- Division of Epigenomics, Aichi Cancer Center Research Institute, Nagoya, Japan
Glioblastomas show heterogeneous histological features. These distinct phenotypic states are thought to be associated with the presence of glioma stem cells (GSCs), which are highly tumorigenic and self-renewing sub-population of tumor cells that have different functional characteristics. Differentiation of GSCs may be regulated by multi-tiered epigenetic mechanisms that orchestrate the expression of thousands of genes. One such regulatory mechanism involves functional non-coding RNAs (ncRNAs), such as microRNAs (miRNAs); a large number of ncRNAs have been identified and shown to regulate the expression of genes associated with cell differentiation programs. Given the roles of miRNAs in cell differentiation, it is possible they are involved in the regulation of gene expression networks in GSCs that are important for the maintenance of the pluripotent state and for directing differentiation. Here, we review recent findings on ncRNAs associated with GSC differentiation and discuss how these ncRNAs contribute to the establishment of tissue heterogeneity during glioblastoma tumor formation.
Introduction
Gliomas are the most common type of malignant primary brain tumor with an incidence of ~5 cases per 100,000 persons (Wen and Kesari, 2008). Glioblastoma multiforme (GBM) is the highest grade glioma (grade 4). Despite advances in treatment using combinations of surgery, radiotherapy, and chemotherapy, GBM confers an average life expectancy of around 14 months from diagnosis (Wen and Kesari, 2008). Accumulating evidence indicates that the presence of a subset of cells with the potential to initiate and maintain growth of gliomas might be crucial for their resistance to conventional therapies (Hadjipanayis and Van Meir, 2009). These cells are designated as glioma stem cells (GSCs; Galli et al., 2004; Singh et al., 2004; Lee et al., 2006; Penuelas et al., 2009; Mazzoleni et al., 2010). GSCs and normal neural stem cells appear to share common features including self-renewal and the capability of differentiating into multiple lineages. Intriguingly, recent studies revealed that in addition to GSCs differentiating into non-GSCs, the reverse process might also occur (Gupta et al., 2011; Natsume et al., 2013). This phenotypic plasticity between the GSC and non-GSC states may be regulated by signals within the tumor microenvironment.
Microenvironmental signals, such as sonic Hedgehog (SHH), Wnt, and Notch, have been shown to regulate the properties of cancer stem cells (Reya and Clevers, 2005; Fan et al., 2010; Takebe et al., 2011). SHH has a critical role in the maintenance of GSCs by regulating so-called “stemness” genes and has also been found to be activated in many high-grade gliomas (Clement et al., 2007; Takezaki et al., 2011). The Wnt/β-catenin pathway has been implicated in the role of GSCs in gliomagenesis through tumor proliferation and invasion (Nager et al., 2012). Notch signaling has been shown to promote GSC self-renewal and to suppress GSC differentiation (Shih and Holland, 2006; Fan et al., 2010; Hu et al., 2011). Genes in the receptor tyrosine kinase (RTK) family mediate several oncogenic growth factor pathways, such as epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR), that have been linked to malignancy, angiogenesis, self-renewal, and multipotency. Recently, it was shown that constitutively activated EGFRvIII expression and loss of the phosphatase and tensin (PTEN) protein in murine neural stem cells results in the formation of glial tumors (Li et al., 2009a). PDGF overexpression has also been implicated in gliomagenesis, and PDGFs can inhibit glial cell differentiation (Fomchenko and Holland, 2007).
Recent advanced technology to identify non-coding RNAs using microarrays or next generation sequencing technologies provide extraordinary abundance of novel data in genome wide-scale and revealed deeper insights into the biology of non-cording RNAs (ncRNAs). More than 90% of the human genome appears to be transcribed and transcription is not limited to protein-coding regions (Birney et al., 2007). Some ncRNAs may play key regulatory and functional roles. Indeed, significant numbers of ncRNAs, such as microRNAs (miRNAs, miRs) and long non-coding RNAs (lncRNAs), are regulated during development in a cell-type specific manner, and are associated with multiple cell functions (Kapranov et al., 2007). miRNAs are the short non-coding endogenous RNAs that post-transcriptionally regulate the expression of a large number of genes (Bartel, 2004). miRNAs play important roles in a wide variety of physiological and pathological processes including tumor formation. Aberrant expression of miRNA can induce tumor suppression or can have an oncogenic effect resulting in tumor formation (Medina and Slack, 2008; Gangaraju and Lin, 2009). lncRNAs are functional ncRNAs that are potentially key regulators not only of cellular differentiation and proliferation, but may also have tumor suppressive or oncogenic functions in many types of cancer (Esteller, 2011; Wapinski and Chang, 2011; Hu et al., 2012; Zhang et al., 2013).
In this review, we provide a summary of the current understanding of miRNAs and lncRNAs in gliomas with a focus on their roles in GSCs.
miRNAs in GSC Differentiation
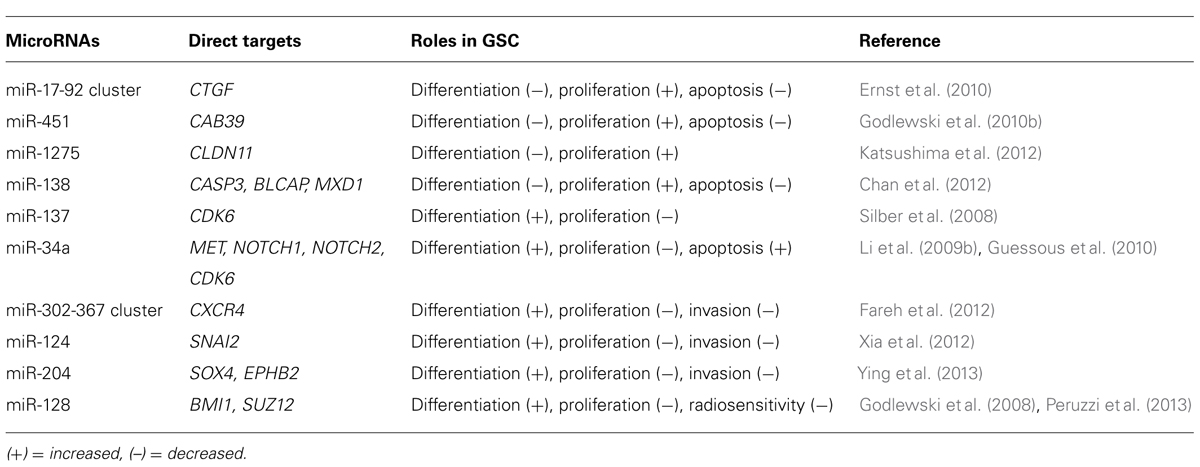
miRNAs are short sequences of 17–25 nucleotides that are not transcribed but have a regulatory function. An RNase III enzyme converts pri-miRNA into pre-miRNA hairpin transcripts that are processed into mature miRNAs and incorporated into a ribonucleoprotein complex called the RNA-induced silencing complex (RISC). The RISC and associated mature miRNA then binds to mRNA and causes a physical block to translation (Ambros and Lee, 2004; Bartel, 2004). Many miRNAs form imperfectly complementary stem-loop structures on the sense strand of the target mRNA. Thus, each miRNA can target multiple mRNA species through recognition of complementary sequences. Upregulation of mature miRNAs may occur as a consequence of transcriptional activation or amplification of the corresponding pre-miRNA locus, whereas downregulation of miRNAs may result from epigenetic silencing or deletion of the corresponding region (Schickel et al., 2008). Although dysregulation of the miRNA-mRNA network has been reported in glioblastoma, little attention has so far been paid to its role in GSCs (Godlewski et al., 2010a). In this section, we describe the information available on the significance of miRNAs in GSCs (Table 1).
miR-17-92 Cluster
The miR-17-92 cluster is thought to be involved in the regulation of GSC differentiation, apoptosis, and proliferation (Ernst et al., 2010). The level of transcripts from miR-17-92 clusters are significantly higher in primary astrocytic tumors than in normal brain tissues and increase significantly with tumor grade progression. A High-level amplification of the miR-17-92 locus has also been found in glioblastoma specimens. Inhibition of miR-17-92 induces apoptosis and decreases cell proliferation in GSCs. mir-17-92 inhibition is also associated with induction of cyclin-dependent kinase inhibitor 1A (CDKN1A), E2F transcription factor 1 (E2F1), PTEN, and connective tissue growth factor (CTGF). Of these, the CTGF gene was shown to be a direct target of miR-17-92 in GSCs.
When GSCs are exposed to the differentiation-promoting conditions, downregulation of the oncogenic miR-17-92 cluster is directly related to the concomitant upregulation of CTGF (Ernst et al., 2010).
miR-124 and miR-137
The initial analysis of miR-124 showed that it promotes neuronal differentiation by targeting the polypyrimidine tract-binding protein 1 (PTBP1) that encodes a global repressor of alternative pre-mRNA splicing; miR reduces the level of PTBP1, which results in an increase in the production of nervous system-specific alternative RNA splicing and promotes the differentiation of progenitor cells to mature neurons (Makeyev et al., 2007). Subsequent analysis showed that both miR-124 and miR-137 are downregulated in high-grade gliomas and up-regulated during adult neural stem cell differentiation (Silber et al., 2008). Transfection of miR-124 or miR-137 inhibits proliferation of GSCs, via suppression of cyclin-dependent protein kinase 6 (CDK6), and induces morphological changes in human GSCs and expression of neuronal differentiation markers. Overexpression of miR-124 has consistently been found to inhibit the CD133+ cell subpopulation of the neurosphere and to downregulate stem cell markers, such as BMI1, Nanog, and Nestin. These effects could be rescued by re-expression of SNAI2, another direct target of miR-124 (Xia et al., 2012).
miR-451
Analysis of the miRNA profiles of GSC (CD133+ cells) and non-GSC (CD133– cells) populations showed that several miRNAs, including miR-451, miR-486, and miR-425, are upregulated in CD133– cells. Transfection of cells with miR-451 has been shown to induce disruption of glioblastoma neurospheres (Gal et al., 2008). Interestingly, this study also showed that SMAD proteins, which are associated with GSC regulation, can upregulate miR-451 by binding to its promoter region. Thus, there is a link between miRNAs and well-known stem cell regulating proteins (Piccirillo et al., 2006). Another interesting finding regarding miR-451 is that its expression level is correlated with glucose concentration. High glucose levels are associated with relatively high levels of miR-451 expression, which promote cell growth; miR-451 expression levels decrease under low glucose conditions, resulting in a reduced rate of cell proliferation but an enhanced rate of cell migration and survival in glioblastomas. This miR-451 effect is mediated by liver kinase B1 (LKB1). These data indicate that tumor cells can survive under metabolic stress conditions and also seek out locations with more favorable growth conditions by migration influenced through an LKB1/AMPK pathway mediated by miR-451 (Godlewski et al., 2010b).
miR-34a
miR-34a is tumor-suppressive and is downregulated in human glioma tissues; miR-34a directly inhibits the expression of c-Met, Notch-1, and Notch-2 in GSCs (Li et al., 2009b). Notch is a critical regulator of cell-fate during development and also of normal stem cell maintenance (Fan et al., 2006; Shih and Holland, 2006; Fan et al., 2010). Activation of the Notch pathway enhances the stemness, proliferation, and radioresistance of GSCs (Wang et al., 2010). Ectopic expression of miR-34a in glioma cells inhibits cell proliferation, survival, and migration. In addition, miR-34a induces GSC differentiation as evidenced by the decreased expression of stem cell markers and increased expression of differentiation markers (Guessous et al., 2010).
miR-128
Two studies have described a link between miR-128 and the polycomb repressor complex (PRC). Two major complexes, PRC1 and PRC2, are recognized as key epigenetic regulators during development (Lund and van Lohuizen, 2004) and are required for maintaining self-renewal and multi-potential capability (Richly et al., 2011). The first study demonstrated that miR-128 has a tumor-suppressive function and that this is downregulated in glioblastoma tissue. miR-128 expression significantly reduces glioma cell proliferation both in vitro and in vivo via downregulation of the oncogene Bmi-1 that is a component of PRC1. In addition, miR-128 inhibits GSC self-renewal (Godlewski et al., 2008). The second study showed that miR-128 directly targets SUZ12, a key component of PRC2. Ectopic expression of miR-128 in GSCs significantly increases their radiosensitivity (Peruzzi et al., 2013). The PRC has been shown to promote normal and cancer stem cell self-renewal and is also implicated in GSC regulation (Abdouh et al., 2009; Suva et al., 2009; Natsume et al., 2013). The findings of these various studies therefore indicate that miR-128 mediates an important epigenetic regulatory pathway in GSCs.
Other miRNAs
Several other miRNAs have been implicated in glioma malignancy. Ectopic expression of the miR-302-367 cluster in GSCs inhibits the CXCR4 pathway resulting in the suppression of stemness signatures, self-renewal, and cell infiltration. Inhibition of the CXCR4 pathway leads to the disruption of the SHH-GLI-NANOG network, which is important for cell self-renewal and tumorigenic properties (Fareh et al., 2012). In both GSCs and non-GSCs, miR-1275 is controlled by a polycomb-mediated silencing mechanism and regulates expression of the oligodendroglial-lineage gene claudin 11 (CLDN11). These data illustrate that miR-1275 is regulated by an epigenetic pathway and that it contributes to the phenotypic diversity of glioblastoma tissues. The increased insight into the roles of these miRs may provide a better understanding of basis for the heterogeneity of glioblastomas in the context of human neurodevelopment (Katsushima et al., 2012). Recently, miR-204 was shown to suppress self-renewal, a stem cell characteristic, and the migration of GSCs by targeting the stemness-governing transcriptional factor SOX4 and the migration-promoting receptor EphB2 (Ying et al., 2013).
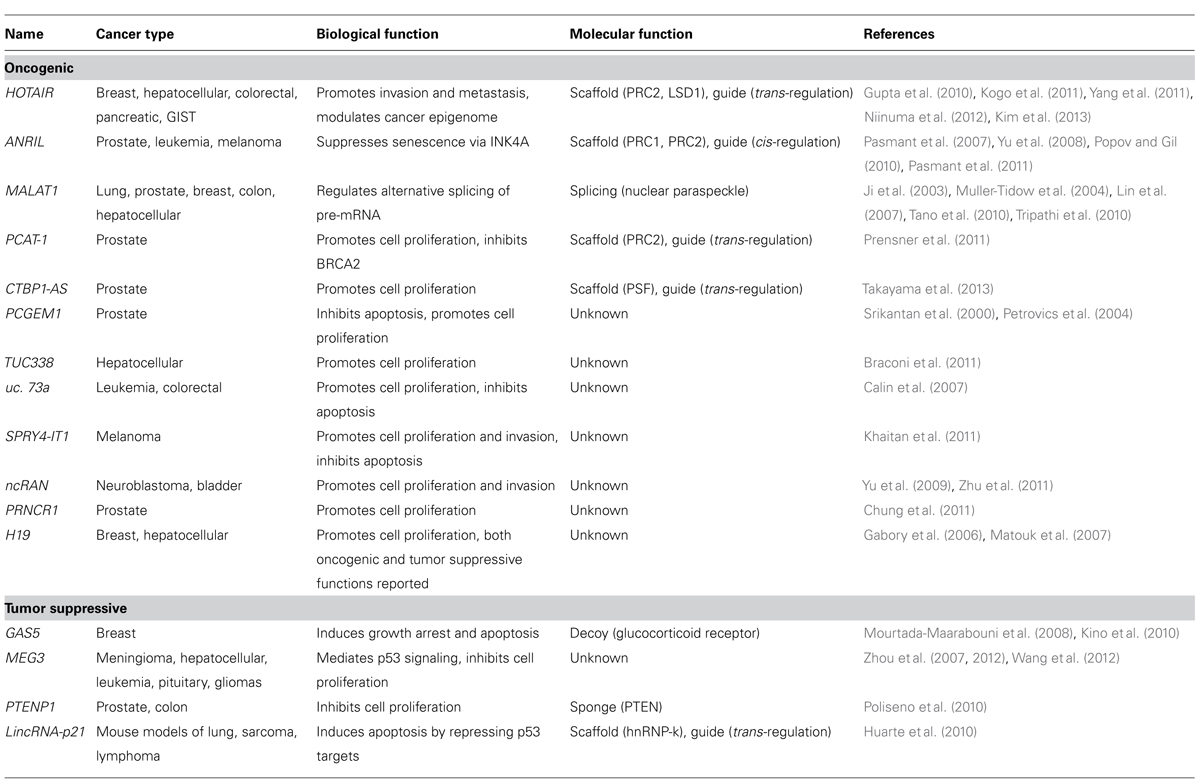
LncRNAs in Cancer
Genome-wide studies showed that there are a large number of ncRNAs, including a group termed lncRNAs (Birney et al., 2007). LncRNAs are generally greater than 200 nucleotides and up to 100 kb in length (Mercer et al., 2009). It is known that lncRNAs are mainly transcribed by RNA polymerase II, are polyadenylated and spliced (Wu et al., 2008; Mercer et al., 2009; Ponting et al., 2009). Approximately 15,000 lncRNAs are estimated to occur in human cells and these are frequently expressed in tissue-specific patterns (Derrien et al., 2012). lncRNAs appear to play important roles in a wide range of biological cellular processes including maintenance of stemness, development, and cell survival (Koziol and Rinn, 2010; Zhang et al., 2013). Currently studies detected a set of lncRNAs in each disease using RNA immunoprecipitation with RNA binding proteins coupled with computational approaches.
Long non-coding RNAs are believed to regulate gene expression through four different pathways (Koziol and Rinn, 2010; Hu et al., 2012). First, lncRNAs can bind to chromatin modifying proteins (which have a scaffold function) and recruit these proteins to target loci. These lncRNA complexes can target genes that are closely situated in the genome (cis-regulation) or genes that are genomically distant (trans-regulation; Nagano et al., 2008; Pandey et al., 2008; Zhao et al., 2008; Gupta et al., 2010; Huarte et al., 2010; Tian et al., 2010; Prensner et al., 2011; Wang et al., 2011). Second, lncRNAs can act as an RNA decoy, that is, they can interact directly with a DNA binding domain to prevent transcription factors interacting with their DNA targets (Kino et al., 2010; Ng et al., 2012). Third, lncRNAs can act as an miRNA sponge, that is, they prevent specific miRNAs from binding to their target mRNAs by competitive binding (Poliseno et al., 2010; Cesana et al., 2011; Karreth et al., 2011). Fourth, lncRNAs can bind to specific combinations of regulatory proteins, such as RNA splicing proteins within ribonucleoprotein complexes (Tripathi et al., 2010; Ng et al., 2012; Schor et al., 2012).
There is increasing evidence to show that a set of lncRNAs is associated with cancer pathogenesis and that these lncRNAs function as regulators in cancer development (Prensner and Chinnaiyan, 2011). lncRNAs that are dysregulated in cancers are listed in Table 2. Below, we provide a brief description of some lncRNAs that are associated with glioma tumorigenesis.
MEG3
Maternally expressed gene 3 (MEG3) is a maternally expressed imprinted gene that can also act as an lncRNA. MEG3 is generally expressed in normal tissues, and its downregulation by aberrant DNA methylation has been found in many types of human cancer (Zhou et al., 2012; Shi et al., 2013). For example, MEG3 expression in glioma tissues is decreased compared to adjacent normal tissues (Wang et al., 2012). The tumor-suppressive role of MEG3 is supported by the fact that it can associate with p53 and that this association is required for p53 activation (Lu et al., 2013). Ectopic expression of MEG3 inhibits cell proliferation and induced cell apoptosis in glioma cell lines (Wang et al., 2012).
CRNDE
Colorectal neoplasia differentially expressed (CRNDE) transcripts are categorized as lncRNAs and have the potential to interact with chromatin-modifying proteins to regulate gene expression through epigenetic changes (Ellis et al., 2012). CRNDE is expressed in the fetal brain and in induced pluripotent stem cells; the level of expression increases during neuronal differentiation but no transcripts can be detected in the adult brain (Lin et al., 2011). Intriguingly, CRNDE is highly expressed in gliomas. The recent study of Ellis et al. demonstrated a direct interaction between CRNDE transcripts and components of PRC2 and the CoREST chromatin-modifying complex. CRNDE provides specific functional scaffolds for regulatory complexes, such as PRC2 and CoREST, and may contribute the maintenance of pluripotent state as well as neuronal differentiation (Ellis et al., 2012).
Concluding Remarks
Following the discovery of cancer stem cells, it became important to elucidate the mechanisms and the environmental cues that control the differentiation of these cells into the diverse array of cell types that form during tumorigenesis. Epigenetic dysregulation has recently been shown to change the balance between differentiation and self-renewal of cortical progenitor cells and, thereby, to alter the rate and developmental timing of neurogenesis (Pereira et al., 2010). Given that cancer is a disease of faulty cellular differentiation, it is likely that aberrant epigenetic mechanisms involving ncRNAs are involved in glioma tumorigenesis. lncRNAs are increasingly important because of their potential for use in clinical diagnosis and treatment. To date, however, the functions of only a few lncRNAs have been elucidated with respect to tumor biology and there are still many aspects that remain to be resolved. Further investigations are required to clarify the functional roles of lncRNAs in order to elucidate the gene regulatory mechanisms in gliomagenesis. Understanding of the interplays between lncRNAs and genomes, which are reversible alterations, may offer a novel opportunity for the development of molecularly targeted therapies. Nevertheless, a better understanding of the glioblastoma core signaling pathways regulated by ncRNAs and other epigenetic mechanisms will undoubtedly provide novel therapeutic targets and strategies with applications in diagnosis and therapy in glioblastoma.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grant from PRESTO of JST, Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
References
Abdouh, M., Facchino, S., Chatoo, W., Balasingam, V., Ferreira, J., and Bernier, G. (2009). BMI1 sustains human glioblastoma multiforme stem cell renewal. J. Neurosci. 29, 8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009
Ambros, V., and Lee, R. C. (2004). Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 265, 131–158. doi: 10.1385/1-59259-775-0131
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Braconi, C., Valeri, N., Kogure, T., Gasparini, P., Huang, N., Nuovo, G. J., et al. (2011). Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 108, 786–791. doi: 10.1073/pnas.1011098108
Calin, G. A., Liu, C. G., Ferracin, M., Hyslop, T., Spizzo, R., Sevignani, C., et al. (2007). Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229. doi: 10.1016/j.ccr.2007.07.027
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chan, X. H., Nama, S., Gopal, F., Rizk, P., Ramasamy, S., Sundaram, G., et al. (2012). Targeting glioma stem cells by functional inhibition of a prosurvival oncomiR-138 in malignant gliomas. Cell Rep. 2, 591–602. doi: 10.1016/j.celrep.2012.07.012
Chung, S., Nakagawa, H., Uemura, M., Piao, L., Ashikawa, K., Hosono, N., et al. (2011). Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 102, 245–252. doi: 10.1111/j.1349-7006.2010.01737.x
Clement, V., Sanchez, P., De Tribolet, N., Radovanovic, I., and Ruiz I Altaba, A. (2007). HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 17, 165–172. doi: 10.1016/j.cub.2006.11.033
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Ellis, B. C., Molloy, P. L., and Graham, L. D. (2012). CRNDE:a long non-coding RNA involved in cancer, neurobiology, and development. Front. Genet. 3:270. doi: 10.3389/fgene.2012.00270
Ernst, A., Campos, B., Meier, J., Devens, F., Liesenberg, F., Wolter, M., et al. (2010). De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 29, 3411–3422. doi: 10.1038/onc.2010.83
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi: 10.1038/nrg3074
Fan, X., Khaki, L., Zhu, T. S., Soules, M. E., Talsma, C. E., Gul, N., et al. (2010). NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells 28, 5–16. doi: 10.1002/stem.254
Fan, X., Matsui, W., Khaki, L., Stearns, D., Chun, J., Li, Y. M., et al. (2006). Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 66, 7445–7452. doi: 10.1158/0008-5472.CAN-06-0858
Fareh, M., Turchi, L., Virolle, V., Debruyne, D., Almairac, F., De-La-Forest Divonne, S., et al. (2012). The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 19, 232–244. doi: 10.1038/cdd.2011.89
Fomchenko, E. I., and Holland, E. C. (2007). Platelet-derived growth factor-mediated gliomagenesis and brain tumor recruitment. Neurosurg. Clin. N. Am. 18, 39–58; viii. doi: 10.1016/j.nec.2006.10.006
Gabory, A., Ripoche, M. A., Yoshimizu, T., and Dandolo, L. (2006). The H19 gene: regulation and function of a non-coding RNA. Cytogenet. Genome Res. 113, 188–193. doi: 10.1159/000090831
Gal, H., Pandi, G., Kanner, A. A., Ram, Z., Lithwick-Yanai, G., Amariglio, N., et al. (2008). MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun. 376, 86–90. doi: 10.1016/j.bbrc.2008.08.107
Galli, R., Binda, E., Orfanelli, U., Cipelletti, B., Gritti, A., De Vitis, S., et al. (2004). Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021. doi: 10.1158/0008-5472.CAN-04-1364
Gangaraju, V. K., and Lin, H. (2009). MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 10, 116–125. doi: 10.1038/nrm2621
Godlewski, J., Newton, H., Chiocca, E., and Lawler, S. (2010a). MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 17, 221–228. doi: 10.1038/cdd.2009.71
Godlewski, J., Nowicki, M. O., Bronisz, A., Nuovo, G., Palatini, J., De Lay, M., et al. (2010b). MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell 37, 620–632. doi: 10.1016/j.molcel.2010.02.018
Godlewski, J., Nowicki, M. O., Bronisz, A., Williams, S., Otsuki, A., Nuovo, G., et al. (2008). Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 68, 9125–9130. doi: 10.1158/0008-5472.CAN-08-2629
Guessous, F., Zhang, Y., Kofman, A., Catania, A., Li, Y., Schiff, D., et al. (2010). microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 9, 1031–1036. doi: 10.4161/cc.9.6.10987
Gupta, P. B., Fillmore, C. M., Jiang, G., Shapira, S. D., Tao, K., Kuperwasser, C., et al. (2011). Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644. doi: 10.1016/j.cell.2011.07.026
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. doi: 10.1038/nature08975
Hadjipanayis, C. G., and Van Meir, E. G. (2009). Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol. Med. 15, 519–530. doi: 10.1016/j.molmed.2009.09.003
Hu, W., Alvarez-Dominguez, J. R., and Lodish, H. F. (2012). Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 13, 971–983. doi: 10.1038/embor.2012.145
Hu, Y. Y., Zheng, M. H., Cheng, G., Li, L., Liang, L., Gao, F., et al. (2011). Notch signaling contributes to the maintenance of both normal neural stem cells and patient-derived glioma stem cells. BMC Cancer 11:82. doi: 10.1186/1471-2407-11-82
Huarte, M., Guttman, M., Feldser, D., Garber, M., Koziol, M. J., Kenzelmann-Broz, D., et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. doi: 10.1016/j.cell.2010.06.040
Ji, P., Diederichs, S., Wang, W., Boing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041. doi: 10.1038/sj.onc.1206928
Kapranov, P., Willingham, A. T., and Gingeras, T. R. (2007). Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8, 413–423. doi: 10.1038/nrg2083
Karreth, F. A., Tay, Y., Perna, D., Ala, U., Tan, S. M., Rust, A. G., et al. (2011). In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395. doi: 10.1016/j.cell.2011.09.032
Katsushima, K., Shinjo, K., Natsume, A., Ohka, F., Fujii, M., Osada, H., et al. (2012). Contribution of microRNA-1275 to Claudin11 protein suppression via a polycomb-mediated silencing mechanism in human glioma stem-like cells. J. Biol. Chem. 287, 27396–27406. doi: 10.1074/jbc.M112.359109
Khaitan, D., Dinger, M. E., Mazar, J., Crawford, J., Smith, M. A., Mattick, J. S., et al. (2011). The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 71, 3852–3862. doi: 10.1158/0008-5472.CAN-10-4460
Kim, K., Jutooru, I., Chadalapaka, G., Johnson, G., Frank, J., Burghardt, R., et al. (2013). HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32, 1616–1625. doi: 10.1038/onc.2012.193
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8. doi: 10.1126/scisignal.2000568
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326. doi: 10.1158/0008-5472.CAN-11-1021
Koziol, M. J., and Rinn, J. L. (2010). RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 20, 142–148. doi: 10.1016/j.gde.2010.03.003
Lee, J., Kotliarova, S., Kotliarov, Y., Li, A., Su, Q., Donin, N. M., et al. (2006). Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403. doi: 10.1016/j.ccr.2006.03.030
Li, L., Dutra, A., Pak, E., Labrie, J. E. III, Gerstein, R. M., Pandolfi, P. P., et al. (2009a). EGFRvIII expression and PTEN loss synergistically induce chromosomal instability and glial tumors. Neurooncology 11, 9–21. doi: 10.1215/15228517-2008-2081
Li, Y., Guessous, F., Zhang, Y., Dipierro, C., Kefas, B., Johnson, E., et al. (2009b). MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576. doi: 10.1158/0008-5472.CAN-09-0529
Lin, M., Pedrosa, E., Shah, A., Hrabovsky, A., Maqbool, S., Zheng, D., et al. (2011). RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE 6:e23356. doi: 10.1371/journal.pone.0023356
Lin, R., Maeda, S., Liu, C., Karin, M., and Edgington, T. S. (2007). A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26, 851–858. doi: 10.1038/sj.onc.1209846
Lu, K. H., Li, W., Liu, X. H., Sun, M., Zhang, M. L., Wu, W. Q., et al. (2013). Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13:461. doi: 10.1186/1471-2407-13-461
Lund, A. H., and van Lohuizen, M. (2004). Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16, 239–246. doi: 10.1016/j.ceb.2004.03.010
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448. doi: 10.1016/j.molcel.2007.07.015
Matouk, I. J., Degroot, N., Mezan, S., Ayesh, S., Abu-Lail, R., Hochberg, A., et al. (2007). The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2:e845. doi: 10.1371/journal.pone.0000845
Mazzoleni, S., Politi, L. S., Pala, M., Cominelli, M., Franzin, A., Sergi Sergi, L., et al. (2010). Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 70, 7500–7513. doi: 10.1158/0008-5472.CAN-10-2353
Medina, P. P., and Slack, F. J. (2008). microRNAs and cancer: an overview. Cell Cycle 7, 2485–2492. doi: 10.4161/cc.7.16.6453
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Mourtada-Maarabouni, M., Pickard, M., Hedge, V., Farzaneh, F., and Williams, G. (2008). GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28, 195–208. doi: 10.1038/onc.2008.373
Muller-Tidow, C., Diederichs, S., Thomas, M., and Serve, H. (2004). Genome-wide screening for prognosis-predicting genes in early-stage non-small-cell lung cancer. Lung Cancer 45(Suppl. 2), S145–S150. doi: 10.1016/j.lungcan.2004.07.979
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R., et al. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720. doi: 10.1126/science.1163802
Nager, M., Bhardwaj, D., Canti, C., Medina, L., Nogues, P., and Herreros, J. (2012). Beta-Catenin signalling in glioblastoma multiforme and glioma-initiating cells. Chemother. Res. Pract. 2012, 192362. doi: 10.1155/2012/192362
Natsume, A., Ito, M., Katsushima, K., Ohka, F., Hatanaka, A., Shinjo, K., et al. (2013). Chromatin Regulator PRC2 Is a Key Regulator of Epigenetic Plasticity in Glioblastoma. Cancer Res. 73, 4559–4570. doi: 10.1158/0008-5472.CAN-13-0109
Ng, S. Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Niinuma, T., Suzuki, H., Nojima, M., Nosho, K., Yamamoto, H., Takamaru, H., et al. (2012). Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72, 1126–1136. doi: 10.1158/0008-5472.CAN-11-1803
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246. doi: 10.1016/j.molcel.2008.08.022
Pasmant, E., Laurendeau, I., Heron, D., Vidaud, M., Vidaud, D., and Bieche, I. (2007). Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969. doi: 10.1158/0008-5472.CAN-06-2004
Pasmant, E., Sabbagh, A., Vidaud, M., and Bieche, I. (2011). ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 25, 444–448. doi: 10.1096/fj.10-172452
Penuelas, S., Anido, J., Prieto-Sanchez, R. M., Folch, G., Barba, I., Cuartas, I., et al. (2009). TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15, 315–327. doi: 10.1016/j.ccr.2009.02.011
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M. W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962. doi: 10.1073/pnas.1002530107
Peruzzi, P., Bronisz, A., Nowicki, M. O., Wang, Y., Ogawa, D., Price, R., et al. (2013). MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neurooncology 15, 1212–1224. doi: 10.1093/neuonc/not055
Petrovics, G., Zhang, W., Makarem, M., Street, J. P., Connelly, R., Sun, L., et al. (2004). Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene 23, 605–611. doi: 10.1038/sj.onc.1207069
Piccirillo, S. G., Reynolds, B. A., Zanetti, N., Lamorte, G., Binda, E., Broggi, G., et al. (2006). Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 444, 761–765. doi: 10.1038/nature05349
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., and Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi: 10.1038/nature09144
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Popov, N., and Gil, J. (2010). Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics 5, 685–690. doi: 10.4161/epi.5.8.12996
Prensner, J. R., and Chinnaiyan, A. M. (2011). The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407. doi: 10.1158/2159-8290.CD-11-0209
Prensner, J. R., Iyer, M. K., Balbin, O. A., Dhanasekaran, S. M., Cao, Q., Brenner, J. C., et al. (2011). Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 29, 742–749. doi: 10.1038/nbt.1914
Reya, T., and Clevers, H. (2005). Wnt signalling in stem cells and cancer. Nature 434, 843–850. doi: 10.1038/nature03319
Richly, H., Aloia, L., and Di Croce, L. (2011). Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2, e204. doi: 10.1038/cddis.2011.84
Schickel, R., Boyerinas, B., Park, S., and Peter, M. (2008). MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27, 5959–5974. doi: 10.1038/onc.2008.274
Schor, I. E., Lleres, D., Risso, G. J., Pawellek, A., Ule, J., Lamond, A. I., et al. (2012). Perturbation of chromatin structure globally affects localization and recruitment of splicing factors. PLoS ONE 7:e48084. doi: 10.1371/journal.pone.0048084
Shi, X., Sun, M., Liu, H., Yao, Y., and Song, Y. (2013). Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 339, 159–166. doi: 10.1016/j.canlet.2013.06.013
Shih, A. H., and Holland, E. C. (2006). Notch signaling enhances nestin expression in gliomas. Neoplasia 8, 1072–1082. doi: 10.1593/neo.06526
Silber, J., Lim, D. A., Petritsch, C., Persson, A. I., Maunakea, A. K., Yu, M., et al. (2008). miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6:14. doi: 10.1186/1741-7015-6-14
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401. doi: 10.1038/nature03128
Srikantan, V., Zou, Z., Petrovics, G., Xu, L., Augustus, M., Davis, L., et al. (2000). PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 12216–12221. doi: 10.1073/pnas.97.22.12216
Suva, M. L., Riggi, N., Janiszewska, M., Radovanovic, I., Provero, P., Stehle, J. C., et al. (2009). EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 69, 9211–9218. doi: 10.1158/0008-5472.CAN-09-1622
Takayama, K. -I., Horie-Inoue, K., Katayama, S., Suzuki, T., Tsutsumi, S., Ikeda, K., et al. (2013). Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 32, 1665–1680. doi: 10.1038/emboj.2013.99
Takebe, N., Harris, P. J., Warren, R. Q., and Ivy, S. P. (2011). Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 8, 97–106. doi: 10.1038/nrclinonc.2010.196
Takezaki, T., Hide, T., Takanaga, H., Nakamura, H., Kuratsu, J., and Kondo, T. (2011). Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci. 102, 1306–1312. doi: 10.1111/j.1349-7006.2011.01943.x
Tano, K., Mizuno, R., Okada, T., Rakwal, R., Shibato, J., Masuo, Y., et al. (2010). MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 584, 4575–4580. doi: 10.1016/j.febslet.2010.10.008
Tian, D., Sun, S., and Lee, J. T. (2010). The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403. doi: 10.1016/j.cell.2010.09.049
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Wang, J., Wakeman, T. P., Lathia, J. D., Hjelmeland, A. B., Wang, X. F., White, R. R., et al. (2010). Notch promotes radioresistance of glioma stem cells. Stem Cells 28, 17–28. doi: 10.1002/stem.261
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Wang, P., Ren, Z., and Sun, P. (2012). Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem. 113, 1868–1874. doi: 10.1002/jcb.24055
Wapinski, O., and Chang, H. Y. (2011). Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361. doi: 10.1016/j.tcb.2011.04.001
Wen, P. Y., and Kesari, S. (2008). Malignant gliomas in adults. N. Engl. J. Med. 359, 492–507. doi: 10.1056/NEJMra0708126
Wu, Q., Kim, Y. C., Lu, J., Xuan, Z., Chen, J., Zheng, Y., et al. (2008). Poly A- transcripts expressed in HeLa cells. PLoS ONE 3:e2803. doi: 10.1371/journal.pone.0002803
Xia, H., Cheung, W. K., Ng, S. S., Jiang, X., Jiang, S., Sze, J., et al. (2012). Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J. Biol. Chem. 287, 9962–9971. doi: 10.1074/jbc.M111.332627
Yang, Z., Zhou, L., Wu, L. M., Lai, M. C., Xie, H. Y., Zhang, F., et al. (2011). Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18, 1243–1250. doi: 10.1245/s10434-011-1581-y
Ying, Z., Li, Y., Wu, J., Zhu, X., Yang, Y., Tian, H., et al. (2013). Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 73, 990–999. doi: 10.1158/0008-5472.CAN-12-2895
Yu, M., Ohira, M., Li, Y., Niizuma, H., Oo, M. L., Zhu, Y., et al. (2009). High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int. J. Oncol. 34, 931–938. doi: 10.3892/ijo_00000219
Yu, W., Gius, D., Onyango, P., Muldoon-Jacobs, K., Karp, J., Feinberg, A. P., et al. (2008). Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206. doi: 10.1038/nature06468
Zhang, H., Chen, Z., Wang, X., Huang, Z., He, Z., and Chen, Y. (2013). Long non-coding RNA: a new player in cancer. J. Hematol. Oncol. 6, 37. doi: 10.1186/1756-8722-6-37
Zhao, J., Sun, B. K., Erwin, J. A., Song, J. J., and Lee, J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756. doi: 10.1126/science.1163045
Zhou, Y., Zhang, X., and Klibanski, A. (2012). MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 48, R45–R53. doi: 10.1530/JME-12-18
Zhou, Y., Zhong, Y., Wang, Y., Zhang, X., Batista, D. L., Gejman, R., et al. (2007). Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742. doi: 10.1074/jbc.M702029200
Keywords: epigenetics, glioma, cancer stem cells, long non-coding RNA, micro RNA
Citation: Katsushima K and Kondo Y (2014) Non-coding RNAs as epigenetic regulator of glioma stem-like cell differentiation. Front. Genet. 5:14. doi: 10.3389/fgene.2014.00014
Received: 06 December 2013; Accepted: 15 January 2014;
Published online: 03 February 2014.
Edited by:
Yoshimasa Saito, Keio University, JapanReviewed by:
Atsushi Natsume, Nagoya University, JapanAlfred Sze-Lok Cheng, Chinese University of Hong Kong, Hong Kong
Copyright © 2014 Katsushima and Kondo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yutaka Kondo, Division of Epigenomics, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan e-mail:eWtvbmRvQGFpY2hpLWNjLmpw
 Keisuke Katsushima
Keisuke Katsushima Yutaka Kondo
Yutaka Kondo