
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Fungal Biol. , 23 February 2024
Sec. Fungi-Plant Interactions
Volume 5 - 2024 | https://doi.org/10.3389/ffunb.2024.1332755
This article is part of the Research Topic Plant-Friendly Microorganisms as a Bio-Barrier Against Pathogens View all articles
 Anna Wockenfuss1
Anna Wockenfuss1 Kevin Chan2
Kevin Chan2 Jessica G. Cooper2
Jessica G. Cooper2 Timothy Chaya2
Timothy Chaya2 Megan A. Mauriello1
Megan A. Mauriello1 Sarah M. Yannarell3
Sarah M. Yannarell3 Julia A. Maresca4†
Julia A. Maresca4† Nicole M. Donofrio2*
Nicole M. Donofrio2*Biological control uses naturally occurring antagonists such as bacteria or fungi for environmentally friendly control of plant pathogens. Bacillus spp. have been used for biocontrol of numerous plant and insect pests and are well-known to synthesize a variety of bioactive secondary metabolites. We hypothesized that bacteria isolated from agricultural soil would be effective antagonists of soilborne fungal pathogens. Here, we show that the Delaware soil isolate Bacillus velezensis strain S4 has in vitro activity against soilborne and foliar plant pathogenic fungi, including two with a large host range, and one oomycete. Further, this strain shows putative protease and cellulase activity, consistent with our prior finding that the genome of this organism is highly enriched in antifungal and antimicrobial biosynthetic gene clusters. We demonstrate that this bacterium causes changes to the fungal and oomycete hyphae at the inhibition zone, with some of the hyphae forming bubble-like structures and irregular branching. We tested strain S4 against Magnaporthe oryzae spores, which typically form germ tubes and penetration structures called appressoria, on the surface of the leaf. Our results suggest that after 12 hours of incubation with the bacterium, fungal spores form germ tubes, but instead of producing appressoria, they appear to form rounded, bubble-like structures. Future work will investigate whether a single antifungal molecule induces all these effects, or if they are the result of a combination of bacterially produced antimicrobials.
A secure food supply relies heavily on managing pathogens and pests that plague major crops. Plant diseases result in a loss of $220 billion/year, according to the Food and Agricultural Organization (Food and Agricultural Organization, 2021), and a recent study demonstrated that as temperatures rise, soilborne fungal plant pathogens will increase in worldwide abundance (Delgado-Baquerizo et al., 2020). A global survey of wheat, soybean, rice, potato and maize indicates between 17 to 30% of these crops will be lost due to plant disease and up to 20% of crops can be lost to fungal diseases post-harvest (Fisher et al., 2012; Savary et al., 2019). While the situation seems dire, plant diseases are routinely managed using myriad techniques including but not limited to naturally occurring genetic plant resistance, fungicides and biocontrol agents. The first two strategies are effective, but simultaneously place selective pressures on the pathogen population to adapt. Evidence of pathogen evolution against plant Resistance (R) genes is demonstrated through studies on the rice blast fungus, Magnaporthe oryzae, via instability of their cognate avirulence proteins; these proteins are recognized by the plant’s R gene, but can accrue mutations, making it unrecognizable and subsequently undetected by the plant (reviewed in (Jia et al., 2016)). Fungicide usage, particularly when they target a single site in the pathogen, has led to development of fungicide resistance (reviewed in (Yin et al., 2023)). Again using M. oryzae as an example, strains of this pathogen have developed resistance to QoI fungicides, which target single sites to inhibit respiration in fungi (Avila-Adame and Köller, 2003; Castroagudín et al., 2015). Naturally occurring genetic resistance and fungicides result in a boom-and-bust cycle of resistance and disease, creating a need for alternative control measures.
Utilizing biological control agents to combat plant disease is predicated on the indirect or direct antagonistic interactions that these agents have against other microbes in their natural environment (reviewed in (Collinge et al., 2022)). Given their multiple modes of action, such as antibiosis, competition for resources and space, and enhancing plant growth, biocontrol agents show promise for avoiding these boom-and-bust cycles. Biological control agents include fungi such as Trichoderma spp., which are widely deployed worldwide (Lorito et al., 2010), and bacteria including but not limited to Bacillus species, which have many of the attributes listed above (reviewed in (Fira et al., 2018; Aloo et al., 2019). There have been 95 peer-reviewed publications in the last 20 years on potential biocontrol agents against the oomycete pathogen, Phytophthora infestans, causal agent of potato late blight (Hashemi et al., 2022). While most do not translate successfully to field applications, the mechanistic knowledge provides invaluable insight into stopping pathogens.
We previously isolated Bacillus (B.) velezensis strain S4 from biochar-amended agricultural soil in Delaware and sequenced its genome (Hempel et al., 2020). We hypothesized that this soil-borne strain of B. velezensis would inhibit the growth of foliar pathogens more than the aggressive, soil-borne plant pathogens Rhizoctonia solani and Sclerotinia sclerotiorum, which are likely to occupy the same niche, and to compete for the same resources (Boer et al., 2005). The prevalence of these two pathogens consistently causing plant diseases in Delaware indicates their success in competing with other microbes (Demissie et al., 2022; Ginn et al., 2023). Further, B. velezensis strains are emerging as promising biocontrol agents; Kjelgaard et al. demonstrated that a B. velezensis strain had potent anti-fungal activity in a high-throughput screen against the cereal pathogen, Fusarium culmorum (Kjeldgaard et al., 2022). In this study, we characterized strain S4 for putative enzyme activity, antimicrobial activity against five fungi and two oomycetes also isolated from Delaware soils, and evaluated its impact on spore germination of the rice blast fungus, Magnaporthe oryzae.
Magnaporthe oryzae (strain 4091), Colletotrichum graminicola, Diaporthe ueckerae, Sclerotinia sclerotiorum, Rhizoctonia solani, Phytophthora nicotianae, and Pythium dissotocum were grown on half-strength Potato Dextrose Agar (PDA; Becton, Dickinson and Company, MD, USA) in a 12:12 light:dark cycle incubator set at 25°C. Magnaporthe oryzae was grown for spore production on half-strength PDA, and subsequently filtered through miracloth (Sigma-Aldrich, MO, USA). All fungal and oomycete strains were isolated from Delaware soils (N. Gregory pers. comm.), except for M. oryzae strain 4091, which was a gift of J. Sweigard.
Bacillus velezensis strain S4. B. velezensis strain S4 was isolated from agricultural soil as described previously (Hempel et al., 2020). For routine growth and transfer in the laboratory, it was grown in solid or liquid LB medium (Fisher Scientific, Pittsburgh, PA, USA) at 28°C. For long-term storage, a liquid culture was amended with glycerol at a final concentration of 10% and maintained at -80°C. Culture supernatant for challenging fungal cultures was prepared by transferring a single colony into LB and growing for 48 hours with shaking at ~30°C, 200 rpm. The culture supernatant was filtered through a 0.2 µm filter to remove whole cells.
Bacillus velezensis strain S10-2-W. This strain was isolated from the same soils as S4 and grown as per S4.
Arabidopsis Col-0 seeds were obtained from the Arabidopsis Biological Resource Center (ABRC). Seed surfaces were decontaminated using 95% ethanol followed by 50% bleach, then rinsed 5X with sterile water. Sterilized seeds were plated on Murashige and Skoog (MS) medium (per liter, 4.4 g MS salt (BioWorld Plant Media, OH, USA), 10 g sucrose, 8 g agar; pH 5.7-5.8. Plates were incubated at 21 ± 2 °C with 12 h/12 h light/dark and illuminated with cool fluorescent light with an intensity of 120 μEm-2s-1 for 20 days.
Bacillus velezensis strain S4 was grown in LB at 30°C for 16 hours at 80 rpm (OD600 nm= 0.5-0.7). Sterile filter papers (8 mm) were briefly dipped into overnight culture with sterile forceps. Each paper was placed in the middle of assay plates to test for enzymatic activity of proteases and lipase (Figures S1A, B); for cellulase activity, the filter paper was briefly dipped in LB or S4 overnight culture and pressed gently onto the center of their respective plates (Figure S1C). Protease activity was tested using skim milk agar (SMA), which includes 100 g skim milk powder (Bectin, Dickson and Co., Franklin Lakes NJ, USA) and 16 g agar per liter (Figure 1) (Baard et al., 2023). Lipase activity was tested using tributyrin agar (TA), which is composed of 12 g agar, 10 mL glyceryl tributyrate (Sigma-Aldrich, St. Louis MO, USA; protocol can be found here: https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/product/documents/417/921/91015dat.pdf), 5 g peptone, 3 g yeast extract, and 6 drops of filter-sterilized 1:1 Tween 20: deionized water per liter (Sigma-Aldrich). Cellulase activity was tested using carboxymethylcellulose agar (CMC), which is composed of carboxymethylcellulose sodium salt 5 g (Sigma-Aldrich, St. Louis MO, USA), sodium nitrate 1 g, potassium chloride 1g, yeast extract 1 g, dipotassium phosphate 0.5 g, magnesium sulfate heptahydrate 0.5 g, and agar 15 g, pH 7.0., respectively (Batubara et al., 2021). After 72 hours, the cellulase plates were flooded with 0.1% Congo Red (10mL/each plate) and left at room temperature for 20 minutes, followed by washing with 1M NaCl (~30 mL). Parallel control plates were made using the same media and technique but using LB broth in place of overnight culture. All plates were incubated at 30°C. The protease and lipase plates observed for 3 days and cellulose plates for 2 days.
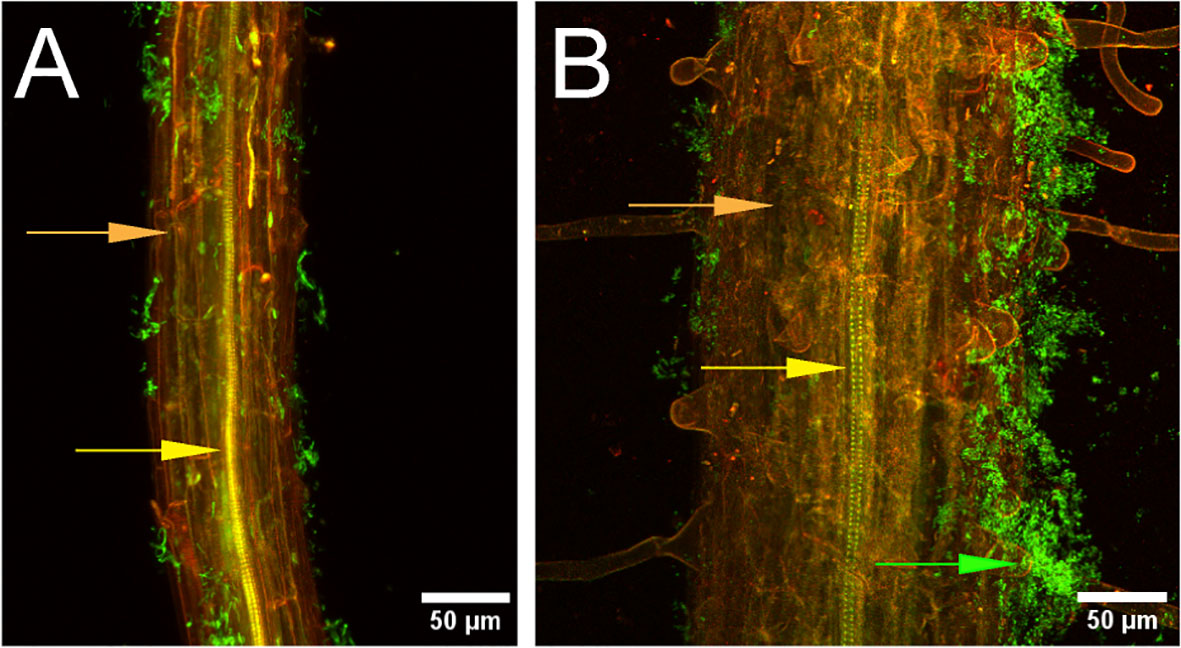
Figure 1 Bacillus velezensis strain S4 aggregates on Arabidopsis thaliana roots. Seedling roots were incubated with suspended Bacillus velezensis strains S10-2-W (A) or S4 (B) cells for 30 hours, then fixed, stained, and imaged. Green fluorescence indicates bacteria (green arrow) on root surface (orange). Xylem is indicated with the yellow arrow.
The genome of B. velezensis strain S4 was previously sequenced (Hempel et al., 2020). To identify gene clusters encoding the synthesis of potential antifungal molecules, the genome was analyzed using AntiSMASH v. 6.0.1 (Blin et al., 2019), with strict detection and default parameters for all other options.
Bacillus velezensis strains S4 and S10-2-W were revived from frozen stocks on solid LB medium and incubated at 28°C overnight. One colony was then transferred to liquid LB and incubated with shaking at 28°C for 12-18 hours, then harvested by centrifugation (4000 × g, 10 min) and washed with sterile water. The OD600 nm was adjusted to 0.02 in 0.5X MS liquid medium (0.5 mM MES, 0.1% sucrose (Bais et al., 2004)).
Five, six-day old Arabidopsis thaliana Col-0 seedlings germinated on MS solid medium were transferred to a magenta box containing 20 mL 0.5X MS liquid medium without touching a mesh support. The mesh was provided to ensure that only roots would grow into the liquid medium. Plants were grown for 12 - 14 d with constant shaking at 90 rpm at 21 ± 2 °C with a 12 h/12 h photoperiod. After 12 d the medium was completely removed and replaced with a suspension of either B. velezensis strain S4 (OD600 nm = 0.02) or S10-2-W in freshly prepared 0.5X MS liquid medium. The magenta boxes were shaken at 90 rpm for 24 h, then incubated for 6 h without shaking to promote biofilm formation (Bais et al., 2004). A. thaliana roots were then carefully removed from the magenta box, rinsed with PBS twice and fixed in 4% formaldehyde. The roots were stained with SYTO®13 (Invitrogen, Molecular Probes, OR, USA) for 5 min, then transferred to sterile water. Images were captured with a 10X Plan-Apochromat objective (numerical aperture 0.45) or 25X C-Apochromat objective (numerical aperture 1.2) on a Zeiss LSM 510 NLO attached to an Axiovert 200M with Zeiss AIM software (Rel. 3.2). Images were acquired with the 488 nm line excitation of an Argon laser using a 505 nm long-pass emission filter.
Bacillus velezensis strain S4 was revived from glycerol stocks on solid LB and incubated overnight, then inoculated into 25 ml of liquid LB (Fisher Scientific, PA, USA) and incubated at 37°C with shaking at 80 rpm for 18 hours. Following the 18-hour incubation period, 500 μl of the overnight culture was transferred into 75 ml of liquid LB (Fisher Scientific, Pittsburgh, PA, USA), incubated at 37°C and 80 rpm for 6 hours, and either used immediately or stored at 10°C for several hours until used.
Assays used half-strength PDA in Fisherbrand 100 mm x 15 mm petri plates (Fisher Scientific, PA, USA). Fisherbrand Q5 9 cm filter paper units (Fisher Scientific, PA, USA) were hole-punched into 0.8 cm circles and autoclaved. On each half-strength PDA plate, one 0.8 cm sterile filter paper disk and one 0.8 cm fungal plug were placed 2 cm away from the edge of the petri plate, directly opposite of each other. 10 μl of the S4 bacterium overnight culture was then pipetted onto the center of the filter paper disk. Plates were sealed with 3M Micropore 1 inch tape (3M, St. Paul, MN, USA) and incubated at 25°C for 14 days. Various time points were used to record the radial measurements of the pathogen growth. Growth inhibition was calculated as [(Rc−Rexp)/Rc] ×100, where Rc represents the longest radius (in cm) of fungal mycelia measured from the center of the fungal plug, and Rexp is the shortest measured radius of fungal mycelia growing towards the bacterium (Chen et al., 2018b).
Half-strength PDA plates were divided into four quadrants. Two sections of the pathogen (2 mm size cube) nearest to B. velezensis strain S4 colony on the diffusible molecule assay plates were cut out with sterile forceps and placed in the center of quadrants one and three (see Figure S2A). From the same plate, an additional two 2 mm cubes of the pathogen were cut from the region where the pathogen is furthest away from the bacteria and placed in the center of quadrants two and four. Two-millimeter cubes were also excised from the control plate (fungus or oomycete without S4) and transferred to half-strength PDA plate that was split into two quadrants. These two cubes were placed in opposing quadrants and monitored alongside the treatment plates for five days.
Half-strength PDA plates were inoculated and incubated at 25°C with a 12h day/night cycle. Images were collected at 5, 6 and 7 days with a Zeiss Axiozoom V16 (Zeiss, Germany) using a Axiocam 512 color camera at 10, 25, 50 and 100x and analyzed using FIJI (Schindelin et al., 2012).
We challenged M. oryzae spores with filtered supernatant from a B. velezensis strain S4 culture, grown to an OD600 nm of 0.579. Fungal spores were isolated from half-strength PDA as described above and adjusted to a concentration of 6 x 105 spores ml-1. Each well of a IBIDI u-bottom 96 well plate (IBIDI USA, Inc, Fitchburg, WI, USA) was inoculated with 100 μL of the spore suspension and 100 μL treatment: sterile water, nystatin (final concentration 20 mg mL-1 or 40 μg mL-1), or diluted, filtered supernatant from a 48 hour culture of B. velezensis strain S4 (1:1, 1:10 or 1:100 dilutions in sterile water). Each treatment was repeated in triplicate. The plate was incubated at 22°C for 2 hours. The wells were then imaged every hour for 16 hours on a CellDiscoverer7 (Zeiss, Germany). Data was acquired using a 20x plan-apochromat objective with a numerical aperture of 0.7. Post analysis acquisition was performed in Zeiss Zen Blue Lite 3.2 (Zeiss, Germany).
We also challenged M. oryzae spores directly with a live culture of B. velezensis strain S4. B. velezensis strain S4 was grown overnight as described above, then diluted 1:1 or 1:100 in sterile water. Each well of a 96-well plate was inoculated with 150 µl of M. oryzae spores (concentration of 5.9 x 105 spores per mL) and 150 µl of sterile water or B. velezensis strain S4 culture (live 1:1, live 1:100, and undiluted, heat killed bacteria). Bacteria were heat-killed by incubating at 95°C for 30 min. Each treatment was repeated in triplicate. The 96-well plate was then incubated at room temperature overnight and placed into the CD7 for imaging. The germ tubes, appressoria, and ungerminated spores were counted manually using Zen 3.2. Three replicates of each treatment were counted with over 700 spores included in statistical analysis.
Statistical analysis was performed using the T-test function in Microsoft Excel comparing the effect of each treatment to the effect of water. Results were considered significant when the null hypothesis was rejected with p-values < 0.05.
Bacillus velezensis strain S4 was analyzed using the AntiSMASH tool (Blin et al., 2019), which identified 12 gene clusters putatively encoding synthesis of antimicrobial or antifungal compounds (Table 1), including clusters for synthesis of bacillaene, bacillibactin, bacilysin, difficidin, fengycin, macrolactin, and mersacidin (all with 100% identity to known gene clusters in Bacillus spp.); plantazolicin and surfactin (91% identity to known gene clusters in Bacillus spp.); and two putative terpenes and a Type III polyketide synthase (T3PKS) with low identity to known gene clusters in other groups of bacteria. Surfactin, bacillaene, fengycin, bacillibactin, and bacilysin have been shown to have antifungal activity in prior work (Koumoutsi et al., 2004; Chen et al., 2009; Um et al., 2013; Hazarika et al., 2019; Dimopoulou et al., 2021). Strain S4 was then assayed on solid media containing different substrates to assess extracellular enzyme activity. The bacterium produced a large clear zone around the inoculation point on skim milk agar, and a small halo on carboxymethylcellulose agar indicating putative protease and cellulose activity, respectively (Figure S1) but suggests a lack of lipase activity under our conditions. While S4 grew on tributyrin agar, there was no zone of clearing observable. The B. velezensis strain S4 genome has 42 annotated proteases, 7 annotated lipases, and 7 annotated enzymes putatively involved in cellulose degradation, including endoglucanase, β-glucosidase, β-glucanase and exoglucanase enzymes (Chen et al., 2018a; Singh et al., 2019; Hempel et al., 2020).
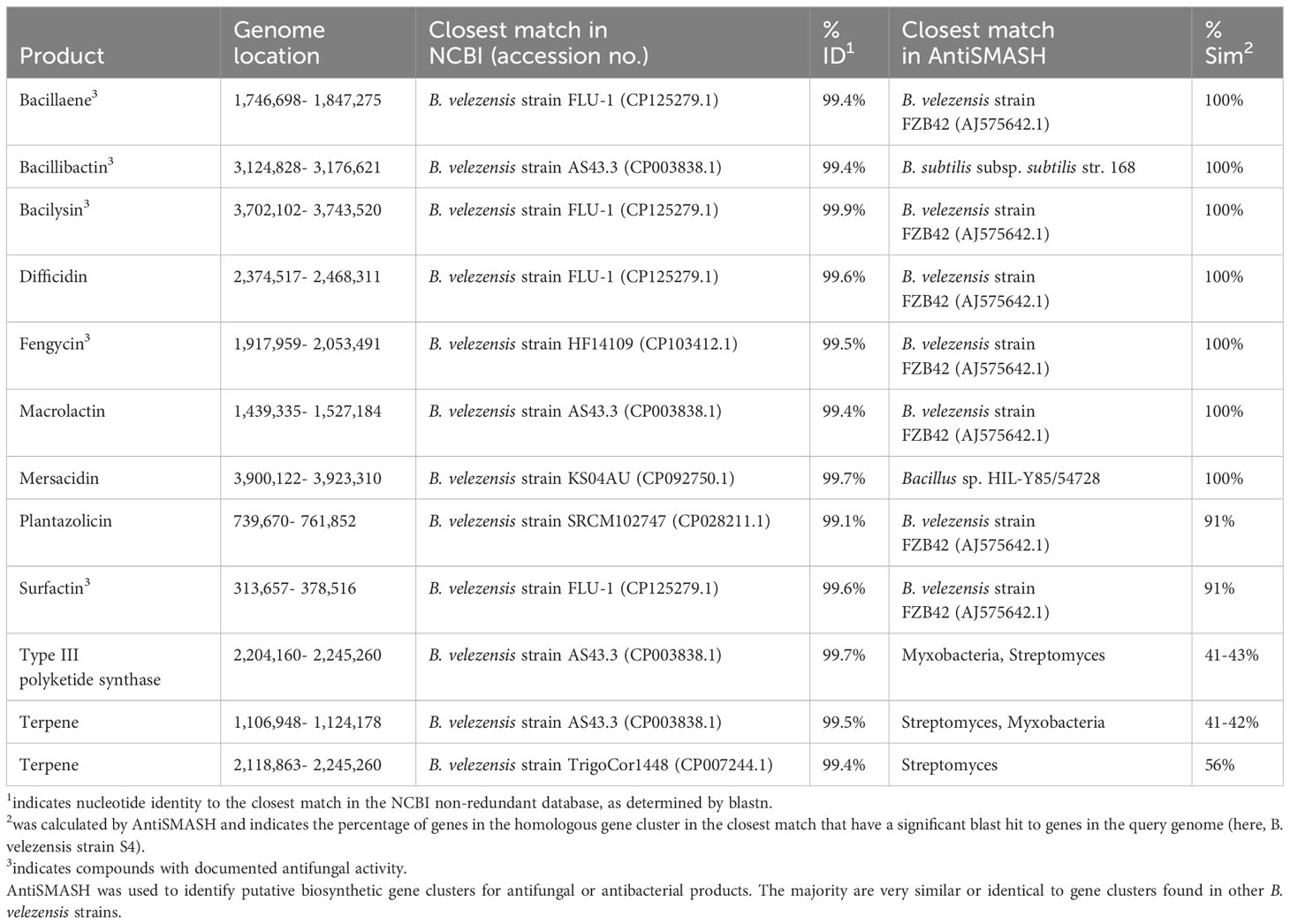
Table 1 Predicted secondary metabolites with antifungal or antibacterial activity identified in the genome of B. velezensis strain S4.
Root assays were performed on Arabidopsis ecotype Col-0 to determine whether strain S4 showed aggregation on root surfaces, potentially indicative of a biofilm formation. It was compared with strain S10-2-W, which was isolated from the same soil as S4, but showed moderate antifungal activity (S.M. Yannarell and J. Maresca, pers. comm.). Results demonstrated that while some bacterial cells from the S10-2-W strain were scattered on the root surface, S4 formed large aggregates. Optical slices (z-stacks) were taken through the root inoculated with S4 and no bacteria were visualized inside the root, indicating they are not entering (data not shown).
We used in vitro antifungal assays to determine whether strain S4 had inhibitory properties against fungi and oomycetes. Previous studies have shown that inhibitory compounds can diffuse through the media, creating a flattened, or inhibition zone, of the fungal colony (Anith et al., 2021; Kjeldgaard et al., 2022). We have access to a collection of soilborne and foliar fungal and oomycete plant pathogens, and except for M. oryzae strain 4091, all were isolated from Delaware plant and soil samples (N. Gregory, pers. comm.). We measured fungal growth along the radius from the center of the fungal plug to the center of the bacterial filter paper, along with the longest radius of the fungal colony. Assays were performed on five fungi and two oomycetes. While growth of all the fungi tested and one oomycete (P. nicotianae) tested was significantly inhibited when compared to the pathogen growing in absence of the bacterium (Figure 2), D. ueckerae and C. graminicola showed the most susceptibility to the diffusible molecules (Table 2). The growth of the water- and soilborne oomycete, P. dissotocum, was not significantly inhibited; however, the hyphae were observably less dense over the bacterial colony than in the rest of the colony or the control (Figure S3).
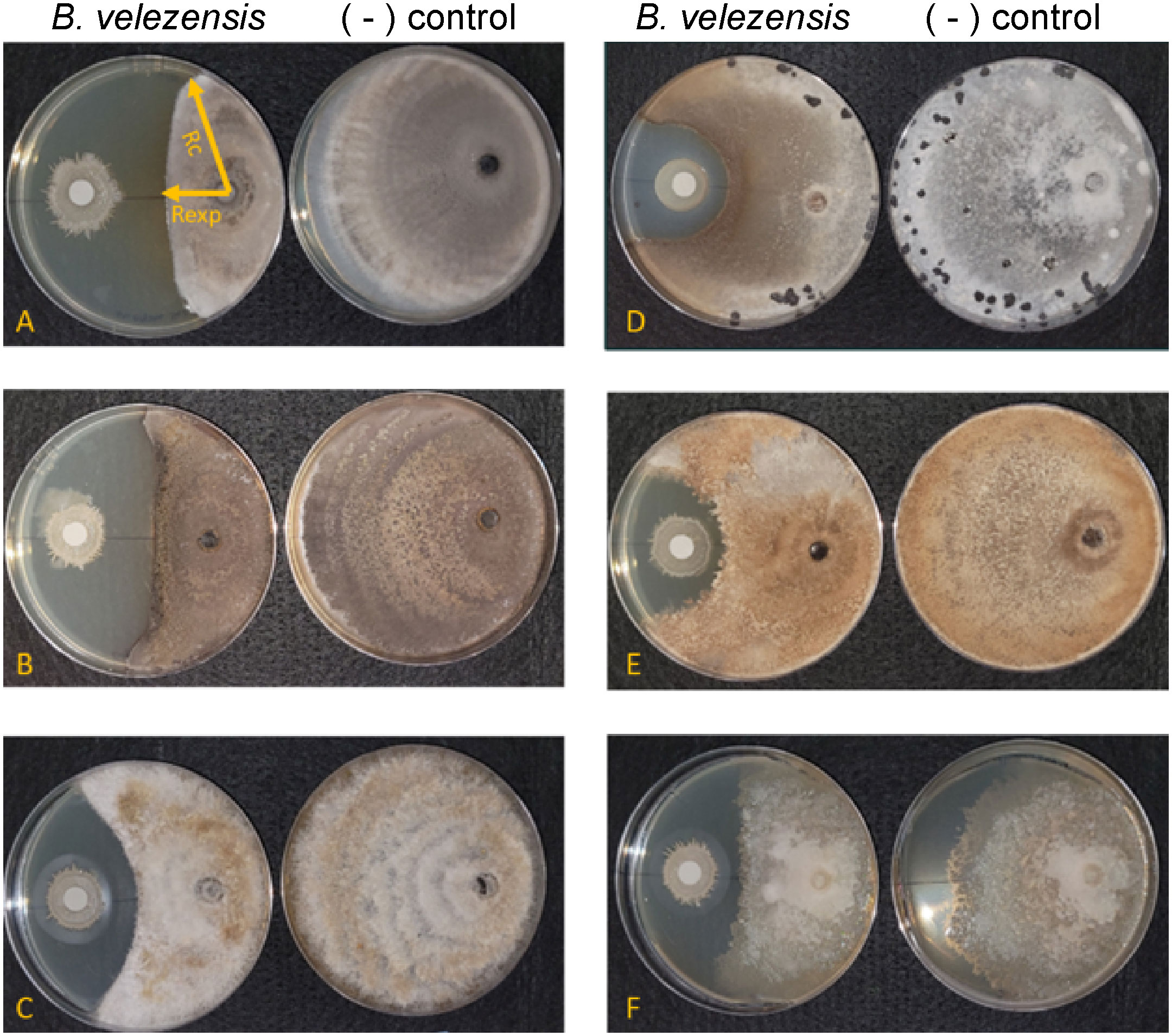
Figure 2 Bacillus velezensis S4 inhibits growth of five fungi and one oomycete. (A) Magnaporthe oryzae; (B) Colletotrichum graminicola; (C) Diaporthe ueckerae; (D) Sclerotinia sclerotiorum; (E) Rhizoctonia solani; (F) Phytophthora nicotianae. For each panel, the left plate is the diffusible molecule assay with the bacterial strain spotted onto a sterile filter paper and the right plate is the fungus growing in the absence of the bacterial cells. Images are representative of six biological replicates.
We then performed recovery assays on fungi and oomycetes from the diffusible molecule assays to determine whether the inhibitory metabolites from strain S4 had fungistatic or fungicidal effects. Two sections from the zone of inhibition, two from the colony not directly in the path of the molecules, and two from the control plates, were plated onto fresh media, grown for five days and imaged (Figure S3). Except for C. graminicola and S. sclerotiorum, strain S4 was clearly present when extracting fungi from the inhibition zone, as it grew out on the quadrant plates and continued to inhibit the fungal colonies. All fungi and oomycetes grew out on recovery plates, indicating that fungal cells are inhibited by exudates secreted by S4, but might not be killed. Alternately, we might have excised some live cells when transferring to the recovery plates.
To begin understanding the mechanism of biocontrol, we first used a dissecting microscope to examine the hyphae in the zone of inhibition for macroscopic changes in structure. In all the fungi, hyphae formed bubble-like structures in the inhibition zone (Figure 3 inset), similar to work from He et al (He et al., 2019) with M. oryzae. No bubbles formed in the hyphae of the oomycete pathogens P. nicotianae and P. dissotocum, however we observed less branching of the hyphae, particularly in P. nicotianae, and some thickened structures that were difficult to identify (Figure 3). The Pythium colony grew over the S4 bacterial colony in the diffusible molecule assay, so growth was not inhibited; however, the hyphae directly over the colony were much less dense than on the rest of the plate, or on the control (Figure S3). Interestingly, we also noted that the S. sclerotiorum colony, usually white except for the sclerotial bodies, were highly pigmented at the zone of inhibition (Figure 2D). In this zone, the hyphae were no longer hyaline, and hyphal tips seemed to terminate in bubble-like structures.
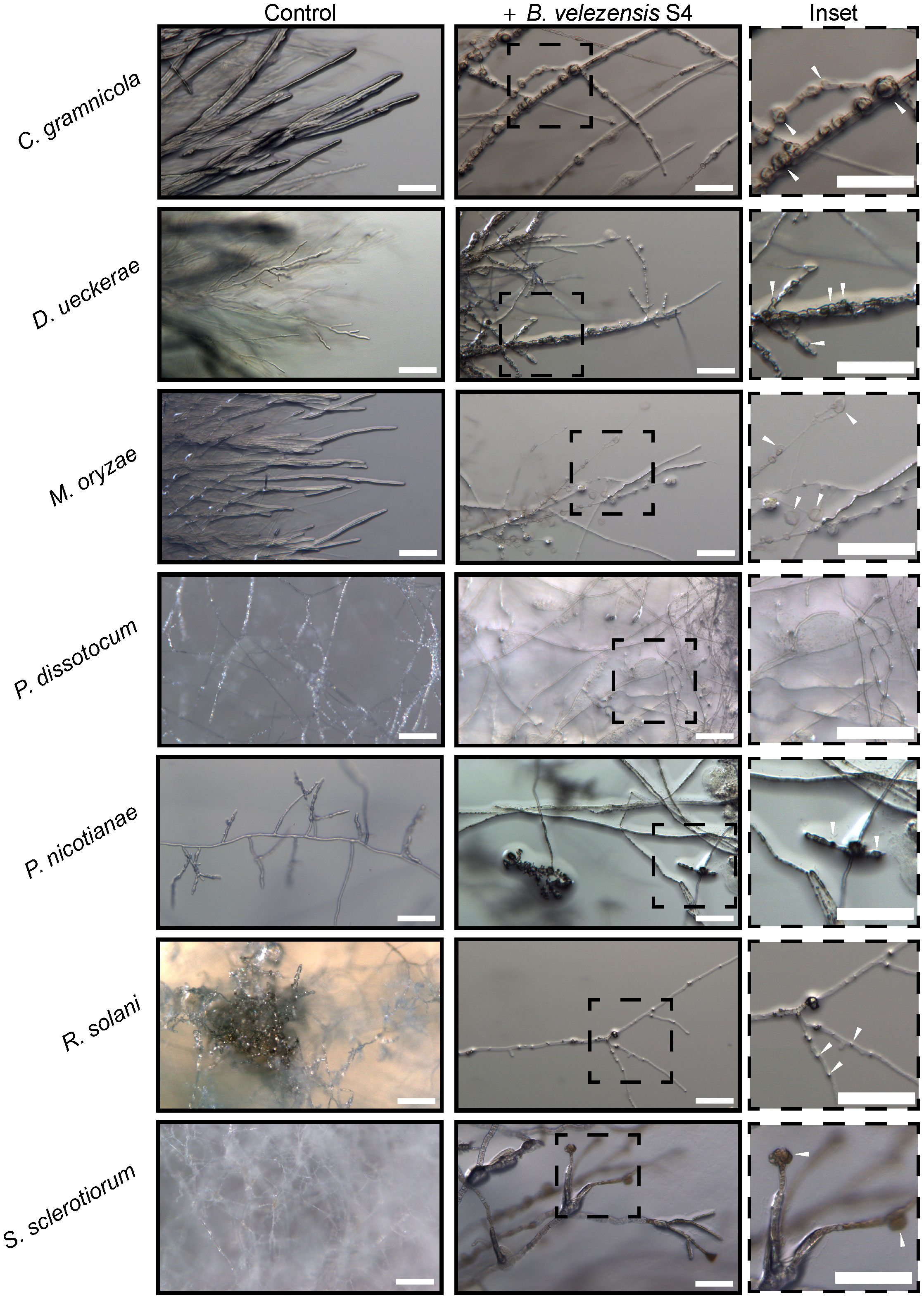
Figure 3 Five fungi and two oomycetes were subjected to Bacillus velezensis S4 small molecules using the diffusible molecule plate assay. Seven days post inoculation, the inhibition zone between microbes was imaged for structural changes at the macroscopic level using a Zeiss Axio Zoom dissecting microscope.
We next examined the impact of strain S4 diffusible molecules on asexual growth of M. oryzae spores. Infection biology of this pathogen is well-studied and begins with an asexual spore, or conidium, landing on an inductive, hydrophobic surface (Choi and Dean, 1997). The conidium then germinates and forms a penetration structure, or appressorium, leading to penetration of the epidermal cells of the plant (Howard et al., 1991). We incubated M. oryzae conidia with water, two dilutions of B. velezensis strain S4, or a heat-killed overnight culture of the bacteria for 20 hours, then imaged germ tube and appressoria formation. Under typical in vitro conditions, a spore will germinate and form a melanized, bulbous appressorium in 9 to 12 hours. Here, the number of ungerminated spores in each treatment was the same. In wells treated only with water, most spores that germinated also formed appressoria, so few conidia with only germ tubes were observed (Figure 4). In the presence of the highest concentration of the bacterium (1:1 dilution), many germ tubes developed but few appressoria formed (Figure 4). More appressoria formed in the wells treated with 1:100 diluted culture of B. velezensis strain S4, with a concomitant reduction in cells with germ tube formation only (Figure 4). Treatment with the heat-killed bacterial culture was not significantly different from the 1:100 dilution.
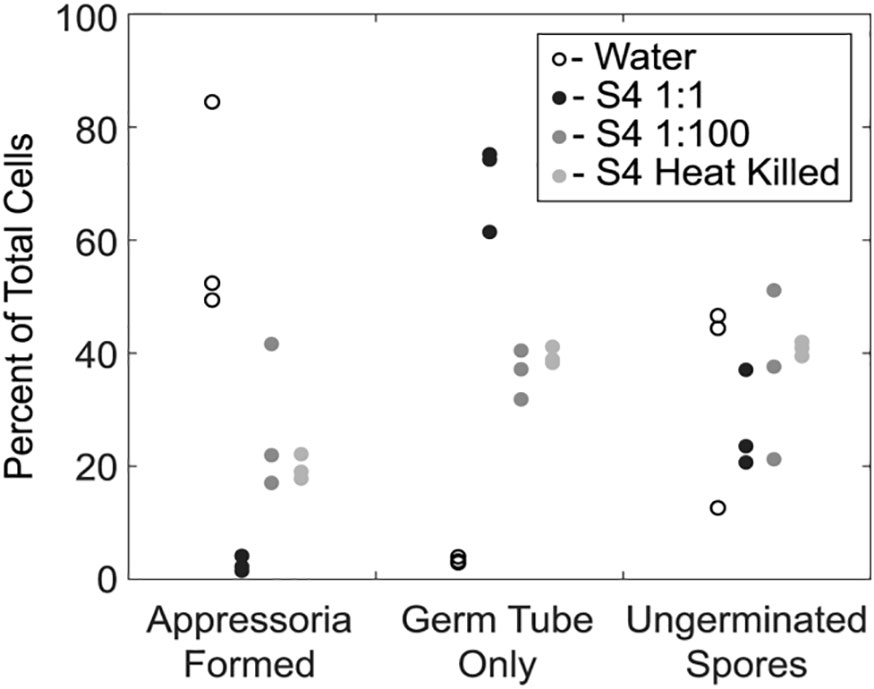
Figure 4 Germination of Magnaporthe oryzae in the presence of S4 shows reduction of appressoria formation. An overnight culture of Bacillus velezensis S4 was added to M. oryzae conidia and examined for differences in germination and appressorial formation. The bacterial culture was added to conidia in a 1:1 and 1:100 dilutions, and as a heat-killed culture. * p-value <0.05, which was considered statistically significant in a one-tailed student’s t-test.
The effects of supernatant from a B. velezensis strain S4 culture (Figure 5) were similar to the effects of diluted live B. velezensis (Figure 4) on M. oryzae spore germination and appressorial formation. Four time points during germination on an inductive surface were examined and revealed bubbles forming at 6 and 12 hours, instead of appressoria (Figure 5). Nystatin, which inhibits appressorium formation but not germination (Eilbert et al., 1999) was used as a control. Together, our observations indicate that the diffusible molecules from strain S4 disrupt normal vegetative growth and conidial germination of M. oryzae, forming bubble-like swellings instead of typical penetration structures on an inductive surface.
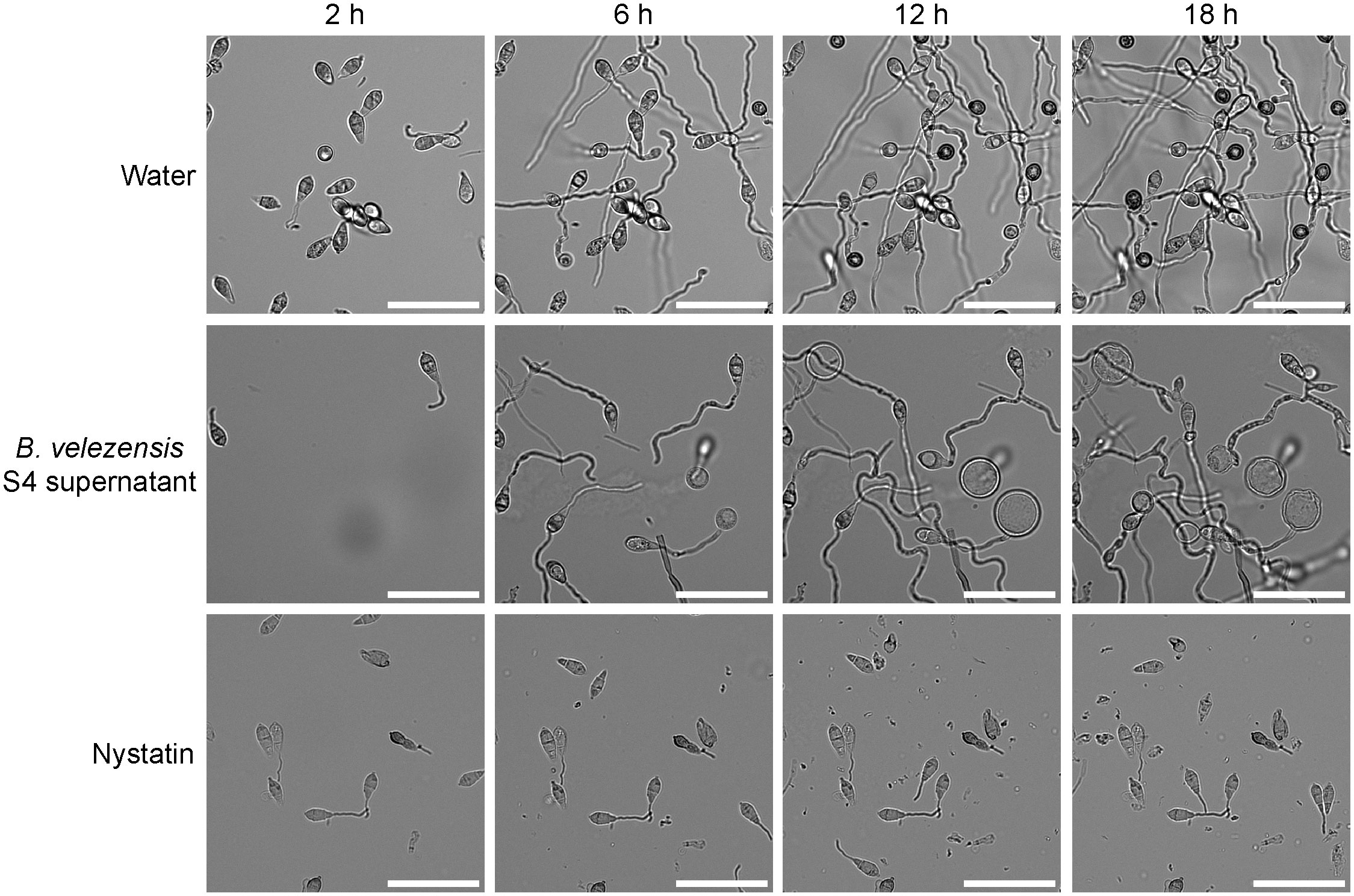
Figure 5 S4 supernatant deforms germinating Magnaporthe oryzae spores. Filtered supernatant from a Bacillus velezensis strain S4 culture was applied to isolated M. oryzae spores and the spores were imaged over time. Appressoria typically begin forming around 6 hours and become fully melanized at 12 hours. In the presence of the S4 supernatant, bubbles appear to differentiate from the germinating spores and no appressoria were formed. Nystatin was used as a germination control. Scale bar equals 50 μm.
We isolated a potential biocontrol bacterium from biochar-amended agricultural soils in Delaware. Bacillus velezensis strain S4 displays potent inhibition of five plant pathogenic fungi, as well as one plant pathogenic oomycete. We predicted that this strain would minimally inhibit growth of soil-borne pathogenic fungi. However, it significantly reduced the growth of Rhizoctonia solani and Sclerotinia sclerotiorum, both found in soil, and was even more effective at inhibiting the foliar fungi C. graminicola and M. oryzae as well as the oomycete P. nicotinianae. Diaporthe ueckerae is an emerging pathogen of particular concern on soybeans, as it is one of several species causing seed decay, however this species has not yet been characterized as foliar or soil-borne (Petrović et al., 2021; A. Koehler, pers. comm.). This specific isolate has not yet been characterized as foliar or soil borne (A. Koehler, pers. comm.). Growth of the soil-borne oomycete Pythium dissotocum was less affected; however, its hyphae did become sparse as it grew over the bacterial colony.
Our results demonstrated strong inhibition of the foliar pathogens C. graminicola and M. oryzae, and weaker but significant inhibition of the aggressive, soilborne pathogens S. sclerotiorum and R. solani. We observed that S. sclerotiorum hyphae, usually hyaline except for its survival structures (sclerotia), was melanized at the inhibition zone and displayed thickened hyphae, many of them terminating in bubble-like structures. The melanin biosynthetic pathway is involved in producing sclerotial bodies (Butler et al., 2009), although one recent study indicates that when the major genes for melanin production are deleted from this fungus, it grows more slowly, is less resistant to UV light, but remains fully pathogenic (Liang et al., 2018). Melanization in the zone of interaction between S. sclerotiorum and B. velezensis suggests that melanin may also play a role in protecting S. sclerotiorum from antagonists. Song et al. isolated a B. velezensis strain from farm soils in South Korea that demonstrated varying levels of inhibition against two different species of S. sclerotiorum and Colletotrichum (Song et al., 2022). The former was melanized, but it is unclear whether their S. sclerotiorum strain has non-pigmented hyphae under normal conditions. S. sclerotiorum is a broad-range, soilborne pathogen known to infect 425 plant species, but likely infects many more (Derbyshire et al., 2022). It has been difficult to manage for many reasons: infection with S. sclerotiorum is unrelated to crop loss in the field, and the environmental variables that impact life stage of S. sclerotiorum in the lab are not relevant in the field (reviewed in (Reich and Chatterton, 2022)).
Rhizoctonia solani is another broad host range, soilborne pathogen. We noted clear inhibition of the pathogen by B. velezensis strain S4, as well as hyphae forming into bubble-like structures. This differs from a recent study deploying B. subtilis strain SL-44 against a R. solani isolate responsible for large losses to pepper plants. Like B. velezensis strain S4, the B. subtilis SL-44 encodes the biosynthesis of fengycin, a noted antifungal compound. Unlike B. velezensis strain S4, their microscopic observations revealed hyphal breakage in R. solani instead of bubbles, with loss of cytosol and cell wall integrity (Wu et al., 2019). A more recent study used a different strain of B. velezensis against multiple isolates of R. solani causing brown patch on turf, and demonstrated that globoid, rounded structures formed at the tips of the hyphae in the inhibition zone (Lee et al., 2023). In contrast, we observed bubble formation behind the hyphal tip, potentially indicating that different strains of B. velezensis will produce different effects on the pathogens, though all may result in biocontrol.
The bubble-like structures we observed in the inhibition zones of all fungi tested, are also consistent with the results of a recent study on biocontrol of M. oryzae (He et al., 2019). There, B. subtilis strain BJ-1 was tested as a biocontrol agent against M. oryzae. The bacteria did not inhibit germination, but did disrupt the cell walls of the hyphae, as well as appressorial development. This is supported by the presence of genes for fengycin production, which has been reported to disrupt hyphal membranes, eventually leading to cell death (Xiao et al., 2021). Furthermore, B. velezensis strain S4 contains the synthetic machinery for bacilysin production, which is known to target cell wall biosynthesis (reviewed in (Andrić et al., 2020). Together, these compounds could account for the bubble-like structures formed within the hyphae, and at the terminal branches of the hyphae in the case of S. sclerotiorum. Indeed, Xiao et al. demonstrated that when the take-all pathogen of wheat was challenged with fengycin, hyphae formed circular structures at the tip (Xiao et al., 2021).
We performed recovery assays from sections of hyphae in the zone of inhibition and found that the fungi recover on fresh media, possibly indicating that strain S4 has a fungistatic rather than a fungicidal activity. However, further studies are needed either that the diffusing molecule(s) do not kill the pathogens, or we are collecting pathogen cells that are not in contact with the S4 exudates. While our search was not exhaustive, we were unable to identify recovery assays in the B. velezensis literature. While these results indicate that strain S4 is fungistatic, future studies will reveal whether fungal and oomycete cells in the inhibition zone are dead or just deformed, but still alive and able to recover.
There has been a recent surge in interest in B. velezensis strains for biocontrol, with several demonstrating both antifungal and plant growth promoting activities. A recent study on the B. velezensis strain FKM10, isolated from an apple orchard in China, suggests that it inhibits apple pathogens, and increases traits like height and fresh weight (Wang et al., 2020). A study from Shin et al. (2021) isolated and characterized a B. velezensis strain from pepper fields in South Korea, which indicate both fungal inhibition and plant growth promoting properties. However, neither study assessed aggregation of B. velezensis on plant roots. Here, we used microscopy to discern whether strain S4 was at least forming aggregates on roots, as other B. subtilis strains with known plant growth promoting and root binding properties do (Rudrappa et al., 2008). We observed large aggregates of S4 on Arabidopsis roots compared with another Bacillus strain that had moderate levels of antifungal activity. We did not observe the bacterium entering plant roots, and further studies are required to determine whether they are forming biofilms and/or whether they have plant growth promoting properties.
Genomic studies of strain S4 reveal an enrichment in potential antibiotic clusters, five of which likely have antifungal activity: surfactin, bacillaene, fengycin, bacillibactin, and bacilysin have been shown to have antifungal activity in prior work (Koumoutsi et al., 2004; Chen et al., 2009; Um et al., 2013; Hazarika et al., 2019; Dimopoulou et al., 2021). We further observed protease and cellulase activity in plate assays, which corresponded with putative protease and cellulose-degrading genes found in the genome. The bioactive compounds could be extracellular enzymes, secondary metabolites, or both, and the B. velezensis strain S4 genome is predicted to encode the synthesis of five small molecules with antifungal activity as well as enzymes capable of protein, triglycerides and cellulose degradation.
Biocontrol has tremendous potential to augment more traditional means of plant disease control, such as fungicide application and genetic resistance. Since biocontrol agents tend to have multiple methods of inhibiting or killing the pathogen, as many strains of B. velezensis do (Chen et al., 2018a; Borriss et al., 2019; Rabbee et al., 2019), evolving genetic resistance to biocontrol is not as straightforward as it would be against a fungicide with a single mode of action. Bardin and Nicot asked whether it is possible for fungi and oomycetes to develop resistance to biocontrol agents (Bardin et al., 2015). The answer will require fully understanding both the mechanisms of inhibition deployed by the biocontrol agent and the responses of the plant pathogens. Changes in hyphal morphology have been demonstrated by multiple labs, including ours, and the next stage will be to examine the genetics of pathogen response, whether the pathogen signals to itself when under duress, and ultra-structural changes within the pathogen cells at the inhibition zone.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AW: Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. KC: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. JC: Formal analysis, Methodology, Visualization, Writing – review & editing. TC: Conceptualization, Investigation, Methodology, Visualization, Writing – review & editing. MM: Conceptualization, Data curation, Methodology, Writing – review & editing. SY: Investigation, Methodology, Visualization, Writing – review & editing. JM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing. ND: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. University of Delaware’s Graduate College and Microbiology Graduate Program provided stipend funding in part for the graduate students AW and MM. KC was funded in part through University of Delaware Undergraduate Research and CANR Unique Strengths summer internship. Microscopy was funded by grants from NIH-NIGMS (P20 GM103446) and NIGMS (P20 GM139760).
We thank Dr. Harsh Bais for assistance with the plant root colonization assays, Noah Savastano from the Bais lab for determining whether S4 enters the root, and Dr. Charles Dhong for assistance with Figure 4. Microscopy access in the Bioimaging Facility at the Delaware Biotechnology Institute was supported by grants from the NIH-NIGMS (P20 GM103446), the NIGMS (P20 GM139760) and the State of Delaware.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2024.1332755/full#supplementary-material
Supplementary Figure 1 | B. velezensis strain S4 shows protease and cellulose activity. (A) skim milk agar; (B) carboxymethylcellulose agar with Congo red staining (light orange around the bacterial colony indicates halo); (C) tributyrin agar. For (A, B), filter papers were inoculated with LB as the control (left plates) and with S4 (right plates) and incubated for 72 hours. For (C), filter papers were dipped in LB control (left plates) or S4 (right plates) and stamped briefly onto the plate, lifted off, and incubated for 48 hours.
Supplementary Figure 2 | Recovery assays show differing amount of hyphal growth. (A) Diagram of recovery assays. Quadrants 2 and 4 are isolated from the side of the plate furthest from the bacterial filter paper, while 1 and 3 are isolated from the zone of inhibition, closest to the bacterial filter paper. For B – G, the left plate shows hyphae isolated from control plates in the absence of S4 and the right plate shows hyphae isolated from antifungal assays. Images were taken 5 days post-inoculation. (B) M. oryzae; (C) C. graminicola; (D) D. uekerae; (E) R. solani; (F) S. sclerotiorum; (G) P. nicotianae
Supplementary Figure 3 | Pythium dissotocum outgrows the bacterial strain. (A) P. dissotocum seven days after plating. The left plate has the B. velezensis S4 bacterial culture on the filter paper, while the right plate only has P. dissotocum. (B) Recovery assay. In quadrants 1 and 3, taken from the inhibition zone (less dense hyphae), bacterial growth is clear, however the oomycete appears largely unaffected, except for some thinner hyphae around the S4 colony.
Aloo B. N., Makumba B. A., Mbega E. R. (2019). The potential of Bacilli rhizobacteria for sustainable crop production and environmental sustainability. Microbiological Res. 219, 26–39. doi: 10.1016/j.micres.2018.10.011
Andrić S., Meyer T., Ongena M. (2020). Bacillus responses to plant-associated fungal and bacterial communities. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01350
Anith K. N., Nysanth N. S., Natarajan C. (2021). Novel and rapid agar plate methods for in vitro assessment of bacterial biocontrol isolates’ antagonism against multiple fungal phytopathogens. Lett. Appl. Microbiol. 73, 229–236. doi: 10.1111/lam.13495
Avila-Adame C., Köller W. (2003). Characterization of spontaneous mutants of Magnaporthe grisea expressing stable resistance to the Qo-inhibiting fungicide azoxystrobin. Curr. Genet. 42, 332–338. doi: 10.1007/s00294-002-0356-1
Baard V., Bakare O. O., Daniel A. I., Nkomo M., Gokul A., Keyster M., et al. (2023). Biocontrol potential of Bacillus subtilis and Bacillus tequilensis against four Fusarium species. Pathogens. 12, 254. doi: 10.3390/pathogens12020254
Bais H. P. P., Fall R., Vivanco J. M. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319. doi: 10.1104/pp.103.028712
Bardin M., Ajouz S., Comby M., Lopez-Ferber M., Graillot B., Siegwart M., et al. (2015). Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00566
Batubara U. M., Mardalisa M., Suparjo S., Maritsa H. U., Pujianto E., Herlini M. (2021). “Isolation and characterization of cellulolytic bacteria diversity in peatland ecosystem and their cellulotylic activities,” in IOP Conf. Series: Earth and Environmental Science. Bristol, Great Britain: IOP Publishing, Vol. 934. doi: 10.1088/1755-1315/934/1/012028
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Boer W., Folman L. B., Summerbell R. C., Boddy L. (2005). Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Borriss R., Wu H., Gao X. (2019). “Secondary Metabolites of the Plant Growth Promoting Model Rhizobacterium Bacillus velezensis FZB42 are Involved in Direct Suppression of Plant Pathogens and in Stimulation of Plant-Induced Systemic Resistance,” in Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms: Discovery and Applications. Eds. Singh H. B., Keswani C., Reddy M. S., Sansinenea E., García-Estrada C. (Springer Singapore, Singapore), 147–168. doi: 10.1007/978-981-13-5862-3_8
Butler M. J., Gardiner R. B., Day A. W. (2009). Melanin synthesis by Sclerotinia sclerotiorum. Mycologia 101, 296–304. doi: 10.3852/08-120
Castroagudín V. L., Ceresini P. C., de Oliveira S. C., Reges J. T. A., Maciel J. L. N., Bonato A. L. V., et al. (2015). Resistance to QoI fungicides is widespread in Brazilian populations of the Wheat Blast Pathogen Magnaporthe oryzae. Phytopathology 105, 284–294. doi: 10.1094/PHYTO-06-14-0184-R
Chen P.-H., Chen R.-Y., Chou J.-Y. (2018b). Screening and evaluation of yeast antagonists for biological control of Botrytis cinerea on strawberry fruits. Mycobiology 46, 33–46. doi: 10.1080/12298093.2018.1454013
Chen L., Gu W., Xu H., Yang G.-L., Shan X.-F., Chen G., et al. (2018a). Complete genome sequence of Bacillus velezensis 157 isolated from Eucommia ulmoides with pathogenic bacteria inhibiting and lignocellulolytic enzymes production by SSF. 3 Biotech. 8, 114. doi: 10.1007/s13205-018-1125-2
Chen X.-H., Koumoutsi A., Scholz R., Borriss R. (2009). More than anticipated - production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J. Mol. Microbiol. Biotechnol. 16, 14–24. doi: 10.1159/000142891
Choi W., Dean R. A. (1997). The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9, 1973–1983. doi: 10.1105/tpc.9.11.1973
Collinge D. B., Jensen D. F., Rabiey M., Sarrocco S., Shaw M. W., Shaw R. H. (2022). Biological control of plant diseases – What has been achieved and what is the direction? Plant Pathol. 71, 1024–1047. doi: 10.1111/ppa.13555
Delgado-Baquerizo M., Guerra C. A., Cano-Díaz C., Egidi E., Wang J.-T., Eisenhauer N., et al. (2020). The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Change 10, 550–554. doi: 10.1038/s41558-020-0759-3
Demissie H. B., Momen B., Everts K. L. (2022). White mold incidence, severity and lima bean (Phaseolus lunatus) yield response to fungicide application timing in the mid-Atlantic region of the United States. Crop Prot. 154, 105890. doi: 10.1016/j.cropro.2021.105890
Derbyshire M. C., Newman T. E., Khentry Y., Owolabi Taiwo A. (2022). The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 23, 1075–1090. doi: 10.1111/mpp.13221
Dimopoulou A., Theologidis I., Benaki D., Koukounia M., Zervakou A., Tzima A., et al. (2021). Direct antibiotic activity of Bacillibactin broadens the biocontrol range of Bacillus amyloliquefaciens MBI600. mSphere 6, e0037621. doi: 10.1128/mSphere.00376-21
Eilbert F., Thines E., Anke H. (1999). Effects of antifungal compounds on conidial germination and on the induction of appressorium formation of Magnaporthe grisea. Z. für Naturforschung C 54, 903–908. doi: 10.1515/znc-1999-1108
Fira D., Dimkić I., Berić T., Lozo J., Stanković S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Fisher M. C., Henk D., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Food and Agricultural Organization (2021). Pathogens, precipitation and produce prices. Nat. Clim. Change 11, 635–635. doi: 10.1038/s41558-021-01124-4
Ginn A. N., Evans T. A., Ernest E. G., Koehler A. M. (2023). First report of Rhizoctonia solani AG4 causing brown bean of lima bean in Delaware. Plant Dis. 107, 214. doi: 10.1094/PDIS-01-22-0118-PDN
Hashemi M., Tabet D., Sandroni M., Benavent-Celma C., Seematti J., Andersen C. B., et al. (2022). The hunt for sustainable biocontrol of oomycete plant pathogens, a case study of Phytophthora infestans. Fungal Biol. Rev. 40, 53–69. doi: 10.1016/j.fbr.2021.11.003
Hazarika D. J., Goswami G., Gautom T., Parveen A., Das P., Barooah M., et al. (2019). Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 19, 71. doi: 10.1186/s12866-019-1440-8
He Y., Zhu M., Huang J., Hsiang T., Zheng L. (2019). Biocontrol potential of a Bacillus subtilis strain BJ-1 against the rice blast fungus Magnaporthe oryzae. Can. J. Plant Pathol. 41, 47–59. doi: 10.1080/07060661.2018.1564792
Hempel P. P., Yao M., Yannarell S., Shevchenko O., Vogt F., Donofrio N., et al. (2020). Complete genome sequence of Bacillus velezensis strain S4, isolated from biochar-treated soil. Microbiol. Resource Announcements 9, e00352–e00320. doi: 10.1128/MRA.00352-20
Howard R. J., Ferrari M. A., Roach D. H., Money N. P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. 88, 11281–11284. doi: 10.1073/pnas.88.24.11281
Jia Y., Zhou E., Lee S., Bianco T. (2016). Coevolutionary dynamics of rice blast resistance gene Pi-ta and Magnaporthe oryzae avirulence gene AVR-Pita 1. Phytopathology® 106, 676–683. doi: 10.1094/PHYTO-02-16-0057-RVW
Kjeldgaard B., Neves A. R., Fonseca C., Kovács Á.T., Domínguez-Cuevas P. (2022). Quantitative high-throughput screening methods designed for identification of bacterial biocontrol strains with antifungal properties. Microbiol. Spectr. 10, e0143321. doi: 10.1128/spectrum.01433-21
Koumoutsi A., Chen X.-H., Henne A., Liesegang H., Hitzeroth G., Franke P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol 186, 1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004
Lee G., Choi H., Liu H., Han Y.-H., Paul N. C., Han G. H., et al. (2023). Biocontrol of the causal brown patch pathogen Rhizoctonia solani by Bacillus velezensis GH1-13 and development of a bacterial strain specific detection method. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1091030
Liang Y., Xiong W., Steinkellner S., Feng J. (2018). Deficiency of the melanin biosynthesis genes SCD1 and THR1 affects sclerotial development and vegetative growth, but not pathogenicity, in Sclerotinia sclerotiorum. Mol. Plant Pathol. 19, 1444–1453. doi: 10.1111/mpp.12627
Lorito M., Woo S. L., Harman G. E., Monte E. (2010). Translational research on Trichoderma: From ‘omics to the field. Annu. Rev. Phytopathol. 48, 395–417. doi: 10.1146/annurev-phyto-073009-114314
Petrović K., Skaltsas D., Castlebury L. A., Kontz B., Allen T. W., Chilvers M. I., et al. (2021). Diaporthe seed decay of soybean [Glycine max (L.) merr.] is endemic in the United States, but new fungi are involved. Plant Dis. 105, 1621–1629. doi: 10.1094/PDIS-03-20-0604-RE
Rabbee M., Ali Md., Choi J., Hwang B., Jeong S., Baek K. (2019). Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 24, 1046. doi: 10.3390/molecules24061046
Reich J., Chatterton S. (2022). Predicting field diseases caused by Sclerotinia sclerotiorum: A review. Plant Pathol. 72, 3–18. doi: 10.1111/ppa.13643
Rudrappa T., Czymmek K. J., Paré P. W., Bais H. P. (2008). Root-secreted Malic acid recruits beneficial soil bacteria. Plant Phys. 148, 1547–1556. doi: 10.1104/pp.108.127613
Savary S., Willocquet L., Pethybridge S. J., Esker P., McRoberts N., Nelson A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Shin J.-H., Park B.-S., Kim H.-Y., Lee K.-H., Kim K. S. (2021). Antagonistic and plant growth-promoting effects of Bacillus velezensis BS1 isolated from rhizosphere soil in a pepper field. Plant Pathol. J. 37, 307–314. doi: 10.5423/PPJ.NT.03.2021.0053
Singh S., Jaiswal D. K., Sivakumar N., Verma J. P. (2019). Developing efficient thermophilic cellulose degrading consortium for glucose production from different agro-residues. Front. Energy Res. 7. doi: 10.3389/fenrg.2019.00061
Song S., Jeon E. K., Hwang C.-W. (2022). Characteristic analysis of soil-isolated Bacillus velezensis HY-3479 and its antifungal activity against phytopathogens. Curr. Microbiol. 79, 357. doi: 10.1007/s00284-022-03060-8
Um S., Fraimout A., Sapountzis P., Oh D.-C., Poulsen M. (2013). The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci. Rep. 3, 3250. doi: 10.1038/srep03250
Wang C., Zhao D., Qi G., Mao Z., Hu X., Du. B., et al. (2020). Effects of Bacillus velezensis FKM10 for promoting the growth of Malus hupehensis Rehd. And inhibiting Fusarium verticillioides. Front. Microbiol. 10, 2889.
Wu Z., Huang Y., Li Y., Dong J., Liu X., Li C. (2019). Biocontrol of Rhizoctonia solani via induction of the defense mechanism and antimicrobial compounds produced by Bacillus subtilis SL-44 on pepper (Capsicum annuum L.). Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02889
Xiao J., Guo X., Qiao X., Zhang X., Chen X., Zhang D. (2021). Activity of fengycin and iturin A isolated from Bacillus subtilis Z-14 on Gaeumannomyces graminis var. tritici and soil microbial diversity. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.682437
Keywords: biocontrol agent, antifungals, Magnaporthe oryzae, appressorial formation, plant pathogens, hyphae
Citation: Wockenfuss A, Chan K, Cooper JG, Chaya T, Mauriello MA, Yannarell SM, Maresca JA and Donofrio NM (2024) A Bacillus velezensis strain shows antimicrobial activity against soilborne and foliar fungi and oomycetes. Front. Fungal Biol. 5:1332755. doi: 10.3389/ffunb.2024.1332755
Received: 03 November 2023; Accepted: 05 February 2024;
Published: 23 February 2024.
Edited by:
Maggie Levy, Hebrew University of Jerusalem, IsraelReviewed by:
Dipali Rani Gupta, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshCopyright © 2024 Wockenfuss, Chan, Cooper, Chaya, Mauriello, Yannarell, Maresca and Donofrio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole M. Donofrio, bmRvbm9mQHVkZWwuZWR1
†Present address: Julia A. Maresca, Department of Chemistry, SUNY-Environmental Science and Forestry, Syracuse, NY, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.