
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Genet. , 20 December 2013
Sec. RNA
Volume 4 - 2013 | https://doi.org/10.3389/fgene.2013.00302
This article is part of the Research Topic The Ccr4-Not complex: an integrator of gene expression that connects nucleus and cytoplasm in eukaryotic cells View all 10 articles
The fungal Ccr4-NOT complex has been implicated in orchestrating gene expression networks that impact on pathways key for virulence in pathogenic species. The activity of Ccr4-NOT regulates cell wall integrity, antifungal drug susceptibility, adaptation to host temperature, and the developmental switches that enable the formation of pathogenic structures, such as filamentous hyphae. Moreover, Ccr4-NOT impacts on DNA repair pathways and genome stability, opening the possibility that this gene regulator could control adaptive responses in pathogens that are driven by chromosomal alterations. Here we provide a synthesis of the cellular roles of the fungal Ccr4-NOT, focusing on pathways important for virulence toward animals. Our review is based on studies in models yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, and two species that cause serious human infections, Candida albicans and Cryptococcus neoformans. We hypothesize that the activity of Ccr4-NOT could be targeted for future antifungal drug discovery, a proposition supported by the fact that inactivation of the genes encoding subunits of Ccr4-NOT in C. albicans and C. neoformans reduces virulence in the mouse infection model. We performed bioinformatics analysis to identify similarities and differences between Ccr4-NOT subunits in fungi and animals, and discuss this knowledge in the context of future antifungal strategies.
Model fungal species, such as the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, have served as powerful genetic models for elucidating the fundamental principles of eukaryotic cellular and molecular biology. In addition to these benign species, the fungal kingdom also contains a number of species that are pathogens of humans, other animals and plants. The pathogenic fungal species have profound effects on the ecosystem, as well as on human health (Brown et al., 2012; Fisher et al., 2012; Kupferschmidt, 2012). In regards to human disease, a small number of species is responsible for most infections, such as Candida sp, the mold Aspergillus fumigatus, and the basidiomyces yeasts belonging to the Cryptococcus species complex (Brown et al., 2012). In most cases, life-threatening disease is seen in hosts with immunocompromised status; therefore fungal infections have become a reason for substantial concern due to the AIDS epidemic and new generation medical procedures that compromise immune function. There are very few treatment options, and mortality from systemic fungal disease remains high in both the developed, and the developing world (Pfaller and Diekema, 2010; Brown et al., 2012). Key mechanisms that impact on the ability of fungi to successfully infect human hosts, survive immune attack, and resist treatments with antifungal drugs include adaptation to growth at host temperature, the ability to mount adaptive responses, genomic and metabolic plasticity, and the capacity to change cell morphology and build and remodel the cell surface, reviewed in Cooney and Klein (2008), Selmecki et al. (2010), Gow and Hube (2012), O'Meara and Alspaugh (2012), Morrow and Fraser (2013). Key regulators have been identified that control these pathogenesis-enabling mechanisms, and many act by modulating gene expression to enable the transitions in cell physiology that favor pathogen survival and virulence. One such regulator is the Ccr4-NOT complex.
Ccr4-NOT is a multisubunit, multifunctional eukaryotic regulator, which has been proposed to serve as a control node for the integration of environmental signals that impinge on cell physiology, as well as acting to coordinate multiple nuclear and cytoplasmic steps in gene expression, reviewed in Collart and Timmers (2004), Collart and Panasenko (2012), Collart et al. (2013). The core Ccr4-NOT complex is composed of nine subunits, which perform roles in gene transcription (initiation and elongation) (Bai et al., 1999; Badarinarayana et al., 2000; Denis et al., 2001; Deluen et al., 2002; Swanson et al., 2003; Qiu et al., 2004; Kruk et al., 2011), posttranscriptional regulation of mRNA stability via poly(A) tail shortening or deadenylation (Tucker et al., 2001, 2002; Chen et al., 2002), as well as several other functions in gene regulation, such as export of mRNAs from the nucleus to the cytoplasm (Kerr et al., 2011), and quality control through interactions with the exosome-dependent pathway (Azzouz et al., 2009a). The architecture is such that the large Not1 subunit serves as a scaffold that brings together the mRNA deadenylases Ccr4 and Caf1 (also known as Pop2) (Bai et al., 1999; Tucker et al., 2001, 2002; Chen et al., 2002), the Not2-5 subunits that mainly function in transcription and contain the Not4 E3 ubiquitin ligase (Collart and Struhl, 1993, 1994; Liu et al., 1998; Bai et al., 1999; Badarinarayana et al., 2000; Hanzawa et al., 2001; Deluen et al., 2002; Panasenko et al., 2006), and Caf40 and Caf130 (Chen et al., 2001). Such complexity of functions in gene regulation, coupled with the fact that Ccr4-NOT activity impacts on the transcriptome in a global way (Cui et al., 2008; Azzouz et al., 2009b; Dagley et al., 2011), means that mutations in the genes encoding Ccr4-NOT subunits lead to pleiotropic phenotypes. In fungi these phenotypes include several that have important implications for virulence of pathogenic species.
Here we provide a synthesis of the knowledge of Ccr4-NOT roles in fungal biology and virulence. We draw on work from S. cerevisiae, as the Ccr4-NOT complex has been mostly studied in this model species, and also review what we know about Ccr4-NOT in pathogenic species. We argue that the pleiotropic functions of this complex are beneficial in the context of fungal pathogenesis in two ways: (i) by studying Ccr4-NOT, we can uncover novel mechanistic links between cellular pathways that impact on fungal virulence, and (ii) the pleiotropic functions could be beneficial for targeting the complex with antifungal molecules—inactivating Ccr4-NOT could simultaneously cripple several functions important for virulence, resulting in more efficient elimination of the pathogen. We therefore hypothesize that the Ccr4-NOT complex could be a potentially attractive target for antifungal drug discovery. The Ccr4-NOT complex is conserved throughout eukaryotes, including between yeast and man. This poses a potential problem, as inhibitors of its activity could be toxic to the host. We performed bioinformatics analysis to revisit the conservation between the fungal and animal Ccr4-NOT subunits, and identified several differences that could be exploited for targeting the fungal complex specifically.
The fungal cell wall is an essential protective structure built from carbohydrates (glucans and chitin), and glycosylated cell wall proteins. The cell wall mediates interactions that enable attachment of fungi to host cells, adherence to medical implants for the formation of drug resistant biofilms, and immune recognition (Van De Veerdonk et al., 2008; Finkel and Mitchell, 2011; Gow and Hube, 2012). The cell wall is not present on human cells, and as such is an attractive target for antifungal drug therapy. The echinocandin drugs, which are used to treat fungal infections, inhibit cell wall synthesis by acting on the enzyme 1,3 ß glucan synthase (Douglas et al., 1997).
Early work in S. cerevisiae has shown that one of the most prominent phenotypes of mutations in Ccr4-NOT relates to defects in the integrity of the cell wall (Betz et al., 2002; Kaeberlein and Guarente, 2002) (Figure 1). Mutants are sensitive to cell wall damaging agents (the echinocandin drug caspofungin, nikkomycin Z that inhibits chitin synthase, calcofluor white, caffeine, SDS), display cell lysis phenotypes, and their growth defects can be suppressed by osmotic stabilization or overexpression of PKC1 that encodes the kinase of the main fungal cell wall stress response pathway (Betz et al., 2002; Kaeberlein and Guarente, 2002; Markovich et al., 2004; Parsons et al., 2004; Dagley et al., 2011). The cell wall phenotypes are particularly obvious for mutations in the mRNA deadenylase subunits of the complex, CCR4 and CAF1/POP2. Also, inactivation of the exonuclease activity of Ccr4 by mutating one of the catalytic residues mimics a complete deletion of the CCR4 gene for caspofungin sensitivity (Dagley et al., 2011). The functions of the NOT sub-module and Caf40 and Caf130 are less clear, but there is evidence of involvement. Inactivation of NOT5 leads to increased levels of expression of genes encoding cell wall functions (Azzouz et al., 2009b), and not4, caf40, and not3 mutants are sensitive to caffeine (Parsons et al., 2004; Kapitzky et al., 2010). Mutations in genes encoding proteins that act together with Ccr4-NOT in mRNA turnover, for example the RNA helicase Dhh1 and the RNA binding protein Puf5 (also called Mpt5), also lead to phenotypes consistent with cell wall defects (Moriya and Isono, 1999; Kaeberlein and Guarente, 2002; Dudley et al., 2005; Stewart et al., 2007; Banuelos et al., 2010; Traven et al., 2010; Lo et al., 2012). This argues that mRNA decay of specific transcripts is key for maintaining fungal cell wall integrity.
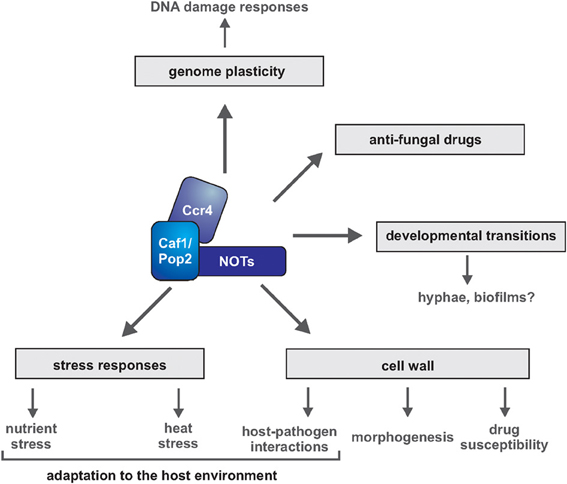
Figure 1. Multiple roles of Ccr4-NOT in pathways important for virulence of human fungal pathogens. Gene regulation by the Ccr4-NOT complex has been implicated in multiple survival mechanisms of fungal pathogens, such as cell wall integrity, filamentous growth and survival of nutrient and temperature stress. Direct assessment of the roles of Ccr4-NOT in fungal biofilm formation and host-pathogen interactions is still to come. Subunits of Ccr4-NOT are required for virulence of C. albicans and C. neoformans in the mouse infection model.
Ccr4 is also important for cell wall integrity in two prominent human fungal pathogens, C. albicans and C. neoformans. In these two pathogenic species, ccr4 mutants display sensitivity to cell wall inhibitors (calcofluor white, congo red, caspofungin), enlarged cells and cell lysis defects (Panepinto et al., 2007; Dagley et al., 2011). In C. albicans, similar phenotypes have also been observed for the deletion of POP2/CAF1 (Dagley et al., 2011). Moreover, the C. albicans the not5 mutant is more sensitive than the wild type to calcofluor white, but more resistant to treatment with the ß-glucanase zymolyase, both phenotypes supportive of alterations in the cell wall (Cheng et al., 2005). C. albicans mutants inactivated for NOT3 display reduced ability to attach to a silicone substrate, which points to changes in cell wall structure and adherence properties (Finkel et al., 2012). In the most direct demonstration roles of Ccr4-NOT in cell wall biogenesis, biochemical analysis of cell wall composition showed that the C. albicans ccr4 and caf1/pop2 mutants display lower relative levels of cell wall ß-glucans (Dagley et al., 2011). This result is consistent with the sensitivity of the mutants to the echinocandin caspofungin, which inhibits 1,3 ß-glucan synthesis. Given that S. cerevisiae and C. neoformans ccr4 mutants are also sensitive to caspofungin (Markovich et al., 2004; Panepinto et al., 2007; Dagley et al., 2011), it is possible that in those yeasts also Ccr4 impacts on the ß-glucan composition of the cell wall.
What are the target genes controlled by Ccr4-NOT to regulate cell wall biology? Several cell wall-related genes display altered expression levels in ccr4 mutants, and some of the observed changes could explain the cell wall ß-glucan phenotypes. These data come from the S. cerevisiae system. Firstly, in the absence of CCR4, KRE6 encoding a ß 1,6-glucan synthesis gene is expressed at lower levels (Betz et al., 2002). Secondly, Ccr4 might be modulating the activity of ß 1,3-glucan synthase (Figure 2). The levels of two regulators of the GTPase Rho1, which is necessary for ß 1,3-glucan synthase activity, are differentially expressed in ccr4 mutants (Ito et al., 2011). The GTP exchange factor ROM2, an activator for Rho1, is down-regulated, while the GTPase activating protein LRG1, a negative regulator of Rho1, is up-regulated. These changes in gene expression likely lead to lower activity of Rho1, which could have a negative impact on ß 1,3-glucan synthase (Figure 2). Rho1 might also have a role in regulating synthesis of ß 1,6-glucans, and is further required for signaling through the PKC-dependent cell wall integrity pathway (Levin, 2011).
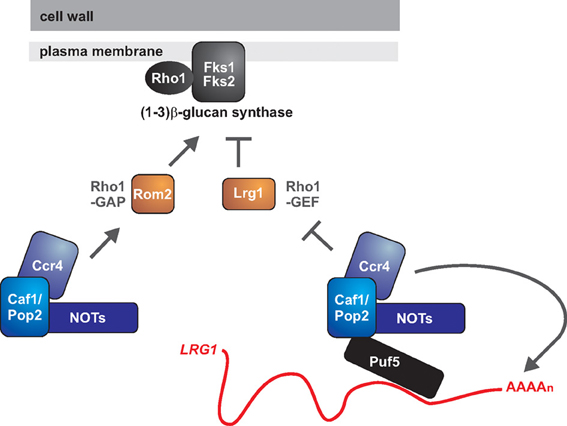
Figure 2. The roles of Ccr4 in fungal cell wall integrity: is Ccr4 a key regulator of glucan synthesis? The GTPase Rho1 is the regulatory subunit of 1,3 ß-glucan synthase and might also have a role in the biogenesis of 1,6 ß-glucan. Rho1 is active in its GTP-bound form. The GTPase activating protein (GAP) Lrg1 negatively regulates Rho1, while the GDP exchange factor (GEF) Rom2 is a positive regulator. Ccr4 has been implicated in repressing the expression of LRG1 and activating the expression of ROM2, with both mechanisms acting to promote Rho1 activity. Ccr4 might be repressing LRG1 via its deadenylase function, and the RNA binding protein Puf5 might be acting to recruit the Ccr4-NOT complex to the LRG1 mRNA. How Ccr4 acts to promote ROM2 expression is presently unclear.
The deadenylase activity of Ccr4 negatively regulates gene expression by causing shortening of the mRNA poly(A) tail, thereby initiating decay (Goldstrohm and Wickens, 2008). Therefore, a direct mRNA target of Ccr4 should be up-regulated in the mutant. We would therefore argue that, of the known affected genes, LRG1 is the most likely direct target of Ccr4 in regards to cell wall phenotypes. Further support for this idea comes from studies of Puf5, an RNA binding protein that binds to 3′ untranslated regions (3′ UTRs) of mRNAs and recruits the Ccr4-NOT complex for deadenylation and translational repression (Goldstrohm et al., 2006). Puf5 also negatively regulates LRG1 levels (Stewart et al., 2007), and it could be doing so via its interaction with Ccr4-NOT, but this needs to be experimentally demonstrated (Figure 2). Whether LRG1 is a relevant Ccr4-target gene in other fungal species remains to be studied.
It is very likely that the effects of Ccr4-NOT on the fungal cell wall are complex, and regulation of multiple, functionally distinct genes might be involved. The C. albicans not3 mutant displays altered levels of several transcripts that encode cell wall proteins (Finkel et al., 2012). Additionally, studies in C. albicans have suggested that the effects of Ccr4 on another pathway, mitochondrial biogenesis, might be linked to its functions in cell wall integrity (Dagley et al., 2011). This, and work that followed from it, has revealed that mitochondrial activity in C. albicans impacts on cell wall integrity and on the ability to respond to treatment with cell wall inhibitors, including the echinocandin drug caspofungin (Dagley et al., 2011; Qu et al., 2012). These studies provide a case in point for our hypothesis that studying the pleoitropic roles of Ccr4-NOT in fungi can reveal unexpected connections and novel players in cellular pathways important in the context of virulence. Studies in S. cerevisiae have revealed respiratory and other mitochondrial defects for several Ccr4-NOT subunits, as well as other factors of the mRNA decay pathway (for example Dhh1) (Betz et al., 2002; Dimmer et al., 2002; Perrone et al., 2005). It is therefore possible that, more generally, Ccr4-NOT and the mRNA turnover machinery link mitochondrial activity with cell wall integrity.
Fungi have the ability to switch between developmental states, which differ in cell morphology, cell surface structure, pathogenicity and interaction with host immunity. One such developmental switch that has been extensively characterized is the yeast to hyphal transition, in which cells change cell morphology from ovoid to elongated filaments (Sudbery, 2011). For C. albicans, the hyphal transition is one of the most recognized pathogenic attributes (Sudbery, 2011).
In S. cerevisiae, pseudohyphal differentiation in diploid strains and agar invasion in haploid strains are two phenotypes that depend on the morphogenetic switch. In large-scale mutant collection screens, not5 mutants were found to display enhanced filamentation on the surface of plates but no invasive growth (Jin et al., 2008), and caf130 mutants were identified as invasion-defective (Ryan et al., 2012). Ccr4 and Pop2 impact on the expression of a key cell surface molecule implicated in invasive and pseudohyphal growth, the adhesin Flo11 (Lo et al., 2012). Ccr4 might be promoting FLO11 transcription by inhibiting the expression of two transcriptional repressors of FLO11, NRG1 and NRG2 (Lo et al., 2012). Low Flo11 levels in the deadenylase mutant alter the cell wall properties, and lead to low adherence to polystyrene (Lo et al., 2012). Cells inactivated for the helicase Dhh1 also display similar phenotypes (Park et al., 2006; Lo et al., 2012). Collectively, these results argue that the mRNA decay pathway plays important roles in linking morphogenesis and cell wall regulation.
Studies in C. albicans support the S. cerevisiae data. Mutations in CCR4, CAF1/POP2, NOT1, NOT3, and NOT5 give rise to cells that are defective in transitioning from yeast to hyphal growth (Cheng et al., 2003; Epp et al., 2010; Dagley et al., 2011). Substrate adherence and hyphal growth are important for the formation of C. albicans biofilms (Finkel and Mitchell, 2011). Given that Ccr4-NOT impacts on both, it is possible that this complex controls the ability of C. albicans to make biofilms.
The target genes of Ccr4-NOT for the filamentous growth pathway are not known. The biogenesis of the cell wall is intimately linked to filament formation. Therefore, the hyphal defects of the ccr4-not mutants could be linked to altered cell wall structure and ß-glucan levels. Consistent with this idea are reports that in C. albicans defects in cell wall ß-glucans lead to reduced ability to form hyphae (Lussier et al., 1998; Herrero et al., 2004; Umeyama et al., 2006). It is also possible that Ccr4-NOT has more specific roles in orchestrating the changes in gene expression programs during the developmental switch from yeast to filaments. Transcriptome-wide approaches to study how Ccr4-NOT impacts on gene regulation during the morphogenetic switch in both S. cerevisiae and C. albicans will answer these questions. Based on current knowledge, we predict that the role of Ccr4-NOT in gene expression is conserved between these two divergent yeasts. However, often, analysis of transcription factor function in different fungal species has revealed a large extent of evolutionary change (Lavoie et al., 2009). How posttranscriptional gene regulation has evolved in fungi is much less understood, and studies of Ccr4-NOT could shed light on this exciting question.
When colonizing the human body, fungal pathogens face numerous stressful conditions, such as nutrient limitation, higher temperature, and low oxygen. In S. cerevisiae, C. neoformans and C. albicans Ccr4-NOT has been implicated in the adaptation to nutrient, temperature and oxygen stress, suggesting this complex is a key player in adaptive response that mediate fungal growth and survival in the host. In this section, we will briefly touch on the S. cerevisiae work, as this has been reviewed before (Denis and Chen, 2003; Collart and Timmers, 2004), and focus more prominently on new knowledge gained in the pathogenic species.
Glucose is the preferred carbon source for yeasts, which they ferment by glycolysis. In S. cerevisiae the Ccr4-NOT complex has important roles for growth in the absence of glucose (so called non-fermentative growth), reviewed in (Denis and Chen, 2003; Collart and Timmers, 2004). Transcriptome profiling by microarrays confirmed a role for Ccr4-NOT in adaptation to glucose depletion, and also more generally in the response to stress (Cui et al., 2008; Azzouz et al., 2009b). A major effect of Ccr4-NOT on stress responsive gene transcription could be mediated by interactions with the SAGA histone acetyltransferase complex (Cui et al., 2008). Ccr4-NOT has also been implicated in regulating the function of stress-responsive transcription factors, such as Msn2 and Skn7 (Lenssen et al., 2002, 2005, 2007). In the niches in the human body, glucose is scarce, and therefore adaptation to alternative carbon sources is important. It remains to be studied how exactly Ccr4-NOT controls the response to non-fermentative conditions in pathogenic fungi. Transcriptome analysis of the ccr4Δ/Δ mutant in C. albicans showed that a large proportion of the differentially expressed genes are related to the function of mitochondria (Dagley et al., 2011), suggesting a role in metabolism. Specific effects on gene expression of mitochondrial functions have also been observed in S. cerevisiae ccr4-not mutants (Cui et al., 2008). Interestingly, the transcriptional profile of the ccr4Δ/Δ mutant of C. albicans correlates with the early response to hypoxia, suggesting a role for Ccr4 in low oxygen conditions (Adnan Sellam and André Nantel, personal communication, manuscript in preparation). Hypoxia is commonly encountered by fungal pathogens in the niches in the human body, and the ability to respond to hypoxia is key for virulence (Grahl et al., 2012).
In C. neoformans, the Ccr4-NOT complex impacts on the expression of a key virulence factor, the laccase LAC1, a multi-copper oxidase required the production of the protective pigment melanin. Work on C. neoformans VAD1, the homolog of S. cerevisiae DHH1, implicated Not1 as a repressor of LAC1 gene expression (Panepinto et al., 2005). The ccr4Δ mutant is defective in laccase activity, but LAC1 mRNA induction is not defective (Bloom and Panepinto, unpublished). These data suggest that the defect in laccase production is posttranscriptional. It appears therefore that in C. neoformans Ccr4-NOT regulates LAC1 via Not1-dependent effects of transcription, and Ccr4-dependent posttranscriptional control. The expression of the LAC1 gene in C. neoformans is tightly regulated by glucose repression, suggesting that the role of Ccr4-NOT in glucose-regulated gene expression is conserved between C. neoformans and S. cerevisiae.
A key stress responsive function of Ccr4 in C. neoformans is the response to temperature stress. This is key for virulence. Mammals, such as us, are normally quite resistant to fungal infections, because most fungi do not grow optimally at our body temperature of 37°C. In order to colonize the human body niches, fungal pathogens need to be able to adapt to growth at 37°C. The C. neoformans ccr4Δ mutant exhibits temperature sensitivity, and is attenuated in virulence (Panepinto et al., 2007). In response to temperature stress in S. cerevisiae, mRNAs that encode ribosomal proteins and other protein synthesis factors are destabilized by Ccr4, suggesting that mRNA decay is a mechanism that promotes stress-responsive re-programming of mRNA pools (Grigull et al., 2004). The destabilization of ribosomal protein mRNAs in response to temperature is also dependent on the RNA polymerase II subunit Rpb4 (Goler-Baron et al., 2008; Harel-Sharvit et al., 2010). The current paradigm is that, under stress conditions, Rbp4 marks transcripts for rapid degradation via co-transcriptional association. In C. neoformans the roles of Ccr4 and Rpb4 in the regulation stress responsive changes in mRNA stability is conserved, suggesting that the coupling of mRNA synthesis and decay is likewise conserved (Bloom et al., 2013). In wild type C. neoformans, a shift from 30°C to 37°C is accompanied by acute and transient changes in mRNA abundance and stability. Ribosomal protein mRNAs are transiently repressed, and ER stress mRNAs are transiently induced (Havel et al., 2011; Bloom et al., 2013). Between 2 and 3 h post-shift, these mRNAs return to pre-shift levels. In the ccr4Δmutant, ribosomal protein mRNAs accumulate and transient repression is absent. Those mRNAs were found to be stable in the ccr4Δ mutant, confirming defects in mRNA degradation as the cause of their up-regulation. Likewise, in a C. neoformans rpb4Δ mutant, mRNAs encoding ribosomal proteins are stabilized, and the transient repression of ribosomal protein transcript mRNA abundance that accompanies temperature adaptation is attenuated (Bloom et al., 2013). Furthermore, in ccr4Δ cells, increased ER stress mRNA abundance persists past 3 h (Havel et al., 2011). The major ER stress sensor is Kar2/BiP, an ER resident Hsp70 family member. In wild type C. neoformans, the KAR2 transcript exhibits distinct changes in mRNA stability in response to temperature stress (Bloom et al., 2013). Under unstressed conditions, the KAR2 mRNA is very stable, but is destabilized following the shift to 37°C, through Rpb4-dependent and -independent mechanisms, and the data is consistent with a role in mRNA degradation limiting both the intensity and the duration of the stress response. The destabilization of KAR2 at peak mRNA abundance will prevent persistent activation of the ER stress response such as that seen in the ccr4Δ mutant. Phenotypically, there is no change in wild type C. neoformans during this adaptation, and growth rate is maintained through the shift. In a ccr4Δ mutant, however, growth is nearly arrested, cell morphology becomes aberrantly large and buds fail to separate (Panepinto, unpublished). Clearly, Ccr4 in C. neoformans is buffering phenotypic change in response to environmental shift.
Pathogenic fungi can acquire macro and micro-chromosomal alterations that might be beneficial to their survival in the host, or in the face of antifungal drug challenge, reviewed in (Selmecki et al., 2010; Morrow and Fraser, 2013). Cells can be generated that display changes in chromosome numbers (aneuploid cells) or even the whole chromosomal set (ploidy changes). Other chromosomal rearrangements, such as translocations, truncations, gene amplifications and loss of heterozygosity (LOH) are also observed. Pathways that impact on genome stability (DNA replication, damage and repair mechanisms) and cell division (cell cycle control mechanisms) are expected to have an impact on the ability of fungal pathogens to adapt by chromosomal alterations. Work in S. cerevisiae and S. pombe implicates Ccr4-NOT in the regulation of genome stability. Data in C. neoformans is consistent with this function, as ccr4 mutant cells are sensitive to DNA damaging agents (Panepinto, unpublished).
Mutations in the Ccr4-NOT complex have been repeatedly found in S. cerevisiae screens that used the deletion collection to find genes that control susceptibility to DNA damaging agents, as well as by researchers using candidate gene approaches (Bennett et al., 2001; Betz et al., 2002; Hanway et al., 2002; Hartman and Tippery, 2004; Parsons et al., 2004; Westmoreland et al., 2004; Mulder et al., 2005; Traven et al., 2005; Woolstencroft et al., 2006; Gaillard et al., 2009). In S. pombe, mutations in Ccr4-NOT result in similar DNA damage response phenotypes (Takahashi et al., 2007; Deshpande et al., 2009). Genetic interactions between genes encoding Ccr4-NOT subunits and genes of the DNA damage checkpoint and repair pathways also support an important role for Ccr4-NOT in DNA damage responses (Traven et al., 2005; Pan et al., 2006; Woolstencroft et al., 2006; Gaillard et al., 2009).
How does Ccr4-NOT activity regulate DNA damage responses? Studies from the S. cerevisiae system suggest a complex scenario, with three possible mechanisms that are not mutually exclusive. Firstly, it seems that Ccr4-NOT has a general (non-gene-specific) roles in transcription-coupled repair (TCR) (Gaillard et al., 2009). The TCR roles could be functionally linked to the function of Ccr4-NOT in transcriptional elongation, but the exact mechanism remains to be understood (Gaillard et al., 2009). Secondly, Ccr4-NOT has gene-specific roles in the response to DNA replication stress: Ccr4-NOT is needed for the induction of ribonucleotide reductase (RNR) genes, (Mulder et al., 2005; Woolstencroft et al., 2006; Kruk et al., 2011). The effects of Ccr4-NOT on the expression of RNR genes are transcriptional and posttranscriptional. Ccr4 might be acting to inhibit the expression of the transcriptional repressor of the RNR genes, CRT1, by shortening its mRNA poly(A) tail (Woolstencroft et al., 2006). In regards to transcriptional control, in absence of Not4 recruitment of RNA polymerase II and the TATA binding protein (TBP) to the promoter of RNR3 in response to HU treatment is defective (Mulder et al., 2005). It has been shown more recently that Ccr4 and Not5 are found at the RNR3 gene in response to MMS, supporting a direct role in transcription of this gene upon DNA damage (Kruk et al., 2011). The third mechanism relates to a co-factor role for Pop2/Caf1 in regulating the subcellular localization of ribonucleotide reductase in response to inhibition of DNA replication (Takahashi et al., 2007). Ccr4 also appears to have some role in this process (Takahashi et al., 2007).
In C. neoformans, multiple DNA damage-related mRNAs were found up regulated in the ccr4Δ strain by microarray analysis (Havel et al., 2011), suggesting conservation of a role for the mRNA decay arm of the Ccr4-NOT complex in genome integrity. C. neoformans lacks an ortholog of CRT1, and induces a basidiomycete-specific ribonucleotide reductase small subunit in response to HU treatment, rather than the retained ascomycete-like gene (Zulkifli et al., 2012). An effect of CCR4 deletion on the regulation of these genes has yet to be determined. Further studies will define the extent of conservation between C. neoformans and the ascomycete yeasts.
The implication of the roles of Ccr4-NOT in genome stability is that in the absence of this complex, genome instability should increase, causing higher chromosomal rearrangements. Increased LOH has been shown in the S. cerevisiae ccr4 and pop2 mutants, but not for all markers tested, and was most pronounced for a locus on chromosome XII (Andersen et al., 2008). The increased LOH was suggested to be linked to the impact of Ccr4 and Pop2 on maintenance of dNTP pools: chromosome XII contains the rDNA repeat, and it was suggested that the stability of this repeat might be sensitive to nucleotide levels (Andersen et al., 2008). Higher chromosomal instability could favor adaptation of pathogens, such as C. albicans and C. neoformans to the host environment and antifungal drug stress. Interesting in this context is a recent report that suggests that in S. pombe and S. cerevisiae, Ccr4-NOT is necessary for ensuring survival of cells that contain chromosomal aneuploidies (Tange et al., 2012). Mutants in the NOT2, NOT3, and CAF40 genes were particularly affected. The deadenylase subunits CCR4 and POP2 also likely have a role, but this could not be precisely determined due to non-specific effects of deadenylase activation in the assays used (Tange et al., 2012). Bone-fide DNA repair genes were also found to be required for growth of aneuploid cells, and it was suggested that the DNA damage response functions of Ccr4-NOT are linked with the roles of the complex in survival of aneuploids (Tange et al., 2012). These results suggest that some forms of chromosomal rearrangements, such as changes to copy numbers, would not be tolerated in the absence of Ccr4-NOT. This could be important in the context of fungal virulence and antifungal drug tolerance, as aneuplodies are observed frequently, and have been shown in some cases to confer increased resistance to drug treatments, reviewed in Selmecki et al. (2010), Morrow and Fraser (2013).
The phenotypes of the mutants argue yes. As discussed above, inactivation of Ccr4-NOT function cripples several pathways important for fungal virulence. Moreover, inactivation of CCR4 in C. albicans and C. neoformans, as well as NOT5 in C. albicans resulted in significantly crippled virulence in mice (Cheng et al., 2005; Panepinto et al., 2007; Dagley et al., 2011). In addition, mutation of CCR4 causes hypersensitivity to the echinocandin drug caspofungin in S. cerevisiae, C. albicans and C. neoformans (Markovich et al., 2004; Panepinto et al., 2007; Dagley et al., 2011), suggesting that targeting Ccr4 could be beneficial for combinatorial therapy with the echinocandins. The polyene amphotericin B and the ergosterol biosynthesis inhibitors from the azole family of drugs are the other two classes of antifungals important in the clinical context. An early report in S. cerevisiae showed sensitivity of the ccr4 mutant to the polyene drug nystatin, which binds to membrane ergosterol (Betz et al., 2002). Understanding how Ccr4-NOT mutants in pathogens respond to azoles and polyenes awaits further experimentation.
Fungi are eukaryotes and many fundamental cellular pathways are conserved with animals. This close evolutionary relationship poses a problem when fungi become our pathogens, because compounds that inhibit fungal cells can cause host toxicity. The Ccr4-NOT complex is a conserved eukaryotic regulator and potentially, targeting subunits of this complex would result in toxic effect in humans. To address this, we performed a detailed bioinformatic analysis of the conservation of the Ccr4-NOT subunits with animals and within the fungal kingdom. Most the key components of the Ccr4-NOT complex (Ccr4, Caf1/Pop2, Not1, Not2, Not3, Not4, Caf40) were already present before the divergence of fungi and animals (Figure 3), and are readily identified in both groups. However, several fungi-specific differences are noteworthy. The animal CNOT10 and CNOT11 were present in the ancestor, but lost in fungi, whereas Not5 and Caf130 are specific for a sub-group of fungi. While previous analysis realized that Not5 and Caf130 are not conserved between S. cerevisiae and animals (Denis and Chen, 2003; Collart and Timmers, 2004), our analysis shows that the acquisition of these two genes is specific for the Saccharomycetes, which contain S. cerevisiae and Candida species, but not other important pathogens such as Aspergillus and Cryptoccous species (Figure 3; Supplemental Table 1). Caf130 shows no relevant similarity to any other protein and is a highly specific invention (we note that occasionally the fungal Caf130 and the animal CNOT10 are listed as homologs (Miller and Reese, 2012), but there is no relevant sequence conservation between the two proteins). Caf130 is fungal-specific, which makes it an interesting candidate for drug development. The function of Caf130 is not clear at the moment, and the S. cerevisiae mutant showed a transcriptional profile similar to the wild type (Azzouz et al., 2009b). However, the caf130 mutant of S. cerevisiae was recently reported to be defective for invasion (Ryan et al., 2012), a phenotype known to affect the virulence potential of C. albicans. It is noteworthy that Caf130 is not present outside the Saccharomycetes, which would limit its usefulness if the aim were a pan-fungal inhibitor. In contrast to Caf130, Not5 shows considerable sequence similarity to Not3 from animals and fungi, and has arisen from a gene duplication event at the base of the Saccharomycetes (Supplemental Figure 1). Our analysis is consistent with previous observations that it is not easily clear whether Not3 or Not5 are more similar to Not3 in organisms with only one copy, as the two genes show similar levels of sequence divergence. Fungi outside the Saccharomycetes, such as Aspergillus and Cryptococcus have only one copy of this gene, which we call NOT3/5. While they are easily identifiable as homologs, the sequence conservation of the Not3/Not5 subunit(s) is not very high between organisms, including between fungi and animals (Supplemental Figure 1). Inactivation of NOT5 has a strong impact on fungal fitness, and in C. albicans the mutant has a cell wall and a virulence defect (Cheng et al., 2005). Collectively, the sequence divergence and roles in virulence suggest that the Not5/Not3 proteins could be targets of interest in the context of antifungal therapy.
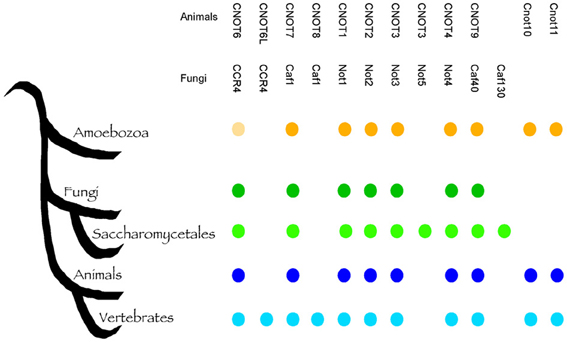
Figure 3. The evolution of the Ccr4-NOT complex. A schematic tree indicating the evolution of the indicated groups based on our current understanding; the presence of the respective members is indicated to emphasize fungi-specific gains (Caf130, Not5), as well as loss (CNOT10, CNOT11) due to their prior presence in the Amoebozoa. Identifiers for sequences supporting this are shown in Supplemental Table 1. The bright shading of Ccr4 in the Amoebozoa indicates the lack of the LRR domain; since this is essential for the binding of Pop2/Caf1 and thus to Not1, this implies that the function of the Ccr4-NOT complex as such could be specific to fungi and animals.
We next investigated the level of sequence conservation for the deadenylase subunits between humans and the fungal pathogens (Cryptococcus, Candida, and Aspergillus; Figure 4). Despite significant differences in length, the Ccr4/Caf1/Not1 interacting domains are conserved between the proteins of fungi and humans (Figures 4A,B) (Basquin et al., 2012). We focused on the sequence regions known to hold the essential sites for the interactions between Ccr4-Caf1 and Caf1-Not1, as determined in a recent structural study (Basquin et al., 2012). The Caf1-Not1 interaction anchors Ccr4 into the Ccr4-NOT complex, and the interactions between these three subunits are essential for deadenylation and growth of S. cerevisiae (Basquin et al., 2012). The high level of conservation of the sites in Ccr4, Pop2/Caf1, and Not1 required for the interactions indicate that functionality of these regions is likely conserved for fungi and animals (Figure 4B). Despite the high similarity, there is a level of variation in the sequences, which could prove beneficial to design compounds to specifically target the fungal complex. We note that loss of the catalytic deadenylase activity of Caf1/Pop2 and the DEDD motif, which is archetypical for this family of deadenylases, is true only for the Saccharomycetes (which include S. cerevisiae and the Candida species), but not the other fungal pathogens (Figure 4C).
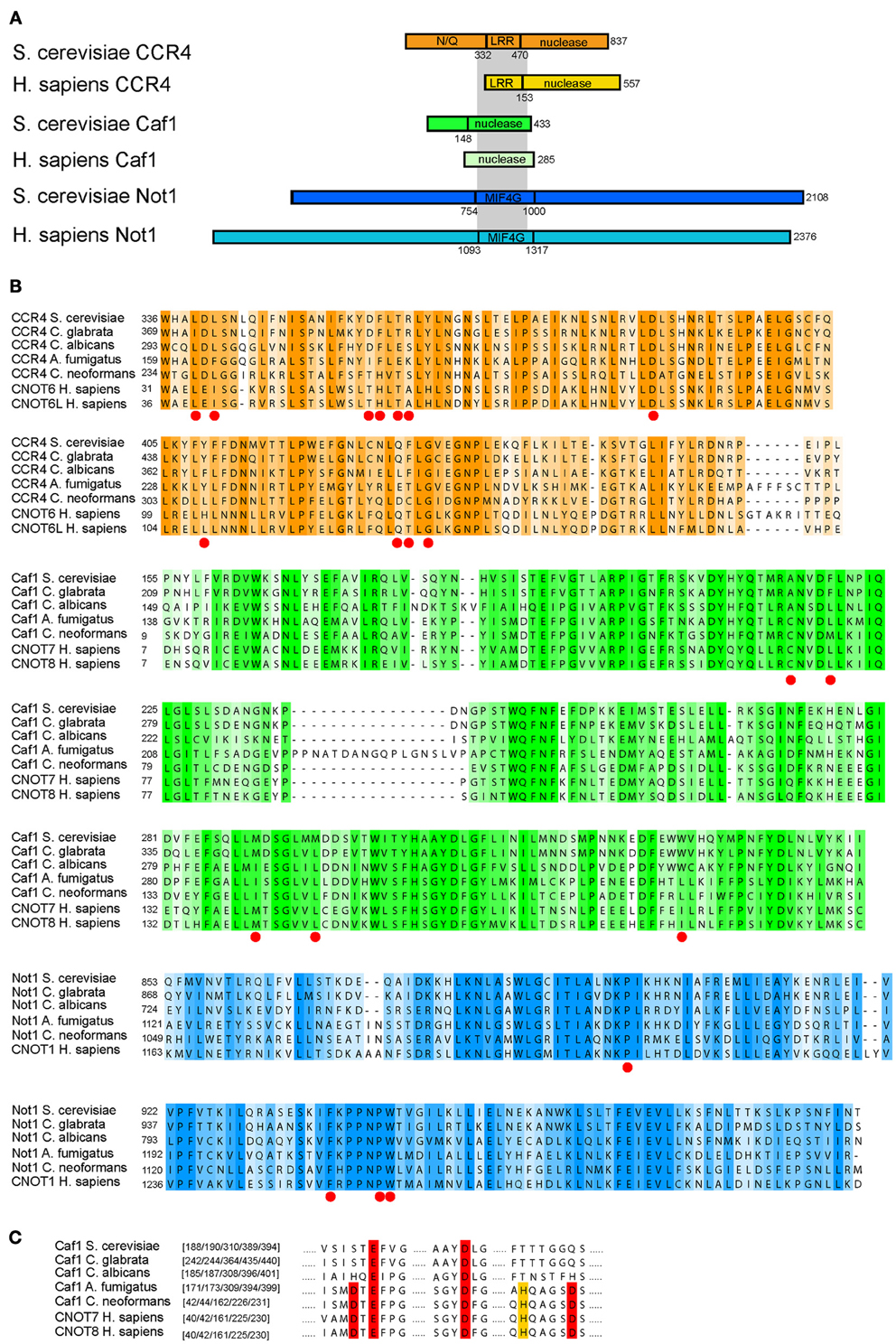
Figure 4. Comparison of Ccr4-Caf1-Not1 interaction domains between humans and pathogenic fungi. (A) A schematic overview of the Ccr4-Caf1-Not1 interactions based on S. cerevisiae, illustrating the interactions between the Ccr4 LRR domain, Caf1, and the Not1 MIF4G fold domain as reported in Basquin et al. (2012) and Petit et al. (2012); the gray shading indicates the interactions. (B) Alignment of the corresponding regions of Ccr4, Caf1, and Not1 in yeast and several fungal pathogens compared to the human homologs. The sites described to be responsible for the function and/or protein-protein interactions in yeast (Clark et al., 2004; Basquin et al., 2012) are indicated by red dots. Color shades represent sequence conservation with a 20% cutoff value as implemented in JalView (Clamp et al., 2004). (C) The DEDD motif of Caf1, as well as the histidine that aids in formation of the cofactor binding pocket (Jonstrup et al., 2007; Horiuchi et al., 2009), are highlighted in red or yellow, respectively. This illustrates the loss of these residues in the Saccharomycetes, but not in other fungal groups. The sequences represent a subset from a full alignment of all sequences. The amino acids are indicated at the start of each block in (B), and the highlighted amino acid residue numbers are given in brackets for each sequence in (C). All alignments are based on the full sequence selection of fungi and animal sequences as given in Supplemental Table 1, and were aligned with muscle (version 3.8.31) (Edgar, 2004), using default settings. Shown sequences were subsequently selected and only-gap sites removed.
The presence of CNOT10 in organisms other than animals (Figure 3) indicates its loss in fungi and potential functional replacement with Caf130, for which no similar proteins outside the Saccharomycetes could be found. This might go in accordance with the differences observed in Not1; whilst the C-terminus, the binding site of Caf1, Ccr4, and Not2-5 (Basquin et al., 2012), is relatively conserved, the N-terminus - which harbors the Caf40 and CNOT10 /CNOT11, and possibly also the Caf130, binding sites (Nasertorabi et al., 2011; Bawankar et al., 2013) - is reduced in fungi compared to animals (Figure 4A). In the Candida group, the proteins lack approximately 180 amino acids at the N-terminal part compared to S. cerevisiae (Supplemental Figure 2 shows an alignment of Not1 sequences from S. cerevisiae, representative fungal pathogens and humans). The Not1 MIF4G region in Not1 contains several sites known to be essential for binding of Caf1 (to which Ccr4 binds) in both fungi and animals (Basquin et al., 2012; Petit et al., 2012) (Figure 4A).
In conclusion, while the Ccr4-NOT complex is generally conserved between yeast and humans, some subunits are fungal-specific, and some sequence variation can be observed in the conserved subunits. These observations suggest that this complex might be explored in the context of antifungal therapy. It is noteworthy that developing antifungal drugs against targets conserved with humans has been done successfully: the target of the commonly used azole drugs, sterol 14 α-demethlyase, is an evolutionarily conserved enzyme required for ergosterol/cholesterol synthesis. However, the inhibition of the fungal enzyme by azoles is higher than inhibition of the human homolog (Parker et al., 2008; Warrilow et al., 2013). It might also be useful to think about Ccr4-NOT in the context of combinatorial therapy with conventional antifungal drugs, such as the echinocandins. One can imagine that the deadenylase activities of Ccr4 could be targeted, and the levels of inhibition adjusted so not to cause adverse effects on human cells, but rather specifically kill fungi in combination with other, fungal-specific drugs.
Ccr4-NOT has been implicated in the ability of C. albicans and C. neoformans to cause disease, despite fundamental differences in the pathogenic processes of these two human pathogens. For example, C. albicans is a human commensal and the temperature of the human host is not sensed as a stressor. In contrast, C. neoformans is an environmental fungus, and the shift from environmental temperature to human core body temperature is sensed as a stressor and involves reprogramming of mRNA pools. Both C. albicans and C. neoformans encounter macrophages. C. neoformans has evolved the ability to survive and even proliferate within the macrophage, whereas C. albicans utilizes the yeast-to-hyphae morphogenetic transition to escape the phagocyte. Though the role of Ccr4 in the survival of C. neoformans or C. albicans in macrophages has not been investigated, the stress sensitivity of the C. neoformans mutant and the lack of morphogenetic capacity of the C. albicans mutant predict that the macrophage-pathogen interaction would lead to elimination of the pathogen. This illustrates how the pleoiotropic phenotypes of the ccr4-not mutants might be beneficial in targeting diverse pathogens, in addition to inactivating several processes important for virulence. Further studies of Ccr4-NOT function in C. albicans and C. neoformans, as well as extension to other pathogenic species, such as the molds and the dimorphs, are likely to be informative for understanding fungal pathogenesis and provide further insight how this complex or a pathway that it regulates could be targeted for improving treatments.
Eva Heinz performed the bioinformatic analysis. John C. Panepinto, Eva Heinz, and Ana Traven wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Trevor Lithgow for help with Figures 1, 2. The work in the Ana Traven lab is supported by grants from the Australian Research Council, the Australian National Health and Medical Research Council and the Monash University Researcher Accelerator Grant. John C. Panepinto is supported by US Public Health Service Grant RO1 AI089920.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2013.00302/abstract
Supplemental Figure 1 | The relationship between Not3, Not5, and CNOT3 with respect to sequence divergence and phylogenetics. (A) Both Not3 and Not5 are present only in the Saccharomycetes. Sequence accession numbers are given in Supplemental Table 1. The alignment was performed by muscle (version 3.8.31) (Edgar, 2004) using default settings. The color shades represent sequence conservation with a 20% cutoff value, and the level of conservation is shown, as implemented in JalView (Clamp et al., 2004). (B) Phylogenetic analysis reveals that the Not3 and Not5 are products of a gene duplication (the duplication event is indicated by a red dot), and show similar levels of sequence divergence, supporting the idea that it is not clear whether the Saccharomycetes Not3 or Not5 is more similar to the single-copy Not3 (CNOT3). The sequence accession numbers are given in Supplemental Table 1. The sequences were aligned with muscle (version 3.8.31) (Edgar, 2004) using default conditions, and divergent regions were removed using trimal (1.2rev59) (Capella-Gutierrez et al., 2009) with the “-automated1” setting. The tree was calculated in MrBayes (version 3.2.1) (Ronquist and Huelsenbeck, 2003) using the amino acid mixture model with 10.000.000 generations, with 25% discarded as burn-in during the consensus tree calculation; support values shown are posterior probabilities.
Supplemental Figure 2 | Divergence of Not1 between fungi and humans. Not1 shows different profiles of conservation, which goes in accordance with the binding domains of conserved (Ccr4, Caf1) and divergent (Caf40, Caf130 vs. CNOT10) binding partners. The alignment was performed by muscle (version 3.8.31) (Edgar, 2004) using default settings, color shades represent sequence conservation with a 20% cutoff value as implemented in JalView (Clamp et al., 2004); accession numbers of all sequences are given in Supplemental Table 1.
Supplemental Table 1 | Table accessions. Accession numbers and sequence data were retrieved from the yeast gene order browser (http://ygob.ucd.ie/) (Byrne and Wolfe, 2005), the Candida gene order browser (http://cgob.ucd.ie/) (Maguire et al., 2013) or the NCBI RefSeq database.
Andersen, M. P., Nelson, Z. W., Hetrick, E. D., and Gottschling, D. E. (2008). A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics 179, 1179–1195. doi: 10.1534/genetics.108.089250
Azzouz, N., Panasenko, O. O., Colau, G., and Collart, M. A. (2009a). The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS ONE 4:e6760. doi: 10.1371/journal.pone.0006760
Azzouz, N., Panasenko, O. O., Deluen, C., Hsieh, J., Theiler, G., and Collart, M. A. (2009b). Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA 15, 377–383. doi: 10.1261/rna.1348209
Badarinarayana, V., Chiang, Y. C., and Denis, C. L. (2000). Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics 155, 1045–1054.
Bai, Y., Salvadore, C., Chiang, Y. C., Collart, M. A., Liu, H. Y., and Denis, C. L. (1999). The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 19, 6642–6651.
Banuelos, M. G., Moreno, D. E., Olson, D. K., Nguyen, Q., Ricarte, F., Aguilera-Sandoval, C. R., et al. (2010). Genomic analysis of severe hypersensitivity to hygromycin B reveals linkage to vacuolar defects and new vacuolar gene functions in Saccharomyces cerevisiae. Curr. Genet. 56, 121–137. doi: 10.1007/s00294-009-0285-3
Basquin, J., Roudko, V. V., Rode, M., Basquin, C., Seraphin, B., and Conti, E. (2012). Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol. Cell 48, 207–218. doi: 10.1016/j.molcel.2012.08.014
Bawankar, P., Loh, B., Wohlbold, L., Schmidt, S., and Izaurralde, E. (2013). NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 10, 228–244. doi: 10.4161/rna.23018
Bennett, C. B., Lewis, L. K., Karthikeyan, G., Lobachev, K. S., Jin, Y. H., Sterling, J. F., et al. (2001). Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29, 426–434. doi: 10.1038/ng778
Betz, J. L., Chang, M., Washburn, T. M., Porter, S. E., Mueller, C. L., and Jaehning, J. A. (2002). Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268, 272–285. doi: 10.1007/s00438-002-0752-8
Bloom, A. L., Solomons, J. T., Havel, V. E., and Panepinto, J. C. (2013). Uncoupling of mRNA synthesis and degradation impairs adaptation to host temperature in Cryptococcus neoformans. Mol. Microbiol. 89, 65–83. doi: 10.1111/mmi.12258
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. doi: 10.1126/science.1222236
Byrne, K. P., and Wolfe, K. H. (2005). The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15, 1456–1461. doi: 10.1101/gr.3672305
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, J., Chiang, Y. C., and Denis, C. L. (2002). CCR4, a 3'-5' poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21, 1414–1426. doi: 10.1093/emboj/21.6.1414
Chen, J., Rappsilber, J., Chiang, Y. C., Russell, P., Mann, M., and Denis, C. L. (2001). Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 314, 683–694. doi: 10.1006/jmbi.2001.5162
Cheng, S., Clancy, C. J., Checkley, M. A., Zhang, Z., Wozniak, K. L., Seshan, K. R., et al. (2005). The role of Candida albicans NOT5 in virulence depends upon diverse host factors in vivo. Infect. Immun. 73, 7190–7197. doi: 10.1128/IAI.73.11.7190-7197.2005
Cheng, S., Nguyen, M. H., Zhang, Z., Jia, H., Handfield, M., and Clancy, C. J. (2003). Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71, 6101–6103. doi: 10.1128/IAI.71.10.6101-6103.2003
Clamp, M., Cuff, J., Searle, S. M., and Barton, G. J. (2004). The Jalview Java alignment editor. Bioinformatics 20, 426–427. doi: 10.1093/bioinformatics/btg430
Clark, L. B., Viswanathan, P., Quigley, G., Chiang, Y. C., McMahon, J. S., Yao, G., et al. (2004). Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 279, 13616–13623. doi: 10.1074/jbc.M313202200
Collart, M. A., and Panasenko, O. O. (2012). The Ccr4–not complex. Gene 492, 42–53. doi: 10.1016/j.gene.2011.09.033
Collart, M. A., Panasenko, O. O., and Nikolaev, S. I. (2013). The Not3/5 subunit of the Ccr4-Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell. Signal. 25, 743–751. doi: 10.1016/j.cellsig.2012.12.018
Collart, M. A., and Struhl, K. (1993). CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12, 177–186.
Collart, M. A., and Struhl, K. (1994). NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8, 525–537. doi: 10.1101/gad.8.5.525
Collart, M. A., and Timmers, H. T. (2004). The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 77, 289–322. doi: 10.1016/S0079-6603(04)77008-7
Cooney, N. M., and Klein, B. S. (2008). Fungal adaptation to the mammalian host: it is a new world, after all. Curr. Opin. Microbiol. 11, 511–516. doi: 10.1016/j.mib.2008.09.018
Cui, Y., Ramnarain, D. B., Chiang, Y. C., Ding, L. H., McMahon, J. S., and Denis, C. L. (2008). Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genomics 279, 323–337. doi: 10.1007/s00438-007-0314-1
Dagley, M. J., Gentle, I. E., Beilharz, T. H., Pettolino, F. A., Djordjevic, J. T., Lo, T. L., et al. (2011). Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol. Microbiol. 79, 968–989. doi: 10.1111/j.1365-2958.2010.07503.x
Deluen, C., James, N., Maillet, L., Molinete, M., Theiler, G., Lemaire, M., et al. (2002). The Ccr4-not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol. Cell. Biol. 22, 6735–6749. doi: 10.1128/MCB.22.19.6735-6749.2002
Denis, C. L., and Chen, J. (2003). The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73, 221–250. doi: 10.1016/S0079-6603(03)01007-9
Denis, C. L., Chiang, Y. C., Cui, Y., and Chen, J. (2001). Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 158, 627–634.
Deshpande, G. P., Hayles, J., Hoe, K. L., Kim, D. U., Park, H. O., and Hartsuiker, E. (2009). Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair (Amst) 8, 672–679. doi: 10.1016/j.dnarep.2009.01.016
Dimmer, K. S., Fritz, S., Fuchs, F., Messerschmitt, M., Weinbach, N., Neupert, W., et al. (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853. doi: 10.1091/mbc.01-12-0588
Douglas, C. M., D'ippolito, J. A., Shei, G. J., Meinz, M., Onishi, J., Marrinan, J. A., et al. (1997). Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-D-glucan synthase inhibitors. Antimicrob. Agents Chemother. 41, 2471–2479.
Dudley, A. M., Janse, D. M., Tanay, A., Shamir, R., and Church, G. M. (2005). A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 1, 2005.0001. doi: 10.1038/msb4100004
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Epp, E., Walther, A., Lepine, G., Leon, Z., Mullick, A., Raymond, M., et al. (2010). Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol. Microbiol. 75, 1182–1198. doi: 10.1111/j.1365-2958.2009.07038.x
Finkel, J. S., and Mitchell, A. P. (2011). Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9, 109–118. doi: 10.1038/nrmicro2475
Finkel, J. S., Xu, W., Huang, D., Hill, E. M., Desai, J. V., Woolford, C. A., et al. (2012). Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525. doi: 10.1371/journal.ppat.1002525
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Gaillard, H., Tous, C., Botet, J., Gonzalez-Aguilera, C., Quintero, M. J., Viladevall, L., et al. (2009). Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 5:e1000364. doi: 10.1371/journal.pgen.1000364
Goldstrohm, A. C., Hook, B. A., Seay, D. J., and Wickens, M. (2006). PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13, 533–539. doi: 10.1038/nsmb1100
Goldstrohm, A. C., and Wickens, M. (2008). Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9, 337–344. doi: 10.1038/nrm2370
Goler-Baron, V., Selitrennik, M., Barkai, O., Haimovich, G., Lotan, R., and Choder, M. (2008). Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 22, 2022–2027. doi: 10.1101/gad.473608
Gow, N. A., and Hube, B. (2012). Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412. doi: 10.1016/j.mib.2012.04.005
Grahl, N., Shepardson, K. M., Chung, D., and Cramer, R. A. (2012). Hypoxia and fungal pathogenesis: to air or not to air? Eukaryotic Cell 11, 560–570. doi: 10.1128/EC.00031-12
Grigull, J., Mnaimneh, S., Pootoolal, J., Robinson, M. D., and Hughes, T. R. (2004). Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24, 5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004
Hanway, D., Chin, J. K., Xia, G., Oshiro, G., Winzeler, E. A., and Romesberg, F. E. (2002). Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. U.S.A. 99, 10605–10610. doi: 10.1073/pnas.152264899
Hanzawa, H., De Ruwe, M. J., Albert, T. K., Van Der Vliet, P.C., Timmers, H. T., and Boelens, R. (2001). The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J. Biol. Chem. 276, 10185–10190. doi: 10.1074/jbc.M009298200
Harel-Sharvit, L., Eldad, N., Haimovich, G., Barkai, O., Duek, L., and Choder, M. (2010). RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143, 552–563. doi: 10.1016/j.cell.2010.10.033
Hartman, J. L. T., and Tippery, N. P. (2004). Systematic quantification of gene interactions by phenotypic array analysis. Genome Biol. 5, R49. doi: 10.1186/gb-2004-5-7-r49
Havel, V. E., Wool, N. K., Ayad, D., Downey, K. M., Wilson, C. F., Larsen, P., et al. (2011). Ccr4 promotes resolution of the endoplasmic reticulum stress response during host temperature adaptation in Cryptococcus neoformans. Eukaryotic Cell 10, 895–901. doi: 10.1128/EC.00006-11
Herrero, A. B., Magnelli, P., Mansour, M. K., Levitz, S. M., Bussey, H., and Abeijon, C. (2004). KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryotic Cell 3, 1423–1432. doi: 10.1128/EC.3.6.1423-1432.2004
Horiuchi, M., Takeuchi, K., Noda, N., Muroya, N., Suzuki, T., Nakamura, T., et al. (2009). Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 284, 13244–13255. doi: 10.1074/jbc.M809250200
Ito, W., Li, X., Irie, K., and Mizuno, T. (2011). RNA-binding protein Khd1 and Ccr4 deadenylase play overlapping roles in the cell wall integrity pathway in Saccharomyces cerevisiae. Eukaryotic Cell 10, 1340–1347. doi: 10.1128/EC.05181-11
Jin, R., Dobry, C. J., McCown, P. J., and Kumar, A. (2008). Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell 19, 284–296. doi: 10.1091/mbc.E07-05-0519
Jonstrup, A. T., Andersen, K. R., Van, L. B., and Brodersen, D. E. (2007). The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 35, 3153–3164. doi: 10.1093/nar/gkm178
Kaeberlein, M., and Guarente, L. (2002). Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160, 83–95.
Kapitzky, L., Beltrao, P., Berens, T. J., Gassner, N., Zhou, C., Wuster, A., et al. (2010). Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol. Syst. Biol. 6, 451. doi: 10.1038/msb.2010.107
Kerr, S. C., Azzouz, N., Fuchs, S. M., Collart, M. A., Strahl, B. D., Corbett, A. H., et al. (2011). The Ccr4-Not complex interacts with the mRNA export machinery. PLoS ONE 6:e18302. doi: 10.1371/journal.pone.0018302
Kruk, J. A., Dutta, A., Fu, J., Gilmour, D. S., and Reese, J. C. (2011). The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 25, 581–593. doi: 10.1101/gad.2020911
Kupferschmidt, K. (2012). Mycology. Attack of the clones.Science 337, 636–638. doi: 10.1126/science.337.6095.636
Lavoie, H., Hogues, H., and Whiteway, M. (2009). Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr. Opin. Microbiol. 12, 655–663. doi: 10.1016/j.mib.2009.09.015
Lenssen, E., Azzouz, N., Michel, A., Landrieux, E., and Collart, M. A. (2007). The Ccr4-not complex regulates Skn7 through Srb10 kinase. Eukaryotic Cell 6, 2251–2259. doi: 10.1128/EC.00327-06
Lenssen, E., James, N., Pedruzzi, I., Dubouloz, F., Cameroni, E., Bisig, R., et al. (2005). The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation–via a newly identified Glc7/Bud14 type I protein phosphatase module–and TFIID promoter distribution. Mol. Cell. Biol. 25, 488–498. doi: 10.1128/MCB.25.1.488-498.2005
Lenssen, E., Oberholzer, U., Labarre, J., De Virgilio, C., and Collart, M. A. (2002). Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43, 1023–1037. doi: 10.1046/j.1365-2958.2002.02799.x
Levin, D. E. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175. doi: 10.1534/genetics.111.128264
Liu, H. Y., Badarinarayana, V., Audino, D. C., Rappsilber, J., Mann, M., and Denis, C. L. (1998). The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17, 1096–1106. doi: 10.1093/emboj/17.4.1096
Lo, T. L., Qu, Y., Uwamahoro, N., Quenault, T., Beilharz, T. H., and Traven, A. (2012). The mRNA decay pathway regulates the expression of the Flo11 adhesin and biofilm formation in Saccharomyces cerevisiae. Genetics 191, 1387–1391. doi: 10.1534/genetics.112.141432
Lussier, M., Sdicu, A. M., Shahinian, S., and Bussey, H. (1998). The Candida albicans KRE9 gene is required for cell wall beta-1, 6-glucan synthesis and is essential for growth on glucose. Proc. Natl. Acad. Sci. U.S.A. 95, 9825–9830. doi: 10.1073/pnas.95.17.9825
Maguire, S. L., Oheigeartaigh, S. S., Byrne, K. P., Schroder, M. S., O'gaora, P., Wolfe, K. H., et al. (2013). Comparative genome analysis and gene finding in Candida species using CGOB. Mol. Biol. Evol. 30, 1281–1291. doi: 10.1093/molbev/mst042
Markovich, S., Yekutiel, A., Shalit, I., Shadkchan, Y., and Osherov, N. (2004). Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 48, 3871–3876. doi: 10.1128/AAC.48.10.3871-3876.2004
Miller, J. E., and Reese, J. C. (2012). Ccr4-Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 47, 315–333. doi: 10.3109/10409238.2012.667214
Moriya, H., and Isono, K. (1999). Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast 15, 481–496.
Morrow, C. A., and Fraser, J. A. (2013). Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin. Cell Dev. Biol. 24, 339–346. doi: 10.1016/j.semcdb.2013.01.008
Mulder, K. W., Winkler, G. S., and Timmers, H. T. (2005). DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 33, 6384–6392. doi: 10.1093/nar/gki938
Nasertorabi, F., Batisse, C., Diepholz, M., Suck, D., and Bottcher, B. (2011). Insights into the structure of the CCR4-NOT complex by electron microscopy. FEBS Lett. 585, 2182–2186. doi: 10.1016/j.febslet.2011.05.071
O'Meara, T. R., and Alspaugh, J. A. (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408. doi: 10.1128/CMR.00001-12
Pan, X., Ye, P., Yuan, D. S., Wang, X., Bader, J. S., and Boeke, J. D. (2006). A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124, 1069–1081. doi: 10.1016/j.cell.2005.12.036
Panasenko, O., Landrieux, E., Feuermann, M., Finka, A., Paquet, N., and Collart, M. A. (2006). The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 281, 31389–31398. doi: 10.1074/jbc.M604986200
Panepinto, J. C., Komperda, K. W., Hacham, M., Shin, S., Liu, X., and Williamson, P. R. (2007). Binding of serum mannan binding lectin to a cell integrity-defective Cryptococcus neoformans ccr4Delta mutant. Infect. Immun. 75, 4769–4779. doi: 10.1128/IAI.00536-07
Panepinto, J., Liu, L., Ramos, J., Zhu, X., Valyi-Nagy, T., Eksi, S., et al. (2005). The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J. Clin. Invest. 115, 632–641. doi: 10.1172/JCI200523048
Park, Y. U., Hur, H., Ka, M., and Kim, J. (2006). Identification of translational regulation target genes during filamentous growth in Saccharomyces cerevisiae: regulatory role of Caf20 and Dhh1. Eukaryotic Cell 5, 2120–2127. doi: 10.1128/EC.00121-06
Parker, J. E., Merkamm, M., Manning, N. J., Pompon, D., Kelly, S. L., and Kelly, D. E. (2008). Differential azole antifungal efficacies contrasted using a Saccharomyces cerevisiae strain humanized for sterol 14 alpha-demethylase at the homologous locus. Antimicrob. Agents Chemother. 52, 3597–3603. doi: 10.1128/AAC.00517-08
Parsons, A. B., Brost, R. L., Ding, H., Li, Z., Zhang, C., Sheikh, B., et al. (2004). Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22, 62–69. doi: 10.1038/nbt919
Perrone, G. G., Grant, C. M., and Dawes, I. W. (2005). Genetic and environmental factors influencing glutathione homeostasis in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 218–230. doi: 10.1091/mbc.E04-07-0560
Petit, A. P., Wohlbold, L., Bawankar, P., Huntzinger, E., Schmidt, S., Izaurralde, E., et al. (2012). The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 40, 11058–11072. doi: 10.1093/nar/gks883
Pfaller, M. A., and Diekema, D. J. (2010). Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36, 1–53. doi: 10.3109/10408410903241444
Qiu, H., Hu, C., Yoon, S., Natarajan, K., Swanson, M. J., and Hinnebusch, A. G. (2004). An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell Biol. 24, 4104–4117.
Qu, Y., Jelicic, B., Pettolino, F., Perry, A., Lo, T. L., Hewitt, V. L., et al. (2012). Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence. Eukaryotic Cell 11, 532–544. doi: 10.1128/EC.05292-11
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryan, O., Shapiro, R. S., Kurat, C. F., Mayhew, D., Baryshnikova, A., Chin, B., et al. (2012). Global gene deletion analysis exploring yeast filamentous growth. Science 337, 1353–1356. doi: 10.1126/science.1224339
Selmecki, A., Forche, A., and Berman, J. (2010). Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryotic Cell 9, 991–1008. doi: 10.1128/EC.00060-10
Stewart, M. S., Krause, S. A., McGhie, J., and Gray, J. V. (2007). Mpt5p, a stress tolerance- and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryotic Cell 6, 262–270. doi: 10.1128/EC.00188-06
Sudbery, P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9, 737–748. doi: 10.1038/nrmicro2636
Swanson, M. J., Qiu, H., Sumibcay, L., Krueger, A., Kim, S. J., Natarajan, K., et al. (2003). A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell Biol. 23, 2800–2820.
Takahashi, S., Kontani, K., Araki, Y., and Katada, T. (2007). Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1-Suc22 assembly. Nucleic Acids Res. 35, 1187–1197. doi: 10.1093/nar/gkm015
Tange, Y., Kurabayashi, A., Goto, B., Hoe, K. L., Kim, D. U., Park, H. O., et al. (2012). The CCR4-NOT complex is implicated in the viability of aneuploid yeasts. PLoS Genet. 8:e1002776. doi: 10.1371/journal.pgen.1002776
Traven, A., Hammet, A., Tenis, N., Denis, C. L., and Heierhorst, J. (2005). Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 169, 65–75. doi: 10.1534/genetics.104.030940
Traven, A., Lo, T. L., Lithgow, T., and Heierhorst, J. (2010). The yeast PUF protein Puf5 has Pop2-independent roles in response to DNA replication stress. PLoS ONE 5:e10651. doi: 10.1371/journal.pone.0010651
Tucker, M., Staples, R. R., Valencia-Sanchez, M. A., Muhlrad, D., and Parker, R. (2002). Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21, 1427–1436. doi: 10.1093/emboj/21.6.1427
Tucker, M., Valencia-Sanchez, M. A., Staples, R. R., Chen, J., Denis, C. L., and Parker, R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386. doi: 10.1016/S0092-8674(01)00225-2
Umeyama, T., Kaneko, A., Watanabe, H., Hirai, A., Uehara, Y., Niimi, M., et al. (2006). Deletion of the CaBIG1 gene reduces beta-1,6-glucan synthesis, filamentation, adhesion, and virulence in Candida albicans. Infect. Immun. 74, 2373–2381. doi: 10.1128/IAI.74.4.2373-2381.2006
Van De Veerdonk, F.L., Kullberg, B. J., Van Der Meer, J.W., Gow, N. A., and Netea, M. G. (2008). Host-microbe interactions: innate pattern recognition of fungal pathogens. Curr. Opin. Microbiol. 11, 305–312. doi: 10.1016/j.mib.2008.06.002
Warrilow, A. G., Parker, J. E., Kelly, D. E., and Kelly, S. L. (2013). Azole affinity of sterol 14alpha-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob. Agents Chemother. 57, 1352–1360. doi: 10.1128/AAC.02067-12
Westmoreland, T. J., Marks, J. R., Olson, J. A. Jr., Thompson, E. M., Resnick, M. A., and Bennett, C. B. (2004). Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryotic Cell 3, 430–446. doi: 10.1128/EC.3.2.430-446.2004
Woolstencroft, R. N., Beilharz, T. H., Cook, M. A., Preiss, T., Durocher, D., and Tyers, M. (2006). Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 119, 5178–5192. doi: 10.1242/jcs.03221
Keywords: Ccr4-NOT, fungal pathogen, cell wall, stress adaptation, Candida albicans, Cryptococcus neoformans
Citation: Panepinto JC, Heinz E and Traven A (2013) The cellular roles of Ccr4-NOT in model and pathogenic fungi—implications for fungal virulence. Front. Genet. 4:302. doi: 10.3389/fgene.2013.00302
Received: 01 October 2013; Paper pending published: 29 October 2013;
Accepted: 09 December 2013; Published online: 20 December 2013.
Edited by:
Martine A. Collart, University of Geneva, SwitzerlandReviewed by:
Zhaohui Wang, University of Texas Southwestern Medical Center, USACopyright © 2013 Panepinto, Heinz and Traven. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John C. Panepinto, Department of Microbiology and Immunology, University at Buffalo, The State University of New York, 117 Biomedical Research Building, 3435 Main Street, Buffalo, NY 14214, USA e-mail:amNwMjVAYnVmZmFsby5lZHU=;
Ana Traven, Department of Biochemistry and Molecular Biology, Building 76, Monash University, Clayton (Melbourne) 3800, Victoria, Australia e-mail:YW5hLnRyYXZlbkBtb25hc2guZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.