- 1Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA
- 2New England Centenarian Study, Section of Geriatrics, Department of Medicine, Boston University School of Medicine, Boston, MA, USA
- 3Division of Statistical Genomics, Washington University School of Medicine, St. Louis, MO, USA
- 4National Institute on Aging, National Institutes of Health, Baltimore, MD, USA
- 5Department of Geriatrics, College of Medicine, Florida State University, Tallahassee, FL, USA
Personality traits have been shown to be associated with longevity and healthy aging. In order to discover novel genetic modifiers associated with personality traits as related with longevity, we performed a genome-wide association study (GWAS) on personality factors assessed by NEO-five-factor inventory in individuals enrolled in the Long Life Family Study (LLFS), a study of 583 families (N up to 4595) with clustering for longevity in the United States and Denmark. Three SNPs, in almost perfect LD, associated with agreeableness reached genome-wide significance (p < 10−8) and replicated in an additional sample of 1279 LLFS subjects, although one (rs9650241) failed to replicate and the other two were not available in two independent replication cohorts, the Baltimore Longitudinal Study of Aging and the New England Centenarian Study. Based on 10,000,000 permutations, the empirical p-value of 2 × 10−7 was observed for the genome-wide significant SNPs. Seventeen SNPs that reached marginal statistical significance in the two previous GWASs (p-value <10−4 and 10−5), were also marginally significantly associated in this study (p-value <0.05), although none of the associations passed the Bonferroni correction. In addition, we tested age-by-SNP interactions and found some significant associations. Since scores of personality traits in LLFS subjects change in the oldest ages, and genetic factors outweigh environmental factors to achieve extreme ages, these age-by-SNP interactions could be a proxy for complex gene–gene interactions affecting personality traits and longevity.
Introduction
Personality traits have been shown to be associated with important health outcomes and longevity (Terracciano et al., 2008). Previous findings suggest that low levels of neuroticism, high levels of conscientiousness, and high levels of extraversion are associated with reduced mortality (Friedman et al., 1993; Wilson et al., 2004, 2005; Weiss and Costa, 2005; Chapman et al., 2011). Centenarian offspring have lower neuroticism and higher extraversion in comparison to published normative data (Givens et al., 2009). We recently assessed domains of agreeableness, conscientiousness, extraversion, neuroticism, and openness to experience using the NEO-five-factor inventory (NEO-FFI) (Costa and McCrae, 1992) in subjects enrolled in the Long Life Family Study (LLFS): a family-based longitudinal study of longevity and healthy aging (Newman et al., 2011). The analysis replicated the association of low neuroticism and high extraversion with longevity and also confirmed differences in distributions of personality scores at different ages.
Heritability estimates of agreeableness, conscientiousness, extraversion, neuroticism, and openness assessed by the NEO-FFI in 6148 Sardinians ranged from 17 to 33% (Pilia et al., 2006), and a recent genome-wide association study (GWAS) and meta-analysis identified many genetic variants associated with personality traits, although just a few reached levels of genome-wide significance (Terracciano et al., 2010; de Moor et al., 2012). This enrichment of associations suggests that personality traits are likely influenced by many genes in a complex manner, each with small effects.
The association between some personality traits and longevity however triggers the question as to whether additional or different genetic variants in long-lived individuals may be associated with longevity-promoting personality traits such as low neuroticism and high extraversion, and as such could contribute to longer life span and health-span. To test this hypothesis, we conducted a GWAS of five domains of NEO-FFI in subjects from the LLFS and examined the results in the context of other studies, Terracciano et al. (2010) and de Moor et al. (2012), and tested gene-by-environment interactions.
Materials and Methods
Study Participants and NEO Five-Factor Inventory
Long Life Family Study
The LLFS is a study of 583 families demonstrating clustering for longevity and healthy aging living in the United States and Denmark. Study eligibility criteria have been described in detail elsewhere (Newman et al., 2011) and enrollment was conducted between 2006 and 2009 from three study centers in the United States (Boston University, University of Pittsburgh, and Columbia University) and in Denmark at the University of Southern Denmark (Sebastiani et al., 2009; Newman et al., 2011). Potential probands were screened for familial longevity using the Family Longevity Selection Score (FLoSS), which scores a family according to birth-year cohort survival probabilities of the proband and siblings (Sebastiani et al., 2009). Family eligibility criteria were a FLoSS >7.0, a proband and at least one living sibling, who did not have dementia, and at least one offspring also willing to participate. The spouses were enrolled as controls of subjects enrolled for familial longevity. Spouses of the offspring generation were recruited as controls while spouses of the proband generation were recruited only if their biological children were enrolled in the study. Enrollment was closed in 2009, and since 2010 subjects have been followed with annual data collections. Phase 1 of the study refers to the initial data collection when the subjects were enrolled and Phase 2 refers to subsequent annual data collections.
NEO-five-factor inventory
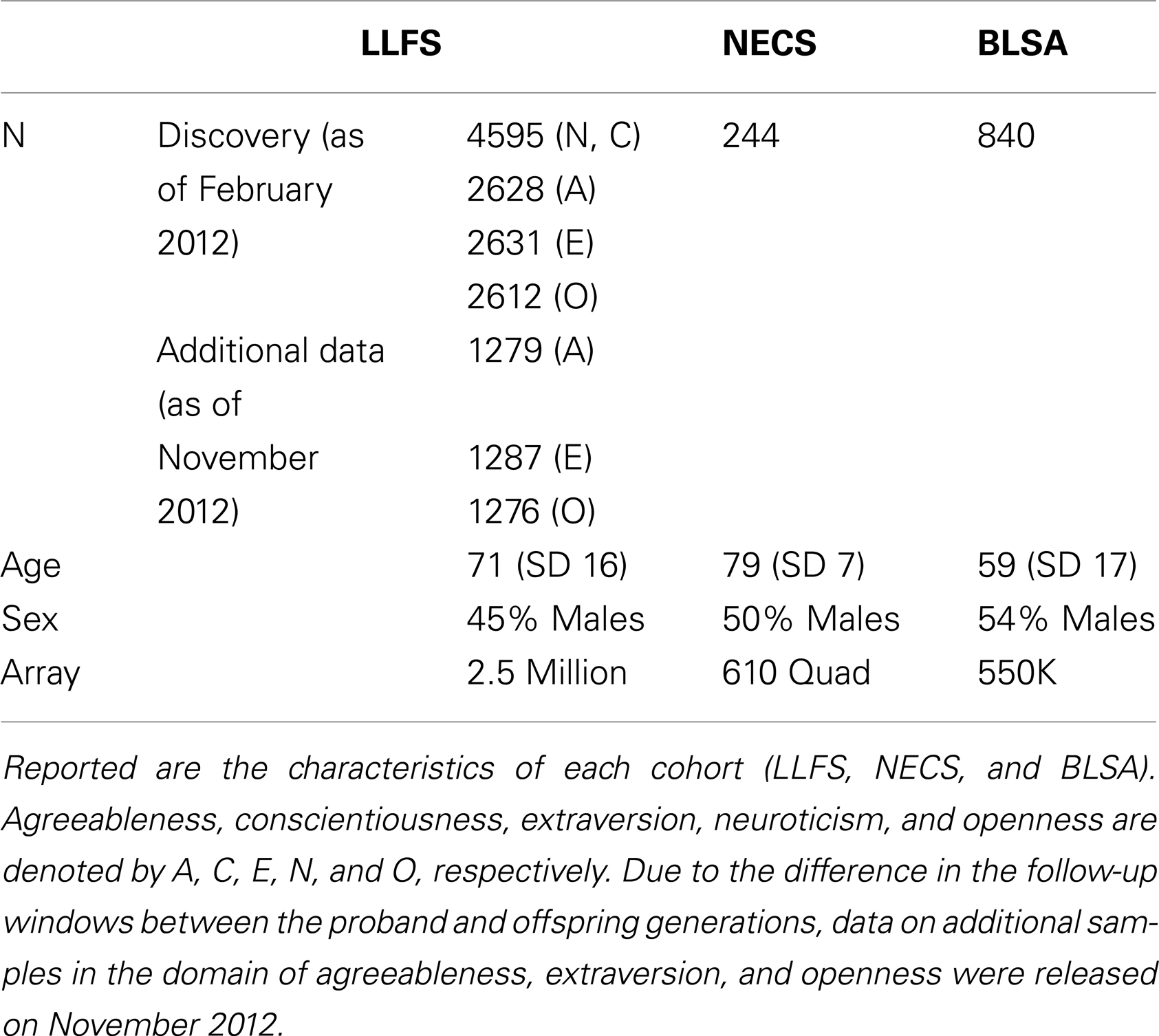
The NEO-FFI is a shortened version of the Revised NEO Personality Inventory (NEO PI-R) and the correlation between the two versions range from 0.75 to 0.89 (Costa and McCrae, 1992). The NEO-FFI consists of 60 items, with 12 items for each of the five domains of personality as compared to the NEO PI-R which has 48 items per domain. Each item in the NEO-FFI is scored using a five-item Likert scale of agreement with each statement. During Phase 1 of the LLFS (2006–2009), only the domains neuroticism and conscientiousness of the NEO-FFI were administered to 4938 participants during a phone interview or an in-home interview. During Phase 2 (2010–present) of the LLFS, all five domains of the NEO-FFI were administered by phone or mail to all living and willing participants, and the data were distributed by the data management and coordinating center in two batches: an initial batch of data from 3032 participants (July 2011) (Andersen et al., 2012a), and an additional batch of data from ∼1300 participants (November 2012). Because of some loss at follow-up and deaths of participants, the sample size for the three domains of agreeableness, extraversion, and openness was ∼4400 (Table 1). The way in which we received the data from the data coordinating center created an independent split of the entire data into two non-overlapping data sets that will be used for internal replication, even though individuals in the two batches of the data are correlated. Note that in the analysis of the two data sets, we fully account for this family correlation. Note also that while the proband generation receives a full follow-up every year, the offspring generation receives a full follow-up every 3 years. As a result of this staggered follow-up window, 99.5% of the samples in the second batch are primarily comprised of subjects in the offspring generation.
New England Centenarian Study
The New England Centenarian Study (NECS) is an ongoing study of exceptional longevity1 that began in 1995 (Andersen et al., 2012b). Approximately 1500 centenarians, 500 offspring, and 150 spouses of offspring have been enrolled and followed annually. Personality data for this study were collected in 2008 from 244 unrelated offspring of centenarians using the NEO-FFI questionnaire for a study of personality traits and exceptional longevity (Givens et al., 2009).
Baltimore Longitudinal Study of Aging
Started in 1958, the Baltimore Longitudinal Study of Aging (BLSA) is an ongoing multidisciplinary study of aging2. The community-dwelling volunteers are assessed at scheduled visits, which include personality assessment. A total of 840 subjects were genotyped (using the Illumina 550K array) and completed the NEO-FFI personality questionnaire at least once. The sample included 46% women and had a mean age of 58.5 years (SD = 17) at the baseline personality assessment.
Genotyping
DNA samples were genotyped at CIDR, and genotypes calls were determined using Illumina recalibrated clusters. The LLFS GWAS SNP data (Illumina Omni 2.5 or 2.5 million SNPs) underwent checking and quality correction centrally at the LLFS data coordinating center (Washington University, St. Louis) where a series of standard procedures were applied. By using GRR package (Abecasis et al., 2001), the GWAS data were checked with the pedigree structure data to verify that the relationships were correct and to avoid sample mismatches. In some cases, direct comparisons of Y and Mitochondrial markers were also applied to verify relationships. Mendelian consistency for all SNPs was assessed with Loki (Heath, 1997) for autosomal markers, PedCheck (O’Connell and Weeks, 1998) for X markers or direct comparison for Y and mitochondrial markers. All Mendel inconsistencies were removed: a sliding threshold depending on minor allele frequency (MAF) was set and if a SNP had inconsistencies below that threshold, the SNP was set to missing in all families with an inconsistency. If the number of families with inconsistencies was above the threshold, the SNP was removed from the analysis. The threshold was set to remove the “worst” ∼0.2% of markers, and ranged from 35 for a MAF of 0.45–0.5 to 1 for a MAF below 0.1. Filters on call rates both by SNP and by individual were also applied as follows: Individuals with a call rate below 97.5% and SNPs with a call rate below 98% had genotype data removed. These steps resulted in 18 individuals and 86,233 autosomal SNPs being removed, as well as 153,363 Mendel inconsistencies set to missing in the families in which they occurred. Approximately two million autosomal SNPs were judged fit for analysis after this filtering.
The NECS DNA samples were genotyped at Boston University using the Illumina Human610-Quad SNP array, with ∼600,000 SNPs. All samples genotyped at Boston University were processed according to the manufacturer’s protocol and BeadStudio Software was used to make genotype calls utilizing the Illumina pre-defined clusters. Samples with less than a 95% call rate were removed and SNPs with a call rate <97.5% were re-clustered. After re-clustering, SNPs with call rates >97.5%, cluster separation score >0.25, excess heterozygosity between −0.10 and 0.10, and MAF >5% were retained in the analysis. We also removed samples with inconsistent sex defined by heterozygosity of the X chromosome that was not consistent with the sex recorded in the database.
Imputation
In LLFS, imputation of un-typed genotypes was performed using MACH (version 1.0.16) (Li et al., 2010) for pre-phasing the genotypic data and MINIMAC (version May 29, 2012) (Howie et al., 2012) for actual imputation with 1000HG genotypic data (version 2010–2011 data freeze, 2012-03-04 haplotypes) including all races as a reference panel. A number of filters before imputing were implemented in the LLFS genotypic data by removing markers that had MAF <1%, HWE p-value <10−6, if LLFS SNPs alleles mismatched with those of 1000HG, and not present in the 1000HG panel. As a result, 38,045,518 variants were imputed and only those with r2 > 0.3 were used for analysis.
Statistical Analysis
Prior to all analyses, raw scores of NEO-FFI in the LLFS and NECS subjects were transformed into sex-specific standardized T-scores with mean = 50 and standard deviation of 10 using the sex-specific means and standard deviations in Table B-4 on page 78 of the NEO PI-R manual (Table S1 in Supplementary Material) (Costa and McCrae, 1992). T-scores between 45 and 55 represent normal values, while T-scores <45 represent lower than normal, and T-scores >55 represent higher than normal values. Lack of departure of the T-score from normality was verified by using the Kurtosis test as reported in Andersen et al. (2012a).
Heritability
Heritability estimates (Table S2 in Supplementary Material) were obtained using variance components analysis implemented in Sequential Oligogenic Linkage Analysis Routines (SOLAR) (Almasy and Blangero, 1998). Under variance components analysis, the total phenotypic variance can be modeled as a sum of an additive genetic component and a non-additive genetic component consisting of environmental factors and measurement errors. The narrow-sense heritability is estimated as the ratio of additive genetic variance to the total phenotypic variance. For each domain, covariates included sex, field centers, and significant polynomial terms of age.
Discovery
Only subjects with completed questionnaires and genotype data (including spouses) and with Caucasian origins were included in the discovery analysis in the LLFS (agreeableness: n = 2628; conscientiousness: n = 4590; extraversion: n = 2631; neuroticism: n = 4595; openness: n = 2612). This discovery set comprised data released in February 2012 (batch 1). Again, due to the difference in the data collection between the proband and offspring generations, the additional data we received on November 2012 (batch 2) were an independent split of the entire data and were used to replicate the top findings from the discovery set in the domains of agreeableness, extraversion, and openness. There were 2759 participants, who had repeated measures on conscientiousness and neuroticism, and the average time interval between the repeated measures was 2.6 years. The agreement between repeated measures of conscientiousness and neuroticism was estimated using the Spearman correlation coefficient (0.67 and 0.66, respectively) and repeated measures were summarized by the average scores and the average ages. No significant trend between age and difference between the two domains was observed, as reported in Andersen et al. (2012a). The association between the five domains of personality and the genotype for each SNP was tested in a linear mixed model with random effects per subject. The random effects were modeled as a multivariate normal distribution with zero mean vector and variance-covariance matrix proportional to the kinship matrix to fully account for familial relations. Covariates included sex, field centers, and significant polynomial terms of age. Significant polynomial terms of age were searched using the model search strategy described in Andersen et al. (2012a). Analyses incorporating the top 10 principal components were also conducted, but the results did not change substantially with this adjustment. All GWAS analyses were performed with R statistical software (version R.2.14) using the “kinship” package. The additive genetic model, which codes the SNP genotype as the number of minor alleles (0, 1, 2), was assumed. SNPs with MAF greater 5% and genotype count >2 were used. To correct for multiple testing, the genome-wide significance threshold of 10−8 was used. Table 2 shows the SNPs that reached genome-wide significance. To assess the chance of a false positive association, 10,000,000 permutation tests were performed for the SNPs that reached genome-wide significance.
Replication in NECS and BLSA
SNPs with p-value <10−5 in the discovery set were sought for replication in the NECS (n = 197) and BLSA (n = 848). We defined replication as SNPs having p-value <0.05 in the replication cohort and consistent direction of effects as in the discovery set. In the NECS, the association between the five domains and the genotype for each SNP was tested using linear regression analysis, adjusting for gender and significant polynomial terms of age, where appropriate, in PLINK (Purcell et al., 2007) software (Table S3 in Supplementary Material). SNPs that were not in the 610 Illumina array were replaced by the closest proxy SNPs in strongest linkage disequilibrium (r2 > 0.8), within a region of 50 kb. Consistency of effects for proxy SNPs was checked by examining the coded allele frequencies of the original SNP and the corresponding proxy SNP. For 53 SNPs we could not find a good proxy. In the BLSA the association analyses were conducted using MERLIN3, and age, sex, and principal components were used as covariates (Table S4 in Supplementary Material).
Replication of findings from other GWASs
All published results from Terracciano et al. (2010) and de Moor et al. (2012) with p-value <10−4 and 10−5 (available through online Supplementary Material), respectively, were tested in the LLFS using the full sample data as of November 2012 and NECS sets. The intention of this replication test was to examine whether any variants that were shown to be significantly associated with personality traits in the previous GWASs also exhibit significant associations in the LLFS and/or NECS. In the case of non-matching SNPs, imputed dosages were used for the LLLF set. The lowest r2, a measure of correlation between the imputed genotype and true genotype, was 0.74 for the SNPs we tested. Proxy SNPs in the NECS set were searched within the region of up to ±50,000 base pairs and r2 > 0.8. Again, consistency of effects for proxy SNPs was checked by examining the coded allele frequencies of the original SNP and the corresponding proxy SNP. Note that only one of the SNPs from Terracciano et al.’s study reached marginal statistical significance in the meta-analysis of de Moor et al.
SNP-by-age interaction
In order to identify SNPs whose effects change with participants’ age on each NEO domain, we tested the significance of a SNP × Age interaction term for those SNPs with significant main effects (p-value <10−6), where age represents the participant’s age at the assessment of NEO Genome-wide testing for interaction would require too much power, as reported in Thomas (2010). Therefore, we chose the p-value of 10−6 to limit the number of significant testing for interaction to 6, 7, 24, 37, and 7 SNPs respectively for agreeableness, conscientiousness, extraversion, neuroticism, and openness, and have some statistical power. Because of the smaller number of tests, a p-value <0.05 was used for statistical significance of the interaction term.
Results
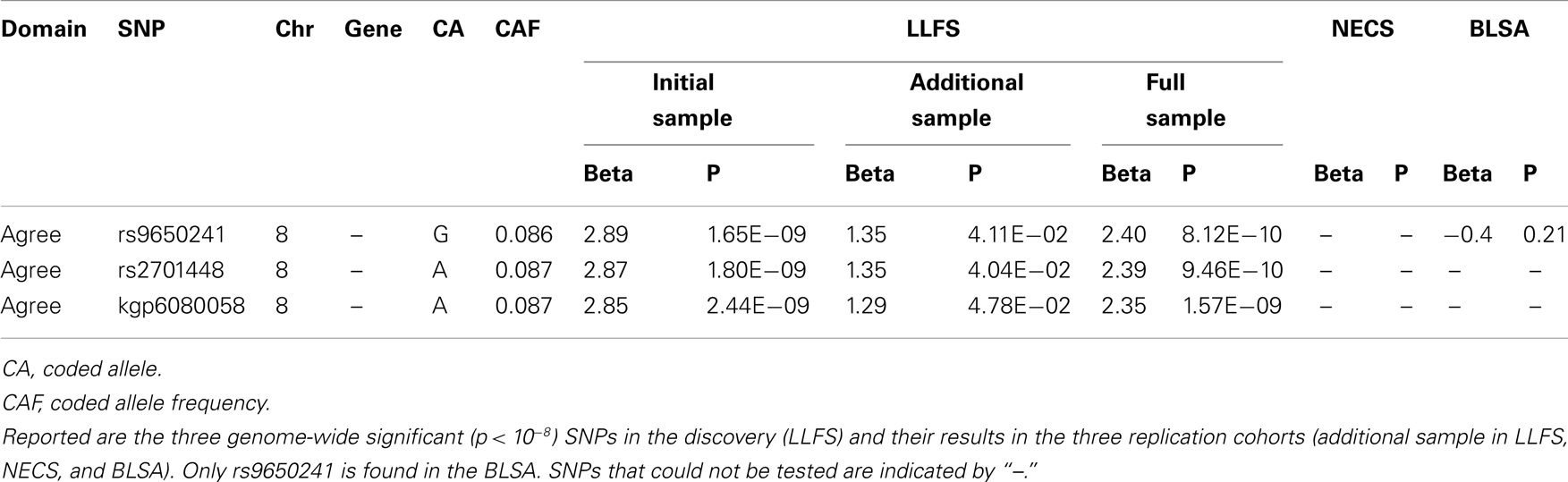
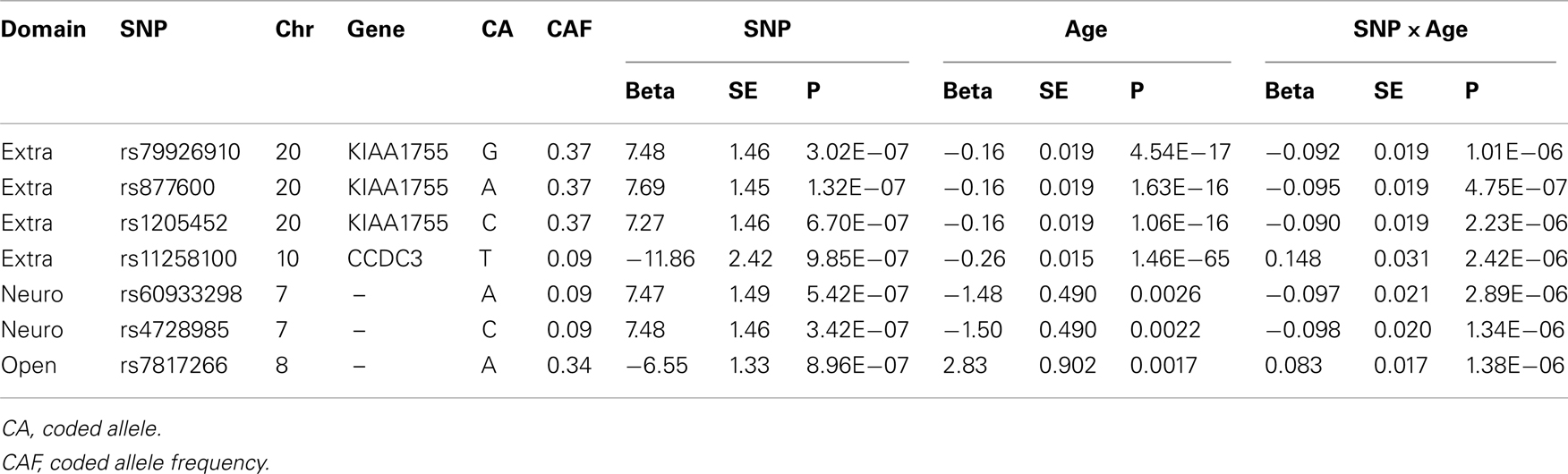
Characteristics of each cohort are summarized in Table 1. The heritability estimates for the five domains and p-values for statistical significance are presented in Table S2 in Supplementary Material and show that all five domains of personality are heritable. Openness was the most heritable (h2 = 49%), while Agreeableness was the least heritable (h2 = 18%). The heritability estimates for conscientiousness, extraversion, and neuroticism were 30, 32, and 25%, respectively. With the exception of the higher heritability of openness, the other estimates were comparable to those reported in Pilia et al. (2006). The QQ-plots and Manhattan plots of the GWAS from the LLFS are shown in Figure 1. Three SNPs reached genome-wide significance (Table 2) in the initial GWAS of the LLFS. The top findings (p-value <10−5) from the LLFS can be found in Table S5 in Supplementary Material (21 SNPs associated with agreeableness; 26 SNPs associated with conscientiousness; 7 SNPs associated with extraversion; 12 SNPs associated with neuroticism; 9 SNPs associated with openness). None of the SNPs that could be tested in the NECS (22 SNPs) or BLSA (24 SNPs) reached statistical significance. Table 3 provides a summary of the replicated results, and Table 4 lists the SNPs in which the genetic effect changes with age. Eighty-one SNPs had significant main effects (p-value <10−6), and were included in the analysis of significant interactions. There were seven SNPs that had significant interactions (four in extraversion, two in neuroticism, and one in openness). Next, results of specific domains are presented.
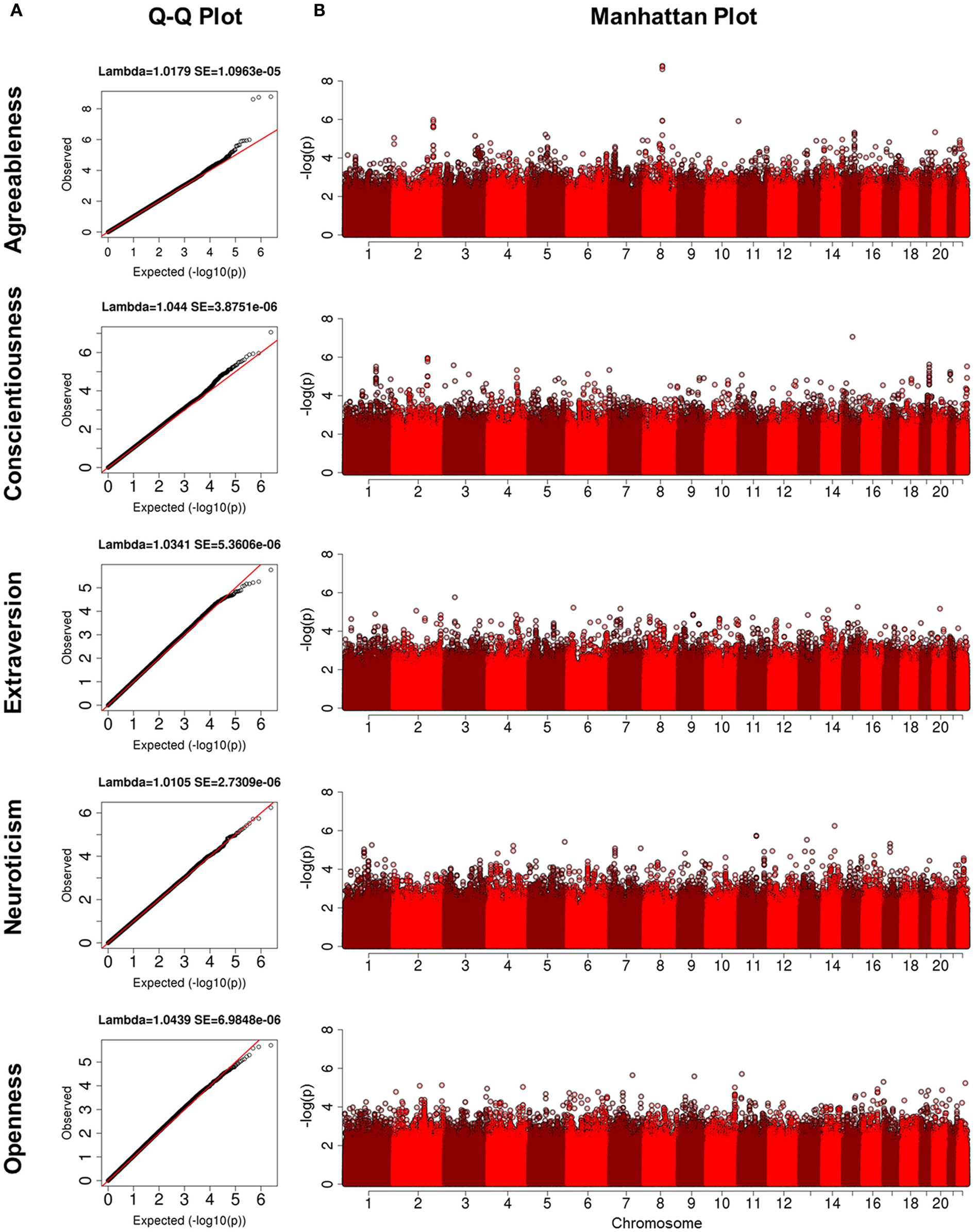
Figure 1. (A) Q-Q plots for five domains of NEO-FFI. The y-axis is the quantiles of observed p-values and the x-axis is the quantiles from the expected distribution; (B) Manahattan plots for five domains of NEO-FFI. The y-axis is the -log(p) and the x-axis is the genomic locations for each SNP ordered by chromosome and base pair positions.
Agreeableness
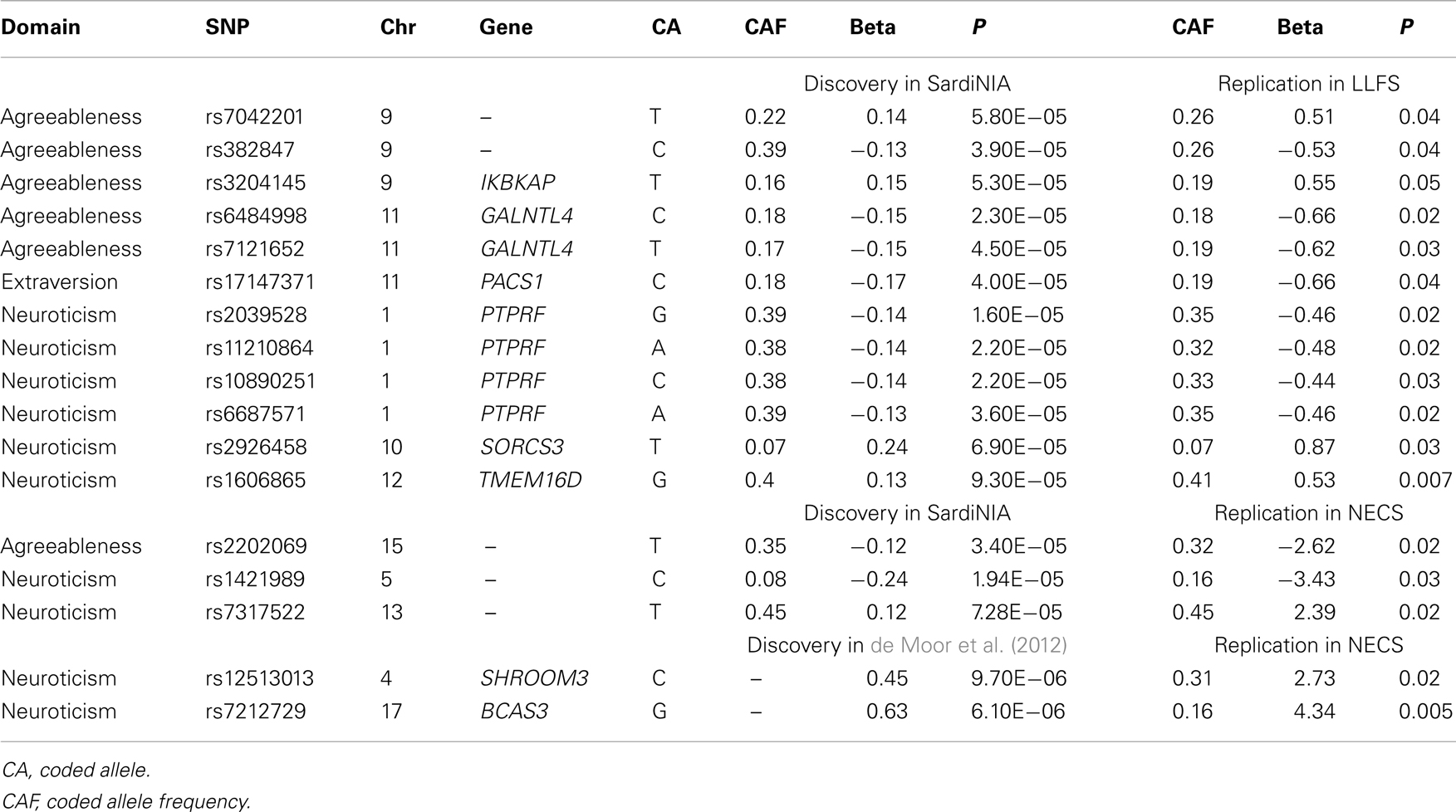
Three SNPs on chromosome 8, which are in almost perfect LD, reached genome-wide significance in the first batch of data and consistent results in the second batch of data in the LLFS with p = 0.04 (Table 2). When analyzed using the combined sample (n = 3907), the statistical significance increased to a p-value of 8.12 × 10−10 for rs9650241 (the most significant SNP). However, these SNPs did not replicate in the BLSA (p-value for rs9650241 was 0.21, and the other two SNPs were not found in their array). These three SNPs were not found in the NECS. From Terracciano et al.’s (2010) findings on agreeableness (114 SNPs with p < 10−4), 109 were found in the LLFS GWAS and 5 replicated, and 20 were found in the NECS GWAS and 1 replicated (Table 3). From de Moor et al.’s (2012) findings on agreeableness (14 SNPs with p < 10−5), 14 were found in the LLFS GWAS, and 2 in the NECS GWAS, but none replicated.
Conscientiousness
No SNP reached genome-wide significance. SNP rs79732200 on chromosome 15 in the gene IGDCC3, reached almost genome-wide significance (p-value 9 × 10−8). However, this SNP was not found in either NECS or BLSA GWAS. From Terracciano et al.’s (2010) findings on conscientiousness (35 SNPs with p < 10−4), 31 were found in the LLFS GWAS, and 6 were found in the NECS GWAS, but none replicated. de Moor et al. (2012) identified rs2576037 in KATNAL2 to be genome-wide significant (4.9 × 10−8), but it did not replicate in the LLFS or NECS GWAS (p = 0.7 and p = 0.5, respectively). From de Moor et al.’s findings on conscientiousness (110 SNPs with p < 10−5), 109 were found in the LLFS GWAS, and 26 were found in the NECS GWAS, but none replicated.
Extraversion
No SNP reached genome-wide significance. From Terracciano et al.’s (2010) findings on extraversion (56 SNPs with p < 10−4), 55 were found in the LLFS GWAS and 7 were found in the NECS GWAS, and 1 replicated in the LLFS GWAS (Table 3). From de Moor et al.’s (2012) findings on extraversion (30 SNPs with p < 10−5 reported), 30 were found in the LLFS GWAS and 7 were found in the NECS GWAS, but none replicated. Four SNPs had significant main effects term (p < 10−6) and interaction term (p < 0.05) (Table 4). Three of these SNPs are in strong LD and are in the gene KIAA1755 and 1 SNP in CCDC3. The plots in Figure S1 in Supplementary Material show that the changes of the effect of the minor allele (A) in SNP rs877600 in KIAA1755 for different ages: carriers of the AG and GG genotype (green and blue lines) tend to score higher than carriers of the AA genotype (red line) in extraversion at younger ages (approximately <80 years) but this trend is reversed in older ages. However, the estimated score remains within “average values” until age 100. The effect of the age × SNP interaction for rs11258100 in CCD3 is different: while carriers of the GG genotype (red line) tend to score lower in extraversion at older ages, in carriers of the genotypes TG and GG the score for extraversion appear to be stable across a wide range of ages.
Neuroticism
No SNP associated with neuroticisms reached genome-wide significance. From Terracciano et al.’s (2010) findings on neuroticism (46 SNPs with p < 10−4), 44 were found in the LLFS GWAS and 6 replicated; 12 were found in the NECS GWAS and 2 replicated (Table 3). From de Moor et al.’s (2012) findings on neuroticism (36 SNPs with p < 10−5), 35 were found in the LLFS GWAS, but none replicated; 3 were found in the NECS GWAS and 2 replicated (Table 3). Two SNPs in strong LD had significant main effects (p < 10−6) and interaction (p < 0.05) terms (Table 4). In Figure S1 in Supplementary Material, subjects with CC genotype (two minor alleles) for SNP rs4782985 have higher neuroticism scores at earlier ages from 40 to 70 than subjects with CT or TT genotypes, but their scores decrease below the normative values after the age of ∼80 years, while subjects with CT or TT genotypes retain the normative values at older ages.
Openness
No SNP associated with openness reached genome-wide significance. From Terracciano et al.’s (2010) findings on openness (62 SNPs with p < 10−4), 57 were found in the LLFS GWAS and 18 were found in the NECS GWAS, but none replicated. de Moor et al. (2012) reported rs1477268 and rs2032794 in RASA1 to be genome-wide significant (p = 2.8 × 10−8 and 3.1 × 10−8, respectively). These SNPs did not replicate in the NECS GWAS with p = 0.83 and p = 0.71, respectively, and did not replicate in the LLFS GWAS with p = 0.69 and p = 0.73, respectively. From de Moor et al.’s findings on openness (39 SNPs with p < 10−5), 39 were found in the LLFS GWAS and 14 were found in the NECS GWAS, but none replicated. In the interaction model, 1 SNP had significant main effects term (p < 10−6) and interaction term (p < 0.05) (Table 4). For this SNP (rs7817266), the three genotype groups show similar trend where their openness scores are higher at earlier ages approximately from 50 to 75 and scores are lower at older ages. However, subjects with AA genotype (two minor alleles) remain within the normative values of openness at all ages, while subjects with AG or GG genotype have openness scores below the normal range after the age of 90.
Discussion
Prior studies have shown that personality traits have a genetic component (Loehlin and Martinb, 2001; Bouchard and McGue, 2003; Pilia et al., 2006) and the heritability estimates derived in the LLFS data confirm these results. Compared to a twin study (Jang and Vernon, 1996) that reported heritability estimates of 41, 53, 61, 41, and 44%, respectively for neuroticism, extraversion, openness, agreeableness, and conscientiousness, LLFS estimates are lower in every domain. Compared to the recent estimates reported by Pilia et al. (2006) from 6148 Sardinians, the estimates of heritability in the LLFS are higher in conscientiousness, extraversion, and openness. The heritability estimate was lower in LLFS for agreeableness, but comparable for neuroticism. These differences may be due to the fact that samples in the two studies are ethnically different (the Sardinian sample was from a genetically isolated population) and phenotypically different. The LLFS is a study of longevity and families were selected for evidence of familial longevity (Newman et al., 2011). Andersen et al. (2012a) showed that all five domains of NEO scores have different distributions at different ages in the LLFS subjects, and this selected population may be enriched for variants that are associated with longevity as well as longevity-promoting personality traits that translate into different heritability estimates.
Intriguingly, the variants associated with agreeableness that reached genome-wide significance in the LLFS did not replicate in the BLSA. The three genome-wide significant SNPs in agreeableness showed consistent results in the two batches of samples of the LLFS. Even though the two data sets are not independent, the fact that these SNPs show significant associations with consistent effects strengthens the validity of this finding. Among the 10,000,000 permutation tests performed, there was 1 permutation that achieved genome-wide significance, which yields an empirical p-value of 2 × 10−7 (North et al., 2002). The analysis using the combined samples yielded improved statistical significance for the three SNPs and two additional SNPs that almost attained genome-wide significance (rs2587559 with p = 4.13 × 10−8 and rs2587561 with p = 5.27 × 10−8), located in the promoter region of TRPA1, which is in a close proximity (within 40,000 bp) to the original variants. This result further corroborates the association between agreeableness and this particular region on chromosome 8, which is linked to tolerance to pain (Doehring et al., 2011). The three genome-wide significant SNPs in the initial GWAS of LLFS have a MAF of around 9%, and there are only 18 subjects out of 2622 with the GG genotype for rs9650241 (the most significant SNP). Therefore, it is difficult to assess the true relationship with the additive coding of SNP genotypes. Under dominant coding (dominant for the G allele) for rs9650241, the statistical significance improves to a p-value of 2.1 × 10−10, compared to 1.7 × 10−9 in an additive model. We observed that the median agreeableness in subjects with the G allele is above the normal range (median T-score = 56.1). The association of this locus with agreeableness is novel in the LLFS, although it did not replicate in the BLSA and it could not be tested in the NECS. Therefore, there is no evidence on whether the same association exists in the general population or this locus is linked to agreeableness through longevity.
There were a total of 75 SNPs (21 SNPs associated with agreeableness; 26 SNPs associated with conscientiousness; 7 SNPs associated with extraversion; 12 SNPs associated with neuroticism; 9 SNPs associated with openness, Table S5 in Supplementary Material) with p-value <10−5 in the GWAS of LLFS. Some of these SNPs were in interesting genes. For example, the most significant SNP in neuroticism (rs177389) is a missense mutation in PAPLN, which changes the aminoacid MET into ARG. This gene was shown to be linked to suicidal ideation during anti-depressant treatment (Laje et al., 2009). On average, carriers of the T allele scored lower in neuroticism; the median scores for the three genotypes (GG, GT, TT) were 43.8, 42.7, and 41.4, respectively, all of which are below the normative value. This SNP could not be replicated in the NECS and BLSA GWAS. Likewise, many of the top findings from LLFS remain to be replicated by other investigators in a larger sample with evidence of longevity.
Interestingly, the LLFS GWAS also replicated findings from other studies (Terracciano et al., 2010; de Moor et al., 2012). Six SNPs in agreeableness, 10 SNPs in neuroticism, and 1 SNP in extraversion replicated in either the LLFS or NECS GWAS (Terracciano et al., 2010; de Moor et al., 2012), although none of the replicated associations passed the Bonferroni correction (p = 0.00015). de Moor et al. reported several SNPs in ARNTL to be associated with agreeableness. In both the LLFS and NECS GWAS, a cluster of SNPs in the same gene were significantly associated with agreeableness at α = 0.05 (Table S6 in Supplementary Material). However, these SNPs were not in LD with those reported in de Moor et al. (highest r2 = 0.234). ARNTL, one of the circadian clock genes, is associated with winter depression and seasonal affective disorder (Partonen et al., 2007) as well as alcohol use disorders (Kovanen et al., 2010). These replicated associations strengthen the original findings in Terracciano et al. (2010), de Moor et al. (2012). Lack of replication of the other findings may be due to different ethnicities, social, and environmental factors.
The significant SNP-by-age interaction terms in Table 4 suggest an interesting hypothesis that it is depicted in Figure 2. It is well known that aging is in part determined by genetic and environmental factors and while genetic factors may explain only 25% of the variability to survive to the mid 1980s (Herskind et al., 1996), the contribution of genes to surviving to older ages is likely much larger (Perls et al., 2000; Sebastiani et al., 2012). Being selected for familial longevity, participants of the LLFS may be enriched for genetic and non-genetic variants that promote longevity, and LLFS subjects who have reached old ages may harbor varying proportions of longevity associated variants. In this hypothetical context, the SNP × age interactions found in this study may actually be surrogates for several gene × gene and gene × environment interactions. This hypothesis is consistent with work in McCrae et al. (2010) that suggested the genetic basis of personality traits is multifactorial, it is likely determined by many interacting genes with individual small effects, and environmental factors are also important factors. However, additional analysis that models the genetic influence on lifespan and personality traits simultaneously is needed to begin to test this hypothesis.
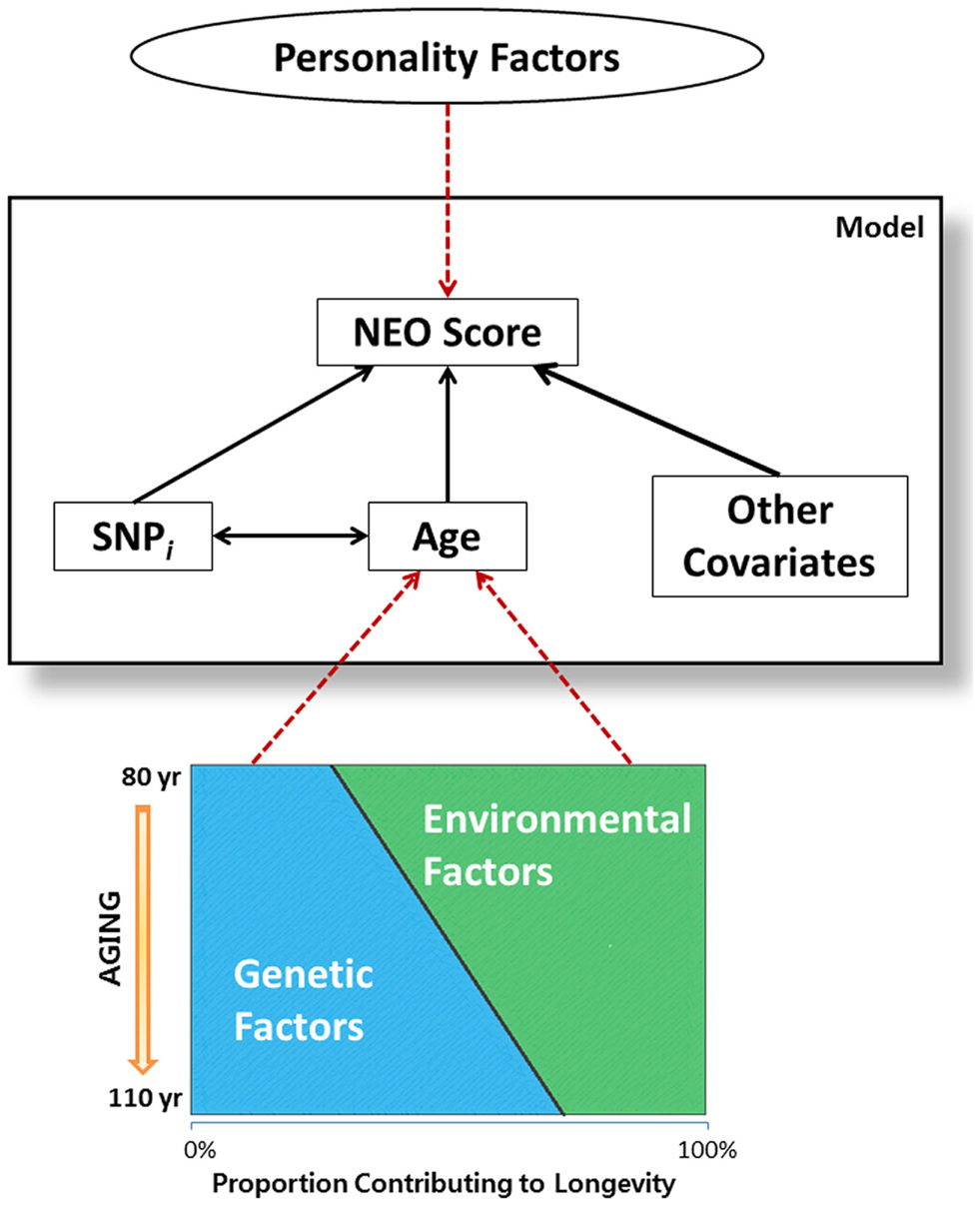
Figure 2. Graphical representation of hypothesis on gene-gene interactions. Attributes inside the large rectangular box are the variables included in the statistical model. Age, which is associated with NEO scores, is reflective of both genetic and environmental factors associated with aging. As genetics factors outweigh the environmental factors in individuals enriched for longevity, SNP-by-age interaction term in the model may imply gene-gene interaction which plays important roles in personality traits, as related to longevity.
Limitations
Our ability to replicate the genome-wide significant associations discovered in the LLFS GWAS was limited by the different arrays that were used to genotype the DNA samples from the BLSA and the NECS. In addition, given that five phenotypes are analyzed, a genome-wide significance threshold of 10−8 may be too liberal and additional replication is necessary to confirm a role of these discovered variants in personality traits.
Conclusion
This study is the first GWAS on five major domains of personality traits assessed by NEO-FFI in a sample enriched for longevity. Our results replicated a few loci identified by others, and confirm that effects of each common genetic variant are modest. As with many complex polygenic traits, genetic framework of personality, as related to longevity, seems multifactorial. In particular, genetic variants that promote longevity may interact with other variants in the establishment of personality traits.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the participants and family members of the Long Life Family Study and the New England Centenarian Study for allowing us to study how they have achieved their longevity. This work was funded by the National Institute on Aging (NIA cooperative agreements U01-AG023755 and U19-AG023122 to T.P. and Intramural Research Program to L.F.), and the National Heart Lung Blood Institute (R21HL114237 to P.S).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Behavioral_and_Psychiatric_Genetics/10.3389/fgene.2013.00065/abstract
Footnotes
References
Abecasis, G. R., Cherny, S. S., Cookson, W. O., and Cardon, L. R. (2001). GRR: graphical representation of relationship errors. Bioinformatics 17, 742–743.
Almasy, L., and Blangero, J. (1998). Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62, 1198–1211.
Andersen, S. L., Sun, J. X., Sebastiani, P., Huntly, J., Gass, J. D., Feldman, L., et al. (2012a). Personality factors in the Long Life Family Study. J. Gerontol. B Psychol. Sci. Soc. Sci. doi:10.1093/geronb/gbs117. [Epub ahead of print].
Andersen, S. L., Sebastiani, P., Dworkis, D. A., Feldman, L., and Perls, T. T. (2012b). Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J. Gerontol. A Biol. Sci. Med. Sci. 67, 395–405.
Bouchard, T. J. Jr., and McGue, M. (2003). Genetic and environmental influences on human psychological differences. J. Neurobiol. 54, 4–45.
Chapman, B. P., Roberts, B., and Duberstein, P. (2011). Personality and longevity: knowns, unknowns, and implications for public health and personalized medicine. J. Aging Res. 2011, 759170.
Costa, P. T. Jr., and McCrae, R. R. (1992). Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor (NEO-FFI) Inventory Professional Manual. Odessa, FL: Psychological Assessment Resources.
de Moor, M. H., Costa, P. T., Terracciano, A., Krueger, R. F., de Geus, E. J., Toshiko, T., et al. (2012). Meta-analysis of genome-wide association studies for personality. Mol. Psychiatry 17, 337–349.
Doehring, A., Kusener, N., Fluhr, K., Neddermeyer, T. J., Schneider, G., and Lotsch, J. (2011). Effect sizes in experimental pain produced by gender, genetic variants and sensitization procedures. PLoS ONE 6:e17724. doi:10.1371/journal.pone.0017724
Friedman, H. S., Tucker, J. S., Tomlinson-Keasey, C., Schwartz, J. E., Wingard, D. L., and Criqui, M. H. (1993). Does childhood personality predict longevity? J. Pers. Soc. Psychol. 65, 176–185.
Givens, J. L., Frederick, M., Silverman, L., Anderson, S., Senville, J., Silver, M., et al. (2009). Personality traits of centenarians’ offspring. J. Am. Geriatr. Soc. 57, 683–685.
Heath, S. C. (1997). Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am. J. Hum. Genet. 61, 748–760.
Herskind, A. M., McGue, M., Holm, N. V., Sorensen, T. I., Harvald, B., and Vaupel, J. W. (1996). The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum. Genet. 97, 319–323.
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J., and Abecasis, G. R. (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959.
Jang, K. L. L. W., and Vernon, P. A. (1996). Heritability of the big five personality dimensions and their facets: a twin study. J. Pers. 64, 577–591.
Kovanen, L., Saarikoski, S. T., Haukka, J., Pirkola, S., Aromaa, A., Lonnqvist, J., et al. (2010). Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol Alcohol. 45, 303–311.
Laje, G., Allen, A. S., Akula, N., Manji, H., John Rush, A., and McMahon, F. J. (2009). Genome-wide association study of suicidal ideation emerging during citalopram treatment of depressed outpatients. Pharmacogenet. Genomics 19, 666–674.
Li, Y., Willer, C. J., Ding, J., Scheet, P., and Abecasis, G. R. (2010). MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834.
Loehlin, J. C., and Martinb, N. G. (2001). Age changes in personality traits and their heritabilities during the adult years: evidence from Australian twin registry samples. Pers. Individ. Dif. 30, 1147–1160.
McCrae, R. R., Scally, M., Terracciano, A., Abecasis, G. R., and Costa, P. T. Jr. (2010). An alternative to the search for single polymorphisms: toward molecular personality scales for the five-factor model. J. Pers. Soc. Psychol. 99, 1014–1024.
Newman, A. B., Glynn, N. W., Taylor, C. A., Sebastiani, P., Perls, T. T., Mayeux, R., et al. (2011). Health and function of participants in the Long Life Family Study: a comparison with other cohorts. Aging 3, 63–76.
North, B. V., Curtis, D., and Sham, P. C. (2002). A note on the calculation of empirical P values from Monte Carlo procedures. Am. J. Hum. Genet. 71, 439–441.
O’Connell, J. R., and Weeks, D. E. (1998). PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266.
Partonen, T., Treutlein, J., Alpman, A., Frank, J., Johansson, C., Depner, M., et al. (2007). Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann. Med. 39, 229–238.
Perls, T., Shea-Drinkwater, M., Bowen-Flynn, J., Ridge, S. B., Kang, S., Joyce, E., et al. (2000). Exceptional familial clustering for extreme longevity in humans. J. Am. Geriatr. Soc. 48, 1483–1485.
Pilia, G., Chen, W. M., Scuteri, A., Orru, M., Albai, G., Dei, M., et al. (2006). Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2:e132. doi:10.1371/journal.pgen.0020132
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575.
Sebastiani, P., Hadley, E. C., Province, M., Christensen, K., Rossi, W., Perls, T. T., et al. (2009). A family longevity selection score: ranking sibships by their longevity, size, and availability for study. Am. J. Epidemiol. 170, 1555–1562.
Sebastiani, P., Solovieff, N., Dewan, A. T., Walsh, K. M., Puca, A., Hartley, S. W., et al. (2012). Genetic signatures of exceptional longevity in humans. PLoS ONE 7:e29848. doi:10.1371/journal.pone.0029848
Terracciano, A., Lockenhoff, C. E., Zonderman, A. B., Ferrucci, L., and Costa, P. T. Jr. (2008). Personality predictors of longevity: activity, emotional stability, and conscientiousness. Psychosom. Med. 70, 621–627.
Terracciano, A., Sanna, S., Uda, M., Deiana, B., Usala, G., Busonero, F., et al. (2010). Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry 15, 647–656.
Thomas, D. (2010). Gene – environment-wide association studies: emerging approaches. Nat. Rev. Genet. 11, 259–272.
Weiss, A., and Costa, P. T. Jr. (2005). Domain and facet personality predictors of all-cause mortality among medicare patients aged 65 to 100. Psychosom. Med. 67, 724–733.
Wilson, R. S., Krueger, K. R., Gu, L., and Bienias, J. L. (2005). Mendes de Leon CF, Evans DA. Neuroticism, extraversion, and mortality in a defined population of older persons. Psychosom. Med. 67, 841–845.
Keywords: neo scores, GWAS, family study, gene-environment interaction, longevity
Citation: Bae HT, Sebastiani P, Sun JX, Andersen SL, Daw EW, Terracciano A, Ferrucci L and Perls TT (2013) Genome-wide association study of personality traits in the Long Life Family Study. Front. Genet. 4:65. doi: 10.3389/fgene.2013.00065
Received: 22 December 2012; Accepted: 09 April 2013;
Published online: 08 May 2013.
Edited by:
Berit Kerner, University of California Los Angeles, USAReviewed by:
Dimitrios Avramopoulos, Johns Hopkins University, USAJudith Ann Badner, University of Chicago, USA
Darlene A. Kertes, University of Florida, USA
Jaime Derringer, University of Colorado, USA
Copyright: © 2013 Bae, Sebastiani, Sun, Andersen, Daw, Terracciano, Ferrucci and Perls. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Harold T. Bae, Department of Biostatistics, Boston University School of Public Health, 801 Massachusetts Avenue, Boston, MA 02118, USA. e-mail:YmFlaEBidS5lZHU=