
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 02 April 2013
Sec. Plant Genetics and Genomics
Volume 4 - 2013 | https://doi.org/10.3389/fgene.2013.00044
This article is part of the Research TopicEpigenetic Mechanisms of Plant Responses to the EnvironmentView all 15 articles
A correction has been applied to this article in:
Corrigendum: Enhancement of RNA-directed DNA methylation of a transgene by simultaneously downregulating a ROS1 ortholog using a virus vector in Nicotiana benthamiana
Expression of Concern: Enhancement of RNA-directed DNA methylation of a transgene by simultaneously downregulating a ROS1 ortholog using a virus vector in Nicotiana benthamiana
Corrigendum: Enhancement of RNA-directed DNA methylation of a transgene by simultaneously downregulating a ROS1 ortholog using a virus vector in Nicotiana benthamiana
Cytosine methylation can be induced by double-stranded RNAs through the RNA-directed DNA methylation (RdDM) pathway. A DNA glycosylase REPRESSOR OF SILENCING 1 (ROS1) participates in DNA demethylation in Arabidopsis and may possibly counteract RdDM. Here, we isolated an ortholog of ROS1 (NbROS1) from Nicotiana benthamiana and examined the antagonistic activity of NbROS1 against virus-induced RdDM by simultaneously inducing RdDM and NbROS1 knockdown using a vector based on Cucumber mosaic virus. Plants were inoculated with a virus that contained a portion of the Cauliflower mosaic virus 35S promoter, which induced RdDM of the promoter integrated in the plant genome and transcriptional silencing of the green fluorescent protein gene driven by the promoter. Plants were also inoculated with a virus that contained a portion of NbROS1, which induced downregulation of NbROS1. Simultaneous induction of RdDM and NbROS1 knockdown resulted in an increase in the level of cytosine methylation of the target promoter. These results provide evidence for the presence of antagonistic activity of NbROS1 against virus-induced RdDM and suggest that the simultaneous induction of promoter-targeting RdDM and NbROS1 knockdown by a virus vector is useful as a tool to enhance targeted DNA methylation.
Nucleotide-sequence-specific interactions mediated by RNA have a role in the control of gene expression via diverse pathways of RNA silencing, which involve either RNA-guided RNA degradation or epigenetic modification of the genome (Baulcombe, 2004; Brodersen and Voinnet, 2006). RNA-guided epigenetic modification was first discovered in transgenic tobacco plants, in which a viroid complementary DNA (cDNA) integrated in the genome was methylated de novo when the plants were infected with the viroid consisting of a self-replicating RNA, a process coined RNA-directed DNA methylation (RdDM; Wassenegger et al., 1994). RdDM can also be induced by viral RNAs (Jones et al., 1998) and transgene-derived RNAs (Mette et al., 2000). In Arabidopsis, 24-nt short interfering RNAs (siRNAs) generated through processing of double-stranded RNAs (dsRNAs) with Dicer-like (DCL) 3 act as a mobile signal and direct RdDM (Molnar et al., 2010). When dsRNAs corresponding to a gene promoter are synthesized, RdDM of the promoter and transcriptional gene silencing (TGS) can be induced. Such promoter-targeted gene silencing has been used to modify gene expression in plants (Jones et al., 2001; Sijen et al., 2001; Cigan et al., 2005; Heilersig et al., 2006; Okano et al., 2008; Kanazawa et al., 2011a,b; Otagaki et al., 2011; Kasai and Kanazawa, 2013) and other organisms (Hawkins and Morris, 2008; Suzuki and Kelleher, 2009).
RdDM induces de novo methylation of cytosine in all sequence contexts (CG, CHG, and CHH, where H is A, C, or T) at the region of siRNA–DNA sequence homology (Matzke et al., 2009). Factors involved in RdDM and TGS have been identified by analyzing mutants of Arabidopsis that are defective in the process. These factors are the canonical RNA silencing machinery that includes DCL and Argonaute (AGO) family proteins, two plant-specific RNA polymerases, Pol IV and Pol V, DOMAINS REARRANGED METHYLTRANSFERASES (DRM1 and DRM2), chromatin-remodeling factors, and several other proteins that can interact with these factors (Matzke et al., 2009; Law and Jacobsen, 2010; Zhang and Zhu, 2011). Cytosine methylation established through RdDM can be maintained through cell division. On the other hand, a family of DNA glycosylases can demethylate cytosine in plants (Chan et al., 2005).
In Arabidopsis, DNA glycosylases of the DEMETER (DME) family demethylate cytosine: a family consisting of DME, DME-LIKE proteins DML2 and DML3, and REPRESSOR OF SILENCING 1 (ROS1; Furner and Matzke, 2011). DNA demethylation by DME occurs during reproductive development and is required for genomic imprinting and seed viability (Choi et al., 2002; Gehring et al., 2006), whereas DNA demethylation by ROS1, DML2, and DML3 occurs in vegetative tissues and protects hundreds of loci from potentially deleterious methylation (Penterman et al., 2007; Zhu et al., 2007).
REPRESSOR OF SILENCING 1 was identified in a screen for mutants with deregulated expression of a repetitive transgene comprising the luciferase gene driven by the RD29A promoter, in which a low level of siRNAs of the RD29A promoter generated from the transgene repeat presumably induces RdDM at both the transgene and endogenous RD29A promoters (Gong et al., 2002). In wild-type plants, ROS1 is thought to demethylate DNA, thereby counteracting the RdDM (Gong et al., 2002; Kapoor et al., 2005).
We previously developed a system that induces RdDM using an RNA virus vector (designated CMV-A1) based on Cucumber mosaic virus (CMV; Kanazawa et al., 2011a,b; Otagaki et al., 2011). Although dozens of virus vectors have been developed for inducing post-transcriptional gene silencing (PTGS; Kanazawa, 2008; Senthil-Kumar and Mysore, 2011), so far, only the CMV vector efficiently induces TGS of not only transgenes but also endogenous genes (Kanazawa et al., 2011a,b). Such efficient induction of TGS is achieved by the function of the virus-encoded 2b protein, which has the ability to facilitate epigenetic modifications through the transport of siRNAs to the nucleus (Kanazawa et al., 2011a). When transgenic Nicotiana benthamiana (line 16c) plants, which express the green fluorescent protein (GFP) gene under the control of Cauliflower mosaic virus (CaMV) 35S promoter, were infected with recombinant CMVs containing a CaMV 35S promoter segment, the level of GFP mRNA decreased and cytosine methylation was induced on the transgene CaMV 35S promoter (Otagaki et al., 2006, 2011). This reduction in GFP expression was accompanied by a decrease in RNA polymerase II bound to the CaMV 35S promoter, which indicates the occurrence of transcriptional repression (Kanazawa et al., 2011a). Using this system, we examined the effect of differences in the length and positions of the promoter dsRNA on the induction of RdDM and found that both the length of dsRNA (above a threshold of 81–91 nt) and the frequency of cytosines at symmetric sites (described in Section “Discussion”) in the region targeted by dsRNA were important for the induction of efficient RdDM and heritable TGS (Otagaki et al., 2011).
Assuming that DNA methylation and demethylation act antagonistically at the promoter region targeted by the CMV-A1 vector, a combination of RdDM and inhibition of DNA demethylase function may enhance cytosine methylation at the target DNA. To test this hypothesis, here we isolated an ortholog of ROS1 in N. benthamiana and examined the effect of knocking-down the ROS1 ortholog on RdDM of the CaMV 35S promoter induced by the virus vector.
Portions of the ROS1 ortholog in N. benthamiana were amplified by reverse transcription-mediated polymerase chain reaction (RT-PCR) using primers designed to anneal regions conserved between Arabidopsis ROS1 and an ortholog of ROS1 in N. tabacum (NtROS1). Then a full-length cDNA of ROS1 ortholog in N. benthamiana (designated NbROS1) was isolated by RT-PCR in combination with 5′- and 3′-rapid amplification of cDNA ends (RACE) techniques. NbROS1 was predicted to encode a protein of 1796 amino acids and contained a putative nuclear localization signal and a conserved DNA glycosylase domain (Figure 1A). A phylogenetic analysis indicated that NbROS1 was most closely related to NtROS1 among proteins belonging to the DME/ROS1 glycosylase family (Figure 1B). We identified two expressed sequence tags (ES887350 and GO612804) that share sequence homology with NbROS1 by employing the BLASTN search program of the Dana Farber Cancer Institute (DFCI) N. benthamiana Gene Index1, which indicates that multiple cognate genes are present in the genome. Quantitative RT-PCR using primers that specifically amplified NbROS1 indicated that the mRNA of NbROS1 gene was present in root, stem, leaf, and inflorescence (Figure 1C), which was consistent with the expression of the AtROS1 gene in both vegetative and reproductive organs (Gong et al., 2002; Ortega-Galisteo et al., 2008).
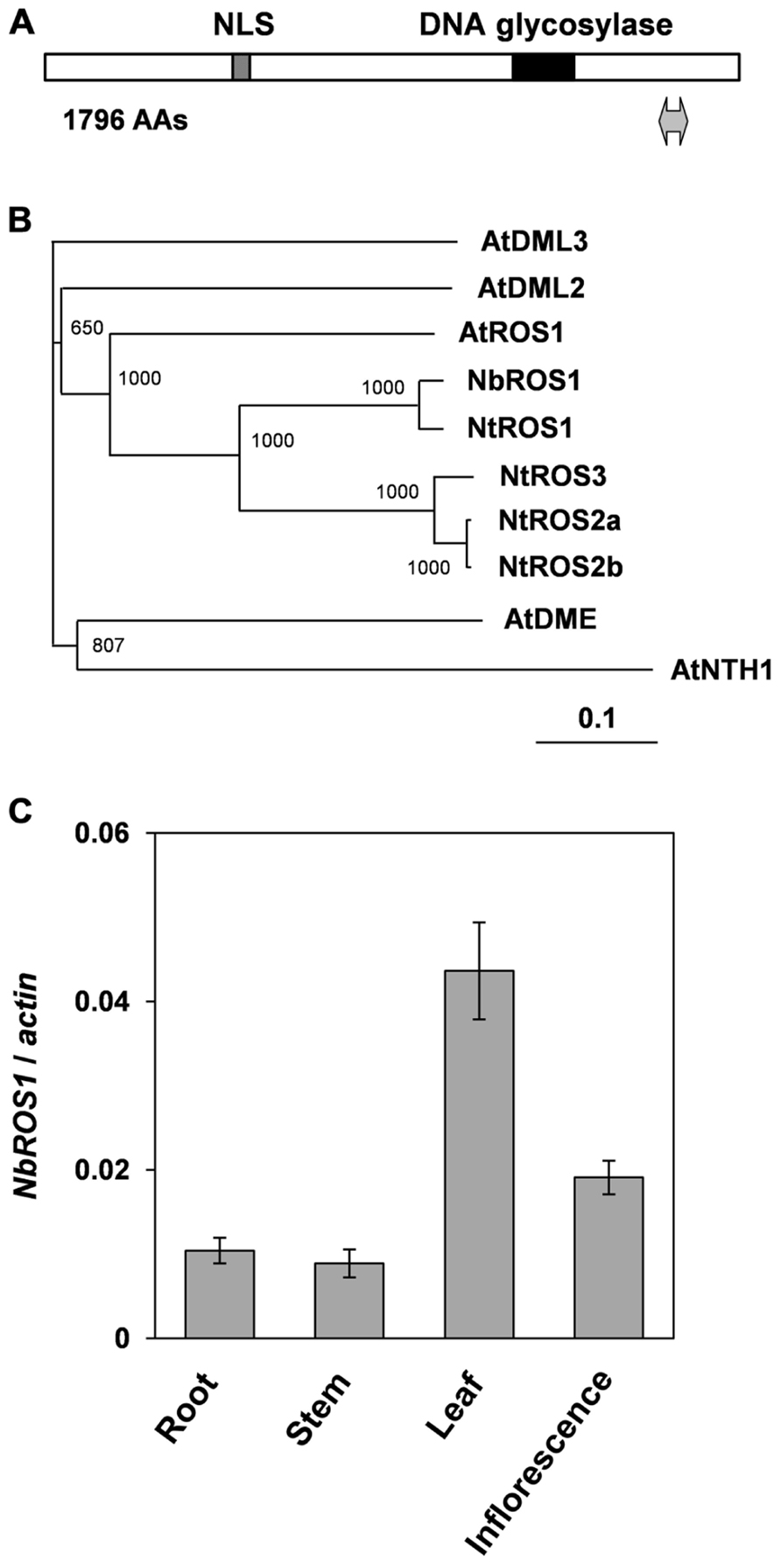
FIGURE 1. Characterization of ROS1 ortholog in N. benthamiana. (A) Structure of NbROS1 protein. Protein size is shown in number of amino acids (AAs). Hatched and black boxes indicate the nuclear localization signal and DNA glycosylase domain, respectively. The region inserted into the CMV-A1 vector is indicated by a thick line with arrowheads at each end. (B) Phylogenetic relationships of DNA glycosylases. The phylogenetic tree was constructed using the neighbor-joining method based on protein sequences deduced from the nucleotide sequences of the genes. Bootstrap values of 1000 replicates are shown. A scale bar represents branch length. Protein sequence of AtNTH1 was used as the outgroup to root the tree. The following sequences were included in the analysis. AtNTH1, Arabidopsis thaliana NTH1 (CAC16135); AtDME, A. thaliana DEMETER (NP_001078527); AtDML2, A. thaliana DEMETER-Like2 (NP_187612); AtDML3, A. thaliana DEMETER-Like3 (NP_195132); AtROS1, A. thaliana ROS1 (Q9SJQ6); NtROS1, Nicotiana tabacum ROS1 (BAF52855); NtROS2a, N. tabacum ROS2a (BAF52856); NtROS2b, N. tabacum ROS2b (BAF52857); NtROS3, N. tabacum ROS3 (BAF52858). (C) Expression of NbROS1. The NbROS1 mRNA level relative to the actin mRNA level was assessed for root, stem, leaf, and inflorescence tissues. Data are the means and standard errors obtained from three replicates.
Because of the wide expression of NbROS1 in tissues including leaves, we induced PTGS of NbROS1 using a virus vector. A 104-nt portion of the NbROS1-coding region (Figure 1A) was inserted into the CMV-A1 vector (Figure 2A), then N. benthamiana plants were infected with the recombinant virus A1:NbROS1. The mRNA level of NbROS1 decreased (Figure 2B) and NbROS1 siRNAs were produced (Figure 2C) in the plants infected with the recombinant virus, which indicated that NbROS1 was downregulated through PTGS.
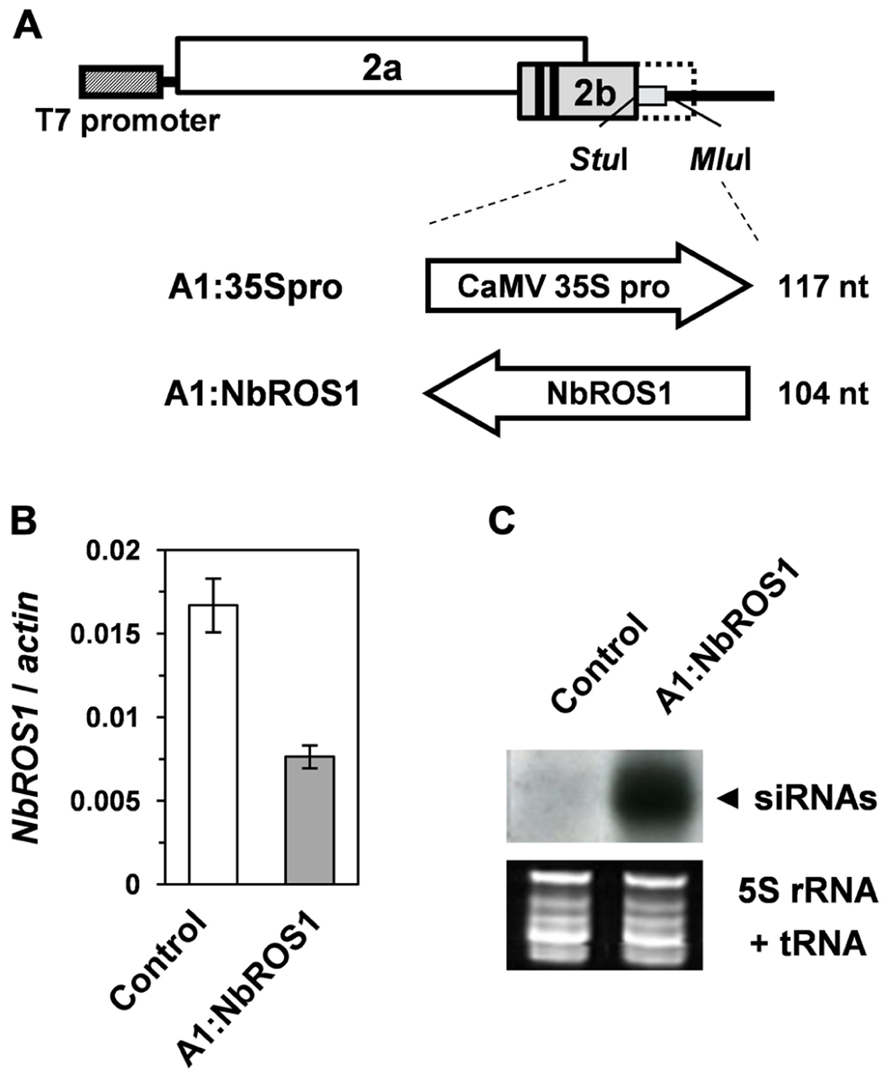
FIGURE 2. Gene silencing using the CMV-A1 vector. (A) Schematic representation of the vector constructs targeting the CaMV 35S promoter or the NbROS1-coding sequence. (B) Changes in mRNA level of NbROS1 as a consequence of infection with virus that contains the NbROS1 insert (A1:NbROS1). The NbROS1 mRNA level was assessed relative to the actin mRNA level in leaf tissues at 18 days post-inoculation (DPI). Data are the means and standard errors obtained from three replicates. (C) Northern blot analysis of low-molecular weight RNAs isolated from leaf tissues of plants infected with the recombinant CMVs at 18 DPI, probed for the NbROS1 gene. Ethidium-bromide-stained 5S rRNA and tRNAs bands are shown below the panel to show that an equal amount of the small RNA fraction was loaded. In the experiments in (B,C), both the control and A1:NbROS1-infected plants were infected with A1:35Spro to eliminate non-specific effects of viral infection on the mRNA level of NbROS1.
Plants of a transgenic N. benthamiana line that carry a transgene containing the CaMV 35S promoter that drives the GFP gene in the genome were infected with a virus containing a portion (-116 to +1 region) of the CaMV 35S promoter (“CMV-A1:-116 to +1”; Otagaki et al., 2011). Here we refer to this construct as “A1:35Spro” for convenience (Figure 2A). Bisulfite sequencing analysis indicated that infection of plants with this virus induces an increase in cytosine methylation of the promoter via RdDM as previously reported (Otagaki et al., 2011; Figures 3A,B).
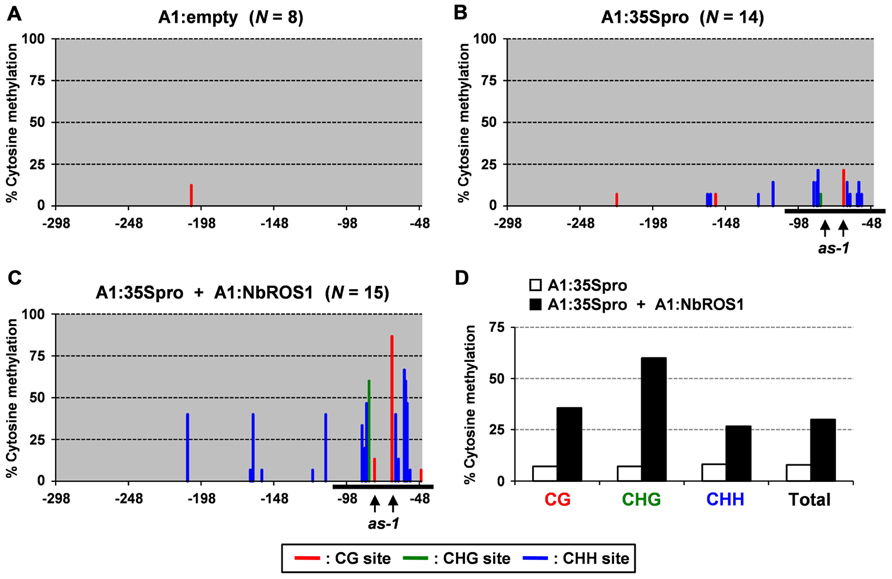
FIGURE 3. Cytosine methylation status of CaMV 35S promoter analyzed by bisulfite sequencing. Cytosine methylation status of the CaMV 35S promoter in plants infected with (A) A1:empty that lacked an insert, (B) A1:35Spro, and (C) both A1:35Spro and A1:NbROS1. The percentages of methylcytosine are shown. Numbers below the line indicate the relative nucleotide positions from the transcriptional start site. Horizontal black lines indicate the targeted region. Red, green, and blue lines indicate frequencies of methylcytosine at the CG, CHG, and CHH sites, respectively. For plants infected with A1:empty, A1:35Spro, and both A1:35Spro and A1:NbROS1, 8, 14, and 15 clones were sequenced, respectively. The positions of two CG sites in the cis-acting as-1 element, to which binding of protein factor(s) is inhibited by cytosine methylation (Kanazawa et al., 2007b), are indicated by vertical arrows. (D) Summary of bisulfite sequencing analysis of CaMV 35S promoter in plants infected with A1:35Spro and those infected with both A1:35Spro and A1:NbROS1.
We next infected plants with both A1:35Spro and A1:NbROS1 to induce RdDM of the CaMV 35S promoter and downregulation of NbROS1, respectively. We found that the level of cytosine methylation in the CaMV 35S promoter was much higher in plants infected with both A1:35Spro and A1:NbROS1 than in plants infected with A1:35Spro alone (Figures 3B,C; Figure A1 in Appendix). Cytosine methylation of the CaMV 35S promoter increased in all sequence contexts, namely, CG, CHG, and CHH (Figure 3D).
Changes in the frequency of cytosine methylation were also analyzed by a method involving digestion of genomic DNA with a methylation-dependent endonuclease followed by PCR amplification (Figure 4). The CaMV 35S promoter region was amplified by PCR from genomic DNA treated with McrBC, an endonuclease that cleaves DNA containing at least two methylcytosines that are preceded by a purine nucleotide and separated each other by 40–80 nt (Sutherland et al., 1992). The level of amplified products of the CaMV 35S promoter in plants infected with both A1:35Spro and A1:NbROS1 was lower than that in plants infected with A1:35Spro alone, the latter of which was lower than that in plants infected with the vector that lacked an insert. These results suggest that RdDM of the promoter was induced in plants infected with A1:35Spro and was enhanced in plants infected with both A1:35Spro and A1:NbROS1.
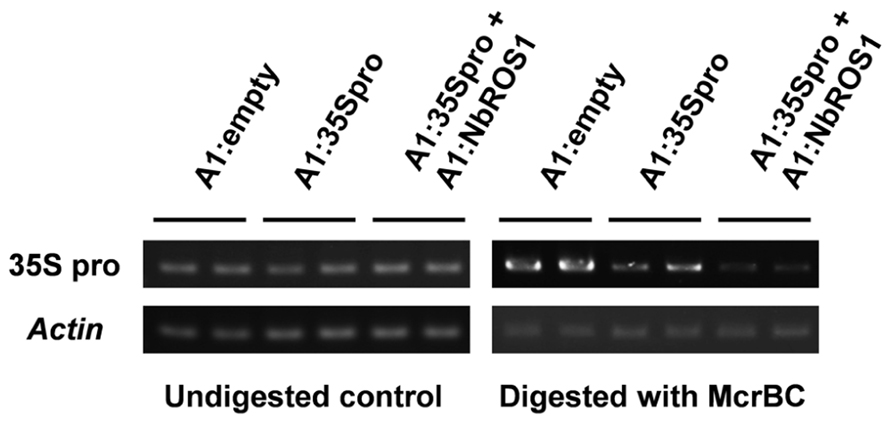
FIGURE 4. Cytosine methylation status of CaMV 35S promoter analyzed by McrBC-PCR. Genomic DNA from plants infected with A1:empty, A1:35Spro, and both A1:35Spro and A1:NbROS1 was digested with McrBC, and was used as a template for amplification of CaMV 35S promoter by PCR. Amplification from undigested DNA and amplification of the actin gene were done as controls.
We analyzed the relative level of viral RNA and production of siRNAs in virus-infected plants. Northern blot analysis indicated that there was no significant difference in the viral accumulation between plants infected with A1:35Spro alone and those infected with both A1:35Spro and A1:NbROS1 (Figure 5A). RT-PCR using a primer that anneals a region adjacent to the cloning site of the vector in combination with an insert-specific primer confirmed that the viral RNAs retained the insert segments in the infected plants (Figure 5B). In addition, no profound difference was detected in the level of siRNAs corresponding to the CaMV 35S promoter between plants infected with A1:35Spro and those infected with both A1:35Spro and A1:NbROS1 by Northern blot analysis (Figure 5C), suggesting that the observed increase in the level of cytosine methylation was not a consequence of a coincidental increase in the level of siRNAs of the targeted promoter by the co-infection. These results suggest that downregulation of NbROS1 facilitates RdDM of the targeted DNA sequence.
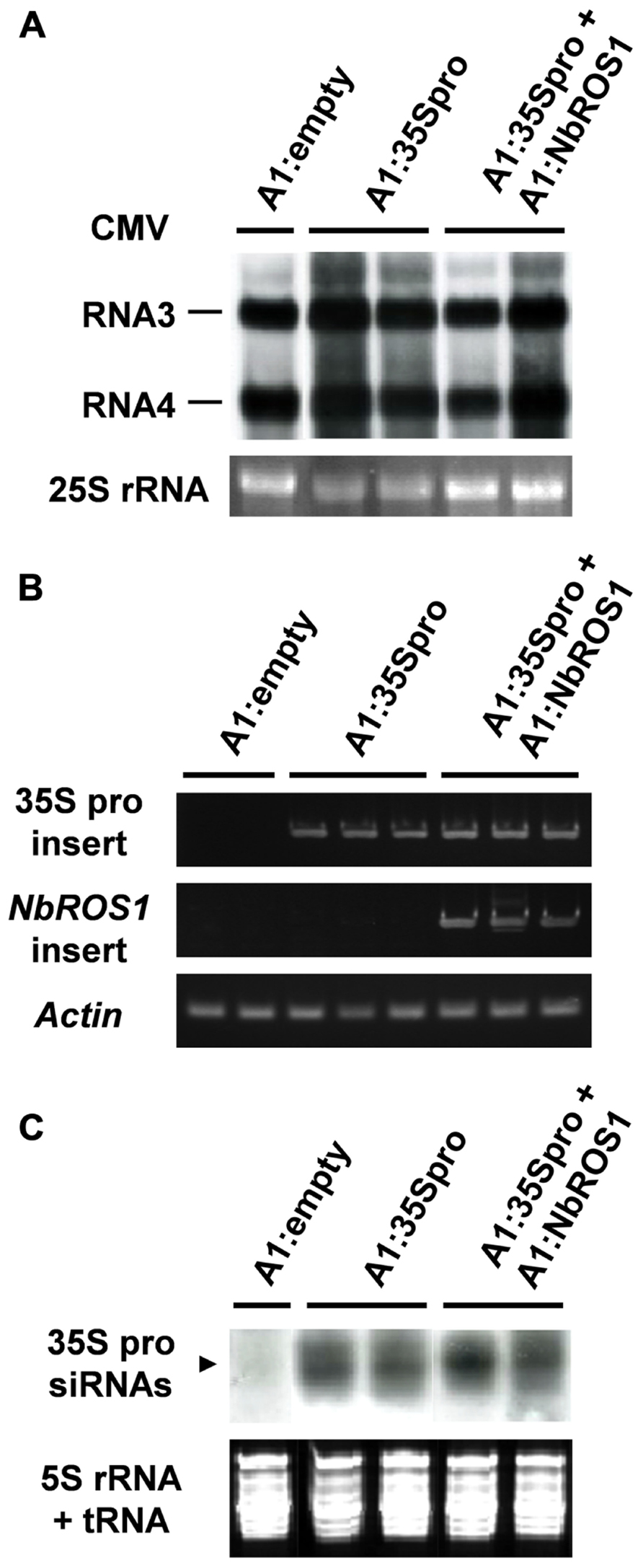
FIGURE 5. Analysis of viral RNAs in plants infected with recombinant CMVs. (A) Northern blot analysis of viral RNAs using a CMV RNA-specific probe. Hybridization signals of CMV RNAs 3 and 4 are shown. Ethidium-bromide-stained 25S RNA bands are shown below the panel to show that an equal amount of RNA was loaded. (B) RT-PCR analysis of viral RNAs. RT-PCR was done using a combination of primers: a primer that anneals a region adjacent to the cloning site of the viral vector and a primer that anneals the CaMV 35S promoter or NbROS1 fragment inserted in the vector. A portion of the actin gene was amplified as a control. (C) Production of siRNAs corresponding to the CaMV 35S promoter in plants infected with recombinant CMVs. Northern blot analysis was done using low-molecular weight RNAs isolated from leaf tissues of the plants infected with the recombinant CMVs at 18 DPI, probed for the CaMV 35S promoter. Ethidium-bromide-stained 5S rRNA and tRNAs bands are shown below the panel to show that an equal amount of the small RNA fraction was loaded. In (A–C), data obtained from two or three individual plants infected with recombinant CMVs are shown.
Despite similar substrate specificity, the DME/ROS1 glycosylases have distinct biological roles, with DME functioning during gametogenesis to establish imprinting and the other family members, including ROS1, functioning in vegetative tissues (Law and Jacobsen, 2010). We found that NbROS1 is expressed in vegetative tissues in N. benthamiana as in Arabidopsis. We also found that the CaMV 35S promoter of the transgene can be a target of ROS1 as previously observed for the RD29A promoter of a transgene (Gong et al., 2002). A recent model for the mechanism(s) of localization of ROS1 protein on a target site involves the sliding of ROS1 protein along DNA (Ponferrada-Marïn et al., 2012), which may fit the notion that ROS1 randomly finds target sites on genomic DNA. The observed demethylation of transgene promoters is consistent with this notion, although whether demethylation by ROS1 occurs at an equal efficiency in different genomic sites is not known.
We have previously reported TGS of the GFP gene driven by the CaMV 35S promoter through RdDM of the promoter (Otagaki et al., 2011). The extent of GFP TGS was affected by the length and cytosine frequency of the promoter segment inserted in the CMV-A1 vector (Otagaki et al., 2011). Both the extent of GFP mRNA reduction and the level of cytosine methylation induced by the CMV-A1:-116 to +1 (A1:35Spro in the present study) was lower than those induced by other constructs of a similarly sized insert (e.g., CMV-A1:-208 to -89; Otagaki et al., 2011). Therefore, we had expected that infection of plants with both A1:35Spro and A1:NbROS1 might result in an increase in both the extent of mRNA reduction and the level of cytosine methylation. However, no profound enhancement of GFP mRNA reduction was brought about by the downregulation of NbROS1 (Figure 6), although an increase in the level of cytosine methylation was induced by the treatment (Figures 3 and 4). These results suggest that the level of cytosine methylation is not necessary tightly linked with the extent of TGS, although TGS induction was always associated with cytosine methylation of the promoter in this system (Otagaki et al., 2011). We have found that the lower level of TGS induction by CMV-A1:-116 to +1 (A1:35Spro) is correlated not only with a less cytosine methylation but also a lower frequency (3.4/100 nt; 4/117 nt) of cytosines at symmetrical sites (CG and CHG) in the target DNA region: the latter value of the other constructs that induced a higher extent of TGS was 5.4–9.8/100 nt (Otagaki et al., 2011). Taking into account such a constraint in terms of sequence composition, a plausible explanation for the lack of additional effect on TGS is that the extent of mRNA reduction by A1:35Spro was already at a maximum irrespective of the expression level of NbROS1. The strategy of co-inoculation with a virus for downregulating a ROS1 ortholog may be useful for enhancing TGS of the target, particularly when the levels of viral propagation and/or production of promoter siRNAs are limited.
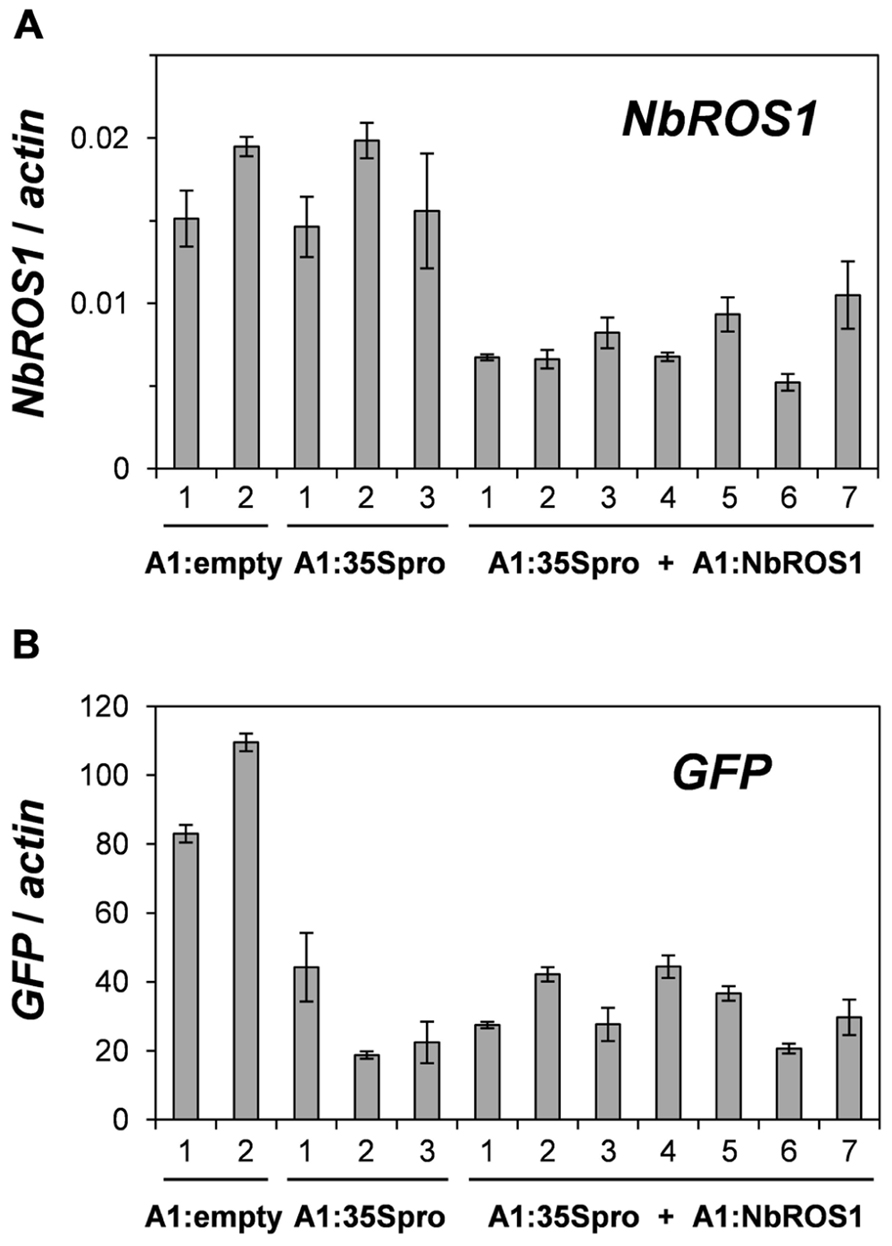
FIGURE 6. Changes in mRNA levels of NbROS1 and GFP genes after infection with CMVs containing a portion of CaMV 35S promoter or NbROS1. The mRNA levels of NbROS1 (A) and GFP (B) were analyzed in plants infected with A1:empty, A1:35Spro, or both A1:35Spro and A1:NbROS1. For these plants, data obtained from 2, 3, and 7 individual plants, respectively, are shown. The mRNA levels were assessed relative to the actin mRNA level in leaf tissues at 18 DPI. Data are means and standard errors obtained from three replicates.
We found that viral RNA levels in plants infected with A1:35Spro were similar to those in plants infected with both A1:35Spro and A1:NbROS1. Despite the co-existence of A1:35Spro with A1:NbROS1, the level of siRNAs derived from A1:35Spro in plants infected with both A1:35Spro and A1:NbROS1 was not different from that in plants infected with A1:35Spro alone (Figure 5C). In this regard, the rate of degradation of A1:NbROS1 RNA may be higher than that of A1:35Spro RNA because degradation of A1:NbROS1 RNA can be amplified by NbROS1 PTGS. This notion is consistent with a previous observation that virus-induced PTGS can lead to elimination of the viral RNA (Ruiz et al., 1998; Vaistij et al., 2002). Although viral RNA levels at different time points remain elusive, the present results suggest that the co-infection does not cause an extensive interference between the viruses, e.g., an enhancement of the degradation of A1:35Spro RNA in trans by siRNAs derived from A1:NbROS1 RNA.
In terms of controlling virus-induced changes, the effect of virus-induced PTGS has been enhanced by placing a target gene segment in an inverted repeat position in a viral vector (Lacomme et al., 2003) and was found to be higher in a mutant of a gene that encodes a protein involved in the 3′ end formation of RNA (Herr et al., 2006). To our knowledge, however, no report for enhancing virus-induced RdDM has been published.
The first process that allows the control of the level of targeted DNA methylation may be the induction phase of RdDM, in which accumulation of siRNAs in the nucleus can affect the level of de novo methylation. In fact, a transgene-derived hairpin RNA that resides in an intron and thus is expected to be retained in the nucleus was found to efficiently trigger RdDM (Dalakouras et al., 2009). In this regard, the CMV-A1 vector originally has the advantage that the 2b protein encoded in the vector facilitates RdDM through the transport of siRNAs to the nucleus (Kanazawa et al., 2011a). Another process that can be modified involves the maintenance of methylation and/or demethylation, which was tested in this study. Here we found that downregulation of NbROS1 enhanced virus-induced RdDM, which consequently provides evidence for antagonistic activity of NbROS1 against virus-induced RdDM. Thus, the present method can be used to control the level of cytosine methylation in a targeted DNA region via RdDM.
Wild-type N. benthamiana plants and plants of transgenic N. benthamiana line 16c (Ruiz et al., 1998) were used for analyses. Plants were grown under a 16-h light/8-h dark cycle at 24°C.
Portions of NbROS1 were amplified from cDNA by PCR using primers designed to anneal regions conserved between AtROS1 (DDBJ/EMBL/GenBank accession AY286009) and NtROS1 (No. AB281587). A cDNA fragment covering the entire coding region of NbROS1 was isolated using PCR in combination with 5′- and 3′-RACE techniques using a SMART RACE cDNA Amplification Kit (Clonetech) according to the manufacturer’s instructions. Primers used for PCR in this study are listed in Table A1 in Appendix. The NbROS1 cDNA sequence data has been deposited in the DDBJ database under accession number AB778815.
Total RNA was isolated from leaf tissues of N. benthamiana plants at 18 days post-inoculation (DPI) as described previously (Otagaki et al., 2006). For analyzing NbROS1 and GFP expression, cDNA synthesis, RT-PCR and quantitative RT-PCR were done as described previously (Kasai et al., 2012). In all PCR experiments, a reaction mixture without reverse transcriptase was used as a control to confirm that no amplification occurred from genomic DNA contaminants in the RNA sample. Amplification of a single DNA species was confirmed by both melting curve analysis of quantitative RT-PCR and gel electrophoresis of PCR products. For analyzing viral RNAs, Northern blot analysis was done as described previously (Otagaki et al., 2006). A CMV RNA-specific probe was prepared by amplifying a portion of the pCY3 plasmid containing the cDNA of CMV-Y RNA 3 (Suzuki et al., 1991). RT-PCR was done as described above except that cDNA was synthesized using a mixture of random 9-nt primers (TaKaRa).
The protein sequences deduced from the nucleotide sequences of DNA glycosylase genes were aligned using the CLUSTAL W Multiple Sequence Alignment Program version 2.12 (Thompson et al., 1994). A phylogenetic tree was constructed using the neighbor-joining (NJ) method (Saitou and Nei, 1987) based on the protein sequences.
For inducing RdDM of the CaMV 35S promoter, the -116 to +1 region (positions are relative to the transcription start site) of the CaMV 35S promoter was amplified by PCR. The amplified fragment was cloned between the StuI and MluI sites of the CMV-A1 vector as described previously (Otagaki et al., 2011). For the downregulation of NbROS1, a 104-bp portion of the NbROS1-coding region was also amplified by PCR and the amplified fragment was cloned into the same site in the vector in the antisense orientation. Plasmids containing full-length cDNA of viral RNA were transcribed in vitro, and leaves of young N. benthamiana plants were dusted with carborundum and rub-inoculated with the transcripts as described previously (Otagaki et al., 2006).
Low-molecular weight RNAs were isolated and siRNAs were detected by gel-blot analysis according to the method of Goto et al. (2003). DIG-labeled RNAs corresponding to the -345 to +1 region of the CaMV 35S promoter (Otagaki et al., 2006, 2011) and the 104-bp portion of the NbROS1-coding region were used as a probe.
DNA was isolated from leaf tissues at 24 DPI using the Nucleon PhytoPure DNA extraction kit (GE Healthcare). Bisulfite treatment of DNA and subsequent PCR amplification were done as described previously (Kanazawa et al., 2007a). As a control to ensure that bisulfite treatment was complete, DNA isolated from Arabidopsis leaves was simultaneously treated. A region of the Arabidopsis ASA1 gene that is not methylated was amplified as previously reported (Kanazawa et al., 2007a). All five cloned sequences of PCR products showed complete conversion of cytosines to thymidines.
DNA (500 ng) was digested with McrBC for overnight and precipitated with ethanol. After centrifugation, the pellet was dissolved in water. One-fifth volume of the DNA solution was used as a template for amplification by PCR. The products of PCR were analyzed by electrophoresis on an agarose gel.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to D. Baulcombe for 16c N. benthamiana plants. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Cultures, Sports, Science and Technology of Japan and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280.
Chan, S. W., Henderson, I. R., and Jacobsen, S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 351–360.
Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42.
Cigan, A. M., Unger-Wallace, E., and Haug-Collet, K. (2005). Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J. 43, 929–940.
Dalakouras, A., Moser, M., Zwiebel, M., Krczal, G., Hell, R., and Wassenegger, M. (2009). A hairpin RNA construct residing in an intron efficiently triggered RNA-directed DNA methylation in tobacco. Plant J. 60, 840–851.
Furner, I. J., and Matzke, M. (2011). Methylation and demethylation of the Arabidopsis genome. Curr. Opin. Plant Biol. 14, 137–141.
Gehring, M., Huh, J. H., Hsieh, T. F., Penterman, J., Choi, Y., Harada, J. J., et al. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506.
Gong, Z., Morales-Ruiz, T., Ariza, R. R., Roldán-Arjona, T., David, L., and Zhu, J. K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814.
Goto, K., Kanazawa, A., Kusaba, M., and Masuta, C. (2003). A simple and rapid method to detect plant siRNAs using nonradioactive probes. Plant Mol. Biol. Rep. 21, 51–58.
Hawkins, P. G., and Morris, K. V. (2008). RNA and transcriptional modulation of gene expression. Cell Cycle 7, 602–607.
Heilersig, B. H., Loonen, A. E., Janssen, E. M., Wolters, A. M., and Visser, R. G. (2006). Efficiency of transcriptional gene silencing of GBSSI in potato depends on the promoter region that is used in an inverted repeat. Mol. Genet. Genomics 275, 437–449.
Herr, A. J., Molnàr, A., Jones, A., and Baulcombe, D. C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 14994–15001.
Jones, A. L., Thomas, C. L., and Maule, A. J. (1998). De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 17, 6385–6393.
Jones, L., Ratcliff, F., and Baulcombe, D. C. (2001). RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11, 747–757.
Kanazawa, A. (2008). RNA silencing manifested as visibly altered phenotypes in plants. Plant Biotechnol. 25, 423–435.
Kanazawa, A., Inaba, J., Shimura, H., Otagaki, S., Tsukahara, S., Matsuzawa, A., et al. (2011a). Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J. 65, 156–168.
Kanazawa, A., Inaba, J., Kasai, M., Shimura, H., and Masuta, C. (2011b). RNA-mediated epigenetic modifications of an endogenous gene targeted by a viral vector: a potent gene silencing system to produce a plant that does not carry a transgene but has altered traits. Plant Signal. Behav. 6, 1090–1093.
Kanazawa, A., O’Dell, M., and Hellens, R. P. (2007a). Epigenetic inactivation of chalcone synthase-A transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol. 48, 638–647.
Kanazawa, A., O’Dell, M., and Hellens, R. P. (2007b). The binding of nuclear factors to the as-1 element in the CaMV 35S promoter is affected by cytosine methylation in vitro. Plant Biol. 9, 435–441.
Kapoor, A., Agarwal, M., Andreucci, A., Zheng, X., Gong, Z., Hasegawa, P. M., et al. (2005). Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr. Biol. 15, 1912–1918.
Kasai, M., and Kanazawa, A. (2013). Induction of RNA-directed DNA methylation and heritable transcriptional gene silencing as a tool to engineer novel traits in plants. Plant Biotechnol. (in press).
Kasai, M., Koseki, M., Goto, K., Masuta, C., Ishii, S., Hellens, R. P., et al. (2012). Coincident sequence-specific RNA degradation of linked transgenes in the plant genome. Plant Mol. Biol. 78, 259–273.
Lacomme, C., Hrubikova, K., and Hein, I. (2003). Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J. 34, 543–553.
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220.
Matzke, M., Kanno, T., Daxinger, L., Huettel, B., and Matzke, A. J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21, 367–376.
Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A., and Matzke, A. J. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201.
Molnar, A., Melnyk, C. W., Bassett, A., Hardcastle, T. J., Dunn, R., and Baulcombe, D. C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875.
Okano, Y., Miki, D., and Shimamoto, K. (2008). Small interfering RNA (siRNA) targeting of endogenous promoters induces DNA methylation, but not necessarily gene silencing, in rice. Plant J. 53, 65–77.
Ortega-Galisteo, A. P., Morales-Ruiz, T., Ariza, R. R., and Roldán-Arjona, T. (2008). Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681.
Otagaki, S., Arai, M., Takahashi, A., Goto, K., Hong, J. S., Masuta, C., et al. (2006). Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant Biotechnol. 23, 259–265.
Otagaki, S., Kawai, M., Masuta, C., and Kanazawa, A. (2011). Size and positional effects of promoter RNA segments on virus-induced RNA-directed DNA methylation and transcriptional gene silencing. Epigenetics 6, 681–691.
Penterman, J., Zilberman, D., Huh, J. H., Ballinger, T., Henikoff, S., and Fischer, R. L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U.S.A. 104, 6752–6757.
Ponferrada-Marïn, M. I., Roldán-Arjona, T., and Ariza, R. R. (2012). Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 40, 11554–11562.
Ruiz, M. T., Voinnet, O., and Baulcombe, D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Senthil-Kumar, M., and Mysore, K. S. (2011). New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 16, 656–665.
Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J. N., et al. (2001). Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11, 436–440.
Sutherland, E., Coe, L., and Raleigh, E. A. (1992). McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225, 327–348.
Suzuki, K., and Kelleher, A. D. (2009). Transcriptional regulation by promoter targeted RNAs. Curr. Top. Med. Chem. 9, 1079–1087.
Suzuki, M., Kuwata, S., Kataoka, J., Masuta, C., Nitta, N., and Takanami, Y. (1991). Functional analysis of deletion mutants of cucumber mosaic virus RNA3 using an in vitro transcription system. Virology 183, 106–113.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Vaistij, F. E., Jones, L., and Baulcombe, D. C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14, 857–867.
Wassenegger, M., Heimes, S., Riedel, L., and Sänger, H. L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76, 567–576.
Zhang, H., and Zhu, J. K. (2011). RNA-directed DNA methylation. Curr. Opin. Plant Biol. 14, 142–147.
Zhu, J., Kapoor, A., Sridhar, V. V., Agius, F., and Zhu, J. K. (2007). The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 17, 54–59.
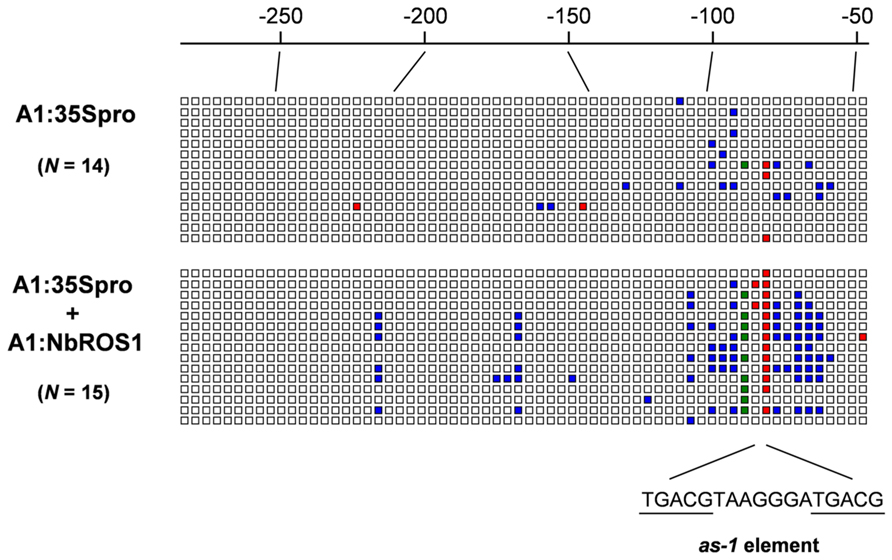
FIGURE A1. Graphical representation of cytosine methylation status of CaMV 35S promoter. Sequencing data for PCR products amplified from bisulfite-treated DNA from plants infected with A1:35Spro (upper) and both A1:35Spro and A1:NbROS1 (lower) have been compiled. For these plants, 14 and 15 clones were sequenced, respectively. Open boxes indicate unmethylated cytosines. Red, green, and blue boxes indicate methylcytosines at the CG, CHG, and CHH sites, respectively. Numbers on the line indicate nucleotide positions relative to the transcription start site of the CaMV 35S promoter. The position of the cis-acting as-1 element, to which binding of protein factor(s) is inhibited by cytosine methylation (Kanazawa et al., 2007b), is shown below the boxes.
Keywords: Cucumber mosaic virus, DNA demethylation, RNA-directed DNA methylation, ROS1, virus-induced gene silencing
Citation: Otagaki S, Kasai M, Masuta C and Kanazawa A (2013) Enhancement of RNA-directed DNA methylation of a transgene by simultaneously downregulating a ROS1 ortholog using a virus vector in Nicotiana benthamiana. Front. Genet. 4:44. doi: 10.3389/fgene.2013.00044
Received: 29 January 2013; Accepted: 12 March 2013;
Published online: 02 April 2013.
Edited by:
Michael Wassenegger, RLP AgroScience GmbH, GermanyReviewed by:
Ming-Bo Wang, Commonwealth Scientific and Industrial Research Organisation, AustraliaCopyright: © 2013 Otagaki, Kasai, Masuta and Kanazawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Akira Kanazawa, Research Faculty of Agriculture, Hokkaido University, Kita 9, Nishi 9, Kita-ku, Sapporo 060-8589, Japan. e-mail:kanazawa@res.agr.hokudai.ac.jp
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.