
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 02 April 2014
Sec. RNA
Volume 5 - 2014 | https://doi.org/10.3389/fgene.2014.00067
This article is part of the Research Topic Molecular Biology of the Transfer RNA Revisited View all 13 articles
Sulfur is an essential element for a variety of cellular constituents in all living organisms. In tRNA molecules, there are many sulfur-containing nucleosides, such as the derivatives of 2-thiouridine (s2U), 4-thiouridine (s4U), 2-thiocytidine (s2C), and 2-methylthioadenosine (ms2A). Earlier studies established the functions of these modifications for accurate and efficient translation, including proper recognition of the codons in mRNA or stabilization of tRNA structure. In many cases, the biosynthesis of these sulfur modifications starts with cysteine desulfurases, which catalyze the generation of persulfide (an activated form of sulfur) from cysteine. Many sulfur-carrier proteins are responsible for delivering this activated sulfur to each biosynthesis pathway. Finally, specific “modification enzymes” activate target tRNAs and then incorporate sulfur atoms. Intriguingly, the biosynthesis of 2-thiouridine in all domains of life is functionally and evolutionarily related to the ubiquitin-like post-translational modification system of cellular proteins in eukaryotes. This review summarizes the recent characterization of the biosynthesis of sulfur modifications in tRNA and the novel roles of this modification in cellular functions in various model organisms, with a special emphasis on 2-thiouridine derivatives. Each biosynthesis pathway of sulfur-containing molecules is mutually modulated via sulfur trafficking, and 2-thiouridine and codon usage bias have been proposed to control the translation of specific genes.
A characteristic structural and functional feature of RNA is post-transcriptional modification. More than 100 forms of naturally occurring chemical modification have been reported to date1,2 (Cantara et al., 2011; Machnicka et al., 2013). The roles of modified nucleosides in tRNA are important and wide-ranging, and include critical roles in biogenesis, structural stability, codon recognition, maintenance of reading frame, and identification elements for the translation machinery (Björk, 1995; Curran, 1998).
The biosynthesis and functions of thionucleosides have been elucidated mainly by using Escherichia coli, Salmonella enterica serovar Typhimurium, and Saccharomyces cerevisiae as model organisms. E. coli tRNAs contain five thionucleosides, 4-thiouridine (s4U) at position 8, 2-thiocytidine (s2C) at position 32, 5-methylaminomethyl-2-thiouridine (mnm5s2U) or 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) at position 34, and 2-methylthio-N6-isopentenyladenosine (ms2i6A) at position 37 (Figure 1). The biosynthesis of these thionucleosides can be divided into two major groups depending on the involvement of iron–sulfur (Fe–S) cluster biosynthesis. The thiouridines s4U8 and (c)mnm5s2U34 are synthesized independently of Fe–S cluster formation, while s2C32 and ms2i6A37 synthesis is dependent upon Fe–S cluster formation, which suggest that Fe–S-containing proteins are present in the latter biosynthesis pathways (Figure 2; Lauhon et al., 2004; Leipuviene et al., 2004).
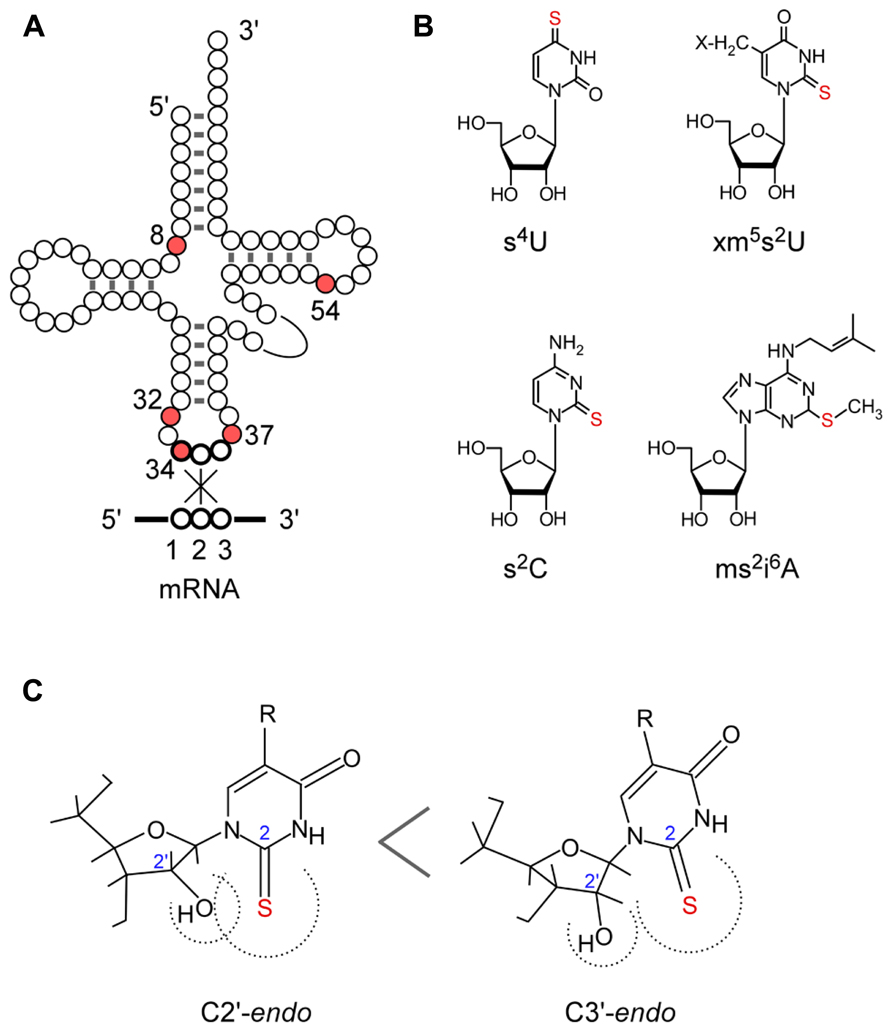
FIGURE 1. Sulfur-containing tRNA modifications. (A) Secondary structure of tRNA and positions of thiolated nucleosides in tRNA. (B) Chemical structure of thiolated nucleosides in E. coli: s4U, 4-thiouridine; s2C, 2-thiocytidine; xm5s2U, 5-methyl-2-thiouridine derivatives; ms2i6A, 2-methylthio-N6-isopentenyladenosine. (C) Conformation of the xm5s2U: C3’ -endo form is preferred because of the steric hindrance of the 2-thio and 2’ -OH groups.
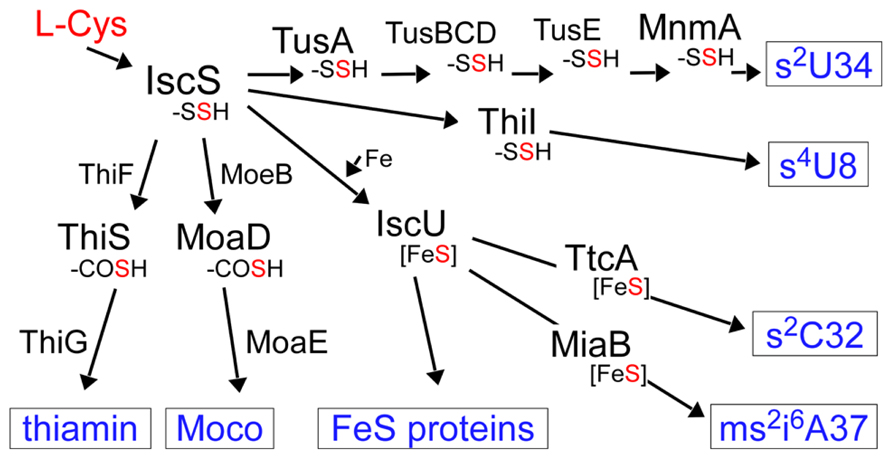
FIGURE 2. Sulfur flow in the biosynthesis of various sulfur-containing molecules in E. coli. The sulfur atom in cysteine is first activated by IscS as a persulfide. The persulfide sulfur on IscS is delivered to acceptor proteins in each pathway. -SSH: persulfide, -COSH: thiocarboxylate, Fe–S: iron–sulfur cluster, Moco: molybdenum cofactor.
The first step of mobilization of sulfur in both pathways starts with the activation of the sulfur atom of cysteine by an enzyme, cysteine desulfurase, IscS (Figure 2). IscS forms an enzyme-bound persulfide (IscS–SSH) using pyridoxal-5’-phospate (PLP) as a cofactor and this activated sulfur is transferred to the next acceptor protein in each pathway. For the biosynthesis of s4U8 and (c)mnm5s2U34, the sulfur atom is transferred to specific sulfur-carrier proteins or a “modification enzyme” (Palenchar et al., 2000; Ikeuchi et al., 2006). The modification enzymes bind and activate target tRNA and catalyze the final step of sulfur transfer to tRNA. For the biosynthesis of s2C32 and ms2i6A37 (Fe–S cluster dependent pathway), the persulfide generated by IscS is transferred to the “scaffold” protein IscU, on which the Fe–S cluster is synthesized, and the Fe–S cluster is then incorporated into the modification enzymes for ms2i6A37 (and maybe also for s2C32; Pierrel et al., 2002, 2003, 2004; Jäger et al., 2004). In ms2i6A synthesis, it was reported that the sulfur atom in the Fe–S cluster is not the sulfur donor (Forouhar et al., 2013); therefore, the ultimate sulfur donor in vivo remains to be determined.
The sulfur atom activated by IscS is also used in molybdenum cofactor (Moco) and thiamin biosynthesis (Figure 2). These are sulfur-containing cofactors whose biosynthesis also includes unique sulfur-carrier proteins. Moco is incorporated into the active sites of many molybdoenzymes, including nitrate reductase, sulfite oxidase, and xanthine dehydrogenase (Schindelin et al., 2001). Moco contains a molybdenum atom and a pterin named molybdpterin (MPT). In MPT biosynthesis, two sulfur atoms are incorporated into precursor Z using a protein-thiocarboxylate as a sulfur donor. Thiamin is an essential cofactor for enzymes involved in carbohydrate and branched-chain amino acid metabolism and is synthesized from thiazole and pyrimidine moieties (Settembre et al., 2003). The sulfur atom of the thiazole ring is added in most bacteria by a system similar to the Moco biosynthetic machinery.
In S. cerevisiae, there are two thiouridines in tRNA, 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34) in cytosolic tRNAs and 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U34) in mitochondrial tRNAs. The biosynthesis pathway of 2-thiouridine in cytosolic tRNA is Fe–S cluster dependent, while the mitochondrial pathway is independent of Fe–S cluster formation (Umeda et al., 2005; Nakai et al., 2007). The biosynthesis of s2U in cytosolic tRNA in the eukaryote utilizes a protein-thiocarboxylate as intermediate sulfur donor. This pathway is functionally and evolutionarily related to the ubiquitin-like post-translational modification system of cellular proteins in eukaryotes and a similar biosynthesis pathway in archaea was reported (Humbard et al., 2010; Miranda et al., 2011).
In some thermophiles, 5-methyl-2-thiouridine (m5s2U) [also called 2-thioribothymidine (s2T)] occurs at position 54 in the T-loop (Figure 1). Intriguingly, the biosynthesis pathway (Shigi et al., 2008) is similar to that of cytosolic s2U34 in eukaryotes, and ubiquitin-like post-translational modification of cellular proteins has recently been discovered also in the bacteria domain (Shigi, 2012).
In this review, I summarize recent advances with respect to the characterization of the biosynthesis mechanisms of sulfur modifications in tRNA, with special reference to 2-thiouridine derivatives. Up to the time of writing, two major pathways for the biosynthesis of 2-thiouridine have been reported. These pathways differ in terms of the types of modification enzyme and the ultimate sulfur donor (Table 1). The novel roles of 2-thiouridine in cellular functions have been revealed by new techniques including genome-wide analyses in some model organisms. Interestingly, each biosynthesis pathway to sulfur-containing molecules has been suggested to be mutually modulated via sulfur trafficking and translational control of specific genes by 2-thiouridine derivatives in tRNAs.
The 2-thiouridine modification at position 34 and 54 plays critical roles in protein synthesis. Position 34 (the wobble base) of tRNAs for Glu, Gln, and Lys are universally modified to 5-methyl-2-thiouridine derivatives (xm5s2U; Figure 1): 5-methylaminomethyl-2-thiouridine (mnm5s2U) and 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) in bacterial tRNAs, 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) in eukaryotic cytosolic tRNAs, cmnm5s2U in yeast mitochondrial tRNA, and 5-taurinomethyl-2-thiouridine (τm5s2U) in mammalian mitochondrial tRNAs (Suzuki, 2005).
The conformation of xm5s2U preferentially takes the C3’-endo form of ribose puckering, because of the steric effect of the bulky 2-thiocarbonyl group toward the 2’-hydroxyl group (Figure 1C; Yokoyama et al., 1985; Agris et al., 1992). The xm5s2U34 base pairs preferentially with purines and prevents misreading of near cognate codons ending in pyrimidines (Agris et al., 1973; Yokoyama et al., 1985; Murphy et al., 2004; Durant et al., 2005; Johansson et al., 2008) and frame shifting (Urbonavicius et al., 2001; Atkins and Björk, 2009; Isak and Ryden-Aulin, 2009; Jäger et al., 2013). The 2-thio group of xm5s2U34 is required for efficient codon recognition on the ribosome (Ashraf et al., 1999; Vendeix et al., 2012; Rodriguez-Hernandez et al., 2013). In addition, the 2-thio group of cmnm5s2U34 in tRNAGluacts as the identity element for specific recognition by glutaminyl-tRNA synthetase (Sylvers et al., 1993; Rodriguez-Hernandez et al., 2013). In human, a defect in mitochondrial translation is induced by the lack of xm5s2U34 modification in mutant mitochondrial tRNALys from patients with myoclonus epilepsy with ragged-red fibers (MERRF; Yasukawa et al., 2000, 2001).
The 2-thio modification of m5s2U (s2T) at position 54 in the T-loop also plays an important role in protein synthesis in high temperature environments. In thermophilic organisms such as Thermus thermophilus and Pyrococcus furiosus, almost all tRNA species are modified to m5U54 and m5s2U54 (Watanabe et al., 1974; Kowalak et al., 1994). The m5s2U54 is also found in the hyperthermophilic bacterium Aquifex aeolicus (Awai et al., 2009). The 2-thiolation content of m5U54 increases with cultivation temperature (Watanabe et al., 1976; Kowalak et al., 1994). As deletion strains of T. thermophilus lacking the 2-thio group of the m5s2U54 modification show a temperature sensitive phenotype, this modification is suggested to be required for survival of the thermophile at high temperature (Shigi et al., 2006a). In the L-shaped tRNA structure, m5s2U54 is buried inside the tertiary core and forms a reverse Hoogsteen base pair with m1A58 and also stacking with G53 and ψ55. In addition, ψ55 and C56 form tertiary base pairs with G18 and G19 in the D-loop, respectively. The rigid conformation of m5s2U54 stabilizes the A-form helix of the D-loop–T-loop interaction, contributing to the thermostability of tRNAs in the thermophile (Watanabe et al., 1974; Horie et al., 1985).
In E. coli, seven proteins are responsible for 2-thiolation of 5-methylaminomethyl-2-thiouridine (mnm5s2U) or 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) in the wobble base of tRNAGluUUC, tRNAGlnUUG, and tRNALysUUU: a cysteine desulfurase (IscS), a modification enzyme (MnmA), and three persulfide carriers (TusA, TusBCD complex, and TusE; Figure 3A; Kambampati and Lauhon, 2003; Ikeuchi et al., 2006; Numata et al., 2006a). The sulfur atom of L-cysteine is first activated by IscS cysteine desulfurase to form an enzyme-bound persulfide. The small sulfur-carrier proteins TusA, TusBCD, and TusE relay this sulfur atom via their active site cysteine residues to MnmA. Tus proteins stimulate sulfur transfer from IscS to the catalytic cysteine of MnmA (Cys199). MnmA is an N-type ATP-pyrophosphatase that possesses the characteristic PP-motif (Bork and Koonin, 1994) and two conserved cysteine residues (Cys102 and Cys199). The reaction mechanism was well documented in a biochemical study based on the crystal structure of the MnmA-tRNA complex (Numata et al., 2006b). MnmA binds the anticodon arm and D-stem regions of tRNA and activates the C2-position of the uracil ring at position 34 as an acyl-adenylated intermediate (tRNA-OAMP). This is then followed by nucleophilic attack by the persulfide sulfur of MnmA-Cys199-SSH, which results in the completion of 2-thiouridine formation.
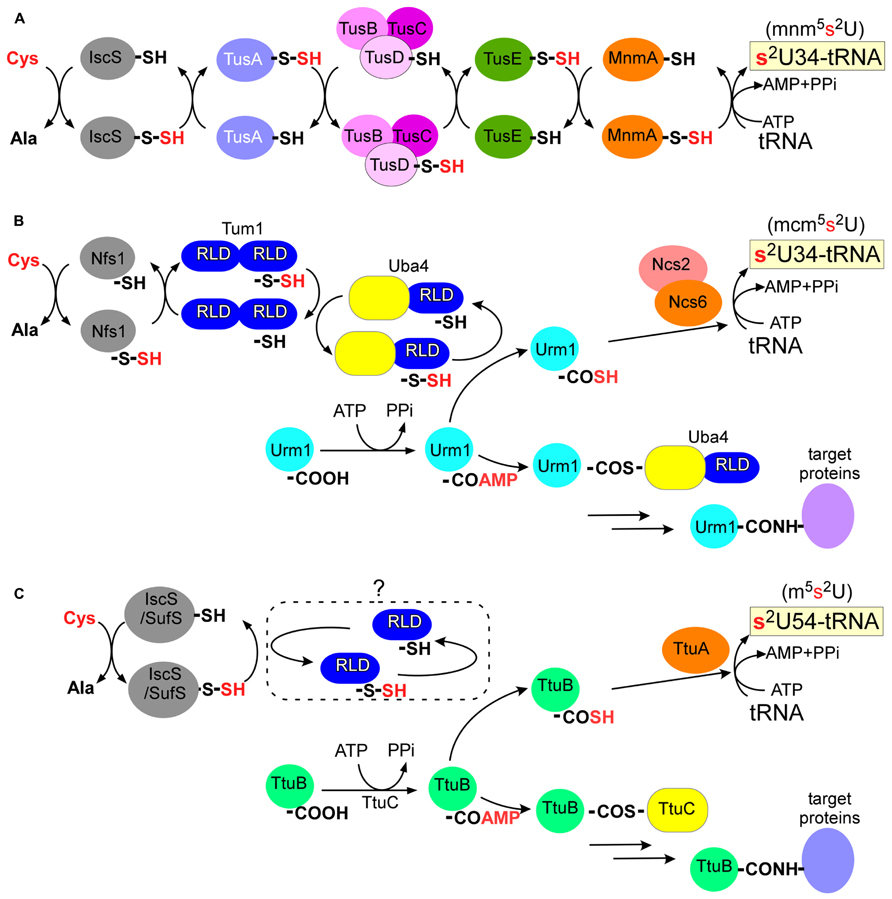
FIGURE 3. Biosynthesis pathway of 2-thiouridine derivatives. (A) In E. coli, Tus proteins relay the persulfide sulfur of IscS to a modification enzyme, MnmA. (B) In S. cerevisiae, Urm1 acts both as sulfur carrier and protein modifier. A thioester conjugate or an acyldisulfide conjugate (not shown) of Urm1 and Uba4 were proposed in urmylation pathway. RLD: rhodanese-like domain. (C) In T. thermophilus, TtuB also acts both as sulfur carrier and protein modifier. An unidentified RLD protein might be involved in sulfur transfer.
Genomic analysis of bacteria revealed that IscS, TusA, and MnmA are mostly conserved, whereas TusBCD and TusE are not found in many organisms (Kotera et al., 2010). This implies that a variation in the sulfur-transfer pathways from IscS to MnmA may exist. In eukaryotic mitochondria, NifS and Mtu1 (homologs of IscS and MnmA, respectively) are responsible for 2-thiolation of cmnm5s2U in yeast and 5-taurinomethyl-2-thiouridine (τ m5s2U) in mammals (Umeda et al., 2005). In eukaryotic mitochondria, the intermediate sulfur carriers remained to be identified.
The Ncs6/Urm1 pathway is responsible for the 2-thiolation of 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) in the wobble base of tRNAGluUUC, tRNAGlnUUG, and tRNALysUUU in the cytosol of eukaryotes (S. cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Homo sapiens; Esberg et al., 2006; Björk et al., 2007; Dewez et al., 2008; Huang et al., 2008; Nakai et al., 2008; Schlieker et al., 2008; Schmitz et al., 2008; Leidel et al., 2009; Noma et al., 2009). A similar pathway was reported subsequently in plants (Leiber et al., 2010; Nakai et al., 2012). With the exception of the first step catalyzed by cysteine desulfurase Nfs1, the eukaryotic pathway is quite different from the MnmA pathway in bacteria described above. The function of Nfs1 is to donate the sulfur to the Fe–S cluster and 2-thiouridine. Formation of 2-thiouridine is dependent on Fe–S cluster biosynthesis (ISC) and cytosolic Fe–S cluster assembly (CIA) machineries in yeast (Nakai et al., 2007). This suggests that the Ncs6/Urm1 pathway depends on Fe–S protein(s), although at the time of writing it remains to be determined which protein(s) possess Fe–S cluster(s).
The Ncs6/Urm1 pathway is composed of at least six proteins including a cysteine desulfurase (Nfs1), a modification enzyme complex (Ncs6/Ncs2), two sulfur carriers (Urm1 and Tum1), and an activation enzyme for Urm1 (Uba4; Figure 3B). The gene names here are those of S. cerevisiae, and homologs of Ncs6/Ncs2 and Uba4 in humans are designated ATPBD3/CTU2 and MOCS3 (molybdenum cofactor synthesis 3), respectively. Tum1 and Uba4 contain rhodanese-like domains (RLDs) bearing conserved cysteine residues. Rhodanese is a widespread sulfur-carrier enzyme that catalyzes sulfur-transfer reactions in various metabolic pathways (Bordo and Bork, 2002). The conserved cysteine residues of RLDs in Tum1 and Uba4 are critical for 2-thiouridine formation in vivo. Tum1 probably directs sulfur flow to 2-thiouridine formation (Noma et al., 2009). The persulfide of Nfs1 is transferred to the RLD of Uba4 mainly via the RLD of Tum1.
Urm1 is a ubiquitin-related modifier and Uba4 is an E1-like Urm1-activating enzyme involved in protein urmylation (see the following; Furukawa et al., 2000). The carboxy-terminus of Urm1 is first activated as an acyl-adenylate intermediate (Urm1-COAMP) and then thiocarboxylated (Urm1-COSH) by a persulfide from the RLD of Uba4 (Figure 3B; Schlieker et al., 2008; Schmitz et al., 2008; Leidel et al., 2009; Noma et al., 2009). The activated thiocarboxylate may be utilized in subsequent reactions for 2-thiouridine formation, which is mediated by a heterodimer complex, Ncs6/Ncs2 (Dewez et al., 2008; Noma et al., 2009). Ncs6 has the PP-motif and many CXXC motifs (Figure 4A; see TtuA/TtuB Pathway for 2-Thiouridine Synthesis in Thermophile tRNAs). Thus, 2-thiolation of mcm5s2U shares a pathway and chemical reactions with protein urmylation. Intriguingly, eukaryotic 2-thiouridine formation employs a thiocarboxylated intermediate as the active form of the sulfur atom, which is a mechanism distinct from bacterial sulfur-relay based on persulfide chemistry.
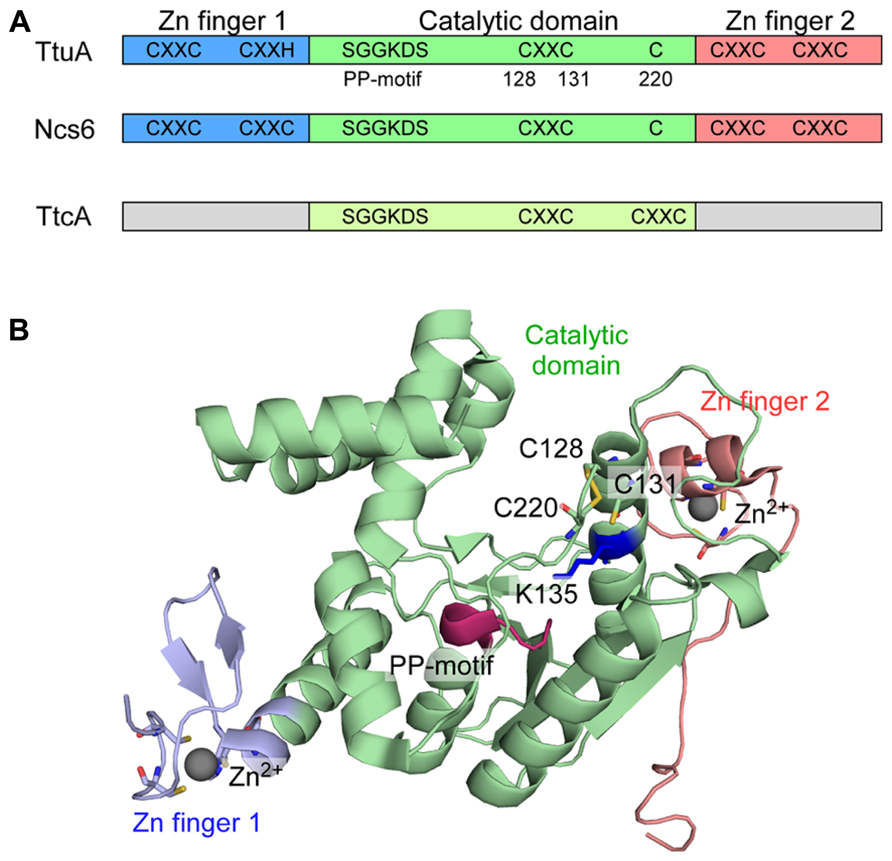
FIGURE 4. TtuA and Ncs6 proteins in 2-thiouridine biosynthesis. (A) Schematic representation of TtcA family proteins. TtuA and Ncs6 belong to group II of the TtcA family. TtuA is composed of two Zn finger domains and a catalytic domain. TtcA is responsible for the formation of 2-thiocytidine34. (B) Structure of P. horikoshii TtuA. The three domains are colored the same as in (A). The PP-motif is shown in red. Three Cys residues that is important for enzyme activity is shown with a stick representation. The target K135 for TtuB conjugation (K137 in T. thermophilus) is shown in a blue stick model.
Ubiquitin (Ub) and ubiquitin-like proteins (Ubls) are post-translational protein modifiers with important roles in proteolysis and the regulation of diverse processes in eukaryotes (Hochstrasser, 2009). The breakdown of the Ub/Ubl system is often associated with the development of various diseases. In the first step of conjugation to target proteins, the conserved C-terminal glycine of Ub/Ubl is acyl-adenylated by an activating enzyme (E1) and covalently linked to a cysteine residue of E1 to form an Ub/Ubl-E1 thioester intermediate. The activated Ub/Ubl is next transferred to a conjugating enzyme (E2). Finally, Ub/Ubl is attached to a lysine residue in the target protein by a ligase (E3; Hochstrasser, 2009).
Proteins homologous to eukaryotic Ub/Ubl and E1s exist in almost all members of bacteria and archaea (Iyer et al., 2006; Burroughs et al., 2009, 2012). Earlier works established that these bacterial proteins function in the biosynthesis of sulfur compounds such as molybdenum cofactor and thiamin (Kessler, 2006). Bacterial Ubls (MoaD and ThiS) are adenylated by cognate E1 homologs (MoeB and ThiF), subsequently bind activated sulfur via their C-termini to form thiocarboxylates, and finally act as sulfur donors (Figure 2; Pitterle and Rajagopalan, 1993; Taylor et al., 1998; Lauhon and Kambampati, 2000; Leimkühler et al., 2001; Zhang et al., 2010). These findings imply an evolutionary link between the eukaryotic Ub/Ubl system and the bacterial sulfur-transfer reaction (Iyer et al., 2006; Hochstrasser, 2009). Urm1 is an ubiquitin-related modifier and Uba4 is an E1-like enzyme involved in protein urmylation in eukaryotes (Furukawa et al., 2000; Figure 3B). A thioester conjugate (Furukawa et al., 2000) or an acyldisulfide conjugate (Van der Veen et al., 2011) of Urm1 and Uba4 were proposed in this process. As Urm1 also functions as a sulfur donor for 2-thiouridine synthesis (see the preceding) and has close sequence and structural homology with bacterial Ubls (Xu et al., 2006), Urm1 is thought to be the most ancient Ubl possessing dual functions of protein modifier and sulfur carrier. The E2 and E3 enzymes for urmylation have not been identified at the time of writing.
Several targets of urmylation have been identified upon cell exposure to an oxidant (Van der Veen et al., 2011), although earlier reports only identified a peroxiredoxin Ahp1 (Goehring et al., 2003a, b). Among them, a modification enzyme complex ATPBD3/CTU2 and an E1-like MOCS3, both of which are required for 2-thiouridine biosynthesis, have been identified. The target residues in these proteins have not been identified and the roles of urmylation of these proteins are unknown at the time of writing; however, regulation of the activities of these enzymes would be possible.
Prior work from our group identified the TtuA/TtuB pathway for the biosynthesis of thiouridine (m5s2U) at position 54 in tRNAs from a thermophilic bacterium T. thermophilus. The TtuA/TtuB pathway includes cysteine desulfurases (IscS or SufS), a modification enzyme (TtuA), a small ubiquitin-like sulfur carrier (TtuB), and an activation enzyme for TtuB (TtuC; Figure 3C; Shigi et al., 2006a, b, 2008). Similar to the eukaryotic Ncs6/Urm1 pathway described above, the C-terminal Gly of TtuB is acyl-adenylated (TtuB-COAMP) by TtuC and is then thiocarboxylated (TtuB-COSH) by cysteine desulfurases (IscS or SufS). The sulfur atom of the thiocarboxylated TtuB is transferred to tRNA by TtuA. This step also requires ATP as a cofactor and TtuA possesses the PP-motif, suggesting that TtuA may activate the target uridine as an acyl-adenylate. The sulfur-transfer activity in the in vitro system requires the addition of cell-free extract and the activity was low, suggesting that there may still be additional factors required for TtuA-mediated sulfur transfer to tRNA.
TtuA and eukaryotic Ncs6 are homologous to each other, and belong to group II of the TtcA family, whose members are characterized by five conserved CXXC(H) motifs and the PP-motif (Figure 4A; Bork and Koonin, 1994; Jäger et al., 2004; Björk et al., 2007). TtcA, which catalyzes 2-thiocytidine (s2C) synthesis (Jäger et al., 2004), has only two CXXC motifs and the PP-motif, and therefore belongs to group I of the TtcA family. The PP-motif is used for ATP binding to adenylate the target nucleotide, and is widely distributed among ATP pyrophosphatases, including modification enzymes MnmA (see the preceding; Numata et al., 2006b), ThiI for 4-thiouridine (s4U) synthesis (Mueller and Palenchar, 1999; Palenchar et al., 2000), and TilS for tRNAIle2 lysidine synthesis (Ikeuchi et al., 2005).
We determined the crystal structure of the TtuA homolog (PH0300) of the archeaon P. horikoshii (Nakagawa et al., 2013). The P. horikoshii genome has two TtuA/Ncs6-like ORFs: one (PH0300) seems to be an ortholog of T. thermophilus TtuA; and the other (PH1680) seems to be an ortholog of eukaryotic Ncs6, based on their sequence homology to TtuA and Ncs6. The P. horikoshii TtuA forms a homodimer, and each subunit contains a catalytic domain and unique N- and C-terminal zinc fingers (Figure 4B). The N-terminal zinc finger is made up of the first and second CXXC/H motifs, where the zinc atom is coordinated by three Cys residues and one His residue. The C-terminal zinc finger is made up of the fourth and fifth CXXC motifs, where the zinc atom is coordinated by four Cys residues.
Interestingly, the catalytic domain of TtuA has much higher structural similarity to that of another tRNA modification enzyme, TilS (tRNAIle2 lysidine synthetase), than to the other type of tRNA 2-thiolation enzyme, MnmA (Figure 4). However, three Cys residues (128, 131, 220 in PhTtuA) are clustered in the putative catalytic site, which are absent in TilS. Cys128 and Cys131 are in the third CXXC motif and Cys220 is also conserved. By in vivo mutational analysis of TtuA in T. thermophilus (Nakagawa et al., 2013), it became apparent that the three conserved cysteine residues and the putative ATP-binding residues are important for TtuA activity, implying a key role for these Cys residues in sulfur transfer from TtuB-COSH to tRNA. A positively charged surface that includes the catalytic site and the two zinc fingers is likely to provide the tRNA binding site. TtuA recognizes the T-loop (Shigi et al., 2002) and Ncs6/Ncs2 is predicted to recognize the anticodon arm. The recognition mechanisms of the different target sites on tRNA require clarification.
Homology modeling suggests that TtuB possesses a Ub/β-grasp fold and TtuC has significant sequence homology with the adenylation domain of eukaryotic E1s (Shigi et al., 2008). These findings suggest that Ub/Ubl homologous conjugation systems also exist in bacteria. A series of in vivo analyses in T. thermophilus revealed that TtuB is covalently attached to target proteins most likely via its C-terminal glycine (Shigi, 2012). TtuC is required for conjugate formation, and TtuC and TtuA are targets for TtuB conjugation. Mass spectrometric analysis combined with in vivo mutational analysis revealed that lysine residues (K137/K226/K229) in TtuA are covalently modified by the C-terminal carboxylate of TtuB. K137 in T. thermophilus TtuA is situated just after the third CXXC motif. In the crystal structure of P. horikoshii TtuA, K137 (K135 in PhTtuA) is situated close to the catalytic center of this enzyme family (Figure 4B; Nakagawa et al., 2013). K137 in TtuA is conserved in related bacteria and archaea, such as Aquifex, Pyrococcus, Thermococcus, and Metanocaldococcus. On the other hand, this position is occupied by a conserved arginine in eukaryotic Ncs6. K226 and K229 in T. thermophilus TtuA are situated just after Cys222 (Cys220 in PhTtuA), although the regions near the two lysine residues were disordered in the PhTtuA structure. However, K226 and K229 are only conserved in T. thermophilus and a few other species, possibly implying species-specific functions of the conjugation. Intriguingly, a deletion mutant of a JAMM [JAB1/MPN/Mov34 metalloenzyme (Ambroggio et al., 2004)] ubiquitin isopeptidase homolog in T. thermophilus showed aberrant TtuB-conjugates of TtuC and TtuA, and a ~50% decrease in the amount of thiouridine in tRNA (Shigi, 2012). These results support the hypothesis that thiouridine synthesis is regulated by TtuB conjugation.
Although the precise chemical structure of the archaeal counterpart remains unknown at the time of writing, the existence of modified uridines at the wobble position in tRNALys and tRNAGlufrom Haloferax volcanii has been reported (Gupta, 1984). The existence of 2-thiouridine in tRNALys from H. valcanii was suggested by APM-electrophoresis (Miranda et al., 2011), a method that can detect sulfur modifications in RNAs (Igloi, 1988). Because the homologs of eukaryotic Ncs6 are widely distributed in archaeal genomes, these proteins may be involved in this modification (Kotera et al., 2010). Genetic analysis in H. valcanii shows that SAMP2 (small archaeal modifier protein 2) and E1-like protein UbaA are required for thiouridine formation in this organism (Miranda et al., 2011). These results indirectly suggest that SAMP2-COSH is formed and used as a sulfur donor for thiouridine formation in archaea, as observed previously in eukaryotes and bacteria. SAMPs are the first example of a ubiquitin-like protein modifier identified other than from a eukaryote (Humbard et al., 2010) and extensive studies show that the archaeal protein modification system resembles that of eukaryotes in many aspects (Van der Veen et al., 2011; Hepowit et al., 2012; Miranda et al., 2014). SAMP2 covalently conjugates too many target proteins including UbaA (a Uba4 homolog), HVO_0580 (a Ncs6 homolog), and HVO_0025 (a Tum1 homolog) (Humbard et al., 2010), implying that the SAMP2 modification also regulates the thiolation machinery.
4-Thiouridine (s4U), is a modified nucleotide of tRNA that is conserved from bacteria to archaea, where it functions as a sensor for near-UV irradiation (Favre et al., 1969; Carre et al., 1974; Ryals et al., 1982). Sulfur transfer in the biosynthesis of s4U has been extensively studied in bacteria (Kambampati and Lauhon, 2000; Mueller et al., 2001; Leipuviene et al., 2004). The persulfide of IscS is transferred to the RLD of ThiI, a PP-motif-containing modification enzyme. Recently, ThiI lacking an RLD domain has been characterized in methanogenic archaea (Liu et al., 2012). It has three cysteine residues (two of which come from a CXXC motif) in the putative catalytic site, and they are all required for persulfide intermediate formation. This may provide a hint about the catalytic mechanism of TtuA/Ncs6, because of the sequence and structural similarities between TtuA and ThiI (Waterman et al., 2006; Nakagawa et al., 2013).
The mobilization of sulfur in biosynthesis pathways of sulfur-containing compounds starts with the activation of the sulfur atom of cysteine by the cysteine desulfurase IscS. IscS forms an enzyme-bound persulfide and this activated sulfur is transferred to the next acceptor protein in each pathway, such as TusA (2-thiouridine in tRNA), ThiI (4-thiouridine in tRNA), IscU (Fe–S cluster), ThiS (thiamin), and MoaD (molybdenum cofactor; Figure 2). It is conceivable that each biosynthesis pathway of sulfur-containing molecules is mutually modulated via competition of sulfur trafficking. An interesting observation with respect this was made during a study of lambda phage infection in E. coli (Maynard et al., 2010, 2012). During viral infection, the normal amount of modified uridine in tRNALysUUU grarantees a normal translation and frameshifting rate for production of the proper ratio of viral gpG and gpGT proteins (gpGT production needs programmed ribosomal frameshifting). Hypomodification of tRNALysUUUcaused by deletion of Tus genes in the host cell leads to increased frameshifting in the translation of viral mRNA of G and T genes, which affects the ratio of viral gpG to gpGT. A lower gpG:gpGT ratio leads to decreased virion production. Another factor lowering infection is overexpression of IscU in the host cell. In this situation, higher sulfur flow from IscS to IscU conversely lowers the sulfur flow to Tus proteins, which leads to hypomodification of tRNALysUUU and abnormal frameshifting, which finally affects the viral infection rate. The competitive binding of TusA and IscU to IscS has been analyzed in detail, based on the structures of the complexes IscS/TusA and IscS/IscU (Shi et al., 2010; Marinoni et al., 2012).
The sharing of a factor downstream of cysteine desulfurase also occurs in this sulfur-transfer network. TusA was originally identified as a sulfur carrier for 2-thiouridine synthesis (Ikeuchi et al., 2006); however, it was later reported to be involved in Moco synthesis in E. coli (Dahl et al., 2013; Kozmin et al., 2013). It has been proposed that the deletion of TusA leads to the overproduction of Fe–S clusters, which finally affects the expression of several genes (Dahl et al., 2013). A study of the link between the MnmA pathway and cellular redox state has recently been reported (Nakayashiki et al., 2013). By screening for mutants sensitive to hydroxyurea (HU) in E. coli, the authors identified mutations in all genes in the MnmA pathway (iscS, mnmA, and tusA-E in 2-thiouridine synthesis at the wobble position). These mutations resulted in a more reduced state, which may have led to a change in the activity of ribonucleotide reductase, an enzyme inhibited by HU. It is possible that a change in sulfur flow to each pathway in mutants of the MnmA pathway led to the reduced cellular state, although the precise mechanism underlying this phenomenon still remains unknown.
There is another interesting case. In E. coli, bacterial Ubls (MoaD and ThiS) are adenylated by cognate E1 homologs (MoeB and ThiF), subsequently bind activated sulfur at their C-termini to form thiocarboxylate, and work as sulfur donors for Moco and thiamin biosynthesis, respectively (Figure 2; Kessler, 2006). In the T. thermophilus genome, there is only one E1 homolog, TtuC. The ttuC mutant cannot synthesize 2-thio modification of m5s2U54; moreover, Moco and thiamin biosynthesis are also defective. TtuC and cysteine desulfurase can activate and thiocarboxylate TtuB, MoaD, and ThiS in vitro. Thus, TtuC is a common E1-like enzyme shared by the biosynthesis pathways of these three sulfur-containing compounds (Shigi et al., 2008). Similarly, in human, MOCS2A (a MoaD homolog) and URM1 are adenylated by an E1-like MOCS3 (Uba4 homolog; Chowdhury et al., 2012). In archaea, E1-like UbaA is involved in the biosynthesis of Moco and thiouridine (Miranda et al., 2011). Interestingly, these E1 homologs are also involved in the post-translational modification of cellular proteins in all domains of life (Furukawa et al., 2000; Miranda et al., 2011; Shigi, 2012).
The inactivation of the genes in the Ncs6/Urm1 pathway results in a pleotropic phenotype that includes increased sensitivity to high temperature, oxidative stress, and rapamycin, a Tor-signaling inhibitor (Furukawa et al., 2000; Goehring et al., 2003a, b; Dewez et al., 2008). These phenotypes can be suppressed by overexpression of tRNALys, tRNAGlu, and tRNAGln, which normally possess mcm5s2U34 (Leidel et al., 2009). The phenotypes are similar to those of mutants of the elongator complex (Esberg et al., 2006), which is essential for modification of the C5 position of U34. These observations provide evidence that the mcm5s2U modification in tRNAs affects global translation, resulting in a pleotropic phenotype.
In S. cerevisiae, pioneering work has addressed the function of tRNA modifications on gene expression. The mcm5 modification of the wobble base of specific tRNAs modulates the expression of a DNA damage response mRNA, whose cognate codons are unusually overrepresented (Begley et al., 2007). A similar observation has been made concerning telomeric gene silencing (Chen et al., 2011). Proteome analysis in S. pombe showed that the amount of a specific group of proteins, including those involved in cell division, was decreased in mutants defective in the mcm5s2U modification (Bauer and Hermand, 2012; Bauer et al., 2012). The genes coding for these proteins have skewed lysine codon usage, such that the AAA codon was overrepresented compared to the AAG codon. The mcm5s2U modification in tRNALysUUU is necessary especially for the efficient translation of mRNAs enriched in the AAA codon. Among the genes affected by the mcm5s2U modification, a central regulator of mitosis and cytokinesis, Cdr2, was identified. The amount of Cdr2, a protein kinase, was recovered by overexpression of tRNALysUUU in the mutant defective in mcm5s2U biosynthesis. In addition, after substituting AAG codons for all AAA codons in cdr2 mRNA, the Cdr2 protein amount was no longer affected by the mcm5s2U modification. This study provides an interesting example of how translational control of a specific group of mRNAs can be affected by tRNA modifications and codon usage. The translation of specific genes in S. pombe and S. cerevisiae has also been reported by two other groups to be controlled by the s2U modification and codon usage (Fernandez-Vazquez et al., 2013; Rezgui et al., 2013).
In S. cerevisiae, 2-thiouridine formation in tRNA-Lys, -Glu, and -Gln is actively downregulated when methionine and cysteine are limiting, which leads to an overall reduction in translational capacity and reduced growth, because Glu, Gln, and especially Lys codons are overrepresented in the genes essential for translation and growth (Laxman et al., 2013). In this case, tRNA thiolation works as a key effector to maintain amino acid homeostasis.
In the course of the characterization of biosynthesis pathways of sulfur-containing modifications, a number of sulfur-carrier proteins have been identified. The sulfur-carrier proteins may have evolved to deliver reactive sulfur atoms to specific targets and avoid non-specific transfer of activated sulfur atoms, which could inactivate other biomolecules. At the time of writing, it is still unclear whether or not the differences in the chemical properties of persulfide and thiocarboxylate result in different biological outcomes.
Each biosynthesis pathway of sulfur-containing molecules is mutually modulated by sulfur trafficking, and translational control of specific genes by 2-thiouridine and codon usage bias is now proposed in some model organisms. It would be interesting to determine whether similar mechanisms exist in higher eukaryotes.
Unexpectedly, the characterization of the biosynthesis of 2-thiouridine has revealed molecular fossils, namely, ancient ubiquitin-like molecules, including Urm1 in eukaryotes, TtuB in bacteria, and SAMP2 in archaea. These proteins have two functions; they function as sulfur carriers for 2-thiouridine synthesis and as protein modifiers. Therefore, these proteins may be evolutionarily intermediates between ancient sulfur-carrier proteins and protein modifiers. It is possible that an adenylated or thiocarboxylated intermediate, formed in the course of 2-thiouridine biosynthesis, was incidentally attached to adjacent proteins at some time in the past. By this post-translational modification, the activities of the attached proteins have probably changed. This was certainly the origin of the post-translational modification of proteins by these Ubls. The primitive function of these conjugates was probably self-regulation. Conjugates of Ubls and the modification enzymes Ncs6/Ncs2 and TtuA have already been detected, but the function of these post-translational modifications remains to be clarified. The resultant conjugates have become to be used as tags for the recognition and regulation of other proteins. By acquiring E2 and E3 enzymes, which can recognize target proteins precisely, the Ub system evolved considerably to become the sophisticated system it is today in eukaryotes.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I would like to thank all collaborators, including Dr. Tsutomu Suzuki (University of Tokyo), Dr. Kimitsuna Watanabe (Tokyo University of Pharmacy and Life Sciences), and Dr. Shigeyuki Yokoyama (RIKEN), as well as members of their laboratories, namely Ms. Yuriko Sakaguchi (University of Tokyo) and Dr. Hirofumi Nakagawa (University of Tokyo and RIKEN). This study was supported in part by KAKENHI Grant (24570173) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Naito Foundation, the Kurata Memorial Hitachi Science and Technology Foundation, and the Takeda Science Foundation.
Agris, P. F., Sierzputowska-Gracz, H., Smith, W., Malkiewicz, A., Sochacka, E., and Nawrot, B. (1992). Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. J. Am. Chem. Soc. 114, 2652–2656. doi: 10.1021/ja00033a044
Agris, P. F., Soll, D., and Seno, T. (1973). Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry 12, 4331–4337. doi: 10.1021/bi00746a005
Ambroggio, X. I., Rees, D. C., and Deshaies, R. J. (2004). JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2:E2. doi: 10.1371/journal.pbio.0020002
Ashraf, S. S., Sochacka, E., Cain, R., Guenther, R., Malkiewicz, A., and Agris, P. F. (1999). Single atom modification (O-- > S) of tRNA confers ribosome binding. RNA 5, 188–194. doi: 10.1017/S1355838299981529
Atkins, J. F., and Björk, G. R. (2009). A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev. 73, 178–210. doi: 10.1128/MMBR.00010-08
Awai, T., Kimura, S., Tomikawa, C., Ochi, A., Ihsanawati, Bessho, Y., et al. (2009). Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 284, 20467–20478. doi: 10.1074/jbc.M109.020024
Bauer, F., and Hermand, D. (2012). A coordinated codon-dependent regulation of translation by Elongator. Cell Cycle 11, 4524–4529. doi: 10.4161/cc.22689
Bauer, F., Matsuyama, A., Candiracci, J., Dieu, M., Scheliga, J., Wolf, D. A., et al. (2012). Translational control of cell division by Elongator. Cell Rep. 1, 424–433. doi: 10.1016/j.celrep.2012.04.001
Begley, U., Dyavaiah, M., Patil, A., Rooney, J. P., DiRenzo, D., Young, C. M., et al. (2007). Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28, 860–870. doi: 10.1016/j.molcel.2007.09.021
Björk, G. R. (1995). “Biosynthesis and function of modified nucleosides,” in Trna: Structure, Biosynthesis, and Function, eds. D. Soll and U. L. RajBhandary (Washington, DC: ASM Press), 165–205.
Björk, G. R., Huang, B., Persson, O. P., and Bystrom, A. S. (2007). A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13, 1245–1255. doi: 10.1261/rna.558707
Bordo, D., and Bork, P. (2002). The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 3, 741–746. doi: 10.1093/embo-reports/kvf150
Bork, P., and Koonin, E. V. (1994). A P-loop-like motif in a widespread ATP pyrophosphatase domain: implications for the evolution of sequence motifs and enzyme activity. Proteins 20, 347–355. doi: 10.1002/prot.340200407
Burroughs, A. M., Iyer, L. M., and Aravind, L. (2009). Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins 75, 895–910. doi: 10.1002/prot.22298
Burroughs, A. M., Iyer, L. M., and Aravind, L. (2012). The natural history of ubiquitin and ubiquitin-related domains. Front. Biosci. (Landmark Ed.) 17:1433–1460.
Cantara, W. A., Crain, P. F., Rozenski, J., McCloskey, J. A., Harris, K. A., Zhang, X., et al. (2011). The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201. doi: 10.1093/nar/gkq1028
Carre, D. S., Thomas, G., and Favre, A. (1974). Conformation and functioning of tRNAs: cross-linked tRNAs as substrate for tRNA nucleotidyl-transferase and aminoacyl synthetases. Biochimie 56, 1089–1101. doi: 10.1016/S0300-9084(74)80097-0
Chen, C., Huang, B., Eliasson, M., Ryden, P., and Bystrom, A. S. (2011). Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 7:e1002258. doi: 10.1371/journal.pgen.1002258
Chowdhury, M. M., Dosche, C., Lohmannsroben, H. G., and Leimkuhler, S. (2012). Dual role of the molybdenum cofactor biosynthesis protein MOCS3 in tRNA thiolation and molybdenum cofactor biosynthesis in humans. J. Biol. Chem. 287, 17297–17307. doi: 10.1074/jbc.M112.351429
Curran, J. F. (1998). “Modified nucleosides in translation,” in Modification and Editing of RNA, eds. H. Grosjean and R. Benne (Washington, DC: ASM Press), 493–516.
Dahl, J. U., Radon, C., Buhning, M., Nimtz, M., Leichert, L. I., Denis, Y., et al. (2013). The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J. Biol. Chem. 288, 5426–5442. doi: 10.1074/jbc.M112.431569
Dewez, M., Bauer, F., Dieu, M., Raes, M., Vandenhaute, J., and Hermand, D. (2008). The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. U.S.A. 105, 5459–5464. doi: 10.1073/pnas.0709404105
Durant, P. C., Bajji, A. C., Sundaram, M., Kumar, R. K., and Davis, D. R. (2005). Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44, 8078–8089. doi: 10.1021/bi050343f
Esberg, A., Huang, B., Johansson, M. J., and Bystrom, A. S. (2006). Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 24, 139–148. doi: 10.1016/j.molcel.2006.07.031
Favre, A., Yaniv, M., and Michelson, A. M. (1969). The photochemistry of 4-thiouridine in Escherichia coli t-RNA Val1. Biochem. Biophys. Res. Commun. 37, 266–271. doi: 10.1016/0006-291X(69)90729-3
Fernandez-Vazquez, J., Vargas-Perez, I., Sanso, M., Buhne, K., Carmona, M., Paulo, E., et al. (2013). Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 9:e1003647. doi: 10.1371/journal.pgen.1003647
Forouhar, F., Arragain, S., Atta, M., Gambarelli, S., Mouesca, J. M., Hussain, M., et al. (2013). Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat. Chem. Biol. 9, 333–338. doi: 10.1038/nchembio.1229
Furukawa, K., Mizushima, N., Noda, T., and Ohsumi, Y. (2000). A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 275, 7462–7465. doi: 10.1074/jbc.275.11.7462
Goehring, A. S., Rivers, D. M., and Sprague, G. F. Jr. (2003a). Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol. Biol. Cell 14, 4329–4341. doi: 10.1091/mbc.E03-02-0079
Goehring, A. S., Rivers, D. M., and Sprague, G. F. Jr. (2003b). Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot. Cell 2, 930–936. doi: 10.1128/EC.2.5.930-936.2003
Gupta, R. (1984). Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 259, 9461–9471.
Hepowit, N. L., Uthandi, S., Miranda, H. V., Toniutti, M., Prunetti, L., Olivarez, O., et al. (2012). Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol. Microbiol. 86, 971–987. doi: 10.1111/mmi.12038
Hochstrasser, M. (2009). Origin and function of ubiquitin-like proteins. Nature 458, 422–429. doi: 10.1038/nature07958
Horie, N., Hara-Yokoyama, M., Yokoyama, S., Watanabe, K., Kuchino, Y., Nishimura, S., et al. (1985). Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 24, 5711–5715. doi: 10.1021/bi00342a004
Huang, B., Lu, J., and Bystrom, A. S. (2008). A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14, 2183–2194. doi: 10.1261/rna.1184108
Humbard, M. A., Miranda, H. V., Lim, J. M., Krause, D. J., Pritz, J. R., Zhou, G., et al. (2010). Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463, 54–60. doi: 10.1038/nature08659
Igloi, G. L. (1988). Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27, 3842–3849. doi: 10.1021/bi00410a048
Ikeuchi, Y., Shigi, N., Kato, J., Nishimura, A., and Suzuki, T. (2006). Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21, 97–108. doi: 10.1016/j.molcel.2005.11.001
Ikeuchi, Y., Soma, A., Ote, T., Kato, J., Sekine, Y., and Suzuki, T. (2005). Molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol. Cell 19, 235–246. doi: 10.1016/j.molcel.2005.06.007
Isak, G., and Ryden-Aulin, M. (2009). Hypomodification of the wobble base in tRNAGlu, tRNALys, and tRNAGln suppresses the temperature-sensitive phenotype caused by mutant release factor 1. J. Bacteriol. 191, 1604–1609. doi: 10.1128/JB.01485-08
Iyer, L. M., Burroughs, A. M., and Aravind, L. (2006). The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 7:R60. doi: 10.1186/gb-2006-7-7-r60
Jäger, G., Leipuviene, R., Pollard, M. G., Qian, Q., and Bjork, G. R. (2004). The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J. Bacteriol. 186, 750–757. doi: 10.1128/JB.186.3.750-757.2004
Jäger, G., Nilsson, K., and Bjork, G. R. (2013). The phenotype of many independently isolated +1 frameshift suppressor mutants supports a pivotal role of the P-site in reading frame maintenance. PLoS ONE 8:e60246. doi: 10.1371/journal.pone.0060246
Johansson, M. J., Esberg, A., Huang, B., Bjork, G. R., and Bystrom, A. S. (2008). Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 28, 3301–3312. doi: 10.1128/MCB.01542-07
Kambampati, R., and Lauhon, C. T. (2000). Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J. Biol. Chem. 275, 10727–10730. doi: 10.1074/jbc.275.15.10727
Kambampati, R., and Lauhon, C. T. (2003). MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42, 1109–1117. doi: 10.1021/bi026536+
Kessler, D. (2006). Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 30, 825–840. doi: 10.1111/j.1574-6976.2006.00036.x
Kotera, M., Bayashi, T., Hattori, M., Tokimatsu, T., Goto, S., Mihara, H., et al. (2010). Comprehensive genomic analysis of sulfur-relay pathway genes.Genome Inform. 24, 104–115. doi: 10.1142/9781848166585_0009
Kowalak, J. A., Dalluge, J. J., McCloskey, J. A., and Stetter, K. O. (1994). The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33, 7869–7876. doi: 10.1021/bi00191a014
Kozmin, S. G., Stepchenkova, E. I., and Schaaper, R. M. (2013). TusA (YhhP) and IscS are required for molybdenum cofactor-dependent base-analog detoxification. Microbiologyopen 2, 743–755. doi: 10.1002/mbo3.108
Lauhon, C. T., and Kambampati, R. (2000). The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275, 20096–20103. doi: 10.1074/jbc.M002680200
Lauhon, C. T., Skovran, E., Urbina, H. D., Downs, D. M., and Vickery, L. E. (2004). Substitutions in an active site loop of Escherichia coli IscS result in specific defects in Fe-S cluster and thionucleoside biosynthesis in vivo. J. Biol. Chem. 279, 19551–19558. doi: 10.1074/jbc.M401261200
Laxman, S., Sutter, B. M., Wu, X., Kumar, S., Guo, X., Trudgian, D. C., et al. (2013). Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154, 416–429. doi: 10.1016/j.cell.2013.06.043
Leiber, R. M., John, F., Verhertbruggen, Y., Diet, A., Knox, J. P., and Ringli, C. (2010). The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22, 1898–1908. doi: 10.1105/tpc.109.073007
Leidel, S., Pedrioli, P. G., Bucher, T., Brost, R., Costanzo, M., Schmidt, A., et al. (2009). Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458, 228–232. doi: 10.1038/nature07643
Leimkühler, S., Wuebbens, M. M., and Rajagopalan, K. V. (2001). Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 276, 34695–34701. doi: 10.1074/jbc.M102787200
Leipuviene, R., Qian, Q., and Bjork, G. R. (2004). Formation of thiolated nucleosides present in tRNA from Salmonella enterica serovar Typhimurium occurs in two principally distinct pathways. J. Bacteriol. 186, 758–766. doi: 10.1128/JB.186.3.758-766.2004
Liu, Y., Zhu, X., Nakamura, A., Orlando, R., Soll, D., and Whitman, W. B. (2012). Biosynthesis of 4-thiouridine in tRNA in the methanogenic archaeon Methanococcus maripaludis. J. Biol. Chem. 287, 36683–36692. doi: 10.1074/jbc.M112.405688
Machnicka, M. A., Milanowska, K., Osman Oglou, O., Purta, E., Kurkowska, M., Olchowik, A., et al. (2013). MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 41, D262–D267. doi: 10.1093/nar/gks1007
Marinoni, E. N., de Oliveira, J. S., Nicolet, Y., Raulfs, E. C., Amara, P., Dean, D. R., et al. (2012). (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51, 5439–5442. doi: 10.1002/anie.201201708
Maynard, N. D., Birch, E. W., Sanghvi, J. C., Chen, L., Gutschow, M. V., and Covert, M. W. (2010). A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. PLoS Genet. 6:e1001017. doi: 10.1371/journal.pgen.1001017
Maynard, N. D., Macklin, D. N., Kirkegaard, K., and Covert, M. W. (2012). Competing pathways control host resistance to virus via tRNA modification and programmed ribosomal frameshifting. Mol. Syst. Biol. 8:567. doi: 10.1038/msb.2011.101
Miranda, H. V., Antelmann, H., Hepowit, N., Chavarria, N. E., Krause, D. J., Pritz, J. R., et al. (2014). Archaeal Ubiquitin-like SAMP3 is Isopeptide-linked to Proteins via a UbaA-dependent Mechanism. Mol. Cell. Proteomics 13, 220–239. doi: 10.1074/mcp.M113.029652
Miranda, H. V., Nembhard, N., Su, D., Hepowit, N., Krause, D. J., Pritz, J. R., et al. (2011). E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 4417–4422. doi: 10.1073/pnas.1018151108
Mueller, E. G., and Palenchar, P. M. (1999). Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis. Protein Sci. 8, 2424–2427. doi: 10.1110/ps.8.11.2424
Mueller, E. G., Palenchar, P. M., and Buck, C. J. (2001). The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J. Biol. Chem. 276, 33588–33595. doi: 10.1074/jbc.M104067200
Murphy, F. V. IV, Ramakrishnan, V., Malkiewicz, A., and Agris, P. F. (2004). The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 11, 1186–1191. doi: 10.1038/nsmb861
Nakagawa, H., Kuratani, M., Goto-Ito, S., Ito, T., Katsura, K., Terada, T., et al. (2013). Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA. Proteins 81, 1232–1244. doi: 10.1002/prot.24273
Nakai, Y., Harada, A., Hashiguchi, Y., Nakai, M., and Hayashi, H. (2012). Arabidopsis molybdopterin biosynthesis protein Cnx5 collaborates with the ubiquitin-like protein Urm11 in the thio-modification of tRNA. J. Biol. Chem. 287, 30874–30884. doi: 10.1074/jbc.M112.350090
Nakai, Y., Nakai, M., and Hayashi, H. (2008). Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 283, 27469–27476. doi: 10.1074/jbc.M804043200
Nakai, Y., Nakai, M., Lill, R., Suzuki, T., and Hayashi, H. (2007). Thio modification of yeast cytosolic tRNA is an iron-sulfur protein-dependent pathway. Mol. Cell. Biol. 27, 2841–2847. doi: 10.1128/MCB.01321-06
Nakayashiki, T., Saito, N., Takeuchi, R., Kadokura, H., Nakahigashi, K., Wanner, B. L., et al. (2013). The tRNA thiolation pathway modulates the intracellular redox state in Escherichia coli. J. Bacteriol. 195, 2039–2049. doi: 10.1128/JB.02180-12
Noma, A., Sakaguchi, Y., and Suzuki, T. (2009). Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 37, 1335–1352. doi: 10.1093/nar/gkn1023
Numata, T., Fukai, S., Ikeuchi, Y., Suzuki, T., and Nureki, O. (2006a). Structural basis for sulfur relay to RNA mediated by heterohexameric TusBCD complex. Structure 14, 357–366. doi: 10.1016/j.str.2005.11.009
Numata, T., Ikeuchi, Y., Fukai, S., Suzuki, T., and Nureki, O. (2006b). Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 442, 419–424. doi: 10.1038/nature04896
Palenchar, P. M., Buck, C. J., Cheng, H., Larson, T. J., and Mueller, E. G. (2000). Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J. Biol. Chem. 275, 8283–8286. doi: 10.1074/jbc.275.12.8283
Pierrel, F., Bjork, G. R., Fontecave, M., and Atta, M. (2002). Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 277, 13367–13370. doi: 10.1074/jbc.C100609200
Pierrel, F., Douki, T., Fontecave, M., and Atta, M. (2004). MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 279, 47555–47563. doi: 10.1074/jbc.M408562200
Pierrel, F., Hernandez, H. L., Johnson, M. K., Fontecave, M., Atta, M., and Bjork, G. R. (2003). MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA-methylthiotransferase. J. Biol. Chem. 278, 29515–29524. doi: 10.1074/jbc.M301518200
Pitterle, D. M., and Rajagopalan, K. V. (1993). The biosynthesis of molybdopterin in Escherichia coli. Purification and characterization of the converting factor. J. Biol. Chem. 268, 13499–13505.
Rezgui, V. A., Tyagi, K., Ranjan, N., Konevega, A. L., Mittelstaet, J., Rodnina, M. V., et al. (2013). tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U.S.A. 110, 12289–12294. doi: 10.1073/pnas.1300781110
Rodriguez-Hernandez, A., Spears, J. L., Gaston, K. W., Limbach, P. A., Gamper, H., Hou, Y. M., et al. (2013). Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position. J. Mol. Biol. 425, 3888–3906. doi: 10.1016/j.jmb.2013.05.018
Ryals, J., Hsu, R. Y., Lipsett, M. N., and Bremer, H. (1982). Isolation of single-site Escherichia coli mutants deficient in thiamine and 4-thiouridine syntheses: identification of a nuvC mutant. J. Bacteriol. 151, 899–904.
Schindelin, H., Kisker, C., and Rajagopalan, K. V. (2001). Molybdopterin from molybdenum and tungsten enzymes. Adv. Protein Chem. 58, 47–94. doi: 10.1016/S0065-3233(01)58002-X
Schlieker, C. D., Van der Veen, A. G., Damon, J. R., Spooner, E., and Ploegh, H. L. (2008). A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 18255–18260. doi: 10.1073/pnas.0808756105
Schmitz, J., Chowdhury, M. M., Hanzelmann, P., Nimtz, M., Lee, E. Y., Schindelin, H., et al. (2008). The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry 47, 6479–6489. doi: 10.1021/bi800477u
Settembre, E., Begley, T. P., and Ealick, S. E. (2003). Structural biology of enzymes of the thiamin biosynthesis pathway. Curr. Opin. Struct. Biol. 13, 739–747. doi: 10.1016/j.sbi.2003.10.006
Shi, R., Proteau, A., Villarroya, M., Moukadiri, I., Zhang, L., Trempe, J. F., et al. (2010). Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8:e1000354. doi: 10.1371/journal.pbio.1000354
Shigi, N. (2012). Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J. Biol. Chem. 287, 17568–17577. doi: 10.1074/jbc.M112.359844
Shigi, N., Sakaguchi, Y., Asai, S., Suzuki, T., and Watanabe, K. (2008). Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 27, 3267–3278. doi: 10.1038/emboj.2008.246
Shigi, N., Sakaguchi, Y., Suzuki, T., and Watanabe, K. (2006a). Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 281, 14296–14306. doi: 10.1074/jbc.M511675200
Shigi, N., Suzuki, T., Terada, T., Shirouzu, M., Yokoyama, S., and Watanabe, K. (2006b). Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 281, 2104–2113. doi: 10.1074/jbc.M510771200
Shigi, N., Suzuki, T., Tamakoshi, M., Oshima, T., and Watanabe, K. (2002). Conserved bases in the TPsi C loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J. Biol. Chem. 277, 39128–39135. doi: 10.1074/jbc.M207323200
Suzuki, T. (2005). “Biosynthesis and function of tRNA wobble modifications,” in Fine-Tuning of RNA Functions by Modification and Editing, ed. H. Grosjean (Heidelberg: Springer), 23–69.
Sylvers, L. A., Rogers, K. C., Shimizu, M., Ohtsuka, E., and Soll, D. (1993). A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 32, 3836–3841. doi: 10.1021/bi00066a002
Taylor, S. V., Kelleher, N. L., Kinsland, C., Chiu, H. J., Costello, C. A., Backstrom, A. D., et al. (1998). Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273, 16555–16560. doi: 10.1074/jbc.273.26.16555
Umeda, N., Suzuki, T., Yukawa, M., Ohya, Y., Shindo, H., Watanabe, K., et al. (2005). Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 280, 1613–1624. doi: 10.1074/jbc.M409306200
Urbonavicius, J., Qian, Q., Durand, J. M., Hagervall, T. G., and Bjork, G. R. (2001). Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873. doi: 10.1093/emboj/20.17.4863
Van der Veen, A. G., Schorpp, K., Schlieker, C., Buti, L., Damon, J. R., Spooner, E., et al. (2011). Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. U.S.A. 108, 1763–1770. doi: 10.1073/pnas.1014402108
Vendeix, F. A., Murphy, F. V. IV, Cantara, W. A., Leszczynska, G., Gustilo, E. M., Sproat, B., et al. (2012). Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol. 416, 467–485. doi: 10.1016/j.jmb.2011.12.048
Watanabe, K., Oshima, T., Saneyoshi, M., and Nishimura, S. (1974). Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GT psi C in tRNA of an extreme thermophile. FEBS Lett. 43, 59–63. doi: 10.1016/0014-5793(74)81105-1
Watanabe, K., Shinma, M., Oshima, T., and Nishimura, S. (1976). Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72, 1137–1144. doi: 10.1016/S0006-291X(76)80250-1
Waterman, D. G., Ortiz-Lombardia, M., Fogg, M. J., Koonin, E. V., and Antson, A. A. (2006). Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 356, 97–110. doi: 10.1016/j.jmb.2005.11.013
Xu, J., Zhang, J., Wang, L., Zhou, J., Huang, H., Wu, J., et al. (2006). Solution structure of Urm1 and its implications for the origin of protein modifiers. Proc. Natl. Acad. Sci. U.S.A. 103, 11625–11630. doi: 10.1073/pnas.0604876103
Yasukawa, T., Suzuki, T., Ishii, N., Ohta, S., and Watanabe, K. (2001). Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 20, 4794–4802. doi: 10.1093/emboj/20.17.4794
Yasukawa, T., Suzuki, T., Ishii, N., Ueda, T., Ohta, S., and Watanabe, K. (2000). Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNA(Lys) with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 467, 175–178. doi: 10.1016/S0014-5793(00)01145-5
Yokoyama, S., Watanabe, T., Murao, K., Ishikura, H., Yamaizumi, Z., Nishimura, S., et al. (1985). Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. U.S.A. 82, 4905–4909. doi: 10.1073/pnas.82.15.4905
Zhang, W., Urban, A., Mihara, H., Leimkuhler, S., Kurihara, T., and Esaki, N. (2010). IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J. Biol. Chem. 285, 2302–2308. doi: 10.1074/jbc.M109.082172
Keywords: post-transcriptional modification, post-translational modification, sulfur, tRNA, ubiquitin
Citation: Shigi N (2014) Biosynthesis and functions of sulfur modifications in tRNA. Front. Genet. 5:67. doi: 10.3389/fgene.2014.00067
Received: 25 February 2014; Accepted: 17 March 2014;
Published online: 02 April 2014.
Edited by:
Akio Kanai, Keio University, JapanCopyright © 2014 Shigi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naoki Shigi, Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, 2-4-7 Aomi, Koto-ku, Tokyo 135-0064, Japan e-mail:bmFva2ktc2hpZ2lAYWlzdC5nby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.