
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet. , 04 December 2013
Sec. Epigenomics and Epigenetics
Volume 4 - 2013 | https://doi.org/10.3389/fgene.2013.00265
This article is part of the Research Topic (Epi)genetic control of human trophoblast invasion View all 7 articles
Specialized cell types of trophoblast cells form the placenta in which each cell type has particular properties of proliferation and invasion. The placenta sustains the growth of the fetus throughout pregnancy and any aberrant trophoblast differentiation or invasion potentially affects the future health of the child and adult. Recently, the field of epigenetics has been applied to understand differentiation of trophoblast lineages and embryonic stem cells (ESC), from fertilization of the oocyte onward. Each trophoblast cell-type has a distinctive epigenetic profile and we will concentrate on the epigenetic mechanism of DNA methyltransferases and TETs that regulate DNA methylation. Environmental factors affecting the mother potentially regulate the DNA methyltransferases in trophoblasts, and so do steroid hormones, cell cycle regulators, such as p53, and cytokines, especially interlukin-1β. There are interesting questions of why trophoblast genomes are globally hypomethylated yet specific genes can be suppressed by hypermethylation (in general, tumor suppressor genes, such as E-cadherin) and how invasive cell-types are liable to have condensed chromatin, as in metastatic cancer cells. Future work will attempt to understand the interactive nature of all epigenetic mechanisms together and their effect on the complex biological system of trophoblast differentiation and invasion in normal as well as pathological conditions.
The epigenetic mechanism of DNA methylation partially regulates trophoblast differentiation and invasion into the endometrium of the uterus in order to establish and maintain a healthy placenta for the growing fetus. The placenta is an essential organ for sustaining the life of the fetus through which nutrients, oxygen exchange, immune barrier protection, and waste disposal are achieved between the fetal and maternal blood circulations (Cross et al., 1994; Arck and Hecher, 2013). Defective placentation and aberrant differentiation and invasion of the trophoblasts potentially results in pathologies of pregnancy, which include pre-eclampsia, intrauterine growth restriction (IUGR) (Maccani and Carmen, 2009; Kokkinos et al., 2010), spontaneous abortion, preterm birth (Khong and Brosens, 2011), placenta acreta, “Hemolysis, Elevated Liver enzymes, Low Platelets” (HELLP) (van Dijk et al., 2012), and choriocarcinoma (Graham and Lala, 1992; Norwitz, 2006). Environmental factors, such as diet and oxygen stress, can adversely affect the epigenetic mechanisms that the embryo relies on to implant and fully grow into a healthy fetus (Yuen et al., 2013). Epigenetic mechanisms partially regulate extra-villous trophoblast differentiation and invasiveness into the endometrium (Figure 1) (Rahnama et al., 2006; Rugg-Gunn, 2012; Chen et al., 2013a). Over the last decade the study of epigenetics has been applied to many complex biological systems, such as trophoblast differentiation, in order to reach an understanding of the mechanisms and the pathways (Cox et al., 2009, 2011; Choi, 2010; Hemberger, 2010; Senner and Hemberger, 2010; Turner et al., 2012; Arnold et al., 2013).
DNA methylation, histone modification and non-coding RNA (ncRNA) and the interactions between them are some of the epigenetic mechanisms that have been studied in the placenta (Fuks et al., 2003; Nelissen et al., 2011). The field of epigenetics is continually expanding and there is recent interest in DNA hydroxymethylation and TET methylcytosine dioxygenases (ten-eleven translocation) (Tahiliani et al., 2009; Dahl et al., 2011; Xu et al., 2011; Zhao and Chen, 2013), and the role of various types of ncRNA, such as micro RNAs (miRNA) (Tsai et al., 2009; Ji et al., 2013; Mouillet et al., 2013) and long intergenic ncRNA (lincRNA) (van Dijk et al., 2012). The three main DNMTs that methylate cytosines in CpG dinucleotides to 5-methylcytosine (5mC) are DNMT1, the maintenance methyltransferase, and the de novo methyltransferases DNMT3A and DNMT3B, which each have gene-specific methylation sites in distinct genomic DNA regions (Hsieh, 1999; Okano et al., 1999; Takeshima et al., 2006). Another member of the DNMT3 family is DNMT3L, which is essential for placental development through establishing maternal gene imprinting (Chédin et al., 2002; Hata et al., 2002; Arima et al., 2006). Imprinting of paternal or maternal genes has an important effect on placenta gene expression and subsequent growth of the fetus (Reik and Walter, 2001; Wang et al., 2013a). DNA methylation of CpGs generally silences a gene by at least three different functions; the binding of transcription factors (Watt and Molloy, 1988), attracting methyl binding domain proteins (MBD) (Boyes and Bird, 1991), and altering chromatin packaging (Ng and Bird, 1999). Moreover, DNMTs have both DNA methylation-independent as well as methylation-dependent functions (Bachman et al., 2001; Fuks et al., 2001; Esteve et al., 2005; Pavlopoulou and Kossida, 2010). There are a few instances when instead of repressing transcription, DNA methylation activates transcription, but how this is achieved is still unknown, but release of inhibitors is most likely (Gellersen and Kempf, 1990; Niesen et al., 2005). TETs are involved in an active DNA demethylation pathway, by converting 5mC to 5-hydroxymethylcytosine (5hmC) (Seisenberger et al., 2013). These epigenetic mechanisms critically regulate the differentiation of cells essential for the development of the placenta and embryo (Table 1) (Ohgane et al., 2002; Rugg-Gunn, 2012).
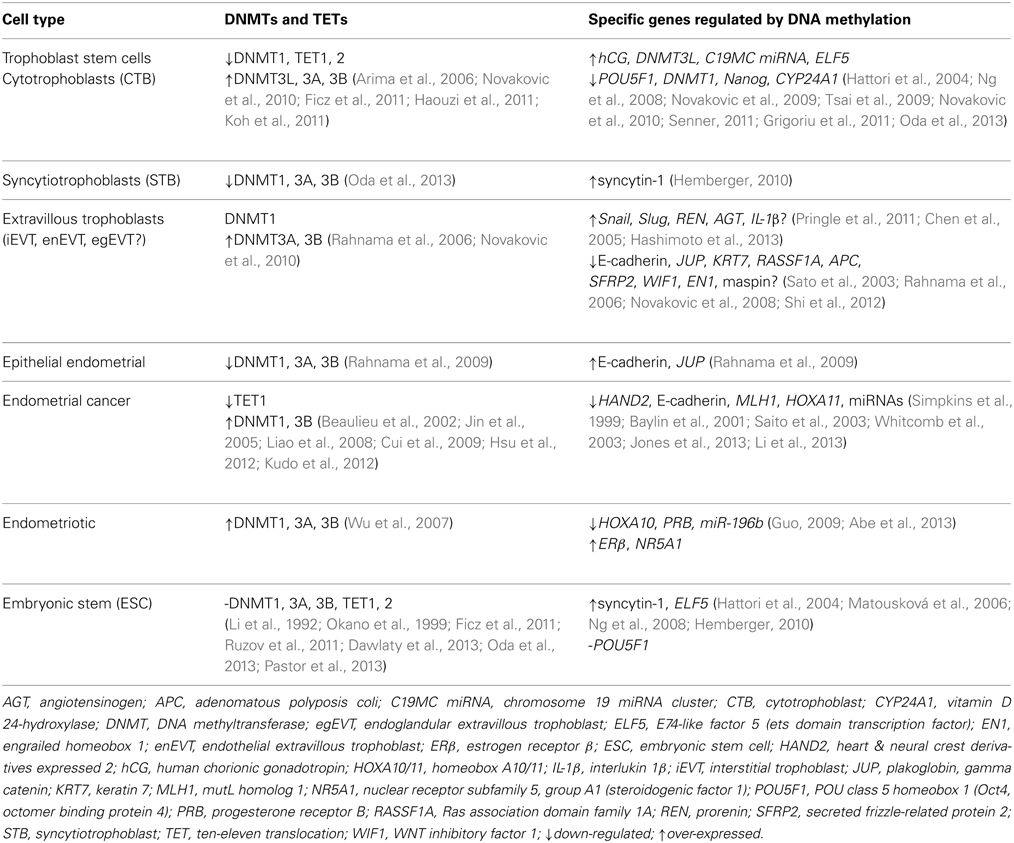
Table 1. DNA methyltransferases (DNMTs) and TETs regulate specific key genes for differentiation and invasion.
DNA methylation is involved in the distinction between the specialized cell lineages of pregnancy beginning from fertilization until term (Hemberger, 2007; Nelissen et al., 2011; Xu et al., 2011). The blastocyst has embryonic stem cell (ESC) lineages in the inner cell mass, which will form the embryo, and trophoblast stem cells in the trophectoderm of extra-embryonic cells, which will develop into the placenta (Bischof and Irminger-Finger, 2005). Histone modifications are thought to establish the lineages of inner cell mass and trophectoderm (Nakanishi et al., 2012) and then DNA methylation profiles “lock in” and maintain the lineages to create a barrier between them, i.e., to block ESC from becoming trophoblast lineage (Ng et al., 2008; Senner, 2011; Oda et al., 2013). Both these lineages have the defining feature of a distinctive global DNA methylation pattern (Senner et al., 2012), so that the genome of the embryo is highly methylated, whilst the genome of the placental trophectoderm lineage of trophoblasts is globally hypomethylated (Hemberger, 2010; Xie et al., 2013). In the trophoblasts, DNMT1 is down-regulated by promoter methylation whereas DNMT3L is up-regulated, and in turn activates both DNMT3A and DNMT3B (Table 1) (Chédin et al., 2002; Hata et al., 2002; Suetake et al., 2004; Chen et al., 2005; Novakovic et al., 2010; Haouzi et al., 2011). A complex biological system, such as differentiating the trophoblast stem cell lineage, builds in redundancy. If DNMT1 is not expressed then DNMT3A and DNMT3B can substitute for DNMT1, if necessary, and maintain the methylation of DNA during cell proliferation (Liang et al., 2002; Walton et al., 2011; Arand et al., 2012). In a mouse triple knockout of DNMT1/3a/3b, extra-embryonic stem cells survived and proliferated whereas ESC either died or with minimal CpG methylation occasionally differentiated into trophoblast-like cells (Li et al., 1992; Jackson et al., 2004; Tsumura et al., 2006; Ng et al., 2008; Sakaue et al., 2010). However, knocking out DNMT3L disrupted the placenta in mice (Hata et al., 2002; Arima et al., 2006; Nelissen et al., 2011). Another feature of trophoblasts is the overall low level of 5hmC in the genome compared with the high level of 5hmC in the ESC genome (Ito et al., 2010; Ruzov et al., 2011).
To differentiate the trophoblast stem cell lineage from the ESC, the pluripotency, transcription factor gene, POU5F1 (OCT4) is hypermethylated and repressed in trophoblast stem cells (Hattori et al., 2004; Li et al., 2007; Zhang et al., 2008; Zafarana et al., 2009; Senner, 2011). There are more hypermethylated promoters in trophoblast stem cells that silence pluripotency genes than in ESC whereas in ESC pluripotency-related genes are generally hypomethylated and expressed (Farthing et al., 2008). POU5F1 up-regulates TET1/2 in ESC to induce a high level of 5hmC and therefore pluripotency (Ruzov et al., 2011), on the other hand depletion of POU5F1 in trophoblast stem cells inhibits TET1/2 gene expression, which decreases 5hmC and pluripotency (Koh et al., 2011). TET1/2 depletion hypermethylates specific genes and if this occurs in ESC then pluripotency-related genes are down-regulated and this tends to differentiate ESC into extra-embryonic cells (Ficz et al., 2011; Koh et al., 2011; Williams et al., 2012). In humans, mutated, inactive TET2 can lead to cancers, such as hematopoietic cancer (Salker et al., 2011; Kudo et al., 2012; Perez et al., 2012), yet TET1/2 knockout mice can survive with reduced 5hmC. Perhaps the TET1/2 knockout mice survive because TET3 was increased and substituted for TET1/2, but the increased methylation compromised imprinted genes, and there was also an increase in extra-embryonic stem cells (Dawlaty et al., 2011, 2013). In a microarray study of TET1 knockout mice, 221 genes, mainly developmental, were down-regulated (Dawlaty et al., 2011). Depleted TET1 in stem cells up-regulate ELF5, the key trophoblast lineage-enforcing gene, which is hypomethylated and expressed in trophoblast stem cells but in ESC ELF5 must be methylated and silenced (Ng et al., 2008; Senner and Hemberger, 2010; Hemberger, 2010; Senner, 2011). If ESC are induced to express ELF5 then ESC can become trophoblast-like (Roper and Hemberger, 2009; Koh et al., 2011).
DNMTs and TETs are involved in the differentiation and regulation of the specialized trophoblast cells in the placenta (Table 1). Trophoblast stem cells are differentiated into proliferating, polarized epithelial cell cytotrophoblasts (CTB) and thence into villous CTB of anchoring villi and non-proliferating syncytiotrophoblasts (STB) (Figure 1) (Bischoff et al., 2000; Bischof and Irminger-Finger, 2005; Ji et al., 2013). The down-regulation of DNMT1, 3A and 3B in CTB hypomethylates the retroviral, fusogenic-protein, syncytin-1, and the consequent high expression of syncytin-1 differentiates the CTBs into multi-nucleated, single cell STB (Blond et al., 2000; Hemberger, 2010). Failure of this differentiation into STBs may result in pre-eclampsia or IUGR, with a different pattern of DNMTs and MBD proteins for each condition (Baczyk et al., 2009; Ruebner et al., 2013). For instance, the over-expression of DNMT3A will inhibit syncytin-1 gene expression and disrupt formation of STBs (Ruebner et al., 2013). The other pathway for CTB progenitor differentiation, from the anchoring villi columns, produces invasive intermediate trophoblasts (Fisher and Damsky, 1993), which invade the endometrium as either interstitial EVTs (iEVT) or endovascular-EVTs (enEVT) (Figure 1) (Pijnenborg et al., 1980). The intermediate EVTs also possibly invade uterine glandular cells as endoglandular EVTs (egEVT) in order to remodel and open up the uterine glands that can then secrete into the inter-villous space (Fitzgerald et al., 2010; Moser et al., 2010). The iEVTs continue to invade through the endometrium into the first third of the myometrium and in the placental bed become mononuclear cell aggregate and multi-nucleated giant cells (Fisher and Damsky, 1993; Al-Lamki et al., 1999). The enEVTs invade into the maternal spiral arteries that have been prepared for EVT invasion by decidual cells and uterine natural killer cells (Lash et al., 2010; Hannon et al., 2012) and by replacing the vascular endothelial and smooth muscle cells (Zhou et al., 1997), remodel these arteries into low pressure, high volume vessels capable of delivering sufficient maternal blood to the fetus through the placenta (Lockwood et al., 1999). The morphology and function of iEVTs and enEVTs are quite different to the STBs (Zhou et al., 1997).
To successfully become invasive EVTs, the CTBs undergo a transition from epithelial to highly invasive mesenchymal phenotype, called EMT (epithelial-mesenchymal transition), similar to invasive, metastatic cancer cells, except that EVTs are under strict spatial and temporal regulation (Bischof and Irminger-Finger, 2005; Perry et al., 2010; Apps et al., 2011). The decidual cells and extracellular matrix (ECM) inhibit the EVTs from invading too deeply (Bischoff et al., 2000). Recently, a microarray of first trimester trophoblasts identified over 3000 differentially regulated genes in the transition from villous CTB to EVT and this included ELF5 that was down-regulated in EVT (Apps et al., 2011). In the EMT transition of epithelial CTBs into EVTs, the epithelial marker genes of E-cadherin and keratin 7 (KRT7) are hypermethylated and down-regulated (Rahnama et al., 2006; Chen et al., 2013a). Similar to invasive cancer cells, the tumor suppressor genes in EVTs are generally methylated, which down-regulates the genes, such as APC, E-cadherin, and possibly maspin (Rahnama et al., 2006; Wong et al., 2008; Shi et al., 2012). Two other EMT differentiation proteins, Snail and Slug, that are also involved in cancer and down-regulate E-cadherin, are up-regulated by hypomethylation in trophoblast cell-lines and differentiate CTBs into EVTs (Table 1) (Chen et al., 2013b). In endometrial cancer, an invasive phenotype is produced by an aberrant over-expression of DNMT1 and DNMT3B that down-regulates E-cadherin (Chan et al., 2003; Saito et al., 2003; Jin et al., 2005; Liao et al., 2008). In a similar epithelial-based invasive pathology, of endometriosis, DNMT1, 3A, 3B are over-expressed in the ectopic endometrial cells (Wu et al., 2007). DNMT1, 3A, 3B hypermethylate and down-regulate PRB, the progesterone receptor B, but there is no change in E-cadherin levels and so there is no EMT differentiation (Shaco-Levy et al., 2008). Concomitantly in endometriosis, DNMT1, 3A, 3B hypermethylate and down-regulate the anti-invasive HOXA10 gene expression in the eutopic endometrium (Chu et al., 2004; Wu et al., 2005; Santamaria et al., 2012).
Perhaps EVTs have chromatin condensation to enable migration and invasion? Although cancer cells generally have partially condensed chromatin between that of normal proliferative and senescent cells (Oh et al., 2013) there is a correlation between increased DNA hypermethylation, and therefore chromatin condensation, and increased invasiveness, which is essential during metastasis for the cancer cell intravasion into and extravasion out of blood vessels (Dufer et al., 2000; Fu et al., 2012). During cell migration the condensed chromatin allows the nuclei to be squeezed to fit through narrow gaps between cells (Gerlitz and Bustin, 2010; Fu et al., 2012). Constitutive heterochromatin is invariably condensed but when necessary the facultative heterochromatin can be condensed for invasion (Trojer and Reinberg, 2007). Chromatin condensation has also been observed in sperm, leukocytes, glioma cells, and neurons (Hammadeh et al., 1998; Gerlitz and Bustin, 2010). If cells are differentiated into invasive phenotypes then there is a tendency for the epigenetic mechanisms of DNA methylation and histone modification to condense the chromatin (Gerlitz and Bustin, 2010). When treatments of the histone methyltransferase inhibitor (5′-deoxy-5′-methyl-thioadenosine), HDAC inhibitors, or TETs were applied to invasive cell types then those cells decondensed the chromatin and invasiveness was inhibited (Gerlitz and Bustin, 2010; Fu et al., 2012; Song et al., 2013). In other cell types over-expression of TET increases 5hmC, which decreases 5mC (hypomethylation) and decondenses chromatin, so possibly the lack of TET expression in EVTs encourages an invasive phenotype by condensing the chromatin (Wanunu et al., 2010; Hsu et al., 2012; Pfeifer et al., 2013).
Trophoblast genomes are globally hypomethylated, predominantly in repetitive DNA elements and intergenic regions, relative to somatic tissue genomes, yet concomitantly specific genes in trophoblasts are hypermethylated (generally tumor suppressor genes) to ensure EMT into EVTs and induce invasiveness similar to metastatic cancer cells (Gama-Sosa et al., 1983; Ehrlich, 2009; Apps et al., 2011; Novakovic and Saffery, 2012; Shen et al., 2012). Specific genes that are down-regulated by DNA methylation in EVTs have been identified and include E-cadherin, plakoglobin (JUP), KRT7, DNMT1, and five Wnt signaling inhibitors APC, SFRP2, RASSF1A, EN1, and WIF1 (Table 1) (Rahnama et al., 2006; Chiu et al., 2007; Novakovic et al., 2008, 2010; Guilleret et al., 2009; Chen et al., 2013a). The increased Wnt signaling stimulates β-catenin accumulation in the nucleus, and that promotes EVT invasiveness (Pollheimer et al., 2006). Sometimes there is hypomethylation and up-regulation of specific genes in EVTs, such as the renin-angiotensin system genes (RAS) (Wang et al., 2012). In the immortalized EVT-like cell-line, HTR8/SVneo, treatments of both cyclic AMP and 5-aza-2′-deoxycytidine (AZA), an inhibitor of DNMTs and DNA methylation, up-regulated prorenin (REN), and angiotensinogen (AGT) (Wang et al., 2013b). RAS genes regulate EVT invasion and vascular remodeling and it has been suggested that aberrant expression could be linked to pre-eclampsia and IUGR (Williams et al., 2010; Pringle et al., 2011).
The question that has long been asked has been how do specific genes become hypermethylated and silenced in cells that otherwise have globally hypomethylated genomes, such as in cancer and now here in trophoblasts (Caiafa and Zampieri, 2005)? Global hypomethylation generally consists of vast hypomethylated regions of the genome that are depleted of genes whereas hypermethylated genes, such as tumor suppressor genes, have methylated promoters (Shen et al., 2012; Varley et al., 2013). A mouse tumor model showed that a functional DNMT3a determined that genomic hypomethylation was restricted to regional rather than the uniformly widespread hypomethylation throughout the genome that occurs in DNMT3a deficient mice (Raddatz et al., 2012). (This difference in DNMT3A expression could explain the possible difference between STB and EVT genomic hypomethylation.) DNA methylation should not be regarded as static in cancer because genomic hypomethylation and hypermethylation of specific genes increase progressively as the tumor increases in malignancy (Jones and Baylin, 2007; Esteller, 2008; Ehrlich, 2009) and even then not all tumor suppressor genes are hypermethylated (Hamilton et al., 2005). Regulation of hypermethylation of specific genes and genomic hypomethylation in cancer, and presumably in trophoblasts, is not dependent solely on DNMTs but involves the combined action with histone modification (Freitag and Selker, 2005; Esteller, 2008; Ehrlich, 2009), ncRNA (Kulis and Esteller, 2010), and TETs (Hsu et al., 2012; Williams et al., 2012). The inactivation of TETs can be involved in the targeted hypermethylation and silencing of specific tumor suppressor genes (Hsu et al., 2012; Williams et al., 2012). There is ample evidence that DNMTs interact with HDACs (Fuks et al., 2000; Bachman et al., 2001; Geiman et al., 2004) and transcription factors (Di Croce et al., 2002; Suzuki et al., 2005; Hervouet et al., 2009; Pavlopoulou and Kossida, 2010; Yuen et al., 2013), often in complexes, to target the methylation of specific genes (Robertson et al., 2000; Fuks et al., 2001; Zhang et al., 2005). Specific genes can be expressed in specific tissues (Schroeder et al., 2013), for example, syncytin-1 is highly expressed and tissue specific to the placenta (Mi et al., 2000; Matousková et al., 2006; Muir et al., 2006) however aberrant hypomethylation and expression of the syncytin-1 gene in the brain has been proposed to contribute to multiple sclerosis (Perron et al., 1997; Mattson and Taub, 2004).
So what factors regulate the DNMTs? DNMTs can be regulated by steroid hormones (Cui et al., 2009; Yamagata et al., 2009; Vincent et al., 2011; Logan et al., 2013) growth factors (Shafiei et al., 2008), cell cycle regulators (Lin et al., 2010), viruses (Flanagan, 2006; Shamay et al., 2006), and cytokines (Karmakar and Das, 2002; Braconi et al., 2010). The cytokines of IL-1β, IL-6, VEGF, and cytokine-activated HIF-1α are involved in trophoblast invasion (Karmakar and Das, 2002; Dubinsky et al., 2010). IL-1β can regulate DNMTs, suppress methylation, reduce E-cadherin, stimulate MMP9 [crucial for EVT invasion (Whiteside et al., 2001)] and positively correlates with invasiveness (Librach et al., 1994; Karmakar and Das, 2004; Hashimoto et al., 2010; Nakano et al., 2013). Conversely, IL-1β can be methylated and regulated at a single CpG, −299 bp (Hashimoto et al., 2013). DNMTs can also both be regulated by and interact with the cell cycle regulators p53/Sp1, MDM, pRb, and E2F (Robertson et al., 2000; Kimura et al., 2003; Esteve et al., 2005; Lin et al., 2010).
The TETs are down-regulated in the placenta, just like in cancer cells, which means 5hmC is reduced in the DNA of cells and the reduction in gene body-associated 5hmC generally inhibits gene expression (Ficz et al., 2011; Lian et al., 2012; Pfeifer et al., 2013). TET1 partly suppresses invasion in cancer by binding to TIMP2/3 and demethylating the promoters to activate TIMP2/3, and in turn down-regulates MMP expression (Hsu et al., 2012). Conversely, depletion of TET1 hypermethylates and silences TIMP2/3 (Hsu et al., 2012). The TETs are the proposed active DNA demethylases that convert 5mC to 5hmC, and then can continue that pathway so that eventually 5mC is converted to cytosine (Seisenberger et al., 2013). There has been a suggestion that oxygen regulation of gene expression is mediated by TET conversion of 5mC to 5hmC (Thalhammer et al., 2011). Recently it has been reported that under redox conditions DNMT3A and DNMT3B, as well as being methylases, can demethylate DNA, such as in cancer (Kangaspeska et al., 2008; Metivier et al., 2008; Chen et al., 2012). Perhaps DNMT3A and DNMT3B can, under hypoxic conditions in the endometrium, aberrantly demethylate DNA in EVTs?
If any of the epigenetic enzymes of DNMTs, TETs, HDACs, and ncRNAs are dysfunctional in EVTs then the differentiation and invasiveness of EVTs may be compromised (Rugg-Gunn, 2012; Fu et al., 2013; Ji et al., 2013; Mouillet et al., 2013). Environmental factors, such as diet or oxygen stress, can disrupt epigenetic regulation of trophoblasts (Novakovic and Saffery, 2012), for example, hypoxia of <1% oxygen in STBs can affect the AP-1 transcription factor and adversely disrupt the function of DNMT1 (Yuen et al., 2013). Poorly functioning trophoblasts fail to provide an adequate blood supply in circulation through the placenta to the fetus and this has been linked to pathologies of pregnancy such as preeclampsia and IUGR (Maccani and Carmen, 2009; Kokkinos et al., 2010). Future work will attempt to understand the interactive nature of all the epigenetic mechanisms together and their effect on the complex biological system of trophoblast differentiation and EVT invasion in normal as well as pathological conditions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abe, W., Nasu, K., Nakada, C., Kawano, Y., Moriyama, M., and Narahara, H. (2013). miR-196b targets c-myc and Bcl-2 expression, inhibits proliferation and induces apoptosis in endometriotic stromal cells. Hum. Reprod. 28, 750–761. doi: 10.1093/humrep/des446
Al-Lamki, R. S., Skepper, J. N., and Burton, G. J. (1999). Are human placental bed giant cells merely aggregates of small mononuclear trophoblast cells? An ultrastructural and immunocytochemical study. Hum Reprod. 14, 496–504. doi: 10.1093/humrep/14.2.496
Apps, R., Sharkey, A., Gardner, L., Male, V., Trotter, M., Miller, N., et al. (2011). Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta 32, 33–43. doi: 10.1016/j.placenta.2010.10.010
Arand, J., Spieler, D., Karius, T., Branco, M. R., Meilinger, D., Meissner, A., et al. (2012). In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 8:e1002750. doi: 10.1371/journal.pgen.1002750
Arck, P. C., and Hecher, K. (2013). Fetomaternal immune cross-talk and its consequences for maternal and offspring's health. Nat. Med. 99, 548–556. doi: 10.1038/nm.3160
Arima, T., Hata, K., Tanaka, S., Kusumi, M., Li, E., Kato, K., et al. (2006). Loss of the maternal imprint in Dnmt3Lmat-/- mice leads to a differentiation defect in the extraembryonic tissue. Dev. Biol. 297, 361–373. doi: 10.1016/j.ydbio.2006.05.003
Arnold, P., Schöler, A., Pachkov, M., Balwierz, P. J., Jørgensen, H., Stadler, M. B., et al. (2013). Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res. 23, 60–73. doi: 10.1101/gr.142661.112
Bachman, K. E., Rountree, M. R., and Baylin, S. B. (2001). Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276, 32282–32287. doi: 10.1074/jbc.M104661200
Baczyk, D., Drewlo, S., Proctor, L., Dunk, C., Lye, S., and Kingdom, J. (2009). Glial cell missing-1 transcription factor is required for the differentiation of the human trophoblast. Cell Death Differ. 16, 719–727. doi: 10.1038/cdd.2009.1
Baylin, S. B., Esteller, M., Rountree, M. R., Bachman, K. E., Schuebel, K., and Herman, J. G. (2001). Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10, 687–692. doi: 10.1093/hmg/10.7.687
Beaulieu, N., Morin, S., Chute, I. C., Robert, M.-F., Nguyen, H., MacLeod, A. R. (2002). An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J. Biol. Chem. 277, 28176–28181. doi: 10.1074/jbc.M204734200
Bischof, P., and Irminger-Finger, I. (2005). The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 37, 1–16. doi: 10.1016/j.biocel.2004.05.014
Bischoff, P., Meisser, A., and Campana, A. (2000). Paracrine and autocrine regulators of trophoblast invasion—A review. Placenta 21(Suppl. A), S55–S60. doi: 10.1053/plac.2000.0521
Blond, J.-L., Lavillette, D., Cheynet, V., Bouton, O., Oriol, G., Chapel-Fernandes, S., et al. (2000). An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74, 3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000
Boyes, J., and Bird, A. (1991). DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64, 1123–1134. doi: 10.1016/0092-8674(91)90267-3
Braconi, C., Huang, N., and Patel, T. (2010). MicroRNA-dependent regulation of, DNA. methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 51, 881–890. doi: 10.1002/hep.23381
Caiafa, P., and Zampieri, M. (2005). DNA methylation and chromatin structure: the puzzling CpG islands. J. Cell. Biochem. 94, 257–265. doi: 10.1002/jcb.20325
Chan, A. O.-O., Lam, S.-K., Wong, B. C.-Y., Wong, W.-M., Yuen, W.-M., Yeung, Y.-H., et al. (2003). Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut 52, 502–506. doi: 10.1136/gut.52.4.502
Chédin, F., Lieber, M. R., and Hsieh, C.-L. (2002). The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. U.S.A. 99, 16916–16921. doi: 10.1073/pnas.262443999
Chen, C.-C., Wang, K.-Y., and Shen, C.-K. J. (2012). The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J. Biol. Chem. 287, 33116–33121. doi: 10.1074/jbc.C112.406975
Chen, Y., Wang, K., and Leach, R. (2013a). 5-Aza-dC treatment induces mesenchymal-to-epithelial transition in 1st trimester trophoblast cell line HTR8/SVneo. Biochem. Biophys. Res. Commun. 432, 116–122. doi: 10.1016/j.bbrc.2013.01.075
Chen, Y., Wang, K., Qian, C.-N., and Leach, R. (2013b). DNA methylation is associated with transcription of Snail and Slug genes. Biochem. Biophys. Res. Commun. 430, 1083–1090. doi: 10.1016/j.bbrc.2012.12.034
Chen, Z.-X., Mann, J. R., Hsieh, C.-L., Riggs, A. D., and Chédin, F. (2005). Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 95, 902–917. doi: 10.1002/jcb.20447
Chiu, R. W. K., Chim, S. S. C., Wong, I. H. N., Wong, C. S. C., Lee, W.-S., To, K. F., et al. (2007). Hypermethylation of RASSF1A in human and rhesus placentas. Am. J. Pathol. 170, 941–950. doi: 10.2353/ajpath.2007.060641
Choi, J. (2010). Systems biology and epigenetic gene regulation. IET Syst. Biol. 4, 289–295. doi: 10.1049/iet-syb.2010.0008
Chu, M., Selam, F., and Taylor, H. (2004). HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol. Ther. 3, 568–572. doi: 10.4161/cbt.3.6.848
Cox, B., Kotlyar, M., Evangelou, A. I., Ignatchenko, V., Ignatchenko, A., Whiteley, K., et al. (2009). Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol. Syst. Biol. 5:279. doi: 10.1038/msb.2009.37
Cox, B., Sharma, P., Evangelou, A. I., Whiteley, K., Ignatchenko, V., Ignatchenko, A., et al. (2011). Translational analysis of mouse and human placental protein and mRNA reveals distinct molecular pathologies in human preeclampsia. Mol. Cell. Prot. 10:M111.012526. doi: 10.1074/mcp.M111.012526
Cross, J., Werb, Z., and Fisher, S. (1994). Implantation and the placenta: key pieces of the development puzzle. Science 266, 1508–1518. doi: 10.1126/science.7985020
Cui, M., Wen, Z., Yang, Z., Chen, J., and Wang, F. (2009). Estrogen regulates DNA methyltransferase 3B expression in Ishikawa endometrial adenocarcinoma cells. Mol. Biol. Rep. 36, 2201–2207. doi: 10.1007/s11033-008-9435-9
Dahl, C., Gronbaek, K., and Guldberg, P. (2011). Advances in DNA methylation: 5-hydroxymethylcytosine revisited. Clin. Chim. Acta 412, 831–836. doi: 10.1016/j.cca.2011.02.013
Dawlaty, M. M., Breiling, A., Le, T., Raddatz, G., Barrasa, M. I., Cheng, A. W., et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323. doi: 10.1016/j.devcel.2012.12.015
Dawlaty, M. M., Ganz, K., Powell Benjamin, E., Hu, Y.-C., Markoulaki, S., Cheng Albert, W., et al. (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175. doi: 10.1016/j.stem.2011.07.010
Di Croce, L., Raker, V. A., Corsaro, M., Fazi, F., Fanelli, M., Faretta, M., et al. (2002). Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295, 1079–1082. doi: 10.1126/science.1065173
Dubinsky, V., Poehlmann, T. G., Suman, P., Gentile, T., Markert, U. R., and Gutierrez, G. (2010). Role of regulatory and angiogenic cytokines in invasion of trophoblastic cells. Am. J. Reprod. Immunol. 63, 193–199. doi: 10.1111/j.1600-0897.2009.00778.x
Dufer, J., Poupon, M. F., and Yatouji, S. (2000). Nuclear DNA content and chromatin pattern of rat rhabdomyosarcoma cell sublines with different metastatic potentials. Anal. Cell. Pathol. 20, 41–48.
Ehrlich, M. (2009). DNA hypomethylation in cancer cells. Epigenomics 1, 239–259. doi: 10.2217/epi.09.33
Esteller, M. (2008). Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159. doi: 10.1056/NEJMra072067
Esteve, P.-O., Chin, H. G., and Pradhan, S. (2005). Human maintenance, DNA. (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc. Natl. Acad. Sci. U.S.A. 102, 1000–1005. doi: 10.1073/pnas.0407729102
Farthing, C. R., Ficz, G., Ng, R. K., Chan, C.-F., Andrews, S., Dean, W., et al. (2008). Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4:e1000116. doi: 10.1371/journal.pgen.1000116
Ficz, G., Branco, M. R., Seisenberger, S., Santos, F., Krueger, F., Hore, T. A., et al. (2011). Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402. doi: 10.1038/nature10008
Fisher, S. J., and Damsky, C. H. (1993). Human cytotrophoblast invasion. Sem. Cell Biol. 4, 183–188. doi: 10.1006/scel.1993.1022
Fitzgerald, J. S., Germeyer, A., Huppertz, B., Jeschke, U., Knöfler, M., Moser, G., et al. (2010). Governing the invasive trophoblast: current aspects on intra- and extracellular regulation. Am. J. Reprod. Immunol. 63, 492–505. doi: 10.1111/j.1600-0897.2010.00824.x
Flanagan, J. M. (2006). Host epigenetic modifications by oncogenic viruses. Br. J. Cancer 96, 183–188. doi: 10.1038/sj.bjc.6603516
Freitag, M., and Selker, E. U. (2005). Controlling DNA methylation: many roads to one modification. Curr. Opin. Genet. Dev. 15, 191–199. doi: 10.1016/j.gde.2005.02.003
Fu, G., Brkiæ, J., Hayder, H., and Peng, C. (2013). MicroRNAs in human placental development and pregnancy complications. Int. J. Mol. Sci. 14, 5519–5544. doi: 10.3390/ijms14035519
Fu, Y., Chin, L. K., Bourouina, T., Liu, A. Q., and VanDongen, A. M. J. (2012). Nuclear deformation during breast cancer cell transmigration. Lab Chip 12, 3774–3778. doi: 10.1039/c2lc40477j
Fuks, F., Burgers, W. A., Brehm, A., Hughes-Davies, L., and Kouzarides, T. (2000). DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24, 88–91. doi: 10.1038/71750
Fuks, F., Burgers, W. A., Godin, N., Kasai, M., and Kouzarides, T. (2001). Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20, 2536–2544. doi: 10.1093/emboj/20.10.2536
Fuks, F., Hurd, P. J., Wolf, D., Nan, X., Bird, A. P., and Kouzarides, T. (2003). The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278, 4035–4040. doi: 10.1074/jbc.M210256200
Gama-Sosa, M. A., Slagel, V. A., Trewyn, R. W., Oxenhandler, R., Kuo, K. C., Gehrke, C. W., et al. (1983). The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 11, 6883–6894. doi: 10.1093/nar/11.19.6883
Geiman, T. M., Sankpal, U. T., Robertson, A. K., Zhao, Y., Zhao, Y., and Robertson, K. D. (2004). DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochem. Biophys. Res. Commun. 318, 544–555. doi: 10.1016/j.bbrc.2004.04.058
Gellersen, B., and Kempf, R. (1990). Human prolactin gene expression: positive correlation between site- specific methylation and gene activity in a set of human lymphoid cell lines. Mol. Endocrinol. 4, 1874–1886. doi: 10.1210/mend-4-12-1874
Gerlitz, G., and Bustin, M. (2010). Efficient cell migration requires global chromatin condensation. J. Cell Sci. 123, 2207–2217. doi: 10.1242/jcs.058271
Graham, C., and Lala, P. (1992). Mechanisms of placental invasion of the uterus and their control. Biochem. Cell Biol. 70, 867–874. doi: 10.1139/o92-135
Grigoriu, A., Ferreira, J. C., Choufani, S., Baczyk, D., Kingdom, J., and Weksberg, R. (2011). Cell specific patterns of methylation in the human placenta. Epigenetics 6, 368–379. doi: 10.4161/epi.6.3.14196
Guilleret, I., Osterheld, M.-C., Braunschweig, R., Gastineau, V., Taillens, S., and Benhattar, J. (2009). Imprinting of tumor-suppressor genes in human placenta. Epigenetics. 4, 62–68. doi: 10.4161/epi.4.1.7471
Guo, S.-W. (2009). Epigenetics of endometriosis. Mol. Hum. Reprod. 15, 587–607. doi: 10.1093/molehr/gap064
Hamilton, D. W., Lusher, M. E., Lindsey, J. C., Ellison, D. W., and Clifford, S. C. (2005). Epigenetic inactivation of the RASSF1A tumour suppressor gene in ependymoma. Cancer Lett. 227, 75–81. doi: 10.1016/j.canlet.2004.11.044
Hammadeh, M. E., Stieber, M., Haidl, G., and Schmidt, W. (1998). Association between sperm cell chromatin condensation, morphology based on strict criteria, and fertilization, cleavage and pregnancy rates in an IVF program. Andrologia 30, 29–35. doi: 10.1111/j.1439-0272.1998.tb01379.x
Hannon, T., Innes, B. A., Lash, G. E., Bulmer, J. N., and Robson, S. C. (2012). Effects of local decidua on trophoblast invasion and spiral artery remodeling in focal placenta creta – An immunohistochemical study. Placenta 33, 998–1004. doi: 10.1016/j.placenta.2012.09.004
Haouzi, D., Dechaud, H., Assou, S., Monzo, C., de Vos, J., and Hamamah, S. (2011). Transcriptome analysis reveals dialogues between human trophectoderm and endometrial cells during the implantation period. Hum. Reprod. 26, 1440–1449. doi: 10.1093/humrep/der075
Hashimoto, K., Kokubun, S., Itoi, E., and Roach, H. I. (2010). Inflammatory cytokines cause loss of DNA methylation together with induction of abnormal gene expression in healthy human articular chondrocytes. J. Bone Joint Surg [Br]. 92-B(Suppl. I), 55.
Hashimoto, K., Otero, M., Imagawa, K., de Andres, M. C., Coico, J. M., Roach, H. I., et al. (2013). Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1 beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J. Biol. Chem. 288, 10061–10072. doi: 10.1074/jbc.M112.421156
Hata, K., Okano, M., Lei, H., and Li, E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993.
Hattori, N., Nishino, K., Ko, Y.-G., Hattori N., Ohgane J., Tanaka, S., et al. (2004). Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 279, 17063–17069. doi: 10.1074/jbc.M309002200
Hemberger, M. (2007). Epigenetic landscape required for placental development. Cell. Mol. Life Sci. 64, 2422–2436. doi: 10.1007/s00018-007-7113-z
Hemberger, M. (2010). Genetic-epigenetic intersection in trophoblast differentiation: implications for extraembryonic tissue function. Epigenetics 5, 24–29. doi: 10.4161/epi.5.1.10589
Hervouet, E., Vallette, F. M., and Cartron, P.-F. (2009). Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics 4, 487–499. doi: 10.4161/epi.4.7.9883
Hsieh, C.-L. (1999). In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19, 8211–8218.
Hsu, C.-H., Peng, K.-L., Kang, M.-L., Chen, Y.-R., Yang, Y.-C., Tsai, C.-H., et al. (2012). TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2, 568–579. doi: 10.1016/j.celrep.2012.08.030
Ito, S., D'Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Jackson, M., Krassowska, A., Gilbert, N., Chevassut, T., Forrester, L., Ansell, J., et al. (2004). Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 24, 8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004
Ji, L., Brkić, J., Liu, M., Fu, G., Peng, C., and Wang, Y.-L. (2013). Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Mol. Aspect Med. 34, 981–1023. doi: 10.1016/j.mam.2012.12.008
Jin, F., Dowdy, S. C., Xiong, Y., Eberhardt, N. L., Podratz, K. C., and Jiang, S.-W. (2005). Up-regulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol. Oncol. 96, 531–538. doi: 10.1016/j.ygyno.2004.10.039
Jones, A., Teschendorff, A. E., Li, Q., Hayward, J. D., Kannan, A., Mould, T., et al. (2013). Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med. 10:e1001551. doi: 10.1371/journal.pmed.1001551
Jones, P. A., and Baylin, S. B. (2007). The epigenomics of cancer. Cell 128, 683–692. doi: 10.1016/j.cell.2007.01.029
Kangaspeska, S., Stride, B., Metivier, R., Polycarpou-Schwarz, M., Ibberson, D., Carmouche, R. P., et al. (2008). Transient cyclical methylation of promoter DNA. Nature 452, 112–115. doi: 10.1038/nature06640
Karmakar, S., and Das, C. (2002). Regulation of trophoblast invasion by, I.L-.1β and TGF- β1. Am. J. Reprod. Immunol. 48, 210–219. doi: 10.1034/j.1600-0897.2002.01151.x
Karmakar, S., and Das, C. (2004). Modulation of ezrin and, E-.cadherin expression by IL-1β and TGF-β1 in human trophoblasts. J. Reprod. Immunol. 64, 9–29. doi: 10.1016/j.jri.2004.04.005
Khong, Y., and Brosens, I. (2011). Defective deep placentation. Best Prac. Res. Clin. Obstet. Gynaecol. 25, 301–311. doi: 10.1016/j.bpobgyn.2010.10.012
Kimura, H., Nakamura, T., Ogawa, T., Tanaka, S., and Shiota, K. (2003). Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acid Res. 31, 3101–3113. doi: 10.1093/nar/gkg406
Koh, K. P., Yabuuchi, A., Rao, S., Huang, Y., Cunniff, K., Nardone, J., et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. doi: 10.1016/j.stem.2011.01.008
Kokkinos, M. I., Murthi, P., Wafai, R., Thompson, E. W., and Newgreen, D. F. (2010). Cadherins in the human placenta – epithelial–mesenchymal transition (EMT) and placental development. Placenta 31, 747–755. doi: 10.1016/j.placenta.2010.06.017
Kudo, Y., Tateishi, K., Yamamoto, K., Yamamoto, S., Asaoka, Y., Ijichi, H., et al. (2012). Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 103, 670–676. doi: 10.1111/j.1349-7006.2012.02213.x
Kulis, M., and Esteller, M. (2010). “DNA methylation and cancer,” in Advances in Genetics, eds H. Zdenko and U. Toshikazu (Waltham, MA: Academic Press), 27–56.
Lash, G. E., Robson, S. C., and Bulmer, J. N. (2010). Review: functional role of uterine natural killer (uNK) cells in human early pregnancy decidua. Placenta 31(Suppl.), S87–S92. doi: 10.1016/j.placenta.2009.12.022
Li, B.-L. W., Lu, C., Lu, J.-J., Qu, T.-T., Yang, Q. Y., and Wan, X.-P. (2013). CpG island hypermethylation-associated silencing of microRNAs promotes human endometrial cancer. Cancer Cell. Int. 13:44. doi: 10.1186/1475-2867-13-44
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi: 10.1016/0092-8674(92)90611-F
Li, J.-Y., Pu, M.-T., Hirasawa, R., Li, B.-Z., Huang, Y.-N., Zeng, R., et al. (2007). Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 27, 8748–8759. doi: 10.1128/MCB.01380-07
Lian, C. G., Xu, Y., Ceol, C., Wu, F., Larson, A., Dresser, K., et al. (2012). Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150, 1135–1146. doi: 10.1016/j.cell.2012.07.033
Liang, G., Chan, M. F., Tomigahara, Y., Tsai, Y. C., Gonzales, F. A., Li, E., et al. (2002). Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22, 480–491. doi: 10.1128/MCB.22.2.480-491.2002
Liao, X., Siu, M. K.-Y., Chan, K. Y.-K., Wong, E. S.-Y., Ngan, H. Y.-S., Chan, Q. K.-Y., et al. (2008). Hypermethylation of RAS effector related genes and DNA methyltransferase 1 expression in endometrial carcinogenesis. Int. J. Cancer 123, 296–302. doi: 10.1002/ijc.23494
Librach, C. L., Feigenbaum, S. L., Bass, K. E., Cui, T. Y., Verastas, N., Sadovsky, Y., et al. (1994). Interleukin-1 beta regulates human cytotrophoblast metalloproteinase activity and invasion in vitro. J. Biol. Chem. 269, 17125–17131.
Lin, R.-K., Wu, C.-Y., Chang, J.-W., Juan, L.-J., Hsu, H.-S., Chen, C.-Y., et al. (2010). Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 70, 5807–5817. doi: 10.1158/0008-5472.CAN-09-4161
Lockwood, C., Krikun, G., and Schatz, F. (1999). The decidua regulates hemostasis in human endometrium. Semin. Reprod. Endocrinol. 17, 45–51. doi: 10.1055/s-2007-1016211
Logan, P. C., Ponnampalam, A. P., Steiner, M., and Mitchell, M. D. (2013). Effect of cyclic AMP and estrogen/progesterone on the transcription of DNA methyltransferases during decidualization of human endometrial stromal cells. Mol. Hum. Reprod. 19, 302–312. doi: 10.1093/molehr/gas062
Maccani, M. A., and Carmen, J. M. (2009). Epigenetics in the placenta. Am. J. Reprod. Immunol. 62, 78–89. doi: 10.1111/j.1600-0897.2009.00716.x
Matousková, M., Blazková, J., Pajer, P., Pavlícek, A., and Hejnar, J. (2006). CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell Res. 312, 1011–1020. doi: 10.1016/j.yexcr.2005.12.010
Mattson, M. P., and Taub, D. D. (2004). Ancient viral protein enrages astrocytes in multiple sclerosis. Nat. Neurosci. 7, 1021–1023. doi: 10.1038/nn1004-1021
Metivier, R., Gallais, R., Tiffoche, C., Le Peron, C., Jurkowska, R. Z., Carmouche, R. P., et al. (2008). Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50. doi: 10.1038/nature06544
Mi, S., Lee, X., Li, X.-P., Veldman, G. M., Finnerty, H., Racie, L., et al. (2000). Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789. doi: 10.1038/35001608
Moser, G., Gauster, M., Orendi, K., Glasner, A., Theuerkauf, R., and Huppertz, B. (2010). Endoglandular trophoblast, an alternative route of trophoblast invasion? Analysis with novel confrontation co-culture models. Hum. Reprod. 25, 1127–1136. doi: 10.1093/humrep/deq035
Mouillet, J.-F., Donker, R. B., Mishima, T., Cronqvist, T., Chu, T., and Sadovsky, Y. (2013). The unique expression and function of miR-424 in human placental trophoblasts. Biol. Reprod. 89, 25, 1–9. doi: 10.1095/biolreprod.113.110049
Muir, A., Lever, A. M. L., and Moffett, A. (2006). Human endogenous retrovirus-W envelope (syncytin) is expressed in both villous and extravillous trophoblast populations. J. Gen. Virol. 87, 2067–2071. doi: 10.1099/vir.0.81412-0
Nakanishi, M. O., Hayakawa, K., Nakabayashi, K., Hata, K., Shiota, K., and Tanaka, S. (2012). Trophoblast-specific DNA methylation occurs after the segregation of the trophectoderm and inner cell mass in the mouse periimplantation embryo. Epigenetics 7, 173–182. doi: 10.4161/epi.7.2.18962
Nakano, K., Boyle, D. L., and Firestein, G. S. (2013). Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J. Immunol. 190, 1297–1303. doi: 10.4049/jimmunol.1202572
Nelissen, E. C. M., van Montfoort, A. P. A., Dumoulin, J. C. M., and Evers, J. L. H. (2011). Epigenetics and the placenta. Hum. Reprod. Update 17, 397–417. doi: 10.1093/humupd/dmq052
Ng, H.-H., and Bird, A. (1999). DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9, 158–163. doi: 10.1016/S0959-437X(99)80024-0
Ng, R. K., Dean, W., Dawson, C., Lucifero, D., Madeja, Z., Reik, W., et al. (2008). Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290. doi: 10.1038/ncb1786
Niesen, M. I., Osborne, A. R., Yang, H., Rastogi, S., Chellappan, S., Cheng, J. Q., et al. (2005). Activation of a methylated promoter mediated by a sequence-specific DNA-binding protein, RFX. J. Biol. Chem. 280, 38914–38922. doi: 10.1074/jbc.M504633200
Norwitz, E. R. (2006). Defective implantation and placentation: laying the blueprint for pregnancy complications. Reprod. Biomed. Online 13, 591–599. doi: 10.1016/S1472-6483(10)60649-9
Novakovic, B., Rakyan, V., Ng, H. K., Manuelpillai, U., Dewi, C., Wong, N. C., et al. (2008). Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Mol. Hum. Reprod. 14, 547–554. doi: 10.1093/molehr/gan046
Novakovic, B., and Saffery, R. (2012). The ever growing complexity of placental epigenetics – Role in adverse pregnancy outcomes and fetal programming. Placenta 33, 959–970. doi: 10.1016/j.placenta.2012.10.003
Novakovic, B., Sibson, M., Ng, H. K., Manuelpillai, U., Rakyan, V., Down, T., et al. (2009). Placenta-specific methylation of the vitamin D 24-hydroxylase gene: implications for feedback autoregulation of active vitamin D levels at the fetomaternal interface. J. Biol. Chem. 284, 14838–14848. doi: 10.1074/jbc.M809542200
Novakovic, B., Wong, N. C., Sibson, M., Ng, H.-K., Morley, R., Manuelpillai, U., et al. (2010). DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. J. Biol. Chem. 285, 9583–9593. doi: 10.1074/jbc.M109.064956
Oda, M., Oxley, D., Dean, W., and Reik, W. (2013). Regulation of lineage specific DNA hypomethylation in mouse trophectoderm. PLoS ONE 8:e68846. doi: 10.1371/journal.pone.0068846
Oh, J. H., Gertych, A., and Tajbakhsh, J. (2013). Nuclear DNA methylation and chromatin condensation phenotypes are distinct between normally proliferating/aging, rapidly growing/immortal, and senescent cells. Oncotarget 4, 474–493.
Ohgane, J., Hattori, N., Oda, M., Tanaka, S., and Shiota, K. (2002). Differentiation of trophoblast lineage is associated with DNA methylation and demethylation. Biochem. Biophys. Res. Commun. 290, 701–706. doi: 10.1006/bbrc.2001.6258
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi: 10.1016/S0092-8674(00)81656-6
Pastor, W. A., Aravind, L., and Rao, A. (2013). TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell. Biol. 14, 341–356. doi: 10.1038/nrm3589
Pavlopoulou, A., and Kossida, S. (2010). Cytosine methyltransferases as tumor markers. Curr. Genomics 11, 568–577. doi: 10.2174/138920210793360916
Perez, C., Martinez-Calle, N., Martin-Subero, J. I., Segura, V., Delabesse, E., Fernandez-Mercado, M., et al. (2012). TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS ONE 7:e31605. doi: 10.1371/journal.pone.0031605
Perron, H., Garson, J. A., Bedin, F., Beseme, F., Paranhos-Baccala, G., Komurian-Pradel, F., et al. (1997). Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 94, 7583–7588. doi: 10.1073/pnas.94.14.7583
Perry, J. K., Lins, R. J., Lobie, P. E., and Mitchell, M. D. (2010). Regulation of invasive growth: similar epigenetic mechanisms underpin tumour progression and implantation in human pregnancy. Clin. Sci. 118, 451–457. doi: 10.1042/CS20090503
Pfeifer, G., Kadam, S., and Jin, S.-G. (2013). 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 6, 10. doi: 10.1186/1756-8935-6-10
Pijnenborg, R., Dixon, G., Robertson, W., and Brosens, I. (1980). Trophoblastic invasion of human decidua from 8 to 18 weeks of pregnancy. Placenta 1, 3–19. doi: 10.1016/S0143-4004(80)80012-9
Pollheimer, J., Loregger, T., Sonderegger, S., Saleh, L., Bauer, S., Bilban, M., et al. (2006). Activation of the canonical wingless/T-cell factor signaling pathway promotes invasive differentiation of human trophoblast. Am. J. Pathol. 168, 1134–1147. doi: 10.2353/ajpath.2006.050686
Pringle, K. G., Tadros, M. A., Callister, R. J., and Lumbers, E. R. (2011). The expression and localization of the human placental prorenin/renin-angiotensin system throughout pregnancy: Roles in trophoblast invasion and angiogenesis? Placenta 32, 956–962. doi: 10.1016/j.placenta.2011.09.020
Raddatz, G., Gao, Q., Bender, S., Jaenisch, R., and Lyko, F. (2012). Dnmt3a protects active chromosome domains against cancer-associated hypomethylation. PLoS Genet. 8:e1003146. doi: 10.1371/journal.pgen.1003146
Rahnama, F., Shafiei, F., Gluckman, P. D., Mitchell, M. D., and Lobie, P. E. (2006). Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology 147, 5275–5283. doi: 10.1210/en.2006-0288
Rahnama, F., Thompson, B., Steiner, M., Shafiei, F., Lobie, P. E., and Mitchell, M. D. (2009). Epigenetic regulation of, E-cadherin controls endometrial receptivity. Endocrinology 150, 1466–1472. doi: 10.1210/en.2008-1142
Reik, W., and Walter, J. (2001). Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32. doi: 10.1038/35047554
Robertson, K. D., Ait-Si-Ali, S., Yokochi, T., Wade, P. A., Jones, P. L., and Wolffe, A. P. (2000). DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25, 338–342. doi: 10.1038/77124
Roper, S., and Hemberger, M. (2009). Defining pathways that enforce cell lineage specification in early development and stem cells. Cell Cycle 8, 1515–1525. doi: 10.4161/cc.8.10.8381
Ruebner, M., Strissel, P. L., Ekici, A. B., Stiegler, E., Dammer, U., Goecke, T. W., et al. (2013). Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region. PLoS ONE 8:e56145. doi: 10.1371/journal.pone.0056145
Rugg-Gunn, P. J. (2012). Epigenetic features of the mouse trophoblast. Reprod. Biomed. Online 25, 21–30. doi: 10.1016/j.rbmo.2012.01.012
Ruzov, A., Tsenkina, Y., Serio, A., Dudnakova, T., Fletcher, J., Bai, Y., et al. (2011). Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 21, 1332–1342. doi: 10.1038/cr.2011.113
Saito, T., Nishimura, M., Yamasaki, H., and Kudo, R. (2003). Hypermethylation in promoter region of E-cadherin gene is associated with tumor dedifferention and myometrial invasion in endometrial carcinoma. Cancer 97, 1002–1009. doi: 10.1002/cncr.11157
Sakaue, M., Ohta, H., Kumaki, Y., Oda, M., Sakaide, Y., Matsuoka, C., et al. (2010). DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr. Biol. 20, 1452–1457. doi: 10.1016/j.cub.2010.06.050
Salker, M. S., Christian, M., Steel, J. H., Nautiyal, J., Lavery, S., Trew, G., et al. (2011). Deregulation of the serum- and glucocorticoid-inducible kinase SGK1 in the endometrium causes reproductive failure. Nat. Med. 17, 1509–1513. doi: 10.1038/nm.2498
Santamaria, X., Massasa, E. E., and Taylor, H. S. (2012). Migration of cells from experimental endometriosis to the uterine endometrium. Endocrinology 153, 5566–5574. doi: 10.1210/en.2012-1202
Sato, N., Fukushima, N., Matsubayashi, H., and Goggins, M. (2003). Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene 23, 1531–1538. doi: 10.1038/sj.onc.1207269
Schroeder, D. I., Blair, J. D., Lott, P., Yu, H. O. K., Hong, D., Crary, F., et al. (2013). The human placenta methylome. Proc. Natl. Acad. Sci. U.S.A. 110, 6037–6042. doi: 10.1073/pnas.1215145110
Seisenberger, S., Peat, J. R., Hore, T. A., Santos, F., Dean, W., and Reik, W. (2013). Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368. doi: 10.1098/rstb.2011.0330
Senner, C. E. (2011). The role of DNA methylation in mammalian development. Reprod. Biomed. Online 22, 529–535. doi: 10.1016/j.rbmo.2011.02.016
Senner, C. E., and Hemberger, M. (2010). Regulation of early trophoblast differentiation - Lessons from the mouse. Placenta 31, 944–950. doi: 10.1016/j.placenta.2010.07.013
Senner, C. E., Krueger, F., Oxley, D., Andrews, S., and Hemberger, M. (2012). DNA methylation profiles define stem cell identity and reveal a tight embryonic–extraembryonic lineage boundary. Stem Cells 30, 2732–2745. doi: 10.1002/stem.1249
Shaco-Levy, R., Sharabi, S., Benharroch, D., Piura, B., and Sion-Vardy, N. (2008). Matrix metalloproteinases 2 and 9, E-cadherin, and β-catenin expression in endometriosis, low-grade endometrial carcinoma and non-neoplastic eutopic endometrium. Eur. J. Obstet. Gynecol. Reprod. Biol. 139, 226–232. doi: 10.1016/j.ejogrb.2008.01.004
Shafiei, F., Rahnama, F., Pawella, L., Mitchell, M. D., Gluckman, P. D., and Lobie, P. E. (2008). DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells. Oncogene 27, 2602–2612. doi: 10.1038/sj.onc.1210917
Shamay, M., Krithivas, A., Zhang, J., and Hayward, S. D. (2006). Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U.S.A. 103, 14554–14559. doi: 10.1073/pnas.0604469103
Shen, X., He, Z., Li, H., Yao, C., Zhang, Y., He, L., et al. (2012). Distinct functional patterns of gene promoter hypomethylation and hypermethylation in cancer genomes. PLoS ONE 7:e44822. doi: 10.1371/journal.pone.0044822
Shi, X., Wu, Y., Liu, H., Gong, X., Du, H., Li, Y., et al. (2012). Effect of epigenetic modification of maspin on extravillous trophoblastic function. J. Huazhong Univ. Sci. Technol. Med. Sci. 32, 879–882. doi: 10.1007/s11596-012-1051-1
Simpkins, S. B., Bocker, T., Swisher, E. M., Mutch, D. G., Gersell, D. J., Kovatich, A. J., et al. (1999). MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum. Mol. Genet. 8, 661–666. doi: 10.1093/hmg/8.4.661
Song, S. J., Poliseno, L., Song Min, S., Ala, U., Webster, K., Ng, C., et al. (2013). MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154, 311–324. doi: 10.1016/j.cell.2013.06.026
Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H., and Tajima, S. (2004). DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816–27823. doi: 10.1074/jbc.M400181200
Suzuki, M., Yamada, T., Kihara-Negishi, F., Sakurai, T., Hara, E., Tenen, D. G., et al. (2005). Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b. Oncogene 25, 2477–2488. doi: 10.1038/sj.onc.1209272
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Takeshima, H., Suetake, I., Shimahara, H., Ura, K., Tate, S.-I., and Tajima, S. (2006). Distinct DNA methylation activity of Dnmt3a and Dnmt3b towards naked and nucleosomal DNA. J. Biochem. 139, 503–515. doi: 10.1093/jb/mvj044
Thalhammer, A., Hansen, A. S., El-Sagheer, A. H., Brown, T., and Schofield, C. J. (2011). Hydroxylation of methylated CpG dinucleotides reverses stabilisation of DNA duplexes by cytosine 5-methylation. Chem. Commun. 47, 5325–5327. doi: 10.1039/c0cc05671e
Trojer, P., and Reinberg, D. (2007). Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28, 1–13. doi: 10.1016/j.molcel.2007.09.011
Tsai, K.-W., Kao, H.-W., Chen, S.-J., and Lin, W.-C. (2009). Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 4, 587–592. doi: 10.4161/epi.4.8.10230
Tsumura, A., Hayakawa, T., Kumaki, Y., Takebayashi, S.-I., Sakaue, M., Matsuoka, C., et al. (2006). Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814. doi: 10.1111/j.1365-2443.2006.00984.x
Turner, A., Lones, M., Fuente, L., Stepney, S., Caves, L., and Tyrrell, A. (2012). “Using artificial epigenetic regulatory networks to control complex tasks within chaotic systems,” in Information Processing in Cells and Tissues, eds M. Lones, S. Smith, S. Teichmann, F. Naef, J. Walker, and M. Trefzer (Berlin; Heidelberg: Springer), 1–11. doi: 10.1007/978-3-642-28792-3_1
van Dijk, M., Thulluru, H. K., Mulders, J., Michel, O. J., Poutsma, A., Windhorst, S., et al. (2012). HELLP babies link a novel lincRNA to the trophoblast cell cycle. J. Clin. Invest. 122, 4003–4011. doi: 10.1172/JCI65171
Varley, K. E., Gertz, J., Bowling, K. M., Parker, S. L., Reddy, T. E., Pauli-Behn, F., et al. (2013). Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567. doi: 10.1101/gr.147942.112
Vincent, Z. L., Farquhar, C. M., Mitchell, M. D., and Ponnampalam, A. P. (2011). Expression and regulation of DNA methyltransferases in human endometrium. Fertil. Steril. 95, 1522.e1–1525.e1 doi: 10.1016/j.fertnstert.2010.09.030
Walton, E. L., Francastel, C., and Velasco, G. (2011). Maintenance of DNA methylation: Dnmt3b joins the dance. Epigenetics 6, 1373–1377. doi: 10.4161/epi.6.11.17978
Wang, X., Miller, D. C., Harman, R., Antczak, D. F., and Clark, A. G. (2013a). Paternally expressed genes predominate in the placenta. Proc. Natl. Acad. Sci. U.S.A. 110, 10705–10710. doi: 10.1073/pnas.1308998110
Wang, Y., Pringle, K. G., and Lumbers, E. R. (2013b). The effects of cyclic AMP, sex steroids and global hypomethylation on the expression of genes controlling the activity of the renin - angiotensin system in placental cell lines. Placenta 34, 275–280. doi: 10.1016/j.placenta.2012.12.018
Wang, Y., Pringle, K. G., Chen, Y. X., Zakar, T., and Lumbers, E. R. (2012). Regulation of the renin-angiotensin system (RAS) in BeWo and HTR-8/SVneo trophoblast cell lines. Placenta 33, 634–639. doi: 10.1016/j.placenta.2012.05.001
Wanunu, M., Cohen-Karni, D., Johnson, R. R., Fields, L., Benner, J., Peterman, N., et al. (2010). Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J. Am. Chem. Soc. 133, 486–492. doi: 10.1021/ja107836t
Watt, F., and Molloy, P. L. (1988). Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 2, 1136–1143. doi: 10.1101/gad.2.9.1136
Whitcomb, B. P., Mutch, D. G., Herzog, T. J., Rader, J. S., Gibb, R. K., and Goodfellow, P. J. (2003). Frequent HOXA11 and THBS2 promoter methylation, and a methylator phenotype in endometrial adenocarcinoma. Clin. Cancer Res. 9, 2277–2287.
Whiteside, E. J., Jackson, M. M., Herington, A. C., Edwards, D. R., and Harvey, M. B. (2001). Matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-3 are key regulators of extracellular matrix degradation by mouse embryos. Biol. Reprod. 64, 1331–1337. doi: 10.1095/biolreprod64.5.1331
Williams, K., Christensen, J., and Helin, K. (2012). DNA methylation: TET proteins - guardians of CpG islands? EMBO Rep. 13, 28–35. doi: 10.1038/embor.2011.233
Williams, P. J., Mistry, H. D., Innes, B. A., Bulmer, J. N., and Broughton Pipkin, F. (2010). Expression of AT1R, AT2R and AT4R and their roles in extravillous trophoblast invasion in the human. Placenta 31, 448–455. doi: 10.1016/j.placenta.2010.02.014
Wong, N. C., Novakovic, B., Weinrich, B., Dewi, C., Andronikos, R., Sibson, M., et al. (2008). Methylation of the adenomatous polyposis coli (APC) gene in human placenta and hypermethylation in choriocarcinoma cells. Cancer Lett. 268, 56–62. doi: 10.1016/j.canlet.2008.03.033
Wu, Y., Halverson, G., Basir, Z., Strawn, E., Yan, P., and Guo, S.-W. (2005). Aberrant methylation at HOXA10 may be responsible for its aberrant expression in the endometrium of patients with endometriosis. Am. J. Obstet. Gynecol. 193, 371–380. doi: 10.1016/j.ajog.2005.01.034
Wu, Y., Strawn, E., Basir, Z., Halverson, G., and Guo, S.-W. (2007). Aberrant expression of deoxyribonucleic acid methyltransferases DNMT1, DNMT3A, and DNMT3B in women with endometriosis. Fertil. Steril. 87, 24–32. doi: 10.1016/j.fertnstert.2006.05.077
Xie, W., Schultz Matthew, D., Lister, R., Hou, Z., Rajagopal, N., Ray, P., et al. (2013). Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148. doi: 10.1016/j.cell.2013.04.022
Xu, Y., Wu, F., Tan, L., Kong, L., Xiong, L., Deng, J., et al. (2011). Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464. doi: 10.1016/j.molcel.2011.04.005
Yamagata, Y., Asada, H., Tamura, I., Lee, L., Maekawa, R., Taniguchi, K., et al. (2009). DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum. Reprod. 24, 1126–1132. doi: 10.1093/humrep/dep015
Yuen, R. K. C., Chen, B., Blair, J. D., Robinson, W. P., and Nelson, D. M. (2013). Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics 8, 192–202. doi: 10.4161/epi.23400
Zafarana, G., Avery, S. R., Avery, K., Moore, H. D., and Andrews, P. W. (2009). Specific knockdown of OCT4 in human embryonic stem cells by inducible short hairpin RNA interference. Stem Cells 27, 776–782. doi: 10.1002/stem.5
Zhang, H. J., Siu, M. K. Y., Wong, E. S. Y., Wong, K. Y., Li, A. S. M., Chan, K. Y. K., et al. (2008). Oct4 is epigenetically regulated by methylation in normal placenta and gestational trophoblastic disease. Placenta 29, 549–554. doi: 10.1016/j.placenta.2008.03.003
Zhang, Q., Wang, H. Y., Marzec, M., Raghunath, P. N., Nagasawa, T., and Wasik, M. A. (2005). STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 102, 6948–6953. doi: 10.1073/pnas.0501959102
Zhao, H., and Chen, T. (2013). Tet family of 5-methylcytosine dioxygenases in mammalian development. J. Hum. Genet. 58, 421–427. doi: 10.1038/jhg.2013.63
Keywords: DNA methyltransferases, trophoblasts, cell differentiation, TETs, placenta, chromatin condensation, hydroxymethylation, epigenetics
Citation: Logan PC, Mitchell MD and Lobie PE (2013) DNA methyltransferases and TETs in the regulation of differentiation and invasiveness of extra-villous trophoblasts. Front. Genet. 4:265. doi: 10.3389/fgene.2013.00265
Received: 06 August 2013; Accepted: 15 November 2013;
Published online: 04 December 2013.
Edited by:
Marie van Dijk, VU University Medical Center, NetherlandsReviewed by:
Lori A. Pile, Wayne State University, USACopyright © 2013 Logan, Mitchell and Lobie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip C. Logan, The Liggins Institute, University of Auckland, 85 Park Road, Private Bag 92019, Auckland 1023, New Zealand e-mail:cGMubG9nYW5AYXVja2xhbmQuYWMubno=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.