- 1Laboratory of Gene Expression Regulation in Development, Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russia
- 2Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russia
Due to advances in genome-wide technologies, consistent distant interactions within chromosomes of higher eukaryotes have been revealed. In particular, it has been shown that enhancers can specifically and directly interact with promoters by looping out intervening sequences, which can be up to several hundred kilobases long. This review is focused on transcription factors that are supposed to be involved in long-range interactions. Available data are in agreement with the model that several known transcription factors and insulator proteins belong to an abundant but poorly studied class of proteins that are responsible for chromosomal architecture.
Introduction
In recent years, considerable progress has been made in understanding chromosome organization (for reviews, see Gibcus and Dekker, 2013; Krijger and de Laat, 2013; Nora et al., 2013; Tanay and Cavalli, 2013). High-resolution chromosome conformation capture techniques have provided evidence that chromosomes in the genomes of human, mouse, and Drosophila are partitioned into a series of discrete topologically associating domains (TADs; Lieberman-Aiden et al., 2009; Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). Their characteristic feature is that regulatory elements within a TAD display extensive long-range interactions with each other but interact far less frequently with regulatory elements located outside their domain. The size of TADs ranges from 10 to 500 kb, with a median of about 100 kb, in Drosophila (Sexton et al., 2012) and from slightly less than 100 kb to several megabases, with a median of 1 Mb, in humans and mice (Dixon et al., 2012; Nora et al., 2012). Within a TAD, numerous local chromatin loops are formed between enhancers, silencers, and promoters, with their length in some cases exceeding 100 kb (Li et al., 2012; Sanyal et al., 2012; Shen et al., 2012; Phillips-Cremins et al., 2013).
Two decades have elapsed since insulators were recognized as a specific class of DNA sequence elements that contribute to organization of independent gene function domains by restricting the enhancer and silencer functions (Ghirlando et al., 2012; Herold et al., 2012; Kirkland et al., 2013). However, although considerable progress has been made in the study of chromosomal architecture, we still do not have a clear mechanistic picture of how long-range interactions between distant regulatory regions are established and maintained through the cell cycle. In the past few years, a concept has been formed that there is a special class of architectural proteins, including some known insulator proteins, that are responsible for global chromosome architecture as well as for local regulation of enhancer–promoter interactions (Maksimenko et al., 2008; Holwerda and de Laat, 2012; Gibcus and Dekker, 2013; Nora et al., 2013; Kyrchanova and Georgiev, 2014). This paper is an attempt to summarize recent progress in understanding the role (function) of transcription factors and insulator proteins as architectural proteins.
Drosophila Transcription Factors That are Involved in Distant Enhancer–Promoter Interactions
Drosophila is a unique model system to study long-distance interactions between regulatory elements. Using transposon-mediated transformation or attP-phage-based integration and manipulation with recombination systems, it is possible to obtain different combinations of the regulatory elements in the same genomic position in order to study the role of particular regulatory elements in reporter expression (for reviews, see Bischof et al., 2007; Venken and Bellen, 2012). These approaches have made it possible to discover several “tethering” elements near promoters that ensure specific long-distance interactions between enhancers and corresponding promoters (Calhoun et al., 2002; Calhoun and Levine, 2003; Akbari et al., 2008; Melnikova et al., 2008). However, the proteins that bind to tethering elements and are responsible for their activity have not yet been identified.
To date, only two known transcription factors, Chip and Zeste, have been considered to be involved in supporting distant enhancer–promoter interactions inDrosophila (Figure 1A). The Chip protein can form dimmers and mediate interactions between different classes of transcription factors (for review, see Matthews and Visvader, 2003). Chip has two domains well-conserved among higher eukaryotes: an amino-terminal homodimerization domain (SID) and a carboxy-terminal LIM interaction domain (LID; Figure 1A). The LID domain interacts with LIM-homeodomain (LIM-HD) and LIM-only (LMO) proteins, which have important roles in cell fate determination, tissue development, and cytoskeletal organization. Recent data show that Chip is responsible for cooperative binding of LIM-HD and GATA proteins onto target promoters and enhancers (Heitzler et al., 2003; Bronstein et al., 2010). There is indirect genetic evidence that Chip supports long-range enhancer–promoter interactions in the cut locus (Morcillo et al., 1997) and achaete-scute complex (Ramain et al., 2000; Heitzler et al., 2003).
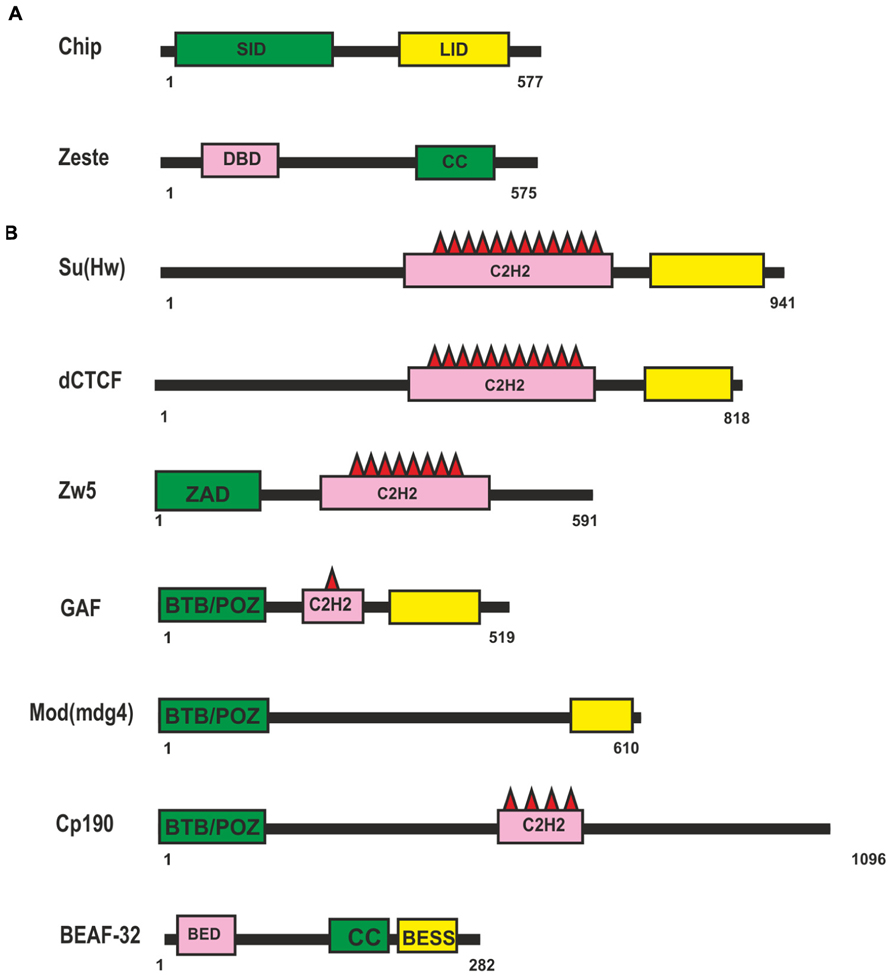
FIGURE 1. Drosophila proteins suggesting to be involved long-distance interactions. (A) Domain organization of the Chip and Zeste proteins. (B) Domain organization of the Drosophila insulator proteins. Self-interaction, protein-protein interaction and DNA-binding domains are shown as green, yellow, and pink boxes respectively. Abbreviations: self-interaction domain (SID), LIM-interaction domain (LID), DNA binding domain (DBD), coiled-coil domain (CC), Zinc-fingers of C2H2 type (C2H2), Zinc-finger Associated Domain (ZAD), BTB/POZ domain, BED-type (BEAF and DREF) zinc finger domain (BED), BEAF, Su(var)3-7, and Stonewall domain (BESS).
A putative role of another protein, Zeste (Figure 1A), in distant interactions has also been evidenced only in genetic studies with transgenic lines (Qian et al., 1992; Laney and Biggin, 1997; Kostyuchenko et al., 2009). Zeste is a sequence-specific DNA-binding protein that binds to the regulatory regions of many genes, including the white and Ubx genes, and stimulates their expression (Benson and Pirrotta, 1988; Chen and Pirrotta, 1993). A unique feature of Zeste is that it binds cooperatively to multiple binding sites as a higher-order homo-oligomer (Chen and Pirrotta, 1993). Zeste oligomerization is the result of interactions mediated by carboxy-terminal leucine zipper motifs. In particular, Zeste binds to the enhancer and promoter of the white gene (Qian et al., 1992). In transgenic lines, Zeste is strongly required for the distant interaction between the eye enhancer and the white promoter across the heterologous yellow gene (Kostyuchenko et al., 2009). At the same time, inactivation of Zeste has no effect on the activity of the eye enhancer when it is located relatively close to the white promoter. The deletion of Zeste binding sites in the upstream promoter region does not affect the basal level of white expression but eliminates Zeste-dependent long-range communication between the enhancer and the promoter. Thus, it appears that Zeste is not required for basal activity of the promoter but contributes to organization of specific enhancer–promoter interactions. However, there is no direct evidence that Zeste itself is sufficient for establishing enhancer–promoter interactions. Therefore, it may well be that additional, as yet unknown transcription factors cooperate with Zeste to support specific enhancer–promoter interactions stimulating white expression.
Drosophila Insulator Proteins are Likely Candidates for Architectural Transcription Factors
Most information about potential transcription factors involved in long-range interactions has been obtained in studies on Drosophila insulators. The Drosophila genome contains many sequences with an insulator function (Herold et al., 2012). The first insulators to be identified were scs and scs’ located at the boundaries of two heat shock 70 genes (Kellum and Schedl, 1991, 1992). Two proteins, Zw5 and BEAF (Figure 1B), bind to scs and scs’, respectively, and partially account for their insulator properties (Zhao et al., 1995; Gaszner et al., 1999). The best characterized insulator consisting of reiterated binding sites for the Su(Hw) protein (Figure 1B) was found in the regulatory region of the gypsy retrotransposon (Holdridge and Dorsett, 1991; Geyer and Corces, 1992). The Su(Hw) protein associates with thousands of genomic sites, with the vast majority of them carrying a single copy of the corresponding sequence (Golovnin et al., 2003; Parnell et al., 2003; Kuhn-Parnell et al., 2008; Soshnev et al., 2012, 2013).
Insulators named Mcp, Fab-6, Fab-7, and Fab-8 were identified at the boundaries of enhancer domains regulating proper activation of the Abd-B gene in the Bithorax complex (Gyurkovics et al., 1990; Barges et al., 2000; Hogga et al., 2001; Schweinsberg et al., 2004; Gruzdeva et al., 2005; Rodin et al., 2007; Iampietro et al., 2008, 2010; Aoki et al., 2012). Binding sites for a Drosophila homolog of vertebrate insulator protein CTCF (dCTCF; Figure 1B) were found in Mcp, Fab-6, and Fab-8 insulators (Moon et al., 2005; Holohan et al., 2007). Other transcription factors – GAF, ELBA, and BEAF-32 – were also found to frequently bind to known Drosophila insulators. In addition, several insulators were described for which DNA binding proteins have not yet been identified (Herold et al., 2012).
As shown in transgenic lines, pairing of two identical insulators can support distant activation of a promoter by an enhancer or yeast GAL4 activator (Cai and Shen, 2001; Muravyova et al., 2001; Kyrchanova et al., 2007; Kyrchanova et al., 2008a, b). The relative orientation of two identical insulators defines the mode of loop formation that either allows or blocks enhancer (GAL4)–promoter interaction (Kyrchanova et al., 2008a,b). This phenomenon is explained by the assumption that when the insulators are located in opposite orientations, the loop configuration is favorable for communication between regulatory elements located beyond the loop (Figure 2). The loop formed by two insulators located in the same orientation juxtaposes two elements located within and beyond the loop. Supposedly, this orientation-dependent interaction is accounted for by at least two insulator-bound proteins that are involved in specific protein–protein interactions.
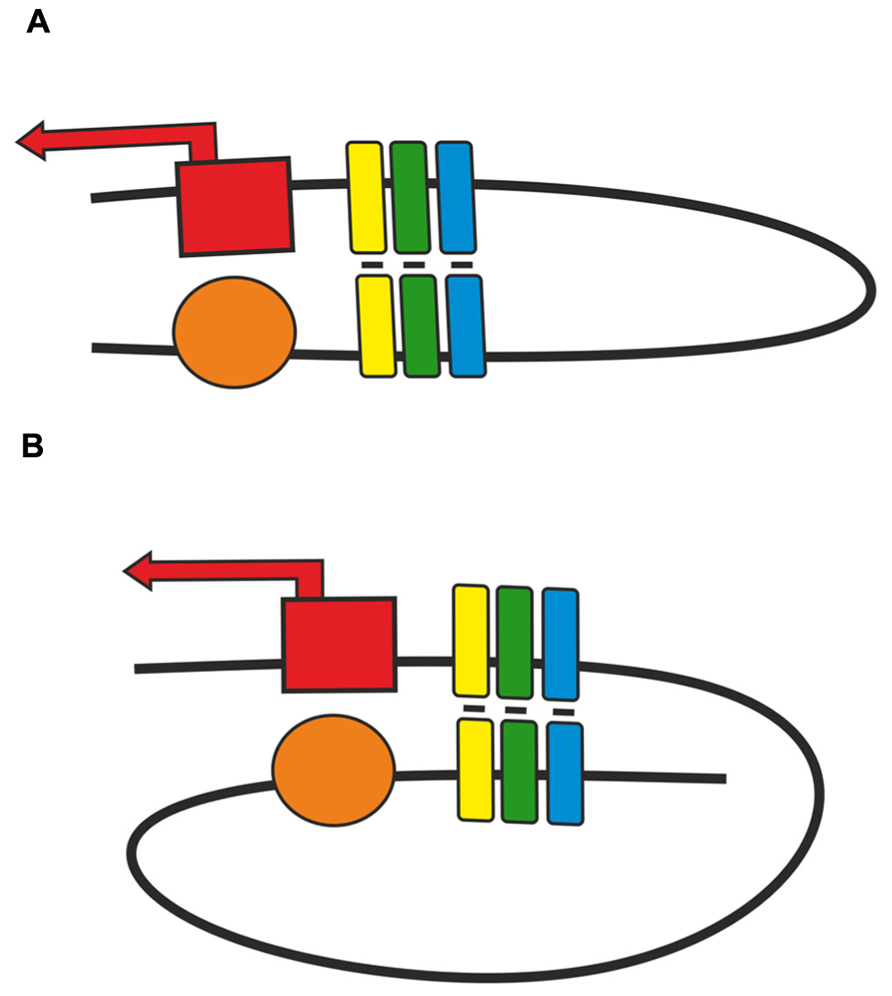
FIGURE 2. Two modes of pairing between two copies of an insulator inserted in either (A) opposite or (B) same orientation. Presumptive proteins responsible for insulator pairing are shown as a cluster of yellow, green, blue boxes. Red box with solid arrow indicates promoter region, orange oval – enhancer element.
It has also been found that two identical insulators can support interactions between regulatory elements located in transgenes inserted at distances up to several megabases from each other (Sigrist and Pirrotta, 1997; Muller et al., 1999; Kravchenko et al., 2005; Vazquez et al., 2006; Li et al., 2011, 2013). The most striking example is the insulator termed Homie that is located between the TER94 promoter and regulatory region of the eve gene (Fujioka et al., 2009). The presence of Homie in a transgene as far as 3.3 Mb away from the endogenous copy facilitates long-range communication between endogenous eve enhancers located near Homie and a promoter placed on the transgene (Fujioka et al., 2009, 2013). These facts suggest that proteins bound to insulators can support very specific distant interactions through the cell cycle. Thus, insulators are good candidates to form interactive boundaries partitioning Drosophila chromosomes into TADs. Indeed, insulator-bound proteins are frequently found at the presumed borders of TADs (Sexton et al., 2012).
To support specific long-range interactions, insulator proteins should have homodimerization domains. Three insulator proteins – Su(Hw), Zw5, and dCTCF (Figure 1B) – contain multiple C2H2 zinc fingers (Kim et al., 1996; Gaszner et al., 1999; Moon et al., 2005). To date, these proteins have not been examined for the presence of dimerization domains. Only Zw5 was found to contain a zinc finger-associated domain (ZAD) specific for insects at the N-terminus (Gaszner et al., 1999; Blanton et al., 2003). More than 90 ZAD-proteins were also identified in the Drosophila genome (Chung et al., 2002), but they have not yet been studied sufficiently. They are characterized by a conserved constellation of four cysteines within the ZAD, which form a zinc-coordinated fold. The crystal structure of the ZAD of Grauzone protein provides evidence that two ZAD molecules interact in a head-to-tail mode to form a dimer, which suggests that ZAD domains of other proteins are also able to self-associate (Jauch et al., 2003). Therefore, the ZAD domain of Zw5 may be involved in distant interactions, but this assumption requires experimental verification.
Moreover, it has been shown that Su(Hw) interacts with the CP190 protein and Mod(mdg4) isoform named Mod(mdg4)-67.2 (Büchner et al., 2000; Gause et al., 2001; Ghosh et al., 2001; Pai et al., 2004; Golovnin et al., 2007), and the dCTCF protein interacts with CP190 (Gerasimova et al., 2007; Mohan et al., 2007).
The GAF, Mod(mdg4)-67.2, and CP190 proteins have the BTB (bric-a-brac, tramtrack, and broad complex)/POZ (poxvirus and zinc finger) domain at the N-terminus. The BTB is a conserved protein–protein interaction motif contained in a variety of transcription factors involved in development, chromatin remodeling, insulator activity, and carcinogenesis (Stogios et al., 2005; Perez-Torrado et al., 2006). All well-studied mammalian BTB domains form obligate homodimers and, rarely, tetramers. The BTB domains of Drosophila GAF and Mod(mdg4)-67.2 factors belong to the “ttk group,” which contains several highly conserved sequences not found in other BTB domains, and exist as higher-order multimers (Zollman et al., 1994; Espinas et al., 1999; Mahmoudi et al., 2002; Bonchuk et al., 2011).
The role of BTB domains and especially of GAF and Mod(mdg4)-67.2 in organization of long-distance interactions either between insulators or between an enhancer and a promoter have been discussed for a long time. Electron microscopic and DNA pull-down experiments have shown that GAF complexes can form a protein link between separate DNA elements in vitro (Katsani et al., 1999; Mahmoudi et al., 2002). Similar results have also been obtained for the Bach1 BTB/POZ protein interaction domain required for the formation of looped DNA structures between different regulatory elements within the human β-globin LCR, as visualized by atomic force microscopy in vitro (Yoshida et al., 1999). As shown by functional in vivo assays, GAF can facilitate gene activation in a heterologous model system such as human 911 cells (Mahmoudi et al., 2002) and yeast (Petrascheck et al., 2005) by acting as an anchor that links the remote GAL4 binding sites to the promoters. However, binding sites for GAF do not support distant interaction between GAL4 activator and the white promoter in Drosophila transgenic lines (Bonchuk et al., 2011), while binding sites for Zw5, dCTCF, or Su(Hw) can support such interactions in the same model system (Kyrchanova et al., 2008a). Thus, there is no conclusive experimental evidence for the ability of GAF to support long-distance interactions in Drosophila. On the other hand, oligomerization of the BTB domains is required for cooperative binding of GAF to many adjacent sites in the same regulatory region (enhancer, insulator, or promoter; Katsani et al., 1999). As a result, GAF can open chromatin regions, thereby allowing the recruitment of other transcription factors to regulatory regions (Leibovitch et al., 2002). A similar role may be played by the self-association domain located at the C-terminus of BEAF protein (Hart et al., 1997; Gilbert et al., 2006).
Biochemical experiments have shown that each BEAF protein (Figure 1B) binds with its N-terminal BED finger domain to specific DNA motif CGATA, while BEAF trimers bind with high affinity to clusters of CGATA motifs (Hart et al., 1997). According to the results of genome-wide analysis, BEAF preferentially binds to such clusters in the promoter regions of active genes and is required for stimulation of their transcription (Emberly et al., 2008; Jiang et al., 2009).
A new insulator complex, named ELBA, recently described in Drosophila (Aoki et al., 2012) is composed of two proteins, Elba1 and Elba2, which share a conserved C-terminal “BEN domain” mediating binding to DNA. The third protein, Elba3, is responsible for “dimerization” of the Elba1-2 BEN domains and is encoded by the gene closely linked to Elba1. In this case, dimerization domain is required for cooperative binding of two BEN domains to corresponding insulator sites. Thus, dimerization domains in many insulator-bound proteins may be essential for effective binding of insulator proteins to chromatin but not for organization of long-distance interactions.
The Su(Hw), Mod(mdg4)-67.2, and CP190 proteins colocalize in discrete foci, named insulator bodies, in the Drosophila interphase cell nucleus (Gerasimova et al., 2000; Pai et al., 2004). Hence, it has been asserted (Gerasimova et al., 2000) that the insulator bodies arise via association of individual Su(Hw)-containing nucleoprotein complexes located at distant chromosomal sites. Hypothetically, a number of Su(Hw) insulators coalesce into an insulator body owing to interactions between the BTB domains of insulator proteins Mod(mdg4)-67.2 and CP190. However, recent results show that the insulator bodies are aggregates of insulator proteins that resemble well-known promyelocytic leukemia nuclear bodies (PML-NB) and stress bodies, which comprise many unrelated proteins (Golovnin et al., 2008, 2012; Schoborg et al., 2013). Thus, there is no direct evidence that Mod(mdg4) and CP190 are important for supporting interactions between insulators located at a large distance from each other. Since inactivation of either CP190 or Mod(mdg4)-67.2 leads to weaker Su(Hw) binding to chromosomes (Pai et al., 2004; Golovnin et al., 2007; Schwartz et al., 2012), it seems likely that the BTB-containing proteins are important for cooperative binding of insulator proteins to their sites and consequent formation of insulator complexes.
In conclusion, it should be noted that some of Drosophila insulator proteins are good candidates to be architectural proteins. However, the mechanisms of and possible protein domains involved in long-distance interactions have not yet been identified.
Cooperation of CTCF with Cohesin in Chromatin Architecture of Mammalian Genome
CTCF (CCCTC-binding factor) is regarded as the main insulator protein in mammals (Ohlsson et al., 2010; Chaumeil and Skok, 2012; Lee and Iyer, 2012; Merkenschlager and Odom, 2013). This protein is ubiquitously expressed across most mammalian tissues (Wendt et al., 2008) and is required for early mouse development (Fedoriw et al., 2004), participating in cell-cycle progression, apoptosis, and cell differentiation (Splinter et al., 2006; Heath et al., 2008; Soshnikova et al., 2010). Many independent experiments on genome-scale mapping of CTCF binding in cells of different mammalian tissues have revealed its preferential binding at the gene-dense regions but with little or no enrichment in promoters (Kim et al., 2007; Chen et al., 2008; Wang et al., 2012; Lee et al., 2012). This protein localizes at the DNase I-hypersensitive sites, open chromatin determinants that are generally common across cell types (Song et al., 2011). There is ample experimental evidence for the role of CTCF in organization of chromatin architecture in particular loci and formation of TADs (Chaumeil and Skok, 2012; Herold et al., 2012; Holwerda and de Laat, 2012; Merkenschlager and Odom, 2013).
Although CTCF is recognized as the main architectural protein, information on the involvement of its domains in long-distance interactions is scarce (Figure 3A). The central part of its molecule contains 11 C2H2 zinc fingers (ZFs), with ZFs 4–7 recognizing the core consensus DNA motif (Nakahashi et al., 2013). Nonconserved flanking DNA sequences are recognized by ZFs 1–2 and ZFs 8–11 clusters, which also stabilize CTCF. This protein is capable of self-association, but domains involved in this process have not been characterized sufficiently (Yusufzai and Felsenfeld, 2004). Pant et al. (2004) obtained evidence for a pairwise interaction between the C-terminal end of one CTCF molecule and the ZF domain of another in vitro. However, the ZF domain of CTCF can also interact with many different proteins, including CHD8, Sin3A, and YB-1 (Chernukhin et al., 2000; Lutz et al., 2000; Ishihara et al., 2006). Therefore, such a ZF-mediated mechanism is unlikely to account for specific long-distance interactions between CTCF binding sites, and further studies are needed to identify CTCF domains responsible for such interactions.
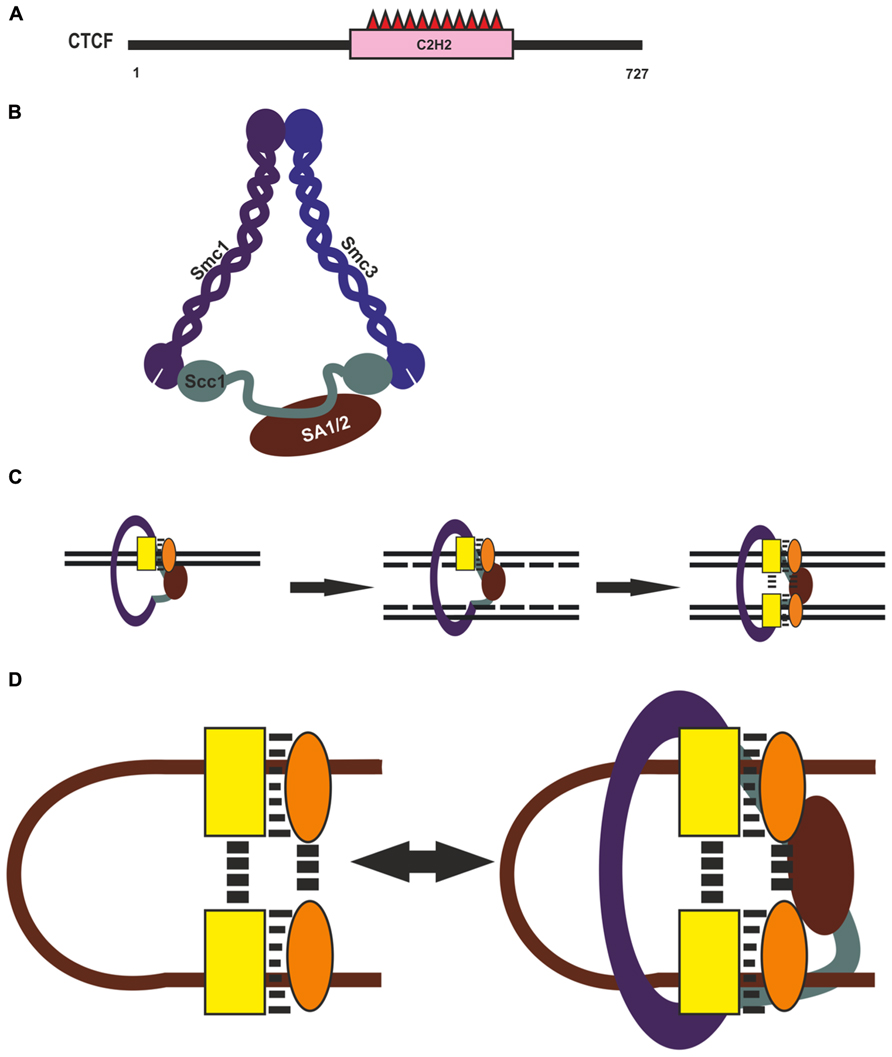
FIGURE 3. CTCF and cohesin complex in chromatin architecture. (A) Domain organization of CTCF. (B) Cohesin complex. Structural Maintenance of Chromosomes (Smc), Sister Chromatid Cohesion (Scc), Stromal Antigen (SA). (C) Possible role of Cohesin in reproducing of CTCF complex on newly synthesized DNA during replication. CTCF and assumptive interaction protein are shown by yellow rectangle and orange oval. (D) Diagram illustrating possible role of CTCF with its partners and Cohesin in organization and supporting of long-distance interactions.
An important role for CTCF-mediated distant interactions has been suggested for the cohesin complex (Holwerda and de Laat, 2012; Lee and Iyer, 2012; Mehta et al., 2013; Merkenschlager and Odom, 2013). This macromolecular complex, conserved from yeast to human, is responsible for the fidelity of chromosome segregation during mitosis and meiosis, keeping the sister chromatids together from S phase to anaphase (for review, see Peters, 2012; Dorsett and Merkenschlager, 2013; Remeseiro and Losada, 2013). The complex is composed of four protein components: two long coiled-coil molecules, Smc1 and Smc3, which form an open-ended heterodimer; Scc1 (Rad21), which bridges its open end; and SA1 (or SA2), which interacts with Scc1 and is external to the Smc1/Smc3/Scc1 trimer (Figure 3B). Thus, the three core subunits of cohesin – Smc1, Smc3, and Scc1 (Rad21) – form a ring-shaped structure, and the SA1 and SA2 proteins interact with it in a mutually exclusive manner. It has been proposed that such complexes mediate chromatid cohesion by trapping the two sister DNA molecules inside the cohesin ring and can have an effect on chromatin structure, forming or stabilizing intrachromatid loops (Figure 3C).
The results of genome-wide analysis of CTCF and cohesin binding show that their patterns largely overlap (Parelho et al., 2008; Wendt et al., 2008). CTCF and subunits of the cohesin complex coprecipitate in the nuclear lysate, and SA2 directly interacts with CTCF in vitro (Xiao et al., 2011), suggesting that SA2 may be responsible for cohesin recruitment to CTCF-binding sites. Another potential participant in the stabilization of CTCF–cohesin interaction is the DEAD-box RNA helicase p68, which functions in association with the steroid receptor RNA activator (SRA; Yao et al., 2010). As shown by genome-wide ChIP-Seq analysis, 22% of p68 peaks are associated with CTCF-binding sites, and 7% of CTCF sites bind p68. The p68/SRA complex interacts with both CTCF and cohesin, and depletion of p68 or SRA results in the loss of cohesin binding to CTCF. On this basis, the authors (Yao et al., 2010) suggest that RNA helicase stabilizes the cohesin–CTCF interaction.
In CTCF-depleted cells, only a small part of cohesin sites is lost, indicating that CTCF is only one of many factors recruiting the cohesin complex to chromatin (Wendt et al., 2008; Hadjur et al., 2009; Nativio et al., 2009). For example, it has been shown that transcription factor Klf4 interacts with the cohesin complex and recruits it to the Oct4 distal enhancer (Wei et al., 2013). A CTCF-independent role for cohesin in transcription regulation was also demonstrated by Schmidt et al. (2010), who revealed cohesin and estrogen receptor co-binding near upregulated genes upon estrogen treatment of MCF-7 cells. Of interest are recent data that cohesin and CTCF contribute differentially to the topological domain architecture (Zuin et al., 2013), which further support the model that many additional transcriptional factors in cooperation with cohesin are involved in organization of long-distance interactions.
Recently cohesin binding has been revealed at most of active regulatory regions (Schaaf et al., 2013; Yan et al., 2013), suggesting that cohesin can support but not organize specific long-distance interactions between CTCF binding sites (Figure 3D). Genome-wide studies have shown that only a minor part of CTCF binding sites are involved in loop formation, which is evidence that additional proteins may participate in this process (Handoko et al., 2011; Dixon et al., 2012; Sanyal et al., 2012). Many CTCF-binding partners that can aid in the diverse functions of CTCF have been reported to date (for reviews, see Herold et al., 2012; Lee and Iyer, 2012). For example, zinc-finger protein Prdm5 interacts with CTCF and colocalizes with it at many genomic sites (Galli et al., 2013). Taken together, these observations suggest that CTCF helps in recruiting additional transcriptional factors that, in turn, might be involved in mediating in specific selective long-distance interactions between CTCF binding sites (Figure 3D).
Cooperation of Mediator and Cohesin in Short-Range Enhancer–Promoter Interactions in Mammals
Cohesin copurifies and colocalizes with the Mediator complex, which binds to most of active promoters and enhancers in eukaryotes (Ebmeier and Taatjes, 2010; Kagey et al., 2010). Mediator is a highly conserved, large multisubunit complex comprising 25 subunits in yeast and 30 or more subunits in higher organisms (for reviews, see Malik and Roeder, 2010; Ansari and Morse, 2013). Several Mediator subunits have been shown to interact with various activators both in yeast and metazoans (Brzovic et al., 2011; Vojnic et al., 2011), with its specific subunits interacting with Pol II subunits and other general transcription factors bound to promoters (Takagi et al., 2006; Esnault et al., 2008; Cai et al., 2010). The classical model suggests that Mediator acts as an adaptor that conveys transcription signals from activators to the general transcription machinery to help initiate transcription by Pol II (Malik and Roeder, 2010; Ansari and Morse, 2013).
It has been shown that DNA looping takes place between enhancers and promoters occupied by the Mediator and cohesin complexes (Kagey et al., 2010; Seitan et al., 2011). Inactivation of cohesin or Mediator components leads to partial loss of enhancer–promoter interactions. On this basis, it has been suggested that Mediator and cohesin together bridge cell-type-specific enhancer–promoter interactions (Phillips-Cremins et al., 2013). The model proposed by these authors is also based on the ability of the large Mediator complex to simultaneously interact with enhancer-bound activators and general transcription factors bound to a promoter. Hence, Mediator may potentially bring together remote enhancers and promoters, while the cohesin complex stabilizes such interactions by forming a ring around an enhancer and a promoter sites. This model is attractive, but it has not yet been supported by direct experimental evidence. On the contrary, there is at least one example demonstrating that the loss of a cohesin-associated site at the one of Myc-mediated enhancers does not lead to the loss of another paired site on the interacting promoter (Yan et al., 2013).
Cohesin also extensively colocalizes with transcription activators (Yan et al., 2013) and Polycomb repressive complex 1 (Schaaf et al., 2013), facilitating the recruitment of these proteins to their sites. Moreover, cohesin is essential for protein complex formation on newly synthesized DNA during replication, since it is responsible for holding the nascent sister chromatids together at regulatory regions (Yan et al., 2013). Such a role of cohesin binding in promoting re-establishment of transcription factors on corresponding regulatory elements during the cell cycle suggest the ability for cohesin to help in recruiting CTCF and some unknown architectural proteins onto newly synthesized DNA during replication (Figure 3C). These proteins organize specific long-distance interactions, which, in turn, are also stabilized with participation of cohesin (Figure 3D). In accordance with this assumption, cohesin depletion in non-cycling mouse thymocytes proved to have no significant effect on preexisting architectural compartments but diminished interactions between some cohesin-bound sites (Seitan et al., 2013).
Insulator–Promoter Interactions in Vertebrates and Drosophila
As shown in several recent studies, CTCF binding sites frequently interact with active promoters, and CTCF may be involved in organization of enhancer–promoter interactions (Handoko et al., 2011; Sanyal et al., 2012). The molecular mechanism of the CTCF–promoter interaction might be explained by the recent finding that CTCF interacts with TAF3, a component of the basal TFIID transcriptional machinery (Liu et al., 2011). In Drosophila, the enhancer-blocking activity of several promoters and insulators depends on general transcription factors that inhibit RNAP II elongation (Chopra et al., 2009). It has been speculated that insulators interact with components of the RNAP II complex at stalled promoters and that the resulting chromatin loops can prevent the inappropriate activation of stalled genes by enhancers associated with the neighboring locus. Drosophila insulators located on the 3′-side of genes interact with promoters, and these interactions are in some cases necessary for the basal activity of the promoters (Erokhin et al., 2011; Kyrchanova et al., 2013). In addition to the possible role of a gene loop in the enhancement of RNAP II recycling and mRNA export, insulators may serve to bring to the promoter the remodeling and histone modification complexes that improve the binding and stabilization of the TFIID complex. In transgenic lines, insulators proved to interact with different promoters, suggesting that insulator proteins can interact with components of general transcription complex assembled on promoters. Insulator protein GAF interacts with TAF3, as does human CTCF (Chopra et al., 2008), which indicates that TAF3 may be a key protein in the TFIID complex that is responsible for nonspecific interaction between insulators and promoters.
Transcription Factors Involved in Erythropoiesis as Possible Organizers of Enhancer–Promoter Interactions
Current knowledge of vertebrate proteins maintaining chromatin loops between enhancers and promoters has come mainly from studies on genes involved in erythropoiesis, the process dependent on lineage-specific transcription factors GATA1, GATA2, Tal1, E2A, FOG, and Klf1 (for review, see Cantor and Orkin, 2002; Anantharaman et al., 2011; Palstra and Grosveld, 2012).
The β-globin locus was the first gene cluster at which long-range (about 40 kb) chromosomal interactions between a distal enhancer, the locus control region (LCR), and the target β-globin promoters during erythropoiesis were described (Carter et al., 2002; Tolhuis et al., 2002). Transcription factor GATA1 was shown to be essential for the induction of most, if not all, erythroid genes (Welch et al., 2004; Fujiwara et al., 2009). The GATA1 protein contains a highly conserved Cys4-type dual zinc finger module (Figure 4), with the zinc fingers located closer to the N- and C-termini being named NF and CF, respectively. The CF is responsible (and sufficient) for high-affinity GATA1 binding to the cognate DNA site (WGATAR); NF is also involved in stabilizing GATA1 binding to DNA, but its main function is to interact with different transcriptional cofactors such as FOG (Tsang et al., 1997; Fox et al., 1999), LMO2 (Osada et al., 1997), SP1 (Merika and Orkin, 1995; Gregory et al., 1996; Imanishi et al., 2010), Klf1 (Merika and Orkin, 1995; Gregory et al., 1996), and many others.
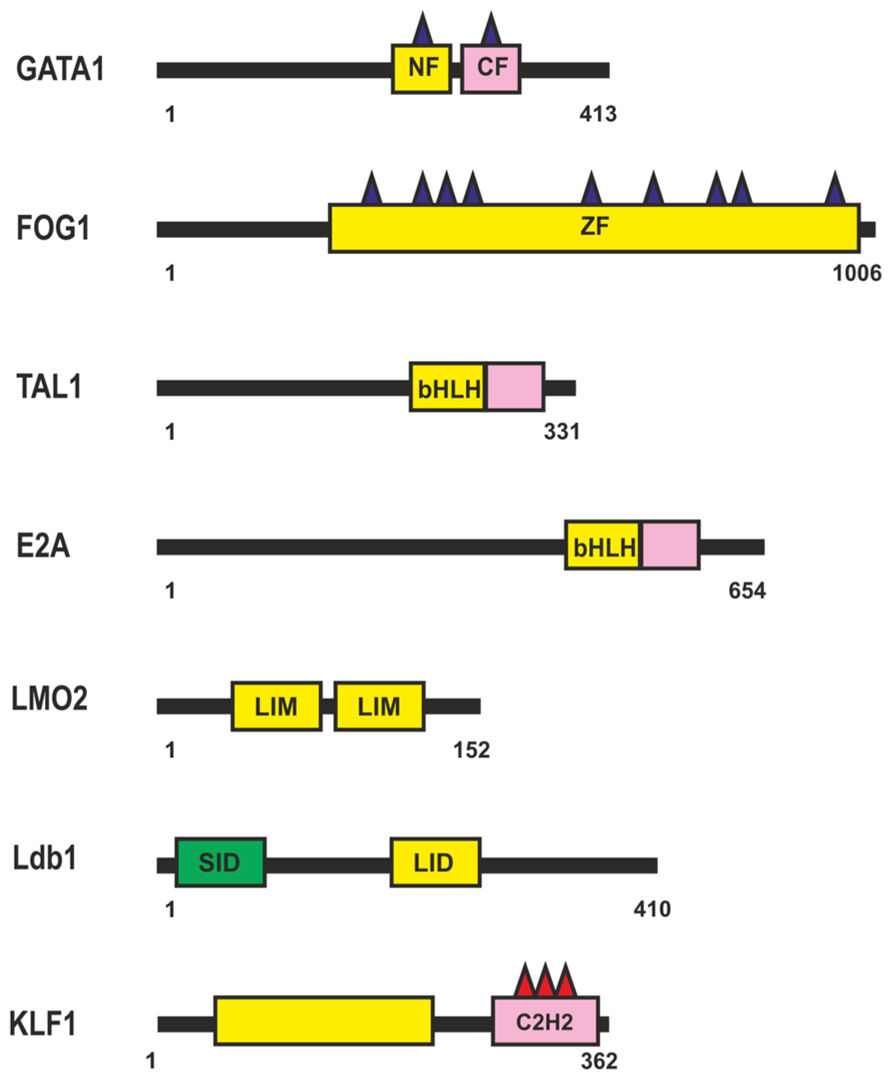
FIGURE 4. Transcription factors involved in regulation of the β-globin locus during erythropoiesis. N-termini C4-type zinc finger domain (NF), C-termini C4-type zinc finger domain (CF), CCHC-type Zinc-Finger domain (ZF), Basic Helix-Loop-Helix domain (bHLH), Lin11, Isl-1, and Mec-3 domain (LIM). For other designations, see Figure 1.
Most GATA1-regulated events require its binding to FOG1 (Figure 4), a coregulator protein containing nine zinc fingers, five of them with the CCHC arrangement of zinc-chelating residues. Four of the FOG1 protein zinc fingers bind GATA1 with a similar modest affinity in vitro, each contributing to the ability of FOG1 to regulate the transcriptional activity of GATA1 (Fox et al., 1999). Thus, a single FOG1 molecule can potentially interact with several GATA1 molecules bound at separate sites. However, previous studies indicate that FOG1 with a single intact GATA1-binding zinc finger is sufficient for erythroid differentiation (Cantor and Orkin, 2002). Therefore, simultaneous binding of many GATA1 molecules appears to be an excess function of FOG1. As a consequence of their interaction, FOG1 and GATA1 mutually facilitate each other’s binding to chromatin and, in particular, to the β-globin gene promoter (Mancini et al., 2012).
TAL1 and E2A (Figure 4) are members of the basic helix-loop-helix (bHLH) family of transcription factors (for review, see Anantharaman et al., 2011). TAL1 heterodimerizes with E2A and binds to canonical DNA sequences, CANNTG, termed E-boxes, each monomer recognizing one-half of the E-box (Massari and Murre, 2000). Many other HLH proteins can also interact with E-box elements in erythroid cell-specific genes, with the specificity of these interactions being in particular determined by nearby bound transcription factors. Tal1 is among the earliest expressed transcription factors important for the specification of hematopoietic cells. Tal1 exists as part of different activator and repressor complexes and is responsible for the activity of many proteins activated during erythropoiesis. Genome-wide analysis of protein-DNA interactions has shown that Tal1 can be recruited to DNA either directly via E-box or in a DNA-binding-independent manner, through interaction with other transcription factors (Kassouf et al., 2010). Dissection of the TAL1-E2A interface shows weak interaction with DNA, suggesting that the complex can bind regulatory regions in cooperation with additional DNA-bound transcription factors (El Omari et al., 2013).
The TAL1:E2A heterodimer interacts with the LMO2 protein and its partner, LDB1 (LIM domain -binding protein 1; Lécuyer and Hoang, 2004). LMO2 (Figure 4) is a versatile adaptor protein that, through interaction with additional regulators, plays a critical role in recruiting complexes to DNA. LMO2 comprises two LIM domains that act as protein-interaction motifs (Wadman et al., 1997). A single LMO2 molecule bridges the DNA-binding proteins GATA1 and TAL1/E2A, thereby creating a stable complex on DNA (Wadman et al., 1997; Wilkinson-White et al., 2011; El Omari et al., 2013). The DNA contacts are made by TAL1/E2A heterodimers and the C-terminal zinc-finger of GATA1. The GATA1 NF binds the C-terminal half of the LIM2 domain of LMO2, leaving LIM1 and the N-terminal half of LMO2 available for contact with TAL1/E2A (Wilkinson-White et al., 2011). The Ldb1 protein (Figure 4) can interact with multiple transcription factors and mediate interactions between them (Matthews and Visvader, 2003). This protein contains the N-terminal self-association domain that forms trimers (Cross et al., 2010), and its C-terminal domain is involved in the interaction with LMO2. The multiprotein complexes containing GATA1, TAL1, E2A, LMO2, and LDB1 proteins (named Ldb1 complexes) bind to a conserved paired motif composed of a consensus E-box and a GATA motif (Figure 5A) with restricted orientation and spacing, CANNTG-N8-10-GATA (Cheng et al., 2009; Soler et al., 2010; Li et al., 2013).
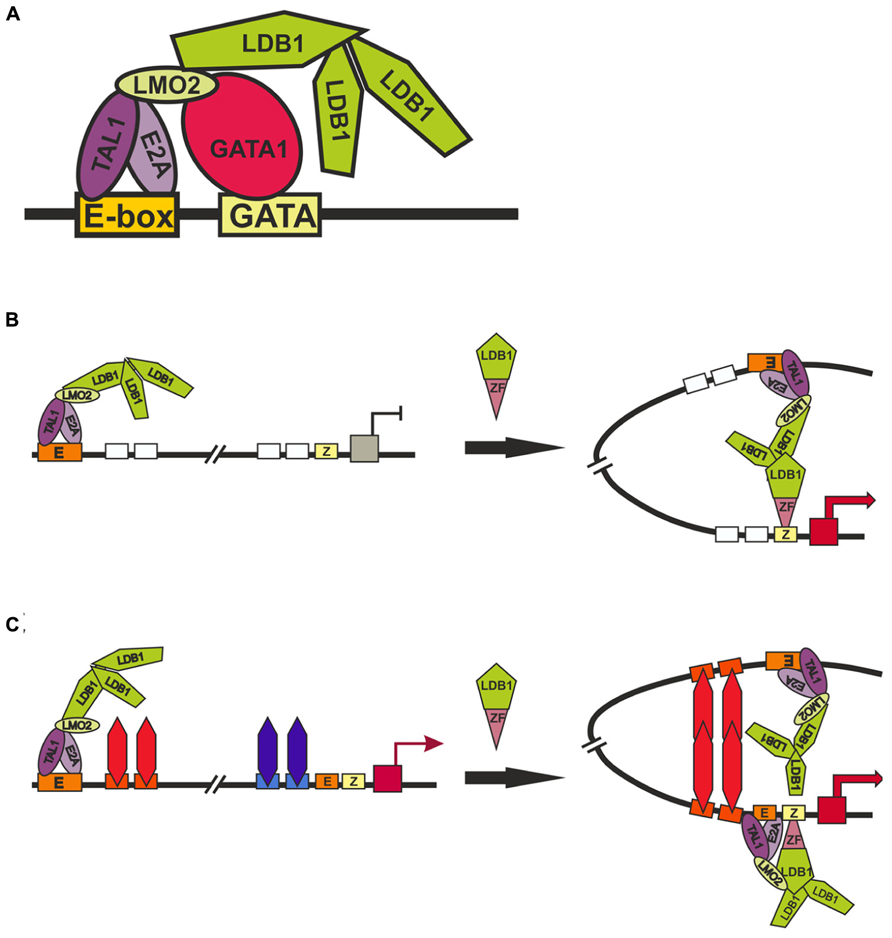
FIGURE 5. Models of long-distance interaction between LCT and the β-globin promoter. (A) Scheme for the formation of the Ldb1 complex at the E-box and GATA-sites. (B) and (C) two alternative models describing role of Ldb1 in organization of distance interactions. Designations: E-box (E), GATA1-binding site (GATA), zinc-finger binding site (Z).
Genome-wide analysis has revealed a high percentage of overlapping binding sites for KLF1 (Figure 4) and the Ldb1 complex near TSS or within the first intron at putative erythroid lineage–specific promoters (Tallack et al., 2012; Li et al., 2013). It is supposed that Klf1 and the Ldb1 complex function cooperatively to regulate transcription of shared target genes during erythropoiesis. In particular the major globin promoter and LCR contain a number of EKLF-binding sites (Perkins, 1999; Bieker, 2001). KLF1 recognizes the CACCC-box motif, which is found in erythroid-specific gene promoters and is required for their activation (Yien and Bieker, 2013). KLF1 contains three similar C2H2 zinc fingers at the C-terminus that comprise its DNA-binding domain. KLF1 interacts with components of the basal transcription machinery, such as the p62 subunit of TFIIH (Mas et al., 2011), and with TAF9 (Sengupta et al., 2009). These interactions are necessary for stabilization of transcription machinery on promoters, the β-globin promoter in particular (Sengupta et al., 2009; Mas et al., 2011). GATA1 can physically interact with KLF1 and exhibits functional synergy with KLF1 at erythroid promoters (Merika and Orkin, 1995; Gregory et al., 1996). KLF1 also interacts with chromatin-modifying and remodeling factors, such as P/CAF, CBP/p300, SWI/SNF complex, and possibly BAF47/BAF155 (Yien and Bieker, 2013). Erythroid cells that lack KLF1 exhibit an aberrant chromatin configuration and altered components at KLF1-dependent target promoters, the β-globin promoter in particular, resulting in histone hypoacetylation, loss of DNase I hypersensitivity, and the absence of CBP, BRG1, TBP, and RNA polymerase II (Pol II; Bottardi et al., 2006). Thus, KLF1 is essential for the formation of erythroid-specific active promoters.
Inactivation of GATA1 and its cofactors – FOG1 (Vakoc et al., 2005), KLF1 (Drissen et al., 2004), and Ldb1 (Song et al., 2007) – proved to strongly reduce the expression of β-globin gene and impair interactions between the LCR and promoter. These results were interpreted as evidence for the involvement of these factors in long-distance interactions between the LCR and the promoter. One of the main problems in interpreting the results of experiments on RNAi-mediated inactivation of tested genes is that these transcription factors are of general importance for stimulating transcription of the genes during erythropoiesis and, in addition, are subject to cross-stimulation (Tallack et al., 2010; Mancini et al., 2012; Li et al., 2013). Thus, the inactivation of any of the factors may lead to changes in the expression of other known and unknown factors that are involved in the distant enhancer–promoter interactions. To overcome such a problem, an elegant model system has been developed that employs artificial zinc fingers to tether Ldb1 to the β-globin promoter in GATA1-null erythroblasts (G1E cells), in which the β-globin locus is inactive (Deng et al., 2012). Since G1E cells lack GATA1, the β-globin promoter is devoid of Ldb1, whereas the LCR retains its activity and ability to bind Ldb1. Experiments with this model has shown that the targeting of Ldb1 or its self-association domain alone to the β-globin promoter substantially activates transcription in the absence of GATA1 and that promoter tethering of Ldb1 provides for the formation of a 40-kb chromatin loop between the LCR and promoter and for transcription activation. According to the authors, their findings support the model that the self-association domain of Ldb1 is an essential rate-limiting effector of GATA1 during chromatin loop formation between the LCR and promoter (Figure 5B). However, since the self-association domain of Ldb1 is relatively weak (Cross et al., 2010), it is difficult to imagine how the binding of one Ldb1-ZF chimeric protein to a single site in the promoter region of the β-globin gene can provide for the establishment of specific interaction with the LCR located at a distance of 40 kb. Indeed, the authors themselves have shown that nonspecific dimerization domains of GAF, lexA, and p65NFkB proteins recruited to the LCR and promoter fail to support this distant interaction (Deng et al., 2012).
Alternatively (Figure 5C), the chimeric Ldb1-ZF protein can possibly substitute for GATA1 by facilitating the recruitment of E2A/TAL1/LMO2 complex to the promoter. This explains why the recruitment of the LMO2-interacting domain of Ldb1 alone proved to be sufficient for partially restoring the expression of β-globin gene and the distant interaction of the promoter with the LCR (Deng et al., 2012). In this case, we assume the existence of some unknown architectural proteins that bind to the LCR and promoter region and support distant interaction between them only when the promoter is active. This model explains the role played in chromatin loop formation by Brg1, the ATPase component of the SWI/SNF nucleosome remodeling complex (Kim et al., 2009), the general transcription factor TFII-I (Ren et al., 2011), and transcription factors such as KLF1, FOG-1, and GATA1 (Drissen et al., 2004; Vakoc et al., 2005, Song et al., 2007). All these factors are required for the formation of active β-globin promoter, with consequent activation of putative architectural proteins that form the chromatin loop with the LCR. In addition to organizing specific distant interactions, these architectural proteins must remain on the regulatory elements during mitosis. In contrast to most other DNA-binding factors, GATA1 remains bound to the subset of its target genes during mitosis (Kadauke et al., 2012). All examined GATA1 cofactors (FOG1, TAL1, Ldb1, and LMO2) vacate mitotic chromatin regardless of whether GATA1 is retained, which indicates that they do not influence GATA1 binding to mitotic chromatin. However, inactivation of GATA1 only partially affects DNaseI hypersensitivity (HS), suggesting that additional unknown factors are involved in the formation of nucleosome-free regions. HS propagation through mitosis is also mediated by a GATA1-independent mechanism. These findings may be regarded as evidence for the existence of not yet identified architectural proteins that form a mitotically stable platform for the binding of GATA1 and reassembly of coregulator complexes at the appropriate genomic locations.
There is ample evidence for possible involvement of several other proteins in organization of long-range interactions, including the transcription factor SP1 that contains C2H2-type zinc finger DNA-binding domain and glutamine-rich dimerization domain (Courey et al., 1989; Mastrangelo et al., 1991; Su et al., 1991), the transcription factor Klf4 (Wei et al., 2013) that interact with many transcription regulators, including Oct4 and Sox2 (Wei et al., 2009), general activator p300/CBP, and repressors such as HDAC and CtBP (Swamynathan, 2010), MAR-binding protein SATB1 (Cai et al., 2006; Gong et al., 2011), TFIIIC (Kirkland et al., 2013), and condensins (D’Ambrosio et al., 2008). In any case however, it would be premature to arrive at any definitive conclusions about the role of these proteins in the chromosome architecture.
Conclusion and Prospects for the Future
Chromatin looping between different types of regulatory elements (promoters, enhancers, silencers, and insulators) is widely observed and appears to be a general mechanism for establishing long-range functional interactions in the genomes of higher eukaryotes. In contrast, distant interactions between regulatory elements in yeast are relatively rare. For example, GAL4 activator can stimulate yeast promoters only when its binding sites are located in relatively close proximity to the promoter, at a distance of no more than a few hundred base pairs (Guarente and Hoar, 1984; Struhl, 1984). Thus, we can postulate that higher eukaryotes possess a special class of architectural proteins responsible for distance interactions, which are absent in the yeast genome. It is important to note that the cohesin and Mediator complexes are highly conserved among all eukaryotes (Ansari and Morse, 2013). In yeast, the Mediator complex is recruited to GAL4 activator sites (Reeves and Hahn, 2005; Ansari and Morse, 2013). Moreover, cohesin is likely to influence transcription in Saccharomyces cerevisiae via interaction with the Mediator complex (Cena et al., 2013). If cohesin and Mediator can support interactions over distances of many kilobases in mammals, it is difficult to explain why homologous proteins in yeast fail to stimulate promoter from the GAL4 activator bound at a distance of only 400–500 bp.
Insulator proteins such as CTCF, Su(Hw), and Zw5 have no homologs in the yeast genome, which makes them probable candidates for organizing distant interactions. To consistently support such interactions in chromosomes, putative architectural proteins should be able to remain bound to chromosomes during mitosis, the process that imposes dramatic and dynamic changes on nuclear organization (Kadauke and Blobel, 2013). In contrast to most transcription factors, the Su(Hw) and dCTCF proteins in Drosophila and CTCF in mammals have predominantly constitutive binding sites in different cell lines and tissues (Chen et al., 2008; Song et al., 2011; Lee et al., 2012; Schwartz et al., 2012; Soshnev et al., 2012, 2013; Wang et al., 2012), suggesting that these transcription factors bind to chromosomes during the cell cycle. Contradictory results concerning the potential binding of CTCF to mitotic chromosomes (Burke et al., 2005; Komura et al., 2007; Wendt et al., 2008) may be explained by difficulties in selecting suitable antibodies for which the recognizable epitope is not occluded due to chromatin compaction during mitosis and mitosis-specific post-translational modifications (Kadauke and Blobel, 2012). Additional experimental approaches are required to elucidate the ability of the insulator proteins to bind to their sites through the cell cycle. Another still unresolved question is how architectural (insulator) proteins can organize specific interactions between distantly located sites. It appears that, to this end, they should have special homodimerization domains. If so, the role of cohesin is limited to supporting already established distant interactions.
The important but as yet unresolved question is as to how long-distance interactions are regulated. Possible roles of non-coding RNAs and different protein modifications in stimulation/repression of such interactions are discussed in several recent publications (Herold et al., 2012; Lee and Iyer, 2012; Li et al., 2013; Merkenschlager and Odom, 2013).
Hopefully, further studies will provide a deeper insight into the mechanisms of specific long-distance interactions, their regulation, and the principles of organization of chromosomal architecture in higher eukaryotes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to N. A. Gorgolyuk for his help in preparing the manuscript. This study was supported by RFBR 12-04-92423-EMBL-a and no. SS-2591.2012.4 (to Pavel Georgiev), by RFBR 12-04-33171-mol-a-ved and by the Presidential Stipend no. SP-1960.2012.4 (to Oksana Maksimenko), Russian Federation Ministry of Education and Science (14.B25.31.0022). The literature regarding transcription factors and insulator proteins is overwhelming, and we apologize to the authors whose studies we have failed to cite.
References
Akbari, O. S., Bae, E., Johnsen, H., Villaluz, A., Wong, D., and Drewell, R. A. (2008). A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development 135, 123–131. doi: 10.1242/dev.010744
Anantharaman, A., Lin, I. J., Barrow, J., Liang, S. Y., Masannat, J., Strouboulis, J., et al. (2011). Role of helix-loop-helix proteins during differentiation of erythroid cells. Mol. Cell. Biol. 31, 1332–1343. doi: 10.1128/MCB.01186-10
Ansari, S. A., and Morse, R. H. (2013). Mechanisms of mediator complex action in transcriptional activation. Cell. Mol. Life Sci. 70, 2743–2756. doi: 10.1007/s00018-013-1265-9
Aoki, T., Sarkeshik, A., Yates, J., and Schedl, P. (2012). Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife 1, e00171. doi: 10.7554/eLife.00171
Barges, S., Mihaly, J., Galloni, M., Hagstrom, K., Muller, M., Shanower, G., et al. (2000). The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127, 779–790.
Benson, M., and Pirrotta, V. (1988). The Drosophila zeste protein binds cooperatively to sites in many gene regulatory regions: implications for transvection and gene regulation. EMBO J. 7, 3907–3915.
Bieker, J. J. (2001). Krüppel-like factors: three fingers in many pies. J. Biol. Chem. 276, 34355–34358. doi: 10.1074/jbc.R100043200
Bischof, J., Maeda, R. K., Hediger, M., Karch, F., and Basler, K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3127. doi: 10.1073/pnas.0611511104
Blanton, J., Gaszner, M., and Schedl, P. (2003). Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17, 664–675. doi: 10.1101/gad.1052003
Bonchuk, A., Denisov, S., Georgiev, P., and Maksimenko, O. (2011). Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J. Mol. Biol. 412, 423–436. doi: 10.1016/j.jmb.2011.07.052
Bottardi, S., Ross, J., Pierre-Charles, N., Blank, V., and Milot, E. (2006). Lineage specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 25, 3586–3595. doi: 10.1038/sj.emboj.7601232
Bronstein, R., Levkovitz, L., Yosef, N., Yanku, M., Ruppin, E., Sharan, R., et al. (2010). Transcriptional regulation by CHIP/LDB complexes. PLoS Genet. 6:e1001063. doi: 10.1371/journal.pgen.1001063
Brzovic, P. S., Heikaus, C. C., Kisselev, L., Vernon, R., Herbig, E., Pacheco, D., et al. (2011). The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953. doi: 10.1016/j.molcel.2011.11.008
Büchner, K., Roth, P., Schotta, G., Krauss, V., Saumweber, H., Reuter, G., et al. (2000). Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155, 141–157.
Burke, L. J., Zhang, R., Bartkuhn, M., Tiwari, V. K., Tavoosidana, G., Kurukuti, S., et al. (2005). CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 24, 3291–3300. doi: 10.1038/sj.emboj.7600793
Cai, H. N., and Shen, P. (2001). Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291, 493–495. doi: 10.1126/science.291.5503.493
Cai, G., Imasaki, T., Yamada, K., Cardelli, F., Takagi, Y., and Asturias, F. J. (2010). Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 17, 273–279. doi: 10.1038/nsmb.1757
Cai, S., Lee, C. C., and Kohwi-Shigematsu, T. (2006). SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288. doi: 10.1038/ng1913
Calhoun, V. C., Stathopoulos, A., and Levine, M. (2002). Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. U.S.A. 99, 9243–9247. doi: 10.1073/pnas.142291299
Calhoun, V. C., and Levine, M. (2003). Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. U.S.A. 100, 9878–9883. doi: 10.1073/pnas.1233791100
Cantor, A. B., and Orkin, S. H. (2002). Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21, 3368–3376. doi: 10.1038/sj.onc.1205326
Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F., and Fraser, P. (2002). Long rangechromatin regulatory interactions in vivo. Nat. Genet. 32, 623–626. doi: 10.1038/ng1051
Cena, A., Skoneczny, M., Chełstowska, A., Kowalec, P., Natorff, R., and Kurlandzka, A. (2013). Cohesin Irr1/Scc3 is likely to influence transcription in Saccharomyces cerevisiae via interaction with Mediator complex. Acta Biochim. Pol. 60, 233–238.
Chaumeil, J., and Skok, J. A. (2012). The role of CTCF in regulating V(D)J recombination. Curr. Opin. Immunol. 24, 153–159. doi: 10.1016/j.coi.2012.01.003
Chen, J. D., and Pirrotta, V. (1993). Stepwise assembly of hyperaggregated forms of Drosophila zeste mutant protein suppresses white gene expression in vivo. EMBO J. 12, 2061–2073.
Chen, X., Xu, H., Yuan, P., Fang, F., Huss, M., Vega, V. B., et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 doi: 10.1016/j.cell.2008.04.043
Cheng, Y., Wu, W., Kumar, S. A., Yu, D., Deng, W., Tripic, T., et al. (2009). Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 19, 2172–2184. doi: 10.1101/gr.098921.109
Chernukhin, I. V., Shamsuddin, S., Robinson, A. F., Carne, A. F., Paul, A., El-Kady, A. I., et al. (2000). Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 275, 29915–29921. doi: 10.1074/jbc.M001538200
Chopra, V. S., Cande, J., Hong, J. W., and Levine, M. (2009). Stalled Hox promoters as chromosomal boundaries. Genes Dev. 23, 1505–1509. doi: 10.1101/gad.1807309
Chopra, V. S., Srinivasan, A., Kumar, R. P., Mishra, K., Basquin, D., and Docquier, M. (2008). Transcriptional activation by GAGA factor is through its direct interaction with dmTAF3. Dev. Biol. 317, 660–670. doi: 10.1016/j.ydbio.2008.02.008
Chung, H. R., Schäfer, U., Jäckle, H., and Böhm, S. (2002). Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep 3, 1158–1162. doi: 10.1093/embo-reports/kvf243
Courey, A. J., Holtzman, D. A., Jackson, S. P., and Tjian, R. (1989). Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59, 827–836. doi: 10.1016/0092-8674(89)90606-5
Cross, A. J., Jeffries, C. M., Trewhella, J., and Matthews, J. M. (2010). LIM domain binding proteins 1 and 2 have different oligomeric states. J. Mol. Biol. 399, 133–144. doi: 10.1016/j.jmb.2010.04.006
D’Ambrosio, C., Schmidt, C. K., Katou, Y., Kelly, G., Itoh, T., Shirahige, K., et al. (2008). Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22, 2215–2227. doi: 10.1101/gad.1675708
Deng, W., Lee, J., Wang, H., Miller, J., Reik, A., Gregory, P. D., et al. (2012). Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244. doi: 10.1016/j.cell.2012.03.051
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Dorsett, D., and Merkenschlager, M. (2013). Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell. Biol. 25, 327–333. doi: 10.1016/j.ceb.2013.02.003
Drissen, R., Palstra, R. J., Gillemans, N., Splinter, E., Grosveld, F., Philipsen, S., et al. (2004). The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18, 2485–2490. doi: 10.1101/gad.317004
Ebmeier, C. C., and Taatjes, D. J. (2010). Activator–Mediator binding regulates Mediator–cofactor interactions. Proc. Natl. Acad. Sci. U.S.A. 107, 11283–11288. doi: 10.1073/pnas.0914215107
El Omari, K., Hoosdally, S. J., Tuladhar, K., Karia, D., Hall-Ponselé, E., Platonova, O., et al. (2013). Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 4, 135–147. doi: 10.1016/j.celrep.2013.06.008
Emberly, E., Blattes, R., Schuettengruber, B., Hennion, M., Jiang, N., and Hart, C. M. (2008). BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 6:2896–2910. doi: 10.1371/journal.pbio.0060327
Erokhin, M., Davydova, A., Kyrchanova, O., Parshikov, A., Georgiev, P., and Chetverina, D. (2011). Insulators form gene loops by interacting with promoters in Drosophila. Development 138, 4097–4106. doi: 10.1242/dev.062836
Esnault, C., Ghavi-Helm, Y., Brun, S., Soutourina, J., Van Berkum, N., Boschiero, C., et al. (2008). Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31, 337–346. doi: 10.1016/j.molcel.2008.06.021
Espinas, M. L., Jimenez-Garcia, E., Vaquero, A., Canudas, S., Bernues, J., and Azorin, F. (1999). The N terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 274, 16461–16469. doi: 10.1074/jbc.274.23.16461
Fedoriw, A. M., Stein, P., Svoboda, P., Schultz, R. M., and Bartolomei, M. S. (2004). Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303, 238–240. doi: 10.1126/science.1090934
Fox, A. H., Liew, C., Holmes, M., Kowalski, K., MacKay, J., and Crossley, M. (1999). Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18, 2812–2822. doi: 10.1093/emboj/18.10.2812
Fujioka, M., Wu, X., and Jaynes, J. B. (2009). A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development 136, 3077–3087. doi: 10.1242/dev.036467
Fujioka, M., Sun, G., and Jaynes, J. B. (2013). The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 9:e1003883. doi: 10.1371/journal.pgen.1003883
Fujiwara, T., O’Geen, H., Keles, S., Blahnik, K., Linnemann, A. K., Kang, Y. A., et al. (2009). Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681. doi: 10.1016/j.molcel.2009.11.001
Galli, G. G., Carrara, M., Francavilla, C., Honnens de Lichtenberg K., Olsen, J. V., Calogero, R. A., et al. (2013). Genomic and proteomic analyses of Prdm5 reveal interactions with insulator binding proteins in embryonic stem cells. Mol. Cell. Biol. 33, 4504–4516. doi: 10.1128/MCB.00545-13
Gaszner, M., Vazquez, J., and Schedl, P. (1999). The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13, 2098–2107. doi: 10.1101/gad.13.16.2098
Gause, M., Morcillo, P., and Dorsett, D. (2001). Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21, 4807–4817. doi: 10.1128/MCB.21.14.4807-4817.2001
Gerasimova, T. I., Byrd, K., and Corces, V. G. (2000). A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6, 1025–1035. doi: 10.1016/S1097-2765(00)00101-5
Gerasimova, T. I., Lei, E. P., Bushey, A. M., and Corces, V. G. (2007). Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell 28, 761–772. doi: 10.1016/j.molcel.2007.09.024
Geyer, P. K., and Corces, V. G. (1992). DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6, 1865–1873. doi: 10.1101/gad.6.10.1865
Ghirlando, R., Giles, K., Gowher, H., Xiao, T., Xu, Z., Yao, H., et al. (2012). Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta 1819, 644–651. doi: 10.1016/j.bbagrm.2012.01.016
Ghosh, D., Gerasimova, T. I., and Corces, V. G. (2001). Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20, 2518–2527. doi: 10.1093/emboj/20.10.2518
Gibcus, J. H., and Dekker, J. (2013). The hierarchy of the 3D genome. Mol. Cell 49, 773–782. doi: 10.1016/j.molcel.2013.02.011
Gilbert, M. K., Tan, Y. Y., and Hart, C. M. (2006). The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173, 1365–1375. doi: 10.1534/genetics.106.056002
Golovnin, A., Biryukova, I., Romanova, O., Silicheva, M., Parshikov, A., Savitskaya, E., et al. (2003). An endogenous Su(Hw) insulator separates the yellow gene from the achaete-scute gene complex in Drosophila. Development 130, 3249–3258. doi: 10.1242/dev.00543
Golovnin, A., Mazur, A., Kopantseva, M., Kurshakova, M., Gulak, P. V., Gilmore, B., et al. (2007). Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila melanogaster. Mol. Cell. Biol. 27, 963–974. doi: 10.1128/MCB.00795-06
Golovnin, A., Melnikova, L., Volkov, I., Kostuchenko, M., Galkin, A. V., and Georgiev, P. (2008). ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 9, 440–445. doi: 10.1038/embor.2008.32
Golovnin, A., Volkov, I., and Georgiev, P. (2012). SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J. Cell Sci. 125, 2064–2074. doi: 10.1242/jcs.100172
Gong, F., Sun, L., Wang, Z., Shi, J., Li, W., Wang, S., et al. (2011). The BCL2 gene is regulated by a special AT-rich sequence binding protein 1-mediated long range chromosomal interaction between the promoter and the distal element located within the 30-UTR. Nucleic Acids Res. 39, 4640–4652. doi: 10.1093/nar/gkr023
Gregory, R. C., Taxman, D. J., Seshasayee, D., Kensinger, M. H., Bieker, J. J., and Wojchowski, D. M. (1996). Functional interaction of GATA1 with erythroid Krüppel-like factor and SP1 at defined erythroid promoters. Blood 87, 1793–1801.
Gruzdeva, N., Kyrchanova, O., Parshikov, A., Kullyev, A., and Georgiev, P. (2005). The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer–promoter communication. Mol. Cell. Biol. 25, 3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005
Guarente, L., and E. Hoar. (1984). Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box.” Proc. Natl. Acad. Sci. U.S.A. 81, 7860–7864. doi: 10.1073/pnas.81.24.7860
Gyurkovics, H., Gausz, J., Kummer, J., and Karch, F. (1990). A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9, 2579–2585.
Hadjur, S., Williams, L. M., Ryan, N. K., Cobb, B. S., Sexton, T., Fraser, P., et al. (2009). Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413. doi: 10.1038/nature08079
Handoko, L., Xu, H., Li, G., Ngan, C. Y., Chew, E., Schnapp, M., et al. (2011). CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638. doi: 10.1038/ng.857
Hart, C. M., Zhao, K., and Laemmli, U. K. (1997). The scs’ boundary element: characterization of boundary element associated factors. Mol. Cell. Biol. 17, 999–1009.
Heath, H., Ribeiro de Almeida, C., Sleutels, F., Dingjan, G., van de Nobelen, S., Jonkers, I., et al. (2008). CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 27, 2839–2850. doi: 10.1038/emboj.2008.214
Heitzler, P., Vanolst, L., Biryukova, I., and Ramain, P. (2003). Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes Dev. 17, 591–596. doi: 10.1101/gad.255703
Herold, M., Bartkuhn, M., and Renkawitz, R. (2012). CTCF: insights into insulator function during development. Development 139, 1045–1057. doi: 10.1242/dev.065268
Hogga, I., Mihaly J., Barges, S., and Karch, F. (2001). Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell 8, 1145–1151. doi: 10.1016/S1097-2765(01)00377-X
Holdridge, C., and Dorsett, D. (1991). Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11, 1894–1900.
Holohan, E. E., Kwong, C., Adryan, B., Bartkuhn, M., Herold, M., Renkawitz, R., et al. (2007). CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3:e112. doi: 10.1371/journal.pgen.0030112
Holwerda, S., and de Laat, W. (2012). Chromatin loops, gene positioning, and gene expression. Front. Genet. 3:217. doi: 10.3389/fgene.2012.00217
Iampietro, C., Cléard, F., Gyurkovics, H., Maeda, R. K., and Karch, F. (2008). Boundary swapping in the Drosophila Bithorax complex. Development 135, 3983–3987. doi: 10.1242/dev.025700
Iampietro, C., Gummalla, M., Mutero, A., Karch, F., and Maeda, R. K. (2010). Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 6:e1001260. doi: 10.1371/journal.pgen.1001260
Imanishi, M., Imamura, C., Higashi, C., Yan, W., Negi, S., Futaki, S., et al. (2010). Zinc finger-zinc finger interaction between the transcription factors, GATA-1 and Sp1. Biochem. Biophys. Res. Commun. 400, 625–630. doi: 10.1016/j.bbrc.2010.08.116
Ishihara, K., Oshimura, M., and Nakao, M. (2006). CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 23, 733–742. doi: 10.1016/j.molcel.2006.08.008
Jauch, R., Bourenkov, G. P., Chung, H. R., Urlaub, H., Reidt, U., and Jäckle, H. (2003). The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure 11, 1393–1402. doi: 10.1016/j.str.2003.09.015
Jiang, N., Emberly, E., Cuvier, O., and Hart, C. M. (2009). Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 29, 3556–3568. doi: 10.1128/MCB.01748-08
Kadauke, S., and Blobel, G. A. (2012). “Remembering” tissue-specific transcription patterns through mitosis. Cell Cycle 11, 3911–3912. doi: 10.4161/cc.22237
Kadauke, S., and Blobel, G. A. (2013). Mitotic bookmarking by transcription factors. Epigenet. Chromat. 6, 6. doi: 10.1186/1756-8935-6-6
Kadauke, S., Udugama, M. I., Pawlicki, J. M., Achtman, J. C., Jain, D. P., Cheng, Y., et al. (2012). Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150, 725–737. doi: 10.1016/j.cell.2012.06.038
Kagey, M. H., Newman, J. J., Bilodeau, S., Zhan, Y., Orlando, D. A., van Berkum, N. L., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. doi: 10.1038/nature09380
Kassouf, M. T., Hughes, J. R., Taylor, S., McGowan, S. J., Soneji, S., Green, A. L., et al. (2010). Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 20, 1064–1083. doi: 10.1101/gr.104935.110
Katsani, K. R., Hajibagheri, M. A., and Verrijzer, C. P. (1999). Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18, 698–708. doi: 10.1093/emboj/18.3.698
Kellum, R., and Schedl, P. (1991). A position-effect assay for boundaries of higher order chromosomal domains. Cell 64, 941–950. doi: 10.1016/0092-8674(91)90318-S
Kellum, R., and Schedl, P. (1992). A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12, 2424–2431.
Kim, J., Shen, B., Rosen, C., and Dorsett, D. (1996). The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol. Cell. Biol. 16, 3381–3392.
Kim, S. I., Bresnick, E. H., and Bultman, S. J. (2009). BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 37, 6019–6027. doi: 10.1093/nar/gkp677
Kim, T. H., Abdullaev, Z. K., Smith, A. D., Ching, K. A., Loukinov, D. I., Green, R. D., et al. (2007). Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245. doi: 10.1016/j.cell.2006.12.048
Kirkland, J. G., Raab, J. R., and Kamakaka, R. T. (2013). TFIIIC bound DNA elements in nuclear organization and insulation. Biochim. Biophys. Acta 1829, 418–424. doi: 10.1016/j.bbagrm.2012.09.006
Komura, J., Ikehata, H., and Ono, T. (2007). Chromatin fine structure of the c-MYC insulator element/DNase I-hypersensitive site I is not preserved during mitosis. Proc. Natl. Acad. Sci. U.S.A. 104, 15741–1576. doi: 10.1073/pnas.0702363104
Kostyuchenko, M., Savitskaya, E., Koryagina, E., Melnikova, L., Karakozova, M., and Georgiev P. (2009). Zeste can facilitate long-range enhancer-promoter communication and insulator bypass in Drosophila melanogaster. Chromosoma 118, 665–674. doi: 10.1007/s00412-009-0226-4
Kravchenko, E., Savitskaya, E., Kravchuk, O., Parshikov, A., Georgiev, P., and Savitsky, M. (2005). Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 25, 9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005
Krijger, P. H., and de Laat, W. (2013). Identical cells with different 3D genomes; cause and consequences? Curr. Opin. Genet. Dev. 23, 191–196. doi: 10.1016/j.gde.2012.12.010
Kuhn-Parnell, E. J., Helou, C., Marion, D. J., Gilmore, B. L., Parnell, T. J., Wold, M. S., et al. (2008). Investigation of the properties of non-gypsy suppressor of hairy-wing-binding sites. Genetics 179, 1263–1273. doi: 10.1534/genetics.108.087254
Kyrchanova, O., Toshchakov, S., Parshikov, A., and Georgiev, P. (2007). Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer–promoter communication. Mol. Cell. Biol. 27, 3035–3043. doi: 10.1128/MCB.02203-06
Kyrchanova, O., Chetverina, D., Maksimenko, O., Kullyev, A., and Georgiev, P. (2008a). Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 36, 7019–7028. doi: 10.1093/nar/gkn781
Kyrchanova, O., Toshchakov, S., Podstreshnaya, Y., Parshikov, A., and Georgiev, P. (2008b). Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 28, 4188–4195. doi: 10.1128/MCB.00229-08
Kyrchanova, O., and Georgiev, P. (2014). Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 588, 8–14. doi: 10.1016/j.febslet.2013.10.039
Kyrchanova, O., Maksimenko, O., Stakhov, V., Ivlieva, T., Parshikov, A., Studitsky, V. M., et al. (2013). Effective blocking of the white enhancer requires cooperation between two main mechanisms suggested for the insulator function. PLoS Genet. 9:e1003606. doi: 10.1371/journal.pgen.1003606
Laney, J. D., and Biggin, M. D. (1997). Zeste-mediated activation by an enhancer is independent of cooperative DNA binding in vivo. Proc. Natl. Acad. Sci. U.S.A. 94, 3602–3604. doi: 10.1073/pnas.94.8.3602
Lécuyer, E., and Hoang, T. (2004). SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32, 11–24. doi: 10.1016/j.exphem.2003.10.010
Lee, B. K., Bhinge, A. A., Battenhouse, A., McDaniell, R. M., Liu, Z., Song, L., et al. (2012). Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 22, 9–24. doi: 10.1101/gr.127597.111
Lee, B. K., and Iyer, V. R. (2012). Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 287, 30906–30913. doi: 10.1074/jbc.R111.324962
Leibovitch, B. A., Lu, Q., Benjamin, L. R., Liu, Y., Gilmour, D. S., and Elgin, S. C. (2002). GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol. 22, 6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002
Li, H. B., Müller, M., Bahechar, I. A., Kyrchanova, O., Ohno, K., Georgiev, P., et al. (2011). Insulators, not polycomb response elements, are required for long-range interactions between polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 31, 616–625. doi: 10.1128/MCB.00849-10
Li, H. B., Ohno, K., Gui, H., and Pirrotta, V. (2013). Insulators target active genes to transcription factories and polycomb-repressed genes to polycomb bodies. PLoS Genet 9:e1003436. doi: 10.1371/journal.pgen.1003436
Li, G., Ruan, X., Auerbach, R. K., Sandhu, K. S., Zheng, M., Wang, P., et al. (2012). Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98. doi: 10.1016/j.cell.2011.12.014
Li, L., Freudenberg, J., Cui, K., Dale, R., Song, S. H., Dean, A., et al. (2013). Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood 121, 4575–4585. doi: 10.1182/blood-2013-01-479451
Li, Y., Huang, W., Niu, L., Umbach, D. M., Covo, S., and Li, L. (2013). Characterization of constitutive CTCF/cohesin loci: a possible role in establishing topological domains in mammalian genomes. BMC Genomics 14:553. doi: 10.1186/1471-2164-14-553
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–393. doi: 10.1126/science.1181369
Liu, Z., Scannell, D. R., Eisen, M. B., and Tjian, R. (2011). Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146, 720–731. doi: 10.1016/j.cell.2011.08.005
Lutz, M., Burke, L. J., Barreto, G., Goeman, F., Greb, H., Arnold, R., et al. (2000). Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 28, 1707–1713. doi: 10.1093/nar/28.8.1707
Mahmoudi, T., Katsani, K. R., and Verrijzer, C. P. (2002). GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 21, 1775–1781. doi: 10.1093/emboj/21.7.1775
Maksimenko, O., Golovnin, A., and Georgiev, P. (2008). Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus. Mol. Cell. Biol. 28, 5469–5477. doi: 10.1128/MCB.00461-08
Malik, S., and Roeder, R. G. (2010).The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772. doi: 10.1038/nrg2901
Mancini, E., Sanjuan-Pla, A., Luciani, L., Moore, S., Grover, A., Zay, A., et al. (2012). FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J. 31, 351–365. doi: 10.1038/emboj.2011.390
Mas, C., Lussier-Price, M., Soni, S., Morse, T., Arseneault, G., Di Lello, P., et al. (2011). Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF). Proc. Natl. Acad. Sci. U.S.A. 108, 10484–10489. doi: 10.1073/pnas.1017029108
Massari, M. E., and Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20, 429–440. doi: 10.1128/MCB.20.2.429-440.2000
Mastrangelo, I. A., Courey, A. J., Wall, J. S., Jackson, S. P., and Hough, P. V. (1991). DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc. Natl. Acad. Sci. U.S.A. 88, 5670–5674. doi: 10.1073/pnas.88.13.5670
Matthews, J. M., and Visvader, J. E. (2003). LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep 4, 1132–1137. doi: 10.1038/sj.embor.7400030
Mehta, G. D., Kumar, R., Srivastava, S., and Ghosh, S. K. (2013). Cohesin: functions beyond sister chromatid cohesion. FEBS Lett. 587, 2299–2312. doi: 10.1016/j.febslet.2013.06.035
Melnikova, L., Kostuchenko, M., Silicheva, M., and Georgiev, P. (2008). Drosophila gypsy insulator and yellow enhancers regulate activity of yellow promoter through the same regulatory element. Chromosoma 117, 137–145. doi: 10.1007/s00412-007-0132-6
Merika, M., and Orkin, S. H. (1995). Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15, 2437–2447.
Merkenschlager, M., and Odom, D. T. (2013). CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152, 1285–1297. doi: 10.1016/j.cell.2013.02.029
Mohan, M., Bartkuhn, M., Herold, M., Philippen, A., Heinl, N., Bardenhagen, I., et al. (2007). The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 26, 4203–4214. doi: 10.1038/sj.emboj.7601851
Moon, H., Filippova, G., Loukinov, D., Pugacheva, E., Chen, Q., Smith, S. T., et al. (2005). CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6, 165–170. doi: 10.1038/sj.embor.7400334
Morcillo, P., Rosen, C., Baylies, M. K., and Dorsett, D. (1997). Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 11, 2729–2740. doi: 10.1101/gad.11.20.2729
Muller, M., Hagstrom, K., Gyurkovics, H., Pirrotta, V., and Schedl P. (1999). The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153, 1333–1356.
Muravyova, E., Golovnin, A., Gracheva, E., Parshikov, A., Belenkaya, T., Pirrotta, V., et al. (2001). Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291, 495–498. doi: 10.1126/science.291.5503.495
Nakahashi, H., Kwon, K. R., Resch, W., Vian, L., Dose, M., Stavreva, D., et al. (2013). A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 3, 1678–1689. doi: 10.1016/j.celrep.2013.04.024
Nativio, R., Wendt, K. S., Ito, Y., Huddleston, J. E., Uribe-Lewis, S., Woodfine, K., et al. (2009). Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5:e1000739. doi: 10.1371/journal.pgen.1000739
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Nora, E. P., Dekker, J., and Heard, E. (2013). Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays 35, 818–828. doi: 10.1002/bies.201300040
Ohlsson, R., Lobanenkov, V., and Klenova, E. (2010). Does CTCF mediate between nuclear organization and gene expression? Bioessays 32, 37–50. doi: 10.1002/bies.200900118
Osada, H., Grutz, G. G., Axelson, H., Forster, A., and Rabbitts, T. H. (1997). LIM-only protein Lmo2 forms a protein complex with erythroid transcription factor GATA-1. Leukemia 3, 307–312.
Pai, C. Y., Lei, E. P., Ghosh, D., and Corces, V. G. (2004). The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16, 737–748. doi: 10.1016/j.molcel.2004.11.004
Palstra, R. J., and Grosveld, F. (2012). Transcription factor binding at enhancers: shaping a genomic regulatory landscape in flux. Front. Genet. 3:195. doi: 10.3389/fgene.2012.00195
Pant, V., Kurukuti, S., Pugacheva, E., Shamsuddin, S., Mariano, P., Renkawitz, R., et al. (2004). Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24, 3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004
Parelho, V., Hadjur, S., Spivakov, M., Leleu, M., Sauer, S., Gregson, H. C., et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433. doi: 10.1016/j.cell.2008.01.011
Parnell, T. J., Viering, M. M., Skjesol, A., Helou, C., Kuhn, E. J., and Geyer, P. K. (2003). An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 100, 13436–13441. doi: 10.1073/pnas.2333111100
Perez-Torrado, R., Yamada, D., and Defossez, P. A. (2006). Born to bind: the BTB protein-protein interaction domain. Bioessays 28, 1194–1202. doi: 10.1002/bies.20500
Perkins, A. (1999). Erythroid Kruppel like factor: from fishing expedition to gourmet meal. Int. J. Biochem. Cell. Biol. 31, 1175–1192. doi: 10.1016/S1357-2725(99)00083-7
Peters, J. M. (2012). The many functions of cohesion – different rings to rule them all? EMBO J. 31, 2061–2063. doi: 10.1038/emboj.2012.90
Petrascheck, M., Escher, D., Mahmoudi, T., Verrijzer, C. P., Schaffner, W., and Barberis, A. (2005). DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 33, 3743–3750. doi: 10.1093/nar/gki689
Phillips-Cremins, J. E., Sauria, M. E., Sanyal, A., Gerasimova, T. I., Lajoie, B. R., Bell, J. S., et al. (2013). Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295. doi: 10.1016/j.cell.2013.04.053
Qian, S., Varjavand, B., and Pirrotta, V. (1992). Molecular analysis of the zeste white interaction reveals a promoter-proximal element essential for distant enhancer–promoter communication. Genetics 131, 79–90.
Ramain, P., Khechumian, R., Khechumian, K., and Arbogast, N., Ackermann, C., and Heitzler, P. (2000). Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol. Cell 6, 781–790. doi: 10.1016/S1097-2765(05)00079-1
Reeves, W. M., and Hahn, S. (2005). Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 25, 9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005
Remeseiro, S., and Losada, A. (2013). Cohesin, a chromatin engagement ring. Curr. Opin. Cell. Biol. 25, 63–71. doi: 10.1016/j.ceb.2012.10.013
Ren, X., Siegel, R., Kim, U., and Roeder, R. G. (2011). Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol. Cell 42, 342–355. doi: 10.1016/j.molcel.2011.04.011
Rodin, S., Kyrchanova, O., Pomerantseva, E., Parshikov, A., and Georgiev, P. (2007). New properties of Drosophila Fab-7 insulator. Genetics 177, 113–121. doi: 10.1534/genetics.107.075887
Sanyal, A., Lajoie, B. R., Jain, G., and Dekker, J. (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–113. doi: 10.1038/nature11279
Schaaf, C. A., Misulovin, Z., Gause, M., Koenig, A., Gohara, D. W., Watson, A., et al. (2013). Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 9:e1003560. doi: 10.1371/journal.pgen.1003560
Schoborg, T., Rickels, R., Barrios, J., and Labrador, M. (2013). Chromatin insulator bodies are nuclear structures that form in response to osmotic stress and cell death. J. Cell. Biol. 202, 261–276. doi: 10.1083/jcb.201304181
Schmidt, D., Schwalie, P. C., Ross-Innes, C. S., Hurtado, A., Brown, G. D., Carroll, J. S., et al. (2010). A CTCF- independent role for cohesion in tissue-specific transcription. Genome Res. 20, 578–588. doi: 10.1101/gr.100479.109
Schwartz, Y. B., Linder-Basso, D., Kharchenko, P. V., Tolstorukov, M. Y., Kim, M., Li, H. B., et al. (2012). Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22, 2188–2198. doi: 10.1101/gr.138156.112
Schweinsberg, S., Hagstrom, K., Gohl, D., Schedl, P., Kumar, R. P., Mishra, R., et al. (2004). The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168, 1371–1384. doi: 10.1534/genetics.104.029561
Seitan, V. C., Hao, B., Tachibana-Konwalski, K., Lavagnolli, T., Mira-Bontenbal, H., Brown, K. E., et al. (2011). A role for cohesin in T cell receptor rearrangement and thymocyte differentiation. Nature 476, 467–471. doi: 10.1038/nature10312
Seitan, V., Faure, A., Zhan, Y., McCord, R., Lajoie, B., Ing-Simmons, E., et al. (2013). Cohesin-based chromatin interactions enable regulated gene expression within pre-existing architectural compartments. Genome Res. 23, 2066–2077. doi: 10.1101/gr.161620.113
Sengupta, T., Cohet, N., Morle, F., and Bieker, J. J. (2009). Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 106, 4213–4218. doi: 10.1073/pnas.0808347106
Sexton, T., Yaffe, E., Kenigsberg, E., Bantignies, F., Leblanc, B., Hoichman, M., et al. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472. doi: 10.1016/j.cell.2012.01.010
Shen, Y., Yue, F., McCleary, D. F., Ye, Z., Edsall, L., Kuan, S., et al. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120. doi: 10.1038/nature11243
Sigrist, C. J. A., and Pirrotta, V. (1997). Chromatin insulator elements block the silencing of a target gene by the Drosophila Polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147, 209–221.
Soler, E., Andrieu-Soler, C., DeBoer, E., Bryne, J. C., Thongjuea, S., Stadhouders, R., et al. (2010). The genome- wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 24, 277–289. doi: 10.1101/gad.551810
Song, S. H., Hou, C., and Dean, A. (2007). A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell 28, 810–822. doi: 10.1016/j.molcel.2007.09.025
Song, L., Zhang, Z., Grasfeder, L. L., Boyle, A. P., Giresi, P. G., Lee, B. K., et al. (2011). Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21, 1757–1767. doi: 10.1101/gr.121541.111
Soshnev, A. A., He, B., Baxley, R. M., Jiang, N., Hart, C. M., Tan, K., et al. (2012). Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 40, 5415–5431. doi: 10.1093/nar/gks225
Soshnev, A. A., Baxley, R. M., Manak, J. R., Tan, K., and Geyer PK. (2013). The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development 140, 3613–3623. doi: 10.1242/dev.094953
Soshnikova, N., Montavon, T., Leleu, M., Galjart, N., and Duboule, D. (2010). Functional analysis of CTCF during mammalian limb development. Dev. Cell 19, 819–830. doi: 10.1016/j.devcel.2010.11.009
Splinter, E., Heath, H., Kooren, J., Palstra, R. J., Klous, P., Grosveld, F., et al. (2006). CTCF mediates long-range chromatin looping and local histone modification in the b-globin locus. Genes Dev. 20, 2349–2354. doi: 10.1101/gad.399506
Stogios, P. J., Downs, G. S., Jauhal, J. J., Nandra, S. K., and Prive, G. G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82. doi: 10.1186/gb-2005-6-10-r82
Struhl, K. (1984). Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc. Natl. Acad. Sci. U.S.A. 81, 7865–7869. doi: 10.1073/pnas.81.24.7865
Su, W., Jackson, S., Tjian, R., and Echols, H. (1991). DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 5, 820–826. doi: 10.1101/gad.5.5.820
Swamynathan, S. K. (2010). Krüppel-like factors: three fingers in control. Hum. Genomics 4, 263–270. doi: 10.1186/1479-7364-4-4-263
Takagi, Y., Calero, G., Komori, H., Brown, J. A., Ehrensberger, A. H., Hudmon, A., et al. (2006). Head module control of mediator interactions. Mol. Cell 23, 355–364. doi: 10.1016/j.molcel.2006.06.007
Tallack, M. R., Whitington, T., Yuen, W. S., Wainwright, E. N., Keys, J. R., Gardiner, B. B., et al. (2010). A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 20, 1052–1063. doi: 10.1101/gr.106575.110
Tallack, M. R., Magor, G. W., Dartigues, B., Sun, L., Huang, S., Fittock, J. M., et al. (2012). Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 22, 2385–2398. doi: 10.1101/gr.135707.111
Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F., and de Laat, W. (2002). Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465. doi: 10.1016/S1097-2765(02)00781-5
Tanay, A., and Cavalli, G. (2013). Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Curr. Opin. Genet. Dev. 23, 197–203. doi: 10.1016/j.gde.2012.12.009
Tsang, A. P., Visvader, J. E., Turner, C. A., Fujiwara, Y., Yu, C., Weiss, M. J., et al. (1997). FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90, 109–119. doi: 10.1016/S0092-8674(00)80318-9
Vakoc, C. R., Letting, D. L., Gheldof, N., Sawado, T., Bender, M. A., Groudine, M., et al. (2005). Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462. doi: 10.1016/j.molcel.2004.12.028
Vazquez, J., Müller, M., Pirrotta, V., and Sedat, J. W. (2006). The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol. Biol. Cell. 17, 2158–2165. doi: 10.1091/mbc.E06-01-0049
Venken, K. J., and Bellen, H. J. (2012). Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and ΦC31 integrase. Methods Mol. Biol. 859, 203–228. doi: 10.1007/978-1-61779-603-6_12
Vojnic, E., Mourao, A., Seizl, M., Simon, B., Wenzeck, L., and Lariviere, L. (2011). Structure and VP16 binding of the mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 18, 404–409. doi: 10.1038/nsmb.1997
Wadman, I. A., Osada, H., Grutz, G. G., Agulnick, A. D., Westphal, H., Forster, A., et al. (1997). The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16, 3145–3157. doi: 10.1093/emboj/16.11.3145
Wang, H., Maurano, M. T., Qu, H., Varley, K. E., Gertz, J., Pauli, F., et al. (2012). Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688. doi: 10.1101/gr.136101.111
Wei, Z., Yang, Y., Zhang, P., Andrianakos, R., Hasegawa, K., Lyu, J., et al. (2009). Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 27, 2969–2978. doi: 10.1002/stem.231
Wei, Z., Gao, F., Kim, S., Yang, H., Lyu, J., An, W., et al. (2013). Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13, 36–47. doi: 10.1016/j.stem.2013.05.010
Welch, J. J., Watts, J. A., Vakoc, C. R., Yao, Y., Wang, H., and Hardison, R. C. (2004). Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147. doi: 10.1182/blood-2004-04-1603
Wendt, K. S., Yoshida, K., Itoh, T., Bando, M., Koch, B., Schirghuber, E., et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801. doi: 10.1038/nature06634
Wilkinson-White, L., Gamsjaeger, R., Dastmalchi, S., Wienert, B., Stokes, P. H., and Crossley, M. (2011). Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc. Natl. Acad. Sci. U.S.A. 108, 14443–14448. doi: 10.1073/pnas.1105898108
Xiao, T., Wallace, J., and Felsenfeld, G. (2011). Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol. Cell. Biol. 31, 2174–2183. doi: 10.1128/MCB.05093-11
Yan, J., Enge, M., Whitington, T., Dave, K., Liu, J., Sur, I., et al. (2013). Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor cites. Cell 154, 801–813. doi: 10.1016/j.cell.2013.07.034
Yao, H., Brick, K., Evrard, Y., Xiao, T., Camerini-Otero, R. D., and Felsenfeld, G. (2010). Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555. doi: 10.1101/gad.1967810
Yien, Y. Y., and Bieker, J. J. (2013). EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol. Cell. Biol. 33, 4–13. doi: 10.1128/MCB.01058-12
Yoshida, C., Tokumasu, F., Hohmura, K. I., Bungert, J., Hayashi, N., Nagasawa, T., et al. (1999). Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells 4, 643–655. doi: 10.1046/j.1365-2443.1999.00291.x
Yusufzai, T. M., and Felsenfeld, G. (2004). The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. U.S.A. 101, 8620–8624. doi: 10.1073/pnas.0402938101
Zhao, K., Hart, C. M., and Laemmli, U. K. (1995). Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81, 879–889. doi: 10.1016/0092-8674(95)90008-X
Zollman, S., Godt, D., Prive, G. G., Couderc, J. L., and Laski, F. A. (1994). The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 91, 10717–10721. doi: 10.1073/pnas.91.22.10717
Keywords: chromatin looping, long-range interactions, chromatin insulators, cis-regulation, transcription, enhancers
Citation: Maksimenko O and Georgiev P (2014) Mechanisms and proteins involved in long-distance interactions. Front. Genet. 5:28. doi: 10.3389/fgene.2014.00028
Received: 11 October 2013; Accepted: 25 January 2014;
Published online: 18 February 2014.
Edited by:
Huabing Li, Yale University School of Medicine, USAReviewed by:
Zdenka Pausova, The Hospital for Sick Children, CanadaSushma S. Iyengar, University of Southern California, USA
Huabing Li, Yale University School of Medicine, USA
Copyright © 2014 Maksimenko and Georgiev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pavel Georgiev, Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, 34/5 Vavilov Street, Moscow 119334, Russia e-mail: georgiev_p@mail.ru
 Oksana Maksimenko
Oksana Maksimenko