
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 28 October 2013
Sec. Behavioral and Psychiatric Genetics
Volume 4 - 2013 | https://doi.org/10.3389/fgene.2013.00213
This article is part of the Research Topic Ion Channels and Mental Illness: Exploring etiology and pathophysiology in major psychiatric disorders View all 9 articles
The sodium channel Nav1.6, encoded by the gene SCN8A, is one of the major voltage-gated channels in human brain. The sequences of sodium channels have been highly conserved during evolution, and minor changes in biophysical properties can have a major impact in vivo. Insight into the role of Nav1.6 has come from analysis of spontaneous and induced mutations of mouse Scn8a during the past 18 years. Only within the past year has the role of SCN8A in human disease become apparent from whole exome and genome sequences of patients with sporadic disease. Unique features of Nav1.6 include its contribution to persistent current, resurgent current, repetitive neuronal firing, and subcellular localization at the axon initial segment (AIS) and nodes of Ranvier. Loss of Nav1.6 activity results in reduced neuronal excitability, while gain-of-function mutations can increase neuronal excitability. Mouse Scn8a (med) mutants exhibit movement disorders including ataxia, tremor and dystonia. Thus far, more than ten human de novo mutations have been identified in patients with two types of disorders, epileptic encephalopathy and intellectual disability. We review these human mutations as well as the unique features of Nav1.6 that contribute to its role in determining neuronal excitability in vivo. A supplemental figure illustrating the positions of amino acid residues within the four domains and 24 transmembrane segments of Nav1.6 is provided to facilitate the location of novel mutations within the channel protein.
SCN8A encodes one of the major voltage-gated sodium channels that regulate the initiation and propagation of action potentials in the nervous system. The sodium channel transmembrane proteins were first purified 30 years ago (Hartshorne and Catterall, 1981; Tamkun and Catterall, 1981) and cDNA clones were isolated shortly thereafter (Noda et al., 1986). The Scn8a gene, encoding the sodium channel Nav1.6, was identified in 1995 by positional cloning of the mouse neurological mutant motor endplate disease (med) (Burgess et al., 1995) and by isolation of a novel sodium channel cDNA from rat brain (Schaller et al., 1995). SCN8A is a member of the gene family comprised of nine evolutionarily related sodium channels with specific roles in neurons and in skeletal muscle and cardiac muscle (Lopreato et al., 2001; Meisler and Kearney, 2005; Meisler et al., 2010; Zakon et al., 2011; Zakon, 2012).
Human SCN8A was mapped to chromosome 12q13 in 1998 (Plummer et al., 1998). The role of SCN8A in human disease was initially investigated by screening for mutations in families segregating inherited disorders such as ataxia, dystonia, and tremor (Trudeau et al., 2006; Sharkey et al., 2009a). These analyses identified only one family with an inherited mutation of SCN8A (Trudeau et al., 2006). Recently, the ability to sequence the entire exome or genome from an individual patient has made it possible to identification of de novo mutations in patients who do not have a family history of disease (Bamshad et al., 2011; Doherty and Bamshad, 2012; Need et al., 2012; Rauch et al., 2012). Using this technology, more than ten mutations of SCN8A have been described during the past year, in patients with epileptic encephalopathy and intellectual disability. This rapid progress indicates that mutations of SCN8A are a previously unrecognized cause of these and possibly other neurological disorders. Here we describe the recently discovered patient mutations and review the unique features of Nav1.6 as a framework for understanding the pathological consequences of human mutations.
The first de novo mutation in SCN8A was discovered in 2012 by whole genome sequencing of a child with an early onset, debilitating epileptic encephalopathy. The clinical picture included developmental delay, features of autism, intellectual disability and ataxia (Veeramah et al., 2012). Afebrile seizures began at 6 months of age, and by 5 years EEG recordings detected short bursts of frontocentrally predominant generalized spike-wave activity, and bifrontal and multifocal spikes. Neither the parents nor an unaffected sibling carried the de novo mutation, p.Asn1768Asp, that was detected in the patient. The biophysical properties of the mutant channel include increase in persistent sodium current, incomplete channel inactivation, and a depolarizing shift in the voltage dependence of steady-state fast-inactivation (Veeramah et al., 2012). Current tracings of cells transfected with mutant channels reveal as much as 20% of maximal current remaining after 100 ms, compared with only 1% in cells transfected with wild-type channel (Figure 1). The elevated persistent current increases the likelihood of premature firing of neurons after subthreshold depolarization. Transfection of mouse hippocampal neurons with the mutant cDNA resulted in increased spontaneous and induced firing characteristic of neuronal hyperexcitability, consistent with the dominant expression of seizures in the heterozygous patient. Increased persistent current is also a common feature of mutations in the channel SCN1A that cause the epileptic encephalopathy Dravet Syndrome (Meisler and Kearney, 2005). Increased activity of Nav1.6 has also been implicated in the seizure-prone Celf4−/− mouse mutant (Sun et al., 2013) and suggested in fibroblast-derived neurons from patients with Dravet syndrome (Liu et al., 2013).
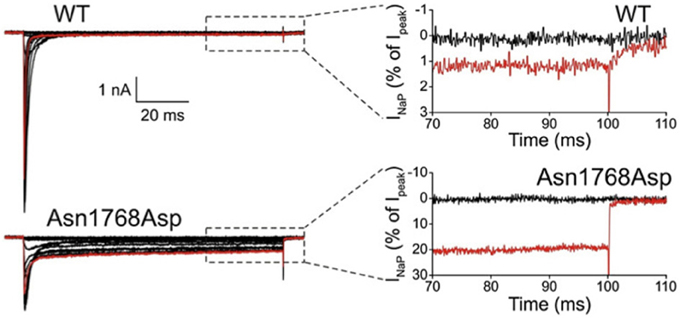
Figure 1. Increased persistent current in SCN8A-p.Asn1768Asp mutant channel. Wildtype and mutant cDNAs were transiently transfected into the neuronal cell line ND7/23. At 100 ms after induction of an action potential, cells expressing the mutant cDNA had 20% persistent current compared with 1% in the wildtype. Cells were held at −120 mV, and a family of step depolarizations (−80 to +60 mV in 5 mV increments) were applied every 5 s. Insets show persistent inward currents (normalized by maximal transient peak currents) from WT and p.Asn1768Asp channels at the end of a 100 ms step depolarization to −80 mV (black, control) and +20 mV (red). [reprinted from Veeramah et al. (2012), with permission].
A second missense mutation, SCN8A-p.Leu1331Val, was identified by targeted resequencing of 65 candidate genes in 500 individuals with epileptic encephalopathy (Carvill et al., 2013). The proband presented with epileptic encephalopathy at 18 months of age, and the mutation was inherited from a mosaic father. Two additional mutations were identified in this study, p.Arg662Cys and p.Arg1872Gln, but family data regarding inheritance was not available (Carvill et al., 2013). The mutation, SCN8A-p.Arg223Gly, was recently identified in child that presented with epileptic encephalopathy at 6 months of age (Kovel et al., submitted). In a screen for de novo mutations in 264 patients with infantile spasms or Lennox-Gastaut syndrome, the SCN8A mutation p.Leu876Gln was found in a child with Lennox-Gastaut (Epi4K Consortium and Epilepsy Phenome/Genome Project, 2013). The locations of the epilepsy-associated mutations are indicated in Figure 2.
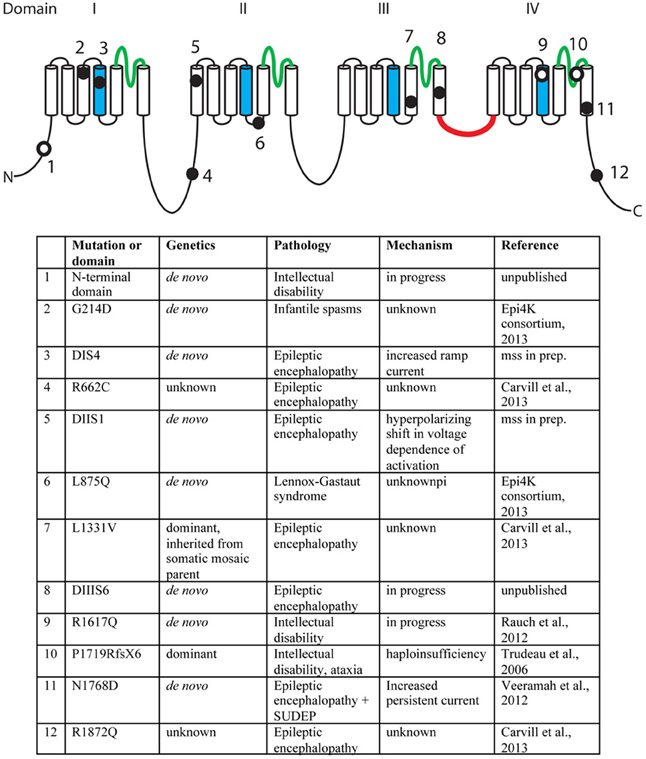
Figure 2. Mutations of human SCN8A. The positions of twelve recently identified mutations of SCN8A are indicated on the backbone of the channel structure. The four homologous domains are labeled with the pore domains in green, the voltage-sensing transmembrane segments (S4) in blue, and the inactivation gate in red. Filled circles, mutations identified in patients with epilepsy. Open circles, mutations identified in patients with cognitive deficits. Unpublished mutations are shown in their approximate positions.
In 2006, we described the heterozygous loss-of-function mutation P1719RfsX1724 that segregated with cognitive deficits in a small family (Trudeau et al., 2006). Heterozygous children in this family were enrolled in special education classes, and heterozygous adults were unable to live independently. In 2012, Rauch and colleagues sequenced the exomes of 51 individuals with severe non-syndromic intellectual disability (Rauch et al., 2012). These patients were offspring of healthy, non-consanguineous parents and presented with intellectual disability, grossly normal motor function, and lack of syndrome-specific abnormality. The de novo missense variant p.Arg1617Gln in the voltage-sensing transmembrane segment of domain 4 of SCN8A was identified in one patient (Figure 2). Four additional de novo missense mutations in SCN8A have been discovered by exome sequencing of patients with intellectual disability (Figure 2). The limited functional data suggest that mutations causing increased channel activity are associated with seizures, while heterozygous loss-of-function of SCN8A predisposes to intellectual disability (Figure 2).
Over the past 18 years, fifteen mutant alleles of mouse Scn8a have been characterized. These include six spontaneous mutants, eight ENU-induced mutations, and one random transgene insertion (Figure 3) (Meisler et al., 2004). Several of these are null mutations with complete loss of Scn8a function. Homozygous null mice exhibit motor defects at 2 weeks of age, including ataxia and tremor, and do not survive beyond 3 weeks (Burgess et al., 1995; Kohrman et al., 1995). Homozygosity for severe hypomorphic alleles such as medJ and nmf58 is viable, but results in ataxia and tremor with progression to muscle weakness and dystonia. Homozygosity for five mildly hypomorphic alleles (medjo, jolting2J, tremorD, clth, 9J) results in tremor, ataxia and reduced body size. These observations suggest that mutations of human SCN8A may be found in the future in patients with movement disorders. Nav1.6 is expressed at a low level in cardiac myocytes, and null mice have prolonged cardiac action potentials, suggesting a possible role in cardiac arrythmias (Noujaim et al., 2012). Homozygous knockout of Scn8a in Purkinje cells results in impaired learning in Morris Water Maze and eyeblink conditioning tests (Woodruff-Pak et al., 2006).
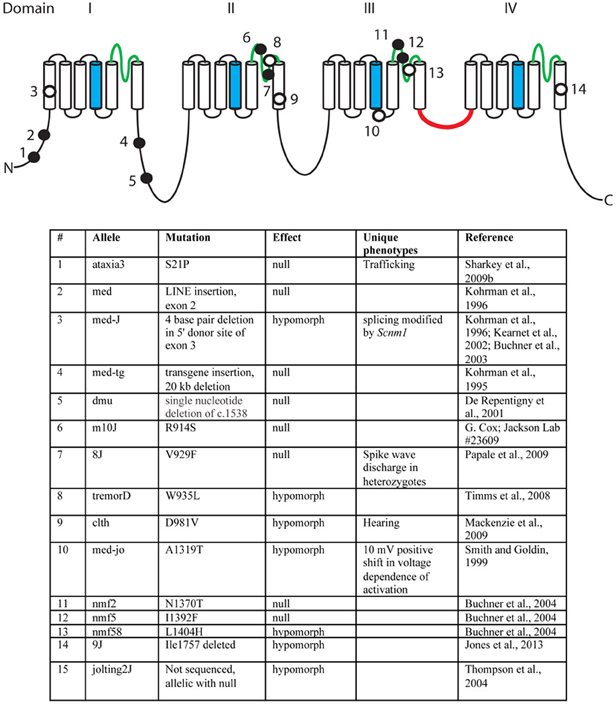
Figure 3. Mutations of mouse Scn8a. Fourteen allelic mutations are shown on the channel backbone as described in Figure 2. Amino acids are numbered according to Genbank AF049617. Filled circles, null alleles; open circles, hypomorphic alleles.
Mice that are heterozygous for loss-of-function mutations exhibit minor abnormalities such as spike-wave discharges suggestive of absence epilepsy (Papale et al., 2009), disrupted sleep architecture (Papale et al., 2010), and behavioral deficits including anxiety (McKinney et al., 2008). Haploinsufficiency of Nav1.6 also reduces susceptibility to genetic- and chemically-induced seizures (Martin et al., 2007, 2010). Scn8amed/+ and Scn8amed−jo/+ heterozygotes have reduced susceptibility to flurothyl and kainic acid induced seizures, and the combination of one mutant allele of Scn8a with Nav1.1 heterozygous or homozygous null mice results in extended lifespan and reduced seizure susceptibility. These observations suggest that reduced expression of Scn8a protects against seizures by decreasing neuronal excitability.
Direct evidence for the in vivo role of Nav1.6 has been advanced by recordings from neurons from several different lines of Scn8a null and conditional null mice developed in our laboratory (Burgess et al., 1995; Levin and Meisler, 2004; Levin et al., 2006) (Table 1). Reduced repetitive firing is consistently observed in cerebellar Purkinje cells, granule neurons, trigeminal mesencephalic neurons, and retinal ganglion cells from Scn8a mutant mice (Raman and Bean, 1997; Raman et al., 1997; Van Wart and Matthews, 2006; Aman and Raman, 2007). Reduced persistent and resurgent current was observed in several types of neurons by multiple investigators (Table 1). In addition to induced firing, spontaneous firing is reduced in Purkinje neurons isolated from null mice (Khaliq et al., 2003). Overall, the work summarized in Table 1 demonstrates that Scn8a is a key determinant of neuronal excitability in vivo.
The role of Scn8a in regulating neuronal excitability may be related to three properties of Nav1.6: its role in persistent and resurgent current, its voltage dependence of activation, and its subcellular localization at the axon initial segment (AIS), the site of initiation of action potentials. Persistent current is a steady-state sodium current that persists after firing and is involved in action potential initiation at membrane voltages near the threshold of firing (Crill, 1996; Smith et al., 1998; Rush et al., 2005; Osorio et al., 2010). Persistent current is important for generation of repetitive firing in neurons such as cerebellar Purkinje cells. In cerebellar Purkinje cells isolated from Scn8a null mice, persistent current was reduced by 70% compared with wild-type littermates (Raman et al., 1997). In tsA-201 kidney cells, the persistent current generated by Nav1.6 is five-fold higher than that generated by Nav1.2 (Chen et al., 2008). The differences in magnitude of persistent current in different types of neurons suggests that this property is modulated by neuron-specific factors (Rush et al., 2005; Chen et al., 2008). Mutations that further increase Nav1.6 persistent current result in epileptogenesis (e.g., Figure 1) (Veeramah et al., 2012).
Resurgent current is a voltage- and time-dependent property in which depolarization after the initial action potential elicits a small, transient current (Hille, 2001). This rapidly reversible form of inactivation allows neurons to fire quickly and repetitively. Resurgent current is thought to contribute to spontaneous firing and multi-peaked action potentials in cerebellar Purkinje cells that are compromised in mutants lacking Nav1.6 (Raman and Bean, 1997; Raman et al., 1997). The β4 sodium channel subunit is involved in generating resurgent current in cerebellar Purkinje neurons and cerebellar granule cell neurons, but the blocking factor appears to vary by neuron type (Raman and Bean, 2001; Grieco et al., 2005; Bant and Raman, 2010).
In transfected DRG neurons, there is a 15 mV leftward shift in voltage dependence of fast activation of Nav1.6 compared to Nav1.2, meaning that Nav1.6 is more activated earlier during depolarization (Rush et al., 2005). Nav1.6 is also less likely to inactivate at higher stimulation frequencies (20–100 Hz) (Rush et al., 2005). In transfected HEK-tsA-201 cells, Nav1.6 displayed a more positive voltage dependence of slow inactivation, passing ~10% more current in the −35 to −25 mV range than Nav1.2 (Chen et al., 2008). These features of Nav1.6 contribute to the positive effect of Nav1.6 on neuronal excitability.
The AIS is the membrane domain at the proximal end of the axon in which sodium channels are highly concentrated, electrical signals from the soma and dendrites are summed, and the threshold for action potential initiation is lowest (Royeck et al., 2008). The channel composition of the AIS appears to determine the firing threshold for different types of neurons (Lorincz and Nusser, 2008). Nav1.6 is highly concentrated in the distal half of the AIS in many neurons, including cerebellar granule cells and cerebellar Purkinje cells (Van Wart and Matthews, 2006; Lorincz and Nusser, 2008; Royeck et al., 2008). In the absence of Nav1.6, there is relocation of Nav1.1 and Nav1.2 to occupy the distal AIS (Van Wart and Matthews, 2006; Xiao et al., 2013). Cultured hippocampal CA1 pyramidal cells from Scn8a-null mice exhibit a 5 mV depolarizing (rightward) shift in the voltage dependence of activation, 60% reduction in persistent current, and 75% reduction in resurgent current (Royeck et al., 2008). This combination renders Scn8a null neurons less excitable than their wild type counterparts, as demonstrated by an 8 mV depolarizing shift in the spike threshold (Royeck et al., 2008).
In cortical pyramidal neurons, action potentials initiate at the distal part of the AIS, where sodium channel concentrations are highest (Van Wart et al., 2007; Kole and Stuart, 2008; Kole et al., 2008). The distal AIS in these cells contains predominantly Nav1.6, while the proximal AIS contains predominantly Nav1.2 (Hu et al., 2009). Step-depolarizations of patched neurons revealed that the activation threshold in the distal AIS was −55 mV, while the activation threshold in the proximal AIS closest to the soma was −43 mV (Hu et al., 2009), consistent with a role for Nav1.6 in lowering the threshold of action potential initiation.
Action potentials are primarily directed down the axon, away from the soma, but backpropagation into the soma occurs at low frequency (Hu et al., 2009). Current injection into the distal AIS does not produce backpropagation, while current injection at the proximal AIS leads to detectable action potentials in the soma (Hu et al., 2009). Thus, localization of Nav1.6 to the distal AIS is associated with a lower threshold for action potential initiation and direction of the action potential away from the soma. Overall, membranes containing Nav1.6 are more excitable than those containing only Nav1.1 and Nav1.2, and loss of Nav1.6 results in a higher threshold for initiation of action potentials (Van Wart and Matthews, 2006).
The SCN8A gene is located on human chromosome 12q13.13 (Plummer et al., 1998) and mouse distal chromosome 15 (Burgess et al., 1995). The 27 exons of SCN8A span 170 kb and encode a protein of 1980 residues (GenBank AF050736). The location of the amino acid residues within the 4 homologous domains and 24 transmembrane segments of Nav1.6 is shown in Figure S1. Nav1.6 protein is concentrated ~1,000-fold in two membrane domains, the AIS and the nodes of Ranvier of myelinated axons (Schaller and Caldwell, 2000; Boiko et al., 2001, 2003; Van Wart and Matthews, 2006; Van Wart et al., 2007; Lorincz and Nusser, 2008, 2010). Nav1.6 is also present at lower abundance in non-myelinated axons, neuronal soma, and dendrites (Krzemien et al., 2000; Lorincz and Nusser, 2010). The full-length SCN8A transcript is highly expressed throughout the brain, with concentration in the cerebellum and olfactory bulb of the rat (Schaller and Caldwell, 2000).
Transcriptional regulation of sodium channel genes is not well characterized. The transcription start sites for Scn8a are located in noncoding exons 70 kb upstream of the translation initiation site (Drews et al., 2005). Exon 1c is highly conserved through evolution and includes potential binding sites for neuronal transcription factors Pou6f1/Brn5, YY1, and REST/NRSF (Drews et al., 2007). Exon 1c and upstream sequences are sufficient to drive neuron-specific expression of LacZ in transgenic mice (Drews et al., 2007).
SCN8A contains two pairs of mutually exclusive, alternative coding exons whose splicing regulates channel function. Exons 5N/5A and 18N/18A encode the S3–S4 transmembrane segments of domain I and domain III, respectively (Plummer et al., 1997). Exon 18N contains an in-frame stop codon and is only expressed in non-neuronal cells (Plummer et al., 1997) including glia (O'Brien et al., 2012a). The neuronal splice factors RBFOX1 and RBFOX2 can activate inclusion of exon 18A in neurons, resulting in neuron-specific expression of the full length, active channel (Gehman et al., 2012; O'Brien et al., 2012a; Zubovic et al., 2012). Splice enhancers and silencers in exons 18A and 18N also contribute to temporal and spatial regulation (Zubovic et al., 2012). Alternative polyadenylation sites are located 4 and 6.5 kb downstream from the translation termination site of Scn8a, generating full-length coding transcripts of 9 and 12 kb (Drews et al., 2005). Transcripts with the shorter and longer 3′ UTR are equally represented in brain RNA and are not known to be associated with specific functions.
The pharmacology of compounds that target voltage-gated sodium channels has recently been reviewed (Eijkelkamp et al., 2012). The epileptic encephalopathies described in this review could in principle be treated with specific inhibitors of Nav1.6. However, the extensive sequence conservation among the neuronal and muscle sodium channels has made it difficult to develop drugs with specificity for a single channel. Two compounds with preferential effects on Nav1.6 have been described. The tetrodotoxin derivative 4,9-anhydrotetrodotoxin inactivates Nav1.6 expressed in Xenopus oocytes at concentrations that have minimal effects on six of the other channels (Rosker et al., 2007). The beta-scorpion toxin Cn2 also binds Nav1.6 specifically (Schiavon et al., 2006); this compound enhanced resurgent current inducing a hyperpolarizing shift in voltage dependence of channel activation in Purkinje slices, indicative of channel activation, while in HEK cells the effect was inhibitory. We have generated a mouse model of epileptic encephalopathy carrying the SCN8A-p.Asn1768Asp mutation that may be useful for future evaluation of drug specificity and effectiveness in vivo.
Voltage-gated sodium channels are components of large, multi-protein complexes that vary between neurons and at specific subcellular domains. The known sites of protein interaction with Nav1.6 are indicated in Figure 4. The N-terminus of Nav1.6 interacts with the light chain of microtubule-associated protein Map1b (Mtap1b), and co-transfection increases current density in transfected cells via increased trafficking of Nav1.6 to the cell surface (O'Brien et al., 2012b). Phosphorylation of Nav1.6 by the stress-activated MAP kinase p38 facilitates binding of E3 ubiquitin ligases and channel degradation (Sudol and Hunter, 2000; Zarrinpar and Lim, 2000; Gasser et al., 2010). Protein kinases PKA and PKC have only a small effect on channel activity (Chen et al., 2008). Ankyrin G binds to the first intracellular loop of Nav1.6 and other neuronal sodium channels (Srinivasan et al., 1988; Davis et al., 1996; Hill et al., 2008), and is essential for targeting and localization of Nav1.6 to nodes of Ranvier (Gasser et al., 2012).
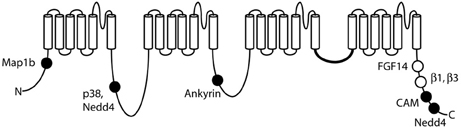
Figure 4. Locations of protein interactions with Nav1.6. Filled circles represent binding sites that have been localized to specific residues of Nav1.6: Map1b (77–80), p38 (553), ankyrin (1089–1122), calmodulin (1902–1912), and Nedd4 (551–554 and 1943–1945). Open symbols, binding sites that have not been mapped to specific residues.
The intracellular fibroblast growth factors FGF11-FGF14 interact with Nav1.6 and other voltage-gated sodium channels (Wittmack et al., 2004; Laezza et al., 2009; Shakkottai et al., 2009; Xiao et al., 2013). FGF13 interacts with the C-terminus in an isoform-dependent manner (Wittmack et al., 2004), which may allow specific sub-populations of neurons to fine-tune firing properties via alternative splicing of FGF13. Fgf14 null mice develop ataxia and ~80% of their cerebellar Purkinje cells lack repetitive firing (Shakkottai et al., 2009). The abundance of Nav1.6 in the AIS is reduced in cerebellar Purkinje cells from Fgf14 null mice, suggesting that FGF14 plays a key role in the organization of a subunits in the AIS (Xiao et al., 2013).
The sodium channel subunits β1 to β4 are small single-transmembrane cell-adhesion molecule proteins that modulate current and surface expression of the α subunit (Patino and Isom, 2010). Studies of mice null for the β1 subunit (Scn1b−/−) suggest that interaction between β1 and Nav1.6 is required for function of Nav1.6 at the distal AIS (Brackenbury et al., 2010). The β4 subunit has been implicated in the generation of resurgent Nav1.6 current in Purkinje neurons (Grieco et al., 2005; Aman et al., 2009), but resurgent current was not generated by co-transfection of β4 and Nav1.6 in HEK cells (Chen et al., 2008; Aman et al., 2009).
The calcium responsive protein calmodulin binds the IQ motif located in the C-terminus of Nav1.6 (residues 1902–1912). Apo-calmodulin accelerates inactivation and Ca2+ increases excitability of Nav1.6 (Herzog et al., 2003). The E3 ubiquitin ligase Nedd4 also binds to the C-terminus of Scn8a at a PXY motif (residues 1943–1945), and the PXpS/pTP motif in the first cytoplasmic loop (residues 551–554) (Abriel et al., 2000; Sudol and Hunter, 2000; Fotia et al., 2004; Ingham et al., 2004; van Bemmelen et al., 2004; Rougier et al., 2005). Both sites are necessary for Nedd4 binding and internalization of Nav1.6 (Gasser et al., 2010). Ubiquitination of Nav1.6 by Nedd4 is thought to target Nav1.6 for degradation and may be part of the neuronal stress response.
These interactions are relevant to the genetics of neurological and psychiatric disorders, since proteins that bind Nav1.6 may be considered candidate genes for the same disorders caused by mutations of Nav1.6. Further, common variants of the interacting proteins may act as modifiers of the severity of SCN8A mutations in patients (Meisler et al., 2010; Meisler and O'Brien, 2012).
Nav1.6 is a major sodium channel in human brain. The features of Nav1.6 that influence neuronal excitability include contributions to persistent and resurgent neuronal currents, low threshold for excitation, and concentration in the AIS. Mutations of Scn8a in the mouse result in movement disorders including ataxia, dystonia, and tremor. Within the past year, de novo mutations of human SCN8A detected by exome sequencing have revealed a role for Nav1.6 in epilepsy and intellectual disability. Hypoactivity and hyperactivity of Nav1.6 are both pathogenic, but with different outcomes: haploinsufficiency is associated with impaired cognition (Trudeau et al., 2006; McKinney et al., 2008; Rauch et al., 2012) while hyperactivity can result in epilepsy (Veeramah et al., 2012). Analysis of additional mutants in the near future should provide insight into structure-function relationships of Nav1.6 and the mechanisms of pathogenesis in neurological disease.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Our work on human and mouse SCN8A is supported by NIH grant R01 NS34509 to Miriam H. Meisler. Janelle E. O'Brien acknowledges support from NIH T32 GM007544 and the Rackham School of Graduate Studies, University of Michigan.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2013.00213/abstract
Figure S1 | Amino acid sequence of human and mouse SCN8A. The predicted human amino acid sequence corresponds to the predominant transcript in adult brain containing exon 5A and exon 18A and using the upstream splice donor site of exon 10B. The approximate locations of transmembrane segments S1–S6 of domains I–IV of the protein are underlined. Human coding sequence, GenBank AF050736; mouse coding sequence, GenBank AF049617. ∇, exon borders; *, protein kinase A consensus sequence; arrow, tyrosine kinase consensus; dots, amino acid identity. Adapted from Plummer et al. (1998).
Abriel, H., Kamynina, E., Horisberger, J. D., and Staub, O. (2000). Regulation of the cardiac voltage-gated Na+ channel (H1) by the ubiquitin-protein ligase Nedd4. FEBS Lett. 466, 377–380. doi: 10.1016/S0014-5793(00)01098-X
Aman, T. K., and Raman, I. M. (2007). Subunit dependence of Na channel slow inactivation and open channel block in cerebellar neurons. Biophys. J. 92, 1938–1951. doi: 10.1529/biophysj.106.093500
Aman, T. K., Grieco-Calub, T. M., Chen, C., Rusconi, R., Slat, E. A., Isom, L. L., et al. (2009). Regulation of persistent Na current by interactions between beta subunits of voltage-gated Na channels. J. Neurosci. 29, 2027–2042. doi: 10.1523/JNEUROSCI.4531-08.2009
Bamshad, M. J., Ng, S. B., Bigham, A. W., Tabor, H. K., Emond, M. J., Nickerson, D. A., et al. (2011). Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755. doi: 10.1038/nrg3031
Bant, J. S., and Raman, I. M. (2010). Control of transient, resurgent, and persistent current by open-channel block by Na channel beta4 in cultured cerebellar granule neurons. Proc. Natl. Acad. Sci. U.S.A. 107, 12357–12362. doi: 10.1073/pnas.1005633107
Boiko, T., Rasband, M. N., Levinson, S. R., Caldwell, J. H., Mandel, G., Trimmer, J. S., et al. (2001). Compact myelin dictates the differential targeting of two sodium channel isoforms in the same axon. Neuron 30, 91–104. doi: 10.1016/S0896-6273(01)00265-3
Boiko, T., Van Wart, A., Caldwell, J. H., Levinson, S. R., Trimmer, J. S., and Matthews, G. (2003). Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. J Neurosci 23, 2306–2313.
Brackenbury, W. J., Calhoun, J. D., Chen, C., Miyazaki, H., Nukina, N., Oyama, F., et al. (2010). Functional reciprocity between Na+ channel Nav1.6 and beta1 subunits in the coordinated regulation of excitability and neurite outgrowth. Proc. Natl. Acad. Sci. U.S.A. 107, 2283–2288. doi: 10.1073/pnas.0909434107
Buchner, D. A., Seburn, K. L., Frankel, W. N., and Meisler, M. H. (2004). Three ENU-induced neurological mutations in the pore loop of sodium channel Scn8a (Na(v)1.6) and a genetically linked retinal mutation, rd13. Mamm. Genome 15, 344–351. doi: 10.1007/s00335-004-2332-1
Buchner, D. A., Trudeau, M., and Meisler, M. H. (2003). SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301, 967–969. doi: 10.1126/science.1086187
Burgess, D. L., Kohrman, D. C., Galt, J., Plummer, N. W., Jones, J. M., Spear, B., et al. (1995). Mutation of a new sodium channel gene, Scn8a, in the mouse mutant ‘motor endplate disease’. Nat. Genet 10, 461–465. doi: 10.1038/ng0895-461
Carvill, G. L., Heavin, S. B., Yendle, S. C., McMahon, J. M., O'Roak, B. J., Cook, J., et al. (2013). Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 45, 825–830. doi: 10.1038/ng.2646
Chen, Y., Yu, F. H., Sharp, E. M., Beacham, D., Scheuer, T., and Catterall, W. A. (2008). Functional properties and differential neuromodulation of Na(v)1.6 channels. Mol. Cell. Neurosci. 38, 607–615. doi: 10.1016/j.mcn.2008.05.009
Crill, W. E. (1996). Persistent sodium current in mammalian central neurons. Annu. Rev. Physiol. 58, 349–362. doi: 10.1146/annurev.ph.58.030196.002025
Cummins, T. R., Dib-Hajj, S. D., Herzog, R. I., and Waxman, S. G. (2005). Nav1.6 channels generate resurgent sodium currents in spinal sensory neurons. FEBS Lett. 579, 2166–2170. doi: 10.1016/j.febslet.2005.03.009
Davis, J. Q., Lambert, S., and Bennett, V. (1996). Molecular composition of the node of Ranvier: identification of ankyrin-binding cell adhesion molecules neurofascin (mucin+/third FNIII domain-) and NrCAM at nodal axon segments. J. Cell Biol. 135, 1355–1367. doi: 10.1083/jcb.135.5.1355
De Repentigny, Y., Cote, P. D., Pool, M., Bernier, G., Girard, S., Vidal, S. M., et al. (2001). Pathological and genetic analysis of the degenerating muscle (dmu) mouse: a new allele of Scn8a. Hum. Mol. Genet. 10, 1819–1827. doi: 10.1093/hmg/10.17.1819
Do, M. T., and Bean, B. P. (2004). Sodium currents in subthalamic nucleus neurons from Nav1.6-null mice. J. Neurophysiol. 92, 726–733. doi: 10.1152/jn.00186.2004
Doherty, D., and Bamshad, M. J. (2012). Exome sequencing to find rare variants causing neurologic diseases. Neurology 79, 396–397. doi: 10.1212/WNL.0b013e3182617170
Drews, V. L., Lieberman, A. P., and Meisler, M. H. (2005). Multiple transcripts of sodium channel SCN8A (Na(V)1.6) with alternative 5′- and 3′-untranslated regions and initial characterization of the SCN8A promoter. Genomics 85, 245–257. doi: 10.1016/j.ygeno.2004.09.002
Drews, V. L., Shi, K., de Haan, G., and Meisler, M. H. (2007). Identification of evolutionarily conserved, functional noncoding elements in the promoter region of the sodium channel gene SCN8A. Mamm. Genome 18, 723–731. doi: 10.1007/s00335-007-9059-8
Eijkelkamp, N., Linley, J. E., Baker, M. D., Minett, M. S., Cregg, R., Werdehausen, R., et al. (2012). Neurological perspectives on voltage-gated sodium channels. Brain 135(Pt 9), 2585–2612. doi: 10.1093/brain/aws225
Enomoto, A., Han, J. M., Hsiao, C. F., and Chandler, S. H. (2007). Sodium currents in mesencephalic trigeminal neurons from Nav1.6 null mice. J. Neurophysiol. 98, 710–719. doi: 10.1152/jn.00292.2007
Epi4K Consortium and Epilepsy Phenome/Genome Project, Allen, A. S., Berkovic, S. F., Cossette, P., Delanty, N., Dlugos, D., Eichler, E. E., et al. (2013). De novo mutations in epileptic encephalopathies. Nature 501, 217–221. doi: 10.1038/nature12439
Fotia, A. B., Ekberg, J., Adams, D. J., Cook, D. I., Poronnik, P., and Kumar, S. (2004). Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4-2. J. Biol. Chem. 279, 28930–28935. doi: 10.1074/jbc.M402820200
Gasser, A., Cheng, X., Gilmore, E. S., Tyrrell, L., Waxman, S. G., and Dib-Hajj, S. D. (2010). Two Nedd4-binding motifs underlie modulation of sodium channel Nav1.6 by p38 MAPK. J. Biol. Chem. 285, 26149–26161. doi: 10.1074/jbc.M109.098681
Gasser, A., Ho, T. S., Cheng, X., Chang, K. J., Waxman, S. G., Rasband, M. N., et al. (2012). An ankyrinG-binding motif is necessary and sufficient for targeting Nav1.6 sodium channels to axon initial segments and nodes of Ranvier. J. Neurosci. 32, 7232–7243. doi: 10.1523/JNEUROSCI.5434-11.2012
Gehman, L. T., Meera, P., Stoilov, P., Shiue, L., O'Brien, J. E., Meisler, M. H., et al. (2012). The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 26, 445–460. doi: 10.1101/gad.182477.111
Grieco, T. M., Malhotra, J. D., Chen, C., Isom, L. L., and Raman, I. M. (2005). Open-channel block by the cytoplasmic tail of sodium channel beta4 as a mechanism for resurgent sodium current. Neuron 45, 233–244. doi: 10.1016/j.neuron.2004.12.035
Hartshorne, R. P., and Catterall, W. A. (1981). Purification of the saxitoxin receptor of the sodium channel from rat brain. Proc. Natl. Acad. Sci. U.S.A. 78, 4620–4624. doi: 10.1073/pnas.78.7.4620
Herzog, R. I., Liu, C., Waxman, S. G., and Cummins, T. R. (2003). Calmodulin binds to the C terminus of sodium channels Nav1.4 and Nav1.6 and differentially modulates their functional properties. J. Neurosci. 23, 8261–8270.
Hill, A. S., Nishino, A., Nakajo, K., Zhang, G., Fineman, J. R., Selzer, M. E., et al. (2008). Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 4:e1000317. doi: 10.1371/journal.pgen.1000317
Hu, W., Tian, C., Li, T., Yang, M., Hou, H., and Shu, Y. (2009). Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nat. Neurosci. 12, 996–1002. doi: 10.1038/nn.2359
Ingham, R. J., Gish, G., and Pawson, T. (2004). The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23, 1972–1984. doi: 10.1038/sj.onc.1207436
Kearney, J. A., Buchner, D. A., De Haan, G., Adamska, M., Levin, S. I., Furay, A. R., et al. (2002). Molecular and pathological effects of a modifier gene on deficiency of the sodium channel Scn8a (Na(v)1.6). Hum. Mol. Genet. 11, 2765–2775. doi: 10.1093/hmg/11.22.2765
Khaliq, Z. M., Gouwens, N. W., and Raman, I. M. (2003). The contribution of resurgent sodium current to high-frequency firing in Purkinje neurons: an experimental and modeling study. J. Neurosci. 23, 4899–4912.
Kohrman, D. C., Harris, J. B., and Meisler, M. H. (1996). Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J. Biol. Chem. 271, 17576–17581. doi: 10.1074/jbc.271.29.17576
Kohrman, D. C., Plummer, N. W., Schuster, T., Jones, J. M., Jang, W., Burgess, D. L., et al. (1995). Insertional mutation of the motor endplate disease (med) locus on mouse chromosome 15. Genomics 26, 171–177. doi: 10.1016/0888-7543(95)80198-U
Kole, M. H., Ilschner, S. U., Kampa, B. M., Williams, S. R., Ruben, P. C., and Stuart, G. J. (2008). Action potential generation requires a high sodium channel density in the axon initial segment. Nat. Neurosci. 11, 178–186. doi: 10.1038/nn2040
Kole, M. H., and Stuart, G. J. (2008). Is action potential threshold lowest in the axon? Nat. Neurosci. 11, 1253–1255. doi: 10.1038/nn.2203
Krzemien, D. M., Schaller, K. L., Levinson, S. R., and Caldwell, J. H. (2000). Immunolocalization of sodium channel isoform NaCh6 in the nervous system. J. Comp. Neurol. 420, 70–83. doi: 10.1002/(SICI)1096-9861(20000424)420:1<70::AID-CNE5>3.0.CO;2-P
Laezza, F., Lampert, A., Kozel, M. A., Gerber, B. R., Rush, A. M., Nerbonne, J. M., et al. (2009). FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 42, 90–101. doi: 10.1016/j.mcn.2009.05.007
Levin, S. I., Khaliq, Z. M., Aman, T. K., Grieco, T. M., Kearney, J. A., Raman, I. M., et al. (2006). Impaired motor function in mice with cell-specific knockout of sodium channel Scn8a (NaV1.6) in cerebellar purkinje neurons and granule cells. J. Neurophysiol. 96, 785–793. doi: 10.1152/jn.01193.2005
Levin, S. I., and Meisler, M. H. (2004). Floxed allele for conditional inactivation of the voltage-gated sodium channel Scn8a (NaV1.6). Genesis 39, 234–239. doi: 10.1002/gene.20050
Liu, Y., Lopez-Santiago, L. F., Yuan, Y., Jones, J. M., Zhang, H., O'Malley, H. A., et al. (2013). Dravet Syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann. Neurol. 74, 128–139. doi: 10.1002/ana.23897
Lopreato, G. F., Lu, Y., Southwell, A., Atkinson, N. S., Hillis, D. M., Wilcox, T. P., et al. (2001). Evolution and divergence of sodium channel genes in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 98, 7588–7592. doi: 10.1073/pnas.131171798
Lorincz, A., and Nusser, Z. (2008). Cell-type-dependent molecular composition of the axon initial segment. J. Neurosci. 28, 14329–14340. doi: 10.1523/JNEUROSCI.4833-08.2008
Lorincz, A., and Nusser, Z. (2010). Molecular identity of dendritic voltage-gated sodium channels. Science 328, 906–909. doi: 10.1126/science.1187958
Mackenzie, F. E., Parker, A., Parkinson, N. J., Oliver, P. L., Brooker, D., Underhill, P., et al. (2009). Analysis of the mouse mutant Cloth-ears shows a role for the voltage-gated sodium channel Scn8a in peripheral neural hearing loss. Genes Brain Behav. 8, 699–713. doi: 10.1111/j.1601-183X.2009.00514.x
Martin, M. S., Dutt, K., Papale, L. A., Dube, C. M., Dutton, S. B., de Haan, G., et al. (2010). Altered function of the SCN1A voltage-gated sodium channel leads to gamma-aminobutyric acid-ergic (GABAergic) interneuron abnormalities. J. Biol. Chem. 285, 9823–9834. doi: 10.1074/jbc.M109.078568
Martin, M. S., Tang, B., Papale, L. A., Yu, F. H., Catterall, W. A., and Escayg, A. (2007). The voltage-gated sodium channel Scn8a is a genetic modifier of severe myoclonic epilepsy of infancy. Hum. Mol. Genet. 16, 2892–2899. doi: 10.1093/hmg/ddm248
Maurice, N., Tkatch, T., Meisler, M., Sprunger, L. K., and Surmeier, D. J. (2001). D1/D5 dopamine receptor activation differentially modulates rapidly inactivating and persistent sodium currents in prefrontal cortex pyramidal neurons. J. Neurosci. 21, 2268–2277.
McKinney, B. C., Chow, C. Y., Meisler, M. H., and Murphy, G. G. (2008). Exaggerated emotional behavior in mice heterozygous null for the sodium channel Scn8a (Nav1.6). Genes Brain Behav. 7, 629–638. doi: 10.1111/j.1601-183X.2008.00399.x
Meisler, M. H., and Kearney, J. A. (2005). Sodium channel mutations in epilepsy and other neurological disorders. J. Clin. Invest. 115, 2010–2017. doi: 10.1172/JCI25466
Meisler, M. H., and O'Brien, J. E. (2012). “Gene interactions and modifiers in epilepsy,” in Jasper's Basic Mechanisms of the Epilepsies [Internet], 4th Edn, eds J. L. Noebels, M. Avoli, M. A. Rogawski, R. W. Olsen, and A. V. Delgado-Escueta (Bethesda, MD: National Center for Biotechnology Information (US)).
Meisler, M. H., O'Brien, J. E., and Sharkey, L. M. (2010). Sodium channel gene family: epilepsy mutations, gene interactions and modifier effects. J. Physiol. Proc. Natl. Acad. Sci. U.S.A. 588(Pt 11), 1841–1848. doi: 10.1113/jphysiol.2010.188482
Meisler, M. H., Plummer, N. W., Burgess, D. L., Buchner, D. A., and Sprunger, L. K. (2004). Allelic mutations of the sodium channel SCN8A reveal multiple cellular and physiological functions. Genetica 122, 37–45. doi: 10.1007/s10709-004-1441-9
Mercer, J. N., Chan, C. S., Tkatch, T., Held, J., and Surmeier, D. J. (2007). Nav1.6 sodium channels are critical to pacemaking and fast spiking in globus pallidus neurons. J. Neurosci. 27, 13552–13566. doi: 10.1523/JNEUROSCI.3430-07.2007
Need, A. C., Shashi, V., Hitomi, Y., Schoch, K., Shianna, K. V., McDonald, M. T., et al. (2012). Clinical application of exome sequencing in undiagnosed genetic conditions. J. Med. Genet. 49, 353–361. doi: 10.1136/jmedgenet-2012-100819
Noda, M., Ikeda, T., Kayano, T., Suzuki, H., Takeshima, H., Kurasaki, M., et al. (1986). Existence of distinct sodium channel messenger RNAs in rat brain. Nature 320, 188–192. doi: 10.1038/320188a0
Noujaim, S. F., Kaur, K., Milstein, M., Jones, J. M., Furspan, P., Jiang, D., et al. (2012). A null mutation of the neuronal sodium channel NaV1.6 disrupts action potential propagation and excitation-contraction coupling in the mouse heart. FASEB J. 26, 63–72. doi: 10.1096/fj.10-179770
O'Brien, J. E., Drews, V. L., Jones, J. M., Dugas, J. C., Barres, B. A., and Meisler, M. H. (2012a). Rbfox proteins regulate alternative splicing of neuronal sodium channel SCN8A. Mol. Cell. Neurosci. 49, 120–126. doi: 10.1016/j.mcn.2011.10.005
O'Brien, J. E., Sharkey, L. M., Vallianatos, C. N., Han, C., Blossom, J. C., Yu, T., et al. (2012b). Interaction of voltage-gated sodium channel Nav1.6 (SCN8A) with microtubule-associated protein Map1b. J. Biol. Chem. 287, 18459–18466. doi: 10.1074/jbc.M111.336024
Osorio, N., Cathala, L., Meisler, M. H., Crest, M., Magistretti, J., and Delmas, P. (2010). Persistent Nav1.6 current at axon initial segments tunes spike timing of cerebellar granule cells. J. Physiol. Annu. Rev. Physiol. 588(Pt 4), 651–670. doi: 10.1113/jphysiol.2010.183798
Papale, L. A., Beyer, B., Jones, J. M., Sharkey, L. M., Tufik, S., Epstein, M., et al. (2009). Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum. Mol. Genet. 18, 1633–1641. doi: 10.1093/hmg/ddp081
Papale, L. A., Paul, K. N., Sawyer, N. T., Manns, J. R., Tufik, S., and Escayg, A. (2010). Dysfunction of the Scn8a voltage-gated sodium channel alters sleep architecture, reduces diurnal corticosterone levels, and enhances spatial memory. J. Biol. Chem. 285, 16553–16561. doi: 10.1074/jbc.M109.090084
Patino, G. A., and Isom, L. L. (2010). Electrophysiology and beyond: multiple roles of Na+ channel beta subunits in development and disease. Neurosci. Lett. 486, 53–59. doi: 10.1016/j.neulet.2010.06.050
Plummer, N. W., Galt, J., Jones, J. M., Burgess, D. L., Sprunger, L. K., Kohrman, D. C., et al. (1998). Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics 54, 287–296. doi: 10.1006/geno.1998.5550
Plummer, N. W., McBurney, M. W., and Meisler, M. H. (1997). Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J. Biol. Chem. 272, 24008–24015. doi: 10.1074/jbc.272.38.24008
Raman, I. M., and Bean, B. P. (1997). Resurgent sodium current and action potential formation in dissociated cerebellar Purkinje neurons. J. Neurosci. 17, 4517–4526.
Raman, I. M., and Bean, B. P. (2001). Inactivation and recovery of sodium currents in cerebellar Purkinje neurons: evidence for two mechanisms. Biophys. J. 80, 729–737. doi: 10.1016/S0006-3495(01)76052-3
Raman, I. M., Sprunger, L. K., Meisler, M. H., and Bean, B. P. (1997). Altered subthreshold sodium currents and disrupted firing patterns in Purkinje neurons of Scn8a mutant mice. Neuron 19, 881–891. doi: 10.1016/S0896-6273(00)80969-1
Rauch, A., Wieczorek, D., Graf, E., Wieland, T., Endele, S., Schwarzmayr, T., et al. (2012). Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380, 1674–1682. doi: 10.1016/S0140-6736(12)61480-9
Rosker, C., Lohberger, B., Hofer, D., Steinecker, B., Quasthoff, S., and Schreibmayer, W. (2007). The TTX metabolite 4,9-anhydro-TTX is a highly specific blocker of the Na(v1.6) voltage-dependent sodium channel. Am. J. Physiol. Cell Physiol. 293, C783–C789. doi: 10.1152/ajpcell.00070.2007
Rougier, J. S., van Bemmelen, M. X., Bruce, M. C., Jespersen, T., Gavillet, B., Apotheloz, F., et al. (2005). Molecular determinants of voltage-gated sodium channel regulation by the Nedd4/Nedd4-like proteins. Am. J. Physiol. Cell Physiol. 288, C692–C701. doi: 10.1152/ajpcell.00460.2004
Royeck, M., Horstmann, M. T., Remy, S., Reitze, M., Yaari, Y., and Beck, H. (2008). Role of axonal NaV1.6 sodium channels in action potential initiation of CA1 pyramidal neurons. J. Neurophysiol. 100, 2361–2380. doi: 10.1152/jn.90332.2008
Rush, A. M., Dib-Hajj, S. D., and Waxman, S. G. (2005). Electrophysiological properties of two axonal sodium channels, Nav1.2 and Nav1.6, expressed in mouse spinal sensory neurones. J. Physiol. 564(Pt 3), 803–815. doi: 10.1113/jphysiol.2005.083089
Schaller, K. L., and Caldwell, J. H. (2000). Developmental and regional expression of sodium channel isoform NaCh6 in the rat central nervous system. J. Comp. Neurol. 420, 84–97. doi: 10.1002/(SICI)1096-9861(20000424)420:1<84::AID-CNE6>3.0.CO;2-9
Schaller, K. L., Krzemien, D. M., Yarowsky, P. J., Krueger, B. K., and Caldwell, J. H. (1995). A novel, abundant sodium channel expressed in neurons and glia. J. Neurosci. 15(5 Pt 1), 3231–3242.
Schiavon, E., Sacco, T., Cassulini, R. R., Gurrola, G., Tempia, F., Possani, L. D., et al. (2006). Resurgent current and voltage sensor trapping enhanced activation by a beta-scorpion toxin solely in Nav1.6 channel. Significance in mice Purkinje neurons. J. Biol. Chem. 281, 20326–20337. doi: 10.1074/jbc.M600565200
Shakkottai, V. G., Xiao, M., Xu, L., Wong, M., Nerbonne, J. M., Ornitz, D. M., et al. (2009). FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol. Dis. 33, 81–88. doi: 10.1016/j.nbd.2008.09.019
Sharkey, L. M., Jones, J. M., Hedera, P., and Meisler, M. H. (2009a). Evaluation of SCN8A as a candidate gene for autosomal dominant essential tremor. Parkinsonism Relat. Disord. 15, 321–323. doi: 10.1016/j.parkreldis.2008.06.010
Sharkey, L. M., Cheng, X., Drews, V., Buchner, D. A., Jones, J. M., Justice, M. J., et al. (2009b). The ataxia3 mutation in the N-terminal cytoplasmic domain of sodium channel Nav1.6 disrupts intracellular trafficking. J. Neurosci. 29, 2733–2741. doi: 10.1523/JNEUROSCI.6026-08.2009
Smith, M. R., and Goldin, A. L. (1999). A mutation that causes ataxia shifts the voltage-dependence of the Scn8a sodium channel. Neuroreport 10, 3027–3031. doi: 10.1097/00001756-199909290-00028
Smith, M. R., Smith, R. D., Plummer, N. W., Meisler, M. H., and Goldin, A. L. (1998). Functional analysis of the mouse Scn8a sodium channel. J. Neurosci. 18, 6093–6102.
Srinivasan, Y., Elmer, L., Davis, J., Bennett, V., and Angelides, K. (1988). Ankyrin and spectrin associate with voltage-dependent sodium channels in brain. Nature 333, 177–180. doi: 10.1038/333177a0
Sudol, M., and Hunter, T. (2000). NeW wrinkles for an old domain. Cell 103, 1001–1004. doi: 10.1016/S0092-8674(00)00203-8
Sun, W., Wagnon, J. L., Mahaffey, C. L., Briese, M., Ule, J., and Frankel, W. N. (2013). Aberrant sodium channel activity in the complex seizure disorder of Celf4 mutant mice. J. Physiol. Ann. Neurol. 591(Pt 1), 241–255. doi: 10.1113/jphysiol.2012.240168
Tamkun, M. M., and Catterall, W. A. (1981). Reconstitution of the voltage-sensitive sodium channel of rat brain from solubilized components. J. Biol. Chem. 256, 11457–11463.
Thompson, D., Ward-Bailey, P. F., Donahue, L. R., Bronson, R. T., Soukup, J., and Davisson, M. T. (2004). A remutation to Scn8a named jolting 2 Jackson. MGI Direct Data Submission.
Trudeau, M. M., Dalton, J. C., Day, J. W., Ranum, L. P., and Meisler, M. H. (2006). Heterozygosity for a protein truncation mutation of sodium channel SCN8A in a patient with cerebellar atrophy, ataxia, and mental retardation. J. Med. Genet. 43, 527–530. doi: 10.1136/jmg.2005.035667
van Bemmelen, M. X., Rougier, J. S., Gavillet, B., Apotheloz, F., Daidie, D., Tateyama, M., et al. (2004). Cardiac voltage-gated sodium channel Nav1.5 is regulated by Nedd4-2 mediated ubiquitination. Circ. Res. 95, 284–91. doi: 10.1161/01.RES.0000136816.05109.89
Van Wart, A., and Matthews, G. (2006). Impaired firing and cell-specific compensation in neurons lacking nav1.6 sodium channels. J. Neurosci. 26, 7172–7180. doi: 10.1523/JNEUROSCI.1101-06.2006
Van Wart, A., Trimmer, J. S., and Matthews, G. (2007). Polarized distribution of ion channels within microdomains of the axon initial segment. J. Comp. Neurol. 500, 339–352. doi: 10.1002/cne.21173
Veeramah, K. R., O'Brien, J. E., Meisler, M. H., Cheng, X., Dib-Hajj, S. D., Waxman, S. G., et al. (2012). De Novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am. J. Hum. Genet. 90, 502–510. doi: 10.1016/j.ajhg.2012.01.006
Wittmack, E. K., Rush, A. M., Craner, M. J., Goldfarb, M., Waxman, S. G., and Dib-Hajj, S. D. (2004). Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J. Neurosci. 24, 6765–6775. doi: 10.1523/JNEUROSCI.1628-04.2004
Woodruff-Pak, D. S., Green, J. T., Levin, S. I., and Meisler, M. H. (2006). Inactivation of sodium channel Scn8A (Na-sub(v)1.6) in Purkinje neurons impairs learning in Morris water maze and delay but not trace eyeblink classical conditioning. Behav. Neurosci. 120, 229–240. doi: 10.1037/0735-7044.120.2.229
Xiao, M., Bosch, M. K., Nerbonne, J. M., and Ornitz, D. M. (2013). FGF14 localization and organization of the axon initial segment. Mol. Cell. Neurosci. 56C, 393–403. doi: 10.1016/j.mcn.2013.07.008
Zakon, H. H. (2012). Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc. Natl. Acad. Sci. U.S.A. 109(Suppl. 1), 10619–10625. doi: 10.1073/pnas.1201884109
Zakon, H. H., Jost, M. C., and Lu, Y. (2011). Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol. Biol. Evol. 28, 1415–1424. doi: 10.1093/molbev/msq325
Zarrinpar, A., and Lim, W. A. (2000). Converging on proline: the mechanism of WW domain peptide recognition. Nat. Struct. Biol. 7, 611–613. doi: 10.1038/77891
Keywords: voltage-gated sodium channels, epilepsy, intellectual disability, SCN8A, Nav1.6, neurogenetics, genetics, exomes
Citation: O'Brien JE and Meisler MH (2013) Sodium channel SCN8A (Nav1.6): properties and de novo mutations in epileptic encephalopathy and intellectual disability. Front. Genet. 4:213. doi: 10.3389/fgene.2013.00213
Received: 14 August 2013; Paper pending published: 14 September 2013;
Accepted: 04 October 2013; Published online: 28 October 2013.
Edited by:
Kathleen D. Askland, Brown University, USACopyright © 2013 O'Brien and Meisler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miriam H. Meisler, Department of Human Genetics, University of Michigan, 4909 Buhl, Ann Arbor, MI 48109-5618, USA e-mail:bWVpc2xlcm1AdW1pY2guZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.