- 1Department of Genetics and Developmental Biology, School of Medicine, University of Connecticut Health Center, Farmington, CT, USA
- 2Department of Molecular Biology, Cell Biology and Biochemistry, Division of Biology and Medicine, Brown University, Providence, RI, USA
Decreased expression of the fly and worm Indy genes extends longevity. The fly Indy gene and its mammalian homolog are transporters of Krebs cycle intermediates, with the highest rate of uptake for citrate. Cytosolic citrate has a role in energy regulation by affecting fatty acid synthesis and glycolysis. Fly, worm, and mice Indy gene homologs are predominantly expressed in places important for intermediary metabolism. Consequently, decreased expression of Indy in fly and worm, and the removal of mIndy in mice exhibit changes associated with calorie restriction, such as decreased levels of lipids, changes in carbohydrate metabolism and increased mitochondrial biogenesis. Here we report that several Indy alleles in a diverse array of genetic backgrounds confer increased longevity.
Introduction
Aging is a complex process that can be modulated by environment and affected by genetic manipulations, such as single gene mutations. Understanding the underlying mechanisms by which single gene mutations extend life span can contribute to our understanding of the process of aging, and allow us to design therapeutic interventions that could postpone age-related decline and extend healthy aging. For example, based on the genetic data shown that down-regulation of the TOR signaling pathway extends longevity of yeast, worms, and fruit flies, experiments were performed that show that rapamycin, a drug that down-regulates the TOR signaling pathway, extends mice and fruit flies longevity (Vellai et al., 2003; Jia et al., 2004; Kapahi et al., 2004; Kaeberlein et al., 2005; Harrison et al., 2009; Bjedov et al., 2010).
Mutations in the Indy (I’m Not Dead Yet) gene extend life span of the fruit fly, Drosophila melanogaster (Rogina et al., 2000; Wang et al., 2009). Similarly, decreased expression of two of the worm Indy homologs extend worm longevity (Fei et al., 2003, 2004). Indy encodes the fly homolog of a mammalian di and tricarboxylate transporter involved in reabsorbing Krebs cycle intermediates, such as citrate, pyruvate, and α-ketoglutarate (Knauf et al., 2002, 2006; Pajor, 2006). Functional characterization of the transporter encoded by the Indy structural gene confirmed that it is a transporter of Krebs cycle intermediates (Inoue et al., 2002; Knauf et al., 2002). Studies in frog oocytes and mammalian cells showed that INDY mediates Na+, K+, and Cl− independent high-affinity flux of dicarboxylates and citrate across the plasma membrane (Inoue et al., 2002; Knauf et al., 2002). Further studies have shown that INDY functions as an anion exchanger of dicarboxylate and tricarboxylate Krebs cycle intermediates (Knauf et al., 2006). Crystal structure of a bacterial INDY homolog from Vibrio cholera (VcINDY) reveals that one citrate and one sodium molecule is bound per protein but the mature transporter is likely found in the form of a dimer (Mancusso et al., 2012).
The fly INDY is most highly expressed in the gut, fat bodies, and oenocytes, all places where intermediary metabolism takes place, suggesting its role in metabolism (Knauf et al., 2002). Similarly, worm homologs (ceNaDC1 and ceNaDC2) are expressed in the intestinal tract (Fei et al., 2003), and the mouse gene mIndy (mINDY; SLC13A5) is predominantly expressed in liver (Birkenfeld et al., 2011). Based on INDY expression and a role in transporting Krebs cycle intermediates it has been hypothesized that decreased INDY activity creates a state similar to calorie restriction (CR). Studies in flies and mice support this hypothesis mainly by showing similarities between the physiology of Indy mutant flies and mIndy knockout mice on high calorie food and control flies and mice on CR (Wang et al., 2009; Birkenfeld et al., 2011).
It has recently been reported that longevity was not extended in worms with decreased levels of the Indy or in fruit flies with one of the alleles utilized by Rogina et al. (2000) and Toivonen et al. (2007). Toivonen et al. (2007), attributed the life span extension in Indy to the genetic background and bacterial infection (Toivonen et al., 2007). Subsequently, it was confirmed that the original Indy206 mutation extends longevity after backcrossing into the yw background but not after backcrossing into the w1118 genetic background (Wang et al., 2009; reviewed in Frankel and Rogina, 2012). Furthermore, it was demonstrated that the results published in Toivonen et al., are most likely due to differences in the caloric content of the food (Toivonen et al., 2007; Wang et al., 2009).
Here we report that the presence of one copy of an Indy206 mutant chromosome extends longevity in several genetic backgrounds when compared to genetically matched controls. In order to further address the issues of Wolbachia contamination we treated the previously reported Indy159 allele, and several new alleles, with tetracycline and backcrossed all of these Indy alleles into a yw genetic background for 10 generations. We determined survivorships of all Indy alleles on standard laboratory diet and found that several new Indy mutant alleles can also extend the longevity of male and female Drosophila. The data presented here further confirm the role of the Indy gene in Drosophila longevity and show the relationship between life span extension and reduction in Indy mRNA.
Results
Mutation in Indy206 Extends Life Span in Different Genetic Backgrounds
In order to further examine if genetic background may contribute to the life span extension of heterozygous Indy mutant flies, we determined the survivorship of Indy heterozygous mutant flies in Hyperkinetic1 (Hk1) and long- and short-lived selected Luckinbill lines (Figures 1A-E) (Kaplan and Trout, 1969; Luckinbill and Clare, 1985). Hk1 is a recessive mutation characterized by hyperactivity and shorter life span of Drosophila. Hyperactivity is due to mutation of the beta (Hk1) subunit of the potassium channel, which causes increased neuronal excitability (Trout and Kaplan, 1970). Hk1 is an X-linked recessive mutation, thus only male flies in those background live shorter (Trout and Kaplan, 1970; Rogina and Helfand, 1995). We used the Hk1 line since it was isolated by an EMS mutagenesis of Canton-S (CS) stock in 1969 and therefore had many years of divergence from the CS background of the original Indy lines. We determined the survivorship of flies heterozygous for Hk1 and either Indy206 or control-2216. The 2216 and 1085 lines that were derived from the same mutagenesis as Indy206, but do not have a P-element insertions in the Indy region were used as control in Rogina et al., 2000. Survivorship analysis revealed that the median life span of male flies with one copy of the Indy mutation in Hk background is 52.0% increased as compared to the control Hk;2216. A similar increase in survivorship of 57.0% was observed in Hk;Indy206 female flies when compared to the control females, Figures 1A,B; Table 1. (Median life span: Hk;Indy206 males = 38.0 days, females = 68.0; Hk;2216 males = 25.0, females = 43.3).
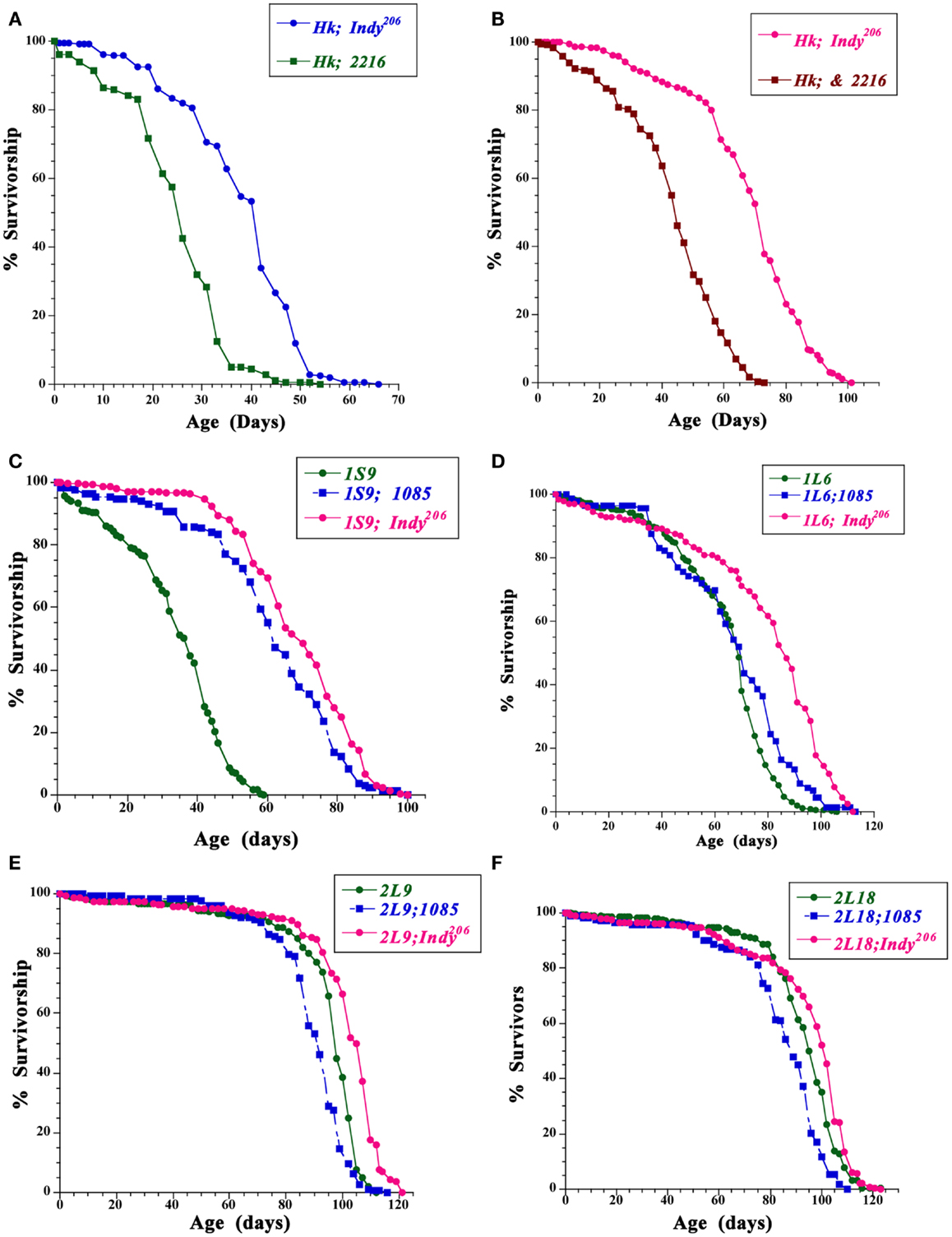
Figure 1. Mutation in the Indy gene extends the life span of male and female Drosophila in different genetic backgrounds. (A,B) Survivorships for male (A) and female (B) heterozygous for the Indy206 (Hk;Indy206), and control (Hk;2216) enhancer-trap line in Hyperkinetic background. (C–F) Survivorship for male homozygous flies for the Luckinbill short live line 1S9 (C), the Luckinbill long-lived line 1L6(D), 2L9 (E), or 2L18 (F) and heterozygous for the Indy206, or 1085 and the Luckinbill 1S9, 1L6, 2L9, or 2L18 lines at 25°C. Between 135–537 flies were used for each life span.
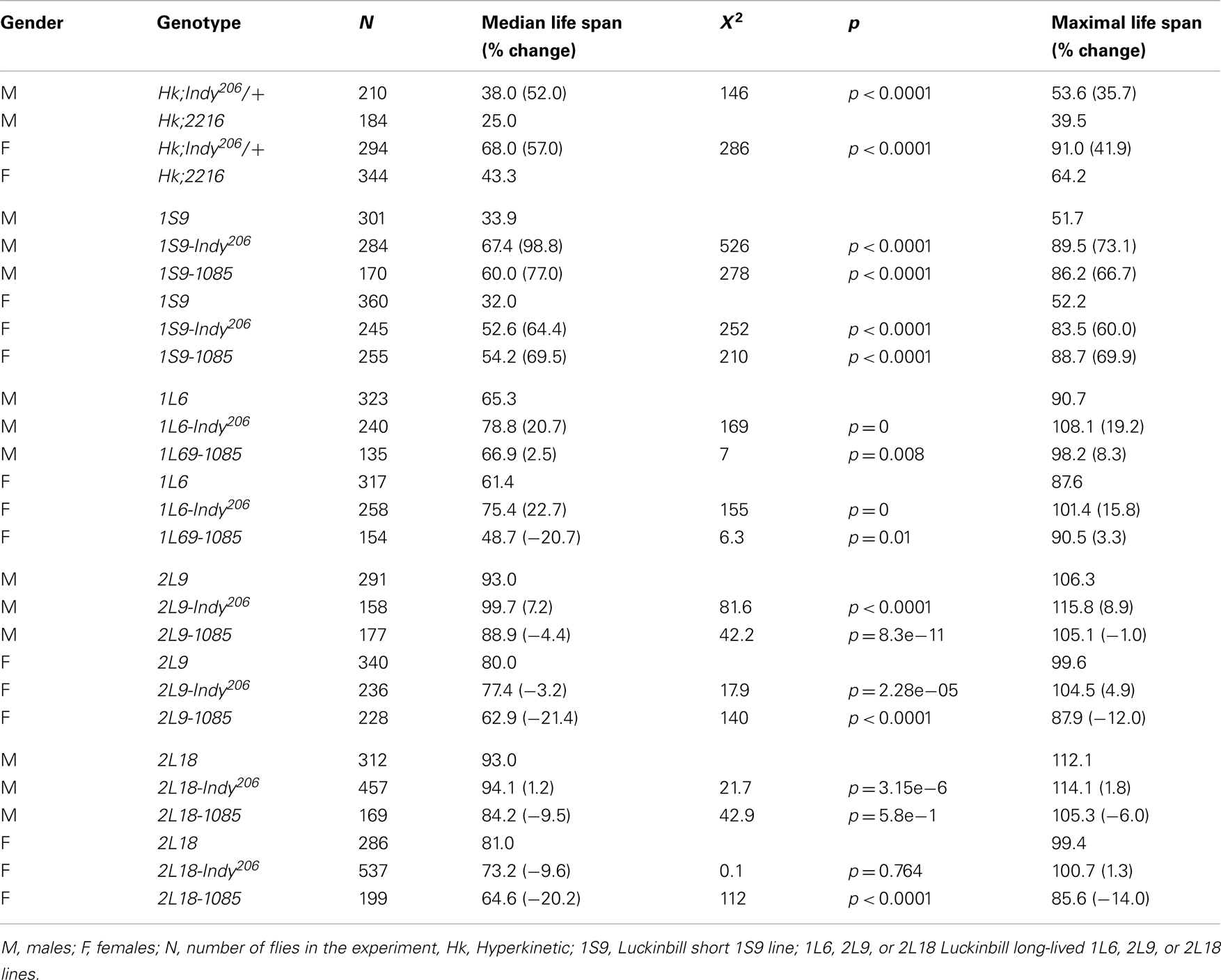
Table 1. Life span of Indy206 heterozygous flies is longer compared to the control flies in different genetic backgrounds.
Indy206 Mutant Heterozygous Flies Live Longer in Luckinbill Short- and Long-Lived Lines Compared to Control Lines
Luckinbill short- and long-lived lines were selected based on reproduction of a population of outbreed Drosophila early or late in life (Luckinbill and Clare, 1985). Selective breeding was carried out for 21 or 29 generations and resulted in a large difference in median longevity between short- and long-lived lines. For instance, median life span of males for the short-lived 1S9 line was 33.9 day, while median longevity for the long-lived line 1L6 = 65.3, 2L9 was 93.0 and 2L18 was 93.0 days. Similar differences in median longevity between short and long-lived line can be seen in females (Median longevity 1S9 = 32.0, 1L6 = 61.4, 2L9 = 80.0, and 2L18 = 81.0 days.) We examined if Indy mutant flies can affect longevity of Luckinbill short- and long-lived line differently as compared to controls and further extend the life of long-lived lines beyond that expected from hybrid vigor. Our data show that the Indy206 mutation increases longevity of both short- and long-lived lines in all conditions, with one exception, the female 1S9;Indy206 flies have a similar median longevity compared to the controls. While, F1 heterozygous males flies from a cross between the control 1085 and the 1S9 short Luckinbill line show the expected life span extension due to hybrid vigor and have a 77% increase in median longevity as compared to the homozygous 1S9 line, F1 heterozygous Indy206;1S9 male flies have much higher increase in median longevity of 98.8% compared to 1S9 homozygous flies, Figure 1C; Table 1. Indy206 mutation further increased longevity of all long-lived Luckbill lines. F1 heterozygote animals from a cross between the Indy206 enhancer-trap line and the laboratory selected long-lived line 1L6 of Luckinbill (Indy206;1L6) live 20.7% longer compared to the homozygous 1L6. In contrast, heterozygous control 1085;1L6, live only 2.5% longer then homozygous 1L6 flies. 2L9 homozygous long-lived Luckinbill line live much longer compared to 1L6 Figure 1D. However, Indy mutant heterozygous flies in 2L9 (Indy206;2L9) still live 7.2% longer compared to the 2L9 homozygous flies Figure 1E. In contrast, F1 heterozygous control males, 1085;2L9 have 4.4% shorter median life span compared to the homozygous 2L9 flies. Median male life span in days: Indy206;2L9 males = 99.7, 1085;2L9 = 88.9, Figure 1E; Table 1. Thus, heterozygous Indy206;2L9 male flies have an increase in life span of 12% over matched controls (2L9;1085), and 7.2% over the homozygote Luckinbill long-lived 2L9 line itself (Table 1). Median life span of female 1085;2L9 is decreased by 21.4% compared to median longevity of homozygous 2L9 female flies, while longevity of Indy206;2L9 females is only 3.2% shorter compared to homozygous flies. (Median female longevity in days: 1085;2L9 = 62.9, Indy206;2L9 = 77.4, Table 1). Heterozygous Indy206 flies in the background of the 2L18 long-lived line do not live significantly longer compared to homozygous 2L18 flies; however, they live significantly longer compared to control 1085;2L18 heterozygous male flies, which live 9.5% shorter compared to 2L18 homozygous male flies. (Median male life span in days: 2L18 = 93.0, Indy206;2L18 = 94.1, 1085;2L18 = 84.2, Figure 1F; Table 1). Similarly, Indy206;2L18 heterozygous females live longer in 2L18 background compared to the controls.
Life Span Extensions in Different Indy Alleles
We have previously reported that five independent Indy mutant alleles extend the life span of male and female Drosophila in wild type CS and yw genetic backgrounds (Rogina et al., 2000; Wang et al., 2009). We have now tested an additional six Indy alleles for their effect on fly longevity (IndyEP3044, IndyEP3366, IndyEY01442, IndyEY01458, IndyEY013297, IndyKG07717). Genomic organization of the Indy locus and position of P-elements insertion in different Indy mutant alleles used in this manuscript is shown in Figure 2A. These six new alleles and three previously tested Indy alleles (Indy206, Indy302, Indy159) and yw control flies, were all treated with tetracycline to eliminate any possible bacterial contamination by Wolbachia. Although the absence of Wolbachia contamination after tetracycline treatment was not confirmed by PCR, we have previously confirmed the absence of Wolbachia after identical treatment (Wang et al., 2009). All of the Indy alleles and one of the control stocks 1085, which has the same genetic background as Indy206and Indy302, were backcrossed into the yw genetic background for 10 generations. We have determined longevity of all Indy alleles as heterozygotes in yw background and calculated median longevity for males and females, Table 2. Representative survivorships of two new Indy alleles are plotted in Figures 2B,C. Heterozygous yw;Indy206/+, yw;Indy302/+, yw;Indy159/+, yw;IndyEY01442/+ yw;IndyEY01458/+, and yw;IndyEY013297/+ male and female flies have a significantly longer life compared to control yw flies. Longevity extension in males with one copy of Indy mutant allele varies from 34.4 to 14.0%, and in females Indy mutant extension range from 29.4 to 10.7%, Table 2. In addition, female, but not male heterozygous yw;IndyEP3044 flies live 9.2% longer compared to the controls. No effect on longevity was observed in male and female heterozygous yw;IndyKG07717/+, yw;Indy3366/+, and male heterozygous yw;Indy3044/+ mutant flies. We determined the levels of Indy mRNA isolated from Head& Thorax of male heterozygous for two of the new Indy alleles (yw;IndyEY01442/+, yw; IndyEP3366/+), one old (yw;Indy206/+), and their genetic control (yw). The levels of Indy mRNA in heterozygous yw;Indy206/+ allele are 51.1% and in heterozygous yw;IndyEY01442/+ allele are 60.6% of the levels of Indy mRNA found in yw flies, Figure 2D. A similar decrease in the levels of Indy mRNA in yw;Indy206/+ was previously reported (Wang et al., 2009). We found only a minor, non-significant decrease in the levels of Indy mRNA in heterozygous yw;IndyEP3366/+ mutant flies. Lack of longevity effect in yw;IndyEP3366/+ allele is most likely due to only a small effect of the P-element insertion on the Indy mRNA levels in yw;IndyEP3366/+mutant flies. Our data show a strong correlation between the level of Indy mRNA and longevity extension.
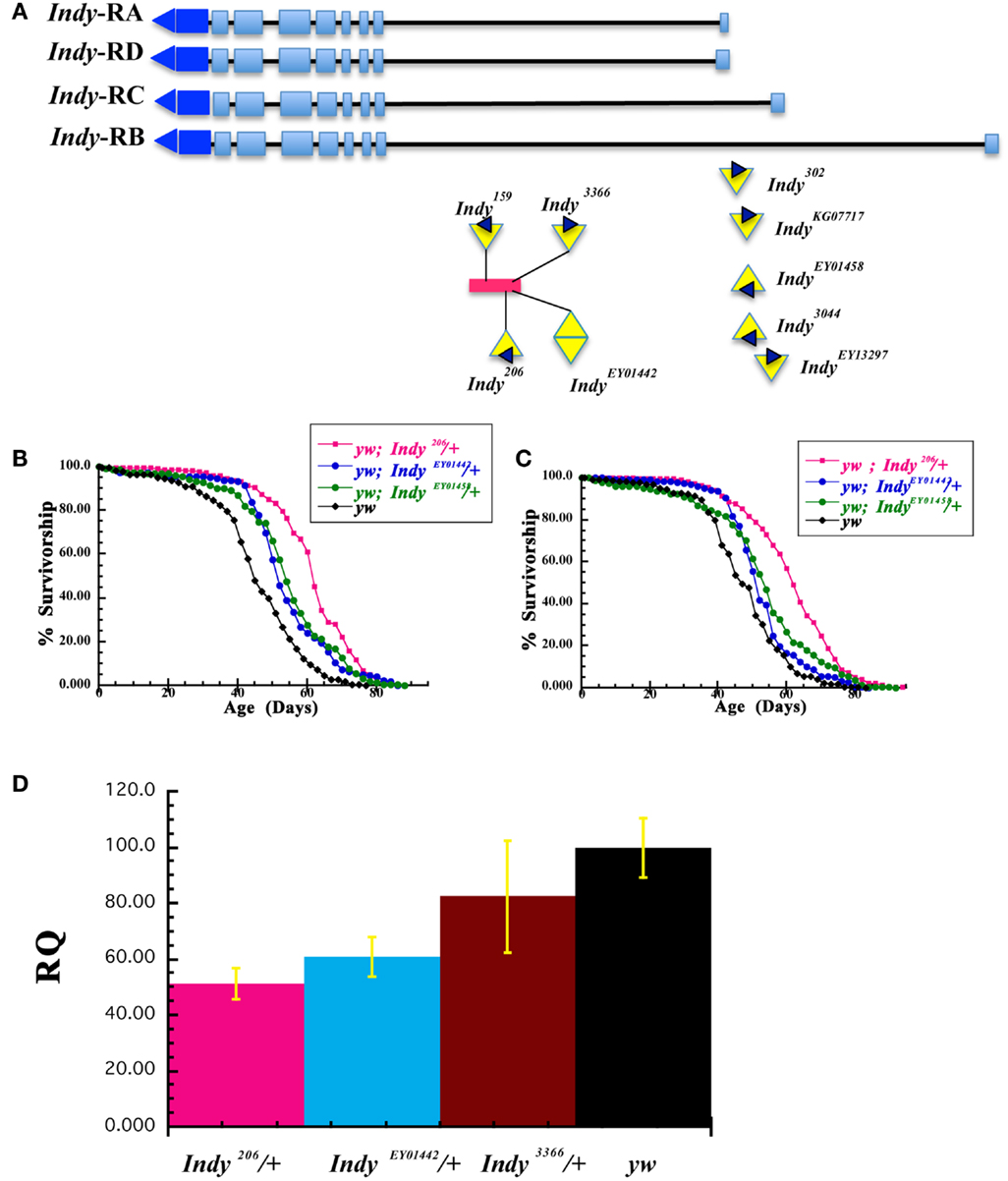
Figure 2. Survivorships of different Indy mutant alleles in yw background. (A) Genomic organization of the Indy locus with insertion sites and orientation of P-element in Indy206, Indy159, IndyEY01442, Indy3366, Indy302, IndyKG07717, IndyEY01458, IndyEP3044, and IndyEY013297 alleles used in this manuscript. Orientation of P-element in IndyEY01442allele is not known. The red rectangle represents the conserved Hoppel transposable element. Indy encodes four putative transcripts (RA, RB, RD, and RC), which have different 5′ exon. (B,C) Life span of males (B) and females (C) heterozygous for Indy206, IndyEY01442, IndyEY01458, and yw on standard laboratory corn diet after 10× backcrossing into the yw. (D) Indy mutants have decreased levels of Indy mRNA. Q-PCR determination of Indy mRNA expression levels in Heads and Thorax of Indy206/+, IndyEY01442/+, Indy33662/+, and yw 20 days old male flies. Experiments were done in two (Indy206/+) or three (IndyEY01442/+, Indy3366/+, and yw) replicates with 2 × 15(Indy206/+), 3 × 40 (Indy3366/+), or 3 × 50 (IndyEY01442/+ and yw) flies in each group.
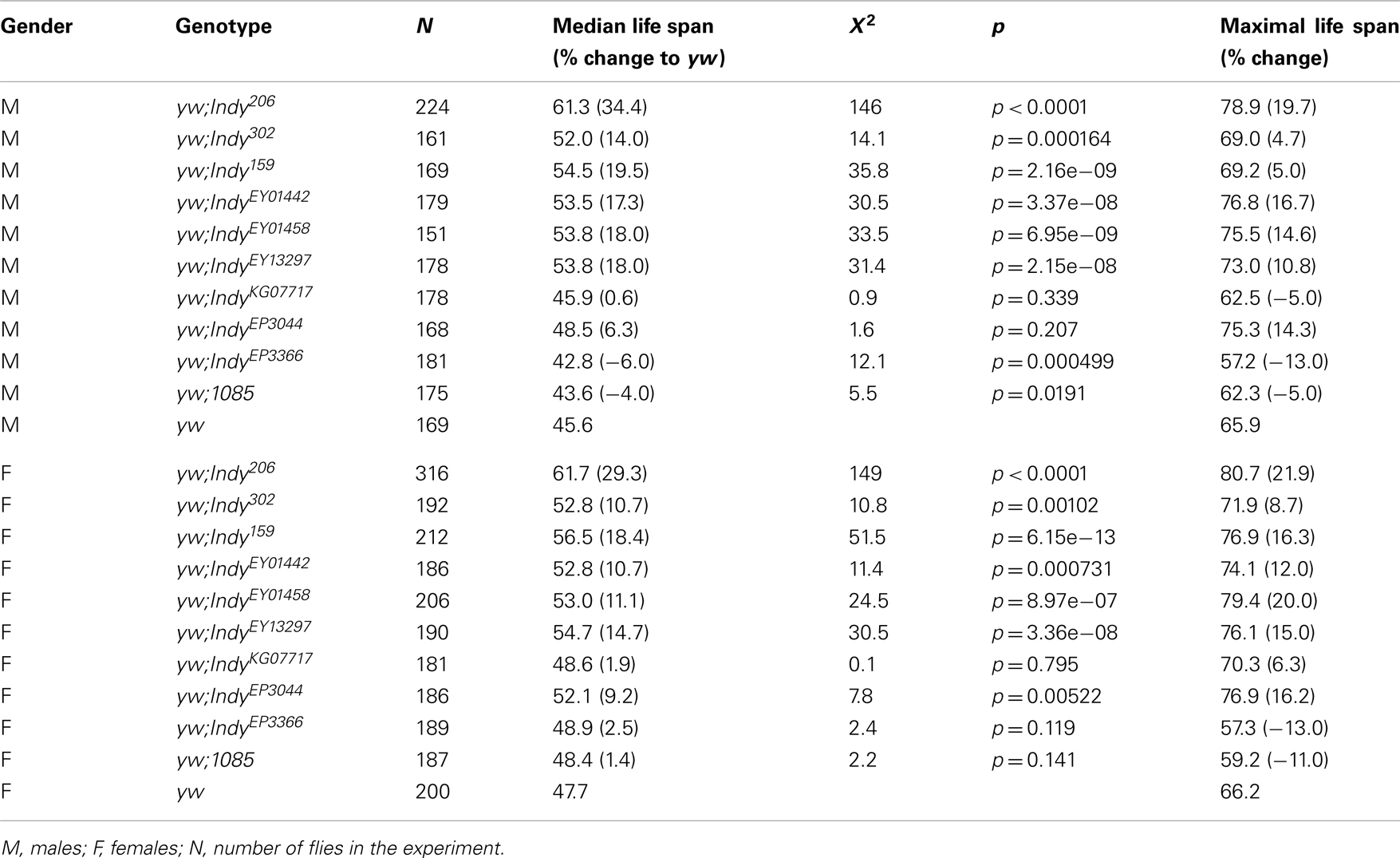
Table 2. Life span of several different Indy mutant alleles as heterozygous is longer compared to the control flies in yw genetic background.
Discussion
We have previously identified and characterized five independent mutations in the Indy gene in Drosophila that cause an increase in average and maximal life span for both male and female fruit flies (Rogina et al., 2000). The original five alleles were derived from three different mutageneses (Boynton and Tully, 1992; Rogina et al., 2000). Life spans of flies carrying one copy of P-element in the Indy gene were compared with their close genetically matched controls, flies from the same mutagenesis without a P-element insertion in the Indy gene. Here we show that Indy206 heterozygous mutant flies also live longer when crossed into three different genetic backgrounds, Hk, short, and long-lived Luckinbill lines as compared to control flies from the same genetic background as Indy also crossed to these three different genetic backgrounds. Luckinbill short and long-lived lines have been generated by selective breeding for early and late female fecundity (Luckinbill and Clare, 1985). Presence of the yw;Indy206 mutant chromosome significantly extends longevity in the background of the Luckinbill short 1S9 line compared to the control line 1085. Moreover, the Indy206 mutation further extends longevity of two long-lived Luckinbill lines and does not cause shortening of life span of 2L18 long-lived line. At the same time, median longevity of control lines when crossed to Luckinbill long-lived lines are significantly shorter compared to homozygous Luckinbill lines. These data show that extension of life span by this Indy allele is not limited to the background of the short-lived lines, but further extends lines already selected for long life span.
We also report extension of longevity by additional Indy mutant alleles. All Indy mutant alleles were treated by tetracycline to prevent any effects of Wolbachia and backcrossed to yw background. Wolbachia infection was proposed as a contributing factor to Indy longevity by Toivonen et al. (2007). IndyEY01442, IndyEY01458, IndyEY013297, IndyKG07717 were generated by the Berkeley Drosophila Genome Project (BDGP) gene disruption project (Bellen et al., 2004). The Indy gene region appears to be a “hot spot” for P-element insertions illustrated by isolation of 5 KG, 28 EY, and 10 EP element insertions in the Indy region (Bellen et al., 2004). P-element insertion in Indy206, Indy159, IndyEY01442, and IndyEP3366 are within the Hoppel element in the first intron of the Indy gene, upstream of the putative translational start site, Figure 2A. The conserved Hoppel element is present in the same position in wild type flies (Rogina et al., 2000). The insertion in Indy302, IndyEY013297, IndyEY01458, IndyKG07717, and IndyEP3044 lines is upstream from putative transcriptional start sites. Indy encodes four putative transcripts, which have different 5′-exons. The positions of P-elements in Indy302, IndyEP3044, IndyEY01458, IndyEY013297, and IndyKG07717 are located close to the three putative transcriptional start sites for three putative Indy transcripts (Indy-RA, Indy-RD, and Indy-RC) and about 5,000 bp upstream from the putative transcriptional start site in Indy-RB. Genomic organization of the Indy locus and positions of P-element insertion in different Indy alleles used in this manuscript are shown in Figure 2A. Positions of additional P-elements insertion can be seen in FlyBase: http://flybase.org/reports/FBgn0036816.html. It was previously shown that the presence of the P-element in Indy206 and Indy302 mutant alleles decreases the levels of Indy mRNA most likely by affecting transcription (Knauf et al., 2006; Wang et al., 2009). The levels of Indy mRNA are decreased about 95% in homozygous Indy206 and about 40% in homozygous Indy302 alleles (Wang et al., 2009). The levels of INDY protein are also dramatically decreased in Indy206 homozygous mutant flies (Knauf et al., 2002). Similarly, here we show that the levels of Indy mRNA are decreased about 39% in the heterozygous IndyEY01442/+ allele and about 49% in the heterozygous Indy206/+ allele compared to the levels of Indy mRNA found in yw flies. No significant decrease in the levels of Indy mRNA were observed in heterozygous Indy3366/+ flies, which correlates with the absence of longevity extension. It is likely that variation in longevity effects of different Indy alleles correlates to actual Indy mRNA levels and differential effects of P-elements on transcription. We found that male flies heterozygous for six Indy alleles have longevity extension ranging from 14.0 to 34.4%. Females heterozygous for seven Indy alleles show similar result having longevity extension ranging from 9.2 to 29.3%. Our data further confirm our hypothesis that the level of Indy expression is central for longevity extension. When the levels of Indy mRNA are decreased approximately 49%, as in Indy206/+ heterozygous mutant flies, there is dramatic longevity extension of 34%. We have previously reported that when the levels of Indy mRNA are radically reduced, as in Indy206 homozygous flies, longevity extension is less than extension of the Indy206/+ heterozygous flies (Wang et al., 2009). A smaller longevity effect of 17% was observed when Indy mRNA levels are moderately reduced, as in IndyEY01442/+. Insignificant reduction of Indy mRNA levels, as in IndyEP3366/+ mutant flies, resulted in no longevity effect. Besides IndyEP3366/+, no longevity extension was found in another one of the new alleles, IndyKG07717. In summary, maximal longevity in Indy mutant flies is associated with optimal reduction of Indy mRNA levels. When Indy levels are too low or close to normal, longevity effects are diminished. Although a recent report attributed life span extension in Indy to hybrid vigor, due to life span evaluation in an incorrect genetic background, and bacterial infection, our data presented here corroborate a link between the Indy mutations and longevity in flies (Toivonen et al., 2007; Wang et al., 2009). The effect of the Indy mutation on longevity was supported by findings that decreased activity of NaDC2, a C. elegans homolog of the Indy gene, extends the life span of worms (Fei et al., 2003, 2004). Similar effects of increased longevity associated with mutations in the fly and the worm Indy gene suggests a possibility of evolutionary conservation and a universal role of INDY in longevity (Fei et al., 2003, 2004).
Several studies have investigated the molecular mechanisms underlying the effects of the Indy mutation on longevity and health span of worms, flies, and mice (Fei et al., 2003; Marden et al., 2003; Neretti et al., 2009; Wang et al., 2009; Birkenfeld et al., 2011). INDY is a plasma membrane transporter that may mediate the movement of dicarboxylic acids through the epithelium of the gut and into organs important in intermediary metabolism and storage (Knauf et al., 2002, 2006). Location of the INDY transporter in the fat body and oenocytes suggest a role in intermediary metabolism and expression in the gut suggests a role in uptake of nutrients. Reductions in INDY activity may alter uptake, utilization, or storage of important nutrients and affect normal metabolism. It has been hypothesized that reductions in Indy activity seen in Indy mutations might be altering the normal energy supply in flies resulting in life span extension through a mechanism similar to CR. CR has been shown to increase life span and delay the onset of age-related symptoms in a broad range of organisms (McCay et al., 1935; Weindruch and Walford, 1988). Consistent with the hypothesis that Indy is important in metabolism is the finding that Indy mutant worms, flies, and mice have disrupted lipid metabolism (Fei et al., 2003; Wang et al., 2009; Birkenfeld et al., 2011). Similarly to CR animals, Indy mutant flies have increased spontaneous physical activity, decreased starvation resistance, weight, egg production, and insulin signaling. Furthermore, wild type flies on CR have significantly decreased levels of Indy mRNA (Wang et al., 2009). Indy homozygous mutant flies live shorter on low calorie foods compared to controls, which is consistent with our hypothesis that Indy mutant flies are already in a state of reduced nutrition on normal food and when food is further reduced, life span is shortened due to starvation (Wang et al., 2009). In addition, Indy mutant flies have increased mitochondrial biogenesis in heads and thoraces similar to CR animals (Neretti et al., 2009). Similarly, mIndy knockout mice have increased mitochondrial biogenesis in the liver. The mechanism of the effect of a decrease in INDY on metabolism is likely from its physiological function as a citrate transporter. Cytosolic citrate is the main precursor for the synthesis of fatty acid, cholesterol, triacylglycerols, and low-density lipoproteins. In addition, cytosolic citrate inhibits glycolysis and fatty acid β-oxidation. Therefore, INDY by affecting the levels of cytosolic citrate may alter glucose and lipid metabolism in a manner that favors longevity. Additional support that Indy mutation mimics CR comes from the findings that mIndy knockout mice are protected against adiposity and insulin resistance when kept on high fat diet (Birkenfeld et al., 2011). The data from worm, fly, and mice studies highlight the importance of INDY in health span and longevity. New Indy alleles described here should provide additional tools to further explore the role of INDY in metabolism and its connection to extended longevity and health.
Materials and Methods
Fly Strains
1S9 a short-lived and 1L6, 2L9, and 2L18 long-lived lines were a kind gift from James W. Curtsinger and originally described in Luckinbill and Clare (1985). Indy206, Indy302, 1085, and 2216 were obtained from Tim Tully (Boynton and Tully, 1992). Indy159 was kind gift from the Bier lab (Bier et al., 1989). IndyEP3044, IndyEP3366, IndyEY01442, IndyEY01458, IndyEY013297, IndyKG07717alleles, and Hk1 were obtained from the Bloomington Stock Center or Exelexis. Heterozygous flies used in survivorship analysis are F1 generations from crosses in which virgin females homozygous for Hk1, short-lived, long-lived Luckinbill lines, or yw were mated to males homozygous for different Indy alleles, or the control lines 1085 or 2216.
Backcrossing Scheme
Indy206, Indy302, Indy159, IndyEP3044, IndyEP3366, IndyEY01442, IndyEY01458, IndyEY013297, IndyKG07717, and 1085 were backcrossed into the yw background. Female virgins from yw were mated with males of different Indy alleles or 1085. Heterozygous females were then backcrossed to yw males for 10 generations.
Food Recipe
We used standard yeast, corn, sucrose food in our experiments: 113 g Sucrose (MP Biomedicals, Fischer Scientific) and 28 g Brewers yeast (MP Biomedicals, Fischer Scientific) was mixed with 643 ml water and autoclaved for 20 min. 49 g corn (MP Biomedicals, Fischer Scientific) and 8.1 g Agar (SciMart) were mixed in 268 ml water and added to the food mixture and autoclaved for 20 min. The food was cooled down with constant mixing. 2.4 g tegosept (Fischer Scientific) dissolved in 10.7 ml 100% EtOH was added when the food temperature was 65°C. Approximately 10 ml food was poured to plastic vials using Fly food dispenser (Fischer Scientific), and vials were covered with Kimwipes and cheese cloth. Once the food was cooled down it was stored at 4°C. Before use the food was warmed up to room temperature.
Life Span
Vials were cleared of adult flies in the morning and the collection of newly eclosed flies occurred in the afternoon. Approximately 20 male and 20 female flies were kept together in a plastic vials with approximately 5–10 ml of a standard cornmeal media (Rogina et al., 2000). Flies were housed in humidity-controlled incubators, maintained at 25°C on a 12 h light: dark cycle. Vials of fresh food were supplied three times weekly (Monday, Wednesday, and Friday) and the number of dead flies was recorded during each passage from old to new vials.
mRNA Isolation Q-PCR Analysis
The standard Chomczynski protocol and Trizol reagent (Gibco BRL) were used to isolate mRNA (Chomczynski and Sacchi, 1987). Male flies at age 20 were placed on a cold block and Head with Thorax were dissected. Three biological replicates of 50 males were used in each isolations of IndyEY01442/+ and yw flies, three biological replicates of 40 Indy3366/+ males and two biological replicates of 15 Indy206/+ males. Q-PCR was performed with Indy and Ankyrin specific primers obtained from Applied Biosystems according to the manufacturers protocol. Ankyrin was used as an endogenous control. The samples were run on the AB 7500 System.
Statistical Analysis
Life span data were analyzed by long-rank tests (http://bioinf.wehi.edu.au/software/russell/logrank/). Maximum life span was calculated as the median life span of the longest surviving 10% of the population.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Suzanne Kowalski and Ryan P. Rogers for their excellent technical help and Dr. Stewart Frankel, Ryan P. Rogers, and Jared Woods for critical reading of the manuscript. This work was supported by a NIA grant AG023088 to Blanka Rogina and by NIA grants AG16667, AG24353, and AG25277 to Stephen L. Helfand.
References
Bellen, J., Lewis, R. W., Liao, G., He, Y., Carlson, J. W., Tsang, G., et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781.
Bier, E., Vaessin, H., Shepherd, S., Lee, K., McCall, K., Barbel, S., et al. (1989). Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 9, 1273–1287.
Birkenfeld, A. L., Lee, H.-Y., Guebre-Egziabher, F., Alves, T. C., Jurczak, M. J., Jornayvaz, F. R., et al. (2011). Deletion of the mammalian INDY homolog mimics aspects of dietary restriction and protects against adiposity and insulin resistance in mice. Cell Metab. 14, 184–195.
Bjedov, I., Toivonen, J. M., Kerr, F., Slack, C., Jacobson, J., Foley, A., et al. (2010). Mechanisms of life span extension by rapamycin in fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46.
Boynton, S., and Tully, T. (1992). Latheo, a new gene involved in associative learning and memory in Drosophila melanogaster, identified from P element mutagenesis. Genetics 131, 655–672.
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159.
Fei, Y. J., Inoue, K., and Ganapathy, V. (2003). Structural and functional characteristics of two sodium-coupled dicarboxylate transporters (ceNaDC1 and ceNaDC2) from Caenorhabditis elegans and their relevance to life span. J. Biol. Chem. 278, 6136–6144.
Fei, Y. J., Liu, J. C., Inoue, K., Zhuang, L., Miyake, K., Miyauchi, S., et al. (2004). Relevance of NAC-2, an Na+-coupled citrate transporter, to life span, body size and fat content in Caenorhabditis elegans. Biochem. J. 379, 191–198.
Frankel, S., and Rogina, B. (2012). Indy mutants: live long and prosper. Front Genet. 3:13. doi:10.3389/fgene.2012.00013
Harrison, D. E., Strong, R., Sharp, Z. D., Nelson, J. F., Astle, C. M., Flurkey, K., et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395.
Inoue, K., Fei, Y. J., Huang, W., Zhuang, L., Chen, Z., and Ganapathy, V. (2002). Functional identity of Drosophila melanogaster Indy as a cation-independent, electroneutral transporter for tricarboxylic acid-cycle intermediates. Biochem. J. 367, 313–319.
Jia, K., Chen, D., and Riddle, D. L. (2004). The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131, 3897–3906.
Kaeberlein, M., Powers, R. W. III, Steffen, K. K., Westman, E. A., Hu, D., Dang, N., et al. (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196.
Kapahi, P., Zid, B. M., Harper, T., Koslover, D., Sapin, V., and Benzer, S. (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14, 885–890.
Kaplan, W. D., and Trout, W. E. III (1969). The behavior of four neurological mutants of Drosophila. Genetics 61, 399–409.
Knauf, F., Mohebbi, N., Teichert, C., Herold, D., Rogina, B., Helfand, S., et al. (2006). The life-extending gene Indy encodes an exchanger for Krebs-cycle intermediates. Biochem. J. 397, 25–29.
Knauf, F., Rogina, B., Jiang, Z., Aronson, P. S., and Helfand, S. L. (2002). Functional characterization and immunolocalization of the transporter encoded by the life-extending gene Indy. Proc. Natl. Acad. Sci. U.S.A. 99, 14315–14319.
Luckinbill, L. S., and Clare, M. J. (1985). Selection for life span in Drosophila melanogaster. Heredity 55, 9–18.
Mancusso, R., Gregorio, G. G., Liu, Q., and Wang, D. N. (2012). Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature 491, 622–626.
Marden, J., Rogina, B., Montooth, K. L., and Helfand, S. L. (2003). Conditional tradeoffs between aging and organismal performance of Indy long-lived mutant flies. Proc. Natl. Acad. Sci. U.S.A. 100, 3369–3373.
McCay, C. M., Crowell, M. F., and Maynard, L. A. (1935). The effect of retarded growth upon the length of life span and upon the ultimate body size. J. Nutr. 10, 63–79.
Neretti, N., Wang, P.-Y., Brodsky, A. S., Nyguyen, H. H., White, K. P., Rogina, B., et al. (2009). Long-lived Indy induces reduced mitochondrial ROS production and oxidative damage. Proc. Natl. Acad. Sci. U.S.A. 106, 2277–2282.
Pajor, A. M. (2006). Molecular properties of the SLC13 family of dicarboxylate and sulfate transporters. Pflugers Arch. 451, 597–605.
Rogina, B., and Helfand, S. L. (1995). Regulation of gene expression is linked to life span in adult Drosophila. Genetics 141, 1043–1048.
Rogina, B., Reenan, R. A., Nielsen, S. P., and Helfand, S. L. (2000). Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290, 2137–2140.
Toivonen, J. M., Walker, G. A., Martinez-Diaz, P., Bjedov, I., Driege, Y., Jacobs, H. T., et al. (2007). No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PLoS Genet. 3:e95. doi:10.1371/journal.pgen.0030095
Trout, W. E., and Kaplan, W. D. (1970). A relation between longevity, metabolic rate, and activity in shaker mutants of Drosophila melanogaster. Exp. Gerontol. 5, 83–92.
Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A. L., Orosz, L., and Müller, F. (2003). Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620.
Keywords: Indy, Drosophila melanogaster, aging and longevity, fruit flies, single gene mutation
Citation: Rogina B and Helfand SL (2013) Indy mutations and Drosophila longevity. Front. Genet. 4:47. doi: 10.3389/fgene.2013.00047
Received: 30 November 2012; Accepted: 14 March 2013;
Published online: 08 April 2013.
Edited by:
Elena G. Pasyukova, Institute of Molecular Genetics of Russian Academy of Sciences, RussiaReviewed by:
Giovanni Cenci, University of L’Aquila, ItalyWilliam Ja, The Scripps Research Institute, USA
Copyright: © 2013 Rogina and Helfand. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Blanka Rogina, Department of Genetics and Developmental Biology, School of Medicine, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, CT 06030-6403, USA. e-mail:Um9naW5hQG5ldXJvbi51Y2hjLmVkdQ==
