, M. T., Gagnier, L., Dunn, C. A., Van De Lagemaat, L. N., and Mager, D. L. (2006). Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2:e2. doi: 10.1371/journal.pgen.0020002
Manikkam, M., Tracey, R., Guerrero-Bosagna, C., and Skinner, M. K. (2012). Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS ONE 7:e46249. doi: 10.1371/journal.pone.0046249
Manikkam, M., Tracey, R., Guerrero-Bosagna, C., and Skinner, M. K. (2013). Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 8:e55387. doi: 10.1371/journal.pone.0055387
Marraffini, L. A., and Sontheimer, E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190. doi: 10.1038/nrg2749
Martis, M. M., Zhou, R., Haseneyer, G., Schmutzer, T., Vrana, J., Kubalakova, M., et al. (2013). Reticulate evolution of the rye genome. Plant Cell 25, 3685–3698. doi: 10.1105/tpc.113.114553
Masaki, T., Qu, J., Cholewa-Waclaw, J., Burr, K., Raaum, R., and Rambukkana, A. (2013). Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell 152, 51–67. doi: 10.1016/j.cell.2012.12.014
Matsunaga, W., Kobayashi, A., Kato, A., and Ito, H. (2012). The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana. Plant Cell Physiol. 53, 824–833. doi: 10.1093/pcp/pcr179
Mccue, A. D., Nuthikattu, S., Reeder, S. H., and Slotkin, R. K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 8:e1002474. doi: 10.1371/journal.pgen.1002474
Meehan, A. M., and Poeschla, E. M. (2010). Chromatin tethering and retroviral integration: recent discoveries and parallels with DNA viruses. Biochim. Biophys. Acta 1799, 182–191. doi: 10.1016/j.bbagrm.2009.10.001
Mercer, T. R., Edwards, S. L., Clark, M. B., Neph, S. J., Wang, H., Stergachis, A. B., et al. (2013). DNase I-hypersensitive exons colocalize with promoters and distal regulatory elements. Nat. Genet. 45, 852–859. doi: 10.1038/ng.2677
Mercer, T. R., and Mattick, J. S. (2013). Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res. 23, 1081–1088. doi: 10.1101/gr.156612.113
Mermoud, J. E., Rowbotham, S. P., and Varga-Weisz, P. D. (2011). Keeping chromatin quiet: how nucleosome remodeling restores heterochromatin after replication. Cell Cycle 10, 4017–4025. doi: 10.4161/cc.10.23.18558
Michalak, M., Barciszewska, M. Z., Barciszewski, J., Plitta, B. P., and Chmielarz, P. (2013). Global changes in DNA methylation in seeds and seedlings of Pyrus communis after seed desiccation and storage. PLoS ONE 8:e70693. doi: 10.1371/journal.pone.0070693
Miller, K. M., Tjeertes, J. V., Coates, J., Legube, G., Polo, S. E., Britton, S., et al. (2010). Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151. doi: 10.1038/nsmb.1899
Miller, M., Zhang, C., and Chen, Z. J. (2012). Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 (Bethesda) 2, 505–513. doi: 10.1534/g3.112.002162
Mirouze, M., Reinders, J., Bucher, E., Nishimura, T., Schneeberger, K., Ossowski, S., et al. (2009). Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461, 427–430. doi: 10.1038/nature08328
Moldovan, D., Spriggs, A., Yang, J., Pogson, B. J., Dennis, E. S., and Wilson, I. W. (2010). Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 61, 165–177. doi: 10.1093/jxb/erp296
Mou, Z., Kenny, A. E., and Curcio, M. J. (2006). Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172, 2157–2167 doi: 10.1534/genetics.105.054072
Mularoni, L., Zhou, Y., Bowen, T., Gangadharan, S., Wheelan, S. J., and Boeke, J. D. (2012). Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 22, 693–703. doi: 10.1101/gr.129460.111
Munoz-Galvan, S., Jimeno, S., Rothstein, R., and Aguilera, A. (2013). Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 9:e1003237. doi: 10.1371/journal.pgen.1003237
Nakaminami, K., Matsui, A., Shinozaki, K., and Seki, M. (2012). RNA regulation in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 149–153. doi: 10.1016/j.bbagrm.2011.07.015
Nakayashiki, H. (2011). The Trickster in the genome: contribution and control of transposable elements. Genes Cells 16, 827–841. doi: 10.1111/j.1365-2443.2011.01533.x
Narlikar, G. J., Sundaramoorthy, R., and Owen-Hughes, T. (2013). Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503. doi: 10.1016/j.cell.2013.07.011
Navarro, B., Pantaleo, V., Gisel, A., Moxon, S., Dalmay, T., Bisztray, G., et al. (2009). Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS ONE 4:e7686. doi: 10.1371/journal.pone.0007686
Neumann, P., Navratilova, A., Koblizkova, A., Kejnovsky, E., Hribova, E., Hobza, R., et al. (2011). Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mob. DNA 2:4. doi: 10.1186/1759-8753-2-4
Neumann, P., Yan, H., and Jiang, J. (2007). The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 176, 749–761. doi: 10.1534/genetics.107.071902
Neves-Costa, A., and Varga-Weisz, P. (2006). The roles of chromatin remodelling factors in replication. Results Probl. Cell Differ. 41, 91–107. doi: 10.1007/400_007
Ng, D. W., Lu, J., and Chen, Z. J. (2012). Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin. Plant Biol. 15, 154–161. doi: 10.1016/j.pbi.2012.01.007
Novikov, A., Smyshlyaev, G., and Novikova, O. (2012). Evolutionary history of LTR retrotransposon chromodomains in plants. Int. J. Plant Genomics 2012:874743. doi: 10.1155/2012/874743
Nuthikattu, S., Mccue, A. D., Panda, K., Fultz, D., Defraia, C., Thomas, E. N., et al. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131. doi: 10.1104/pp.113.216481
Omarov, R. T., and Scholthof, H. B. (2012). Biological chemistry of virus-encoded suppressors of RNA silencing: an overview. Methods Mol. Biol. 894, 39–56. doi: 10.1007/978-1-61779-882-5_3
Osley, M. A., Tsukuda, T., and Nickoloff, J. A. (2007). ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 618, 65–80. doi: 10.1016/j.mrfmmm.2006.07.011
Ou, X., Zhang, Y., Xu, C., Lin, X., Zang, Q., Zhuang, T., et al. (2012). Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 7:e41143. doi: 10.1371/journal.pone.0041143
Ozhuner, E., Eldem, V., Ipek, A., Okay, S., Sakcali, S., Zhang, B., et al. (2013). Boron stress responsive microRNAs and their targets in barley. PLoS ONE 8:e59543. doi: 10.1371/journal.pone.0059543
Palomera-Sanchez, Z., and Zurita, M. (2011). Open, repair and close again: chromatin dynamics and the response to UV-induced DNA damage. DNA Repair (Amst.) 10, 119–125. doi: 10.1016/j.dnarep.2010.10.010
Pang, M., Woodward, A. W., Agarwal, V., Guan, X., Ha, M., Ramachandran, V., et al. (2009). Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol. 10:R122. doi: 10.1186/gb-2009-10-11-r122
Pantaleo, V. (2011). Plant RNA silencing in viral defence. Adv. Exp. Med. Biol. 722, 39–58. doi: 10.1007/978-1-4614-0332-6_3
Partridge, J. F. (2008). Centromeric chromatin in fission yeast. Front. Biosci. 13, 3896–3905. doi: 10.2741/2977
Pask, A. J., Papenfuss, A. T., Ager, E. I., Mccoll, K. A., Speed, T. P., and Renfree, M. B. (2009). Analysis of the platypus genome suggests a transposon origin for mammalian imprinting. Genome Biol. 10:R1. doi: 10.1186/gb-2009-10-1-r1
Peaston, A. E., Evsikov, A. V., Graber, J. H., De Vries, W. N., Holbrook, A. E., Solter, D., et al. (2004). Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606. doi: 10.1016/j.devcel.2004.09.004
Pelaez, P., and Sanchez, F. (2013). Small RNAs in plant defense responses during viral and bacterial interactions: similarities and differences. Front. Plant Sci. 4:343. doi: 10.3389/fpls.2013.00343
Perez-Quintero, A. L., Quintero, A., Urrego, O., Vanegas, P., and Lopez, C. (2012). Bioinformatic identification of cassava miRNAs differentially expressed in response to infection by Xanthomonas axonopodis pv. manihotis. BMC Plant Biol. 12:29. doi: 10.1186/1471-2229-12-29
Plate, I., Hallwyl, S. C., Shi, I., Krejci, L., Muller, C., Albertsen, L., et al. (2008). Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283, 29077–29085. doi: 10.1074/jbc.M804881200
Poot, R. A., Bozhenok, L., Van Den Berg, D. L., Hawkes, N., and Varga-Weisz, P. D. (2005). Chromatin remodeling by WSTF-ISWI at the replication site: opening a window of opportunity for epigenetic inheritance? Cell Cycle 4, 543–546. doi: 10.4161/cc.4.4.1624
Price, B. D., and D'andrea, A. D. (2013). Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354. doi: 10.1016/j.cell.2013.02.011
Qin, Y., Duan, Z., Xia, X., and Yin, W. (2011). Expression profiles of precursor and mature microRNAs under dehydration and high salinity shock in Populus euphratica. Plant Cell Rep. 30, 1893–1907. doi: 10.1007/s00299-011-1096-9
Queen, K. J., Shi, M., Zhang, F., Cvek, U., and Scott, R. S. (2013). Epstein-Barr virus-induced epigenetic alterations following transient infection. Int. J. Cancer 132, 2076–2086. doi: 10.1002/ijc.27893
Quintero, A., Perez-Quintero, A. L., and Lopez, C. (2013). Identification of ta-siRNAs and cis-nat-siRNAs in cassava and their roles in response to cassava bacterial blight. Genom. Proteomics Bioinform. 11, 172–181. doi: 10.1016/j.gpb.2013.03.001
Rahavi, M. R., Migicovsky, Z., Titov, V., and Kovalchuk, I. (2011). Transgenerational adaptation to heavy metal salts in Arabidopsis. Front. Plant Sci. 2:91. doi: 10.3389/fpls.2011.00091
Raja, P., Jackel, J. N., Li, S., Heard, I. M., and Bisaro, D. M. (2014). Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 88, 2611–2622. doi: 10.1128/JVI.02305-13
Raman, V., Simon, S. A., Romag, A., Demirci, F., Mathioni, S. M., Zhai, J., et al. (2013). Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus, Magnaporthe oryzae. BMC Genomics 14:326. doi: 10.1186/1471-2164-14-326
Ramesh, S. V., Ratnaparkhe, M. B., Kumawat, G., Gupta, G. K., and Husain, S. M. (2014). Plant miRNAome and antiviral resistance: a retrospective view and prospective challenges. Virus Genes. 48, 1–14. doi: 10.1007/s11262-014-1038-z
Rechavi, O., Minevich, G., and Hobert, O. (2011). Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147, 1248–1256. doi: 10.1016/j.cell.2011.10.042
Redon, C. E., Dickey, J. S., Bonner, W. M., and Sedelnikova, O. A. (2009). gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Adv. Space Res. 43, 1171–1178. doi: 10.1016/j.asr.2008.10.011
Ren, Y., Chen, L., Zhang, Y., Kang, X., Zhang, Z., and Wang, Y. (2012). Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct. Integr. Genomics 12, 327–339. doi: 10.1007/s10142-012-0271-6
Ren, Y., Chen, L., Zhang, Y., Kang, X., Zhang, Z., and Wang, Y. (2013). Identification and characterization of salt-responsive microRNAs in Populus tomentosa by high-throughput sequencing. Biochimie 95, 743–750. doi: 10.1016/j.biochi.2012.10.025
Rigal, M., and Mathieu, O. (2011). A “mille-feuille” of silencing: epigenetic control of transposable elements. Biochim. Biophys. Acta 1809, 452–458. doi: 10.1016/j.bbagrm.2011.04.001
Robert, T., Vanoli, F., Chiolo, I., Shubassi, G., Bernstein, K. A., Rothstein, R., et al. (2011). HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 471, 74–79. doi: 10.1038/nature09803
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., and Bonner, W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868.
Rongrui, L., Na, H., Zongfang, L., Fanpu, J., and Shiwen, J. (2014). Epigenetic mechanism involved in the HBV/HCV-related hepatocellular carcinoma tumorigenesis. Curr. Pharm. Des. 20, 1715–1725. doi: 10.2174/13816128113199990533
Ruiz-Ferrer, V., and Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. doi: 10.1146/annurev.arplant.043008.092111
Ryan, D. P., and Owen-Hughes, T. (2011). Snf2-family proteins: chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 15, 649–656. doi: 10.1016/j.cbpa.2011.07.022
Sabin, L. R., and Cherry, S. (2013). Small creatures use small RNAs to direct antiviral defenses. Eur. J. Immunol. 43, 27–33. doi: 10.1002/eji.201243201
Sani, E., Herzyk, P., Perrella, G., Colot, V., and Amtmann, A. (2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14:R59. doi: 10.1186/gb-2013-14-6-r59
Savva, Y. A., Jepson, J. E., Chang, Y. J., Whitaker, R., Jones, B. C., St. Laurent, G., et al. (2013). RNA editing regulates transposon-mediated heterochromatic gene silencing. Nat. Commun. 4:2745. doi: 10.1038/ncomms3745
Schmidt, C. K., and Jackson, S. P. (2013). On your mark, get SET(D2), go! H3K36me3 primes DNA mismatch repair. Cell 153, 513–515. doi: 10.1016/j.cell.2013.04.018
Seeber, A., Hauer, M., and Gasser, S. M. (2013). Nucleosome remodelers in double-strand break repair. Curr. Opin. Genet. Dev. 23, 174–184. doi: 10.1016/j.gde.2012.12.008
Sha, A., Zhao, J., Yin, K., Tang, Y., Wang, Y., Wei, X., et al. (2014). Virus-based microRNA silencing in plants. Plant Physiol. 164, 36–47. doi: 10.1104/pp.113.231100
Shapiro, J. A. (2011). Evolution: a View from the 21st Century. Upper Saddle River, NJ: FT Press Science.
Sharma, A. (2013). Transgenerational epigenetic inheritance: focus on soma to germline information transfer. Prog. Biophys. Mol. Biol. 113, 439–446. doi: 10.1016/j.pbiomolbio.2012.12.003
Sharma, A., Larue, R. C., Plumb, M. R., Malani, N., Male, F., Slaughter, A., et al. (2013a). BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. U.S.A. 110, 12036–12041. doi: 10.1073/pnas.1307157110
Sharma, A., Wolfgruber, T. K., and Presting, G. G. (2013b). Tandem repeats derived from centromeric retrotransposons. BMC Genomics 14:142. doi: 10.1186/1471-2164-14-142
Shi, L., and Oberdoerffer, P. (2012). Chromatin dynamics in DNA double-strand break repair. Biochim. Biophys. Acta 1819, 811–819. doi: 10.1016/j.bbagrm.2012.01.002
Shivaprasad, P. V., Dunn, R. M., Santos, B. A., Bassett, A., and Baulcombe, D. C. (2012). Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 31, 257–266. doi: 10.1038/emboj.2011.458
Shpiz, S., Kwon, D., Rozovsky, Y., and Kalmykova, A. (2009). rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 37, 268–278. doi: 10.1093/nar/gkn960
Shu, L., and Hu, Z. (2012). Characterization and differential expression of microRNAs elicited by sulfur deprivation in Chlamydomonas reinhardtii. BMC Genomics 13:108. doi: 10.1186/1471-2164-13-108
Shuai, P., Liang, D., Zhang, Z., Yin, W., and Xia, X. (2013). Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics 14:233. doi: 10.1186/1471-2164-14-233
Sienski, G., Donertas, D., and Brennecke, J. (2012). Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–980. doi: 10.1016/j.cell.2012.10.040
Sijen, T., and Plasterk, R. H. (2003). Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426, 310–314. doi: 10.1038/nature02107
Skinner, M. K., Haque, C. G., Nilsson, E., Bhandari, R., and Mccarrey, J. R. (2013). Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and the subsequent germ line. PLoS ONE 8:e66318. doi: 10.1371/journal.pone.0066318
Slaughter, A., Daniel, X., Flors, V., Luna, E., Hohn, B., and Mauch-Mani, B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158, 835–843. doi: 10.1104/pp.111.191593
Slotkin, R. K. (2010). The epigenetic control of the Athila family of retrotransposons in Arabidopsis. Epigenetics 5, 483–490. doi: 10.4161/epi.5.6.12119
Sokolov, M. V., Dickey, J. S., Bonner, W. M., and Sedelnikova, O. A. (2007). gamma-H2AX in bystander cells: not just a radiation-triggered event, a cellular response to stress mediated by intercellular communication. Cell Cycle 6, 2210–2212. doi: 10.4161/cc.6.18.4682
Soubry, A., Hoyo, C., Jirtle, R. L., and Murphy, S. K. (2014). A paternal environmental legacy: evidence for epigenetic inheritance through the male germ line. Bioessays 36, 359–371. doi: 10.1002/bies.201300113
Stenberg, P., and Saura, A. (2013). Meiosis and its deviations in polyploid animals. Cytogenet. Genome Res. 140, 185–203. doi: 10.1159/000351731
Sun, G., Stewart, C. N. Jr. Xiao, P., and Zhang, B. (2012). MicroRNA expression analysis in the cellulosic biofuel crop switchgrass (Panicum virgatum) under abiotic stress. PLoS ONE 7:e32017. doi: 10.1371/journal.pone.0032017
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05.001
Suzuki, S., Ono, R., Narita, T., Pask, A. J., Shaw, G., Wang, C., et al. (2007). Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3:e55. doi: 10.1371/journal.pgen.0030055
Svetlova, M. P., Solovjeva, L. V., and Tomilin, N. V. (2010). Mechanism of elimination of phosphorylated histone H2AX from chromatin after repair of DNA double-strand breaks. Mutat. Res. 685, 54–60. doi: 10.1016/j.mrfmmm.2009.08.001
Takahashi, K. (2014). Influence of bacteria on epigenetic gene control. Cell. Mol. Life Sci. 71, 1045–1054. doi: 10.1007/s00018-013-1487-x
Tang, Z., Zhang, L., Xu, C., Yuan, S., Zhang, F., Zheng, Y., et al. (2012). Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 159, 721–738. doi: 10.1104/pp.112.196048
Tayale, A., and Parisod, C. (2013). Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenet. Genome Res. 140, 79–96. doi: 10.1159/000351318
Testori, A., Caizzi, L., Cutrupi, S., Friard, O., De Bortoli, M., Cora, D., et al. (2012). The role of transposable elements in shaping the combinatorial interaction of transcription factors. BMC Genomics 13:400. doi: 10.1186/1471-2164-13-400
Tian, Y., Yang, W., Song, J., Wu, Y., and Ni, B. (2013). Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell. Biol. 33, 2810–2816. doi: 10.1128/mcb.00205-13
Tian, Z., Yu, Y., Lin, F., Sanmiguel, P. J., Wing, R. A., Mccouch, S. R., et al. (2011). Exceptional lability of a genomic complex in rice and its close relatives revealed by interspecific and intraspecific comparison and population analysis. BMC Genomics 12:142. doi: 10.1186/1471-2164-12-142
Tomas, D., Bento, M., Viegas, W., and Silva, M. (2012). Involvement of disperse repetitive sequences in wheat/rye genome adjustment. Int. J. Mol. Sci. 13, 8549–8561. doi: 10.3390/ijms13078549
Tsukahara, S., Kawabe, A., Kobayashi, A., Ito, T., Aizu, T., Shin-I, T., et al. (2012). Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes Dev. 26, 705–713. doi: 10.1101/gad.183871.111
Vago, R., Leva, V., Biamonti, G., and Montecucco, A. (2009). DNA ligase I and Nbs1 proteins associate in a complex and colocalize at replication factories. Cell Cycle 8, 2600–2607. doi: 10.4161/cc.8.16.9352
Van Attikum, H., and Gasser, S. M. (2005). ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle 4, 1011–1014. doi: 10.4161/cc.4.8.1887
Van Rij, R. P., Saleh, M. C., Berry, B., Foo, C., Houk, A., Antoniewski, C., et al. (2006). The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995. doi: 10.1101/gad.1482006
Vandegehuchte, M. B., and Janssen, C. R. (2013). Epigenetics in an ecotoxicological context. Mutat. Res. doi: 10.1016/j.mrgentox.2013.08.008. [Epub ahead of print].
Vanegas, M., Llano, M., Delgado, S., Thompson, D., Peretz, M., and Poeschla, E. (2005). Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 118, 1733–1743. doi: 10.1242/jcs.02299
Vanitharani, R., Chellappan, P., and Fauquet, C. M. (2005). Geminiviruses and RNA silencing. Trends Plant Sci. 10, 144–151. doi: 10.1016/j.tplants.2005.01.005
Vos, L. J., Famulski, J. K., and Chan, G. K. (2006). How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem. Cell Biol. 84, 619–639. doi: 10.1139/o06-078
Wang, B., Li, Y., Shao, C., Tan, Y., and Cai, L. (2012). Cadmium and its epigenetic effects. Curr. Med. Chem. 19, 2611–2620. doi: 10.2174/092986712800492913
Wang, G. P., Ciuffi, A., Leipzig, J., Berry, C. C., and Bushman, F. D. (2007). HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17, 1186–1194. doi: 10.1101/gr.6286907
Wang, H., Chai, Y., Chu, X., Zhao, Y., Wu, Y., Zhao, J., et al. (2009). Molecular characterization of a rice mutator-phenotype derived from an incompatible cross-pollination reveals transgenerational mobilization of multiple transposable elements and extensive epigenetic instability. BMC Plant Biol. 9:63. doi: 10.1186/1471-2229-9-63
Wang, N., Wang, H., Wang, H., Zhang, D., Wu, Y., Ou, X., et al. (2010). Transpositional reactivation of the Dart transposon family in rice lines derived from introgressive hybridization with Zizania latifolia. BMC Plant Biol. 10:190. doi: 10.1186/1471-2229-10-190
Wang, T., Chen, L., Zhao, M., Tian, Q., and Zhang, W. H. (2011). Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367. doi: 10.1186/1471-2164-12-367
Wang, X., Wu, R., Lin, X., Bai, Y., Song, C., Yu, X., et al. (2013a). Tissue culture-induced genetic and epigenetic alterations in rice pure-lines, F1 hybrids and polyploids. BMC Plant Biol. 13:77. doi: 10.1186/1471-2229-13-77
Wang, Z. H., Zhang, D., Bai, Y., Zhang, Y. H., Liu, Y., Wu, Y., et al. (2013b). Genomewide variation in an introgression line of rice-Zizania revealed by whole-genome re-sequencing. PLoS ONE 8:e74479. doi: 10.1371/journal.pone.0074479
Watanabe, T., Nozawa, T., Aikawa, C., Amano, A., Maruyama, F., and Nakagawa, I. (2013). CRISPR regulation of intraspecies diversification by limiting IS transposition and intercellular recombination. Genome Biol. Evol. 5, 1099–1114. doi: 10.1093/gbe/evt075
Weber, B., Heitkam, T., Holtgrawe, D., Weisshaar, B., Minoche, A. E., Dohm, J. C., et al. (2013). Highly diverse chromoviruses of Beta vulgaris are classified by chromodomains and chromosomal integration. Mob. DNA 4:8. doi: 10.1186/1759-8753-4-8
Weiner, A., Chen, H. V., Liu, C. L., Rahat, A., Klien, A., Soares, L., et al. (2012). Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 10:e1001369. doi: 10.1371/journal.pbio.1001369
Williams, R. S., Williams, J. S., and Tainer, J. A. (2007). Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520. doi: 10.1139/O07-069
Winkler, D. D., and Luger, K. (2011). The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374. doi: 10.1074/jbc.R110.180778
Wolfgruber, T. K., Sharma, A., Schneider, K. L., Albert, P. S., Koo, D. H., Shi, J., et al. (2009). Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet. 5:e1000743. doi: 10.1371/journal.pgen.1000743
Wu, R., Wang, X., Lin, Y., Ma, Y., Liu, G., Yu, X., et al. (2013). Inter-species grafting caused extensive and heritable alterations of DNA methylation in solanaceae plants. PLoS ONE 8:e61995. doi: 10.1371/journal.pone.0061995
Xiao, J., Song, C., Liu, S., Tao, M., Hu, J., Wang, J., et al. (2013). DNA methylation analysis of allotetraploid hybrids of red crucian carp (Carassius auratus red var.) and common carp (Cyprinus carpio L.). PLoS ONE 8:e56409. doi: 10.1371/journal.pone.0056409
Xie, W., Gai, X., Zhu, Y., Zappulla, D. C., Sternglanz, R., and Voytas, D. F. (2001). Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21, 6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001
Xin, M., Wang, Y., Yao, Y., Song, N., Hu, Z., Qin, D., et al. (2011). Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11:61. doi: 10.1186/1471-2229-11-61
Xin, M., Wang, Y., Yao, Y., Xie, C., Peng, H., Ni, Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. doi: 10.1186/1471-2229-10-123
Xiong, Z., Gaeta, R. T., and Pires, J. C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. U.S.A. 108, 7908–7913. doi: 10.1073/pnas.1014138108
Xu, L., Wang, Y., Zhai, L., Xu, Y., Wang, L., Zhu, X., et al. (2013). Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. J. Exp. Bot. 64, 4271–4287. doi: 10.1093/jxb/ert240
Xu, Y., Ayrapetov, M. K., Xu, C., Gursoy-Yuzugullu, O., Hu, Y., and Price, B. D. (2012a). Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733. doi: 10.1016/j.molcel.2012.09.026
Xu, Y., Huang, L., Fu, S., Wu, J., and Zhou, X. (2012b). Population diversity of rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphgax striatellus. PLoS ONE 7:e46238. doi: 10.1371/journal.pone.0046238
Xu, Y., and Price, B. D. (2011). Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10, 261–267. doi: 10.4161/cc.10.2.14543
Xu, Z., Zhong, S., Li, X., Li, W., Rothstein, S. J., Zhang, S., et al. (2011). Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 6:e28009. doi: 10.1371/journal.pone.0028009
Yaakov, B., and Kashkush, K. (2011a). Massive alterations of the methylation patterns around DNA transposons in the first four generations of a newly formed wheat allohexaploid. Genome 54, 42–49. doi: 10.1139/g10-091
Yaakov, B., and Kashkush, K. (2011b). Methylation, transcription, and rearrangements of transposable elements in synthetic allopolyploids. Int. J. Plant Genomics 2011:569826. doi: 10.1155/2011/569826
Yaakov, B., and Kashkush, K. (2012). Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol. Biol. 80, 419–427. doi: 10.1007/s11103-012-9957-3
Yang, L., Jue, D., Li, W., Zhang, R., Chen, M., and Yang, Q. (2013). Identification of MiRNA from eggplant (Solanum melongena L.) by small RNA deep sequencing and their response to Verticillium dahliae infection. PLoS ONE 8:e72840. doi: 10.1371/journal.pone.0072840
Yang, N., and Kazazian, H. H. Jr. (2006). L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13, 763–771. doi: 10.1038/nsmb1141
Yang, X., Xie, Y., Raja, P., Li, S., Wolf, J. N., Shen, Q., et al. (2011). Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7:e1002329. doi: 10.1371/journal.ppat.1002329
Yang, X., Yu, Y., Jiang, L., Lin, X., Zhang, C., Ou, X., et al. (2012). Changes in DNA methylation and transgenerational mobilization of a transposable element (mPing) by the topoisomerase II inhibitor, etoposide, in rice. BMC Plant Biol. 12:48. doi: 10.1186/1471-2229-12-48
Yao, Y., Danna, C. H., Zemp, F. J., Titov, V., Ciftci, O. N., Przybylski, R., et al. (2011). UV-C-irradiated Arabidopsis and tobacco emit volatiles that trigger genomic instability in neighboring plants. Plant Cell 23, 3842–3852. doi: 10.1105/tpc.111.089003
Yao, Y., Kathiria, P., and Kovalchuk, I. (2013). A systemic increase in the recombination frequency upon local infection of Arabidopsis thaliana plants with oilseed rape mosaic virus depends on plant age, the initial inoculum concentration and the time for virus replication. Front. Plant Sci. 4:61. doi: 10.3389/fpls.2013.00061
Yao, Y., and Kovalchuk, I. (2011). Abiotic stress leads to somatic and heritable changes in homologous recombination frequency, point mutation frequency and microsatellite stability in Arabidopsis plants. Mutat. Res. 707, 61–66. doi: 10.1016/j.mrfmmm.2010.12.013
Yap, K. L., and Zhou, M. M. (2011). Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 50, 1966–1980. doi: 10.1021/bi101885m
Ye, Y. H., Woolfit, M., Huttley, G. A., Rances, E., Caragata, E. P., Popovici, J., et al. (2013). Infection with a virulent strain of disrupts genome wide-patterns of cytosine methylation in the mosquito. PLoS ONE 8:e66482. doi: 10.1371/journal.pone.0066482
Yin, Z., Li, Y., Han, X., and Shen, F. (2012). Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae-inoculated cotton roots. PLoS ONE 7:e35765. doi: 10.1371/journal.pone.0035765
Youngson, N. A., Kocialkowski, S., Peel, N., and Ferguson-Smith, A. C. (2005). A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481–490. doi: 10.1007/s00239-004-0332-0
Yu, S., Teng, Y., Waters, R., and Reed, S. H. (2011). How chromatin is remodelled during DNA repair of UV-induced DNA damage in Saccharomyces cerevisiae. PLoS Genet. 7:e1002124. doi: 10.1371/journal.pgen.1002124
Yu, Y., Deng, Y., Reed, S. H., Millar, C. B., and Waters, R. (2013a). Histone variant Htz1 promotes histone H3 acetylation to enhance nucleotide excision repair in Htz1 nucleosomes. Nucleic Acids Res. 41, 9006–9019. doi: 10.1093/nar/gkt688
Yu, Y., Yang, X., Wang, H., Shi, F., Liu, Y., Liu, J., et al. (2013b). Cytosine methylation alteration in natural populations of Leymus chinensis induced by multiple abiotic stresses. PLoS ONE 8:e55772. doi: 10.1371/journal.pone.0055772
Zeng, Q. Y., Yang, C. Y., Ma, Q. B., Li, X. P., Dong, W. W., and Nian, H. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 12:182. doi: 10.1186/1471-2229-12-182
Zhai, L., Liu, Z., Zou, X., Jiang, Y., Qiu, F., Zheng, Y., et al. (2013). Genome-wide identification and analysis of microRNA responding to long-term waterlogging in crown roots of maize seedlings. Physiol. Plant. 147, 181–193. doi: 10.1111/j.1399-3054.2012.01653.x
Zhan, X., Wang, B., Li, H., Liu, R., Kalia, R. K., Zhu, J. K., et al. (2012). Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 18198–18203. doi: 10.1073/pnas.1216199109
Zhang, W., Gao, S., Zhou, X., Chellappan, P., Chen, Z., Zhou, X., et al. (2011). Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 75, 93–105. doi: 10.1007/s11103-010-9710-8
Zhang, W., Lee, H. R., Koo, D. H., and Jiang, J. (2008a). Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20, 25–34. doi: 10.1105/tpc.107.057083
Zhang, X., Ge, X., Shao, Y., Sun, G., and Li, Z. (2013). Genomic change, retrotransposon mobilization and extensive cytosine methylation alteration in Brassica napus introgressions from two intertribal hybridizations. PLoS ONE 8:e56346. doi: 10.1371/journal.pone.0056346
Zhang, Y., and Mager, D. L. (2012). Gene properties and chromatin state influence the accumulation of transposable elements in genes. PLoS ONE 7:e30158. doi: 10.1371/journal.pone.0030158
Zhang, Z., Lin, H., Shen, Y., Gao, J., Xiang, K., Liu, L., et al. (2012). Cloning and characterization of miRNAs from maize seedling roots under low phosphorus stress. Mol. Biol. Rep. 39, 8137–8146. doi: 10.1007/s11033-012-1661-5
Zhang, Z., Wei, L., Zou, X., Tao, Y., Liu, Z., and Zheng, Y. (2008b). Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann. Bot. 102, 509–519. doi: 10.1093/aob/mcn129
Zheng, X., Chen, L., Li, M., Lou, Q., Xia, H., Wang, P., et al. (2013). Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 8:e80253. doi: 10.1371/journal.pone.0080253
Zheng, Y., Ao, Z., Jayappa, K. D., and Yao, X. (2010). Characterization of the HIV-1 integrase chromatin- and LEDGF/p75-binding abilities by mutagenic analysis within the catalytic core domain of integrase. Virol. J. 7:68. doi: 10.1186/1743-422x-7-68
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. doi: 10.1104/pp.112.208702
Zhu, H., and Guo, H. (2012). The role of virus-derived small interfering RNAs in RNA silencing in plants. Sci. China Life Sci. 55, 119–125. doi: 10.1007/s11427-012-4281-3
Zhu, J., Gaiha, G. D., John, S. P., Pertel, T., Chin, C. R., Gao, G., et al. (2012). Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2, 807–816. doi: 10.1016/j.celrep.2012.09.008
Zhu, Y., Rowley, M. J., Bohmdorfer, G., and Wierzbicki, A. T. (2013). A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell 49, 298–309. doi: 10.1016/j.molcel.2012.11.011
Zhuang, Y., Zhou, X. H., and Liu, J. (2014). Conserved miRNAs and their response to salt stress in wild Eggplant Solanum linnaeanum roots. Int. J. Mol. Sci. 15, 839–849. doi: 10.3390/ijms15010839
Zucchi, F. C., Yao, Y., and Metz, G. A. (2012). The secret language of destiny: stress imprinting and transgenerational origins of disease. Front. Genet. 3:96. doi: 10.3389/fgene.2012.00096
Keywords: mutation, evolution, natural genetic engineering, mobile DNA, viruses, mobile genetic elements, non-coding RNA
Citation: Shapiro JA (2014) Epigenetic control of mobile DNA as an interface between experience and genome change. Front. Genet. 5:87. doi: 10.3389/fgene.2014.00087
Received: 27 February 2014; Accepted: 01 April 2014;
Published online: 25 April 2014.
Edited by:
Ilaria Negri, University of Turin, ItalyReviewed by:
Jennifer Cropley, Victor Chang Cardiac Research Insitute, AustraliaMauro Mandrioli, University of Modena and Reggio Emilia, Italy
Copyright © 2014 Shapiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James A. Shapiro, Department of Biochemistry and Molecular Biology, University of Chicago, GCIS W123B, 979 E. 57th Street, Chicago, IL 60637, USA e-mail:anNoYUB1Y2hpY2Fnby5lZHU=
 James A. Shapiro
James A. Shapiro