- 1 Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, Tokyo, Japan
- 2 Department of Computational Biology, Graduate School of Frontier Sciences, The University of Tokyo, Chiba, Japan
RNA interference (RNAi) is a mechanism through which small interfering RNA (siRNA) induces sequence-specific posttranscriptional gene silencing. RNAi is commonly recognized as a powerful tool not only for functional genomics but also for therapeutic applications. Twenty-one-nucleotide-long siRNA suppresses the expression of the intended gene whose transcript possesses perfect complementarity to the siRNA guide strand. Hence, its silencing effect has been assumed to be extremely specific. However, accumulated evidences revealed that siRNA could downregulate unintended genes with partial complementarities mainly to the seven-nucleotide seed region of siRNA. This phenomenon is referred to as off-target effect. We have revealed that the capability to induce off-target effect is strongly correlated to the thermodynamic stability in siRNA seed-target duplex. For understanding accurate target gene function and successful therapeutic application, it may be critical to select a target gene-specific siRNA with minimized off-target effect. Here we present our siRNA design software for a target-specific RNAi. In addition, we also introduce the software programs open to the public for designing functional siRNAs.
Introduction
RNA interference (RNAi) is a broadly used technique by which small interfering RNA (siRNA) downregulates a specific target gene with perfect complementary sequence, and promised to use in therapeutic application for human diseases (Castanotto and Rossi, 2009; Ketting, 2011). Although human has more than 20,000 genes, it is desirable to use siRNA which is highly functional and has no effects on any genes other than its specific target. In this article, we review an optimized method to design siRNA based on the mechanism of RNAi. In addition, we introduce the websites open to the public for selecting siRNA sequences.
Optimized Design of siRNA
Duplexes of 21-nucleotide (nt) RNA with 2 nt 3′ overhangs (siRNA) is usually used for RNAi experiments. Upon delivery into the cells, siRNAs are incorporated into the RNA-induced silencing complex (RISC) as a double-stranded RNA. RISC is the effector complex containing Argonaute protein (Ago) with slicer activity (Hammond et al., 2001; Martinez et al., 2002). The siRNA guide strand containing the thermodynamically less stable 5′-end is preferentially retained by RISC (Khvorova et al., 2003; Schwarz et al., 2003; Ui-Tei et al., 2004). The passenger strands of most of the double-stranded siRNAs loaded onto RISC are cleaved by Ago2 protein and degraded (Matranga et al., 2005; Rand et al., 2005; Leuschner et al., 2006). The retained guide strand pairs target mRNA with perfectly complementary sequence, and represses it by cleavage by Ago2 protein at nucleotide position 10 of siRNA guide strand (Elbashir et al., 2001; Hammond et al., 2001; Martinez et al., 2002). However, an accumulated evidence from genome-wide experiments indicate that a great number of mRNAs with partial complementarities to the guide strand are also reduced (Jackson et al., 2003, 2006; Lim et al., 2005; Birmingham et al., 2006; Ui-Tei et al., 2008). This phenomenon is referred to as seed-dependent off-target effect and preferably observed in mRNA 3′ UTRs. The target recognition mechanism of this off-target effect is known to be similar to that of miRNA-mediated gene silencing (Lewis et al., 2005; Lim et al., 2005; Grimson et al., 2007). The transcripts with sequences complementary to the seed region positioned 2–8 from the 5′ terminal are mainly reduced. The seed region is known to be situated on the surface of Ago in a quasi-helical form to serve as the entry or nucleation site for small RNAs in the RISCs (Ma et al., 2005; Yuan et al., 2005). Thus, the seed region first identifies the target mRNAs, and subsequently form perfect base-pairing with intended target mRNA and induce RNAi by Ago2.
Based on the mechanism of RNAi, a target gene-specific siRNA is considered to be selectable according to the following three steps.
STEP 1: Selection of Functional siRNA Sequence
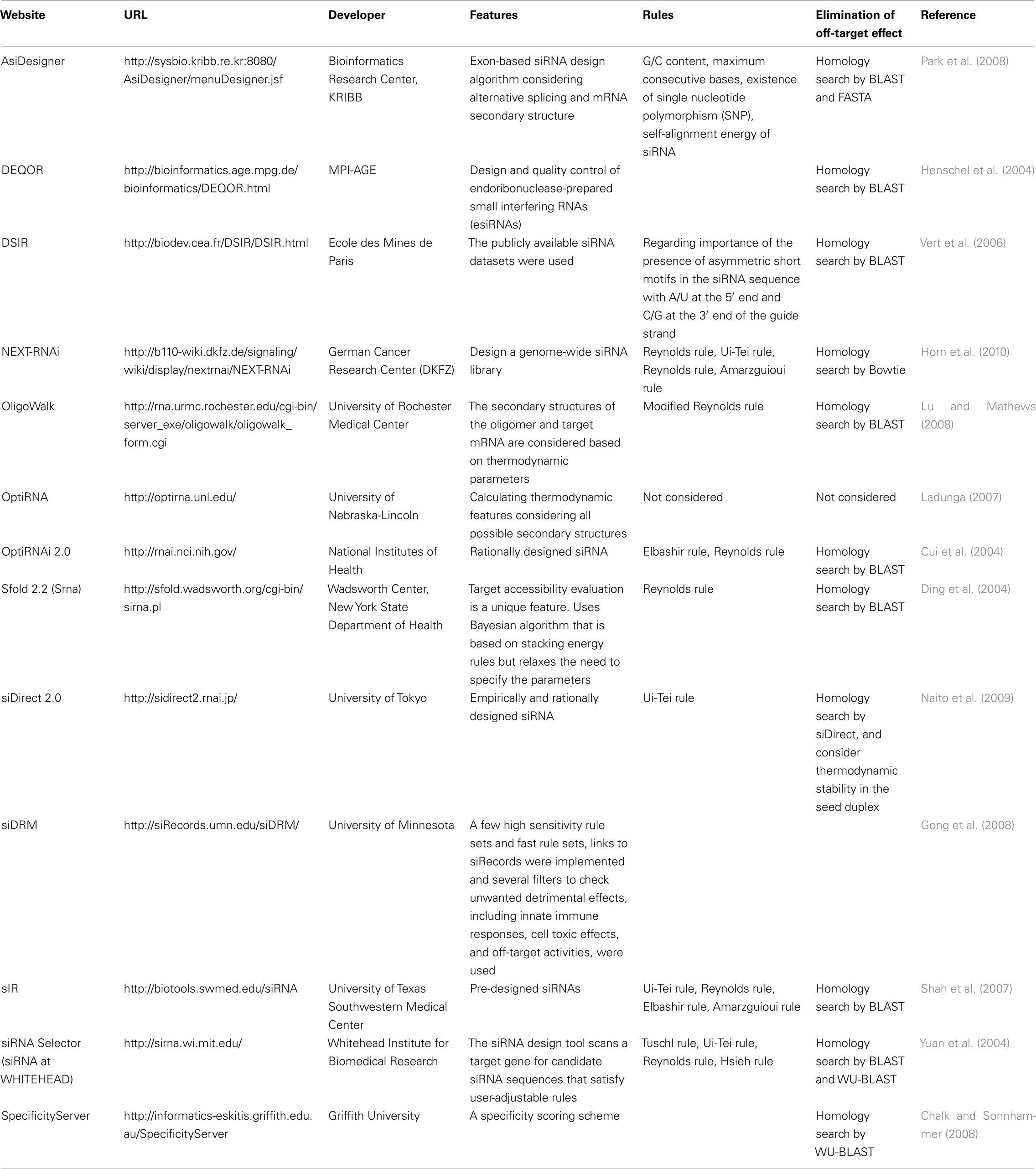
The knockdown efficiencies of siRNAs are revealed to be highly dependent on their sequences. We advocated the empirically based rule which prescribed the characteristics of highly functional siRNAs (Ui-Tei et al., 2004), such rule is called as Ui-Tei rule (Figure 1). The siRNA selected by Ui-Tei rule satisfy the following four conditions simultaneously: (1) A or U at position 1 from 5′ terminus of siRNA guide strand, (2) G or C at position 19, (3) AU richness (AU ≥4) in positions 1–7, and (4) no long GC stretch ≥10. Except for (4), our rule indicated that the functional siRNA has asymmetrical stability in 5′ and 3′ terminals. Our experimental validation using luciferase reporter assay showed that 98% of the siRNAs satisfying above conditions reduced the expression of luciferase reporter below 33% (Ui-Tei et al., 2004). Other groups also demonstrated the rules of highly functional siRNAs referred to as Reynolds rule (Reynolds et al., 2004) and Amarzguioui rule (Amarzguioui and Prydz, 2004) as summarized in Figure 1. These rules also clearly showed that functional siRNAs are asymmetrical: an RNA strand with unstable 5′ terminal was effective as a guide strand. Furthermore, common in these rules, 5′ terminus of functional siRNA guide strand was preferable to be A or U. It was revealed later that the results reflected the structural features of human Ago2 (Frank et al., 2010). The crystal structure of a MID (middle) domain from human AGO2 and NMR titration experiments showed that nucleotide monophosphates, AMP and UMP, bind with up to 30-fold higher affinity than either CMP or GMP, providing structural evidence for nucleotide-specific interactions in the MID domain of eukaryotic AGO proteins.
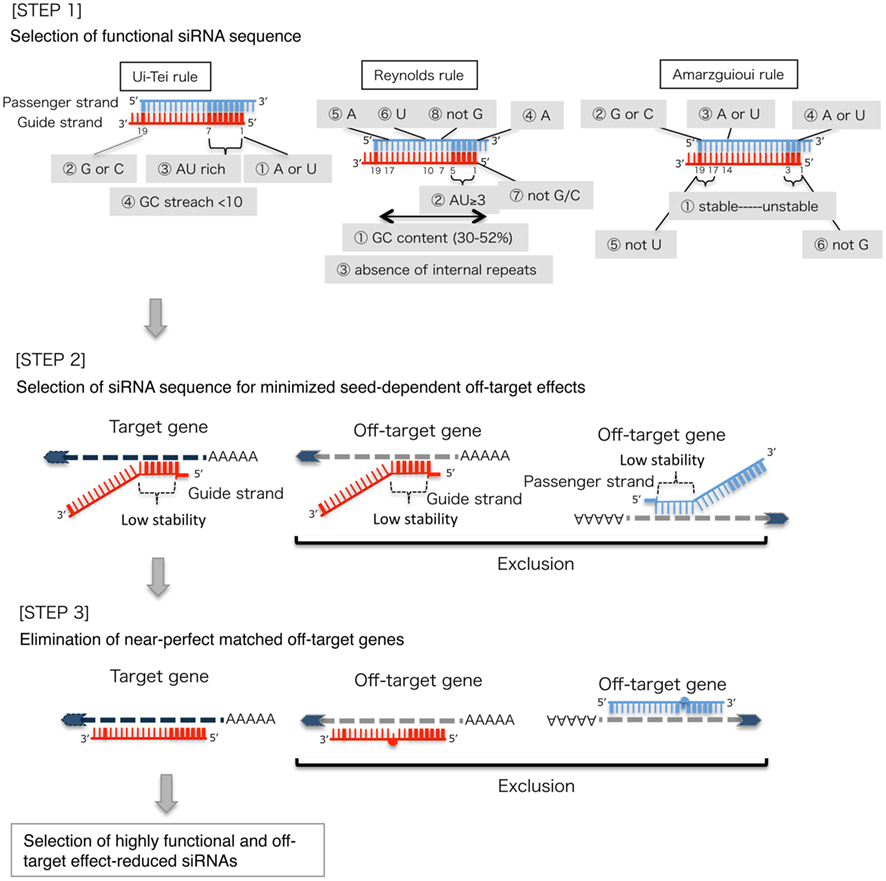
Figure 1. Schematic representation for selecting functional and off-target effect-reduced siRNAs. Selection of highly functional siRNA by Ui-Tei, Reynolds, Amarzguioui rules, or the combination of them (STEP 1). Selection of siRNAs with low stability in the seed-target duplexes (STEP 2). Elimination of siRNAs with near-perfect matched sequences to non-target genes (STEP 3). In each rule, the nucleotide position indicates the number of nucleotide counted from 5′ terminal of the guide strand.  :
:  A/U at position 1.
A/U at position 1.  G/C at position 19.
G/C at position 19.  4 to 7 A/Us in positions 1-7.
4 to 7 A/Us in positions 1-7. 
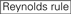 :
:  GC content (30%-52%).
GC content (30%-52%).  A/U ≥ 3 at positions 1-5.
A/U ≥ 3 at positions 1-5.  Absence of internal repeats.
Absence of internal repeats. 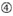 A at position 1.
A at position 1. 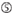 A at position 17.
A at position 17. 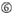 U at position 10.
U at position 10. 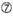 Not G/C at position 1.
Not G/C at position 1.  Not G at position 7.
Not G at position 7. 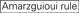 :
:  An asymmetry in the stability of the duplex ends (measured as the A/U differential of the three terminal basepairs at either end of the duplex).
An asymmetry in the stability of the duplex ends (measured as the A/U differential of the three terminal basepairs at either end of the duplex).  G or C at position 19.
G or C at position 19.  A or U at position 1.
A or U at position 1. 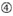 A or U at position 14.
A or U at position 14. 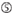 Not U at position 19.
Not U at position 19. 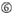 Not G at position 1.
Not G at position 1.
STEP 2: Selection of siRNA Sequence with Reduced Off-Target Effects
To avoid seed-dependent off-target effects, one approach may be to select the siRNA guide strand whose seed sequence is not complementary to any sequences in the 3′ UTR of all non-targeted genes. However, this approach is proved to be impossible, because human siRNAs with the most infrequent seven-nt seed sequence still had seed-complementarities with several non-targeted mRNAs. So, we have looked for the rules that govern the capability of siRNAs to induce seed-dependent off-target effect, and revealed that the efficiency of off-target effect is highly correlated to the thermodynamic stability of the duplex formed between the seed region of siRNA guide strand and its target mRNA (Ui-Tei et al., 2008). The melting temperature (Tm), one of the thermodynamic parameters for the formation of RNA duplex, showed strong positive correlation with the induction of seed-dependent off-target effects. Thus, selecting the siRNAs with low Tm of the seed-target duplex should minimize seed-dependent off-target silencing (Figure 1). The Tm of 21.5°C may serve as the benchmark, which discriminates the almost off-target-free seed sequences from the off-target-positive ones. Furthermore, since the off-target effect may be caused by not only by the guide strand but also by the passenger strand, siRNAs whose seed-target Tm is sufficiently low for both strands are favorable.
STEP 3: Elimination of Near-Perfect Matched Off-Target Genes
Even when the Tm value of the seed-target duplex is sufficiently low, the target gene silencing can still take place if the non-seed region is completely complementary. Therefore, in the third step, siRNAs that have near-perfect matches to any other non-targeted transcripts were eliminated (Figure 1).
siRNA Design Software
We presented siRNA design software, siDirect 2.0 (http://siDirect2.RNAi.jp/; Figure 2), which provides functional, target-specific siRNA design software according to the procedures mentioned above (Naito et al., 2009). In default parameter, siRNAs satisfying Ui-Tei rule can be selected. When the candidate functional siRNAs could form seed-target duplexes with Tm values below 21.5°C, and their 19-nt regions spanning positions 2–20 of both strands have at least two mismatches to any other non-targeted transcripts, siDirect 2.0 can design at least one qualified siRNA for >94% of human mRNA sequences in RefSeq.

Figure 2. Screen views of siDirect 2.0 siRNA design software. (A) Top page (http://siDirect2.RNAi.jp/). (B) Optional parameters for siRNA design. (C) Result page. (D) Detailed list of off-target candidates with near-perfect matches. The alignment between each off-target transcript and the siRNA sequence visualizes the positions of mismatches.
Other software to select functional siRNAs were open to public as shown in Table 1. In many of those, Ui-Tei rule (Ui-Tei et al., 2004), Reynolds rule (Reynolds et al., 2004), Amarzguioui rule (Amarzguioui and Prydz, 2004), Tuschl rule (Elbashir et al., 2002), and the combination of them were frequently and widely used. To eliminate the near-perfect matched non-target genes, BLAST search was used for homology search in several software. However, since BLAST search is not so accurate for short sequences like siRNAs, siDirect, WU-BLAST, and Bowtie, which are highly accurate homology search engine for short sequences are often used. Among them, siDirect 2.0 may bring the most accurate results. Furthermore, some of the software consider the additional features, such as mRNA secondary structure (Ladunga, 2007; Lu and Mathews, 2008), alternative splicing (Park et al., 2008), or the motif sequence by which the immune response by a RNA virus might occur (Gong et al., 2008). These features are not considered in siDirect 2.0. So, to take into account these features, siRNAs commonly selected by siDirect 2.0 and the other appropriate software programs may produce optimum result.
In addition, it is practically important consideration to use two or more siRNAs targeting different sites in an intended target gene, since the knockdown effects of an intended target gene are supposed to be common but the off-target effects are likely to be different between siRNAs.
Conclusion
The efficacy of each siRNA is known to be widely varies depending on its sequence in mammalian cells, and only a limited fraction of randomly designed siRNAs is functional. Moreover, off-target silencing effects arise when the siRNA has partial complementarity in the seed region with unintended genes. Here, based on the RNAi machinery, we described the rational design of functional, off-target effect-reduced siRNAs, which are expected to knockdown a target gene-specifically.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partially supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and the Cell Innovation Project (MEXT), and Core Research Project for Private University; matching fund subsidy to Kumiko Ui-Tei.
References
Amarzguioui, M., and Prydz, H. (2004). An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 316, 1050–1058.
Birmingham, A., Anderson, E. M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., Baskerville, S., Maksimova, E., Robinson, K., Karpilow, J., Marshall, W. S., and Khvorova, A. (2006). 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204.
Castanotto, D., and Rossi, J. J. (2009). The promises and pitfalls of RNA-interference-based therapeutics. Nature 457, 426–433.
Chalk, A. M., and Sonnhammer, E. L. L. (2008). siRNA specificity searching incorporating mismatch tolerance data. Bioinformatics 24, 1316–1317.
Cui, W., Ning, J., Naik, U. P., and Duncan, M. K. (2004). OptiRNAi, and RNAi design tool. Comput. Methods Programs Biomed. 75, 67–73.
Ding, Y., Chan, C. Y., and Lawrence, C. E. (2004). Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 32, W135–W141.
Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498.
Elbashir, W. M., Harborth, J., Weber, K., and Tuschl, T. (2002). Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26, 199–213.
Frank, F., Sonenberg, N., and Nagar, B. (2010). Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822.
Gong, W., Ren, Y., Zhou, H., Wang, Y., Kang, S., and Li, Tongbin. (2008). siDRM: and effective and generally applicable online siRNA design tool. Bioinformatics 24, 2405–2406.
Grimson, A., Farh, K. K., Johnston, W. K., Garrett-Engele, P., Lim, L. P., and Bartel, D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105.
Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R., and Hannon, G. J. (2001). Argonaute 2 link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150.
Henschel, A., Buchholz, F., and Habermann, B. (2004). DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 32, W113–W120.
Horn, T., Sandmann, T., and Boutros, M. (2010). Design and evaluation of genome-wide libraries for RNA interference screens. Genome Biol. 11, R61.
Jackson, A. L., Bartz, S. R., Shelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P. S. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637.
Jackson, A. L., Burchard, J., Schelter, J., Chau, B. M., Cleary, M., Lim, L., and Linsley, P. S. (2006). Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187.
Khvorova, A., Reynolds, A., and Jayasena, S. D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216.
Ladunga, I. (2007). More complete gene silencing by fewer siRNAs: transparent optimized design and biophysical signature. Nucleic Acids Res. 35, 433–440.
Leuschner, P. J., Ameres, S. L., Kueng, S., and Martinez, J. (2006). Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 7, 314–320.
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20.
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S., and Johnson, J. M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773.
Lu, Z. J., and Mathews, D. H. (2008). Efficient siRNA selection using hybridization thermodynamics. Nucleic Acid Res. 36, 640–647.
Ma, J.-B., Yuan, Y. R., Meister, G., Pei, Y., Tuschl, T., and Patel, D. J. (2005). Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus piwi protein. Nature 434, 666–670.
Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. (2002). Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574.
Matranga, C., Tomari, Y., Shin, C., Bartel, D. P., and Zamore, P. D. (2005). Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620.
Naito, Y., Yoshimura, J., Morishita, S., and Ui-Tei, K. (2009). siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10, 392.
Park, Y.-K., Park, S.-M., Choi, Y.-C., Lee, D., Won, M., and Kim, Y. J. (2008). AsiDesigner: exon-based siRNA design server considering alternative splicing. Nucleic Acid Res. 36, W97–W103.
Rand, T. A., Petersen, S., Du, F., and Wang, X. (2005). Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629.
Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S., and Khvorova, A. (2004). Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330.
Schwarz, D. S., Hutvangner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P. D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208.
Shah, J. K., Garner, H. R., White, M. A., Shames, D. S., and Minna, J. D. (2007). sIR: siRNA information resource, a web-based tool for siRNA sequence design and analysis and an open access siRNA database. BMC Bioinformatics 8, 178. doi:10.1186/1471-2105-8-178
Ui-Tei, K., Naito, Y., Nishi, K., Juni, A., and Saigo, K. (2008). Thermodynamic stability and Watoson-Crick base pairing in the seed duplex are major determinants of the efficiency of the sRNA-based off-target effect. Nucleic Acids Res. 36, 7100–7109.
Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., Ueda, R., and Saigo, K. (2004). Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32, 936–948.
Vert, J.-P., Foveau, N., Lajaunie, C., and Vandenbrouck, Y. (2006). An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 7, 520. doi:10.1186/1471-2105-7-520
Yuan, B., Latek, R., Rossbach, M., Tuschl, T., and Lewitter, F. (2004). siRAN selection server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32, W130–W134.
Keywords: siRNA, seed region, off-target effect, thermodynamic stability
Citation: Naito Y and Ui-Tei K (2012) siRNA design software for a target gene-specific RNA interference. Front. Gene. 3:102. doi: 10.3389/fgene.2012.00102
Received: 26 April 2012; Accepted: 20 May 2012;
Published online: 11 June 2012.
Edited by:
Akio Kanai, Keio University, JapanReviewed by:
Rintaro Saito, Keio University, JapanKengo Sato, Keio University, Japan
Motoaki Wakiyama, RIKEN, Japan
Copyright: © 2012 Naito and Ui-Tei. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Yuki Naito, Genome Information Biology, Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
†Present address: Yuki Naito, Database Center for Life Science, Research Organization of Information and Systems, Tokyo, Japan. e-mail:eS1uYWl0b0BSTkFpLmpw