- Max Planck Research Group for Gene Expression and Signaling, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany
The extracellular environment is a complex entity comprising of the extracellular matrix (ECM) and regulatory molecules. It is highly dynamic and under cell-extrinsic stress, transmits the stressed organism’s state to each individual ECM-connected cell. microRNAs (miRNAs) are regulatory molecules involved in virtually all the processes in the cell, especially under stress. In this review, we analyse how miRNA expression is regulated downstream of various signal transduction pathways induced by changes in the extracellular environment. In particular, we focus on the muscular dystrophy-associated cell adhesion molecule dystroglycan capable of signal transduction. Then we show how exactly the same miRNAs feedback to regulate the extracellular environment. The ultimate goal of this bi-directional signal transduction process is to change cell behavior under cell-extrinsic stress in order to respond to it accordingly.
Each individual organism in its lifetime has to cope with multiple kinds of stress that occur when body’s homeostasis is shifted from its optimal state. Development, puberty, changing environmental conditions, injuries, diseases, and aging – all influence the organism as a whole no matter which cell or system was the first to encounter the stressor. Organism’s systemic reaction to stress is achieved because cells are not isolated from each other. Rather, they communicate through direct cell–cell contacts or via the extracellular matrix (ECM).
In this review, in the beginning we introduce the ECM and the cell-extrinsic stress-induced signaling molecules. We define the extracellular environment, focusing on how it can encode for stress signals. We then discuss how a cell can read the encoded signals, focusing on cell adhesion molecule dystroglycan (Dg) capable of outside-in signal transduction. In the last two parts of the review, we show how a cell can change levels of intracellular microRNAs (miRNAs) in response to outside stress signals, and how these miRNAs can then modulate the cell behavior by targeting components of the extracellular environment.
ECM as a Reservoir for Cell-Extrinsic Stress-Induced Signaling Molecules
The ECM is composed of three major types of structural components: insoluble collagen fibers, viscous proteoglycans, and soluble multiadhesive proteins. Examples of multiadhesive proteins are laminin and fibronectin that connect collagens and proteoglycans to the cell receptors. Metabolic enzymes responsible for the restructuring of the ECM, like matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs), can be deposited outside the cell (Yu and Woessner, 2000; Yu et al., 2000) and thus can be considered as the fourth, enzymatic, component of the ECM (Table 1).
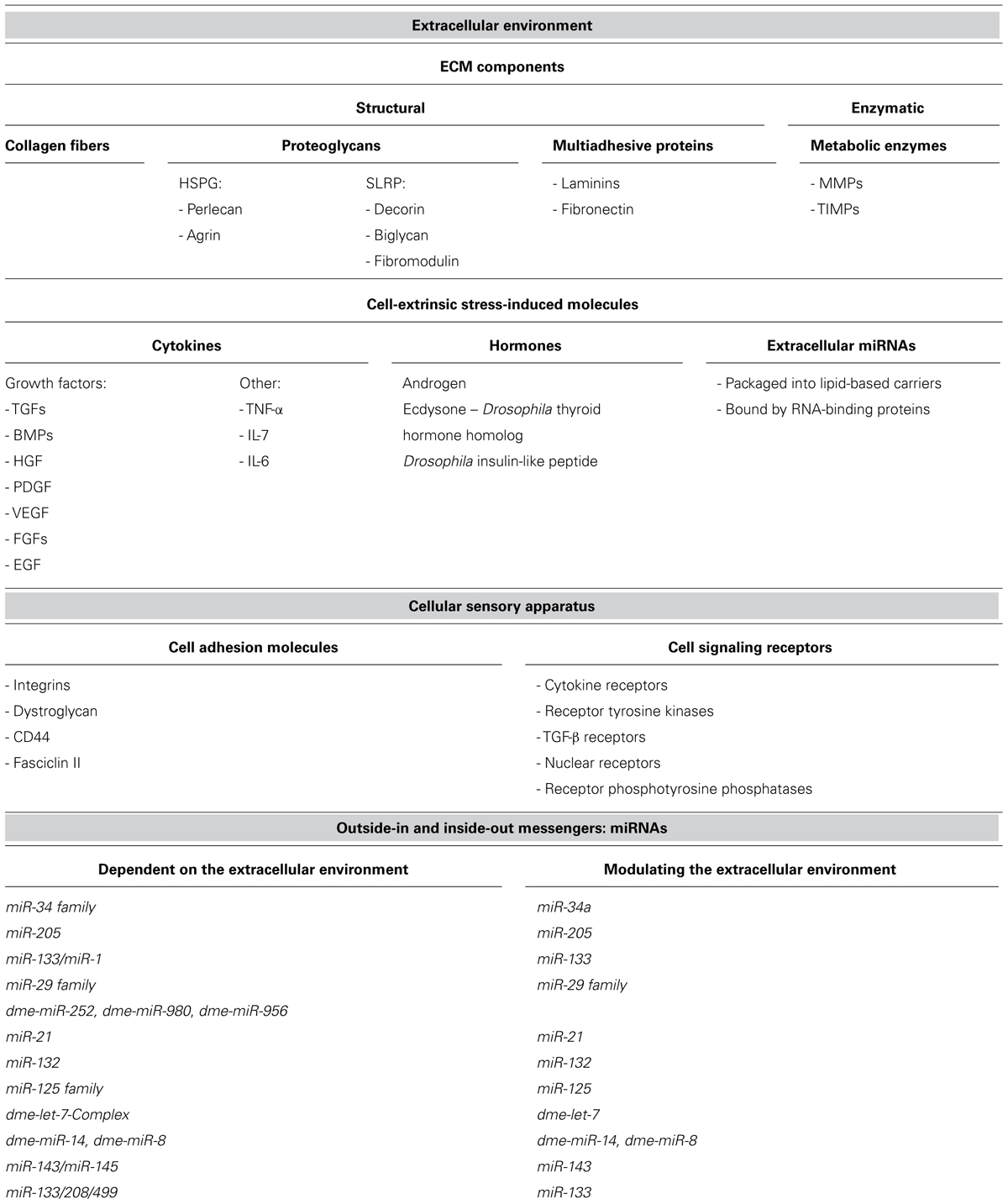
TABLE 1. The components of the extracellular environment, cellular sensory apparatus, and miRNAs, mentioned in the review.
The three-dimensional net of ECM components acts as a reservoir and a scaffold for a diversity of regulatory molecules: cytokines (including various growth factors and interleukins), hormones, and extracellular miRNAs. These molecules constitute the cell-extrinsic stress-induced signaling components, as they warn different cells of the organism about changes in the environmental conditions such as injuries, pathogens, and other external stressors that endanger the organism’s homeostasis and welfare.
The Extracellular Environment
Extracellular environment can be defined as a combination of the ECM and the extracellular regulatory molecules. Specific composition of the ECM of each ECM-connected cell defines the types of regulatory molecules found in close proximity to the cell. For example, different types of cytokines can be bound to different types of ECM proteins, contributing to where the cytokine exerts its function in mediating a specific cell fate or activity, as we now discuss.
Collagen-Binding Cytokines
Procollagen of type IIB is synthesized and deposited into the ECM by differentiated chondrocytes. Immature chondrogenic progenitors synthesize the other splice variant of procollagen – type IIA that has an additional cysteine-rich domain. This domain was shown to bind growth factors, such as transforming growth factor-β1 (TGF-β1), bone morphogenetic proteins bone morphogenetic protein-2 (BMP-2) and BMP-4 (Zhu et al., 1999; Larraín et al., 2000). Hence, the type of ECM procollagen defines the presence of differentiation factors and thus cell’s responsiveness to them.
Types of collagen deposited by cells also impose spatial constraints on the biological activity of platelet-derived and hepatocyte growth factors (PDGF and HGF; Somasundaram and Schuppan, 1996; Schuppan et al., 1998). PDGF and HGF trigger mitosis in mesenchymal cells and hepatocytes respectively. Thus, the ECM collagenous composition serves as an important clue for growth factors binding to the appropriate target cell types.
Fibronectin-Binding Cytokines
Cell-to-ECM adhesion protein fibronectin can also modulate effects of various signaling molecules, as for example of vascular endothelial growth factor (VEGF) that promotes migration and proliferation of endothelial cells. Fibronectin has separate cell-binding and VEGF-binding domains, and only when it binds VEGF and a cell simultaneously, the VEGF stimulation is significantly enhanced (Wijelath et al., 2006). Thus, the degree of endothelial cell response to VEGF can be regulated by the ECM composition.
Another domain of fibronectin was shown to interact with tumor necrosis factor-α (TNF-α) – initiator and regulator of inflammatory reactions. When bound to fibronectin, TNF-α enhances adhesion of activated immune cells to this glycoprotein, suggesting that ECM-bound TNF-α may recruit and direct immune cells to the sites of inflammation (Hershkoviz et al., 1994). In addition, interleukin-7 (IL-7) has been shown to modulate adhesive properties of immune cells in the context of the ECM composition. IL-7 binding to fibronectin augments adhesion of resting T-cells to fibronectin (Ariel et al., 1997). Hence, the ECM environment in which immune reactions take place modulates effects of cytokines on immune cells.
HSPG-Binding Cytokines
Heparan sulfate proteoglycans (HSPGs) are cell surface and ECM molecules composed of a protein core to which heparan sulfate chains are attached. Sulfate chains can bind basic fibroblast growth factors (FGF) for storage and protection from proteolytic degradation (Saksela et al., 1988). Bound FGFs can be released in bioactive form by partial proteolysis of the protein core or through digestion of heparan sulfate moieties (Ishai-Michaeli et al., 1990; Saksela and Rifkin, 1990). However, in order to interact with their cell receptors basic FGFs have to be bound by heparan sulfate chains (Rapraeger et al., 1991; Yayon et al., 1991).
Perlecan and agrin are HSPGs involved in modulation of FGFs signaling in the processes of bone formation and neurite outgrowth respectively. Perlecan secreted by chondrocytes localizes to the growth plate of the developing long bones; FGF18 bound to perlecan enhances FGF receptor 3 signaling to control proper cartilage/bone transition zone formation (Chuang et al., 2010). Agrin in neuronal basal laminae binds FGF2 and probably enhances FGF2 affinity to its cellular receptor resulting in enhancement of FGF2 neurite outgrowth stimulation (Kim et al., 2003).
Small leucine-rich proteoglycans (SLRPs) represent a different type of proteoglycan-growth factor interactions. The core proteins of SLRPs decorin, biglycan and fibromodulin were shown to bind to isoforms of TGF-β (Hildebrand et al., 1994), sequestering TGF-β into the ECM for signaling.
Extracellular miRNAs
In addition to cytokines, the extracellular environment also contains hormones and miRNAs. Recent studies have suggested that these extracellular miRNAs can act as regulatory molecules. miRNAs are small non-coding regulatory RNA molecules. In most cases, they act as negative regulators of protein translation by binding to the 3′UTRs of the target mRNA molecules, subjecting them for silencing or degradation (He and Hannon, 2004). miRNAs are encoded by the genome of each organism. They undergo the maturation process first in the nucleus and then in the cytoplasm. Extracellular miRNAs are expressed by a cell but they are secreted into the extracellular environment to act on other cells. They are different from intracellular miRNAs, which act on the same cell that have expressed the miRNA (reviewed in Rayner and Hennessy, 2013).
Components of the ECM and ECM-tethered and free cell-extrinsic stress-induced signaling molecules (cytokines, hormones, and extracellular miRNAs) together constitute the extracellular environment, surrounding each individual cell (summarized in Table 1). This environment is highly dynamic and changes in response to different stimuli. The profile of the signaling molecules may be altered or the composition of the ECM may change, which will also bring a change in the quantity and distribution of certain ECM-associated signaling molecules. Thus, the extracellular environment represents the state of the organism, translated into the language that each individual cell can understand: in the language of cell-extrinsic stress-induced signaling molecules.
How do Extracellular Signals Regulate miRNA Expression
In general, the cell can interpret the extracellular signals because it has various receptors that enable the ECM-bound cell to communicate with the outside world by modulating signal transduction. There are two main types of receptors: (1) cell signaling receptors responsible for binding and transducing signals from cell-extrinsic stress-induced signaling molecules, and (2) cell adhesion molecules that bind and respond to the ECM components.
Cell signaling receptors, such as cytokine receptors (CR), receptor tyrosine kinases (RTK), TGF-β receptors, nuclear receptors (NRs), and receptor phosphotyrosine phosphatases among others, bind growth factors, interleukins, and other cytokines, steroid and protein hormones. Each type of receptor activates specific signaling pathways inside the cell but often pathways activated by different types of receptors overlap. For example, the Ras-mitogen-activated protein (MAP) kinase pathway, and the PI-3 kinase pathway are common to more than one type of receptor (Figure 1). Since many miRNAs are encoded by their own genes, the pathways activated by cell signaling receptors can regulate expression of miRNA-coding genes as well as of protein-coding genes.
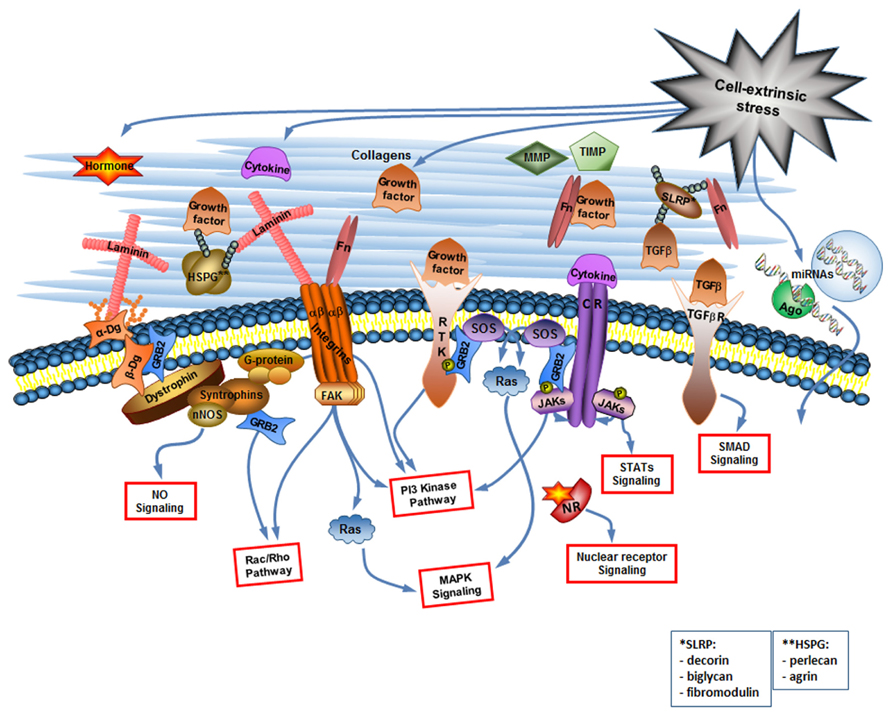
FIGURE 1. The cell-extrinsic stress signaling brings about the changes in the extracellular environment that are read by the cellular sensory apparatus. The extracellular environment is a complex but organized entity composed of structural and enzymatic ECM proteins [collagens, proteoglycans, multiadhesive proteins like laminin and fibronectin (Fn), MMPs and TIMPs] and cell-extrinsic stress-induced signaling molecules including cytokines (considering growth factors as a type of cytokine), hormones, and extracellular miRNAs. The cellular sensory apparatus includes the ECM receptors and receptors for signaling molecules. Integrins and dystroglycan (Dg) are the ECM receptors. They do not only connect cell to its matrix but are also involved in the cell signaling. Multiple receptors for signaling molecules [including receptor tyrosine kinases (RTK), cytokine receptors (CR), TGF-β receptors, and nuclear receptors (NR)] transduce changes in the extracellular environment into the various signaling pathways inside the cell. Extracellular miRNAs can probably be taken up by the cell via yet unknown mechanism. NO, nitric oxide, Ago, argonaute.
Most prominent cell adhesion molecules are the proteins of the integrin family that bind to collagen, perlecan, laminin, and fibronectin. Intracellular integrin signaling can act either through modulation of signaling from growth receptors or through direct activation of intracellular signaling events. Directly activated by integrins are the MAP kinase pathway, the PI3 kinase (PI3K) pathway, and the small GTPases of the Rho family (Keely et al., 1998; Giancotti, 1999; Figure 1). Although integrin signaling has a potential to regulate miRNA expression profile (Gerson et al., 2012), we would like to focus here on another cell adhesion molecule Dg capable of regulating miRNA expression. Dg is a part of the Dystrophin Glycoprotein Complex, associated with a group of fatal inherited diseases muscular dystrophies (Campbell, 1995; Cohn, 2005; Goddeeris et al., 2013). No cure exists for these neuromuscular degenerative diseases, and we believe that better understanding of Dg role in cell-ECM communication will aid in the development of future therapeutics.
Dystroglycan as a Cell Adhesion Molecule
Dystroglycan is a major non-integrin ECM binding receptor. α- and β-Dgs are translated in mammals from the same mRNA transcript and are then separated by posttranslational cleavage and modifications to form a membrane complex with extracellular α-Dg tightly bound to transmembrane β-Dg (Barresi and Campbell, 2006; Figure 1).
α-Dystroglycan can bind to different ECM proteins: laminin, agrin, and perlecan in muscle, and neurolexin in brain (Barresi and Campbell, 2006). Importance of the α-Dg association with the ECM is highlighted by the experimental data that mice with mutation in the gene encoding Dg do not develop into adults due to improper formation of the early embryonic basement membrane (Williamson et al., 1997). In Dg-null murine muscle, α7B integrin receptor subunit is selectively upregulated suggesting that certain types of integrins can partially compensate for the absence of Dg in basement membrane assembly and maintenance (Côté et al., 2002).
Transmembrane β-Dg binds inside the cell to dystrophin in skeletal muscles and to alternative proteins of the dystrophin locus or to an autosomal homolog of dystrophin, utrophin, in non-muscle tissues (Barresi and Campbell, 2006). β-Dg also binds the adaptor protein growth factor receptor-bound protein 2 (Grb2) both in muscle and in brain (Yang et al., 1995). Dystrophin localizes to the plasma membrane another type of adaptor proteins – syntrophins with different syntrophin isoforms being expressed in muscle and neuronal tissues (Bhat et al., 2013).
The interaction partners of Dg and dystrophin, as well as the Dg–dystrophin signaling are extensively studied in mammalian muscle cell culture and in muscle tissue, as well as on the Drosophila muscular dystrophy model (Shcherbata et al., 2007). In the genetic interaction screen on Drosophila, we identified new muscle specific partners of Dg and dystrophin, of which some are implicated in mechanical and stress-induced pathways (Kucherenko et al., 2011). Loss of Dg or dystrophin in Drosophila resulted in altered cellular levels of reactive oxygen species, suggesting the function of these proteins in regulation of homeostasis (Marrone et al., 2011b). In muscle cell culture, phosphorylation of syntrophins due to laminin-1 binding to α-Dg was shown to increase the association between syntrophins and Grb2, resulting in the initiation of Rac1 signaling (Zhou et al., 2006). In skeletal muscle, syntrophins also bind heterotrimeric G-protein. This binding together with laminin attachment to Dg complex is necessary for activation of the PI3K/Akt signaling pathway (Xiong et al., 2009). Interestingly, recent data also links the Dg–dystrophin signaling via syntrophins to the expression of miRNAs in muscle progenitors. Syntrophins can localize neuronal nitric oxide synthase (nNOS) to the muscle cell sarcolemma leading to the production of nitric oxide as a second messenger that nitrosylates histone deacetylase 2 (HDAC2) influencing expression of certain genes, including miRNAs, important for muscle cell differentiation and maintenance (Cacchiarelli et al., 2010).
The nervous system expression of Dg together with dystrophin and syntrophins suggests the role for Dg in the nervous tissue. Importance of the cell-to-ECM adhesion via Dg in the nervous system is supported by the fact that mice with astrocytes-specific deletion of Dg show discontinuities in the pial surface basal lamina (Moore et al., 2002). In the genetic interaction screen on Drosophila brain, we showed that Dg and dystrophin interact with proteins involved in actin cytoskeleton remodeling, which is essential for cell homeostasis (Marrone et al., 2011a). Moreover, defects in the Dg glycosylation that disrupt its association with the ECM cause numerous human diseases, such as muscle-eye-brain disease, Walker–Warburg syndrome, forms of congenital muscular dystrophies, symptoms of which include prominent neurological abnormalities (Cohn, 2005).
Hence, integrins, Dg, and a variety of signaling receptors for the cell-extrinsic stress-induced signaling molecules constitute the major cellular sensory apparatus, allowing the cell to read information encoded by the extracellular environment (Table 1; Figure 1). Signaling pathways activated by the cellular sensory apparatus have a potential to regulate intracellular miRNA expression, examples of which we discuss in the next section.
miRNAs Dependent on the Extracellular Environment
The importance of extracellular environment for regulation of miRNA expression profile was highlighted in the experiments with Matrigel. Matrigel is a 3D cell culture medium composed of the protein mixture secreted by the mouse sarcoma cells, resembling the basement membrane (Kleinman and Martin, 2005). Human cancer cells cultured on the Matrigel have a significantly different miRNA expression profile compared to cells cultured on plastic (Price et al., 2012). Hence, miRNA cellular levels depend on the composition of the extracellular environment. In the similar experimental setup, it was shown that p53 expression levels and nuclear localization are enhanced in human cells cultured on Matrigel (Li et al., 2003). Taking into account that certain miRNAs – miR-34 family members and miR-205– can be directly regulated by p53 (Shi et al., 2008; Piovan et al., 2012), these results again suggest that cellular miRNA profile can change in response to the extracellular environmental composition.
miRNA expression profile can be regulated in the ECM-dependent manner at the transcription level through (a) regulation of epigenetic marks or (b) through regulation of transcription factors’ activity. Additionally, cellular miRNA expression profile can be altered through direct incorporation of extracellular miRNAs.
Epigenetic Modifications
miRNA expression can be regulated through changes in the chromatin modifications. For example, inhibition of histone deacetylases (HDACs) was shown to alter miRNA levels in breast cancer cell line (Scott et al., 2006). Many pathways can lead to HDACs inhibition. One of the signaling mechanisms via HDAC2 inhibition can be suggested to modulate levels of specific miRNAs in muscle progenitor cells depending on the extracellular environment. This pathway involves dystrophin and nNOS localization to plasma membrane by syntrophins. nNOS causes nitric oxide production, S-nitrosylation and subsequent inhibition of HDAC2 that directly controls expression of miR-133/miR-1 and miR-29 (Cacchiarelli et al., 2010). Since dystrophin is tightly bound to Dg, and Dg is an important ECM binding molecule, it can be hypothesized that Dg–dystrophin–syntrophin–nNOS–HDAC2 pathway can be modulated from the extracellular space in response to stimuli to alter miRNA levels.
Though this pathway was described on muscle progenitor cells, it may be important for tuning miRNA expression in non-muscle tissues as well. Dg, dystrophin and syntrophin isoforms were shown to be expressed in non-muscle tissues, like in brain, liver, and kidney (Tinsley et al., 1994; Bhat et al., 2013). Moreover, we have identified on whole-fly Drosophila RNA extracts a group of Dg and dystrophin-dependent stress-response miRNAs. Levels of these miRNAs change under temperature stress in wild type flies. On the contrary, in flies with no functional Dg or dystrophin, no change in the expression of these miRNAs is observed. At least three of such Dg- and dystrophin-dependent stress-response Drosophila miRNAs –miR-252, miR-980, and miR-956– depend also on the levels of syntrophin 1 that is a Drosophila homolog of mammalian syntrophins capable of nNOS localization (Alessi et al., 2006; Marrone et al., 2012). Since miR-252, miR-980, and miR-956 are suggested to have a nervous system expression, the Dg–dystrophin–syntrophin–NOS–HDAC pathway may exist additionally in non-muscle tissues and contribute to the miRNA expression changes under stress.
Modifications Via Transcription Factors
The change in the miRNA transcription due to external stimulation can occur because of the modulation of the availability and activity of certain transcription factors. For example, TGF-β signaling acts via direct activation of cytosolic Smad transcription factors, and miR-29 was shown in human cell culture to be regulated downstream of TGF-β2 pathway (Luna et al., 2011). Interestingly, this same miRNA is involved in the muscle cell differentiation and maintenance program and, as discussed above, is controlled by the Dg–dystrophin–syntrophin–nNOS–HDAC2 pathway.
Various immune-response cytokines act via direct activation of signal transducer and activator of transcription (STAT) factors where STAT dimer is translocated into the nucleus upon phosphorylation by JAK receptors in response to extracellular signaling. miR-21 – a tumor suppressor miRNA associated with many types of cancer – has been shown to be regulated by this signaling pathway since the promoter of the miR-21 gene can be directly bound by STAT3 transcription factor (Löffler et al., 2007).
Many cytokines via Ras-mitogen-activated protein kinase (MAPK) signaling can activate cAMP-response element binding protein (CREB), which binds to promoters and regulates transcription of various genes. The expression of one of the CREB-activated genes in rat neuronal cells miR-132 is turned on in response to neurotrophins stimulation (Vo et al., 2005). Growth factor signaling activates several pathways and kinase cascades, including MAPK signaling cascade, leading to activation of transcription factors specific to the cell type and external stimulus. For example, miR-125 expression is reduced in human lung cancer cell culture due to epidermal growth factor (EGF) stimulation, though it is not yet clear which transcription factors are involved in this response (Wang et al., 2009).
Many steroid hormones belong to the NR family. Inactive NRs are cytoplasmic. Upon hormone binding, they translocate into the nucleus and serve as transcription factors. miRNAs directly activated by NRs exist. For example, human miR-125b-2 contains a functional androgen-responsive element upstream of its gene locus (Shi et al., 2007). In Drosophila, miR-125 belongs to an evolutionary conserved let-7-Complex of three miRNAs, miR-100, let-7, and miR-125 (Pasquinelli et al., 2000). Ecdysteroid hormone signaling was shown to upregulate transcription of let-7-Complex but repress transcription of other Drosophila miRNAs miR-14 and miR-8 (Varghese and Cohen, 2007; Garbuzov and Tatar, 2010; Chawla and Sokol, 2012; Jin et al., 2012; Kucherenko et al., 2012). Such NR-responsive miRNAs can react very fast to hormone stimulation without multiple intermediate activation steps required.
Direct Input from the ECM
In addition to the ECM-dependent modulation of signaling pathways that leads to changes in the intracellular miRNA profile, extracellular miRNAs can be directly incorporated by cells. miRNAs can be prominently secreted from certain tissues, transported via blood by lipid- or protein-carriers and can then be taken up by recipient cells in different tissues (Rayner and Hennessy, 2013). For example, in human endothelial cells atheroprotective shear stress significantly upregulated expression of miR-143/miR-145 cluster. miR-143/miR-145 were then found enriched in exosomes of endothelial cells, secreted by them and taken up by co-cultured smooth muscle cells. This prevented smooth muscle cells from de-differentiation (Hergenreider et al., 2012). Hence, in the organism miR-143/miR-145 secreted from the endothelial cells in response to atheroprotective shear stress may travel via blood to the smooth muscle cells and protect them from malfunction.
miRNAs can be secreted from inflamed tissues, signaling that a possible threat for the whole organism exists. Analysis of plasma from patients allowed to suggest that miR-133/208/499 are secreted from myocardium following acute myocardial infarction (AMI) and circulate in blood packaged into exosomes or microvesicles (Corsten et al., 2010; De Rosa et al., 2011; Olivieri et al., 2013).
miRNAs take part in virtually all critical cellular processes (He and Hannon, 2004), ranging from stem cell division, maintenance and differentiation (Hatfield et al., 2005; Qi et al., 2009; Mathieu and Ruohola-Baker, 2013) to aging of the organism (Inukai and Slack, 2013). We have now summarized some evidence that the constantly tuned and adjusted miRNA profile of cells is regulated downstream of many classical ECM-connected pathways, as well as of a newly discovered ECM-connected Dg–dystrophin–syntrophin–nNOS–HDAC2 pathway (Figure 2).
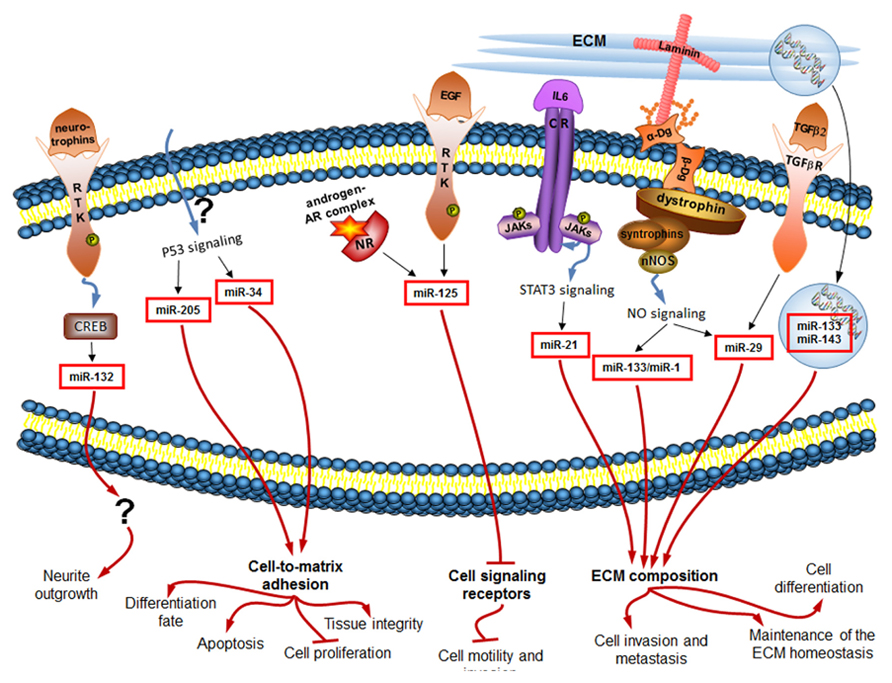
FIGURE 2. miRNAs regulated downstream of signaling induced by changes in the extracellular environment (black arrows) influence in turn the ECM composition and the cellular sensory apparatus, and ultimately the cell behavior (red arrows). Only mammalian miRNAs and pathways are shown.
Altered miRNA Profile Changes Cell Behavior
miRNAs differentially expressed in response to ECM-dependent signaling may have various functions in the cell. Interestingly, a proportion of those ECM-regulated miRNAs was shown to feedback to alter the ECM composition or the cellular sensory apparatus (Figure 2). In this section, we would like to highlight how each of the introduced in the previous section miRNAs modulates cell behavior by acting either on the components of the extracellular environment or on the cell adhesion molecules/cell signaling receptors (Table 1).
Modulation of Cell Behavior Via Extracellular Environment
The ECM is a remarkably complex still organized entity. Its careful regulation is essential for proper functioning of cells and subsequently tissues, organs and the whole organism. miRNAs can regulate the ECM composition. For example, the above discussed TGF-β2 signaling-dependent miR-29 family members were shown to play a role in the maintenance of the ECM homeostasis. Expression of miR-29 members in human cell culture resulted in significant reduction of the ECM components, such as laminin, fibronectin, collagen I, collagen IV, and SPRC (secreted protein, acidic, and rich in cysteine; Villarreal et al., 2011). miR-133, which is secreted from diseased myocardium following AMI, was shown in the experiments in rats and in human cell culture to directly target 3′UTR of the pro-α1 chains of type I collagen, changing the ECM properties of the recipient tissues (Castoldi et al., 2012). Interestingly, miR-133 is also one of the miRNAs activated by the Dg–dystrophin–syntrophin–nNOS–HDAC2 pathway in muscle progenitor cells, and it is known there to inhibit cell proliferation via inhibition of the serum response factor, and to induce cell differentiation via HDAC4 inhibition [as summarized in (Marrone and Shcherbata, 2011)].
Fibronectin is a high-molecular weight ECM glycoprotein that binds integrin receptors modulating their signaling activity thus playing an important role in cell adhesion, growth, differentiation, and migration. Fibronectin mRNA can be targeted by miRNAs. In human cell culture experiments miR-143 was shown to directly target the 3′UTR of the fibronectin type III domain containing protein 3B – molecule that regulates cell motility. By repressing fibronectin expression, high levels of miR-143 induce invasive and metastatic behavior of liver tumors in mice (Zhang et al., 2009). It is interesting to note, that miR-143 is one of the miRNAs found in the extracellular space packaged into exosomes (Hergenreider et al., 2012). Since miR-143 is an onco-miRNA, cancer cells may communicate and promote each other’s metastatic and invasive properties via secretion of this circulating miRNA.
For proper ECM maintenance, availability and activity of ECM modulating enzymes MMPs and TIMPs are of utmost significance. miRNAs were shown to control their expression. Elevated levels of the IL-6 dependent oncogenic miR-21 were shown to inhibit MMP inhibitors, promoting MMP activity, cancer invasion, and metastasis of glioblastoma in mouse model (Gabriely et al., 2008). Another study on human tissues suggested that miR-21 induces inhibition of TIMP3, and additionally of programmed cell death 4 protein, accounting for increased invasiveness and reduced apoptosis in cholangiocarcinomas (Selaru et al., 2009).
Signaling molecules are a part of the extracellular environment, and miRNA signaling can target such molecules. For example, expression of the insulin-like peptide in Drosophila, secreted by insulin-producing cells in the fly brain and involved in control of energy homeostasis, is regulated by miR-14. It was recently reported that miR-14 directly targets in insulin-producing cells a negative regulator of insulin-like peptide gene expression. Thus, ecdysteroid-dependent miR-14 provides a link between steroid hormone signaling and insulin secretion to allow nutrient-independent insulin-production control (Varghese et al., 2010). Another Drosophila miRNA miR-8, repressed by ecdysteroid signaling, is involved in innate immune homeostasis control. It keeps antimicrobial peptides’ expression by fly innate immune system organ, fat body, at low basal level. Hence, in pathogen-free flies miR-8 is suggested to keep immune system from firing and thus to prevent autoimmune reactions (Choi and Hyun, 2012).
Modulation of Cell Behavior Via Cell Adhesion Molecules and Signaling Receptors
As important as the extracellular environment is for cell behavior, proper cell functioning depends also on appropriate cell-to-ECM attachment and signaling. miRNAs play a role in regulation of cell adhesion and signaling as well. For example, it was shown that silencing of the p53-induced miR-205 in human prostatic cell line reduces secretion of all laminin-332 subunits and integrin-β4 – critical cell-to-ECM adhesion molecules in control of tissue integrity. Hence, loss of miR-205 in prostate cancer results in basement membrane discontinuities (Guess and Quaranta, 2009; Gandellini et al., 2012). Another p53-induced miR-34a can directly target CD44 in human cancer cell lines. CD44 is a glycoprotein expressed on cell surfaces and capable of ECM binding, playing a role in cell migration and adhesion. miR-34a-induced decrease in CD44 levels was shown to inhibit prostate cancer regeneration and metastasis (Liu et al., 2011). In another work on mice, directed delivery of miR-34a to cancer cells with nanovector resulted in induced apoptosis, decreased proliferation, and ultimately inhibition of tumor growth (Pramanik et al., 2011).
Proper cell adhesion is important not only for stable differentiated state of the cell as opposed to de-differentiation during cancerogenesis, but also for the early development and initial differentiation of the cell. Neuronal differentiation is a good model for emphasizing the importance of cell adhesion in the complex process of nervous system development. When neurons differentiate, they send their axons to defined places and establish contacts with specific cells and ECM components due to tightly controlled cell adhesion process. miRNAs fine-tune this process. For example, in rat cortical neurons miR-132 expression in response to stimulation with neurotrophins results in neurite outgrowth (Vo et al., 2005). Ecdysteroid induced miRNA let-7 in the developing Drosophila brain controls via a cytokine-dependent transcription factor expression levels of the neural cell adhesion molecule Fasciclin II, ultimately regulating neurons’ differentiation fate (Kucherenko et al., 2012; discussed in Kucherenko and Shcherbata, 2013). Interestingly, in the differentiated neuron the target of let-7 is a negative regulator of ecdysteroid signaling. Thus, let-7 is involved in the positive feedback loop to enhance its own expression level.
miR-125 gives an example of a miRNA that regulates expression of cell signaling receptors. In human cancer cell line, miR-125 members are inhibited due to EGF stimulation (Wang et al., 2009). Interestingly, miR-125 overexpression results in the reduction of the transcript and protein levels of the EGF receptors themselves, significantly reducing cell motility and invasion (Scott et al., 2007). Hence, miR-125 is also involved in the positive feedback loop down-regulating its own inhibitor, and leading to inhibition of certain properties of cancerous cells.
Concluding Remarks
The extracellular environment is a complex system of ECM components and signaling molecules. It undergoes constant rearrangements following different kinds of stress. Cells have developed a sensitive apparatus to respond to the changes of the extracellular environment by tuning the expression of genes, including the miRNA-coding genes. Interestingly, some of those miRNAs feedback to affect the ECM and cellular sensory apparatus with the ultimate goal being, to change the cell behavior. This further widens the idea of the “dynamic reciprocation” between the cell and its extracellular environment under stress (Bissell et al., 1982).
Although it is the extreme cases of miRNAs influencing cell behavior (like invasion, metastasis and apoptosis) that are mostly studied, we can hypothesize that under mild stress, miRNAs together with other effector molecules in an affected cell coordinate their work to return the organism to its initial homeostasis. The stronger and longer the stress is – the harder it is for the cell to constantly shift back to its initial state. Then, a new altered homeostasis is obtained. The new homeostasis represents the stress-induced state of the cell and though in most cases favorable, it can sometimes lead to cell transformations resulting in, for example, cancer. Under extreme levels of stress, a cell may enter apoptosis or turn into anastasis (Figure 3).
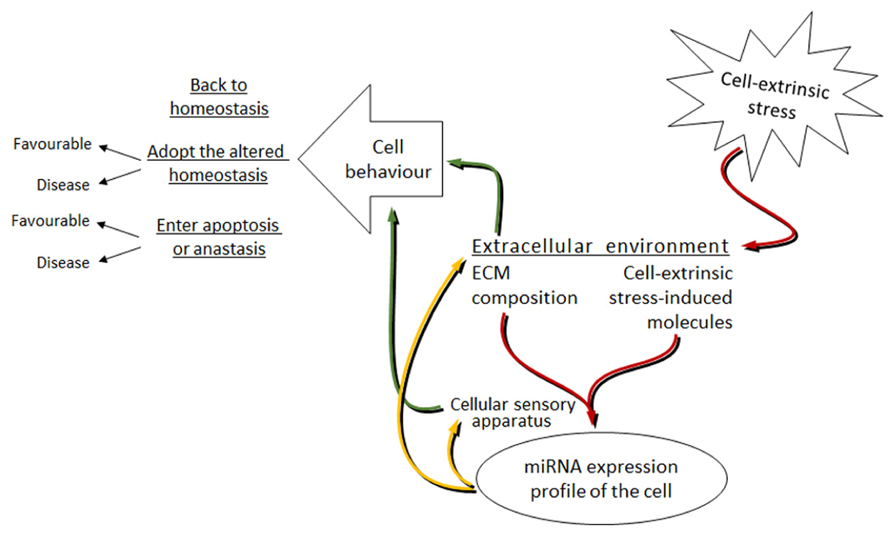
FIGURE 3. Proposed model of the miRNAs involvement in the bi-directional signal transduction between the cell and extracellular environment under cell-extrinsic stress. Red arrows: cell-extrinsic stress changes the extracellular environment of cells in the organism. The ECM composition and the cell-extrinsic stress-induced molecules, as parts of the altered extracellular environment, signal via the cellular sensory apparatus to change the miRNA expression profile of cells. Yellow arrows: altered miRNA levels target the cellular sensory apparatus and the extracellular environment. Green arrows: this leads to the changes in cell behavior. The mode of cell behavior is different dependent on the strength and length of the applied cell-extrinsic stress.
In this review, we focused on the ECM bound cells and on their reaction toward the cell-extrinsic stress. The general picture is much more complex. Many cells do not have contacts to the ECM and organize into three-dimensional structures due to multiple cell-to-cell contacts. In addition to cell-extrinsic stress, cell-intrinsic stress, independent from extracellular environmental signaling, can lead to the changes in cell behavior. In those cases, the cellular transcriptome, including the miRNA profile, will be the one to change first. It may then be translated into the changes in the cell behavior and at the same time into the changes in the ECM composition and cell adhesion. Both may again change the cellular transcriptome and miRNA profile, and ultimately fine-tune the cell behavior.
With the growing field of miRNA research, further studies will identify new ECM-dependent pathways or will prove that canonical pathways regulate miRNA expression. As miRNAs seem to be ubiquitous to all cellular processes, for many of them a role in the regulation of the ECM composition and the cellular sensory apparatus will be attributed. This will firmly consolidate the role of miRNAs in the bi-directional signal transduction process between the cell and its exterior.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to researchers whose studies we were unable to cite due to the space limitation of this review. We would like to thank all members of Halyna Shcherbata lab, Kanika Vanshylla, Anita Smarandache, and Sumana Sharma for careful reading and valuable comments on the manuscript, and Max Planck Society for financial support. We are grateful to Qiagen for the inspiration in drawing Figures 1 and 2.
References
Alessi, A., Bragg, A. D., Percival, J. M., Yoo, J., Albrecht, D. E., Froehner, S. C., et al. (2006). gamma-Syntrophin scaffolding is spatially and functionally distinct from that of the alpha/beta syntrophins. Exp. Cell Res. 312, 3084–3095. doi: 10.1016/j.yexcr.2006.06.019
Ariel, A., Hershkoviz, R., Cahalon, L., Williams, D. E., Akiyama, S. K., Yamada, K. M., et al. (1997). Induction of T cell adhesion to extracellular matrix or endothelial cell ligands by soluble or matrix-bound interleukin-7. Eur. J. Immunol. 27, 2562–2570. doi: 10.1002/eji.1830271015
Barresi, R., and Campbell, K. P. (2006). Dystroglycan: from biosynthesis to pathogenesis of human disease. J. Cell Sci. 119, 199–207. doi: 10.1242/jcs.02814
Bhat, H. F., Adams, M. E., and Khanday, F. A. (2013). Syntrophin proteins as Santa Claus: role(s) in cell signal transduction. Cell. Mol. Life Sci. 70, 2533–2554. doi: 10.1007/s00018-012-1233-9
Bissell, M. J., Hall, H. G., and Parry, G. (1982). How does the extracellular matrix direct gene expression? J. Theor. Biol. 99, 31–68. doi: 10.1016/0022-5193(82)90388-5
Cacchiarelli, D., Martone, J., Girardi, E., Cesana, M., Incitti, T., Morlando, M., et al. (2010). MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 12, 341–351. doi: 10.1016/j.cmet.2010.07.008
Campbell, K. P. (1995). Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell 80, 675–679. doi: 10.1016/0092-8674(95)90344-5
Castoldi, G., Di Gioia, C. R. T., Bombardi, C., Catalucci, D., Corradi, B., Gualazzi, M. G., et al. (2012). MiR-133a regulates collagen 1A1: potential role of miR-133a in myocardial fibrosis in angiotensin II-dependent hypertension. J. Cell. Physiol. 227, 850–856. doi: 10.1002/jcp.22939
Chawla, G., and Sokol, N. S. (2012). Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development 139, 1788–1797. doi: 10.1242/dev.077743
Choi, I. K., and Hyun, S. (2012). Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev. Comp. Immunol. 37, 50–54. doi: 10.1016/j.dci.2011.12.008
Chuang, C. Y., Lord, M. S., Melrose, J., Rees, M. D., Knox, S. M., Freeman, C., et al. (2010). Heparan sulfate-dependent signaling of fibroblast growth factor 18 by chondrocyte-derived perlecan. Biochemistry 49, 5524–5532. doi: 10.1021/bi1005199
Cohn, R. D. (2005). Dystroglycan: important player in skeletal muscle and beyond. Neuromuscul. Disord. 15, 207–217. doi: 10.1016/j.nmd.2004.11.005
Corsten, M. F., Dennert, R., Jochems, S., Kuznetsova, T., Devaux, Y., Hofstra, L., et al. (2010). Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 3, 499–506. doi: 10.1161/CIRCGENETICS.110.957415
Côté, P. D., Moukhles, H., and Carbonetto, S. (2002). Dystroglycan is not required for localization of dystrophin, syntrophin, and neuronal nitric-oxide synthase at the sarcolemma but regulates integrin alpha 7B expression and caveolin-3 distribution. J. Biol. Chem. 277, 4672–4679. doi: 10.1074/jbc.M106879200
De Rosa, S., Fichtlscherer, S., Lehmann, R., Assmus, B., Dimmeler, S., and Zeiher, A. M. (2011). Transcoronary concentration gradients of circulating microRNAs. Circulation 124, 1936–1944. doi: 10.1161/CIRCULATIONAHA.111.037572
Gabriely, G., Wurdinger, T., Kesari, S., Esau, C. C., Burchard, J., Linsley, P. S., et al. (2008). MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 28, 5369–5380. doi: 10.1128/MCB.00479-08
Gandellini, P., Profumo, V., Casamichele, A., Fenderico, N., Borrelli, S., Petrovich, G., et al. (2012). miR-205 regulates basement membrane deposition in human prostate: implications for cancer development. Cell Death Differ. 19, 1750–1760. doi: 10.1038/cdd.2012.56
Garbuzov, A., and Tatar, M. (2010) Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly (Austin) 4, 306–311. doi: 10.4161/fly.4.4.13008
Gerson, K. D., Shearstone, J. R., Maddula, V. S. R. K., Seligmann, B. E., and Mercurio, A. M. (2012). Integrin β 4 regulates SPARC protein to promote invasion. J. Biol. Chem. 287, 9835–9844. doi: 10.1074/jbc.M111.317727
Giancotti, F. G. (1999). Integrin signaling. Science 285, 1028–1033. doi: 10.1126/science.285.5430.1028
Goddeeris, M. M., Wu, B., Venzke, D., Yoshida-Moriguchi, T., Saito, F., Matsumura, K., et al. (2013). LARGE glycans on dystroglycan function as a tunable matrix scaffold to prevent dystrophy. Nature 503, 136–140. doi: 10.1038/nature12605
Guess, C. M., and Quaranta, V. (2009). Defining the role of laminin-332 in carcinoma. Matrix Biol. 28, 445–455. doi: 10.1016/j.matbio.2009.07.008
Hatfield, S. D., Shcherbata, H. R., Fischer, K. A., Nakahara, K., Carthew, R. W., and Ruohola-Baker, H. (2005). Stem cell division is regulated by the microRNA pathway. Nature 435, 974–978. doi: 10.1038/nature03816
He, L., and Hannon, G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. doi: 10.1038/nrg1379
Hergenreider, E., Heydt, S., Tréguer, K., Boettger, T., Horrevoets, A. J. G., Zeiher, A. M., et al. (2012). Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 14, 249–256. doi: 10.1038/ncb2441
Hershkoviz, R., Cahalon, L., Miron, S., Alon, R., Sapir, T., Akiyama, S. K., et al. (1994). TNF-alpha associated with fibronectin enhances phorbol myristate acetate- or antigen-mediated integrin-dependent adhesion of CD4+ T cells via protein tyrosine phosphorylation. J. Immunol. 153, 554–565.
Hildebrand, A., Romarís, M., Rasmussen, L. M., Heinegård, D., Twardzik, D. R., Border, W. A., et al. (1994). Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem. J. 302, 527–534.
Inukai, S., and Slack, F. (2013). MicroRNAs and the genetic network in aging. J. Mol. Biol. 425, 3601–3608. doi: 10.1016/j.jmb.2013.01.023
Ishai-Michaeli, R., Eldor, A., and Vlodavsky, I. (1990). Heparanase activity expressed by platelets, neutrophils, and lymphoma cells releases active fibroblast growth factor from extracellular matrix. Cell Regul. 1, 833–842.
Jin, H., Kim, V. N., and Hyun, S. (2012). Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 26, 1427–1432. doi: 10.1101/gad.192872.112
Keely, P., Parise, L., and Juliano, R. (1998). Integrins and GTPases in tumour cell growth, motility and invasion. Trends Cell Biol. 8, 101–106. doi: 10.1016/S0962-8924(97)01219-1
Kim, M. J., Cotman, S. L., Halfter, W., and Cole, G. J. (2003). The heparan sulfate proteoglycan agrin modulates neurite outgrowth mediated by FGF-2. J. Neurobiol. 55, 261–277. doi: 10.1002/neu.10213
Kleinman, H. K., and Martin, G. R. (2005). Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15, 378–386. doi: 10.1016/j.semcancer.2005.05.004
Kucherenko, M. M., Barth, J., Fiala, A., and Shcherbata, H. R. (2012). Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 31, 4511–4523. doi: 10.1038/emboj.2012.298
Kucherenko, M. M., Marrone, A. K., Rishko, V. M., Magliarelli, H. de. F., and Shcherbata, H. R. (2011). Stress and muscular dystrophy: a genetic screen for dystroglycan and dystrophin interactors in Drosophila identifies cellular stress response components. Dev. Biol. 352, 228–242. doi: 10.1016/j.ydbio.2011.01.013
Kucherenko, M. M., and Shcherbata, H. R. (2013). Steroids as external temporal codes act via miRNAs and cooperate with cytokines in differential neurogenesis. Fly (Austin) 7, 8–7. doi: 10.4161/fly.25241
Larraín, J., Bachiller, D., Lu, B., Agius, E., Piccolo, S., and De Robertis, E. M. (2000). BMP-binding modules in chordin: a model for signalling regulation in the extracellular space. Development 127, 821–830.
Li, L.-N., Wang, D.-R., Sato, M., Kojima, N., Imai, K., Higashi, N., et al. (2003). Extracellular matrix-regulated p53 expression and nuclear localization in cultured Detroit 562 cells derived from pharyngeal carcinoma. Arch. Histol. Cytol. 66, 419–428. doi: 10.1679/aohc.66.419
Liu, C., Kelnar, K., Liu, B., Chen, X., Calhoun-Davis, T., Li, H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211–215. doi: 10.1038/nm.2284
Löffler, D., Brocke-Heidrich, K., Pfeifer, G., Stocsits, C., Hackermüller, J., Kretzschmar, A. K., et al. (2007). Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 110, 1330–1333. doi: 10.1182/blood-2007-03-081133
Luna, C., Li, G., Qiu, J., Epstein, D. L., and Gonzalez, P. (2011). Cross-talk between miR-29 and transforming growth factor-betas in trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci. 52, 3567–3572. doi: 10.1167/iovs.10-6448
Marrone, A. K., Edeleva, E. V., Kucherenko, M. M., Hsiao, N. H., and Shcherbata, H. R. (2012). Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol. 13:26. doi: 10.1186/1471-2121-13-26
Marrone, A. K., Kucherenko, M. M., Rishko, V. M., and Shcherbata, H. R. (2011a). New dystrophin/dystroglycan interactors control neuron behavior in Drosophila eye. BMC Neurosci. 12:93. doi: 10.1186/1471-2202-12-93
Marrone, A. K., Kucherenko, M. M., Wiek, R., Göpfert, M. C., and Shcherbata, H. R. (2011b). Hyperthermic seizures and aberrant cellular homeostasis in Drosophila dystrophic muscles. Sci. Rep. 1, 47. doi: 10.1038/srep00047
Marrone, A. K., and Shcherbata, H. R. (2011). Dystrophin orchestrates the epigenetic profile of muscle cells via miRNAs. Front. Genet. 2:64. doi: 10.3389/fgene.2011.00064
Mathieu, J., and Ruohola-Baker, H. (2013). Regulation of stem cell populations by microRNAs. Adv. Exp. Med. Biol. 786, 329–351. doi: 10.1007/978-94-007-6621-1_18
Moore, S. A., Saito, F., Chen, J., Michele, D. E., Henry, M. D., Messing, A., et al. (2002). Deletion of brain dystroglycan recapitulates aspects of congenital muscular dystrophy. Nature 418, 422–425. doi: 10.1038/nature00838
Olivieri, F., Antonicelli, R., Capogrossi, M. C., and Procopio, A. D. (2013). Circulating microRNAs (miRs) for diagnosing acute myocardial infarction: an exciting challenge. Int. J. Cardiol. 167, 3028–3029. doi: 10.1016/j.ijcard.2012.11.103
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. doi: 10.1038/35040556
Piovan, C., Palmieri, D., Di Leva, G., Braccioli, L., Casalini, P., Nuovo, G., et al. (2012). Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol. Oncol. 6, 458–472. doi: 10.1016/j.molonc.2012.03.003
Pramanik, D., Campbell, N. R., Karikari, C., Chivukula, R., Kent, O. A., Mendell, J. T., et al. (2011). Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 10, 1470–1480. doi: 10.1158/1535-7163.MCT-11-0152
Price, K. J., Tsykin, A., Giles, K. M., Sladic, R. T., Epis, M. R., Ganss, R., et al. (2012). Matrigel basement membrane matrix influences expression of microRNAs in cancer cell lines. Biochem. Biophys. Res. Commun. 427, 343–348. doi: 10.1016/j.bbrc.2012.09.059
Qi, J., Yu, J.-Y., Shcherbata, H. R., Mathieu, J., Wang, A. J., Seal, S., et al. (2009). microRNAs regulate human embryonic stem cell division. Cell Cycle 8, 3729–3741. doi: 10.4161/cc.8.22.10033
Rapraeger, A. C., Krufka, A., and Olwin, B. B. (1991). Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708. doi: 10.1126/science.1646484
Rayner, K. J., and Hennessy, E. J. (2013). Extracellular communication via microRNA: lipid particles have a new message. J. Lipid Res. 54, 1174–1181. doi: 10.1194/jlr.R034991
Saksela, O., Moscatelli, D., Sommer, A., and Rifkin, D. B. (1988). Endothelial cell-derived heparan sulfate binds basic fibroblast growth factor and protects it from proteolytic degradation. J. Cell Biol. 107, 743–751. doi: 10.1083/jcb.107.2.743
Saksela, O., and Rifkin, D. B. (1990). Release of basic fibroblast growth factor-heparan sulfate complexes from endothelial cells by plasminogen activator-mediated proteolytic activity. J. Cell Biol. 110, 767–775. doi: 10.1083/jcb.110.3.767
Schuppan, D., Schmid, M., Somasundaram, R., Ackermann, R., Ruehl, M., Nakamura, T., et al. (1998). Collagens in the liver extracellular matrix bind hepatocyte growth factor. Gastroenterology 114, 139–152. doi: 10.1016/S0016-5085(98)70642-0
Scott, G. K., Goga, A., Bhaumik, D., Berger, C. E., Sullivan, C. S., and Benz, C. C. (2007). Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 282, 1479–1486. doi: 10.1074/jbc.M609383200
Scott, G. K., Mattie, M. D., Berger, C. E., Benz, S. C., and Benz, C. C. (2006). Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 66, 1277–1281. doi: 10.1158/0008-5472.CAN-05-3632
Selaru, F. M., Olaru, A. V., Kan, T., David, S., Cheng, Y., Mori, Y., et al. (2009). MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 49, 1595–1601. doi: 10.1002/hep.22838
Shcherbata, H. R., Yatsenko, A. S., Patterson, L., Sood, V. D., Nudel, U., Yaffe, D., et al. (2007). Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 26, 481–493. doi: 10.1038/sj.emboj.7601503
Shi, X.-B., Tepper, C. G., and deVere White, R. W. (2008). Cancerous miRNAs and their regulation. Cell Cycle 7, 1529–1538. doi: 10.4161/cc.7.11.5977
Shi, X.-B., Xue, L., Yang, J., Ma, A.-H., Zhao, J., Xu, M., et al. (2007). An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 19983–19988. doi: 10.1073/pnas.0706641104
Somasundaram, R., and Schuppan, D. (1996). Type I, II, III, IV, V, and VI collagens serve as extracellular ligands for the isoforms of platelet-derived growth factor (AA, BB, and AB). J. Biol. Chem. 271, 26884–26891. doi: 10.1074/jbc.271.43.26884
Tinsley, J. M., Blake, D. J., Zuellig, R. A., and Davies, K. E. (1994). Increasing complexity of the dystrophin-associated protein complex. Proc. Natl. Acad. Sci. U.S.A. 91, 8307–8313. doi: 10.1073/pnas.91.18.8307
Varghese, J., and Cohen, S. M. (2007). microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 21, 2277–2282. doi: 10.1101/gad.439807
Varghese, J., Lim, S. F., and Cohen, S. M. (2010). Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 24, 2748–2753. doi: 10.1101/gad.1995910
Villarreal, G., Oh, D.-J., Kang, M. H., and Rhee, D. J. (2011). Coordinated regulation of extracellular matrix synthesis by the microRNA-29 family in the trabecular meshwork. Invest. Ophthalmol. Vis. Sci. 52, 3391–3397. doi: 10.1167/iovs.10-6165
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., et al. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431. doi: 10.1073/pnas.0508448102
Wang, G., Mao, W., Zheng, S., and Ye, J. (2009). Epidermal growth factor receptor-regulated miR-125a-5p – a metastatic inhibitor of lung cancer. FEBS J. 276, 5571–5578. doi: 10.1111/j.1742-4658.2009.07238.x
Wijelath, E. S., Rahman, S., Namekata, M., Murray, J., Nishimura, T., Mostafavi-Pour, Z., et al. (2006). Heparin-II domain of fibronectin is a vascular endothelial growth factor-binding domain: enhancement of VEGF biological activity by a singular growth factor/matrix protein synergism. Circ. Res. 99, 853–860. doi: 10.1161/01.RES.0000246849.17887.66
Williamson, R. A., Henry, M. D., Daniels, K. J., Hrstka, R. F., Lee, J. C., Sunada, Y., et al. (1997). Dystroglycan is essential for early embryonic development: disruption of Reichert’s membrane in Dag1-null mice. Hum. Mol. Genet. 6, 831–841. doi: 10.1093/hmg/6.6.831
Xiong, Y., Zhou, Y., and Jarrett, H. W. (2009). Dystrophin glycoprotein complex-associated Gbetagamma subunits activate phosphatidylinositol-3-kinase/Akt signaling in skeletal muscle in a laminin-dependent manner. J. Cell. Physiol. 219, 402–414. doi: 10.1002/jcp.21684
Yang, B., Jung, D., Motto, D., Meyer, J., Koretzky, G., and Campbell, K. P. (1995). SH3 domain-mediated interaction of dystroglycan and Grb2. J. Biol. Chem. 270, 11711–11714. doi: 10.1074/jbc.270.20.11711
Yayon, A., Klagsbrun, M., Esko, J. D., Leder, P., and Ornitz, D. M. (1991). Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848. doi: 10.1016/0092-8674(91)90512-W
Yu, W. H., and Woessner, J. F. (2000). Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7). J. Biol. Chem. 275, 4183–4191. doi: 10.1074/jbc.275.6.4183 t
Yu, W. H., Yu, S., Meng, Q., Brew, K., and Woessner, J. F. (2000). TIMP-3 binds to sulfated glycosaminoglycans of the extracellular matrix. J. Biol. Chem. 275, 31226–31232. doi: 10.1074/jbc.M000907200
Zhang, X., Liu, S., Hu, T., Liu, S., He, Y., and Sun, S. (2009). Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50, 490–499. doi: 10.1002/hep.23008
Zhou, Y. W., Thomason, D. B., Gullberg, D., and Jarrett, H. W. (2006). Binding of laminin alpha1-chain LG4-5 domain to alpha-dystroglycan causes tyrosine phosphorylation of syntrophin to initiate Rac1 signaling. Biochemistry 45, 2042–2052. doi: 10.1021/bi0519957
Keywords: miRNAs, extrinsic stress, ECM composition, bi-directional signal transduction, dystroglycan
Citation: Edeleva EV and Shcherbata HR (2013) Stress-induced ECM alteration modulates cellular microRNAs that feedback to readjust the extracellular environment and cell behavior. Front. Genet. 4:305. doi: 10.3389/fgene.2013.00305
Received: 20 September 2013; Accepted: 16 December 2013;
Published online: 31 December 2013.
Edited by:
Patrick McGowan, University of Toronto, CanadaReviewed by:
Vincent Edward Sollars, Marshall University, USADonald Palmer, University of London, UK
Copyright © 2013 Edeleva and Shcherbata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Halyna R. Shcherbata, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany e-mail:aGFseW5hLnNoY2hlcmJhdGFAbXBpYnBjLm1wZy5kZQ==
 Evgeniia V. Edeleva
Evgeniia V. Edeleva Halyna R. Shcherbata
Halyna R. Shcherbata