
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
GENERAL COMMENTARY article
Front. Genet., 21 May 2012
Sec. Toxicogenomics
Volume 3 - 2012 | https://doi.org/10.3389/fgene.2012.00088
A commentary on
Applications of next-generation sequencing in fish ecotoxicogenomics
by Mehinto, A. C., Martyniuk, C. J., Spade, D. J., and Denslow, N. D. (2012). Front. Genet. 3:62. doi: 10.3389/fgene.2012.00062
Ecotoxicants are elements expelled into the environment through human actions and having the potential to affect ecosystems at relatively low concentrations (Connell et al., 1999). Aquatic environments are often exposed by ecotoxicants through several routes. As fish is the most diverse group of vertebrates, knowing the mode of actions of ecotoxicants on fish helps in assessing the well-being of the marine environment. Such a study is also important as fish can serve as a warning of possible impacts on mankind (Zelikoff, 1998). Fish can also be a good model organism for examining ecotoxicants of concern to wellness of mankind. The goal of ecotoxicogenomics is to study effects that ecotoxicants elicit upon the environment using high-throughput genomic approaches. To this end, genomics approaches have revolutionized biomedical research to study biological problems at high-throughput levels that were never possible before. Applying genomics approaches to the fish ecotoxicology is the promise of next-generation sequencing technologies. Mehinto et al. (2012) summarized the recent developments and future prospects of current NGS platforms and bioinformatics tools that can be utilized to study the transcriptome of non-model fish species. With the genomic integration, multi-“omics” approaches will be employed to predict biological contexts including molecular functions, pathways, biological processes, relationship, and compounds. This will enable a speedy and comprehensive manner for us to traverse the levels of our understanding of fish ecotoxicogenomics from base pairs to genes to isoforms or variants and to narrow down molecular targets or develop testable new hypotheses.
In the last decade, several studies focused in finding biomarkers of environmental exposure to ecotoxicants (Benninghoff and Williams, 2008; Jayapal et al., 2010). The future goal would be to catalog these chemicals with known mode of action to predict the genomic signature in animals with unknown exposure. However, most of the experiments of the last decade were study-specific and with limited numbers of genes. This is mainly because of genome information is accessible only to selected organisms. Currently, with access to the next-generation sequencing technologies, it is expected to supplement and even replace array based technologies in the near future (Mehinto et al., 2012). One such method mainly used in the fish toxicogenomics is RNA-seq (Fraser et al., 2011). It has several advantages over microarrays as it does not require reference genome and is able to detect isoform abundance and alternative splicing events. Microarrays are restricted to specific probes on the arrays, whereas RNA-seq is capable of non-targeted transcript detection.
Methylation sequencing is another method shown to have the potential to be exploited as early predictors of environmentally induced epigenetic change. Studying methylation status using NGS technology can unravel biomarkers of potential oncogenic, damaging environments for aquatic species. This area of research is one of the less studied in the marine biology. Recent studies have applied methyl-seq in non-model organism and showed link between environment, epigenetics, and cancer (Mirbahai et al., 2011). It is evident from these studies that epigenetic mechanisms are disrupted by environmental toxicants in aquatic species. Thus, it is important in deciphering the effect of these toxicants and its role in diseases of aquatic organisms.
Current NGS application in fish ecotoxicogenomics is also extended to identification of novel microRNAs (miRNAs) to understand marine biology. MicroRNAs are small, single-stranded RNAs of found in both plant and animal cells. miRNAs are non-protein-coding RNAs and they are shown to play vital role in variety of cellular processes including cell proliferation, differentiation, and developmental stages. To understand these and other biological processes, several studies are applying high-throughput miRNA sequencing (miRNAome) based identification and quantification (Soares et al., 2009). Such miRNAome studies will find novel miRNAs signifying their specific role in the physiological or pathological conditions. One such recent study using miRNA sequencing showed the relationship between host miRNA expression and S. eriocheiris infections in crustaceans (Ou et al., 2012). The study also reports that differentially regulated miRNAs might be essential in host-pathogen interactions. The main goal in ecotoxicogenomics is to be able to predict and prevent damaging effects on the environment. Thus, it is important to understand the molecular mechanisms of action of ecotoxicants. From the aspect of toxicity, knowing the mechanism can help in assessing the risk of toxicity. Expanding fish ecotoxicogenomics program to miRNAome approach will provide additional mechanistic insight into actions of toxicants. Besides ecotoxicity and risk assessment, this research is expected to provide a substrate for development of additional biomarkers for safety and efficacy. Identification of biomarkers for ecotoxicity screening may lead to the development of simple and high-throughput toxicologic assays, which could be field-deployable. That in turn, would allow one to deduce how organisms respond to changes in the external environment.
With accessibility to NGS technology, the future research should aim to integrate leading-edge genomic platforms to develop new tools for studying the molecular mechanisms underlying diseases and ecotoxic risk. The overall integrated genomics approach for ecotoxicity assessment and their possible outcomes are schematically illustrated in Figure 1. It is a viable way ahead for applying the integrated genomic approach for ecotoxicological risk assessment to predict environmental risks, compound/agent characterization, and elucidating their molecular mechanism of action.
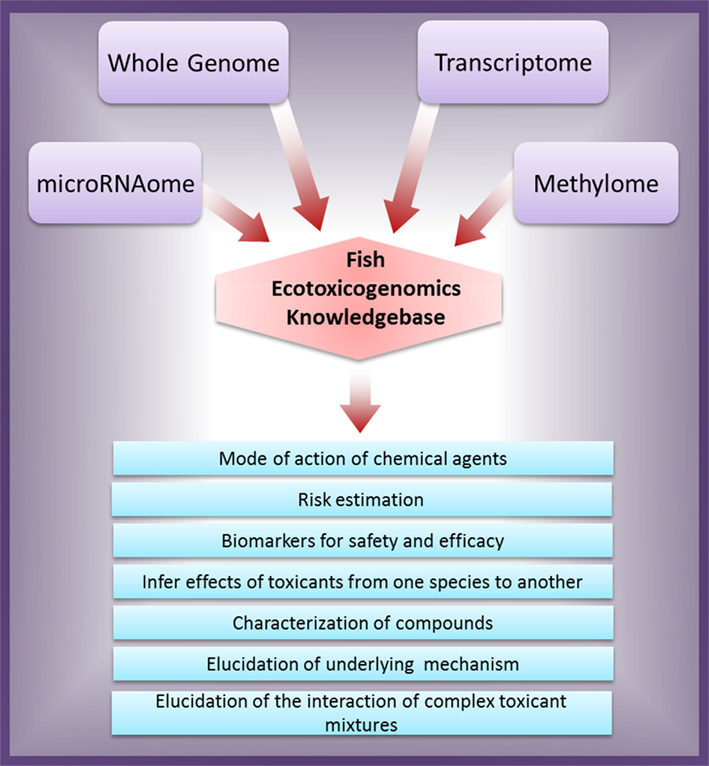
Figure 1. Schematic representation of integrated genomics approach for ecotoxicity assessment and their possible outcomes.
Benninghoff, A. D., and Williams, D. E. (2008). Identification of a transcriptional fingerprint of estrogen exposure in rainbow trout liver. Toxicol. Sci. 101, 65–80.
Connell, D., Lam, P., Richardsonm, B., and Wu, R. (1999). Introduction to Ecotoxicology. Oxford: Blackwell Science.
Fraser, B. A., Weadick, C. J., Janowitz, I., Rodd, H., and Hughes, K. A. (2011). Sequencing and characterization of the guppy (Poecilia reticulata) transcriptome. BMC Genomics 12, 202. doi: 10.1186/1471-2164-12-202
Jayapal, M., Bhattacharjee, R. N., Melendez, A. J., and Hande, M. P. (2010). Environmental toxicogenomics: a post-genomic approach to analysing biological responses to environmental toxins. Int. J. Biochem. Cell Biol. 42, 230–240.
Mehinto, A. C., Martyniuk, C. J., Spade, D. J., and Denslow, N. D. (2012). Applications of next-generation sequencing in fish ecotoxicogenomics. Front. Genet. 3:62. doi: 10.3389/fgene.2012.00062
Mirbahai, L., Williams, T. D., Zhan, H., Gong, Z., and Chipman, J. K. (2011). Comprehensive profiling of zebrafish hepatic proximal promoter CpG island methylation and its modification during chemical carcinogenesis. BMC Genomics 12, 1–16. doi: 10.1186/1471-2164-12-3
Ou, J., Meng, Q., Li, Y., Xiu, Y., Du, J., Gu, W., Wu, T., Li, W., Ding, Z., and Wang, W. (2012). Identification and comparative analysis of the Eriocheir sinensis microRNA transcriptome response to Spiroplasma eriocheiris infection using a deep sequencing approach. Fish Shellfish Immunol. 32, 345–352.
Soares, A. R., Pereira, P. M., Santos, B., Egas, C., Gomes, A. C., Arrais, J., Oliveira, J. L., Moura, G. R., and Santos, M. A. (2009). Parallel DNA pyrosequencing unveils new zebrafish microRNAs. BMC Genomics 10, 195. doi: 10.1186/1471-2164-10-195
Citation: Jayapal M (2012) Integration of next-generation sequencing based multi-omics approaches in toxicogenomics. Front. Gene. 3:88. doi: 10.3389/fgene.2012.00088
Received: 30 April 2012; Accepted: 02 May 2012;
Published online: 21 May 2012.
Copyright: © 2012 Jayapal. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence:bWpheWFwYWxAa2F1LmVkdS5zYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.